Abstract
The N-glycans attached to proteins contain various GlcNAc branches, the aberrant formation of which correlates with various diseases. N-Acetylglucosaminyltransferase-IVa (GnT-IVa or MGAT4A) and Gnt-IVb (or MGAT4B) are isoenzymes that catalyze the formation of the β1,4-GlcNAc branch in N-glycans. However, the functional differences between these isozymes remain unresolved. Here, using cellular and UDP-Glo enzyme assays, we discovered that GnT-IVa and GnT-IVb have distinct glycoprotein preferences both in cells and in vitro. Notably, we show that GnT-IVb acted efficiently on glycoproteins bearing an N-glycan premodified by GnT-IV. To further understand the mechanism of this reaction, we focused on the noncatalytic C-terminal lectin domain, which selectively recognizes the product glycans. Replacement of a nonconserved amino acid in the GnT-IVb lectin domain with the corresponding residue in GnT-IVa altered the glycoprotein preference of GnT-IVb to resemble that of GnT-IVa. Our findings demonstrate that the C-terminal lectin domain regulates differential substrate selectivity of GnT-IVa and GnT-IVb, highlighting a new mechanism by which N-glycan branches are formed on glycoproteins.
Keywords: glycobiology, glycoprotein biosynthesis, glycosylation, glycosyltransferase, GnT-IV, substrate specificity
Abbreviations: α1-antitrypsin, alpha-1-antitrypsin; α1AGP, alpha-1-acid glycoprotein; BACE1, β-site amyloid precursor protein cleaving enzyme-1; BSA, bovine serum albumin; CBB, Coomassie brilliant blue; cDNA, complementary DNA; CHX, cycloheximide; DKO, double KO; DSA, Datura stramonium agglutinin; ER, endoplasmic reticulum; FACS, fluorescence-activated cell sorting; GnT, N-acetylglucosaminyltransferase; HEK293, human embryonic kidney 293 cell line; HRP, horseradish peroxidase; IgG, immunoglobulin G; MS, mass spectrometry; PNGaseF, peptide N-glycanase F; RCA, Ricinus communis agglutinin; SSA, Sambucus sieboldiana agglutinin; TBS, Tris-buffered saline; TBS-T, TBS with Tween-20
Glycosylation, one of the most common post-translational modifications in mammals, regulates various protein functions (1). Glycans on proteins are biosynthesized in the endoplasmic reticulum (ER) and the Golgi apparatus by stepwise and competitive actions of various glycosyltransferases (2). Accordingly, a considerable variety of glycan structures are synthesized, which affects protein folding, activity, trafficking, and stability, thereby regulating many physiological phenomena (3). Furthermore, abnormal glycosylation is often associated with the development and exacerbation of various diseases, including cancer (4), chronic obstructive pulmonary disease (5), diabetes (6), and Alzheimer’s disease (7). Therefore, understanding how protein glycosylation is regulated and dysregulated in cells is important for elucidating pathogenesis and developing new therapies.
Among several common classes of animal glycans, we have focused on N-glycans, which are highly abundant and widespread in many species (8). N-Glycan biosynthesis starts with attaching a common triglucosylated glycan (Glc3Man9GlcNAc2) to the asparagine in the consensus sequence N-X-S/T in the ER (8, 9). After glycosidase-mediated processing in the ER, many Golgi-resident glycosyltransferases further modify N-glycan structures in species-, cell type-, protein-, and even site-specific manners (8). A unique structural feature of N-glycan is the variable number of GlcNAc branches that are biosynthesized by the specific N-acetylglucosaminyltransferases-I–V (GnT-I–V, also designated as MGAT1–5) in mammals (10, 11). Because the expression levels of each N-glycan branch on specific glycoproteins are highly related to various diseases, such as cancer, diabetes, and Alzheimer’s disease (12), GnTs are potential drug targets against these diseases. However, the mechanisms defining how these branching enzymes act on their specific target proteins in cells are largely unresolved. Moreover, the presence of isozymes (GnT-IVa and GnT-IVb), which apparently have similar activity (13) (Fig. 1A), adds further challenges for understanding how each N-glycan branch is biosynthesized in cells. Therefore, clarifying protein selectivity of each branching enzyme and the functional differences of the isozymes for branching is pivotal in elucidating the complex biosynthetic mechanisms of N-glycans.
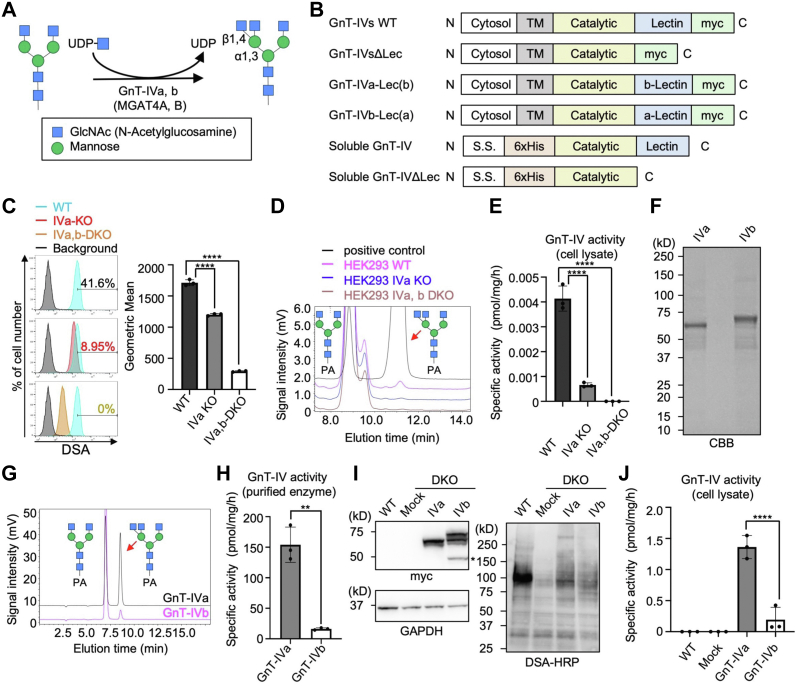
Figure 1.
In vitro and intracellular activity of GnT-IVa and GnT-IVb.A, schematic model of the GlcNAc transfer reaction catalyzed by GnT-IVa and GnT-IVb (MGAT4A and MGAT4B). B, plasmid constructs used in this study. C, FACS analysis of HEK293 WT (blue), HEK293 GnT-IVa KO (red), and HEK293 DKO cells (orange) with DSA lectin (left). The right graph shows the geometric means (n = 3, means ± SD, ∗∗∗∗p < 0.0001, Tukey's multiple comparisons test). D, the endogenous GnT-IV activity in HEK293 WT, GnT-IVa KO, and DKO cells was measured by incubating the lysates with the PA-labeled acceptor sugar (GnGnbi-PA) and analyzing by HPLC. E, the specific activity of GnT-IV in the HEK293 WT, GnT-IVa KO, and DKO cell lysates calculated by the peak area in (D) (n = 3, means ± SD, ∗∗∗∗p < 0.0001, Tukey's multiple comparisons test). F, soluble GnT-IVa and GnT-IVb were expressed in COS7 cells and purified from the media using a Ni2+ column. Purified GnT-IVa and GnT-IVb were separated by SDS-PAGE and visualized by CBB staining. G, enzyme activity of the purified GnT-IVa and GnT-IVb was measured by incubating the enzymes with GnGnbi-PA and analyzing by HPLC. H, the specific activity of the purified GnT-IVa and GnT-IVb was calculated by the peak area in (G) (n = 3, means ± SD, ∗∗p < 0.01, unpaired t test). I, proteins from mock-treated HEK293 WT cells and DKO cells transfected with an empty vector (mock) or a plasmid for expression of GnT-IVa or GnT-IVb were subjected to SDS-PAGE and blotted with anti-myc antibody (upper left), anti-GAPDH antibody (lower left), or HRP-conjugated DSA (right). J, lysates of mock-treated HEK293 WT cells and DKO cells transfected with an empty vector (mock) or a plasmid for expression of GnT-IVa or GnT-IVb were reacted with GnGnbi-PA and analyzed by HPLC (n = 3, means ± SD, ∗∗∗∗p < 0.0001, Tukey's multiple comparisons test). CBB, Coomassie brilliant blue; DKO, double KO; DSA, Datura stramonium agglutinin; FACS, fluorescence-activated cell sorting; GnT, N-acetylglucosaminyltransferase; HEK293, human embryonic kidney 293 cell line; HRP, horseradish peroxidase.
Unlike other GnTs, only GnT-IV forms a large family. GnT-IV is composed of four homologous family members: GnT-IVa (MGAT4A), GnT-IVb (MGAT4B), GnT-IVc (MGAT4C, also known as GnT-VI), and GnT-IVd (MGAT4D, also known as GnT-1IP), and GnT-IVs constitute the CAZy GT54 family (14, 15, 16, 17). GnT-IVa and GnT-IVb isoenzymes have GnT-IV activity to transfer GlcNAc from UDP-GlcNAc to the α1,3-Man arm of N-glycan via a β1,4-linkage (Fig. 1A). Previous enzyme assays toward various oligosaccharides showed that these two isozymes have almost the same acceptor glycan specificity, with GnT-IVa exhibiting higher activity (18). No GnT-IV-like activity (addition of β1,4-GlcNAc on the α1,3 arm) has so far been demonstrated for GnT-IVc or GnT-IVd. In fish and birds, GnT-IVc, also known as GnT-VI, has a different activity, transferring GlcNAc to the Manα1–6 arm (19). Expressed human GnT-IVc, also designated GnT-IV homolog, was shown to have neither GnT-IV activity nor GnT-VI activity (16). On the other hand, GnT-IVd appears to act as an inhibitor of GnT-I in the Golgi (17), rather than having any enzymatic function. Furthermore, double deficiency of GnT-IVa and GnT-IVb in mice completely abolishes both GnT-IV activity and expression of β1,4-GlcNAc on N-glycans in tissues (20), demonstrating that GnT-IVa and GnT-IVb are responsible for the biosynthesis of this branch in mammals. However, the difference in the molecular function between GnT-IVa and GnT-IVb is poorly understood. As to tissue distribution, GnT-IVb is ubiquitously expressed, whereas GnT-IVa expression is specific to some tissues, especially the pancreas (6, 15). GnT-IVa-deficient mice have impaired glucose-stimulated insulin secretion from pancreatic β-cells because of the reduced cell surface residency of GLUT2, leading to type 2-diabetic phenotypes such as abnormal glucose tolerance (6). Consistently, human pancreatic β-cells from type 2 diabetes patients also exhibit transcriptional downregulation of GnT-IVa and disordered glucose-stimulated insulin secretion (21), suggesting that branch formation on specific proteins by GnT-IVa in the pancreas may be a therapeutic target of diabetes. In contrast, our knowledge about the roles of GnT-IVb in cells and tissues is sparse.
Like many Golgi glycosyltransferases, GnT-IV enzymes commonly have a type-II transmembrane protein structure, consisting of a short cytoplasmic tail, a membrane-spanning region, a stem region, and a large C-terminal region (18) (Fig. 1B). We recently revealed that GnT-IVa, unlike other glycosyltransferases acting on N-glycans, uniquely has a lectin domain at its C terminus. We solved the crystal structure of the GnT-IVa lectin domain and found its structural similarity to a bacterial GlcNAc-binding lectin (22). Furthermore, this lectin domain specifically recognizes the product glycans having β1–4 GlcNAc branch synthesized by the catalytic domain, and the recognition of product glycans by this lectin domain is critical to the full enzymatic activity toward an oligosaccharide (22). However, the importance of the lectin domain in GnT-IVb and the functional roles of lectin domains from GnT-IVa and GnT-IVb toward glycoprotein substrates remain unknown.
In this study, we investigated the difference in activity toward glycoprotein substrates between GnT-IVa and GnT-IVb and found that these two isozymes have clearly distinct glycoprotein preferences. Furthermore, the lectin domain was found to be required for glycan biosynthesis in cells and a key for substrate selectivity toward glycoproteins. Our findings provide new insights into how these isozymes differentially biosynthesize the GlcNAc branch in cells.
Results
Different glycoprotein selectivity of GnT-IVs in cells
Because the tissue distribution patterns of GnT-IVa and GnT-IVb are different (20), we first examined whether the expression of GnT-IVa and GnT-IVb is exclusive to each other or both enzymes simultaneously function in the same cells. We generated human embryonic kidney 293 (HEK293) GnT-IVa single KO (IVaKO) and GnT-IVa/GnT-IVb-double KO (DKO) cells (22) to discriminate these two possibilities and compared the levels of GnT-IV products by flow cytometry. For glycan detection, we used Datura stramonium agglutinin (DSA), which recognizes β1,4-linked oligomers of GlcNAc (23). Single KO of GnT-IVa was found to reduce the signals of DSA partially, and the remaining signals were further reduced by additive deletion of GnT-IVb (Fig. 1C). This observation indicates that GnT-IVa and GnT-IVb function as glycosyltransferases within the same cells. To confirm that cellular GnT-IV activity is abolished in DKO cells, we incubated the cell lysates with a fluorescence-labeled biantennary N-glycan, GnGnbi-PA, as an acceptor substrate, and the reaction mixtures were analyzed by reversed-phase HPLC (Fig. 1D), as reported previously (24). The enzyme products were successfully detected in WT cells, and the endogenous GnT-IV activity was reduced in GnT-IVa single KO cells and completely disappeared in DKO cells, as expected (Fig. 1E). These findings suggest that GnT-IVa and GnT-IVb are not simply expressed in different cells but have distinct functions within one cell.
We next compared in vitro and intracellular activity of GnT-IVa and GnT-IVb to define the differences in function between GnT-IVa and GnT-IVb. We expressed recombinant soluble GnT-IVa and GnT-IVb in COS7 cells and purified these two proteins (Fig. 1, B and F). These enzymes were incubated with the acceptor substrate GnGnbi-PA to measure in vitro catalytic activity. Enzyme activity was detected for both GnT-IVa and GnT-IVb, and the specific activity of GnT-IVa was noticeably higher than GnT-IVb (Fig. 1, G and H), which is consistent with a previous report (18). We next examined the differences of these two enzymes in biosynthetic activity in cells. This was achieved by expressing full-length myc-tagged GnT-IVa or GnT-IVb (Fig. 1B) in DKO cells and analyzing the cell lysates by Western and lectin blotting. Anti-myc blotting showed that GnT-IVa and GnT-IVb were expressed at very similar levels (Fig. 1I, left). The band with a molecular weight of ∼50 kDa (Fig. 1I, upper left, asterisk) is derived from GnT-IVb and less than the theoretical molecular weight of full-length GnT-IVb (63.3 kDa), suggesting that GnT-IVb undergoes proteolysis or degradation more readily than GnT-IVa in cells. Compared with the mock-treated DKO cells, exogenous expression of either GnT-IVa or GnT-IVb increased the signals of DSA (Fig. 1I, right), indicating that this experimental system allows us to evaluate the intracellular biosynthetic activity of GnT-IVa and GnT-IVb toward glycoproteins in cells. Remarkably, the band patterns of DSA were slightly different between these two enzymes. In particular, the larger glycoproteins (over 150 kDa) were efficiently modified by GnT-IVa, whereas proteins at ∼75 kDa were modified by GnT-IVb. This observation suggests that these two enzymes act on at least a few distinct acceptor proteins in cells while also sharing substrate proteins. In vitro enzyme assays toward GnGnbi-PA using the cell lysates as enzyme sources showed that the enzyme activity of full-length GnT-IVa toward glycan substrates was again much higher than GnT-IVb (Fig. 1J), which is the same as the purified truncated enzymes (Fig. 1H). Collectively, these data indicate that the two isozymes GnT-IVa and GnT-IVb have not only different in vitro activity toward glycans but distinct protein selectivity in cells.
Intracellular localization of GnT-IVa and GnT-IVb
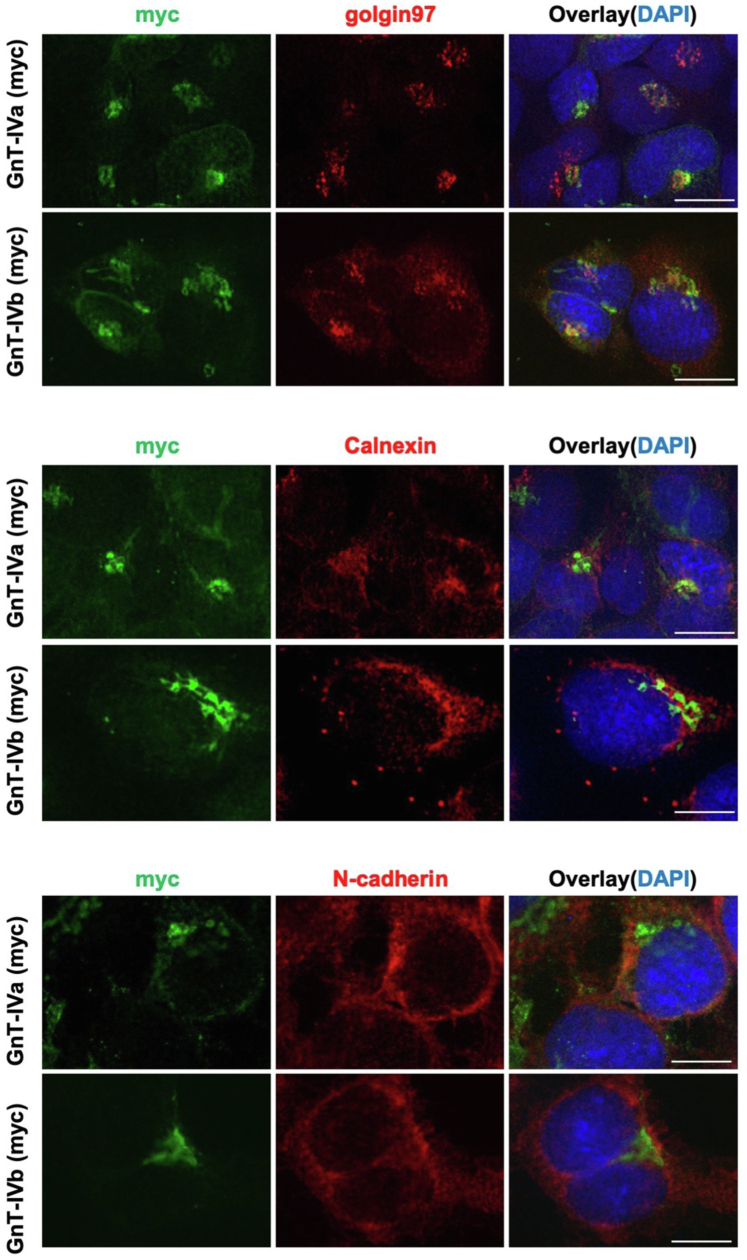
As a possible mechanism by which GnT-IVa and GnT-IVb act on different proteins in cells, we hypothesized that GnT-IVa and GnT-IVb are differentially localized in cells or selectively recognize their respective target polypeptides in the catalytic reactions. These possibilities were tested by first examining the intracellular localization of GnT-IVs. Immunofluorescence staining of myc-tagged GnT-IVa and GnT-IVb expressed in HEK293–DKO cells showed that GnT-IVa and GnT-IVb largely overlapped with the Golgi marker Golgin-97 but not with the ER marker (calnexin) and the plasma membrane marker (N-cadherin) (Fig. 2), indicating that these glycosyltransferases predominantly localize to the Golgi apparatus. The apparent absence of distribution in other organelles for both enzymes indicates that the different protein selectivities of GnT-IVs in cells are probably not caused by their different subcellular localizations.
Figure 2.
Intracellular localization of GnT-IVa and GnT-IVb. DKO cells transfected with a plasmid for expression of GnT-IVa or GnT-IVb were stained with anti-myc (green), antibodies for marker proteins (Golgin-97 for the Golgi, calnexin for the ER, or N-cadherin for the plasma membrane) (red), and DAPI (blue). The scale bar represents 10 μm. DAPI, 4′,6-diamidino-2-phenylindole; DKO, double KO; ER, endoplasmic reticulum; GnT, N-acetylglucosaminyltransferase.
Different specificity toward glycoprotein substrates between GnT-IVa and GnT-IVb
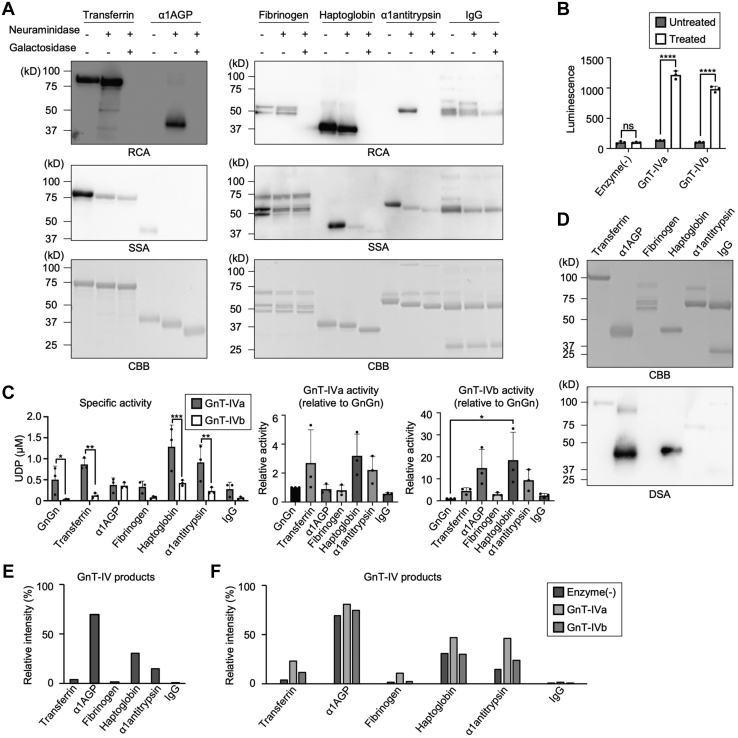
We next examined whether the enzyme activity of GnT-IVa and GnT-IVb on glycoproteins is influenced by a polypeptide moiety in acceptors. As our HPLC-based enzyme assays can only be applied to small substrates such as glycans, we performed UDP-Glo assays to examine the GnT-IV activity toward glycoprotein substrates. In UDP-Glo assays, the level of UDP released from UDP-GlcNAc by a glycosyltransferase reaction is measured. Therefore, any kind of acceptor substrates (oligosaccharides and glycoproteins) is acceptable in this assay system (25). We selected six human plasma glycoproteins as model substrates, transferrin apoform, alpha-1-acid glycoprotein (α1AGP), fibrinogen, haptoglobin, alpha-1-antitrypsin (α1-antitrypsin), and immunoglobulin G (IgG). The summary of the glycan structures of these glycoproteins reported in the literature (26) is shown in Table S1. All glycoprotein substrates were initially treated with neuraminidase and β-galactosidase to remove terminal sialic acid and galactose residues because GnT-IVs cannot react with an N-glycan whose GlcNAc is capped with galactose or sialic acid (18). Sambucus sieboldiana agglutinin (SSA) lectin and Ricinus communis agglutinin (RCA) lectin were used to confirm the removal of sialic acid and galactose, respectively. SSA recognizes α2,6-sialic acid (27), and RCA recognizes terminal galactose exposed by eliminating sialic acid (28). Reactivity of all glycoproteins with RCA was drastically reduced after treatment with neuraminidase and β-galactosidase (Fig. 3A, top panels). Furthermore, Coomassie brilliant blue (CBB) staining showed that these glycoproteins in an SDS-PAGE gel were step-wisely shifted to lower molecular weights by the glycosidase treatments (Fig. 3A, bottom panels). These data strongly suggest that GlcNAc residues were exposed in N-glycans of the prepared glycoprotein substrates and potentially serve as GnT-IV acceptors. Indeed, we found that in the UDP-Glo assay, the reactivity of GnT-IVa and GnT-IVb toward α1AGP became detectable after the glycosidase treatments (Fig. 3B). These findings suggest that the prepared substrates are suitable for analyzing GnT-IV activity toward glycoproteins.
Figure 3.
The in vitro activity of soluble GnT-IVa and GnT-IVb toward an oligosaccharide and various glycoproteins.A, glycoprotein substrates for the UDP-Glo assay were pretreated with neuraminidase and β-galactosidase. The treated proteins were stained with RCA, SSA, or CBB. B, the UDP-Glo assay was performed using purified soluble GnT-IVa or GnT-IVb and glycosidase-treated or untreated α1AGP (n = 3, means ± SD, ∗∗∗∗p < 0.0001; ns, not significant, Holm–Sidak's multiple comparisons test). C, comparison of the in vitro activity of GnT-IVa and GnT-IVb toward an oligosaccharide and various glycoproteins using the UDP-Glo assay (n = 3, means ± SD, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Holm–Sidak's multiple comparisons test [left]; n = 3, means ± SD, ∗p < 0.05, Tukey's multiple comparisons test [middle and light]). D, the glycosidase-treated proteins were stained with CBB or DSA. E, sum of the signal intensities of GnT-IV product N-glycans (#2c, 3, 5c, 6, and 11d in Table S2) in the sialidase- and galactosidase-treated six glycoprotein substrates without incubating with GnT-IV from LC–MS analysis. F, the sialidase- and galactosidase-treated six glycoprotein substrates were incubated with GnT-IVa or GnT-IVb in vitro. Sum of the signal intensities of GnT-IV product N-glycans from LC–MS analysis is shown. α1AGP, alpha-1-acid glycoprotein; CBB, Coomassie brilliant blue; DSA, Datura stramonium agglutinin; GnT, N-acetylglucosaminyltransferase; RCA, Ricinus communis agglutinin; SSA, Sambucus sieboldiana agglutinin.
Glycosylation efficiency among the glycoproteins was compared by using the same amount of N-glycan (100 pmol) in each reaction and taking into consideration the number of N-glycans in the glycoproteins. Using the prepared substrates and purified soluble GnT-IVa and GnT-IVb, the enzyme activity toward the oligosaccharide substrate (GnGnbi-PA) and six glycoprotein substrates (α1AGP, transferrin, fibrinogen, haptoglobin, α1-antitrypsin, and IgG) was measured. Consistent with the aforementioned results in the HPLC-based assay, the enzyme activity of GnT-IVa toward GnGnbi-PA was much higher than that of GnT-IVb in this assay system (Fig. 3C, left graph, GnGn). The activity of GnT-IVa toward glycoprotein substrates was higher than that of GnT-IVb for all the glycoproteins tested, but intriguingly, GnT-IVa and GnT-IVb showed distinct glycoprotein selectivity (Fig. 3C, middle and right graphs). In detail, GnT-IVa exhibited high activity toward asialoagalacto forms of transferrin, haptoglobin, and α1-antitrypsin and showed signals comparable to or less than that of the oligosaccharide GnGnbi-PA for the poor glycoprotein substrates (α1AGP, fibrinogen, and IgG). In contrast, GnT-IVb displayed higher activity toward all the glycoproteins than GnGnbi-PA, with a markedly higher preference for haptoglobin and α1AGP. These data demonstrate that the two enzymes have distinct glycoprotein preferences in the catalytic reactions and that efficient glycosylation by GnT-IVb depends strongly on the polypeptide moiety.
Next, we investigated the mechanism by which GnT-IVs selectively act on glycoproteins in their enzyme reactions. Both GnT-IVa and GnT-IVb have C-terminal lectin domains, and we recently revealed that the specific binding of the GnT-IVa lectin domain to the enzyme product β1,4-GlcNAc is essential for efficient enzyme activity (22). Based on these findings, we hypothesized that the N-glycan structures of the acceptor glycoproteins, particularly the presence of the β1,4-GlcNAc branch, influence the activity of GnT-IVs toward glycoproteins. Therefore, we stained the six prepared glycoprotein substrates with DSA lectin, which recognizes GnT-IV products (Fig. 3D). As a result, glycosidase-treated α1AGP and haptoglobin, the two best substrates for GnT-IVb, were highly reactive with DSA, compared with the other glycoproteins. These results suggest that GnT-IVb may preferentially act on glycoproteins that already possess GnT-IV-modified glycan(s).
To directly analyze the glycan structures of the substrate glycoproteins, we released N-glycans from these glycoproteins with or without GnT-IV reactions and analyzed their structures by LC–MS (Fig. S1 and Table S2). To distinguish isomeric GlcNAc branch structures, we also analyzed the fetuin-derived standard asialoagalacto-N-glycans, which were reacted with purified GnT-III, GnT-IVa, or GnT-V (Fig. S2). Consistent with the results of DSA lectin blotting (Fig. 3D), α1AGP and haptoglobin were confirmed to already have the high levels of GnT-IV products in their N-glycans without incubation with GnT-IVa or GnT-IVb (Fig. 3E). Furthermore, we detected the increased GnT-IV products by mass spectrometry (MS) after incubation with GnT-IVa or GnT-IVb for all the glycoproteins (Fig. 3F), confirming that β1,4-linked GlcNAc was formed on these glycoproteins by GnT-IVa or GnT-IVb. Although MS is not completely quantitative, the poor substrates for both GnT-IVa and GnT-IVb (fibrinogen and IgG) in UDP-Glo assays also showed the lower levels of GnT-IV products when analyzed by MS.
Intracellular activity of GnT-IVs toward a multiglycosylated model protein
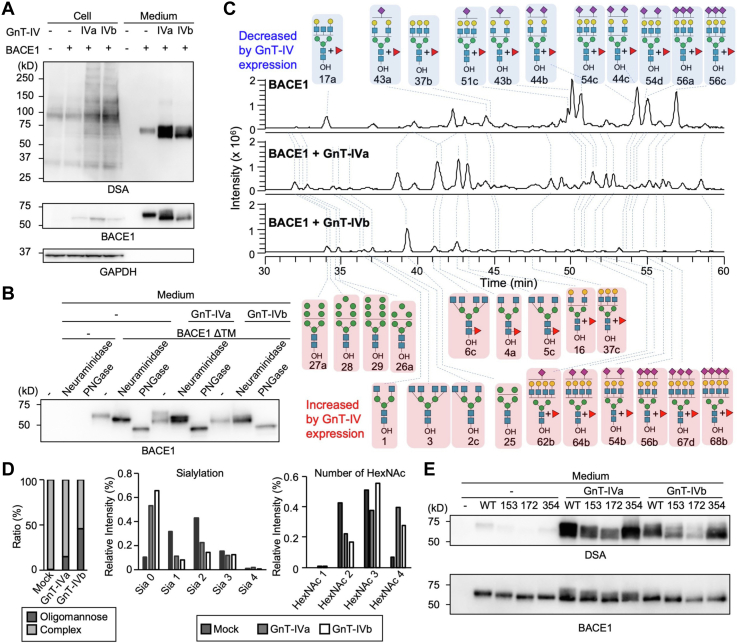
We coexpressed a multiglycosylated protein with GnT-IVa or GnT-IVb to further investigate the different activity toward glycoproteins in cells. As a model glycoprotein, we used β-site amyloid precursor protein cleaving enzyme-1 (BACE1), which has four N-glycans (N153, N172, N223, and N354) with multiple branches and sialic acids when expressed in cultured cells (29). A truncated soluble His-tagged form of BACE1 (BACE1ΔTM) was coexpressed with GnT-IVa or GnT-IVb in DKO cells, and the cell lysates and secreted BACE1 purified with Ni2+ beads were analyzed by Western and lectin blotting. Compared with the sole expression of BACE1, exogenous coexpression of either GnT-IVa or GnT-IVb increased the DSA reactivity of secreted BACE1 (Fig. 4A), demonstrating that GnT-IVa and GnT-IVb modified BACE1 glycans in cells. Interestingly, coexpression of either GnT-IVa or GnT-IVb gave rise to two BACE1 bands in the medium with faster and slower mobility in the SDS-PAGE gel than solely expressed BACE1. Furthermore, in the GnT-IVb-expressing sample, the lower band was dominant over the upper band, whereas BACE1 coexpressed with GnT-IVa showed comparable levels of these two bands (Fig. 4A). This result suggested that GnT-IVa and GnT-IVb differentially modify BACE1 in cells.
Figure 4.
Differential actions of GnT-IVa and GnT-IVb toward BACE1 in cells.A, cell lysates (cell) and secreted BACE1 purified with Ni2+ beads (medium) were analyzed by Western and lectin blotting. Proteins from HEK293 DKO cells transfected with the plasmids for expression of GnT-IVa, GnT-IVb, and BACE1, or empty vector, were subjected to SDS-PAGE and blotted with HRP-conjugated DSA (upper), anti-BACE1 antibody (middle), and anti-GAPDH antibody (lower). B, secreted BACE1 purified with Ni2+ beads was treated with neuraminidase or PNGaseF, subjected to SDS-PAGE, and blotted with the anti-BACE1 antibody. C, LC–MS total ion chromatograms (TICs) of N-glycans derived from secreted BACE1 with or without coexpression of GnT-IVa or GnT-IVb in HEK293 DKO cells. Pink, glycans increased by coexpression with GnT-IVa or GnT-IVb; blue, glycans decreased by coexpression with GnT-IVa or GnT-IVb. D, left, sum of the signal intensities of oligomannose glycans and hybrid or complex glycans from LC–MS analysis is shown. Middle, sum of the signal intensities of non-, mono-, di-, tri-, and tetrasialylated N-glycans from LC–MS analysis is shown. Right, sum of the signal intensities of N-glycans with 1, 2, 3, and 4 HexNAc residues (HexNAc residues in chitobiose were excluded) from LC–MS analysis is shown. E, secreted BACE1 WT and its N-glycosylation site mutants (N153S, N172S, and N354S) purified with Ni2+ beads were subjected to SDS-PAGE and blotted with HRP-conjugated DSA (upper) and the anti-BACE1 antibody (lower). BACE1, β-site amyloid precursor protein cleaving enzyme-1; DKO, double KO; DSA, Datura stramonium agglutinin; GnT, N-acetylglucosaminyltransferase; HEK293, human embryonic kidney 293 cell line; HRP, horseradish peroxidase; PNGaseF, peptide N-glycanase F.
We hypothesized that these two BACE1 bands observed by GnT-IV expression are different glycoforms. This hypothesis was tested by treating secreted proteins with neuraminidase and peptide N-glycanase F (PNGaseF) to remove terminal sialic acids and all N-glycans, respectively (Fig. 4B). The two BACE1 bands observed by GnT-IV expression were both sensitive to PNGaseF, and the mobility of BACE1 after PNGaseF treatment was almost the same regardless of GnT-IV expression, suggesting that the two BACE1 bands observed by GnT-IV expression are only different in N-glycan structures from solely expressed BACE1. In contrast, in the GnT-IV-expressing samples, only the upper band was neuraminidase sensitive, and the lower band was not, whereas BACE1 without GnT-IV expression was sensitive to neuraminidase (Fig. 4B). These results indicated that BACE1 is normally sialylated in DKO cells and that the expression of GnT-IVs generates both sialylated and unsialylated forms of BACE1. Moreover, the unsialylated form is dominant when modified by GnT-IVb but not by GnT-IVa. To confirm these findings, we conducted N-glycomic analysis for BACE1 using LC–MS. BACE1 was expressed with or without GnT-IV and purified, and its N-glycans were released and analyzed (Figs. 4C and S3; Table S3). As a result, the amounts of sialylated N-glycans among total complex N-glycans were reduced by GnT-IV expression (Fig. 4D, middle), consistent with the results in Figure 4B. Furthermore, the number of HexNAc was increased by GnT-IV expression as expected (Fig. 4D, right), which was likely attributed to the biosynthesis of the GlcNAc branch by coexpressed GnT-IV. More surprisingly, the rates of oligomannose glycans in BACE1 N-glycans were increased by GnT-IV expression (Fig. 4D, left), particularly by GnT-IVb. This finding suggests that an unknown mechanism exists by which Golgi enzyme GnT-IVs affect an earlier step in N-glycan biosynthesis in the ER or cis-Golgi.
Next, we examined the activity of GnT-IVa and GnT-IVb toward BACE1 WT and N-glycosylation site mutants. Because the expression level of BACE1 N223S was extremely low for an unknown reason (Fig. S4), we expressed the other three mutants (N153S, N172S, and N354S) with GnT-IVa or GnT-IVb. The DSA signals on secreted BACE1 and its mutants coexpressed with GnT-IVa were all higher than those coexpressed with GnT-IVb. The DSA signal on BACE1 N153S and N172S mutants was markedly weaker than WT for both GnT-IVa and GnT-IVb (Fig. 4E). This suggests that these two sites are the preferred sites for GnT-IVa and GnT-IVb, with N172 being the most preferred site. Taken together, these findings indicate that GnT-IVa and GnT-IVb have similar glycosylation site preferences toward BACE1 glycoprotein, but these two enzymes differentially modify BACE1 N-glycans in cells.
Lectin domain is important for glycoprotein selectivity
As shown in Figure 3, C–E, the activity of GnT-IVa and GnT-IVb on glycoproteins likely depends on the glycan structures of the acceptor glycoproteins. We also recently reported that the C-terminal lectin domain is essential for GnT-IVa activity (22). Based on these findings, we reasoned that the lectin domains of GnT-IVs contribute to the selective recognition of acceptor glycoproteins.
The importance of the lectin domain for the activity of GnT-IVa and GnT-IVb was confirmed by preparing constructs, GnT-IVsΔLec, that lacked the lectin domain (Fig. 1B), and these constructs were expressed in DKO cells (Fig. 5A). In vitro assays of enzyme activity of the cell lysates toward GnGnbi-PA clearly showed that the activity of GnT-IVsΔLec was almost abolished (Fig. 5B). This suggests that the lectin domain is needed for the enzymatic activity of both GnT-IVa and GnT-IVb. It is possible that loss of activity of the mutants is caused by conformational changes affecting the catalytic activity, but we found that GnT-IVΔLec mutants were still mainly localized in the Golgi apparatus (Fig. S5A). Furthermore, soluble truncated forms of GnT-IVΔLec with a signal sequence were successfully secreted into the media (Fig. S5B), and their activity was shown to be greatly less than WT enzymes (Fig. S5C). These data suggested that the lectin domain deletion mutants maintain folding at least partially while dramatically losing activity.
Figure 5.
Lectin domain regulates protein selectivity of GnT-IV.A, proteins from mock-treated HEK293 WT and DKO cells transfected with an empty vector (mock) or a plasmid to express myc-tagged GnT-IVa, GnT-IVb, GnT-IVaΔLec, or GnT-IVbΔLec were subjected to SDS-PAGE and blotted with the anti-myc antibody or anti-GAPDH antibody. B, cell lysates of mock-treated HEK293 WT and DKO cells transfected with an empty vector (mock) or a plasmid for expression of GnT-IVa, GnT-IVb, GnT-IVaΔLec, or GnT-IVbΔLec were reacted with GnGnbi-PA and analyzed by HPLC (n = 3, means ± SD, ∗∗p < 0.01, Tukey's multiple comparisons test). C, proteins from mock-treated HEK293 WT and DKO cells transfected with an empty vector (mock) or a plasmid for expression of GnT-IVa, GnT-IVb, GnT-IVa-Lec(b), or GnT-IVb-Lec(a) were subjected to SDS-PAGE and blotted with the anti-myc antibody or anti-GAPDH antibody. D, lysates of mock-treated HEK293 WT and DKO cells transfected with an empty vector (mock) or a plasmid for expression of GnT-IVa, GnT-IVb, GnT-IVa-Lec(b), or GnT-IVb-Lec(a) were reacted with GnGnbi-PA and analyzed by HPLC (n = 3, means ± SD, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Tukey's multiple comparisons test). E, the structure of the lectin domain of GnT-IVa modeled with GlcNAc. The residues conserved between GnT-IVa and GnT-IVb are shown in red. F, soluble GnT-IVb, GnT-IVb F413H, or GnT-IVb I456Q was expressed in COS7 cells and purified from the media using a Ni2+ column. Purified soluble GnT-IVb, GnT-IVb F413H, or GnT-IVb I456Q was separated by SDS-PAGE and visualized by CBB staining. G, the specific activity of the purified GnT-IVb, GnT-IVb F413H, and GnT-IVb I456Q toward GnGnbi-PA was analyzed by HPLC (n = 3, means ± SD, a significant p value was not observed, Tukey's multiple comparisons test). H, comparison of the in vitro activity of GnT-IVb, GnT-IVb F413H, and GnT-IVb I456Q toward an oligosaccharide and various glycoproteins (same as Fig. 3) using the UDP-Glo assay (n = 3, means ± SD, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, Tukey's multiple comparisons test). CBB, Coomassie brilliant blue; DKO, double KO; GnT, N-acetylglucosaminyltransferase; HEK293, human embryonic kidney 293 cell line.
Next, to examine whether the lectin domain is mutually compatible between GnT-IVa and GnT-IVb, two GnT-IV mutants (GnT-IVa-Lec(b) and GnT-IVb-Lec(a)) in which the lectin domains were swapped with each other (Fig. 1B), were expressed in DKO cells (Fig. 5C). In vitro enzyme assays toward GnGnbi-PA showed that both mutants have much weaker activity than WT enzymes (Fig. 5D), indicating that the whole lectin domains of GnT-IVa and GnT-IVb are not interchangeable. We therefore sought to modify the functions of lectin domain by point mutation, and potential functional differences of the lectin domains from GnT-IVa and GnT-IVb were examined. Close inspection of the structure of the GnT-IVa lectin domain around the putative GlcNAc-binding site (Fig. 5E) and sequence alignment of the lectin domains (Fig. S6) revealed that most of the key residues around the putative GlcNAc-binding site are conserved between GnT-IVa and GnT-IVb (Fig. 5E, red). Nonetheless, H397 and Q440 in GnT-IVa are replaced with F413 and I456 in GnT-IVb, respectively. We hypothesized that conversion of these amino acids of GnT-IVb to the corresponding residues of GnT-IVa might modify the functions of the GnT-IVb lectin domain, resulting in GnT-IVa-like enzymatic activity. Recombinant soluble GnT-IVb F413H and I456Q mutants were expressed in COS7 cells and purified (Fig. 5F), and the activity of these mutants toward GnGnbi-PA and the glycoproteins was measured by the HPLC-based assay (Fig. 5G) and UDP-Glo (Fig. 5H) to test this hypothesis. We found no statistically significant differences in the activity of F413H and I456Q toward the oligosaccharide from WT IVb. Surprisingly, however, the F413H mutant showed enhanced activity toward particular glycoprotein substrates, such as haptoglobin, α1-antitrypsin, and transferrin (Fig. 5H), which were the best three substrates for GnT-IVa (Fig. 3C). Taken together, the glycoprotein preference of GnT-IVb can be modified by engineering its lectin domain, suggesting that the lectin domain plays an important role in determining acceptor preference against glycoproteins.
Discussion
In this study, we found that two N-glycan branching isozymes, GnT-IVa and GnT-IVb, have distinct glycoprotein preferences. Furthermore, the C-terminal lectin domain of GnT-IV contributes to the selectivity against glycoprotein substrates. Several glycosyltransferase isozymes, particularly those directly acting on polypeptides, have been reported to display different glycoprotein selectivity. For example, N-acetylgalactosaminyltransferases, O-fucosyltransferases (POFUT1,2), and oligosaccharyltransferase have different specificities toward acceptor polypeptides among the isozymes (30, 31, 32). In contrast, it remains unclear whether and how the glycosyltransferase isozymes for branching or capping of glycans also show glycoprotein selectivity. In this study, we revealed different glycoprotein preferences of GnT-IV isozymes for the first time, providing new clues for understanding how N-glycan branches are differentially formed on various glycoproteins.
As expected, we found that GnT-IVa and GnT-IVb are predominantly localized in the Golgi apparatus. However, it is still possible that these two enzymes have subtly distinct localization within the Golgi apparatus, which may influence different protein selectivity of GnT-IVs. Further investigation that details where GnT-IVa and GnT-IVb exist in the Golgi cisternae is warranted and requires the use of super-resolution or electron microscopy. Moreover, the appearance of a likely degradation product of GnT-IVb (Fig. 1I, upper left, asterisk) suggests that GnT-IVb is more unstable than GnT-IVa in cells and readily targeted to the degradation pathway. To confirm this notion, degradation rates of GnT-IVa and GnT-IVb were compared by cycloheximide (CHX) chase experiments, and we observed that GnT-IVb was more rapidly degraded than GnT-IVa as expected (Fig. S7A). We also tested whether the GnT-IVb smaller band is detected in the medium, because most glycosyltransferases are cleaved near the transmembrane border and secreted (33). As a result, the smaller band was not detected in the medium (Fig. S7B), suggesting that the smaller band is a degradation product but not a soluble secreted form. Detailed mechanisms that define the different stability between GnT-IVa and GnT-IVb in cells should be characterized, and the effect of this stability on the intracellular activity of GnT-IVs should also be examined.
We raise two possible explanations for the different preferences of GnT-IVa and GnT-IVb toward glycoprotein substrates. The first is that GnT-IVa and GnT-IVb actively recognize a polypeptide moiety or a combination of peptide and glycan. The other explanation is that glycosylation by GnT-IVs may take place only on glycoproteins or glycosylation sites with less steric hindrance. Consistent with this notion, IgG, whose Fc region possibly causes steric hindrance, was a poor substrate for both GnT-IVa and GnT-IVb. Notably, GnT-IVb showed much higher activity toward all glycoproteins than free GnGn, indicating that GnT-IVb prefers glycoprotein to glycan as a substrate. Moreover, GnT-IVb displayed higher activity toward haptoglobin and α1AGP than toward the other glycoproteins tested (Fig. 3C), and these two glycoproteins reacted strongly with DSA (Fig. 3D). Therefore, GnT-IVb prefers glycoproteins that GnT-IV has already modified. We propose that the former explanation is more likely based on these findings. Although we previously reported that the GnT-IVa lectin domain binds to β1,4-GlcNAc specifically (22), specificity and avidity of glycan binding by the GnT-IVb lectin domain have not been clarified. In addition, as the tertiary structures of the catalytic domains of GnT-IVa and GnT-IVb have also not been solved, elucidating the structures of GnT-IVs complexed with substrates would uncover how these enzymes recognize substrate glycoproteins. Furthermore, forming a protein complex with other molecules may also influence acceptor recognition by GnT-IVs. GnT-IVb was reported to interact with the UDP-GlcNAc transporter (34), and searching for interacting partners of GnT-IVs represents an interesting issue to understand their activity in cells further. In addition, considering the preference of GnT-IVb for glycoproteins that already possess GnT-IV-modified glycans, it is possible that there is an order of actions of GnT-IVa and GnT-IVb in cells. If both GnT-IVa and GnT-IVb are simultaneously expressed, GnT-IVb could modify glycoproteins that GnT-IVa premodified.
We found that deletion or swapping of the lectin domain almost abolished activity (Fig. 5, B and C). Furthermore, mutation (F413H) of a nonconserved amino acid in the putative GlcNAc-binding site in GnT-IVb enhanced the activity only toward particular glycoproteins (Fig. 5H). These findings indicate that the lectin domain plays important roles in determining the acceptor preference against glycoproteins, and the nonconserved residues in the lectin domain may be crucial for defining the interaction with glycans or glycoproteins. It is important to examine in the future whether and how the glycan-binding activity of the lectin domain and its binding specificity regulate substrate protein selectivity of GnT-IVa and GnT-IVb. Although it is unclear how the lectin domain regulates the activity and specificity of the catalytic domain, we surmise that specific binding to a glycan on a glycoprotein through the lectin domain may induce a conformational change of the catalytic domain that leads to this domain being more reactive. Besides GnT-IVa and GnT-IVb, several glycosyltransferases also have a lectin domain. UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase have a C-terminal lectin domain, which binds GalNAc, the product of the catalytic domain, thereby modulating acceptor peptide selectivity (35, 36). In addition, the lectin domain of protein O-linked mannose β1,2-N-acetylglucosaminyltransferase 1 also binds to the β-linked GlcNAc moiety in O-mannosyl glycans, a product of protein O-linked mannose β1,2-N-acetylglucosaminyltransferase 1 (37). These studies raised a possibility that the lectin domains of glycosyltransferases bind the products, thereby regulating glycoprotein selectivity.
In addition to the distinct glycoprotein preferences of GnT-IVa and GnT-IVb, we also demonstrated that GnT-IVa and GnT-IVb differentially modify N-glycans of a single glycoprotein (BACE1) in cells (Fig. 4, A–D). In particular, the expression of GnT-IVb predominantly produced the unsialylated glycoforms. BACE1 is normally sialylated without the expression of GnT-IVa and GnT-IVb, whereas BACE1 was more modified with oligomannosidic glycans upon GnT-IVb overexpression. This suggests that overexpressed GnT-IVb or the produced β1,4-GlcNAc interferes with the actions of enzymes for early steps in N-glycan biosynthesis in the ER and cis-Golgi. Further investigation on the effects of GnT-IV expression on the activity and localization of these enzymes is required to explore these possible inhibitory mechanisms. Our present study also showed that GnT-IVa and GnT-IVb have a preferred site within a single glycoprotein (N172 on BACE1). We have also reported that another N-glycan branching enzyme GnT-III prefers different glycosylation sites (N153 and N223) in BACE1 (29). Thus, these findings indicate that each branching enzyme modifies different sites within a single glycoprotein.
In conclusion, we demonstrated functional differences between GnT-IVa and GnT-IVb isozymes. Our data showed that these isozymes have different glycoprotein selectivity, which is at least partially regulated by their lectin domains, providing insights into how complex N-glycans are differentially synthesized on glycoproteins. We recently reported that the N-terminal region of the luminal noncatalytic domain of another branching enzyme, GnT-V, plays a critical role in recognizing glycoprotein substrates (38). These findings, together with the present study, suggest that for some glycosyltransferases dedicated to glycan extension and capping, noncatalytic domains control acceptor selectivity. In the future, identifying GnT-IVa- and GnT-IVb-specific physiological substrates should lead to understanding the mechanisms responsible for differential protein selectivity by GnT-IVa and GnT-IVb. Together with our activity assay data, the expression levels of GnT-IVa and GnT-IVb can predict a dominant GnT-IV enzyme for modification of a certain glycoprotein. For example, we showed that GnT-IVb has comparable and weaker (approximately one-third of GnT-IVa) activity toward α1AGP and haptoglobin, respectively, compared with GnT-IVa (Fig. 3C); however, others’ transcriptomic data (39) indicate that human MGAT4B mRNA is six times more highly expressed than MGAT4A in the liver, the organ where these glycoproteins are produced. This suggests that GnT-IVb is the primary β1,4-GlcNAc-branching enzyme modifying these glycoproteins in vivo.
Experimental procedures
Reagents
The following antibodies and lectins were used: anti-myc (mouse, clone 4A6; Millipore; catalog no.: 05-724), anti-BACE1 (mouse, clone 1A11, generous gift from Dr Bart De Strooper (40)), anti-GAPDH (mouse, clone 6C5; Merck Millipore; catalog no.: MAB374), anti-Golgin-97 (rabbit, clone D8P2K, Cell Signaling Technology; catalog no.: 13192S), anticalnexin (rabbit, abcam; catalog no.: ab22595), anti-N-cadherin (rabbit, abcam; catalog no.: ab18203), horseradish peroxidase (HRP)–conjugated antimouse IgG (GE Healthcare; catalog no.: NA931V), HRP-conjugated anti-rabbit IgG (GE Healthcare, Amersham; catalog no.: NA934V), DSA (J-Chemical; catalog no.: J105), SSA letin (J-Chemical; catalog no.: J118), biotinylated RCA (Vector Laboratories; catalog no.: B-1085), Alexa546-conjugated anti-rabbit IgG (Invitrogen; catalog no.: A10040), and Alexa488-conjugated antimouse IgG (Invitrogen; catalog no.: A21202). DSA and SSA were conjugated to HRP using a Peroxidase Labeling Kit-NH2 (Dojindo), according to the manufacturer’s protocol.
Plasmid construction
Primers used in this study are listed in Table S4. pcDNA6 myc-His A/mouse GnT-IVa (UniProt ID: Q812G0-2) was constructed, as described previously (22). Complementary DNA (cDNA) encoding mouse GnT-IVb was amplified by PCR using the mouse brain cDNA library as a template and cloned into pCR4Blunt-TOPO. The cDNA fragment of mouse GnT-IVb was digested with EcoRI/XhoI and inserted into EcoRI/XhoI sites of pcDNA6 myc-His A. Construction of the expression plasmids for GnT-IVaΔLec or GnT-IVbΔLec, in which the lectin domain (GnT-IVa, Asn382 to Asp493; GnT-IVb, Asn398 to Asp548) was removed, involved amplifying the DNA fragment encoding from the N terminus to Val381 (GnT-IVa) or Val397 (GnT-IVb) by PCR using pcDNA6 myc-His A/mouse GnT-IVa or pcDNA6 myc-His A/mouse GnT-IVb as a template. PCR products were digested with NotI and XhoI or EcoRI and XhoI and then ligated to the NotI–XhoI sites of pcDNA6/myc-His A or EcoRI–XhoI sites of pcDNA6/myc-His A. The cDNA encoding the GnT-IVb lectin domain (Asn398 to Asp548) was amplified by PCR using pcDNA6 myc-His A/mouse GnT-IVb as a template to construct the plasmid for GnT-IVa-Lec(b). The amplified fragment was ligated to pcDNA6 myc-His A/mouse GnT-IVaΔLec, which had been digested with XhoI, using NEBuilder HiFi DNA Assembly Master Mix, according to the manufacturer’s protocol. Similarly, for the construction of GnT-IVb-Lec(a), the cDNA encoding the GnT-IVa lectin domain (Asn382 to Asp493) was amplified by PCR using pcDNA6 myc-His A/mouse GnT-IVa as a template. The amplified fragment was ligated to pcDNA6 myc-His A/mouse GnT-IVbΔLec, which had been digested with XhoI, using the NEBuilder HiFi DNA Assembly Master Mix, according to the manufacturer’s protocol. The plasmid for the 6× His-tagged soluble GnT-IVa (pcDNA-IH/GnT-IVa) and pcDNA-IH/GnT-IVaΔlec was constructed, as described previously (22). For the construction of pcDNA-IH/GnT-IVb, the DNA fragment encoding the catalytic region (Ser61 to C terminus) was amplified by PCR using pcDNA6 myc-His A/mouse GnT-IVb as a template and ligated to the EcoRV–XhoI sites of pcDNA-IH. For construction of pcDNA-IH/GnT-IVbΔlec, the DNA fragment encoding the catalytic region (Ser61 to Gly377) was amplified by PCR using pcDNA-IH/GnT-IVb as a template and ligated to the EcoRV–XhoI sites of pcDNA-IH. The plasmid for myc-His-tagged BACE1ΔTM (pcDNA6 myc-His A/human BACE1ΔTM) was constructed, as described previously (29). The plasmids for the N-glycosylation site mutants of BACE1 were constructed using pcDNA6 myc-His A/human BACE1ΔTM as a template and the QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies), according to the manufacturer’s protocol. The plasmids for the point mutants of GnT-IVb (F413H and I456Q) were constructed using pcDNA-IH/GnT-IVb as a template and the QuikChange Lightning Site-Directed Mutagenesis Kit, according to the manufacturer’s protocol.
Cell culture
COS7, HEK293, and HEK293 GnT-IVa/GnT-IVb-DKO cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and 50 μg/ml kanamycin under 5% CO2 conditions at 37 °C. The HEK293-DKO cell clone was established by a CRISPR system, as described previously (22).
Plasmid transfection
According to the manufacturer's protocol, cells at 70 to 80% confluency on a 6-cm dish were transfected with 2 μg of plasmids using Lipofectamine 3000 reagent (Thermo Fisher Scientific). Cells were collected 48 h after transfection and used for subsequent experiments. For expression of recombinant soluble GnT-IVs, polyethyleneimine MAX (Polyscience) was used, as described later (see “Purification of recombinant proteins” section).
Fluorescence-activated cell sorting analysis
Cells were washed with PBS twice and collected by cell scrapers, followed by precipitation by centrifugation at 1400g for 3 min. The cells were washed with fluorescence-activated cell sorting (FACS) buffer (1% bovine serum albumin [BSA] and 0.1% NaN3 in PBS) once and stained with FITC-conjugated D. stramonium lectin (purchased from Vector Laboratories; catalog no.: FL-1181) diluted in FACS buffer (1:200 dilution). Cells were then washed with PBS twice and analyzed with a FACS Melody cell sorter (BD Biosciences). The collected data were analyzed by FlowJo software (BD Biosciences).
Western and lectin blotting and CBB staining
Cells were washed with PBS and collected by centrifugation at 410g for 5 min. The cells were washed again with PBS and centrifuged at 13,800g for 1 min. The cells were lysed with lysis buffer (50 mM Tris–HCl [pH 7.4], 150 mM NaCl, 1% Nonidet P-40, Protease Inhibitor Cocktail Set V [EDTA free] [Fujifilm]) and sonicated. The protein concentrations of the cell lysates were measured using the Pierce Bicinchoninic Acid Protein Assay Kit (Thermo Fisher Scientific), according to the manufacturer’s protocol. Lysates or protein solution samples for SDS-PAGE analysis were prepared by mixing these samples with Laemmli 5× SDS sample buffer and boiling the samples at 95 °C for 5 min. The same amount of protein was loaded into each lane of the SDS-PAGE gel, and the proteins were resolved by 5 to 20% SDS-PAGE. For CBB staining of the gel, the proteins were stained with GelCode Blue Safe Protein Stain (Thermo Fisher Scientific) and visualized using FUSION-SOLO 7s EDGE (Vilber–Lourmat). The proteins separated in the gel were transferred to nitrocellulose membranes using a semidry blotter for Western blotting. The membranes were blocked with 5% skim milk in Tris-buffered saline (TBS) containing 0.1% Tween-20 (TBS-T) and incubated with primary antibodies diluted with 5% skim milk in TBS-T overnight at 4 °C. After washing with TBS-T for 5 min three times, the membranes were incubated with HRP-conjugated secondary antibodies at room temperature for 1 h. For blotting with HRP–lectins, membranes were blocked with 1% BSA in TBS-T overnight at 4 °C, followed by incubation at room temperature for 1 h with HRP-conjugated lectins that were diluted with 1% BSA in TBS-T. For blotting with biotinylated lectins, membranes were blocked with TBS-T overnight at 4 °C, followed by incubation at room temperature for 30 min with biotinylated lectins diluted with TBS-T. After washing with TBS, the membranes were incubated with the VECTASTAIN ABC Standard kit (Vector Laboratories) (1:400 dilution in TBS-T) at room temperature for 30 min. Proteins were detected with the Western Lightning Plus-ECL (PerkinElmer) using FUSION-SOLO 7s EDGE.
Purification of recombinant proteins
COS7 cells were cultured on 15-cm dishes to obtain soluble His-tagged GnT-IVa and GnT-IVb and their ΔLec mutants. Cells were transfected with the plasmids using polyethyleneimine MAX (Polyscience) when confluency reached 70 to 80%. After 6 h, the medium was replaced with Opti-MEM I, followed by further incubation for 72 h. Soluble His-tagged GnT-IVs were purified from the media using a Ni2+ column. After washing the column with 10 mM phosphate buffer (pH 7.4) containing 20 mM imidazole and 0.5 M NaCl, the recombinant proteins were eluted with 10 mM phosphate buffer (pH 7.4) containing 0.5 M NaCl and 0.5 M imidazole (elution buffer), followed by desalting using a NAP-5 gel filtration column (GE Healthcare). The eluates were diluted with 50 mM Mes (pH 7.7) buffer to ensure the enzyme concentrations were the same between GnT-IVa and GnT-IVb and were used directly as the enzyme sources.
GnT-IV activity assay using HPLC
GnT-IV activity toward an oligosaccharide substrate was measured using HPLC, as described previously (24), with slight modifications. In brief, the fluorescence-labeled oligosaccharide GnGnbi-PA (pyridylamine) was used as the substrate, and cell lysates or purified soluble His-tagged GnT-IVs were used as the enzyme source. The enzyme source was incubated at 37 °C in 10 μl reaction buffer, which contained 10 μM acceptor substrate, 20 mM UDP-GlcNAc, 25 mM Mes (pH 7.7), 0.5% (v/v) Triton X-100, 5 mg/ml BSA, and 7.5 mM MnCl2. The GnT-IV reaction was stopped by boiling at 95 °C for 5 min, and 40 μl of water was added to the mixture. After centrifugation at 16,000g for 3 min, the supernatant was analyzed by reversed-phase HPLC equipped with an ODS column (TSKgel ODS-80TM, TOSOH Bioscience, 4.6 × 150 mm). HPLC analysis was conducted in the isocratic mode with buffers A (20 mM ammonium acetate buffer [pH 4.0]) and B (1% 1-butanol in buffer A) mixed in a proportion of five to one.
CHX chase
After 24 h of transfection, CHX (Fujifilm) was added to the culture medium at 100 μg/ml. After incubation for 0, 2, 4, 6, and 24 h, the cells were collected and the cell lysates were subjected to Western blotting.
Immunofluorescence staining
Cells seeded on an 8-well glass chamber slide were transfected with 0.1 μg of plasmid using Lipofectamine 3000 transfection reagent. After 48 h, the cells were washed with PBS and fixed with 4% paraformaldehyde/PBS for 15 min at room temperature. The cells were washed with PBS and then permeabilized by incubating with PBS containing 0.1% Nonidet P-40 and 3% BSA for 30 min. After washing with PBS, the cells were incubated with primary antibodies for 1 h at room temperature, followed by incubation with Alexa488- or Alexa546-conjugated secondary antibodies and 4′,6-diamidino-2-phenylindole. Fluorescence signals were visualized using a BZX-800 all-in-one fluorescence microscope (KEYENCE).
Preparation of the acceptor substrate for the UDP-Glo assay
Human transferrin apoform (transferrin) (Wako), human α1AGP (Sigma; catalog no.: G9885), human fibrinogen (Fujifilm; catalog no.: 061-03691), human haptoglobin (Sigma; catalog no.: H3536), human α1-antitrypsin (Sigma; catalog no.: A9024), and human IgG (Wako; catalog no.: 143-09501) were purchased and used for the assays. One milligram of α1AGP, transferrin, fibrinogen, haptoglobin, α1-antitrypsin, or IgG was digested with 3.8 munit/μl of Arthrobacter ureafaciens neuraminidase (Nacalai Tesque) and 1.2 unit/μl of Streptococcus pneumoniae β-galactosidase (New England Biolabs) in 87 mM sodium acetate buffer (pH 4.5) at 37 °C overnight.
LC–MS glycan analysis
Commercially available six human serum glycoproteins (transferrin, α1AGP, fibrinogen, haptoglobin, α1antitrypsin, and IgG) treated with neuraminidase and galactosidase and a soluble form of BACE1 purified from HEK293 culture media using Ni2+-sepharose were subjected to SDS-PAGE. After transfer to poly(vinylidene fluoride) membrane and protein staining with DirectBlue71, the bands corresponding to the six glycoproteins and BACE1 were excised. N-Glycans from these glycoproteins were released and analyzed as described previously (38, 41) with some modifications. After release and reduction, N-glycan alditols were separated on a carbon column (5 μm HyperCarb, 1 mm I.D. × 100 mm; Thermo Fisher Scientific) using a Vanquish HPLC pump (Thermo Fisher Scientific; flow rate: 50 μl/min, column oven: 40 °C). The eluate was continuously introduced into an electrospray ionization source (LTQ Orbitrap XL; Thermo Fisher Scientific). MS spectra were obtained in the negative ion mode using Orbitrap MS (mass range m/z 500 to m/z 2500), and MS/MS spectra were obtained using ion trap MS. Monoisotopic masses were assigned with possible monosaccharide compositions using the GlycoMod software tool (mass tolerance for precursor ions is ±0.006 Da; https://web.expasy.org/glycomod/). Xcalibur software, version 2.2 (Thermo Fisher Scientific) was used to show the total ion chromatogram, base peak chromatogram, extracted ion chromatogram and to analyze MS and MS/MS data. The standard N-glycans having defined GlcNAc branches were prepared from bovine fetuin N-glycans whose structures were already determined (42, 43). The desialylated and degalactosylated biantennary and triantennary N-glycan alditols were prepared from bovine fetuin (from fetal bovine serum; Sigma; catalog no.: F2379-100MG) as described previously. The N-glycans (233 pmol per reaction) were incubated with purified recombinant GnT-III, GnT-IVa, or GnT-V derived from COS7 cells (44) with 0.2 mM UDP-GlcNAc for 3 h at 37 °C.
GnT-IV activity assay using UDP-Glo
The UDP-Glo assay was performed using a UDP-Glo Glycosyltransferase Assay kit (Promega), as described previously (25). Briefly, the GnT-IV reaction was conducted in a 96-well white plate by incubating purified GnT-IVs with acceptor asialoagalacto substrates for 2 h at room temperature in 10 μl of the reaction mixture that contained 10 mM ultrapure UDP-GlcNAc (supplied in the kit), an acceptor substrate, 25 mM Mes (pH 7.7), 0.5% (v/v) Triton X-100, 5 mg/ml BSA, and 7.5 mM MnCl2. The concentrations of the acceptor substrates were defined so that 100 pmol of N-glycans were added per well based on the number of N-glycans they have (GnGnbi-PA, 1; transferrin, 2; α1AGP, 5; fibrinogen, 6; haptoglobin, 4; α1-antitrypsin, 3; and IgG, 2). After the reaction, 15 μl of the GnT-IV buffer (25 mM Mes [pH 7.7], 0.5% [v/v] Triton X-100, 5 mg/ml BSA, and 7.5 mM MnCl2) was added to each well followed by adding 25 μl of the UDP detection reagent (supplied in the kit). Then, the plate was incubated in the dark for 1 h at room temperature. Chemiluminescence signals were measured using a SYNERGY H1 microplate reader (BioTek). The duplicate measurements for each sample were performed.
Analysis of BACE1 glycans by Western blotting
HEK293 DKO cells were cultured on 6-cm dishes and transfected with the plasmids (see “Plasmid transfection” section) to obtain recombinant soluble BACE1 and its mutants. After 4 h, the medium was replaced with Opti-MEM I. After 48 h, the media containing soluble His-tagged BACE1 and its mutants were incubated with Ni2+-Sepharose 6 Fast Flow (GE Healthcare) for 3 h or more at 4 °C with gentle rotation. After washing the beads three times with 10 mM phosphate buffer (pH 7.4) containing 20 mM imidazole and 0.5 M NaCl, the recombinant proteins were eluted with 10 mM phosphate buffer (pH 7.4) containing 0.5 M NaCl and 0.5 M imidazole. The eluates were mixed with Laemmli 5× SDS sample buffer and boiled at 95 °C for 5 min, followed by SDS-PAGE analysis. For neuraminidase treatment, the protein-bound beads were incubated with 5 U/μl of Clostridium perfringens neuraminidase (New England BioLabs) in TBS at 37 °C for 2 h. For PNGaseF treatment, the beads were first incubated with TBS containing 0.5% SDS at 95 °C for 5 min. After diluting with four volumes of TBS containing Nonidet P-40 (0.5% at final concentration), the beads were incubated with 40 mU/μl PNGaseF of Flavobacterium meningosepticum (New England Biolabs) at 37 °C for 2 h.
Structural representation
Atomic structure of mouse GnT-IVa lectin domain in unliganded form (Protein Data Bank code: 7VMT) was recently determined (22). The docking model of GlcNAc complex was built by using HADDOCK Web server (45). The structural figure was depicted by PyMOL (The PyMOL Molecular Graphics System, version 2.0; Schrödinger, LLC).
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8 software (GraphPad Software, Inc). Unpaired t test was used for comparison between two groups, and Tukey’s test or Holm–Sidak’s test was used for comparison between three or more groups.
Data availability
Glycomic raw data for glycan-structure analysis have been deposited to the GlycoPOST (announced ID: GPST000277). All the other data are contained within the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Ms Chizuko Yonekawa, Yuko Tokoro, and Emiko Mori (Gifu University) for their technical help. We also thank Dr Bart De Strooper (University College London) for providing the anti-BACE1 antibody. We thank Edanz (https://jp.edanz.com/ac) for editing a draft of this article.
Author contributions
Y. K. conceptualization; N. O., Masamichi Nagae, Miyako Nakano, and T. H. investigation; N. O. writing–original draft; Y. K. writing–review & editing; Y. K. supervision; Y. K. project administration; Y. K. funding acquisition.
Funding and additional information
This work was partially supported by a Grant-in-Aid for Scientific Research (B) to Y. K. (grant no.: 20H03207), a Leading Initiative for Excellent Young Researchers project (to Y. K.) from the Japan Society for the Promotion of Science, a CREST grant (grant no.: 18070267; to Y. K.) and a FOREST (Fusion Oriented REsearch for disruptive Science and Technology) grant (grant no.: JPMJFR215Z; to Y. K.) from Japan Science and Technology Agency, and a grant from the Takeda Science Foundation to Y. K.
Edited by Chris Whitfield
Supporting information
References
- 1.Varki A. Biological roles of glycans. Glycobiology. 2017;27:3. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moremen K.W., Tiemeyer M., Nairn A.V. Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao Y.-Y., Takahashi M., Gu J.-G., Miyoshi E., Matsumoto A., Kitazume S., et al. Functional roles of N-glycans in cell signaling and cell adhesion in cancer. Cancer Sci. 2008;99:1304–1310. doi: 10.1111/j.1349-7006.2008.00839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mereiter S., Balmaña M., Campos D., Gomes J., Reis C.A. Glycosylation in the era of cancer-targeted therapy: where are we heading? Cancer Cell. 2019;36:6–16. doi: 10.1016/j.ccell.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Wang X., Inoue S., Gu J., Miyoshi E., Noda K., Li W., et al. Dysregulation of TGF-beta1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15791–15796. doi: 10.1073/pnas.0507375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohtsubo K., Takamatsu S., Minowa M.T., Yoshida A., Takeuchi M., Marth J.D. Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell. 2005;123:1307–1321. doi: 10.1016/j.cell.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 7.Kizuka Y., Kitazume S., Fujinawa R., Saito T., Iwata N., Saido T.C., et al. An aberrant sugar modification of BACE1 blocks its lysosomal targeting in Alzheimer’s disease. EMBO Mol. Med. 2015;7:175–189. doi: 10.15252/emmm.201404438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanley P., Taniguchi N., Aebi M. In: Essentials of Glycobiology. Varki A., Cummings R.D., Esko J.D., Hart G.W., editors. Cold Spring Harbor; New York, NY: 2015. N-Glycans. [PubMed] [Google Scholar]
- 9.Cherepanova N., Shrimal S., Gilmore R. N-linked glycosylation and homeostasis of the endoplasmic reticulum. Curr. Opin. Cell Biol. 2016;41:57. doi: 10.1016/j.ceb.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brockhausen I., Narasimhan S., Schachter H. The biosynthesis of highly branched N-glycans: studies on the sequential pathway and functional role of N-acetylglucosaminyltransferases I, II, III, IV, V and VI. Biochimie. 1988;70:1521–1533. doi: 10.1016/0300-9084(88)90289-1. [DOI] [PubMed] [Google Scholar]
- 11.Boscher C., Dennis J.W., Nabi I.R. Glycosylation, galectins and cellular signaling. Curr. Opin. Cell Biol. 2011;23:383–392. doi: 10.1016/j.ceb.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Kizuka Y., Taniguchi N. Enzymes for N-glycan branching and their genetic and nongenetic regulation in cancer. Biomolecules. 2016;6:25. doi: 10.3390/biom6020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagae M., Yamaguchi Y., Taniguchi N., Kizuka Y. 3D structure and function of glycosyltransferases involved in N-glycan maturation. Int. J. Mol. Sci. 2020;21:437. doi: 10.3390/ijms21020437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minowa M.T., Oguri S., Yoshida A., Hara T., Iwamatsu A., Ikenaga H., et al. cDNA cloning and expression of bovine UDP-N-acetylglucosamine: alpha1, 3-D-mannoside beta1,4-N-acetylglucosaminyltransferase IV. J. Biol. Chem. 1998;273:11556–11562. doi: 10.1074/jbc.273.19.11556. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida A., Minowa M.T., Takamatsu S., Hara T., Ikenaga H., Takeuchi M.T. A novel second isoenzyme of the human UDP-N-acetylglucosamine:alpha1,3-D-mannoside beta1,4-N-acetylglucosaminyltransferase family: cDNA cloning, expression, and chromosomal assignment. Glycoconj. J. 1998;15:1115–1123. doi: 10.1023/a:1006951519522. [DOI] [PubMed] [Google Scholar]
- 16.Sakamoto Y., Taguchi T., Honke K., Korekane H., Watanabe H., Tano Y., et al. Molecular cloning and expression of cDNA encoding chicken UDP-N-acetyl-D-glucosamine (GlcNAc): GlcNAcbeta 1-6(GlcNAcbeta 1-2)- manalpha 1-R[GlcNAc to man]beta 1,4N-acetylglucosaminyltransferase VI. J. Biol. Chem. 2000;275:36029–36034. doi: 10.1074/jbc.M005860200. [DOI] [PubMed] [Google Scholar]
- 17.Huang H.H., Hassinen A., Sundaram S., Spiess A.N., Kellokumpu S., Stanley P. GnT1IP-L specifically inhibits MGAT1 in the Golgi via its luminal domain. Elife. 2015;4 doi: 10.7554/eLife.08916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oguri S., Yoshida A., Minowa M.T., Takeuchi M. Kinetic properties and substrate specificities of two recombinant human N-acetylglucosaminyltransferase-IV isozymes. Glycoconj. J. 2006;23:473–480. doi: 10.1007/s10719-006-6216-3. [DOI] [PubMed] [Google Scholar]
- 19.Taguchi T., Ogawa T., Inoue S., Inoue Y., Sakamoto Y., Korekane H., et al. Purification and characterization of UDP-GlcNAc: GlcNAcbeta 1-6(GlcNAcbeta 1-2)Manalpha 1-R [GlcNAc to Man]-beta 1, 4-N-acetylglucosaminyltransferase VI from hen oviduct. J. Biol. Chem. 2000;275:32598–32602. doi: 10.1074/jbc.M004673200. [DOI] [PubMed] [Google Scholar]
- 20.Takamatsu S., Antonopoulos A., Ohtsubo K., Ditto D., Chiba Y., Le D.T., et al. Physiological and glycomic characterization of N-acetylglucosaminyltransferase-IVa and -IVb double deficient mice. Glycobiology. 2010;20:485–497. doi: 10.1093/glycob/cwp200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohtsubo K., Chen M.Z., Olefsky J.M., Marth J.D. Pathway to diabetes through attenuation of pancreatic beta cell glycosylation and glucose transport. Nat. Med. 2011;17:1067–1076. doi: 10.1038/nm.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagae M., Hirata T., Tateno H., Sushil K.M., Manabe N., Osada N., et al. Discovery of a lectin domain that regulates enzyme activity in N-acetylglucosaminyltransferase-IVa (MGAT4A) Commun. Biol. 2022;5 doi: 10.1038/s42003-022-03661-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crowley J.F., Goldstein I.J., Arnarp J., Lönngren J. Carbohydrate binding studies on the lectin from Datura stramonium seeds. Arch. Biochem. Biophys. 1984;231:524–533. doi: 10.1016/0003-9861(84)90417-x. [DOI] [PubMed] [Google Scholar]
- 24.Takamatsu S., Korekane H., Ohtsubo K., Oguri S., Park J.Y., Matsumoto A., et al. N-Acetylglucosaminyltransferase (GnT) assays using fluorescent oligosaccharide acceptor substrates: GnT-III, IV, V, and IX (GnT-Vb) Methods Mol. Biol. 2013;1022:283–298. doi: 10.1007/978-1-62703-465-4_21. [DOI] [PubMed] [Google Scholar]
- 25.Hirata T., Nagae M., Osuka R.F., Mishra S.K., Yamada M., Kizuka Y. Recognition of glycan and protein substrates by N-acetylglucosaminyltransferase-V. Biochim. Biophys. Acta Gen. Subj. 2020;1864:129726. doi: 10.1016/j.bbagen.2020.129726. [DOI] [PubMed] [Google Scholar]
- 26.Clerc F., Reiding K.R., Jansen B.C., Kammeijer G.S.M., Bondt A., Wuhrer M. Human plasma protein N-glycosylation. Glycoconj. J. 2016;33:309–343. doi: 10.1007/s10719-015-9626-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibuya N., Tazaki K., Song Z.W., Tarr G.E., Goldstein I.J., Peumans W.J. A comparative study of bark lectins from three elderberry (Sambucus) species. J. Biochem. 1989;106:1098–1103. doi: 10.1093/oxfordjournals.jbchem.a122972. [DOI] [PubMed] [Google Scholar]
- 28.Bhattacharyya L., Haraldsson M., Brewer C.F. Precipitation of galactose-specific lectins by complex-type oligosaccharides and glycopeptides: studies with lectins from Ricinus communis (agglutinin I), Erythrina indica, Erythrina arborescens, Abrus precatorius (agglutinin), and Glycine max (soybean) Biochemistry. 1988;27:1034–1041. doi: 10.1021/bi00403a028. [DOI] [PubMed] [Google Scholar]
- 29.Kizuka Y., Nakano M., Kitazume S., Saito T., Saido T.C., Taniguchi N. Bisecting GlcNAc modification stabilizes BACE1 protein under oxidative stress conditions. Biochem. J. 2016;473:21–30. doi: 10.1042/BJ20150607. [DOI] [PubMed] [Google Scholar]
- 30.Kong Y., Joshi Hi.J., Schjoldager K.T.B.G., Madsen T.D., Gerken T.A., Vester C.M.B., et al. Probing polypeptide GalNAc-transferase isoform substrate specificities by in vitro analysis. Glycobiology. 2015;25:55–65. doi: 10.1093/glycob/cwu089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holdener B.C., Haltiwanger R.S. Protein O-fucosylation: structure and function. Curr. Opin. Struct. Biol. 2019;56:78–86. doi: 10.1016/j.sbi.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shrimal S., Cherepanova N.A., Gilmore R. Cotranslational and posttranslocational N-glycosylation of proteins in the endoplasmic reticulum. Semin. Cell Dev. Biol. 2015;41:71–78. doi: 10.1016/j.semcdb.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hobohm L., Koudelka T., Bahr F.H., Truberg J., Kapell S., Schacht S.S., et al. N-terminome analyses underscore the prevalence of SPPL3-mediated intramembrane proteolysis among Golgi-resident enzymes and its role in Golgi enzyme secretion. Cell. Mol. Life Sci. 2022;79:185. doi: 10.1007/s00018-022-04163-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khoder A.F., Sosicka P., Escriva C.M., Hassinen A., Glumoff T., Olczak M., et al. N-acetylglucosaminyltransferases and nucleotide sugar transporters form multi-enzyme-multi-transporter assemblies in Golgi membranes in vivo. Cell. Mol. Life Sci. 2019;76:1821–1832. doi: 10.1007/s00018-019-03032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubota T., Shiba T., Sugioka S., Furukawa S., Sawaki H., Kato R., et al. Structural basis of carbohydrate transfer activity by human UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10) J. Mol. Biol. 2006;359:708–727. doi: 10.1016/j.jmb.2006.03.061. [DOI] [PubMed] [Google Scholar]
- 36.Fritz T.A., Hurley J.H., Trinh L.B., Shiloach J., Tabak L.A. The beginnings of mucin biosynthesis: the crystal structure of UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase-T1. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15307–15312. doi: 10.1073/pnas.0405657101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuwabara N., Manya H., Yamada T., Tateno H., Kanagawa M., Kobayashi K., et al. Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of α-dystroglycan. Proc. Natl. Acad. Sci. U. S. A. 2016;113:9280–9285. doi: 10.1073/pnas.1525545113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osuka R.F., Hirata T., Nagae M., Nakano M., Shibata H., Okamoto R., et al. N-acetylglucosaminyltransferase-V requires a specific non-catalytic luminal domain for its activity toward glycoprotein substrates. J. Biol. Chem. 2022;298:101666. doi: 10.1016/j.jbc.2022.101666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y.F., Aoki K., Akase S., Ishihara M., Liu Y.S., Yang G., et al. Global mapping of glycosylation pathways in human-derived cells. Dev. Cell. 2021;56:1195–1209.e7. doi: 10.1016/j.devcel.2021.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou L., Chávez-Gutiérrez L., Bockstael K., Sannerud R., Annaert W., May P.C., et al. Inhibition of beta-secretase in vivo via antibody binding to unique loops (D and F) of BACE1. J. Biol. Chem. 2011;286:8677–8687. doi: 10.1074/jbc.M110.194860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakano M., Saldanha R., Göbel A., Kavallaris M., Packer N.H. Identification of glycan structure alterations on cell membrane proteins in desoxyepothilone B resistant leukemia cells. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M111.009001. M111.009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pabst M., Bondili J.S., Stadlmann J., Mach L., Altmann F. Mass + retention time = structure: a strategy for the analysis of N-glycans by carbon LC-ESI-MS and its application to fibrin N-glycans. Anal. Chem. 2007;79:5051–5057. doi: 10.1021/ac070363i. [DOI] [PubMed] [Google Scholar]
- 43.Green E.D., Adelt G., Baenziger J.U., Wilson S., van H.H. The asparagine-linked oligosaccharides on bovine fetuin. Structural analysis of N-glycanase-released oligosaccharides by 500-megahertz 1H NMR spectroscopy. J. Biol. Chem. 1988;263:18253–18268. [PubMed] [Google Scholar]
- 44.Vibhute A.M., Tanaka H., Mishra S.K., Osuka R.F., Nagae M., Yonekawa C., et al. Structure-based design of UDP-GlcNAc analogs as candidate GnT-V inhibitors. Biochim. Biophys. Acta Gen. Subj. 2022;1866:130118. doi: 10.1016/j.bbagen.2022.130118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Z.G.C.P., Rodrigues J.P.G.L.M., Trellet M., Schmitz C., Kastritis P.L., Karaca E., et al. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Glycomic raw data for glycan-structure analysis have been deposited to the GlycoPOST (announced ID: GPST000277). All the other data are contained within the article.