Abstract
Type 2 diabetes mellitus, a metabolic disorder characterized by abnormally elevated blood sugar, poses a growing social, economic, and medical burden worldwide. The skeletal muscle is the largest metabolic organ responsible for glucose homeostasis in the body, and its inability to properly uptake sugar often precedes type 2 diabetes. Although exercise is known to have preventative and therapeutic effects on type 2 diabetes, the underlying mechanism of these beneficial effects is largely unknown. Animal studies have been conducted to better understand the pathophysiology of type 2 diabetes and the positive effects of exercise on type 2 diabetes. However, the complexity of in vivo systems and the inability of animal models to fully capture human type 2 diabetes genetics and pathophysiology are two major limitations in these animal studies. Fortunately, in vitro models capable of recapitulating human genetics and physiology provide promising avenues to overcome these obstacles. This review summarizes current in vitro type 2 diabetes models with focuses on the skeletal muscle, interorgan crosstalk, and exercise. We discuss diabetes, its pathophysiology, common in vitro type 2 diabetes skeletal muscle models, interorgan crosstalk type 2 diabetes models, exercise benefits on type 2 diabetes, and in vitro type 2 diabetes models with exercise.
I. INTRODUCTION
Diabetes mellitus refers to a group of metabolic diseases that are characterized by elevated levels of blood glucose known as hyperglycemia. More than 400 × 106 people worldwide have diabetes.1 The U.S. accounts for approximately 34 × 106 (about 1 in 10) and roughly 90% of them are type 2 diabetes (T2D).1 Type 1 diabetes occurs when the immune system of the body attacks and destroys β-cells, the insulin-producing cells in the pancreas. This results in the buildup of sugar level in the bloodstream due to little to no insulin, which promotes cell glucose uptake. T2D occurs when the pancreatic β-cells do not produce enough insulin and the body responds poorly to insulin, known as insulin resistance, and takes in less sugar.1 Diverse genetic and environmental factors lead to the loss of pancreatic β-cell mass and function.2 When β-cells are unable to fully compensate for decreased insulin sensitivity, T2D results.3
Skeletal muscle is the largest metabolic organ and the most prominent site for the disposal of ingested glucose in healthy individuals.4 Hence, the well-being of the skeletal muscle is essential for glucose homeostasis in the body. In insulin-resistant states, insulin-stimulated glucose uptake and related insulin signaling in the skeletal muscle are significantly impaired.4 Typically, insulin resistance precedes T2D and the earliest site of insulin resistance is the skeletal muscle.2 In addition, many factors secreted by the skeletal muscle, including proteins and peptides (myokines), metabolites, microRNAs (miRNAs), and exosomes, mediate the interaction between the skeletal muscle and other organs such as the pancreas, adipose tissues, and the liver in the development of T2D. Thus, a deeper understanding of the pathophysiological molecular mechanisms underlying skeletal muscle in the progression of T2D is crucial in finding better treatments for the patients.
Moreover, the skeletal muscle contributes to numerous preventative and therapeutic effects of exercise on diabetes. The studies found that exercise increases glucose uptake in the skeletal muscle.5–8 In addition, regular exercise yields reduced basal and glucose-stimulated insulin levels, improved mitochondrial function, and increased muscle mass, insulin activity, and free fatty acid oxidation.5,9–11 In the recent decades, studies also found that contracting the skeletal muscles secrete protein factors or myokines that may be anti-inflammatory and have the potential to be beneficial to diabetes where chronic inflammation is a major underlying pathophysiology.
Animal studies have been used to study the role of the skeletal muscle tissues in the pathophysiology of T2D and the positive effects of muscle contraction on T2D. In vivo T2D rodent models, including monogenic and polygenic obese models, diet-induced obese models, and transgenic nonobese models, are widely used to study the onset and progression of T2D and exercise benefits in diabetes.12 Each model is useful in investigating the genetic, environmental, or endocrine factors present in the evolution of T2D. However, in vivo systems are highly complex, making the precise mechanistic understanding difficult, in addition to the ethical considerations of human or animal experimentations and their tremendous costs. No single model captures all human T2D phenotypes and genetics, and it is challenging to interpret and extrapolate the results from animal models to humans.13
The advent of in vitro models using patient-derived T2D muscle cells or induced pluripotent stem cells (iPSCs) has enabled researchers to recapitulate T2D in a dish with human genetics and disease phenotypes.14–20 In particular, iPSCs are advantageous because they could serve as unlimited cell source and are minimally invasive to obtain. These in vitro models of T2D are important avenues to better understand the cellular and molecular processes involved in the disease. They allow researchers to study the precise and various experimental conditions without the full complexity of in vivo physiology. In addition, genetic modification can easily be leveraged to investigate the functional roles of genes in diseases. Moreover, in vitro models have shown to capture the effects of secreted factors that directly link muscle, liver, and adipose tissues and dissect casual T2D-related interorgan connections. In addition, the advent of more sophisticated in vitro T2D and various in vitro exercise protocols have enabled detailed mechanistic studies on the benefits of exercise on the disease.
In this review, we discuss diabetes mellitus, its known pathophysiology, and the various in vitro models used to model T2D, all with a focus on the skeletal muscle, which is the primary site of glucose disposal. We then talk about interorgan crosstalk in T2D development as well as the exercise benefits on alleviating diabetes mellitus. Finally, we discuss existing in vitro T2D skeletal muscle models capturing both interorgan crosstalk and exercise.
II. PATHOPHYSIOLOGICAL FACTORS IN T2D SKELETAL MUSCLES
Below we will focus on the biological mechanisms underlying T2D-related impairment of the skeletal muscle (Fig. 1), since T2D accounts for 90% of diabetic occurrence and the skeletal muscle is the largest tissue responsible for glucose homeostasis in the body.
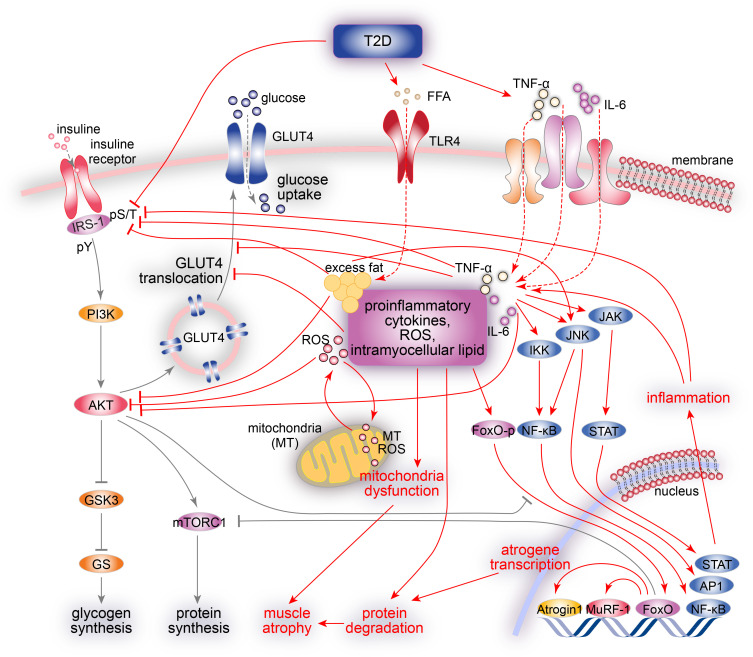
FIG. 1.
A summary of T2D pathophysiology in the skeletal muscle. In the healthy skeletal muscle (black arrows), insulin stimulates intracellular glucose metabolism by tyrosine phosphorylation (pY) of insulin receptors and IRSs. IRS proteins activate the PI3K/AKT pathway and promote many insulin actions via the activation of AKT by (1) increasing glucose influx via the translocation of GLUT4, (2) promoting protein synthesis via mTORC1 activation, (3) preventing the transcription of FoxO-dependent atrogene via FoxO phosphorylation, and (4) promoting glycogen synthesis via GSK3 inactivation. In the T2D skeletal muscle (red arrows), the inhibitory Ser/Thr phosphorylation (pS/T) of IRS-1 impairs its tyrosine phosphorylation (pY), resulting in the development of insulin resistance by impairing PI3K/AKT signaling, decreasing GLUT4 translocation and glucose transport, and impairing glucose phosphorylation and synthesis. In addition, altered phosphorylation of IRS-1 and reduced PI3K activity exacerbates muscle atrophy via the inhibition of mTORC1 signaling and activation of FoxO-dependent atrogene transcription (Atrogin1 and MuRF1). Increased ROSs, intramyocellular lipids, and proinflammatory cytokines (TNF-α and IL-6) lead to mitochondria dysfunction, lipotoxicity, and chronic inflammation via the activation of JNK, IKK/NF-κB, and JAK–STAT stress kinase pathways, resulting in Ser/Thr phosphorylation (pS/T) of insulin receptors and IRS proteins, further contributing to insulin resistance.
A. Glucose and insulin dysregulation
The key phenotype of diabetic patients is the abnormal elevations of glucose and insulin concentrations. The glucose level of healthy individuals is typically maintained at a steady state. The level rises post-ingestion but would return to pre-prandial concentrations within 2 h.21,22 In T2D patients, however, the glucose concentration is at higher levels both before and after ingestion. The impaired glucose homeostasis prompts the β-cells in the pancreas to produce excess insulin to enhance cellular glucose uptake. When the body becomes less sensitive to insulin, also known as insulin-resistance, both elevated glucose and insulin concentrations persist. Normally, insulin level rises to a peak within 30 min of ingestion in the body. In T2D patients, however, insulin secretion is delayed, and peak insulin level does not occur until 2 h after ingestion.21,22 The insulin concentration stays abnormally elevated for the following several hours.
Insulin resistance in the skeletal muscle is considered the most important extra-pancreatic factor in the T2D pathophysiology.23 In the healthy skeletal muscle, insulin stimulates intracellular glucose metabolism by activating the insulin receptor and subsequently inducing second messengers that activate a cascade of phosphorylation–dephosphorylation reactions4 (Fig. 1). When the insulin receptors are activated by insulin binding, the receptors recruit receptor substrates, including insulin-receptor substrate-1 (IRS-1), and subsequently phosphorylate IRS-1 on multiple tyrosine residues. The tyrosine-phosphorylated IRS-1 recruits and activates phosphatidylinositol 3-kinase (PI3K), further phosphorylating and activating protein kinase B (PKB)/AKT (Ak strain transforming). AKT is a central intermediate for many insulin actions (Fig. 1). First, the activation of AKT leads to increased glucose influx into muscle cells via the activation of the glucose transport system (glucose transporter 4, GLUT4) and the translocation of GLUT4 to the plasma membrane. Second, AKT promotes protein synthesis via the activation of mammalian target of rapamycin complex 1 (mTORC1) pathway. Third, AKT phosphorylates Forkhead box O (FoxO), which leads to the exclusion of FoxO from the nucleus, and thus, prevents the transcription of FoxO-dependent atrogene. Fourth, AKT also inactivates glycogen synthase kinase 3 (GSK3) and activates glycogen synthase (GS), inducing glycogen synthesis. Other AKT involvements in the insulin signaling pathway are reviewed in detail in the literature.4,24 This PI3K/AKT pathway is responsible for most metabolic actions of insulin, but mitogen-activated protein kinase (MAPK) is another insulin signaling pathway that involves insulin receptors and IRS-1. MAPK activates the cascade of extracellular signal-regulated kinases (ERK) and promotes cell growth and differentiation.4
The role of insulin to promote glucose uptake is drastically reduced in the T2D skeletal muscle.4,24 Importantly, both lean and obese T2D individuals show a significant decrease in insulin-stimulated glucose uptake in their skeletal muscle.25 In the T2D skeletal muscle, tyrosine phosphorylation of IRS-1 is severely damaged, which contributes to lowered insulin sensitivity. Tyrosine phosphorylation of IRS-1 induces the insulin signaling pathway while serine (Ser) and threonine (Thr) phosphorylation of IRS-1 inhibit the insulin signaling pathway.24 An increase in Ser/Thr phosphorylation impairs the tyrosine phosphorylation of IRS-1, which further reduces the association of the p85 regulatory subunit of PI3K activity with IRS-1 and decreases PI3K activation. As a result, this impaired IRS-1 and PI3K signaling leads to the development of insulin resistance in the skeletal muscle by (1) decreasing glucose transport via impaired translocation and decreased activity of glucose transporters (e.g., glucose transporter 4, GLUT4) and (2) diminishing glucose phosphorylation, and (3) impairing glycogen synthesis activity4 (Fig. 1).
B. Muscle atrophy and fiber type redistribution
The muscle atrophy seen in diabetes is an imbalance of protein synthesis and protein degradation, as in all muscle atrophy. Reduced insulin signaling leads to lower protein synthesis signaling, such as AKT signaling, higher protein degradation activities, and higher ubiquitin-proteasome pathways.26 Altered phosphorylation of IRS-1 and reduced PI3K activity could exacerbate muscle atrophy by reducing the activation of AKT. The reduced AKT activation could enhance protein degradation via FoxO-dependent atrogene transcription, while simultaneously decreasing protein synthesis via inhibition of the downstream mTORC1 signaling.27 Reduction of the AKT activity in muscle promotes protein degradation by directly increasing muscle atrophy F-box (MAFbx) and Muscle RING finger 1 (MuRF1), the representative E3 ligases of muscle atrophy28 (Fig. 1).
In T2D patients, the fraction of slow muscle fiber and the fiber's expression of GLUT4, the most abundant glucose transporter in the skeletal muscle, were found to be lower compared with either obese or healthy subjects.29,30 Slow-twitch fibers are more insulin-sensitive and responsive compared with fast-twitch fibers.31 Thus, such changes may contribute to the reduction in insulin-stimulated glucose uptake in the skeletal muscle of T2D, especially since GLUT4 expression is normally higher in slow fibers compared with fast fibers.32 To make matter worse, muscle atrophy further worsens insulin resistance, resulting in a vicious feedback loop in the skeletal muscle sugar uptake.
C. Excessive fat supply and impaired fat oxidation
Body weight is highly related to diabetes, and several studies show that obesity increases the risk of diabetes.33–36 Obese individuals without a family history of T2D have a 35%–50% decrease in whole body insulin-mediated glucose uptake.3,37 In these patients, there are higher amounts of non-esterified fatty acids, which contribute to insulin resistance and T2D.38,39 Independent of excessive fat supply to the skeletal muscle, T2D patients also show impaired muscle fat oxidation,40–43 which indicates an impaired ability of mitochondria to oxidize fat.
The increases in intramyocellular lipids and other lipotoxic metabolites (e.g., ceramide) play a causal role in the development of the skeletal muscle insulin resistance.4 An increasing level of free fatty acids (FFAs) in the body results in the accumulation of intramyocellular lipids, such as diacylglycerol and ceramides. Lipotoxicity caused by the accumulation of lipid intermediates can inhibit insulin signaling via the reduction in GLUT4 transporters on the myocyte membrane,44–46 leading to impaired glucose utilization, reduced lipid oxidation, and decreased muscle function27,47–51 (Fig. 1). Ceramide is found to inhibit AKT signaling, which plays an important role in glucose homeostasis and protein synthesis. AKT signaling inhibition due to excess lipids leads to decreased glucose transport and glycogen synthesis, contributing to abnormally elevated blood glucose.27 In addition, ceramides induce insulin resistance via c-Jun N-terminal Kinase (JNK) activation.52 Palmitate, a fatty acid, also causes insulin resistance in the skeletal muscle by promoting endoplasmic reticulum stress, cytokine production, and JNK activation.53
D. Chronic inflammation
Chronic inflammation is not only an important contributor to T2D but also accelerates and exacerbates T2D. The skeletal muscle in the proinflammatory state is common in diabetic patients. The levels of cytokines, including Tumor Necrosis Factor-α (TNF-α) and interleukin 6 (IL-6), are upregulated in the skeletal muscle.54,55 TNF-α activates nuclear factor kappa B (NF-κB) and increases protein degradation via MuRF-1-dependent signal pathways. Also, TNF-α induces insulin resistance by stimulating JNK and inhibitor of nuclear factor kappa-B kinase subunit beta (IKKβ)/NF-κB pathways and increasing serine/threonine phosphorylation of IRS-156 (Fig. 1). IL-6 promotes protein degradation and insulin resistance by reducing the expression of both GLUT4 and IRS-1 and activating the Janus kinase/signal transducer and activator of transcription proteins (JAK/STAT)54,55 (Fig. 1). Specific factors and signaling pathways are often correlated with each other. For example, the IKKβ/NF-κB activation can induce the secretion of proinflammatory cytokines, which leads to further stimulation of IKKβ/NF-κB signaling.57 Moreover, both NF-κB and JNK were found to induce phosphorylation of insulin receptor substrate, leading to impaired downstream insulin signaling. Studies in humans and mice showed that the impairment in insulin-stimulated muscle glucose transport is associated with disruptions in the translocation of GLUT4, the most abundant glucose transporter isoform in the skeletal muscle, to the muscle cell surface58–62 (Fig. 1).
The interplay between inflammation and fat infiltration causes glucose dysregulation and impaired insulin signaling, leading to T2D. Excess fat and reactive oxygen species (ROS) play a role in activating JNK and NF-κB, which are important protein and transcription factors involved in cellular stress and inflammation.63 Abnormally elevated free fatty acids (FFAs) induce adipocytes to produce proinflammatory cytokines, such as TNF-α, IL-1β, and interferon-gamma (IFN-γ), which contribute to both local and systemic insulin resistance observed in the skeletal muscle.60–62,64–66
E. Mitochondria dysfunction
The mitochondrial dysfunction is associated with T2D development.1 In the skeletal muscles of insulin-resistant individuals, mitochondrial dysfunction has also been observed. The accumulation of ROS in the mitochondria is one of the proposed mechanisms linking mitochondrial dysfunction to insulin resistance.67 The genes involved in oxidative metabolism are down-regulated in T2D tissues: peroxisome proliferator-activated receptor-gamma coactivator (PGC-1α), an important gene involved in mitochondrial biogenesis, was also found to be downregulated in subjects with T2D.67–69 The phosphocreatine re-synthesis rate is also diminished.70 Elevated electron donors stemming from excess nutrient and catabolism increase electron supply to the mitochondrial electron transport chain, which results in excess ROS production, inducing cellular damage, insulin resistance, and mitochondrial damage58,59 (Fig. 1).
Obesity is associated with mitochondria dysfunction in T2D tissues. The oxidative capacity of mitochondria was found to decrease in obese individuals compared with lean subjects, leading to reductions in lipid metabolism and mitochondrial respiration.40,71,72 During the insulin resistance state, mitochondrial fat oxidation is decreased and the influx of free fatty acid into the skeletal muscle is enhanced.4 Lipotoxicity caused by the accumulation of lipid intermediates leads to excessive mitochondrial fission via the activation of a mitochondrial fission protein, dynamin-related protein 1 (DRP1) activation.73 Both the development of ROS and excessive lipid accumulation result in mitochondrial damage and further enable the removal of damaged mitochondria through mitophagy (selective mitochondrial autophagy).74
F. Altered myokine secretion
Contracting the skeletal muscle secretes protein factors, named myokines, which can exert their effects in an autocrine, paracrine, or endocrine fashion. Though very little is known about the functions and the mechanistic actions of these myokines, many of them play a positive role in energy metabolism.75 However, some of the myokine secretions are altered in diseased states, such as in T2D. For example, plasma FGF21 was found to be increased in T2D patients compared with normal controls.76 FGF21 enhances the skeletal muscle glucose uptake and may be a compensatory mechanism to elevated blood glucose in T2D individuals. Another study showed that conditioned media from diabetic myotubes treated with high glucose, insulin, and lipids significantly reduced insulin-stimulated insulin secretion in β-cells, suggesting altered myokines in diabetic and metabolically challenged skeletal muscle.77 Other studies have shown that insulin-resistant or diabetic myotubes secreted elevated levels of IL-6, IL-8, IL-15, and TNF-α.78,79 IL-8 was further investigated and found to be involved in the reduced capillarization observed in the skeletal muscle of T2D patients.80 A deeper understanding of the myokines dysregulated in T2D has the potential to help discover more effective therapeutics.
III. IN VITRO T2D SKELETAL MUSCLE MODEL
A. Animal models and the motivation for in vitro models
Different animal models have been developed to reproduce key characteristics of T2D such as insulin resistance and β-cell dysfunction. However, there is no single animal model that can encompass all of the clinical complications and pathophysiological mechanisms in human T2D. Thus, the choice of the model should depend on what aspect of the disease is being investigated. The details of currently available T2D animal models were reviewed in the literature.12,81–84 Below we provide a short summary of these models.
1. Rodent T2D models
The rodent model has proven to be a reliable model to study the genetic or environmental factors contributing to T2D. Rodent models have the obvious advantages such as lower cost and feasibility of conducting longitudinal studies compared to large animal models. T2D rodent models can be categorized into two groups: obese and nonobese models. The obese models can be generated by (1) naturally occurring mutations or genetic manipulation (monogenic and polygenic T2D models) and (2) high fat diet (diet-induced obese T2D model).
2. Monogenic obese T2D models
Monogenic models of T2D with obesity are commonly used in the research. In these models, the obesity induced T2D is caused by monogenic mutation in leptin or leptin receptor, rendering leptin signaling dysfunctional. Leptin induces satiety, and thus, the lack of functional leptin causes hyperphagia and obesity followed by hyperlipidemia, hyperinsulinemia, and hyperglycemia.12
3. Polygenic obese T2D models
Polygenic T2D models provide a more accurate model to mirror the complicated human T2D conditions such as the interplay of obesity and glucose homeostasis and diabetic complications. Among polygenic models, KK mice,12,85 and NoncNZO10/LtJ mice82 models demonstrated insulin resistance in skeletal muscles. However, there are no wild-type controls.
4. Diet-induced obese T2D model
Models of high fat feeding (58% of energy derived from fat) have developed to model the diet-induced metabolic changes in the development of T2D. Since the obesity is caused by environmental factors rather than single genetic mutation, diet-induced obesity model better captures insulin resistance in skeletal muscles caused by increased circulating concentrations of free fatty acids.86 However, both genetic and high-fat feeding-induced mouse models do not capture the same islet pathology as humans. In particular, diabetes in the mouse models was caused by a failure to adequately increase the β-cell mass, which is one of the secondary acquired metabolic abnormalities in humans.81
5. Nonobese T2D models
Nonobese T2D models have been developed such as Goto–Kakizaki rats and hIAPP mice, since not all T2D patients are obese. Goto–Kakizaki model can mimic glucose intolerance and defective glucose-induced insulin secretion, but this abnormal glucose metabolism is caused by aberrant β-cell mass. Transgenic mice (hIAPP mice) expressing human islet amyloid polypeptide (hIAPP) have been created to closely mimic the pathogenic effects of hIAPP on β-cell destruction in humans.
6. Large animal T2D models
Larger animal T2D models, such as cats, dogs, pigs, and non-human primates, have been created for T2D research. In particular, cat T2D models were found to closely mimic insulin resistance and subsequent β-cell loss and islet amyloidosis in humans. Among all the animal models, a main challenge remains—the animals are not able to fully recapitulate the human genetics and disease phenotypes with varying degrees of insulin resistance and β-cell destruction.
In vitro models of T2D are important avenues to a better understanding of the cellular and molecular processes involved in the disease. They not only allow researchers to capture human genetics and pathophysiology but also enable the study of precise and various experimental conditions without the full complexity of in vivo physiology. In addition, genetic modification can easily be leveraged to investigate the functional roles of genes in diseases in controlled and simplified environments. Several strategies have been utilized to study the underlying pathophysiology of T2D in vitro. These methods include exposing cultured myotubes to elevated systemic concentrations of glucose, insulin, fatty acids, and inflammatory cytokines to mimic the pathophysiology of diabetes. Recent advances in T2D in vitro modeling include growing myotubes derived from diabetic individuals using both muscle stem cells and induced pluripotent stem cells. In the following sections, we discuss each of these methods in further details (Tables I and II).
TABLE I.
A summary of in vitro T2D models.
| Cell source | Method | Concentration | Impact on myoblasts | Impact on myotubes | References |
|---|---|---|---|---|---|
| C2C12 | Elevated glucose | 60 mM | ↓Myogenesis, ↓insulin sensitivity, ↓AKT signaling | 87 | |
| C2C12 | Elevated glucose | 10–25 mM | ↓Myogenesis, ↓insulin sensitivity, ↑mitochondrial fragmentation, ↑ROS | ↑ROS, ↑protein degradation, ↑apoptosis | 88 |
| C2C12 | Elevated insulin | 50 nM | ↓Insulin sensitivity, ↓AKT signaling, ↓mRNAs of GLUT4 and PGC1-α | 91, 92 | |
| C2C12 | Elevated glucose + insulin | Glucose, 15 mM Insulin, 50 nM | ↑Myogenesis | 91, 92 | |
| Human myotubes | Elevated glucose + insulin | High glucoseInsulin, 100 nM | ↓Insulin sensitivity, ↓AKT signaling | 95 | |
| C2C12 | Hyperlipidemia | Palmitate, 0.25 mM for 2 h | ↑Apoptotic signaling, ↑mitochondrial fragmentation | 89 | |
| C2C12 | Hyperlipidemia | Palmitate, 0.1–0.6 mM for 24 h | ↓Insulin sensitivity, ↓AKT signaling | 101, 102 | |
| C2C12 | Hyperlipidemia | Palmitate, 0.5 mM for 24 h | ↓Myokine irisin | 105 | |
| L6 | Hyperlipidemia | 0.75 mM for 2–16 h | ↓GLUT4 expression, ↑NF-κB, ↑TNF-α | 103, 104 | |
| Human myotubes | Hyperlipidemia | Palmitate, 0.5 mM for 24 h | ↑Mitochondrial fragmentation, ↑IL-6 | 106 | |
| Human myotubes | Hyperlipidemia + inflammation | Palmitate, 0.5 mM for 48 h TNF-α, 10 ng/ml for 24 h | Dysregulated enhancers | 107 | |
| C2C12 | Inflammation | TNF-α, 10 ng/ml for 1 h | ↓Insulin sensitivity | 108 | |
| C2C12 | Inflammation | TNF-α, 10 ng/ml for 16 h | ↓AMPK signaling | 66 | |
| T2D subjects | ↓Myogenesis, ↑IL-1β, dysregulated autophagy, dysregulated miRNAs | ↑IL-1β, NF-κB, IL-6/8/15, TNF-α dysregulated autophagy, ↓glycogen synthesis, ↓glucose oxidation, ↓insulin sensitivity, ↑ceramide and glycosphingolipids metabolism, ↓lipolysis and hormone-sensitive lipase | 14–17, 79, 112, 117–119 | ||
| iPSCs from T2D subjects | ↓Insulin sensitivity, ↓mitochondrial oxidation, | Dysregulated phosphoproteome | 18–20 |
TABLE II.
A comparison of in vitro T2D models.
| Method | Effect | Advantage |
|---|---|---|
| High glucose and insulin treatment | • Impaired myogenesis | Ability to capture the T2D mechanisms underlying glucose and insulin dysregulation in the skeletal muscle |
| • Impaired glucose metabolism and insulin signaling | ||
| • Increased protein degradation and ubiquitin-proteasome stimulation | ||
| • Increased ROS and apoptosis | ||
| High lipids and proinflammatory cytokines treatment | • Impaired glucose metabolism and insulin signaling | Ability to capture the T2D mechanisms underlying high fat infiltration and inflammation |
| • Increased apoptosis | ||
| • Impaired mitochondrial metabolism | ||
| Incorporation of T2D subject-derived myoblasts and myotubes | • Altered gene expression of myogenic transcription factors | Ability to recapitulate human-specific genetic and metabolic phenotypes |
| • Impaired myogenesis | ||
| • Downregulated microRNAs | ||
| • Elevated expression of inflammation genes | ||
| • Dysregulated expression of autophagy genes | ||
| • Impaired glucose metabolism and insulin signaling | ||
| • Increased lipid accumulation | ||
| Incorporation of T2D-induced pluripotent stem cells | • Impaired glucose metabolism and insulin signaling | Patient-specific T2D modeling |
| • Dysregulated phosphoproteome | ||
| • Impaired mitochondrial metabolism |
B. High glucose and insulin treatment
To better understand how abnormal elevations of glucose and insulin contribute to cell and tissue dysfunction, researchers have exposed cultured myotubes to elevated glucose and/or insulin. Below we summarize the findings of these studies on how elevated glucose and/or insulin affect myoblasts and myotubes.
C2C12 myoblasts differentiated in high glucose or insulin media showed impaired myogenesis and metabolism. The myoblasts had a significantly lower ability to form myotubes when they are differentiated in medium containing 60 mM glucose compared with differentiation in medium containing 25 mM glucose.87 The high glucose also reduced both basal and insulin-stimulated GLUT4 expressions and glucose uptakes. The impaired myogenesis and glucose metabolism were shown to be related to reductions in the expressions of myogenesis-related genes, namely, myoD and myogenin, and decreased AKT signaling, which is important for glucose uptake and metabolism.87 A much older study showed that even a concentration of 25 mM glucose in culture medium desensitized the insulin-stimulated glucose uptake in C2C12 myoblasts compared with a concentration of 5.5 mM, which is around the normal physiologic glucose level in human and mice at a fasting state.88 C2C12 myoblasts cultured in 25 mM glucose also showed higher mitochondrial fragmentation and elevated ROS compared with myoblasts cultured in 5.6 mM glucose.89,90 Furthermore, C2C12 myoblasts differentiated in 15 mM glucose medium displayed reduction in myogenesis seen in lowered fusion and expressions of myoD, myogenin, and myosin heavy chain compared with myoblasts cultured in basal medium.91 Interestingly, C2C12 myoblasts cultured in both high glucose (15 mM) and insulin (50 nM) showed increased fusion, myogenin, and a drop in myostatin compared with myoblasts cultured in 15 mM glucose only or basal medium.91,92 However, C2C12 myoblasts differentiated in the presence of elevated insulin was found to have reduced AKT signaling, lowered insulin sensitivity, and decreased mRNA expressions of genes related to glucose uptake and mitochondrial biogenesis, such as GLUT4 and PGC1-α.93
In addition to myoblasts, C2C12 myotubes in media supplemented with 10 or 25 mM glucose displayed increased protein degradation, ubiquitin-proteasome stimulation, ROS, and apoptosis.94 Human myotubes showed impaired metabolism and insulin signaling when exposed to high insulin levels. Human myotubes cultured in high glucose medium with 3 days of 100 nM insulin exposure showed impaired insulin-stimulated glucose uptake and blunted insulin (IRS-1) and AKT signaling.95
Exposing murine myoblasts or myotubes to high glucose or insulin in vitro has deepened our understanding of the mechanisms underlying glucose and insulin dysregulation in the skeletal muscle. This allows us to parse through the deleterious effects of abnormally elevated glucose and/or insulin in the skeletal muscle from the effects of other T2D pathogenic mechanisms, such as fat infiltration and inflammation. Below we discuss papers focusing on elucidating the effect of surplus lipids and inflammation in the skeletal muscle.
C. High lipids and proinflammatory cytokines treatment
The plasma free fatty acid levels are unusually elevated in most diabetic patients and high intramyocellular lipid is associated with insulin resistance.96 Fat infusion in healthy humans revealed that elevated fat reduced glucose uptake and glycogen synthesis in a dose-dependent manner.97 In addition to impaired glucose transport and glycogen synthesis, rats infused with elevated lipids also showed reduced insulin signaling.98 Earlier studies found that saturated fatty acids, such as palmitate, has been correlated with reduced insulin sensitivity and impaired metabolism, whereas increases in certain unsaturated fatty acids have been shown to not have deleterious effects on metabolism.99–101 Despite the past few decades of research, the underlying mechanisms of how increased lipids and inflammation lead to insulin resistance and diabetes are still incompletely understood. Researchers have built in vitro models that aimed to recapitulate the pathologic lipid elevation and inflammation in the skeletal muscle to deepen our understanding of T2D pathophysiology. Below we summarize their studies and findings.
In limited hyperlipidemic studies on myoblasts, one showed that C2C12 myoblasts exposed to 0.25 mM palmitate for 2 h displayed an increase in apoptotic signaling and mitochondrial impairment, including an increase in mitochondrial fragmentation and mitochondrial membrane potential.89 The increased apoptotic signaling and mitochondrial impairment are correlated with an increased reactive oxygen species level.89
Similarly on myoblasts, elevated palmitate or proinflammatory cytokines have deleterious effects on myotubes. C2C12 myotubes treated with 0.1–0.6 mM of palmitate for 24 h showed decreased insulin-stimulated glycogen synthesis, glucose oxidation, glucose uptake, and AKT signaling.101,102 Furthermore, L6 and myotubes exposed to 0.75 mM palmitate for 2–16 h displayed a reduction in GLUT4 protein expression and an increase in NF-κB and TNF-α activities, indicative of inflammation.103,104 In therapeutic studies, the inhibition of protein kinase C (PKC) was found to eliminate palmitate-induced TNF-α expression and restore GLUT4 mRNA reduction, suggesting that targeting PKC could potentially treat fatty acid-induced insulin resistance.104 Other studies found that the treatment of 0.5 mM palmitate for 24 h on C2C12 myotubes negatively regulated the expression of myokine irisin and that knockdown of Smad3 alleviated the inhibitory effect of palmitate. The study suggests that palmitate could induce insulin resistance through Smad3-mediated down-regulation of irisin.105 Interestingly, not all saturated fatty acids induce metabolic dysfunction in myotubes. Recent studies in human myotubes showed that 0.5 mM palmitic acid, but not lauric acid, induced mitochondrial fragmentation and inflammatory cytokine IL-6.106 As human myotubes are exposed to 0.5 μM palmitate for 48 h or 10 ng/ml TNF-α for 24 h, another study identified dysregulated enhancers that overlap with genetic loci implicated in metabolic disease using a chromatin conformation assay. In addition to genes with known roles in metabolism, the study identified novel targets that have not been linked to human metabolic diseases.107
Studies mimicking the skeletal muscle inflammation found that 1 h of 10 ng/ml TNF-α exposure to C2C12 myotubes decreased insulin-stimulated IRS-1 tyrosine phosphorylation, leading to lowered insulin-stimulated glucose uptake.108 Moreover, 16 h of 10 ng/ml TNF-α exposure was found to suppress the adenosine monophosphate-activated protein kinase (AMPK) activity, which is important in fatty-acid oxidation, thus, worsening the insulin resistance.66
Overall, the in vitro models have shed lights on dysregulated molecular mechanisms implicated in the skeletal muscle cultured in hyperlipidemic and inflammatory conditions. The models hold promise in parsing out the complex genetic and environmental factors contributing to diabetes. Genetic studies, including sequencing, genetic knockouts, and enhancer mapping, will help identify potential novel targets to better treat diabetes. The models can act as preclinical drug screening platforms and narrow down effective targets to be tested in animals or humans.
The myoblast cell line (C2C12 and L6)-based in vitro models demonstrated the capability to model insulin resistance and T2D-related metabolic conditions because myoblast cell lines preserve key insulin signaling pathways, including IRS-1, PI3K, AMPK, mTOR, and AKT signaling pathways, and high level expression of GLUT and MYH proteins.109 However, though the widespread use of C2C12 has expanded our knowledge of T2D pathophysiology, the cell line lacks the ability to fully replicate the complex genetic makeup and metabolic dysfunction in humans. Below we discuss in vitro models built with myotubes derived from T2D patients and their advantages.
D. Incorporation of T2D subject-derived myoblasts and myotubes
Myoblasts and myotubes derived from individuals with T2D not only have the advantage of capturing human disease phenotypes and the diverse epigenetic backgrounds but also provide unique platforms for personalized drug discovery and a deeper understanding of the human molecular basis of disease. Earlier studies had shown that myotubes derived from T2D patients retain diabetic phenotypes such as impaired glucose uptake,110 reduced lipid oxidation,111 and increased inflammatory markers.14 Furthermore, in vitro systems allow gene knockdowns of human cells to investigate the functional roles of genes implicated in T2D. The studies below highlight the recent work conducted in in vitro human systems.
Compared with those of healthy individuals, myoblasts from T2D patients were shown to have altered gene expression of myogenic transcription factors, deteriorated myogenic differentiation, and increased expression of proinflammatory cytokine, IL-1β.15 T2D myoblasts also showed dysregulated expression of non-canonical autophagy genes, including VAMP8 and TP53INP1 compared with healthy control.16 Recently, VPS39 was found to be downregulated in T2D myoblasts and its knockdown in human myoblasts showed that VPS39 deficiency is implicated in impaired autophagy, abnormal epigenetic reprogramming, and dysregulation of myogenic differentiation.17 Moreover, studies on microRNAs (miRNAs) showed that miRNA-23b/27b was downregulated in myoblasts derived from T2D individuals compared with healthy control. Knocking down of the miRNA in myoblasts obtained from healthy donors impaired the myogenic capacity of the myoblasts.112 In addition, a subpopulation showing distinct miRNA expression was identified in myoblasts derived from T2D individuals compared with healthy controls. These characteristic miRNAs are implicated in the regulation of glucose transport and AKT pathway.113 miRNAs are increasingly recognized as important regulators of metabolism and are dysregulated in metabolic diseases, including T2D.114–116 The studies discussed above showed that diabetic abnormalities in the skeletal muscle exist even at the progenitor stage.
Unsurprisingly, many of the impairments found in T2D myoblasts are also present in myotubes derived from patient myoblasts. T2D myotubes showed elevated expressions of inflammation, such as NF-κB14 and IL-1β,15 and dysregulated expressions of autophagy genes, including DRAM1, VAMP8, and ATG7.16 Moreover, T2D myotubes displayed dysregulated intramyocellular glucose metabolism. Myotubes derived from obese T2D exhibited lowered rates of glycogen synthesis and glucose oxidation compared with myotubes derived from nondiabetic individuals.117 T2D myotubes showed suppressed insulin-stimulated glucose uptake.14 In addition, more than 200 genes were found to be significantly differentially expressed in T2D compared with obesity.118 Specifically, metabolism associated with ceramide and glycosphingolipids was upregulated in myocytes derived from T2D subjects compared with the control.118 The studies also found that lipolysis and hormone-sensitive lipase were lowered in T2D myotubes.119 This could contribute to the pathologic accumulation of lipids in the skeletal muscle. Myokines were also found to increase in myotubes derived from T2D patients compared with control and many of them are inflammatory, including IL-6, IL-8, IL-15, and TNF-α.79 This could contribute to the chronic inflammation observed in T2D patients. Both the accumulation of lipids and chronic inflammation lead to insulin resistance. Together, these findings showed that patient-derived T2D in vitro models do capture many of the diabetic phenotypes observed in vivo.
The main advantages of studying T2D in myoblasts and myotubes derived from individuals are the ability to recapitulate human genetic and metabolic phenotypes, perform high throughput drug screening, and conduct gene knockouts to better understand the molecular basis underlying the disease. However, the main challenges of harvesting satellite cells and myoblasts from human subjects are limited cell numbers and cells losing regenerative capacity after extended in vitro culture. Fortunately, induced pluripotent stem cells (iPSCs) are a promising solution to overcome these challenges and their use in T2D disease modeling is discussed in the following section.
E. Integration of T2D-induced pluripotent stem cells
Induced pluripotent stem cells (iPSCs) allow the generation of stem cells from blood or skin cells.120 Utilizing either small molecule differentiation or direct reprogramming, the iPSCs are then differentiated into myogenic progenitor cells that are capable of fusing and becoming multinucleated myotubes. They offer similar advantages to myoblasts harvested from patients in providing unique patient-specific disease models and gene editing to better understand the molecular basis of healthy and diabetic patients. In addition, iPSCs models provide the benefits of minimally-invasive harvest of patient cells and having unlimited cell source without losing the regenerative capacity of the cells. Below we discuss several recent T2D-related studies using iPSCs.
Interestingly, RNA-seq analysis conducted on iPSCs clones derived from insulin-resistant and insulin-sensitive subjects was able to identify differentially expressed genes in the groups.18 A set of genes that regulate critical aspects of insulin sensitivity was identified and their functional relevance for insulin responsiveness in immortalized human myogenic cells was confirmed.18 Moreover, myogenic cells derived from iPSCs of T2D patients retained defects, such as altered insulin signaling, reduced insulin-stimulated glucose uptake, and lowered mitochondrial oxidation, as observed in the muscle tissues of T2D individuals.19 While insulin resistance is a major disease phenotype of T2D and many studies have focused on insulin signaling, the study found that a number of phosphoproteome dysregulated in T2D are insulin-independent, including proteins involved in mRNA processing, vesicular trafficking, and chromatin remodeling.19 In addition, myotubes derived from iPSCs of individuals with mutations in the insulin receptor also captured the impairment in insulin signaling, insulin-stimulated glucose uptake, and metabolic gene expressions.20
Although iPSCs can address challenges associated with patient-derived myoblasts and hold much promise in furthering our understanding of disease physiology, a few major obstacles remain. The current iPSC differentiation protocols still need improvement to control the differentiation direction of cells more precisely. The maturity level of myofibers derived from iPSCs also needs improvement to make the in vitro models more suitable for the study of muscle diseases.
IV. INTERORGAN CROSSTALK IN T2D DEVELOPMENT
The interorgan crosstalk contributes to the development of T2D. There are at least three levels of crosstalk regulation between organs: organokines, hormones, and metabolites.121–125 Many factors secreted by the skeletal muscle, including proteins and peptides (myokines), metabolites, microRNAs (miRNAs), and exosomes, mediate the interaction between the skeletal muscle and other organs via an endocrine signaling and regulates metabolic health (Fig. 2). These muscle-secreted factors show potential to mediate the function and survival of β-cells and isolated islets during exercise and the insulin resistance process.123 The muscle-secreted factors promote browning and thermogenesis in white adipose tissue and alter adipocyte metabolism.126 Inversely, other organokines, including adipokines secreted by adipose tissue and hepatokines released by the liver, are also involved in the regulation of insulin sensitivity in the muscle tissues. Metabolites, adipokines, and hepatokines cause metabolic changes that can induce ROS production, inflammation, and cell death and contribute to T2D development.121 Many of these factors are regulated by exercise, and we summarized exercise effects on T2D skeletal muscle in Sec. VI. In addition, other factors including excessive ROS and lipid accumulation have been linked to the T2D-related interorgan crosstalk. For example, liver lipid accumulation, a key pathogenic factor of nonalcoholic fatty liver disease, induces insulin resistance in hepatic, adipose, and muscle tissues, thus, increasing the risk of T2D.127,128 Also, mitochondrial dysfunction in T2D tissues induces an increase in ROS and oxidative damage to muscle, liver, and adipose tissues. The reduction in lipolysis in adipose tissue induces increased plasma free fatty acid and uptake by liver and muscle, leading to lipotoxicity-induced insulin resistance.129 Taken together, these findings suggest the central role of interorgan crosstalk in the development of T2D.
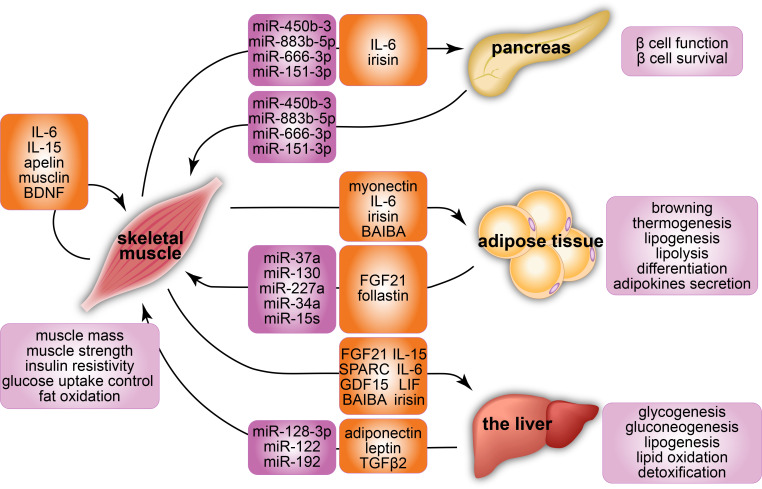
FIG. 2.
A summary of interorgan crosstalk via microRNAs and myokines with a focus on the skeletal muscle. The microRNAs and myokines are key factors secreted by the skeletal muscle that mediate the interaction between skeletal muscle and other organs, including the pancreas, adipose tissue, and the liver. The crosstalk of the skeletal muscle (1) enhances muscle mass and strength, insulin resistivity, glucose uptake control, and fat oxidation in skeletal muscles; (2) improves the function and survival of β-cells; (3) induces browning, thermogenesis, lipogenesis, lipolysis, differentiation, and adipokines secretion in adipose tissues; and (4) promotes glycogenesis, gluconeogenesis, lipogenesis, lipid oxidation, and detoxification in the liver.
V. IN VITRO T2D SKELETAL MUSCLE MODEL FOR INTERORGAN CROSSTALK STUDIES
In vitro skeletal models have been used to study the interorgan crosstalk in the development of T2D. The nature of blood-borne factors makes it difficult to dissect the effects of secreted factors that directly link muscle, liver, and adipose tissues, using in vivo animal models. To circumvent this, in vitro models have been used to study the effects of interorgan crosstalk in the development of T2D by (1) using conditioned media secreted from in vitro muscles, (2) co-culturing skeletal muscles with other tissues, and (3) overexpressing targeted organokine genes in the muscles. These in vitro interorgan approaches show the potential to dissect causal T2D-related interorgan connections and identify target functions and tissues of organokines, hormones, and metabolites.
A. Crosstalk between skeletal muscle and islets
In vitro studies suggest that muscle-secreted factors influence islets functions. The conditioned media collected from skeletal muscle cells or myotubes affected glucose-stimulated insulin secretion of pancreatic islets. C57BL6/J mouse islets and Wistar rat β-cells were found to have a higher level of glucose-stimulated insulin secretion after being incubated with human primary myotubes.130 The study also revealed that insulin-treated myotubes alone did not affect glucose-stimulated insulin secretion, proving that insulin secretion was mediated by the crosstalk between the β-cells and myotubes, not that the hormones secreted by the myotubes themselves. Similarly in other study, both normal and diabetic INS-1 832/3 pseudoislets treated with conditioned media secreted by C2C12 myotubes was found to have an acute increase in their insulin production, along with their enhanced mitochondrial oxidation levels.131 When INS-1 cells were cultured in the conditioned media collected from T2D patient-derived myotubes in another study by Ryan et al.,77 the T2D skeletal muscle media demonstrated an impaired ability to protect glucose-stimulated insulin secretion from metabolic inflammation challenge induced by increased glucose, insulin, and palmitate level. It was also found that the adverse effect of T2D patient-derived myotube conditioned media on glucose-stimulated insulin secretion was mediated via multiple signaling pathway, including p38 MAPK, PI3K, and PKC pathways. Merz et al.132 extended this idea to suggest that p21-activated kinase 1 (PAK1) of skeletal muscles and its circulating derivatives enhance pancreatic β-cells function in insulin secretion, as observed from β-cells cultured in PAK1-enriched conditioned media compared to isolated β-cells. In addition, a study of Rutti et al. used human skeletal muscle cells and identified two myokines, angiogenin and osteoprotegerin that can prevent TNF-α-induced apoptosis of β-cells.133
B. Crosstalk between skeletal muscle and adipose tissue
The crucial role of the adipocyte-skeletal muscle crosstalk in regulating pathophysiology of T2D has been consistently demonstrated in in vitro models. Pandurangan et al. found that co-culturing C2C12 and 3T3-L1 preadipocyte cells using a transwell insert altered the expression levels of calpains, caspases, and heat shock proteins (HSPs) in both cells, confirming a crosstalk between muscle cells and adipocytes in in vitro setting.134 This muscle-adipose tissue crosstalk was also observed in 3T3-L1 cells treated with electrically stimulated C2C12 myotubes conditioned media.126 Myotube conditioned media promoted adipogenesis and lipid metabolism by increasing PPARγ2 and PPARγ-regulated gene expression in adipocytes.126 Nintou et al. confirmed the browning effects of contracting myotubes on adipose cells by enhancing the expression of thermogenic proteins (UCP1 and IL-6) in the co-cultured C2C12 myotubes and 3T3-L1 adipocytes under electrical stimulation.135
C. Crosstalk between skeletal muscle and the liver
In coherence with adipose tissue and islets, skeletal muscles also engage in crosstalk with the liver. A recent in vitro nonalcoholic fatty liver disease model demonstrated the pathogenic link between fatty liver disease and T2D.128 De Chiara et al. demonstrated that the treatment of the conditioned media collected from fatty hepatocytes impaired myotube differentiation by altered gene expression of myogenic transcription factors in a 3D C2C12 myotube model.128 Moreover, this in vitro platform successfully confirmed the beneficial effect of albumin, a plasma protein produced by the liver in response to a high lipid challenge on the skeletal muscle tissue. Albumin pretreatment rescued C2C12 myotubes from the impaired myotube differentiation caused by fatty hepatocytes.128 Recently, hepatokines secreted by the liver have been identified for their roles in metabolic control in the muscle–liver axis.136 Seo et al. identified apolipoprotein J (ApoJ) as a novel hepatokine targeting muscle glucose metabolism and insulin sensitivity.137 It was revealed that the deletion of hepatic ApoJ causes shutdown of ApoJ to lipoprotein receptor-related protein-2 (LRP2) signaling pathway, which in turn knocks down LRP2 in C2C12 muscle cells to cause a significant reduction in insulin receptor internalization/endocytosis induced by insulin. The final result was a significant reduction of glucose uptake in the skeletal cell. The study showed that the interaction between the liver and muscle cells is needed for the insulin-dependent IR internalization; therefore, the crosstalk plays a crucial role in insulin sensitivity of the cells.137 Leukocyte cell-derived chemotaxin 2 (LECT2) was also found to be an energy-sensing hepatokine in the muscle-liver axis.138 The deletion of LECT2 increased insulin sensitivity in the skeletal muscle, while expression of endogenous LECT2 and treatment of recombinant LECT2 protein simultaneously in C2C12 myotubes impaired insulin signaling via increased JNK phosphorylation.138 Both ApoJ and LECT2 protein originating from hepatocytes are identified as a key regulator in development of insulin resistance in the skeletal muscle, suggesting them as potential therapeutic targets for the antidiabetic treatment.
VI. EXERCISE BENEFITS IN T2D
Exercise is widely known to have a plethora of preventative and therapeutic benefits in diabetes139–143 (Fig. 3). Studies found that exercise increases glucose uptake in the skeletal muscle during and transiently post-exercise.5–8 The initial glucose uptake during exercise was found to be insulin-independent, where the increasing demand for glucose and oxygen drives up AMP concentration, which activates PI3K and AMPK signaling leading to GLUT4 translocation.5 Shortly after exercise, the insulin-dependent glucose uptake kicks in, where IRS-1 is phosphorylated, leading to AKT signaling and GLUT4 translocation.5 Exercise also results in a higher concentration of Ca2+ in the muscle fiber, leading to increased GLUT4 expression, which positively correlates with exercise intensity and duration.5,144,145 Moreover, regular exercise yields reduced basal and glucose-stimulated insulin levels, improved mitochondrial function, and increased muscle mass, GLUT4 concentration, insulin activity, and free fatty acid oxidation.5,9–11 Exercise can revert glucose uptake to near-normal level in insulin resistant individuals.5,144,146,147 In addition, the expression of PGC1-α, a marker of mitochondrial biogenesis, increases in exercise.148,149 Not surprisingly, physical activity is an important determinant of insulin sensitivity.5,150
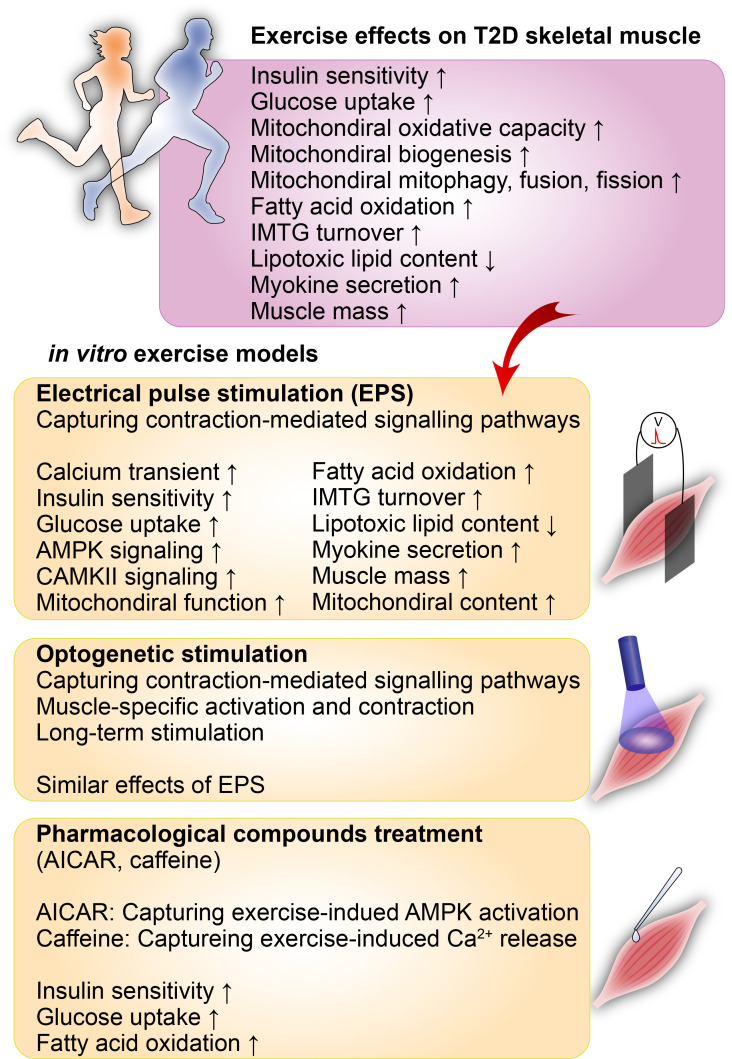
FIG. 3.
In vitro exercise models for the study of T2D pathophysiology. Various in vitro exercise models have enabled mechanistic studies on numerous preventative and therapeutic effects of exercise on diabetes. Electrical pulse stimulation (EPS) of cultured myotubes has been commonly used to capture muscle-contraction-mediated signaling pathways. The optogenetic stimulation model provides an alternative method to activate contraction-mediated signaling pathways and enable prolonged stimulation and muscle maturation. Pharmacological compounds such as AICAR and AMPK have been used to study the effects of exercise on T2D in in vitro T2D models. AICAR has been used to mimic the exercise-induced activation of AMPK. In contrast, caffeine treatment has been applied to capture exercise-induced Ca2+ release from the sarcoplasmic reticulum and the downstream effects.
In the recent decades, studies found that contracting skeletal muscles secrete protein factors or myokines that may be anti-inflammatory and have the potential to be beneficial to various diseases, including diabetes, where chronic inflammation is a major underlying pathophysiology. For example, IL-6 secretion rises after exercise and is associated with decreases in proinflammatory cytokine, TNF-α.149,151 IL-6 was also found to induce the production of anti-inflammatory cytokines, IL-1 receptor antagonist and IL-10, in blood mononuclear cells.152,153 Moreover, IL-6 is known to stimulate lipolysis.153 Both IL-6 and another myokine, Brain-derived neurotrophic factor (BDNF), are involved in fat oxidation.153 Despite our current knowledge, the beneficial effects of myokines and exercise in diabetes are still largely unknown. Below we discuss in vitro models built to better mimic and understand exercise and diabetes.
VII. IN VITRO SKELETAL MUSCLE MODEL OF EXERCISE AND T2D
To simulate exercise in vivo, electrical pulse stimulation (EPS) of cultured myotubes has been commonly used to induce muscle contractions in vitro154–156 to capture muscle-contraction-mediated signaling pathways157 (Fig. 3). EPS is known to increase glucose uptake, mitochondrial biogenesis, and fatty acid oxidation in skeletal muscles, similar to the adaptive changes observed during exercise in vivo.158,159 In the past decade, EPS has also been utilized to elucidate how diabetic patients respond to exercise. Studies showed that short-term, high-frequency EPS-induced acute, strength training-like upregulation of glucose uptake, lactate production, mTOR, AKT, and decreased adenosine triphosphate (ATP) and phosphocreatine content.158–160 In contrast, longer-term, low-frequency EPS-induced endurance training-like upregulation of AMPK, PGC1-α, mitochondrial biogenesis, and glucose and fatty acid oxidation.159,161–164 Many studies have also shown that exercise has differential effects on obese or diabetic verses healthy individuals. For example, EPS was found to improve insulin sensitivity and glucose oxidative capacity but not lipid oxidation in myotubes from obese subjects with or without T2D compared with lean nondiabetic subjects.165,166 EPS increased AMPK activation in healthy myotubes but the effect is less pronounced in myotubes from obese individuals.166 Myotubes derived from obese donors with and without T2D showed reduced lipid (intramuscular triglyceride, IMTG) turnover and fat oxidation rate post-EPS compared with those derived from lean subjects.167 EPS also has differential effects on mitochondrial dynamics on obese verses lean individuals.168
Optogenetic stimulation has been developed to provide an alternative method to induce muscle contraction (Fig. 3). This is achieved by genetic expression of light-sensitive ion channels in muscle cells. Optical activation of light-sensitive ion channels in muscle cells allows for optical control of muscle activation and contraction. Thus, optogenetic stimulation of muscles eliminates potential chemical-leaching associated with EPS electrodes. Also, optogenetic stimulation enables muscle-specific activation and contraction by expressing the light-sensitive channels in muscle tissues via skeletal muscle-specific protomers.169 Pulsed illumination of ChR2 expression C2C12 myotubes demonstrated optogenetically stimulated muscle contraction170 and prolong stimulation enhanced the muscle maturation.171 Although the optogenetic method can induce muscle contraction and is a valuable addition to exclude any nonspecific effects caused by EPS, the application of optogenetic stimulation method on T2D models has not reported yet.
Several pharmacological compounds such as 5-aminoimidazole-4-carboxamide-1-β-dribofuranoside (AICAR) and caffeine, have been used to study the effects of exercise on T2D in in vitro T2D models. AICAR has been used to mimic the exercise-induced activation of AMPK, while caffeine treatment has been applied to capture exercise-induced Ca2+ release from the sarcoplasmic reticulum and the downstream effects157 (Fig. 3). In particular, AICAR treatment activated key exercise-induced insulin signaling pathways such as enhanced insulin responsiveness and GLUT4 expression and translocation.157 Importantly, AICAR has also been used in in vitro human myotubes to investigate AICAR effects on the AMPK activation between healthy donor and T2DM-derived myotubes.172–174 But since the pharmacological compounds treatment do not induce muscle contraction, there is lack of several effects of in vivo exercise, including unchanged ATP content and myotube lactate production.157
These in vitro exercise models elucidate how exercise is beneficial to the skeletal muscle and how the positive effects may be reduced in T2D. A deeper understanding of how T2D compromises the benefits of exercise will help researchers find more effective therapeutics for diabetic patients.
VIII. FUTURE PERSPECTIVES
Although numerous efforts have been made to develop in vitro T2D skeletal muscle models, recent advances in the development of 3D tissue-level microphysiological systems175 are now bringing in vitro T2D skeletal muscle models to the next level. First, the longer-term culture and improved tissue maturation of 3D models compared to the traditional 2D monolayers muscle models will allow researchers to better understand mechanisms in the pathogenesis of T2D, a chronic progressive metabolic disorder. Second, the close resemblance of the in vivo microenvironment of the skeletal muscle in 3D models will facilitate the in-depth study of T2D-mediated pathogenetic extracellular matrix (ECM) remodeling mechanisms such as increased collagen and reduced collagen integrity,176 upregulated matrix metalloproteases, and altered integrin signaling.177 Third, 3D tissue models have capability to better capture beneficial effects of muscle contraction on T2D muscles due to their in vivo tissue-like stiffness compared to 2D monolayer cultured on rigid substrates. These 3D models will be useful to investigate mechanisms responsible for exercise-induced myofiber injury in T2D muscle tissues.177 Fourth, the microfluidics-based 3D tissue multiorgan systems will allow the study of interorgan crosstalk in T2D development by emulating cross-communication between skeletal muscles and other organs.178 Also, the integration of multiomics technology, including secretomics,179 metabolomics,180–182 and exosomics,183,184 into the multi-organ T2D models will uncover the identity of myokines, lipids, metabolites, and exosomes responsible for the crosstalk and elucidate their targets and therapeutic functions. Also, although the dynamics nature of these factors makes it difficult to detect, the integration of in situ sensing technology185,186 into 3D tissue-based microphysiological systems will identify the factors with therapeutic potentials and delineate the mechanistic roles of these factors. Finally, 3D tissue constructs will provide novel therapeutic approaches to the T2D treatment. The recent study187 demonstrated that the implantation of in vitro 3D fibrin gel-based skeletal muscle constructs that overexpressed the GLUT4 transporter in diet-induced obesity T2D mice significantly improved glucose homeostasis and insulin sensitivity, even though the implanted muscle tissue was only 1.1% of the total weight of the abdominal muscle of the mice. The authors also pointed out that this systemic effect is mediated by myokines. Thus, this work suggests that the genetically engineered 3D skeletal muscle may serve as a delivery tool of myokines, lipids, metabolites, and exosomes for the novel T2D supportive therapy. The continued development of in vitro T2D models will enable researchers to reverse-engineer closely in vivo T2D signaling dynamics closely in in vitro setting and help to develop novel therapies with the precision mechanisms for T2D.
IX. SUMMARY
Advances in the skeletal muscle, T2D, and exercise in vitro models have enabled us to study more precisely the underlying pathophysiology in diabetes, the therapeutic effects of exercise, and how these effects may be compromised in T2D. More recent T2D and exercise studies are focused on in vitro human myotubes derived either from the myoblasts or iPSCs from T2D subjects. These models hold promise in parsing out the complex genetic and environmental factors contributing to diabetes. In addition, in vitro T2D models have demonstrated the potentials to emulate how interorgan crosstalk contributes to the development of T2D and how exercise is mechanistically beneficial to T2D. More sophisticated genetic studies and proteomics investigations in T2D in vitro models will help identify potential novel targets to better treat diabetes in the near future.
ACKNOWLEDGMENTS
This work was supported by the Parker H. Petit Institute Interdisciplinary Research Grant Program. S.-J.P. was funded by Georgia Institute of Technology, Emory University School of Medicine, Emory School of Medicine Bridge Funding Program, and American Heart Association Career Development Award (No. 857583). C.Y.S. was funded by Cell and Tissue Engineering NIH Biotechnology Training Grants (Grant Nos. T32 GM-008433 and T32 GM145735).
AUTHOR DECLARATIONS
Conflict of Interest
The authors have no conflicts to disclose.
Author Contributions
Christina Y. Sheng: Formal analysis (lead); Investigation (lead); Resources (lead); Writing – original draft (lead). Young Hoon Son: Writing – original draft (supporting); Writing – review & editing (supporting). Jeongin Jang: Writing – original draft (supporting); Writing – review & editing (supporting). Sung-Jin Park: Conceptualization (lead); Funding acquisition (lead); Project administration (lead); Supervision (lead); Visualization (lead); Writing – original draft (lead); Writing – review & editing (lead).
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1. Galicia-Garcia U., Benito-Vicente A., Jebari S., Larrea-Sebal A., Siddiqi H., Uribe K. B., Ostolaza H., and Martin C., Int. J. Mol. Sci. 21(17), 6275 (2020). 10.3390/ijms21176275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abdelalim E. M., Cell Mol. Life Sci. 78(6), 2459–2483 (2021). 10.1007/s00018-020-03710-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kahn S. E., Hull R. L., and Utzschneider K. M., Nature 444(7121), 840–846 (2006). 10.1038/nature05482 [DOI] [PubMed] [Google Scholar]
- 4. Abdul-Ghani M. A. and DeFronzo R. A., J. Biomed. Biotechnol. 2010, 476279. 10.1155/2010/476279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Whillier S., Adv. Exp. Med. Biol. 1228, 137–150 (2020). 10.1007/978-981-15-1792-1 [DOI] [PubMed] [Google Scholar]
- 6. O'Gorman D. J., Karlsson H. K., McQuaid S., Yousif O., Rahman Y., Gasparro D., Glund S., Chibalin A. V., Zierath J. R., and Nolan J. J., Diabetologia 49(12), 2983–2992 (2006). 10.1007/s00125-006-0457-3 [DOI] [PubMed] [Google Scholar]
- 7. Ross R., Diabetes Care 26(3), 944–945 (2003). 10.2337/diacare.26.3.944 [DOI] [PubMed] [Google Scholar]
- 8. DiMenna F. J. and Arad A. D., BMC Sports Sci. Med. Rehabil. 10, 21 (2018). 10.1186/s13102-018-0110-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kirwan J. P., del Aguila L. F., Hernandez J. M., Williamson D. L., O'Gorman D. J., Lewis R., and Krishnan R. K., J. Appl. Physiol. 88(2), 797–803 (2000). 10.1152/jappl.2000.88.2.797 [DOI] [PubMed] [Google Scholar]
- 10. Sigal R. J., Kenny G. P., Wasserman D. H., and Castaneda-Sceppa C., Diabetes Care 27(10), 2518–2539 (2004). 10.2337/diacare.27.10.2518 [DOI] [PubMed] [Google Scholar]
- 11. Stanford K. I. and Goodyear L. J., Adv. Physiol. Educ. 38(4), 308–314 (2014). 10.1152/advan.00080.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. King A. J., Br. J. Pharmacol. 166(3), 877–894 (2012). 10.1111/j.1476-5381.2012.01911.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Srinivasan K. and Ramarao P., Indian J. Med. Res. 125(3), 451–472 (2007). [PubMed] [Google Scholar]
- 14. Green C. J., Pedersen M., Pedersen B. K., and Scheele C., Diabetes 60(11), 2810–2819 (2011). 10.2337/db11-0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakamura S., Yonekura S., Shimosato T., and Takaya T., Front. Physiol. 12, 679152 (2021). 10.3389/fphys.2021.679152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Henriksen T. I., Wigge L. V., Nielsen J., Pedersen B. K., Sandri M., and Scheele C., Sci. Rep. 9(1), 8169 (2019). 10.1038/s41598-019-44535-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davegardh C., Sall J., Benrick A., Broholm C., Volkov P., Perfilyev A., Henriksen T. I., Wu Y., Hjort L., Brons C., Hansson O., Pedersen M., Wurthner J. U., Pfeffer K., Nilsson E., Vaag A., Stener-Victorin E., Pircs K., Scheele C., and Ling C., Nat. Commun. 12(1), 2431 (2021). 10.1038/s41467-021-22068-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carcamo-Orive I., Henrion M. Y. R., Zhu K., Beckmann N. D., Cundiff P., Moein S., Zhang Z., Alamprese M., D'Souza S. L., Wabitsch M., Schadt E. E., Quertermous T., Knowles J. W., and Chang R., PLoS Comput. Biol. 16(12), e1008491 (2020). 10.1371/journal.pcbi.1008491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Batista T. M., Jayavelu A. K., Wewer Albrechtsen N. J., Iovino S., Lebastchi J., Pan H., Dreyfuss J. M., Krook A., Zierath J. R., Mann M., and Kahn C. R., Cell Metab. 32(5), 844–859 (2020). 10.1016/j.cmet.2020.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iovino S., Burkart A. M., Warren L., Patti M. E., and Kahn C. R., Proc. Natl. Acad. Sci. U. S. A. 113(7), 1889–1894 (2016). 10.1073/pnas.1525665113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Butler P. C. and Rizza R. A., Diabetes 40, 73 (1991). 10.2337/diab.40.1.73 [DOI] [PubMed] [Google Scholar]
- 22. Rizza R. A., Diabetes 59(11), 2697–2707 (2010). 10.2337/db10-1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petersen K. F. and Shulman G. I., Am. J. Cardiol. 90(5), 11–18 (2002). 10.1016/S0002-9149(02)02554-7 [DOI] [Google Scholar]
- 24. Boucher J., Kleinridders A., and Kahn C. R., Cold Spring Harbor Perspect. Biol. 6(1), a009191 (2014). 10.1101/cshperspect.a009191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Butterfield W. J. H. and Whichelow M. J., Diabetologia 1(1), 43–53 (1965). 10.1007/BF01338715 [DOI] [Google Scholar]
- 26. Wang X., Hu Z., Hu J., Du J., and Mitch W. E., Endocrinology 147(9), 4160–4168 (2006). 10.1210/en.2006-0251 [DOI] [PubMed] [Google Scholar]
- 27. Huang X., Liu G., Guo J., and Su Z., Int. J. Biol. Sci. 14(11), 1483–1496 (2018). 10.7150/ijbs.27173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stitt T. N., Drujan D., Clarke B. A., Panaro F., Timofeyva Y., Kline W. O., Gonzalez M., Yancopoulos G. D., and Glass D. J., Mol. Cell 14(3), 395–403 (2004). 10.1016/S1097-2765(04)00211-4 [DOI] [PubMed] [Google Scholar]
- 29. Gaster M., Staehr P., Beck-Nielsen H., Schrøder H. D., and Handberg A., Diabetes 50, 1324 (2001). 10.2337/diabetes.50.6.1324 [DOI] [PubMed] [Google Scholar]
- 30. Deshmukh A. S., Murgia M., Nagaraj N., Treebak J. T., Cox J., and Mann M., Mol. Cell Proteomics 14(4), 841–853 (2015). 10.1074/mcp.M114.044222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bianchi L. and Volpato S., Acta Diabetol. 53(6), 879–889 (2016). 10.1007/s00592-016-0880-y [DOI] [PubMed] [Google Scholar]
- 32. Gaster M., Staehr P., Handberg A., Schroder D., and Beck-Nielsen H., Am. J. Physiol. Endocrinol. Metab. 278, E910 (2000). 10.1152/ajpendo.2000.278.5.E910 [DOI] [PubMed] [Google Scholar]
- 33. DeFronzo R. A., Int. J. Obes. 6, 73–82 (1982). [PubMed] [Google Scholar]
- 34. Lemay A., Turcot L., Dechene F., Dodin S., and Forest J. C., Menopause 17(2), 321–325 (2010). 10.1097/gme.0b013e3181b7c521 [DOI] [PubMed] [Google Scholar]
- 35. Wedick N. M., Snijder M. B., Dekker J. M., Heine R. J., Stehouwer C. D., Nijpels G., and van Dam R. M., Obesity 17(8), 1609–1614 (2009). 10.1038/oby.2008.666 [DOI] [PubMed] [Google Scholar]
- 36. Schrauwen P., Proc. Nutr. Soc. 66(1), 33–41 (2007). 10.1017/S0029665107005277 [DOI] [PubMed] [Google Scholar]
- 37. Bogardus C., Lillioja S., Mott D., Reaven G. R., Kashiwagi A., and Foley J. E., J. Clin. Invest. 73(3), 800–805 (1984). 10.1172/JCI111274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wondmkun Y. T., Diabetes, Metab. Syndr. Obes. 13, 3611–3616 (2020). 10.2147/DMSO.S275898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Palomer X., Pizarro-Delgado J., Barroso E., and Vazquez-Carrera M., Trends Endocrinol. Metab. 29(3), 178–190 (2018). 10.1016/j.tem.2017.11.009 [DOI] [PubMed] [Google Scholar]
- 40. Kelley D. E., Goodpaster B., Wing R. R., and Simoneau J. A., Am. J. Physiol. 277(6), E1130–E1141 (1999). 10.1152/ajpendo.1999.277.6.E1130 [DOI] [PubMed] [Google Scholar]
- 41. Hulver M. W., Berggren J. R., Cortright R. N., Dudek R. W., Thompson R. P., Pories W. J., MacDonald K. G., Cline G. W., Shulman G. I., Dohm G. L., and Houmard J. A., Am. J. Physiol.-Endocrinol. Metab. 284(4), E741–E747 (2003). 10.1152/ajpendo.00514.2002 [DOI] [PubMed] [Google Scholar]
- 42. Steinberg G. R., Parolin M. L., Heigenhauser G. J., and Dyck D. J., Am. J. Physiol. Endocrinol. Metab. 283(1), 187–192 (2002). 10.1152/ajpendo.00542.2001 [DOI] [PubMed] [Google Scholar]
- 43. Turcotte L. P., Swenberger J. R., Zavitz Tucker M., and Yee A. J., Diabetes 50(6), 1389–1396 (2001). 10.2337/diabetes.50.6.1389 [DOI] [PubMed] [Google Scholar]
- 44. Chavez J. A. and Summers S. A., Arch. Biochem. Biophys. 419(2), 101–109 (2003). 10.1016/j.abb.2003.08.020 [DOI] [PubMed] [Google Scholar]
- 45. Itani S. I., Ruderman N. B., Schmieder F., and Boden G., Diabetes 51(7), 2005–2011 (2002). 10.2337/diabetes.51.7.2005 [DOI] [PubMed] [Google Scholar]
- 46. Montell E., Turini M., Marotta M., Roberts M., Noé V., Ciudad C. J., Macé K., and Gómez-Foix A. M., Am. J. Physiol. Endocrinol. Metab. 280(2), E229–E237 (2001). 10.1152/ajpendo.2001.280.2.E229 [DOI] [PubMed] [Google Scholar]
- 47. Holland W. L., Brozinick J. T., Wang L. P., Hawkins E. D., Sargent K. M., Liu Y., Narra K., Hoehn K. L., Knotts T. A., Siesky A., Nelson D. H., Karathanasis S. K., Fontenot G. K., Birnbaum M. J., and Summers S. A., Cell Metab. 5(3), 167–179 (2007). 10.1016/j.cmet.2007.01.002 [DOI] [PubMed] [Google Scholar]
- 48. Holland W. L., Knotts T. A., Chavez J. A., Wang L.-P., Hoehn K. L., and Summers S. A., Nutr. Rev. 65(6), 39–46 (2007). 10.1301/nr.2007.jun.S39-S46 [DOI] [PubMed] [Google Scholar]
- 49. Hilton T. N., Tuttle L. J., Bohnert K. L., Mueller M. J., and Sinacore D. R., Phys. Ther. 88(11), 1336–1344 (2008). 10.2522/ptj.20080079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lowell B. B. and Shulman G. I., Science 307(5708), 384–387 (2005). 10.1126/science.1104343 [DOI] [PubMed] [Google Scholar]
- 51. Goodpaster B. H., Carlson C. L., Visser M., Kelley D. E., Scherzinger A., Harris T. B., Stamm E., and Newman A. B., J. Appl. Physiol. 90(6), 2157–2165 (2001). 10.1152/jappl.2001.90.6.2157 [DOI] [PubMed] [Google Scholar]
- 52. Schenk S., Saberi M., and Olefsky J. M., J. Clin. Invest. 118(9), 2992–3002 (2008). 10.1172/JCI34260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shi H., Kokoeva M. V., Inouye K., Tzameli I., Yin H., and Flier J. S., J. Clin. Invest. 116(11), 3015–3025 (2006). 10.1172/JCI28898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wood N., Straw S., Scalabrin M., Roberts L. D., Witte K. K., and Bowen T. S., ESC Heart Failure 8(1), 3–15 (2021). 10.1002/ehf2.13121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reyna S. M., Ghosh S., Tantiwong P., Meka C. S., Eagan P., Jenkinson C. P., Cersosimo E., Defronzo R. A., Coletta D. K., Sriwijitkamol A., and Musi N., Diabetes 57(10), 2595–2602 (2008). 10.2337/db08-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Perry B. D., Caldow M. K., Brennan-Speranza T. C., Sbaraglia M., Jerums G., Garnham A., Wong C., Levinger P., Asrar Ul Haq M., Hare D. L., Price S. R., and Levinger I., Exercise Immunol. Rev. 22, 94–109 (2016). [PMC free article] [PubMed] [Google Scholar]
- 57. Chen L., Chen R., Wang H., and Liang F., Int. J. Endocrinol. 2015, 508409. 10.1155/2015/508409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Anderson E. J., Lustig M. E., Boyle K. E., Woodlief T. L., Kane D. A., Lin C. T., Price J. W. III, Kang L., Rabinovitch P. S., Szeto H. H., Houmard J. A., Cortright R. N., Wasserman D. H., and Neufer P. D., J. Clin. Invest. 119(3), 573–581 (2009). 10.1172/JCI37048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cross C. E., Halliwell B., Borish E. T., Pryor W. A., Ames B. N., Saul R. L. et al. , Ann. Intern. Med. 107, 526 (1987). 10.7326/0003-4819-107-4-526 [DOI] [PubMed] [Google Scholar]
- 60. Shepherd P. and Kahn B., N. Engl. J. Med. 341, 248 (1999). 10.1056/NEJM199907223410406 [DOI] [PubMed] [Google Scholar]
- 61. Evans P. L., McMillin S. L., Weyrauch L. A., and Witczak C. A., Nutrients 11(10), 2432 (2019). 10.3390/nu11102432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huang C., Somwar R., Patel N., Niu W., Török D., and Klip A., Diabetes 51(7), 2090–2098 (2002). 10.2337/diabetes.51.7.2090 [DOI] [PubMed] [Google Scholar]
- 63. Shoelson S. E., Lee J., and Goldfine A. B., J. Clin. Invest. 116(7), 1793–1801 (2006). 10.1172/JCI29069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zatterale F., Longo M., Naderi J., Raciti G. A., Desiderio A., Miele C., and Beguinot F., Front. Physiol. 10, 1607 (2019). 10.3389/fphys.2019.01607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hotamisligil G. S., Shargill N. S., and Spiegelman B. M., Science 259(5091), 87–91 (1993). 10.1126/science.7678183 [DOI] [PubMed] [Google Scholar]
- 66. Steinberg G. R., Michell B. J., van Denderen B. J., Watt M. J., Carey A. L., Fam B. C., Andrikopoulos S., Proietto J., Gorgun C. Z., Carling D., Hotamisligil G. S., Febbraio M. A., Kay T. W., and Kemp B. E., Cell Metab. 4(6), 465–474 (2006). 10.1016/j.cmet.2006.11.005 [DOI] [PubMed] [Google Scholar]
- 67. Sergi D., Naumovski N., Heilbronn L. K., Abeywardena M., O'Callaghan N., Lionetti L., and Luscombe-Marsh N., Front. Physiol. 10, 532 (2019). 10.3389/fphys.2019.00532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mootha V. K., Lindgren C. M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., and Groop L. C., Nat. Genet. 34(3), 267–273 (2003). 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- 69. Patti M. E., Butte A. J., Crunkhorn S., Cusi K., Berria R., Kashyap S., Miyazaki Y., Kohane I., Costello M., Saccone R., Landaker E. J., Goldfine A. B., Mun E., DeFronzo R., Finlayson J., Kahn C. R., and Mandarino L. J., Proc. Natl. Acad. Sci. U. S. A. 100(14), 8466–8471 (2003). 10.1073/pnas.1032913100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schrauwen-Hinderling V. B., Kooi M. E., Hesselink M. K. C., Jeneson J. A. L., Backes W. H., van Echteld C. J. A., van Engelshoven J. M. A., Mensink M., and Schrauwen P., Diabetologia 50(1), 113–120 (2007). 10.1007/s00125-006-0475-1 [DOI] [PubMed] [Google Scholar]
- 71. Kim J. Y., Hickner R. C., Cortright R. L., Dohm G. L., and Houmard J. A., Am. J. Physiol. Endocrinol. Metab. 279(5), 1039–1044 (2000). 10.1152/ajpendo.2000.279.5.E1039 [DOI] [PubMed] [Google Scholar]
- 72. Phielix E., Schrauwen-Hinderling V. B., Mensink M., Lenaers E., Meex R., Hoeks J., Kooi M. E., Moonen-Kornips E., Sels J. P., Hesselink M. K., and Schrauwen P., Diabetes 57(11), 2943–2949 (2008). 10.2337/db08-0391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jheng H. F., Tsai P. J., Guo S. M., Kuo L. H., Chang C. S., Su I. J., Chang C. R., and Tsai Y. S., Mol. Cell Biol. 32(2), 309–319 (2012). 10.1128/MCB.05603-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. da Silva Rosa S. C., Nayak N., Caymo A. M., and Gordon J. W., Physiol. Rep. 8(19), e14607 (2020). 10.14814/phy2.14607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Laurens C., Bergouignan A., and Moro C., Front. Physiol. 11, 91 (2020). 10.3389/fphys.2020.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mashili F. L., Austin R. L., Deshmukh A. S., Fritz T., Caidahl K., Bergdahl K., Zierath J. R., Chibalin A. V., Moller D. E., Kharitonenkov A., and Krook A., Diabetes Metab. Res. Rev. 27(3), 286–297 (2011). 10.1002/dmrr.1177 [DOI] [PubMed] [Google Scholar]
- 77. Ryan A. J., Ciaraldi T. P., and Henry R. R., Front. Physiol. 10, 1608 (2019). 10.3389/fphys.2019.01608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bouzakri K., Plomgaard P., Berney T., Donath M. Y., Pedersen B. K., and Halban P. A., Diabetes 60(4), 1111–1121 (2011). 10.2337/db10-1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ciaraldi T. P., Ryan A. J., Mudaliar S. R., and Henry R. R., PLoS One 11(7), e0158209 (2016). 10.1371/journal.pone.0158209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Amir Levy Y., Ciaraldi T. P., Mudaliar S. R., Phillips S. A., and Henry R. R., Am. J. Physiol. Endocrinol. Metab. 309(1), E22–E34 (2015). 10.1152/ajpendo.00513.2014 [DOI] [PubMed] [Google Scholar]
- 81. Cefalu W. T., ILAR J. 47(3), 186–198 (2006). 10.1093/ilar.47.3.186 [DOI] [PubMed] [Google Scholar]
- 82. Leiter E. H., Methods Mol. Biol. 560, 1–17 (2009). 10.1007/978-1-59745-448-3 [DOI] [PubMed] [Google Scholar]
- 83. Ostler J. E., Maurya S. K., Dials J., Roof S. R., Devor S. T., Ziolo M. T., and Periasamy M., Am. J. Physiol. Endocrinol. Metab. 306(6), E592–E605 (2014). 10.1152/ajpendo.00277.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fang J. Y., Lin C. H., Huang T. H., and Chuang S. Y., Nutrients 11(3), 530 (2019). 10.3390/nu11030530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Clee S. M. and Attie A. D., Endocr. Rev. 28(1), 48–83 (2007). 10.1210/er.2006-0035 [DOI] [PubMed] [Google Scholar]
- 86. Mühlhäusler B. S., Toop C., and Gentili S., Methods Mol. Biol. 2076, 43–69 (2020). 10.1007/978-1-4939-9882-1 [DOI] [PubMed] [Google Scholar]
- 87. Luo W., Ai L., Wang B. F., and Zhou Y., Biomed. Pharmacother. 120, 109498 (2019). 10.1016/j.biopha.2019.109498 [DOI] [PubMed] [Google Scholar]
- 88. Mangnall D., Bruce C., and Fraser R. B., Biochem. Soc. Trans. 21, 438S (1993). 10.1042/bst021438s [DOI] [PubMed] [Google Scholar]
- 89. Dohl J., Foldi J., Heller J., Gasier H. G., Deuster P. A., and Yu T., Life Sci. 211, 238–244 (2018). 10.1016/j.lfs.2018.09.041 [DOI] [PubMed] [Google Scholar]
- 90. Liu X., Heras G., Lauschke V. M., Mi J., Tian G., and Gastaldello S., Exp. Cell Res. 395(2), 112234 (2020). 10.1016/j.yexcr.2020.112234 [DOI] [PubMed] [Google Scholar]
- 91. Grzelkowska-Kowalczyk K., Wieteska-Skrzeczynska W., Grabiec K., and Tokarska J., Cell Biol. Int. 37(1), 29–35 (2013). 10.1002/cbin.10004 [DOI] [PubMed] [Google Scholar]
- 92. Grabiec K., Gajewska M., Milewska M., Blaszczyk M., and Grzelkowska-Kowalczyk K., J. Endocrinol. Invest. 37(3), 233–245 (2014). 10.1007/s40618-013-0007-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Turner M. C., Player D. J., Martin N. R. W., Akam E. C., and Lewis M. P., J. Cell Biochem. 119(7), 5686–5695 (2018). 10.1002/jcb.26748 [DOI] [PubMed] [Google Scholar]
- 94. Russell S. T., Rajani S., Dhadda R. S., and Tisdale M. J., Exp. Cell Res. 315(1), 16–25 (2009). 10.1016/j.yexcr.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 95. Turner M. C., Martin N. R. W., Player D. J., Ferguson R. A., Wheeler P., Green C. J., Akam E. C., and Lewis M. P., J. Mol. Endocrinol. 64(3), 125–132 (2020). 10.1530/JME-19-0169 [DOI] [PubMed] [Google Scholar]
- 96. McGarry J. D., Diabetes 51, 7–18 (2002). 10.2337/diabetes.51.1.7 [DOI] [PubMed] [Google Scholar]
- 97. Boden G., Chen X., Ruiz J., White J. V., and Rossetti L., J. Clin. Invest. 93, 2438 (1994). 10.1172/JCI117252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Griffin M. E., Marcucci M. J., Cline G. W., Bell K., Barucci N., Lee D., Goodyear L. J., Kraegen E. W., White M. F., and Shulman G. I., Diabetes 48, 1270 (1999). 10.2337/diabetes.48.6.1270 [DOI] [PubMed] [Google Scholar]
- 99. Storlien L. H., Jenkins A. B., Chisholm D. J., Pascoe W. S., Khouri S., and Kraegen E. W., Diabetes 40, 280 (1991). 10.2337/diab.40.2.280 [DOI] [PubMed] [Google Scholar]
- 100. Storlien L. H., Kraegen E. W., Chisholm D. J., Ford G. L., Bruce D. G., and Pascoe W. S., Science 237(4817), 885–888 (1987). 10.1126/science.3303333 [DOI] [PubMed] [Google Scholar]
- 101. Hirabara S. M., Curi R., and Maechler P., J. Cell Physiol. 222(1), 187–194 (2010). 10.1002/jcp.21936 [DOI] [PubMed] [Google Scholar]
- 102. Yang M., Wei D., Mo C., Zhang J., Wang X., Han X., Wang Z., and Xiao H., Lipids Health Dis. 12(1), 104 (2013). 10.1186/1476-511X-12-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ebersbach-Silva P., Poletto A. C., David-Silva A., Seraphim P. M., Anhe G. F., Passarelli M., Furuya D. T., and Machado U. F., Lipids Health Dis. 17(1), 64 (2018). 10.1186/s12944-018-0714-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Jove M., Planavila A., Sanchez R. M., Merlos M., Laguna J. C., and Vazquez-Carrera M., Endocrinology 147(1), 552–561 (2006). 10.1210/en.2005-0440 [DOI] [PubMed] [Google Scholar]
- 105. Guo Q., Wei X., Hu H., Yang D., Zhang B., Fan X., Liu J., He H., Oh Y., Wu Q., Zhang Y., Wang C., Liu C., and Gu N., Biochem. Biophys. Res. Commun. 520(3), 619–626 (2019). 10.1016/j.bbrc.2019.10.077 [DOI] [PubMed] [Google Scholar]
- 106. Sergi D., Luscombe-Marsh N., Naumovski N., Abeywardena M., and O'Callaghan N., Front. Nutr. 8, 663838 (2021). 10.3389/fnut.2021.663838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Williams K., Ingerslev L. R., Bork-Jensen J., Wohlwend M., Hansen A. N., Small L., Ribel-Madsen R., Astrup A., Pedersen O., Auwerx J., Workman C. T., Grarup N., Hansen T., and Barres R., Nat. Commun. 11(1), 2695 (2020). 10.1038/s41467-020-16537-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. del Aguila L. F., Claffey K. P., and Kirwan J. P., Am. J. Physiol. 276(5), E849–E855 (1999). 10.1152/ajpendo.1999.276.5.E849 [DOI] [PubMed] [Google Scholar]
- 109. Wong C. Y., Al-Salami H., and Dass C. R., J. Pharm. Pharmacol. 72(12), 1667–1693 (2020). 10.1111/jphp.13359 [DOI] [PubMed] [Google Scholar]
- 110. Gaster M., Petersen I., Hojlund K., Poulsen P., and Beck-Nielsen H., Diabetes 51, 927 (2002). 10.2337/diabetes.51.4.921 [DOI] [PubMed] [Google Scholar]
- 111. Gaster M., Rustan A. C., Aas V., and Beck-Nielsen H., Diabetes 53, 542 (2004). 10.2337/diabetes.53.3.542 [DOI] [PubMed] [Google Scholar]
- 112. Henriksen T. I., Davidsen P. K., Pedersen M., Schultz H. S., Hansen N. S., Larsen T. J., Vaag A., Pedersen B. K., Nielsen S., and Scheele C., Mol. Metab. 6(7), 770–779 (2017). 10.1016/j.molmet.2017.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Henriksen T. I., Heywood S. E., Hansen N. S., Pedersen B. K., Scheele C. C., and Nielsen S., Front. Physiol. 9, 883 (2018). 10.3389/fphys.2018.00883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M., Heim M. H., and Stoffel M., Nature 474(7353), 649–653 (2011). 10.1038/nature10112 [DOI] [PubMed] [Google Scholar]
- 115. Kornfeld J. W., Baitzel C., Konner A. C., Nicholls H. T., Vogt M. C., Herrmanns K., Scheja L., Haumaitre C., Wolf A. M., Knippschild U., Seibler J., Cereghini S., Heeren J., Stoffel M., and Bruning J. C., Nature 494(7435), 111–115 (2013). 10.1038/nature11793 [DOI] [PubMed] [Google Scholar]
- 116. Gallagher I. J., Scheele C., Keller P., Nielsen A. R., Remenyi J., Fischer C. P., Roder K., Babraj J., Wahlestedt C., Hutvagner G., Pedersen B. K., and Timmons J. A., Genome Med. 2, 9 (2010). 10.1186/gm130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zou K., Turner K., Zheng D., Hinkley J. M., Kugler B. A., Hornby P. J., Lenhard J., Jones T. E., Pories W. J., Dohm G. L., and Houmard J. A., Am. J. Physiol.-Cell Physiol. 319(6), C1011–C1019 (2020). 10.1152/ajpcell.00157.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Varemo L., Henriksen T. I., Scheele C., Broholm C., Pedersen M., Uhlen M., Pedersen B. K., and Nielsen J., Genome Med. 9(1), 47 (2017). 10.1186/s13073-017-0432-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kase E. T., Feng Y. Z., Badin P. M., Bakke S. S., Laurens C., Coue M., Langin D., Gaster M., Thoresen G. H., Rustan A. C., and Moro C., Biochim. Biophys. Acta 1851(9), 1194–1201 (2015). 10.1016/j.bbalip.2015.03.005 [DOI] [PubMed] [Google Scholar]
- 120. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., and Yamanaka S., Cell 131(5), 861–872 (2007). 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 121. Sanches J. M., Zhao L. N., Salehi A., Wollheim C. B., and Kaldis P., “ Pathophysiology of type 2 diabetes and the impact of altered metabolic interorgan crosstalk,” FEBS J. (published online 2021). 10.1111/febs.16306 [DOI] [PubMed] [Google Scholar]
- 122. Gancheva S., Jelenik T., Álvarez-Hernández E., and Roden M., Physiol. Rev. 98(3), 1371–1415 (2018). 10.1152/physrev.00015.2017 [DOI] [PubMed] [Google Scholar]
- 123. Barlow J. P. and Solomon T. P., Am. J. Physiol. Endocrinol. Metab. 314(4), E297–E307 (2018). 10.1152/ajpendo.00353.2017 [DOI] [PubMed] [Google Scholar]
- 124. Aoi W. and Tanimura Y., Front. Endocrinol. 12, 697204 (2021). 10.3389/fendo.2021.697204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Romero A. and Eckel J., Cells 10(8), 2082 (2021). 10.3390/cells10082082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Tamura K., Goto-Inoue N., Miyata K., Furuichi Y., Fujii N. L., and Manabe Y., PLoS One 15(8), e0237095 (2020). 10.1371/journal.pone.0237095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Targher G., Corey K. E., Byrne C. D., and Roden M., Nat. Rev. Gastroenterol. Hepatol. 18(9), 599–612 (2021). 10.1038/s41575-021-00448-y [DOI] [PubMed] [Google Scholar]
- 128. De Chiara F., Ferret-Miñana A., Fernández-Costa J. M., Senni A., Jalan R., and Ramón-Azcón J., Biomedicines 10(5), 958 (2022). 10.3390/biomedicines10050958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Goedeke L., Perry R. J., and Shulman G. I., Annu. Rev. Pharmacol. Toxicol. 59, 65–87 (2019). 10.1146/annurev-pharmtox-010716-104727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Mizgier M. L., Cataldo L. R., Gutierrez J., Santos J. L., Casas M., Llanos P., Contreras-Ferrat A. E., Moro C., Bouzakri K., and Galgani J. E., J. Diabetes Res. 2017, 1328573. 10.1155/2017/1328573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Barlow J. and Solomon T. P. J., Metabolism 91, 1–9 (2019). 10.1016/j.metabol.2018.11.004 [DOI] [PubMed] [Google Scholar]
- 132. Merz K. E., Tunduguru R., Ahn M., Salunkhe V. A., Veluthakal R., Hwang J., Bhattacharya S., McCown E. M., Garcia P. A., Zhou C., Oh E., Yoder S. M., Elmendorf J. S., and Thurmond D. C., Front. Endocrinol. 13, 821849 (2022). 10.3389/fendo.2022.821849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Rutti S., Dusaulcy R., Hansen J. S., Howald C., Dermitzakis E. T., Pedersen B. K., Pinget M., Plomgaard P., and Bouzakri K., Sci. Rep. 8(1), 10072 (2018). 10.1038/s41598-018-28117-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Pandurangan M., Jeong D., Amna T., Van Ba H., and Hwang I., In Vitro Cell. Dev. Biol.-Animal 48(9), 577–582 (2012). 10.1007/s11626-012-9550-8 [DOI] [PubMed] [Google Scholar]
- 135. Nintou E., Karligiotou E., Vliora M., Fatouros I. G., Jamurtas A. Z., Sakellaridis N., Dimas K., and Flouris A. D., Life 11(11), 1227 (2021). 10.3390/life11111227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Jensen-Cody S. O. and Potthoff M. J., Mol. Metab. 44, 101138 (2021). 10.1016/j.molmet.2020.101138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Seo J. A., Kang M. C., Yang W. M., Hwang W. M., Kim S. S., Hong S. H., Heo J. I., Vijyakumar A., de Moura L. P., Uner A., Huang H., Lee S. H., Lima I. S., Park K. S., Kim M. S., Dagon Y., Willnow T. E., Aroda V., Ciaraldi T. P., Henry R. R., and Kim Y. B., Nat. Commun. 11(1), 2276 (2020). 10.1038/s41467-020-16305-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Lan F., Misu H., Chikamoto K., Takayama H., Kikuchi A., Mohri K., Takata N., Hayashi H., Matsuzawa-Nagata N., Takeshita Y., Noda H., Matsumoto Y., Ota T., Nagano T., Nakagen M., Miyamoto K.-I., Takatsuki K., Seo T., Iwayama K., Tokuyama K., Matsugo S., Tang H., Saito Y., Yamagoe S., Kaneko S., and Takamura T., Diabetes 63(5), 1649–1664 (2014). 10.2337/db13-0728 [DOI] [PubMed] [Google Scholar]
- 139. Church T. S., Blair S. N., Cocreham S., Earnest P. C. et al. , JAMA 304, 2253 (2010). 10.1001/jama.2010.1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Karstoft K., Winding K., Knudsen S. H., Nielsen J. S., Thomsen C., Pedersen B. K., and Solomon T. P., Diabetes Care 36(2), 228–236 (2013). 10.2337/dc12-0658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Wallberg-Henriksson H. and Zierath J. R., Nat. Rev. Endocrinol. 11(4), 198–200 (2015). 10.1038/nrendo.2015.24 [DOI] [PubMed] [Google Scholar]
- 142. Losa-Reyna J., Baltasar-Fernandez I., Alcazar J., Navarro-Cruz R., Garcia-Garcia F. J., Alegre L. M., and Alfaro-Acha A., Exp. Gerontol. 115, 114–121 (2019). 10.1016/j.exger.2018.11.022 [DOI] [PubMed] [Google Scholar]
- 143. Martinez-Velilla N., Casas-Herrero A., Zambom-Ferraresi F., Saez de Asteasu M. L., Lucia A., Galbete A., Garcia-Baztan A., Alonso-Renedo J., Gonzalez-Glaria B., Gonzalo-Lazaro M., Apezteguia Iraizoz I., Gutierrez-Valencia M., Rodriguez-Manas L., and Izquierdo M., JAMA Intern. Med. 179(1), 28–36 (2019). 10.1001/jamainternmed.2018.4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Rohling M., Herder C., Stemper T., and Mussig K., J. Diabetes Res. 2016, 2868652. 10.1155/2016/2868652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Rose A. J., Kiens B., and Richter E. A., J. Physiol. 574(Pt 3), 889–903 (2006). 10.1113/jphysiol.2006.111757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Zierath J. R., He L., Gumà A., Odegoard Wahlström E., Klip A., and Wallberg-Henriksson H., Diabetologia 39(10), 1180–1189 (1996). 10.1007/BF02658504 [DOI] [PubMed] [Google Scholar]
- 147. Stephens F. B. and Tsintzas K., Biochem. Soc. Trans. 46(1), 111–118 (2018). 10.1042/BST20170198 [DOI] [PubMed] [Google Scholar]
- 148. Stepto N. K., Benziane B., Wadley G. D., Chibalin A. V., Canny B. J., Eynon N., and McConell G. K., PLoS One 7(12), e53080 (2012). 10.1371/journal.pone.0053080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Yang D., Yang Y., Li Y., and Han R., Ann. Nutr. Metab. 74(4), 313–321 (2019). 10.1159/000500110 [DOI] [PubMed] [Google Scholar]
- 150. Balkau B., Mhamdi L., Oppert J. M., Nolan J., Golay A., Porcellati F., Laakso M., and Ferrannini E., Diabetes 57(10), 2613–2618 (2008). 10.2337/db07-1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Karstoft K. and Pedersen B. K., Immunol. Cell Biol. 94(2), 146–150 (2016). 10.1038/icb.2015.101 [DOI] [PubMed] [Google Scholar]
- 152. Steensberg A., Fischer C. P., Keller C., Møller K., and Pedersen B. K., Am. J. Physiol. Endocrinol. Metab. 285(2), E433–E437 (2003). 10.1152/ajpendo.00074.2003 [DOI] [PubMed] [Google Scholar]
- 153. Pedersen B. K., Eur. J. Clin. Invest. 47(8), 600–611 (2017). 10.1111/eci.12781 [DOI] [PubMed] [Google Scholar]
- 154. Tarum J., Folkesson M., Atherton P. J., and Kadi F., Exp. Physiol. 102(11), 1405–1413 (2017). 10.1113/EP086581 [DOI] [PubMed] [Google Scholar]
- 155. Nikolic N., Gorgens S. W., Thoresen G. H., Aas V., Eckel J., and Eckardt K., Acta Physiol. 220(3), 310–331 (2017). 10.1111/apha.12830 [DOI] [PubMed] [Google Scholar]
- 156. Vepkhvadze T. F., Vorotnikov A. V., and Popov D. V., Biochemistry 86(5), 597–610 (2021). 10.1134/S0006297921050084 [DOI] [PubMed] [Google Scholar]
- 157. Carter S. and Solomon T. P. J., Pflugers Arch. 471(3), 413–429 (2019). 10.1007/s00424-018-2210-4 [DOI] [PubMed] [Google Scholar]
- 158. Aas V., Torblå S., Andersen M. H., Jensen J., and Rustan A. C., Ann. N. Y. Acad. Sci. 967, 506–515 (2002). 10.1111/j.1749-6632.2002.tb04309.x [DOI] [PubMed] [Google Scholar]
- 159. Nikolic N., Bakke S. S., Kase E. T., Rudberg I., Halle I. F., Rustan A. C., Thoresen G. H., and Aas V., PLoS One 7(3), e33203 (2012). 10.1371/journal.pone.0033203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Valero-Breton M., Warnier G., Castro-Sepulveda M., Deldicque L., and Zbinden-Foncea H., Front. Bioeng. Biotechnol. 8, 565679 (2020). 10.3389/fbioe.2020.565679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Pourteymour S., Eckardt K., Holen T., Langleite T., Lee S., Jensen J., Birkeland K. I., Drevon C. A., and Hjorth M., Mol. Metab. 6(4), 352–365 (2017). 10.1016/j.molmet.2017.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Fujita H., Nedachi T., and Kanzaki M., Exp. Cell Res. 313(9), 1853–1865 (2007). 10.1016/j.yexcr.2007.03.002 [DOI] [PubMed] [Google Scholar]
- 163. Lambernd S., Taube A., Schober A., Platzbecker B., Gorgens S. W., Schlich R., Jeruschke K., Weiss J., Eckardt K., and Eckel J., Diabetologia 55(4), 1128–1139 (2012). 10.1007/s00125-012-2454-z [DOI] [PubMed] [Google Scholar]
- 164. Nedachi T., Fujita H., and Kanzaki M., Am. J. Physiol. Endocrinol. Metab. 295(5), E1191–E1204 (2008). 10.1152/ajpendo.90280.2008 [DOI] [PubMed] [Google Scholar]
- 165. Feng Y. Z., Nikolic N., Bakke S. S., Kase E. T., Guderud K., Hjelmesaeth J., Aas V., Rustan A. C., and Thoresen G. H., Am. J. Physiol. Cell Physiol. 308(7), C548–C556 (2015). 10.1152/ajpcell.00314.2014 [DOI] [PubMed] [Google Scholar]
- 166. Park S., Turner K. D., Zheng D., Brault J. J., Zou K., Chaves A. B., Nielsen T. S., Tanner C. J., Treebak J. T., and Houmard J. A., J. Physiol. 597(2), 449–466 (2019). 10.1113/JP276990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Løvsletten N. G., Rustan A. C., Laurens C., Thoresen G. H., Moro C., and Nikolić N., Appl. Physiol. Nutr. Metab. 45(2), 169–179 (2020). 10.1139/apnm-2019-0265 [DOI] [PubMed] [Google Scholar]
- 168. Kugler B. A., Deng W., Francois B., Anderson M., Hinkley J. M., Houmard J. A., Gona P. N., and Zou K., Med. Sci. Sports Exercise 53(6), 1151–1160 (2021). 10.1249/MSS.0000000000002580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Gundelach L. A., Huser M. A., Beutner D., Ruther P., and Bruegmann T., Pflugers Arch. 472(5), 527–545 (2020). 10.1007/s00424-020-02387-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Asano T., Ishizua T., and Yawo H., Biotechnol. Bioeng. 109(1), 199–204 (2012). 10.1002/bit.23285 [DOI] [PubMed] [Google Scholar]
- 171. Asano T., Ishizuka T., Morishima K., and Yawo H., Sci. Rep. 5, 8317 (2015). 10.1038/srep08317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Costford S. R., Bajpeyi S., Pasarica M., Albarado D. C., Thomas S. C., Xie H., Church T. S., Jubrias S. A., Conley K. E., and Smith S. R., Am. J. Physiol. Endocrinol. Metab. 298(1), E117–E126 (2010). 10.1152/ajpendo.00318.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Kitzmann M., Lantier L., Hebrard S., Mercier J., Foretz M., and Aguer C., Biochim. Biophys. Acta 1812(4), 423–430 (2011). 10.1016/j.bbadis.2010.12.007 [DOI] [PubMed] [Google Scholar]
- 174. Nascimento E. B., Osler M. E., and Zierath J. R., Am. J. Physiol. Endocrinol. Metab. 305(11), E1408–E1414 (2013). 10.1152/ajpendo.00212.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Khodabukus A., Prabhu N., Wang J., and Bursac N., Adv. Healthcare Mater. 7(15), e1701498 (2018). 10.1002/adhm.201701498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Kang L., Ayala J. E., Lee-Young R. S., Zhang Z., James F. D., Neufer P. D., Pozzi A., Zutter M. M., and Wasserman D. H., Diabetes 60(2), 416–426 (2011). 10.2337/db10-1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Ahmad K., Lee E. J., Moon J. S., Park S. Y., and Choi I., Cells 7(10), 148 (2018). 10.3390/cells7100148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Danku A. E., Dulf E. H., Braicu C., Jurj A., and Berindan-Neagoe I., Front. Bioeng. Biotechnol. 10, 840674 (2022). 10.3389/fbioe.2022.840674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Song P., Kwon Y., Joo J. Y., Kim D. G., and Yoon J. H., Int. J. Mol. Sci. 20(16), 3893 (2019). 10.3390/ijms20163893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Klein M. S. and Shearer J., J. Diabetes Res. 2016, 3898502. 10.1155/2016/3898502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Arneth B., Arneth R., and Shams M., Int. J. Mol. Sci. 20(10), 2467 (2019). 10.3390/ijms20102467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Chen Z. Z. and Gerszten R. E., Circ. Res. 126(11), 1613–1627 (2020). 10.1161/CIRCRESAHA.120.315898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Xiao Y., Zheng L., Zou X., Wang J., Zhong J., and Zhong T., J. Extracell. Vesicles 8(1), 1625677 (2019). 10.1080/20013078.2019.1625677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Liu J., Sun X., Zhang F. L., Jin H., Yan X. L., Huang S., Guo Z. N., and Yang Y., Front. Endocrinol. 11, 596811 (2020). 10.3389/fendo.2020.596811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Wu J., Dong M., Rigatto C., Liu Y., and Lin F., npj Digital Med. 1, 7 (2018). 10.1038/s41746-017-0014-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Glieberman A. L., Pope B. D., Zimmerman J. F., Liu Q., Ferrier J. P., Kenty J. H. R., Schrell A. M., Mukhitov N., Shores K. L., Tepole A. B., Melton D. A., Roper M. G., and Parker K. K., Lab Chip 19(18), 2993–3010 (2019). 10.1039/C9LC00253G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Beckerman M., Harel C., Michael I., Klip A., Bilan P. J., Gallagher E. J., LeRoith D., Lewis E. C., Karnieli E., and Levenberg S., Sci. Adv. 7(42), eabg3947 (2021). 10.1126/sciadv.abg3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.