Significance
Viruses must make use of a small number of genes to evade host detection and hijack host systems that are critical to replicate the viral genome. Bacteriophage T7 and HIV-2 encode unrelated proteins that target related host proteins. Gp1.2 from T7 inhibits Dgt catalytic activity, while Vpx from HIV-2 targets SAMHD1 for proteasomal degradation. The fact that two unrelated viruses independently evolved means to limit the action of this class of enzymes highlights the importance of doing so for viral fitness. Our structural and biochemical data reveal how T7 Gp1.2 binds to and inactivates Dgt. We propose a new model for Gp1.2 function in vivo that overcomes longstanding limitations.
Keywords: cryo-EM, enzyme inhibition, allosteric regulation, NMR, dGTP triphosphohydrolase
Abstract
Levels of the cellular dNTPs, the direct precursors for DNA synthesis, are important for DNA replication fidelity, cell cycle control, and resistance against viruses. Escherichia coli encodes a dGTPase (2′-deoxyguanosine-5′-triphosphate [dGTP] triphosphohydrolase [dGTPase]; dgt gene, Dgt) that establishes the normal dGTP level required for accurate DNA replication but also plays a role in protecting E. coli against bacteriophage T7 infection by limiting the dGTP required for viral DNA replication. T7 counteracts Dgt using an inhibitor, the gene 1.2 product (Gp1.2). This interaction is a useful model system for studying the ongoing evolutionary virus/host “arms race.” We determined the structure of Gp1.2 by NMR spectroscopy and solved high-resolution cryo-electron microscopy structures of the Dgt–Gp1.2 complex also including either dGTP substrate or GTP coinhibitor bound in the active site. These structures reveal the mechanism by which Gp1.2 inhibits Dgt and indicate that Gp1.2 preferentially binds the GTP-bound form of Dgt. Biochemical assays reveal that the two inhibitors use different modes of inhibition and bind to Dgt in combination to yield enhanced inhibition. We thus propose an in vivo inhibition model wherein the Dgt–Gp1.2 complex equilibrates with GTP to fully inactivate Dgt, limiting dGTP hydrolysis and preserving the dGTP pool for viral DNA replication.
Escherichia coli encodes a 2′-deoxyguanosine-5′-triphosphate (dGTP) triphosphohydrolase (dGTPase; Dgt), encoded by the dgt gene, which catalyzes the hydrolysis of dGTP into 2′-deoxyguanosine (dG) and triphosphate (PPPi) (1). The enzyme is highly specific for dGTP, although it is able to hydrolyze other canonical and noncanonical dNTPs with lower efficiency (2). Deletion of the dgt gene was reported to lead to a twofold increase in the cellular dGTP pool (3). More recently, deletion of the dgt gene was shown to yield a mutator phenotype, whereby certain base pair substitution mutations occur at higher frequencies than in the wild-type strain (4). Such a mutator phenotype is consistent with other observations indicating that cellular dNTP levels are critical for controlling DNA polymerase fidelity and cellular mutation rates (5).
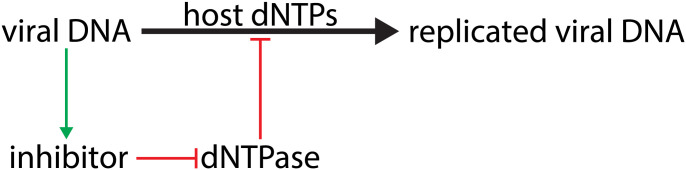
dNTP pool maintenance is also important for cells as a potential means to protect themselves against infection by invading DNAs, like viruses (Fig. 1). Viruses replicate their genomes using the dNTPs available within the cell, so host proteins like dNTP triphosphohydrolases (dNTPase) can limit infection by restricting the available dNTP pool (6, 7). In turn, some viruses have evolved dNTPase inhibitors to counteract this cellular defense. A relevant example of such virus/host interactions is between the E. coli Dgt and the 1.2 gene product (Gp1.2) of bacteriophage T7, a system that was initially discovered and investigated in detail by the Richardson group (3, 8, 9). An E. coli strain expressing elevated levels of Dgt due to an up-mutation in the dgt promoter (optA1) (3) can no longer be infected by T7 if it is missing a functional Gp1.2 (8). It was then discovered that Gp1.2 is a direct inhibitor of E. coli Dgt and that the inability of the mutant 1.2 phage to replicate in this strain was caused by its failure to maintain the necessary dGTP levels for its DNA replication (9). More precisely, phage T7 breaks down the host DNA following infection to secure an elevated supply of DNA precursors; but the loss of Gp1.2 leads to a 200-fold reduced dGTP level when replicating in the optA1 strain, leading to premature termination of phage DNA replication (8). Thus, dGTP limitation is a clear mechanism by which E. coli can defend itself against invading DNAs, while the phage has acquired a defense response in the form of a specific inhibitor. As such, this system is an interesting example of the established evolutionary “arms race” between hosts and viruses (10). A detailed understanding of such host versus invader interactions is also of practical interest, both for cases where viral infections need to be avoided and for other cases when viral action is desirable, like in phage therapies where phages are used to treat bacterial infections.
Fig. 1.
Host cells encode dNTPase enzymes, which can limit viral DNA replication by restricting the pool of DNA precursors (dNTPs). To counteract this effect, some viruses preserve the dNTP pool by encoding dNTPase inhibitors.
Recently, significant advances have been made in understanding the structure and function of E. coli Dgt. The enzyme is a hexameric homolog of the tetrameric human SAMHD1 (11), a dNTPase that was also implicated as a restriction factor against viruses such as HIV (6). While SAMHD1 activity is stimulated via allosteric dNTP binding sites, E. coli Dgt binds tightly to single-stranded DNA (12), which stimulates its hydrolysis activity (11). DNA binding to Dgt is also relevant during the interaction with Gp1.2, because Gp1.2 fails to fully inhibit Dgt that is already bound to nucleic acids (9). The Dgt–Gp1.2 complex can still bind to dGTP or GTP, but the ligands are not hydrolyzed (13). There remain questions about how Gp1.2 can inhibit Dgt in vivo when its inhibitory activity is limited by dGTP binding to Dgt (9). The dual binding of Gp1.2 to both the free and substrate-bound Dgt suggests a mixed-type inhibition (9). More recently, GTP was found to inhibit Dgt activity on its own, while a GTP-bound structure revealed GTP binding to the active site in place of dGTP (14). These data support a competitive mode of inhibition for GTP.
In the present work, we use NMR to obtain the structure of the Gp1.2 protein and describe high-resolution cryo-electron microscopy (cryo-EM) structures of the Dgt–Gp1.2 complex and of ternary complexes bound to either dGTP or GTP. The structures reveal how Gp1.2 recognizes and inhibits Dgt. Gp1.2 binds near the active site in a conformation that leaves the site partly open to solution so that nucleotide ligands can still equilibrate with the enzyme. When the ligands bind to the inhibited active site, they interact directly with the N terminus of Gp1.2 to form a stable ternary complex in a conformation that is refractory for catalysis. Importantly, enzymatic assays demonstrate that Gp1.2 and GTP combine to enhance Dgt inhibition, with the ternary complex remaining fully inhibited even in the presence of excess dGTP. Together, the structural and kinetic data provide insights into the function of Gp1.2 during infection of E. coli by T7 bacteriophage. We propose that Gp1.2 evolved to bind to the GTP-bound Dgt, with the Dgt–Gp1.2–GTP complex being the fully inhibited state of Dgt in vivo.
Results
NMR Structure of Gp1.2.
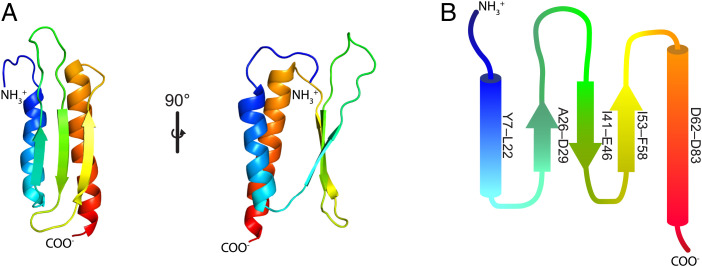
To characterize the structural and mechanistic basis for the Gp1.2 inhibition of Dgt, we began by determining the structure of Gp1.2 (Fig. 2 and SI Appendix, Table S1). The Gp1.2 sequence shows no significant homology to other proteins in structural databases, so it was not possible to predict its structure based on sequence. In view of the small size of Gp1.2 (∼10 kDa), NMR spectroscopy was chosen to determine its solution structure. Using triple resonance NMR methods (Materials and Methods), a structure was obtained revealing a somewhat barrel-shaped fold with a three-stranded antiparallel beta sheet covered on one side by two parallel alpha helices (Fig. 2).
Fig. 2.
NMR structure of the T7 bacteriophage protein Gp1.2 (PDB ID: 2MDP). (A) overview of Gp1.2 structure colored as a rainbow from the N terminus (blue) to the C terminus (red). The structure consists of a bundle of two alpha helices and a three-stranded antiparallel beta sheet. State 4 of the NMR ensemble is shown. Images were generated in PyMOL. (B) Secondary structure diagram colored as in A. The start and end residues are listed at the right of each alpha helix or beta strand.
A search for similar structures with the program DALI (15) revealed mammalian proteins of a similar size that are related to the double-stranded RNA binding domain fold (e.g., Protein Data Bank [PDB] ID: 2KHX) (16). The most significant DALI matches were E. coli proteins that inhibit ribosome activity, including the hibernation-promoting factor (HPF) and related YfiA. However, there was a significant difference between HPF and Gp1.2 with regard to the angle between the alpha helices (SI Appendix, Fig. S1, compare red to purple); the measurement of residual dipolar couplings confirms the precision of the interhelical angle (SI Appendix, Fig. S2B). This angle was useful for identifying the correct orientation of Gp1.2 when docking the NMR model into our cryo-EM maps.
Cryo-EM Structures of Dgt–Gp1.2 Complexes.
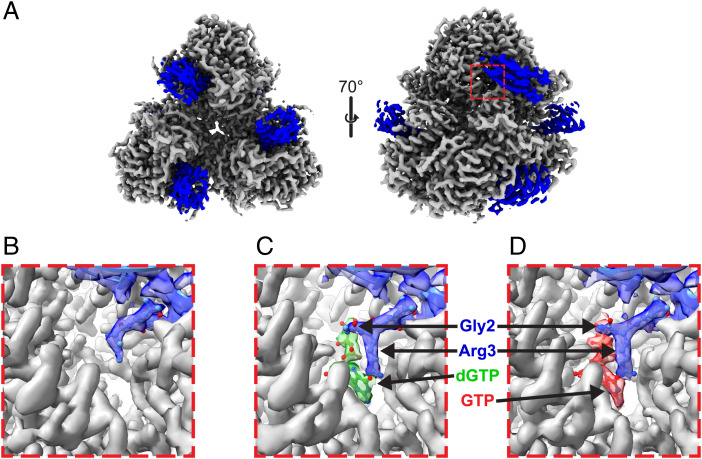
Given the large size of the Dgt hexamer (>300 kDa), we next turned to cryo-EM to determine structures of the Dgt–Gp1.2 complex (Fig. 3 and SI Appendix, Table S2). We produced three high-resolution structures: 1) a Dgt–Gp1.2 complex (2.85 Å), 2) a Dgt–Gp1.2–dGTP ternary complex (2.96 Å), and 3) a Dgt–Gp1.2–GTP ternary complex (2.54 Å). We are able to fully visualize each nucleotide ligand in the relevant structures. GTP was included in these studies because the Gp1.2-inhibited state of Dgt was reported, intriguingly, to be a tight GTP-binding protein (13). GTP was also recently recognized as an inhibitor of Dgt (14).
Fig. 3.
Cryo-EM maps of E. coli Dgt in complex with T7 bacteriophage protein Gp1.2 with or without (d)GTP ligands. (A) Dgt–Gp1.2 complex resolved to 2.85 Å. The Dgt hexamer is shown in gray, and the Gp1.2 molecules are in blue. Images were generated in ChimeraX. (B–D) Zoom-in views of the Dgt–Gp1.2 active-site density, highlighting the insertion of the Gp1.2 N terminus into the Dgt active site with or without ligands, as indicated. DeepEMhancer postprocessed maps are presented for the Dgt density, while the Gp1.2 and ligand densities were generated using the Phenix autosharpen tool. Images were generated in Chimera.
The density for Gp1.2 in our data is of good quality where it interacts with Dgt but is discontinuous in other parts of the protein that lack obvious contact and seem to be “hanging” out into the solvent (SI Appendix, Fig. S3). Therefore, it is possible that Gp1.2 binding to Dgt is conformationally flexible, resulting in the discontinuous density in these regions of our maps. The discontinuous regions of the Gp1.2 maps also correspond to the lowest local resolutions within the structures (SI Appendix, Fig. S4). Likewise, the loops that were disordered in the NMR structure (SI Appendix, Fig. S2) remain disordered in the Dgt-bound structures (SI Appendix, Fig. S3). Nevertheless, the Gp1.2 density is complete enough for us to dock the NMR structure into the maps. The backbone trace of the three-stranded beta sheet is mostly visible. Thus, by aligning the beta sheet, we were then able to use the angle between the Gp1.2 helices (see above) to identify the correct orientation of Gp1.2. The Gp1.2 structure remains largely unchanged after binding to Dgt (SI Appendix, Fig. S5).
The Gp1.2 helix α1 binds at the interface of two Dgt monomers (Fig. 3A and SI Appendix, Fig. S6), with the N terminus inserted directly into the active site of one (Fig. 3). While the N terminus of Gp1.2 was disordered in the NMR structure, it is well ordered when bound to Dgt in the cryo-EM maps (SI Appendix, Fig. S5). The density for Gp1.2 begins with G2, consistent with previous N-terminal protein sequencing results that showed G2 is the N terminus of Gp1.2 when expressed in E. coli (17). The genetically encoded N-terminal methionine is likely removed by methionine aminopeptidase (18). There is complete density for G2/R3 in the structures with ligands (Fig. 3 C and D) but only partial density for them in the structure lacking ligands (Fig. 3B). The structures, when processed without symmetry imposed, showed six Gp1.2 molecules bound per hexamer and one dGTP or GTP per active site (SI Appendix, Fig. S6). We subsequently processed the maps with D3 symmetry imposed, which improved resolution without significantly affecting the overall positions within the complex. Given the symmetry within the structures and for the sake of brevity, we will focus on the monomer A active site and the Gp1.2 bound between monomers A and E.
Gp1.2 Recognizes Dgt with Specific Interactions.
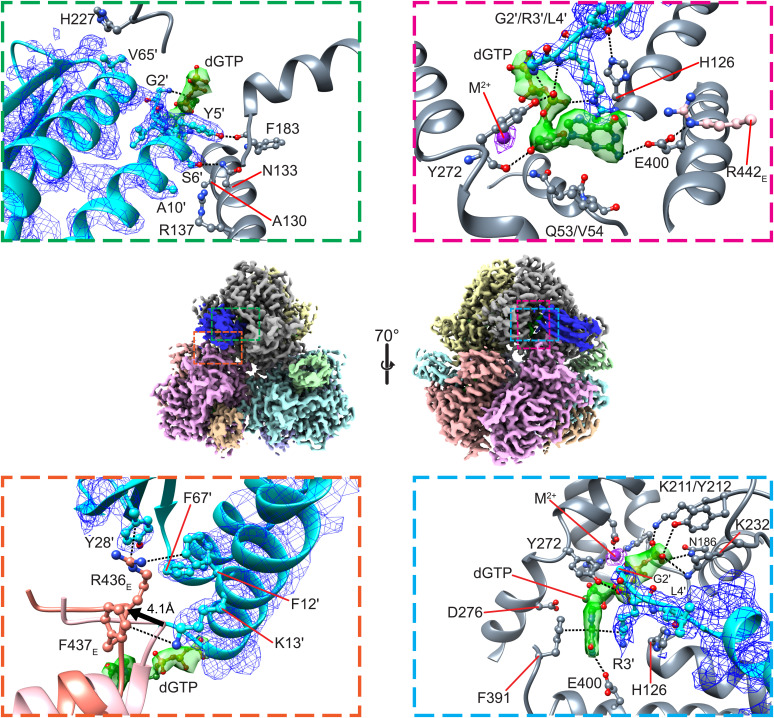
The cryo-EM structures of the Dgt–Gp1.2 complexes provide structural insight into the modes of binding and inhibition. Fig. 4 shows Gp1.2 binding near the active site of monomer A at its interface with monomer E. It has a significantly larger interaction surface with monomer A (∼630 Å2) than with monomer E (∼240 Å2). A portion of monomer E is displaced by >4 Å to accommodate the Gp1.2 helix α1, reducing the interface between A and E by ∼80 Å2 (Fig. 4, Bottom Left). Gp1.2 binding in this conformation leaves the active site of Dgt mostly open to solution (SI Appendix, Fig. S6 B and C), suggesting that ligands should be able to equilibrate with the inhibited active site.
Fig. 4.
Interactions between Dgt, Gp1.2, and dGTP. Middle, overview of the Dgt–Gp1.2–dGTP structure. Dgt and Gp1.2 are colored by monomer, while the dGTPs are colored green. Top Left, close-up of hydrogen-bond interactions between Gp1.2 and Dgt monomer A. Bottom Left, close-up of cation–π interactions between Gp1.2 and Dgt monomer E. The apo-Dgt (PDB ID: 4XDS, colored salmon) is aligned by the monomer A active-site histidine-aspartate (HD) motif. A portion of monomer E is displaced by >4 Å (as measured by the R436E C-α) to accommodate Gp1.2 helix α1. Top Right, close-up of interactions between Gp1.2, Dgt monomer A, and dGTP. Bottom Right, point of view rotated ∼90° from the image in the Top Right. The Middle images were generated in ChimeraX, while the other images were generated in Chimera.
Dgt is recognized by Gp1.2 by specific interactions at two distinct interfaces (Fig. 4, Left). The mode of binding for Gp1.2 is apparently independent of ligand binding, and each of the interactions described below is present in all three of the Dgt–Gp1.2 complexes. Gp1.2 recognizes monomer A primarily with van der Waals interactions, including Y5′ (a prime indicates a Gp1.2 residue when referring to Dgt–Gp1.2 complexes) with K232A and P233A, S6′ with A130A, G7′ with H182A, A10′ with R137A, and V65′ with H227A, although there are also specific hydrogen bonds (Fig. 4, Top Left). The Y5′ sidechain hydrogen bonds with the backbone carbonyl of F193A, and S6′ hydrogen bonds with the N133A sidechain. In contrast, it recognizes monomer E through a series of cation–π interactions (Fig. 4, Bottom Left). F12′ and Y28′ form stacking interactions with the guanidinium of R436E, while K13′ forms a cation–π interaction with F437E.
The Gp1.2 N Terminus Traps the Active Site in a Noncatalytic Conformation.
The cryo-EM grids yielding the ternary Dgt–Gp1.2–(d)GTP structures were obtained by adding the GTP or dGTP ligands after first preequilibrating Dgt–Gp1.2, so the observed presence of ligands in our structures indicates that they were able to enter the active site of the preformed protein complex. After ligand binding, the Gp1.2 N terminus becomes visibly ordered and makes direct interactions with both the active site and substrate. These interactions, described in the next paragraph, prevent both the alignment of the scissile bond with the nucleophilic water and block the catalytic acid (H126) from protonating the leaving group. The interactions observed in the structures therefore provide a clear mechanistic explanation for potent Dgt inhibition by Gp1.2.
The Gp1.2 N-terminal amino group makes ionic hydrogen bonds with the (d)GTP phosphates (Figs. 4 and 5), while the sidechain of R3′ makes an ionic hydrogen bond with the α-phosphate proRP oxygen (Fig. 5). These interactions yield a significantly displaced triphosphate conformation relative to normal dGTP-bound structure of Dgt (14), whereby the α-phosphate is displaced by >2.5 Å (Fig. 5 and SI Appendix, Fig. S7). This conformation also forces the scissile bond between the α-phosphorous atom and the 5′-oxygen out of alignment with the active site metal ions. In view of the proposed catalytic mechanism for Dgt (SI Appendix, Fig. S8), which is proposed to be conserved with SAMHD1 (19, 20), this alternate substrate conformation by itself is expected to be sufficient to inhibit the Dgt reaction by preventing the nucleophilic attack by the hydrolytic metal-bound water/hydroxide. However, additionally, R3′ forms an extended cation–π stacking interaction with the guanine ring and residues F391A and H126A (SI Appendix, Fig. S7). To accommodate the R3′ sidechain, the dGTP guanine ring is displaced by ∼2 Å and F391A by 1.6 Å, while H126A is sterically hindered from accessing the substrate. H126A is further held in this conformation by a hydrogen bond with the L4′ backbone carbonyl. Given that H126A is the acid proposed to protonate the 5′-oxygen of the 2′-deoxyguanosine leaving group (14, 19), the combination of the triphosphate displacement and the steric blockage of H126 provides sufficient explanation for potent inhibition of Dgt by Gp1.2.
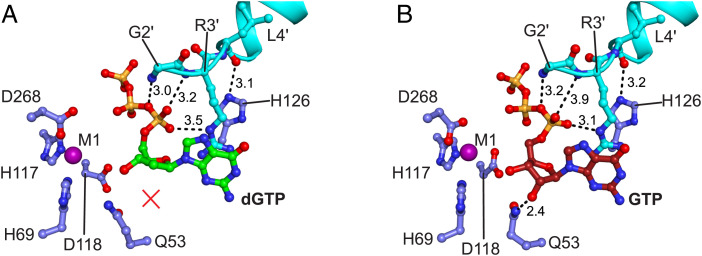
Fig. 5.
The Gp1.2 N terminus interacts with dGTP/GTP ligands and critical Dgt residues in the active site. Image generated in PyMOL. (A) Active site of the Dgt–Gp1.2–dGTP structure, with Dgt residues shown in blue, Gp1.2 in cyan, and dGTP in green and all colored by heteroatom. Dashed lines are the interactions with Gp1.2 that replace productive substrate interactions. dGTP lacks the interaction with Q53 (red X) in the ternary complex. (B) Active site of the Dgt–Gp1.2–GTP structure, with Dgt residues shown in blue, Gp1.2 in cyan, and GTP in red and all colored by heteroatom. Dashed lines are the interactions between Gp1.2 and GTP equivalent to the interactions in A, as well as the productive interaction between Q53 and the GTP 2′-OH.
Interestingly, the sidechain of Y272A, which normally interacts with the proSP oxygen of the β-phosphate, interacts in the inhibited state instead with the proSP oxygen of the α-phosphate (Fig. 4, Top Right). This repositioning of Y272A breaks the helicity of the helix on which it resides, and the Y272 backbone carbonyl now interacts with the 3′-OH of the (d)GTP (Fig. 4, Top Right). It is noteworthy that dGTP and GTP are found in near identical conformations in the inhibited complexes, including a similar set of interactions with Gp1.2 (Fig. 5 and SI Appendix, Fig. S7). The main difference is that GTP maintains the interactions between its 2′-OH and active-site residues Q53A/D276A, while dGTP necessarily lacks these interactions (Fig. 5). This structural observation is consistent with findings that the Dgt–Gp1.2 complex binds to GTP more tightly than dGTP (13).
Gp1.2 Is a Mixed-Type Inhibitor and GTP Is a Competitive Inhibitor of Dgt.
Given that GTP forms a ternary complex with Dgt and Gp1.2 (ref. 13 and this work) but also inhibits Dgt activity on its own (14), we next sought to establish whether Gp1.2 and GTP may inhibit Dgt synergistically. We began by establishing the mode of inhibition for each inhibitor individually. It was previously reported by the Richardson group that inhibition by Gp1.2 requires a kinetically slow step and is complete only after the complex has preequilibrated for 5 to 10 min (9). We therefore preincubated the Dgt–Gp1.2 complexes for at least 20 min before initiating all of the assays below that include Gp1.2. While this assay design limits our ability to draw conclusions about the effects of the ligands on the kinetics of inhibition by Gp1.2, the experiments nevertheless provide important information about the binding equilibria of Gp1.2 and GTP and their combinatorial effects on Dgt inhibition.
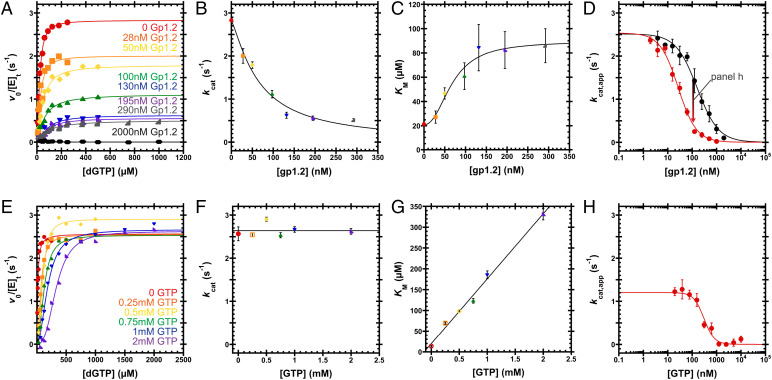
Gp1.2 yields both a decreased kcat and an increased KM and thus is a mixed-type inhibitor, contributing both competitive (affecting KM) and noncompetitive (affecting Vmax) effects by interacting with both the free enzyme (E) and the enzyme–substrate (ES) complex (Fig. 6 A–C and SI Appendix, Table S3). These results directly confirm inferences about the mode of inhibition that were made from previous observations that 1) Gp1.2 lowers the Vmax, 2) Gp1.2 binds to E, and 3) inhibition is reversible (9). Unlike a pure noncompetitive inhibitor, which binds equally to both E and the ES complex, a mixed-type inhibitor binds to E and ES with different affinities. This yields different effects on the substrate’s KM that depend on the relationship of the two affinities. In this case, the increasing KM for dGTP implies that Gp1.2 binds more tightly to E than ES. This is consistent with previous data indicating that dGTP binding to Dgt limits inhibition by Gp1.2 (13).
Fig. 6.
Inhibition of Dgt by Gp1.2 and GTP. (A) v0/[E]t versus [dGTP] curves varying Gp1.2 (see labels). A single experiment was performed for each condition. Eq. 1 is fit to the data. (B and C) Gp1.2 displays a mixed-type inhibition, wherein kcat decreases and KM increases with increasing Gp1.2. Data are the kcat or KM and SE from fitting Eq. 1 to the data in A. Eq. 2 is fit to the data in B to yield IC50 = 66 ± 8 nM and nH = 1.3 ± 0.2. Eq. 3 is fit to the data in C to yield IC50 = 70 ± 10 nM and nH = 2.3 ± 0.9. (D) Titrating Gp1.2 against Dgt under kcat,app conditions (black, 250 µM dGTP) or Dgt with 1 mM GTP (red, 2 mM dGTP). Data are the mean and SEM of three independent experiments. Eq. 4 is fit to the data. Gp1.2 inhibits Dgt to completion with IC50 = 160 ± 20 nM and nH = 1.0 ± 0.1. With 1 mM GTP, Gp1.2 inhibits with IC50 = 27 ± 3 nM and nH = 1.2 ± 0.1. (E) v0/[E]t versus [dGTP] curves at various GTP concentrations (see labels). A single experiment was performed for each condition. Eq. 1 is fit to the data. (F and G) GTP displays a competitive mode of inhibition, wherein kcat is independent of GTP concentration, while KM is linearly dependent on GTP concentration. Data are the kcat or KM and SE from fitting Eq. 1 to the data in E. The line in F represents the average of the kcat values: 2.6 ± 0.2 s−1. Fitting Eq. 5 to the data in G, the inhibitor binding constant (KJ,0; Fig. 6) is determined to be 120 ± 50 µM. (H) Titrating GTP against Dgt (∼25 nM) bound to 100 nM Gp1.2 under kcat,app conditions (2 mM dGTP). Data are the mean and SEM of three independent experiments. Eq. 4 is fit to the data. GTP inhibits Dgt–Gp1.2 to completion with IC50 = 310 ± 50 nM and nH = 2.1 ± 0.5.
GTP increases the dGTP KM but leaves the kcat unaffected and thus acts as a competitive inhibitor of Dgt (Fig. 6 E–G and SI Appendix, Table S3). This competitive behavior is consistent with a recent demonstration that GTP is able to bind directly to the Dgt active site and to inhibit enzyme turnover (14). The slope of the dependence of KM on the concentration of a competitive inhibitor (Fig. 6G) is proportional to both the substrate KM in the absence of inhibitor (KM,0) and the binding constant for the inhibitor itself; the GTP-binding constant that we derive from this slope is 120 ± 50 µM. This value is consistent with previous KM measurements for GTP hydrolysis by Dgt (110 to 150 µM) (1, 21). Importantly, this level of affinity means that Dgt will readily bind GTP in vivo, where GTP is in the high micromolar to low millimolar range (22–24). Interestingly, this also implies that GTP will affect the effective KM for dGTP in vivo and that its value will be significantly higher than the in vitro value (≤20 µM) in the absence of GTP (1, 2, 11, 21).
GTP and Gp1.2 Combinatorially Inhibit Dgt.
After establishing the kinetic parameters for inhibition by Gp1.2 and GTP individually, we tested for a possible synergistic effect of the two inhibitors, because the ternary Dgt complex with the two inhibitors could be relevant for Gp1.2 action in vivo. This was done by titrating each inhibitor against the complex of Dgt with the other, followed by a final challenge with a high concentration of dGTP substrate (Fig. 6 D and H and SI Appendix, Table S3). Titrating Gp1.2 in the absence of GTP yielded a Gp1.2 half maximal inhibitory concentration (IC50) of 160 ± 20 nM (Fig. 6D, black). The conditions for this assay approximate kcat conditions (kcat,app) because the chosen concentration of dGTP (250 μM) is in significant excess over the uninhibited KM. When we included 1 mM GTP in this reaction (Fig. 6D, red), again ensuring kcat,app conditions by including 2 mM dGTP substrate, the Gp1.2 IC50 was significantly lowered to 27 ± 3 nM. Note that the lowest IC50 that can be measured for an inhibitor is the enzyme concentration (25), which in this case corresponds to ∼25 nM Dgt active sites. Thus, given this experimental limitation, we can say that the Gp1.2 IC50 for the Dgt–GTP complex is at most 27 ± 3 nM. Therefore, the presence of GTP enhances inhibition by Gp1.2 by at least 5.9-fold.
We next titrated GTP against the Dgt–Gp1.2 complex (Fig. 6H). We used the same Dgt concentration as above and included 100 nM Gp1.2, which had yielded a partially inhibited state of Dgt in the absence of GTP (Fig. 6D). We used 2 mM dGTP substrate for these assays, which should maintain kcat,app conditions throughout the GTP titration. Under these conditions, GTP inhibits the Dgt–Gp1.2 complex to completion, with IC50 = 310 ± 50 nM (Fig. 6H). It is not usually possible to directly calculate both KI values from a single IC50 measurement in most mixed-inhibition systems (26). Nevertheless, IC50 is typically greater than or equal to the KI. Thus, Gp1.2 enhances the affinity of GTP for the Dgt active site by at least 390-fold compared with the KI for GTP alone (120 ± 50 µM). This remarkably strong inhibitory effect of GTP in the presence of Gp1.2 suggests that the inhibitors act synergistically.
Discussion
Our results provide structural and biochemical insights into the mechanism of E. coli Dgt inhibition by Gp1.2 of bacteriophage T7. The biochemical assays confirm that Gp1.2 acts as a mixed-type inhibitor and that GTP is a competitive inhibitor. Importantly, we show that the two inhibitors can inhibit Dgt turnover in a combinatorial manner, wherein each individual inhibitor effect is enhanced by the other. These data are borne out by the cryo-EM structures of the Dgt–Gp1.2 complexes, which show that Gp1.2 makes extensive interactions with the Dgt active site as well as the (d)GTP ligands, positioning them in a noncatalytic conformation.
When dGTP and GTP are bound to Dgt alone, they bind in quite different conformations. However, in their ternary complexes, they are bound by a similar set of interactions. When dGTP binds to Dgt alone, its 3′-OH interacts with the conserved Q53A and D276A residues, while its α- and γ-phosphates make direct interactions to the active-site metal ions to productively align the scissile bond for hydrolysis (SI Appendix, Fig. S7A) (14, 19). By contrast, for the case of GTP, Q53A and D276A interact instead with the 2′-OH (SI Appendix, Fig. S7E) (14). This conformation repositions the 3′-OH nearer to the active-site metal, displacing the phosphates and hence preventing hydrolysis. Interestingly, this inactive conformation occupied by GTP is similar to the Gp1.2-bound conformations of both GTP and dGTP in the ternary structures (SI Appendix, Fig. S7). In this light, Gp1.2 appears to force the dGTP substrate to bind in a conformation that resembles the nonproductive conformation of the GTP-bound enzyme.
The differential effects of Gp1.2 on dGTP and GTP are most apparent when comparing the Gp1.2-bound structures to the Gp1.2-free structures (SI Appendix, Fig. S7). dGTP is significantly displaced after Gp1.2 binding, with the α-phosphate moved by >2.5 Å and the guanine ring displaced by ∼2 Å to accommodate the Gp1.2 R3′ sidechain. By contrast, the GTP α-phosphate is only displaced by 1.6 Å and the guanine ring by only 0.6 Å. These differences in the ligands’ relative displacement exist despite R3′ being bound in a nearly identical conformation with either ligand. The observation that the GTP 2′-OH retains its contacts with Q53A and D276A in the Gp1.2-inhibited conformation, while dGTP necessarily lacks these interactions, also helps to explain previous observations that GTP binds much more tightly to the Dgt–Gp1.2 complex than dGTP (13). Thus, it appears that Gp1.2 evolved to specifically bind to the Dgt with GTP in the active site. This hypothesis is strongly supported by the combinatorial effects of Gp1.2 and GTP on inhibiting Dgt, inhibiting turnover in vitro in the nanomolar concentration range even in the presence of millimolar dGTP (Fig. 6).
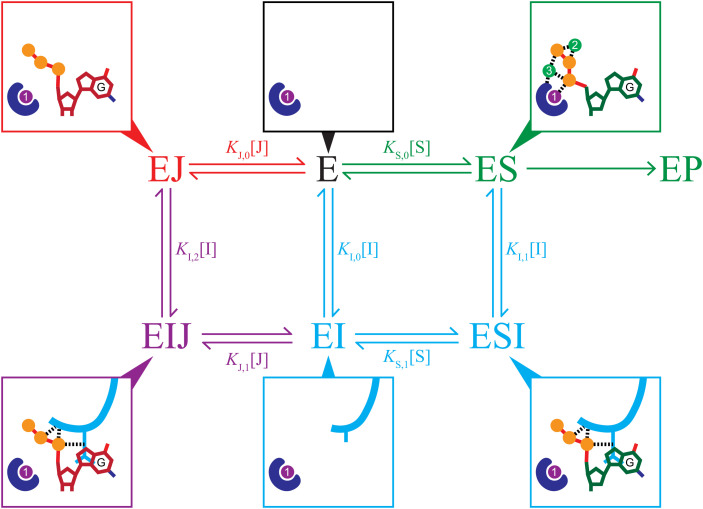
Given everything above, we propose a Gp1.2 inhibition model, as outlined in Fig. 7. In this model, Gp1.2 binds to Dgt and equilibrates into a ternary complex with GTP (EIJ), and this ternary complex represents the effective inhibited state of Dgt in vivo (Fig. 7). This model addresses longstanding questions about how Gp1.2 can inhibit Dgt in vivo, despite requiring a slow equilibration step that is apparently blocked if substrate binding occurs first (8, 9). As described above, GTP is normally in the high micromolar to low millimolar range in cells, so the low micromolar binding constant for GTP implies that Dgt active sites will often be occupied by GTP. Furthermore, the GTP concentration will be in vast excess of its IC50 value for Dgt–Gp1.2 (Fig. 6H), indicating that the complex will be fully saturated with GTP. The competitive GTP inhibition data (Fig. 6G) indicate that the effective KM for dGTP in vivo will be above 100 µM, so the enzyme active sites will be significantly less occupied with dGTP than the in vitro KM might suggest. Importantly, complete inhibition of Dgt–Gp1.2 by GTP occurs in the nanomolar range despite the reactions including millimolar dGTP, implying that GTP easily outcompetes dGTP for the Gp1.2-inhibited Dgt active site. The dGTP concentration in the reactions is also well above the low micromolar concentrations expected in the cell, so the results also suggest that the Dgt–Gp1.2–GTP ternary complex will remain stably bound in vivo. Thus, after infection by T7, the nascent Gp1.2 protein will encounter and interact with Dgt active sites in at least three ligand-bound states (E, ES, and EJ; see Fig. 7). As the Dgt–Gp1.2 complex equilibrates, most should end up in the ternary complex with GTP (EIJ) due to both the extremely tight binding of GTP to the Dgt–Gp1.2 complex (Fig. 6H) and the fact that the dGTP off rate from the complex is faster than the GTP off rate (13).
Fig. 7.
An inhibition scheme combining a mixed-type inhibitor (I = Gp1.2, cyan) with a competitive inhibitor (J = GTP, red). The blue shape represents the active-site HD motif, and the purple and green circles are the proposed catalytic metal ions (SI Appendix, Fig. S5) (20). Product release and any possible conformational change steps are excluded for simplicity. The surrounding boxes show minimal schematics of the active site in the various states of the scheme with or without Gp1.2 (cyan), dGTP (green carbons), or GTP (red carbons). In a mixed-type inhibition scheme (cyan), inhibitor (I) binds to either E or ES, yielding inhibited complexes (EI or ESI) that prevent turnover to product (P). The increasing KM observed in Fig. 6C indicates a competitive-like mixed-type inhibition, wherein the inhibitor binds more tightly to E (i.e., KI,0 < KI,1). In a competitive inhibition scheme (red), inhibitor (J) binds only to E and prevents it from binding to substrate (S), therefore preventing turnover to product (P). Because Gp1.2 and GTP use different inhibition modes and form productive interactions in a ternary complex with Dgt (EIJ, purple), inhibition by both is combinatorial.
Given the proposed inhibition model, the slow equilibration of Gp1.2 into the inhibited ternary complex, which apparently occurs over the course of several minutes (9), is not likely to be a limiting factor for T7 fitness and may not be subject to major selective pressure. The delayed inhibition should be easily tolerated because the levels of the four dNTPs do not significantly rise until ∼10 min after T7 infection begins (7). This slow increase in the dNTP pool is related to the mechanism that T7 uses to increase the pool for its own DNA production. T7 encodes nucleases (endonuclease Gp3 and exonuclease Gp6) that degrade the host chromosome, releasing the nucleotides as monophosphates. These dNMPs are then converted by kinases into the dNTPs for use by the T7 DNA polymerase. The endonuclease is expressed to high levels during the first 15 min of infection (27). Because this also correlates with the delayed increase in the dNTP levels, slow inhibition of Dgt by Gp1.2 should not limit the T7 DNA synthesis during infection. So long as the GTP concentration remains in excess of the nanomolar concentration range, Gp1.2 will equilibrate slowly into the inhibited ternary complex with Dgt and GTP during the first minutes of an infection and then remain trapped in that state as dGTP is produced from the dGMP liberated from the host chromosome.
Like phage T7 carrying the Dgt inhibitor Gp1.2, HIV-2 carries a SAMHD1 inhibitor, Vpx, that interferes with SAMHD1-mediated restriction (6, 28). The two inhibitors do not use the same mechanism, as Vpx relieves restriction by triggering proteasomal degradation of SAMHD1 (6, 28), and they are not related by sequence. Thus, viruses have independently evolved inhibitors of dNTPase homologs on at least two occasions, suggesting that the ability to override host controls on the nucleotide pool is critical for these viruses to replicate. An outstanding question is whether these are the only two instances of viruses evolving inhibitors of Dgt/SAMHD1 homologs. Given the wide distribution of the dNTPase proteins (29) and the long evolutionary time that viruses have been infecting their hosts, it remains possible that other viruses have evolved other novel dNTPase inhibitors to gain control of the host nucleotide pool to replicate their genomes.
Materials and Methods
Expression and Purification of Dgt.
The expression and purification of E. coli Dgt was performed with a modified protocol 3 from our previous report (11) to obtain a DNA- and metal-free E. coli Dgt for kinetic studies. First, all buffers included 25 mM Tris (pH 8) instead of switching to 25 mM sodium/potassium phosphate (pH 8) for the nickel-nitrilotriacetic acid (Ni2+-NTA) column. Next, after removal of the N-terminal 6×His-tag by enteropeptidase (enterokinase light chain, NEB) and subsequent separation of the protease by a Ni2+-NTA column, Dgt was made metal free by overnight dialysis in our storage buffer also containing ethylenediaminetetraacetic acid (EDTA; 25 mM Tris pH 8, 75 mM sodium citrate, 15 mM EDTA, and 1 mM β-mercaptoethanol). After this, the protein was dialyzed twice for >2 h each into two separate batches of a metal-free storage buffer (25 mM Tris pH 8, 75 mM sodium citrate, and 1 mM β-mercaptoethanol). A separate portion of this purification that was not made metal free was used instead for the cryo-EM grid preparation, with Dgt aliquots stored in storage buffer (25 mM Tris pH 8, 75 mM sodium citrate, 10 mM MgCl2, and 1 mM β-mercaptoethanol).
Expression and Purification of Gp1.2.
BL21-DE3 cells were transformed with pDest-566 (Invitrogen) carrying the phage T7 1.2 gene for isotopic labeling; Rosetta-DE3 cells were likewise transformed for unlabeled growth. The cells were grown in either 15N- or 13C-15N-M9 minimal medium (15NH4Cl) or lysogeny broth (LB) to an optical density (OD600) of ∼1.0, induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and expressed overnight at 25 °C. Cells were pelleted (5,000 × g for 20 min at 4 °C), resuspended in buffer A (50 mM sodium/potassium phosphate pH 6.5, 100 mM NaCl, and protease inhibitors), sonicated, and centrifuged to separate the soluble lysate (30,000 × g for 10 min at 4 °C). The protein was purified using immobilized metal affinity chromatography. The lysate was applied to a 5-mL Ni column. The column was washed with buffer A plus 20 mM imidazole and eluted with buffer A plus 300 mM imidazole. Imidazole was removed using a 3-kDa concentrator to exchange to buffer B (50 mM Tris pH 7.7 and 150 mM NaCl). The Gp1.2 was then concentrated to 1 mL, and the affinity tag was cleaved using 250 µL of 1 mg/mL Tobacco Etch Virus (TEV) protease and incubated overnight at 4 °C. The cleaved Gp1.2 was further purified using a Superdex S75 gel filtration column with an isocratic elution in buffer B. Peak fractions were concentrated and exchanged to 1× PBS (137 mM NaCl, 3 mM KCl, 4.2 mM Na2HPO4, and 0.15 mM KH2PO4, pH 7.4) for unlabeled samples and 1/3× PBS with 1 µM EDTA for isotope-labeled samples.
Structural Determination of Gp1.2 by NMR.
All measurements were performed at 25 °C on a 600-MHz Varian Inova NMR spectrometer equipped with a cryogenically cooled triple resonance probe. The assignment of resonances and nuclear Overhauser effect (NOE) studies of Gp1.2 were determined by standard triple resonance NMR methods similar to that described previously (30). Briefly, backbone and sidechain atoms were assigned by scalar coupling experiments starting with amide-detected experiments followed by sidechain total correlation spectroscopy. Experiments used are listed in PDB entry 2MDP. Residual dipolar couplings (RDC) were measured by comparing J-couplings in the presence and absence of bacteriophage Pf1. The calculation of the ensemble of structures used initial models from CS-Rosetta (31) followed by automated assignment of the NOE spectra with CYANA (32). After multiple iterations, the top 20 structures were refined with X-PLOR-NIH in explicit solvent (33). An example of NOE assignments determined by CYANA is shown in SI Appendix, Fig. S2C. This figure shows long-range NOEs between the aromatic sidechain of Y78 and the methyl group of L22, which helps constrain the positions of helices 1 and 2. Data and structure quality factors are included in SI Appendix, Table S1.
The variation in the angle phi in the ensemble of NMR structures is used as a pseudoorder parameter (34). The high S(phi) values show that the loop between helix α1 and strand β1 is well ordered, whereas the lower S(phi) values for the other loops and nonsecondary structure elements, such as the N terminus, indicate reduced order (SI Appendix, Fig. S2A). The measured residual dipolar couplings provide orientational restraints for the structure calculation (SI Appendix, Fig. S2B) (35). The error bars are the SD in the experimental data versus the fit to the structures. These are small, indicating that the structure is a good fit to the data. Another feature of the data is that the H-N RDC values for both helices are around −5 Hz, indicating that they are likely to be parallel or antiparallel in orientation. Thus, the angle between helix α1 and the beta sheet in the NMR model is reasonably precise.
Oligomeric State of Gp1.2 in Solution.
The 1H-15N heteronuclear single quantum coherence (HSQC) spectra of U-[15N] Gp1.2 showed strong, well-resolved resonances at concentrations of ≤0.2 mM, but linewidth increased significantly at higher concentrations, suggesting significant concentration-dependent aggregation. To evaluate whether Gp1.2 was monomeric at low concentrations, pulsed gradient spin-echo experiments were performed to measure the diffusion rate of Gp1.2 at 0.2 mM. SI Appendix, Fig. S9, compares the diffusion rates measured for 15N-labeled Gp1.2, RNase H, and NuiA using the isotope-filtered gradient diffusion experiment of Nesmelova et al. (36). The data are plotted so that the slope of the line corresponds to the rate of diffusion. The calculated radii of gyration from the monomeric structures of Gp1.2, RNase H (37), and NuiA (38) are 14.3, 14.8, and 16.7 Å, respectively, which directly correlate with the observed slopes. Therefore, Gp1.2 is most likely a monomer at low concentrations because it is diffusing faster than the slightly larger RNase H. The measured diffusion constant for Gp1.2 was best fit to 1.5 × 10−6 cm2/s, which corresponds to a Stokes radius of 16.3 Å. Assuming a hydration shell of 1.6 to 3.0 Å, the radius of gyration (Rg) value is reasonable for the monomeric structure of Gp1.2.
Cryo-EM Sample Preparation and Grid Setting.
Dgt and Gp1.2 aliquots were thawed on ice and then mixed for a final ratio of 1.25 to 1 Gp1.2 to Dgt monomer concentration and incubated at room temperature for at least 20 min before setting grids, regardless of ligands. The GTP ligand (final concentration of 1 mM) was added immediately after the proteins were mixed, and the complex was incubated together before setting grids. The dGTP ligand (final concentration of 1 mM) was mixed only after the proteins had been preincubated for ∼20 min to limit hydrolysis, and the complex was incubated together for >5 min before setting grids. Three microliters of the mixtures was applied to glow-discharged UltrAuFoil R 1.2/1.3 300 mesh grids (Quantifoil Micro Tools GmbH). Grids were then blotted 2.5 to 3.5 s and vitrified in liquid ethane using an Automatic Plunge Freezer EM GP (Leica).
Cryo-EM Data Collection.
Movies of Dgt–Gp1.2 complexes were collected with a Talos Arctica electron microscope (Thermo Fisher Scientific) operating at 200 keV using a K2 Summit direct electron detector (Gatan). Movies were collected in counting mode at a nominal magnification of ×45,000, corresponding to a physical pixel size of 0.932 Å/pixel. Overall, 1,638 (Dgt–Gp1.2), 1,108 (Dgt–Gp1.2–GTP), or 1,206 (Dgt–Gp1.2–dGTP) movies were collected with 60 frames/movie. Defocus values ranged from −0.50 to −2.5 μm. Movies were recorded over 8.4 s, resulting in a fluence of ∼54 e−/Å2 and flux of 5.6 e−/pixel/s.
Cryo-EM Data Processing.
All data were processed in RELION (39) using normal procedures (SI Appendix, Table S2 and Fig. S10) using a workflow similar to what we described previously for the Leeuwenhoekiella blandensis dGTPase (20). Briefly, movie frames were aligned using MotionCor2 (40). Contrast transfer functions (CTFs) were estimated using CTFFIND-4.1 (41). The CTF-corrected micrographs were manually inspected for quality, with 1,404 (Dgt–Gp1.2), 1,004 (Dgt–Gp1.2–GTP), and 1,037 (Dgt–Gp1.2–dGTP) used for the remaining steps. Particles were picked using Laplacian-of-Gaussian autopicking, extracted with 4× binning (3.728 Å/pixel), and submitted to two-dimensional (2D) classification with alignment (T = 2). Two-dimensional (2D) classes with good signal to noise and visible secondary structure were used to generate a 15-Å initial model aligned to D3 symmetry for the Dgt–Gp1.2–GTP structure. The particles were then reextracted without binning and with recentering and submitted to three-dimensional (3D) autorefinement, with the Dgt–Gp1.2–GTP initial model (D3 symmetry imposed) as a reference map for all three structures.
Preliminary refinement and postprocessing without symmetry (C1) indicated that there were six Gp1.2 molecules per hexamer and six ligands bound in the active sites of the ternary complexes, thus indicating fully saturated complexes. In subsequent steps, we imposed D3 symmetry during refinement, which significantly improved resolution without significantly changing the model. The structures from this first autorefinement were of poor quality, with resolution estimates >4 Å and a geometric appearance with incoherent protein connectivity. The refined particles were submitted to 2D classification without alignment. We noted several thousand bad “particles” in each dataset that had made it through the initial 2D classification steps, which were removed. Subsequent refinement produced improved maps (<4 Å with good secondary structure). These maps were then submitted to CTF refinement, Bayesian polishing, and a second CTF refinement. Three-dimensional autorefinement and postprocessing were run after each CTF refinement and polishing step to monitor for improvements in map quality.
After the final postprocessing step, the maps had Fourier shell correlation (FSC) cross-correlation resolution estimates based on the 0.143 cutoff of 2.85 Å (Dgt–Gp1.2), 2.54 Å (Dgt–Gp1.2–GTP), and 2.96 Å (Dgt–Gp1.2–dGTP). Postprocessed maps were generated from the final half-maps in RELION (39), autosharpened maps in PHENIX (42), and the DeepEMhancer postprocessing artificial neural network (43). The maps were validated, and FSC resolution estimates were made using MTRIAGE (44). The local resolution distribution of the maps was estimated using RELION (45). The apo-Dgt crystal structure and the Gp1.2 NMR structure were docked into each map for real-space refinement in PHENIX (42). Final models were obtained by iterative rounds of manual inspection and building in Coot (46) and real-space refinement in PHENIX (42). Structures were validated with MolProbity (47). The PISA server was used to perform the interface analysis presented in the results on the Dgt–Gp1.2 structure (48). Figures were prepared using PyMOL (Schrödinger, LLC), Chimera (49), or ChimeraX (50), as indicated in the figure legends.
Inhibition Studies of E. coli Dgt by Gp1.2 and GTP.
We recently adapted our previously published enzyme-coupled assay (51) to a 96-well plate format, as described for the L. blandensis dGTPase (20). Briefly, all reactions in this study were performed in a reaction buffer containing 100 mM Tris pH 8, 5 mM MgCl2, 125 µM MnCl2, and 5 mM inorganic phosphate pH 7. Hydrolysis of dGTP to dG and PPPi is coupled to a spectroscopic change by inclusion of purine nucleoside phosphorylase (PNP; Sigma) and xanthine oxidase (XOD; Sigma), yielding a maximum difference in the absorbance spectrum at 297 nm (51). All reactions were run in half-area, flat-bottom, ultraviolet-transparent Corning microplates (Sigma, CLS3679) with the change in absorbance at 297 nm monitored in real time in a SpectraMax M5 microplate reader (Molecular Devices). We used a previously determined experimental conversion factor to convert the change in absorbance to the product concentration in our plate assays (20). This conversion factor is the slope of the linear relationship of ΔA297 endpoints from a dilution series of guanine reacted with XOD (1.5 × 10−3 ΔA297/µM for the 40-µL reaction volume used in this study).
For Dgt initial rate (v0) versus concentration of dGTP ([dGTP]) curves (Fig. 6 A and E), substrate stocks were serially diluted into two rows. Each row included a zero-substrate well. One row served as no-enzyme controls and was diluted 1:1 with reaction buffer. We used the zero-substrate well from the control row as a plate blank. The reactions in the second row were initiated using a multichannel pipette by 1:1 addition of an enzyme mixture (∼50 nM Dgt monomer, 100 mU/mL PNP, and 1 U/mL XOD) in reaction buffer. The enzyme mixture was prepared by preincubating Dgt in reaction buffer at 37 °C for 20 min and adding PNP and XOD and distributing the mixture into the plate. For curves including Gp1.2, the Dgt–Gp1.2 complex was preincubated at 37 °C and then otherwise handled as above. For curves including GTP, the GTP was included in the dGTP wells. For the kcat,app curves (Fig. 6 D and H), the dGTP substrate was constant at either 250 µM or 2 mM, as indicated. Gp1.2 or GTP was serially diluted into wells containing fixed concentrations of Dgt and the other inhibitor (as indicated) and preincubated at 25 °C in the plate reader for >20 min before initiating reactions as above.
Data processing was handled as described previously (20). Briefly, the no-enzyme control wells were used to correct for changes in baseline absorbance across the substrate dilution series. We then subtracted the no-substrate well to correct for an upward drift in A297 that was observed in all enzyme-containing wells. The data were then converted into product concentrations using the conversion factor described above. Initial velocities (v0, nM/s) were determined by fitting a line to the linear region of the concentration versus time curves for each condition. We controlled for enzyme concentration by dividing by the total Dgt monomer concentration to yield rate constants for each condition (v0/[E]t, s−1). A Michaelis–Menten equation with a Hill coefficient (n) was fit to the v0/[E]t versus dGTP plots (Eq. 1). Eq. 2 was fit to the kcat versus Gp1.2 curve, whereby kcat,0 is the rate constant for Dgt activity without Gp1.2, IC50 is the concentration of Gp1.2 required for half inhibition, and n is a Hill coefficient. Eq. 3 was fit to the KM versus Gp1.2 curve, whereby KM,0 is the dGTP KM without Gp1.2, ΔKM is the amplitude, IC50 is the concentration of Gp1.2 required for half inhibition, and n is a Hill coefficient. Eq. 4, which is a modified version of Eq. 2, was fit to the kcat,app versus Gp1.2 curves with and without 1 mM GTP. Eq. 5 was fit to the linear relationship of the dGTP KM with GTP. The equation was used to obtain the binding constant for GTP to Dgt (KJ,0) from the dGTP KM absent a competitive inhibitor (KM,0).
| [1] |
| [2] |
| [3] |
| [4] |
| [5] |
Supplementary Material
Acknowledgments
This work was supported, in part, by the Intramural Research Program of the NIH; National Institute of Environmental Health Sciences (NIEHS; 1ZIAES102906, 1ZICES103326, and 1ZIAES101905 to R.E.L., M.J.B., and R.M.S., respectively). The NMR dataset was collected using a 600 MHz Varian at the NIEHS. The EM datasets were collected using a Talos Arctica at NIEHS. We thank Drs. Niketa Bhawsinghka and Andrew Sikkema for carefully reading this manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2123092119/-/DCSupplemental.
Data, Materials, and Software Availability
The Gp1.2 NMR assignments and structural models have been deposited in the Biological Magnetic Resonance Data Bank (BMRB) and the PDB under accession codes BMRB ID 19498 (52) and PDB ID 2MDP (53), respectively. The cryo-EM maps and associated atomic models have been deposited in the Electron Microscopy Data Bank and the PDB under accession codes EMD-26360 (54) and PDB ID 7U65 (55) (Dgt–Gp1.2), EMD-26361 (56) and PDB ID 7U66 (57) (Dgt–dGTP–Gp1.2), and EMD-26362 (58) and PDB ID 7U67 (59) (Dgt–GTP–Gp1.2). All other data are included in the manuscript and/or supporting information.
References
- 1.Kornberg S. R., Lehman I. R., Bessman M. J., Simms E. S., Kornberg A., Enzymatic cleavage of deoxyguanosine triphosphate to deoxyguanosine and tripolyphosphate. J. Biol. Chem. 233, 159–162 (1958). [PubMed] [Google Scholar]
- 2.Seto D., Bhatnagar S. K., Bessman M. J., The purification and properties of deoxyguanosine triphosphate triphosphohydrolase from Escherichia coli. J. Biol. Chem. 263, 1494–1499 (1988). [PubMed] [Google Scholar]
- 3.Quirk S., Bhatnagar S. K., Bessman M. J., Primary structure of the deoxyguanosine triphosphate triphosphohydrolase-encoding gene (dgt) of Escherichia coli. Gene 89, 13–18 (1990). [DOI] [PubMed] [Google Scholar]
- 4.Gawel D., Hamilton M. D., Schaaper R. M., A novel mutator of Escherichia coli carrying a defect in the dgt gene, encoding a dGTP triphosphohydrolase. J. Bacteriol. 190, 6931–6939 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar D., et al. , Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 39, 1360–1371 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laguette N., et al. , SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tal N., et al. , Bacteria deplete deoxynucleotides to defend against bacteriophage infection. Nat. Microbiol. 7, 1200–1209 (2022). [DOI] [PubMed] [Google Scholar]
- 8.Myers J. A., Beauchamp B. B., Richardson C. C., Gene 1.2 protein of bacteriophage T7. Effect on deoxyribonucleotide pools. J. Biol. Chem. 262, 5288–5292 (1987). [PubMed] [Google Scholar]
- 9.Huber H. E., Beauchamp B. B., Richardson C. C., Escherichia coli dGTP triphosphohydrolase is inhibited by gene 1.2 protein of bacteriophage T7. J. Biol. Chem. 263, 13549–13556 (1988). [PubMed] [Google Scholar]
- 10.Safari F., et al. , The interaction of phages and bacteria: The co-evolutionary arms race. Crit. Rev. Biotechnol. 40, 119–137 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Singh D., et al. , Structure of Escherichia coli dGTP triphosphohydrolase: A hexameric enzyme with DNA effector molecules. J. Biol. Chem. 290, 10418–10429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wurgler S. M., Richardson C. C., DNA binding properties of the deoxyguanosine triphosphate triphosphohydrolase of Escherichia coli. J. Biol. Chem. 268, 20046–20054 (1993). [PubMed] [Google Scholar]
- 13.Nakai H., Richardson C. C., The gene 1.2 protein of bacteriophage T7 interacts with the Escherichia coli dGTP triphosphohydrolase to form a GTP-binding protein. J. Biol. Chem. 265, 4411–4419 (1990). [PubMed] [Google Scholar]
- 14.Barnes C. O., et al. , The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity. Proc. Natl. Acad. Sci. U.S.A. 116, 9333–9339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm L., Sander C., The FSSP database: Fold classification based on structure-structure alignment of proteins. Nucleic Acids Res. 24, 206–209 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mueller G. A., et al. , Solution structure of the Drosha double-stranded RNA-binding domain. Silence 1, 2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers J. A., Beauchamp B. B., White J. H., Richardson C. C., Purification and characterization of the gene 1.2 protein of bacteriophage T7. J. Biol. Chem. 262, 5280–5287 (1987). [PubMed] [Google Scholar]
- 18.Ben-Bassat A., et al. , Processing of the initiation methionine from proteins: Properties of the Escherichia coli methionine aminopeptidase and its gene structure. J. Bacteriol. 169, 751–757 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris E. R., et al. , Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis. Nat. Commun. 11, 3165 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klemm B. P., et al. , High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP. J. Biol. Chem. 298, 102073 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beauchamp B. B., Richardson C. C., A unique deoxyguanosine triphosphatase is responsible for the optA1 phenotype of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 85, 2563–2567 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bochner B. R., Ames B. N., Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 257, 9759–9769 (1982). [PubMed] [Google Scholar]
- 23.Buckstein M. H., He J., Rubin H., Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 190, 718–726 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett B. D., et al. , Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams J. W., Morrison J. F., The kinetics of reversible tight-binding inhibition. Methods Enzymol. 63, 437–467 (1979). [DOI] [PubMed] [Google Scholar]
- 26.Burlingham B. T., Widlanski T. S., An intuitive look at the relationship of Ki and IC50: A more general use for the Dixon plot. J. Chem. Educ. 80, 214 (2003). [Google Scholar]
- 27.Center M. S., Studier F. W., Richardson C. C., The structural gene for a T7 endonuclease essential for phage DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 65, 242–248 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hrecka K., et al. , Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu C. H., et al. , Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification. Nat. Commun. 12, 731 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loeffler P. A., et al. , Structural studies of the PARP-1 BRCT domain. BMC Struct. Biol. 11, 37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen Y., Vernon R., Baker D., Bax A., De novo protein structure generation from incomplete chemical shift assignments. J. Biomol. NMR 43, 63–78 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrmann T., Güntert P., Wüthrich K., Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Clore G. M., Schwieters C. D., How much backbone motion in ubiquitin is required to account for dipolar coupling data measured in multiple alignment media as assessed by independent cross-validation? J. Am. Chem. Soc. 126, 2923–2938 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharya A., Tejero R., Montelione G. T., Evaluating protein structures determined by structural genomics consortia. Proteins 66, 778–795 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Lipsitz R. S., Tjandra N., Residual dipolar couplings in NMR structure analysis. Annu. Rev. Biophys. Biomol. Struct. 33, 387–413 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Nesmelova I. V., Idiyatullin D., Mayo K. H., Measuring protein self-diffusion in protein-protein mixtures using a pulsed gradient spin-echo technique with WATERGATE and isotope filtering. J. Magn. Reson. 166, 129–133 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Pari K., Mueller G. A., DeRose E. F., Kirby T. W., London R. E., Solution structure of the RNase H domain of the HIV-1 reverse transcriptase in the presence of magnesium. Biochemistry 42, 639–650 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Kirby T. W., et al. , The nuclease A inhibitor represents a new variation of the rare PR-1 fold. J. Mol. Biol. 320, 771–782 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez-Garcia R., et al. , DeepEMhancer: A deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afonine P. V., et al. , New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 74, 814–840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh D., Schaaper R. M., Hochkoeppler A., A continuous spectrophotometric enzyme-coupled assay for deoxynucleoside triphosphate triphosphohydrolases. Anal. Biochem. 496, 43–49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mueller G. A., London R. E., Schaaper R. M., BMRB Entry 19498, gp1.2. Biological Magnetic Resonance Data Bank. https://bmrb.io/data_library/summary/index.php?bmrbId=19498. Deposited 16 September 2013. [Google Scholar]
- 53.Mueller G. A., London R. E., The bacteriophage T7 encoded inhibitor (gp1.2) of E. coli dGTP triphosphohydrolase. Protein Data Bank. https://www.rcsb.org/structure/2MDP. Deposited 16 September 2013. [Google Scholar]
- 54.Klemm B. P., Hsu A. L., Borgnia M. J., Schaaper R. M., Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-26360. Deposited 3 March 2022. [Google Scholar]
- 55.Klemm B. P., Hsu A. L., Borgnia M. J., Schaaper R. M., Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2. Protein Data Bank. https://www.rcsb.org/structure/7U65. Deposited 3 March 2022. [Google Scholar]
- 56.Klemm B. P., Dillard L. B., Borgnia M. J., Schaaper R. M., Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and dGTP. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-26361. Deposited 3 March 2022. [Google Scholar]
- 57.Klemm B. P., Dillard L. B., Borgnia M. J., Schaaper R. M., Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and dGTP. Protein Data Bank. https://www.rcsb.org/structure/7U66. Deposited 3 March 2022. [Google Scholar]
- 58.Klemm B. P., Hsu A. L., Borgnia M. J., Schaaper R. M., Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and GTP. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-26362. Deposited 3 March 2022. [Google Scholar]
- 59.Klemm B. P., Hsu A. L., Borgnia M. J., Schaaper R. M., Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and GTP. Protein Data Bank. https://www.rcsb.org/structure/7U67. Deposited 3 March 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Gp1.2 NMR assignments and structural models have been deposited in the Biological Magnetic Resonance Data Bank (BMRB) and the PDB under accession codes BMRB ID 19498 (52) and PDB ID 2MDP (53), respectively. The cryo-EM maps and associated atomic models have been deposited in the Electron Microscopy Data Bank and the PDB under accession codes EMD-26360 (54) and PDB ID 7U65 (55) (Dgt–Gp1.2), EMD-26361 (56) and PDB ID 7U66 (57) (Dgt–dGTP–Gp1.2), and EMD-26362 (58) and PDB ID 7U67 (59) (Dgt–GTP–Gp1.2). All other data are included in the manuscript and/or supporting information.