Significance
In different individuals, the same mutation can lead to different phenotypes due to genetic background effects. This is commonly observed in various systems, including many human diseases. While isolated examples of such background effects have been observed, a systematic view across a large number of individuals is still lacking. Here, we surveyed genetic background effects associated with gene loss-of-function mutations across a population of natural isolates of the yeast Saccharomyces cerevisiae. We found that ∼15% of genes can display a background-dependent fitness change. Genes related to mitochondrial functions are significantly overrepresented, and showed reversed patterns of fitness gain or loss with genes involved in transcription and chromatin remodeling as well as in nuclear–cytoplasmic transport, suggesting a potential functional rewiring.
Keywords: background effect, fitness variability, transposition saturation, yeast, Saccharomyces cerevisiae
Abstract
In natural populations, the same mutation can lead to different phenotypic outcomes due to the genetic variation that exists among individuals. Such genetic background effects are commonly observed, including in the context of many human diseases. However, systematic characterization of these effects at the species level is still lacking to date. Here, we sought to comprehensively survey background-dependent traits associated with gene loss-of-function (LoF) mutations in 39 natural isolates of Saccharomyces cerevisiae using a transposon saturation strategy. By analyzing the modeled fitness variability of a total of 4,469 genes, we found that 15% of them, when impacted by a LoF mutation, exhibited a significant gain- or loss-of-fitness phenotype in certain natural isolates compared with the reference strain S288C. Out of these 632 genes with predicted background-dependent fitness effects, around 2/3 impact multiple backgrounds with a gradient of predicted fitness change while 1/3 are specific to a single genetic background. Genes related to mitochondrial function are significantly overrepresented in the set of genes showing a continuous variation and display a potential functional rewiring with other genes involved in transcription and chromatin remodeling as well as in nuclear–cytoplasmic transport. Such rewiring effects are likely modulated by both the genetic background and the environment. While background-specific cases are rare and span diverse cellular processes, they can be functionally related at the individual level. All genes with background-dependent fitness effects tend to have an intermediate connectivity in the global genetic interaction network and have shown relaxed selection pressure at the population level, highlighting their potential evolutionary characteristics.
The same mutation might show different phenotypic effects across genetically distinct individuals due to standing genomic variation (1–8). Such background effects have been described across species and impact the phenotype–genotype relationship, including in the context of health and disease. Indeed, they have been observed in multiple human Mendelian disorders, where individuals carrying the same causal mutation can display a wide range of clinical symptoms, including variable severity and age of onset (1, 7, 9–12). The underlying origin of these background effects may be both intrinsic, namely due to interactions between the causal variant and other genetic modifiers (9–11), and/or extrinsic, namely due to environmental factors (12, 13). To date, a handful of modifier genes have been found associated with human disorders, most notably in cystic fibrosis (11, 12). However, such examples remain rare and anecdotal due to the low number of sample cases in most human Mendelian diseases.
In recent years, several large-scale surveys in different model organisms, such as the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe and the nematode Caenorhabditis elegans, and various human cell lines highlighted the broad influence of genetic backgrounds on the phenotypic outcomes associated with loss-of-function mutations (14–22). In yeast, a study comparing systematic gene deletion collections in two laboratory strains, Σ1278b and S288C, showed that ∼1% of all genes (57/5,100) can display background-dependent gene essentiality, that is, where the deletion of the same gene can be lethal in one background but not the other (14). Several origins underlying such gene essentiality have been identified, including genetic interactions between the mitochondrial genome and/or viral elements with the nuclear genome (23) as well as genetic interactions between the primary gene deletion and background-specific modifiers (24). While gene essentiality may be the most severe manifestation associated with loss-of-function mutations, gain- and loss-of-fitness variation related to genetic backgrounds or environmental conditions were also found in yeast (5, 25). For example, about 20% of yeast genes showed background-dependent fitness variation under a wide range of growth conditions, including the presence of various drugs, osmotic stress, and nutrient sources in four genetically diverse isolates (25). However, all these studies only include a limited number of genetic backgrounds and therefore cannot accurately reflect the extent of the background effect at the species level.
Recently, a large collection of 1,011 S. cerevisiae isolates originating from various ecological and geographical sources has been completely sequenced (26), representing an incomparable resource to systematically study the effects of genetic backgrounds at the species level. Several strategies have been developed in S. cerevisiae to explore the impact of loss-of-function mutations, including systematic gene deletions using homologous recombination (14), gene disruption using the CRISPR-Cas9 editing systems (27), repeated backcrosses (25), and transposon mutagenesis (28–30). Among these strategies, transposon mutagenesis based on random excision and insertions is particularly attractive for exploring in parallel a large number of genetically diverse individuals. This method relies on transposition events via a carrier plasmid, which allow for the generation of millions of mutants carrying genomic insertions leading to loss-of-function mutations (30). Due to the random insertion patterns in each genetic background, these methods do not depend on sequence homology, as is the case for traditional PCR-based gene deletions and CRISPR-Cas9–related strategies (27, 31), and they do not present the risk of inadvertently introducing exogenous genomic regions, as is potentially the case for backcross-based strategies (25).
Here, we selected over a hundred natural isolates broadly representative of the diversity of the S. cerevisiae species, and performed transposon saturation analyses using the Hermes transposition system (29). We generated, sequenced, and analyzed large pools of transposon insertion mutants and constructed a logistic model to predict the fitness effects of gene loss of function based on the insertion densities in and around each annotated gene. Comparing the fitness prediction between the different isolates and the S288C reference, we identified 632 genes with predicted background-dependent fitness effects, corresponding to ∼15% of the genome. Overall, they are functionally related, with members of the same protein complex or biological process showing similar variability in each genetic background. They also tend to show an intermediate level of integration in genetic networks compared with nonessential and essential genes, and might be under positive or relaxed purifying selection at the population level.
Results
Generation of Loss-of-Function Mutant Collections Using the Hermes Transposon System.
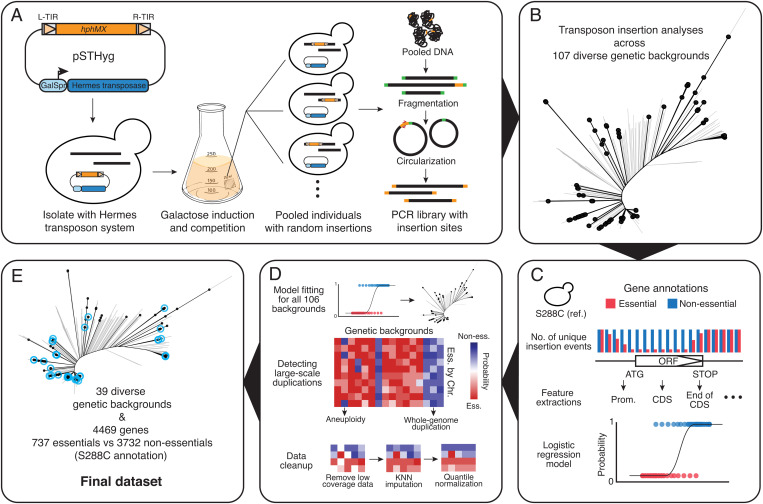
To gain insight into fitness variation associated with loss-of-function mutations across different S. cerevisiae genetic backgrounds, we performed transposon saturation assays in various natural isolates using the Hermes transposon system. The Hermes transposon system has previously been adapted in yeast to allow the selection of random insertion events in liquid culture, which makes this system particularly suitable for parallel analyses of a large number of genetically diverse individuals (29). This system is based on a centromeric plasmid, which contains the Hermes transposase under the control of a modified galactose inducible promoter, GalS, as well as a transposon carrying a selectable marker (Fig. 1A). Briefly, for any strain of interest, the plasmid is first transformed into stable haploid cells and then propagated in media containing galactose to induce excision and reinsertion of the transposon at random locations in the genome, thereby generating a large pool of individuals with hundreds of thousands of insertions along the genome (Fig. 1A). After a recovery phase in rich medium, the genome of this pool of mutants is recovered, and then fragmentated and circularized (Fig. 1A). Using PCR with outward-facing primers specifically targeting the transposon, a library that exclusively contains the insertion sites can be constructed and then sequenced using standard Illumina methods (Fig. 1A). In principle, transposon insertions that cause severe fitness defects, for example those occurring in essential genes, will not be recovered due to the competitive disadvantage compared with events occurring in genes which are not essential. Analysis of insertion patterns along the genomes of different individuals therefore provides a proxy for fitness variation related to loss-of-function mutations.
Fig. 1.
Summary of the Hermes transposon saturation procedure. (A) A centromeric plasmid carrying the Hermes transposase and a transposon containing a hygromycin resistance marker (HygMX) are transformed into a haploid isolate background. Random transposon insertions are induced and selected. The mutant pool is then recovered and a PCR library that contains only the insertion sites is constructed and sequenced. (B) Distribution of the selected 107 isolates across the species. The neighbor-joining tree was constructed using biallelic SNPs in the 1,011 yeast collection (26). Selected strains are highlighted in black. (C) A logistic model was constructed using insertion profiles in the reference strain S288C. Gene essentiality annotations were used as a binary classifier, excluding those annotated as involved in galactose metabolism, respiration, and slow growth. (D) The logistic model was applied to insertion patterns in the remaining 106 isolates. Large-scale genome duplications were detected by looking at fitness predictions for all annotated essential genes along each chromosome. Low-coverage regions were removed and then imputed using the k-nearest-neighbor method. The imputed fitness matrix was then quantile-normalized. (E) The final dataset after imputation consists of 39 isolates and 4,469 genes. Strains included in the final dataset are highlighted in blue.
In addition to the S288C reference strain, we selected 106 isolates originating from various ecological and geographical sources that are broadly representative of the species diversity (Fig. 1B and Dataset S1). Stable haploid variants of this set of isolates have been generated previously (32, 33) and are all capable of growing in galactose medium. We have adapted the initial version of the Hermes transposon plasmid to carry a hygromycin resistance marker instead of nourseothricin to ensure compatibility with the selected strains, which may carry either a KanMX or NatMX marker at the HO locus. Transposon insertion profiles for each isolate were obtained as described (Fig. 1A). We observed a marked variability in terms of insertion efficiency across different genetic backgrounds, ranging from ∼100 to ∼300,000 unique insertion sites (SI Appendix, Fig. S1A and Dataset S1). No discernible correlation between the genetic origin of the isolates and the transposon insertion efficiency was observed (Dataset S1). We then compared the insertion preferences between the S288C reference strain and the 106 natural isolates (SI Appendix, Fig. S1B). Insertion densities for known sequence motifs (29) were conserved across the different genetic backgrounds (SI Appendix, Fig. S1B).
Using insertion profiles and the annotation of gene essentiality in the S288C reference, we analyzed the average insertion patterns in the promoters (−500 bp to ATG, 100-bp window), the coding DNA sequences (CDSs), and the terminators (STOP to +500 bp, 100-bp window) for all annotated essential vs. nonessential genes. Compared with nonessential genes, the number of insertions for essential genes drops from −100 bp prior to the CDS and extends to −100 bp prior to the stop codon (before the terminator region), with on average ∼3 times fewer insertions within the CDS (SI Appendix, Fig. S1 C–E). This pattern is consistent with the results obtained in previous studies using the Hermes system (29).
Modeling Based on Insertion Patterns to Identify Genes with Background-Dependent Fitness Effects.
We constructed a logistic model that simultaneously takes into account transposon insertions that have occurred in the genes and surrounding regions using the insertion profiles from the reference strain S288C and the corresponding gene essentiality annotations as a binary classifier (Materials and Methods, Fig. 1C, and Dataset S2). We applied this model to the insertion profiles of all 106 diverse isolates and the reference S288C (Fig. 1D). For each annotated open reading frame (ORF) (approximately a total of 6,300 ORFs), a probability was calculated based on the model, ranging from a value of 1, corresponding to most likely nonessential, to 0, corresponding to most likely essential. Genomic regions with low insertion densities contribute to overall low predictive powers (SI Appendix, Fig. S2 A and B), which were subsequently removed. Due to the variability of insertion efficiency across strains (SI Appendix, Fig. S1A), removal of regions with low insertion density has led to entire strain backgrounds with few interpretable genes. By maximizing the number of both strains and genes that remained after data imputation (SI Appendix, Fig. S2 C and D), a total of 52 backgrounds and probability predictions for 4,469 genes were retained for subsequent analyses (Dataset S3).
Large-scale genome duplications, including aneuploidies and endoreduplications, are frequently observed in yeast experimental evolution (34–36). Such events may hamper the accuracy of the modeled fitness effect in the context of the transposon insertion assay, as genes in the duplicated region will all appear to be fit due to insertions in a single copy of the gene. We searched for signals of large-scale genome duplications by examining all annotated essential genes along the chromosomes in our set of isolates (Fig. 1D and SI Appendix, Fig. S3A). We detected endoreduplication events in 7 out of 52 strains where all chromosomes appeared to be duplicated based on the high predicted probability values for all essential genes (SI Appendix, Fig. S3A). In another set of six strains, essential genes showed an intermediate- to high-probability prediction but not high enough to be confidently classified as nonessential. These six strains were then confirmed as a mixture of haploid and diploid cells using flow cytometry. In addition to these whole-genome events, we also detected three strains with an aneuploidy of chromosome I (ACT, BKL, and ACV), one strain with an aneuploidy of chromosome XII (CPG), and one strain with an aneuploidy of chromosome XIV (CQA). These aneuploidies were not present in the original isolate, with the exception of chromosome I aneuploidies in ACT and ACV strains, highlighting the dynamics of genome instability in different genetic backgrounds. All 13 strains with whole-genome endoreduplication were entirely removed from the dataset. We also excluded aneuploid chromosomes from the analysis (Dataset S3).
Next, we looked specifically at the probability predictions in the reference S288C. The final set of 4,469 genes includes 3,732 and 737 that were annotated nonessential and essential, respectively. Among the genes annotated as nonessential, ∼180 were predicted to be likely essential in our data (Dataset S3), of which more than 70% correspond to slow-growth or galactose-specific fitness defect genes. For example, the hexose transporters HXT6/7 and genes involved in galactose metabolism are all predicted to be likely essential, as expected by using our transposition saturation strategy (SI Appendix, Fig. S3B). On the other hand, 26 genes annotated as essential were predicted to be likely nonessential, with a predicted probability >0.8 (Dataset S3). Among these, we found FUR1, HIP1, and SSY5, consisting of amino acid transporters that are only essential in the multiauxotrophic BY4741 background, isogenic to the prototrophic S288C we used in our study (Dataset S3). We have also found genes where the essentiality concerns only part of the ORF, namely the essential domains, as has also been observed in previous studies using this transposon saturation strategy (28) (SI Appendix, Fig. S3C). Notably, we found the RET2 and SRP14 genes, which are also among the essential genes specific to the S288C background compared with Σ1278b in systematic gene deletion collections (14). Indeed, these domain essential effects are recaptured in our dataset when comparing insertion patterns between S288C and Σ1278b (SI Appendix, Fig. S3D). In fact, background-specific essential genes between S288C and Σ1278b that did not display severe fitness defects when deleted in the nonessential background (24), including S288C-specific essential genes (RET2, UBC1, and SRP14) and Σ1278b-specific essential genes (SKI8, TMA108, and AAT2), all showed domain essential effects and are all recaptured in our data (SI Appendix, Fig. S3D).
Overall, the predicted probability based on our logistic model can serve as a reasonable proxy for fitness variation related to loss-of-function mutations. Modeled fitness (predicted probability for nonessentiality) is more accurate at predicting nonessential/high-fitness cases than essential/low-fitness cases, which may in part be due to the fact that some nonessential genes are de facto slow growers in the context of our experimental conditions, and in part due to bias in transposon insertion densities for some genomic regions across genetic backgrounds. Essentialities related to specific domains can be recaptured by the raw insertion patterns but not by our modeled fitness values (SI Appendix, Fig. S3 C and D). However, this effect is inherent to the transposon saturation system and should not lead to differential fitness effect prediction in different genetic backgrounds. In principle, biases related to low insertion densities for genomic regions and/or protein domains as mentioned above will likely lead to false negative calls for background-dependent fitness predictions and will not impact the specificity of the model. Our final dataset consists of 39 isolates from various origins and predicted fitness for 4,469 genes, which is analyzed in more detail (Fig. 1E and Dataset S3). Compared with the initial set of 106 isolates, the final of 39 isolates is still representative of the species diversity, although some of the most divergent groups, such as isolates from French Guiana and China, are underrepresented (Fig. 1E).
Fitness Variability Associated with Loss-of-Function Mutations Distinguishes Rare and Common Effects across Genetic Backgrounds.
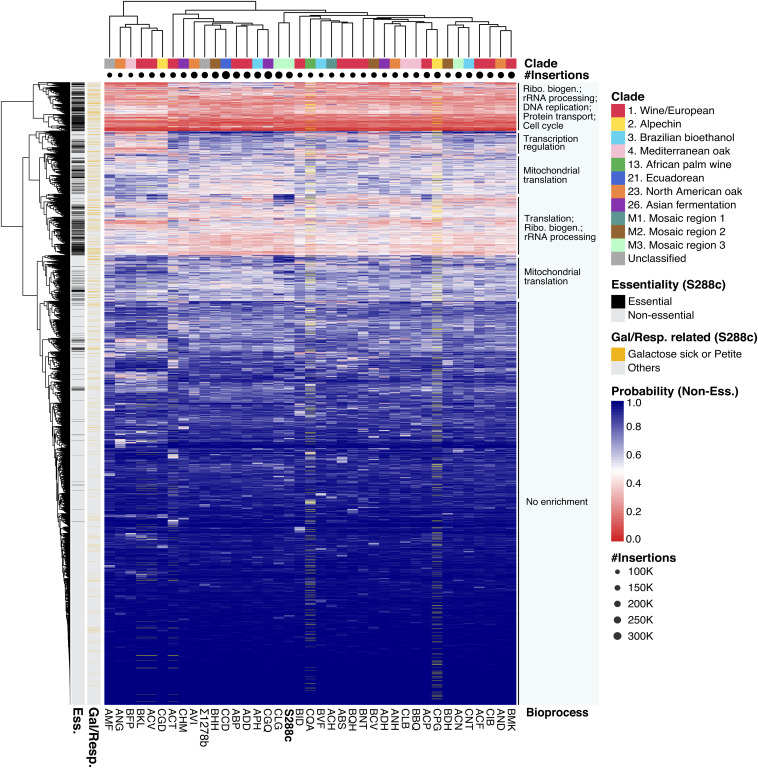
We first performed a hierarchical clustering based on the predicted fitness values of 4,469 genes across the 39 genetic backgrounds (Fig. 2). Profile similarity based on the predicted fitness effects did not correlate with the genetic origins of the isolates (Fig. 2). Genes that are consistently essential in different isolates clustered together and are enriched for essential biological processes, including ribosome biogenesis, ribosomal RNA (rRNA) processing, DNA replication, protein transport, and the cell cycle (Fig. 2). Genes that are consistently nonessential in all backgrounds formed a large cluster without significant enrichment for any specific biological process. Interestingly, several clusters of genes with variable fitness effects were identified, displaying modular switches from fit to nonfit phenotypes across the entire population. Gene enrichment analyses revealed genes involved in mitochondrial translation, transcription regulation, and general translational processes (Fig. 2). A large proportion of these genes with population-wide fitness variation consists of nuclear-encoded mitochondrial genes involved in respiration, which were expected to show a selective disadvantage in our pool of mutants that must grow on galactose. This observation suggests that such general fitness variability may be environment-related rather than background-specific per se. However, other biological processes in addition to respiration and mitochondrial functions have also been enriched, such as transcription regulation (Fig. 2), for which the impact of environment vs. genetic background on their fitness variability remains unclear.
Fig. 2.
Hierarchical clustering of 4,469 fitness predictions across 39 genetic backgrounds. The distance matrix was calculated using the Euclidean distance method. The genetic origin of each isolate was color-coded, and the total insertion numbers per isolate are represented by dot size under the origin color code. Genes annotated as essential in the reference S288C are highlighted in black, and genes annotated as either galactose- or respiration-related are highlighted in yellow on the side bars. Genes within duplicated chromosomes were removed (yellow bars on heatmap). Biological processes that are enriched in subclusters are annotated.
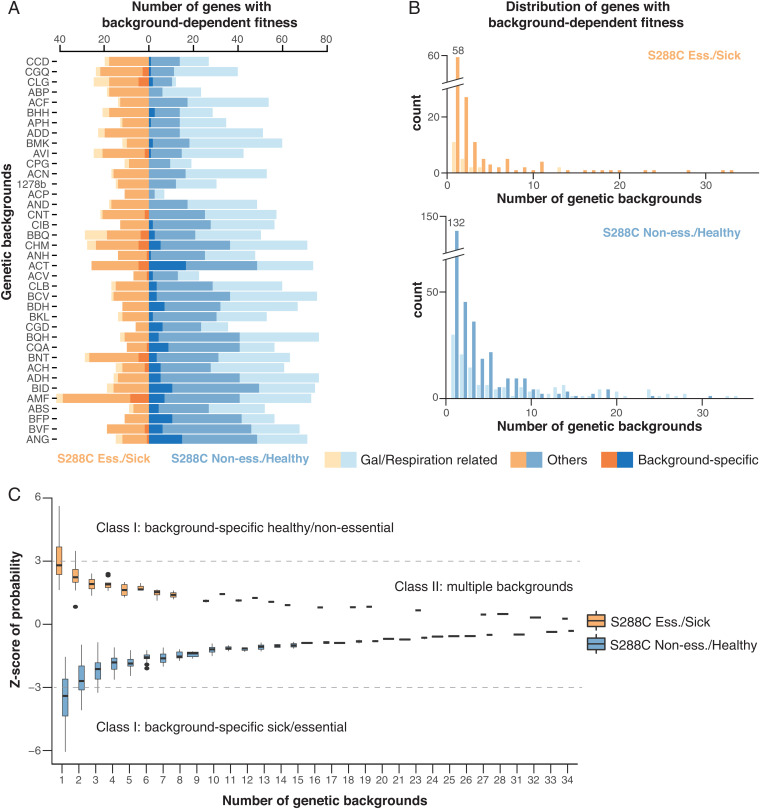
To further characterize the background-dependent fitness variation, we systematically compared the predicted fitness values for each gene in a given isolate with the predictions of the reference strain S288C. A differential fitness score for each gene in each background was calculated by subtracting the predicted fitness value in a given strain from the corresponding fitness prediction in the reference S288C. A minimum absolute value of the differential fitness score of 0.5 was considered significant, which corresponds to a bona fide reverse in the direction of being predicted as essential or nonessential according to our logistic model. In total, 632 genes were identified with marked fitness variation, with 458 and 174 showing a loss of fitness (S288C healthy and background sick) and a gain of fitness (S288C sick and background healthy) compared with the reference, respectively. The number of identified differential fitness genes ranges from 8 (ACP) to 88 (BQH) for loss-of-fitness cases (with a median of 61), and from 6 (CGD) to 42 (AMF) for gain-of-fitness cases (with a median of 16) (Fig. 3A). A total of 163 out of all 632 hits are related to respiration and mitochondrial functions, representing ∼20 to ∼60% of loss-of-fitness hits depending on the genetic background (Fig. 3A). Furthermore, these respiration-related genes tend to impact more backgrounds on average than non–respiration-related hits (Fig. 3B). These observations echoed what was shown with hierarchical clustering where mitochondrial-related genes were highly enriched in clusters with modular fitness variation in several backgrounds (Fig. 2). Again, due to the overrepresentation of these respiration-related genes and their continuous fitness variation in the population, we suspect that these hits are likely to be impacted by the environment (i.e., pooled competition in galactose media) in addition to any specific genetic backgrounds.
Fig. 3.
Number and distribution of background-dependent fitness variation genes. (A) Number of hits detected in each genetic background. Genes annotated as galactose- or respiration-related and background-specific genes are color-coded as indicated. Strains are sorted according to the total number of insertions. (B) The number of genetic backgrounds impacted by the detected hits. (B, Top) Gain-of-fitness genes compared with S288C. (B, Bottom) Loss-of-fitness genes compared with S288C. (C) Z-statistic distribution for hits that impact different numbers of genetic backgrounds. A cutoff of |z statistics| >3 is indicated with dashed lines.
We then calculated the z statistics for all variable fitness hits to distinguish those that only impact few backgrounds (class I) from the others that impact multiple backgrounds (class II) (Dataset S4). In principle, a low z statistic indicates variability involving many strains whereas cases that are truly specific to some genetic backgrounds should be outliers with a high z-statistic score (|z| > 3) (Fig. 3C). Of the set of 632 genes, we found 179 that are background-specific, which mainly impact a single genetic background (Dataset S4). These background-specific genes are rarer compared with the other group, with a median of five identified per isolate of both loss- and gain-of-fitness types combined (Fig. 3A). Genes related to respiration and mitochondrial functions are not overrepresented in this group (23/179 vs. 691/4,469 in the background, Fisher’s exact test, P = 0.82). For the class I group, no significant enrichment for any biological processes or molecular functions has been identified. By contrast, respiration-related genes are significantly overrepresented in the class II group (140/453 vs. 691/4,469, Fisher’s exact test, P = 1.6e-10, odds ratio [OR] = 2). On average, this group of genes impacts ∼6 genetic backgrounds.
Common Fitness Variation across Multiple Backgrounds Reveals Functional Rewiring.
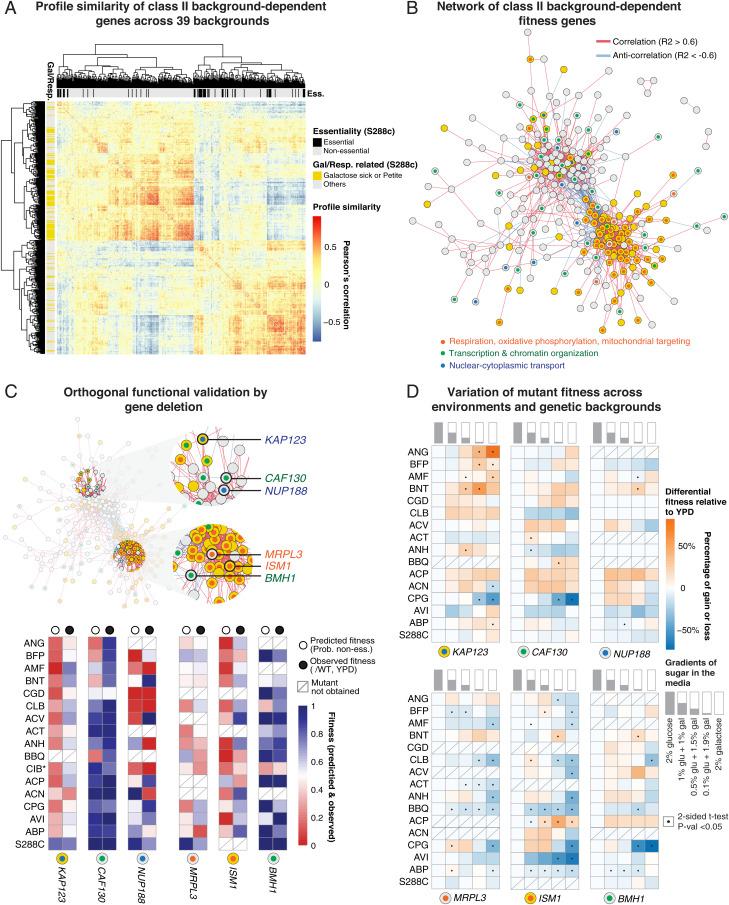
The overrepresentation of respiration-related genes in the 453 class II hits led us to hypothesize that this group of genes might be impacted by the experimental environment in addition to the genetic backgrounds. While a large fraction of these hits corresponds to genes involved in respiration, the majority of this group is involved in other biological processes. To explore the functional relationships within this group, we calculated the pairwise correlations between these genes using predicted fitness values across the 39 strain backgrounds (Fig. 4A). We constructed a network based on the profile similarities where the edges correspond to a Pearson’s correlation >0.6 (correlation) or <−0.6 (anticorrelation) (Fig. 4B and SI Appendix, Fig. S4). In total, 292 out of the 453 environment-related hits exceeded our stringent correlation cutoffs (SI Appendix, Fig. S4). The profile similarity and network structure revealed two main subnetworks, which are correlated within the subgroup but are anticorrelated between subgroups (Fig. 4 A and B). The first subgroup contains mainly respiration-related genes, in particular genes involved in mitochondrial translation (Fig. 4A and SI Appendix, Fig. S4A), which are anticorrelated with genes involved in transcription regulation and chromatin remodeling (SPT7, SPT8, SWC4, SWC5, ARP6, ARP7, SIN3, RKR1, YAF9, UME1, NGG1, and CAF130, for example) as well as genes involved in nuclear–cytoplasmic protein transfer (KAP120, KAP122, KAP123, NUP57, NUP100, NUP188, POM152, NIC96, and MLP1, for example) (SI Appendix, Fig. S4A). Many of these correlations have been found between members of the same protein complexes. Several members of the transcription and nuclear transport subgroup are also annotated as related to respiration (deletion leads to absence of respiration) although they are not directly involved in mitochondrial function, such as SIN3, a general chromatin remodeler, and KAP123, a karyopherin responsible for nuclear import of ribosomal proteins. In addition to this large network, several small networks have also been detected (SI Appendix, Fig. S4 B–D), including PMT1, PMT2, and GET2, which are involved in endoplasmic reticulum–related glycosylation and are known to have physical interactions. Functional enrichments in the anticorrelated subgroups suggest a potential “rewiring” between mitochondrial translation and transcription regulation/nuclear transport, where modular switches of fitness effects associated with gene loss of function may occur in different genetic backgrounds.
Fig. 4.
Correlation analyses for class II hits. (A) Pairwise profile similarity based on predicted fitness across 39 backgrounds. Distance matrix was based on pairwise Pearson’s correlation. Gene essentiality annotations are indicated on the Upper side bar and genes annotated as involved in galactose/respiration are indicated on the Left side bar. (B) Network based on profile similarity among class II hits. Genes annotated as involved in galactose/respiration are colored in yellow. Positive correlations (>0.6) are represented as red edges and negative correlations (<−0.6) are represented as blue edges. Enriched processes are indicated with colored dots. The complete network with annotated gene names can be found in SI Appendix, Fig. S4. (C) Functional validation using gene deletion. Candidate hits selected for deletion are highlighted and labeled. The predicted fitness (logistic probability of nonessential) and the observed fitness values (colony size ratio after tetrad dissection) for 88 strain/gene combinations varying from 0 to 1 are shown. Strain names are shown on the y axis. *CIB has wrinkles colony phenotype and was excluded for subsequent phenotyping using robotic pinning. (D) Variation of mutant fitness across environments and genetic backgrounds. For each mutant, the fitness (colony ratio mutant/wild type [WT]) variation was measured across five media conditions with different combinations of glucose and galactose. The proportions of these two sugar types are schematically represented and labeled. The relative fitness gain or loss was calculated by subtracting the fitness for a given condition by the fitness observed on YPD (first column, 2% glucose). Significant gain or loss is indicated with a black dot. Two-sided t test, P < 0.05.
We performed an orthogonal validation by gene deletion in a subset of cases across 17 out of the 39 isolates in the final dataset (Fig. 4C). We selected six candidate genes spanning the three main enriched biological processes, specifically the MRPL3 and ISM1 genes for mitochondrial translation, the CAF130 and BMH1 genes for transcription regulation, and the KAP123 and NUP188 genes involved in nuclear–cytoplasmic transport. Two of the candidates, KAP123 and ISM1, are known to have reduced fitness in galactose media when deleted in the reference strain S288C (Fig. 4 B and C). By using isogenic diploid strains, we obtained 88 deletion mutants out of the total 102 gene/strain combinations. We measured the fitness of the mutants by calculating the ratio between the colony size of haploid segregants that carry the mutation and the wild-type segregants after tetrad dissection on YPD complete medium (Fig. 4C, SI Appendix, Fig. S5A, and Dataset S4). Overall, observed fitness values for deletion mutants are significantly correlated with predicted fitness based on the logistic probability (Pearson’s correlation = 0.4, P = 0.0001; SI Appendix, Fig. S5B). At the gene level, the correlation between the predicted and observed fitness values can be variable, with the largest discrepancies observed for respiration/galactose–related genes, in particular for the KAP123, MRPL3, and ISM1 genes (Fig. 4C). Such discrepancies are more or less expected, as they could echo the potential environmental effect we observed. We therefore tested the 88 mutants and wild-type strains on media containing various gradients of glucose and/or galactose and systematically measured the relative fitness gain or loss compared with YPD (Fig. 4D and Dataset S4). Significant fitness gain or loss was observed in ∼45% of strain/mutant pairs (40 out of 88) in at least one media condition containing variable concentrations of galactose (Fig. 4D). The genes with the most discrepancies between the predicted and observed fitness values on YPD, namely the KAP123, MRPL3, and ISM1 genes, display greater environment-related fitness variability (on average, nine gene/strain combinations are variable against four in the other cases) (Fig. 4D). Moreover, this environment-related fitness variability also depends on the strain background. For example, for the strain CPG, loss of fitness of more than 50% was observed on galactose media compared with YPD for five out of the six genes tested, while no significant change was observed for any of the genes tested in the S288C reference background (Fig. 4D). These results highlight the robustness of our transposon insertion data and fitness predictions. In addition, they demonstrate the unequivocal impacts of environment, genetic background, and interplay between the two on the fitness of loss-of-function mutations.
Functional Insights into Genes with Background-Dependent Fitness Effects.
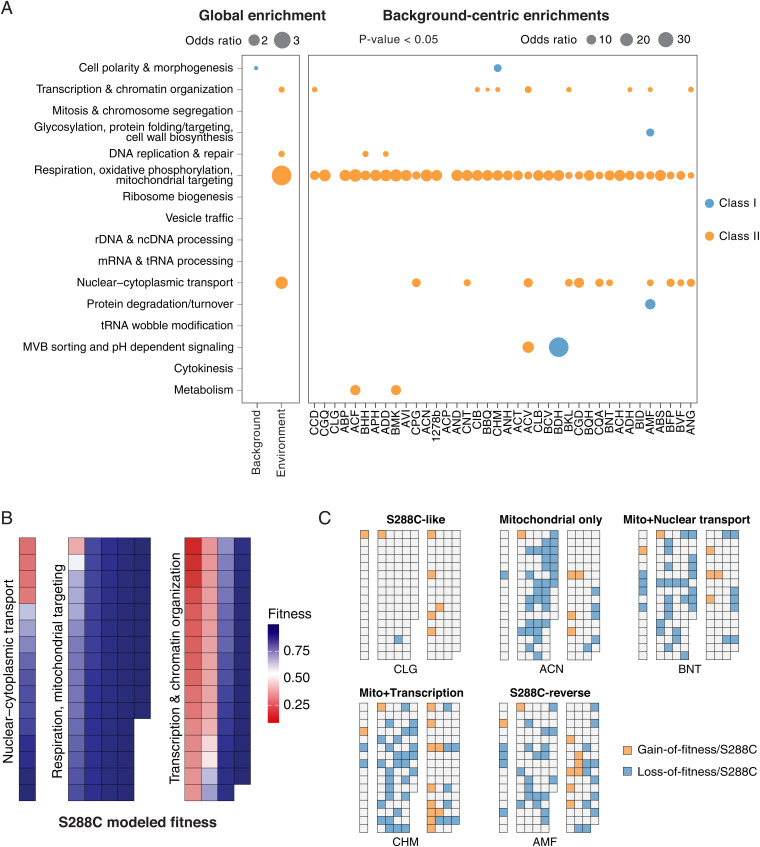
To further explore the functional enrichments of fitness variation genes at the strain level, we annotated genes in our dataset into 16 functional neighborhoods according to SAFE (37) and looked for enrichment in different neighborhoods (Fig. 5A). For each neighborhood, we calculated the OR of enrichment based on the number of hits annotated in the neighborhood vs. the total number of hits, with the size of the neighborhood and the total number of genes as background (one-sided Fisher’s exact test). Globally, background-specific hits (class I, 179 genes) are not enriched for most processes except for cell polarity (OR = 1.49, P = 0.026). Hits impacting multiple backgrounds (class II, 453 genes) are enriched for respiration and mitochondrial functions (OR = 3.77, P = 4.16e-17), as well as transcription and chromatin regulation (OR = 1.53, P = 0.002), nuclear–cytoplasmic transport (OR = 2.14, P = 0.004), and DNA repair (OR = 1.55, P = 0.01) (Fig. 5A and Dataset S4). When looking at the same neighborhood enrichment at the strain level, class II hits are enriched for mitochondrial functions in most genetic backgrounds, with the exception of the ACP and CLG strains, the latter of which has a predicted fitness profile that was most similar to the reference S288C (Fig. 2). A large fraction of isolates showed significant enrichments for transcription and chromatin regulation as well as nuclear–cytoplasmic transport (Fig. 5A). These enrichments are consistent with the rewiring hypothesis based on the profile similarity network analysis (Fig. 4B). Indeed, by specifically looking at the annotated genes in these functional neighborhoods, we observed various degrees of rewiring depending on the backgrounds (Fig. 5 B and C). In the reference S288C, loss of function for annotated genes in these three neighborhoods showed either high- or low-fitness predictions (Fig. 5B and SI Appendix, Fig. S5B), while in other genetic backgrounds, these predictions may be reversed as gain- or loss-of-fitness hits compared with S288C, with profiles ranging from similar to S288C (CLG) to almost completely reversed (AMF) (Fig. 5C). Most notably, such rewiring could include either only mitochondrial-related genes, or with one or more processes related to either transcription and chromatin regulation or nuclear–cytoplasmic transport (Fig. 5C). Depending on the genetic background, different sets of genes within the same functional neighborhood could be involved, highlighting the dynamics of such rewiring (SI Appendix, Fig. S5C).
Fig. 5.
Functional enrichments and rewiring for genes with predicted background-dependent fitness. (A) Enrichments across 16 functional neighborhoods defined by SAFE (37). Dot sizes represent ORs between the number of hits in a given neighborhood and the total number of hits detected, with the size of the neighborhood vs. the total number of genes in the dataset as background, using one-sided Fisher’s exact test. Global enrichment for class I (blue) and class II (orange) hits (Left) and strain-centric enrichments are presented (Right). Enrichments with a P < 0.05 are shown. Backgrounds highlighted by dashed lines correspond to example rewiring diagrams in C. (B) Predicted fitness for genes annotated in respiration/mitochondrial targeting, transcription and chromatin organization, and nuclear–cytoplasmic transport in the reference S288C. Genes in different processes are arranged by descending order of the modeled fitness. A detailed annotated version of this diagram can be found in SI Appendix, Fig. S5A. (C) Example rewiring diagrams in other backgrounds compared with the reference S288C. A switch from healthy to sick (loss of fitness) is indicated in blue and a switch from sick to healthy (gain of fitness) is indicated in orange for any given gene in a given background. The diagrams for all 38 isolates are shown in SI Appendix, Fig. S5B.
Compared with class II genes that impact multiple backgrounds, the class I genes are background-specific, rare, and tend to show little functional enrichment, as expected. However, in cases where multiple genes are detected in the same genetic background, some enrichments emerge (Fig. 5A and Dataset S4). For example, in the strain BDH, eight background-specific genes were detected with three annotated into one of the 16 functional neighborhoods, and two of which are involved in multivesicular body (MVB) sorting and pH-dependent signaling (RIM8 and RIM101). Both genes are nonessential in S288C but predicted as loss-of-fitness in the BDH background (Fig. 5A). In the strain AMF, 16 background-specific genes were detected with 11 annotated, among which 2 were involved in protein degradation and turnover (VID28 and PRE3) and 3 were involved in glycosylation and cell-wall biogenesis (OST1, OPI3, and FAB1). These observations demonstrate that genes with background-specific fitness variation, while rare, can be functionally related and may involve multiple members of the same protein complex or biological process.
Finally, as previously posited (6), genes exhibiting background-dependent fitness variation tend to show an intermediate level of connectivity in terms of genetic interactions (SI Appendix, Fig. S6A and Dataset S5) and an intermediate functional similarity between interacting gene pairs compared with genes that are consistently nonessential or essential (SI Appendix, Fig. S6B). Both class I and class II hits have the same pattern. Interestingly, background-specific genes display higher nonsynonymous-to-synonymous substitution rates (dN/dS) than essential genes (SI Appendix, Fig. S6C), indicating potential positive or relaxed purifying selection on these genes at the population level. Overall, genes with background-dependent fitness effects tend to be diverse yet can be functionally related within a single genetic background, and may display distinct evolutionary characteristics compared with genes that are consistently essential or nonessential across different backgrounds.
Discussion
To have a better insight into the background-dependent fitness variation associated with gene loss of function, we explored a large number of natural yeast isolates using a transposon saturation strategy. We modeled fitness by considering transposon insertion densities within gene coding sequence and surrounding regions. The comparison of the modeled fitness between different isolates and the reference S288C allowed the identification of 632 genes displaying background-dependent phenotypes. The majority of these cases (71,7%), denoted as class II, showed a gradient of fitness prediction across the population and is at least partly related to the environment. By contrast, background-specific cases, denoted as class I, tend to be rare. Within the same isolate background, both class I and class II hits tend to be functionally related.
The impact of the environment on the class II genes can be supported by two main observations. First, this set of genes was highly enriched for respiration and mitochondrial functions, which is consistent with a fitness loss under prolonged growth in media with galactose as the sole carbon source. Indeed, mitochondrial-related genes were also found to be background-dependent in a previous study involving four different isolates under conditions with nonfermentable carbon sources (25). Second, these genes showed a continuous fitness variation across the population, which is not directly correlated with the corresponding genetic diversity observed. We performed orthogonal functional validations in a subset of class II cases by gene deletion. The results showed that, while some predicted background-dependent fitness variation can be recapitulated in standard media, the effect of the environment, specifically galactose in this case, is unequivocal and may accentuate the observed growth phenotypes.
For all the class II cases, further analyses highlighted that genes involved in two biological processes, namely transcription and chromatin remodeling, as well as nuclear–cytoplasmic transport, are anticorrelated with genes involved in mitochondrial translation in terms of their predicted fitness profiles. These anticorrelations indicate a modular change in the relative fitness of genes involved in these processes, and may suggest coherent functional rewiring in a background-dependent manner. However, whether such rewiring is exclusively related to the growth conditions or could represent a general background-dependent effect remains difficult to disentangle due to the experimental settings required for transposon saturation analyses.
In a recent large-scale analysis of environment-dependent genetic interactions, it has been shown that most interactions specific to an environmental condition are in fact part of the global genetic interaction network that was exacerbated or attenuated in the tested condition (38). Compared with genetic interactions between pairs of gene deletion mutants, the background-dependent gene loss-of-function phenotype could be considered as an interaction between the loss-of-function gene and background-specific modifiers, which are expected to share general properties with deletion mutants. Indeed, we tested the gene deletion phenotype for six environment-related genes across 17 isolates and significant correlation was observed between the predicted fitness values (galactose media) and the observed fitness on standard rich media, YPD. Furthermore, depending on the genetic background, the effect of environment on the fitness variation of gene loss of function can be highly variable, suggesting that some of the observed effects could be well-conserved beyond a specific experimental condition.
Although the transposon saturation strategy can be used for various natural isolates , this method also presents some limitations. Among all the isolates initially tested, only about half showed a reasonable level of insertion efficiency, highlighting the unexpected variability of transposon activity between different individuals. This variability results in an underestimate of the number of genes associated with background-dependent phenotypes. In addition, loss-of-function phenotypes that are related to specific protein domains but not the entire ORF are difficult to identify, unless the insertion efficiency is extremely high. Furthermore, correlating the insertion density in and around a given gene to the actual loss-of-function fitness phenotype in a biological context will inevitably result in some biases no matter how sophisticated the mathematical model used, which again will likely lead to false negatives in the data. Finally, the Hermes system, like all currently available saturation systems, requires the step of a transposon induction in the presence of galactose (30). This competition effect in this specific carbon source may complicate downstream analysis, as the effects of environment vs. genetic background can be difficult to unravel. New strategies that take into account these factors are still needed in order to get a more precise view of background-dependent gene loss-of-function phenotypes at the species level.
Materials and Methods
Strains and Growth Conditions.
A total of 106 isolates were selected from the 1,011 S. cerevisiae collection (26). A prototrophic haploid strain, FY5, isogenic to the reference strain S288C, was also included. Haploid segregants derived from the 106 natural isolates were obtained after HO deletion and tetrad dissection (32, 33). Detailed descriptions of the strains can be found in Dataset S1. Strains were maintained at 30 °C using YPD (1% yeast extract; 2% peptone, 2% dextrose) in liquid culture or solid plates (2% agar). Transposon activity was induced in YPGal (1% yeast extract; 2% peptone, 2% galactose) with hygromycin B (200 µg/mL). Sporulation was induced on solid plates containing 1% potassium acetate and 2% agar.
Ploidy Control.
Ploidy was estimated by flow cytometry. Cells in exponential growth phase were washed in water and then 70% ethanol and sodium-citrate buffer (50 mM, pH 7.5), followed by RNase A treatment (500 µg/mL). To avoid cell aggregates, each sample was sonicated, and then the DNA was labeled with propidium iodide (16 µg/mL), a fluorescent intercalating agent. DNA content was then quantified using the 488-nm excitation laser of the Accuri C6 Plus flow cytometer (BD Biosciences).
Cell Transformation.
Cells in exponential growth phase were chemically transformed using the EZ-Yeast Transformation Kit (MP Biomedicals). We incubated cells for 30 min at 42 °C with EZ-Transformation solution, carrier DNA, and either 100 ng pSTHyg plasmid or 1 µg PCR fragment. After regeneration in YPD, cells were spread on solid YPD plates supplemented with hygromycin B and incubated at 30 °C until transformants appeared.
Construction of the pSTHyg Plasmid.
In order to be compatible with our isolates already carrying either a nourseothricin or kanamycin resistance cassette, the nourseothricin cassette of the pSG36 plasmid (29) was replaced by a hygromycin B resistance cassette. The pSG36 plasmid was amplified in two fragments by PCR excluding the natMX cassette, and then assembled with the hphMX cassette amplified from the p41 plasmid (Addgene, 58547) with overlapping regions using Gibson assembly. The new plasmid, pSTHyg, was amplified in Escherichia coli and extracted using the GeneJET Plasmid Miniprep Kit (Thermo Scientific). The construction was verified using enzymatic digestion with KpnI and PvuI.
Generation of Transposon Insertion Mutant Pools.
The protocol is adapted and modified from Gangadharan et al. (29). Each natural isolate was grown in liquid YPD medium and chemically transformed with 100 ng pSTHyg plasmid as described. From the selective transformation plates, a single clone was picked and grown in 30 mL YPD supplemented with hygromycin B under agitation at 30 °C until saturation (∼24 h). Cells were then diluted at an optical density (OD) of 0.05 in 50 mL of YPGal supplemented with hygromycin B to activate the transposase and induce the transposition for 72 h at 30 °C. Two successive dilutions were then performed for 24 h at an OD of 0.5 in 100 mL YPD and then YPD was supplemented with hygromycin B to enrich for cells with the transposon inserted in their genome. The final 100-mL culture was centrifuged and water-washed and 500-µL aliquots of cells were frozen at −20 °C.
Sequencing Library Preparation.
In order to sequence the genomic regions with a transposon insertion, the genomic DNA (gDNA) of the pool of cells carrying insertion events was extracted using the MasterPure Yeast DNA Purification Kit (Lucigen). Cells were lysed using a lysis solution supplemented with zymolyase 20T (1.5 mg/mL). Proteins and cellular debris were removed with MPC Protein Precipitation Reagent and several RNase A treatments were performed to eliminate RNA. gDNA was then precipitated with ethanol. The pellet was washed twice with 70% ethanol and resuspended in 80 µL water. The gDNA sample integrity was controlled on a 1% agarose gel and quantified by NanoDrop and Qubit using the Qubit dsDNA BR Assay Kit (Invitrogen). gDNA (2 × 2 µg) was digested in parallel with 50 U DpnII (NEB, R0543L) and NlaIII (NEB, R0125L) in 50 µL for 16 h at 37 °C. The enzymatic reactions were inactivated for 20 min at 65 °C and DNA fragments were ligated with 25 Weiss units of T4 ligase (Thermo Scientific, EL0011) in a total volume of 400 µL for 6 h at 22 °C. Circular DNA was then precipitated overnight at −20 °C with ethanol, salt (3 M NaOAc, pH 5.2), and glycogen. After a 70% ethanol wash, the DNA pellet was resuspended in 50 µL water. The junction between the genomic region and the transposon insertion site was amplified on both DpnII- and NlaIII-digested and recircularized gDNA by PCR using outward-facing primers targeting the transposon. The PCR products were controlled on a 1% agarose gel and displayed variable sizes centered around 750 bp. NanoDrop and Qubit using the Qubit dsDNA BR Assay Kit (Invitrogen) quantifications were then used to pool the same amount of NlaIII- and DpnII-digested PCR products. For each sample, at least 6 µg at a minimum of 30 ng/µL was then sent to the Beijing Genomics Institute for sequencing. In total, each sequencing run provided 1 Gb of 100-bp paired-end reads using Illumina HiSeq 4000 or DNBSeq technologies.
Determination of Transposon Insertion Sites.
The reads that contained the amplified part of the transposon were selected, the corresponding 57-bp sequence was trimmed with Cutadapt (39), and the reads corresponding to the plasmid were discarded. The cleaned reads were mapped specifically for each isolate by imputing the corresponding background-specific single-nucleotide polymorphisms (SNPs) into the reference genome (26) with BWA (40). The genomic position of an insertion site was defined as the first base pair aligned on the genome after the transposon region. For each insertion site, the number of reads and their orientation were obtained.
Modeling the Fitness Effect of Gene Loss of Function Based on Transposon Insertion Profiles.
The number of insertions in the promoter region (−100 bp to ATG), the beginning of the coding region (−100 bp to +100 bp from ATG), the coding region, and the end of the coding region (−100 bp to +100 bp from the stop codon) were normalized as insertion densities per 100 bp. Gene essentiality annotations were obtained from SGD (phenotype “inviable”) exclusively for annotations with gene deletion in the S288C background. Respiration-related gene annotations were obtained from SGD with the phenotype “respiration: absent” after gene deletion in S288C. Galactose-specific loss of fitness was determined as in Costanzo et al. (38), with a stringent cutoff of <−0.2. A logistic model was constructed using the glm() function from the R package “stats,” using insertion densities in the reference strain S288C, the promoter region (−100 bp to ATG), beginning of the coding region (−100 bp to +100 bp from ATG), the coding region, and the end of the coding region (−100 bp to +100 bp from the stop codon), raw insertion number in the coding region and gene sizes as predictors, and essentiality annotations as a binary classifier. Genes displaying a slow-growth phenotype (41), genes with differential fitness defects in galactose media (38), as well as genes showing respiration defects were excluded. Genes that are localized in regions with low insertion densities, namely fewer than 3 insertions in the terminator region (STOP to +300 bp) and fewer than 50 insertions in a 10-kb region surrounding the gene (−5 kb before ATG and +5 kb after STOP), were also excluded. A total of 4,600 genes were included in the model corresponding to 867 essential genes and 3,737 nonessential genes (Dataset S2). Tenfold cross-validation was performed using the R package “caret,” with trainControl() and train() functions, method = “glm,” and family = “binomial.” Cross-validation results showed an average accuracy of 0.88 with a Kappa of 0.57 and an F1 score of 0.73 (Dataset S2). The predictive value for nonessential labels is 0.91, contrasting a lower predictive value of 0.70 for essential labels, indicating a better accuracy in predicting nonessential genes using this model. This lower predictive power for essential genes is more or less expected, as the absence or low numbers of insertions could be linked to the overall low insertion density in certain genomic regions, which is independent of gene essentiality. Imputations for missing values were performed using the function impute.knn() in the R package “impute,” with k = 10, rowmax = 50%, and colmax = 80%. Quantile normalization of the imputed matrix was performed using the normalize.quantiles() function in the R package “preprocessCore.” All fitness prediction data can be found in Dataset S3.
Validation of the Phenotypic Consequence of BMH1 Gene Loss of Function.
Stable haploid isolates FY5 and CIB were diploidized using the pHS2 plasmid (Addgene, 81037) containing the HO gene encoding the endonuclease responsible for mating-type switching and a hygromycin resistance cassette. The BMH1 gene was replaced with a hygromycin B resistance cassette in the diploid isolates. Sporulation was induced on a potassium acetate medium in diploid isolates heterozygous for BMH1 gene deletion. Around 20 resulting tetrads were then dissected on YPD using an MSM 400 micromanipulator (Singer Instruments). Each spore grew for 48 h at 30 °C and the colony size was captured with the camera of the colony picker, PIXL (Singer Instruments). Colony size measurements were then analyzed using custom R scripts.
Validation Using Gene Deletion and Subsequent Phenotyping.
We developed plasmid constructs for direct homologous gene deletion in diploid isolates using CRISPR-Cas9 as described previously (24). A fragment carrying the natMX marker bordered by ∼200 bp of sequence homologous to the targeted genes was cloned onto a plasmid backbone containing spCas9 and a guide RNA, the URA3 marker, the yeast CEN6 sequence fused to an autonomous replication sequence, as well as an ampicillin resistance marker and an E. coli replication origin site from the standard pBluescript SK II (+). A total of 17 diploid isolates that are ura3Δ0 but otherwise isogenic to the haploid isolates used in the transposon mutagenesis were selected for functional validation using gene deletion. A total of six genes were selected, specifically the MRPL3 and ISM1 genes (mitochondrial translation), CAF130 and BMH1 (transcription regulation), and NUP188 and KAP123 genes (nuclear–cytoplasmic transport). For each gene, the deletion plasmid was constructed and transformed into all 17 isolates, and deletion mutants were selected as previously described (24). For each mutant, five tetrads were dissected and grown for 48 h on YPD at 30 °C. The plates were then imaged and the colony sizes for both the wild-type segregant and deletion mutants were determined. To test the growth phenotype across multiple media with different concentrations of glucose and galactose, the haploid deletion mutants and wild-type isolates were arrayed onto solid YPD square plates in a 384 format, with two intraplate replicates and two interplate replicates. The colonies were then pinned onto five different media with increased concentrations of galactose relative to glucose, namely YP 2% glucose (YPD), YP 1% glucose + 1% galactose, YP 0.5% glucose + 1.5% galactose, YP 0.1% glucose + 1.9% galactose, and 2% YP galactose. The plates were incubated at 30 °C and imaged at 16-, 24-, 40-, and 48-h time points. The colony sizes were then calculated. Detailed data can be found in Dataset S4.
Supplementary Material
Acknowledgments
We thank Agnès Michel for helpful suggestions throughout the project. This work was supported by the European Research Council (ERC Consolidator Grant 772505). It is also part of Interdisciplinary Thematic Institutes (ITI) Integrative Molecular and Cellular Biology (IMCBio), as part of the ITI 2021-to-2028 program of the University of Strasbourg, CNRS, and Inserm, supported by IdEx Unistra (ANR-10-IDEX-0002) and L’École Universitaire de Recherche (EUR) (IMCBio ANR-18-EUR-0016) under the framework of the French Investments for the Future Program. J.S. is a Member of the Institut Universitaire de France.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. D.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2204206119/-/DCSupplemental.
Data, Materials, and Software Availability
All sequencing data related to this study have been deposited in the European Nucleotide Archive (accession no. PRJEB45777) (42).
References
- 1.Cooper D. N., Krawczak M., Polychronakos C., Tyler-Smith C., Kehrer-Sawatzki H., Where genotype is not predictive of phenotype: Towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum. Genet. 132, 1077–1130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandler C. H., Chari S., Tack D., Dworkin I., Causes and consequences of genetic background effects illuminated by integrative genomic analysis. Genetics 196, 1321–1336 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sackton T. B., Hartl D. L., Genotypic context and epistasis in individuals and populations. Cell 166, 279–287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chow C. Y., Bringing genetic background into focus. Nat. Rev. Genet. 17, 63–64 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Mullis M. N., Matsui T., Schell R., Foree R., Ehrenreich I. M., The complex underpinnings of genetic background effects. Nat. Commun. 9, 3548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou J., van Leeuwen J., Andrews B. J., Boone C., Genetic network complexity shapes background-dependent phenotypic expression. Trends Genet. 34, 578–586 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen R., et al. , Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat. Biotechnol. 34, 531–538 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Fournier T., Schacherer J., Genetic backgrounds and hidden trait complexity in natural populations. Curr. Opin. Genet. Dev. 47, 48–53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow C. Y., Kelsey K. J. P., Wolfner M. F., Clark A. G., Candidate genetic modifiers of retinitis pigmentosa identified by exploiting natural variation in Drosophila. Hum. Mol. Genet. 25, 651–659 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinberg M. H., Sebastiani P., Genetic modifiers of sickle cell disease. Am. J. Hematol. 87, 795–803 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorfman R., Modifier gene studies to identify new therapeutic targets in cystic fibrosis. Curr. Pharm. Des. 18, 674–682 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Cutting G. R., Modifier genes in Mendelian disorders: The example of cystic fibrosis. Ann. N. Y. Acad. Sci. 1214, 57–69 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams R. A., Mamotte C. D. S., Burnett J. R., Phenylketonuria: An inborn error of phenylalanine metabolism. Clin. Biochem. Rev. 29, 31–41 (2008). [PMC free article] [PubMed] [Google Scholar]
- 14.Dowell R. D., et al. , Genotype to phenotype: A complex problem. Science 328, 469 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D.-U., et al. , Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 28, 617–623 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blomen V. A., et al. , Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Boutros M., et al. ; Heidelberg Fly Array Consortium, Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303, 832–835 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Hart T., et al. , High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Vu V., et al. , Natural variation in gene expression modulates the severity of mutant phenotypes. Cell 162, 391–402 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Wang T., et al. , Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamath R. S., et al. , Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Paaby A. B., et al. , Wild worm embryogenesis harbors ubiquitous polygenic modifier variation. eLife 4, e09178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards M. D., Symbor-Nagrabska A., Dollard L., Gifford D. K., Fink G. R., Interactions between chromosomal and nonchromosomal elements reveal missing heritability. Proc. Natl. Acad. Sci. U.S.A. 111, 7719–7722 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou J., Tan G., Fink G. R., Andrews B. J., Boone C., Complex modifier landscape underlying genetic background effects. Proc. Natl. Acad. Sci. U.S.A. 116, 5045–5054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galardini M., et al. , The impact of the genetic background on gene deletion phenotypes in Saccharomyces cerevisiae. Mol. Syst. Biol. 15, e8831 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peter J., et al. , Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadhu M. J., et al. , Highly parallel genome variant engineering with CRISPR-Cas9. Nat. Genet. 50, 510–514 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michel A. H., et al. , Functional mapping of yeast genomes by saturated transposition. eLife 6, e23570 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gangadharan S., Mularoni L., Fain-Thornton J., Wheelan S. J., Craig N. L., DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo. Proc. Natl. Acad. Sci. U.S.A. 107, 21966–21972 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Opijnen T., Levin H. L., Transposon insertion sequencing, a global measure of gene function. Annu. Rev. Genet. 54, 337–365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharon E., et al. , Functional genetic variants revealed by massively parallel precise genome editing. Cell 175, 544–557.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou J., et al. , The hidden complexity of Mendelian traits across natural yeast populations. Cell Rep. 16, 1106–1114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fournier T., et al. , Extensive impact of low-frequency variants on the phenotypic landscape at population-scale. eLife 8, e49258 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harari Y., Ram Y., Rappoport N., Hadany L., Kupiec M., Spontaneous changes in ploidy are common in yeast. Curr. Biol. 28, 825–835.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Venkataram S., et al. , Development of a comprehensive genotype-to-fitness map of adaptation-driving mutations in yeast. Cell 166, 1585–1596.e22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson M. S., et al. , Phenotypic and molecular evolution across 10,000 generations in laboratory budding yeast populations. eLife 10, e63910 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costanzo M., et al. , A global genetic interaction network maps a wiring diagram of cellular function. Science 353, aaf1420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Costanzo M., et al. , Environmental robustness of the global yeast genetic interaction network. Science 372, eabf8424 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10 (2011). [Google Scholar]
- 40.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giaever G., et al. , Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Caudal E., et al. , Loss-of-function mutation survey revealed that genes with background-dependent fitness are rare and functionally related in yeast. European Nucleotide Archive. https://www.ebi.ac.uk/ena/browser/view/PRJEB45777?show=reads. Deposited 30 October 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data related to this study have been deposited in the European Nucleotide Archive (accession no. PRJEB45777) (42).