Abstract
Mycobacterium tuberculosis can persist in an altered physiological state for many years after initial infection, and it may reactivate to cause active disease. An analogous persistent state, possibly consisting of several different subpopulations of bacteria, may arise during chemotherapy; this state is thought to be responsible for the prolonged period required for effective chemotherapy. Using two models of drug-induced persistence, we show that both microaerophilic stationary-phase M. tuberculosis treated with a high dose of rifampin in vitro and pyrazinamide-induced persistent bacteria in mice are nonculturable yet still contain 16S rRNA and mRNA transcripts. Also, the in vitro persistent, plate culture-negative bacteria incorporate radioactive uridine into their RNA in the presence of rifampin and can rapidly up-regulate gene transcription after the replacement of the drug with fresh medium and in response to heat shock. Our results show that persistent M. tuberculosis has transcriptional activity. This finding provides a molecular basis for the rational design of drugs targeted at persistent bacteria.
Tuberculosis is an important infectious disease, with over 1 billion people subclinically infected and the 3 million new cases each year resulting in 7% of the total number of deaths (20). Case finding and treatment are the main control measures, but chemotherapy takes 6 months to achieve a cure because bacteria persist during chemotherapy and cause relapse (2 to 5% in 5 years) after the end of treatment (14). These persistent bacteria are usually drug sensitive at relapse, and so their resistance to chemotherapy is phenotypic (17) (or tolerant) rather than genetic. It is suggested that an altered physiological state of persistent Mycobacterium tuberculosis accounts for its tolerance to drugs as well as the ability to survive in the host for many years. Persistence is likely to be a combined effect of both the immune system and bacterial physiology, resulting in what is generally referred to as a latent state (1). A key question is whether the metabolism of persistent bacteria is switched off with no cell division (spore-like) or, alternatively, is active (30).
The first major advance in the detection of quiescent M. tuberculosis was the development of the Cornell mouse model (25). In this model, animals are infected with M. tuberculosis and treated for 3 months with chemotherapy which renders the tissues sterile; then steroids are administered, which leads to relapse in a high proportion of the mice. Immediately after the end of antibiotic therapy, the persistent bacteria are invisible by microscopy and are uncultivable, but about 105 genome equivalents of M. tuberculosis DNA per ml of tissue in the organs have been detected by DNA amplification (4). The presence of DNA could be indicative of dead bacilli or, possibly, of live bacilli which can multiply when the host immune system becomes weakened by steroids. Since DNA can be detected in dead bacteria (9, 16, 24), targeting genomic DNA is not suitable for the determination of metabolic activity and viability of bacteria.
It has been suggested that the presence of rRNA could be an indicator of viability in M. smegmatis (33), Escherichia coli (26, 32), and Staphylococcus aureus (26), although the correlation of CFU counts with levels of 16S rRNA is delayed, often by several days. Most mRNAs are very unstable, with a short half-life (18, 19). Thus, the presence of mRNA is usually indicative of the continuous transcription of a gene. Patel et al. (31) used reverse transcription (RT)-PCR to examine the viability of heat-killed M. leprae and detected dnaK mRNA only in living cells. Recently, Hellyer et al. (15) found that exposure of log-phase M. tuberculosis to isoniazid and rifampin for 24 h reduced the levels of fbpB (85B, α-antigen) mRNA to <4 and <0.01%, respectively, of those in the drug-free control. A recent study has also shown that mRNA for the 85 antigen is present in vivo in persistent organisms (29). Similarly, in other bacteria, mRNA is not detected in cells killed by heat and other experimental treatments (32). Because of its short half-life, mRNA rapidly disappears from dead cells, and so its detection provides a useful indicator of cellular viability and metabolic activity.
It has been suggested by Wayne (34) that M. tuberculosis can adapt to microaerophilic or anaerobic conditions and can persist in a stationary phase for long periods of time. However, this microaerophilic stationary-phase culture model contains different populations of persistent organisms (35). To study populations of persisters which are tolerant to antibiotics, we have used both (i) an in vitro microaerophilic stationary-phase model (34) in which we treat M. tuberculosis with high-dose rifampin and (ii) a pyrazinamide-induced model in mice (the Cornell model) (25). Rifampin and pyrazinamide were chosen since they are the two antituberculosis drugs with the greatest ability to kill persisting bacilli during the treatment of pulmonary tuberculosis (27, 28). We show here that persistent bacteria have transcriptional activity which indicates an active, though possibly restricted, metabolism.
MATERIALS AND METHODS
M. tuberculosis growth; estimation of viability and incorporation of [3H]uridine after rifampin treatment.
M. tuberculosis was grown in 7H9 medium containing 0.05% Tween 80 supplemented with 10% albumin dextrose complex (Difco Laboratories, Detroit, Mich.) without shaking for 100 days. To obtain evenly dispersed cultures prior to experimental treatment, clumps in the cultures were broken up by vortexing the cultures in the presence of 2-mm-diameter glass beads (Philip Harris Scientific, Staffordshire, United Kingdom) for 5 min, followed by sonication in a water bath sonicator (Branson Ultrasonic B.V.) for 5 min. After addition of rifampin at a final concentration of 100 μg ml−1, the cultures were incubated at 37°C for 5 days. Viability was estimated initially and at daily intervals. The cells were washed by centrifugation three times with phosphate-buffered saline and finally resuspended to the original volume in 7H9 broth. For CFU counts, three 100-μl samples from the appropriate serial 10-fold dilutions were plated onto three one-third segments of plates of 7H11 agar medium supplemented with oleic albumin dextrose complex (Difco). Before rifampin treatment, the plates were usually inoculated from the 10−3 and 10−4 dilution. After addition of rifampin, the neat washed culture (1 ml) was plated. Colonies were counted after incubation of the plates in plastic bags for 3 weeks at 37°C. Broth counts were performed as serial 10-fold dilutions from each of which triplicate 1-ml samples were added to 9 ml of 7H9 medium. At 10-day intervals over a 2-month period of incubation at 37°C, the broth cultures were examined for visible turbidity. Growth of M. tuberculosis in turbid tubes was confirmed by colonial morphology on 7H11 agar plates. The most probable number of viable bacilli was then estimated from the patterns of positive and negative tubes (13). The absence of bacteria other than mycobacteria from the washed cultures was shown by plating on blood agar medium. For incorporation of radioactive uridine, 10 ml of the culture for each time point was washed to remove the remaining rifampin, then resuspended in 10 ml of 7H9 medium, and incubated with [3H]uridine (10 μCi ml−1) for 20 h. [3H]uridine (10 μCi ml−1) was also added to the washed culture, which was further incubated in the presence of rifampin for 20 h. RNA was extracted as described below; [3H]uridine incorporation into total RNA was determined as counts per minute of trichloroacetic acid-precipitated RNA.
Cornell mouse model.
M. tuberculosis was grown in BALB/c mice weighing 18 to 20 g which were infected intravenously with 2 × 105 CFU of a mouse-passaged, virulent H37Rv strain of M. tuberculosis. Spleens and lungs were removed rapidly after sacrifice, and sterile autopsy was performed at a time point designated week −2; portions of the organs were immediately frozen in liquid N2 for subsequent RNA extraction. CFU counts of viable M. tuberculosis were estimated by grinding half of the spleen and lungs in 5 ml of sterile water in motor-driven polytetrafluorethylene-glass grinders. CFU counts were estimated from serial 10-fold dilutions of the homogenate on plates of selective 7H11 medium (Difco), incubated at 37°C for 3 to 4 weeks. At 2 weeks after infection (week 0), a sample of four mice yielded 2 × 107 to 5 × 107 CFU/lung or spleen. Treatment was then given for 14 weeks with 1,000 mg of pyrazinamide and 25 mg of isoniazid per kg of body weight in the pelleted diet, after which the sterile state was reached. Six mice were sacrificed at week 7 to monitor CFU counts. Another eight mice were then sacrificed at week 14; and the organ homogenates were cultured in selective Kirchner liquid medium for 4 weeks, with subsequent subculturing onto selective Löwenstein-Jensen slopes for a further 4 weeks. At each time point, one-fifth portions of half of each of the organs were immediately frozen in liquid N2 for subsequent RNA extraction. Culture media were made selective for M. tuberculosis by the addition of 200 U of polymyxin B ml−1, 100 μg of carbenicillin ml−1, 20 μg of trimethoprim ml−1, and 10 μg of amphotericin B ml−1. After a further 8 weeks during which each of the remaining mice received 0.5 mg of hydrocortisone acetate together with chloramphenicol in the drinking water at 50 mg/kg, 21 of 23 mice yielded positive spleen and lung cultures.
RNA extraction.
Total RNA was extracted from in vitro cultures by the method of Mangan et al. (23). Mycobacterial RNA extraction from organs was performed on individual portions of each organ that had been separately stored frozen in liquid nitrogen by the method of Butcher et al. (2), but employing a prior differential tissue lysis using a reciprocal shaker and 1-mm beads (Hybaid Ltd., Teddington, United Kingdom) (23). RNA was treated with RNase-free DNase I (Life Technologies) to remove contaminating genomic DNA.
Nucleotide sequence accession number.
Gene annotation was according to Cole et al. (3). GenBank accession numbers and nucleotide positions are given for the genes from which PCR primer pairs were designed to be in the coding regions: 16K (AL021899, nucleotides [nt] 16796 to 17230; hspX, Rv2031c), 16S (X52917, nt 1471844 to 1473380; rrnS), sigA (Z96072; nt 21832 to 23418; Rv2703), sigB (Z96072; nt 26458 to 27429; Rv2710), dnaK (Z95324; nt 1526 to 1800; Rv0350), and rpoB (Z95972, nt 9875 to 13393; Rv0667). The sequences of the oligonucleotides are as follows: 16K, 5′-GAAGACGAGATGAAAGAGGGG-3′ and 5′-GTAAGAATGCCCTTGTCGTAGG-3′; 16S, 5′-GCCTGGGAAACTGGGTCTAA-3′ and 5′-TCTCCACCTACCGTCAATCC-3′; rpoB, 5′-GTTCGGGGAGATGGAGTGCT-3′ and 5′-CGTTGCGGGACAGATTGATT-3′; sigB, 5′-AGATCAACGACCTGCTGGAA-3′ and 5′-GGGACAGCCCGAATAGTTTG-3′; sigA, 5′-CCAGCACGAAGCCGCAAC-3′ and 5′-TCATCCCAGACGAAATCACC-3′; dnaK, 5′-ATTGTGCACGTCACCGCCAA-3′ and 5′-ACCGCGGCATCAACCTTGTT-3′. The sensitivity of these primers was determined by measuring their ability to detect M. tuberculosis chromosomal DNA in a DNA PCR. A series of 10-fold dilutions of known concentrations of M. tuberculosis DNA was subjected to PCR amplification with each pair of primers. The sensitivity of the primers was defined as the endpoint of DNA concentration at which the primers could produce product.
RT of RNA samples and PCR.
Total RNA was reverse transcribed essentially as described previously (2) in a total volume of 20 μl containing 0.5 mM each dATP, dCTP, dGTP and dTTP, 2.5 μM reverse primer, 5 mM dithiothreitol, 40 U of RNAsin (Promega), 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, and 200 U of Superscript II (Life Technologies). The RNA was denatured at 85°C for 5 min and then chilled on ice. After addition of the reaction mixture, the RT reaction was carried out at 42°C for 60 min. PCR was performed in a final volume of 50 μl which contained 10 μl of cDNA template, using Taq DNA polymerase (Promega). The reaction mixture contained 1 μM each primer. PCR amplification was carried out for 40 cycles (94°C for 1 min, 58°C for 2 min, and 72°C for 3 min). Each RT-PCR was repeated twice or, for some genes, three times. A limited choice of genes was available due to severely limited amounts of input RNA from these models of persistence. Each RT reaction mixture contained the equivalent RNA from one-fifth of one lung. A 20-μl PCR sample from each reaction was subjected to electrophoresis on a 1.5% agarose gel containing ethidium bromide. Non-reverse-transcribed PCR controls indicated absence of contaminating genomic DNA and that PCR products derived from mRNA.
RESULTS
Isolation of in vitro persistent M. tuberculosis.
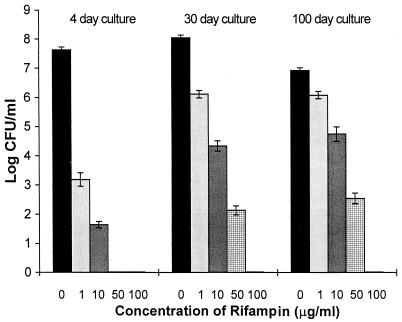
To establish the experimental conditions necessary to select subpopulations of in vitro microaerophilic stationary-phase bacteria that persist during exposure to antibiotic, several preliminary experiments were performed to determine the optimal concentration of rifampin required, duration of exposure, and age of culture (Fig. 1). The log-phase (4-day) cultures (10 ml, 3.9 × 107 CFU ml−1; used as a control for viability) and stationary-phase (30-day [10 ml, 1.12 × 108 CFU ml−1] and 100-day [10 ml, 8.5 × 106 CFU ml−1]) cultures were incubated with 1, 10, 50, and 100 μg of rifampin ml−1, for 5 days. Then the cultures were washed and resuspended in 7H9 broth to the original volume. Viability (CFU) was examined with 1 ml of each 10-ml culture as described in Materials and Methods. As shown in Fig. 1, rifampin at 50 μg ml−1 significantly reduced the CFU counts of stationary-phase cultures and reduced the CFU counts of the log-phase culture to zero. Treatment with 100 μg of rifampin ml−1 resulted in CFU counts of zero in all plate cultures.
FIG. 1.
Effects of different concentrations of rifampin on the viability of log-phase and stationary-phase cultures of M. tuberculosis H37Rv. Log-phase (4-day) and stationary-phase (30- and 100-day) cultures were incubated with 1, 10, 50, and 100 μg of rifampin ml−1 for 5 days. After washing of the cultures, viability was estimated by examination of CFU counts. Values shown are averages of three independent experiments.
To investigate whether all populations of the bacilli in these cultures were killed by 100 μg of rifampin ml−1, resuscitation with fresh 7H9 broth was carried out as described in Materials and Methods in an effort to recover any remaining viable bacilli. As shown in Table 1, broth dilution counts revealed that 0.085, 85, and 850 cells per ml remained after rifampin treatment of 4-, 30-, and 100-day cultures, respectively, indicating that a significant subpopulation from the 100-day cultures persists in the presence of rifampin.
TABLE 1.
Broth dilution counts of rifampin survivors according to age of initial cultures
| Age of initial culture (days) | No. of turbid tubes observed (dilutions)a | Broth count (cells ml−1)b |
|---|---|---|
| 4 | 1 (100), 0 (10−1), 0 (10−2) | 0.085 |
| 30 | 3 (10−1), 2 (10−2), 0 (10−3) | 85 |
| 100 | 3 (10−2), 2 (10−3), 0 (10−4) | 8.51 × 102 |
After exposure to rifampin (100 μg ml−1) for 5 days. These representative results have been confirmed in five independent experiments. Starting culture was 10 ml of 2 × 107 ml−1.
Estimated as described by Fisher and Yates (13).
To confirm that the organisms detected in the broth dilutions in Table 1 were derived from viable, persistent M. tuberculosis rather than contamination by other bacteria, samples from all broth dilutions were plated onto blood agar plates, which were incubated overnight. No other bacteria were grown. Also, the samples from all dilutions were plated onto selective 7H11 agar plates at the end of the 2 months of incubation. All positive broth cultures produced typical M. tuberculosis colonies, containing acid-fast organisms, on the agar plates after 3 weeks of incubation. Negative broth cultures showed no CFU counts. These results indicated that 100 μg of rifampin ml−1 killed all actively replicating bacilli because only a few bacilli survived in log-phase cultures (0.085 from 10 ml of 5 × 107 ml−1 [Table 1]). However, in the stationary-phase cultures treated with rifampin at 100 μg ml−1, although a 5-log killing was observed, nearly 3 logs of bacteria remained viable by broth counts (851 from 10 ml of 5 × 107 ml−1 [Table 1]). The surviving bacilli may be representative of organisms persisting after chemotherapy in vivo. These persistent bacteria are plate count negative but broth dilution count positive.
Identification of phenotypic resistance of persisters to rifampin.
To distinguish between phenotypic and genotypic resistance, we washed the 100-day stationary-phase persisting bacteria three times after rifampin treatment. The washed cultures were diluted 10-fold, and 1 ml of each dilution, including the neat culture, was added to 9 ml of 7H9 broth containing 0.1 μg of rifampin ml−1. A normal MIC for M. tuberculosis in 7H9 medium is approximately 0.1 μg ml−1. Controls were performed by adding 1 ml of each dilution to 9 ml of 7H9 broth without the drug. After 2 months of incubation, no growth was observed in rifampin-containing cultures (n = 3), indicating that the persistent bacilli were sensitive to rifampin after they emerged from stationary phase and started to grow in aerated, fresh broth. Persistent bacilli resuscitated in the absence of rifampin (day 100 [Table 1]) showed viable bacilli by broth dilution counts. These bacteria were negative for rifampin resistance mutations in rpoB as detected both by RT-PCR followed by sequencing and hybridization with oligonucleotide probes specific for the rifampin resistance-associated mutations (Immunogenetics N.V., Rijswijk, The Netherlands) (data not shown). These data indicate that the resistance is phenotypic in this model. This model is operationally defined as a 100-day culture of M. tuberculosis incubated with 100 μg of rifampin ml−1 for 5 days.
RT-PCR analysis of persistent bacilli in the in vitro persistent model.
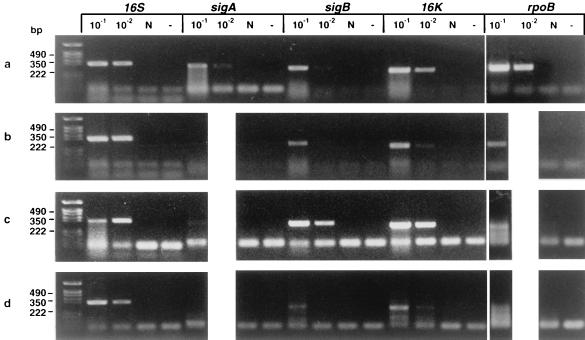
RT-PCR for the following genes was performed: rpoB (encodes the rifampin target, RNA polymerase subunit B), hspX (16-kDa protein [α-crystallin homolog]), rrnS (16S rRNA), and sigA and sigB (two ς70 homologs) (8). sigB may be functionally analogous to stationary-phase sigma factor gene rpoS of E. coli (21) and to M. tuberculosis sigF (6), which is transcribed in the stationary growth phase, but the sequences are quite divergent. Stationary-phase M. tuberculosis is associated with increased expression of the α-crystallin 16-kDa protein (37). Prior to RT-PCR, we determined the sensitivity of primers for the genes described in Materials and Methods. sigA-specific primers are able to detect 107 M. tuberculosis gene copies, the 16S rRNA primer can detect 105 gene copies, and the rest of the primers can detect 106 gene copies. Thus, RT-PCR cannot generate products with the specific primers from the samples containing fewer than 105, 106, or 107 copies of templates, respectively. Total RNA was extracted from the same volume of cultures before and after rifampin treatment in the in vitro persistent model, and this was used for RT-PCR. Figure 2a shows that all of these genes are transcribed in vitro in stationary-phase bacilli. After 5 days of rifampin treatment, which reduced the CFU to zero (Fig. 2b), the 16S rRNA was not diminished and transcripts for rpoB, 16K, and sigB could still be detected, albeit at reduced levels, while the transcript for sigA was not seen. The sequences of the sigA, sigB, and hspX PCR products were verified as M. tuberculosis in origin by commercial sequencing (Cambridge BioScience Ltd., Cambridge, United Kingdom).
FIG. 2.
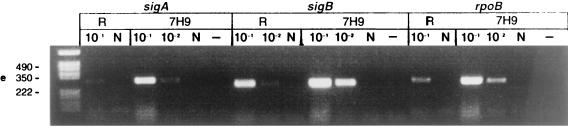
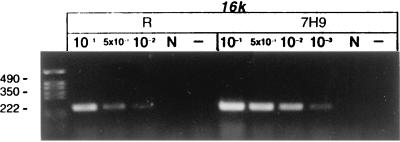
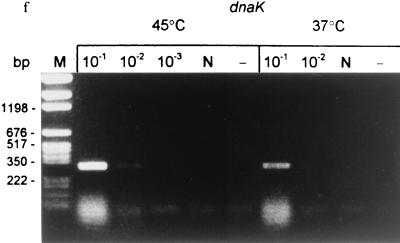
Detection of mRNA in M. tuberculosis H37Rv by RT-PCR before and after treatment with antituberculosis agents in vitro and in vivo. (a) RNA was extracted from 100-day cultures before rifampin treatment (106 CFU per RT reaction). (b) After rifampin (100 μg/ml) treatment for 5 days (0 CFU; same volume of culture used for RT as in panel a). (c) RNA was extracted from infected mice before antituberculosis drug treatment; week 0 (106 CFU per RT). (d) After antituberculosis drug treatment in mice; week 14 (0 CFU; same volume of tissue used for RT as in panel c). (e) The 100-day cultures were treated with rifampin (100 μg/ml) for 5 days. The cells were washed three times to remove rifampin and then incubated with 7H9 broth for 12 h. RT-PCR was performed with sigA, sigB, rpoB, and 16K primers before (R) and after (7H9) resuscitation with 7H9 medium. (f) The 100-day cultures were treated with rifampin for 5 days, washed free of rifampin, and immediately heat shocked at 45°C for 30 min compared to incubation at 37°C for 30 min. Detected genes are shown above lanes; the PCR product sizes are as follows: sigA, 306 bp; sigB, 273 bp; rpoB, 307 bp; dnaK, 275 bp; 16K (hspX), 242 bp; 16S, 336 bp. 10−1, 5 × 10−1, 10−2, and 10−3 indicate that 10-, 50-, 100-, and 1,000-fold-diluted cDNAs were used for PCR. N, DNase I-treated RNA but no RT enzyme. −, PCR control with each primer and 10 μl of water instead of template. The following limits of detection (gene copies) were estimated by DNA PCR: 16S, 105 (equivalent to 100 CFU of a log-phase growth culture); sigA, 107; sigB, 106; 16K, 106; rpoB, 106; dnaK, 106 (equivalent to ∼104 CFU).
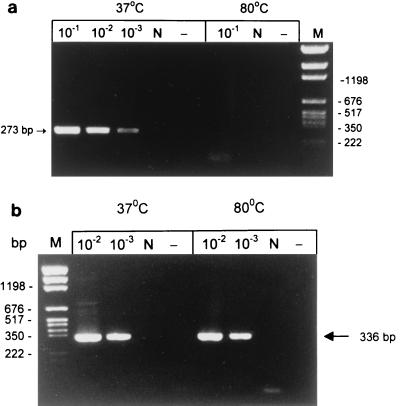
To provide further evidence that the mRNA present in persistent bacilli was derived from metabolically active bacilli rather than from dead cells, RNA was extracted from a 100-day culture which was first killed by heating at 80°C for 30 min followed by incubation of the cultures at room temperature for 24 h to ensure complete degradation of the mRNA. RT-PCR was performed to detect hspX, sigA, and sigB mRNAs and 16S rRNA. No hspX, sigA, or sigB mRNA was detected in the heat-killed culture; however, the 16S rRNA was still detectable, indicating its relative stability. Results for sigB mRNA and 16S rRNA are shown in Fig. 3. No viable counts were obtained from the heat-killed culture, as determined by both plate counts and broth dilution counts. RT-PCR was also performed with RNA extracted from H2O2 (20 mM for 10 h)- and acetic acid (pH 3.5 for 10 h)-killed bacilli. No mRNA was detected in these killed bacteria, while 16S rRNA remained unchanged (data not shown). These results confirmed that mRNA is a good correlate of viability compared to rRNA and that mRNA detected in rifampin-treated cultures was derived from living persistent organisms.
FIG. 3.
mRNA as a correlate of viability, analyzed by limiting dilution RT-PCR of heat-killed and viable 100-day cultures of M. tuberculosis. (a) sigB mRNA; (b) 16S rRNA. Lanes N and −, as in Fig. 2; lane M, molecular weight markers.
Persistent bacilli are environmentally responsive.
Further confirmation that persistent bacilli are alive and retain the potential for rapid metabolic responses to environmental change came from the examination of mRNA from rifampin-treated cultures to which fresh 7H9 medium was added. As shown in Fig. 2e, the levels of the hspX, rpoB, and sigA mRNAs increased 5- to 10-fold after 12 h of resuscitation. In Fig. 2e, strong sigA and sigB signals were observed at 10−1 dilution, but the signal at the 10−2 dilution was unexpectedly low. This emphasizes that RT-PCR is, at best, only semiquantitative. RT-PCR was performed to detect dnaK mRNA after the bacilli were exposed to elevated temperature. Total RNA was extracted from the washed cultures before and immediately after heat shock at 45°C for 30 min. As shown in Fig. 2f, dnaK mRNA was detectable from the persistent bacilli at 37°C. In a temperature shift from 37 to 45°C, dnaK mRNA increased 10-fold, indicating that these persistent bacilli were responsive to heat shock.
Incorporation of [3H]uridine by persistent bacilli.
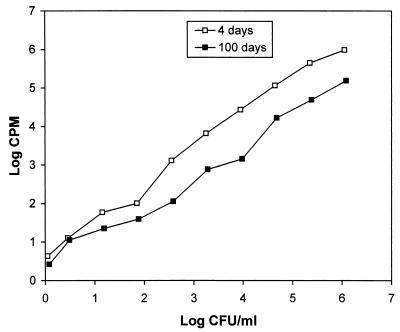
We performed an assay to measure the incorporation of [3H]uridine into RNA as a correlate of metabolic and transcriptional activity in persistent bacilli. So as to be able to accurately compare uridine uptake rates between low numbers of persisting bacteria in a rifampin-treated culture with the uptake rate of rapidly growing bacteria, the quantitative relationship between bacterial numbers and uridine incorporation was determined. Log-phase (4 days with an initial count of 1.12 × 106 CFU ml−1) and stationary-phase (100 days with an initial count of 1.21 × 106 CFU ml−1) cultures were serially diluted in fivefold steps and incubated in the absence of rifampin with [3H]uridine (10 μCi ml−1) for 20 h (Fig. 4). [3H]uridine incorporation was proportionally reduced as the cell numbers decreased as shown in Fig. 4. Above bacterial cell densities of 2 log10 CFU ml−1, the incorporation of [3H]uridine into RNA by 100-day cultures is approximately 15% of the 4-day culture values for the same number of viable organisms (CFU counting). This indicates active but reduced metabolism in stationary-phase bacteria.
FIG. 4.
Kinetic analysis of [3H]uridine incorporation by log-phase and stationary-phase M. tuberculosis. Log-phase (4-day) and stationary-phase (100-day) cultures were serially diluted fivefold. [3H]uridine was added to the cultures, and RNA was extracted after 20 h of incubation. The results were confirmed in three independent experiments.
To measure uridine uptake rates in the subpopulation of rifampin-treated persisting organisms, we performed a series of experiments (Table 2) in which rifampin was added to a 100-day culture (6.6 × 105 CFU ml−1) at a final concentration of 100 μg ml−1 for 5 days. At 1-day intervals, a sample of the culture was washed and incubated with [3H]uridine (10 μCi ml−1). [3H]uridine was also added to a 5-day rifampin-treated culture in the presence of the drug; heat-killed (80°C for 30 min) cultures were included as background control. Viability was estimated by CFU counts and broth dilution counts. As shown in Table 2, after 1 to 5 days of rifampin treatment, the CFU counts were reduced to zero, but about 103 cells ml−1 (2.93 log10 at 95% confidence limits; 2.29 to 3.51) remained from 3.8 × 107 cells ml−1 (7.45 log10; 6.17 to 8.73) at day 5, as estimated by triplicate broth counts. This result agrees with the 100-day culture viability data in Table 1 and confirms the existence of a subpopulation of persisters in this model.
TABLE 2.
Incorporation of [3H]uridine into M. tuberculosis after addition of rifampin
| Days in rifampin | Plate count (CFU ml−1) | Broth count (cells ml−1) | [3H]uridine (mean cpm ± SD)a |
|---|---|---|---|
| 0 | 6.6 × 105 | 3.8 × 107 | 74,682 ± 311 |
| 1 | 0 | 7.89 × 102 | 2,228 ± 33 |
| 2 | 0 | 1.73 × 103 | 2,316 ± 15 |
| 3 | 0 | 1.73 × 103 | 2,430 ± 23 |
| 4 | 0 | 3.8 × 103 | 2,318 ± 18 |
| 5 | 0 | 1.73 × 103 | 2,388 ± 31 |
| 5b | 0 | 1.73 × 103 | 518 ± 9c |
| 0 | 0 (heat killed) | 0 | 180 ± 11c |
Mean of six representative experiments with triplicate counting, except for the heat-killed test, in which case results were confirmed in three independent experiments.
The culture was incubated with [3H]uridine (10 μCi ml−1) in the presence of rifampin for 20 h, and RNA was extracted as described in Materials and Methods.
P < 0.0001.
In the presence of rifampin, the persistent bacilli incorporated 338 cpm of [3H]uridine (518 − 180 cpm background [heat-killed control]) (Table 2). The ratio between the [3H]uridine incorporation of persisters (at 5 days) and the incorporation of log-phase organisms (from the linear range in Fig. 4) is 338/968,500 = 3.490 × 10−4. The corresponding ratio between the broth counts of persisters and the plate counts of log-phase organisms is 1.73 × 103/1.12 × 106 = 1.545 × 10−3. This indicates that the persisters were incorporating uridine at about one-quarter [3.490/(1.545 × 10) = 0.23] of the rate of the same number of log-phase bacteria. This agrees fairly well with data derived from Fig. 4, in which the incorporation of [3H]uridine into stationary-phase cultures was about 15% of that in log-phase cultures. Thus, the rifampin persisters have uridine uptake rates similar to those of the bulk of the stationary-phase bacilli and represent a subpopulation of bacilli in stationary-phase cultures.
Broth counts and CFU counts are equivalent in log-phase cultures but vary by nearly 1.7 logs in 100-day cultures (Table 2). This difference in 100-day cultures probably reflects presence of the subpopulation of persisters (described above) that are CFU negative. After resuscitation of persisters with fresh 7H9 medium, irrespective of the duration of treatment with rifampin (1 to 5 days), the incorporation of [3H]uridine by persisters increased by 82%, from 338 to 2,156 cpm (average of 2,336, minus background of 180), a transcriptional activity similar to that for log-phase growth. This indicated that the persisters were viable, with reduced metabolic and transcriptional activity, and rapidly reinitiated growth in fresh broth, as measured both by increased uridine uptake and increased levels of mRNA (Fig. 2e).
RT-PCR analysis of persistent bacilli in the Cornell model.
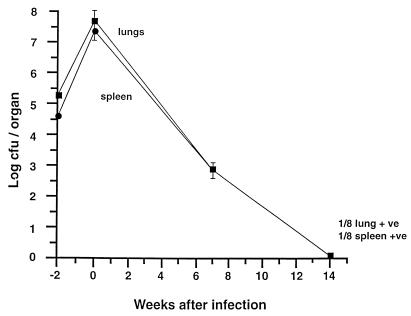
To establish whether a similar situation exists in vivo, we chose the murine Cornell model. In our experiments (Fig. 5), the lungs and spleens of the mice had high plate viable counts before antibiotic treatment and low counts after 44 days of treatment. They reached a sterile state with negative lung and spleen cultures on solid and liquid media at 14 weeks, when treatment was completed. Since it was difficult to measure the concentration of bacterial RNA extracted from mouse lungs and spleens which were M. tuberculosis culture negative after 14 weeks of chemotherapy, we used RNA extracted from equal volumes of mouse organs before and after chemotherapy for RT-PCR. RT-PCR of RNA from acutely infected lungs (5 × 107 CFU/lung [Fig. 5]) removed before treatment revealed transcripts for rpoB, sigA, sigB, 16K, and 16S rRNA (Fig. 2c). Significantly, bacterial mRNA for sigB, rpoB, and 16K was detected in lungs after 14 weeks of chemotherapy, and the level of 16S rRNA was reduced, as seen by RT-PCR at 10−2 dilution (Fig. 2d), compared to the prechemotherapy level. The sigA transcripts were not detectable in either in vitro persisters or mouse persisters (Fig. 2c and d, lanes sigA 10−1), probably due to low numbers of bacteria (see Discussion). The rpoB lanes showed multiple low-molecular-weight bands with no detectable correct-size band. Also in Fig. 2c, the 16S signal was stronger in 10−2 dilution than that in 10−1 dilution. These aberrant results may be due to the interference of residual mouse RNA with the RT-PCR. RT-PCR was also performed with the RNA extracted from spleens; the patterns of RT-PCR amplification were similar to those of the lungs. Here only the lung data are presented. All lungs and spleens used for RT-PCR after the 14-week chemotherapy treatment were preconfirmed as culture negative determined by CFU counts and broth inoculation counts. RT-PCR was also performed with RNA extracted from noninfected mouse lungs as a control for cross-reaction with mouse RNA. No PCR products were amplified (data not shown). The sequence of the hspX PCR product was verified as described above. All controls were performed to ensure that RT-PCR products derived from mRNA and not from residual genomic DNA in the RNA preparation (Fig. 2, lanes N). The entire in vivo experiment was undertaken twice with the same results each time; in the second experiment, the RT-PCR was performed blind by two different workers who obtained the same results.
FIG. 5.
Viability of M. tuberculosis H37Rv in vivo before and after treatment with antituberculosis agents in the mouse Cornell model. The results of a single experiment are shown, with viability expressed as log CFU counts per organ (spleen or lung). Mice were infected intravenously at week −2, and the infection allowed to progress for 2 weeks prior to treatment with pyrazinamide and isoniazid for 14 weeks (weeks 0 to 14). At week 14, 2 mice yielded a positive organ culture, one for lung only (1/8 lung +ve) and one for spleen only (1/8 spleen +ve). The remaining 6 mice at week 14 + 1 were all culture negative, and their spleens and lungs were frozen in one-fifth portions for RNA extraction and RT-PCR analysis. Not shown is the bacteriological relapse in 21 of 23 mice after steroid treatment given at week 14 + 8, indicating restoration of culturability and hence rescue from dormancy.
DISCUSSION
We have used two models of phenotypic persistence in M. tuberculosis to measure mRNA transcripts by RT-PCR in order to investigate the transcriptional activity and hence the physiological state of persistent bacilli. In the first model, the organisms are cultured long term in a microaerophilic gradient (34), where the stationary-phase organisms are viable; they are resistant to the MIC of rifampin (0.1 μg ml−1). While log-phase bacilli are very sensitive to antituberculosis agents, stationary-phase bacteria are partially tolerant to these drugs (7, 27, 35), and a subpopulation of persistent bacilli are not killed by any known antituberculous agents (28). The rationale for the in vitro approach taken in this study was to treat microaerophilic stationary-phase cultures with rifampin, which kills the replicating bacilli; the organisms which remain after chemotherapy (∼0.005%) we term persisters. To start with, we established the correct conditions for the assay, in particular, the concentration of rifampin and the duration of chemotherapy. To avoid the generation of genetic resistance by selective pressure (12) during the experiments, we used high doses of the antibiotic for short periods of time. Rifampin is particularly suitable since it is bactericidal and starts killing M. tuberculosis within an hour of exposure. Importantly, it is one of the most active sterilizing drugs for chemotherapy of tuberculosis (28). We treated the stationary-phase bacteria with a high level of rifampin (100 μg ml−1) which reduces plate counts to zero but results in a small number of persisting organisms which are detectable and quantifiable (13) by broth dilution counting. The second model is murine chronic tuberculosis, which is induced by chemotherapy (Cornell model [25]). Mice are infected with M. tuberculosis and then treated for 14 weeks with high-dosage pyrazinamide (one of the most active sterilizing drugs) and isoniazid, which reduce the broth and plate counts in the spleens and lungs to zero (Fig. 5). However, mice in this sterile state contain large amounts of M. tuberculosis DNA, which is estimated to be equivalent to 105 bacteria per organ (5). Upon treatment of mice with steroids, spleens and lungs become culture positive again, indicating reactivation from a nonculturable and drug-insensitive (tolerant) state. Both models mirror chemotherapy and subsequent relapse of tuberculosis in humans.
In the in vitro persistent model, RNA was detected from bacilli which survived the treatment with rifampin. The mRNA detected was not from dead bacilli since it disappeared from dead organisms (Fig. 3). We have previously measured the half-lives of mRNA in both log-phase and long-term stationary-phase M. tuberculosis cultures and shown them to be growth phase independent, between 2 and 3 min for 16K and sigB and >40 min for sigA (18, 19). We also compared the kinetics of loss of RT-PCR signals between 4-day and 30-day cultures. There was no difference of RT-PCR signal loss between log-phase and stationary-phase cultures (data not shown). Thus, it is unlikely that the RT-PCR results (Fig. 2b) are accounted for by long-term stable mRNA. These results suggest that ongoing transcriptional (metabolic) activity is retained in persistent organisms. Although sigA mRNA is constitutively transcribed and has a long half-life (19), it is not detected in persistent bacilli. This result is most likely due to the low sensitivity of the primers in specimens with low bacilliary load so that the sigA mRNA transcripts become undetectable. The limit of detection by DNA PCR for sigA was 107 genome equivalents, at least a logfold less sensitive than for the other mRNA transcripts (see the legend to Fig. 2). Alternatively, the transcription of sigA might be turned off in these persistent organisms. The data cannot distinguish between these possibilities at such low bacillary counts. Importantly, the persistent organisms incorporated [3H]uridine into RNA even in the presence of a high concentration of rifampin, which confirmed the RT-PCR data and indicated that active transcription took place in these persistent bacilli but at a reduced rate.
The persisters were very responsive to changes in their environment. Upon resuscitation of 100-day rifampin-treated cultures with fresh 7H9 medium, transcription of the bacilli increased within 12 to 20 h, as demonstrated by 5- to 10-fold increases in mRNA by RT-PCR (Fig. 2e) and by 5-fold increases in incorporation of [3H]uridine into M. tuberculosis RNA (Table 2). The transcription of dnaK increased in a temperature shift from 37 to 45°C (Fig. 2f). These results indicate that the in vitro persisters not only were metabolically active but also retained their potential for environmental responsiveness.
Whether bacilli which persist in human tuberculosis infection and in murine chronic animal models are metabolically active has been a crucial open question for many years (10, 11). Although PCR amplification of dormant M. tuberculosis DNA in the Cornell model indicated the presence of ∼105 genome equivalents per lung or spleen (5), it has not been possible to distinguish between dead bacilli and living organisms. A recent study (29) has shown mRNA for 85 antigen can be detected in viable but none culturable bacilli in the Cornell model, indicating that the persistent bacteria are transcriptionally active. Here we show that some of the M. tuberculosis mRNA (hspX and sigB) was detectable in mouse tissues which were negative for tubercle bacilli by CFU counts and broth inoculation counts after 14 weeks of chemotherapy (the sterile state). It is surprising to find any mRNA at all after 14 weeks of chemotherapy since mRNA is highly labile and depleted within killed bacilli within a very short time (18, 19). Since it was not experimentally possible to measure the half-lives of these mRNAs or the incorporation of [3H]uridine into RNA in the infected mouse, we cannot discriminate between active transcription of unstable mRNA and stable mRNAs in Cornell model persisters. Nevertheless, the data suggest that substantial numbers of nonculturable persistent bacilli are present in infected tissues postchemotherapy. These bacteria may be metabolically active because they contain mRNA and rRNA. The data for the Cornell model parallel those for the in vitro long-term stationary microaerophilic model and argue strongly for the hypothesis that M. tuberculosis can adapt to a nonculturable yet metabolically active state in which mRNA is detected and low levels of active transcription may be measured. We propose that some form of an adaptive gene expression response, yet to be characterized, allows a subpopulation of bacteria to persist within the host without being killed and that this state confers resistance to high concentrations of antimycobacterial agents. Thus, persisting M. tuberculosis may exist in some physiological form in which limited metabolism occurs, presumably with little or no cell turnover, accounting for tolerance to conventional antibiotic treatment. The mechanism of tolerance to rifampin (a transcriptional inhibitor) has not been investigated. It has been shown that tubercle bacilli incubated under long-term microaerophilic and anaerobic conditions developed a very thick cell wall outer layer (4). A change in drug permeability may be one of the possible mechanisms of bacterial tolerance to antibiotics. In addition, the use of alternative sigma factors (such as SigB) in the persistent state might confer differential sensitivity of the RNA polymerase holoenzyme to rifampin as shown for E. coli (36), thus allowing continued but modified transcription. In this state, active transcription continues since labile mRNA is detectable and uridine incorporation into RNA continues even in the presence of rifampin. These persisting bacilli also maintain their environmental responsiveness (Fig. 2e and f) necessary for regrowth.
The recent demonstration that HSP-65 DNA vaccination could suppress bacteriological relapse in M. tuberculosis-infected mice after 12 weeks of chemotherapy (22) and the relapse rate for tuberculosis of 2 to 5% per annum in AIDS patients suggests an important role for immunity in the maintenance of persistence. The experiments described here provide an approach to study gene expression associated with persistence as well as to investigate the mechanisms of antibiotic tolerance (phenotypic resistance) in persisters. Furthermore, since antimicrobial agents require some level of bacterial metabolism to be effective, a scientific rationale has now been provided for the development of chemotherapeutic agents which inhibit transcription in persisters. Such agents should be able to kill organisms persisting in the presence of rifampin or pyrazinamide, the most actively sterilizing of current drugs (28), and therefore substantially shorten the course of the chemotherapy of human tuberculosis. They might also be useful for the chemoprophylaxis of latent bacilli, which currently infect one-third of the world's population.
ACKNOWLEDGMENTS
We thank the British Medical Research Council for financial support (grants G9328099, G9814061, and G9525858) and The Sanger Centre for bioinformatics support prior to completing and making available the M. tuberculosis genome sequence.
REFERENCES
- 1.Bloom B R, McKinney J D. The death and resurrection of tuberculosis. Nat Med. 1999;5:872–874. doi: 10.1038/11309. [DOI] [PubMed] [Google Scholar]
- 2.Butcher P D, Mangan J A, Monahan I M. Intracellular gene expression: analysis of RNA from mycobacteria in macrophages using RT-PCR. Methods Mol Biol. 1998;101:285–306. doi: 10.1385/0-89603-471-2:285. [DOI] [PubMed] [Google Scholar]
- 3.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham A F, Spreadbury C L. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton α-crystallin homolog. J Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Wit D, Wootton M, Dhillon J, Mitchison D A. The bacterial DNA content of mouse organs in the Cornell model of dormant tuberculosis. Tuber Lung Dis. 1995;76:555–562. doi: 10.1016/0962-8479(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 6.DeMaio J, Zhang Y, Ko C, Young D B, Bishai W R. A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1996;93:2790–2794. doi: 10.1073/pnas.93.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickinson J M, Mitchison D A. Experimental models to explain the high sterilizing activity of rifampicin in the chemotherapy of tuberculosis. Am Rev Respir Dis. 1981;123:367–371. doi: 10.1164/arrd.1981.123.4.367. [DOI] [PubMed] [Google Scholar]
- 8.Doukhan L, Predich M, Nair G, Dussurget O, Mandic-Mulec I, Cole S T, Smith D R, Smith I. Genomic organization of the mycobacterial sigma gene cluster. Gene. 1995;165:67–70. doi: 10.1016/0378-1119(95)00427-8. [DOI] [PubMed] [Google Scholar]
- 9.Dupray E, Caprais M P, Derrien A, Fach P. Salmonella DNA persistence in natural seawaters using PCR analysis. J Appl Microbiol. 1997;82:507–510. doi: 10.1046/j.1365-2672.1997.00143.x. [DOI] [PubMed] [Google Scholar]
- 10.Gangadharam P R J. Mycobacterial dormancy. Tuber Lung Dis. 1995;76:477–479. doi: 10.1016/0962-8479(95)90521-9. [DOI] [PubMed] [Google Scholar]
- 11.Grange J M. The mystery of the mycobacterial ‘persistor.’. Tuber Lung Dis. 1992;73:249–251. doi: 10.1016/0962-8479(92)90128-7. [DOI] [PubMed] [Google Scholar]
- 12.Guillemot D, Carbon C, Balkau B, Geslin P, Lecoeur H, Vauzelle-Kervroedan F, Bouvenot G, Eschwege E. Low dosage and long treatment duration of beta-lactam: risk factors for carriage of penicillin-resistant Streptococcus pneumoniae. JAMA. 1998;279:365–370. doi: 10.1001/jama.279.5.365. [DOI] [PubMed] [Google Scholar]
- 13.Fisher R A, Yates F. Statistical tables for biological, agricultural and medical research, Table VIII2. Edinburgh, United Kingdom: Oliver and Boyd; 1963. p. 66. [Google Scholar]
- 14.Fox W. Whither short-course chemotherapy? Br J Dis Chest. 1981;75:331–357. doi: 10.1016/0007-0971(81)90022-x. [DOI] [PubMed] [Google Scholar]
- 15.Hellyer T J, DesJardin L E, Hehman G L, Cave M D, Eisenach K D. Quantitative analysis of mRNA as a marker for viability of Mycobacterium tuberculosis. J Clin Microbiol. 1999;37:290–295. doi: 10.1128/jcm.37.2.290-295.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herman L. Detection of viable and dead Listeria monocytogenes by PCR. Food Microbiol. 1997;14:103–110. [Google Scholar]
- 17.Hong Kong Tuberculosis Treatment Services and East African and British Medical Research Councils. First-line chemotherapy in the retreatment of bacteriological relapses of pulmonary tuberculosis following a shortcourse regimen. Lancet. 1976;1:162–163. [PubMed] [Google Scholar]
- 18.Hu Y, Coates A R M. Transcription of the stationary-phase-associated hspX gene of Mycobacterium tuberculosis is inversely related to synthesis of the 16-kilodalton protein. J Bacteriol. 1999;181:1380–1387. doi: 10.1128/jb.181.5.1380-1387.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y, Coates A R M. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J Bacteriol. 1999;181:469–476. doi: 10.1128/jb.181.2.469-476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kochi A. The global tuberculosis situation and the new control strategy of the World Health Organisation. Tubercle. 1991;72:1–6. doi: 10.1016/0041-3879(91)90017-m. [DOI] [PubMed] [Google Scholar]
- 21.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 22.Lowrie D B, Tascon R E, Bonata V L D, Lima V M F, Faccioli L H, Stravropoulos E, Colston M J, Hewinson R G, Moelling K, Silva C L. Therapy of tuberculosis in mice by DNA vaccination. Nature. 1999;400:269–271. doi: 10.1038/22326. [DOI] [PubMed] [Google Scholar]
- 23.Mangan J A, Sole K M, Mitchison D A, Butcher P D. An effective method of RNA extraction from bacteria refractory to disruption, including mycobacteria. Nucleic Acids Res. 1997;25:675–676. doi: 10.1093/nar/25.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masters C I, Shallcross J A, Mackey B M. Effect of stress treatments on the detection of Listeria monocytogenes and enterotoxigenic Escherichia coli by the polymerase chain reaction. J Appl Bacteriol. 1994;77:73–79. doi: 10.1111/j.1365-2672.1994.tb03047.x. [DOI] [PubMed] [Google Scholar]
- 25.McCune R M, Feldmann F M, McDermott W. Microbial persistence. II. Characteristics of the sterile state of tubercle bacilli. J Exp Med. 1966;123:469–486. doi: 10.1084/jem.123.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKillip J L, Jaykus L-A, Drake M. rRNA stability in heat-killed and UV-irradiated enterotoxigenic Staphylococcus aureus and Escherichia coli O157:H7. Appl Environ Microbiol. 1998;64:4264–4268. doi: 10.1128/aem.64.11.4264-4268.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchison D A, Selkon J B. The bactericidal activities of antituberculous drugs. Am Rev Tuberc. 1956;74(Suppl.):109–116. doi: 10.1164/artpd.1956.74.2-2.109. [DOI] [PubMed] [Google Scholar]
- 28.Mitchison D A. Role of individual drugs in the chemotherapy of tuberculosis. Int J Tuberc Lung Dis. 2000;4:796–806. [PubMed] [Google Scholar]
- 29.Pai S R, Actor J K, Sepulveda E, Hunter R L, Jr, Jagannath C. Identification of viable and non-viable Mycobacterium tuberculosis in mouse organs by directed RT-PCR for antigen 85B mRNA. Microb Pathog. 2000;28:335–342. doi: 10.1006/mpat.2000.0353. [DOI] [PubMed] [Google Scholar]
- 30.Parrish N M, Dick J D, Bishai W R. Mechanisms of latency in Mycobacterium tuberculosis. Trends Microbiol. 1998;6:107–112. doi: 10.1016/s0966-842x(98)01216-5. [DOI] [PubMed] [Google Scholar]
- 31.Patel B K R, Banerjee D K, Butcher P D. Determination of M. leprae viability by the polymerase chain reaction amplification of 71kDa heat shock protein mRNA. J Infect Dis. 1993;168:799–800. doi: 10.1093/infdis/168.3.799. [DOI] [PubMed] [Google Scholar]
- 32.Sheridan G E C, Masters C I, Shallcross J A, Mackey B M. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl Environ Microbiol. 1998;64:1313–1318. doi: 10.1128/aem.64.4.1313-1318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van der Vliet G M, Schepers P, Schukkink R A, Van Gemen B, Klatser P R. Assessment of mycobacterial viability by RNA amplification. Antimicrob Agents Chemother. 1994;38:1959–1965. doi: 10.1128/aac.38.9.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wayne L G. Dynamics of submerged growth of Mycobacterium tuberculosis under aerobic and microaerophilic conditions. Am Rev Respir Dis. 1976;114:807–811. doi: 10.1164/arrd.1976.114.4.807. [DOI] [PubMed] [Google Scholar]
- 35.Wayne L G, Sramek H A. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:2054–2058. doi: 10.1128/aac.38.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wegrzyn A, Szalewska-Palasz A, Blaszczak A, Liberek K, Wegrzyn G. Differential inhibition of transcription from sigma70- and sigma32-dependent promotors by rifampicin. FEBS Lett. 1998;440:172–174. doi: 10.1016/s0014-5793(98)01449-5. [DOI] [PubMed] [Google Scholar]
- 37.Yuan Y, Crane D D, Barry C E. Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial α-crystallin homolog. J Bacteriol. 1996;178:4484–4492. doi: 10.1128/jb.178.15.4484-4492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]