Abstract
Drug resistance by pathogenic microbes has emerged as a matter of great concern to mankind. Microorganisms such as bacteria and fungi employ multiple defense mechanisms against drugs and the host immune system. A major line of microbial defense is the biofilm, which comprises extracellular polymeric substances that are produced by the population of microorganisms. Around 80% of chronic bacterial infections are associated with biofilms. The presence of biofilms can increase the necessity of doses of certain antibiotics up to 1000-fold to combat infection. Thus, there is an urgent need for strategies to eradicate biofilms. Although a few physicochemical methods have been developed to prevent and treat biofilms, these methods have poor efficacy and biocompatibility. In this review, we discuss the existing strategies to combat biofilms and their challenges. Subsequently, we spotlight the potential of enzymes, in particular, polysaccharide degrading enzymes, for biofilm dispersion, which might lead to facile antimicrobial treatment of biofilm-associated infections.
Keywords: microbial infections, biofilm dispersion, enzymes, polysaccharides, glycoside hydrolases
Abbreviations: 5-FU, 5-fluorouracil; AHL, N-acyl homoserine lactone; AMP, antimicrobial peptide; CF, cystic fibrosis; eDNA, extracellular DNA; EPS, extracellular polymeric substance; GH, glycoside hydrolase; PDB, Protein Data Bank; PDT, photodynamic therapy; PNAG, poly N-acetyl glucosamine; PL, polysaccharide lyase; QS, quorum sensing; ROS, reactive oxygen species
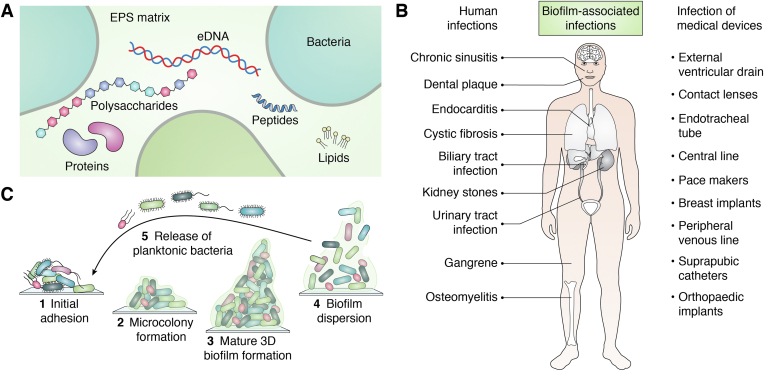
The emergence of multiple drug resistance in pathogenic microbes has led to poor management of bacterial and fungal diseases in both humans and animals. Microorganisms employ multiple defense mechanisms against the host immune system and antimicrobials, which lead to establishment of microbial pathogenesis. Biofilm formation is one of the leading reasons for the emergence of multidrug-resistant bacteria and fungi that affects human health and the world economy severely (1, 2, 3, 4). These microorganisms have two distinct modes of survival: motile and sedentary. Biofilms are representative of bacteria and fungi in the sedentary state of their life cycle as opposed to a motile state. Bacteria and fungi have an innate tendency to cling onto both biotic and abiotic surfaces and ensconce themselves in a self-produced slimy matrix, known as extracellular polymeric substances (EPSs), which mainly comprise polysaccharides, extracellular DNAs (eDNAs), lipids, and proteins (Fig. 1A) (5, 6, 7).
Figure 1.
Biofilms and their impact on human health.A, the major components of the EPS matrix of biofilm. B, biofilm-associated infections in humans. C, the life cycle of microbial biofilm. EPS, extracellular polymeric substance.
Biofilms in bacterial species differ in their EPS composition, the production of which is orchestrated by various genes. Biofilms can increase the necessity of certain antibiotics up to 1000-fold to be effective against microorganisms and provide a protective niche to microorganisms against the host’s immune defense (8, 9, 10, 11, 12). Biofilm-producing microorganisms exhibit altered physiology such as stunted growth, changed phenotype, overexpression of various resistance determinants including efflux pumps, and alterations in membrane and biofilm matrix composition (13, 14). The situation becomes more complex and severe due to the presence of persisters, a small subpopulation of cells that can tolerate exceedingly high doses of antibiotics (15). Biofilms are omnipresent and can be found in human body, natural, and man-made environments. A large number of both gram-positive and gram-negative bacteria are known to be associated with biofilm-mediated infections (16). Reports suggest that biofilm-producing microorganisms are involved in nearly 80% of chronic bacterial infections (1, 17, 18). A few well-known biofilm-associated bacterial infections are urinary tract infections, lung infections, cystic fibrosis (CF), and chronic tonsillitis. Medical implants and devices such as pacemakers, catheters, prosthetics, contact lenses, dentures, etc., are also highly susceptible to biofilm formation (Fig. 1B) (6, 19). Like bacteria, fungi also tend to form biofilms and flourish in aggregated communities enclosed in an extracellular matrix (20, 21). Over 65% of human fungal infections involve biofilm (21). Among fungi-related biofilm-mediated infections, Candida species are the ubiquitous etiologic agents. However, other yeasts and filamentous fungi like Cryptococcus, Saccharomyces, Pneumocystis, Aspergillus, Trichosporon, Blastoschizomyces, Malassezia, Coccidioides, etc. are known to infect individuals with indwelling medical devices such as catheters, pacemakers, and implants (22, 23, 24, 25). Introduction of invasive medical implants into the patient's body forms an appropriate condition for fungi to adhere and form biofilms. Fungi are eukaryotic with complex cellular systems, which add to the difficulty in diagnosing and treating biofilm-mediated fungal infections. The situation is further aggravated by amplified cell density of sessile populations with altered physiological states and the expression of a specific array of genes (26). A recent study found that preformed biofilms were unaffected by high concentrations of most antifungal agents. For example, even newly adherent cells can grow, proliferate, and form biofilms in the presence of high concentrations of fluconazole (a common antifungal medicine that is used to prevent and cure fungal and yeast infections) (27). As a consequence, biofilm-mediated fungal infections are associated with high mortality rates.
Biofilms can be monomicrobial and polymicrobial, consisting of various bacterial strains, fungi, algae, etc., which results in a highly complex structure (5). Biofilm formation occurs in five stages: (1) migration and adhesion of cells on a suitable substratum; (2) microcolony formation by EPS secretion and cell aggregation; (3) multiple layer formation over the microbial colonies and their maturation; (4) attaining maximum cell growth and cell density; and (5) release of the mature and healthy cells to nearby sites for spreading the infection further (Fig. 1C) (28, 29, 30, 31).
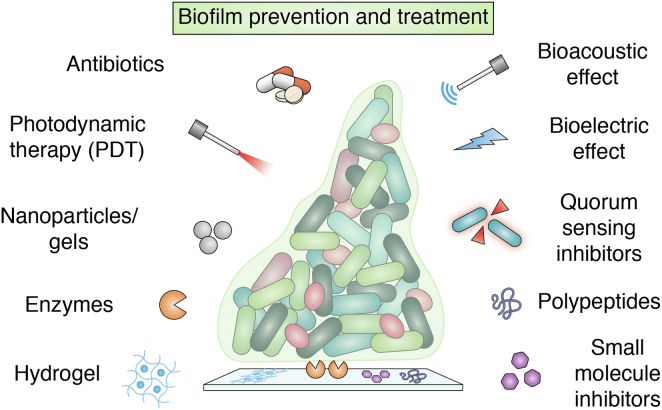
Since the first report of biofilms in the seventeenth century (32), research on biofilm formation by microorganisms has steadily increased as their contribution to pathogenicity is increasingly recognized. The current strategies to treat biofilms include both inhibition of biofilm formation and eradication of preformed biofilms. Biofilm formation can be inhibited by preventing the initial attachment of cells to surfaces or by arresting their maturation. Initial adherence of cells can be blocked either by modifying the attachment surface or preconditioning it with antimicrobials, which have been widely explored (33, 34, 35). In contrast, eradication of the preformed biofilms is more challenging as the recalcitrant polymer matrix must be degraded, leading the biofilm to disperse. Strategies that have been developed against biofilm-associated infection include administering antimicrobial, inhibition of quorum sensing (QS), and photodynamic therapy (Figs. 2 and 3). However, the major limitation of the existing methods is their poor biocompatibility. The quest for biocompatible antibiofilm agents has spurred interest in utilizing enzymes as potential therapeutic agents to treat biofilm-associated infections.
Figure 2.
Methods to prevent and treat biofilms.
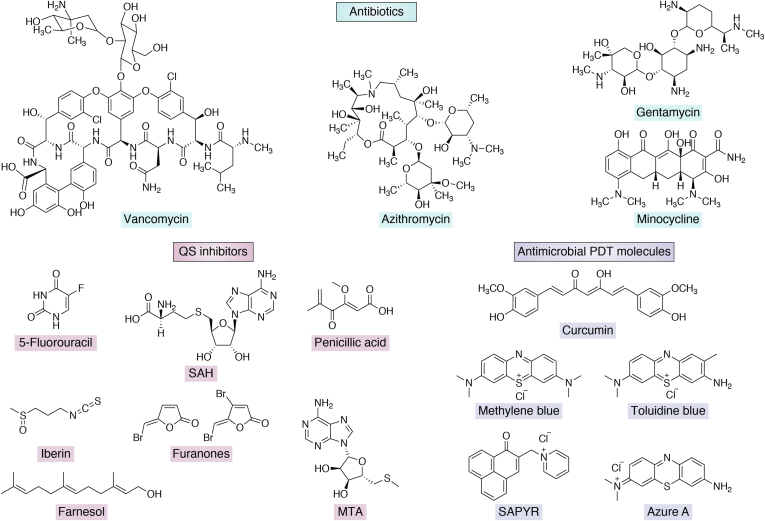
Figure 3.
Examples of small molecules used for the treatments of biofilm-associated infections.
In this review, we briefly summarize the utility and limitations of the existing methods to combat biofilms; more details on these approaches can be found in various recent review articles (5, 9, 28, 36, 37, 38). Our main focus is on enzymatic biofilm dispersion, which has garnered substantial interest in recent years. We discuss an emerging enzyme-based strategy targeting one of the major components of the EPS matrix, the polysaccharides, for efficient dispersion of biofilms under physiological conditions.
Existing methods to combat biofilms
Antimicrobial coating/surface modification
Minimizing the initial microbial adherence to a substratum is a promising strategy to impede biofilm development. Adhesion depends on features such as surface roughness, charge, topography, stiffness, and environmental factors such as hydrodynamics (39). Disruption of the initial adhesion can be achieved through surface modification employing antimicrobial coatings, which is a widely explored method against biofilms (40). For instance, coating albumin on polyethylene/polypropylene surface of medical devices significantly reduces bacterial adhesion (41). However, albumin coating requires additional chemical modification using functionalized cyclodextrins to facilitate the adsorption of albumin (42). Modification of intubations and prostheses made of silicones grafted with C1 and C8 alkyl side chains exhibited ∼92% reduction in biofilm formation by Candida albicans in vitro (43). Similarly, coating the surface with cationic peptides Histatin5 and its synthetic variants led to a 93% reduction in biofilm formation by C. albicans in vitro (44, 45).
Nanoparticles were also found to be effective in reducing the surface adhesion of bacteria (46). Metal nanoparticles possess a positive charge that facilitates their electrostatic attraction to the negatively charged bacterial membrane and, therefore, can perturb the bacterial cells prone to adhere to the metal surface. Reports suggest that silver nanoparticles disrupt the bacterial cell membrane, trigger reactive oxygen species (ROS) generation, and thereby denature proteins, damage DNA, and affect the respiratory enzymes (47). Silver nanoparticle-coated medical implants such as urinary catheters and silver-loaded hydrogels are currently in use. Likewise, selenium, titanium, copper oxide, iron oxide, molybdenum, and curcumin nanoparticles/quantum particles have been explored (48, 49, 50, 51, 52). Although this strategy looks promising, it suffers from a lack of longevity in the application. Additionally, applying the metallic coating often requires elevated temperatures that may damage the nonmetallic implant materials (53).
Antimicrobial peptides (AMPs) have also been employed as surface coating agents against biofilm formation. AMPs exhibit broad spectrum antibacterial activities against both the gram-positive and gram-negative bacteria. AMPs target the functionality and stability of membrane and can kill microbes in the planktonic state, sessile state, and even in dormancy by pore formation (54, 55). Prominently, melimine has shown a broad spectrum activity targeting methicillin-resistant Staphylococcus aureus, Pseudomonas aeruginosa, fungi, protozoa, etc. (56) Various studies have demonstrated successful covalent immobilization of AMPs on medical implants and materials such as glass, titanium oxide, resin beads, and silicone surfaces. Nevertheless, the high production costs of AMPs and poor optimization of their properties have limited their clinical applications and commercialization (57).
Recent studies have shown that implants coated with biodegradable polymers and hydrogels could prevent the initial stages of biofilm formation. PEG and hyaluronan-based hydrogels are widely explored, which efficiently encapsulate and systematically release antibiotics (58, 59). Controlled release of antibiotics to the implant site helps to minimize side effects. Even though antimicrobial agents play a significant role as a preventive measure by reducing the frequency with which implants need to be replaced, there remain challenges associated with the stability and longevity of coatings, toxicity elicited by them, and integration of surrounding indigenous tissues with the implant (60). Antimicrobial coatings can unpredictably promote biofilm formation due to the deposition of initial dead microorganisms killed by the coating (61). Further, the slow release of antibiotics can lead to the development of bacterial resistance (62).
QS inhibition
Another approach to inhibiting biofilm formation involves targeting the bacterial communication system known as QS that regulates a wide variety of bacterial physiology such as motility, virulence, symbiosis, competence, conjugation, sporulation, antibiotic production, as well as biofilm formation (63). After attaining the threshold cell density, bacteria produce N-acyl homoserine lactones (AHLs) and furanones (among gram-negative isolates), or autoinducer peptides (among gram-positive isolates), or autoinducer-2 (among some gram-positive and some gram-negative isolates), which help in modulating specific gene expression for the EPS production and expression of virulence factors (64, 65). AHLs are the primary QS molecules in gram-negative bacteria, which are produced by cognate AHL synthases. When the bacterial population increases and AHL molecules exceed their threshold concentration, specific receptors belonging to the class of DNA-binding transcription factors sense it and alter gene expression (66). Autoinducer peptides are signaling molecules released by membrane transporters. As their concentration rises, they bind to the corresponding histidine kinases, which phosphorylate downstream response regulators and ultimately change the expression of target genes (67). Although QS is not a prerequisite for bacterial growth, its quenching can be an excellent antivirulence strategy (68) and therefore help in inhibiting biofilm formation. Various enzymes, peptide-based agents, and small molecules have been explored extensively as QS inhibitors (69, 70). Specific lactonases and acylases were shown to degrade AHLs (71, 72). Analogs of the precursors of AHLs such as 5′-methylthioadenosine, S-adenosyl-homocysteine (Fig. 3), butyryl-SAM, and sinefungin have been employed against P. aeruginosa (73, 74). Further, the transition state analogs of 5′-methylthioadenosine have been shown to interrupt the QS of Vibrio cholerae and Escherichia coli (75). Investigators have also explored QS signaling antagonizing analogs such as N-octanoyl cyclopentylamides, N-nonanoyl cyclopentylamides, and N-decanoyl cyclopentylamides that exhibited anti-QS activity. Interestingly, N-decanoyl cyclopentylamides was also reported to exhibit strong antibiofilm activity (76). However, only a few of these quorum-quenching and antagonizing molecules have reached clinical trials owing to their ineffectiveness against the preformed, mature biofilms (77, 78). Azithromycin is used as a QS inhibitor for treating pulmonary transplanted and CF patients in clinical trials (79, 80, 81). In vitro, it disrupts the QS in P. aeruginosa strains; however, in vivo it was not effective (82). Natural QS inhibitors such as garlic extracts were also ineffective in clinical trials on patients (83). 5-Fluorouracil (5-FU), which is used as an anticancer drug, has been shown to prevent the expression of QS-regulated virulence factors in P. aeruginosa; therefore, its use as a coating agent for catheters was suggested in the clinical trials (84). 5-FU inhibits thymidylate synthetase, thereby inhibiting DNA replication. Studies suggest that 5-FU has various side effects on patients, and the molecule can affect the cardiovascular system (85). However, recently, Sedlmayer et al. have reported anti-QS activity of 5-FU against methicillin-resistant S. aureus in the murine model (86).
Bioelectric effect
Bacterial cells are sensitive to electric fields; in fact, electric fields have been used for making competent cells. In combination with antibiotics, electrical methods such as applying DC voltages, low AC currents, and pulsed electric fields have proved synergistic against biofilms produced by P. aeruginosa and S. aureus in vitro (87). Electric field applied in conjunction with QS inhibitors inhibited E. coli biofilm growth and showed propitious synergetic effects with gentamycin (88). These methods were even found effective in preventing planktonic bacteria from initiating biofilm formation. In spite of showing promising results in in vitro studies (77, 89), the bioelectric effect is minimally tested in animals and calls for further in vivo experiments before any conclusions on its utility can be made.
Bioacoustic effect
Ultrasonication can improve in the permeability of the biofilm to antibiotics (90, 91, 92), thereby reducing the amount of antibiotics required to inhibit bacterial growth. Ultrasonication combined with antibiotics substantially reduced biofilms formation by E. coli, Staphylococcus epidermidis, and P. aeruginosa in catheters (11, 77). The acoustic ultrasonic wave acts as a repulsive force by interfering with the attachment of planktonic bacteria to the surface. Most of the in vitro studies were performed in a sonication bath with the biofilm formed on implants. In vivo studies were carried out by administering antibiotics along with the ultrasound treatment using a small portable transmitter (93). The major challenge for in vivo studies is limited application time, which results in inefficient therapy, especially with indwelling medical devices.
Antimicrobial photodynamic therapy
Antimicrobial photodynamic therapy (PDT) is a noninvasive therapeutic approach that employs photosensitizers combined with near-infrared lasers and oxygen to suppress biofilm-associated infections (94, 95). PDT involves two steps: first a photosensitizer is administered and then light irradiation generates photoexcited molecules that react with ambient oxygen to generate ROS. The ROS react with the proteins and eDNA present in the EPS and thereby disrupt the biofilm. The photosensitizer molecules are chosen based on their ability to bind to microbial cells, as efficient attachment is important for antibiofilm activity—a selection of these molecules is depicted in Figure 3. It is possible that the photosensitizer molecules are sequestered by EPS or partially penetrate through the biofilm or bind to microbial cells. In vitro, antimicrobial PDT has shown promising efficacy in biofilm eradication. Further, PDT has proven efficient in in vivo models such as implants inserted in mice, biofilm-associated wounds, burn injuries, etc. Clinical studies suggest that PDT can treat oral biofilm formed on teeth with negligible side effects (96). However, overaccumulated photosensitizer on untargeted areas can cause burns, redness, pain, and swelling. Therefore, this technique is mainly applied where the topical or local administration of photosensitizer is possible, rather than by intravenous or oral systemic administration (95).
Enzymatic degradation of polysaccharides—an emergent approach of biofilm dispersion
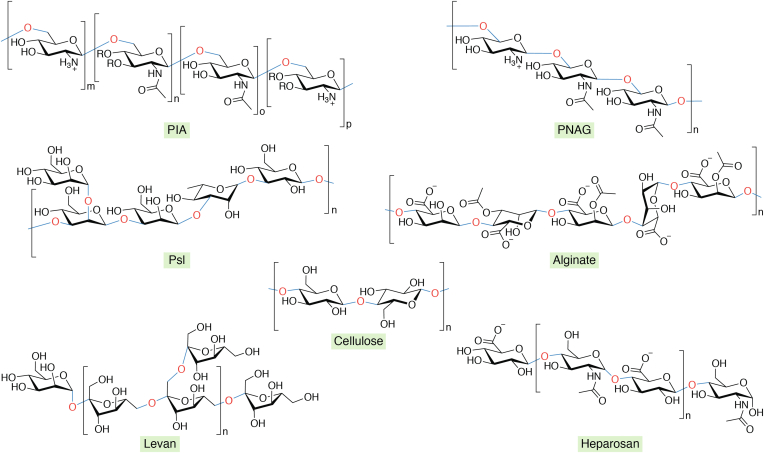
Success in the war against drug-resistant microbes invariably requires effective dispersion of biofilms under physiological conditions. The EPS matrix contributes up to ∼90% of the total biofilm. The dissolution of the EPS matrix of mature biofilm to gain access to the indwellers (microbes that live inside the biofilm) is an emerging area of biofilm research. Polysaccharides are the major component of the EPS matrix, which helps microbes to adhere to a variety of different surfaces. Further, polysaccharides provide immense strength and inertness to the biofilm and thereby protect the microbial population from desiccation, filter out antimicrobials, act as nutrient reservoirs, and facilitate formation of a suitable structured microenvironment for microbes to persist (97). For instance, the primary polysaccharide associated with S. aureus and S. epidermidis biofilms is polysaccharide intercellular adhesion, a partially de-N-acetylated homopolymer composed of β-1,6-linked 2-amino-2-deoxy-D-glucopyranosyl residues (98). Bacteria can also modulate the degree of N-acetylation and O-succinylation depending upon their requirements (99). Polysaccharide intercellular adhesion is referred to as poly N-acetyl glucosamine (PNAG) in gram-negative bacteria, which lacks the machinery for O-succinylation (100, 101). Likewise, P. aeruginosa biofilms contain three different polysaccharides, namely Pel, Psl, and alginate. The chemical structure and composition of Pel were unclear for a long time. Recent studies have suggested that Pel is a cationic exopolysaccharide with partially de-N-acetylated 1,4-linked N-acetylgalactosamine (102, 103, 104). The nonmucoid and mucoid isolates of P. aeruginosa secrete Psl, which is a polymer of pentasaccharide repeating units consisting of D-mannose, D-glucose, and L-rhamnose (103, 105). Mucoid P. aeruginosa isolates produce Psl and alginate that is a random linear polymer of acetylated 1,4-linked β-D-mannuronic acid and its C5 epimer α-L-guluronic acid (106, 107). Pel and Psl are found in the sputum of individuals with CF, and these exopolysaccharides facilitate aggregation of P. aeruginosa in CF airways, which helps in biofilm formation that results in chronic infection (103). Further, capsular polysaccharides contain repeating monosaccharides and are extensively hydrated, mainly involved in the biofilms of the members of Enterobacteriaceae such as Klebsiella pneumoniae and E. coli. Besides, in their biofilms, both cellulose and colanic acid or M antigen are present (108). The biofilms of Streptococcus mutans are primarily composed of the neutral homopolymer of β-D-fructans with irregular and extensive branching, commonly termed levans (97). A list of the well-characterized polysaccharides associated with the biofilms produced by various pathogenic bacteria and fungi is shown in Table 1 (97, 102, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126). Further, the structures of a few of these polysaccharides are depicted in Figure 4.
Table 1.
Various pathogenic microbes and the polysaccharides present in their biofilms
| Pathogenic microbes | Major polysaccharides in the biofilm |
|---|---|
| Salmonella enterica | Cellulose (β-1,4) |
| Pseudomonas aeruginosa | Alginate (β-1,4) and (α-1,4), Pel (1,4), Psl (α-1,2) (β-1,3), (α -1,3) |
| Streptococcus mutans | Glucans (α-1,6), levan (β-D-fructans) |
| Staphylococcus epidermidis | (β-1,6), 2-deoxy-2-amino-D-glucopyranosyl residues, PNAG (β-1,6)a |
| Staphylococcus aureus | PNAG (β-1,6)a |
| Escherichia coli (Uro-pathogen) | PNAG (β-1,6), Cellulose (β-1,4), colanic acid (α-1,4 & β-1,3) |
| Klebsiella pneumoniae | PNAG (β-1,6) |
| Enterobacter spp. | Colanic acid (α-1,4 & β-1,3), N-acetylheparosan |
| Proteus mirabilis | N-acetyl-D-glucosamine (β-1,4), N-acetyl-L-fucosamine (α-1,3), D-glucuronic acid (α-1,3) |
| Bacterioides fragilis | {,3) α-D-AATGalp(1,4)[ β-D-Galf(1,3)] α-D-GalpNAc(1,3) β-D-Galp(1, }b |
| Serratia marcescens | Stewartan, Emulsan, polysaccharide B, capsular polysaccharide |
| Aspergillus fumigatus | Galactosaminogalactan (partially deacetylated heteropolymer of α -1,4-linked galactose and N-acetyl galactosamine) |
| Candida albicans | α-1,2 branched and α-1,6 mannans |
In gram-positive bacteria PNAG is known as polysaccharide intercellular adhesion (PIA) that has O-succinylated groups.
AATGal: Acetamido-amino-2,4,6-trideoxygalactose. The galactopyranosyl residue has a pyruvate substituent. D-Galp: D-galactopyranose; D-GalpNAc: N-acetyl-D-galactopyranosamine; D-Galf: D-galactofuranose.
Figure 4.
Structures of the polysaccharides found in the EPS matrix of biofilms produced by various pathogenic bacteria.O-succinylation of PIA is indicated by the R group. EPS, extracellular polymeric substance; PIA, polysaccharide intercellular adhesion.
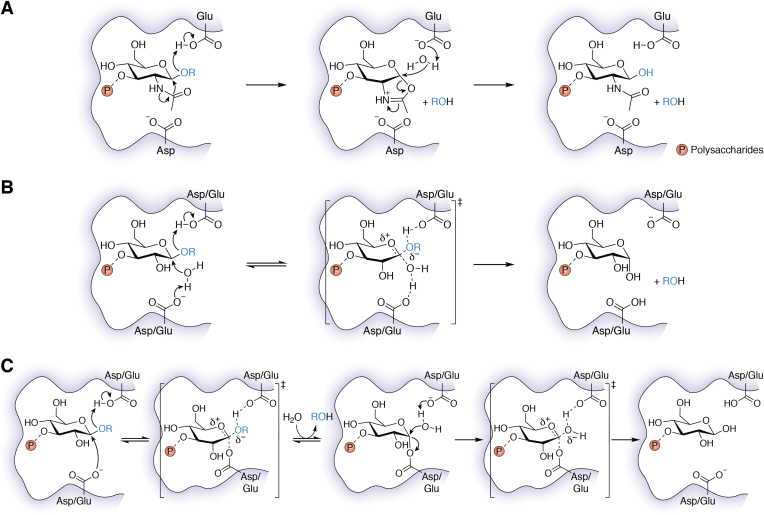
Degradation of the polysaccharides could weaken the biofilm substantially and render the sessile microbial population easily accessible to antimicrobial, thereby ensuring better clearance of the microbes from the infected areas. Interestingly, indwellers of biofilm are known to produce saccharolytic enzymes to initiate biofilm dispersal events in response to nutrient deprivation (127). It has been found that the gram-negative bacterium Actinobacillus actinomycetemcomitans produces a glycoside hydrolase (GH) enzyme, Dispersin B, for the dispersal of self-produced mature biofilm (128). Dispersin B is a β-N-acetylglucosaminidase enzyme that catalyzes cleavage of the β-(1,6)-linked PNAG, which is a major component of the exopolysaccharides of biofilms produced by E. coli, Pseudomonas fluorescens, Actinobacillus pleuropneumoniae (gram-negative bacteria), as well as S. epidermidis and S. aureus (gram-positive bacteria). Binding of the polysaccharide substrate to the active site of Dispersin B is assisted by a set of conserved residues: one Glu, three Trp, and one Tyr (129). The site of cleavage of the glycosidic bond (endo- or exo-) of the polysaccharide by Dispersin B depends on the nature of the substrate (129, 130, 131). The active site architecture of the enzyme (Protein Data Bank [PDB] ID: 1YHT) (132) displays a conserved Asp-Glu dyad, where the Glu residue performs the general acid-base catalysis to hydrolyze the glycosidic bond through an oxazolinium intermediate (Fig. 5A). The Asp residue assists the N-acetyl group for nucleophilic attack. The ability of Dispersin B to disrupt biofilms has stimulated several attempts to utilize a similar strategy for effective dispersion of biofilms.
Figure 5.
Plausible mechanisms of cleavage of glycosidic linkages of polysaccharides in biofilm by GH enzymes. A, the mechanism of Dispersin B, which involves oxazolinium intermediate during the catalysis. B, the inverting mechanism of GH, which follows a concerted pathway. C, the retaining mechanism of GH, which involves the formation of an enzyme-substrate covalent intermediate. In all cases, conserved Asp/Glu residues are involved in acid-base catalysis. The orange circle with P stands for polysaccharides. GH, glycoside hydrolase.
Sourjik et al. have engineered an E. coli strain by introducing Dispersin B into the microorganism, which could disrupt the biofilm containing PNAG (133). In an alternative strategy, Collin et al. have developed a T7 phage that induces a gene encoding an EPS degrading enzyme to the phage genome. The engineered phage led to efficient dispersal of E. coli biofilm and bacterial cell lysis (134). However, the specificity of bacteriophage targeting the host systems and release of toxins upon bacterial lysis makes this approach unpromising for clinical applications. Further, quick clearance by phagocytes in the human body makes phage therapy challenging and limited to the laboratory scale disruption of biofilms. Staphylococcal species and P. aeruginosa are the common bacterial strains found in most biofilm-related infections (77). Recently, Baker et al. have shown that nanomolar concentrations of GHs (PelA and PslG) present in the periplasm of P. aeruginosa can disrupt biofilm formed by the microorganism in vitro (135, 136). They have also shown that treatment of Aspergillus fumigatus (an opportunistic fungal pathogen) biofilms with GHs Sph3 or PelA significantly improved the activity of the antifungal agents such as posaconazole, amphotericin B, and caspofungin. Both Sph3 and PelA were found to be noncytotoxic. The intratracheal administration of Sph3 was well tolerated and led to a substantial reduction of fungal outgrowth within the lungs of a neutropenic mouse model (136). Asker et al. have shown that PslG can be uniformly immobilized to commercially available medical grade polyethylene, polyurethane, and polydimethylsiloxane catheter tubing. Moreover, the chemically conjugated PslG was found substantially reduce the formation of P. aeruginosa biofilm both in vitro and in vivo (137, 138). Rumbaugh et al. have investigated the effect of two commercially available GHs, α-amylase from Bacillus subtilis and cellulase from Aspergillus niger, on biofilms produced by S. aureus and P. aeruginosa. Their investigation suggests that the enzymes could disrupt the polymicrobial biofilm produced by the two microorganisms (139). Structural investigation of α-amylase from B. subtilis in the polysaccharide bound form (PDB ID: 1BAG) demonstrates the involvement of three conserved active site acidic residues, two Asp and one Glu, forming a triangle around the cleavage site (140). While one conserved residue Glu and Asp serves as the general acid and base for catalysis, the other conserved Asp residue has a vital role in substrate binding. Similar to α-amylase, cellulase harbors two conserved Glu residues at the active site (PDB ID: 5I77) for catalysis (141).
Both the α-amylase and cellulase are well-known GHs that have been employed for various industrial applications. α-Amylase catalyzes cleavage of the α-(1,4) glycosidic bonds of starch, glycogen, and several other oligosaccharides (140). Cellulase catalyzes hydrolysis of the β-(1,4) glycosidic bond, which is one of the most commonly found linkages in the exopolysaccharides present in biofilms formed by various pathogenic strains such as S. aureus, Salmonella enterica, P. aeruginosa, E. coli, etc. (142) Besides, preclinical studies suggest that cellulase and α-amylase treatment can enhance antibiotic intervention to clear the infection in a murine surgical excision wound model (18). In another study, the Rumbaugh group found six GHs that could disrupt monomicrobial and polymicrobial biofilms produced by S. aureus and P. aeruginosa (143). These pioneer studies demonstrate that GHs could be generally effective for biofilm dispersal. Although the mechanisms of these enzymes have not been investigated in the context of biofilm dispersion, it can be presumed that the enzymes follow the general acid-base catalysis involving conserved Asp/Glu residues to cleave the glycosidic linkages of the polysaccharides present in the biofilm (Fig. 5, B and C) (144, 145, 146).
Jee et al. have demonstrated that the combination of α-amylase and protease (bovine pancreas type-I) significantly inhibits the biofilm growth of E. coli, S. aureus, and methicillin-resistant S. aureus (147). Polysaccharide lyase (PL) is another class of enzymes that cleaves uronic acid–containing polysaccharides using a β-elimination mechanism (148) and thereby disrupt the EPS matrix of the biofilm. Again, cleavage enhances the effect of antibiotics and phagocytosis in the eradication of biofilm-associated infections (149). Recently, a PL, isolated from K. pneumoniae bacteriophage (SH-KP152226), was shown to degrade biofilm produced by multidrug-resistant K. pneumoniae (150). Alginate lyases from the PL class of enzymes have also been explored for the disruption of P. aeruginosa biofilms (151). Although various reports indicate the combinatorial therapy of antibiotics with alginate lyase effectively reduces the minimum inhibitory concentration of multiple antibiotics (149), Lamppa et al. have shown that alginate lyase–mediated biofilm dispersion and the synergy of antibiotic are not coupled with the enzyme activity (152). Although alginate lyases manifest biofilm-dispersive properties, there is controversy regarding the antibiofilm effect of this enzyme. DNase I is another class of enzyme known to degrade biofilms. Disruption of eDNA of the biofilm matrix increases the vulnerability of the biofilm to antibiotics as the negative charge of eDNA is known to sequester cationic antibiotics (153). DNase I, sold as Dornase alfa, is clinically prescribed as a mucolytic agent for CF patients. It acts on the DNA of sputum, helping to reduce its viscosity and allowing it to be cleared (154). Nevertheless, the efficacy of the treatment in the CF patients is more related to compliance, as the enzyme needs to be nebulized and inhaled over 1 h, which is uncomfortable for the patients. In vitro studies suggest that DNase I treatment might be beneficial in preventing the establishment of chronic P. aeruginosa infection of the CF lung by inhibiting biofilm formation (155). Recently Pirlar et al. have demonstrated that the synergistic action of DNase I and trypsin helped to disperse the polymicrobial biofilm of S. aureus and P. aeruginosa in the wound-like medium, which resulted in a 2.5-fold reduction in the minimum concentration of antibiotic needed for biofilm eradication (156).
Nevertheless, investigations suggest that enzymes, particularly the polysaccharide degrading enzymes, might hold a key to the effective dispersion of biofilms produced by many pathogenic microbes. The chemical structures and the nature of the glycosidic linkages of various polysaccharides present in the biofilms of a large number of microbes have been explored (Table 1). Therefore, selectively targeting biofilms using GH enzymes might be feasible. A list of these enzymes used for biofilm dispersion is shown in Table 2 (135, 139, 157, 158, 159, 160, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 171, 172, 173, 174, 175, 176, 177, 178, 179).
Table 2.
Reported glycoside hydrolases for biofilm dispersion
| Glycoside hydrolase | Targeting glycosidic linkage | Monosaccharide composition of the polysaccharides |
|---|---|---|
| Dispersin B | GlcNAc -(β-1,6)- GlcNAc |
N-acetyl-D-glucosamine |
| α-Amylase | Glc-(α-1,4)-Glc | D-glucose |
| Cellulase | Glc-(β-1,4)-Glc | D-glucose |
| PslG | Manp-(β-1,3)- Manpa | D-mannose, D-glucose and L-rhamnose |
| PelA | (1,4)b | Partially de-N-acetylated N-acetylgalactosamine |
| β-Mannosidase | Man-(β-1,4)-Man | D-mannose |
| α-Mannosidase | Man-(α-1,3)-Man | D-mannose |
| PgaB | GlcN-(β-1,6)-GlcN | D-glucosamine |
| Ega3 | GalN-(α-1,4)-GalN | D-galactosamine |
| Sph3 | GalNAc-(α-1,4)-GalNAc | N-acetyl-D-galactosamine |
| CcsZ | Glc-(β-1,4)-Glc | D-glucose |
| PssZ | ManN-(β-1,4)-ManN | D-mannosamine |
The targeting glycosidic linkage of PslG is predicted.
PelA enzyme works on Pel, a linear cationic polysaccharide composed of partially de-N-acetylated 1,4 linked N-acetylgalactosamine.
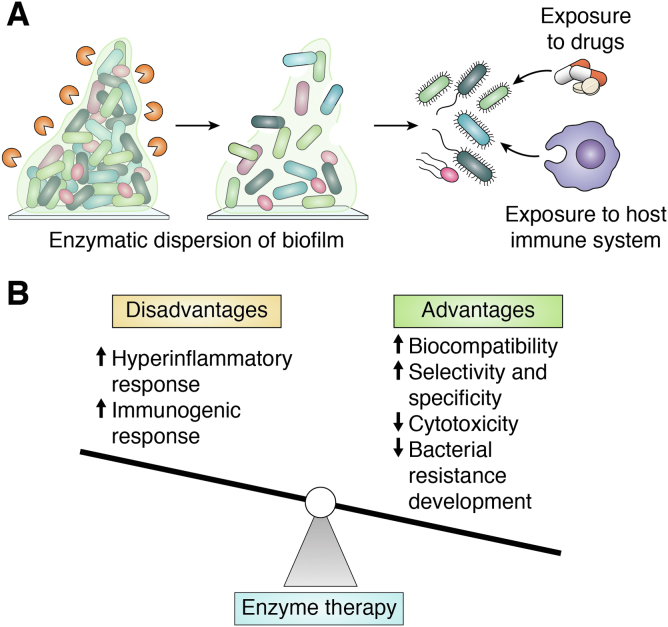
The pilot studies of biofilm dispersion (vide supra) by various GH enzymes serve as the proof of concept that GH enzymes have substantial potential for biofilm dispersion. The ability of GH enzymes to disperse mature biofilms both in vitro and in vivo makes them unique from other physicochemical methods (180). Many GHs have been studied from thermophilic fungi due to interest in these enzymes for recalcitrant biomass degradation at high temperatures for biofuel production. However, such enzymes function poorly under physiological conditions, restricting their ability to effectively degrade the EPS matrix's polysaccharides in biofilms. Therefore, mesophilic GH enzymes, preferably from mammalian microbiome, which would catalyze biofilm dispersion under physiological conditions with minimal immunogenic effect, are urgently needed to combat biofilm-related infections. The CAZy database holds information on thousands of carbohydrate-active enzymes from various microorganisms, and the recent surge in the metagenomic analysis of biomass-degrading microbes has tremendously increased the entries of novel GHs in the CAZy database (181). Such analysis also includes the discovery of numerous saccharolytic enzymes from ruminant animals such as cows and goats that have cellulose-rich diets (182, 183, 184). This observation suggests that some of the most active mesophilic cellulolytic enzymes from the mammalian microbiome may serve as potential candidates for effective dispersion of biofilms. Such enzymes can play a pivotal role in degrading biofilm by which the microbes become planktonic, which then can be killed by the low dose of drugs. EPS-degrading enzymes thus may be of potential therapeutic value in treating microbial infections. These enzymes may also be useful to dislodge microbial colonies from the implants, which otherwise can make the implant nonfunctional. It is important to note that enzymes are typically noncytotoxic to sessile microbes; instead, they will disperse the biofilm by disrupting the EPS matrix and provide easy access of the drugs or the host immune system to the indwellers (Fig. 6A). Therefore, broad spectrum antimicrobial activity could be achieved by combining these enzymes with antimicrobial drugs. Such combinatorial therapy has shown promising results in the efficient treatment of chronic wound infections (27, 139). Nonetheless, a few challenges associated with the microbial enzyme therapy need to be addressed: these include the immunogenicity, short in vivo half-life of the enzymes, cost, and applicability of enzymes in the clinic. Immunogenicity toward foreign proteins is unavoidable in the host systems, which may require further protein modification such as PEGylation (185, 186), development of chimeric protein system (187, 188), or chemical modifications (189) to nullify the off-target effects. PEG is a nontoxic and amphipathic polymer, widely used for modulating enzyme activity and pharmacokinetics to protect from immunoreactivity, immunogenicity, and in vivo degradation of the enzymes (190). In addition, enzyme encapsulation and modification with bioconjugates such as antibodies, peptides, hormones, vitamins, and DNA are under study to enhance the in vivo stability and reduce immunogenicity and clearance (191, 192). Cost, however, may not be a major factor for utilizing enzymes for biofilm dispersion. Heterologous overexpression and purification of enzymes using microorganisms such as E. coli and yeast has revolutionized the field of protein biochemistry. The process gives an opportunity for strain manipulation, overexpression and faster protein production, easy scale up, recovery, and purification (193). In conjunction with the inexpensive gene synthesis, the approach has become an economically viable option for protein production.
Figure 6.
Enzymatic dispersion of biofilms: advantages and disadvantages.A, enzymatic dispersion of biofilm can lead to facile access of the drugs and the host immune system to the indwellers. B, advantages and disadvantages of enzyme therapy. The disadvantages of enzyme therapy can be dealt with established methods. The black arrows indicate the likely increase or decrease of various responses and factors related to the enzyme therapy.
Enzyme therapy on mixed species biofilm is still a challenge due to the substrate-specific nature of most enzymes. Thus, efficient dispersal of biofilm requires multienzyme formulations capable of degrading polysaccharides, eDNA, and QS molecules present in the EPS matrix (194, 195). Moreover, the enzymatic unmasking of biofilm can cause huge microbial exposure to the nearby tissues, which may elicit hyperinflammatory responses. These responses may vary by organism and the site of infection; however, the combination of enzymes with antimicrobials can successfully overcome these challenges (139). A great advantage of implementing enzyme therapy for biofilm eradication is that it is very unlikely that the biofilm-forming pathogens will rapidly develop resistance to the enzymes since the targets of the enzymes are the polysaccharides of the EPS matrix and not the microorganisms. It is noteworthy that bacteria can modify the molecular mass of polysaccharides and can introduce nonsugar residues by expressing enzymes such as pyruvyl or acetyl transferases (196). Also, they can change the comonomer composition by employing epimerases (197). Further, they can alter elastomeric properties by covalently crosslinking the polymers. Nevertheless, GHs mainly target the glycosidic linkages, so these modifications will not affect the inherent efficacy of the enzymes directly but may restrict the access of the enzyme to the linkages. However, such limitations may be overcome through protein engineering. Nevertheless, the high selectivity, specificity, and biocompatibility of the enzymes make them superior to other biofilm therapeutics (Fig. 6B).
Enzyme therapies have been successfully employed in various fields, such as the medical and food industries. For example, collagenase is used for dispersion of ulcers and burns and is also used for the neurosurgical treatment of Dupuytren’s contracture (198, 199). Lipases from microbial sources are used to treat gastrointestinal disturbance and dyspepsia (200), and bovine pancreatic enzymes are used for the treatment of pancreatic cancer, chronic pancreatitis, and CF (201, 202). Even though the preclinical studies of enzymatic biofilm dispersion are promising, (203) only limited demonstrations of the safety and efficacy of the enzymes in animal models are currently available, and more in vivo studies are needed to bring them to clinical trials.
Conclusion and perspectives
With recent advancements in our understanding of the mechanisms of microbial infection, clinical solutions have been employed to treat patients, yet biofilm-associated infections loom as a grave concern to human health. Biofilm-associated infections develop slowly, and the symptoms show up at a much later phase of infection. Facile disruption of biofilms without adverse side effects is highly desired for effective antimicrobial treatments of biofilm-associated infections. Although current approaches such as antimicrobial coating, bioelectric effect, bioacoustic effect, and photodynamic therapy have shown promise in vitro, only in a very few cases biocompatibility has been demonstrated in vivo. A major limitation of the monoantimicrobial and combinatorial antimicrobial drug therapy is the need for a much higher dose at the biofilm-associated infection site. The dose required is often significantly more than the allowed therapeutic dose, leading to severe hepatotoxicity and nephrotoxicity. The challenges associated with the existing methods have motivated scientists to innovate more effective solutions to biofilms. In this review, we have highlighted one such approach—the enzymatic dispersion of biofilms, which is an emerging avenue of research. In our assessment, GH enzymes, preferably from the mammalian microbiome, which target polysaccharides of the EPS matrix, hold great potential for developing first-in-kind enzyme therapy for dispersion of mature biofilm under physiological conditions. The specificity and lower chances of rejection by the human body may give enzymes an upper hand over the existing physicochemical strategies.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We acknowledge Prof. Neil Marsh, University of Michigan, Ann Arbor, USA for the kind help with editing this manuscript.
Author contributions
D. D. and R. R. conceptualization; D. D., R. R., A. K. S., S. S., and D. C. writing–original draft preparation; D. D., R. R., A. K. S., S. S., and D. C. writing–reviewing and editing; D. D. and D. C. supervision.
Funding and additional information
D. D. acknowledges the Science and Engineering Research Board (CRG/2019/000943) and Indian Institute of Science, Bangalore, India (SR/MHRD-18-0021) for support. D. C. acknowledges TATA Innovation Fellowship, DAE-SRC Fellowship and DBT-IISc IOE for support. R. R. acknowledges the Prime Minister's Research Fellowship, Government of India for support. A. K. S. acknowledges Institute of Eminence-CV Raman Fellowship for support.
Edited by Chris Whitfield
References
- 1.Sharma D., Misba L., Khan A.U. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control. 2019;8:76. doi: 10.1186/s13756-019-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aslam B., Wang W., Arshad M.I., Khurshid M., Muzammil S., Rasool M.H., et al. Antibiotic resistance: a rundown of a global crisis. Infect. Drug Resist. 2018;11:1645–1658. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forsberg K., Woodworth K., Walters M., Berkow E.L., Jackson B., Chiller T., et al. Candida auris: the recent emergence of a multidrug-resistant fungal pathogen. Med. Mycol. 2019;57:1–12. doi: 10.1093/mmy/myy054. [DOI] [PubMed] [Google Scholar]
- 4.Fair R.J., Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Medicin. Chem. 2014;6:25–64. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleming D., Rumbaugh K.P. Approaches to dispersing medical biofilms. Microorganisms. 2017;5:15. doi: 10.3390/microorganisms5020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall-Stoodley L., Costerton J.W., Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 7.Flemming H.C., Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 8.Fux C.A., Costerton J.W., Stewart P.S., Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Wu H., Moser C., Wang H.-Z., Høiby N., Song Z.-J. Strategies for combating bacterial biofilm infections. Int. J. Oral Sci. 2015;7:1–7. doi: 10.1038/ijos.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penesyan A., Nagy S.S., Kjelleberg S., Gillings M.R., Paulsen I.T. Rapid microevolution of biofilm cells in response to antibiotics. NPJ Biofilms Microbiomes. 2019;5:34. doi: 10.1038/s41522-019-0108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srivastava S., Bhargava A. Biofilms and human health. Biotechnol. Lett. 2016;38:1–22. doi: 10.1007/s10529-015-1960-8. [DOI] [PubMed] [Google Scholar]
- 12.Ceri H., Olson M.E., Stremick C., Read R.R., Morck D., Buret A. The calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 1999;37:1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderl J.N., Franklin M.J., Stewart P.S. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 2000;44:1818–1824. doi: 10.1128/aac.44.7.1818-1824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert P., Allison D.G., McBain A.J. Biofilms in vitro and in vivo: do singular mechanisms imply cross-resistance? J. Appl. Microbiol. 2002;92(Suppl):98s–110s. [PubMed] [Google Scholar]
- 15.Lewis K. Persister cells. Annu. Rev. Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 16.Lebeaux D., Ghigo J.M., Beloin C. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 2014;78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jamal M., Ahmad W., Andleeb S., Jalil F., Imran M., Nawaz M.A., et al. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018;81:7–11. doi: 10.1016/j.jcma.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 18.Fleming D., Rumbaugh K. The consequences of biofilm dispersal on the host. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-29121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nandakumar V., Chittaranjan S., Kurian V.M., Doble M. Characteristics of bacterial biofilm associated with implant material in clinical practice. Polym. J. 2013;45:137–152. [Google Scholar]
- 20.Ramage G., Rajendran R., Sherry L., Williams C. Fungal biofilm resistance. Int. J. Microbiol. 2012;2012 doi: 10.1155/2012/528521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez L.R., Fries B.C. Fungal biofilms: relevance in the setting of human disease. Curr. Fungal Infect. Rep. 2010;4:266–275. doi: 10.1007/s12281-010-0035-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cannizzo F.T., Eraso E., Ezkurra P.A., Villar-Vidal M., Bollo E., Castellá G., et al. Biofilm development by clinical isolates of Malassezia pachydermatis. Med. Mycol. 2007;45:357–361. doi: 10.1080/13693780701225767. [DOI] [PubMed] [Google Scholar]
- 23.Cushion M.T., Collins M.S., Linke M.J. Biofilm formation by Pneumocystis spp. Eukaryot. Cell. 2009;8:197–206. doi: 10.1128/EC.00202-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Antonio D., Parruti G., Pontieri E., Di Bonaventura G., Manzoli L., Sferra R., et al. Slime production by clinical isolates of Blastoschizomyces capitatus from patients with hematological malignancies and catheter-related fungemia. Eur. J. Clin. Microbiol. Infect. Dis. 2004;23:787–789. doi: 10.1007/s10096-004-1207-4. [DOI] [PubMed] [Google Scholar]
- 25.Davis L.E., Cook G., Costerton J.W. Biofilm on ventriculo-peritoneal shunt tubing as a cause of treatment failure in coccidioidal meningitis. Emerg. Infect. Dis. 2002;8:376–379. doi: 10.3201/eid0804.010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierce C.G., Uppuluri P., Tristan A.R., Wormley F.L., Jr., Mowat E., Ramage G., et al. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 2008;3:1494–1500. doi: 10.1038/nport.2008.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tits J., Cammue B.P.A., Thevissen K. Combination therapy to treat fungal biofilm-based infections. Int. J. Mol. Sci. 2020;21:8873. doi: 10.3390/ijms21228873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verderosa A.D., Totsika M., Fairfull-Smith K.E. Bacterial biofilm eradication agents: a current review. Front. Chem. 2019;7:824. doi: 10.3389/fchem.2019.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armbruster C.R., Parsek M.R. New insight into the early stages of biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 2018;115:4317–4319. doi: 10.1073/pnas.1804084115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crouzet M., Le Senechal C., Brözel V.S., Costaglioli P., Barthe C., Bonneu M., et al. Exploring early steps in biofilm formation: set-up of an experimental system for molecular studies. BMC Microbiol. 2014;14:253. doi: 10.1186/s12866-014-0253-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koo H., Allan R.N., Howlin R.P., Stoodley P., Hall-Stoodley L. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017;15:740–755. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leewenhoeck A. An abstract of a letter from Mr. Anthony Leevvenhoeck at Delft, dated Sep. 17. 1683. Containing some microscopical observations, about animals in the scurf of the teeth, the substance call'd worms in the nose, the cuticula consisting of scales. Philos. Trans. R. Soc. 1684;14:568–574. [Google Scholar]
- 33.Tang P., Zhang W., Wang Y., Zhang B., Wang H., Lin C., et al. Effect of superhydrophobic surface of titanium on Staphylococcus aureus adhesion. J. Nanomater. 2011;2011 [Google Scholar]
- 34.Pogodin S., Hasan J., Baulin V.A., Webb H.K., Truong V.K., Phong Nguyen T.H., et al. Biophysical model of bacterial cell interactions with nanopatterned cicada wing surfaces. Biophys. J. 2013;104:835–840. doi: 10.1016/j.bpj.2012.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S., Jung U.T., Kim S.-K., Lee J.-H., Choi H.S., Kim C.-S., et al. Nanostructured multifunctional surface with antireflective and antimicrobial characteristics. ACS Appl. Mater. Inter. 2015;7:326–331. doi: 10.1021/am506254r. [DOI] [PubMed] [Google Scholar]
- 36.Rumbaugh K.P., Sauer K. Biofilm dispersion. Nat. Rev. Microbiol. 2020;18:571–586. doi: 10.1038/s41579-020-0385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang Y., Geng M., Bai L. Targeting biofilms therapy: current research strategies and development hurdles. Microorganisms. 2020;8:1222. doi: 10.3390/microorganisms8081222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berhe N., Tefera Y., Tintagu T. Review on biofilm formation and its control options. Int. J. Adv. Res. Biol. Sci. 2017;4:122–133. [Google Scholar]
- 39.Zheng S., Bawazir M., Dhall A., Kim H.E., He L., Heo J., et al. Implication of surface properties, bacterial motility, and hydrodynamic conditions on bacterial surface sensing and their initial adhesion. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.643722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narayana S.V.V.S., Srihari S.V.V. A review on surface modifications and coatings on implants to prevent biofilm. Regen. Eng. Transl. Med. 2019;6:330–346. [Google Scholar]
- 41.Anderson J.M., Rodriguez A., Chang D.T. Foreign body reaction to biomaterials. Semin. Immunol. 2008;20:86–100. doi: 10.1016/j.smim.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nava-Ortiz C.A., Burillo G., Concheiro A., Bucio E., Matthijs N., Nelis H., et al. Cyclodextrin-functionalized biomaterials loaded with miconazole prevent Candida albicans biofilm formation in vitro. Acta Biomater. 2010;6:1398–1404. doi: 10.1016/j.actbio.2009.10.039. [DOI] [PubMed] [Google Scholar]
- 43.Tournu H., Van Dijck P. Candida biofilms and the host: models and new concepts for eradication. Int. J. Microbiol. 2012;2012 doi: 10.1155/2012/845352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Prijck K., De Smet N., Coenye T., Schacht E., Nelis H.J. Prevention of Candida albicans biofilm formation by covalently bound dimethylaminoethylmethacrylate and polyethylenimine. Mycopathologia. 2010;170:213–221. doi: 10.1007/s11046-010-9316-3. [DOI] [PubMed] [Google Scholar]
- 45.De Prijck K., De Smet N., Rymarczyk-Machal M., Van Driessche G., Devreese B., Coenye T., et al. Candida albicans biofilm formation on peptide functionalized polydimethylsiloxane. Biofouling. 2010;26:269–275. doi: 10.1080/08927010903501908. [DOI] [PubMed] [Google Scholar]
- 46.Mu H., Tang J., Liu Q., Sun C., Wang T., Duan J. Potent antibacterial nanoparticles against biofilm and intracellular bacteria. Sci. Rep. 2016;6 doi: 10.1038/srep18877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chernousova S., Epple M. Silver as antibacterial agent: ion, nanoparticle, and metal. Angew. Chem. Int. Ed. 2013;52:1636–1653. doi: 10.1002/anie.201205923. [DOI] [PubMed] [Google Scholar]
- 48.Singh A.K., Prakash P., Singh R., Nandy N., Firdaus Z., Bansal M., et al. Curcumin quantum dots mediated degradation of bacterial biofilms. Front. Microbiol. 2017;8:1517. doi: 10.3389/fmicb.2017.01517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vahdati M., Tohidi Moghadam T. Synthesis and characterization of selenium nanoparticles-lysozyme nanohybrid system with synergistic antibacterial properties. Sci. Rep. 2020;10:510. doi: 10.1038/s41598-019-57333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azizi-Lalabadi M., Ehsani A., Divband B., Alizadeh-Sani M. Antimicrobial activity of Titanium dioxide and Zinc oxide nanoparticles supported in 4A zeolite and evaluation the morphological characteristic. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-54025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arakha M., Pal S., Samantarrai D., Panigrahi T.K., Mallick B.C., Pramanik K., et al. Antimicrobial activity of iron oxide nanoparticle upon modulation of nanoparticle-bacteria interface. Sci. Rep. 2015;5 doi: 10.1038/srep14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh A.K., Mishra H., Firdaus Z., Yadav S., Aditi P., Nandy N., et al. MoS2-Modified curcumin nanostructures: the novel theranostic hybrid having potent antibacterial and antibiofilm activities against multidrug-resistant hypervirulent Klebsiella pneumoniae. Chem. Res. Toxicol. 2019;32:1599–1618. doi: 10.1021/acs.chemrestox.9b00135. [DOI] [PubMed] [Google Scholar]
- 53.Prasad K., Bazaka O., Chua M., Rochford M., Fedrick L., Spoor J., et al. Metallic biomaterials: current challenges and opportunities. Materials. 2017;10:884. doi: 10.3390/ma10080884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang L., Harroun T.A., Weiss T.M., Ding L., Huang H.W. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 2001;81:1475–1485. doi: 10.1016/S0006-3495(01)75802-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raheem N., Straus S.K. Mechanisms of action for antimicrobial peptides with antibacterial and antibiofilm functions. Front. Microbiol. 2019;10:2866. doi: 10.3389/fmicb.2019.02866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yasir M., Dutta D., Kumar N., Willcox M.D.P. Interaction of the surface bound antimicrobial peptides melimine and Mel4 with Staphylococcus aureus. Biofouling. 2020;36:1019–1030. doi: 10.1080/08927014.2020.1843638. [DOI] [PubMed] [Google Scholar]
- 57.Wimley W.C., Hristova K. Antimicrobial peptides: successes, challenges and unanswered questions. J. Membr. Biol. 2011;239:27–34. doi: 10.1007/s00232-011-9343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson C.T., Wroe J.A., Agarwal R., Martin K.E., Guldberg R.E., Donlan R.M., et al. Hydrogel delivery of lysostaphin eliminates orthopedic implant infection by Staphylococcus aureus and supports fracture healing. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E4960–E4969. doi: 10.1073/pnas.1801013115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bayer I.S. Hyaluronic acid and controlled release: a review. Molecules. 2020;25:2649. doi: 10.3390/molecules25112649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hickok N.J., Shapiro I.M. Immobilized antibiotics to prevent orthopaedic implant infections. Adv. Drug Deliv. Rev. 2012;64:1165–1176. doi: 10.1016/j.addr.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahmed W., Zhai Z., Gao C. Adaptive antibacterial biomaterial surfaces and their applications. Mater. Today Bio. 2019;2 doi: 10.1016/j.mtbio.2019.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van de Belt H., Neut D., Schenk W., van Horn J.R., van der Mei H.C., Busscher H.J. Gentamicin release from polymethylmethacrylate bone cements and Staphylococcus aureus biofilm formation. Acta Orthop. Scand. 2000;71:625–629. doi: 10.1080/000164700317362280. [DOI] [PubMed] [Google Scholar]
- 63.Miller M.B., Bassler B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 64.Gupta P., Sarkar S., Das B., Bhattacharjee S., Tribedi P. Biofilm, pathogenesis and prevention--a journey to break the wall: a review. Arch. Microbiol. 2016;198:1–15. doi: 10.1007/s00203-015-1148-6. [DOI] [PubMed] [Google Scholar]
- 65.Oppenheimer-Shaanan Y., Steinberg N., Kolodkin-Gal I. Small molecules are natural triggers for the disassembly of biofilms. Trends Microbiol. 2013;21:594–601. doi: 10.1016/j.tim.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 66.Li Z., Nair S.K. Quorum sensing: how bacteria can coordinate activity and synchronize their response to external signals? Protein Sci. 2012;21:1403–1417. doi: 10.1002/pro.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Preda V.G., Sandulescu O. Communication is the key: biofilms, quorum sensing, formation and prevention. Discoveries. 2019;7 doi: 10.15190/d.2019.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang K., Zhang X.-H. Quorum quenching agents: resources for antivirulence therapy. Mar. Drugs. 2014;12:3245–3282. doi: 10.3390/md12063245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou L., Zhang Y., Ge Y., Zhu X., Pan J. Regulatory mechanisms and promising applications of quorum sensing-inhibiting agents in control of bacterial biofilm formation. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.589640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paluch E., Rewak-Soroczynska J., Jedrusik I., Mazurkiewicz E., Jermakow K. Prevention of biofilm formation by quorum quenching. Appl. Microbiol. Biotechnol. 2020;104:1871–1881. doi: 10.1007/s00253-020-10349-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dong Y.H., Zhang X.F., An S.W., Xu J.L., Zhang L.H. A novel two-component system BqsS-BqsR modulates quorum sensing-dependent biofilm decay in Pseudomonas aeruginosa. Commun. Integr. Biol. 2008;1:88–96. doi: 10.4161/cib.1.1.6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin Y.H., Xu J.L., Hu J., Wang L.H., Ong S.L., Leadbetter J.R., et al. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 2003;47:849–860. doi: 10.1046/j.1365-2958.2003.03351.x. [DOI] [PubMed] [Google Scholar]
- 73.Parsek M.R., Val D.L., Hanzelka B.L., Cronan J.E., Greenberg E.P. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4360–4365. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Subhadra B., Kim D.H., Woo K., Surendran S., Choi C.H. Control of biofilm formation in healthcare: recent advances exploiting quorum-sensing interference strategies and multidrug efflux pump inhibitors. Materials (Basel) 2018;11:1676. doi: 10.3390/ma11091676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gutierrez J.A., Crowder T., Rinaldo-Matthis A., Ho M.C., Almo S.C., Schramm V.L. Transition state analogs of 5'-methylthioadenosine nucleosidase disrupt quorum sensing. Nat. Chem. Biol. 2009;5:251–257. doi: 10.1038/nchembio.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ishida T., Ikeda T., Takiguchi N., Kuroda A., Ohtake H., Kato J. Inhibition of quorum sensing in Pseudomonas aeruginosa by N-acyl cyclopentylamides. Appl. Environ. Microbiol. 2007;73:3183–3188. doi: 10.1128/AEM.02233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khatoon Z., McTiernan C.D., Suuronen E.J., Mah T.F., Alarcon E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon. 2018;4 doi: 10.1016/j.heliyon.2018.e01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muhammad M.H., Idris A.L., Fan X., Guo Y., Yu Y., Jin X., et al. Beyond risk: bacterial biofilms and their regulating approaches. Front. Microbiol. 2020;11:928. doi: 10.3389/fmicb.2020.00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tateda K., Comte R., Pechere J.C., Kohler T., Yamaguchi K., Van Delden C. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2001;45:1930–1933. doi: 10.1128/AAC.45.6.1930-1933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zeng J., Zhang N., Huang B., Cai R., Wu B., E S., et al. Mechanism of azithromycin inhibition of HSL synthesis in Pseudomonas aeruginosa. Sci. Rep. 2016;6 doi: 10.1038/srep24299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Swatton J.E., Davenport P.W., Maunders E.A., Griffin J.L., Lilley K.S., Welch M. Impact of azithromycin on the quorum sensing-controlled proteome of Pseudomonas aeruginosa. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Munir S., Shah A.A., Shahid M., Manzoor I., Aslam B., Rasool M.H., et al. Quorum sensing interfering strategies and their implications in the management of biofilm-associated bacterial infections. Braz. Arch. 2020 doi: 10.1590/1678-4324-2020190555. [DOI] [Google Scholar]
- 83.Smyth A.R., Cifelli P.M., Ortori C.A., Righetti K., Lewis S., Erskine P., et al. Garlic as an inhibitor of Pseudomonas aeruginosa quorum sensing in cystic fibrosis--a pilot randomized controlled trial. Pediatr. Pulmonol. 2010;45:356–362. doi: 10.1002/ppul.21193. [DOI] [PubMed] [Google Scholar]
- 84.Walz J.M., Avelar R.L., Longtine K.J., Carter K.L., Mermel L.A., Heard S.O. Anti-infective external coating of central venous catheters: a randomized, noninferiority trial comparing 5-fluorouracil with chlorhexidine/silver sulfadiazine in preventing catheter colonization. Crit. Care Med. 2010;38:2095–2102. doi: 10.1097/CCM.0b013e3181f265ba. [DOI] [PubMed] [Google Scholar]
- 85.Focaccetti C., Bruno A., Magnani E., Bartolini D., Principi E., Dallaglio K., et al. Effects of 5-fluorouracil on morphology, cell cycle, proliferation, apoptosis, autophagy and ROS production in endothelial cells and cardiomyocytes. PLoS One. 2015;10 doi: 10.1371/journal.pone.0115686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sedlmayer F., Woischnig A.K., Unterreiner V., Fuchs F., Baeschlin D., Khanna N., et al. 5-Fluorouracil blocks quorum-sensing of biofilm-embedded methicillin-resistant Staphylococcus aureus in mice. Nucl. Acids Res. 2021;49:e73. doi: 10.1093/nar/gkab251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Giladi M., Porat Y., Blatt A., Wasserman Y., Kirson Eilon D., Dekel E., et al. Microbial growth inhibition by alternating electric fields. Antimicrob. Agents Chemother. 2008;52:3517–3522. doi: 10.1128/AAC.00673-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Subramanian S., Gerasopoulos K., Guo M., Sintim H.O., Bentley W.E., Ghodssi R. Autoinducer-2 analogs and electric fields - an antibiotic-free bacterial biofilm combination treatment. Biomed. Microdevices. 2016;18:95. doi: 10.1007/s10544-016-0120-9. [DOI] [PubMed] [Google Scholar]
- 89.Del Pozo J.L., Rouse M.S., Patel R. Bioelectric effect and bacterial biofilms. A systematic review. Int. J. Artif. Organs. 2008;31:786–795. doi: 10.1177/039139880803100906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cai Y., Wang J., Liu X., Wang R., Xia L. A review of the combination therapy of low frequency ultrasound with antibiotics. Biomed. Res. Int. 2017;2017 doi: 10.1155/2017/2317846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rediske A.M., Roeder B.L., Brown M.K., Nelson J.L., Robison R.L., Draper D.O., et al. Ultrasonic enhancement of antibiotic action on Escherichia coli biofilms: an in vivo model. Antimicrob. Agents Chemother. 1999;43:1211–1214. doi: 10.1128/aac.43.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hazan Z., Zumeris J., Jacob H., Raskin H., Kratysh G., Vishnia M., et al. Effective prevention of microbial biofilm formation on medical devices by low-energy surface acoustic waves. Antimicrob. Agents Chemother. 2006;50:4144–4152. doi: 10.1128/AAC.00418-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kopel M., Degtyar E., Banin E. Surface acoustic waves increase the susceptibility of Pseudomonas aeruginosa biofilms to antibiotic treatment. Biofouling. 2011;27:701–711. doi: 10.1080/08927014.2011.597051. [DOI] [PubMed] [Google Scholar]
- 94.Liu Y., Qin R., Zaat S.A.J., Breukink E., Heger M. Antibacterial photodynamic therapy: overview of a promising approach to fight antibiotic-resistant bacterial infections. J. Clin. Transl. Res. 2015;1:140–167. [PMC free article] [PubMed] [Google Scholar]
- 95.Hu X., Huang Y.Y., Wang Y., Wang X., Hamblin M.R. Antimicrobial photodynamic therapy to control clinically relevant biofilm infections. Front. Microbiol. 2018;9:1299. doi: 10.3389/fmicb.2018.01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carrera E.T., Dias H.B., Corbi S.C.T., Marcantonio R.A.C., Bernardi A.C.A., Bagnato V.S., et al. The application of antimicrobial photodynamic therapy (aPDT) in dentistry: a critical review. Laser Phys. 2016;26:123001. doi: 10.1088/1054-660X/26/12/123001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Limoli D.H., Jones C.J., Wozniak D.J. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectr. 2015;3:1–19. doi: 10.1128/microbiolspec.MB-0011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rohde H., Frankenberger S., Zahringer U., Mack D. Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur. J. Cell. Biol. 2010;89:103–111. doi: 10.1016/j.ejcb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 99.Nguyen H.T.T., Nguyen T.H., Otto M. The staphylococcal exopolysaccharide PIA - biosynthesis and role in biofilm formation, colonization, and infection. Comput. Struct. Biotechnol. J. 2020;18:3324–3334. doi: 10.1016/j.csbj.2020.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Whitfield G.B., Marmont L.S., Howell P.L. Enzymatic modifications of exopolysaccharides enhance bacterial persistence. Front. Microbiol. 2015;6:471. doi: 10.3389/fmicb.2015.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arciola C.R., Campoccia D., Ravaioli S., Montanaro L. Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects. Front. Cell. Infect. Microbiol. 2015;5:7. doi: 10.3389/fcimb.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jennings L.K., Storek K.M., Ledvina H.E., Coulon C., Marmont L.S., Sadovskaya I., et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. U. S. A. 2015;112:11353–11358. doi: 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jennings L.K., Dreifus J.E., Reichhardt C., Storek K.M., Secor P.R., Wozniak D.J., et al. Pseudomonas aeruginosa aggregates in cystic fibrosis sputum produce exopolysaccharides that likely impede current therapies. Cell. Rep. 2021;34 doi: 10.1016/j.celrep.2021.108782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Le Mauff F., Razvi E., Reichhardt C., Sivarajah P., Parsek M.R., Howell P.L., et al. The Pel polysaccharide is predominantly composed of a dimeric repeat of alpha-1,4 linked galactosamine and N-acetylgalactosamine. Commun. Biol. 2022;5:502. doi: 10.1038/s42003-022-03453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ma L., Jackson K.D., Landry R.M., Parsek M.R., Wozniak D.J. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 2006;188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hay I.D., Ur Rehman Z., Moradali M.F., Wang Y., Rehm B.H.A. Microbial alginate production, modification and its applications. Microb. Biotechnol. 2013;6:637–650. doi: 10.1111/1751-7915.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jones C.J., Wozniak D.J. Psl produced by mucoid Pseudomonas aeruginosa contributes to the establishment of biofilms and immune evasion. mBio. 2017;8 doi: 10.1128/mBio.00864-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lembre P., Lorentz C., Di Martino P. IntechOpen; London: 2012. Exopolysaccharides of the Biofilm Matrix: A Complex Biophysical World; pp. 371–392. [Google Scholar]
- 109.Zogaj X., Nimtz M., Rohde M., Bokranz W., Römling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 110.Solano C., García B., Valle J., Berasain C., Ghigo J.-M., Gamazo C., et al. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 2002;43:793–808. doi: 10.1046/j.1365-2958.2002.02802.x. [DOI] [PubMed] [Google Scholar]
- 111.Ghafoor A., Hay I.D., Rehm B.H.A. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 2011;77:5238–5246. doi: 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bai S., Chen H., Zhu L., Liu W., Yu H.D., Wang X., et al. Comparative study on the in vitro effects of Pseudomonas aeruginosa and seaweed alginates on human gut microbiota. PLoS One. 2017;12 doi: 10.1371/journal.pone.0171576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Passos da Silva D., Matwichuk M.L., Townsend D.O., Reichhardt C., Lamba D., Wozniak D.J., et al. The Pseudomonas aeruginosa lectin LecB binds to the exopolysaccharide Psl and stabilizes the biofilm matrix. Nat. Commun. 2019;10:2183. doi: 10.1038/s41467-019-10201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.O'Gara J.P. Ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 2007;270:179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- 115.Spiliopoulou A.I., Krevvata M.I., Kolonitsiou F., Harris L.G., Wilkinson T.S., Davies A.P., et al. An extracellular Staphylococcus epidermidis polysaccharide: relation to polysaccharide intercellular adhesin and its implication in phagocytosis. BMC Microbiol. 2012;12:76. doi: 10.1186/1471-2180-12-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Idrees M., Sawant S., Karodia N., Rahman A. Staphylococcus aureus biofilm: morphology, genetics, pathogenesis and treatment strategies. Int. J. Environ. Res. Public Health. 2021;18:7602. doi: 10.3390/ijerph18147602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sharma G., Sharma S., Sharma P., Chandola D., Dang S., Gupta S., et al. Escherichia coli biofilm: development and therapeutic strategies. J. Appl. Microbiol. 2016;121:309–319. doi: 10.1111/jam.13078. [DOI] [PubMed] [Google Scholar]
- 118.Benincasa M., Lagatolla C., Dolzani L., Milan A., Pacor S., Liut G., et al. Biofilms from Klebsiella pneumoniae: matrix polysaccharide structure and interactions with antimicrobial peptides. Microorganisms. 2016;4:26. doi: 10.3390/microorganisms4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Singh A.K., Yadav S., Chauhan B.S., Nandy N., Singh R., Neogi K., et al. Classification of clinical isolates of Klebsiella pneumoniae based on their in vitro biofilm forming capabilities and elucidation of the biofilm matrix chemistry with special reference to the protein content. Front. Microbiol. 2019;10:669. doi: 10.3389/fmicb.2019.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vu B., Chen M., Crawford R.J., Ivanova E.P. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules. 2009;14:2535–2554. doi: 10.3390/molecules14072535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Beynon L.M., Dumanski A.J., McLean R.J., MacLean L.L., Richards J.C., Perry M.B. Capsule structure of Proteus mirabilis (ATCC 49565) J. Bacteriol. 1992;174:2172–2177. doi: 10.1128/jb.174.7.2172-2177.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Coyne M.J., Tzianabos A.O., Mallory B.C., Carey V.J., Kasper D.L., Comstock L.E. Polysaccharide biosynthesis locus required for virulence of Bacteroides fragilis. Infect. Immun. 2001;69:4342–4350. doi: 10.1128/IAI.69.7.4342-4350.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kumar M., Kumar M., Pandey A., Thakur I.S. Genomic analysis of carbon dioxide sequestering bacterium for exopolysaccharides production. Sci. Rep. 2019;9:4270. doi: 10.1038/s41598-019-41052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gravelat F.N., Beauvais A., Liu H., Lee M.J., Snarr B.D., Chen D., et al. Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal beta-glucan from the immune system. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sheppard D.C., Howell P.L. Biofilm exopolysaccharides of pathogenic fungi: lessons from bacteria. J. Biol. Chem. 2016;291:12529–12537. doi: 10.1074/jbc.R116.720995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pierce C.G., Vila T., Romo J.A., Montelongo-Jauregui D., Wall G., Ramasubramanian A., et al. The Candida albicans biofilm matrix: composition, structure and function. J. Fungi. 2017;3:14. doi: 10.3390/jof3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kaplan J.B. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 2010;89:205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kaplan J.B., Ragunath C., Ramasubbu N., Fine D.H. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J. Bacteriol. 2003;185:4693–4698. doi: 10.1128/JB.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Manuel S.G., Ragunath C., Sait H.B., Izano E.A., Kaplan J.B., Ramasubbu N. Role of active-site residues of dispersin B, a biofilm-releasing beta-hexosaminidase from a periodontal pathogen, in substrate hydrolysis. FEBS J. 2007;274:5987–5999. doi: 10.1111/j.1742-4658.2007.06121.x. [DOI] [PubMed] [Google Scholar]
- 130.Wang S., Breslawec A.P., Alvarez E., Tyrlik M., Li C., Poulin M.B. Differential recognition of deacetylated PNAG oligosaccharides by a biofilm degrading glycosidase. ACS Chem. Biol. 2019;14:1998–2005. doi: 10.1021/acschembio.9b00467. [DOI] [PubMed] [Google Scholar]
- 131.Breslawec A.P., Wang S., Li C., Poulin M.B. Anionic amino acids support hydrolysis of poly-beta-(1,6)-N-acetylglucosamine exopolysaccharides by the biofilm dispersing glycosidase Dispersin B. J. Biol. Chem. 2021;296 doi: 10.1074/jbc.RA120.015524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ramasubbu N., Thomas L.M., Ragunath C., Kaplan J.B. Structural analysis of dispersin B, a biofilm-releasing glycoside hydrolase from the periodontopathogen Actinobacillus actinomycetemcomitans. J. Mol. Biol. 2005;349:475–486. doi: 10.1016/j.jmb.2005.03.082. [DOI] [PubMed] [Google Scholar]
- 133.Ghalsasi V., Sourjik V. Engineering Escherichia coli to disrupt poly-N-acetylglucosamine containing bacterial biofilms. Curr. Synth. Syst. Biol. 2016;4 doi: 10.4172/2332-0737.1000130. [DOI] [Google Scholar]
- 134.Lu T.K., Collins J.J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11197–11202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Baker P., Hill P.J., Snarr B.D., Alnabelseya N., Pestrak M.J., Lee M.J., et al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci. Adv. 2016;2:e1501632. doi: 10.1126/sciadv.1501632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Snarr B.D., Baker P., Bamford N.C., Sato Y., Liu H., Lehoux M., et al. Microbial glycoside hydrolases as antibiofilm agents with cross-kingdom activity. Proc. Natl. Acad. Sci. U. S. A. 2017;114:7124–7129. doi: 10.1073/pnas.1702798114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Asker D., Awad T.S., Raju D., Sanchez H., Lacdao I., Gilbert S., et al. Preventing Pseudomonas aeruginosa biofilms on indwelling catheters by surface-bound enzymes. ACS Appl. Bio. Mater. 2021;4:8248–8258. doi: 10.1021/acsabm.1c00794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Asker D., Awad T.S., Baker P., Howell P.L., Hatton B.D. Non-eluting, surface-bound enzymes disrupt surface attachment of bacteria by continuous biofilm polysaccharide degradation. Biomaterials. 2018;167:168–176. doi: 10.1016/j.biomaterials.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 139.Fleming D., Chahin L., Rumbaugh K. Glycoside hydrolases degrade polymicrobial bacterial biofilms in wounds. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.01998-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fujimoto Z., Takase K., Doui N., Momma M., Matsumoto T., Mizuno H. Crystal structure of a catalytic-site mutant alpha-amylase from Bacillus subtilis complexed with maltopentaose. J. Mol. Biol. 1998;277:393–407. doi: 10.1006/jmbi.1997.1599. [DOI] [PubMed] [Google Scholar]
- 141.Yan J., Liu W., Li Y., Lai H.L., Zheng Y., Huang J.W., et al. Functional and structural analysis of Pichia pastoris-expressed Aspergillus Niger 1,4-beta-endoglucanase. Biochem. Biophys. Res. Commun. 2016;475:8–12. doi: 10.1016/j.bbrc.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 142.Karatan E., Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 2009;73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Redman W.K., Welch G.S., Williams A.C., Damron A.J., Northcut W.O., Rumbaugh K.P. Efficacy and safety of biofilm dispersal by glycoside hydrolases in wounds. Biofilm. 2021;3 doi: 10.1016/j.bioflm.2021.100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Davies G., Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 145.Vuong T.V., Wilson D.B. Glycoside hydrolases: catalytic base/nucleophile diversity. Biotechnol. Bioeng. 2010;107:195–205. doi: 10.1002/bit.22838. [DOI] [PubMed] [Google Scholar]
- 146.Ardevol A., Rovira C. Reaction mechanisms in carbohydrate-active enzymes: glycoside hydrolases and glycosyltransferases. Insights from ab initio quantum mechanics/molecular mechanics dynamic simulations. J. Am. Chem. Soc. 2015;137:7528–7547. doi: 10.1021/jacs.5b01156. [DOI] [PubMed] [Google Scholar]
- 147.Jee S.-C., Kim M., Sung J.-S., Kadam A.A. Efficient biofilms eradication by enzymatic-cocktail of pancreatic protease type-I and bacterial α-amylase. Polymers. 2020;12:3032. doi: 10.3390/polym12123032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Linhardt R.J., Galliher P.M., Cooney C.L. Polysaccharide lyase. Appl. Biochem. Biotechnol. 1987;12:135–176. doi: 10.1007/BF02798420. [DOI] [PubMed] [Google Scholar]
- 149.Alipour M., Suntres Z.E., Omri A. Importance of DNase and alginate lyase for enhancing free and liposome encapsulated aminoglycoside activity against Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2009;64:317–325. doi: 10.1093/jac/dkp165. [DOI] [PubMed] [Google Scholar]
- 150.Wu Y., Wang R., Xu M., Liu Y., Zhu X., Qiu J., et al. A novel polysaccharide depolymerase encoded by the phage SH-KP152226 confers specific activity against multidrug-resistant Klebsiella pneumoniae via biofilm degradation. Front. Microbiol. 2019;10:2768. doi: 10.3389/fmicb.2019.02768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Blanco-Cabra N., Paetzold B., Ferrar T., Mazzolini R., Torrents E., Serrano L., et al. Characterization of different alginate lyases for dissolving Pseudomonas aeruginosa biofilms. Sci. Rep. 2020;10:9390. doi: 10.1038/s41598-020-66293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lamppa J.W., Griswold K.E. Alginate lyase exhibits catalysis-independent biofilm dispersion and antibiotic synergy. Antimicrob. Agents Chemother. 2013;57:137–145. doi: 10.1128/AAC.01789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Okshevsky M., Regina V.R., Meyer R.L. Extracellular DNA as a target for biofilm control. Curr. Opin. Biotechnol. 2015;33:73–80. doi: 10.1016/j.copbio.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 154.Jones A.P., Wallis C. Dornase alfa for cystic fibrosis. Cochrane Database Syst. Rev. 2010;3:CD001127. doi: 10.1002/14651858.CD001127.pub2. [DOI] [PubMed] [Google Scholar]
- 155.Whitchurch Cynthia B., Tolker-Nielsen T., Ragas Paula C., Mattick John S. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 156.Fanaei Pirlar R., Emaneini M., Beigverdi R., Banar M., van Leeuwen W.B., Jabalameli F. Combinatorial effects of antibiotics and enzymes against dual-species Staphylococcus aureus and Pseudomonas aeruginosa biofilms in the wound-like medium. PLoS One. 2020;15 doi: 10.1371/journal.pone.0235093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fazekas E., Kandra L., Gyémánt G. Model for β-1,6-N-acetylglucosamine oligomer hydrolysis catalysed by DispersinB, a biofilm degrading enzyme. Carbohydr. Res. 2012;363:7–13. doi: 10.1016/j.carres.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 158.Gawande P.V., Leung K.P., Madhyastha S. Antibiofilm and antimicrobial efficacy of DispersinB®-KSL-W peptide-based wound gel against chronic wound infection associated bacteria. Curr. Microbiol. 2014;68:635–641. doi: 10.1007/s00284-014-0519-6. [DOI] [PubMed] [Google Scholar]
- 159.Itoh Y., Wang X., Hinnebusch B.J., Preston J.F., 3rd, Romeo T. Depolymerization of beta-1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 2005;187:382–387. doi: 10.1128/JB.187.1.382-387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Izano E.A., Sadovskaya I., Vinogradov E., Mulks M.H., Velliyagounder K., Ragunath C., et al. Poly-N-acetylglucosamine mediates biofilm formation and antibiotic resistance in Actinobacillus pleuropneumoniae. Microb. Pathog. 2007;43:1–9. doi: 10.1016/j.micpath.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Izano E.A., Wang H., Ragunath C., Ramasubbu N., Kaplan J.B. Detachment and killing of aggregatibacter actinomycetemcomitans biofilms by dispersin B and SDS. J. Dent. Res. 2007;86:618–622. doi: 10.1177/154405910708600707. [DOI] [PubMed] [Google Scholar]
- 162.Kaplan J.B., Ragunath C., Velliyagounder K., Fine D.H., Ramasubbu N. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 2004;48:2633–2636. doi: 10.1128/AAC.48.7.2633-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Waryah C.B., Wells K., Ulluwishewa D., Chen-Tan N., Gogoi-Tiwari J., Ravensdale J., et al. In vitro antimicrobial efficacy of tobramycin against Staphylococcus aureus biofilms in combination with or without DNase I and/or dispersin B: a preliminary investigation. Microb. Drug Resist. 2016;23:384–390. doi: 10.1089/mdr.2016.0100. [DOI] [PubMed] [Google Scholar]
- 164.Yakandawala N., Gawande P.V., LoVetri K., Cardona S.T., Romeo T., Nitz M., et al. Characterization of the poly-β-1,6-N-acetylglucosamine polysaccharide component of Burkholderia biofilms. Appl. Environ. Microbiol. 2011;77:8303–8309. doi: 10.1128/AEM.05814-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Craigen B., Dashiff A., Kadouri D.E. The use of commercially available alpha-amylase compounds to inhibit and remove Staphylococcus aureus biofilms. Open Microbiol. J. 2011;5:21–31. doi: 10.2174/1874285801105010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kalpana B.J., Aarthy S., Pandian S.K. Antibiofilm activity of α-amylase from Bacillus subtilis S8-18 against biofilm forming human bacterial pathogens. Appl. Biochem. Biotechnol. 2012;167:1778–1794. doi: 10.1007/s12010-011-9526-2. [DOI] [PubMed] [Google Scholar]
- 167.Watters C.M., Burton T., Kirui D.K., Millenbaugh N.J. Enzymatic degradation of in vitro Staphylococcus aureus biofilms supplemented with human plasma. Infect. Drug Resist. 2016;9:71–78. doi: 10.2147/IDR.S103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Baker P., Whitfield G.B., Hill P.J., Little D.J., Pestrak M.J., Robinson H., et al. Characterization of the Pseudomonas aeruginosa glycoside hydrolase PslG reveals that its levels are critical for psl polysaccharide biosynthesis and biofilm formation. J. Biol. Chem. 2015;290:28374–28387. doi: 10.1074/jbc.M115.674929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yu S., Su T., Wu H., Liu S., Wang D., Zhao T., et al. PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res. 2015;25:1352–1367. doi: 10.1038/cr.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Banar M., Emaneini M., Satarzadeh M., Abdellahi N., Beigverdi R., Leeuwen W.B.v., et al. Evaluation of mannosidase and trypsin enzymes effects on biofilm production of Pseudomonas aeruginosa isolated from burn wound infections. PLoS One. 2016;11:e0164622. doi: 10.1371/journal.pone.0164622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.McCleary B.V., Matheson N.K. Action patterns and substrate-binding requirements of β-d-mannanase with mannosaccharides and mannan-type polysaccharides. Carbohydr. Res. 1983;119:191–219. [Google Scholar]
- 172.Suits M.D., Zhu Y., Taylor E.J., Walton J., Zechel D.L., Gilbert H.J., et al. Structure and kinetic investigation of Streptococcus pyogenes family GH38 alpha-mannosidase. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Little D.J., Pfoh R., Le Mauff F., Bamford N.C., Notte C., Baker P., et al. PgaB orthologues contain a glycoside hydrolase domain that cleaves deacetylated poly-β(1,6)-N-acetylglucosamine and can disrupt bacterial biofilms. PLoS Pathog. 2018;14:e1006998. doi: 10.1371/journal.ppat.1006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bamford N.C., Le Mauff F., Subramanian A.S., Yip P., Millán C., Zhang Y., et al. Ega3 from the fungal pathogen Aspergillus fumigatus is an endo-α-1,4-galactosaminidase that disrupts microbial biofilms. J. Biol. Chem. 2019;294:13833–13849. doi: 10.1074/jbc.RA119.009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bamford N.C., Le Mauff F., Van Loon J.C., Ostapska H., Snarr B.D., Zhang Y., et al. Structural and biochemical characterization of the exopolysaccharide deacetylase Agd3 required for Aspergillus fumigatus biofilm formation. Nat. Commun. 2020;11:2450. doi: 10.1038/s41467-020-16144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Le Mauff F., Bamford N.C., Alnabelseya N., Zhang Y., Baker P., Robinson H., et al. Molecular mechanism of Aspergillus fumigatus biofilm disruption by fungal and bacterial glycoside hydrolases. J. Biol. Chem. 2019;294:10760–10772. doi: 10.1074/jbc.RA119.008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bamford N.C., Snarr B.D., Gravelat F.N., Little D.J., Lee M.J., Zacharias C.A., et al. Sph3 is a glycoside hydrolase required for the biosynthesis of galactosaminogalactan in Aspergillus fumigatus. J. Biol. Chem. 2015;290:27438–27450. doi: 10.1074/jbc.M115.679050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Scott W., Lowrance B., Anderson A.C., Weadge J.T. Identification of the Clostridial cellulose synthase and characterization of the cognate glycosyl hydrolase, CcsZ. PLoS One. 2020;15 doi: 10.1371/journal.pone.0242686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Koseoglu V.K., Heiss C., Azadi P., Topchiy E., Guvener Z.T., Lehmann T.E., et al. Listeria monocytogenes exopolysaccharide: origin, structure, biosynthetic machinery and c-di-GMP-dependent regulation. Mol. Microbiol. 2015;96:728–743. doi: 10.1111/mmi.12966. [DOI] [PubMed] [Google Scholar]
- 180.Gawande P.V., Clinton A.P., LoVetri K., Yakandawala N., Rumbaugh K.P., Madhyastha S. Antibiofilm efficacy of DispersinB(®) wound spray used in combination with a silver wound dressing. Microbiol. Insights. 2014;7:9–13. doi: 10.4137/MBI.S13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Helbert W., Poulet L., Drouillard S., Mathieu S., Loiodice M., Couturier M., et al. Discovery of novel carbohydrate-active enzymes through the rational exploration of the protein sequences space. Proc. Natl. Acad. Sci. U. S. A. 2019;116:6063–6068. doi: 10.1073/pnas.1815791116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang L., Zhang G., Xu H., Xin H., Zhang Y. Metagenomic analyses of microbial and carbohydrate-active enzymes in the rumen of holstein cows fed different forage-to-concentrate ratios. Front. Microbiol. 2019;10:649. doi: 10.3389/fmicb.2019.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wang G., Luo H., Meng K., Wang Y., Huang H., Shi P., et al. High genetic diversity and different distributions of glycosyl hydrolase family 10 and 11 Xylanases in the goat rumen. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hess M., Sczyrba A., Egan R., Kim T.W., Chokhawala H., Schroth G., et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 185.Jevsevar S., Kunstelj M., Porekar V.G. PEGylation of therapeutic proteins. Biotechnol. J. 2010;5:113–128. doi: 10.1002/biot.200900218. [DOI] [PubMed] [Google Scholar]
- 186.Harris J.M., Chess R.B. Effect of pegylation on pharmaceuticals. Nat. Rev. Drug Discov. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 187.Baldo B.A. Chimeric fusion proteins used for therapy: indications, mechanisms, and safety. Drug Saf. 2015;38:455–479. doi: 10.1007/s40264-015-0285-9. [DOI] [PubMed] [Google Scholar]
- 188.Iyengar A.R.S., Gupta S., Jawalekar S., Pande A.H. Protein chimerization: a new frontier for engineering protein therapeutics with improved pharmacokinetics. J. Pharmacol. Exp. Ther. 2019;370:703–714. doi: 10.1124/jpet.119.257063. [DOI] [PubMed] [Google Scholar]
- 189.Boutureira O., Bernardes G.J. Advances in chemical protein modification. Chem. Rev. 2015;115:2174–2195. doi: 10.1021/cr500399p. [DOI] [PubMed] [Google Scholar]
- 190.de la Fuente M., Lombardero L., Gomez-Gonzalez A., Solari C., Angulo-Barturen I., Acera A., et al. Enzyme therapy: current challenges and future perspectives. Int. J. Mol. Sci. 2021;22:9181. doi: 10.3390/ijms22179181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Arun Richard Chandrasekaran A.R., Levchenko O. DNA Nanocages. Chem. Mater. 2016;28:5569–5581. [Google Scholar]
- 192.Arslan F.B., Ozturk Atar K., Calis S. Antibody-mediated drug delivery. Int. J. Pharm. 2021;596 doi: 10.1016/j.ijpharm.2021.120268. [DOI] [PubMed] [Google Scholar]
- 193.Liu X., Kokare C. Biotechnology of Microbial Enzymes. 2017. Chapter 11 - microbial enzymes of use in industry; pp. 267–298. [Google Scholar]
- 194.Yata V.K., Banerjee S., Ghosh S.S. Folic acid conjugated-bio polymeric nanocarriers: synthesis, characterization and in vitro delivery of prodrug converting enzyme. Adv. Sci. Eng. Med. 2014;6:388–392. [Google Scholar]
- 195.Thallinger B., Prasetyo E.N., Nyanhongo G.S., Guebitz G.M. Antimicrobial enzymes: an emerging strategy to fight microbes and microbial biofilms. Biotechnol. J. 2013;8:97–109. doi: 10.1002/biot.201200313. [DOI] [PubMed] [Google Scholar]
- 196.Wells D.H., Goularte N.F., Barnett M.J., Cegelski L., Long S.R. Identification of a novel pyruvyltransferase using (13)C solid-state nuclear magnetic resonance to analyze rhizobial exopolysaccharides. J. Bacteriol. 2021;203 doi: 10.1128/JB.00403-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kim H.S., Lee M.A., Chun S.J., Park S.J., Lee K.H. Role of NtrC in biofilm formation via controlling expression of the gene encoding an ADP-glycero-manno-heptose-6-epimerase in the pathogenic bacterium, vibrio vulnificus. Mol. Microbiol. 2007;63:559–574. doi: 10.1111/j.1365-2958.2006.05527.x. [DOI] [PubMed] [Google Scholar]
- 198.Coleman S., Gilpin D., Kaplan F.T., Houston A., Kaufman G.J., Cohen B.M., et al. Efficacy and safety of concurrent collagenase clostridium histolyticum injections for multiple Dupuytren contractures. J. Hand Surg. Am. 2014;39:57–64. doi: 10.1016/j.jhsa.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 199.Cuylits N. [News in the treatment of dupuytren's disease: from surgery to collagenase's injection. Rev. Med. Brux. 2013;34:283–286. [PubMed] [Google Scholar]
- 200.Hasan F., Shah A.A., Hameed A. Industrial applications of microbial lipases. Enzyme Microb. Technol. 2006;39:235–251. [Google Scholar]
- 201.Sikkens E.C., Cahen D.L., Kuipers E.J., Bruno M.J. Pancreatic enzyme replacement therapy in chronic pancreatitis. Best Pract. Res. Clin. Gastroenterol. 2010;24:337–347. doi: 10.1016/j.bpg.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 202.Lohr J.M., Hummel F.M., Pirilis K.T., Steinkamp G., Korner A., Henniges F. Properties of different pancreatin preparations used in pancreatic exocrine insufficiency. Eur. J. Gastroenterol. Hepatol. 2009;21:1024–1031. doi: 10.1097/MEG.0b013e328328f414. [DOI] [PubMed] [Google Scholar]
- 203.Ostapska H., Raju D., Lehoux M., Lacdao I., Gilbert S., Sivarajah P., et al. Preclinical evaluation of recombinant microbial glycoside hydrolases in the prevention of experimental invasive aspergillosis. mBio. 2021;12 doi: 10.1128/mBio.02446-21. [DOI] [PMC free article] [PubMed] [Google Scholar]