Abstract
Asparagine synthetase (ASNS) catalyzes synthesis of asparagine (Asn) and Glu from Asp and Gln in an ATP-dependent reaction. Asparagine synthetase deficiency (ASNSD) results from biallelic mutations in the ASNS gene. Affected children exhibit congenital microcephaly, continued brain atrophy, seizures, and often premature mortality. However, the underlying mechanisms are unclear. This report describes a compound heterozygotic ASNSD child with two novel mutations in the ASNS gene, c.1118G>T (paternal) and c.1556G>A (maternal), that lead to G373V or R519H ASNS variants. Structural mapping suggested that neither variant participates directly in catalysis. Growth of cultured fibroblasts from either parent was unaffected in Asn-free medium, whereas growth of the child’s cells was suppressed by about 50%. Analysis of Asn levels unexpectedly revealed that extracellular rather than intracellular Asn correlated with the reduced proliferation during incubation of the child’s cells in Asn-free medium. Our attempts to ectopically express the G373V variant in either HEK293T or JRS cells resulted in minimal protein production, suggesting instability. Protein expression and purification from HEK293T cells revealed reduced activity for the R519H variant relative to WT ASNS. Expression of WT ASNS in ASNS-null JRS cells resulted in nearly complete rescue of growth in Asn-free medium, whereas we observed no proliferation for the cells expressing either the G373V or R519H variant. These results support the conclusion that the coexpression of the G373V and R519H ASNS variants leads to significantly reduced Asn synthesis, which negatively impacts cellular growth. These observations are consistent with the ASNSD phenotype.
Keywords: amino acid, brain metabolism, inborn error of metabolism, neurotransmitter, epilepsy, genetic disease
Abbreviations: AAR, amino acid response; ASNS, asparagine synthetase; ASNSD, asparagine synthetase deficiency; cDNA, complementary DNA; FBS, fetal bovine serum; ISR, integrated stress response; JRS, Jensen rat sarcoma; PDB, protein database; qRT-PCR, quantitative real-time PCR
Human asparagine synthetase (ASNS) catalyzes the synthesis of asparagine (Asn) and glutamate (Glu) from the substrates aspartate (Asp) and glutamine (Gln) in an ATP-dependent amidotransferase reaction (1, 2). The human ASNS gene consists of 13 exons covering about 35 kb and is located at chromosome 7 region 7q21.3 (3, 4, 5). Transcription from the ASNS gene is highly regulated in response to a wide variety of cell stresses, in particular amino acid deprivation and endoplasmic reticulum stress (reviewed in (2, 6)). The human ASNS enzyme consists of 561 amino acid residues with an approximate molecular mass of 64 kDa. The crystal structure of human ASNS was recently reported (protein database [PDB]: 6GQ3) (7) and is homologous to that for Escherichia coli ASNS B (PDB: 1CT9) (1, 8). The C-terminal domain (residues 209–561) is composed of several α-helices and a 5-stranded, parallel β-sheet that contains the ATP- and Asp-binding sites. The reaction catalyzed by the C-terminal domain leads to activation of the Asp carboxyl group by an ATP-dependent process to yield an enzyme bound β-aspartyl-AMP intermediate. Within the distinct N-terminal domain (residues 1–208), Gln binds with the carboxamide group oriented toward the interface of the two domains to allow the glutaminase activity to generate an ammonia group that diffuses through a predicted intramolecular tunnel to react with the β-aspartyl-AMP intermediate, producing Asn (1, 7, 8, 9).
ASNS deficiency (ASNSD) was first described in 2013 based on four families with homozygosity or compound heterozygosity for mutations in the ASNS gene (10). The affected children shared the features of severe encephalopathy, congenital microcephaly, progressive brain atrophy, early onset seizures, axial hypotonia, severe appendicular spasticity, and often, mortality at a premature age (reviewed in (6, 11, 12)). Since the initial report, about 50 independent ASNS protein variants have been identified (summarized by (13, 14, 15)). The exact mechanisms that cause the overt symptoms of the disease are not well understood, and at present, the disease can only be diagnosed through DNA sequencing. Some, but not all, affected individuals have a measurable decrease in the amount of Asn in their serum or cerebrospinal fluid, and thus, this analysis is not a definitive screening approach for suspected cases. Based on animal growth studies, Asn is considered a “nonessential” or “dispensable” amino acid, but at the level of individual cells, an ASNS enzyme deficiency that is sufficiently large leads to extracellular Asn dependence. Our previous characterizations of fibroblasts from ASNSD patients revealed that lowering the extracellular Asn concentration results in a significant decrease in cell proliferation (16, 17). Depending on the severity of the mutation, fibroblasts from some asymptomatic heterozygotic parents also exhibited a decline in proliferation in the absence of sufficient extracellular Asn.
The present results describe a female child with ASNSD who inherited two novel mutations in the ASNS gene, c.1118G>T (paternal) and c.1556G>A (maternal), that lead to aberrant proteins containing G373V or R519H substitutions, respectively. The child has a history of developmental delay, microcephaly, and epilepsy typical of other ASNSD patients. Growth of the child’s fibroblasts was suppressed in medium lacking Asn and measurement of enzyme activity revealed that the R519H variant exhibited reduced function, whereas the G373V variant was too unstable to purify. Transduction with WT ASNS resulted in nearly complete rescue of growth in ASNS-null Jensen rat sarcoma (JRS) cells, but expression of either variant produced little or no growth in the absence of medium-supplied Asn. Collectively, these results support the conclusion that the compound heterozygotic coexpression of the G373V and R519H ASNS variants leads to a significant defect in the cellular ability to produce sufficient Asn, which has a deleterious impact on cellular growth.
Results
Diagnosis and clinical assessment
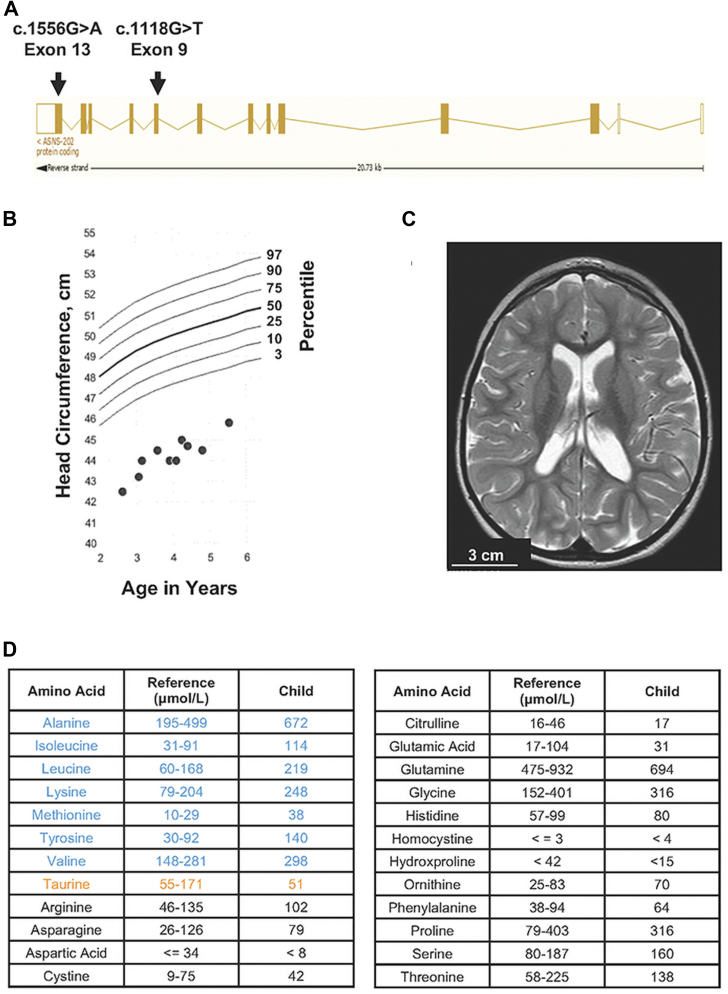
This study describes a female child who has biallelic mutations in the ASNS gene. When whole exome sequencing was performed, mutations in the ASNS gene were identified with c.1118G>T paternally inherited and c.1556G>A maternally inherited (Fig. 1A). The c.1118G>T mutation results in the substitution of valine for glycine at position 373. The G373V variant was not listed in the National Center for Biotechnology Information (NCBI) dbSNP database, and no minor allele frequency was noted in the Exome Sequencing Project cohort, Exome Aggregation Consortium (ExAC), or 1000 Genome Project database. The c.1556G>A mutation results in the substitution of a histidine for arginine at position 519. The R519H variant was listed in the NCBI dbSNP database (rs568570377). According to several mutation prediction software analyses on PROVEAN, PolyPhen-2, and MutationTaster, both variants were predicted to be “deleterious” and “probably damaging.” Neither variant was listed in the Human Gene Mutation Database or reported in peer-reviewed literature. There were no additional predicted deleterious variants reported in the exome.
Figure 1.
Clinical assessment of the child with biallelic mutations in the ASNS gene. A, the ensemble.org transcript ENST00000394308.8 ASNS-202 diagram for ASNS was adapted to show the exon location of both c.1118G>T and c.1556G>A mutations. B, head circumference up to age 6 years. Head circumference in centimeters is the y-axis on the left and the percentile is shown on the right. C, T2 weighted axial MRI scan demonstrating diffuse failure of development of cerebral white matter and microcephaly. D, serum amino acid analysis of the child at age 4. Amino acid levels that were higher than the reference range (μmol/L) are shown in blue, those that were below the range are in orange, and those that were within the range are in black. ASNS, asparagine synthetase.
The child is the first born of nonconsanguineous parents and was delivered at 40 weeks gestation after an uncomplicated pregnancy. At birth, Apgar scores were within the normal range, 8 at 1 min and 9 at 5 min. Head circumference at birth was 32.0 cm (third percentile) and has remained at or below the third percentile (Fig. 1B). The length at birth was 19 in and weight was 7 lbs 9 oz. The father has a head circumference that is small but still within the normal range at 52.5 cm and the mother’s head circumference is 57.0 cm. There is no family history of seizures or additional neurologic concerns. The child was relatively healthy for the first year of life, but parents became concerned about her development at 10 months. Brain MRI at 10 months of age showed microcephaly with diffuse supratentorial white matter volume loss, which primarily involved the central and periventricular white matter (Fig. 1C). She began having seizures at 13 months and she was referred to a genetic specialist at 3 years old for cryptogenic localization-related epilepsy, microcephaly, and global developmental delay. The child’s seizures responded quickly to valproic acid, which is often found to be beneficial with other genetic epilepsies. The patient was noted to have dysmorphic features including microcephaly, triangular facies, widely spaced teeth, and bilateral brachydactyly. She was also noted to have some behavioral concerns such as hand flapping and appeared to be overly happy. At 4 years old, her head circumference was 44.0 cm (<1 percentile), weight was 32 lbs, and height was 36 in. She was noted to have a 7 to 10 word vocabulary, could pull to a stand, and could walk with assistance. Prior to clinical whole exome sequencing, studies revealed normal values for single nucleotide polymorphism microarray, Angelman Syndrome methylation studies, and sequencing of the ubiquitin protein ligase E3A gene. Serum amino acid analysis revealed elevated levels of alanine, isoleucine, leucine, lysine, methionine, tyrosine, and valine, whereas taurine was just below the reference range (Fig. 1D). It is noteworthy that the levels of the ASNS substrates Asp and Gln were normal, as were the products Glu and Asn.
In contrast to most of the ASNSD patients described previously, this child, 9 years old at the time of this study, continues to make developmental progress both with regard to fine and gross motor skills, despite still being microcephalic below the second percentile. She remains nonverbal but is interactive, sociable, and can indicate needs with simple gestures such as pointing and nodding/shaking of the head. The child has been able to walk without assistance for about 2 years now.
Computer modeling of novel human ASNS variants
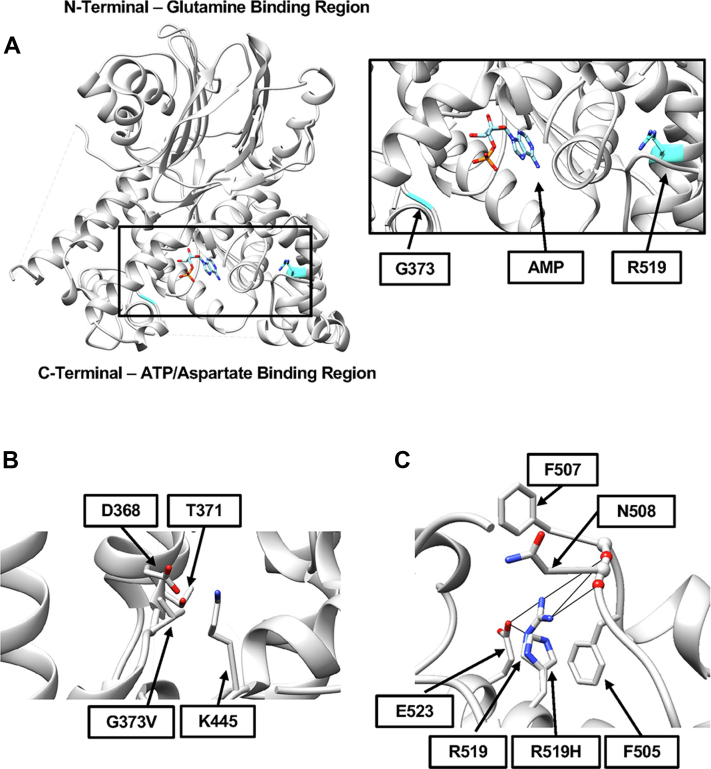
Both amino acids G373 and R519 are highly conserved in ASNS across species, from human to yeast to E. coli. In humans, the first methionine is removed during protein processing, so the cysteine-2 is the first amino acid in the functional enzyme (1). With methionine present, as in E. coli AS-B, both Gln and ammonia can be nitrogen donors, but in the human protein, with the terminal cysteine, only Gln can be the donor. Figure 2 illustrates in silico models of the human ASNS protein based on the crystal structure published by Zhu et al. (7). The locations of residues glycine-373 (G373) and arginine-519 (R519) within the C-terminal domain are shown in Figure 2A. Neither of these residues is located within the active site, which is identified by the location of an AMP molecule, as determined by the crystal structure of the E. coli ASNS-B (PDB: 1CT9) (8). Amino acid G373 resides within a linker region between two helices and R519 is located within a helix near the outer surface of the protein. Indeed, no direct interaction of G373 with other residues is identified in the in silico model, although substitution with the most probable valine rotamer leads to a prediction of possible steric hindrance with residues D368, T371, and K445 (Fig. 2B). The R519 sidechain is predicted to hydrogen bond with the backbone of residues F507 (2.9 and 2.8 Å), N508 (3.0 Å), and E523 (3.4 and 2.9 Å) (Fig. 2C). Modeling of the R519H variant (Fig. 2C) suggests that this substitution may disrupt the ASNS structure because the hydrogen bonds with F507, N508, and E523 are lost, and histidine at this position may cause steric interference with F505.
Figure 2.
In silico modeling of variants within the ASNS protein structure.A, the locations of amino acids G373 and R519 (shown in cyan) were modeled using the human ASNS crystal structure (7). The insert shows a close-up of the C-terminal ATP/Asp-binding region illustrating the distance from the AMP-binding site for G373 (a minimum of 8.9 Å) and R519 (a minimum of 24 Å). B, steric clashes are predicted between the most probable G373V rotamer and residues D368, T371, and K445. C, the region surrounding the R519H variant is shown to illustrate the possible loss of hydrogen bonding and possible steric hindrance. For all panels, nitrogen atoms are shown in blue, oxygen atoms in red, and phosphorous in orange. ASNS, asparagine synthetase.
Amino acid–dependent gene regulation is intact
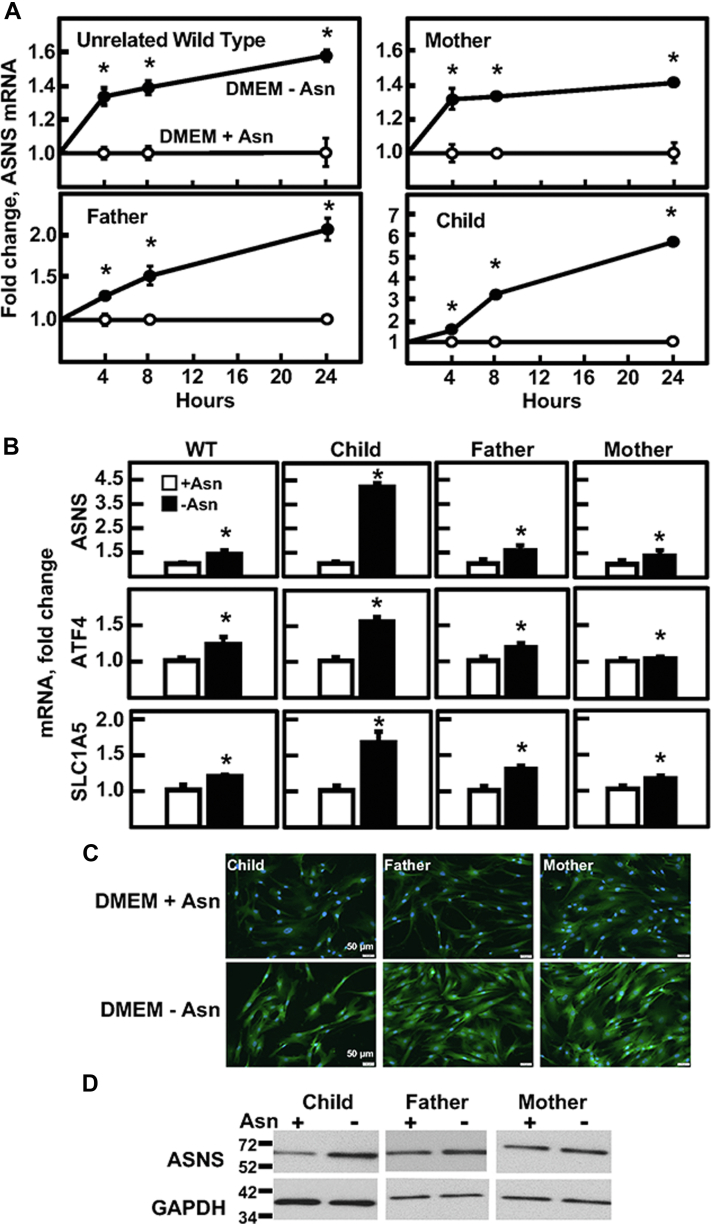
One of the hallmarks of the mammalian ASNS gene is increased transcription in response to endoplasmic reticulum stress or following amino acid limitation of cells that triggers a signal transduction pathway referred to as the amino acid response (AAR) (reviewed in (2, 6, 18, 19)). Both of these stress-induced pathways lead to increased expression of the transcription factor ATF4 (20, 21). Furthermore, both of the stress-induced pathways also lead to increased translation of the ATF4 mRNA by a process referred to as the integrated stress response (ISR), despite a concurrent suppression of general translation (22, 23, 24, 25). Skin fibroblasts from each member of the family were analyzed for the AAR-associated expression of ASNS relative to an unrelated WT human fibroblast cell line (Fig. 3). Incubation of the cells for 0 to 24 h in Dulbecco’s modified Eagle’s medium (DMEM) ± Asn medium revealed that induction of ASNS mRNA was detectable for all family members (Fig. 3A). Interestingly, the level of induction was similar to WT for the maternal- and paternal-derived cells. However, the values of mRNA abundance in the DMEM-Asn condition for the child’s cells were significantly greater (p ≤ 0.05) when compared to either parent’s value at the same time point. To establish that the increase in ASNS mRNA was the result of the integrated stress response, it was observed that mRNA abundance for activating transcription factor 4 (ATF4) itself and the SLC1A5/ASCT2 amino acid transporter, targets of the AAR and ISR, were also increased after incubation of the cells in DMEM ± Asn for 24 h (Fig. 3B). To determine if the ASNS protein expression was uniformly represented within the cell population, anti-ASNS immunostaining of the fibroblasts after 24 h in DMEM ± Asn medium showed the basal expression of ASNS when cells are maintained in complete DMEM medium, as well as the typical AAR-associated increase in ASNS protein expression after incubation of the cells in medium lacking Asn (Fig. 3C, bottom panels). Cell extracts from similarly treated cells were subjected to immunoblotting to detect ASNS abundance (Fig. 3D). While the induction of ASNS protein was modest for the samples from the father and mother, a greater increase was observed for the child. Collectively, these data support the conclusion that the regulated transcription of the ASNS gene and subsequent translation to protein are functional in all family members.
Figure 3.
Amino acid–regulated gene expression in cultured ASNSD fibroblasts. Primary cultures of fibroblasts from the child, both parents, and a nonrelated WT fibroblast cell line were cultured in DMEM ± Asn for 0–24 h. A, ASNS mRNA was measured by qRT-PCR of total RNA and the fold change was calculated for all fibroblast cell lines. The ASNS mRNA values were normalized to the mRNA abundance of GAPDH in the same sample. B, the RNA samples described for panel (A) were analyzed by qRT-PCR for the mRNA abundance of ASNS, ATF4, and the ASCT2 amino acid transporter SLC1A5, each normalized to the GAPDH mRNA content of each sample. For panels (A) and (B), the data are shown as the averages ± SDs for assays in triplicate. Each experiment was repeated at least once with different batches of cells to establish reproducibility. Where not shown, the SD bars are within the symbol. Based on a two-tailed Student’s t test, an asterisk indicates a p value of ≤ 0.05. C, to analyze the uniformity of the ASNS protein content across all cells, immunofluorescence was performed for the fibroblast cell line from each family member after a 24 h incubation in DMEM ± Asn. Immunoblotting was performed for each sample to identify the ASNS protein based on size, with GAPDH used as a control (D). The data shown in panels (C) and (D) are from a representative experiment, and each experiment was repeated at least once to confirm reproducibility. ASNS, asparagine synthetase; ASNSD, ASNS deficiency; qRT-PCR, quatitative real-time PCR.
Impact of ASNS variants on cell proliferation
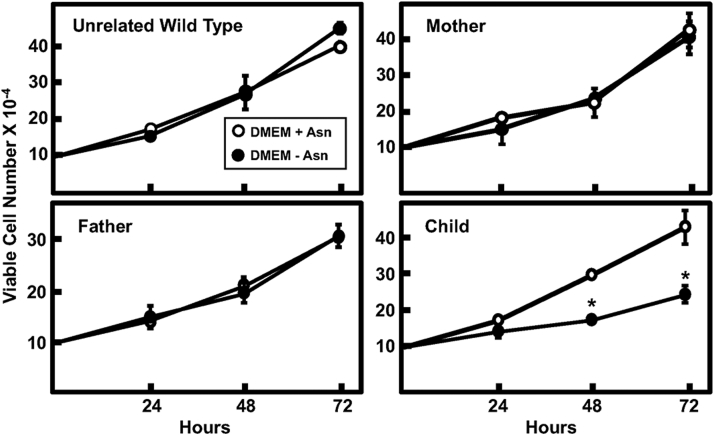
To determine the impact of variant expression on cell proliferation, the fibroblast line of each family member and an unrelated WT were cultured in DMEM ± Asn and viable cell counts determined at 24 h intervals over a 72 h period (Fig. 4). The proliferation studies revealed that both the paternal (G373V) and maternal (R519H) variant-expressing fibroblasts grew at similar rates to an unrelated WT cell line, even in the absence of Asn in the culture medium (Fig. 4). In contrast, the growth of the affected child’s fibroblasts, expressing both variant proteins, grew at a substantially slower rate in the absence of medium-supplied Asn (Fig. 4). Collectively, these data indicate that coexpression of these two variants, in the absence of WT activity, negatively impacts cell proliferation.
Figure 4.
Impact of ASNS variant expression on fibroblast proliferation. Fibroblasts from the child and both parents, along with a nonrelated WT fibroblast cell line, were incubated in DMEM ± Asn for 24, 48, and 72 h. The results shown are viable cell numbers only and were measured using an automated cell counter and an associated trypan blue exclusion method. The data are presented as the averages ± SDs of viable cell numbers in triplicate wells for each time point and culture medium condition. Each experiment was repeated with different batches of cells to establish reproducibility. Where not shown, the SD bars are within the symbol. An asterisk indicates that the DMEM-Asn value was p ≤ 0.05 compared to DMEM + Asn condition. ASNS, asparagine synthetase.
ASNS substrate and product levels in fibroblasts incubated in Asn-free medium
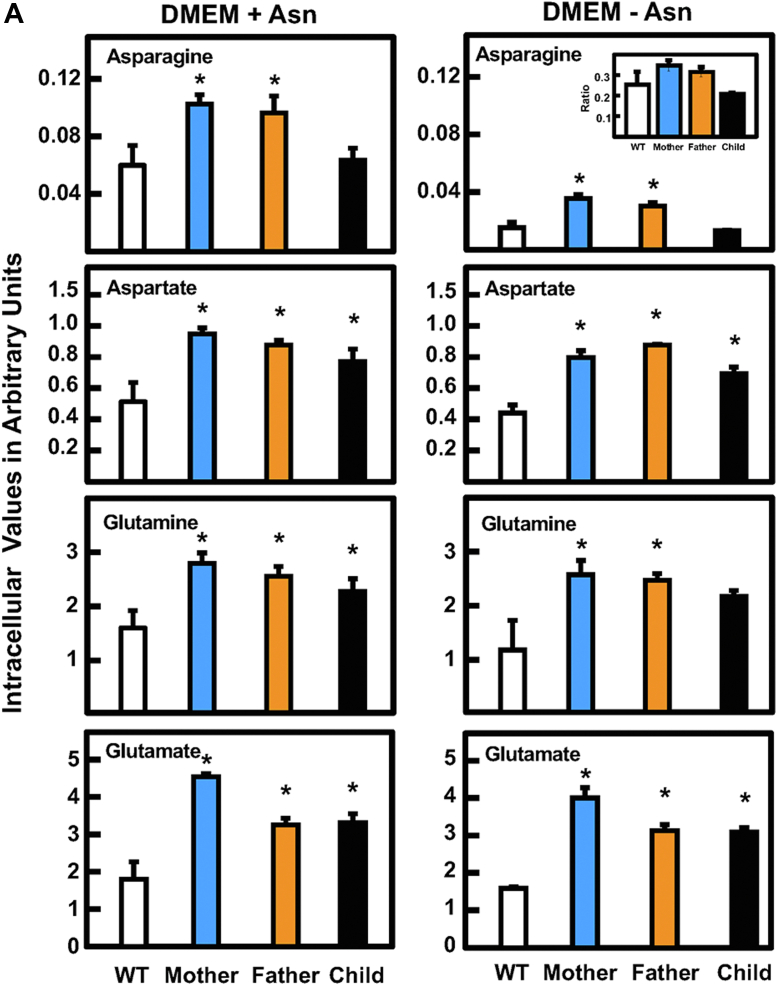
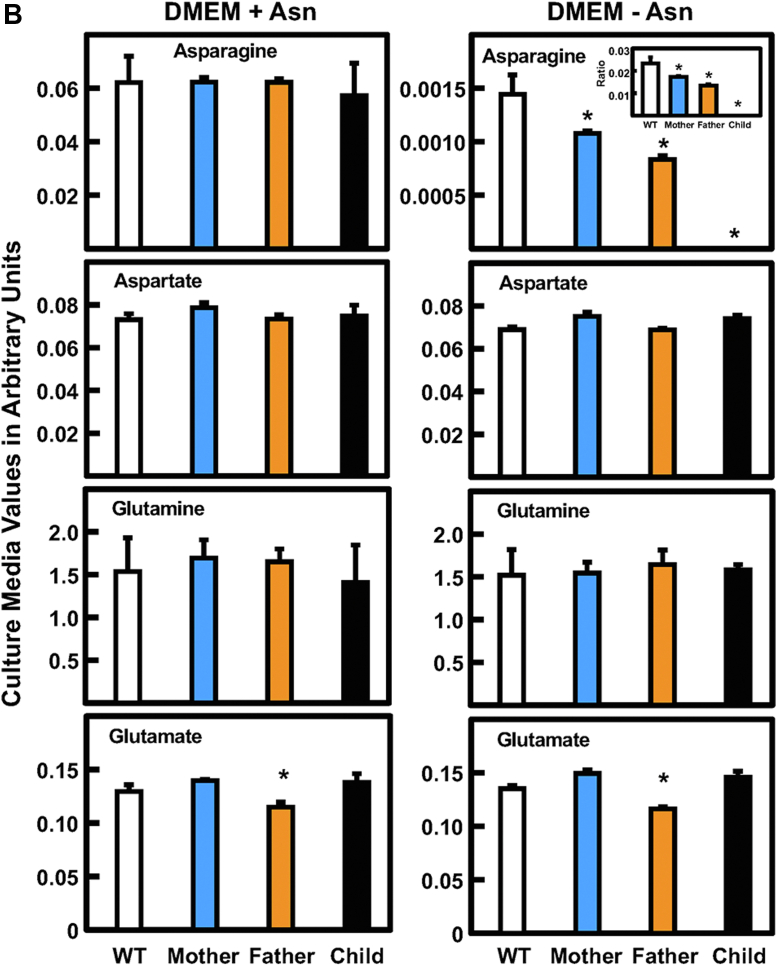
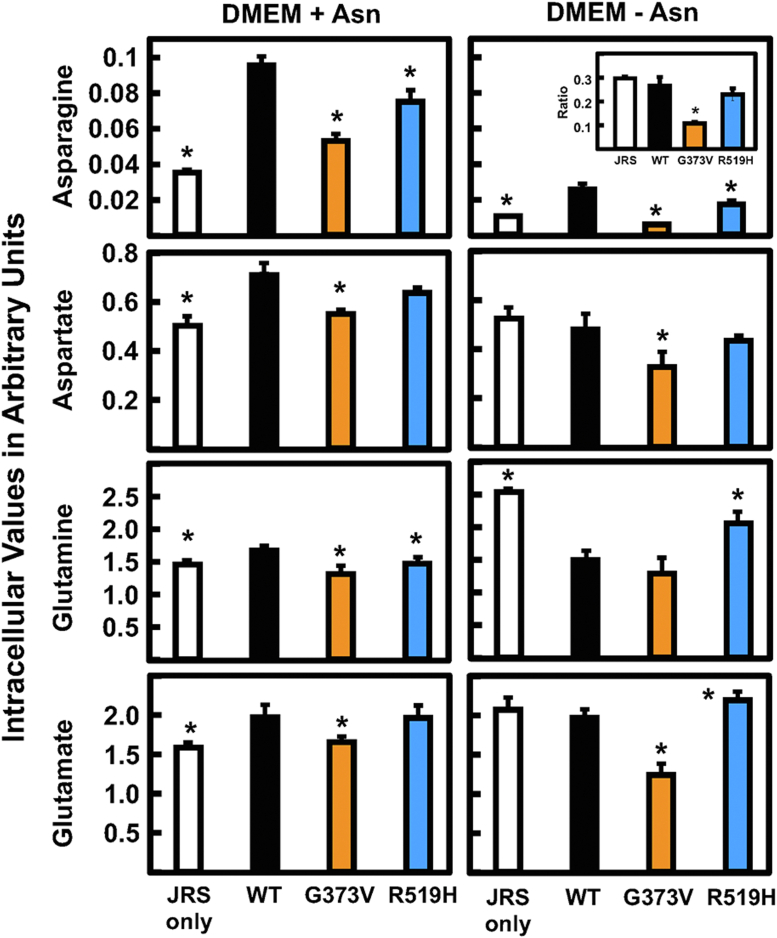
Analysis of intracellular Asn, Asp, Gln, and Glu abundance was investigated in cultured fibroblasts from the ASNSD family. After incubation of the cells in DMEM with or without Asn for 24 h, the cellular content of Asp, Gln, and Glu were elevated relative to WT fibroblasts (Fig. 5A). In contrast, incubation in Asn-free medium caused a decline in Asn in all four cell lines. Given the difference in cell proliferation between the parents and the child, it is noteworthy that the cellular Asn in the child’s cells was less than that for either parent. The basis for the decreased amount of Asn in the WT cells incubated in Asn-free medium relative to the complete DMEM is unclear. To provide a broader picture of metabolism for these four amino acids, the extracellular levels were also analyzed (Fig. 5B). The analysis showed that Asp, Gln, and Glu were largely unchanged regardless of incubation of the cells with or without medium Asn. However, medium Asn content substantially declined for all four cell lines after culture in the absence of Asn. Relative to WT, there was a significant decrease in extracellular Asn for either parent, and the Asn content of the child’s cells was at or below the GC-MS limits of detection (Fig. 5B). The observation that extracellular Asn content was better correlated with the cell proliferation for the ASNSD fibroblasts than intracellular levels was also observed in separate experiments for two different ASNSD families and using LC-MS for the amino acid analysis (data not shown).
Figure 5.
Analysis of substrates and products for the ASNS reaction in fibroblasts incubated in the presence or absence of Asn. Primary cultures of fibroblasts from the ASNSD family and a nonrelated WT fibroblast cell line were incubated in DMEM ± Asn for 24 h. Cell extracts (A) and the extracellular media (B) were collected for amino acid analysis by GC-MS. The results for Asn, Asp, Gln, and Glu are presented as “arbitrary units,” calculated as the averages of triplicate determinations ± SDs of the relative amount of each amino acid per 106 cells. To better illustrate the changes in Asn, the ratios of the values for DMEM-Asn/DMEM+Asn were calculated and are shown as an insert. Where not shown, the SD marks are within the bar. An asterisk indicates that the p value was ≤ 0.05 compared to the value obtained for the WT cells. ASNS, asparagine synthetase.
Analysis of growth after variant expression in ASNS-null JRS cells
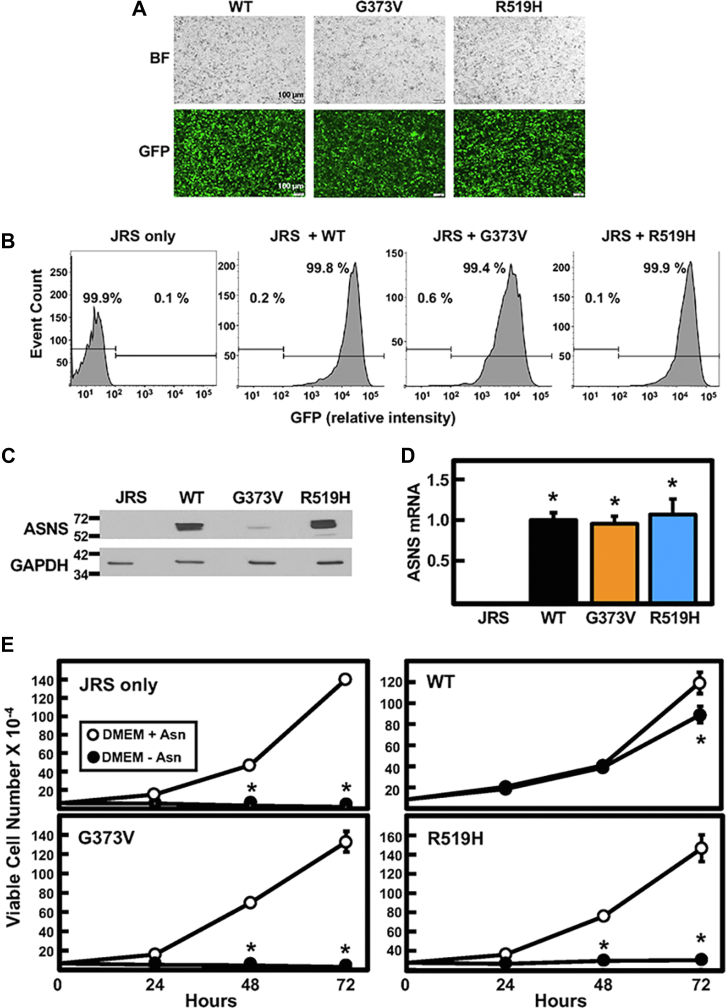
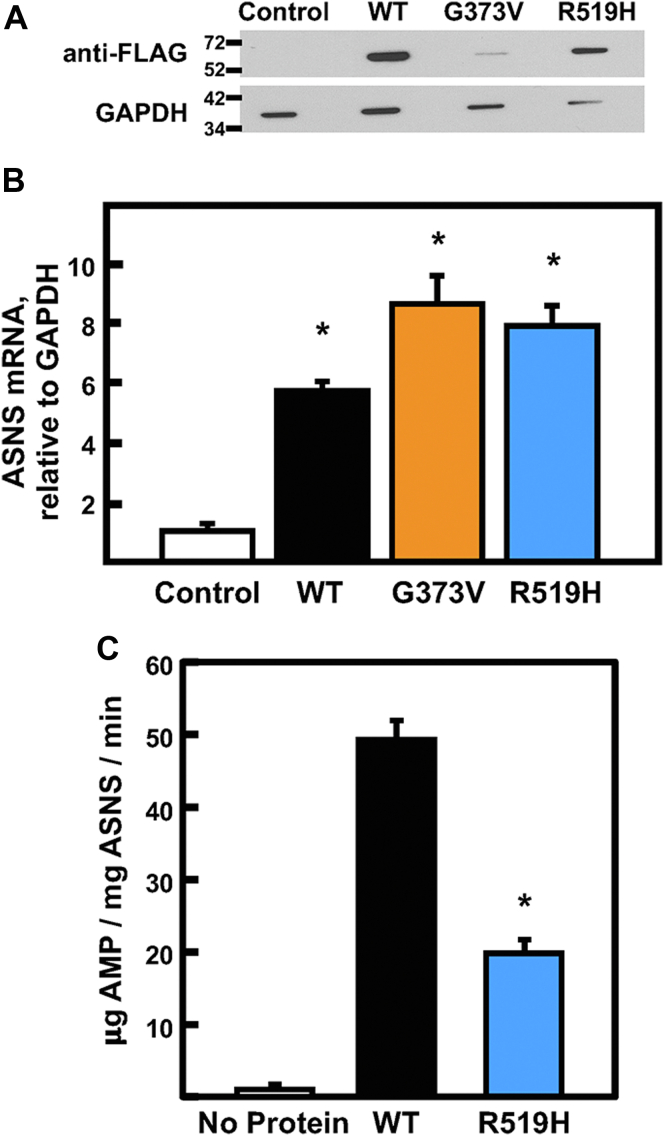
At the cellular level, analysis of a mutation for an individual allele is difficult in the presence of a functional WT allele. Therefore, ectopic expression in an ASNS-null cell line was used to study the impact of each ASNS variant, independent of the presence of WT ASNS or another variant. JRS cells do not express ASNS due to hypermethylation of the ASNS gene (26). Consequently, the JRS cells behave as Asn auxotroph and do not survive in Asn-free culture medium. Stable JRS cell lines, each independently expressing WT, G373V, or R519H ASNS, were generated after transduction with the appropriate ASNS-encoding retrovirus. Based on fluorescent images of GFP, all three cell lines displayed a similar level of transduction (Fig. 6A), and flow cytometry confirmed that the transduction efficiency was greater than 99% (Fig. 6B). Immunoblotting of protein extracts revealed that the parental JRS cells had no detectable ASNS protein, as expected (Fig. 6C). The abundance of ASNS protein for WT and R519H ASNS was similar and readily apparent, but even after a long exposure of the blot, only a small amount of the G373V variant protein was detected. To determine if the lower G373V expression was due to a defect in transcription, the mRNA levels of each cell line were analyzed for ASNS mRNA abundance by quantitative real-time PCR (qRT-PCR) but there were no significant differences (Fig. 6D). These results suggest that the G373V protein may be unstable. The impact of the individual ASNS variants on cell proliferation was determined by measuring the viable cell counts after incubation in DMEM ± Asn for 0 to 72 h (Fig. 6E). To remove any weakly attached, dead cells, the cells were rinsed in ice-cold PBS prior to collection for counting and the small number of trypan blue-staining adherent cells were excluded from the data such that only viable cell numbers are reported. As expected, the parental JRS ASNS-null cells showed no growth in the absence of Asn, and the cells expressing WT ASNS showed a significant rescue of growth in DMEM-Asn. Cells stably expressing either G373V or R519H variant showed little to no growth in medium lacking Asn (Fig. 6E).
Figure 6.
Impact of ectopic expression of ASNS variants in ASNS-null JRS cells. Stable cell lines were generated from ASNS-null JRS cells transduced to express WT, G373V, or R519H variant ASNS protein. A, GFP was coexpressed with the ASNS and shows the relative expression of the transduced plasmids. The top panels show the bright field image (BF). B, flow cytometry was used to quantitate the percent of cells in the population that expressed GFP for nontransduced JRS cells (“JRS only”) or cells transduced with vector encoding WT, G373V, or R519H ASNS. C, immunoblotting of total cell extracts from the stable cell lines described in panel (B) was used to detect relative variant protein abundance, and (D) qRT-PCR analysis was used to measure ASNS mRNA levels. E, cell proliferation of the stable cell lines described in panel (B) was analyzed after incubation in DMEM ± Asn for 24, 48, and 72 h. The results are presented as viable cell numbers only measured with an automated cell counter using a trypan blue exclusion method. For panels (D) and (E), the data are shown as the averages ± SDs for assays in triplicate. Each experiment was repeated at least once with different batches of cells to establish reproducibility. For panel (D), an asterisk indicates a p value of ≤ 0.05 compared to JRS only. For panel (E), where not shown, the SD bars are within the symbol and an asterisk indicates a p value of ≤ 0.05 compared to the DMEM + Asn condition. ASNS, asparagine synthetase; JRS, Jensen rat sarcoma.
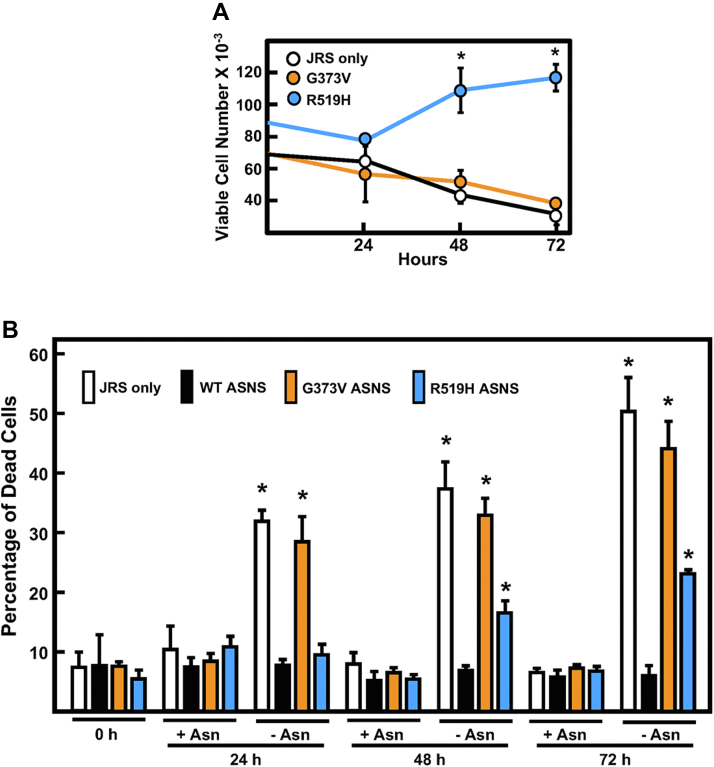
Given the scale of Figure 6E, it is difficult to readily see the differences in viable cell number for those cells incubated in Asn-free DMEM medium. When only the DMEM-Asn data from Figure 6E are graphed to expand the scale (Fig. 7A), it reveals that the viable cell number expressing the R519H variant was maintained or slightly increased over 72 h. In contrast, the number of viable cells expressing G373V decline in a manner similar to the parental JRS cells (Fig. 7A). To assess the degree of cell death for each variant, a time course in DMEM ± Asn, analogous to that shown in Figure 6E, was repeated, but during cell collection, the rinsing step was eliminated and instead the nonadherent cells in the culture medium were combined with the attached cells and the percent trypan blue stained cells was assessed. The results reveal that in the absence of supplied Asn, cell death for the G373V variant was extensive and nearly the same as that for the parental JRS cells (Fig. 7B). An increased amount of cell death was also observed for the R519H but to a lesser degree. Collectively, the results of Figures 6 and 7, obtained in an ASNS-null cell line that provides a uniform genetic background, indicate that the reduction of growth in the child’s fibroblasts is likely resulting from the lack of ASNS activity rather than an independent or additional defect in cell function.
Figure 7.
Effect of Asn deprivation on cell death for each ASNS variant. Panel (A) replots the DMEM-Asn only data shown in Figure 6E to expand the y-axis and illustrate the differences between the control “JRS only” cells, which express no ASNS, and JRS cell lines stably expressing either the G373V or R519H variant. B, after incubation in DMEM ± Asn for the indicated time, the media containing dead/floating cells were saved in tubes and the attached cells were collected by trypsinization and added to the respective media tube. After cell collection by centrifugation, the percent dead cells were determined by trypan blue staining and cell counting. For all panels, the data are shown as the averages ± SDs of assays in triplicate. Where not shown, the SD bars are within the symbol. Each experiment was repeated at least once with different batches of cells to establish reproducibility. An asterisk indicates a p value of ≤ 0.05 compared to the control “JRS only” cells. ASNS, asparagine synthetase; JRS, Jensen rat sarcoma.
ASNS substrate and product levels in variant-expressing JRS cells
The impact of WT, G373V, or R519H ASNS expression on the JRS cellular levels of ASNS substrates and products was determined after incubation of the cells in DMEM ± Asn for 24 h (Fig. 8). The results indicate that cellular Asn content declined after incubation of WT-expressing cells in Asn-free medium, but the decline for cells expressing either G373V or R519H was even greater than WT. The amount of intracellular Asn for the R519H cells was higher than that for JRS control cells lacking ASNS activity, whereas Asn in the G373V cells was below the JRS control. This later result is consistent with the G373V protein instability in the JRS cells (Fig. 6C) and the net cell death of G373V-expressing cells incubated in Asn-free medium (Fig. 7A).
Figure 8.
ASNS substrate and product abundance in JRS cells expressing ASNS variants. Stable cell lines were generated from ASNS-null JRS cells transduced to express WT, G373V, or R519H variant ASNS protein. Each cell line was incubated in DMEM ± Asn for 24 h followed by collection of cell extracts and amino acid analysis by GC-MS. The results for Asn, Asp, Gln, and Glu are presented as “arbitrary units,” calculated as the averages of triplicate determinations ± SDs of the relative amount of each amino acid per 106 cells. To illustrate the relative changes for Asn more clearly, the ratios of the values for DMEM-Asn/DMEM+Asn were calculated for each cell line and are shown as an insert. Where not shown, the SD marks are within the bar and an asterisk indicates that the p value was ≤ 0.05 compared to the value obtained for the cells expressing WT ASNS. ASNS, asparagine synthetase; JRS, Jensen rat sarcoma.
Enzyme activity of purified ASNS protein
To directly assay the enzymatic activity of purified G373V and R519H ASNS proteins in comparison to WT, they were stably expressed in HEK293T cells and then purified on a FLAG affinity gel. This protocol yielded about 10 to 15 μg of protein from a 150 mm culture dish for the WT and R519H clones. However, as observed for the JRS cells (Fig. 6C), despite repeated attempts testing alternate vector constructs, several different complementary DNA (cDNA) delivery systems, and cell selection protocols, the level of G373V protein was never sufficient to assay activity (Fig. 9A). To demonstrate that the lack of G373V protein was not the result of poor infection or transcription, ASNS mRNA content was measured and observed to be similar for WT, G373V, and R519H (Fig. 9B). The WT and R519H variant ASNS proteins were purified and assayed for ASNS enzymatic activity, which revealed that the R519H ASNS variant had a 60% reduction relative to WT (Fig. 9C).
Figure 9.
Enzyme activity of purified ASNS.A, immunoblot analysis and (B) mRNA abundance of FLAG-tagged WT, G373V, and R519H ASNS stably expressed in HEK293T cells. “Control” samples were from nontransfected HEK293T cells. C, ASNS enzymatic activity of the purified ASNS WT or R519H protein was assayed by measuring AMP production. The background value was established by measuring enzyme activity in the absence of added protein. For both panels (B) and (C), the data are shown as the averages ± SDs of assays in triplicate and an asterisk indicates a p value of ≤ 0.05 compared to the control cells (B) or the WT enzymatic activity (C). Each of these experiments was repeated at least once with independently isolated protein preparations to establish reproducibility. ASNS, asparagine synthetase.
Discussion
The present study describes a family that expresses two novel mutations, c.1118G>T (paternal) and c.1556G>A (maternal), within the human ASNS gene that result in G373V and R519H proteins, respectively. ASNSD is a rare autosomal recessive disorder that can arise from homozygosity or, as in this case of the present child, compound heterozygosity. Depending on the mutation(s) present, ASNSD presents with a wide spectrum of severity. The child described here is a 9-year-old female with physical and mental developmental delay, well-controlled seizures, and congenital microcephaly. Many of the most severely affected ASNSD patients succumb to the disease within the first few years of life. The child described here is a 9-year-old who continues to make some developmental progress with regard to fine and gross motor skills, despite still being microcephalic below the second percentile.
To date, more than 50 unique mutations in the ASNS gene have been reported in association with ASNSD diagnoses (15). Most are missense mutations that lead to single amino acid substitutions, but mutations leading to frame shifts have also been reported. The amino acid substitutions are spread randomly across the protein sequence (15). Using the crystal structure of human ASNS (7), it was determined that amino acids G373 and R519 are both located within the ASNS C-terminal domain. Neither is located directly within the active site, but both are predicted to lead to steric interference, alteration of the protein structure, and possibly, protein instability. The immunoblotting data from both the JRS and HEK293T cells suggests that the G373V variant is unstable.
Isolation of skin fibroblasts from the family allowed documentation of ASNS expression, function, and variant impact on cellular growth. Both of these newly identified mutations were located within the coding region of the gene. Although one would not expect such mutations to alter transcription from the ASNS gene, the regulatory sequences that control ASNS transcription are not fully described so basal and stress-induced expression of ASNS was analyzed. Measurement of mRNA and protein abundance, as well as the induction in response to the ISR (22, 23, 24, 25), documented that well-known amino acid–regulated transcription and translation of the ASNS gene were intact (reviewed in (2, 6, 19)). Growth analysis of primary fibroblasts isolated from each family member showed that proliferation of cells from either heterozygotic parent was not suppressed whether medium Asn was present or not. In contrast, fibroblasts from the compound heterozygotic child exhibited about a 50% reduction in proliferation when the cells were maintained in medium lacking Asn.
To our knowledge, no published study has directly measured the Asn content of an ASNSD patient’s cells or tissue. The present results document that when challenged by incubation in Asn-free medium, the decline in intracellular Asn levels in fibroblasts from the child and the parents was similar to that which occurs in WT cells. Conversely, measurement of the extracellular Asn content revealed that the decline in the parents was significantly greater than that for WT cells, and the amount of extracellular Asn for the child’s cells was at or below the limit of detection by GC-MS. The observation that extracellular Asn rather than intracellular Asn declines was unexpected. In addition to the data shown here, two independent cell cultures for the present family were analyzed by LC-MS using a metabolomics core facility, and those studies showed that the intracellular Asn levels were unchanged after incubation of the family’s cells in medium lacking Asn, while the extracellular Asn levels reflected the growth pattern (data not shown). To test whether or not this response was unique to the fibroblasts from this family, the LC-MS analysis was repeated using fibroblast lines from an independent family in which different ASNS variants are expressed, maternal (T337I), paternal (G289A), and a compound heterozygotic child (T337I,G289A) (16). The results from the second family were qualitatively similar to those obtained for the present family (data not shown). Possible explanations for the correlation between cell proliferation and extracellular amino acid levels include modulation of plasma membrane transporter proteins or flux of Asn through bidirectional exchange transporters, either of which could modify the signaling mechanisms associated with the AAR, UPR, or mTOR pathways, all of which sense amino acids. Further investigation of this phenomenon will require a detailed analysis of intracellular metabolic flux and amino acid transporter functions.
The ASNS gene in JRS cells is hypermethylated and no protein is expressed (26), thus providing an ASNS-null cell in which to investigate the consequences of each mutation in the absence of other alleles or compounding cellular differences. Ectopic expression of the WT and R519H variant in the JRS cells documented that WT ASNS rescued their ability to grow in the absence of medium-supplied Asn, but proliferation of the R519H-JRS cells was minimal in the absence of medium Asn. Transduction of the JRS cells with a plasmid encoding the G373V variant resulted in a markedly low level of ASNS protein, despite an mRNA expression level equal to the WT transduced cells. This inability to detect the G373V variant after overexpression is taken as evidence of protein instability. For both R519H and G373V, the degree of cell death in Asn-free medium was significantly greater than for the WT-expressing cells. Thus, the dependence on medium-supplied Asn for the viability and proliferation of JRS cells expressing either R519H or G373V variant alone argues that the Asn dependence of the child’s fibroblasts is the result of insufficient ASNS activity, not the result of some independent or secondary genetic defect associated with the fibroblasts themselves.
Zhu et al. (7) measured ASNS enzymatic activity for two previously reported ASNS variants and discovered that T337I (numbered T336I in the original paper) exhibited little or no activity. This result is consistent with our previous report of significantly reduced growth of fibroblasts from a heterozygotic parent expressing T337I when incubated in Asn-free medium (16). Matsumoto et al. (27) investigated the effect of ASNS mutations associated with four Japanese ASNSD patients initially described by Yamamoto et al. (28). For three of the variants, there was a good correlation between reduced enzymatic activity and an inability to support the growth of ASNS KO cells in Asn-free medium (27). In the present report, a novel method was used to measure ASNS enzymatic activity for specific variant proteins by purification of FLAG-tagged ASNS protein from HEK293T stable cell lines. The enzymatic activity of the purified ASNS protein was analyzed by measuring the Gln-dependent production of AMP. This protocol requires only 100 ng of ASNS protein, a 30 min reaction time, and is highly reproducible. As observed for the JRS cells, despite employing multiple approaches to achieve expression, it was not possible to obtain sufficient G373V protein for enzymatic analysis, even after inhibition of the proteasomal degradation system. Ruzzo et al. (10) were also unsuccessful in blocking ASNS variant turnover by inhibition of the proteasomal degradation pathway. In contrast, the FLAG-tagged WT or R519H constructs led to abundant expression, and the enzymatic analysis documented that the activity of the R519H variant was reduced by about 60%. These data are consistent with the structural models suggesting that the R519H substitution leads to loss of hydrogen bonding that occurs within the native protein and steric hindrance caused by histidine at the 519 position. Thus, for the child described in this report, it appears that little or no G373V protein may exist due to instability and that the R519H variant has a 60% reduction in enzyme activity. Together, these observations are consistent with the approximately 50% reduction of cell proliferation observed for the child’s fibroblasts.
In summary, the present data document two novel mutations in the ASNS gene that lead to coexpression of variants G373V and R519H in the affected child. Several lines of evidence support the interpretation that the G373V and R519H ASNS variants cause the clinical outcome observed, including (1) the apparent instability of the G373V variant, (2) the reduced enzymatic activity of the R519H variant, (3) the inability of either variant to rescue growth of ASNS-null JRS cells, and (4) the lack of cell proliferation by the child’s fibroblasts in the absence of extracellular Asn. These results provide further insight into our understanding of ASNSD and describe new approaches to study the disease in the form of ASNS ectopic expression in JRS ASNS-null cells, measurement of cellular Asn production by GC-MS, and direct enzymatic analysis after ASNS protein variant purification.
Experimental procedures
Exome sequencing, genetic, and metabolite analysis
Whole exome sequencing was performed on the child’s and parental genomic DNA using the NimbleGen V3 targeted sequence capture method to enrich the whole exome (Roche). Sequencing was done using the Illumina HiSeq2500 system with 125 bp paired-ended reads at a minimum coverage of 10× of 95% of the target regions. The exome DNA sequences were mapped and compared to the human genome build UCSC hg19 reference sequence using the GATK Appistry 2013.2 and Alamut Batch 1.4.0 (SOPHiA Genetics) software packages (29). Assessment of quality and coverage for targeted coding exons of known protein coding genes was >97% at 10×. Genes of interest were generated based on the following key phenotypes: microcephaly, developmental delay, intractable seizures, hypotonia, intellectual disability, and triangular facies. Exome analysis filtered thousands of genetic variants in the child and used parental data to help in the filtering process. A subset of these variants was identified as clinically relevant. Sanger sequencing was performed to confirm all potential positive variants. The child’s serum amino acid levels were quantified at age 4 as part of the patient’s clinical care at Cincinnati Children’s Hospital Medical Center by the clinical laboratory.
Protein modeling
The available human ASNS protein model (Uniprot ID: P08243) was used as a template in UCSF Chimera software (version 1.15) (30). The recently crystallized human ASNS (PDB: 6GQ3) (7) was overlayed with E. coli AS-B structure (PDB: 1CT9) to confirm the similarity in structure and allow for human ASNS modeling in the C-terminal active site based on the location of AMP in the E. coli AS-B crystal (8). The human crystal structure sequence does not include the initial methionine residue, thus, G373 and R519 are residue locations G372 and R518, respectively. G373V and R519H variants were introduced with the rotamers tool, and the most probable rotamer was selected based on backbone-dependent rotamer library predictions (31). Predictions of hydrogen bonding were performed with the FindHBond tool set to default H bond constraints provided by the Chimera software. Predictions of clashing residues were obtained through the Find Clashes/Contacts tool set to parameters for default clash criteria. Predicted hydrogen bonds and clashes were confirmed by manual structure inspection and used to infer potential variant impacts on enzyme stability and activity.
Cell culture conditions
Fibroblast cell lines were developed from skin samples collected from the affected child and the parents. An unrelated human adult dermal fibroblast cell line was obtained from ATCC (#PCS-201-012) and serves as a WT cell line. HEK293T (#CRL-3216) and JRS (#CCL-45) cells were obtained from ATCC. All cell lines were cultured in high glucose Dulbecco’s modified Eagle’s medium (DMEM) (Corning #10-013-CV) supplemented with 10% fetal bovine serum (FBS, Bio-Techne #S11550), ABAM (streptomycin, penicillin G, and amphotericin B), 1× nonessential amino acids (Corning #25-025-CL), and 2 mM Gln. To generate DMEM without Asn, 10% dialyzed FBS was used (Bio-Techne #S12850) and the 1× nonessential amino acids supplement was replaced with 0.1 mM glycine, L-alanine, L-Asp, L-Glu, L-proline, and L-serine individually added to the DMEM. Cell seeding densities are listed for each experimental protocol. Cell nutritional basal state was obtained 12 to 18 h before all experiments by changing to fresh complete DMEM media. For all time course cell treatments, the appropriate media were changed every 24 h.
RNA purification and qRT-PCR
For mRNA studies, fibroblasts were seeded in 6-well plates at a density of 1.5 × 105 cells per well. RNA was isolated using TRIzol Reagent (Invitrogen #15596026) per the manufacturer’s protocol. A 1 μg RNA aliquot was converted to cDNA with the qScript cDNA synthesis kit (Quantabio #95047) and then 10% of the cDNA was utilized for each qRT-PCR reaction. qRT-PCR reactions were performed with cDNA, 300 nM forward primer, 300 nM reverse primer, and SYBR Green PCR Master Mix (Applied Biosystems #4309155) in a Bio-Rad CFX-Connect Real-Time System with cycling parameters of 1× 95 °C for 10 min, 40× 95 °C for 15 s and 60 °C for 1 min with a SYBR measurement, 1× 95 °C for 15 s, and a melting curve from 55 °C to 98 °C increasing 0.5 °C every 5 s. Primers used for GAPDH mRNA were forward 5′ -TTGGTATCGTGGAAGGACTC- 3′ and reverse 5′ -ACAGTCTTCTGGGTGGCAGT- 3′. Primers used for ASNS mRNA were forward 5′ -GCAGCTGAAAGAAGCCCAAGT- 3′ and reverse 5′ -TGTCTTCCATGCCAATTGCA- 3′. Primers for ATF4 were forward 5′-GGGACAGATTGGATGTTGGAGA-3′ and reverse 5′-ACCCAACAGGGCATCCAAGT-3’. Primers for SLC1A5 (ASCT2) were forward 5′-TCCTCTTCACCCGCAAAAACCC-3′ and reverse 5′-CCACGCCATTATTCTCCTCCAC-3’. Analysis was performed by the ΔΔCt method (32).
Immunostaining and immunoblotting
Fibroblasts were seeded at 5 × 104 cells per chamber in 8-well Millicell EZ-slides (Millipore Sigma #PEZGS0816). After concluding the experimental protocol, cells were washed with PBS, fixed with 4% paraformaldehyde for 15 min at room temperature (RT), washed with PBS, permeabilized 10 min with 0.1% Triton X-100 in PBS containing 1% bovine serum albumin (Fisher #BP-9703-100), washed with PBS, and then incubated for 1.5 h with 1:100 anti-ASNS rabbit polyclonal antibody (ProteinTech #14681-1-AP). The primary antibody was removed, and the chambers were incubated with 1:500 anti-rabbit Alexa Fluor 488 (Abcam #ab150077) for 1.5 h in the dark. The secondary antibody was removed and slides were mounted with DAPI Fluoromount-G (SouthernBiotech #0100-20). Images were taken on an Olympus IX 73 inverted microscope with an X-Cite LED fluorescence light source. DAPI manual imaging exposure was 488 μs and ASNS was 23 ms with a 4× gain set for both.
For immunoblotting, after treatment for the indicated time in DMEM with or without Asn, cells were scraped for collection in 1 ml medium, collected by centrifugation, washed with PBS, and resuspended in radioimmunoprecipitation assay buffer of the following composition: 150 mM NaCl, 5 mM EDTA pH 8.0, 50 mM Tris pH 8.0, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 1× protease inhibitors (Roche #04693159001). Resuspended pellets were left at 4 °C for 15 min and then centrifuged at 13,000g for 15 min to remove cell debris. Protein quantification was done by Bradford assay (Bio-Rad #5000006), and 20 μg of protein was subjected to gel electrophoresis and immunoblotting. Primary ASNS mouse mAb (33) was diluted 1:500 and the mouse GAPDH antibody (Santa Cruz #sc32233) was diluted to 1:8000 in Tris-buffered saline with Tween-20 containing 5% Carnation nonfat dry milk. The secondary antimouse horseradish peroxidase antibody (Bio-Rad #170-6516) was diluted 1:5000 and added for 1 h at RT. FLAG primary antibody (Cell Signaling #147935) was used at 1:1000 dilution and detected with anti-rabbit horseradish peroxidase –conjugated antibody (Bio-Rad #170-6515) at 1:10,000 dilution. Films were scanned into digital format and analyzed using ImageJ software (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, https://imagej.nih.gov/ij/, 1997-2018).
Analysis of ASNS substrates and products in cultured cells
Fibroblasts or JRS cells were plated at 1 × 106 cells per plate in 10 cm dishes. Cells were allowed to reattach and acclimate for 24 h and then incubation in DMEM ± Asn was initiated. After 24 h, the treatment media was aspirated into 15 ml tubes and frozen at −80 °C. The attached cells were washed with ice-cold PBS, incubated for 5 min in 0.25% trypsin to detach from the plate, and collected into 15 ml tubes. A 50 μL aliquot of the cell suspension was taken for cell counting, and the remaining cell suspension was pelleted at 300g for 5 min at 4 °C. The cell pellets were washed twice in ice-cold PBS, and final pellets were flash frozen in liquid nitrogen and stored frozen at −80 °C until processing. Cell pellets and media samples were processed and analyzed by GC-MS exactly as in published methods (34). In brief, cell pellets were spiked with DL-norleucine internal standard, extracted with acetonitrile:isopropanol:water (3:3:2 by volume), dried with a speedvac, reconstituted in acetonitrile:water (1:1 by volume), and dried in GC-MS reaction v-vials. Without any extraction, 20 μl of media were dried down in GC-MS reaction v-vials. Dried cell extracts and media samples were incubated with 50 μl of 2% MOX reagent for 1.5 h at 30 °C, followed by the addition of 50 μl of MTBSTFA + TBDMS and a 1 h incubation at 60 °C. V-vials were centrifuged and derivatized samples were loaded into GC sample vial inserts. GC-MS data acquisition was accomplished with a Thermo Scientific Single Quadrupole Mass Spectrometer (ISQ) and Gas Chromatograph (Trace 1310). The GC was outfitted with a Restek 30 m dimethyl/diphenyl polysiloxane RTX-5MS column (0.25 mm ID and 0.25 μm film) and a 10 m guard column. Oven temperature was increased from 60 °C (held for 60 s) to 325 °C at a rate of 10 °C/min with a final 5 min hold time. The ion source was set to a temperature of 230 °C and helium (Ultra High Purity) was utilized as a carrier gas. Amino acid peak areas were processed with XCalibur Quan Browser software (ThermoFisher Scientific) and normalized to the peak area of DL-norleucine internal standard.
Retroviral infection of ASNS-null JRS cells
The WT C-terminally FLAG-tagged ASNS sequence and stop codon from the Sino Biological (#HG-16454-CF) plasmid was subcloned into the MSCV-PIG retroviral expression plasmid between the BglII and XhoI restriction sites by Genscript. The ASNS MSCV-PIG plasmid was then mutated by Genscript to generate the independent 1118G>T and 1556G>A ASNS mutant-encoding plasmids. The MSCV-PIG plasmid itself encodes for puromycin selection and expresses IRES-driven GFP. For retrovirus packaging, a 10 μg aliquot of the MSCV-PIG plasmid, containing either WT-ASNS ASNS or an ASNS mutant, and a 10 μg aliquot of pCL-ECO plasmid (IMGENEX) were cotransfected into a 100 mm plate of HEK293T cells at a 1 μg:8 μg ratio of DNA to polyethyleneimine. At 8 h after transfection, the medium was changed to fresh DMEM with 4% FBS, and the transfection efficiency was checked by GFP fluorescence 36 h post-transfection. The medium containing the virus was collected 36 h post-transduction, centrifuged at 800g for 3 min to pellet debris, filtered through a 0.45 μm syringe filter, mixed with polybrene at a ratio of 1 ml medium to 4 μl polybrene, and then added to JRS cells. The transduction was repeated every 24 h for a total of three times. Cells were maintained in fresh DMEM medium containing 1 μg/ml puromycin for 3 days of selection. Stable expression was confirmed by GFP fluorescence and ASNS mRNA and protein abundance.
Flow cytometry to assess trans-gene expression in JRS cells
The transduced JRS cells stably expressing WT ASNS or the individual G373V or R519H variants were collected during growth phase culture for analysis by flow cytometry to quantify percentage of cells expressing GFP. Nontransduced JRS cells were used to set the gate based on exclusion of autofluorescence inherent to the JRS cell line. The cells were resuspended in PBS and processed on a BD LSRFortessa instrument to measure at least 10,000 events.
Cell growth analysis
Fibroblasts were plated at 5 × 104 cells per well in 6-well plates. Following incubation in DMEM ± Asn for the indicated time, cells were rinsed with ice-cold PBS to remove dead cells and debris, and then, the remaining cells were detached with 0.25% trypsin and collected for counting. For incubations of 48 or 72 h, fresh medium was provided every 24 h. Viable cell counts were determined using the Applied Biosystems Countess II FL automated cell counter per the manufacturer’s trypan blue staining protocol. The percentage of viable cells was routinely greater than 99%. To measure the cell death for JRS cells expressing ASNS individual variants by trypan blue staining, the normal cell counting protocol was adapted to retain the dead cells. The rinsing step with ice-cold PBS was eliminated and instead the culture medium, containing any detached dead cells, was aspirated into a 15 ml tube. The attached cells were independently collected by trypsin and then added to the 15 ml tube containing the previously collected medium. After centrifugation at 300g for 5 min, the cells were resuspended in 200 μL PBS, and a Countess II FL automated cell counter was used to measure the total and trypan blue stained cells by the manufacturer’s recommended protocol.
Generation of stable HEK293T cell lines expressing ASNS variants
A C-terminal FLAG-tagged WT ASNS expression construct was obtained from Sino Biological (#HG16454-CF). Constructs for the 1118 C>T (G373V) and 1156G>A (R519H) mutations were generated by Genscript and confirmed by sequencing. Expression plasmids were transfected into HEK293T cells with X-tremeGENE 9 transfection reagent (Roche #06 365 787 001) and stable lines were selected through growth in DMEM containing 100 μg/ml Hygromycin B (EMD Millipore #400052-5ML) for 14 days. The relative expression level of each ASNS variant was confirmed by immunoblotting with anti-FLAG antibody (Cell Signaling #147935).
FLAG-tagged ASNS protein affinity-purification
HEK293T stable lines independently expressing WT ASNS or the variant ASNS proteins were maintained in culture with 50 μg/ml of hygromycin B until 80% confluent. FLAG-tagged ASNS proteins were purified with EZview Red anti-FLAG M2 affinity gel (Millipore Sigma #F2426) per the manufacturer’s protocol. The recommended lysis buffer was modified to contain only 0.1% Triton X-100 to minimize any effect on enzyme activity, and the binding step was extended to overnight to increase yield. Purified proteins were confirmed with SDS-PAGE and Coomassie brilliant blue R-250 staining (Bio-Rad 1#61-0400). Quantification of purified protein was determined by the Bradford assay (Bio-Rad #5000006) and then aliquots were stored at −80 °C until use. Aliquots were only thawed once.
ASNS enzyme assay
The Promega AMP-Glo Assay (#V5011) was adapted to detect the AMP produced by purified WT or variant ASNS proteins. A 100 ng aliquot of protein was determined by Bradford dye assay and combined in a reaction with final concentrations of 50 mM Tris, 10 mM L-Gln, 10 mM L-Asp, 1 mM ATP, 0.75 mM DTT, 0.05 mg/ml bovine serum albumin, and 10 mM MgCl2. After the incubation for 30 min at RT, it was diluted 1:10 with 50 mM Tris to lower the residual ATP concentration (needed to prevent excess ATP interference with AMP determination), and 25 μl of this dilution was transferred to a white 96-well plate for use with the AMP-Glo assay kit per the manufacturer’s protocol for a 25 μl starting volume. The AMP, detected by luminescence for each reaction, is proportional to the ASNS activity. Each assay was performed in triplicate to establish the variability, and each experiment was repeated at least once with a new protein preparation to establish reproducibility. Significant differences between WT and variant ASNS proteins were determined with the Student’s t test.
Statistical analysis
Data containing SD bars were obtained as assays in triplicate for a given experiment to determine the variability, and each experiment was repeated at least once with an independent batch of cells to assess reproducibility. The data are shown as the averages ± SDs and were analyzed by Student’s two-tailed t test. Where indicated with an asterisk, the differences have a p value of ≤ 0.05 relative to the indicated control. For immunoblots and GFP staining images, the data represent individual experiments, which were repeated at least once to ensure reproducibility.
Data availability
Data are available upon request by contacting the corresponding author.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank Dr Harry Nick, University of Florida, for the use of the inverted fluorescent microscope. The authors gratefully acknowledge this family, and other ASNSD families, for their commitment toward research and increased awareness of this disease.
Author contributions
M. S. K., M. E. M., R. M., and S. J. S. methodology; M. S. K., M. E. M., S. J. S., M. C. C., F. Y., Z. Q., L. B. B., and R. M. formal analysis; S. J. S., M. C. C., F. Y., K. C. R., and D. N. F. investigation; Z. Q., L. B. B, M. E. M., and R. M. resources; M. S. K., M. C. C., and S. J. S. writing–original draft; M. S. K., M. E. M., S. J. S., M. C. C., F. Y., Z. Q., L. B. B., R. M., K. C. R., and D. N. F. writing–review & editing.
Funding and additional information
Funding for M. S. K. was from the National Institutes of Health, Institute of Child Health and Human Development (HD110576). Funding for M. E. M. was from the National Institutes of Health, Institute of Diabetes and Digestive and Kidney Diseases (DK132254, DK105346). Funding for L. B. B. was from the National Science Foundation, Molecular and Cellular Biosciences (1817869). Funding for Z. Q. was from National Institutes of Health, National Heart, Lung, and Blood Institute (HL131444).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Ronald Wek
References
- 1.Richards N.G., Kilberg M.S. Asparagine synthetase chemotherapy. Annu. Rev. Biochem. 2006;75:629–654. doi: 10.1146/annurev.biochem.75.103004.142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balasubramanian M.N., Butterworth E.A., Kilberg M.S. Asparagine synthetase: regulation by cell stress and involvement in tumor biology. Am. J. Physiol. Endocrinol. Metab. 2013;304:E789–E799. doi: 10.1152/ajpendo.00015.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greco A., Gong S.S., Ittmann M., Basilico C. Organization and expression of the cell cycle gene, ts 11 , that encodes asparagine synthetase. Mol. Cell Biol. 1989;9:2350–2359. doi: 10.1128/mcb.9.6.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y.P., Lambert M.A., Cairney A.E.L., Wills D., Ray P.N., Andrulis I.L. Molecular structure of the human asparagine synthetase gene. Gene. 1989;4:259–265. doi: 10.1016/0888-7543(89)90329-7. [DOI] [PubMed] [Google Scholar]
- 5.Heng H.H.Q., Shi X.M., Scherer S.W., Andrulis I.L., Sui L.C.T. Refined localization of the asparagine synthetase gene ASNA) to chromosome 7, region q 21.3,and characterization of teh somatic cell hybrid line 4AF/106/KO15. Cytogenet. Cell Genet. 1994;66:135–138. doi: 10.1159/000133685. [DOI] [PubMed] [Google Scholar]
- 6.Lomelino C.L., Andring J.T., McKenna R., Kilberg M.S. Asparagine synthetase: function, structure, and role in disease. J. Biol. Chem. 2017;292:19952–19958. doi: 10.1074/jbc.R117.819060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu W., Radadiya A., Bisson C., Wenzel S., Nordin B.E., Martinez-Marquez F., et al. High-resolution crystal structure of human asparagine synthetase enables analysis of inhibitor binding and selectivity. Commun. Biol. 2019;2:345. doi: 10.1038/s42003-019-0587-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larsen T.M., Boehlein S.K., Schuster S.M. Three-dimensional structure of Escherichia coli asparagine synthetase B: a short journey from substrate to product. Biochemistry (Mosc) 1999;38:16146–16157. doi: 10.1021/bi9915768. [DOI] [PubMed] [Google Scholar]
- 9.Tesson A.R., Soper T.S., Ciustea M., Richards N.G. Revisiting the steady state kinetic mechanism of glutamine-dependent asparagine synthetase from Escherichia coli. Arch. Biochem. Biophys. 2003;413:23–31. doi: 10.1016/s0003-9861(03)00118-8. [DOI] [PubMed] [Google Scholar]
- 10.Ruzzo E.K., Capo-Chichi J.M., Ben-Zeev B., Chitayat D., Mao H., Pappas A.L., et al. Deficiency of asparagine synthetase causes congenital microcephaly and a progressive form of encephalopathy. Neuron. 2013;80:429–441. doi: 10.1016/j.neuron.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sprute R., Ardicli D., Oguz K.K., Malenica-Mandel A., Daimaguler H.S., Koy A., et al. Clinical outcomes of two patients with a novel pathogenic variant in ASNS: response to asparagine supplementation and review of the literature. Hum. Genome. 2019;6:24. doi: 10.1038/s41439-019-0055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radha Rama Devi A., Naushad S.M. Molecular diagnosis of asparagine synthetase (ASNS) deficiency in two Indian families and literature review of 29 ASNS deficient cases. Gene. 2019;704:97–102. doi: 10.1016/j.gene.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 13.Abdel-Salam G.M.H., Abdel-Hamid M.S. Asparagine synthetase deficiency with intracranial hemorrhage can mimic molybdenum cofactor deficiency. Neuropediatrics. 2021;52:201–207. doi: 10.1055/s-0040-1718917. [DOI] [PubMed] [Google Scholar]
- 14.Alharby E., Faqeih E.A., Saleh M., Alameer S., Almuntashri M., Pastore A., et al. Clinical, molecular, and biochemical delineation of asparagine synthetase deficiency in Saudi cohort. Genet. Med. 2020;22:2071–2080. doi: 10.1038/s41436-020-0919-x. [DOI] [PubMed] [Google Scholar]
- 15.Liu L., Wang J., Li H., Dong Y., Li Y., Xia L., et al. An intractable epilepsy phenotype of ASNS novel mutation in two patients with asparagine synthetase deficiency. Clin. Chim. Acta. 2022;531:331–336. doi: 10.1016/j.cca.2022.04.989. [DOI] [PubMed] [Google Scholar]
- 16.Palmer E.E., Hayner J., Sachdev R., Cardamone M., Kandula T., Morris P., et al. Asparagine Synthetase Deficiency causes reduced proliferation of cells under conditions of limited asparagine. Mol. Genet. Metab. 2015;116:178–186. doi: 10.1016/j.ymgme.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sacharow S.J., Dudenhausen E.E., Lomelino C.L., Rodan L., El Achkar C.M., Olson H.E., et al. Characterization of a novel variant in siblings with asparagine synthetase deficiency. Mol. Genet. Metab. 2018;123:317–325. doi: 10.1016/j.ymgme.2017.12.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kilberg M.S., Balasubramanian M., Fu L., Shan J. The transcription factor network associated with the amino acid response in Mammalian cells. Adv. Nutr. 2012;3:295–306. doi: 10.3945/an.112.001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wortel I.M.N., van der Meer L.T., Kilberg M.S., van Leeuwen F.N. Surviving stress: modulation of ATF4-mediated stress responses in normal and malignant cells. Trends Endocrinol. Metab. 2017;28:794–806. doi: 10.1016/j.tem.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siu F., Bain P.J., LeBlanc-Chaffin R., Chen H., Kilberg M.S. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 2002;277:24120–24127. doi: 10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- 21.Dey S., Savant S., Teske B.F., Hatzoglou M., Calkhoven C.F., Wek R.C. Transcriptional repression of ATF4 by C/EBPbeta differentially regulates the integrated stress response. J. Biol. Chem. 2012;287:21936–21949. doi: 10.1074/jbc.M112.351783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fusakio M.E., Willy J.A., Wang Y., Mirek E.T., Al Baghdadi R.J., Adams C.M., et al. Transcription factor ATF4 directs basal and stress-induced gene expression in the unfolded protein response and cholesterol metabolism in the liver. Mol. Biol. Cell. 2016;27:1536–1551. doi: 10.1091/mbc.E16-01-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikonorova I.A., Mirek E.T., Signore C.C., Goudie M.P., Wek R.C., Anthony T.G. Time-resolved analysis of amino acid stress identifies eIF2 phosphorylation as necessary to inhibit mTORC1 activity in liver. J. Biol. Chem. 2018;293:5005–5015. doi: 10.1074/jbc.RA117.001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wek R.C. Role of eIF2alpha kinases in translational control and adaptation to cellular stress. Cold Spring Harb. Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misra J., Holmes M.J., E T.M., Langevin M., Kim H.G., Carlson K.R., et al. Discordant regulation of eIF2 kinase GCN2 and mTORC1 during nutrient stress. Nucl. Acids Res. 2021;49:5726–5742. doi: 10.1093/nar/gkab362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugiyama R.H., Arfin S.M., Harris M. Properties of asparagine synthetase in asparagine-independent variants of Jensen rat sarcoma cells induced by 5-azacytidine. Mol. Cell. Biol. 1983;3:1937–1942. doi: 10.1128/mcb.3.11.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto H., Kawashima N., Yamamoto T., Nakama M., Otsuka H., Ago Y., et al. In vitro functional analysis of four variants of human asparagine synthetase. J. Inherit. Metab. Dis. 2021;44:1226–1234. doi: 10.1002/jimd.12408. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto T., Endo W., Ohnishi H., Kubota K., Kawamoto N., Inui T., et al. The first report of Japanese patients with asparagine synthetase deficiency. Brain Dev. 2017;39:236–242. doi: 10.1016/j.braindev.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 29.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 31.Shapovalov M.V., Dunbrack R.L., Jr. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure. 2011;19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Sheng S., Moraga D.A., Van Heeke G., Schuster S.M. High-level expression of human asparagine synthetase and production of monoclonal antibodies for enzyme purification. Prot. Exp. Purif. 1992;3:337–346. doi: 10.1016/1046-5928(92)90010-t. [DOI] [PubMed] [Google Scholar]
- 34.Mahar R., Chang M.C., Merritt M.E. Measuring NQO1 bioactivation using [(2)H7]Glucose. Cancers (Basel) 2021;13:4165. doi: 10.3390/cancers13164165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available upon request by contacting the corresponding author.