Significance
The innate immune response is the first line of defense against invading pathogens. Virus infection rapidly activates the intracellular signaling pathways, resulting in antiviral and inflammatory responses. The inflammatory response is beneficial for the early protection against the virus infection; however, unregulated inflammation is harmful to the host. We report a cellular antiinflammatory mechanism that inhibits the virus-induced inflammatory gene expression. Using cellular and mouse models, we reveal IRF3, a critical component of innate antiviral immunity, inhibits the activity of NF-κB, the pro-inflammatory transcription factor. Mechanistically, IRF3 binds the NF-κB–p65 subunit and prevents its translocation to the nucleus. It appears, therefore, that we have uncovered a previously unknown function for IRF3 in regulating viral inflammation.
Keywords: IRF3, NF-κB, viral inflammation, antiviral, innate immunity
Abstract
Interferon (IFN) regulatory factor 3 (IRF3) is a transcription factor activated by phosphorylation in the cytoplasm of a virus-infected cell; by translocating to the nucleus, it induces transcription of IFN-β and other antiviral genes. We have previously reported IRF3 can also be activated, as a proapoptotic factor, by its linear polyubiquitination mediated by the RIG-I pathway. Both transcriptional and apoptotic functions of IRF3 contribute to its antiviral effect. Here, we report a nontranscriptional function of IRF3, namely, the repression of IRF3-mediated NF-κB activity (RIKA), which attenuated viral activation of NF-κB and the resultant inflammatory gene induction. In Irf3−/− mice, consequently, Sendai virus infection caused enhanced inflammation in the lungs. Mechanistically, RIKA was mediated by the direct binding of IRF3 to the p65 subunit of NF-κB in the cytoplasm, which prevented its nuclear import. A mutant IRF3 defective in both the transcriptional and the apoptotic activities was active in RIKA and inhibited virus replication. Our results demonstrated IRF3 deployed a three-pronged attack on virus replication and the accompanying inflammation.
The type-I interferon (IFN) system is a critical innate antiviral mechanism in vertebrates (1–7). Virus infection causes rapid induction of type-I IFNs (e.g., IFN-β) by activating the pattern recognition receptors, such as Toll-like receptors (TLRs), RIG-I–like receptors (RLRs), cyclic GMP–AMP synthase (cGAS), and stimulator of IFN genes (STING), which recognize specific viral nucleic acids in distinct cellular compartments (8–19). By recognizing the viral nucleic acids, the pattern-recognition receptors trigger intracellular signaling pathways to activate the downstream transcription factors IFN regulatory factor 3 (IRF3) and NF-κB, thus causing the transcriptional induction of IFNs and the IFN-stimulated genes (ISGs) (20–24). The secreted IFNs act via IFN receptor to activate the JAK/STAT signaling on infected and uninfected cells to amplify the ISGs and protect them from the subsequent viral infection (1, 25, 26). The ISGs, either singly or in combination with others, interfere with specific steps of the viral life cycle to inhibit the virus replication (5, 7, 27, 28).
IRF3, a ubiquitously expressed protein, is an essential transcription factor for inducing IFN and antiviral genes (21, 24, 29–31). IRF3 deficiency causes susceptibility to a wide range of viral infections, due to the lack of antiviral genes (29, 32–35). In addition to the widely accepted notion that IRF3 performs cellular functions exclusively via its transcriptional activity, we uncovered a nontranscriptional IRF3 (nt-IRF3) function: RLR-induced IRF3-mediated pathway of apoptosis (RIPA), which causes the apoptotic killing of the virus-infected cells (35–41). IRF3 functions require activation by phosphorylation for the transcriptional activity but by polyubiquitination for RIPA (24, 35, 41). For transcriptional function, IRF3, upon phosphorylation in the cytoplasm, translocates to the nucleus in virus-infected cells (21, 31, 42). In RIPA, IRF3 is polyubiquitinated and translocates to the mitochondrial membrane (35, 36, 39, 41, 43). We engineered pathway-specific IRF3 mutants activated for either transcriptional or RIPA functions, suggesting that the two branches operate independently of each other (35, 36). Using knock-in mice expressing a transcriptionally inactive Irf3 mutant (Irf3-S1), we showed nt-Irf3 contributes to the innate antiviral response (35). Recently, we used the knock-in mice to demonstrate the functions of nt-Irf3 also regulate fatty liver diseases (44, 45). RIPA-like activities have been implicated in viral and nonviral disease models, strengthening the physiological significance of nt-IRF3 functions (46–52).
It has become clear that virus-induced inflammatory responses, in addition to viral load, contribute significantly to viral pathogenesis. The inflammatory responses are critical in the early stages of infection to prevent viral spread; moreover, the infiltrating immune cells recruited to the site of inflammation are primary responders to limit the viral replication and spread. Later in the infection, however, many cellular mechanisms are in place to regulate viral inflammation and control undesired inflammatory responses. IRFs induce antiinflammatory genes and crosstalk with other signaling pathways to regulate viral inflammatory responses (30, 53–55). In addition to triggering IRF3-dependent antiviral responses, virus-infected cells activate NF-κB, the transcription factor that induces proinflammatory genes. Both IRF3 and NF-κB are activated by partially overlapping signaling pathways and induce shared and distinct genes. Previous studies have shown potential crosstalk between the IRFs, NF-κB, and STATs, all activated by common signaling pathways (54–56). A potential interaction between IRF3 and the components of the NF-κB signaling pathway has been evaluated for hepatic inflammatory diseases (45, 57); however, the molecular details of IRF3 and NF-κB crosstalk in the context of viral infection, which rapidly activates these transcription factors, are unclear. Here, we present a function for IRF3, independent of transcriptional or RIPA activities, interacting with the NF-κB subunit, thereby inhibiting the inflammatory gene expression. We refer to the antiinflammatory function of IRF3 as the repression of IRF3-mediated NF-κB activity (RIKA). RIKA, in addition to virus infection, also inhibits inflammatory gene expression by nonviral stimuli. Furthermore, we show RIKA contributes to the antiviral functions of IRF3 in cells and mice.
Results
Increased Inflammatory Gene Expression in Sendai Virus–Infected Irf3−/− Mice.
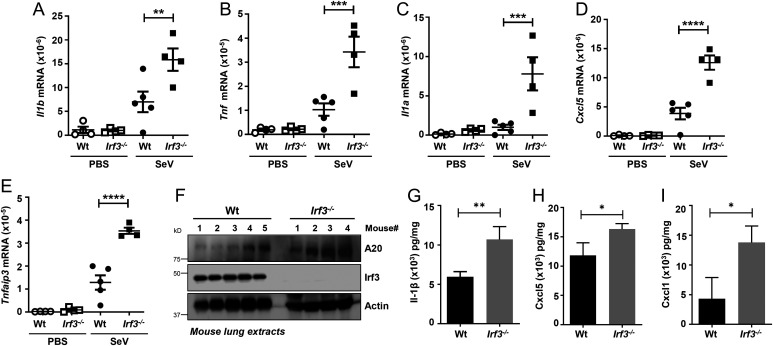
We reported Irf3−/− mice are highly susceptible to Sendai virus (SeV)-induced respiratory pathogenesis (34, 35). As expected, the Irf3−/− mice exhibited increased levels of viral replication, measured by viral messenger RNA (mRNA) and genome levels (SI Appendix, Fig. S1 A and B). In addition to viral load, however, virus-induced inflammatory responses contribute significantly to viral pathogenesis (53, 55). We therefore evaluated whether Irf3−/− mice displayed increased inflammatory gene expression in response to SeV infection. In the SeV-infected Irf3−/− mouse lungs, we observed significantly enhanced levels of a panel of inflammatory target genes, (e.g., Il1b, Tnf, Il1a, Cxcl5, Tnfaip3) compared with the wild-type (Wt) control mice (Fig. 1 A–E). The protein levels of Tnfaip3/A20, which contributes to viral pathogenesis (58, 59), were enhanced in the Irf3−/− mouse lungs (Fig. 1F). We further examined additional inflammatory cytokines (e.g., interleukin-1β, Cxcl5, Cxcl1), which were up-regulated in Irf3−/− mouse lung extracts, compared with the Wt mice (Fig. 1 G–I). The increased inflammatory gene expression in the Irf3−/− mice was associated with the infiltration of immune cells and tissue damage. Hematoxylin and eosin staining indicated the SeV-infected Irf3−/− lung sections had increased infiltrating cells compared with the Wt control (SI Appendix, Fig. S1C). Together, the increased susceptibility of the Irf3−/− mice to SeV pathogenesis was accompanied by an elevated inflammatory response.
Fig. 1.
Increased inflammatory gene expression in Irf3−/− mice upon SeV infection. (A–I) Wt or Irf3−/− mice were infected intranasally with SeV (or treated with the vehicle PBS, as indicated). At 5 d postinfection, the lungs were analyzed for the mRNA levels of Il1b, Tnf, Il1a, Cxcl5, and Tnfaip3 by qRT-PCR (A–E), the protein expression of Tnfaip3/A20 by immunoblot (F), or the cytokine levels by ELISA (G–I). The data represent mean ± SEM; n = 3–5 for each mouse genotype and for each condition, as shown, *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.0001.
IRF3 Inhibits Virus-Induced Inflammatory Gene Induction in Mouse Macrophages.
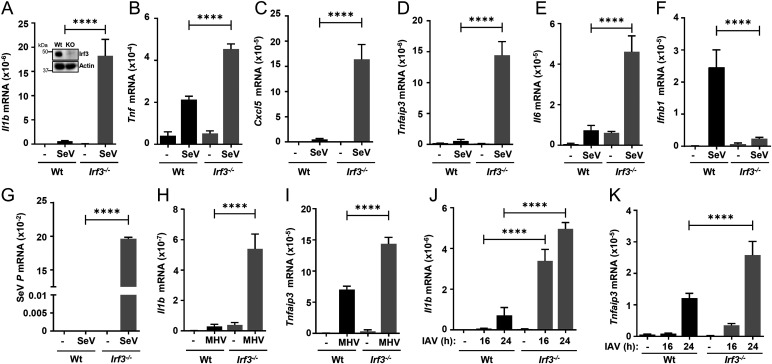
To investigate the cellular mechanism of IRF3-mediated regulation of inflammatory response, we used the Wt and Irf3−/− immortalized bone marrow–derived mouse macrophages (iBMDMs) (Fig. 2A, inset), the critical cell types involved in inflammatory gene expression. SeV infection led to elevated induction of the inflammatory target genes (e.g., Il1b, Il6, Tnf, Tnfaip3, Cxcl5, Cxcl1), in Irf3−/− iBMDMs, compared with the Wt cells (Fig. 2 A–E and SI Appendix, Fig. S2A). As expected, the induction of Ifnb1, which requires the transcriptional activity of Irf3, was inhibited in Irf3−/− cells (Fig. 2F). Viral gene expression, also as expected, was significantly enhanced in the absence of Irf3 (Fig. 2G). To evaluate whether Irf3 regulates viral inflammation in other virus models, we used two additional respiratory viruses, mouse hepatitis virus (MHV), a murine coronavirus, and influenza A virus (IAV). MHV- and IAV-induced inflammatory genes were also elevated in the Irf3−/− iBMDMs, compared with the Wt cells (Fig. 2 H–K and SI Appendix, Fig. S2 B–D). As expected, Irf3−/− iBMDMs had elevated MHV and IAV viral mRNA levels compared with the Wt cells (SI Appendix, Fig. S2 E and F). We further examined whether IRF3 inhibits the induction of antiinflammatory genes. We tested the induction of IL10 and IL13, the expression of which was not elevated in IRF3-deficient cells (SI Appendix, Fig. S3 A–C); therefore, Irf3−/− mouse macrophages showed enhanced virus-induced inflammatory gene expression.
Fig. 2.
Increased inflammatory gene expression in Irf3−/− mouse macrophages upon virus infection. Wt or Irf3−/− iBMDMs (immunoblot of Irf3 expression; inset in A) were infected with SeV (for 4 h postinfection [hpi]; A–G), MHV (for 16 hpi; Hand I), or IAV (for 16 and 24 hpi; J and K), and the mRNA levels of Il1b, Tnf, Cxcl5, Tnfaip3, Il6, Ifnb1 (A–F, H–K), or viral transcript (G) were analyzed by qRT-PCR. The results are representative of three experiments; the data represent mean ± SEM. KO, knockout, ****P < 0.0001.
IRF3 Inhibits Virus-Induced Inflammatory Gene Induction in Human Cells.
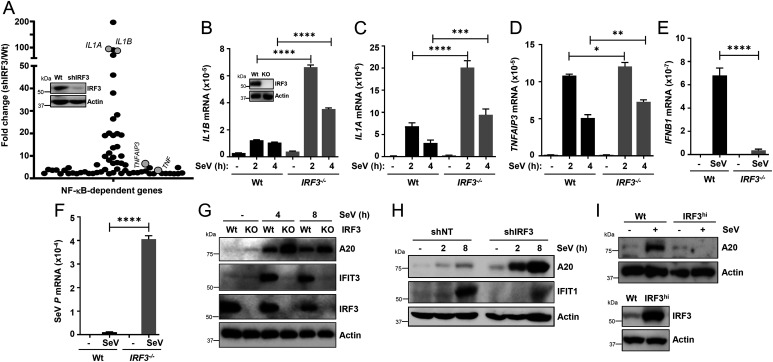
To evaluate the generality of IRF3-mediated inhibition of inflammatory target genes, we performed microarray analyses in SeV-infected Wt and IRF3 knockdown (KD; shIRF3) HT1080 cells (Fig. 3A, inset), which we have used extensively to study IRF3 functions (35–38, 60, 61). Because virus-activated NF-κB primarily contributes to cellular inflammatory responses, we performed targeted analyses of the microarray results for the NF-κB–dependent genes (62, 63). The analyses revealed elevated levels of several NF-κB–dependent genes in the shIRF3 cells compared with the Wt control (Fig. 3A and SI Appendix, Tables S1 and S2). To validate the microarray results, we generated IRF3−/− HT1080 cells using the CRISPR/Cas9 approach [Fig. 3B, inset (64)]. A number of the NF-κB–induced genes (e.g., IL1A, IL1B, TNFAIP3/A20), which we used as readouts in iBMDMs, were up-regulated in the microarray analyses upon SeV infection (Fig. 3 B–D). As expected, in IRF3−/− cells, virus-induced IFNB1 expression was inhibited (Fig. 3E) and viral mRNA expression was elevated (Fig. 3F). To validate the gene expression results at the protein levels, we used immunoblot analyses for TNFAIP3/A20 as a readout of the NF-κB target. SeV-induced A20 expression was elevated in IRF3−/− (Fig. 3G) and HT1080/shIRF3 (Fig. 3H) cells compared with the Wt control. As expected, the expression of IRF3-target genes (e.g., IFIT3 [Fig. 3G] and IFIT1 [Fig. 3H]) was inhibited in the absence of IRF3. In a reciprocal strategy, we used an IRF3-overexpressed cell line, a physiologically relevant approach, because of the wide range of IRF3 levels among various cell types (29, 65). For this purpose, we used the U4C cell line, which is defective in IFN-signaling (60, 66). IRF3 overexpression (IRF3hi) strongly repressed A20 protein induction in SeV-infected U4C cells (Fig. 3I). Together, these findings indicate IRF3 inhibited virus-induced inflammatory gene expression without the requirement of the IFN signaling in mouse and human cells as well as in vivo, using the antiinflammatory activity of IRF3 (RIKA).
Fig. 3.
IRF3 inhibits virus-induced inflammatory gene induction in human cells. (A) Graphical presentation of the microarray analyses of the NF-κB–dependent genes in Wt and shIRF3 (immunoblot of IRF3 expression; inset in A) HT1080 cells after SeV infection (2 h postinfection), are as described in Methods; the genes are listed in SI Appendix, Tables S1 and S2. The microarray results are from duplicate samples for each condition. (B–F) Wt or IRF3−/− (immunoblot of IRF3 expression; inset in B) HT1080 cells were infected with SeV (B–F), and the mRNAs of inflammatory target genes (B–E) or viral mRNA (F) were analyzed by qRT-PCR. (G) Wt and IRF3−/− (knockout [KO]) HT1080 cells were infected with SeV for the indicated time, and the cell lysates were analyzed for TNFAIP3/A20, IFIT1, and IRF3 by immunoblot. (H) The IRF3 shIRF3 HT1080 cells were infected with SeV for the indicated time, and the cell lysates were analyzed for TNFAIP3/A20 and IFIT1 by immunoblot. (I) Wt and IRF3hi U4C cells were infected with SeV and analyzed for TNFAIP3/A20 (Upper panel) and IRF3 (Lower panel) by immunoblot. NT, nontargeting. The results are representative of three experiments; the data represent mean ± SEM. SeV P, SeV P gene, *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.0001.
RIKA Inhibits Inflammatory Gene Expression in Response to Nonviral Stimuli.
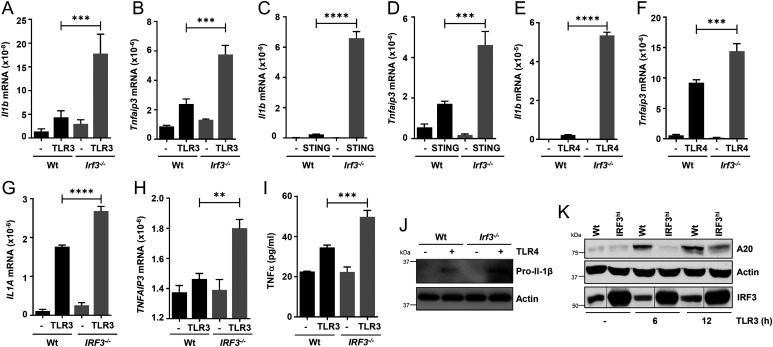
IRF3 and NF-κB are activated by partially overlapping cellular signaling pathways; therefore, their crosstalk has been investigated in various disease models. Previous studies have indicated IRF3 interacts with IKKβ, an upstream kinase that activates NF-κB, as well as the p65 subunit of NF-κB (henceforth, NF-κB–p65), to dampen high-fat diet (HFD)-induced hepatic inflammation (45, 57). How a HFD activates IRF3 and NF-κB is unclear. To evaluate whether RIKA inhibits inflammatory genes in response to nonviral stimuli, we used TLR and STING agonists, known activators of IRF3 and NF-κB. In Irf3−/− iBMDMs, TLR3 stimulation by double-stranded RNA (polyinosinic–polycytidylic acid [polyI:C]) caused increased induction of Il1b and Tnfaip3 (Fig. 4 A and B). Similarly, stimulation by STING (cGAMP) as well as TLR4 (lipopolysaccharide [LPS]) signaling also displayed an inhibition of inflammatory target genes by RIKA (Fig. 4 C–F). Like the iBMDMs, TLR3 stimulation in IRF3−/− HT1080 cells caused increased expression of inflammatory genes (Fig. 4 G and H). The levels of these inflammatory genes were further evaluated at the protein levels for Tnfα (by enzyme-linked immunosorbent assay [ELISA]) (Fig. 4I) and pro–Il-1β (by immunoblot) (Fig. 4J). As expected, in IRF3hi cells, TLR3 stimulation caused strong inhibition of A20 protein expression (Fig. 4K). TLR2 stimulation, which does not activate TRIF or IRF3, did not, however, cause elevated induction of Il1b or Tnf genes in Irf3−/− iBMDMs (SI Appendix, Fig. S3 D and E). Therefore, RIKA inhibited inflammatory target genes in response to viral and nonviral stimuli in human and mouse cells.
Fig. 4.
IRF3 inhibits inflammatory gene expression in response to nonviral stimuli. (A–F) Wt or Irf3−/− iBMDMs were stimulated with polyI:C (TLR3; A and B), cGAMP (STING; C and D), or LPS (TLR4; E and F) for 4 h, and the mRNAs of Il1b and Tnfaip3 were analyzed by qRT-PCR. (G and H) Wt or IRF3−/− HT1080 cells were stimulated with polyI:C (TLR3; G and H) for 4 h, and the mRNAs of Il1a and Tnfaip3 were analyzed by qRT-PCR. (I) Wt and IRF3−/− (knockout) HT1080 cells were stimulated with polyI:C (TLR3) for 8 h, and the supernatants were analyzed for TNFα by ELISA. (J) Wt or Irf3−/− iBMDMs were treated with LPS (TLR4) for 4 h and analyzed for pro–IL-1β by immunoblot. (K) Wt and IRF3hi HT1080 cells were infected with SeV and analyzed for TNFAIP3/A20 by immunoblot. The results are representative of three experiments; the data represent mean ± SEM. **P < 0.005, ***P < 0.0005, ****P < 0.0001.
IRF3 Inhibits Nuclear Translocation of NF-κB to Trigger RIKA.
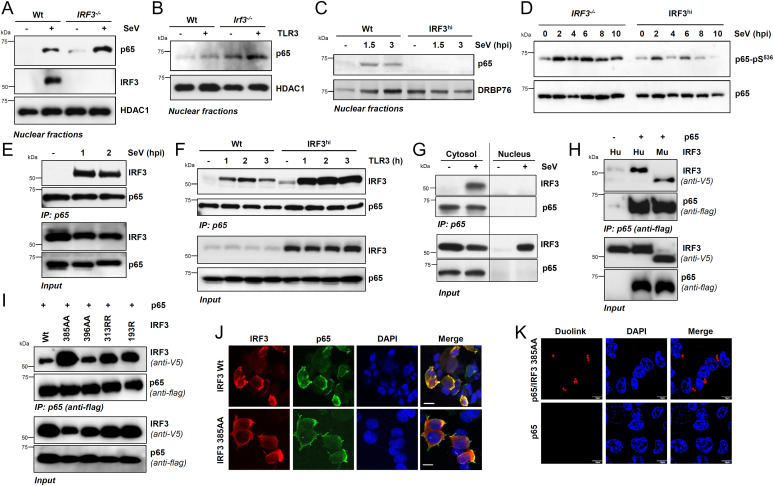
To investigate the molecular mechanism of RIKA, we tested the protein levels of NF-κB–p65/RelA and IκBα, two key players in the NF-κB signaling pathway. IRF3 deficiency did not cause changes in the expression of NF-κB–p65 or IκBα protein levels (SI Appendix, Fig. S4A). We further ruled out that IRF3 regulates the NF-κB–p65 protein expression in SeV-infected cells; IRF3−/− or shIRF3 HT1080 cells did not significantly increase NF-κB–p65 protein expression compared with their Wt control (SI Appendix, Fig. S4 B and C). Furthermore, the IRF3−/− cells did not accumulate the inflammatory genes, due to a lack of apoptotic cell death. SeV infection did not trigger apoptosis in Wt or IRF3−/− cells, as indicated by the absence of cleaved PARP (C-PARP) (SI Appendix, Fig. S4D); SeV-induced apoptosis requires a longer time, as we showed earlier (35, 36). Therefore, we focused on the role of IRF3 in the NF-κB signaling pathway, primarily using the SeV-infection model. IRF3 and NF-κB, the inactive cytosolic proteins, are activated by partially overlapping signaling pathways (e.g., RLR, TLR) and translocate to the nucleus (20–22, 67). In IRF3−/− HT1080 cells, the SeV-induced nuclear translocation of NF-κB–p65 was elevated, as analyzed by both fractionation (Fig. 5A and SI Appendix, Fig. S5 A and B) and confocal microscopy (SI Appendix, Fig. S5 C and D). Nuclear translocation of NF-κB–p65 was also increased in HT1080/shIRF3 cells upon SeV infection (SI Appendix, Fig. S5 E and F). Similarly, in Irf3−/− iBMDMs, TLR3 stimulation caused increased NF-κB–p65 translocation to the nucleus, compared with the Wt cells (Fig. 5B and SI Appendix, Fig. S5 G and H). In the reciprocal strategy, IRF3hi cells displayed reduced p65 translocation to the nucleus (Fig. 5C and SI Appendix, Fig. S5I). The activation of NF-κB–p65 requires its phosphorylation on Ser536, which was inhibited by IRF3 (Fig. 5D and SI Appendix, Fig. S5 J–L). Therefore, our results suggest IRF3 inhibited the nuclear translocation of NF-κB–p65 in human and mouse cells, likely by preventing its phosphorylation.
Fig. 5.
IRF3 interacts with NF-κB–p65, independent of transcriptional or RIPA functions, and inhibits NF-κB activation. (A) Wt and IRF3−/− HT1080 cells were either mock-infected or infected with SeV for 2 h when the nuclear fractions were analyzed for NF-κB–p65 and IRF3 by immunoblot. HDAC1 is a marker for nuclear fractions. (B) Wt and Irf3−/− iBMDMs were stimulated with polyI:C (TLR3) for 2 h, when the nuclear fractions were analyzed for NF-κB–p65 by immunoblot. (C) Wt or IRF3hi HT1080 cells were infected with SeV for the indicated time, when the nuclear fractions were analyzed for NF-κB–p65 by immunoblot; DRBP76 is a marker for the nuclear fractions. (D) IRF3−/− and IRF3hi HT1080 cells were infected with SeV for the indicated time, and phospho-p65 (on Ser536) was analyzed by immunoblot. (E) HT1080 cells infected with SeV for the indicated time were subjected to co-IP analyses for the endogenous NF-κB–p65 and IRF3 proteins using ExactaCruz. (F) Wt and IRF3hi HT1080 cells stimulated with polyI:C (TLR3) for the indicated time were subjected to co-IP analyses for NF-κB–p65 and IRF3 proteins, as in E. (G) The cytosolic and nuclear fractions, isolated from the SeV-infected Wt and IRF3hi HT1080 cells, were subjected to co-IP analyses for NF-κB–p65 and IRF3, as in E. (H and I) HEK293T cells, cotransfected with Flag–NF-κB–p65 and V5-IRF3 Wt (H) human (Hu), murine (Mu), or IRF3 mutants (I), as indicated, were infected with SeV for 2 h and subjected to co-IP analyses for the Flag–NF-κB–p65 and V5-IRF3, as indicated. (J) HEK293T cells, cotransfected with NF-κB–p65 and V5-IRF3 Wt or IRF3 385AA mutant, were infected with SeV and immunostained with anti-Flag and anti-V5 antibodies and analyzed by confocal microscopy. (K) HEK293T cells, cotransfected with NF-κB–p65 and V5-IRF3 385AA mutant, were infected with SeV and immunostained with anti-Flag and anti-V5 antibodies, and analyzed by proximal ligation assay. The results are representative of three experiments; the data represent mean ± SEM. Scale bar, 10 µm. IP, immunoprecipitation.
IRF3 and NF-κB–p65 Interact in the Cytosolic Compartment.
Because IRF3 inhibits the nuclear translocation of the NF-κB–p65 subunit, we examined whether IRF3 interacts with, and sequesters, NF-κB–p65 in the cytosolic compartment. Previous studies have indicated IRF3/NF-κB–p65 interaction (45, 68); however, the molecular details of the interaction are unclear, particularly in the context of viral infection. We performed coimmunoprecipitation (co-IP) analyses of endogenous NF-κB–p65 and IRF3 proteins; strong interaction of IRF3 and NF-κB–p65 was observed upon SeV infection (Fig. 5E) and TLR3 stimulation (Fig. 5F, Wt cells). Furthermore, the IRF3/NF-κB–p65 interaction in TLR3-stimulated cells was enhanced in IRF3hi cells, which displayed IRF3:p65 interaction in unstimulated cells (Fig. 5F), as also noted for NF-κB–p65 nuclear translocation (Fig. 5 A and B and SI Appendix, Fig. S5E), suggesting the two proteins also interact before any stimulation. The interaction was enhanced, however, upon virus infection or TLR3 stimulation. To examine whether the IRF3/NF-κB–p65 interaction takes place in the cytosol, we performed co-IP analyses in the cytosolic and nuclear fractions of IRF3hi cells. The IRF3/NF-κB–p65 interaction of endogenous proteins was observed in the cytosolic but not in the nuclear fractions of SeV-infected cells (Fig. 5G). These results suggest a model that IRF3 binds to NF-κB–p65 in the cytosol to sequester it from translocating to the nucleus. To validate these results genetically, we used IRF3 mutants to examine their interaction with NF-κB–p65. We ectopically expressed epitope-tagged IRF3 and NF-κB–p65 in HEK293T cells and performed a co-IP assay upon SeV infection, the condition that caused optimal IRF3/NF-κB–p65 binding, and both human and mouse IRF3 proteins coimmunoprecipitated with NF-κB–p65 (Fig. 5H). Using this assay, we tested the critical IRF3 mutants, which are defective in either transcriptional (SS385AA, SS396AA) or RIPA (KK313RR, K193R) activities (35, 36). The results indicated IRF3 does not require activation by either transcriptional or RIPA pathways for binding to NF-κB–p65, as evaluated by co-IP, confocal microscopy, and proximal ligation assay, using both human and mouse IRF3 proteins (Fig. 5 I–K and SI Appendix, Fig. S6). Importantly, the IRF3 mutants defective in nuclear translocation (SS385AA, SS396AA) also interacted with NF-κB–p65 (Fig. 5 I–K and SI Appendix, Fig. S6), further confirming IRF3/NF-κB–p65 interaction was cytosolic. Thus, IRF3 interacted with the NF-κB–p65 in the cytosol to inhibit its nuclear translocation.
IRF3 and NF-κB–p65 Interact Directly via Specific Domains.
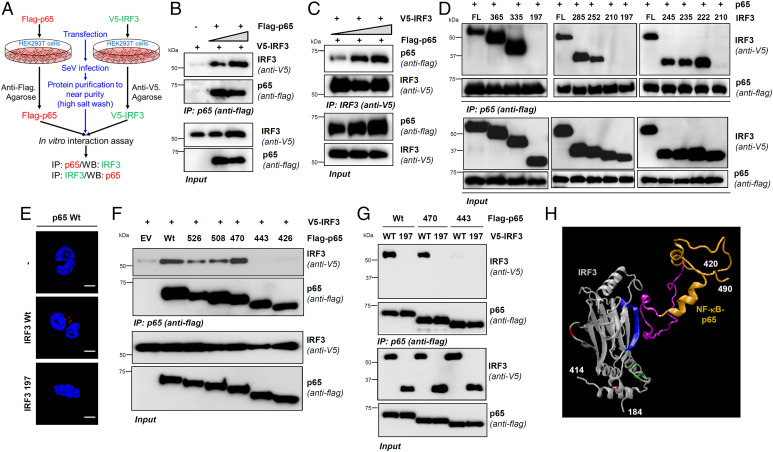
To investigate whether IRF3 and NF-κB–p65 interact directly in virus-infected cells or conduit proteins bridge them, we developed a cell-free interaction assay using partially purified IRF3 and NF-κB–p65 proteins individually from SeV-infected HEK293T cells. Using stringent conditions (e.g., a high-salt wash to remove the interacting partners), we isolated the recombinant proteins in near purity from SeV-infected cells (Fig. 6A). This approach we previously used to study IRF3/BAX direct interaction (36) and, recently, for additional binding partners (69). The results indicate IRF3 and NF-κB–p65, isolated from SeV-infected cells, interacted in vitro (Fig. 6 B and C). We used a co-IP assay to map the interacting domains between the two proteins. We designed a series of C-terminal deletion mutants of IRF3 (SI Appendix, Fig. S7A), guided by the three-dimensional (3D) protein model, to ensure the structural integrity of the mutants, and set up the co-IP assay in HEK293T cells, as in Fig. 5H. Our results revealed the IRF3 domain 210–222 was required for interacting with NF-κB–p65 (Fig. 6 D and E). As expected, this domain is distinct from those needed for activating RIPA or transcriptional functions of IRF3 (SI Appendix, Fig. S7 B and C). Similarly, we designed C-terminal deletion mutants of NF-κB–p65 (SI Appendix, Fig. S7D) and set up co-IP with Wt IRF3. The results indicate the NF-κB–p65 domain 443–470 was required for IRF3 interaction (Fig. 6F). To add rigor to the interaction assay, we included a noninteracting IRF3 mutant (1–197), which did not bind to either the full-length or the truncated NF-κB–p65 proteins (Fig. 6G). Importantly, these interacting domains are highly conserved across various species, adding strength and physiological relevance to IRF3/NF-κB–p65 interaction (SI Appendix, Fig. S7 E and F). These domain-mapping analyses led us to a model for the IRF3:NF-κB–p65 complex with at least one binding interface (Fig. 6H). Together, our results demonstrated a direct interaction between IRF3 and NF-κB–p65 via specific interacting motifs.
Fig. 6.
IRF3 and NF-κB interact directly via specific domains. (A–C) V5-IRF3 or Flag–NF-κB–p65 were ectopically expressed in HEK293T cells, isolated to near purity, and subjected to cell-free interaction assay, as indicated in A, and the complex was analyzed by co-IP (B and C). (D) HEK293T cells, cotransfected with Wt or the C-terminal deletion mutants of V5-IRF3 and Flag-NF-κB–p65, were infected with SeV for 2 h and subjected to co-IP analyses. (E) HEK293T cells expressing either Flag–NF-κB–p65 or Flag–NF-κB–p65 with V5-IRF3 (Wt or 1–197) were analyzed by proximity ligation assay. (F and G) HEK293T cells, cotransfected with full-length or the deletion mutants of Flag–NF-κB–p65 (F) and V5-IRF3 (Wt in F; Wt and 1–197 in G) were infected with SeV for 2 h and subjected to co-IP analyses. (H) A 3D structural model showing the domains and residues of IRF3 (blue: RIKA; red: RIPA; green: transcription) and NF-κB–p65 (gold and pink: IRF3-binding motif) involved in RIKA. The 3D protein structures of IRF3 and NF-κB–p65 were adapted from Protein Data Bank (PDB) templates 1j2f.2.A and 2lww.1, respectively. EV, empty vector. The results are representative of three experiments; the data represent mean ± SEM. Scale bar, 5 µm. IP, immunoprecipitation.
Irf3 Mutant, Defective in Transcriptional and RIPA Functions but Active in RIKA, Inhibits Inflammatory Gene Expression and Contributes to the Antiviral Function of IRF3.
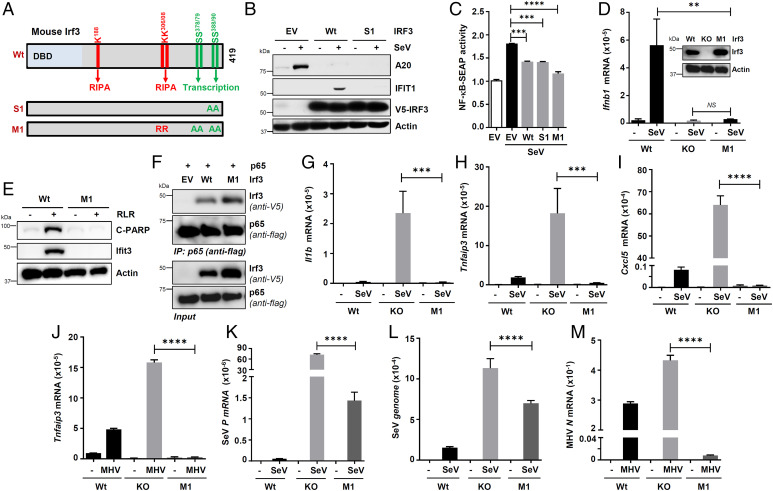
To examine the functional relevance of IRF3/NF-κB–p65 interaction, we used the Irf3-S1 mutant (SS388AA), which is transcriptionally inactive but active in RIPA (Fig. 7A) (35). When expressed in HT1080/shIRF3 cells, the transcriptionally inactive IRF3-S1 mutant, which interacted with NF-κB–p65 (Fig. 5I and SI Appendix, Fig. S6), inhibited TNFAIP3/A20 protein induction, similar to the Wt IRF3, and was expectedly inactive in IFIT1 induction (Fig. 7B). We further confirmed these results in NF-κB–reporter RAW-Blue mouse macrophages expressing NF-κB–dependent secreted embryonic alkaline phosphatase (SEAP) (70). SeV-induced NF-κB–SEAP expression was inhibited by Irf3-Wt or Irf3-S1 (Fig. 7C). Furthermore, we engineered a new Irf3-M1 mutant, which was inactive in both transcriptional and RIPA functions (Fig. 7A), and also inhibited the NF-κB–SEAP activity (Fig. 7C). This result led us to characterize the Irf3-M1 mutant by expressing stably in Irf3−/− iBMDMs at a level similar to endogenous Irf3 (Wt in Fig. 7D, inset). We confirmed Irf3-M1 is transcriptionally inactive; SeV-induced Ifnb1 mRNA levels (Fig. 7D) and RLR-induced Ifit3 protein levels (Fig. 7E) were strongly inhibited. As expected, Irf3-M1 was RIPA-incompetent, assessed by the absence of C-PARP, an apoptotic marker, in RLR-stimulated macrophages (Fig. 7E). Irf3-M1 interacted with NF-κB–p65, similar to Irf3-Wt, as examined by co-IP (Fig. 7F) and confocal microscopy (SI Appendix, Fig. S8A). Irf3-M1 also interacted with the endogenous p65 in SeV-infected iBMDMs (SI Appendix, Fig. S8 A and B). As a result, Irf3-M1 inhibited the induction of Il1b, Tnfaip3, and Cxcl5 in SeV-infected iBMDMs (Fig. 7 G–I). We further validated the functional results in human cells by ectopically expressing Irf3-M1 and Irf3-M2 (SI Appendix, Fig. S8C), which are inactive in transcription, and RIPA in IRF3−/− cells. Similar to Irf3-Wt, both Irf3-M1 and Irf3-M2 mutants suppressed SeV-induced A20 protein expression (SI Appendix, Fig. S8D). In addition to SeV, MHV-induced Tnfaip3 levels were also inhibited by Irf3-M1 (Fig. 7J). Finally, we evaluated the relative contribution of RIKA to the antiviral activity of IRF3 and used the Irf3-M1 iBMDMs. The Irf3−/− iBMDMs, as expected, expressed higher levels of SeV RNA as compared with the Wt cells (Fig. 7 K and L). The RIKA-selective Irf3-M1 mutant suppressed the SeV replication compared with the Irf3−/− cells (Fig. 7 K and L). We further validated these results using MHV; the Irf3-M1 mutant was sufficient to inhibit MHV replication in Irf3−/− iBMDMs (Fig. 7M). In addition to mounting an antiinflammatory response, RIKA, therefore, contributes to the overall antiviral functions of IRF3.
Fig. 7.
IRF3 mutants, active in RIKA but inactive in transcriptional and RIPA, inhibits virus replication and inflammatory gene expression. (A) A cartoon showing mouse Irf3 and its critical residues and domains required for specific functions and the pathway-specific mutants (Irf3-S1 and Irf3-M1). DBD, DNA-binding domain, A, Ala, R, Arg. (B) HT1080/shIRF3 cells, lentivirally expressing Wt or S1 mutant of IRF3, were analyzed for A20 and IFIT1 induction upon SeV infection, by immunoblot 8 h postinfection (hpi). (C) RAW-Blue cells were transfected with Wt or Irf3 mutants (S1 or M1), and the NF-κB–SEAP activity in the culture supernatants was measured upon SeV infection (24 hpi). (D) Wt, Irf3−/−, and Irf3-M1 iBMDMs (Irf3 expression is shown in the inset) were infected with SeV, and Ifnb1 induction was analyzed by qRT-PCR 4 hpi. (E) Wt and Irf3-M1 iBMDMs were transfected with polyI:C for 16 h, when the cell lysates were analyzed for Ifit3 and C-PARP by immunoblot. (F) HEK293T cells, cotransfected with Flag–NF-κB–p65 and V5-Irf3 (Wt or M1), were infected with SeV for 2 h and subjected to co-IP analyses for the Flag–NF-κB–p65 and V5-Irf3. (G–J) Wt, Irf3−/−, and Irf3-M1 iBMDMs were infected with SeV for 4 h (G–I) or MHV for 16 h (J), and the inflammatory target genes were analyzed by qRT-PCR. (K–M) Wt, Irf3−/−, and Irf3-M1 iBMDMs were infected with SeV for 4 h (K and L) or MHV for 16 h (M), and the viral replication was analyzed by qRT-PCR. EV, empty vector. The results are representative of three experiments; the data represent mean ± SEM. IP, immunoprecipitation; KO, knockout, SeV P, SeV P gene, **P < 0.005, ***P < 0.0005, ****P < 0.0001.
Discussion
The optimal physiological function of IRF3 in innate antiviral defense depends on its transcriptional and nontranscriptional (nt) activities (24, 35, 44, 45, 64, 71). In addition to the widely studied role of IRF3 as a transcription factor, we demonstrated a proapoptotic function for nt-IRF3, RIPA, which protects the host from viral pathogenesis and contributes to fatty liver diseases (35, 41, 44, 45). Here, we reveal a function for nt-IRF3, RIKA, which contributes to the antiviral responses of IRF3 and protection against viral pathogenesis, presumably by inhibiting viral inflammation (SI Appendix, Fig. S9). RIKA is independent of the activation of IRF3 in either transcriptional or RIPA pathways. For RIKA, IRF3 interacts with NF-κB–p65 and sequesters it in the cytosol, thus preventing it from translocating to the nucleus, inhibiting the inflammatory gene expression. IRF3 interacts directly with NF-κB–p65, using a domain distinct from the transcriptional or RIPA-activating domains and, therefore, can independently protect against inflammatory diseases. IRF3 may also interact with other NF-κB subunits for its antiinflammatory functions, and the exploration of whether other NF-κB subunits possess the IRF3-interacting motif of NF-κB–p65 will help illuminate such possibilities. We noted IRF3 and NF-κB–p65 formed a complex in unstimulated cells, and the interaction was enhanced upon virus infection or TLR stimulation; therefore, the optimal interaction of IRF3 with NF-κB–p65 is dependent on upstream signaling. It is not clear, however, whether IRF3, or NF-κB–p65, or both proteins require activation, leading to changes in conformation or subcellular localization that favor optimal interaction to trigger RIKA. Both IRF3 and NF-κB–p65 proteins can be activated by a wide range of stimuli, including microbial infections, ethanol, HFD, endoplasmic reticulum stress, reactive oxygen species, cytokines, and damage-associated molecular patterns. Whether some or all of these stimuli activate RIKA will reveal the requirement of any upstream signaling. Our results from TLR2-induced gene expression suggest the role of upstream activation for RIKA, and the molecular details will be investigated in the future. Whether TLR2 stimulation does not provide the activation signal for IRF3 or activates p65 differentially will be explored in future studies.
Because IRF3 and NF-κB are activated by partially overlapping signaling pathways, their potential crosstalk has been studied in inflammatory diseases [e.g., hepatic inflammation (45, 57)], as well as in viral infection (54). These studies lead us to speculate IRF3 may employ multiple mechanisms to dampen the host inflammatory response. In a previous study, IRF3 was shown to interact with IKKβ, an upstream kinase that activates p65, and this interaction prevents hepatic inflammation in response to HFD (57). The exact IKKβ-interacting domain of IRF3 is unclear; however, multiple interacting partners of IRF3 in controlling inflammatory response may have evolved to benefit the host. Future studies will reveal whether IRF3 also interacts with IKKβ in virus-infected cells to inhibit virus-induced inflammatory gene expression. We have previously shown IRF3-NF-κB–p65 interaction can also prevent hepatic inflammation (45). The nt-Irf3 mutant (Irf3-S1) also inhibits HFD-induced inflammation in cells and mice; therefore, IRF3 interacts with IKKβ and NF-κB–p65 to regulate the hepatic inflammatory responses. IRF3, however, may interact with IKKβ and NF-κB–p65 in a cell type–dependent manner. Both Wt and IRF3-S1 interact with LUBAC, which catalyzes, in addition to Irf3, the linear ubiquitination of NEMO/IKKγ, an upstream kinase of the NF-κB signaling pathway. It was consequently unclear whether NEMO mediates IRF3-p65 interaction in the hepatic inflammation model. To address this, we engineered a mutant Irf3 (M1) without the LUBAC-binding sites. Such a mutant interacts with NF-κB–p65 and inhibits virus-induced inflammatory gene expression. Using cell-free interaction assays, our study further established that IRF3 and NF-κB–p65, isolated from virus-infected cells, can interact, and we further narrowed down the interacting sites between the two proteins. An earlier study used recombinant IRF3 and NF-κB–p65 proteins to evaluate their direct interaction, further supporting our results (68). IRF3 and NF-κB–p65, activated by common upstream signaling pathways, are translocated to the nucleus to transcribe shared and distinct sets of genes. Interaction of activated IRF3 and NF-κB–p65, presumably mediated by cAMP-responsive element-binding protein-binding protein (CBP), has been studied in TLR4 signaling, which activates the interferon stimulation response element promoter (72). Moreover, TLR4 signaling also transcriptionally induces a subset of genes (e.g., Scyb9), which requires the interaction of IRF3 and NF-κB–p65 on their promoters (68). The nuclear receptor, GR, inhibits the IRF3/NF-κB–p65 interaction to block the TLR4-induced inflammatory response. Whether GR can disrupt the cytosolic IRF3-NF-κB–p65 complex to elicit inflammatory responses will be investigated in the future.
The viral load is commonly considered a major contributing factor determining the outcome of viral pathogenesis; however, the virus-induced inflammatory responses, particularly in respiratory viruses that trigger lung inflammation, cause detrimental viral diseases. Infections by the IAV, SARS-CoV, trigger rapid lung inflammation to promote viral pathogenesis. Many cellular mechanisms are in place to dampen such undesired inflammatory responses and thereby control the viral pathogenesis. IRF3 provides a key antiviral mechanism against HSV-1 pathogenesis; HSV-1–infected mouse brains exhibit enhanced inflammatory gene expression (73). These studies further support our results indicating that IRF3 controls viral inflammation to prevent pathogenesis. These mechanisms not only regulate the viral infection but can also be expanded to other diseases. For example, IRF7 has been shown to control allergic airway inflammation by regulating the functions of innate lymphoid cells (74). Irf7−/− mice display increased numbers of inflammatory cytokines (e.g., Ccl2) in the Sindbis virus–infected brain, contributing to the viral pathogenesis (75). Increased levels of Tnf, Ccl2, Ccl3, and Ccl4 have been observed in HSV-1–infected Irf7−/− mouse brains (76). IRF7 has been shown to control RSV-induced inflammation in the epithelial cell compartment (77). IRF5 has also been shown to regulate pulmonary inflammation by controlling the inflammatory cytokine expression (78). Whether multiple IRFs can regulate inflammation simultaneously or in a cell- and time-dependent manner will require future investigation. In addition to preventing viral pathogenesis, the cellular antiinflammatory mechanisms may contribute to the antiviral response by directly inhibiting viral replication in the infected cells. The Irf3-M1 mutant, in the absence of transcriptional and RIPA activities, was able to inhibit viral replication. Because NF-κB signaling pathways benefit the virus replication (79), the RIKA branch of IRF3 may also be antiviral to other viruses. The RIKA branch, moreover, may inhibit virus replication by suppressing the proviral inflammatory genes.
It is unclear why IRF3 functions in multiple pathways to protect against viral pathogenesis. We speculate the multiple functions of IRF3 provide the host with independent options to utilize them in specific cell compartments in response to specific pathogens or against inflammatory stimuli. Viruses often evade the immune response by antagonizing specific pathways. Numerous viral proteins have been shown to antagonize the transcriptional and the RIPA activity of IRF3 (80–82). It is also tempting to speculate the viruses have coevolved to evade specific functions of IRF3, and the host can fight back using the additional options. Herpesviruses encode viral IRFs, and it will be interesting to investigate in the future whether these viral proteins can compete with IRF3 for antagonizing NF-κB functions. In summary, our study uncovered a function for IRF3 that contributes to host antiviral and antiinflammatory responses. Although we focused on viral pathogenesis, we speculate RIKA may protect against other inflammatory diseases. A number of inflammatory diseases, including sepsis, are modulated by IRF3, and the role of RIKA in these diseases will be evaluated in the future. Moreover, NF-κB is a critical transcription factor for numerous cellular functions, independent of regulating inflammatory genes, and RIKA may have roles in those functions of NF-κB as well.
Methods
Cell Lines and Viruses.
All cell lines used in this study were maintained in the authors’ laboratory. The human cell lines HT1080 (CCL-121, ATCC), HEK293T (CRL-3216, ATCC), and mouse cells Wt and Irf3−/− iBMDMs (NR-9456, NR-15635, BEI), L929, and RAW-Blue cells (45) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum, penicillin, and streptomycin. The HT1080-derived IRF3−/−, shIRF3, and IRF3hi cell lines were generated as described before (35, 36, 64). SeV Cantell and 52 strains were obtained from Charles River Laboratories. H1N1 IAV strain PR8 was kindly provided by Jacob Yount (Ohio State University). Recombinant murine coronavirus strain icA59 (MHV-A59) was obtained from BEI (NR-43000) and propagated in L929, per the product information sheet from BEI Resources.
Cloning and Mutant Generation.
The expression vectors for human and mouse IRF3 with different epitope tags, as well as NF-κB–p65, were described previously (34–36, 38, 42, 44, 45). Wt human and mouse IRF3 from pLVX-IRES-puromycin (Clontech) were used for the generation of deletion and mutant constructs. The deletion constructs of human IRF3 and NF-κB–p65 were generated by standard molecular cloning methods (35, 36). The mouse IRF3 mutants S1, M1, and M2 were generated by overlap extension PCR and by previously described procedures (35, 36). All the clones and mutants were verified by sequencing by MCLAB.
Antibodies.
The following antibodies were obtained as indicated: Actin (Sigma-Aldrich #5441); A20 (Cell Signaling Technology [CST] #5630); C-PARP (CST #9541); DRBP76 (BD Biosciences); Flag (CST #2368 and Sigma-Aldrich #F1804); HA (CST #3724); HDAC1(CST #34589 and Santa Cruz Biotechnology [SCBT] #sc-81598); NF-κB signaling proteins (CST #9936, 3033, 8242); V5 (CST #13202 and Thermo Fisher #R960-25); IRF3 [SCBT #33641 and as described before (36)]; IL-1β (CST #12242); and IFIT [as described before (36)].
Microarray Analyses.
HT1080 (Wt) cells and the shIRF3/HT1080 (KD) cells were infected with SeV, the total RNA was isolated and treated with DNase I, and the RNA was further purified by using the RNAeasy kit (Qiagen). The purified RNA was then analyzed in an Illumina Mouse Ref-8 gene array, and the data analysis was carried out by using Illumina Genome Studio V2011.1. We selected the mRNAs, which were induced at least two-fold by SeV, and quantified the fold change (KD-SeV/Wt-SeV) or fold induction [(KD-SeV/KD-mock)/(Wt-SeV/Wt-mock)] (as presented in Fig. 3A and SI Appendix, Tables S1 and S2). Targeted analyses were performed for the expression of NF-κB–dependent genes (62, 63).
Activation of Intracellular Signaling and Virus Infection.
The cells were stimulated with polyI:C (25 μg/mL, TLR3) or LPS (1 μg/mL, TLR4), or Pam3CSK4 (0.25 and 1 µg/mL, TLR2) or transfected with polyI:C (2 μg/mL, RLR) or cyclic di-GMP (1 μg/mL, STING) using Lipofectamine 2000 for the indicated period of time to activate the specific intracellular signaling pathways, as described in the figure legends. For virus-infection studies, the cells were infected with SeV (Cantell), IAV, or MHV at the multiplicity of infection of 5 in serum-free DMEM for 2 h, after which the cells were washed and replaced with the normal growth medium. The virus-infected cells were analyzed at the indicated time for cellular or viral gene expression by qRT-PCR or immunoblot, as described in the figure legends.
RNA Isolation and qRT-PCR Analyses.
Total RNA was isolated using TRIzol (Thermo Fisher #15596026 and Sigma #T9424). The DNase (Promega)-treated RNA was used to prepare complementary DNA by random hexamers using the ImProm-II Reverse Transcription Kit (Promega), following the manufacturer’s instructions. The complementary DNA was analyzed using Radiant SYBR Green PCR mix (Alkali Scientific Inc.) in Roche LightCycler 96 instrument, and data were analyzed with the LightCycler 480 Software, version 1.5. The expression levels of the mRNAs were normalized to 18S ribosomal RNA, and the relative expression of each gene to the 18S ribosomal RNA was plotted using GraphPad Prism 9 software. Primers used for qRT-PCR analyses are listed in SI Appendix, Table S3.
Proximity ligation assay and Confocal Microscopy.
The infected cells on the coverslips were fixed in 4% paraformaldehyde (Electron Microscopy Sciences #15710), permeabilized in 0.2% Triton X-100 (Fisher Scientific #9002–93-1), and immunostained with anti-V5 and anti-Flag antibody and harvested for duolink assay (DUO92008-3, DUO92004, DUO92002, Sigma-Aldrich), using manufacturer’s instructions. For confocal microscopy, the cells were incubated with Alexa Fluor–conjugated secondary antibodies (Invitrogen #A-11004) after overnight incubation with primary antibodies. The coverslips were mounted on the microscopy slides using VectaShield/DAPI (H-1200, Vector Laboratories) and analyzed using an Olympus confocal microscope and Olympus Fluoview FV1000 software. The images were further processed using Adobe Photoshop software. The nuclear translocation of NF-κB–p65 was quantified by Pearson’s correlation coefficient, calculated using Image J software.
Immunoblot Analyses.
For immunoblot analyses, the cells were lysed in 50 mM Tris buffer, pH 7.4, containing 150 mM NaCl, 0.1% Triton X-100, 1 mM sodium orthovanadate, 10 mM of sodium fluoride, 10 mM of β-glycerophosphate, 5 mM sodium pyrophosphate and protease inhibitors (Roche), by keeping them on ice for 30 min, followed by centrifugation to clear the lysates. Proteins in the cell lysates were quantified using Bradford reagent (BIO-RAD #500–0006), and equal amounts of total proteins from the cell lysates were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblot.
Co-IP and Cell Fractionation Analyses.
For co-IP of IRF3 and NF-κB–p65, the cells were lysed in EPPS buffer containing protease and phosphatase inhibitors (Roche), using repeated freeze–thaw cycles, and the cell lysates were centrifuged to isolate the supernatants containing the proteins of interest. The immunoprecipitation reactions were performed using anti–V5- or anti–Flag-conjugated agarose beads, using previously described procedures (35, 36). For co-IPs of endogenous IRF3 and NF-κB–p65, ExactaCruz reagents (SCBT) were used as described before (34). After immunoprecipitation, the beads were washed twice with lysis buffer and once with radioimmunoprecipitation assay buffer. The bound proteins were eluted from the beads by elution with Flag peptide (Sigma #F4799) or boiling with 2× sodium dodecyl sulfate (SDS) buffer for 3 min, and the eluates were analyzed on a 10% SDS-PAGE. For isolation of nuclear and cytoplasmic fractions, previously described procedures were followed (35, 36, 42).
Cell-Free Protein Interaction Assay.
Cell-free protein interaction assays were performed using our previously described procedures (36, 69). Briefly, the HEK293T cells expressing V5-tagged IRF3) and Flag-tagged NF-κB–p65 (Flag.NF-κB–p65) were lysed separately in EPPS buffer containing protease inhibitors by repetitive freeze–thaw cycles, and the cell lysates were immunoprecipitated with anti–V5- or anti–Flag-conjugated agarose beads overnight at 4 °C. The beads were washed extensively with EPSS buffer containing 300 mM NaCl by rotating for at least 10 min each. Flag.NF-κB–p65 protein was eluted with Flag peptide, and the eluate was used to incubate with anti-V5 beads bound with IRF3 for 2 h at room temperature with shaking. Elution with V5 peptide and incubation with Flag beads were performed for the reciprocal assay. Respective beads were washed twice with EPPS buffer and once with radioimmunoprecipitation assay buffer before performing final elution for bound proteins by boiling in 2× SDS sample loading buffer (BIO-RAD #161–0737). The interacting proteins were analyzed by SDS-PAGE and immunoblotting.
Molecular-Docking Analyses.
The IRF3 and NF-κB–p65 3D structures were adapted from templates 1j2f.2.A and 2lww.1, respectively, using SWISS-MODEL. The templates were selected and modeled based on sequence similarity and were used for the docking for the complex generation. The molecular modeling for the IRF3–NF-κB–p65 complex generation was performed using Z-DOCK, version 3.0.2. The interacting amino acid residues of IRF3 and NF-κB–p65, identified using biochemical analyses, were used for binding, and noninteracting residues were used for blocking while providing input amino acids for docking. The top 10 predictions from Z-DOCK output results were analyzed, and the most likely conformation was selected according to the interaction based on binding affinity and optimal binding arrangement, as a 3D complex was used as a representative in accordance with experimental data.
NF-κB–SEAP Assay.
RAW-Blue cells (Invivogen #raw-sp) were used for transfection with the IRF3 (Wt or mutants, as indicated in figure legends) plasmids, and 24 h after SeV infection, the cell supernatants were collected to analyze the SEAP activity, using the manufacturer’s instructions (Invivogen), as described previously (45). The SEAP activity of IRF3 and its mutants were normalized to that of the empty vector, which was considered 1.0.
ELISA.
The extracts of the SeV-infected mouse lungs were prepared by homogenization of the tissues in the lysis buffer described in immunoblot analyses, and supernatants separated from tissue debris were subjected to quantification using the Bradford assay. Similar amounts from the tissue lysates were used to measure cytokine levels using ELISA, as indicated in the figure legends. Cells were incubated with SeV for the specified time for adsorption to facilitate infection and kept in serum-free conditions during the postinfection time. The serum-free culture supernatants were collected at mentioned intervals postinfection and used to measure secreted cytokine levels per instructions provided by commercial kits (PEPROTECH for human TNFα; R&D Biosystems for mouse Il1β, Cxcl1, and Cxcl5).
Mice and Virus Infection.
The Wt and Irf3−/− mice, on a C57BL/6 genetic background, were used for the studies, as described before (34, 35). For virus infection, 120,000 pfu of SeV 52 strain in 35 μL of endotoxin-free PBS were intranasally administered to isoflurane-anesthetized 8- to 10-wk-old mice. The mice were monitored daily for their body weight loss and disease symptoms, and the lungs were harvested 5 d postinfection for further analyses. qRT-PCR, immunoblot, and histology were performed using published procedures (34, 35, 42).
Quantification and Statistical Analyses.
Western blot band-intensity measurements were made using Image J for densitometric analyses. The statistical analyses were performed using GraphPad Prism 9 software and Microsoft Excel for Windows 10. The P values were calculated using two-tailed, unpaired Student’s t tests (when comparing two groups) or one-way ANOVA (when comparing more than two groups), based on the number of sets for comparison. P < 0.05 was considered statistically significant. The results presented here are the representatives of at least three independent experiments.
Supplementary Material
Acknowledgments
This work was supported partly by the NIH (Grants AI155545, AI165521, AA026017, and AI153496 [to S. Chattopadhyay], and AA027456 [to L.E.N., G.C.S., and S. Chattopadhyay]); Medical Research Society (S. Chattopadhyay), and the University of Toledo College of Medicine and Life Sciences (S. Chattopadhyay and R.C.). We thank Adolfo García-Sastre, Jacob Yount, and Travis Taylor for the critical reagents and resources used in the study. The following reagents were obtained through BEI Resources, National Institute of Allergy and Infectious Diseases, NIH: macrophage cell line derived from wild-type mice (NR-9456), Irf3 knockout mice (NR-15635), and recombinant murine coronavirus strain icA59 (NR-43000). We thank Kale Bandy for help with editing the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2121385119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Samuel C. E., Antiviral actions of interferons. Clin. Microbiol. Rev. 14, 778–809, (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White C. L., Sen G. C., “Interferons and antiviral actions” inCellular Signaling and Innate Immune Responses to RNA Virus Infections, Brasier A. R., Garcia-Sastre A., Lemon S. M., Eds. (ASM Press, Washington, DC, 2008), pp. 91–106. [Google Scholar]
- 3.Biron C. A., Sen G. C., “Innate responses to viral infections” in Fields Virology, Knipe D. M., Howley P. M., Eds. (Lippincott, Williams and Wilkins, Philadelphia, PA, ed. 5, 2006), pp. 249–278. [Google Scholar]
- 4.Fensterl V., Sen G. C., Interferons and viral infections. Biofactors 35, 14–20 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Schoggins J. W., Interferon-stimulated genes: What do they all do? Annu. Rev. Virol. 6, 567–584 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Reich N. C., Too much of a good thing: Detrimental effects of interferon. Semin. Immunol. 43, 101282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fensterl V., Chattopadhyay S., Sen G. C., No love lost between viruses and interferons. Annu. Rev. Virol. 2, 549–572 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chattopadhyay S., Sen G. C., Tyrosine phosphorylation in Toll-like receptor signaling. Cytokine Growth Factor Rev. 25, 533–541 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barber G. N., STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 35, 88–93 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Cai X., Chiu Y. H., Chen Z. J., The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol. Cell 54, 289–296 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Bruns A. M., Horvath C. M., Antiviral RNA recognition and assembly by RLR family innate immune sensors. Cytokine Growth Factor Rev. 25, 507–512 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan Y. K., Gack M. U., RIG-I-like receptor regulation in virus infection and immunity. Curr. Opin. Virol. 12, 7–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kell A. M., Gale M. Jr., RIG-I in RNA virus recognition. Virology 479-480, 110–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loo Y. M., Gale M. Jr., Immune signaling by RIG-I-like receptors. Immunity 34, 680–692 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malathi K., Dong B., M. Gale, Jr, Silverman R. H., Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448, 816–819 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Der S. D., Zhou A., Williams B. R., Silverman R. H., Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc. Natl. Acad. Sci. U.S.A. 95, 15623–15628 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grandvaux N., et al. , Transcriptional profiling of interferon regulatory factor 3 target genes: Direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76, 5532–5539 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geiss G., et al. , A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J. Biol. Chem. 276, 30178–30182 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa H., Ma Z., Barber G. N., STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiscott J., Convergence of the NF-κB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 18, 483–490 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Hiscott J., Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. 282, 15325–15329 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Honda K., Taniguchi T., IRFs: Master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6, 644–658 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Sadler A. J., Williams B. R., Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8, 559–568 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glanz A., et al. , Transcriptional and non-transcriptional activation, posttranslational modifications, and antiviral functions of interferon regulatory factor 3 and viral antagonism by the SARS-coronavirus. Viruses 13, 575 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borden E. C., et al. , Interferons at age 50: Past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6, 975–990 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazear H. M., Schoggins J. W., Diamond M. S., Shared and distinct functions of type I and type III interferons. Immunity 50, 907–923 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian G., et al. , A new mechanism of interferon’s antiviral action: Induction of autophagy, essential for paramyxovirus replication, is inhibited by the interferon stimulated gene, TDRD7. PLoS Pathog. 14, e1006877 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian G., et al. , The interferon-inducible protein TDRD7 inhibits AMP-activated protein kinase and thereby restricts autophagy-independent virus replication. J. Biol. Chem. 295, 6811–6822 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanai H., et al. , Revisiting the role of IRF3 in inflammation and immunity by conditional and specifically targeted gene ablation in mice. Proc. Natl. Acad. Sci. U.S.A. 115, 5253–5258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jefferies C. A., Regulating IRFs in IFN driven disease. Front. Immunol. 10, 325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petro T. M., IFN regulatory factor 3 in health and disease. J. Immunol. 205, 1981–1989 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Ikushima H., Negishi H., Taniguchi T., The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harb. Symp. Quant. Biol. 78, 105–116 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Sato M., et al. , Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity 13, 539–548 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Chattopadhyay S., et al. , Inhibition of viral pathogenesis and promotion of the septic shock response to bacterial infection by IRF-3 are regulated by the acetylation and phosphorylation of its coactivators. MBio 4, e00636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chattopadhyay S., Kuzmanovic T., Zhang Y., Wetzel J. L., Sen G. C., Ubiquitination of the transcription factor IRF-3 activates RIPA, the apoptotic pathway that protects mice from viral pathogenesis. Immunity 44, 1151–1161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chattopadhyay S., et al. , Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J. 29, 1762–1773 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chattopadhyay S., Yamashita M., Zhang Y., Sen G. C., The IRF-3/Bax-mediated apoptotic pathway, activated by viral cytoplasmic RNA and DNA, inhibits virus replication. J. Virol. 85, 3708–3716 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chattopadhyay S., et al. , Role of interferon regulatory factor 3-mediated apoptosis in the establishment and maintenance of persistent infection by Sendai virus. J. Virol. 87, 16–24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chattopadhyay S., Sen G. C., IRF-3 and Bax: A deadly affair. Cell Cycle 9, 2479–2480 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chattopadhyay S., Sen G. C., dsRNA-activation of TLR3 and RLR signaling: Gene induction-dependent and independent effects. J. Interferon Cytokine Res. 34, 427–436 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chattopadhyay S., Sen G. C., RIG-I-like receptor-induced IRF3 mediated pathway of apoptosis (RIPA): A new antiviral pathway. Protein Cell 8, 165–168 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chattopadhyay S., et al. , EGFR kinase activity is required for TLR4 signaling and the septic shock response. EMBO Rep. 16, 1535–1547 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White C. L., Chattopadhyay S., Sen G. C., Phosphatidylinositol 3-kinase signaling delays sendai virus-induced apoptosis by preventing XIAP degradation. J. Virol. 85, 5224–5227 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanz-Garcia C., et al. , The non-transcriptional activity of IRF3 modulates hepatic immune cell populations in acute-on-chronic ethanol administration in mice. J. Hepatol. 70, 974–984 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanz-Garcia C., et al. , Nontranscriptional activity of interferon regulatory factor 3 protects mice from high-fat diet-induced liver injury. Hepatol. Commun. 3, 1626–1641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petrasek J., et al. , STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc. Natl. Acad. Sci. U.S.A. 110, 16544–16549 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sze A., et al. , Host restriction factor SAMHD1 limits human T cell leukemia virus type 1 infection of monocytes via STING-mediated apoptosis. Cell Host Microbe 14, 422–434 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Zierhut C., et al. , The cytoplasmic DNA sensor cGAS promotes mitotic cell death. Cell 178, 302–315.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iracheta-Vellve A., et al. , Endoplasmic reticulum stress-induced hepatocellular death pathways mediate liver injury and fibrosis via stimulator of interferon genes. J. Biol. Chem. 291, 26794–26805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Paolo N. C., Doronin K., Baldwin L. K., Papayannopoulou T., Shayakhmetov D. M., The transcription factor IRF3 triggers “defensive suicide” necrosis in response to viral and bacterial pathogens. Cell Rep. 3, 1840–1846 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakhaei P., et al. , IκB kinase ε-dependent phosphorylation and degradation of X-linked inhibitor of apoptosis sensitizes cells to virus-induced apoptosis. J. Virol. 86, 726–737 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng Z., Lemon S. M., Innate immunity to enteric hepatitis viruses. Cold Spring Harb. Perspect. Med. 9, a033464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brasier A. R., Zhao Y., Measurement of the innate immune response in the airway. Adv. Exp. Med. Biol. 795, 233–254 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Czerkies M., et al. , Cell fate in antiviral response arises in the crosstalk of IRF, NF-κB and JAK/STAT pathways. Nat. Commun. 9, 493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian B., et al. , BRD4 couples NF-κB/RelA with airway inflammation and the IRF-RIG-I amplification loop in respiratory syncytial virus infection. J. Virol. 91, e00007–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Indukuri H., et al. , Ikkepsilon regulates viral-induced interferon regulatory factor-3 activation via a redox-sensitive pathway. Virology 353, 155–165 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Wang X. A., et al. , Interferon regulatory factor 3 constrains IKKβ/NF-κB signaling to alleviate hepatic steatosis and insulin resistance. Hepatology 59, 870–885 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Maelfait J., et al. , A20 (Tnfaip3) deficiency in myeloid cells protects against influenza A virus infection. PLoS Pathog. 8, e1002570 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maelfait J., et al. , A20 deficiency in lung epithelial cells protects against influenza A virus infection. PLoS Pathog. 12, e1005410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peters K., Chattopadhyay S., Sen G. C., IRF-3 activation by Sendai virus infection is required for cellular apoptosis and avoidance of persistence. J. Virol. 82, 3500–3508 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sears N., Sen G. C., Stark G. R., Chattopadhyay S., Caspase-8-mediated cleavage inhibits IRF-3 protein by facilitating its proteasome-mediated degradation. J. Biol. Chem. 286, 33037–33044 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pahl H. L., Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18, 6853–6866 (1999). [DOI] [PubMed] [Google Scholar]
- 63.Shelest E., Kel A. E., Göessling E., Wingender E., Prediction of potential C/EBP/NF-kappaB composite elements using matrix-based search methods. In Silico Biol. 3, 71–79 (2003). [PubMed] [Google Scholar]
- 64.Glanz A., et al. , High throughput screening of FDA-approved drug library reveals the compounds that promote IRF3-mediated pro-apoptotic pathway inhibit virus replication. Viruses 12, 442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chow K. T., et al. , Differential and overlapping immune programs regulated by IRF3 and IRF5 in plasmacytoid dendritic cells. J. Immunol. 201, 3036–3050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elco C. P., Guenther J. M., Williams B. R., Sen G. C., Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-κB, and interferon but not Toll-like receptor 3. J. Virol. 79, 3920–3929 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Freaney J. E., Kim R., Mandhana R., Horvath C. M., Extensive cooperation of immune master regulators IRF3 and NFκB in RNA Pol II recruitment and pause release in human innate antiviral transcription. Cell Rep. 4, 959–973 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ogawa S., et al. , Molecular determinants of crosstalk between nuclear receptors and Toll-like receptors. Cell 122, 707–721 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McGowan J., et al. , 14-3-3ζ-TRAF5 axis governs interleukin-17A signaling. Proc. Natl. Acad. Sci. U.S.A. 117, 25008–25017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abdulkhalek S., Guo M., Amith S. R., Jayanth P., Szewczuk M. R., G-protein coupled receptor agonists mediate Neu1 sialidase and matrix metalloproteinase-9 cross-talk to induce transactivation of TOLL-like receptors and cellular signaling. Cell. Signal. 24, 2035–2042 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Glanz A., et al. , Autophagic degradation of IRF3 induced by the small-molecule auranofin inhibits its transcriptional and proapoptotic activities. J. Biol. Chem. 297, 101274 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wietek C., Miggin S. M., Jefferies C. A., O’Neill L. A., Interferon regulatory factor-3-mediated activation of the interferon-sensitive response element by Toll-like receptor (TLR) 4 but not TLR3 requires the p65 subunit of NF-κ. J. Biol. Chem. 278, 50923–50931 (2003). [DOI] [PubMed] [Google Scholar]
- 73.Menachery V. D., Pasieka T. J., Leib D. A., Interferon regulatory factor 3-dependent pathways are critical for control of herpes simplex virus type 1 central nervous system infection. J. Virol. 84, 9685–9694 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He J., et al. , IRF-7 is a critical regulator of type 2 innate lymphoid cells in allergic airway inflammation. Cell Rep. 29, 2718–2730.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Schultz K. L. W., Troisi E. M., Baxter V. K., Glowinski R., Griffin D. E., Interferon regulatory factors 3 and 7 have distinct roles in the pathogenesis of alphavirus encephalomyelitis. J. Gen. Virol. 100, 46–62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Canivet C., et al. , Both IRF3 and especially IRF7 play a key role to orchestrate an effective cerebral inflammatory response in a mouse model of herpes simplex virus encephalitis. J. Neurovirol. 24, 761–768 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Fang L., et al. , Ataxia telangiectasia mutated kinase mediates NF-κB serine 276 phosphorylation and interferon expression via the IRF7-RIG-I amplification loop in paramyxovirus infection. J. Virol. 89, 2628–2642 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Byrne A. J., et al. , A critical role for IRF5 in regulating allergic airway inflammation. Mucosal Immunol. 10, 716–726 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bais S. S., et al. , Chandipura virus utilizes the prosurvival function of RelA NF-κB for its propagation. J. Virol. 93, e00081-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hilton L., et al. , The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J. Virol. 80, 11723–11732 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xia H., et al. , Evasion of type I interferon by SARS-CoV-2. Cell Rep. 33, 108234 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hardy S., Jackson B., Goodbourn S., Seago J., Classical swine fever virus Npro antagonises IRF3 to prevent IFN-independent TLR3 and RIG-I-mediated apoptosis. J. Virol. 95, e01136-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.