Abstract
The role of the carboxy terminus of the Escherichia coli cell division protein FtsA in bacterial division has been studied by making a series of short sequential deletions spanning from residue 394 to 420. Deletions as short as 5 residues destroy the biological function of the protein. Residue W415 is essential for the localization of the protein into septal rings. Overexpression of the ftsA alleles harboring these deletions caused a coiled cell phenotype previously described for another carboxy-terminal mutation (Gayda et al., J. Bacteriol. 174:5362–5370, 1992), suggesting that an interaction of FtsA with itself might play a role in its function. The existence of such an interaction was demonstrated using the yeast two-hybrid system and a protein overlay assay. Even these short deletions are sufficient for impairing the interaction of the truncated FtsA forms with the wild-type protein in the yeast two-hybrid system. The existence of additional interactions between FtsA molecules, involving other domains, can be postulated from the interaction properties shown by the FtsA deletion mutant forms, because although unable to interact with the wild-type and with FtsAΔ1, they can interact with themselves and cross-interact with each other. The secondary structures of an extensive deletion, FtsAΔ27, and the wild-type protein are indistinguishable when analyzed by Fourier transform infrared spectroscopy, and moreover, FtsAΔ27 retains the ability to bind ATP. These results indicate that deletion of the carboxy-terminal 27 residues does not alter substantially the structure of the protein and suggest that the loss of biological function of the carboxy-terminal deletion mutants might be related to the modification of their interacting properties.
FtsA is an essential cell division protein of Escherichia coli that is widely conserved in bacteria. Together with ftsZ, which codes for a GTPase analog of the eukaryotic tubulin, ftsA forms one of the most frequently conserved gene pairs among the cell division genes in the eubacteria. Based on sequence homology it has been proposed that FtsA belongs to the sugar kinase/hsp70/actin superfamily (4). This superfamily comprises several proteins with a common two-domain topology and the ability to bind and hydrolyze ATP. FtsA binds to columns of ATP-agarose and can be isolated from cells either as a phosphorylated or a nonphosphorylated form (29), but so far no other biochemical function has been described for this protein.
FtsA is present both in the cytoplasm and in the cytoplasmic membrane (29), where it forms a structural part of the septum (32). It has been proposed that FtsA is a component of a membrane-associated complex (septator or divisome), which would include periplasmic, transmembrane, and cytoplasmic proteins acting coordinately to perform septation (27, 35). Genetic analysis suggests that FtsA may interact, directly or indirectly, with other cell division proteins, such as FtsZ, PBP3, FtsQ, and FtsN (9, 10, 24, 33, 34). The FtsZ/FtsA ratio is important for cell division, and the localization of FtsA to a central position along the cell length depends on the formation of the FtsZ ring (1, 8, 11, 15, 23). Accordingly, the interaction between FtsA and FtsZ from different bacteria has been established using the yeast two-hybrid system (12, 25, 38, 40). In addition the presence of functional FtsA is required for the localization of FtsK, PBP3 (also known as FtsI), FtsQ, and FtsN to the septal ring (2, 7, 37, 41), suggesting that FtsA may indeed be a part of a multiprotein complex (27).
Interaction of FtsA with itself has been suggested based on the ability of ftsA104 (coding for FtsA104: T215A) to complement efficiently two ftsA(Ts) mutations (ftsA2 and ftsA3), but with less efficiency an amber one (ftsA16) (29), and has been recently reported by Yan et al. (40) using the two-hybrid system. Involvement of the protein carboxy terminus in the interaction was suspected from previous results showing that overproduction of a carboxy-terminally truncated form of FtsA causes the formation of long fibers in the cells that also show a curved morphology phenotype (17).
To study the role of the carboxy terminus in the function of FtsA a series of mutants with sequential deletions spanning from 1 to 27 residues was constructed. We have found that this region is essential for the biological activity and for the correct self-interaction of the protein and that the residue W415 is essential for the correct localization of FtsA into septal rings.
MATERIALS AND METHODS
Bacterial and yeast strains, media, and growth conditions.
E. coli strain DH5α [F− endA1 hsdR17 supE44 thi1 recA1 gyrA relA1 Δ(lacZYA-argF) U169, (Φ80lacZΔM15) (19)] was used as host for cloning. Strain OV2 [F− ilv leu thyA (deo), his, ara(Am), lac125(Am) galKu42(Am) galE trp(Am) tsx(Am) tyrT] [also known as supFA81(Ts)] was used as wild type for ftsA (31). Strains OV16 [as OV2 ftsA16(Am) (13)] and D2 [as OV2 ftsA2(Ts) leu+ (31)] were used for cell division complementation assays. BL21(DE3) pLysS [F− ompT hsdSB (rB− mB−) gal dcm (DE3); obtained from Novagen] was used for expression of His-tagged FtsA forms. Luria-Bertani (LB) broth (28) and LB agar, supplemented with antibiotics when required (ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 50 μg/ml; and rifampin, 25 μg/ml) were used for routine cultures of E. coli at 37°C (except when indicated otherwise). Strains carrying ftsA(Ts) alleles used for complementation assays were grown in nutrient broth medium (Oxoid no. 2 nutrient broth) supplemented with thymine (50 μg/ml) (NBT) and antibiotics when required and incubated at the permissive (30°C) or restrictive (42°C) temperatures. Complementation tests were done as described by Sánchez et al (29). VIP386 is BL21(DE3) harboring pMFV12 (expressing an N-terminal fusion of ftsA+ to His tag under the control of a T7 promoter contained in a pET28a plasmid purchased from Novagen; construction and strain obtained from M. J. Ferrándiz). VIP516 is BL21(DE3) harboring pSRV2 (expressing an N-terminal fusion of ftsAΔ27 to His tag under the control of a T7 promoter contained in a pET28a plasmid).
Yeast two-hybrid experiments were performed with Saccharomyces cerevisiae strain SFY526 (MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112 canr gal4-542 gal80-538 URA3::GAL1-lacZ) (3). Yeast strains were grown in YEPD or SD medium (supplemented with required amino acids and glucose) described in the Matchmaker kit manual (Clontech).
Cell parameter measurements, photography, and immunofluorescence microscopy.
Cultures were grown at the permissive temperature in liquid media in a shaking water bath so that balanced growth was maintained for several doublings (not less than four) before the beginning of the experiment. The optical density at 600 nm (OD600) was measured using a Shimadzu UV-1203 spectrophotometer; the optical density was always kept below 0.40 to 0.50 by appropriate dilution with prewarmed medium. Particles were fixed in 0.75% formaldehyde and counted using a ZM Coulter counter, with a 30-μm-diameter orifice connected to a C1000 Channelyzer (both from Coulter Electronics). Cells were spread on thin agar-azide layers and photographed under phase-contrast optics using a Sensys charge-coupled device camera (Photometrics) coupled to a Zeiss Axiolab HBO 50 microscope. The software used for image capture was IPLab Spectrum, and Adobe Photoshop 5.0.2 software was used for processing.
For immunofluorescence microscopy cells grown as indicated above were prepared as described by Addinall and Lutkenhaus (1). The final lysozyme concentration used was 8 μg/ml, and the permeabilization time was 1 min. The primary antibody used for FtsA immunolocalization was MVM1, a polyclonal antiserum raised against FtsA (29) previously purified by membrane affinity. Cy3-conjugated anti-rabbit serum (Amersham Pharmacia Biotech) was used as the secondary antibody. Cells were observed by fluorescence microscopy using a Zeiss Axiolab HBO 50 microscope, with a 100× immersion oil lens. Images were captured with a Sensys charge-coupled device camera (Photometrics), fitted with an HQ:CY3 filter (excitation, 545/30 nm; emission, 610/75 nm; beam splitter, 565LP).
Plasmid constructions and DNA manipulation.
Plasmid DNA isolation, cloning techniques, and transformation procedures were done as described by Sambrook et al. (28). Restriction endonucleases and other enzymes were purchased from and used as recommended by Roche Molecular Biochemicals. DNA was sequenced by the dideoxynucleotide method using Sequenase version 2.0 (U.S. Biochemicals). Two-hybrid system cloning vectors (pGAD424 and pGBT9) were obtained from Clontech.
Plasmid pLYV29 contains the 3′-truncated ftsAΔ27 allele (with an ochre codon spontaneously inserted instead of the Glu-394 coding triplet, thus rendering an FtsA protein lacking the last 27 residues), cloned into the vector pJF119HE (16). Plasmid pLYV32 contains the ftsAΔ23Φ36 allele (resulting from a spontaneous frameshift in the 3′ end of the gene, rendering an FtsA protein with the last carboxy terminal 23 residues replaced by a different 36-residue sequence), cloned into pJF119EH (as pJF119HE but with the multicloning site in the opposite orientation). Plasmids pLYV33, -34, -35, and -41 encode different carboxy-terminal deletion derivatives of FtsA, (lacking the last 13, 6, 5, and 1 residues respectively), cloned into pJF119EH. Plasmid pLYV30 contains the wild-type ftsA gene cloned into pJF119HE. These deletions were obtained by PCR-mediated mutagenesis as follows. The 3′ ftsA coding region was obtained from pZAQ (39) by PCR amplification with the upstream primer MF11 (5′-GGGGTGAACGACCTCGAA-3′) and the following downstream primers (respectively): LY23 (5′-ATAGTCGACTTACGAGCCAACTGATGCTGT-3′), which inserts a stop codon instead of the TGG coding for Trp-408 and a SalI restriction site immediately downstream; LY24 (5′-ATAGTCGACTTAACTATTGAGTCGCTTGATC-3′), which inserts a stop codon instead of the TGG coding for Trp-415 together with a SalI restriction site immediately downstream; LY25 (5′-ATAGTCGACTTACCAACTATTGAGTCGCTTG-3′), which inserts a stop codon instead of the CTG coding for Leu-416 and a SalI restriction site immediately downstream; and LY28 (5′-ATAGTCGACTTACTCTTTTCGCAGCCAACT-3′), which inserts a stop codon instead of the TTT coding for Phe-420 together with a downstream SalI restriction site. The PCR-amplified fragments were digested with AscI and SalI and were used to replace the 3′ region of ftsAΔ23Φ36 in pLYV32. The presence of all the mutations was confirmed by DNA sequencing.
Plasmids for the complementation assays were constructed as follows. The 3′ end fragments of the partially deleted ftsA alleles (from the AscI site to the end of the gene) were obtained from plasmids pLYV29, -33, -34, -35 and -41 by digestion with appropriate enzymes and were cloned into pMSV20 (29) in place of the wild-type 3′ region to give plasmids pLYV57, -58, -59, -60, and -61, respectively. This places the corresponding deletion mutant alleles downstream from an ftsQ sequence fragment (starting at a NdeI site). This sequence context working together with any potential readthrough transcription from the plasmid promoters at the copy number of the vector (a pBR322 derivative) produces levels of each protein that fall within the level range found in strains expressing the wild-type ftsA+ from the chromosome (from 0.90 to 1.14 at 30°C, and from 1.36 to 1.73 at 42°C, relative to the values measured in OV2).
Plasmids for the GAL4 two-hybrid assays were constructed as follows. The ftsA+ coding sequence was obtained from pZAQ (39) by PCR amplification with the upstream and downstream primers MF24 (5′-AATCGCATATGATCAAGGCGACGGACA-3′) and LY20 (5′-CAGGTCGACCGTAATCATCGTCGGCCTC-3′), which incorporate the restriction sites NdeI and SalI, respectively. The amplified fragment was cloned into pGBT9 and pGAD424 containing amino acids 1 to 147 of the DNA-binding domain of GAL4 and amino acids 768 to 881 from the activation domain of GAL4 respectively, yielding plasmids pLYV44 and pLYV43. These plasmids were used as templates for the construction of the GAL4-ftsA truncated allele fusions by replacing the wild-type 3′ end with the corresponding 3′-truncated regions obtained by digestion of pLYV29, -32, -33, -34, -35, and -41 with AscI and SalI.
To construct the His-FtsA+ fusion the coding sequence of ftsA+ was amplified from pZAQ using oligonucleotides MF24 (which introduces an NdeI restriction site overlapping with the ftsA initiation codon, 5′-AATCGCATATGATCAAGGCGACGGACA-3′) and MF19 (which is just downstream of an EcoRI site in the 3′ extreme of ftsA, 5′-CATCGGTATTTACCGCGAAG-3′). The PCR product was digested with NdeI and EcoRI and cloned into the vector pET28a (Novagen) to yield plasmid pMFV12. Construction of plasmid pSRV2, used to produce His-FtsAΔ27, involved ligation of a 360-bp AscI-EcoRI fragment from pMFV5 to a similarly digested pMVF12. pMFV5 contains the ftsA106 allele in which the following changes are present: M167T, T215D, and an ochre codon at position 394. The D210A ftsA mutation was constructed by inverse PCR using oligonucleotide JM1 (5′-ATAGCGACGACGCAGAC-3′) as a mutagenic primer and JM2 (5′-CGGTGGTGGTACAATGG-3′) and pMFV12 as template DNA. These constructions were checked by sequencing.
Yeast two-hybrid assays.
Yeast strains were transformed using the Li acetate method (18) and selected in SD medium supplemented with the required amino acids and glucose at 30°C. Qualitative β-galactosidase assays were performed using Whatman no. 5 filters as described in the Matchmaker kit manual (Clontech). For color development, filters were incubated up to 16 h at 30°C (although no changes were usually observed after 3 h). For quantitative analysis of the interactions β-galactosidase assays were done in liquid cultures at exponential growth phase using ONPG (o-nitrophenyl-β-d-galactopyranoside) as the substrate as described by Miller (26). Cell lysis was achieved by two freeze (dry ice-ethanol) and thaw (37°C) cycles. The values presented are the average of at least three independent experiments, carried out in duplicate each time.
Expression and purification of wild-type and carboxy-terminally truncated FtsA proteins used in Fourier transform infrared (FTIR) and circular dichroism (CD) spectroscopy measurements.
E. coli strains VIP386 (His-FtsA+) and VIP516 (His-FtsAΔ27) were grown at 37°C in LB medium supplemented with antibiotics when required (50 μg of kanamycin and/or chloramphenicol per ml) to an OD600 of 0.4. Overexpression of the proteins was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Thirty minutes after addition of the inducer, rifampin (25 μg/ml) was added to the cell culture. Growth was continued for 1 or 2 h depending on the strain. Cells were harvested by centrifugation and resuspended in ice-cold buffer A (5 mM imidazole, 10 mM HEPES [pH 7.9]). The bacteria were lysed by sonic disruption and centrifuged for 15 min at 10,000 rpm at 4°C (10,800 × g) (Sorvall SA-300 rotor). The His-tagged proteins recovered in the supernatant were purified by metal affinity chromatography on a cobalt column (TALON resin; Clontech) equilibrated with buffer A. The proteins were eluted with a step gradient of imidazole ranging from 30 to 200 mM in a 10 mM HEPES, pH 7.9, buffer. Integrity and purity of proteins were checked by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis (SDS–12% PAGE), and fractions containing pure proteins were pooled and dialyzed against 2 mM HEPES, pH 7.9, before analysis.
Overlay assays.
Sample proteins were blotted to nitrocellulose membranes (Schleicher and Schuell) using a slot blot filtration manifold (Amersham Pharmacia Biotech AB). The membranes were blocked by incubation for 1 h with 3% skimm milk in blotting buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.05% IGEPAL CA-630). Purified His-FtsA+ was biotinylated in vitro with d-biotin-N-hydroxysuccinimide ester (Roche Molecular Biochemicals) following the manufacturer's instructions. The nitrocellulose membranes were washed with blotting buffer and then incubated for 1 h at room temperature with biotinylated His-FtsA+ (0.1 μg/ml) in blotting buffer. The membranes were then washed and incubated for 1 h with streptavidin-peroxidase conjugate (1:10,000; Roche Molecular Biochemicals). After extensive washing, biotinylated FtsA bound to the membranes was detected using the BM chemiluminiscence blotting substrate (POD; Roche Molecular Biochemicals).
Photoaffinity labeling.
Proteins were purified by Ni-nitrilotriacetic acid chromatography. For labeling, the imidazole elution buffer was changed in a G-50 column (Amersham Pharmacia Biotech AB) to labeling buffer (50 mM Tris [pH 8.0], 50 mM NaCl, 5 mM MgCl2, 2 mM dithiothreitol). Three micrograms of protein was mixed with 2 μCi of 8-azido[α-32P]ATP (20 Ci/mmol; ICN Biochemicals) in 20 μl. After a 5-min incubation in the dark the samples were placed on a paraffin film and irradiated for 10 min with a UVP model GL-25 UV lamp at a distance of 10 cm from the samples. After irradiation, samples were mixed with 50 μl of Ni-nitrilotriacetic acid in binding buffer, washed twice, and mixed with SDS-PAGE loading buffer containing 10 mM EDTA. Labeled proteins were analyzed by SDS-PAGE and autoradiography of the dried gels.
ATR-FTIR spectroscopy.
Attenuated total reflection (ATR)-FTIR spectra were recorded at 20°C on a Bruker IFS-55 spectrometer, equipped with a liquid nitrogen-cooled mercury cadmium telluride detector with a nominal resolution of 2 cm−1. The spectrometer was continuously purged with dry air. The proteins were spread on a germanium ATR plate with an aperture angle of 45° (50 by 20 by 2 mm; Harrick EJ2121) by slowly evaporating the sample under nitrogen. A total of 1,024 scans were averaged to improve the signal/noise ratio and corrected for the spectrum of the clean ATR plate.
CD spectroscopy.
Spectra of FtsA and FtsAΔ27 were recorded in 2 mM HEPES, pH 7.9, at room temperature, using a Jasco J-710 spectropolarimeter in quartz cells with a 0.02-cm path length. The spectra were monitored from 185 to 260 nm, using a scan speed of 50 nm/min, a response time of 2 s, a band width of 1.0 nm, and a resolution of 2 data points/nm. Eight spectra were averaged and subsequently corrected for the spectrum of the buffer.
Analysis of secondary structure.
The determination of protein secondary structure was based on a multivariate statistical analysis method of band shape recognition (Oberg et al., unpublished data). This analysis used a commercially available partial least-squares package to determine fractional secondary structure composition (PLSPlus 2.1 for Spectra Calc; Galactic Industries, Salem, N.H.). A reference set (RaSP50) was generated by collecting ATR-FTIR and CD spectra of 50 proteins with known X-ray structures selected to represent a wide range of helix and sheet compositions as well as 60 different protein domain folds. To analyze combined CD and IR data, hybrid spectra were built by placing side-by-side CD (185 to 260 nm) and IR (1,720 to 1,500 cm−1) spectra in a single array. Before analysis, all spectra were normalized so that the PLS algorithm was forced to use band shapes, rather than absolute intensities. Cross validation procedures using the RaSP50 protein spectra have shown root mean square structure determination errors of ±4.5% for α-helix and ±6.3% for β-sheet conformations.
RESULTS
Deletion mutants of FtsA lacking five or more residues from the carboxy terminus are not biologically active in E. coli.
To study the role of the carboxy terminus of E. coli FtsA on its function, a series of sequential ftsA deletion mutants comprising codons coding for 1, 5, 6, 13, and 27 amino acid residues from the carboxy terminus were constructed (Fig. 1). A spontaneous mutant in which the last 23 residues have been replaced by a 36-residue unrelated amino acid sequence, FtsAΔ23Φ36, was also studied. The functionality of these mutants was analyzed by complementation assays of strains containing either a thermosensitive ftsA mutation (D2) or an amber mutation in ftsA in a temperature-sensitive suppressor background (OV16). Strains D2 and OV16 were transformed with plasmids expressing the deletion or wild-type ftsA alleles at levels similar to the expression of ftsA from the chromosome (pLYV57, -58, -59, -60, -61, and pMSV20), and their ability to grow on NBT plates at 30 or 42°C was tested. Western blot analysis using an affinity-purified antibody raised against FtsA indicated that, when produced under the same gene expression conditions, the cellular levels of all the truncated forms were similar to those of the wild-type protein at both growth temperatures (see Materials and Methods); consequently, the possibility that expression from an artificial system, or in vivo thermal instability of the mutant forms can cause unspecific side effects on viability can be excluded. The results of the complementation analysis show that only ftsA+ and ftsAΔ1 are able to rescue the two strains (Table 1). The ftsAΔ5 allele in its turn is able to partially complement ftsA2 but not ftsA16. No complementation of any of the two mutations was observed when the ftsAΔ6, ftsΔ13, ftsΔ27, and ftsΔ23Φ36 were tested. We conclude that deletion of five or more FtsA carboxy-terminal residues yields proteins that are not functional in vivo, suggesting that this part of the molecule has an important role on the biological activity of FtsA.
FIG. 1.
Sequence of the carboxy terminus of FtsA. The top part of the figure shows the alignment of the carboxy end of the published FtsA sequences from 25 bacterial species, including E. coli, obtained using the program ClustalW. The residues have been highlighted accordingly to their average BLOMSUM62 scores as follows: black for maximum (3.0)-score residues, medium gray for mid-range (1.5)-score residues, and light grey for low (0.5)-score residues. Similar residues according to BLOMSUM62 are shaded as the most conserved one. The bottom part of the figure shows the sequences of the E. coli FtsA carboxy end derived from the wild type and from the deleted ftsA alleles used in this work. The alien residues in FtsAΔ23Φ36 appear in italics. Shading codes are as described above; data were provided by L. Sánchez and A. Valencia.
TABLE 1.
Complementation assays of ftsA point mutations by ftsA carboxy-terminal deletion mutantsa
| Allele | D2 (ftsA2) | OV16 (ftsA16) |
|---|---|---|
| ftsA+ | + | + |
| ftsAΔ1 | + | + |
| ftsAΔ5 | +/− | − |
| ftsAΔ6 | − | − |
| ftsAΔ13 | − | − |
| ftsAΔ27 | − | − |
| ftsAΔ23Φ36 | − | − |
D2 and OV16 strains containing plasmids in which the different carboxy-terminal deletion mutant alleles were placed under the control of their natural control sequences were plated in NBT and incubated at 42°C, and growth was observed after 16 h. Twenty-five colonies of each strain were analyzed as described in Materials and Methods. Symbols: +, all the colonies produced confluent growth after 14 h; +/−, some of the clones produced isolated colonies or low-density growth; −, no growth after 14 h of incubation.
The same plasmids were used to measure the effect of expressing the ftsA deletion alleles at basal levels on the growth rate, cell morphology, and cell length of E. coli strain OV16 grown at 30 or 42°C. The results (not shown) indicate that the truncated forms of FtsA, although present in OV16 at detectable levels, did not negatively affect either the increases in optical density and particle numbers, or the cell shape or length. At 30 or 42°C the presence of plasmids containing either ftsAΔ1 or ftsA+ restored the length of the OV16 cells to wild-type (OV2) values [note that OV16 cells at 42°C are filaments while at 30°C are some 25% longer than the wild-type OV2 at the same temperature, due to the lower amounts of FtsA provided by the combination of an amber mutation, ftsA16, with a low-efficiency temperature-sensitive tyrT suppressor, supFA81(Ts) (6)]. On the other hand the presence of plasmids containing any of the other ftsA deletion mutants had no effect in shortening the length of OV16 cells. These results indicate that the FtsA forms do not exert a trans dominant effect over the wild type, and that, except FtsAΔ1, they are not active in cell division.
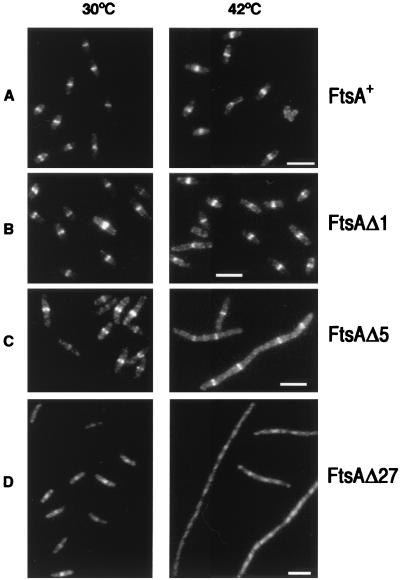
To analyze the subcellular localization of the mutant proteins by immunofluorescence, strain OV16 grown at 42°C, which produces negligible amounts of FtsA from the chromosome (less than 1%) was used. The analysis of these cells showed that the wild-type and Δ1 proteins correctly localize, forming a septal ring in the cell center (Fig. 2A and B). The mutated FtsAΔ5 was still able to form rings, but in agreement with the lack of complementation of this mutation in OV16 they were not functional, as the cells did not divide and formed long filaments (Fig. 2C). The mutated FtsAΔ6, FtsΔ13, and FtsΔ27 were not able to localize in the cell center (Fig. 2D shows the results for FtsAΔ27).
FIG. 2.
Localization of FtsA in E. coli cells expressing different carboxy-terminally truncated forms of FtsA. OV16 strains containing plasmids pLYV29 (ftsAΔ27), pLYV35 (ftsAΔ5) or pLYV41 (ftsAΔ1) were grown at 30°C until an OD600 of 0.15 was reached. At time zero each culture was divided into two portions; one portion was transferred to 42°C, and the other was left at 30°C. Samples were withdrawn after 2 h, processed for immunostaining with purified MVM1 FtsA antiserum, and observed as described in the text. All the frames were corrected to minimize background differences using the Photoshop Levels adjustment. The reference bar marks 5 μm. (A) OV16/pLYV30 (FtsA+); (B) OV16/pLYV41 (FtsAΔ1); (C) OV16/pLYV35 (FtsAΔ5); (D) OV16/pLYV29 (FtsAΔ27). Note the different scale in the D frames.
Effect of the overexpression of FtsA carboxy terminus deletion mutants on cell morphology.
Increasing amounts of FtsA cause cell filamentation, a phenotype that has been interpreted as the result of an imbalance in the FtsA/FtsZ ratio (8, 11). However, the overproduction of FtsA carboxy-terminal truncations causes a peculiar phenotype, in which the cells coil forming C-shaped (short) and coiled (longer) cells (CC phenotype). This phenotype has already been observed by Gayda et al. (17) in wild-type cells containing a form of FtsA lacking the last 28 residues encoded by a plasmid.
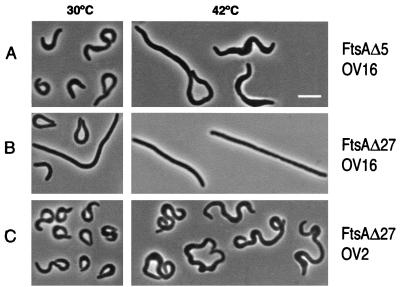
As expression of the deletion mutants at low levels did not alter the cell shape, we cloned them in expression vector pJF119 under the control of the Ptac promoter and analyzed their phenotypes upon induction with 50 μM IPTG. Overproduction of FtsAΔ1 in strain OV16 at either 30 or 42°C generated straight filaments similar to those obtained when overexpressing the wild-type protein (data not shown). Overexpression of the mutant ftsAΔ5, on the other hand, generated a pronounced CC phenotype (Fig. 3A) at either permissive or restrictive temperatures. Induction of the ftsAΔ6, ftsAΔ13, and ftsAΔ27 mutants produced the CC phenotype only at 30°C, while at 42°C (when no more than 1% of the wild-type levels of FtsA+ are produced from the chromosomal gene) they formed straight filaments (Fig. 3B shows the results for ftsAΔ27). To find out whether this was due to temperature sensitivity of the mutant proteins or to the lack of functional septa, they were overexpressed in E. coli OV2, the parental ftsA+ strain of OV16. In this case all the mutants showed the curly phenotype at both 30 and 42°C. These results suggested that in ftsAΔ6, ftsAΔ13, and ftsAΔ27, as well as in ftsAΔ23Φ36, the phenotype depends on the presence of the full-length protein, while in ftsAΔ5 the phenotype is independent of it.
FIG. 3.
Morphology of E. coli cells expressing two different carboxy-terminal deletion mutant forms of FtsA. OV2 and OV16 strains containing plasmid pLYV35 (ftsAΔ5) or pLYV29 (ftsAΔ27) were grown at 30°C until an OD600 of 0.15 was reached. At time zero they were split in four portions; two of them were transferred to 42°C, while the remaining two were left at 30°C. After 15 min IPTG (50 μM) was added to one portion at each temperature. Samples were withdrawn 2 h later, fixed, and observed as described in the text. Only the induced samples are shown. The reference bar marks 5 μm. (A) OV16/pLYV35; (B) OV16/pLYV29; (C) OV2/pLYV29.
A carboxy-terminally truncated FtsA protein, FtsAΔ27, retains its secondary structure and its ability to bind ATP.
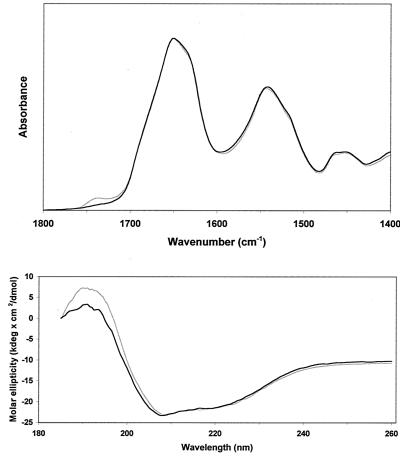
To test if the carboxy-terminal deletions had any effect on the structure of FtsA we analyzed the secondary structure of His-FtsA+ and His-FtsAΔ27 (the protein missing the longest carboxy terminus stretch among the ones used in this study). ATR-FTIR and CD spectra were recorded in 2 mM HEPES, pH 7.9. No significant difference was detected between the spectra of both proteins (Fig. 4). Their secondary structure was determined by a new method based on the recognition of the spectral bandshapes (K. Oberg, J.-M. Ruysschaert, and E. Goormaghtigh, unpublished data). The input spectra are made of a combination of ATR-FTIR and CD spectra, and the secondary structure fractions are determined by similarity of the hybrid patterns to the reference set made of 50 selected proteins. The data reported in Table 2 show no obvious change in the secondary structure of the protein resulting from the truncation of the carboxy terminus. We must stress here that the variations observed are in the range of the structure evaluation method error and therefore are not related to any structural modification of the His-FtsAΔ27 polypeptidic chain with respect to the wild type.
FIG. 4.
ATR-FTIR (top) and CD (bottom) spectra of His-FtsA+ (grey) and the carboxy-terminal deletion mutant form His-FtsAΔ27 (black) in 2 mM HEPES, pH 7.9.
TABLE 2.
Secondary structure of wild-type His-FtsA+ and carboxy-terminal deletion form His-FtsAΔ27 determined by a combination of ATR-FTIR and CD spectroscopy
| Protein | Helix (%) | Sheet (%) | Turn (%) | Other (%) |
|---|---|---|---|---|
| His-FtsA+(n = 5) | 28.6 | 26.6 | 11.8 | 33.9 |
| His-FtsAΔ27 (n = 3) | 25.9 | 27.1 | 12.2 | 35.0 |
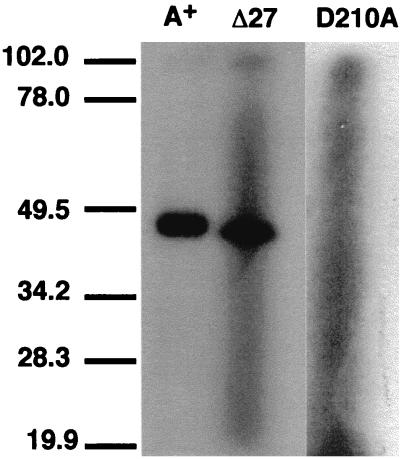
The binding of 8-azido[α-32P]ATP to purified His-FtsA+ and His-FtsAΔ27 was tested as a clue to the structural integrity of the ATP binding site. FtsAΔ27 was able to bind ATP to an extent similar to that of the wild type, showing that the carboxy terminus deletion does not affect the structure of the ATP-binding site (Fig. 5). As a negative control the His-FtsA D210A mutant—which, by analogy to other members of the same protein family (4, 21), was predicted to not bind ATP—was used. As expected, it did not bind ATP (Fig. 5).
FIG. 5.
Photoafinity labeling of His-FtsA+ and His-FtsAΔ27 with the photoreactive ATP analog 8-azido[α-32P]ATP. The SDS-PAGE separation of the material obtained after the photoactivation of the cross-linking reagent was performed and developed as described in the text. The reaction mixture loaded in the lane marked as A+ contained His-FtsA+, while His-FtsAΔ27 was contained in the one loaded in lane Δ27. Lane D210A is a negative control (see text for details). The position of molecular mass markers (in kilodaltons) is indicated at the left.
These experiments showed that the ATP-binding site of His-FtsAΔ27 is functional and its secondary structure is nearly identical to that of the wild-type protein. From these results it can be concluded that the inability of ftsAΔ27 (the more extensive truncation analyzed) to complement ftsA2 or ftsA16 is not likely to be caused by a major disruption of the structure of the resultant protein.
Role of the carboxy-terminus in interaction of FtsA molecules.
The different phenotypes produced by the mutants in OV2 and OV16 at 42°C suggested that the FtsA molecules might establish interactions with one another. To test this possibility the yeast two-hybrid system (14) was used. The wild-type ftsA gene was fused to the GAL4-binding and activation domain sequences, and the resulting plasmids were transformed into the yeast reporter strain SFY526. Control experiments showed that FtsA+ alone does not activate transcription from the GAL4 promoter. But when both fusion genes were assayed together, blue colonies were obtained in filter assays (Table 3), and 10.9 ± 1.8 Miller units of β-galactosidase activity were measured in liquid culture assays, demonstrating that FtsA+ molecules interact in the yeast two-hybrid.
TABLE 3.
Interaction between ftsA alleles revealed by yeast two-hybrid assaya
| DNA binding domain fused to: | Activation domain fused to:
|
||||||
|---|---|---|---|---|---|---|---|
| FtsA+ | FtsAΔ1 | FtsAΔ5 | FtsAΔ6 | FtsAΔ13 | FtsAΔ27 | FtsAΔ23Φ36 | |
| FtsA+ | + (10.9) | + (12.4) | − (<1) | − (<1) | − (<1) | − (<1) | − (<1) |
| FtsAΔ1 | + (14.7) | + (21.2) | − | NDb | ND | − | ND |
| FtsAΔ5 | − (<1) | − (<1) | + (37.6) | + (10.3) | + | + | + |
| FtsAΔ6 | − (<1) | ND | + (26.7) | + (11.4) | + | + | + |
| FtsAΔ13 | − (<1) | ND | + | + | + (15.6) | + | + |
| FtsAΔ27 | − (<1) | − | + | + | + | + (29.0) | + |
| FtsAΔ23Φ36 | − (<1) | ND | + | + | + | + | + (17.0) |
Transformants of yeast strain SFY526 containing fusions of the different ftsA alleles to the indicated GAL4 domains were grown on top of filters which were then soaked in buffer Z containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) to reveal β-galactosidase activity. Symbols: +, blue color; −, absence of blue color. Values in parentheses are the average of three measurements obtained by determining the β-galactosidase activity (expressed as Miller units) present in cultures in the exponential growth phase.
ND, not done.
The role of the carboxy terminus of FtsA on this interaction was also investigated. For this purpose the deletion mutants were subcloned in the two-hybrid vectors fused to both GAL4 domains. Combinations of the resulting hybrid plasmids together with the plasmids carrying the wild-type FtsA fusions were then introduced into the yeast strain SFY526. Cotransformants were assayed for β-galactosidase activity (Table 3). Control experiments showed that none of the FtsA truncated hybrids were capable of activating the expression of the reporter gene by itself (data not shown). When only the last amino acid (the highly conserved Phe-420) was removed from the carboxy terminus, the resulting FtsAΔ1 protein was still able to interact both with the wild-type form and also with itself to a similar extent as the wild-type form did, suggesting that Phe-420 is dispensable for the interaction as it is for the biological role of FtsA.
When assayed in combination with the full-length FtsA+ hybrids, the rest of the FtsA hybrids formed white colonies, indicating that deletions of five residues or more from the FtsA carboxy terminus are sufficient to impair the interaction with the wild-type protein. On the other hand, each truncated protein was able to interact with itself, and with the other deleted forms, except with FtsAΔ1 (Table 3). This result, although unexpected, indicated that the hybrid proteins were correctly synthesized in yeast, were efficiently transported into the nucleus, and maintained a tertiary structure proficient for the interaction.
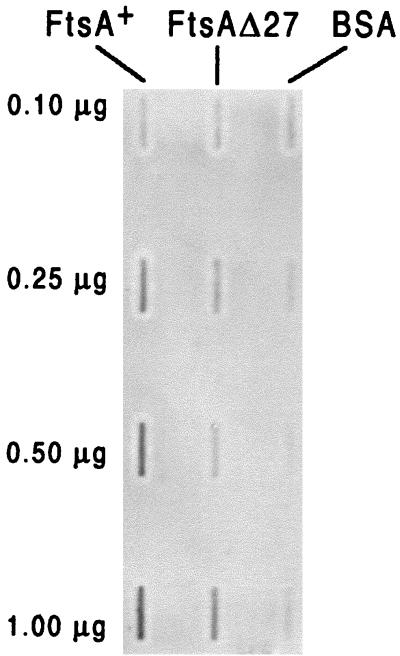
In an alternative approach His-tagged FtsA+ was purified to study the interaction in vitro. His-FtsA+ was bound to a nitrocellulose membrane using a slot blot apparatus. A fraction of the protein was biotinylated in vitro and incubated with the membrane for 1 h. After several washes the membrane was incubated with a streptavidin-peroxidase conjugate and developed by chemiluminiscence (Fig. 6). While the biotinylated protein did not bind to bovine serum albumin used as a control, it bound strongly to His-FtsA+, confirming the results obtained with the yeast two-hybrid system. His-FtsAΔ27 shows a decrease in the relative affinity of the interaction with the soluble biotinylated His-FtsA+ (Fig. 6), resulting in a binding too weak to be observed in the yeast two-hybrid assay.
FIG. 6.
Interaction of His-FtsA+ and His-FtsAΔ27 measured by an overlay assay. Different quantities (as indicated) of purified His-FtsA+ and His-FtsAΔ27 were blotted to nitrocellulose membranes using a slot blot filtration manifold. Bovine serum albumin (BSA) was included as a control. The filters were then blocked to prevent further binding and incubated with in vitro-biotinylated His-FtsA+. Binding of biotinylated protein to the filter-bound proteins was detected by incubating with streptavidin-peroxidase conjugate and developing with a chemiluminiscence detection reagent.
DISCUSSION
Several mutants producing carboxy terminus truncations of the FtsA cell division protein that can be grouped in three categories have been constructed. The first one, FtsAΔ1, behaves in all respects like the wild-type protein, showing that the last Phe residue, even though it is conserved in many sequences, is dispensable. Next, FtsAΔ5 is able to localize into mid-cell rings but does not complement ftsA mutations and, when overproduced, generates coiled cells independently of the presence of the full-length protein. Finally, FtsAΔ6, FtsAΔ13, FtsAΔ27, and FtsAΔ23Φ36, which are not functional, do not localize into septal rings, and produce coiled cells only in the presence of a full-length FtsA, but not in its absence (Fig. 3). All together, these results indicate that residues 415 to 419 are necessary for the biological function of FtsA, which is in agreement with the results of Ma et al. (23), who, by using green fluorescent protein fusions, mapped the intracellular localization domains of FtsA to both termini of the protein, and moreover, W415 is essential for the localization of the protein into septal rings.
When overproducing FtsAΔ6, FtsAΔ13, FtsAΔ27, or FtsAΔ23Φ36 in strains OV2 or OV16 at 30°C (with FtsA+ levels sufficient to support both normal growth and division) cell morphology is strikingly altered to a CC phenotype, while no such phenotype is observed after overproduction of the same proteins in OV16 at 42°C (containing no more than 1% of the FtsA+ levels). This result suggests that an interaction between the truncated proteins and a full-length protein might be necessary for the production of the CC phenotype. FtsA belongs to a protein family (4, 29) that includes members that are able to establish self-interactions. In the case of actin this interaction is essential for its biological function, i.e., the formation of the actin filament that is a major component of the cytoskeleton of all eukaryotic cells (30).
The yeast two-hybrid system (14) has already been used to detect interactions among some of the components of the bacterial cell division machinery, including those of the MinCDE system (20), and those of FtsZ with SulA (20), ZipA (25), SpoIIE (22), or FtsA (12, 24, 25, 38, 40). More recently, Yan et al. (40) have reported the self-interaction of E. coli FtsA using the two-hybrid system, a result that has been confirmed in the present work. Using a protein overlay assay (5) we have also detected in vitro the interaction of FtsA molecules. In these assays biotinylated FtsA was used at a concentration of 2.5 nM in the liquid phase. As this concentration is much lower than the intracellular concentration of FtsA, estimated to be around 100 nM when assuming that 150 molecules are contained in each E. coli cell (36), the observed interaction falls well within the expected physiological range.
As discussed by Yan et al. (40) this interaction might be unique to E. coli FtsA, because in the yeast two-hybrid system, the Staphylococcus aureus FtsA molecules do not interact. But given that the strength of the interaction measured for E. coli FtsA falls in the lower range of the two-hybrid assay (11 Miller units for the wild-type proteins), it is possible that if the S. aureus FtsA interaction is slightly weaker than the E. coli one, it might fall below the significant range of the assay.
When tested in the two-hybrid system, deletions of five or more residues impaired the interaction with the wild-type protein, showing that the carboxy terminus of the protein is important for the self-interaction. Although they did not interact in the yeast two-hybrid assay we could nevertheless detect binding of biotinylated FtsA+ to FtsAΔ27 when using the protein overlay assay. A plausible interpretation of these results is that the deletion produces a decrease in the binding affinity between the wild type and the mutant protein, weakening the interaction below the detection threshold of the two-hybrid assay. This might be the reason also for the CC phenotype being expressed only when the truncated proteins are produced at high levels. These alterations of the interaction properties observed in carboxy-terminally truncated FtsAΔ proteins (lacking five residues or more) are accompanied by the simultaneous loss of their function in E. coli division, as evidenced by their inability to complement ftsA mutants D2 and OV16.
Each truncated protein species was able to interact with itself and with other truncated FtsA proteins with the exception of FtsAΔ1. This shows that truncation of the carboxy terminus does not result in an extreme change in the protein structure, as the mutated proteins can still self-interact. Furthermore, a comparative FTIR-CD spectroscopy analysis of the soluble His-FtsA+ and His-FtsAΔ27, the more extensive of the truncated FtsA forms analyzed, showed that their secondary structures are similar (Table 2 and Fig. 4). This result indicates that the deletion of the 27 last carboxy-terminal residues, although important for the biological activity, does not affect significantly the structure of the protein. Accordingly, FtsAΔ27 retained the ability to bind ATP (Fig. 5), indicating that the structure of the ATP-binding site remains intact.
The FtsAΔ6, FtsAΔ13, and FtsAΔ27 proteins cannot localize into septal rings, and they express a coiled phenotype only in the presence of FtsA+. FtsAΔ5, on the other hand, is able to localize into rings and produces coiled cells independently of the presence of FtsA+. These data suggest that both a correct localization of the protein in the septal ring and a correct interaction between FtsA molecules are important for cell division. Alterations in the interaction of FtsA might be responsible for the distorted cell shape observed in these mutants.
We can postulate that the FtsA molecule contains domains able to establish interactions (evidence to include the carboxy terminus among them is shown) and can speculate that removal of a carboxy-terminus segment exposes one or more domains, normally masked in the wild-type protein, that may be able to establish a different and stronger interaction. Alternatively the deletion of the carboxy terminus may affect the properties of the resultant protein to modify its transport into the yeast nucleus, resulting in an alteration of the molecules available for the establishment of the interaction. This trivial explanation would be more difficult to reconcile with the apparent conservation of the protein structure in the deleted forms. In any case the loss of interaction with the wild type is accompanied by a loss of function, but the exact nature of the FtsA interactions and the molecular mechanisms responsible for the impairment of the biological function observed when the carboxy terminus of FtsA is deleted remain, nevertheless, to be discovered.
ACKNOWLEDGMENTS
We thank Alfonso Valencia and Luis Sánchez for computer analysis of the FtsA sequence, and María José Ferrándiz for the construction of pMFV plasmids. The excellent technical assistance of Pilar Palacios is acknowledged.
This work has been jointly financed in the laboratories of M.V. and J.-M.R. by EC DGXII project BIO4-CT96-0122. Additional funds granted to M.V. were from project BIO97-1246 from Plan Nacional de I+D (Ministerio de Educación y Cultura, Spain). J.M. and S.R. were supported by fellowships from the Comunidad Autónoma de Madrid.
REFERENCES
- 1.Addinall S G, Lutkenhaus J. FtsA is localized to the septum in an FtsZ-dependent manner. J Bacteriol. 1996;178:7167–7172. doi: 10.1128/jb.178.24.7167-7172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinall S G, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 3.Bartel P L, Chien C-T, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. BioTechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- 4.Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin and hsp70 heat shock proteins. Proc Natl Acad Sci USA. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr D W, Scott J D. Blotting and band-shifting: techniques for studying protein-protein interactions. Trends Biochem Sci. 1992;17:246–249. doi: 10.1016/0968-0004(92)90402-u. [DOI] [PubMed] [Google Scholar]
- 6.Celis J E, Hooper M L, Smith J D. Amino acid acceptor stem of E. coli suppressor tRNATyr is a site of synthetase recognition. Nat New Biol. 1973;244:261–264. doi: 10.1038/newbio244261a0. [DOI] [PubMed] [Google Scholar]
- 7.Chen J C, Weiss D S, Ghigo J M, Beckwith J. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J Bacteriol. 1999;181:521–530. doi: 10.1128/jb.181.2.521-530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai K, Xu Y, Lutkenhaus J. Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts) J Bacteriol. 1993;175:3790–3797. doi: 10.1128/jb.175.12.3790-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Descoteaux A, Drapeau G R. Regulation of cell division in Escherichia coli K-12: probable interactions among proteins FtsQ, FtsA, and FtsZ. J Bacteriol. 1987;169:1938–1942. doi: 10.1128/jb.169.5.1938-1942.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dewar S J, Begg K J, Donachie W D. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol. 1992;174:6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Din N, Quardokus E M, Sackett M J, Brun Y V. Dominant carboxy-terminal deletions of FtsZ that affect its ability to localise in Caulobacter and its interaction with FtsA. Mol Microbiol. 1998;27:1051–1063. doi: 10.1046/j.1365-2958.1998.00752.x. [DOI] [PubMed] [Google Scholar]
- 13.Donachie W D, Begg K J, Lutkenhaus J F, Salmond G P C, Martínez-Salas E, Vicente M. Role of the ftsA gene product in control of Escherichia coli cell division. J Bacteriol. 1979;140:338–394. doi: 10.1128/jb.140.2.388-394.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–247. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 15.Flärdh K, Palacios P, Vicente M. Cell division genes ftsQAZ in Escherichia coli require distant cis-acting signals upstream of ddlB for full expression. Mol Microbiol. 1998;30:305–316. doi: 10.1046/j.1365-2958.1998.01064.x. [DOI] [PubMed] [Google Scholar]
- 16.Fürste J P, Pansegrau W, Frank R, Blöcker H, Scholtz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 17.Gayda R C, Henk M C, Leong D. C-shaped cells caused by expression of an ftsA mutation in Escherichia coli. J Bacteriol. 1992;174:5362–5370. doi: 10.1128/jb.174.16.5362-5370.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, Cao C, Lutkenhaus J. Interaction between FtsZ and inhibitors of cell division. J Bacteriol. 1996;178:5080–5085. doi: 10.1128/jb.178.17.5080-5085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurley J. The sugar kinase/heat-shock protein 70/actin superfamily: implications of conserved structure for mechanism. Annu Rev Biophys Biomol Struct. 1996;25:137–162. doi: 10.1146/annurev.bb.25.060196.001033. [DOI] [PubMed] [Google Scholar]
- 22.Lucet I, Feucht A, Yudkin M D, Errington J. Direct interaction between the cell division protein FtsZ and the cell differentiation protein SpoIIE. EMBO J. 2000;19:1467–1475. doi: 10.1093/emboj/19.7.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma X, Ehrhardt D W, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma X, Sun Q, Wang R, Singh G, Jonietz E L, Margolin W. Interactions between heterologous FtsA and FtsZ proteins at the FtsZ ring. J Bacteriol. 1997;179:6788–6797. doi: 10.1128/jb.179.21.6788-6797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma X, Margolin W. Genetic and functional analyses of the conserved carboxy-terminal core domain of Escherichia coli FtsZ. J Bacteriol. 1999;181:7531–7544. doi: 10.1128/jb.181.24.7531-7544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 27.Nanninga N. Morphogenesis of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:110–129. doi: 10.1128/mmbr.62.1.110-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sánchez M, Valencia A, Ferrándiz M J, Sander C, Vicente M. Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J. 1994;13:4919–4925. doi: 10.1002/j.1460-2075.1994.tb06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheterline P, Clayton J, Sparrow J. Actin. Protein Profile. 1995;2(L):1–103. [PubMed] [Google Scholar]
- 31.Tormo A, Martínez-Salas E, Vicente M. Involvement of the ftsA gene product in late stages of the Escherichia coli cell cycle. J Bacteriol. 1980;141:806–813. doi: 10.1128/jb.141.2.806-813.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tormo A, Vicente M. The ftsA gene product participates in formation of the Escherichia coli septum structure. J Bacteriol. 1984;157:779–784. doi: 10.1128/jb.157.3.779-784.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tormo A, Ayala J A, de Pedro M A, Aldea M, Vicente M. Interaction of FtsA and PBP3 proteins in the Escherichia coli septum. J Bacteriol. 1986;166:985–992. doi: 10.1128/jb.166.3.985-992.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vicente M, Errington J. Structure, function and controls in microbial division. Mol Microbiol. 1996;20:1–7. doi: 10.1111/j.1365-2958.1996.tb02482.x. [DOI] [PubMed] [Google Scholar]
- 35.Vicente M, Palacios P, Dopazo A, Garrido T, Pla J, Aldea M. On the chronology and topography of bacterial cell division. Res Microbiol. 1991;142:253–257. doi: 10.1016/0923-2508(91)90038-c. [DOI] [PubMed] [Google Scholar]
- 36.Wang H C, Gayda R C. High-level expression of the FtsA protein inhibits cell septation in Escherichia coli K-12. J Bacteriol. 1990;172:4736–4740. doi: 10.1128/jb.172.8.4736-4740.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Khattar M K, Donachie W D, Lutkenhaus J. FtsI and FtsW are localized to the septum in Escherichia coli. J Bacteriol. 1998;180:2810–2816. doi: 10.1128/jb.180.11.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Huang J, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP and FtsA. J Bacteriol. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward J E, Jr, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985;42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- 40.Yan K, Pearce K H, Payne D J. A conserved residue at the extreme carboxy-terminus of FtsZ is critical for the FtsA-FtsZ interaction in Staphylococcus aureus. Biochem Biophys Res Commun. 2000;270:387–392. doi: 10.1006/bbrc.2000.2439. [DOI] [PubMed] [Google Scholar]
- 41.Yu X C, Tran A H, Sun Q, Margolin W. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol. 1998;180:1296–1304. doi: 10.1128/jb.180.5.1296-1304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]