Significance
Anatomical and molecular studies have revealed that in many animal nervous systems, neuronal cell types can often be subclassified into highly related subtypes with only small phenotypic differences. We decipher here the regulatory logic of such cell type diversification processes. We show that identity features of neurons that are highly similar to one another are controlled by master regulatory transcription factors and that phenotypic differences between related cell types are controlled by downstream acting transcription factors that promote or antagonize the ability of master regulatory factors to control unique identity features. Our findings help explain how neuronal cell types diversify and suggest hypothetical scenarios for neuronal cell-type evolution.
Keywords: C. elegans, homeobox, transcriptional control, neuronal identity, homeosis
Abstract
The classification of neurons into distinct types reveals hierarchical taxonomic relationships that reflect the extent of similarity between neuronal cell types. At the base of such taxonomies are neuronal cells that are very similar to one another but differ in a small number of reproducible and select features. How are very similar members of a neuron class that share many features instructed to diversify into distinct subclasses? We show here that the six very similar members of the Caenorhabditis elegans IL2 sensory neuron class, which are all specified by a homeobox terminal selector, unc-86/BRN3, differentiate into two subtly distinct subclasses, a dorsoventral subclass and a lateral subclass, by the toggle switch–like action of the sine oculis/SIX homeobox gene unc-39. unc-39 is expressed only in the lateral IL2 neurons, and loss of unc-39 leads to a homeotic transformation of the lateral into the dorsoventral class; conversely, ectopic unc-39 expression converts the dorsoventral subclass into the lateral subclass. Hence, a terminal selector homeobox gene controls both class- as well as subclass-specific features, while a subordinate homeobox gene determines the ability of the class-specific homeobox gene to activate subtype-specific target genes. We find a similar regulatory mechanism operating in a distinct class of six motor neurons. Our findings underscore the importance of homeobox genes in neuronal identity control and invite speculations about homeotic identity transformations as potential drivers of evolutionary novelty during cell-type evolution in the brain.
Understanding developmental programs in the nervous system necessitates the precise delineation and classification of neuronal cell types (1). Single cell transcriptomic techniques have ushered in a new era of cell-type classification in the nervous system of all animal species (2, 3). While classic anatomical and functional studies have revealed that neuronal cell types can be subdivided into distinct subtypes (4, 5), the inclusion of molecular criteria, and particularly recent scRNA transcriptome data, have begun to generate much finer-grained opportunities for cell-type classification (1–3, 6). Such classification reveals taxonomic relationships with hierarchical structures that reflect the extent of similarity between neuronal cell types (1, 6). For example, single cell profiling of retinal bipolar cells, which all share certain anatomical, functional, and molecular features, identified novel subtypes and established hierarchical relationships between subtypes of previously known retinal bipolar cells (7). Similarly, recent comprehensive single cell transcriptomic analysis of other major parts of the brain (e.g., cortex and hippocampus) revealed a plethora of hierarchical, multitier subdivisions of specific inhibitory and excitatory neuron classes (3, 8, 9). The hierarchical categorization of relationships of cellular identities poses an intriguing question, particularly at the bottom of such clustering diagrams: What are the molecular mechanisms by which cells that share a broad range of phenotypic identity features diversify into subtypes that express subtly distinct features? We address this problem here in the nervous system of the nematode Caenorhabditis elegans, where anatomical (10) and scRNA-based molecular atlases (11) have unearthed multiple example of subtypologies of highly related neuronal cell types.
The six polymodal C. elegans IL2 sensory neurons, arranged as three symmetrically arranged neuron pairs (a dorsal, lateral, and ventral pair; Fig. 1A) have been classified together due to their characteristic cell body position, neurite projections, association with the inner labial sensilla, and their shared synaptic connectivity (10, 12) (Fig. 1B). All six IL2 neurons stereotypically innervate the IL1 sensory-motor neurons and OLQ sensory neurons, the RIH interneuron, and the RME and URA motorneurons, a pattern shared by no other neuron class (Fig. 1B). However, only the lateral IL2 neuron pair, but not the dorsal or ventral pairs (from here on referred to as dorsoventral IL2 neurons) makes reciprocal chemical synapses with the dopaminergic ADE sensory neurons (Fig. 1B). In addition, the growth of extensive dendritic branches occurring during entry in a diapause arrest stage, when these neurons acquire specific mechanosensory functions (13), is mostly restricted to the dorsoventral IL2 neurons (14). Reporter gene studies as well as recent scRNA profiling have corroborated the notion that while all six IL2 neurons share the expression of a large number of genes, the dorsoventral IL2 and lateral IL2 neurons also differentially express a small number of genes (11, 15, 16) (Fig. 1C). We investigate here how these subtype differentiation events are brought about. We show that the two IL2 subtypes represent two distinct, interconvertible states. Each state can be switched to the other by gain or loss of the sine oculis/SIX-type homeobox gene unc-39, which is normally expressed only in the lateral IL2 neurons; its removal leads to a homeotic identity transformation of IL2 lateral identity into dorsoventral IL2 identity, while ectopic expression of unc-39 in the dorsoventral IL2 neurons switches them to the IL2 lateral identity. We find that another sine oculis/SIX homeobox gene, ceh-32, fulfills a similar role in subtype diversification of an unrelated, but also radially symmetric head motor neuron class.
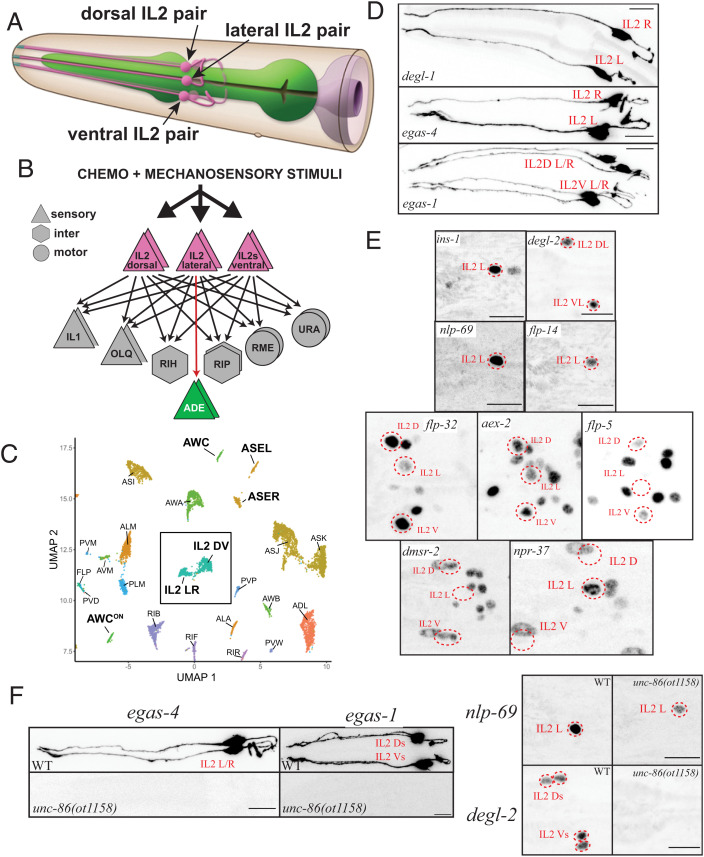
Fig. 1.
IL2 neurons are composed of distinct subclasses, all specified by the unc-86 terminal selector. (A) Schematic drawing of IL2 neurons, based on electron microscopical reconstruction (10). Reproduced with permission from ref. 58. (B) Synaptic connectivity of IL2 neurons. Data extracted from refs. 10 and 59. (C) Single cell transcriptome analysis showing similarity and differences among IL2 neuron subtypes, relative to other neuron classes. Reproduced from ref. 11. (D and E) Expression pattern analysis using reporter alleles or reporter transgenes support IL2 subtype classification. (D) Subtype-specific, cytoplasmically localized gfp promoter fusions expressed from integrated transgenes that show expression in the lateral IL2 subtype (degl-1: otIs825, egas-4: otIs833), or the dorsoventral IL2 subtype (egas-1: otIs846). (E) Nuclear-localized reporter alleles for subtype-specific markers. Lateral IL2 subtype: ins-1(syb5452[ins-1::SL2::gfp::H2B]), flp-14(syb3323[flp-14::T2A::3xNLS::gfp]), nlp-69(syb4512[nlp-69::SL2::gfp::H2B]), and npr-37(syb4440[npr-37::SL2::gfp::H2B]). Dorsoventral IL2 subtype: degl-2(syb5229[degl-2::SL2::gfp::H2B]), flp-32(syb4374[flp-32::SL2::gfp::H2B]), aex-2(syb4447[aex-2::SL2::gfp::H2B]), flp-5(syb4513syb4513[flp-5::SL2::gfp::H2B]), and dmsr-2(syb4514 [dmsr-2::SL2::gfp::H2B]). For several of the more broadly expressed markers, NeuroPAL was used for cell identification (ID) (images in SI Appendix, Fig. S2A). (F) IL2 subtype marker genes are unc-86 dependent. In an unc-86(ot1158) null allele, both lateral IL2 markers [nlp-69(syb4512) and egas-4/otIs833), and dorsoventral IL2 markers [degl-2(syb5229), egas-1/otIs846, flp-32(syb4374), and flp-5(syb4513)] are affected. Quantification is in SI Appendix, Fig. S2B. Images and quantification for flp-32 and flp-5 regulation by unc-86 are shown in SI Appendix, Fig. S2 C and D.
Results
Defining Subtypes of the IL2 Sensory Neuron Class.
Single cell transcriptional profiling of the entire, mature nervous system of C. elegans (11) revealed that all six IL2 neurons display a plethora of commonalities among the six neurons, supporting their initially anatomically based classification into a unique class (10) (Fig. 1C). Nevertheless, a distinctive set of molecules is differentially expressed between the dorsoventral pairs and the lateral pair (SI Appendix, Fig. S1) (11). Since the IL2 neurons are possible mechanoreceptors, at least in the dauer stage (13), it is of particular note that scRNA profiling revealed distinctive expression patterns of the DEG/ENaC/ASIC family of ion channels, several of which (e.g., MEC-4 and MEC-10) are known mechanoreceptors in other cellular contexts (SI Appendix, Fig. S1) (17). Specifically, two DEG/ENaC/ASIC family members, asic-2 and del-4, are expressed in all six IL2 neurons, while five unusual DEG/ENaC/ASIC family members show a highly patterned expression in the IL2 subtypes (SI Appendix, Fig. S1). Three DEG/ENaC/ASIC proteins characterized by the existence of an additional EGF domain (the EGAS proteins) (18) are expressed either only in the dorsoventral IL2 neurons (egas-1 and egas-3) or only in the lateral IL2 neurons (egas-4). Similarly, two small transmembrane proteins with an DEG/ENaC/ASIC domain also show expression in either the dorsoventral IL2 neurons (F58G6.8) or the lateral IL2s (Y57G11C.44) (11). We named these two proteins DEGL-1 (Y57G11C.44) and DEGL-2 (F58G6.8), for “degenerin channel-like.” We generated promoter fusions for three of these five unusual, subtype-specific, putative mechanoreceptors (egas-1, egas-4, and degl-1) and generated a reporter allele by CRISPR/Cas9 genome engineering for another one (degl-2). These reporters confirmed their expression in either the lateral or dorsoventral IL2 subtypes, as predicted by the scRNA analysis (Fig. 1 D and E).
In addition, as per our scRNA data (11), several neuropeptides and neuropeptide receptors also show IL2 subtype–specific expression (SI Appendix, Fig. S1). We recapitulate the scRNA data by generating reporter alleles (via CRISPR/Cas9 genome engineering) for four neuropeptides (flp-32, flp-5, flp-14, and nlp-69), two neuropeptide receptor (dmsr-2 and npr-37), and one insulin-like peptide (ins-1). We found that these genes are indeed all expressed in a subtype-specific manner in either lateral (flp-14, nlp-69, ins-1, and npr-37) or dorsoventral IL2 neurons (flp-32, flp-5, and dmsr-2, aex-2) (Fig. 1 D and E and SI Appendix, Fig. S2A). Taken together, the dorsal and ventral pair form one subclass of IL2 neurons and the lateral pair another subclass.
Subclass-Specific Features Are Controlled by the Class-Specific Terminal Selector unc-86.
The identity of all six IL2 neurons is specified by the BRN3-type POU homeobox gene unc-86, a phylogenetically conserved, terminal selector transcription factor (19–21). UNC-86/BRN3 is expressed in all six IL2 neurons and is required for the expression of all tested molecular features of the IL2 neurons (20–22), as well as for their unique dendritic morphology (14). The specificity of action of UNC-86/BRN3, which is expressed in multiple other neuron types as well (22), is determined by the IL2-neuron–specific collaboration of UNC-86 with the transcription factors CFI-1 and SOX-2 (20, 23).
We tested whether subtype-specific IL2 markers also require the unc-86 terminal selector, or, alternatively, whether such subtype features are regulated independently of unc-86-dependent “pan-class” features (20). Crossing two lateral IL2 markers (egas-4 and nlp-69) and four dorsoventral IL2 markers (egas-1, degl-2, flp-32, and flp-5) with an unc-86 null mutant allele generated by CRISPR/Cas9 genome engineering revealed that each tested marker requires unc-86 for expression in either the lateral or dorsoventral IL2 (Fig. 1F and SI Appendix, Fig. S2 B–D). This observation 1) demonstrates that unc-86/BRN3 is critical not only for controlling pan-class features but also for IL2 subtype diversification and 2) suggests that other factors must either promote and/or restrict the ability of UNC-86 to control expression of IL2 subtype–specific genes.
unc-39, a SIX4/5 Homolog, Is Continuously Expressed in the Lateral IL2 Subtypes.
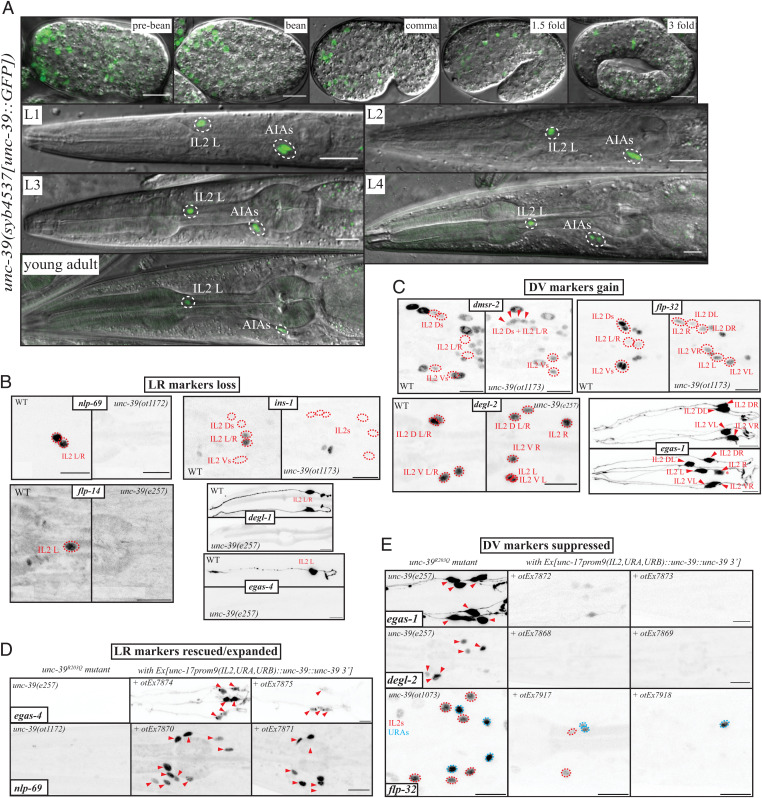
Through the nervous system–wide expression analysis of all GFP-tagged homeodomain proteins, we have found that each C. elegans neuron class expresses a unique combination of homeodomain proteins (24). Using a fosmid-based reporter transgene, we found that one of the homeodomain proteins, the SIX4/5 ortholog UNC-39 (25), is selectively expressed in the lateral IL2, but not the dorsoventral IL2 neurons (24). We sought to confirm this expression pattern by inserting gfp at the C terminus of the unc-39 locus by CRISPR/Cas9 genome engineering (Fig. 2A). Expression of the gfp-tagged locus, unc-39(syb4537), is observed in the lateral IL2 (but not dorsoventral IL2) neurons throughout all larval and adult stages (Fig. 2A). In the embryo, expression is also restricted to the lateral IL2 neurons and never observed in the dorsoventral IL2 neurons (26). The only other cell type that expresses the unc-39 reporter allele throughout larval and adult stages is the AIA interneuron class. There are additional sites of expression in the embryo, consistent with unc-39 affecting proper development of other cell types (25, 27). Expression of unc-39 in the lateral IL2 neurons is eliminated in unc-86 mutants at all stages, but not in the AIA neurons, where unc-86 is not expressed (SI Appendix, Fig. S3A).
Fig. 2.
Molecular IL2 subtype diversification is controlled by unc-39. (A) Expression pattern of the unc-39(syb4537[unc-39::gfp]) reporter allele across embryonic and larval stages. (B) Lateral IL2 marker expression is lost in unc-39R203Q mutant animals (either canonical e257 allele or CRISPR/Cas9 genome engineered ot1172 or ot1173 alleles with identical nucleotide change). Markers are nlp-69(syb4512[nlp-69::SL2::gfp::H2B]), ins-1(syb5452[ins-1::SL2::gfp::H2B]), flp-14(syb3323[flp-14::SL2::gfp::H2B]), degl-1 (otIs825), and egas-4 (otIs833). NeuroPAL images for cell identification and quantification are in SI Appendix, Fig. S4 B and C. (C) Dorsoventral IL2 markers dmsr-2(syb4514[dmsr-2::SL2::gfp::H2B]), flp-32(syb4374[flp-32::SL2::gfp::H2B]), degl-2(syb5229[degl-2::SL2::gfp::H2B]), and egas-1(otIs846) are gained in all IL2 neurons of unc-39R203Q mutant animals (either canonical e257 allele or CRISPR/Cas9 genome engineered ot1173 allele with identical nucleotide change). We counted gain of expression as an all or none phenotype, but the converted lateral IL2 sometimes can be dimmer (in the Bottom Right subpanel, egas-1, the asterisk marks such a case). NeuroPAL images for cell ID and quantification are in SI Appendix, Fig. S4 B and C. Note that the IL2 are often displaced (Fig. 3A) in unc-39R203Q mutant animals. In the unc-39(ok2137) null mutant animals, the front part of the anterior ganglion moves past the anterior bulb of the pharynx (SI Appendix, Fig. S4D). (D) Ectopic expression of unc-39 in the IL2, URA, and URB neurons in an unc-39R203Q mutant background (e257, ot1172, or ot1173 allele) results in rescue of mutant phenotype in the lateral IL2 neurons and conversion of dorsal/lateral IL2 neurons. In each subpanel, the unc-39R203Q control is shown on the Left and two independent extrachromosomal array lines on the Right. Two lateral IL2 markers [nlp-69(syb4512) rescue lines otEx7870 and otEx7871, egas-4/otIs833 otEx7874, and otEx7875] that are lost in the hypomorph are now partially rescued and even expanded in more than just the two lateral IL2s or even the six IL2 (nlp-69, 10 cells in the Middle panel vs. 6 on the Right). (E) Three dorsoventral IL2 markers [degl-2(syb5229) rescue lines otEx7868 otEx7869, egas-1/otIs846 rescue lines otEx7872 and otEx7873, and flp-32(syb4374) rescue lines otEx7917 and otEx7918] that are found in all the IL2 in the hypomorphic mutant animals are repressed in all subtypes upon unc-39 misexpression. unc-39 is able to suppress an unc-86–dependent marker like flp-32(syb4374) in all IL2s but also the relatively similar URAs, which all share unc-86 as identity regulators. In the flp-32(ot1073) image a ventral view is provided. Quantification is in SI Appendix, Fig. S4 E and F.
Continuous expression of a transcription factor throughout the life of a cell is often an indication of autoregulation (28, 29). We tested whether unc-39 autoregulates its own expression by first defining cis-regulatory elements required for continuous expression in the IL2 and AIA neurons. We found that neuronal unc-39 expression in the lateral IL2 neurons and the AIA neurons is recapitulated by a 2.3-kb region directly upstream of the unc-39 coding region (SI Appendix, Fig. S3B). Expression of this promoter fusion construct is down-regulated in the unc-39 mutant animals (SI Appendix, Fig. S3B). We deleted a predicted homeodomain binding site within this region into the endogenous unc-39(syb4537) reporter allele using CRISPR/Cas9 genome engineering and also observed loss of expression in both IL2 and to a lesser extent in AIA (SI Appendix, Fig. S3 C and D). This regulatory mutation is not the result of a failure to initiate unc-39 expression in IL2 and AIA, since these mutant animals display wild-type (WT)–like expression of the gfp-tagged unc-39 locus until the first larval stage, after which expression fades away (SI Appendix, Fig. S3C). Since maintained expression genetically depends on unc-39, as well as on a predicted homeodomain binding site in its promoter, we suspect that the predicted homeodomain binding site is an UNC-39 binding site. We conclude that like in other well-studied cases (28, 29), unc-39 expression is initiated by an upstream regulatory factor (unc-86) and then maintained through autoregulation.
unc-39 Is Required for IL2 Subtype Diversification.
To investigate the functional effects of unc-39, we relied mainly on the canonical unc-39(e257) loss-of-function allele, which contains a R203Q mutation in the SIX domain of unc-39 (25). To avoid linkage issues , we introduced the same mutation in different reporter strain background using the CRISPR/Cas9 system. We find that unc-39R203Q mutant animals display a loss of the expression of all five tested lateral specific markers, egas-4, degl-1, ins-1, flp-14, and nlp-69 (Fig. 2B). Conversely, unc-39R203Q mutant animals display a concomitant gain of all four tested dorsoventral IL2 marker expression (egas-1, flp-32, dmsr-2, and degl-2) in the lateral IL2 neurons of unc-39 mutants (Fig. 2C), indicating that the lateral IL2 neurons have adopted the identity of the dorsoventral IL2 neurons.
unc-39 selectively affects IL2 subtype specification, since we observe no effects on the expression of identity features that are expressed by all six IL2 neurons. Specifically, expression of the vesicular transporter unc-17 and the kinesin klp-6, both controlled by unc-86 in all six IL2 neurons (20), is not affected in unc-39(e257) mutant animals or the unc-39(ok2137) null allele (SI Appendix, Fig. S4D).
unc-39 Is Sufficient to Transform IL2 Subtype Identities.
To ask whether unc-39 is not only required but also sufficient to induce lateral IL2 subtype identity and to suppress dorsoventral IL2 identity, we ectopically expressed unc-39 in the dorsoventral IL2 neurons, using a pan-IL2 driver, an enhancer fragment from the unc-17 locus (30). We verified that this driver is unaffected in unc-39 null and hypomorphic animals (SI Appendix, Fig. S4D). In an unc-39(e257) background, this transgene was not only able to rescue the loss of the lateral IL2 marker genes egas-4 and nlp-69 in the lateral IL2 neuron, but was also able to induce egas-4 and nlp-69 expression in the dorsoventral IL2 subtype (Fig. 2D). Conversely, ectopic expression of the dorsoventral IL2 markers in the lateral IL2 neurons, observed in unc-39 mutants, is not only suppressed by this transgene, but expression of these dorsoventral IL2 subtype markers are repressed in the dorsoventral IL2 subtypes as well (Fig. 2E). Together, these findings demonstrate that unc-39 is not only required, but also sufficient to promote lateral IL2 identities.
In animals that express unc-39 under the unc-17prom9 driver, we also noted occasional ectopic expression of the lateral IL2 marker nlp-69 in a few additional neurons, which are likely the URA and URB neurons, which also express the unc-17prom9 driver (30). unc-86 is known to work as a putative terminal selector in the URA and URB neurons (20). We therefore surmise that unc-39, in conjunction with unc-86, is able to induce IL2 lateral identity in other cellular contexts as well. Vice versa, ectopic unc-39 expression is also sufficient to suppress dorsoventral IL2 marker genes that normally happen to be expressed in other neurons. Specifically, the neuropeptide-encoding flp-32 marker is normally strongly expressed in the unc-86–dependent URA and dorsoventral IL2, and dimly in the lateral IL2 neurons. Ectopic unc-39 expression with the unc-17prom9 driver is able to almost fully suppress flp-32 expression in these neurons (Fig. 2E), to an even lower levels than the already weak wild-type lateral IL2 expression (Fig. 1E). Taken together unc-39 is sufficient to activate and to repress specific target genes not just in the context of the IL2 neurons, but also in other cellular contexts that normally require the unc-86 homeobox gene for their proper differentiation.
Transformation of Other Phenotypic Features of IL2 Subtypes in unc-39 Mutants.
A transformation of lateral IL2 to dorsoventral IL2 identity in unc-39 mutants is not only indicated by molecular markers, but is also further supported by an analysis of IL2 morphology. First, we note that in unc-39(e257) mutants, lateral IL2 cell bodies can become closely associated with dorsoventral IL2 cell bodies and their dendritic projections are often fasciculated with the dorsoventral IL2 dendrites (Fig. 3A and SI Appendix, Fig. S5A). Moreover, the positional stereotypy of the lateral IL2 soma is transformed to the much more variable dorsoventral IL2 soma position that we had previous described (31).
Fig. 3.
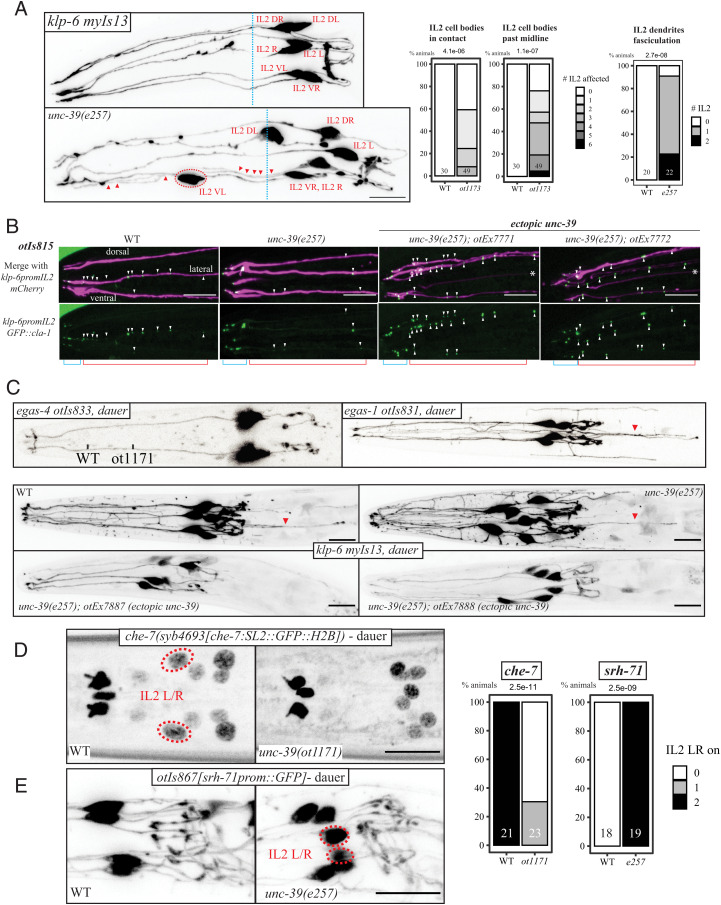
Morphological IL2 subtype diversification is controlled by unc-39. (A) The neurites of all six IL2 are labeled by the unc-39 independent marker myIs13 (klp-6prom::GFP). Left: In WT animals, IL2 cell bodies are usually adjacent to the anterior bulb of the pharynx and their processes run in six fascicles alongside. In the unc-39 hypomorphic alleles ot1171 and e257 (both carrying the R203Q change), the lateral IL2 neurons are often out of position, fully overtaking the midline of the anterior bulb (red circled neuron past the blue dash line). Their dendrites join the the dorsal or ventral tracts (red triangles, see SI Appendix, Fig. S4A). Right: Quantification of neurite fasciculation defects (abnormally close tracts). We also considered cell body position defect like anteriorly displaced IL2 (crossing the blue midline) or IL2 soma entering into contact with each other, using NeuroPAL transgene otIs669 in wild type and unc-39R203Q animals. (B) A cla-1 reporter transgene, that uses the klp-6 driver to label the neurite with mCherry and expresses a GFP-tagged short CLA-1 protein isoform (otIs815) is found in the endings of all six IL2 dendrites (blue segment), but also further along the dendrites (red segment; punctae marked with white triangles) of the lateral IL2 neurons. This second group punctae (along the dendrite) are eliminated in the unc-39R203Q hypomorph. Upon misexpression of unc-39 these punctae frequently appear in multiple dendrites. See quantifications in SI Appendix, Fig. S5B. (C) Dauer arborization changes as shown by myIs13[klp-6prom::GFP]; worms are SDS selected. Top: In wild-type animals, IL2 neurons display dauer-specific arborization, with increased branching in the dorsoventral subtypes compared to the lateral subtypes. We show dauer pictures of our subtype-specific intregrated cytoplasmic reporter, egas-4(otIs833) Top Left in ventral position, egas-1 (otIs831) Top Right. Bottom: IL2 branching imaged with myIs13 cytoplasmic reporter shows increased branching of the lateral IL2 neurons of unc-39(e257) animals, which is difficult to quantify, while overexpressing unc-39 using the unc-17prom::gfp driver in all IL2 neurons (transgenic lines otEx7887 and otEx7888) strongly suppress this arborization.The backward projecting neurites we saw are labeled with red triangles. See quantifications in SI Appendix, Fig. S5C. (D) Expression of the innexin che-7 reporter allele syb4693 is gained in dauer in the lateral IL2 in WT worms, but not in unc-39R203Q mutants. (E) An srh-71 GPCR reporter transgene (otIs867) is expressed in all IL2 in wild-type worms and lost from the lateral IL2 when they go into dauer. In unc-39R203Q mutant dauers (ot1171 and e257 alleles), expression remains in the lateral IL2 neurons while expression is eliminated in wild-type animals.
We serendipitously discovered another subcellular feature that distinguishes lateral from dorsoventral IL2 neurons: All the IL2 neurons localize the synaptic active zone marker CLA-1/clarinet at the dendritic endings in the nose of the worm, but the lateral IL2 neurons also show CLA-1 punctae localized along the dendrite (Fig. 3B). Synaptic vesicle machinery had previously been observed in ciliary endings of other sensory neurons (32–34), but had not been examined in the IL2 neurons to date. Dendritic CLA-1 localization is likely unrelated to extracellular vesicle (EV) release by IL2 neurons, since we find it to be unaffected in klp-6(ky611) mutants, which lack the kinesin that transports EVs (35). In unc-39(e257) mutants, CLA-1 localization along the length of the lateral IL2 dendrites is reduced if not eliminated, comparable to what is normally seen only in the dorsoventral IL2 subtypes in wild-type animals. Conversely, ectopic unc-39 expression in all six IL2 neurons results in CLA-1 signals being observed along the length of all IL2 dendrites, comparable to what is normally only observed in the lateral IL2 neurons (Fig. 3B). We conclude that CLA-1 dendritic localization is differentially regulated in the two IL2 subtypes and that unc-39 controls this feature as well.
Upon entry into the dauer stage, the six IL2 neurons adopt further subtype-specific features. On a morphological level, the dorsoventral, but not lateral IL2 neurons, develop extensive dendritic branches (14) (Fig. 3C). unc-39(e257) animals form SDS-resistant dauer normally, but the lateral IL2 neurons now seem to display dendritic branches, i.e., adopt features normally exclusive to the dorsoventral IL2 neurons (Fig. 3C). These branches appear to merge with the IL2 dorsoventral branches. Conversely, ectopic expression of unc-39 in all six IL2 neurons almost completely suppresses branch formation in the dorsoventral IL2 neuron, making them resemble the branchless lateral IL2 neurons.
We have also noticed that in the dauer-stage animals dorsoventral IL2 neurons appear to project neurites posterior to the cell body, as visualized with the cytoplasmic and subtype-specific egas-1 and egas-4 markers (Fig. 3C). While ectopic projections are hard to score in unc-39 mutant animals, they obviously disappear upon ectopic expression of unc-39 in all IL2 neurons. This observation further corroborates the notion that unc-39 controls subtype-specific morphological features of the IL2 neurons (Fig. 3C).
Entry into the dauer stage also leads to IL2 subtype–specific changes in gene expression. These include the dauer-specific induction of a GFP-tagged electrical synapse protein, CHE-7, exclusively in the lateral IL2 neurons (36). Conversely, the GPCR encoding gene srh-71, which, under replete conditions, is expressed in all six IL2 neurons, becomes selectively repressed in the lateral IL2 neurons (37). Both of these molecular remodeling events fail to occur in unc-39 mutant dauers (Fig. 3 D and E). A che-7 transcriptional CRISPR reporter fails to be properly induced in lateral IL2 neurons and, conversely, a srh-71 reporter construct is derepressed in the lateral IL2 neurons of unc-39 mutant dauer animals (Fig. 3E). After dauer recovery, srh-71 is turned off in wild-type animals, while in unc-39 mutant animals, srh-71 expression persists (SI Appendix, Fig. S5D). Taken together, both molecular as well as morphological data are consistent with what can be referred to as a “homeotic identity transformation” of the lateral IL2 to the dorsoventral IL2 neurons in unc-39 mutants.
Nested Homeobox Gene Function Also Controls Subtype Diversification in the RMD Neck Motor Neuron Class.
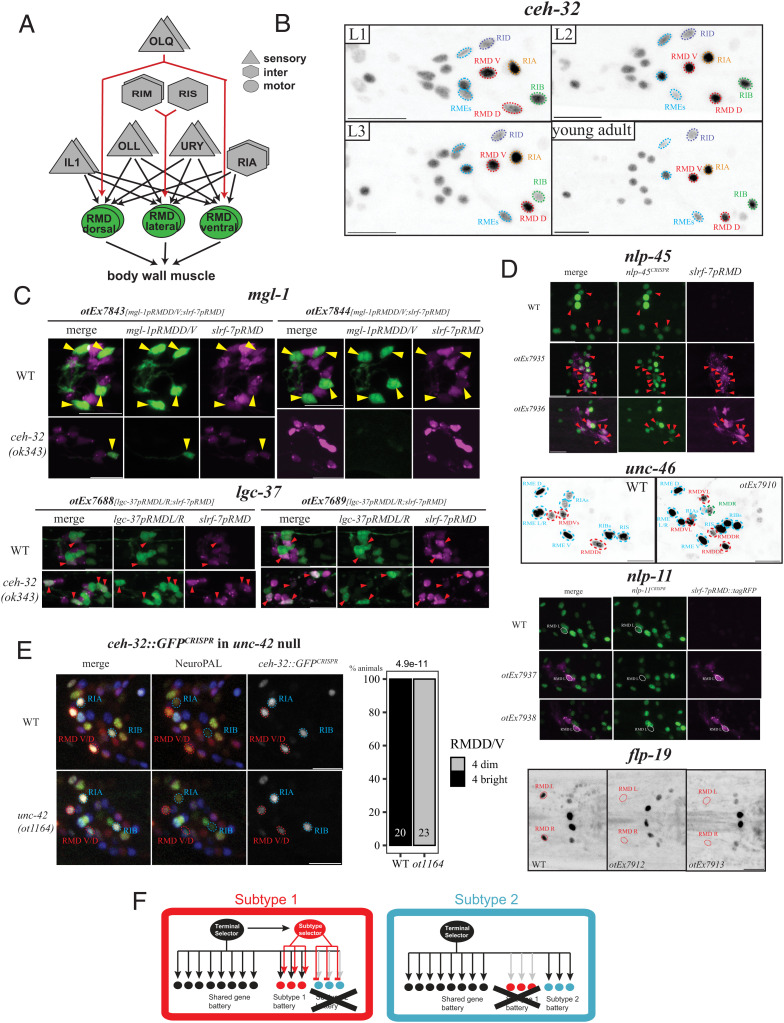
Our functional analysis of the IL2 neurons reveals a nested set of homeobox gene function: a homeobox terminal selector controls all identity features of a neuron and a subordinate, subtype-specific homeobox gene differentiates the identity of neuronal subtypes. Does the concept of nested homeobox gene function apply in other neuron classes as well? Intriguingly, our genome- and nervous-system–wide expression pattern analysis of homeodomain proteins revealed that another member of sine oculis/SIX homeodomain protein family, the SIX3/6-like CEH-32 protein, is expressed in a subtype-specific manner in yet another radially symmetric class of neurons, the RMD neck motorneurons (24) (Fig. 4A). A CRISPR/Cas9-engineered gfp reporter allele is expressed throughout all larval and adult stages only in the dorsoventral RMD neuron subclass, not the lateral RMD subclass (Fig. 4B).
Fig. 4.
Another sine oculis/SIX-type homeobox gene diversifies another radially symmetric neuron class. (A) RMD neck motor neuron overview. Other connectivity difference between RMD subtypes exist (e.g., dopamine neurons; or lateral neuron-specific feedback to RIA) (10) but are not illustrated here for clarity. (B) A GFP-tagged ceh-32 reporter allele (ot1040) is expressed across all larval stages in the ventral and dorsal RMD subtypes (circled in red), located, respectively, dorsally and ventrally. ceh-32 expression is never observed in the lateral RMD. The ceh-32 expressing neuron classes that are closest to the RMD (RIA/RIB/RID/RME) are labeled. (C) Loss-of-function experiments. RMD subtype marker expression in ceh-32(ok343) null mutant animals. slrf-7prom::TagRFP is present on all transgenes to label overall RMD fate. Top: expression of a mgl-1 reporter, expressed in dorsoventral RMD (yellow triangles), is lost in two different lines (otEx7843 and otEx7844). Bottom: a lgc-37 reporter, expressed in lateral RMD (red triangles) is ectopically expressed in more than just two RMDs, as seen in two different lines (otEx7688 and otEx7689). The number of slrf-7(+) neurons is not affected (expression in the dorsoventral RMDs appears brighter in ceh-32 mutants). Quantifications are found in SI Appendix, Fig. S6C. (D) Gain-of-function experiments. ceh-32 is misexpressed in all six RMD neurons using the slrf-7 driver. The dorsoventral subtype markers nlp-45 (reporter allele ot1032) and unc-46 (fosmid otIs568) can be found in more than the usual four expected RMDs (using ceh-32 misexpressing arrays otEx7935 and otEx7936 for nlp-45 reporter allele, and otEx7909 for unc-46 fosmid line; another line, otEx7910, had no effect). In the case of nlp-45, the cells expressing both nlp-45 and a slrf-7p::tagRFP marker as part of the otEx7935/7936 misexpression arrays are shown with red triangles. The lateral RMD subtype markers nlp-11 (reporter allele syb4759) and flp-19 (reporter allele syb3278) are repressed by pan-RMD expression of ceh-32 (expressed with arrays otEx7937 and otEx7938 in the nlp-11 reporter allele and otEx7912 and otEx7913 in the flp-19 reporter allele). In the nlp-11 reporter allele background, we also used slrf-7p::tagRFP as part of the misexpression array to help identify the RMD neurons. The lateral RMDs are circled in white. For flp-19, we show ventral images, the lateral RMDs are circled in red and would be anterior to the two bright cells in the middle (AIAs). Quantifications are found in SI Appendix, Fig. S6D. (E) In an unc-42(ot1164) null allele, the ceh-32 reporter allele ot1040 becomes dimmer in both dorsal and ventral RMDs. We quantified the RMDD/V (circled red) expression as bright vs. dim, using the brightness of RIA and RIB (circled blue) as reference. Those neurons do not express unc-42 and stay unchanged in unc-42 null mutant animals. P values are done via Fisher two-sided test. (F) Summary. Terminal selectors operate in a nested regulatory configuration in which they regulate the expression of subtype terminal selectors that specify subtype identity. Loss of a subtype selector results in homeotic identity transformation to another subtype identity. Subtype selectors promote the ability of a terminal selector to activate specific subtype identity features (“subtype 1 battery”) and antagonize the ability of a terminal selector to activate specific subtype identity features (“subtype 2 battery”). Open questions are how the subtype specificity of subtype regulators are controlled and whether subtype selectors act directly on target genes (as indicated here) or through intermediary factors.
The RMD neck motor neuron class is entirely distinct from the IL2 sensory neuron class, in terms of location in a different ganglion, function (motor vs. sensory neuron), morphology (i.e., neurite projection patterns), synaptic connectivity and molecular composition. But like the IL2 neuron class, the RMD class is also composed of three bilateral neuron pairs, a dorsal, lateral, and ventral pair that share a plethora of anatomical, as well as molecular features among each other (10, 11, 15) (Fig. 4A). Similar to the IL2 neuron case, all six RMD neurons are genetically specified by a homeobox-type terminal selector gene, the Prop1-like unc-42 gene, which controls the expression of essentially all known terminal identity features of the RMD neurons (38). Yet, despite many similarities, the dorsoventral and the lateral pairs do show differences in synaptic connectivity, as well as molecular composition (10, 11, 15) (Fig. 4A). For example, the metabotropic glutamate receptor mgl-1 has been found to be expressed in the dorsoventral RMD pairs, but not the lateral pair (20, 39), while the GABA receptor subunit lgc-37 is selective expressed in the lateral, but not dorsoventral RMD pairs (11, 20, 39, 40). Through the CRISPR/Cas9-mediated engineering of reporter alleles, we validated other genes that scRNA analysis found to be subtype-specifically expressed, including several”neuropeptide-encoding genes (flp-19, nlp-11, and nlp-45) (Fig. 4D).
Armed with these markers of subtype identity, we asked whether CEH-32 parallels the subtype diversification function of UNC-39 in the context of the RMD neurons. We found that expression of a reporter transgenes that monitors expression of mgl-1 in the dorsoventral RMD neuron is lost in ceh-32 mutant animals (Fig. 4C and SI Appendix, Fig. S6A). Conversely, the lgc-37 gene, normally restricted to the lateral RMD neurons, becomes derepressed in the dorsoventral RMD neurons (Fig. 4C and SI Appendix, Fig. S6A). In contrast, ceh-32 does not affect expression of a pan-RMD marker, C42D4.1/srlf-7 (Fig. 4C and SI Appendix, Fig. S7).
To test for sufficiency of ceh-32 function, we expressed ceh-32 in all six RMD neurons using the promoter of the pan-RMD C42D4.1/slrf-7 promoter. Such transgenic animals showed gain of several dorsoventral subtype markers in the lateral RMD neurons and a concomitant loss of left/right subtype markers (Fig. 4D and SI Appendix, Fig. S6B). Taken together, our data indicate a homeotic identity change of RMD subtypes that is akin to what we observed in the IL2 neurons upon manipulation of unc-39 expression.
The pan-RMD terminal selector unc-42 affects both the subtype-specific markers, as well as the pan-RMD markers (38, 40, 41) (SI Appendix, Fig. S8). Moreover, ceh-32 expression in the dorsoventral RMD neurons is diminished in unc-42 mutants (Fig. 4E). Hence, as in the case of the IL2 neurons, a subtype-specific since oculis/SIX-type homeodomain protein controls subtype-specific identity features, while a pan-class homeobox terminal selector controls both pan-class identity features as well as subtype-specific features.
Discussion
An analysis of the taxonomic relationship between neuronal cell types in many different animal brains reveals closely related neuronal subtypes at the terminal branch point of such taxonomies that share a multitude of anatomical and molecular features but differ in a select number of phenotypic properties. In theory, one can envision that such very closely related cell types are independently specified by entirely distinct regulatory programs. In the cases that we describe here, this is not the case. We rather uncover, in two distinct cellular contexts (IL2 neuron class and RMD neuron class), a nested regulatory architecture that specifies differences between closely related neuronal cell types (Fig. 4F). Shared, as well as subtype-specific features of a neuronal cell type are controlled by a terminal selector transcription factor that is expressed in all subtypes (e.g., UNC-86 in all IL2 subtypes and UNC-42 in all RMD subtypes) (Fig. 4F). Subtype specificity of select identity features are dictated by “subtype terminal selectors” that modulate the ability of a terminal selector to control the expression of specific target genes (Fig. 4F). Depending on the target genes, a subtype terminal selector like UNC-39 may either assist the ability of a class-specific terminal selector like UNC-86 to bind DNA and/or to activate transcription of subtype-specific features (e.g., lateral IL2 markers) or, on other target genes, prevent the terminal selector from doing so (e.g., dorsoventral IL2 markers) (Fig. 4F). Given that the diversification of neuronal cell types into closely related subtypes is observed in many different contexts in invertebrate and vertebrate brains (3, 7–9), we envision the nested gene regulatory principle described here to be broadly applicable, with similarities of two cell types being defined by the same terminal selector, but diversified by subtype terminal selectors.
In both distinct cellular contexts in which we described such a nested regulatory architecture, both the terminal selectors as well as the subtype selectors are homeobox genes, thereby further corroborating the importance of homeobox genes in neuronal identity control (24). A nested, homeobox-dependent regulatory architecture that involves also sine oculis/SIX family proteins may also exist in other nervous systems. For example, vertebrate cone photoreceptor differentiation requires a terminal selector–type function of the CRX and OTX2 proteins, broad regulators of all cone photoreceptors (42) and, in zebrafish, the subordinate control of a distinct type of cone photoreceptors by the SIX6/7 proteins (43, 44). Non-homeobox genes can function as subtype selectors as well. One recent example is the FEZF1 Zn finger transcription factor that acts as a subtype selector to control a binary fate choice between closely related on and off starburst amacrine cells in the vertebrate retina (45).
The subtype identity transformation phenotypes of the sine oculis/SIX-type homeobox gene mutants that we describe here are conceptually reminiscent of body segment identity transformation in HOX cluster mutants. In fact, we had previously described that C. elegans HOX genes also act in terminal motor neuron differentiation to diversify motor neuron subtype identities along the anterior/posterior axis (46). Viewing neuronal subtype identity transformations as “homeotic” places them into the rich framework of evolutionary thought that started with William Bateson’s definition of homeosis in 1894 and further elaborated on by Richard Goldschmidt (47–49). While it remains debatable whether homeotic identity changes of entire body parts have been drivers of evolution (50, 51), homeotic identity transformations of smaller units, i.e., individual cell types, can be more easily envisioned to have played a role in the evolution of the composition and complexity of tissue types (48). Specifically, it is conceivable that neuronal subtypes evolved from a homogeneous, terminal selector–controlled state in which a group of neurons all shared the same features. The gain of expression of a subtype selector in a subgroup of these cells may have then enabled the modulation of the expression of a subset of the shared identity features of this group of neurons.
Materials and Methods
Strains and Mutant Alleles.
A complete list of mutant and transgenic strains can be found in SI Appendix, Table S1. Due to linkage issues (nlp-69, che-7, and NeuroPAL integrant otIs669), we regenerated the e257 allele, a G > A mutation that alters arginine 203 to glutamine in the SIX domain in several different strain backgrounds, using CRISPR/Cas9 genome engineering following ref. 53. We use the following repair template (bold: sgRNA, italics: G > A mutation): CGTGGCAAAGAACTGAATCCAGTGGAAAAATATCaGCTGAGACGAAAGTTTCCGGCTCCGAAAACAATTTGG. The G > A edit eliminates the PAM site, the edited allele is identical to e257.
The unc-39 binding site mutant ot1193 was generated via the same CRISPR protocol, with the guide CC939 and repair template shown in SI Appendix, Fig. S3B.
We generated in the background of the ceh-32 reporter allele ot1040 a novel unc-42 null allele, unc-42(ot1164), using the same CRISPR protocol as described above, deleting the entire coding gene of unc-42. We used the two following guides, GCTCATtgtgtgagtgaaag and tctcactgatagactaatgt, and the following repair oligo:
ttcggtcacccctcactttccacatttctccgctttgttggtacgtacatttacaaaaagacaacggttt.
Mixes using the described amount, were injected as simple arrays except for the ceh-32 and unc-39 rescue strains, which were injected as complex. pBluescript DNA as an injection filler to reach 100 ng/μL for simple injection mixes, and OP50 genomic DNA for complex mixes. Integration was done via gamma radiation, with the strains being outcrossed at least three times.
The coinjection marker inx-18promAVG::TagRFP is from ref. 54. The inx-6prom(2TAAT-deletion)::GFP coinjection marker is from ref. 55 and labels the procorpus of the pharynx.
Reporter Construction.
Reporter alleles for che-7, dmsr-2, degl-2, flp-5, flp-14, flp-19, flp-32, ins-1, nlp-69, and nlp-11 (see strain list) were generated using CRISPR/Cas9, inserting a SL2::gfp::h2b cassette at the C terminus of the respective gene. These strains, as well as the unc-39 reporter allele, syb4537, a direct fusion of gfp to the C terminus of unc-39, were generated by Sunybiotech.
For srh-71, we reinjected the srh-71 PCR fusion mix from ref. 23, yielding an otEx7765 pha-1 rescue line that was integrated.
Promoter fusions for the egas1, egas-4, degl-1, and unc-39 genes were Gibson cloned into the pPD95.75 backbone, using either the SphI/XmaI multiple cloning sites for promoters and KpnI/ApaI for coding sequence/3′ UTR. Primer sequences are shown below:
-
•
egas-1 1422bp promoter: fwd aaccagtgtaccgtccatctg rev atgatttataaggacgttagggaagtttg
-
•
egas-4 278bp promoter: fwd caaatcaaaaacaggtctcatgtacatac rev ttcttgactgaatcctaataggaaattc
-
•
degl-1 230bp promoter: fwd aacaacgctaacaagtaaccagatg rev aaaatttacaaaacactgagcttgtgc
-
•
slrf-7 570bp promoter: fwd gattcgcggaactctagtaaattgaatc rev cctgaaatcaaccatttccatcaagaag
-
•
unc-39 2260bp promoter: fwd gaagagtgtcgaatattgcagcag rev tggaatactactcatcttgcaaacgttc
-
•
unc-39 1754bp CDS and 3′ UTR: fwd ATGACAGACCATCCGCCAATTG rev tgccaacattgattgatctctacg
-
•
ceh-32 3694bp CDS and 3′ UTR: fwd atgttcactccagaacagttcac rev tattttgcagattcgcattacttcgg.
Misexpression Approaches.
The unc-39 and ceh-32 CDS and 3′ UTR fragment described above were misexpressed in all six IL2 or RMD neurons, respectively, using the unc17prom9 driver for the IL2 neurons (30) and, for the RMD neurons, a 570 bp of the 5′ region of the C42D4.1/srlf-7 gene, which encodes a small secreted protein that is a member of a large family of SXP/RAL2-related proteins, characterized by an ANIS5 Pfam domain (PF02520). scRNA analysis had shown expression in the RMD neurons (11) and this pattern was confirmed with promoter fusions (56). We find that 570 bp of the D4.1/srlf-7 promoter drive expression of TagRFP or GFP in all six RMD neurons, with occasional expression in the SAA neurons (SI Appendix, Fig. S2).
Microscopy and Image Processing.
Worms were anesthetized using 100 mM sodium azide (NaN3) and mounted on 5% agarose pads on glass slides. Z-stack images (40× for L4 animals, 0.7-μm steps, 63× for L1 with 0.4 steps) were acquired using a Zeiss confocal microscope (LSM880) or Zeiss compound microscope (Imager Z2) using ZEN software. Maximum intensity projections of 2 to 30 slices were generated with ImageJ software (57). All scale bars are 10 μm. We show our reporters as inverted gray, with the same range between genotypes.
In the case of NeuroPAL images, we split the channels as the GFP signal channel in gray, and the NeuroPAL coloring channels (mTabBFP2,CyOFP,mNeptune). We adjusted when needed the gamma to 0.5 but for the latter group of channels only. Images are shown as GFP signal in inverted gray in the main figures, and for each of those the merge/GFP in gray/Neuropal colors are shown in supplemental figures. Depth coloring for SI Appendix, Fig. S4A was done in Fiji via the image/hyperstacks/temporal color-code command.
In the case of dauer experiments, we grew gravid adults for 6 to 8 d at 25 °C, washed the plates with 1% SDS into multiple tubes, and left the worms to incubate under gentle agitation for 20 min at least and 2 h at most. Before imaging, we washed off worms in M9 one plate at a time and replated the living dauer and nondauer carcasses on an uncoated plate, imaging for 1 h or so before switching to a new tube of worms in SDS.
Data Analysis and Statistics.
All statistics and plots were done in R. We used the R tidyverse package collection and the ggplot2 graph library. We created bar plots with “geom_bar,” organized scoring data into contingency tables, and used the two-sided Fisher test for P values for most experiments except for misexpression experiments. In those case we plotted neuron counts with “geom_beeswarm,” and used a Wilcoxon rank sum test to compare each misexpression line to the relevant control (unc-39 hypomorph, ceh-32 WT). We adjusted P values for multiple testing via Holm’s method. We did not determine sample sizes in advance, but used as baseline 30 to 40 worms when scoring on a dissecting scope, and for confocal/microscope scoring we stopped at 10 to 15 worms for fully penetrant phenotypes and 15 to 25 otherwise. Extrachromosomal lines were scored as 10 to 15 worms, with a control and at least two lines. On the graphs, the number on each bar shows the number of worms scored per genotype.
Supplementary Material
Acknowledgments
We thank Chi Chen for generating transgenic lines, Tessa Tekieli for an unc-86 null allele and pictures of flp-5 crossed to it, Maryam Majeed for generating the IL2 cla-1 line integrated here and a flp-3 IL1 reporter, Robert Fernandez for providing multiple neuropeptide reporter alleles already crossed to NeuroPAL, Molly Reilly for generating the ceh-32 fosmid rescue, Eduardo Leyva-Díaz for unc-42 CRISPR deletion allele reagents and comments on the manuscript, Surojit Sural for help and advice on dauer experiments, and Berta Vidal for a srh-71 PCR fusion injection mix. We thank the O.H. laboratory members and Nathan Schroeder for advice and feedback. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (grant P40 OD010440). This work was funded by NIH grant R21NS106843 and R01NS110391 and by the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2206817119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Zeng H., Sanes J. R., Neuronal cell-type classification: Challenges, opportunities and the path forward. Nat. Rev. Neurosci. 18, 530–546 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Poulin J. F., Tasic B., Hjerling-Leffler J., Trimarchi J. M., Awatramani R., Disentangling neural cell diversity using single-cell transcriptomics. Nat. Neurosci. 19, 1131–1141 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Network B. I. C. C.; BRAIN Initiative Cell Census Network (BICCN), A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cajal R. y., Histologie du Système Nerveux de l’Homme et des Vertébrés (Maloine, Paris, 1911). [Google Scholar]
- 5.Masland R. H., The fundamental plan of the retina. Nat. Neurosci. 4, 877–886 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Arendt D., Bertucci P. Y., Achim K., Musser J. M., Evolution of neuronal types and families. Curr. Opin. Neurobiol. 56, 144–152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shekhar K., et al. , Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308–1323.e30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Z., et al. , A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241.e26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cembrowski M. S., Spruston N., Heterogeneity within classical cell types is the rule: Lessons from hippocampal pyramidal neurons. Nat. Rev. Neurosci. 20, 193–204 (2019). [DOI] [PubMed] [Google Scholar]
- 10.White J. G., Southgate E., Thomson J. N., Brenner S., The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1–340 (1986). [DOI] [PubMed] [Google Scholar]
- 11.Taylor S. R., et al. , Molecular topography of an entire nervous system. Cell 184, 4329–4347.e23 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ward S., Thomson N., White J. G., Brenner S., Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans.?2UU. J. Comp. Neurol. 160, 313–337 (1975). [DOI] [PubMed] [Google Scholar]
- 13.Lee H., et al. , Nictation, a dispersal behavior of the nematode Caenorhabditis elegans, is regulated by IL2 neurons. Nat. Neurosci. 15, 107–112 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Schroeder N. E., et al. , Dauer-specific dendrite arborization in C. elegans is regulated by KPC-1/Furin. Curr. Biol. 23, 1527–1535 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobert O., Glenwinkel L., White J., Revisiting neuronal cell type classification in Caenorhabditis elegans. Curr. Biol. 26, R1197–R1203 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Wang J., et al. , Cell-specific transcriptional profiling of ciliated sensory neurons reveals regulators of behavior and extracellular vesicle biogenesis. Curr. Biol. 25, 3232–3238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Sheikh U., Kang L., Mechano-gated channels in C. elegans. J. Neurogenet. 34, 363–368 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Hobert O., The neuronal genome of Caenorhabditis elegans. WormBook, 10.1895/wormbook.1.161.1 (2013). Accessed 23 August 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leyva-Díaz E., Masoudi N., Serrano-Saiz E., Glenwinkel L., Hobert O., Brn3/POU-IV-type POU homeobox genes-Paradigmatic regulators of neuronal identity across phylogeny. Wiley Interdiscip. Rev. Dev. Biol. 9, e374 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Zhang F., et al. , The LIM and POU homeobox genes ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron types. Development 141, 422–435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaham S., Bargmann C. I., Control of neuronal subtype identity by the C. elegans ARID protein CFI-1. Genes Dev. 16, 972–983 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finney M., Ruvkun G., The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63, 895–905 (1990). [DOI] [PubMed] [Google Scholar]
- 23.Vidal B., et al. , C. elegans SoxB genes are dispensable for embryonic neurogenesis but required for terminal differentiation of specific neuron types. Development 142, 2464–2477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reilly M. B., Cros C., Varol E., Yemini E., Hobert O., Unique homeobox codes delineate all the neuron classes of C. elegans. Nature 584, 595–601 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanowitz J. L., et al. , UNC-39, the C. elegans homolog of the human myotonic dystrophy-associated homeodomain protein Six5, regulates cell motility and differentiation. Dev. Biol. 272, 389–402 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Ma X., et al. , A 4D single-cell protein atlas of transcription factors delineates spatiotemporal patterning during embryogenesis. Nat. Methods 18, 893–902 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Lim M. A., et al. , Neuroendocrine modulation sustains the C. elegans forward motor state. eLife 5, e19887 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertrand V., Hobert O., Linking asymmetric cell division to the terminal differentiation program of postmitotic neurons in C. elegans. Dev. Cell 16, 563–575 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leyva-Díaz E., Hobert O., Transcription factor autoregulation is required for acquisition and maintenance of neuronal identity. Development 146, dev177378 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serrano-Saiz E., et al. , Modular organization of Cis-regulatory control information of neurotransmitter pathway genes in Caenorhabditis elegans. Genetics 215, 665–681 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yemini E., et al. , NeuroPAL: A multicolor atlas for whole-brain neuronal identification in C. elegans. Cell 184, 272–288.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ojeda Naharros I., et al. , Loss-of-function of the ciliopathy protein Cc2d2a disorganizes the vesicle fusion machinery at the periciliary membrane and indirectly affects Rab8-trafficking in zebrafish photoreceptors. PLoS Genet. 13, e1007150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Datta P., et al. , Accumulation of non-outer segment proteins in the outer segment underlies photoreceptor degeneration in Bardet-Biedl syndrome. Proc. Natl. Acad. Sci. U.S.A. 112, E4400–E4409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazelova J., Ransom N., Astuto-Gribble L., Wilson M. C., Deretic D., Syntaxin 3 and SNAP-25 pairing, regulated by omega-3 docosahexaenoic acid, controls the delivery of rhodopsin for the biogenesis of cilia-derived sensory organelles, the rod outer segments. J. Cell Sci. 122, 2003–2013 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J., et al. , C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr. Biol. 24, 519–525 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattacharya A., Aghayeva U., Berghoff E. G., Hobert O., Plasticity of the electrical connectome of C. elegans. Cell 176, 1174–1189.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vidal B., et al. , An atlas of Caenorhabditis elegans chemoreceptor expression. PLoS Biol. 16, e2004218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berghoff E. G., et al. , The Prop1-like homeobox gene unc-42 specifies the identity of synaptically connected neurons. eLife 10, e64903 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greer E. R., Pérez C. L., Van Gilst M. R., Lee B. H., Ashrafi K., Neural and molecular dissection of a C. elegans sensory circuit that regulates fat and feeding. Cell Metab. 8, 118–131 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gendrel M., Atlas E. G., Hobert O., A cellular and regulatory map of the GABAergic nervous system of C. elegans. eLife 5, e17686 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun H., Hobert O., Temporal transitions in the post-mitotic nervous system of Caenorhabditis elegans. Nature 600, 93–99 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corbo J. C., et al. , CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res. 20, 1512–1525 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogawa Y., et al. , Six6 and Six7 coordinately regulate expression of middle-wavelength opsins in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 116, 4651–4660 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogawa Y., Corbo J. C., Partitioning of gene expression among zebrafish photoreceptor subtypes. Sci. Rep. 11, 17340 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng Y. R., et al. , Binary fate choice between closely related interneuronal types is determined by a Fezf1-dependent postmitotic transcriptional switch. Neuron 105, 464–474.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kratsios P., et al. , An intersectional gene regulatory strategy defines subclass diversity of C. elegans motor neurons. eLife 6, e25751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bateson W., Materials for the Study of Variation, Treated with Especial Regard to Discontinuity in the Origin of Species (Macmillan, London, 1894). [Google Scholar]
- 48.Arlotta P., Hobert O., Homeotic transformations of neuronal cell identities. Trends Neurosci. 38, 751–762 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Goldschmidt R., The Material Basis of Evolution (Yale University Press, New Haven, CT, 1940). [Google Scholar]
- 50.Akam M., Hox genes, homeosis and the evolution of segment identity: No need for hopeless monsters. Int. J. Dev. Biol. 42, 445–451 (1998). [PubMed] [Google Scholar]
- 51.Theissen G., Saltational evolution: Hopeful monsters are here to stay. Theory Biosci. 128, 43–51 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Reilly M. B., et al. , Widespread employment of conserved C. elegans homeobox genes in neuronal identity specification. PLoS Genetics, in press (2022). [DOI] [PMC free article] [PubMed]
- 53.Dokshin G. A., Ghanta K. S., Piscopo K. M., Mello C. C., Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics 210, 781–787 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oren-Suissa M., Bayer E. A., Hobert O., Sex-specific pruning of neuronal synapses in Caenorhabditis elegans. Nature 533, 206–211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhattacharya A., Hobert O., A new anterior pharyngeal region specific fluorescent co-transformation marker. MicroPubl. Biol. 10.17912/micropub.biology.000084 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lorenzo R., Onizuka M., Defrance M., Laurent P., Combining single-cell RNA-sequencing with a molecular atlas unveils new markers for Caenorhabditis elegans neuron classes. Nucleic Acids Res. 48, 7119–7134 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schindelin J., Rueden C. T., Hiner M. C., Eliceiri K. W., The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 82, 518–529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hall D. H., Altun Z., C. Elegans Atlas (Cold Spring Harbor Laboratory Press, 2007). [Google Scholar]
- 59.Cook S. J., et al. , Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 571, 63–71 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.