Abstract
Catabolic and anabolic processes are finely coordinated in microorganisms to provide optimized fitness under varying environmental conditions. Understanding this coordination and the resulting physiological traits reveals fundamental strategies of microbial acclimation. Here, we characterized the system-level physiology of Methanococcus maripaludis, a niche-specialized methanogenic archaeon, at different dilution rates ranging from 0.09 to 0.003 h−1 in chemostat experiments under phosphate (i.e., anabolic) limitation. Phosphate was supplied as the limiting nutrient, while formate was supplied in excess as the catabolic substrate and carbon source. We observed a decoupling of catabolism and anabolism resulting in lower biomass yield relative to catabolically limited cells at the same dilution rates. In addition, the mass abundance of several coarse-grained proteome sectors (i.e., combined abundance of proteins grouped based on their function) exhibited a linear relationship with growth rate, mostly ribosomes and their biogenesis. Accordingly, cellular RNA content also correlated with growth rate. Although the methanogenesis proteome sector was invariant, the metabolic capacity for methanogenesis, measured as methane production rates immediately after transfer to batch culture, correlated with growth rate suggesting translationally independent regulation that allows cells to only increase catabolic activity under growth-permissible conditions. These observations are in stark contrast to the physiology of M. maripaludis under formate (i.e., catabolic) limitation, where cells keep an invariant proteome including ribosomal content and a high methanogenesis capacity across a wide range of growth rates. Our findings reveal that M. maripaludis employs fundamentally different strategies to coordinate global physiology during anabolic phosphate and catabolic formate limitation.
Subject terms: Archaeal physiology, Environmental microbiology
Introduction
Microorganisms tightly coordinate catabolic and anabolic processes to fine-tune allocation of cellular resources and to modulate growth in response to the varying environment. Escherichia coli, for example, responds to catabolic (e.g., carbon substrate-) or anabolic (e.g., ammonia-) limitations by increasing the expression of functionally relevant proteome sectors to alleviate the limitation [1, 2]. Despite complex regulatory details, a microbial proteome can be partitioned into coarse-grained functional sectors, which allows effective, quantitative characterization of their strategy of global resource allocation [1–3]. Proteome sectors that comprise up to half of the proteome by mass exhibit linear relations with growth rate under different limitations in E. coli [1, 2, 4, 5]. These growth-dependent proteome changes modulate different aspects of cell physiology, including metabolism [6], growth yield [7], cell composition [8], and cell size [9]. In marked contrast to E. coli, our recent study on a metabolically specialized, methane-producing archaeon (i.e., methanogen), Methanococcus maripaludis, showed a cellular proteome and cellular composition invariant of growth rate under catabolic limitation [10]. The difference in the resource allocation strategies by different prokaryotes reveals versatility of physiological acclimation to environmental changes. A system-level understanding of physiological regulation can help to form predictive phenotypic models and guide design of bioprocesses [1].
Phosphorus is an essential nutrient for all known life including microorganisms. The assimilable form of inorganic phosphate (PO43−, Pi) is frequently found at low, limiting concentrations in natural environments [11]. Phosphate limitation has been reported for microbial growth in freshwater [12, 13], seawater [14], wastewater [15], and drinking water [16], as well as in clinical settings [17, 18]. How cells coordinate catabolic and anabolic functions and other physiological processes in response to changes in limiting phosphate (anabolic) flux, however, is underexplored [19].
In this study, we investigated the physiology of the methanogen Methanococcus maripaludis at various growth rates under anabolic limitation with phosphate as the limiting nutrient. The hydrogenotrophic (hydrogen-utilizing), anaerobic M. maripaludis has an ecophysiological strategy that differs from metabolically versatile model microorganisms such as E. coli. We previously showed that M. maripaludis maintains a constant proteome and cellular composition, including ribosome number, over a two orders of magnitude range of catabolically limited growth rates controlled by formate (i.e., electron donor) flux [10]. This observation raised the question whether M. maripaludis also adopts a different strategy in response to anabolic limitation compared to model microorganisms like E. coli. For example, does M. maripaludis modulate its ribosome number in response to growth rate under anabolically limited growth conditions? Using the established experimental platform of anoxic chemostats, we studied the effect of phosphate limitation on M. maripaludis’ physiology at different growth rates.
Materials and methods
Growth conditions, chemostat setup, and metabolite determination
M. maripaludis strain MM901 [20] was grown in anoxic glycylglycine-buffered medium with formate as the energy substrate at 30 °C as previously described [10]. Briefly, bioreactor (Eppendorf Bioflo 120, Hamburg, Germany) and medium bottles were pressurized to 3 psi with N2 gas (NF, 99.997%, Linde, Dublin, Ireland) to maintain an anoxic environment. Peristaltic pumps were used to maintain continuous in- and out-flow of medium. Each chemostat experiment started with a fresh cryostock inoculum to prevent accumulation of adaptive mutations and was operated for approximately 6 retention times before sampling and analysis. We used dilution rates of 0.0875 ± 0.0029 h−1 (“fast”, 38% of μmax), 0.0195 ± 0.0015 h−1 (“intermediate”, 8% of μmax), and 0.0034 ± 0.0001 h−1 (“very slow”, 1% of μmax) in biological triplicates. While the dilution rate in a chemostat is numerically different from growth rate (dilution rate = growth rate – death rate), we previously described a slow M. maripaludis cell lysis rate during chemostat cultivation of 0.002 h−1 that is not dependent on dilution rate [10]. Therefore, we use the terms dilution rate and growth rate interchangeably and refer to them as “fast”, “intermediate”, and “very slow”. Under phosphate limitation, 60 µM sodium phosphate was supplied as the sole source of phosphorus, while formate was supplied in excess in the form of 200 mM sodium formate (fast growth), 200 mM sodium formate + 200 mM formic acid (intermediate growth) or 200 mM sodium formate + 400 mM formic acid (very slow growth). Additional electron donor at slower growth rates was necessary to keep the cells phosphate limited, due to the reduction in growth yield on formate at lower growth rates caused by the increased impact of maintenance energy and cell lysis at lower growth rates [10]. Formic acid was supplied instead of formate because it does not cause net consumption of protons - in other words, does not affect reactor pH - when metabolized to methane. The low pH, formic acid-containing growth medium was only supplied to actively growing cultures so that it would be immediately consumed without decreasing reactor pH.
During chemostat experiments, formate, methane, OD, and pH were monitored daily to check steady-state operation. Formate concentration was determined by high performance liquid chromatography (HPLC) with an Aminex HPX-87H column (Bio-Rad, Hercules, CA, USA). Methane and hydrogen were measured by an 8610C gas chromatograph (SRI Instruments, Torrance, CA, USA) as previously described [10].
Proteomic analysis
Biological triplicate samples were harvested via centrifugation for mass spectrometry and proteomic analysis as described previously [10]. Proteome sectors were quantified by adding the mass fraction of corresponding proteins determined by spectral counting and individual proteins were quantified by relative quantification against a 15N-labeled reference [10]. Growth rate dependence was determined using a linear model in R (version 4.0.2) to test for significant non-zero slopes (p values were adjusted for multiple comparisons using the Benjamini–Hochberg false discovery rate correction [21]). Statistical analysis of pairwise comparison between proteins under formate and phosphate limitation was done in Perseus (version 1.6.14.0) [22]. The mass spectrometry proteomics data are available on MassIVE under accession number MSV000087621.
Capacity for methanogenesis
Aliquots of cultures were transferred anoxically (using syringes and needles pre-flushed in the H2S-enriched headspace of an anoxic serum bottle containing a 1 M sodium sulfide solution) from the chemostat to Hungate tubes flushed with N2 with addition of 150 mM sodium formate, 1 mM sodium sulfide, as well as 0.8 mM K2HPO4 and/or 5 µg mL−1 translation inhibitor pseudomonic acid when indicated. Methane production and OD600 increase were immediately monitored by the SRI 8610C gas chromatograph and a WPA CO8000 Cell Density Meter (Biochrom, Cambridge, United Kingdom), respectively, for 5–6 h to calculate the initial specific methanogenesis rate [10].
Determination of cell size and cellular components
Cell size and chromosomal copy number per cell were determined by flow cytometry as previously described [10]. Briefly, cells fixed in 3% glutaraldehyde + paraformaldehyde were diluted in tris-ethylenediaminetetraacetic acid (TE) buffer with 0.25% Tween-20, digested by RNAse A, and stained by PicoGreen dye (Invitrogen, Waltham, MA, USA). Cell size was calibrated using microspheres from a flow cytometry size calibration kit (Invitrogen). Chromosome copy number was calibrated using rifampicin-treated E. coli cells, which contain either 1 or 2 copies of chromosomes [10].
Total RNA was extracted using a phenol:chloroform-based chemical separation method, and quantified by Bioanalyzer (Agilent, Santa Clara, CA, USA) using the luciferase RNA (Promega, Madison, WI, USA) as internal standard as described before [10].
To convert cell density measured at OD600 to cell count, cell numbers were determined at various cell densities by counting at least 1000 cells using a Petroff-Hausser Counting Chamber (Hausser Scientific, Horsham, PA, USA) and the resulting linear calibration curve (cell number = 1.53 * 109 * OD600 – 2.2 * 108, Supplementary Fig. S1) was used for conversion.
Quantification of polysome fraction
As described before [10], cultures were quickly harvested and stored at −80 °C in lysis buffer (20 mM tris-HCl pH 8.0, 10 mM MgCl2, 100 mM NH4Cl, 0.4% Triton X-100, 400 U/mL RNaseOUT, 2 mM dithiothreitol (DTT), 100 U/mL RNase-free DNase I) before further analysis. After lysis by two freeze-thaw cycles, 80–200 μg RNA of lysate were loaded to 10–50% linear sucrose gradients (20 mM Tris-HCl pH 7.5, 10 mM MgCl2, 100 mM NH4Cl, and 2 mM DTT) and centrifuged at 221,652 × g for 2 h at 4 ˚C. Fractions containing 30 S, 50 S, free 70 S, and monosome ribosomes were pooled and separated from the polysome fraction. RNA from different fractions was measured as described above to calculate polysome fractions.
Results
Phosphate limitation affected cellular energy allocation
Under phosphate limitation, M. maripaludis decouples catabolic and anabolic processes [23, 24]. In preliminary experiments, we determined in batch cultures that a phosphate concentration of 60 µM became limiting for M. maripaludis growth above an OD600 of ca. 0.5. Phosphate limitation was confirmed by an observed increase of cell density upon addition of phosphate, but not upon addition of formate. When implementing phosphate limitation in the chemostat, cultures were supplied with 60 µM phosphate and with as much formate as necessary to reach the OD threshold where they were phosphate limited. These concentrations were empirically determined to be 200 mM at fast, 400 mM at intermediate, and 600 mM at very slow growth rates (formate in excess of 200 mM was supplied in the form of formic acid to keep the cultures at a stable pH).
The decoupling of catabolic and anabolic processes in phosphate-limited M. maripaludis cells lead to a reduction in growth yield (cell production measured as OD600 per mol substrate) compared to formate limitation (Supplementary Fig. S2, also see Supplementary Dataset S1 for accompanying data of the chemostat runs). The extent of this difference is affected by the amount of additional electron donor that was added, as we observed that phosphate-limited cultures were able to consume electron donor without further increasing biomass.
Several key proteome sectors correlated with growth rates in phosphate-limited cells
The proteome was quantified using proteomic mass spectrometry with three independent biological replicates for each condition. A total of 1185 proteins were detected, corresponding to 69% of the predicted open reading frames, and of these, 774 (65%) were quantified against an 15N-labeled reference (see Supplementary Dataset S2 for an overview of mass fraction, relative quantification and functional classification of M. maripaludis proteins), which is comparable in coverage to the previous study [10]. We employed regression analysis on both coarse-grained functional proteome sectors [10] and single proteins to compare their abundance at different growth rates and conditions. When comparing this dataset to the previous formate limitation dataset, proteins of the phosphate uptake operon (pst operon, MMP1095–1099) were, as expected, significantly induced under phosphate-limited conditions, especially at higher growth rates (Supplementary Fig. S3) confirming cells were indeed phosphate-limited.
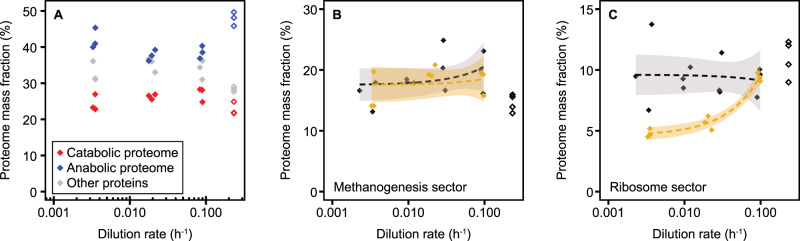
Both the catabolic and anabolic proteome sectors were largely invariant with growth rate under phosphate-limited conditions (Fig. 1A). The catabolic proteome is mainly composed of the enzymes of the methanogenesis pathway (the largest functional sector), which comprise 18% ± 2% of the total proteome by mass, independent of growth rate (Fig. 1B). Several other proteome sectors, however, did exhibit statistically significant increases or decreases as a function of growth rate under phosphate limitation (Supplementary Fig. S4), which is in marked contrast to their invariance in formate-limiting growth [10]. Most prominently, the ribosome sector, which is the largest anabolic proteome sector, decreased by about half from 9.4% ± 0.3% of the total proteome at fast growth to 4.9% ± 0.3% of the total proteome at very slow growth (Fig. 1C). Similarly, the ribosome biogenesis sector (containing 33 genes involved in rRNA modification and ribosome assembly and maturation) decreased from 1.95% ± 0.13% to 1.55% ± 0.04% (Supplementary Fig. S4). Other anabolic proteome sectors involved in synthesis of RNA (transcription, purine/pyrimidine metabolism) showed a significant inverse trend by increasing with decreasing growth rate (Supplementary Fig. S4). Similar trends have been observed before for formate-limited conditions, although the trends did not pass the significance test (Supplementary Fig. S4) [10]. Besides these, the incomplete reductive TCA cycle and queuosine-archaeosine biosynthesis sectors were significantly negatively correlated with growth rate (Supplementary Fig. S4). Significant positive correlations were found for the V-type ATPase (from 4.57% ± 0.16% at fast to 1.90% ± 0.04% at very slow growth) as well as the motility/chemotaxis sector consisting of 12 proteins of the archaellum [25, 26] and 7 putative chemotaxis proteins (from 2.22% ± 0.23% at fast to 0.42% ± 0.18% at very slow growth) (Supplementary Fig. S4). Both trends were also observed under formate-limited conditions but only the latter was significant (Supplementary Fig. S4).
Fig. 1. Proteome allocation at different growth rates.
Allocation of the catabolic and anabolic proteome (A) and the respective largest functional sub-sectors, methanogenesis (B) and ribosomes (C), in relation to growth rate. In B, C, mass fraction of total proteome is shown for phosphate-limited chemostat growth (gold, this study) with data under formate limitation [10] shown for comparison (black, closed symbols). In all three plots, results from an exponential phase batch culture sample are shown as open symbols, but not used for regression analysis. The ribosome sector under phosphate-limited conditions correlates positively with growth rate (significantly non-zero slope of linear regression, p < 0.05 after correction for multiple comparisons). Shaded areas depict 5% confidence level intervals.
At the single protein level, 27 proteins correlated positively and 2 proteins negatively with growth rate, mostly belonging to ribosome (11 proteins), V-type ATPase and motility/chemotaxis (4 proteins each), and other sectors (10 proteins) (Supplementary Table S1).
Overall, M. maripaludis has a more static proteome even under phosphate-limited conditions with only approximately 10% of the proteome being altered over a wide range of growth rates compared to E. coli where up to 50% of the proteome is remodeled [1].
Methanogenesis capacity was affected by phosphate limitation
To test whether cells growing at different growth rates retained the capacity for constant methanogenesis rates, as was the case under formate limitation [10], we assayed chemostat-grown cells for their intrinsic capacity of methanogenesis by supplying excess formate (150 mM). In the chemostat, the rate of methane formation correlated linearly with the dilution rate (or, the medium supply) as expected (Fig. 2A, gold color). We then transferred these cells from chemostat to batch culture in Hungate tubes with excess formate and followed their immediate methane production rate with and without protein synthesis inhibitor (Fig. 2A, red color). These rates represent the intrinsic methanogenesis capacity of the cells under chemostat-growing conditions. The addition of excess phosphate during those batch experiments did not lead to an increase in methanogenesis capacity (Supplementary Fig. S5).
Fig. 2. Methane production rates and methanogenesis capacities at different growth rates.
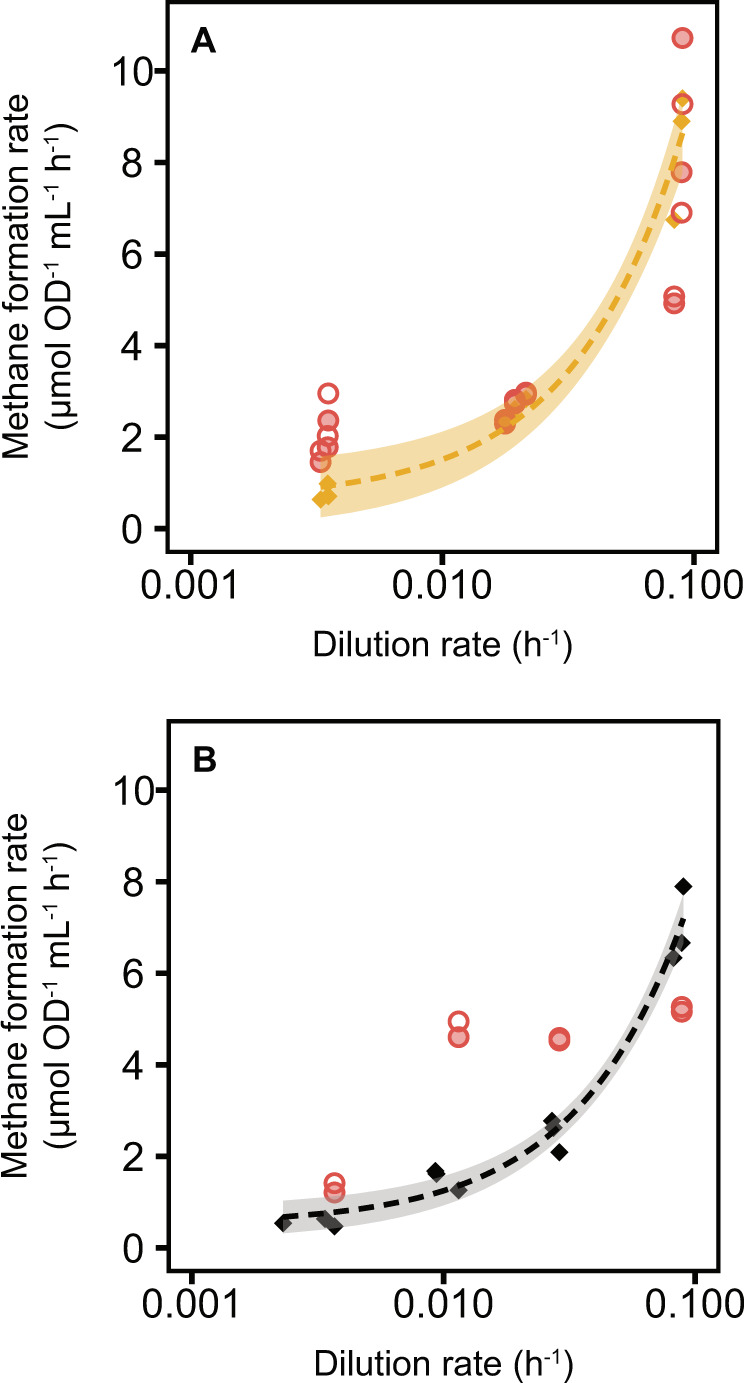
A Methane production rates of M. maripaludis under phosphate-limited conditions of cells growing in the chemostat (gold diamonds) and after being transferred to Hungate tubes with excess formate (150 mM) for 6 h (red circles, x-axis indicates dilution rate before transfer). Specific rates of in situ methane formation linearly correlated with dilution rate (dashed line, shaded area depicts 5% confidence level interval). Specific methanogenesis rates of transferred cells (methanogenesis capacity) were measured with (filled circles) and without (open circles) addition of pseudomonic acid, an antibiotic that inhibits protein synthesis. B Analog graph to A under formate-limited conditions, re-plotted from Müller et al. 2021 [10].
We observed a significant decrease in methanogenesis capacity independent of new protein synthesis that correlated with the growth rate and was similar to the in situ methanogenesis rates observed in the chemostats (Fig. 2A). The proteome analysis predicted an invariant high catabolic capacity at all growth rates due to the growth-rate invariant methanogenesis proteome fraction (Fig. 1B), suggesting a phosphate limitation-induced regulation of metabolic activity that is independent of gene expression. This finding is in marked contrast with our previous observation of methanogenesis capacity in formate-limited growth (Fig. 2B). During formate limitation, M. maripaludis cells maintained a high methanogenesis capacity at medium and slow growth rates consistent with the invariant proteome [10] (Fig. 1B).
Cellular composition and ribosomal activity were altered under phosphate limitation
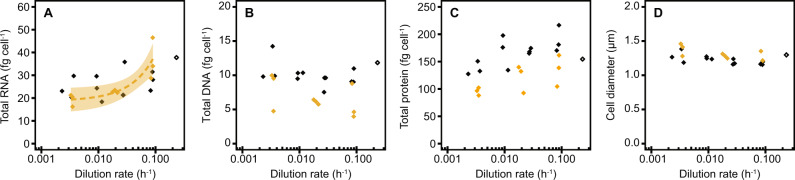
We determined the macromolecular composition (i.e., RNA, DNA, and protein contents) of M. maripaludis at different growth rates. Consistent with our proteomic data, RNA content of cells significantly correlated with growth rates (Fig. 3A). RNA content decreased from 36 ± 9 fg cell−1 at fast (0.0875 ± 0.0029 h−1) to 19 ± 3 fg cell−1 at very slow (0.0034 ± 0.0001 h−1) dilution rates. DNA and protein content did not significantly correlate with growth rate (Fig. 3B). Yet, the average DNA content of 3.7 ± 1.2 chromosomal copies per cell (6.6 ± 2.2 fg cell−1) is significantly lower than when M. maripaludis is formate-limited (5.6 ± 0.8 chromosomal copies per cell, 10.0 ± 1.4 fg cell−1). The protein content of 117 ± 26 fg cell−1 is also significantly lower than during formate-limited growth (176 ± 23 fg cell−1) (Fig. 3C). Despite an overall lower macromolecular content, cell size of M. maripaludis was slightly, yet significantly, larger at phosphate-limited (1.31 ± 0.08 µm) compared to formate-limited (1.22 ± 0.06 µm) conditions based on flow cytometry and did not significantly correlate with growth rate (Fig. 3D).
Fig. 3. Macromolecular composition and cell size of M. maripaludis at different growth rates, showing data from chemostat-grown cells (filled diamonds) under phosphate- (gold, this study) and formate- (black, [10]) limited conditions and exponential phase batch culture samples (open diamonds).
A RNA content, B DNA content, C protein content, D cell size. Data from exponential phase batch culture is not used for regression analysis, shaded area depicts 95% confidence level interval.
We further investigated the activity of ribosomes by measuring the fraction of polysomes using sucrose gradient ultracentrifugation. The polysome fraction decreased from 59% ± 7% to 28% ± 9% from fast to slow growth (Fig. 4). In comparison with formate-limited cells, the polysome fraction of phosphate-limited cells was noticeably higher. Together with lower ribosome content, this showed that a higher fraction of the ribosomes was engaged in translation to reach the same growth rate relative to formate limitation and compensate for the lower ribosome content, albeit with lower protein content per cell under phosphate limitation (Figs. 3C, 4).
Fig. 4. Ribosome activity of M. maripaludis at different growth rates, showing data from chemostat-grown cells (filled diamonds) under phosphate- (gold, this study) and formate- (black, [10]) limited conditions and exponential phase batch culture samples (open diamonds).
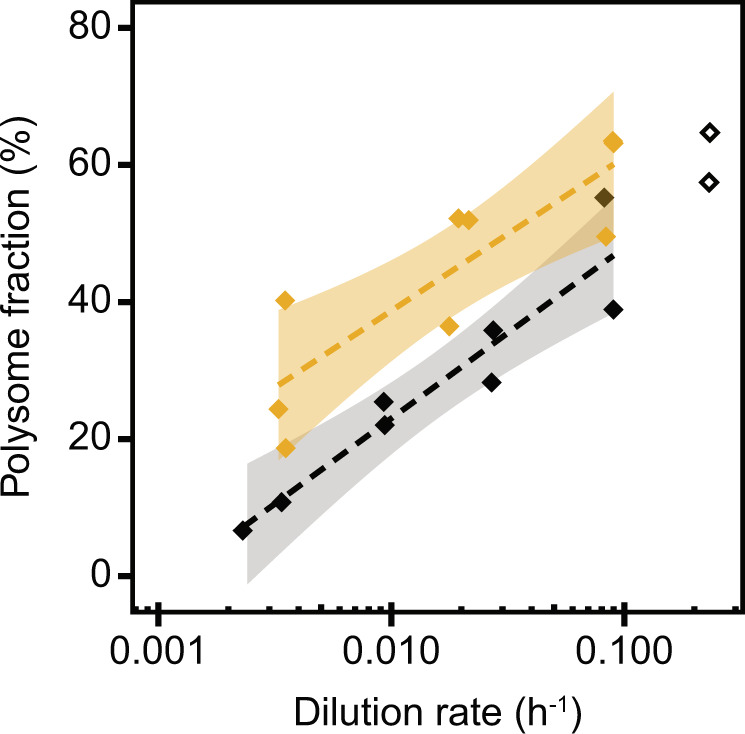
The polysome fraction correlated logarithmically with dilution rate under both phosphate- (y = 9.73 ln(x) + 84, R² = 0.75, p < 0.01) and formate- (y = 11.54 ln(x) + 76, R² = 0.93, p < 0.01) limited conditions. Shaded areas depict 5% confidence level intervals. Data from exponential phase batch culture are not used for regression analysis.
Discussion
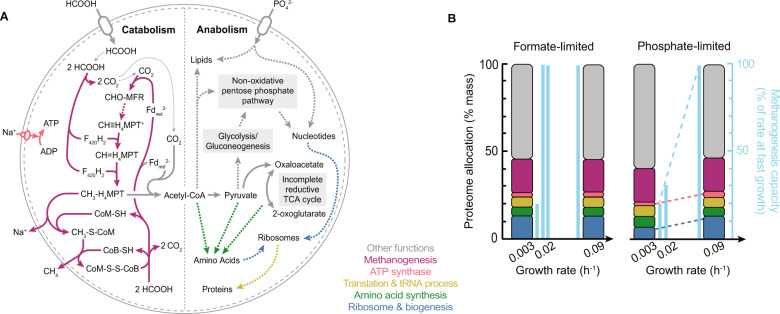
We present here a system-level characterization of physiological traits of M. maripaludis at different growth rates under anabolic phosphate limitation. M. maripaludis is a metabolically specialized methanogen [27] with a catabolic pathway that cannot be truncated. The reduced ferredoxin required for the initial reduction of CO2 is generated in the last step of methanogenesis by the heterodisulfide reductase, ultimately rendering methanogenesis a cyclic pathway also known as the Wolfe cycle [28] (Fig. 5A). M. maripaludis also has a simpler carbon flow between the catabolic pathway and the carbon assimilation pathways. Carbon assimilation proceeds mostly by combining the methyl-group from methyl-H4MPT generated in the methanogenesis pathway with CO2-derived CO in the carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) complex to produce acetyl-CoA [29, 30] (Fig. 5A). This routing of carbon from the catabolic methanogenesis cycle necessitates anaplerotic formation of reduced ferredoxin, for example, by an Eha-hydrogenase [31]. These metabolic features make M. maripaludis an interesting case for studying proteome allocation strategy in comparison to versatile microorganisms such as E. coli.
Fig. 5. Metabolic pathways and proteome allocation in M. maripaludis.
Summary of (A) catabolic (methanogenesis) and key anabolic metabolic pathways of M. maripaludis when growing on formate as the energy substrate (modified from [31, 47–50]), colors of arrows indicate the proteome sectors the pathways are assigned to, dashed arrows indicate there are intermediate steps not shown; (B) proteome allocation (left-hand axis, color for functional sector matched the color of pathways in A) and methanogenesis capacity (right-hand axis, bars in light blue) of M. maripaludis at different growth rates when catabolically (formate-) [10] vs. anabolically (phosphate-) limited. Representative illustrations of proteome allocation are shown only for the fastest and slowest growth rates investigated. Dashed lines indicate growth rate dependence.
We previously discovered that under catabolic limitation, M. maripaludis exhibits an invariant proteome allocation and invariant macromolecular cell composition at different growth rates [10] (Fig. 5B), contrary to what has been observed in many microorganisms including E. coli [2, 3, 32]. To our surprise, when anabolic flux is limiting due to phosphate limitation, the mass fraction of several proteome sectors, in particular the ribosomal sector, was growth rate-dependent in M. maripaludis. The ribosomal change with growth rate has been observed at the transcriptional level, i.e., increase in rRNA abundance based on microarray analysis, in M. maripaludis in a previous study of two growth rates [33]. In comparison to formate limitation, phosphate-limited cells produced 66% of protein with roughly half the number of ribosomes at slower growth (Fig. 1), but with a higher fraction of ribosomes engaged, likely compensating for the lower ribosome content (Fig. 4). Such strategy is an acclimation to phosphate limitation as RNA is phosphorus-rich and ribosomal RNA is the most abundant cellular RNA. This also indicates that ribosome content is not coupled to growth rate per se, but might reflect a more specific response of the cell to acclimate to environmental conditions. Therefore, the invariant ribosome level across growth rates observed under formate-limitation may reflect an acclimation of cells to resume growth quickly when the energy substrate is replenished [19], while the high phosphate requirement of a large unused ribosome pool might be prohibitive under phosphate limitation. These ribosomal regulation traits are similar to E. coli where cells exhibit lower ribosome levels when phosphorus-limited compared to when carbon- or nitrogen-limited and the ribosome level positively correlates with growth rate [19]. In contrast, M. maripaludis has similar ribosomal expression in phosphate- vs. formate-limited cells during fast growth while E. coli shows consistently lower ribosomal levels at a wide range of growth rates under phosphate-limited conditions (Fig. 1).
Apart from ribosome expression, the overall proteome responds more dynamically to growth rate changes under phosphate-limited conditions compared to the static composition under formate limitation. Several growth rate-dependent proteome sectors showed similar trends, but the trends were less pronounced under catabolic limitation for sectors such as transcription, purine/pyrimidine metabolism, motility/chemotaxis, and V-type ATP synthase. In contrast, other sectors seem to be uniquely regulated under anabolic phosphate limitation, including ribosome, ribosome biosynthesis, incomplete reductive TCA cycle, and queuosine-archaeosine biosynthesis sectors. Among them, transcription, purine/pyrimidine metabolism, tRNA modification, and queuosine-archaeosine biosynthesis sectors are expressed at higher level at slower growth, which may counteract the lower phosphate flux as overexpression may shift the limiting phosphate to other pathways.
Phosphate limitation affects M. maripaludis by inducing a reduction of methanogenesis capacity and a decoupling of catabolism and anabolism. Decoupling of catabolism from biomass formation has also been reported for phosphorus limitation in Methanosarcina barkeri and Methanobacterium thermoautotrophicum [34, 35], indicating that this might be a widespread feature in methanogens. The observed invariant methanogenesis proteome level in M. maripaludis suggests that the rate of methanogenesis is downregulated at lower growth rates independent of transcription/translation, for example, by post-translational modifications (PTMs). The exact mechanism of regulating activity of methanogenesis, however, warrants further investigation. In addition, our data indicates a lower macromolecular content (especially DNA and protein) per cell in phosphate-limited cells compared to formate-limited cells at all growth rates. Macromolecular content (especially RNA) also decreased with growth rate under phosphate limitation. Cell size remained similar in all conditions, which indicates a lack of cell size regulation.
Phosphorus is essential for genetic inheritance, energy metabolism, and membrane integrity. Many bacteria developed complex genetic regulation - phosphate regulons under a two-component master regulator PhoRB - to acclimate to its scarcity [36–39]. Nevertheless, a similar PhoRB-mediated regulation has not been reported in archaea [40]. In M. maripaludis, we observed induction of an ABC-type (Pst-like) phosphate transporter (MMP1095–1099) under phosphate limitation (Supplementary Fig. S3), consistent with previous studies [33, 41, 42]. A previous study reported two other putative phosphate transporters, MMP0666 (Na+/Pi cotransporter), MMP1199 (PhoU homolog) and their respective operons [42]. Both are present at low to nondetectable levels in our dataset and are not significantly differentially expressed in comparison to formate-limited condition. Besides the pst operon, we did not find homologs of genes for alkaline phosphatase, polyphosphate kinase, or cyclic-2,3-diphosphoglycerate synthetase in M. maripaludis from genome or literature search, even though these enzymes are common targets of phosphate regulons and several methanogens were found to regulate these enzymes when phosphate-stressed [35, 43–45]. Understanding how the various phosphate-dependent pathways are differentially regulated in M. maripaludis at different growth rates under phosphate limitation will require future investigation [46].
In summary, our characterization revealed a distinct strategy of M. maripaludis for anabolic limitation compared to catabolic limitation where cells apparently have different priorities for resource allocation (Fig. 5B). Under phosphate limitation when ribosome production becomes resource-costly, M. maripaludis down-regulates ribosome expression at slow growth compared to keeping it at a constant level during catabolic limitation. In addition, phosphate limitation limits the catabolic capacity of M. maripaludis - while catabolic limitation does not - at a wide range of growth rates, indicating potential non-transcriptional/translational cross-regulation of catabolic and anabolic activities. Such a regulation allows the cells to readily increase catabolism only when doing so will allow for new growth, but not when other factors (such as phosphate) are growth-limiting. Our findings showcase the presence of a different microbial lifestyle in response to different environments and call for a wider survey of microbial physiology for deducing general principles.
Supplementary information
Acknowledgements
We thank Dr. Vadim Patsalo for recording and analyzing the proteomic mass spectrometry data. This work is supported by grants from the US Army Research Office (W911NF2010111 to AMS) and National Science Foundation/University of Southern California, Center for Dark Energy Biosphere Investigations (OCE-0939564), and a grant from the National Institutes of Health (R35-GM136412 to JRW).
Author contributions
WG, ALM, and AMS conceived and designed the experiments. WG and ALM performed the experiments and analyzed the data. All authors contributed materials/analysis tools. All authors contributed to the writing of the manuscript.
Data availability
The mass spectrometry proteomics data are available in the MassIVE database (https://massive.ucsd.edu) under accession number MSV000087621.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Wenyu Gu, Albert L. Müller.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01278-9.
References
- 1.Hui S, Silverman JM, Chen SS, Erickson DW, Basan M, Wang J, et al. Quantitative proteomic analysis reveals a simple strategy of global resource allocation in bacteria. Mol Syst Biol. 2015;11:784. doi: 10.15252/msb.20145697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.You C, Okano H, Hui S, Zhang Z, Kim M, Gunderson CW, et al. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature. 2013;500:301–6. doi: 10.1038/nature12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. Interdependence of cell growth and gene expression: origins and consequences. Science. 2010;330:1099–102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- 4.Makman RS, Sutherland EW. Adenosine 3’,5’-phosphate in Escherichia Coli. J Biol Chem. 1965;240:1309–14. [PubMed] [Google Scholar]
- 5.Kochanowski K, Okano H, Patsalo V, Williamson J, Sauer U, Hwa T. Global coordination of metabolic pathways in Escherichia coli by active and passive regulation. Mol Syst Biol. 2021;17:1–14. doi: 10.15252/msb.202010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basan M, Hui S, Okano H, Zhang Z, Shen Y, Williamson JR, et al. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature. 2015;528:99–104. doi: 10.1038/nature15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsson C, Von Stockar U, Marison I, Gustafsson L. Growth and metabolism of Saccharomyces cerevisiae in chemostat cultures under carbon-, nitrogen-, or carbon- and nitrogen-limiting conditions. J Bacteriol. 1993;175:4809–16. doi: 10.1128/jb.175.15.4809-4816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai X, Zhu M, Warren M, Balakrishnan R, Patsalo V, Okano H, et al. Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat Microbiol. 2016;2:1–9. doi: 10.1038/nmicrobiol.2016.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basan M, Zhu M, Dai X, Warren M, Sévin D, Wang Y, et al. Inflating bacterial cells by increased protein synthesis. Mol Syst Biol. 2015;11:836. doi: 10.15252/msb.20156178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Müller AL, Gu W, Patsalo V, Deutzmann JS, Williamson JR, Spormann AM. An alternative resource allocation strategy in the chemolithoautotrophic archaeon Methanococcus maripaludis. Proc Natl Acad Sci USA. 2021;118:1–8. doi: 10.1073/pnas.2025854118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smil V. Phosphorus in the environment: natural flows and human Interferences. Annual Review of Energy and the Environment; Palo Alto. 2000;25:53.
- 12.Schindler DW. Evolution of phosphorus limitation in lakes. Science. 1977;195:260–2. doi: 10.1126/science.195.4275.260. [DOI] [PubMed] [Google Scholar]
- 13.Müller S, Mitrovic SM. Phytoplankton co-limitation by nitrogen and phosphorus in a shallow reservoir: progressing from the phosphorus limitation paradigm. Hydrobiologia. 2015;744:255–69. [Google Scholar]
- 14.Garcia NS, Bonachela JA, Martiny AC. Interactions between growth-dependent changes in cell size, nutrient supply and cellular elemental stoichiometry of marine. Synechococcus ISME J. 2016;10:2715–24. doi: 10.1038/ismej.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arne Alphenaar P, Sleyster R, De Reuver P, Ligthart GJ, Lettinga G. Phosphorus requirement in high-rate anaerobic wastewater treatment. Water Res. 1993;27:749–56. [Google Scholar]
- 16.Miettinen IT, Vartiainen T, Martikainen PJ. Phosphorus and bacterial growth in drinking water. Appl Environ Microbiol. 1997;63:3242–5. doi: 10.1128/aem.63.8.3242-3245.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shropshire H, Jones RA, Aguilo-Ferretjans MM, Scanlan DJ, Chen Y. Proteomics insights into the Burkholderia cenocepacia phosphorus stress response. Environ Microbiol. 2021;23:5069–86. doi: 10.1111/1462-2920.15451. [DOI] [PubMed] [Google Scholar]
- 18.Jones RA, Shropshire H, Zhao C, Murphy A, Lidbury I, Wei T, et al. Phosphorus stress induces the synthesis of novel glycolipids in Pseudomonas aeruginosa that confer protection against a last-resort antibiotic. ISME J. 2021;15:3303–14. doi: 10.1038/s41396-021-01008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li SHJ, Li Z, Park JO, King CG, Rabinowitz JD, Wingreen NS, et al. Escherichia coli translation strategies differ across carbon, nitrogen and phosphorus limitation conditions. Nat Microbiol. 2018;3:939–47. doi: 10.1038/s41564-018-0199-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa KC, Wong PM, Wang T, Lie TJ, Dodsworth JA, Swanson I, et al. Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc Natl Acad Sci USA. 2010;107:11050–5. doi: 10.1073/pnas.1003653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 22.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–19. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 23.De Poorter LMI, Geerts WJ, Keltjens JT. Coupling of Methanothermobacter thermautotrophicus methane formation and growth in fed-batch and continuous cultures under different H2 gassing regimens. Appl Environ Microbiol. 2007;73:740–9. doi: 10.1128/AEM.01885-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schönheit P, Moll J, Thauer RK. Growth parameters (Ks, μmax, Ys) of Methanobacterium thermoautotrophicum. Arch Microbiol. 1980;127:59–65. doi: 10.1007/BF00403508. [DOI] [PubMed] [Google Scholar]
- 25.Chaban B, Ng SYM, Kanbe M, Saltzman I, Nimmo G, Aizawa SI, et al. Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis. Mol Microbiol. 2007;66:596–609. doi: 10.1111/j.1365-2958.2007.05913.x. [DOI] [PubMed] [Google Scholar]
- 26.Albers SV, Jarrell KF. The archaellum: how Archaea swim. Front Microbiol. 2015;6:1–12. doi: 10.3389/fmicb.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitman WB, Shieh J, Sohn S, Caras DS, Premachandran U. Isolation and characterization of 22 mesophilic methanococci. Syst Appl Microbiol. 1986;7:235–40. [Google Scholar]
- 28.Kaster AK, Moll J, Parey K, Thauer RK. Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc Natl Acad Sci USA. 2011;108:2981–6. doi: 10.1073/pnas.1016761108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shieh J, Whitman WB. Autotrophic acetyl coenzyme A biosynthesis in Methanococcus maripaludis. J Bacteriol. 1988;170:3072–9. doi: 10.1128/jb.170.7.3072-3079.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Major TA, Liu Y, Whitman WB. Characterization of energy-conserving hydrogenase B in Methanococcus maripaludis. J Bacteriol. 2010;192:4022–30. doi: 10.1128/JB.01446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lie TJ, Costa KC, Lupa B, Korpole S, Whitman WB, Leigh JA. Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis. Proc Natl Acad Sci USA. 2012;109:15473–8. doi: 10.1073/pnas.1208779109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brauer M, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, et al. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–67. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendrickson EL, Liu Y, Rosas-Sandoval G, Porat I, Söll D, Whitman WB, et al. Global responses of Methanococcus maripaludis to specific nutrient limitations and growth rate. J Bacteriol. 2008;190:2198–205. doi: 10.1128/JB.01805-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Archer DB. Uncoupling of methanogenesis from growth of Methanosarcina barkeri by phosphate limitation. Appl Environ Microbiol. 1985;50:1233–7. doi: 10.1128/aem.50.5.1233-1237.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seely RJ, Fahrney DE. Levels of cyclic-2,3-diphosphoglycerate in Methanobacterium thermoautotrophicum during phosphate limitation. J Bacteriol. 1984;160:50–54. doi: 10.1128/jb.160.1.50-54.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tommassen J, de Geus P, Lugtenberg B, Hackett J, Reeves P. Regulation of the pho regulon of Escherichia coli K-12: cloning of the regulatory genes phoB and phoR and identification of their gene products. J Mol Biol. 1982;157:265–74. doi: 10.1016/0022-2836(82)90233-9. [DOI] [PubMed] [Google Scholar]
- 37.Hulett FM. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol Microbiol. 1996;19:933–9. doi: 10.1046/j.1365-2958.1996.421953.x. [DOI] [PubMed] [Google Scholar]
- 38.Novak R, Cauwels A, Charpentier E, Tuomanen E. Identification of a Streptococcus pneumoniae gene locus encoding proteins of an ABC phosphate transporter and a two-component regulatory system. J Bacteriol. 1999;181:1126–33. doi: 10.1128/jb.181.4.1126-1133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kočan M, Schaffer S, Ishige T, Sorger-Herrmann U, Wendisch VF, Bott M. Two-component systems of Corynebacterium glutamicum: deletion analysis and involvement of the phoS-phoR system in the phosphate starvation response. J Bacteriol. 2006;188:724–32. doi: 10.1128/JB.188.2.724-732.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wende A, Furtwängler K, Oesterhelt D. Phosphate-dependent behavior of the archaeon Halobacterium salinarum strain R1. J Bacteriol. 2009;191:3852–60. doi: 10.1128/JB.01642-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akinyemi TS, Shao N, Lyu Z, Drake IJ, Liu Y, Whitman WB. Tuning gene expression by phosphate in the methanogenic archaeon Methanococcus maripaludis. ACS Synth Biol. 2021;10:3028–39. doi: 10.1021/acssynbio.1c00322. [DOI] [PubMed] [Google Scholar]
- 42.Xia Q, Wang T, Hendrickson EL, Lie TJ, Hackett M, Leigh JA. Quantitative proteomics of nutrient limitation in the hydrogenotrophic methanogen Methanococcus maripaludis. BMC Microbiol. 2009;9:1–10. doi: 10.1186/1471-2180-9-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paula FS, Chin JP, Schnürer A, Müller B, Manesiotis P, Waters N, et al. The potential for polyphosphate metabolism in Archaea and anaerobic polyphosphate formation in Methanosarcina mazei. Sci Rep. 2019;9:1–12. doi: 10.1038/s41598-019-53168-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.König H, Nusser E, Stetter KO. Glycogen in Methanolobus and Methanococcus. FEMS Microbiol Lett. 1985;28:265–9. [Google Scholar]
- 45.Rudnick H, Hendrich S, Pilatus U, Blotevogel KH. Phosphate accumulation and the occurrence of polyphosphates and cyclic 2,3-diphosphoglycerate in Methanosarcina frisia. Arch Microbiol. 1990;154:584–8. [Google Scholar]
- 46.Shalvarjian KE, Nayak DD. Transcriptional regulation of methanogenic metabolism in archaea. Curr Opin Microbiol. 2021;60:8–15. doi: 10.1016/j.mib.2021.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–91. [DOI] [PubMed]
- 48.Goyal N, Zhou Z, Karimi IA. Metabolic processes of Methanococcus maripaludis and potential applications. Micro Cell Fact. 2016;15:1–19. doi: 10.1186/s12934-016-0500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simpson PG, Whitman WB. Anabolic pathways in methanogens. In: Methanogenesis. Boston, MA: Springer; 1993. p. 445–72.
- 50.Dai X, Zhu M. Coupling of ribosome synthesis and translational capacity with cell growth. Trends Biochem Sci. 2020;45:681–92. doi: 10.1016/j.tibs.2020.04.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data are available in the MassIVE database (https://massive.ucsd.edu) under accession number MSV000087621.