Abstract
The skeleton of reef-building coral harbors diverse microbial communities that could compensate for metabolic deficiencies caused by the loss of algal endosymbionts, i.e., coral bleaching. However, it is unknown to what extent endolith taxonomic diversity and functional potential might contribute to thermal resilience. Here we exposed Goniastrea edwardsi and Porites lutea, two common reef‐building corals from the central Red Sea to a 17-day long heat stress. Using hyperspectral imaging, marker gene/metagenomic sequencing, and NanoSIMS, we characterized their endolithic microbiomes together with 15N and 13C assimilation of two skeletal compartments: the endolithic band directly below the coral tissue and the deep skeleton. The bleaching-resistant G. edwardsi was associated with endolithic microbiomes of greater functional diversity and redundancy that exhibited lower N and C assimilation than endoliths in the bleaching-sensitive P. lutea. We propose that the lower endolithic primary productivity in G. edwardsi can be attributed to the dominance of chemolithotrophs. Lower primary production within the skeleton may prevent unbalanced nutrient fluxes to coral tissues under heat stress, thereby preserving nutrient-limiting conditions characteristic of a stable coral-algal symbiosis. Our findings link coral endolithic microbiome structure and function to bleaching susceptibility, providing new avenues for understanding and eventually mitigating reef loss.
Subject terms: Microbial ecology, Microbial ecology
Introduction
The health and ecological success of reef-building corals are supported by a wide range of symbiotic microorganisms, prompting scientists to consider this consortium as a discrete biological unit—the coral holobiont [1, 2]. The past thirty years of research have provided important insights into the diversity, biogeography, and functional roles of the microorganisms associated with corals. The most well-known coral symbionts are dinoflagellates from the Symbiodiniaceae family [3], which fulfill most corals’ energy requirements [4]. These microalgae are the photosynthetic engines of the reef, powering the metabolically expensive process of coral calcification [4]. In addition to Symbiodiniaceae, coral tissues also harbor an array of protists, fungi, bacteria, archaea, and viruses, which play numerous roles in coral health [5, 6]. These other microorganisms have been implicated in the cycling of essential nutrients [7–11], production of antimicrobial and antioxidant compounds [12–14], protection against UV radiation [15], and coral larval settlement [16, 17]. Although interspecies interactions occurring in the coral tissue and mucus are the focus of many studies, these compartments are usually less than 3 mm thick and, metaphorically speaking, represent the tip of the iceberg with respect to the volume of a coral [18, 19]. Indeed, the vast expanse of the underlying skeleton harbors an abundance of endolithic microorganisms, but the functional roles of these endoliths and their contribution to the health of the coral holobiont remain poorly understood [18, 19].
The most common photosynthetic organisms in the coral skeleton belong to a taxonomically diverse group of filamentous green algae from the genus Ostreobium [20, 21], which form dense bands below the tissue of many coral species [22]. Members of this genus can rapidly colonize the skeleton of coral juveniles following their settlement [23]. Under normal conditions, these photosynthetic algae receive very low light levels [24] and are well adapted to photosynthesize in near-darkness [25]. However, when corals bleach following environmental stress, the expulsion of the intracellular Symbiodiniaceae cells from coral tissues results in an increase in the amount of light reaching Ostreobium, enabling these endolithic algae to rapidly photo-acclimate and bloom [26–28]. Endoliths can directly interact with the coral tissue through the transfer of photosynthates and constitute a source of energy or nutrients for the host during environmental stress [29–31]. In addition, the prokaryotic communities inhabiting the coral skeleton are diverse and spatially structured, exhibiting high levels of intra-colony heterogeneity [32, 33]. These endoliths likely contribute to nitrogen, sulfur, and carbon cycling [34], and may be an important source of limiting nutrients for the overlying coral tissue [35]. Endolithic communities therefore potentially play an underacknowledged but fundamental role in the health of their coral hosts.
Unlike the microorganisms associated with coral tissues, endolithic communities seem less sensitive to specific environmental perturbations, such as ocean acidification [32]. However, with the exception of stimulated endolithic photosynthesis during coral bleaching, the response of the endolithic communities to heat stress in terms of community structure, functional potential, and nutrient cycling is currently unknown. Since 2015, more than 75% of reefs worldwide have been affected by mass bleaching events [36], and nearly half of the coral cover has been lost since the mid-80s on the largest reef system in the world, the iconic Great Barrier Reef [37, 38]. Given the escalating impact of marine heatwaves and rising sea surface temperature on coral reef assemblages [39], understanding the microbial processes underpinning coral health, including the overlooked endolithic communities, constitutes an important research priority.
Here we combine metagenomics with high-resolution chemical imaging (NanoSIMS) to characterize the metabolic capacity and putative contribution of endolithic microbes to coral holobiont functioning during heat stress. We compared the metabolic dynamics under heat stress of two species of massive reef-building corals, Goniastrea edwardsi and Porites lutea, which are abundant in the central Red Sea and common across the Indo-Pacific [40]. Both species are among the most environmentally resilient corals in the Indo-Pacific [41], and in particular, the genus Porites is regarded to have high thermal tolerance [42, 43]. These two species have different skeletal properties that likely affect the structure and functions of their endolithic communities with G. edwardsi exhibiting a denser and less porous skeleton compared to P. lutea [44]. We aimed to (i) taxonomically and functionally compare the endolithic communities of these two corals species; (ii) determine whether experimentally induced heat stress differentially impacts these communities taxonomically and functionally, in particular with regards to pathways involved in energy acquisition and cycling; and (iii) quantify the impact of heat stress on carbon and nitrogen assimilation by endolithic microbes.
Methods
Coral sampling & heat stress experiment
Corals were collected using SCUBA from a sheltered inshore reef off KAUST (Abu Shoosha reef: 22°18’16.3”N, 39°02’57.7”E, on 04 February 2019), from approximately 5–8 m of water depth. In total, six colonies of P. lutea and G. edwardsi were collected. All collected coral colonies showed prominent bands in their skeleton. Coral colonies were transferred to the Coastal and Marine Resources Core Lab (CMOR) at KAUST and fragmented directly after collection. Two fragments (one for each treatment: “control” and “heat stress”) were generated for each of the six colonies, resulting in a total of 24 fragments. As both collected coral species displayed massive growth forms, the skeletal areas that became exposed after fragmentation were sealed off with two-component epoxy putty to ensure that biotic and abiotic conditions of the skeleton were not altered by the direct contact with seawater or increases in light availability. All fragmented corals were maintained in a shared 100 L aquarium for a four-day acclimation phase and subsequently distributed into individual flow-through aquaria (100 L each; each aquarium containing one colony fragment of each species) just before the experiment. All aquaria were continuously supplied with sand-filtered seawater freshly collected from a near-shore reef and equipped with 600 W aquaria heaters (Schego, Offenbach, Germany) and Radion light systems (Ecotech Marine Inc., Bethlehem, PA). During the initial acclimation, seawater temperatures and light levels used during the experiment mimicked the ambient conditions recorded at nearby shallow reefs at the depth of coral collection: 27 °C and 750 µmol photons m−2 s−1 (at peak irradiation) in a 12:12 h light:dark cycle. After the initial acclimation phase, seawater temperature was ramped up to 33 °C in half of the aquaria over the course of 3 days (i.e., an incremental increase by 1 °C every 12 h; subsequently referred to as “heat stress” treatment), while the other half of the aquaria was maintained at a constant 27 °C (“control” treatment). Heat stress temperature corresponds to the absolute summer maximum temperature recorded in 2017 [45] and the control corresponds to the measured in situ temperature when corals were collected. The temperature treatments were maintained until Porites sp. fragments were visibly bleached at 33 °C, which occurred after 17 days (14 days at maximum temperature). Of note, G. edwardsi did not exhibit any signs of bleaching at that time.
Stable isotope labelling
On day 17 of the experiment, corals were collected from both temperature treatments and subjected to an incubation period of 24 h in 330 ml beakers filled with isotope-enriched artificial seawater (n = 6 per species and treatment). Artificial seawater composition (salinity = 39 PSU, pH = 8.1, 492.5 mM NaCl, 46.23 mM MgCl2, 10.8 mM Na2SO4, 9.0 mM CaCl2, 7.9 mM KCl) was adapted after Harrison et al. [46] to mimic Red Sea conditions and supplemented with 3 mM 13C-bicarbonate (H13CO3−) and 5 mM 15N-ammonium (15NH4+) for the isotope labeling incubations. During the incubation, corals from heat stress and control treatments were kept in temperature-controlled water baths equipped with Radion light systems (Ecotech Marine Inc., Bethlehem, PA) to maintain temperature and light regimes according to treatment conditions. Throughout the incubation, the seawater in the beakers was constantly stirred at 300 rpm using magnetic stirring bars to homogenously mix the water volume. For each coral species, one fragment was placed in artificial seawater without any isotopic label (isotope label replaced with unlabeled counterparts) to act as control for subsequent NanoSIMS analyses.
Coral sectioning/sampling for next-generation sequencing and NanoSIMS
After the end of the 24 h labelling incubations, coral fragments were collected and immediately sectioned into slices using an electric rotating diamond saw blade. Separate slices from each fragment were used for (a) Next-Generation Sequencing (NGS), (b) NanoSIMS analyses, and (c) spectral imaging of endolithic bands (n = 6 for each application). Sections were kept on ice until further processing. Using a Dremel 4000 rotary tool (Dremel, Prospect, IL), individual sections were trimmed for subsequent sampling. For nucleotide extractions, individual fragments were cut from each slice from (a) coral tissue (defined here as “Tissue”), (b) visible green or pink endolithic bands (defined here as “endolithic band”), and (c) white skeleton without any visible endolithic colonization (defined here as “skeleton”). For NanoSIMS imaging, one fragment per sample (approximately 15 × 5 × 2 mm) containing the coral tissue as well as the underlying skeleton and endolithic communities was collected. Between individual coral fragments and sections, the milling blade was thoroughly rinsed with 70% ethanol to avoid carry-over of tissue and/or microbes between sections. Sections for NGS sequencing were immediately ground manually to a fine powder over liquid nitrogen using a cryogrinder mortar and pestle. Powdered samples were transferred into 2 ml Eppendorf tubes, snap-frozen in liquid nitrogen, and stored at −80 °C until further processing. Sections for NanoSIMS were immediately transferred to fixation buffer (2.5% glutaraldehyde and 1% formaldehyde in PBS-sucrose buffer, pH 7.5) and stored for 24 h at 4 °C.
Photographic documentation and hyperspectral imaging analysis
For each coral sample, one slice was used for conventional photography and hyperspectral imaging analysis. For spectral imaging, sections were illuminated under constant light by two full-spectrum lights (Specim IQ accessory pro kit). Hyperspectral images were recorded with a tripod-mounted Specim IQ camera (Specim, Spectral Imaging LTD, Oulu, Finland) following calibration using the provided white reference tile. Recorded representative spectra were extracted from the coral tissue, the endolithic band, and the underlying coral skeleton for each of the sections using the IQ studio software. Selected spectral regions were statistically compared based on the distance between their cumulative distributions using a Kolmogorov-Smirnov (KS) test (which provides a measure of dissimilarity between the distributions).
DNA extraction and sequencing library preparation
Between 50–100 mg of each powdered sample was weighed directly into a clean 1.5 ml Eppendorf tube. Initial lysis of samples was conducted in 360 µl of ATL buffer + 20 µl of Proteinase K (20 mg/ml) per reaction by incubation at 56 °C and 900 rpm overnight. DNA extractions were performed using the Qiagen QIAamp Investigator Kit (Qiagen, Hilden, Germany) following manufacturer’s instructions for DNA extraction from bone. In the final elution step, DNA was eluted from Qiagen membranes in 25 µl of ATE buffer and stored at −20 °C until further processing. Negative controls (i.e., no template DNA PCR and no sample DNA extraction) were performed to account for contaminants in bacterial community analysis. Quantification of DNA was conducted using a Qubit 2-Fluorometer (Invitrogen, Waltham, MA).
Amplicon and metagenomic library preparation
Amplicon libraries were prepared using the Nextera XT Index Kit (Illumina, San Diego, CA) according to manufacturer’s instructions (Illumina). In brief, each 10 µl PCR reaction contained the Qiagen multiplex PCR kit (Qiagen, Hilden, Germany), 10 ng of DNA, and primers at a final concentration of 0.3 μM. Cycling conditions were 95 °C for 15 min, followed by 27 cycles of 95 °C for 30 sec, 55 °C for 90 sec, 72 °C for 30 sec, with a final extension of 72 °C for 10 min. The hyper-variable region V5-V6 of the bacterial 16 S rRNA gene was amplified using the primers 784 F (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGGATTAGATACCCTGGTA-3′) and 1061R (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCRRCACGAGCTGACGAC- 3′; Illumina overhang adaptor sequences are underlined) [47, 48]. Samples were amplified in triplicates which were combined before cleaning with ExoProStar 1-step (GE Healthcare, Pollards Wood, UK), indexed for 8 cycles, and confirmed on a 1% agarose gel. Samples were normalized using the SequalPrep Normalization Plate Kit (Invitrogen, Waltham, MA). Equimolar ratios of each sample were pooled and quantified using the Qubit dsDNA HS assay with the Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, USA). Pooled libraries were re-quantified using the qPCR KAPA Biosystems library quantification on an ABI HT7900 system (Applied Biosystems, Waltham, MA) and sequenced on the MiSeq platform (Illumina, San Diego, US) with 20% PhiX using the 2 × 301 bp paired-end V3 chemistry at the KAUST BioScience Core Lab (BCL) sequencing facility.
An average of 160 ng of genomic DNA per sample were used as an input to prepare metagenomic libraries using the Ovation Ultralow Library System V2 (Tecan, Männedorf, Switzerland). Briefly, genomic DNA was sheared to fragment sizes of approximately 250–300 bp using the Covaris M2 (90 s, 20 °C, 20% duty factor). Fragments were end-repaired, bidirectionally ligated to sequencing adaptors, and amplified with adaptor-specific primers using 8 PCR cycles. The quality of the libraries was checked on a Bioanalyzer DNA 1000 Chip (Agilent Technologies, Santa Clara, CA), followed by quantification using the Qubit HS DNA system (Invitrogen, Waltham, MA). Libraries were pooled using equimolar ratios to a final concentration of 10 nM. Pooled libraries were re-quantified using the qPCR KAPA Biosystems library quantification on an ABI HT7900 system (Applied Biosystems, Waltham, MA) and paired-end sequenced (2 × 151 bp) on the HiSeq 4000 (Illumina, San Diego, US) platform at 1.8 pM with 1% PhiX at the KAUST BCL sequencing facility.
Sequencing data analysis
16S rRNA gene amplicon sequencing
Amplicon sequence variants (ASVs) were inferred using DADA2 v1.21.0 [49]. In brief, forward and reverse sequence reads were truncated at the 3′ end at 290 and 210 base pairs, respectively. Reads with expected errors >2 or with the presence of ambiguous bases were discarded. A denoising step was done using the “pool=T” option to increase the resolution of singletons and doubletons. ASVs inferred from individual read pairs were merged and checked for chimeras. Taxonomic annotation was done using the SILVA database (version 138) [50]. Sequence reads statistics are found in Table S1 and ASV raw counts are found in Table S2. Relative abundances of the most abundant 10 prokaryotic families were generated using ggplot2 v3.3.5 [51]. PCA analyses were calculated from Euclidean distances of centered-log-ratio (clr)-transformed ASV counts using phyloseq v1.34.0 [52], and differences between bacterial communities were tested using PERMANOVA as implemented in the R package Vegan v2.5 [53]. For alpha diversity estimation, ASV counts were aggregated to families, and the number of observed families was calculated based on rarefied abundances to a sample count of 1966. The identification of differentially abundant ASVs was done using the Analysis of Composition of Microbiomes with Bias Correction (ANCOM-BC) function [54]. ANCOM-BC v1.0.5 function was applied on ASVs absolute counts to compare species and skeletal compartments samples using the False Discovery Rate (FDR) method to correct for multiple testing. A feature (i.e., ASV) was considered enriched in a specific condition when it was significantly more abundant than the reference (i.e., “control”) based on the ANCOM-BC analysis (i.e., FDR-adjusted p values were <0.05).
Gene-centric metagenomic analysis
Sequencing reads were adapter- and quality- (bases with quality scores <20) trimmed using Trimmomatic v.0.38 [55]; reads shorter than 50 nt were removed from the analysis. Assemblies produced by MEGAHIT v1.1.1-2 [56], metaSPAdes v3.13.0 [57], and IDBA v1.1.3-1 [58] were evaluated using QUAST v5.0.2 [59]. The best assembly was produced by MEGAHIT (Table S4) using K-mer lengths of 141. Open reading frames (ORF) were identified using Prodigal v2.6.3 [60], and total read counts to each ORF were estimated using Salmon v0.7.2 [61]. Contigs shorter than 500 bp were excluded from downstream analysis. Taxonomic annotation was done using Kaiju v1.7.3 [62] using the minimum exact matches (MEM) mode with a word length of 11 against the NCBI nr database that includes archaea, bacteria, viruses, and microbial eukaryotes. Functional annotation of ORFs against the KEGG database was done using KOfamscan v 1.3.0 [63], and the best match per contig was chosen based on the lowest E-value (considering only E-values <0.001) and highest bit score (considering only scores >100). ORFs were aggregated into KOs and L3 pathways by adding counts from contigs annotated to the same KO/L3 pathways. A total of 15,213 KOs affiliated to 457 KEGG L3 pathways were annotated, but data analysis and inferences were only made with pathways annotated to metabolism (KEGG map ko00001, 3975 annotated KOs and 160 L3 pathways). Family and metabolism KO alpha diversity was estimated by the Shannon index [64] on rarefied abundances to a sample count of 698,219 and 686,534, respectively, using Vegan [53]. Generalized linear mixed models were fit to test for differences in alpha diversity between species, treatments, and compartments, accounting for a random effect of the genotype using the lmer function of the lme4 v3.1 package [65]. Differentially abundant KOs, L3 pathways, and taxonomic families between coral species and treatments were identified using ANCOM-BC [54]. A feature was considered enriched in a specific condition when it was significantly more abundant than the reference (i.e., “control”) based on the ANCOM-BC analysis (i.e., FDR-adjusted p values <0.05). Ordination plots were computed from Euclidean distances of clr-transformed counts and represented in principal component analysis (PCAs) to account for the compositionality of the data according to Aitchison’s methodology [66]. Functional signatures that best predict species and skeletal compartments were identified using the Selbal algorithm v0.1 based on compositional balances (i.e., ratios of relative abundances) [67]. Selbal was run on KEGG L3 pathways involved in C metabolism, and the predictions were tested by a cross-validation procedure.
Genome-centric metagenomics analysis
In addition to the gene-centric metagenomic analysis, we did metagenomic binning by mapping trimmed reads to the best assembly using Bowtie2 v2.4.1 [68]. Mapping files were used as input files for Metabat2 2.11.1 [69], CONCOCT2 [70], and maxbin2 [71] to calculate tetranucleotide frequencies and differential contig coverage. The resulting bins were evaluated using checkM v1.1.2 [72], and the best binning results among the three software were chosen using MetaWRAP v1.2.1 [73]. Bins with completeness >75% and contamination <10% were considered metagenome-assembled genomes (MAGs). In addition, bins were dereplicated at 95% average nucleotide identity (ANI) using dRep v3.2.2 [74]. Taxonomic placement of MAGs was done based on ANI with the “classify_wf” workflow of GTDB-tk v1.3.0 [75] using the Genome Taxonomy Database (GTDB) release 95 [76]. Functional annotation was done using KOfamscan v 1.3.0 [63], Prokka v1.13 [77], and METABOLIC v2.0 [78]. Completeness of KEGG modules was evaluated using EnrichM v0.4.3 (available at: https://github.com/geronimp/enrichM). Phylogenomic analysis was done using the UBCG pipeline v0.1 [79]. Differential abundance analysis was done using ANCOM-BC using clr-transformed mapped reads to each MAG, results were represented on heatmaps indicating differentially abundant MAGs between species.
Sample embedding and preparation for SEM and NanoSIMS
Coral cross sections for NanoSIMS were preserved in fixation buffer (2.5% glutaraldehyde and 1% formaldehyde in PBS-sucrose buffer, pH 7.5) for 24 h at 4 °C. After the fixation, samples were transferred to 3× PBS-sucrose buffer at 4 °C until embedding.
Initially, the entire coral cross sections (n = 6 per treatment) were embedded into resin. Coral cross sections were dehydrated with increasing concentrations of ethanol (50%, 70%, 90%, 100%), followed by acetone (100%), and then infiltrated with increasing concentrations of Araldite 502 resin (20%, 30%, 40%, 50%, 60%, 75%, 100%; Sigma-Aldrich) under vacuum over a total of 48 h. Coral cross sections were then finally transferred into 1-inch circular molds (3 biological replicates were added to each mold; Figure S1A-B), covered with 5 mm of Araldite 502 resin (100%), and cured at 60 °C under vacuum for 24 hours. The resin-encased coral cross sections were polished down to 1 µm using a RotoPol-35 (Struers, Denmark). Resin blocks were then coated with 5 nm of gold, mounted on SEM specimen holders and mapped using a Zeiss EVO Scanning Electron Microscope (Zeiss, Germany). For each sample, three areas (120 × 80 µm) from each microenvironment (coral tissue, endolithic band, and skeleton) were imaged in high resolution with SEM to be targeted with NanoSIMS imaging. Following SEM, resin blocks were then transferred to a 1-inch sample holder for NanoSIMS analysis.
NanoSIMS analysis
The NanoSIMS-50 (Cameca, France) at the Centre for Microscopy, Characterisation and Analysis (CMCA) at the University of Western Australia was used for all subsequent analyses. The NanoSIMS-50 allows simultaneous collection and counting of five isotopic species, which enables the determination of 13C/12C and 15N/14N ratios. Enrichments of the rare isotopes 13C and 15N were confirmed by an increase in their isotopic ratios above natural abundance values recorded in controls (13C/12C: 0.0110 ± 0.0001 and 15N/14N: 0.0037 ± 0.0001, n = 190 cells). NanoSIMS analysis was undertaken by rastering a 2.5 pA Cs+ beam (~100 nm diameter) across defined 35 µm2 sample areas (256×256 pixels) with a dwell time of 30 ms per pixel. The isotope ratio values are represented hereafter using a colour-coded transform (hue saturation intensity (HSI)) showing natural abundance levels in blue and grading to high enrichment in pink. Images were processed and analyzed using Fiji (http://fiji.sc/Fiji) with the Open-MIMS plug-in (http://nrims.harvard.edu/software). All images were dead-time corrected. Quantitative data were extracted from the mass images through manually drawn regions of interest.
Given that our initial embedding of full coral sections did not yield usable NanoSIMS data (see Results; Figure S1H-O), we used an alternative method to prepare the samples for NanoSIMS. Specifically, fixed coral cross sections were further sub-divided into tissue and endolithic band using a 38 mm diamond cutting wheel and decalcified in 0.5 M EDTA solution over 10 days (solution was changed once after 5 days). Decalcified samples were post-fixed in 1% OsO4 (1× in PBS, final pH 7.5) for 2 h and dehydrated with increasing concentrations of ethanol (50%, 70%, 90%, 100%), followed by acetone (100%). Following this procedure, samples were infiltrated with increasing concentrations of SPURR resin (25%, 50%, 75%, 100%) and cured at 65 °C overnight. Finally, samples were sectioned (200 nm) using an ultramicrotome (Leica Microsystems), mounted on silicon wafers, and coated with 5 nm of gold (for NanoSIMS analysis). For electron microscopy, 100 nm thick sections were mounted onto copper TEM finder grids (coated in formvar—Emgrid, Australia).
Results
To assess the putative functional roles of endolithic microbiomes in coral bleaching, we characterized their taxonomic and metabolic diversity, together with measurements of N and C assimilation during heat stress. For this, we exposed fragments of G. edwardsi and P. lutea to a 17-day heat-stress experiment and combined hyperspectral imaging, 16S rRNA gene amplicon sequencing, metagenomics, and NanoSIMS imaging on different skeletal compartments (Fig. 1).
Fig. 1. Experimental setup.
Coral fragments from G. edwardsi and P. lutea colonies (n = 6) were subjected to a heat stress experiment followed by a 24 h incubation with stable isotope tracers before sample collection and sectioning. Subsequently, we measured the hyperspectral profile of each section and conducted 16S rRNA gene amplicon sequencing, metagenomics, and NanoSIMS analyses to characterize the endolithic microbial communities and their associated metabolic potential as well as differential 13C and 15N assimilation under heat stress.
G. edwardsi and P. lutea have heterogenous skeletal banding patterns
Banding patterns visibly differed in their thickness and position (i.e., distance from tissues) between coral species but were also heterogeneous among fragments of the same coral species and often within the same colony (Figs. 2A, B, 2L, M, and S2). We sampled both the conspicuous (green or pink) endolithic band (found between 0 and 25 mm beneath the tissue in G. edwardsi, and 0 and 10 mm beneath the tissue in P. lutea) and the deep white skeleton underneath (with no visible endolithic colonization, approximately sampled 10 mm beneath the endolithic band). In addition, conspicuous pink bands were observed in two replicate colonies of P. lutea.
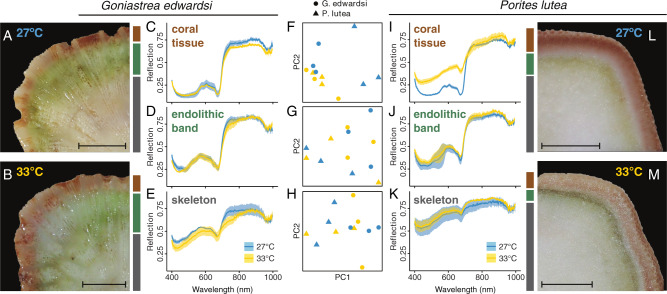
Fig. 2. Hyperspectral imaging of coral fragments from G. edwardsi and P. lutea under heat stress.
Representative picture of skeleton banding patterns of a G. edwardsi colony at A control (27 °C), B heat-stress (33 °C), and hyperspectral profiles of C tissue, D endolithic band, and E skeleton. Hyperspectral profiles of P. lutea I tissue, J endolithic band, and K skeleton. Exemplary picture of skeleton banding patterns of a P. lutea colony at L control (27 °C) and M heat-stress (33 °C). Principal component analysis of the hyperspectral profiles of the F coral tissue, G endolithic band and H skeleton under control (27 °C) and heat stress conditions (33 °C). Means over all samples ± 95% CI. Scale bar: 1 cm.
Hyperspectral profiles were indistinguishable between the tissues of G. edwardsi and P. lutea (Fig. 2C–K and S3). For all three compartments (i.e., coral tissue, endolithic band, skeleton), reflectance spectra followed patterns expected for chlorophyll-containing organisms, including reflectance minima in the blue and red spectra, higher reflectance in the green to orange domain, and a substantial increase in reflectance between 680 to 720 nm (commonly referred to as the red edge) followed by a plateau in the near-infrared domain [80] (Fig. 2C–E and 2I–K). However, some differences were apparent in the endolithic bands between the two species: P. lutea’s reflectance was significantly higher in the red edge (650 to 700 nm) and infrared (900–1000 nm) (KS test, p < 0.01; Figure S3). In addition, the reflectance of the skeleton was significantly higher in P. lutea compared to G. edwardsi across the entire 400 to 1000 nm range (KS test, p < 0.01), with a less pronounced red edge in P. lutea indicative of reduced chlorophyll concentrations.
After 17 days of heat stress, P. lutea fragments were visibly bleached, while G. edwardsi showed no evidence of symbiont loss (Figs. 2A, B and 2L, M). In congruence with the bleaching phenotype, hyperspectral imaging revealed contrasting optical properties between treatments in the tissues of P. lutea, in which heat-treated samples had a substantial and significant increase in reflectance in the 400 to 700 nm range and a less pronounced red edge (KS test, p < 0.01, Figs. 2C and 2I) likely reflecting the loss of chlorophyll-containing Symbiodiniaceae cells. No differences between heat-treated and control samples were observed in the skeleton or endolithic band spectra in P. lutea (Fig. 2J, K). However, reflectance significantly declined during heat stress in the skeleton of G. edwardsi in 400–500 and 700–800 nm ranges (KS test, p < 0.01, Fig. 2E). While the underlying causes of this shift are unknown, a similar decrease in the reflection in the 700–800 nm range has been previously described in plants during fungal pathogen infections [80, 81].
Endolithic microbiomes of G. edwardsi are taxonomically more diverse
We used a metagenomic gene-centric approach (i.e., based on abundances of individual genes) to quantitatively compare the microbial taxonomic and functional diversity in the endolithic band and skeleton of the two coral species under ambient temperature. In both species, metagenomic sequences derived from the endolithic bands and skeletons were mostly of bacterial origin (~75%), followed by microbial eukaryotes (~5%), archaea (<1%), and viruses (<1%) (Figure S4). The unclassified fraction represented ~20% of the community, mainly derived from the coral host. At higher taxonomic ranks (Phylum/Class), taxonomic composition between coral species and skeletal compartments was overall similar (Fig. 3A–D), but significant differences existed at the family level between the endolithic band and the skeleton communities between both coral species (PERMANOVA, padj < 0.05, Fig. 3F, S5A–D, and S6). However, no significant differences were determined between skeletal compartments of the same coral species (Fig. 3F and Table S5). Family alpha diversity was also significantly higher in G. edwardsi than in P. lutea in both compartments (lmer, padj < 0.05), and the skeleton was more diverse than the endolithic band in both species (lmer, padj < 0.01, Fig. 3E and Table S6).
Fig. 3. Taxonomic diversity of G. edwardsi and P. lutea metagenomes.
Bar plots represent the relative abundance of the 10 most abundant taxonomic classes of A Archaea, B Bacteria, C Microbial eukaryotes, and D viruses in G. edwardsi (Gon) and P. lutea (Por). When annotation did not reach the class level, the highest taxonomic rank annotated was used. Less abundant classes were aggregated to the “Others” category. E Shannon diversity of all taxonomic families found in metagenomes. Significant differences in alpha diversity between species and skeletal compartments are denoted by letters above the boxes (Table S6). F Ordination plots of microbial composition at the family level in G. edwardsi and P. lutea. G Maximum likelihood phylogenetical inference of MAGs recovered from G. edwardsi and P. lutea metagenomes. A set of 92 house-keeping genes [79] were used for the reconstruction. H Relative abundance of MAGs clustered to the phylum level with respect to the total binned fraction. Sections G and H share color coding.
As the bacterial fraction represented most of the metagenomic reads, we further assessed bacterial diversity by 16S rRNA gene amplicon sequencing of tissue, endolithic band, and skeleton. The 16S rRNA analysis showed that alpha diversity at the family level consistently increased from tissue to endolithic band to skeleton (Figure S7C) and confirmed that significant differences exist in the composition of bacterial families between tissues and skeletal compartments (Figure S7A and Table S3). Nevertheless, some of the main features of bacterial diversity that were identified by metagenomics were overlooked by 16S rRNA gene sequencing (Fig. 3B, S7B, and S7D, E). A total of 6,491 ASVs assigned to 220 bacterial families were present in the 16S rRNA gene amplicon dataset, in comparison to 492 bacterial families identified through the gene-centric metagenomics approach (Figure S7D). Such discrepancies are common and, in the particular case of our study, may be due to the limited coverage of specific bacterial taxa by the 16S rRNA gene amplicon primers, but other factors (e.g., 16S rRNA secondary structures, gene copy number, coverage in databases) can also play a role [82, 83]. Despite the higher number of bacterial families identified using metagenomics, the most abundant bacterial families were consistent between metagenomes and amplicon data, and included members of the Desulfobacteraceae and Rhodobacteraceae (Figures S5B and S7B). In addition to bacteria, archaea, microbial eukaryotes, and viruses were also present and diverse in endolithic communities found in the metagenomic dataset. Specifically, we identified a total of: (i) 48 archaeal families, dominated by members of the classes Halobacteria, Methanomicrobia, and Thermoplasmata, in both coral species and skeletal compartments (Fig. 3A and S5A); (ii) 643 families of microbial eukaryotes, in which Ostreobiaceae (Ulvophyceae) and Symbiodiniaceae (Dinophyceae) were the two most abundant families in both species and compartments, representing on average 12.5% and 10.6% of the total microbial eukaryotic diversity of G. edwardsi and P. lutea, respectively (Fig. 3C, S5C). Notably, Fungi represented almost 50% of the microbial eukaryotic community and were mainly represented by a large diversity of Ascomycota (from the classes Sordariomycetes, Dothideomycetes, Eurotiomycetes and Saccharomycetes), Basidiomycota (class Agaricomycetes), and Mucoromycota (class Mucoromycetes) (Fig. 3C). Other low-abundant members of the microbial eukaryotic community belonged to the phyla Apicomplexa, Euglenozoa, and Chytridiomycota; (iii) 124 viral families, with the 10 most abundant families representing over 75% of the total viral community and mostly include members of the Caudovirales class such as Myoviridae, Podoviridae, and Siphoviridae (Fig. 3D, S5D).
Overall, 639 families (48.9%) belonging to bacteria, archaea, micro-eukaryotes, and viruses were differentially abundant between coral species in the endolithic band and 444 (34%) in the skeleton. In both compartments, more families were enriched in G. edwardsi in comparison to P. lutea (326 versus 313 in the endolithic band and 263 versus 181 in the skeleton; Figure S8 and Table S7). Most differences between coral species were explained by an enrichment in bacterial families of the Desulfobacterota (formerly belonging to the class of Deltaproteobacteria), including Desulfobacteraceae, Desulfohalobiaceae, and Desulfonatronaceae in the endolithic band and skeleton of G. edwardsi, while the skeletal compartments of P. lutea were enriched with families of the phylum Cyanobacteria, including Cyanobacteriaceae, Prochloraceae, and Microcoleaceae (Figure S9). The differential abundance analysis also revealed that Desulfurococcaceae and Ferroplasmaceae (Archaea), Phycodnaviridae and Picobirnaviridae (Viruses), and several families of the Sordariomycetes (Fungi) were enriched in G. edwardsi in comparison to P. lutea, while Cuniculiplasmataceae and Acidilobaceae (Archaea), Secoviridae and Mimiviridae (Viruses), and several members of the class Dothideomycetes (Eukaryota) were enriched in P. lutea in comparison to G. edwardsi (Table S7, Figure S8).
In addition to the gene-centric approach, we used a genome-centric approach to characterize the metabolic potential of bacterial populations present in the endolithic band and deep skeleton. We recovered a total of 75 bacterial Metagenome-Assembled Genomes (MAGs; completeness >75% and contamination <10%) from the skeletal compartments of G. edwardsi and P. lutea [84] that represented in average 6.91% of the metagenomes across samples (Figure S10 and Table S8). Differential abundance analysis identified 30 MAGs enriched in G. edwardsi (15 only in the endolithic band and 15 in the endolithic band and skeleton) and 21 MAGs enriched in P. lutea (8 only in the skeleton and 13 in the skeleton and endolithic band, Figure S11 and Table S8). The majority of MAGs were affiliated to the phylum Proteobacteria (Fig. 3G) and represented the most abundant fraction of the binned community (Fig. 3H). Besides members of the Proteobacteria, MAGs enriched in G. edwardsi were mainly affiliated to the phyla Desulfobacterota, Bacterioidota, and Firmicutes, while MAGs enriched in P. lutea were mainly affiliated to the phyla Cyanobacteria, Myxococcota, and Planctomycetota (Fig. 3H and S11).
Taken together, our metagenomic approach revealed that endolithic microbial diversity differed between coral species but not within compartments of the same coral host. Endolithic communities were more diverse in G. edwardsi than in P. lutea, with differentially more abundant Deltaproteobacteria in G. edwardsi and Cyanobacteria in P. lutea
Endolithic microbiomes of G. edwardsi are functionally more diverse and redundant
We assessed the metabolic diversity of endolithic microbiomes using a gene-centric approach based on the composition of KEGG Orthologs (KOs) involved in metabolism (KEGG map 01100) between coral species and skeletal compartments. Metabolic diversity was significantly different between species in the endolithic band and the skeleton (PERMANOVA, padj < 0.05), but not significantly different between compartments of the same species (Fig. 4A and Table S9). Species clustering of KOs was evident in ordination plots of overall metabolism (Fig. 4A) and C metabolism (Fig. 4B), while no clustering by species was observed in ordination plots based on N metabolism (Fig. 4C). G. edwardsi was metabolically more diverse than P. lutea in both compartments, and the skeleton was more diverse than the endolithic band in both species (Fig. 4D and Table S10).
Fig. 4. Metagenomic functional diversity in skeletal compartments of G. edwardsi and P. lutea.
Ordination plots based on the composition of KEGG orthologs (KOs) involved in A overall metabolism, B carbon metabolism, and C nitrogen metabolism. D Shannon diversity of metabolism KOs. Significant differences between species and skeletal compartments are denoted by letters above the boxes. E Volcano plots depicting differentially abundant KEGG orthologs (KO) involved in metabolism between G. edwardsi and P. lutea. Each dot represents a KO and differentially abundant KOs are colored by metabolic processes. KOs that are not differentially abundant between species are colored in black. Dashed red lines represent thresholds for FDR-adjusted p value < 0.05 and effect sizes (1 and −1) represented by the Beta statistic obtained from ANCOM-BC. F Differentially abundant carbon metabolism KEGG L3 pathways between G. edwardsi and P. lutea. Clr-transformed pathway counts were plotted and z-normalized by KEGG L3 pathways (rows). Left bars indicate the effect size represented by the W ANCOM-BC statistic and colors represent the coral species in which the process was enriched. Colored boxes represent the affiliation to KEGG L2 pathways.
A higher number of KOs and L3 KEGG pathways were enriched in G. edwardsi in comparison to P. lutea (Fig. 4E and Tables S7 and S10). L3 KEGG pathways enriched in G. edwardsi in comparison to P. lutea, were mainly associated with carbohydrate (21.1%), amino acid (14.0%), and cofactors and vitamin metabolism (12.3%) (Fig. 4F). Contrastingly, P. lutea was enriched in processes involved in the biosynthesis of other secondary metabolites (30.8%), glycan biosynthesis and metabolism (30.8%), and lipid metabolism (15.4%) compared to G. edwardsi (Fig. 4F). We performed a Selbal analysis to identify functional signatures predictive of coral species for each compartment. “Carbon fixation in photosynthetic organisms” was the process with the largest discriminatory effect between coral species and was predictive of the P. lutea endolithic band and skeleton microbiomes, while “Ascorbate and aldarate metabolism” was predictive of G. edwardsi endolithic band microbiomes and “Galactose metabolism” was predictive of G. edwardsi skeleton microbiomes (Figure S12). Of note, the “Carbon fixation in photosynthetic organisms” pathway includes reactions that fix CO2 to organic compounds by the reductive pentose phosphate cycle (i.e., Calvin cycle), unlike the “Carbon fixation pathways in prokaryotes” that includes pathways used by autotrophic bacteria and archaea.
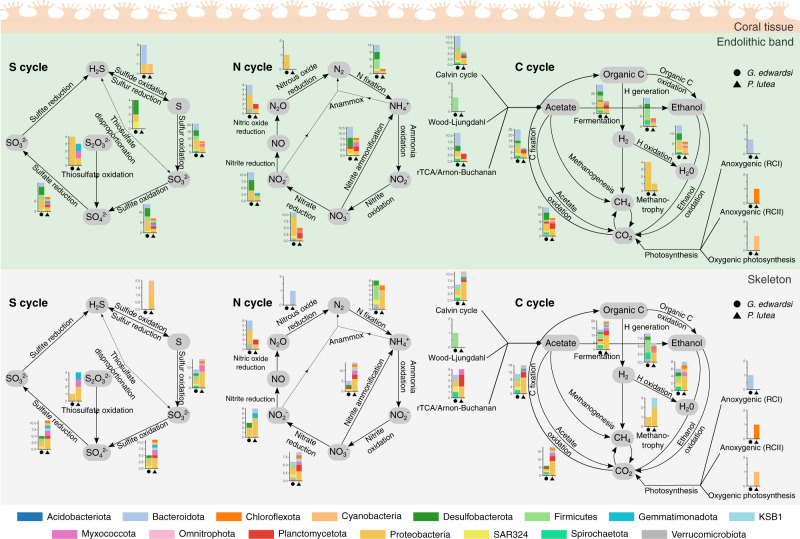
We investigated the prevalence of different prokaryotic carbon fixation pathways as well as their relative contribution in each coral species and compartment (Figure S13). Carbon fixation pathways in G. edwardsi were dominated by the reductive acetyl-CoA pathway (Wood-Ljungdahl pathway), followed by the reductive citric acid cycle (Arnon-Buchanan cycle), the Calvin-Benson cycle, and the 3-hydroxypropionate bi-cycle (Figures S13C-F). In P. lutea the reductive pentose phosphate cycle (i.e., Calvin-Benson cycle) of eukaryotic and cyanobacterial origin was prevalent, representing more than 50% of the relative contribution to total carbon fixation genes in the endolithic band (Figure S13B, S13F). Bacterial MAGs revealed that prokaryotic carbon fixation pathways had a broader phylogenetical distribution in G. edwardsi compared to P. lutea. In G. edwardsi, members from 6 phyla (Bacteroidota, Desulfobacterota, Firmicutes, Proteobacteria, SAR324, and Spirochaetota) harbored at least 70% of the genes involved in carbon fixation in prokaryotes, while in P. lutea only 1 phylum (Proteobacteria) harbored the genes involved in the Arnon-Buchanan cycle (Fig. 5 and S14).
Fig. 5. Taxonomic distribution of central N, S, and C pathways in endolithic metagenome-assembled genomes (MAGs).
For each MAG, only pathways with more than 70% completeness were considered. Barplots show the number of MAGs from each phylum that were assigned to each reaction in G. edwardsi (circles) and P. lutea (triangles).
MAGs enriched in G. edwardsi revealed a higher functional redundancy than in P. lutea. Indeed, 92% of the processes found in the endolithic band were present in more MAGs in G. edwardsi than in P. lutea and each of these processes was represented on average by 2.6 times more taxonomic groups in G. edwardsi compared to P. lutea. Conversely, 83% of the processes found in the skeleton were present in more MAGs in P. lutea than in G. edwardsi and each of these processes was represented on average by 1.8 times more taxonomic groups in P. lutea compared to G. edwardsi. (Fig. 5). Processes including photosynthesis, nitrogen fixation, and sulfate reduction were enriched in the binned fraction of P. lutea in comparison to G. edwardsi, whereas methanotrophy, thiosulfate metabolism, sulfur oxidation, and nitrate reduction were enriched in G. edwardsi’s binned fraction in comparison to P. lutea. Anoxygenic photosynthesis was enriched in the skeleton of both species and nitrite ammonification was enriched in the endolithic band of both species (Fig. 5, S14, and S15).
In summary, both gene-centric and genome-centric metagenomic results revealed that G. edwardsi harbors endolithic microbiomes with greater and more redundant functional diversity than P. lutea. These findings also indicate that genes involved in carbon metabolism, in particular carbon fixation, differed between coral hosts, with anaerobic prokaryotic carbon fixation being more abundant in G. edwardsi and aerobic carbon fixation (i.e., Calvin-Benson cycle) being more abundant in P. lutea.
Taxonomic and functional diversity of skeletal compartments under heat stress
There was no significant difference in the overall taxonomic composition or alpha diversity between control and heat stress samples in either G. edwardsi or P. lutea (Figure S16A, B). Despite the lack of significant differences in the overall microbiome composition between treatments, we identified differentially abundant families between treatments (Table S7). Microbial communities associated with the skeleton of G. edwardsi under heat stress changed the most compared to controls, with 268 differentially abundant prokaryotic and eukaryotic families identified, indicating a higher microbial response to heat in that compartment (Figure S17). These differentially abundant families included members of marine basidiomycete and ascomycete Fungi, specifically the classes Agaricomycetes (n = 37) and Sordariomycetes (n = 20), respectively, which significantly decreased with heat stress, while bacterial families affiliated to the Desulfobacterota phyla (n = 7), including Desulfomicrobiaceae and Desulfuromonadaceae, significantly increased (Table S7). In line with this result, ASVs belonging to the phylum Desulfobacterota and Gammaproteobacteria class were also enriched in the 16S rRNA gene amplicon dataset during heat stress (Table S7).
Similarly, we did not detect significant differences in the overall microbial functional composition or alpha diversity of KEGG orthologs between control and heat stress endolithic microbiomes (Figure S16C, D and Tables S9,10), consistently with fewer differentially abundant KOs and pathways between treatments (Figure S18 and Table S7). P. lutea was enriched with KOs involved in glycolysis/gluconeogenesis and oxidative phosphorylation in comparison to G. edwardsi. In contrast, G. edwardsi had a more diverse metabolic response to heat stress, which included KOs involved in methanogenesis (i.e., formation of methane from CO2), prokaryotic carbon fixation (i.e., 3-Hydroxypropionate bi-cycle and the reductive citrate cycle, acetyl-CoA), antenna proteins (i.e., phycocyanin synthesis and phycobilisome light-harvesting complex), and sulfur metabolism (i.e., thiosulfate metabolism and dissimilatory sulfate reduction) (Figure S18).
P. lutea assimilates more carbon and nitrogen
Given the differences in metabolic potential identified between both coral species, we employed nanoscale secondary ion mass spectrometry (NanoSIMS) to determine whether carbon and nitrogen assimilation differed in the skeletal compartments of both coral species. Entire coral cross sections were initially analyzed, but the carbonated skeleton had a negative impact on the NanoSIMS analysis, resulting in a very low signal for all the isotopic species of interest (12C, 13C, 14N, and 15N). Although low conductivity of biomineralized carbonates has previously been reported [85], the patterns observed here were very pronounced (Figures S1H–O), and the data acquired using this approach were not usable. Specifically, we observed a complete lack of NanoSIMS signal from skeletal surfaces and greatly attenuated signals (together with spatial artifacts, as shown in Figure S1H) in neighboring skeletal pores infiltrated with resin. We, therefore, used an alternative approach to prepare the samples for NanoSIMS, decalcifying separately the tissue and endolithic compartment sections followed by resin embedding of each compartment.
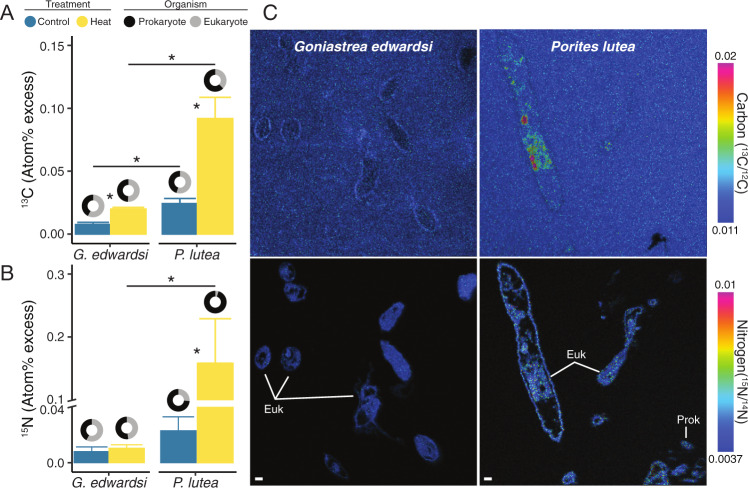
Results derived from this second approach revealed that after incubation with 13C-bicarbonate and 15N-ammonium, endolithic microorganisms in P. lutea were on average 3.2- and 2.9-times more enriched in 13C and 15N, respectively, than those in G. edwardsi (Fig. 6A, B, Table S11). Endolithic microorganisms were characterized as eukaryotes if cells exceeded 3 μm and as prokaryotes if cells were smaller than that threshold. Filamentous endolithic eukaryotes in P. lutea, potentially belonging to Ostreobiaceae (Ulvophyceae) or Aspergillaceae (Eurotiomycetes; Fig. 6C), were on average 2.8-times more enriched in 13C compared to those in G. edwardsi (Kruskal-Wallis test, p < 0.01; Figure S19). Similarly, endolithic prokaryotes in P. lutea were on average 3.1-times more enriched in 13C than those in G. edwardsi (Kruskal–Wallis test, p < 0.01; Figure S19). Conversely, 15N assimilation was barely detectable and not statistically different between coral species when eukaryotes and prokaryotes were considered separately. In addition, coral tissue in P. lutea exhibited a significantly larger enrichment in 13C but a significantly lower enrichment in 15N compared to G. edwardsi (Kruskal–Wallis test, p < 0.01; Figure S19), although the enrichment of their Symbiodiniaceae was not statistically different.
Fig. 6. Carbon and Nitrogen uptake by endolithic microorganisms under heat stress.
A 13C and B 15N assimilation in G. edwardsi and P. lutea. Donut charts represent the proportion of 13C and 15N assimilated by prokaryotes or eukaryotes. C Representative NanoSIMS images showing 13C and 15N assimilation in prokaryotic and eukaryotic cells. Scale bar: 1 µm.
Although only subtle differences were identified in microbial functional diversity between control and heat stress treatments using our metagenomic approach, the assimilation of 13C-bicarbonate by microorganisms from the endolithic band was significantly higher under heat stress in both coral species compared to control conditions (Kruskal-Wallis test, p < 0.01). Indeed, a 2.6- and a 3.8-fold increase in carbon assimilation were recorded in endoliths from G. edwardsi and P. lutea, respectively (Fig. 6A). During heat stress, the endoliths in P. lutea assimilated 4.6-fold more 13C than in G. edwardsi (Fig. 6A). Although heat stress had no effect on 15N assimilation in G. edwardsi (Fig. 6B), endoliths in heat-stressed P. lutea assimilated 82 times more 15N than in control colonies and 184 times more than G. edwardsi at 33 °C (Fig. 6B). More specifically, eukaryotic endoliths in P. lutea assimilated 12.4 times more 15N, while prokaryotic cells assimilated 88.2-times more under heat stress than in the control fragments (Kruskal-Wallis test, p < 0.01; Figure S19). This large increase in 15N assimilation in P. lutea endoliths under stress is in stark contrast with the relatively low and similar levels of assimilation in the endoliths harbored by the control colonies of both species. Meanwhile, 13C and 15N assimilation both significantly decreased under heat stress in the coral tissue from P. lutea (Kruskal–Wallis test, p < 0.01; Figure S19C).
To sum up, endolithic microorganisms in P. lutea assimilated more carbon and nitrogen than those from G. edwardsi under ambient conditions, and these differences were exacerbated further by heat stress. NanoSIMS data therefore indicated that differences in metabolic potential between the endolithic communities of the two coral species contributed to drastic dissimilarities of nutrient uptake, highlighting the putative contribution of endolithic microbiomes to heat stress susceptibility, as further discussed below.
Discussion
Coral bleaching is the primary driver of global reef loss, prompting an urgent need to advance understanding of the mechanisms underlying bleaching susceptibility to mitigate the effects of ocean warming [5, 38]. We are only beginning to understand the role of the endolithic microbiome in coral holobiont functioning and its putative role in mitigating the effects of bleaching, despite the coral skeleton being the largest microbial compartment by means of volume [18]. Previous studies have shown that endolithic microbes can supply the coral tissues with nutrients [29, 30], suggesting that these endolithic communities may act as a “rescue compartment” to supply fixed carbon to compensate for the loss of Symbiodiniaceae during bleaching. Still, we remain naïve with regard to a definitive link between endolithic microbiome structure, functional potential, and coral bleaching susceptibility. To overcome methodological limitations that hamper the exploration of skeletal compartments, we integrated emerging technologies, i.e., single-cell imaging of nutrient assimilation with metagenomic interrogation to characterize endolithic microbial functioning and their potential contribution to bleaching susceptibility.
Metabolic strategies in endolithic microbiomes of two coral species with different bleaching susceptibilities
Endolithic microbiomes were distinct between coral species (i.e., P. lutea and G. edwardsi), but did not differ across endolithic compartments within the same species (Figs. 3F and 4A, Table S5 and S9). The lack of microbial differences between compartments was attributed to high variability across replicates. Therefore, we encourage future studies to include a greater number of replicates to tease apart microbiome differences between the different compartments. In contrast to previous approaches based on ribosomal gene amplicon sequencing [20, 86], our comparative metagenomic approach allowed for a direct comparison across phylogenetically distant groups of endoliths and revealed that bacterial communities accounted for the majority (~75%) of the DNA content in metagenomic samples derived from coral skeleton (Figure S4). Coral host genomic material typically accounts for the majority of the sequenced DNA in tissues [87, 88], but we believe the low host coverage in our study is due to the low coral biomass compared to microbial cells in the skeleton, together with our targeted sampling of the skeletal compartments. One of the most striking observations was the low abundance (<1% of the metagenomic community) of the common microalgal family Ostreobiaceae, often-considered a prominent member of the endolithic band (Fig. 3C). This is consistent with previous reports of low amplicon read numbers affiliated to this taxon [20, 32]. Our findings suggest that differences in endolithic microbiomes between the two coral species were largely explained by the bacterial composition, with G. edwardsi’s endolithic microbiomes being enriched with families belonging to the strictly anaerobic Desulfobacterota, including Desulfobacteraceae and Desulfovibrionaceae, and P. lutea’s endolithic microbiomes enriched with aerobic cyanobacterial families including Cyanobacteriaceae and Phormidesmiaceae (Fig. 3B and S5B). The prevalence of strictly anaerobic bacteria co-occurring with diverse microalgal assemblages in G. edwardsi’s endolithic microbiomes has previously been reported in skeletal compartments [34, 89–91] and supports the existence of micro-niches that protect strict anaerobes from strong daily oxygen oscillations ranging from 10% to 210% air saturation [92].
Functional profiles, particularly those related to carbon metabolism, were largely driven by the relative proportion of anaerobic vs aerobic bacterial taxa. In this regard, the functional signatures that best discriminated endolithic microbiomes were characterized by genes involved in ascorbate metabolism in G. edwardsi and genes involved in the Calvin-Benson cycle in P. lutea (Figure S12). Ascorbate metabolism includes the ROS-scavenging enzyme ascorbate peroxidase, involved in protection against O2 toxicity and reported to be an important mechanism for alleviating photodamage in eukaryotic microalgae, including Symbiodiniaceae, and higher plants during high light intensities [93–95]. These findings, while preliminary, raise the possibility that endolithic microbiomes may be more effective at alleviating heat-induced oxidative stress in G. edwardsi than in P. lutea. On the other hand, P. lutea skeletal compartments were enriched with genes associated with the Calvin-Benson cycle, whereas G. edwardsi’s skeletal compartments were enriched in three carbon fixation pathways exclusive to prokaryotes (Figure S13F). While the Calvin-Benson cycle is the most important contributor to autotrophic CO2 fixation in the ocean, prokaryotic CO2 fixation has also shown to be of high significance, especially in symbiosis in extreme environments [96]. In contrast to the Calvin-Benson cycle, most prokaryotic carbon fixation reactions found in the skeletal compartments of G. edwardsi are oxygen-sensitive and can only be carried out in microaerophilic (i.e., Arnon-Buchanan cycle) or strict anaerobic conditions (i.e., Wood–Ljungdahl pathway). These pathways require less ATP to sustain growth [97, 98], but contain energetically challenging reactions that require a low redox potential and high inorganic carbon concentrations [99]. As a result of such energetic constraints, primary production could be lower in anaerobic chemolithotrophy compared to the Calvin-Benson cycle. This aligns with the higher 13C assimilation measured in the skeleton of P. lutea, which indicates greater levels of primary production.
It is now widely accepted that higher functional diversity and redundancy (i.e., the capacity of one species to functionally compensate for the loss of another) [100, 101] are critical to ecosystem functioning and resilience [102, 103]. We found that endolithic microbiomes in G. edwardsi’s were overall more diverse (taxonomically and functionally) as well as functionally more redundant in comparison to endolithic microbiomes in P. lutea (Figs. 3E and 4D). This suggests that G. edwardsi’s endolithic microbiomes may have a greater repertoire of mechanisms to respond to environmental disturbances, while being less likely to disrupt holobiont homeostasis as a consequence of the loss of microbial functional groups. In this context, we argue that the inability to maintaining holobiont functionality is presumably a crucial aspect of bleaching susceptibility, as evidenced by the dependence of the coral host on organic carbon translocated from its photosynthetic algal symbionts [104]. Besides bleaching susceptibility, microbiomes with a diverse functional potential have been proposed as an adaptive trait enabling corals to persist in highly polluted waters [105]. For instance, endolithic communities capable of utilizing diverse metabolic substrates (e.g., pollutants or excess nutrients) could contribute to maintaining low nutrient concentrations in the overlaying coral tissue. Thus, functionally diverse endolithic communities may increase holobiont environmental resilience, potentially explaining previous reports that identified corals from the genus Goniastrea as environmentally resilient [106, 107]. Our results also argue that higher metabolic diversity (as estimated in G. edwardsi) does not necessarily translate into higher metabolic productivity, but that metabolic versatility might be a critical trait affecting holobiont functioning under stress.
Heat stress stimulates endolithic metabolism
After 17 days of heat stress, P. lutea visibly bleached whereas G. edwardsi showed no visible signs of stress, such as symbiont loss. Besides the apparent differences in bleaching susceptibility, taxonomic and functional profiles were only marginally affected by heat stress and a relatively low number of differentially abundant families were identified between control and heat stress samples (Figures S16, S17). Interestingly, some of these families contributed considerably to the observed differences between coral species (e.g., members of the Desulfobacterota, Figure S9), indicating that heat stress may have exacerbated differences in endolithic communities between these two coral species and implying the existence of species-specific microbial responses to thermal stress. These findings also highlight the stability of these endolithic communities, consistent with the fairly stable environment in which they live and align with prior evidence supporting the stability of endolithic communities under natural gradients of high pCO2 [32]. Our results therefore suggest that endolithic communities respond to heat stress by increasing cellular metabolism, whereas microbial community composition remains unaffected, as shown previously for coral reef microbial communities under environmental change [108, 109].
Interestingly, we showed that 13C assimilation was enhanced by heat stress in the endolithic band of both coral species, but only P. lutea assimilated significantly more 15N during heat stress (Fig. 6A-B). Higher endolithic 15N assimilation in heat-stressed P. lutea may be due to increased access to seawater nutrients as a result of Symbiodiniaceae expulsion from the coral tissue. This could in turn stimulate the growth of endolithic microbes, which may explain previous reports of rapid endolithic proliferation following bleaching events [29, 110].
Putative role of the endolithic microbiome in holobiont stress tolerance
Coral hosts have been proposed to be capable of promoting specific microbial associations to rapidly alleviate environmental stress [111–113], but very little is known about the microbial traits that support host acclimatization to environmental stress, and which trade-offs occur between stress tolerance and holobiont fitness. Our findings show that two coral species with different skeletal physical properties harbor distinct endolithic microbiomes, resulting in contrasting metabolic strategies that may differentially affect bleaching susceptibility (Fig. 7). The bleaching-resistant G. edwardsi was associated with a higher taxonomic and functional endolithic diversity dominated by generalist bacteria with higher metabolic flexibility, which resulted in functionally redundant endolithic microbiomes and lower microbial C and N assimilation. In contrast, skeletal compartments of the bleaching-sensitive P. lutea harbored a lower taxonomic and functional endolithic diversity dominated by specialist bacteria such as Cyanobacteria and, concomitantly, higher C and N assimilation. We hypothesize that the lower productivity observed in G. edwardsi’s endoliths, attributed to the dominance of chemolithotrophs, may result in a constant translocation of C or N of lower magnitude from the skeleton to the host tissues, which may be beneficial in maintaining N limitation, a central feature of the stable coral-algal symbiosis [7, 104]. In contrast, the higher N assimilation observed in P. lutea’s endoliths during heat stress could result in increased N translocation and subsequent excess in the tissues. N excess has previously been linked with coral bleaching [114–116], and is known to rapidly disrupt the coral–algal symbiosis during heat stress [104].
Fig. 7. Proposed model on the effects of endolithic microbiome functional traits in holobiont nutrient cycling and bleaching susceptibility.
Endolithic microbiomes of the bleaching-resistant G. edwardsi were characterized by greater taxonomic diversity in conjunction with greater functional diversity and redundancy, which contrasted with endolithic microbiomes associated with the bleaching-sensitive P. lutea. C and N assimilation was greater in P. lutea than G. edwardsi (represented by black arrows), likely explained by the dominance of aerobic photoautotrophy over chemolithotrophy, respectively. Less C and N are expected to be translocated from the skeleton to the host tissues in G. edwardsi than in P. lutea (red arrows).
We propose a functional link between endolithic microbiome composition, metabolic activity, and bleaching susceptibility, in which the dominance of endolithic photoautotrophs can promote the translocation of excess nutrients to the tissues, which may contribute to Symbiodiniaceae expulsion. Further experimental work in other coral species is required to establish whether the dominance of photoautotrophs is linked to a higher C and N metabolism and whether increased endolithic metabolism is indeed linked to the coral bleaching response. Our findings raise important questions in the context of nutrient cycling and the implications of changes in endolithic metabolism during heat stress. For instance, it would be prudent to elucidate whether highly productive endolithic communities could support the ecological resilience of corals, in particular in the context of overcoming periods of elevated temperature stress. Likewise, it would be important to determine whether less productive endolithic communities favor the re-establishment of the coral-algal symbiosis following bleaching. To better understand whether and how microbes contribute to coral resilience, we must move beyond profiling microbial taxonomic diversity that cannot adequately capture inherent functional potentials. Identifying microbial functional traits that underpin holobiont resilience will be of major importance as they can inform microbiome manipulation approaches to counter the ongoing global loss of coral reefs.
Supplementary information
Acknowledgements
We would like to thank the Coastal & Marine Resources Core Lab (CMOR) for their assistance and support in field and wet laboratory operations. We further thank the KAUST BioScience Core Lab (BCL), in particular, we thank Xiang Zhao, Sadhasivam Perumal, and Emanuele Palescandolo for their assistance provided during library preparation and sequencing. This publication was supported by the Competitive Research Grant (CRG) URF/1/3400-01-01 from the King Abdullah University of Science and Technology (KAUST) to CRV, MP, and J-BR.
Author contributions
NR, CP, JBR, MP, and CRV designed the experiment. AC, NR, CP, JB, PG, MP, and JBR, collected the data. AC, NR, and JBR analysed the data. AC, JBR, and CRV interpreted the results. AC, JBR, and CRV wrote the manuscript with input from all co-authors.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
Scripts used are deposited in the GitHub repository https://github.com/ajcardenasb/Coral_endoliths; MAG assemblies are available from Cárdenas and Voolstra [84]. Raw sequencing data are deposited in the NCBI Sequence Read Archive (SRA) under BioProject PRJNA757245 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA757245).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Anny Cárdenas, Jean-Baptiste Raina.
Contributor Information
Anny Cárdenas, Email: ajcardenasb1@gmail.com.
Jean-Baptiste Raina, Email: jean-baptiste.raina@uts.edu.au.
Christian R. Voolstra, Email: christian.voolstra@uni-konstanz.de
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01283-y.
References
- 1.Pogoreutz C, Voolstra CR, Rädecker N, Weis V, Cardenas A, Raina J-B. The coral holobiont highlights the dependence of cnidarian animal hosts on their associated microbes. In Bosch TCG, Hadfield MG, editors. Cellular Dialogues in the Holobiont. CRC Press; 2020. pp. 91–118. 10.1201/9780429277375-7
- 2.Rohwer F, Seguritan V, Azam F, Knowlton N. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser. 2002;243:1–10. [Google Scholar]
- 3.LaJeunesse TC, Parkinson JE, Gabrielson PW, Jeong HJ, Reimer JD, Voolstra CR, et al. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr Biol. 2018;28:2570–80. doi: 10.1016/j.cub.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Muscatine L, Porter JW. Reef corals: mutualistic symbioses adapted to nutrient-poor environments. Bioscience. 1977;27:454–60. [Google Scholar]
- 5.Christian R, Voolstra DJ, Suggett RS, Peixoto JE, Parkinson KM, Quigley CB, et al. Extending the natural adaptive capacity of coral holobionts. Nature Reviews Earth & Environment. 2021;2:747–762. doi: 10.1038/s43017-021-00214-3. [DOI] [Google Scholar]
- 6.Bourne DG, Morrow KM, Webster NS. Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu Rev Microbiol. 2016;70:317–40. doi: 10.1146/annurev-micro-102215-095440. [DOI] [PubMed] [Google Scholar]
- 7.Rädecker N, Pogoreutz C, Voolstra CR, Wiedenmann J, Wild C. Nitrogen cycling in corals: the key to understanding holobiont functioning? Trends Microbiol. 2015;23:490–7. doi: 10.1016/j.tim.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Matthews JL, Raina JB, Kahlke T, Seymour JR, van Oppen MJ, Suggett DJ. Symbiodiniaceae‐bacteria interactions: rethinking metabolite exchange in reef‐building corals as multi‐partner metabolic networks. Environ Microbiol. 2020;22:1675–87. doi: 10.1111/1462-2920.14918. [DOI] [PubMed] [Google Scholar]
- 9.Kimes NE, Van Nostrand JD, Weil E, Zhou J, Morris PJ. Microbial functional structure of Montastraea faveolata, an important Caribbean reef‐building coral, differs between healthy and yellow‐band diseased colonies. Environ Microbiol. 2010;12:541–56. doi: 10.1111/j.1462-2920.2009.02113.x. [DOI] [PubMed] [Google Scholar]
- 10.Neave MJ, Apprill A, Ferrier-Pagès C, Voolstra CR. Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl Environ Micro. 2016;100:8315–24. doi: 10.1007/s00253-016-7777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neave MJ, Michell CT, Apprill A, Voolstra CR. Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci Rep. 2017;7:1–12. doi: 10.1038/srep40579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krediet CJ, Ritchie KB, Alagely A, Teplitski M. Members of native coral microbiota inhibit glycosidases and thwart colonization of coral mucus by an opportunistic pathogen. ISME J. 2013;7:980–90. doi: 10.1038/ismej.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raina J-B, Tapiolas D, Motti CA, Foret S, Seemann T, Tebben J, et al. Isolation of an antimicrobial compound produced by bacteria associated with reef-building corals. PeerJ. 2016;4:e2275. doi: 10.7717/peerj.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diaz JM, Hansel CM, Apprill A, Brighi C, Zhang T, Weber L, et al. Species-specific control of external superoxide levels by the coral holobiont during a natural bleaching event. Nat Commun. 2016;7:1–10. doi: 10.1038/ncomms13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunlap WC, Shick JM. Ultraviolet radiation‐absorbing mycosporine‐like amino acids in coral reef organisms: a biochemical and environmental perspective. J Phycol. 1998;34:418–30. [Google Scholar]
- 16.Webster NS, Smith LD, Heyward AJ, Watts JE, Webb RI, Blackall LL, et al. Metamorphosis of a scleractinian coral in response to microbial biofilms. Appl Environ Microbiol. 2004;70:1213–21. doi: 10.1128/AEM.70.2.1213-1221.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gómez-Lemos LA, Doropoulos C, Bayraktarov E, Diaz-Pulido G. Coralline algal metabolites induce settlement and mediate the inductive effect of epiphytic microbes on coral larvae. Sci Rep. 2018;8:1–11. doi: 10.1038/s41598-018-35206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pernice M, Raina J-B, Rädecker N, Cárdenas A, Pogoreutz C, Voolstra CR. Down to the bone: the role of overlooked endolithic microbiomes in reef coral health. ISME J. 2020;14:325–34. doi: 10.1038/s41396-019-0548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricci F, Marcelino VR, Blackall LL, Kühl M, Medina M, Verbruggen H. Beneath the surface: community assembly and functions of the coral skeleton microbiome. Microbiome. 2019;7:1–10. doi: 10.1186/s40168-019-0762-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcelino VR, Verbruggen H. Multi-marker metabarcoding of coral skeletons reveals a rich microbiome and diverse evolutionary origins of endolithic algae. Sci Rep. 2016;6:1–9. doi: 10.1038/srep31508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verbruggen H, Marcelino VR, Guiry MD, Cremen MCM, Jackson CJ. Phylogenetic position of the coral symbiont Ostreobium (Ulvophyceae) inferred from chloroplast genome data. J Phycol. 2017;53:790–803. doi: 10.1111/jpy.12540. [DOI] [PubMed] [Google Scholar]
- 22.Del Campo J, Pombert J-F, Šlapeta J, Larkum A, Keeling PJ. The ‘other’coral symbiont: Ostreobium diversity and distribution. ISME J. 2017;11:296–9. doi: 10.1038/ismej.2016.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massé A, Domart-Coulon I, Golubic S, Duché D, Tribollet A. Early skeletal colonization of the coral holobiont by the microboring Ulvophyceae Ostreobium sp. Sci Rep. 2018;8:1–11. doi: 10.1038/s41598-018-20196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halldal P. Photosynthetic capacities and photosynthetic action spectra of endozoic algae of the massive coral Favia. Biol Bull. 1968;134:411–24. [Google Scholar]
- 25.Fork D, Larkum A. Light harvesting in the green alga Ostreobium sp., a coral symbiont adapted to extreme shade. Mar Biol. 1989;103:381–5. [Google Scholar]
- 26.Fine M, Steindler L, Loya Y. Endolithic algae photoacclimate to increased irradiance during coral bleaching. Mar Freshw Res. 2004;55:115–21. [Google Scholar]
- 27.Fine M, Roff G, Ainsworth T, Hoegh-Guldberg O. Phototrophic microendoliths bloom during coral “white syndrome”. Coral Reefs. 2006;25:577–81. [Google Scholar]
- 28.Galindo-Martínez CT, Weber M, Avila-Magaña V, Enríquez S, Kitano H, Medina M, et al. The role of the endolithic alga Ostreobium spp. during coral bleaching recovery. Sci Rep. 2022;12:1–12. doi: 10.1038/s41598-022-07017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fine M, Loya Y. Endolithic algae: an alternative source of photoassimilates during coral bleaching. Proc R Soc B Biol Sci. 2002;269:1205–10. doi: 10.1098/rspb.2002.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlichter D, Zscharnack B, Krisch H. Transfer of photoassimilates from endolithic algae to coral tissue. Naturwissenschaften. 1995;82:561–4. [Google Scholar]
- 31.Sangsawang L, Casareto BE, Ohba H, Vu HM, Meekaew A, Suzuki T, et al. 13C and 15N assimilation and organic matter translocation by the endolithic community in the massive coral Porites lutea. R Soc Open Sci. 2017;4:171201. doi: 10.1098/rsos.171201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcelino VR, Morrow KM, van Oppen MJ, Bourne DG, Verbruggen H. Diversity and stability of coral endolithic microbial communities at a naturally high pCO2 reef. Mol Ecol. 2017;26:5344–57. doi: 10.1111/mec.14268. [DOI] [PubMed] [Google Scholar]
- 33.Marcelino VR, Van Oppen MJ, Verbruggen H. Highly structured prokaryote communities exist within the skeleton of coral colonies. ISME J. 2018;12:300–3. doi: 10.1038/ismej.2017.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang S-H, Tandon K, Lu C-Y, Wada N, Shih C-J, Hsiao SS-Y, et al. Metagenomic, phylogenetic, and functional characterization of predominant endolithic green sulfur bacteria in the coral Isopora palifera. Microbiome. 2019;7:1–13. doi: 10.1186/s40168-018-0616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrer L, Szmant A, editors. Nutrient regeneration by the endolithic community in coral skeletons. Proceedings of the 6th International Coral Reef Symposium; 1988: AIMS Townsville, Australia.
- 36.Eakin CM, Devotta D, Heron S, Connolly S, Liu G, Geiger E, et al. The 2014-17 global coral bleaching event: The most severe and widespread coral reef destruction. Research Square. 2022 doi: 10.21203/rs.3.rs-1555992/v1. [DOI] [Google Scholar]
- 37.Hughes TP, Kerry JT, Álvarez-Noriega M, Álvarez-Romero JG, Anderson KD, Baird AH, et al. Global warming and recurrent mass bleaching of corals. Nature. 2017;543:373–7. doi: 10.1038/nature21707. [DOI] [PubMed] [Google Scholar]
- 38.Hughes TP, Anderson KD, Connolly SR, Heron SF, Kerry JT, Lough JM, et al. Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science. 2018;359:80–3. doi: 10.1126/science.aan8048. [DOI] [PubMed] [Google Scholar]
- 39.Hughes TP, Kerry JT, Baird AH, Connolly SR, Dietzel A, Eakin CM, et al. Global warming transforms coral reef assemblages. Nature. 2018;556:492–6. doi: 10.1038/s41586-018-0041-2. [DOI] [PubMed] [Google Scholar]
- 40.Veron J, Stafford-Smith M, Corals of the World, Volumes 1-3. Australian Institute of Marine Science. Odyssey Publishing; 2000.
- 41.Brown B, Dunne R, Phongsuwan N, Patchim L, Hawkridge J. The reef coral Goniastrea aspera: a ‘winner’becomes a ‘loser’during a severe bleaching event in Thailand. Coral Reefs. 2014;33:395–401. [Google Scholar]
- 42.Klepac C, Barshis D. Reduced thermal tolerance of massive coral species in a highly variable environment. Proc R Soc B Biol Sci. 2020;287:20201379. doi: 10.1098/rspb.2020.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicolas R, Evensen CR, Voolstra M, Fine G, Perna C, Buitrago-López A, et al. Empirically derived thermal thresholds of four coral species along the Red Sea using a portable and standardized experimental approach. Coral Reefs. 2022;41:239–52. doi: 10.1007/s00338-022-02233-y. [DOI] [Google Scholar]
- 44.Madin JS, Anderson KD, Andreasen MH, Bridge TC, Cairns SD, Connolly SR, et al. The Coral Trait Database, a curated database of trait information for coral species from the global oceans. Sci Data. 2016;3:1–22. doi: 10.1038/sdata.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roth F, Karcher DB, Rädecker N, Hohn S, Carvalho S, Thomson T, et al. High rates of carbon and dinitrogen fixation suggest a critical role of benthic pioneer communities in the energy and nutrient dynamics of coral reefs. Funct Ecol. 2020;34:1991–2004. [Google Scholar]
- 46.Harrison PJ, Waters RE, Taylor F. A broad spectrum artificial sea water medium for coastal and open ocean phytoplankton. J Phycol. 1980;16:28–35. [Google Scholar]
- 47.Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bayer T, Neave MJ, Alsheikh-Hussain A, Aranda M, Yum LK, Mincer T, et al. The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl Environ Microbiol. 2013;79:4759–62. doi: 10.1128/AEM.00695-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wickham H. ggplot2. Wiley Interdiscip Rev Comput Stat. 2011;3:180–5. [Google Scholar]
- 52.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dixon P. The vegan package. J Veg Sci. 2003;14:927–30. [Google Scholar]
- 54.Lin H, Peddada SD. Analysis of compositions of microbiomes with bias correction. Nat Commun. 2020;11:1–11. doi: 10.1038/s41467-020-17041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li D, Luo R, Liu C-M, Leung C-M, Ting H-F, Sadakane K, et al. MEGAHIT v1. 0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 57.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824–34. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peng Y, Leung HC, Yiu S-M, Chin FY. Meta-IDBA: a de Novo assembler for metagenomic data. Bioinformatics. 2011;27:i94–i101. doi: 10.1093/bioinformatics/btr216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–5. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:1–11. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–9. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun. 2016;7:1–9. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, et al. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2020;36:2251–2. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hill MO. Diversity and evenness: a unifying notation and its consequences. Ecology. 1973;54:427–32. [Google Scholar]
- 65.Bates D, Sarkar D, Bates MD, Matrix L. The lme4 package. R Package Version. 2007;2:74. [Google Scholar]
- 66.Mandal S, Van Treuren W, White RA, Eggesbø M, Knight R, Peddada SD. Analysis of composition of microbiomes: a novel method for studying microbial composition. Micro Ecol Health Dis. 2015;26:27663. doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rivera-Pinto J, Egozcue JJ, Pawlowsky-Glahn V, Paredes R, Noguera-Julian M, Calle ML. Balances: a new perspective for microbiome analysis. mSystems. 2018;3:e00053–18. doi: 10.1128/mSystems.00053-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7:e7359. doi: 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alneberg J, Bjarnason BS, De Bruijn I, Schirmer M, Quick J, Ijaz UZ, et al. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11:1144–6. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 71.Wu Y-W, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32:605–7. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 72.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uritskiy GV, DiRuggiero J, Taylor J. MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6:1–13. doi: 10.1186/s40168-018-0541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olm MR, Brown CT, Brooks B, Banfield JF. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017;11:2864–8. doi: 10.1038/ismej.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020;36:1925–7. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 77.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 78.Zhou Z, Tran P, Briester AM, Liu Y, Kieft K, Cowley ES, et al. METABOLIC: high-throughput profiling of microbial genomes for functional traits, metabolism, biogeochemistry, and community-scale functional networks. Microbiome. 2022;10:33. doi: 10.1186/s40168-021-01213-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Na S-I, Kim YO, Yoon S-H, Ha S-M, Baek I, Chun J. UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol. 2018;56:280–5. doi: 10.1007/s12275-018-8014-6. [DOI] [PubMed] [Google Scholar]
- 80.Morel J, Jay S, Féret J-B, Bakache A, Bendoula R, Carreel F, et al. Exploring the potential of PROCOSINE and close-range hyperspectral imaging to study the effects of fungal diseases on leaf physiology. Sci Rep. 2018;8:1–13. doi: 10.1038/s41598-018-34429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Calamita F, Imran HA, Vescovo L, Mekhalfi ML, La, Porta N. Early identification of root rot disease by using hyperspectral reflectance: the case of pathosystem Grapevine/Armillaria. Remote Sens. 2021;13:2436. [Google Scholar]
- 82.Brumfield KD, Huq A, Colwell RR, Olds JL, Leddy MB. Microbial resolution of whole-genome shotgun and 16S amplicon metagenomic sequencing using publicly available NEON data. PLoS One. 2020;15:e0228899. doi: 10.1371/journal.pone.0228899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khachatryan L, de Leeuw RH, Kraakman ME, Pappas N, Te Raa M, Mei H, et al. Taxonomic classification and abundance estimation using 16S and WGS—A comparison using controlled reference samples. Forensic Sci Int Genet. 2020;46:102257. doi: 10.1016/j.fsigen.2020.102257. [DOI] [PubMed] [Google Scholar]
- 84.Cardénas A, Voolstra C. 75 Coral Endolith Bacterial Genomes (MAGs) from Red Sea corals Goniastrea edwardsi and Porites lutea (Version 1) [Data set] Zenodo. 2021 doi: 10.5281/zenodo.5606932. [DOI] [Google Scholar]
- 85.Branson O, Bonnin EA, Perea DE, Spero HJ, Zhu Z, Winters M, et al. Nanometer-scale chemistry of a calcite biomineralization template: Implications for skeletal composition and nucleation. Proc Natl Acad Sci. 2016;113:12934–9. doi: 10.1073/pnas.1522864113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sauvage T, Schmidt WE, Suda S, Fredericq S. A metabarcoding framework for facilitated survey of endolithic phototrophs with tufA. BMC Ecol. 2016;16:1–21. doi: 10.1186/s12898-016-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wegley L, Edwards R, Rodriguez‐Brito B, Liu H, Rohwer F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol. 2007;9:2707–19. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- 88.Robbins S, Song W, Engelberts J, Glasl B, Slaby BM, Boyd J, et al. A genomic view of the microbiome of coral reef demosponges. ISME J. 2021;15:1641–54. doi: 10.1038/s41396-020-00876-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang S-Y, Lu C-Y, Tang S-L, Das RR, Sakai K, Yamashiro H, et al. Effects of ocean acidification on coral endolithic bacterial communities in Isopora palifera and Porites lobata. Front Mar Sci. 2020;7:603293. [Google Scholar]
- 90.Yang SH, Lee ST, Huang CR, Tseng CH, Chiang PW, Chen CP, et al. Prevalence of potential nitrogen‐fixing, green sulfur bacteria in the skeleton of reef‐building coral Isopora palifera. Limnol Oceanogr. 2016;61:1078–86. [Google Scholar]
- 91.Cai L, Zhou G, Tian R-M, Tong H, Zhang W, Sun J, et al. Metagenomic analysis reveals a green sulfur bacterium as a potential coral symbiont. Sci Rep. 2017;7:1–11. doi: 10.1038/s41598-017-09032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kühl M, Holst G, Larkum AW, Ralph PJ. Imaging of oxygen dynamics within the endolithic algal community of the massive coral Porites Lobata. J Phycol. 2008;44:541–50. doi: 10.1111/j.1529-8817.2008.00506.x. [DOI] [PubMed] [Google Scholar]
- 93.Roberty S, Bailleul B, Berne N, Franck F, Cardol P. PSI Mehler reaction is the main alternative photosynthetic electron pathway in Symbiodinium sp., symbiotic dinoflagellates of cnidarians. N. Phytol. 2014;204:81–91. doi: 10.1111/nph.12903. [DOI] [PubMed] [Google Scholar]
- 94.Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, et al. Regulation and function of ascorbate peroxidase isoenzymes. J Exp Bot. 2002;53:1305–19. [PubMed] [Google Scholar]
- 95.Roberty S, Fransolet D, Cardol P, Plumier J-C, Franck F. Imbalance between oxygen photoreduction and antioxidant capacities in Symbiodinium cells exposed to combined heat and high light stress. Coral Reefs. 2015;34:1063–73. [Google Scholar]
- 96.Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R, et al. Hydrogen is an energy source for hydrothermal vent symbioses. Nature. 2011;476:176–80. doi: 10.1038/nature10325. [DOI] [PubMed] [Google Scholar]
- 97.McCollom T, Amend J. A thermodynamic assessment of energy requirements for biomass synthesis by chemolithoautotrophic micro‐organisms in oxic and anoxic environments. Geobiology. 2005;3:135–44. [Google Scholar]
- 98.Heijnen J, Van Dijken J. In search of a thermodynamic description of biomass yields for the chemotrophic growth of microorganisms. Biotechnol Bioeng. 1992;39:833–58. doi: 10.1002/bit.260390806. [DOI] [PubMed] [Google Scholar]
- 99.Bar-Even A, Noor E, Milo R. A survey of carbon fixation pathways through a quantitative lens. J Exp Bot. 2012;63:2325–42. doi: 10.1093/jxb/err417. [DOI] [PubMed] [Google Scholar]
- 100.Schulze E-D, Mooney HA, Biodiversity and ecosystem function: Springer Science & Business Media; 2012.
- 101.Lawton JH, Brown VK, Redundancy in ecosystems. Biodiversity and Ecosystem Function: Springer; 1994. p. 255–70.
- 102.Mori AS, Furukawa T, Sasaki T. Response diversity determines the resilience of ecosystems to environmental change. Biol Rev. 2013;88:349–64. doi: 10.1111/brv.12004. [DOI] [PubMed] [Google Scholar]
- 103.Nyström M. Redundancy and response diversity of functional groups: implications for the resilience of coral reefs. Ambio. 2006;35:30–5. [PubMed] [Google Scholar]
- 104.Rädecker N, Pogoreutz C, Gegner HM, Cárdenas A, Roth F, Bougoure J, et al. Heat stress destabilizes symbiotic nutrient cycling in corals. Proc Natl Acad Sci. 2021;118:e2022653118. doi: 10.1073/pnas.2022653118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ziegler M, Grupstra CG, Barreto MM, Eaton M, BaOmar J, Zubier K, et al. Coral bacterial community structure responds to environmental change in a host-specific manner. Nat Commun. 2019;10:1–11. doi: 10.1038/s41467-019-10969-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dikou A, Van, Woesik R. Survival under chronic stress from sediment load: spatial patterns of hard coral communities in the southern islands of Singapore. Mar Pollut Bull. 2006;52:1340–54. doi: 10.1016/j.marpolbul.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 107.Hennige SJ, Smith DJ, Walsh S-J, McGinley MP, Warner ME, Suggett DJ. Acclimation and adaptation of scleractinian coral communities along environmental gradients within an Indonesian reef system. J Exp Mar Biol Ecol. 2010;391:143–52. [Google Scholar]
- 108.Cárdenas A, Neave MJ, Haroon MF, Pogoreutz C, Rädecker N, Wild C, et al. Excess labile carbon promotes the expression of virulence factors in coral reef bacterioplankton. ISME J. 2018;12:59–76. doi: 10.1038/ismej.2017.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cárdenas A, Ye J, Ziegler M, Payet JP, McMinds R, Thurber RV, et al. Coral-associated viral assemblages from the Central Red Sea align with host species and contribute to holobiont genetic diversity. Front Microbiol. 2020;11:572534. doi: 10.3389/fmicb.2020.572534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McCook GD-PLJ. The fate of bleached corals: patterns and dynamics of algal recruitment. Mar Ecol Prog Ser. 2002;232:115–28. [Google Scholar]
- 111.Reshef L, Koren O, Loya Y, Zilber‐Rosenberg I, Rosenberg E. The coral probiotic hypothesis. Environ Microbiol. 2006;8:2068–73. doi: 10.1111/j.1462-2920.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- 112.Voolstra CR, Ziegler M. Adapting with microbial help: microbiome flexibility facilitates rapid responses to environmental change. BioEssays. 2020;42:2000004. doi: 10.1002/bies.202000004. [DOI] [PubMed] [Google Scholar]
- 113.Rosenberg E, Zilber-Rosenberg I. The hologenome concept of evolution after 10 years. Microbiome. 2018;6:1–14. doi: 10.1186/s40168-018-0457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wiedenmann J, D’Angelo C, Smith EG, Hunt AN, Legiret F-E, Postle AD, et al. Nutrient enrichment can increase the susceptibility of reef corals to bleaching. Nat Clim Change. 2013;3:160–4. [Google Scholar]
- 115.DeCarlo TM, Gajdzik L, Ellis J, Coker DJ, Roberts MB, Hammerman NM, et al. Nutrient-supplying ocean currents modulate coral bleaching susceptibility. Sci Adv. 2020;6:eabc5493. doi: 10.1126/sciadv.abc5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pogoreutz C, Rädecker N, Cardenas A, Gärdes A, Voolstra CR, Wild C. Sugar enrichment provides evidence for a role of nitrogen fixation in coral bleaching. Glob Change Biol. 2017;23:3838–48. doi: 10.1111/gcb.13695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Scripts used are deposited in the GitHub repository https://github.com/ajcardenasb/Coral_endoliths; MAG assemblies are available from Cárdenas and Voolstra [84]. Raw sequencing data are deposited in the NCBI Sequence Read Archive (SRA) under BioProject PRJNA757245 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA757245).