Abstract
Clear cell renal cell carcinoma (ccRCC) originates from renal tubular epithelial cells and is the most common pathological renal cell carcinoma type with the worst prognosis. The relationship between the expression, prognosis and mechanism of ccRCC and the E2F family remains challenging. In the present study, RNA sequencing and clinical data of ccRCC from The Cancer Genome Atlas and two datasets, GSE36895 and GSE53757, from the Gene Expression Omnibus were used to identify the role of the E2F family in ccRCC. A total of 10 groups of tumor tissues and paired-normal tissues from patients with ccRCC were verified by reverse transcription-quantitative PCR. the expression, tumor grade and stage, prognosis and regulatory mechanism of the E2F family in ccRCC were analyzed. It was found that the expression levels of E2F1 to 4 and 6 to 8 were higher in ccRCC tissues than in normal tissues, whereas the expression level of E2F5 was lower in the former than in the latter. The expression levels of E2F1 to 8 were correlated with tumor stage and grade. Low expression of E2F1 to 5 and 7 to 8 was significantly associated with longer overall survival, disease-specific survival and progression-free survival times. The data revealed that the E2F family rarely has genetic mutations. The expression of E2F1, E2F2, E2F5, E2F7 and E2F8 was significantly correlated with DNA methylation, and E2F1 to E2F7 were significantly correlated with copy number and the data showed that the expression of E2Fs was significantly correlated with the cell cycle. The results of the present study suggested that E2F family genes may be potential targets for ccRCC molecular diagnosis and targeted therapy.
Keywords: clear cell renal cell carcinoma, E2F family, expression, prognosis, cell cycle
Introduction
Renal cell carcinoma (RCC) is a common malignant tumor in the human genitourinary system that includes numerous different pathological subtypes (1,2). The most common subtype is clear cell renal cell carcinoma (ccRCC). ccRCC originates from renal tubular epithelial cells and accounts for ~60-85% of RCCs (3,4). The prevalence of ccRCC in men is higher than that in women, and most patients are over 60 years old. It is often asymptomatic in the early stage or only has vague systemic symptoms such as fever and fatigue. The typical clinical symptoms are hematuria, pain and a palpable mass in the kidney area. It affects either kidney at an equal rate. At present, the treatment methods are limited, mainly radical nephrectomy, but relapse and metastasis easily occur after surgery and the fatality rate is high (5). The etiology of ccRCC remains unclear and possible related factors include genetics, smoking, obesity, hypertension and antihypertensive drug therapy (6). Therefore, it is very important to explore the potential biomarkers and therapeutic targets of ccRCC.
The E2F family encodes extremely important nuclear transcription factors involved in regulating the cell cycle (7,8). It was first discovered by Kovesdi et al (9) in 1986 during studies of adenovirus. There are numerous members of the E2F family. The ones that have been discovered include E2F1 to E2F8. According to the protein structure and function and transcription characteristics, they are divided into transcription promotion factors (E2F1 to E2F3) and transcription suppressors (E2F4 to E2F8) (10). Clinical studies have found that E2F family proteins are closely related to the occurrence, development, proliferation and apoptosis of gastric, lung, liver, esophageal, prostate, bladder and ovarian cancer and other malignant tumors (7,11). In addition, E2F family proteins exhibit complex and diverse biological functions in different tumors and their expression levels are not consistent in different tumors (11). However, the expression pattern of E2F family proteins in ccRCC and their relationship with the prognosis remains unclear.
To the best of our knowledge, bioinformatics analysis has yet to be applied to explore the role of E2F family in ccRCC. In the present study, RNA sequencing (RNA-Seq) data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) were downloaded to explore the expression characteristics of E2F family proteins in ccRCC and their relationship with prognosis. Finally, the results of bioinformatics analysis we reverified with clinical samples from surgical operations. The present study may provide a new understanding of E2F family proteins in ccRCC and help to interpret the mechanisms underlying their functions.
Materials and methods
Raw data
RNA-Seq and clinical data (from 530 tumor tissues and 72 normal tissues) of ccRCC from TCGA were downloaded from https://portal.gdc.cancer.gov/. The RNA-Seq data were reported as fragments per kilobase million. The datasets GSE36895 (12) (containing 72 tumor tissues and 72 normal tissues) and GSE53757 (13) (containing 29 tumor tissues and 23 normal tissues) were downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo).
Reverse transcription-quantitative (RT-q) PCR
A total of 10 groups of tumor tissues and paired-normal tissues were obtained from 10 patients who underwent radical resection at The Affiliated Suqian First People's Hospital of Nanjing Medical University between January 2020 and August 2021. The patients were diagnosed as ccRCC by imaging and pathological examination, and did not receive chemotherapy or radiotherapy before operation. All experimental procedures were approved (approval no. 2018-SL-0026) by the Ethics Committee of The Affiliated Suqian First People's Hospital of Nanjing Medical University (Suqian, China). Written informed consent was provided by all patients prior to the study. All patients (age range, 55–68 years; median age, 62 years; seven men and three women) were diagnosed with ccRCC by laboratory examination and imaging examination. The clinical information of the patients is provided in Table SI. The mRNA expression of the E2Fs was examined using RT-qPCR. Total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) from the tissues. Total RNA was converted into cDNA, and quantitated using a Fastking One Step Reverse Transcription and Fluorescence Quantitative Kit (Tiangen Biotech Co., Ltd.) as a fluorophore according to the manufacturer's protocol. The primers were designed by online tool ‘primerBank’ (https://pga.mgh.harvard.edu/primerbank). β-actin was used as the internal reference gene. The primer sequences of E2Fs are presented in Table SII. Expression levels of mRNAs relative to β-actin were determined using the 2-ΔΔCq method (14).
Definitions of clinical survival and recurrence types
Raw counts of RNA-sequencing data (level 3) of ccRCC from TCGA were downloaded from https://portal.gdc.cancer.gov/. A total of 3 types of clinical survival and recurrence outcomes were selected in the present study: overall survival (OS), disease-specific survival (DSS), progression-free survival (PFS) and disease-free survival (DFS) (15). Survival analysis with the log-rank test was used to compare the survival difference between the normal and tumor groups. The hazard ratio (HR) was used to indicate the risk difference between the two groups.
Mutation and methylation analysis
The data were downloaded from TCGA (https://portal.gdc.cancer.gov/) and cBioPortal (https://www.cbioportal.org/datasets), copy number and mutation analysis were performed using cBioPortal (https://www.cbioportal.org/) (15). MeV software (http://projects/mev-tm4) was used to create heatmaps. Pearson's correlation analysis and mapping were performed using R 4.1.1 (https://www.r-project.org/). P<0.05 indicates a significant correlation.
Enrichment analysis
Gene enrichment analysis was performed to determine the correlation between E2F expression and the cell cycle. The data were downloaded from TCGA. Gene set enrichment analysis (https://www.gsea-msigdb.org/gsea) was used to perform gene enrichment analysis in three datasets, REACTOME, (Kyoto Encyclopedia of Genes and genomes (KEGG) and Gene Ontology (GO). A normalized enrichment score (NES)>0 means E2F expression is positively correlated with the cell cycle, and a false discovery rate (FDR) <0.05 was considered to indicate a statistically significant difference.
Statistical analysis
The Mann-Whitney U test was performed using GraphPad Prism 9 software (GraphPad Software, Inc.) and R 4.1.1 was used to determine E2F expression between normal tissues and tumor tissues. Kaplan-Meier survival curves and log-rank tests were used to evaluate the effect of E2F expression on survival. The Kruskal-Wallis test was used to detect the differences between groups in normal tissue + stage/grade and stage/grade. P<0.05 was considered to indicate a statistically significant difference.
Results
E2Fs expression in patients with ccRCC
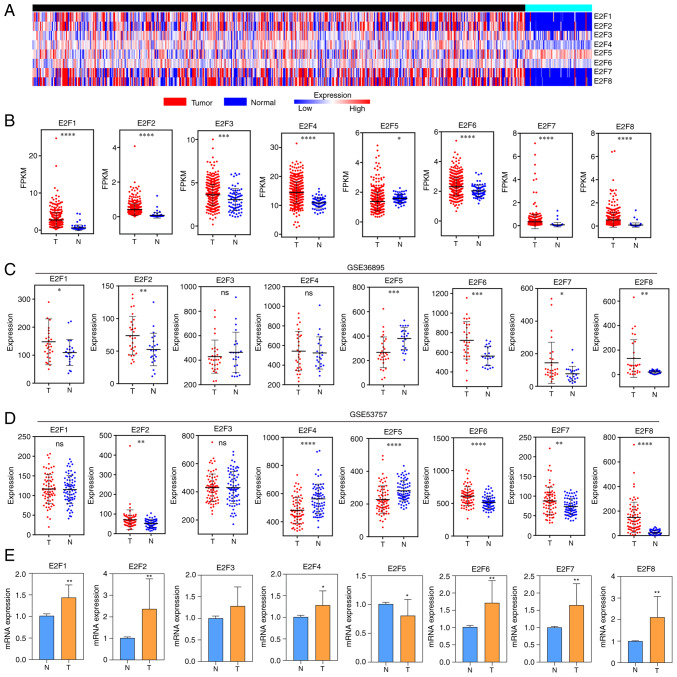
RNA-Seq data from 530 tumors and 72 normal tissue samples from the TCGA dataset were analyzed (Fig. 1A). In data of TCGA, compared with normal tissues, E2F1, E2F2, E2F3, E2F4, E2F6, E2F7 and E2F8 were overexpressed in cancer tissues, and E2F5 was expressed at low levels in cancer tissues (Fig. 1B). In dataset GSE36985, the expression levels of E2F1, E2F2, E2F6, E2F7 and E2F8 in tumor tissues were higher than those in normal tissues, and the expression of E2F5 was lower than that in normal tissues. There was no statistically significant difference between the two groups of E2F3 and E2F4 (Fig. 1C). In dataset GSE53757, the expression of E2F2, E2F6, E2F7 and E2F8 in tumor tissues was higher than that in normal tissues, and the expression of E2F4 and E2F5 was lower than that in normal tissues. There was no statistically significant difference between the two groups of E2F1 and E2F3 (Fig. 1D). In addition, the RT-qPCR verification results of clinical specimens collected from ccRCC surgery revealed that compared with normal tissues, E2F5 expression in cancer tissues was low, and the E2F1, E2F, E2F4, E2F6, E2F7 and E2F8 were overexpressed (Fig. 1E).
Figure 1.
Expression of the E2F family in ccRCC. (A) Heatmap displaying the expression of E2Fs in ccRCC using the total tumor (n=530) and normal (n=72) ccRCC data from TCGA. (B) Scatter plot displaying the expression of tumor tissues (n=530) and normal tissues (n=72) from TCGA. (C) Scatter plot displaying the expression of tumor tissues (n=72) and normal tissues (n=72) from dataset GSE36895. (D) Scatter plot displaying the expression of tumor tissues (n=29) and normal tissues (n=23) from dataset GSE53757. (E) Reverse transcription-quantitative PCR showing the expression of E2Fs in 10 paired ccRCC and normal samples. Data are presented as the mean ± SEM. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. ccRCC, clear cell renal cell carcinoma; TCGA, The Cancer Genome Atlas.
E2F family expression and pathological status of patients with ccRCC
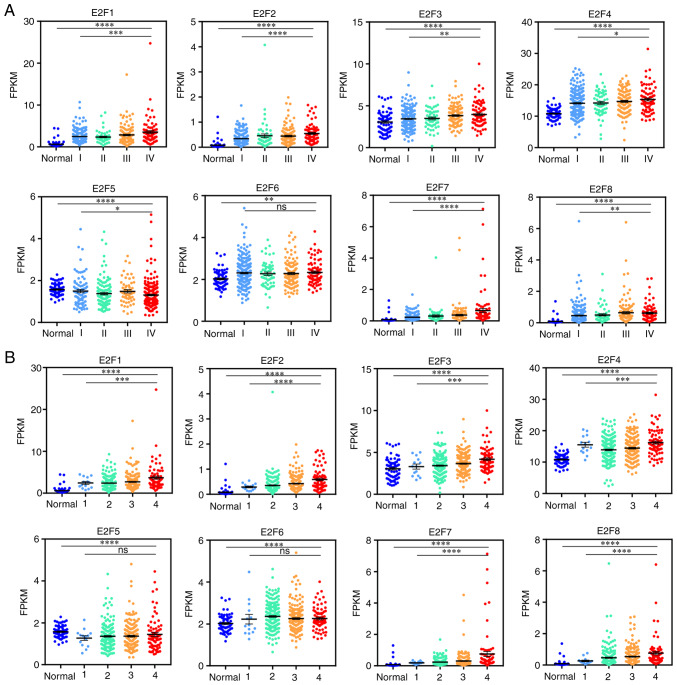
The relationship of E2F family expression with stage and grade was analyzed, respectively in TCGA data. The expression of E2Fs is significantly different in the staging and grading of patients with ccRCC. The higher the pathological stage was, the higher the expression of E2F1, E2F2, E2F3, E2F4, E2F6, E2F7 and E2F8 and the lower the expression of E2F5 (Fig. 2A). The results of pathological grading were similar to the results of staging (Fig. 2B).
Figure 2.
Relationship between E2F expression and pathological status of ccRCC. (A) Scatter plot displaying the expression of E2Fs in normal tissues and stages I–IV of ccRCC. (B) Scatter plot displaying the expression of E2Fs in normal tissues and grade 1–4 ccRCC. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. ccRCC, clear cell renal cell carcinoma.
Prognostic value of E2Fs in patients with ccRCC
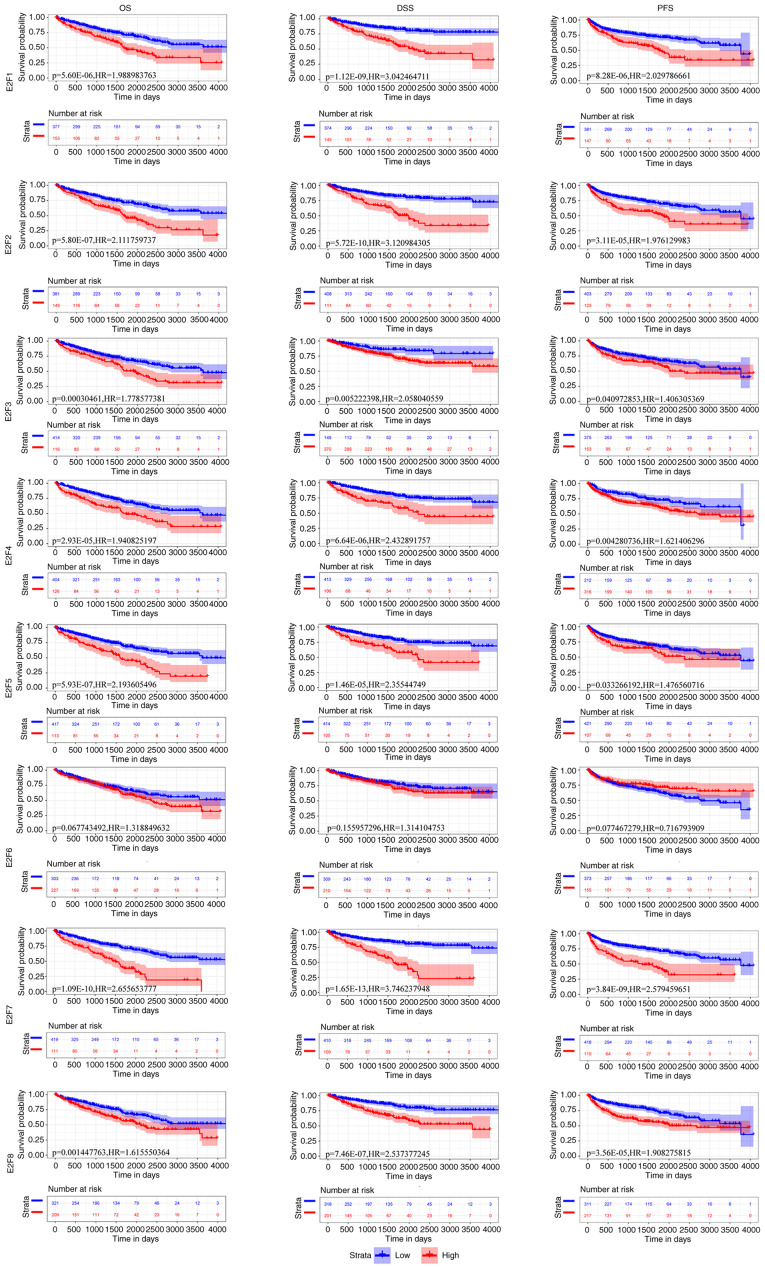
To evaluate the clinical significance of the expression of E2Fs in the survival of patients with ccRCC, E2F expression was assessed in 530 ccRCC clinical samples (≤4,000 days of follow-up) from TCGA for OS, DSS and PFS. Low expression of E2F1, E2F2, E2F3 E2F4, E2F5, E2F7 and E2F8 were significantly associated with a longer OS time (HR=1.989, P<0.0001; HR=2.111, P<0.0001; HR=1.779, P=0.000; HR=1.941, P<0.0001; HR=2.194, P<0.0001; HR=2.656, P<0.0001; HR=1.616, P=0.001), a longer DSS time (HR=3.042, P<0.0001; HR=3.121, P<0.0001; HR=2.058, P=0.005; HR=2.433, P<0.0001; HR=2.355, P<0.0001; HR=3.746, P<0.0001; HR=2.537, P<0.0001) and a longer PFS time (HR=2.030, P<0.0001; HR=1.976, P<0.0001; HR=1.406, P=0.041; HR=1.621, P=0.004; HR=1.477, P=0.033; HR=2.579, P<0.0001; HR=1.908, P<0.0001) in patients with ccRCC, respectively (Fig. 3). However, low expression of E2F6 was not associated with OS (HR=1.319; P=0.068), DSS (HR=1.314; P=0.156) or PFS (HR=0.717; P=0.077) in patients with ccRCC (Fig. 3). However, it was found that the expression of E2F1-E2F8 was not significantly associated with DFS. These results suggested that the mRNA expression levels of E2F1, E2F2, E2F3 E2F4, E2F5, E2F7 and E2F8 may be useful for the prediction of survival of patients with ccRCC.
Figure 3.
Expression of E2Fs was associated with a favorable prognosis in patients with ccRCC. Kaplan-Meier plots depicting the OS, DSS and PFS of patients with high and low expression of E2Fs. All patients with cervical squamous cell carcinoma were included in the TCGA database. OS, overall survival; DSS, disease-specific survival; PFS, progression-free survival; HR, hazard ratio.
Mechanism of E2Fs imbalance in patients with ccRCC
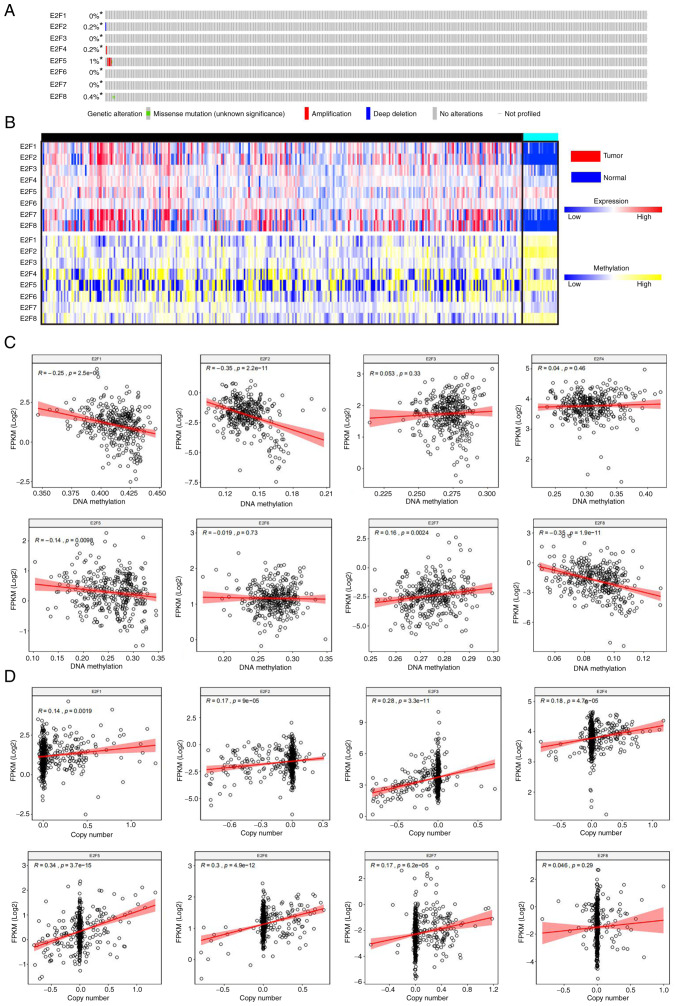
The mutation rates of E2F1-E2F8 were 0, 0.2, 0, 0.2, 1, 0, 0 and 0.4%, respectively (Fig. 4A). The expression of E2F1, E2F2, E2F5 and E2F8 was significantly negatively correlated with DNA methylation (r=−0.25, P<0.0001; r=−0.35, P<0.0001; r=−0.14, P<0.0001; r=−0.35, P<0.0001, respectively), and the expression of E2F7 was significantly positively correlated with DNA methylation (r=−0.16, P=0.0024). The expression of E2F3, E2F4 and E2F6 was not significantly correlated with DNA methylation (r=0.053, P=0.033; r=0.04, P=0.46; r=−0.019, P=0.73) (Fig. 4B and C). The expression of E2F1-E2F7 was significantly positively correlated with copy number (r=0.14, P=0.002; r=0.17, P<0.0001; r=0.28, P<0.0001; r=0.18, P<0.0001; r=0.34, P<0.0001; r=0.30, P<0.0001; r=0.17, P<0.0001, respectively), and the expression of E2F8 was not significantly correlated with copy number (r=0.046, P=0.29) (Fig. 4B and D). The results revealed that the mutation rate of E2Fs in patients with ccRCC is low. E2F1, E2F2 and E2F8 expression may be increased due to hypomethylation in tumors, and E2F4, E2F6 and E2F7 expression may be increased due to an increased copy number in tumors.
Figure 4.
Mechanism of E2Fs dysregulation in patients with ccRCC. (A) Mutation rate of the E2F family in patients with ccRCC. (B) Heatmap displaying E2F expression levels and DNA methylation levels in ccRCC tissues and normal tissues. (C) Association between E2F expression levels and DNA methylation levels in ccRCC tissues. (D) Association between E2F expression level and copy number in ccRCC tissues. ccRCC, clear cell renal cell carcinoma.
E2Fs expression with cell cycle
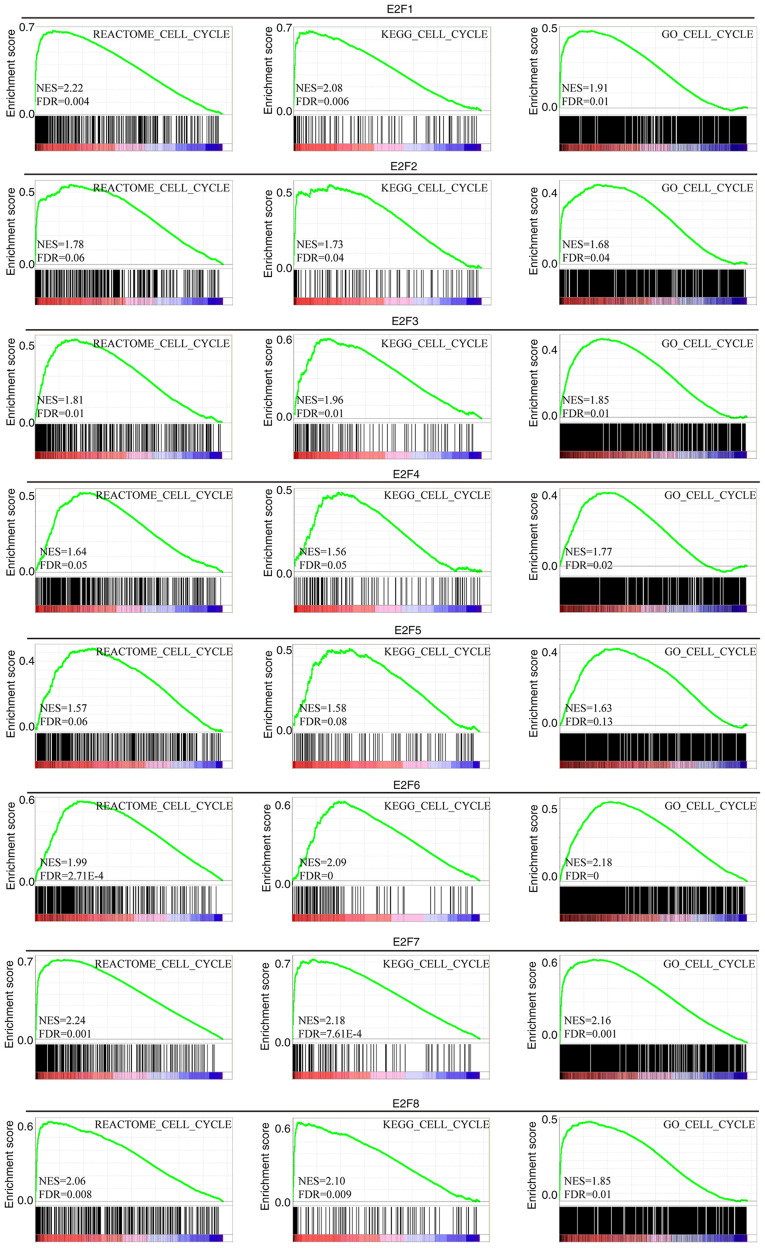
The results demonstrated that the expression of E2F1, E2F3, E2F6, E2F7 and E2F8 in REACTOME was significantly positively correlated with the cell cycle (NES=2.22, FDR=0.004; NES=1.81, FDR=0.01; NES=1.99, FDR<0.001; NES=2.24, FDR=0.001; NES=2.06, FDR=0.008) (Fig. 5). The expression of E2F1, E2F2, E2F3, E2F6, E2F7 and E2F8 in KEGG was significantly positively correlated with the cell cycle (NES=2.08, FDR=0.006; NES=1.73, FDR=0.04; NES=1.96, FDR=0.01; NES=2.09, FDR=0.00; NES=2.18, FDR<0.0001; NES=2.10, FDR=0.009) (Fig. 5). The expression of E2F1, E2F2, E2F3, E2F4, E2F6, E2F7 and E2F8 in GO was significantly positively correlated with the cell cycle (NES=1.91, FDR=0.01; NES=1.68, FDR=0.04; NES=1.85, FDR=0.01; NES=1.77, FDR=0.02; NES=2.18, FDR=0.00; NES=2.16, FDR=0.001; NES=1.85, FDR=0.01) (Fig. 5).
Figure 5.
Association between E2F expression and the cell cycle. Gene set enrichment analysis showing cell cycle signature enrichment analysis in three datasets: REACTOME, KEGG and GO. KEGG, Kyoto Encyclopedia of Genes and genomes; GO, Gene Ontology.
Discussion
Studies have shown that targeted therapy can significantly improve the survival rate of patients with metastatic ccRCC (16). However, ccRCC currently has fewer therapeutic targets in clinical practice, and it is necessary to identify and explore more therapeutic targets to provide a reference for clinical treatment.
E2F is a group of genes encoding transcription factors in higher eukaryotes. According to their different functions, they can be divided into transcription activators and transcription suppressors. To date, 8 E2F family protein members have been identified in mammals, namely, E2F1, E2F2, E2F3, E2F4, E2F5, E2F6, E2F7 and E2F8 (7,11,17). Among them, E2F1 is the only transcription factor among the eight protein family members that has the dual functions of mediating apoptosis and regulating cell proliferation. E2F1 and E2F3a are transcriptional activators in the E2F family of proteins that bind to target genes and participate in the regulation of the cell cycle. E2F4 can participate in the regulation of normal cell proliferation, differentiation, apoptosis and other physiological processes. E2F3 to E2F5 can act as transcription inhibitors after binding to pocket proteins related to retinoblastoma proteins (11). E2F6 inhibits DNA damage-induced apoptosis (8). E2F7 and E2F8 are new members of the E2F family of proteins discovered in recent years. They are atypical family factors and can work in concert with hypoxia-inducible factors to jointly regulate the transcription of vascular endothelial growth factor. E2Fs have been reported to have roles in a variety of cancer types, such as breast (18), liver (19) and gastric cancer (20), since they can regulate numerous cellular functions related to cell cycle progression. Although certain E2F family members have been confirmed to play promising roles in ccRCC, the distinct roles of E2Fs in the development, progression and metastasis of ccRCC remain to be elucidated. In the present study, the expression, mutation and prognostic values of different E2Fs in patients with ccRCC were analyzed.
E2F1 is the most frequently studied gene in the E2F family. E2F1 is an important transcription-promoting factor located on human chromosome 20q11, ~11 kb in size, mainly comprised of six introns and seven exons, and it can encode proteins with a size of more than 400 amino acids (10,21). E2F1 has a very obvious tissue specificity. It can form a heterodimer with retinoblastoma and bind to the corresponding DNA sequence, thereby enhancing or inhibiting the activity of E2F1. The transcription factor E2F1 can regulate biological processes such as the cell cycle, cell proliferation, cell apoptosis and cell differentiation (10). In the present study, E2F1 was highly expressed in cancer tissues and its high expression was closely related to a worse tumor grade and staging and a poor prognosis. In ccRCC, the expression of E2F1 is also significantly positively correlated with the cell cycle.
E2F2 and E2F3 are involved in regulation of the cell cycle and are highly expressed in breast cancer tissues (18). At present, little is known about the expression and role of E2F2 in ccRCC. In the present study, it was revealed that the expression of E2F2 in ccRCC tissues is higher than that in normal tissues, and this expression is significantly related to the tumor stage and grade of patients with ccRCC. In addition, in all patients with ccRCC, high expression of E2F2 was significantly associated with a poor OS, DSS and PFS, which appeared to be consistent with the role of E2F2 as an oncogene. Notably, high expression of E2F3 in cancer tissues was observed in TCGA, but this phenomenon was not identified in the two GEO datasets or the clinical sample validation.
The present study showed that E2F4 may have carcinogenic effects and the high expression of E2F4 is related to poor prognostic factors of ccRCC, such as high TNM stage and high grade. Furthermore, it has been reported that the expression of E2F4 in prostate (22) and breast cancer (18) is higher than that in normal tissues. Although E2F4 is traditionally classified as a cell cycle inhibitor, its pro-proliferation and anti-apoptotic activities have been confirmed in various human cell lines. Notably, gene enrichment analysis in the present study revealed that the positive correlation between E2F4 and the cell cycle was not significant, and the possible relationship of E2F4 with the early stages of carcinogenesis needs to be clarified.
E2F5 has different expression levels in different types of tumors. Studies have reported that E2F5 is overexpressed in glioblastoma (23) and prostate cancer (24). However, E2F5 was downregulated in MCF7 human breast cancer cells, significantly impairing cell proliferation, migration and invasion in vitro and increasing cell cycle arrest in the G0/G1 phase (25). In the present study, it was proved that the expression of E2F5 in ccRCC tissues is lower than that in normal tissues. According to previous studies, the expression of E2F6 in breast cancer tissues is lower than that in normal tissues, but this expression was not correlated with tumor stage in patients with breast cancer (18,26). In the present study, it was identified that the expression of E2F6 in ccRCC tissues was higher than that in normal tissues, and the expression level was significantly positively correlated with the copy number but not significantly correlated with the prognosis of patients with ccRCC. Studies have found that E2F7 and E2F8 are unique inhibitory genes that have a vital inhibitory effect on cell proliferation (27–29). However, the present study demonstrated that E2F7 and E2F8 are highly expressed in ccRCC tissues, suggesting that they may play different biological roles in different cell types.
The change of DNA methylation status is an important factor in tumorigenesis. This change includes the decrease of the overall methylation level of the genome and the abnormal increase of the local methylation level of CpG island, resulting in the instability of the genome and the low expression of tumor suppressor genes (30). It was revealed that E2F1, E2F2 and E2F8 expression may be increased due to hypomethylation in tumors, and E2F4, E2F6 and E2F7 expression may be increased due to an increased copy number in tumors. In addition, the present study showed that the expression of E2F family in ccRCC was significantly positively correlated with cell cycle.
In conclusion, except for the E2F3 and E2F5, all E2F family members are highly expressed in ccRCC tissues, and their expression is closely related to the pathological status and survival prognosis of patients with ccRCC, but different members of the family have differences in different tissue samples. The present findings suggested that E2F family genes may be potential targets for molecular diagnosis and targeted therapy of ccRCC, transcriptional E2F1-5, 7, and 8 were potential prognostic markers for the improvement of ccRCC survival and prognostic accuracy. Targeted therapy against single or combined E2F family proteins may improve the therapeutic effect and patient outcomes. It is expected that the findings of the present study will contribute to available knowledge, improve treatment designs, and enhance the accuracy of prognosis for patients with ccRCC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the Medical Research Project of Jiangsu Provincial Health Commission (grant no. Z2020074), the Six Talent Peaks Project in Jiangsu (grant no. WSN-342), the Jiangsu Provincial Medical Youth Talent (grant no. QNRC2016480), Suqian Guiding Science and Technology Project (grant no. Z2018174) and the Science and Technology Project of Suqian (grant no. S201720).
Availability of data and materials
The datasets generated and analyzed during the current study are available in TCGA (portal.gdc.cancer.gov) and GEO (ncbi.nlm.nih.gov/geo/).
Authors' contributions
ZL, YS and HG conceptualized and designed the research and performed bioinformatics analysis. YW and XYZ performed the experiments. JS, XY, XCZ, HL and XJY analyzed and interpreted the data. ZL and JS drafted and edited the manuscript. XY and XCZ supervised the project. ZL, YS and HG confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved (approval no. 2018-SL-0026) by the Ethics Committee of The Affiliated Suqian First People's Hospital of Nanjing Medical University (Suqian, China). Written informed consent was provided by all patients prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Gray RE, Harris GT. Renal cell carcinoma: Diagnosis and management. Am Fam Physician. 2019;99:179–184. [PubMed] [Google Scholar]
- 2.Linehan WM, Ricketts CJ. The cancer genome atlas of renal cell carcinoma: Findings and clinical implications. Nat Rev Urol. 2019;16:539–552. doi: 10.1038/s41585-019-0211-5. [DOI] [PubMed] [Google Scholar]
- 3.Makhov P, Joshi S, Ghatalia P, Kutikov A, Uzzo RG, Kolenko VM. Resistance to systemic therapies in clear cell renal cell carcinoma: Mechanisms and management strategies. Mol Cancer Ther. 2018;17:1355–1364. doi: 10.1158/1535-7163.MCT-17-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucarelli G, Loizzo D, Franzin R, Battaglia S, Ferro M, Cantiello F, Castellano G, Bettocchi C, Ditonno P, Battaglia M. Metabolomic insights into pathophysiological mechanisms and biomarker discovery in clear cell renal cell carcinoma. Expert Rev Mol Diagn. 2019;19:397–407. doi: 10.1080/14737159.2019.1607729. [DOI] [PubMed] [Google Scholar]
- 5.Tegos T, Tegos K, Dimitriadou A, Dimitriadis G. Current and emerging first-line systemic therapies in metastatic clear-cell renal cell carcinoma. J BUON. 2019;24:1340–1353. [PubMed] [Google Scholar]
- 6.Schodel J, Grampp S, Maher ER, Moch H, Ratcliffe PJ, Russo P, Mole DR. Hypoxia, hypoxia-inducible transcription factors, and renal cancer. Eur Urol. 2016;69:646–657. doi: 10.1016/j.eururo.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Wang X, Xu L, Zhang J, Cao H. Integrated analysis of the E2F transcription factors across cancer types. Oncol Rep. 2020;43:1133–1146. doi: 10.3892/or.2020.7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennycook BR, Vesela E, Peripolli S, Singh T, Barr AR, Bertoli C, de Bruin RAM. E2F-dependent transcription determines replication capacity and S phase length. Nat Commun. 2020;11:3503. doi: 10.1038/s41467-020-17146-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovesdi I, Reichel R, Nevins JR. Role of an adenovirus E2 promoter binding factor in E1A-mediated coordinate gene control. Proc Natl Acad Sci USA. 1987;84:2180–2184. doi: 10.1073/pnas.84.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ertosun MG, Hapil FZ, Nidai OO. E2F1 transcription factor and its impact on growth factor and cytokine signaling. Cytokine Growth Factor Rev. 2016;31:17–25. doi: 10.1016/j.cytogfr.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Kent LN, Leone G. The broken cycle: E2F dysfunction in cancer. Nat Rev Cancer. 2019;19:326–338. doi: 10.1038/s41568-019-0143-7. [DOI] [PubMed] [Google Scholar]
- 12.Zheng L, Dou X, Song H, Gao R, Tang X. TRPV1 acts as a tumor suppressor and is associated with immune cell infiltration in clear cell renal cell carcinoma: Evidence from integrated analysis. J Cancer. 2020;11:5678–5688. doi: 10.7150/jca.45918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang W, Zhang M, Wang Q, Gu D, Huang Z, Wang H, Xiang Y, Xia Q, Cui Z, Jin X. The SLC family are candidate diagnostic and prognostic biomarkers in clear cell renal cell carcinoma. Biomed Res Int. 2020;2020:1932948. doi: 10.1155/2020/1025178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Y, Fang YC, Li J. PD-L1 expression levels on tumor cells affect their immunosuppressive activity. Oncol Lett. 2019;18:5399–5407. doi: 10.3892/ol.2019.10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Yan C, Ye H. Overexpression of MUC16 predicts favourable prognosis in MUC16-mutant cervical cancer related to immune response. Exp Ther Med. 2020;20:1725–1733. doi: 10.3892/etm.2020.8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Saez O, Borau PG, Alonso-Gordoa T, Molina-Cerrillo J, Grande E. Targeting HIF-2 alpha in clear cell renal cell carcinoma: A promising therapeutic strategy. Crit Rev Oncol Hematol. 2017;111:117–123. doi: 10.1016/j.critrevonc.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 17.Attwooll C, Denchi EL, Helin K. The E2F family: Specific functions and overlapping interests. EMBO J. 2004;23:4709–4716. doi: 10.1038/sj.emboj.7600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun CC, Li SJ, Hu W, Zhang J, Zhou Q, Liu C, Li LL, Songyang YY, Zhang F, Chen ZL, et al. Comprehensive analysis of the expression and prognosis for E2Fs in human breast cancer. Mol Ther. 2019;27:1153–1165. doi: 10.1016/j.ymthe.2019.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Jiao Y, Li Y, Fu Z, Hou L, Chen Q, Cai Y, Jiang P, He M, Yang Z. OGDHL expression as a prognostic biomarker for liver cancer patients. Dis Markers. 2019;2019:9037131. doi: 10.1155/2019/9037131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei WY, Yan LH, Wang XT, Li L, Cao WL, Zhang XS, Zhan ZX, Yu H, Xie YB, Xiao Q. E2F-1 overexpression inhibits human gastric cancer MGC-803 cell growth in vivo. World J Gastroenterol. 2015;21:491–501. doi: 10.3748/wjg.v21.i2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang Z, Lin M, Li C, Liu H, Gong C. A comprehensive review of the roles of E2F1 in colon cancer. Am J Cancer Res. 2020;10:757–768. [PMC free article] [PubMed] [Google Scholar]
- 22.Yin H, Lowery M, Glass J. In prostate cancer C/EBPalpha promotes cell growth by the loss of interactions with CDK2, CDK4, and E2F and by activation of AKT. Prostate. 2009;69:1001–1016. doi: 10.1002/pros.20947. [DOI] [PubMed] [Google Scholar]
- 23.Fang DZ, Wang YP, Liu J, Hui XB, Wang XD, Chen X, Liu D. MicroRNA-129-3p suppresses tumor growth by targeting E2F5 in glioblastoma. Eur Rev Med Pharmacol Sci. 2018;22:1044–1050. doi: 10.26355/eurrev_201802_14387. [DOI] [PubMed] [Google Scholar]
- 24.Li SL, Sui Y, Sun J, Jiang TQ, Dong G. Identification of tumor suppressive role of microRNA-132 and its target gene in tumorigenesis of prostate cancer. Int J Mol Med. 2018;41:2429–2433. doi: 10.3892/ijmm.2018.3421. [DOI] [PubMed] [Google Scholar]
- 25.Xu H, Fei D, Zong S, Fan Z. MicroRNA-154 inhibits growth and invasion of breast cancer cells through targeting E2F5. Am J Transl Res. 2016;8:2620–2630. [PMC free article] [PubMed] [Google Scholar]
- 26.Lafta IJ. E2F6 is essential for cell viability in breast cancer cells during replication stress. Turk J Biol. 2019;43:293–304. doi: 10.3906/biy-1905-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno E, Toussaint MJM, van Essen SC, Bongiovanni L, van Liere EA, Koster MH, Yuan R, van Deursen JM, Westendorp B, de Bruin A. E2F7 is a potent inhibitor of liver tumor growth in adult mice. Hepatology. 2021;73:303–317. doi: 10.1002/hep.31259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kent LN, Rakijas JB, Pandit SK, Westendorp B, Chen HZ, Huntington JT, Tang X, Bae S, Srivastava A, Senapati S, et al. E2f8 mediates tumor suppression in postnatal liver development. J Clin Invest. 2016;126:2955–2969. doi: 10.1172/JCI85506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo L, Jones MC, Liu Y, Yv S, Zhu Y, Guo Y. Cross-cultural validation of the student nurse stress index scale: A descriptive survey targeting student nurses in China. J Affect Disord. 2019;251:31–38. doi: 10.1016/j.jad.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 30.Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in cancer and aging. Cancer Res. 2016;76:3446–3450. doi: 10.1158/0008-5472.CAN-15-3278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available in TCGA (portal.gdc.cancer.gov) and GEO (ncbi.nlm.nih.gov/geo/).