Abstract
Class A tick evasins are natural chemokine-binding proteins that block the signaling of multiple chemokines from the CC subfamily through their cognate receptors, thus suppressing leukocyte recruitment and inflammation. Development of tick evasins as chemokine-targeted anti-inflammatory therapeutics requires an understanding of the factors controlling their chemokine recognition and selectivity. To investigate the role of the evasin N-terminal region for chemokine recognition, we prepared chimeric evasins by interchanging the N-terminal regions of four class A evasins, including a newly identified evasin, EVA-RPU02. We show through chemokine binding analysis of the parental and chimeric evasins that the N-terminal region is critical for chemokine binding affinity and selectivity. Notably, we found some chimeras were unable to bind certain cognate chemokine ligands of both parental evasins. Moreover, unlike any natural evasins characterized to date, some chimeras exhibited specific binding to a single chemokine. These results indicate that the evasin N terminus interacts cooperatively with the “body” of the evasin to enable optimum chemokine recognition. Furthermore, the altered chemokine selectivity of the chimeras validates the approach of engineering the N termini of evasins to yield unique chemokine recognition profiles.
Keywords: inflammation, chemokine, evasin, binding, cooperativity
Abbreviations: BRET, bioluminescence resonance energy transfer; cAMP, cyclic AMP; DSF, differential scanning fluorimetry; PEI, polyethylenimine; SPR, surface plasmon resonance
The central hallmark of inflammation is the recruitment of leukocytes from the bloodstream to the affected tissues. This process is regulated by chemokines, a family of proteins expressed in response to tissue injury or infection. Chemokines bind and activate chemokine receptors expressed on the surface of leukocytes, resulting in leukocyte chemotaxis to the affected tissues and a variety of downstream responses, including innate and adaptive immunity and tissue repair (1, 2).
There are ∼50 chemokines in humans that are classified as CCL, CXCL, CX3CL, or XCL (L indicates ligand) based on the arrangement of the first two of four conserved cysteine residues in their sequences (3). Chemokine receptors are G protein–coupled receptors, classified as CCR, CXCR, CX3CR, or XCR (R indicates receptor), named according to the chemokines to which they respond. However, the chemokine–chemokine receptor network exhibits bidirectional promiscuity, whereby each receptor typically responds to several chemokines, and each chemokine typically activates multiple receptors. This complexity is likely to be one factor contributing to the failure of receptor-specific antagonists in clinical trials (4). Thus, it is attractive to consider alternative approaches for the development of anti-inflammatories, such as simultaneously suppressing the activity of several chemokines or receptors.
The simultaneous inhibition of several chemokines is used naturally by viruses, helminths, and ticks, which all secrete armories of molecules, including chemokine-binding proteins, to subvert the host immune response (5). In particular, ticks are hematophagous ectoparasites that can remain undetected by their hosts for several days to weeks. Ticks from several genera secrete salivary proteins, called evasins (designated by the prefix ‘EVA’), that each bind to multiple chemokines, preventing the activation of the cognate chemokine receptors and suppressing the inflammatory response to the tick bite during acquisition of a blood meal (6, 7). The earliest-discovered evasins, EVA-1, EVA-3, and EVA-4, have all exhibited promising anti-inflammatory activities in murine models of various inflammatory diseases (8, 9, 10, 11, 12). Therapeutic applications require evasins that selectively bind to the relevant chemokines in specific diseases, while minimizing the off-target suppression of homeostatic or other beneficial chemokines. Therefore, it is necessary to either discover natural evasins with the desired selectivity (if they exist) or to engineer natural evasins to target the relevant subset of chemokines.
Two distinct structural families of evasins are known. Class A evasins (including EVA-1 and EVA-4) usually contain eight conserved cysteine residues, which form four intramolecular disulfide bonds, and bind exclusively to CC chemokines. Class B evasins (including EVA-3) contain six conserved cysteines, forming three disulfide bonds, and bind selectively to CXC chemokines (7). Within each class, each evasin exhibits a unique spectrum of chemokine binding selectivity, ranging from quite high selectivity (∼5 chemokine targets) to broad spectrum activity (>15 chemokine targets) (13, 14, 15). Here, we focus on class A evasins, for which the structural basis of chemokine recognition is beginning to be understood (16, 17, 18). Structures of EVA-1 bound to CCL3 (18) and EVA-P974 bound to CCL7, CCL17, and a CCL8-CCL7 chimera, supported by mutational data (16), showed that the specificity of class A evasins for CC chemokines (rather than other chemokine subfamilies) is governed by structurally constrained backbone-backbone hydrogen bonds. In contrast, binding preference among CC chemokines is largely controlled by the interactions of side chains in the flexible N- and C-terminal regions of the evasin. Thus, modification of the terminal regions may enable engineering of evasins to target chemokines of interest.
In this study, we found that a new class A evasin, EVA-RPU02 from Rhipicephallus pulchellus (zebra tick or yellow back tick), exhibits similar chemokine inhibitory selectivity to that of EVA-1, despite having substantial sequence differences from EVA-1. To investigate the influence of the N terminus on chemokine selectivity, we characterized several chimeric evasins. The results showed that the chemokine binding affinity and selectivity can be altered by interchanging the N-terminal regions between the evasins. However, they also provide evidence for cooperativity in chemokine binding between the evasin N terminus and other structural regions, highlighting the challenges in rational engineering to target chemokines of interest.
Results
Expression and purification of EVA-RPU02
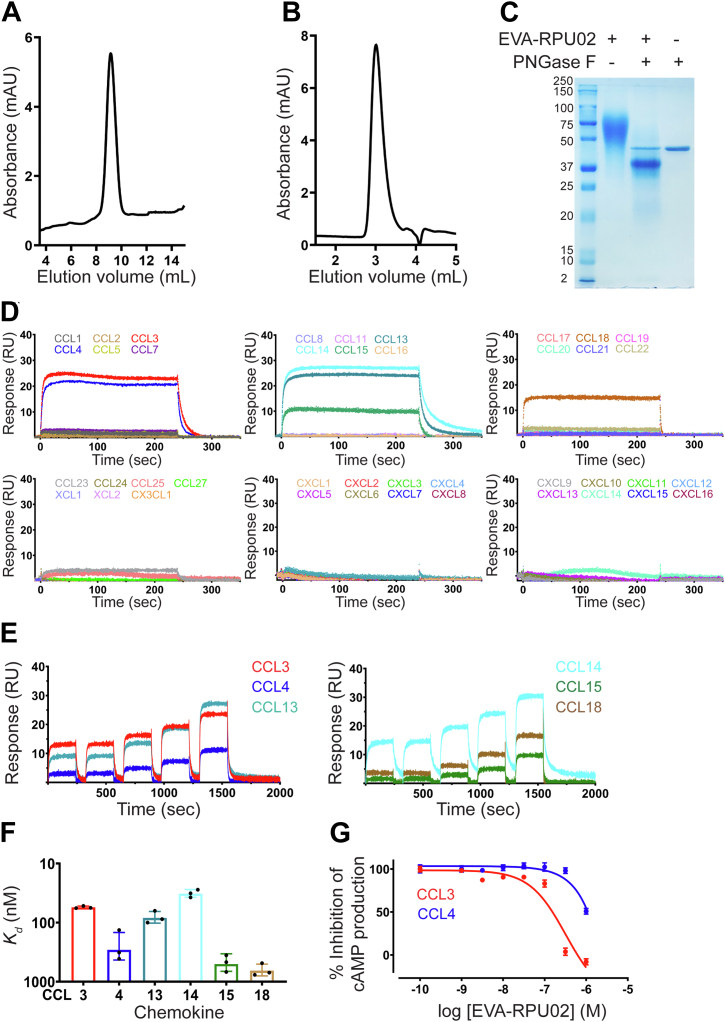
In expression trials of several putative evasins identified in previous bioinformatics searches, we found that EVA-RPU02 (referred to hereafter as EVA-R) was amenable to expression in HEK293 cells and chromatographic purification. Purified EVA-R displayed a single, sharp peak by analytical HPLC (Fig. 1A). However, it eluted as a relatively broad peak corresponding to ∼40 kDa by analytical size-exclusion chromatography (Figs. 1B and S1) and migrated in the molecular weight range ∼40 to 80 kDa by SDS-PAGE (Fig. 1C), which is substantially higher than the predicted molecular mass of ∼15.3 kDa. Treatment with PNGase F led to substantial reduction of the apparent molecular weight (Fig. 1C). Although glycan removal was incomplete, as expected (15, 19, 20, 21), these results indicated that the expressed protein was heavily glycosylated, which is consistent with the presence of up to nine potential N-linked glycosylation sites in the protein sequence (see below; Fig. 2A).
Figure 1.
EVA-R expressed in mammalian cells binds CC chemokines and inhibits their function.A and B, analytical reversed-phase high performance liquid chromatogram (A) and analytical size-exclusion chromatogram (B) of purified EVA-R. C, Coomassie blue–stained SDS-polyacrylamide gel showing purified EVA-R before and after PNGase F treatment: lane 1, protein molecular weight markers (labeled in kDa); lane 2, EVA-R after purification by size-exclusion chromatography; lane 3, purified EVA-R after treatment with PNGase F; lane 4, PNGase F alone. D, representative surface plasmon resonance sensorgrams showing binding of immobilized EVA-R to six CC chemokines (CCL3, CCL4, CCL13, CCL14, CCL15, and CCL18) but not to other CC or any CXC, CX3C, or XC chemokines, at 500 nM concentration. E, representative surface plasmon resonance sensorgrams showing binding of immobilized EVA-R to five different concentrations of the indicated chemokines (31.25, 63.5, 125, 250, and 500 nM). F, binding affinities of EVA-R for CC chemokines, measured by SPR (mean ± SD from three independent experiments). G, concentration response curves showing the inhibition of chemokines CCL3 and CCL4 by EVA-R. FlpInCHO cells stably expressing CCR5 and transfected with the cAMP biosensor CAMYEL, were treated with coelenterazine-H (5 μM, 10 min), followed by forskolin (10 μM, 10 min), followed by CCL3 (60 nM) or CCL4 (80 nM), either alone or pre-incubated with the indicated concentrations of EVA-R. Inhibition of forskolin-induced cAMP production was detected 10 min after chemokine addition. Data are presented as mean ± SEM from three independent experiments. cAMP, cyclic AMP; SPR, surface plasmon resonance.
Figure 2.
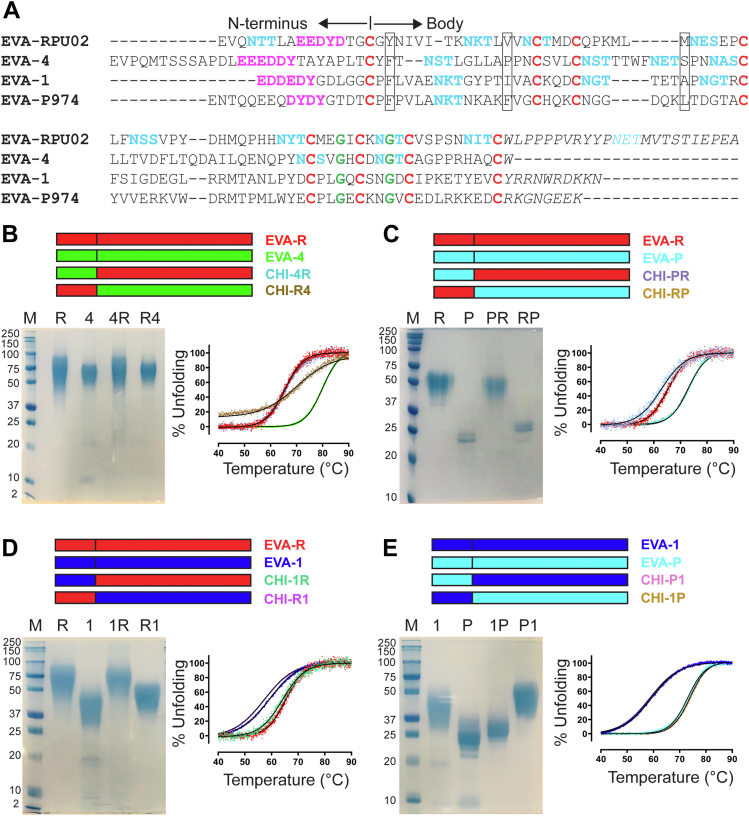
Design, purification, and thermal stability of class A evasins and chimeras.A, Sequence alignment of EVA-R, EVA-4, EVA-1, and EVA-P, highlighting features of class A evasins conserved cysteine (red) and glycine (green) residues; putative tyrosine sulfation sites (magenta); potential N-linked glycosylation sites (cyan); EVA-P residues (and the corresponding residues in other evasins) that form a hydrophobic pocket (boxed); and C-terminal regions (italics). B–E, top panels, schematic representations of parental and chimeric evasins, indicating the evasins from which the N-terminal region and body of each chimera was derived. (B–E, bottom left panels) SDS-PAGE of wildtype and chimeric evasins. Mammalian expressed and purified wildtype evasins and chimeras were subjected to SDS-PAGE under nonreducing conditions and stained with colloidal Coomassie blue. Protein marker (M), EVA-R (R), EVA-4 (4), EVA-P (P), EVA-1 (1), CHI-4R (4R), CHI-R4 (R4), CHI-PR (PR), CHI-RP (RP), CHI-1R (1R), CHI-R1 (R1), CHI-1P (1P), and CHI-P1 (P1). (B–E, bottom right panels) Cooperative thermal unfolding curves of parental and chimeric evasins obtained by nano-differential scanning fluorimetry (nano-DSF).
EVA-R selectively binds and inhibits CC chemokines
We screened the binding of EVA-R to all available human chemokines (22 CC, 16 CXC, 1 CX3C, and 2 XC) using surface plasmon resonance (SPR) at a single chemokine concentration (500 nM; Fig. 1D). As expected for class A evasins, we did not observe binding of EVA-R to any of the CXC, CX3C, or XC chemokines tested. For the six CC chemokines that bound (CCL3, CCL4, CCL13, CCL14, CCL15, and CCL18), SPR at multiple concentrations showed that the affinities (Kd values) ranged from ∼30 to 650 nM (Fig. 1, E and F and Table 1), with CCL14 (Kd = 32.4 ± 4.9 nM) and CCL3 (Kd = 55.1 ± 2.5 nM) exhibiting the highest affinities. Notably, the association and dissociation rate constants showed substantial differences among the binding chemokines (Table 1).
Table 1.
EVA-R chemokine binding parameters
| Chemokines | ka [M-1s-1] | kd [s-1] | Kd [nM] |
|---|---|---|---|
| CCL3 | (8.2 ± 1.9) × 105 | (4.6 ± 1.3) × 10−2 | 55.1 ± 2.5 |
| CCL4 | (2.9 ± 0.4) × 105 | (8.1 ± 3.9) × 10−2 | 288 ± 141 |
| CCL13 | (2.5 ± 1.7) × 105 | (2.3 ± 1.7) × 10−2 | 84 ± 19 |
| CCL14 | (5.8 ± 3.3) × 105 | (2.4 ± 1.9) × 10−2 | 32.4 ± 4.9 |
| CCL15 | (1.4 ± 0.3) × 105 | (6.9 ± 1.8) × 10−2 | 503 ± 166 |
| CCL18 | (4.9 ± 3.1) × 105 | (35 ± 26) × 10−2 | 651 ± 147 |
The kinetic parameters, association rate constant (ka) and dissociation rate constant (kd), and the equilibrium dissociation constants (Kd) of EVA-R to those chemokines exhibiting binding. The values are presented as mean ± SD obtained from three independent experiments (n = 3).
We next examined the ability of EVA-R to inhibit the functions of chemokines in a cell-based receptor-activation assay. Upon activation by chemokines, chemokine receptors give rise to inhibition of cyclic AMP (cAMP) synthesis, which can be detected in CHO cells co-expressing the relevant chemokine receptor and a cAMP biosensor. In this assay, the chemokines CCL3 and CCL4 inhibited cAMP synthesis via activation of their receptor CCR5. This effect was suppressed by EVA-R in a concentration-dependent manner (Fig. 1G). Consistent with their binding affinities, EVA-R was more potent for inhibition of CCL3 (IC50 = 300 ± 2 nM) compared to CCL4 (estimated IC50 ∼1 μM). Collectively, these data show that EVA-R, a class A evasin, not only selectively binds to chemokines but also inhibits their receptor-mediated functions.
Evasins exhibit common sequence features but unique spectra of chemokine binding selectivity
We compared the chemokine-binding selectivity of EVA-R to those of three other well-characterized class A evasins. EVA-1 has been reported to bind to only three of 24 CC chemokines tested, whereas EVA-4 and EVA-P974 (referred to hereafter as EVA-P) have been reported to bind to 17 and 14 CC chemokines, respectively (16, 18, 22). We expressed these evasins using HEK293 cells, purified them, and verified their chemokine binding selectivity using SPR (Table 2, Figs. S2, and S3). Notably, in these experiments, we took advantage of the biotinylated N-terminal Avi-tag to immobilize the evasin to the SPR chip in a specific orientation, whereas previous studies have either used SPR with randomly oriented evasins (9, 13) or biolayer interferometry (15), which is faster than SPR but suffers from lower sensitivity (23). Nevertheless, the observed chemokine binding selectivities and affinities were in good agreement with those reported previously, with minor variation possibly due to the details of the biophysical methods used (9, 13, 15).
Table 2.
Chemokine binding affinities (Kd, nM) of class A evasins (EVA-R, EVA-4, EVA-P, and EVA-1)
| Chemokines | Equilibrium dissociation constants Kd [nM] of WT evasins |
|||
|---|---|---|---|---|
| EVA-R | EVA-4 | EVA-P | EVA-1 | |
| CCL1 | - | 376 ± 63 | 15 ± 2 | - |
| CCL2 | - | 236 ± 132 | 4.1 ± 1.1 | - |
| CCL3 | 55.1 ± 2.5 | 0.2 ± 0.1 | 0.6 ± 0.1 | 0.21 ± 0.08 |
| CCL4 | 288 ± 141 | 158 ± 47 | 1.8 ± 0.1 | 60.7 ± 5.7 |
| CCL5 | - | 6.1 ± 0.1 | - | - |
| CCL7 | - | 0.8 ± 0.1 | 4.4 ± 0.3 | - |
| CCL8 | - | 0.4 ± 0.1 | 0.03 ± 0.01 | - |
| CCL11 | - | 20 ± 3 | 0.3 ± 0.1 | - |
| CCL13 | 84 ± 19 | 253 ± 17 | 0.5 ± 0.1 | - |
| CCL14 | 32.4 ± 4.9 | 1.1 ± 0.2 | 1.8 ± 0.6 | 92 ± 10 |
| CCL15 | 503 ± 166 | 240 ± 40 | 299 ± 244 | - |
| CCL16 | - | 4.1 ± 1.5 | 0.1 ± 0.1 | - |
| CCL17 | - | 283 ± 2 | 23 ± 15 | - |
| CCL18 | 651 ± 147 | 0.4 ± 0.1 | 0.6 ± 0.1 | 192 ± 27 |
| CCL22 | - | 8.5 ± 0.1 | 1.8 ± 0.1 | - |
| CCL23 | - | 7.4 ± 0.1 | - | - |
| CCL24 | - | 55 ± 2 | - | - |
The equilibrium dissociation constant (Kd) of four class A evasins for their cognate chemokines. A hyphen indicates nonmeasurable binding at 500 nM. The values are presented as mean ± SD obtained from three independent experiments (n = 3).
To interrogate the features of these evasins that may contribute to their differences in chemokine selectivity, we compared their aligned sequences (Fig. 2A). All four evasins contain the expected eight conserved cysteine and two conserved glycine residues, which stabilize the evasin structural core. In addition, they contain 1 to 9 potential N-linked glycosylation sites, consistent with differences in SDS-PAGE mobility (Fig. S2), but unlikely to influence chemokine binding (13, 15, 24). Previous studies have shown that residues within the N-terminal regions (before the first conserved Cys residue) of EVA-1 and EVA-P play roles in chemokine binding (16, 18). The N-terminal regions of all four evasins in the present study contain one or two predicted tyrosine sulfation sites. In the case of EVA-P, sulfation at these sites has been reported to enhance chemokine binding affinity (21). However, the N-terminal regions vary considerably in amino acid composition and length, as well as the separation between the predicted tyrosine sulfation sites and the first conserved Cys residue. We therefore postulated that the variations between the N-terminal regions of these evasins are responsible, in part, for their distinct chemokine recognition profiles.
Design, production, and stability of evasin chimeras
To test the hypothesis that the N-terminal regions of class A evasins make substantial contributions to their chemokine recognition profiles, we prepared a series of chimeric proteins in which the N-terminal regions were swapped between these evasins (Fig. 2, B–E). Specifically, we swapped the N-terminal region of EVA-R with those of EVA-4 (Fig. 2B), EVA-P (Fig. 2C), and EVA-1 (Fig. 2D) and also swapped the N-terminal regions of EVA-1 and EVA-P (Fig. 2E). Chimeric evasins were named using the format CHI-XY, in which X represents the evasin from which the N-terminal region was derived, and Y represents the evasin from which the rest of the chimera (the “body” of the evasin) was derived. For example, CHI-4R consists of the N terminus of EVA-4 and the body of EVA-R. Each chimeric evasin was expressed and purified from HEK293 cells. For each chimera, the homogeneity and molecular weight range indicated by SDS-PAGE (Fig. 2, B–E, bottom left panels) were similar to that of the parental evasin from which the body of the chimera was derived. This is consistent with most of the predicted N-linked glycosylation sites being located in the body of each evasin (Fig. 2A).
Considering that the N-terminal regions of evasins are likely to be disordered (in the absence of chemokines), correct folding of the chimeric evasins is expected to give rise to structural stability dominated by the body of the evasin. To test this, we determined the thermal stability of each chimera and each wildtype evasin using differential scanning fluorimetry (DSF) (Fig. 2, B–E, bottom right panels). The wildtype evasins exhibited well-defined unfolding profiles in the range ∼35 to 90 °C, with midpoints (Tm values) of 79.9 ± 0.1 °C (EVA-4), 73.1 ± 0.1 °C (EVA-P), 65.4 ± 0.4 °C (EVA-R), and 59.2 ± 0.1 °C (EVA-1), in decreasing order of stability (Table 3). With the sole exception of CHI-R4, all the chimeric evasins exhibited very similar unfolding profiles and Tm values to the wildtype evasin from which the body of the chimera was derived (Fig. 2, B–E, bottom right panels; Table 3). For example, CHI-1P (Tm = 73.8 ± 0.1 °C) exhibits similar stability to EVA-P (Tm = 73.0 ± 0.1 °C), whereas CHI-P1 (Tm = 58.6 ± 0.1 °C) exhibits similar stability to EVA-1 (Tm = 59.2 ± 0.1 °C) (Fig. 2E). Although CHI-R4 also had a cooperative unfolding curve (Fig. 2B), this curve had a smaller slope and substantially lower midpoint (Tm = 70.1 ± 0.2 °C) compared to EVA-4 (Tm = 79.9 ± 0.1 °C), suggesting that CHI-R4 may be partially misfolded.
Table 3.
Thermal stability of chimeric evasins
| Evasins | Tm (°C) | |
|---|---|---|
| Parental evasin | EVA-R | 65.4 ± 0.4 |
| Chimeras containing the body of EVA-R | CHI-4R | 65.5 ± 0.1 |
| CHI-PR | 62.9 ± 0.4 | |
| CHI-1R | 64.1 ± 0.2 | |
| Parental evasin | EVA-4 | 79.9 ± 0.1 |
| Chimera containing the body of EVA-4 | CHI-R4 | 70.1 ± 0.2 |
| Parental evasin | EVA-P | 73.0 ± 0.1 |
| Chimeras containing the body of EVA-P | CHI-RP | 72.9 ± 0.1 |
| CHI-1P | 73.8 ± 0.1 | |
| Parental evasin | EVA-1 | 59.2 ± 0.1 |
| Chimeras containing the body of EVA-1 | CHI-R1 | 57.4 ± 0.6 |
| CHI-P1 | 58.6 ± 0.1 | |
Melting midpoint (Tm) of parental and chimeric evasins obtained from cooperative thermal unfolding curves. The values are presented as mean ± SD obtained from three independent experiments (n = 3).
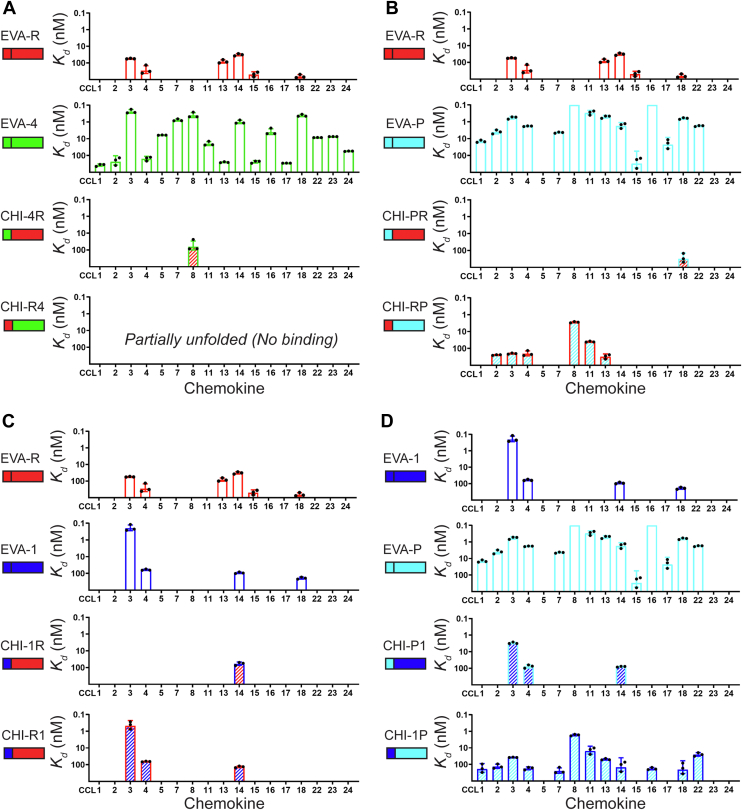
The DSF results establish that seven of the eight chimeras are correctly folded, enabling us to interpret their chemokine binding affinities to understand the contributions of the N-terminal region and the body of each evasin. We therefore used SPR to screen the binding of each chimera to all available chemokines. As expected, all the chimeras failed to bind detectably to CXC, XC, or CX3C chemokines, whereas most of the chimeras bound to subsets of CC chemokines. We determined Kd values for binding of each chimeric evasin to each chemokine (Fig. 3, A–D and Table 4).
Figure 3.
Chimeric evasins display distinct chemokine binding selectivity profiles. Bar diagrams showing binding affinity (Kd) of CC chemokines to: A, EVA-R, EVA-4 and their chimeras; B, EVA-R, EVA-P, and their chimeras; C, EVA-R, EVA-1, and their chimeras; and D, EVA-1, EVA-P, and their chimeras. In each panel, the bar diagrams for the relevant wildtype proteins are repeated for ease of direct comparison to the chimeras. Bars represent mean ± SD of three independent measurements. The Kd for binding of EVA-P to CCL8 and CCL16 is tighter than the measurable limit (∼0.1 nM).
Table 4.
Chemokine binding affinities (Kd, nM) of chimeric evasins
|
Kd (nM) of chimeric evasins |
|||||||
|---|---|---|---|---|---|---|---|
| Chemokine | CHI-4R | CHI-PR | CHI-RP | CHI-1R | CHI-R1 | CHI-P1 | CHI-1P |
| CCL1 | - | - | - | - | - | - | 191 ± 104 |
| CCL2 | - | - | 244 ± 12 | - | - | - | 134 ± 39 |
| CCL3 | - | - | 203 ± 15 | - | 0.5 ± 0.2 | 2.9 ± 0.3 | 37 ± 1 |
| CCL4 | - | - | 207 ± 67 | - | 65 ± 3 | 78 ± 18 | 174 ± 35 |
| CCL7 | - | - | - | - | - | - | 250 ± 90 |
| CCL8 | 64 ± 35 | - | 2.8 ± 0.2 | - | - | - | 1.6 ± 0.2 |
| CCL11 | - | - | 40 ± 3 | - | - | - | 15 ± 8 |
| CCL13 | - | - | 313 ± 99 | - | - | - | 49 ± 5 |
| CCL14 | - | - | - | 60 ± 14 | 134 ± 15 | 79 ± 5 | 149 ± 110 |
| CCL16 | - | - | - | - | - | - | 182 ± 28 |
| CCL18 | - | 315 ± 171 | - | - | - | - | 206 ± 147 |
| CCL22 | - | - | - | - | - | - | 25 ± 6 |
Values shown are the mean ± SD obtained from three independent experiments. A hyphen indicates no measurable binding at 500 nM concentration.
Chemokine binding by chimeras of EVA-R and EVA-4
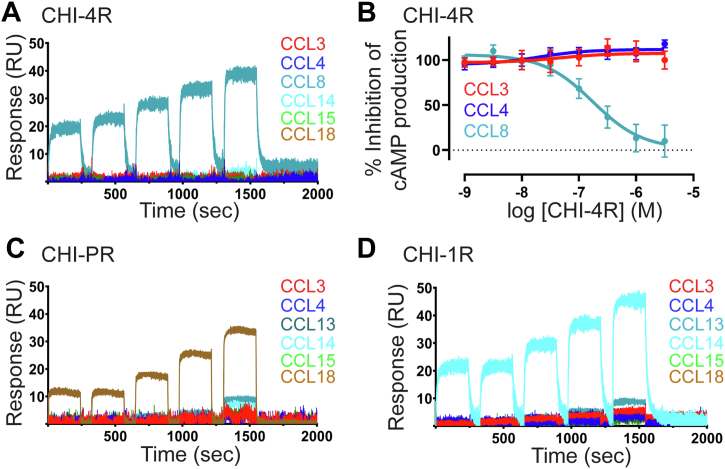
As noted above, EVA-4 displayed a broad spectrum of chemokine recognition, whereas EVA-R had a relatively narrow spectrum of activity. Thus, N-terminal swap chimeras of these two evasins could provide information about the role of the N terminus in broad-spectrum versus narrow-spectrum chemokine recognition. Chimera CHI-R4 did not exhibit measurable binding to any CC chemokines, possibly because it is not correctly folded (see above). Interestingly, CHI-4R bound to CCL8 with quite high affinity (Kd ∼60 nM) but did not bind detectably to any other CC chemokines (at 500 nM concentration), indicating at least 10-fold selectivity for CCL8 over other CC chemokines (Figs. 3A and 4A). Consistent with this selective binding, CHI-4R also selectively inhibited CCL8 (but not CCL3 or CCL4) function (Figs. 4B and S4). To our knowledge, this is the first time an evasin has been engineered to specifically bind and inhibit a single chemokine. The selectivity of CHI-4R differs substantially from those of both the parental evasins, suggesting that it does not result from simple additivity between the chemokine binding interactions of the evasin body and N-terminal regions. Instead, chemokine recognition appears to involve cooperativity between these two regions of the evasins (see Discussion).
Figure 4.
Chimeric evasins selectively bind and inhibit chemokine function.A, C, and D, representative surface plasmon resonance sensorgrams showing the selective binding of immobilized chimeric evasins to chemokines (31.25, 63.5, 125, 250, and 500 nM); CHI-4R (A) bound selectively to CCL8; CHI-PR (C) bound selectively to CCL18; and CHI-1R (D) bound selectively to CCL14. B, concentration response curves showing the selective inhibition of CCL8, but not of CCL3 and CCL4, by CHI-4R; experimental details are the same as for Figure 1G.
Chemokine binding by chimeras of EVA-R and EVA-P
Considering that EVA-P bound to all the same chemokines as EVA-R, as well as a number of additional chemokines, we predicted that the chimeras CHI-PR and CHI-RP would, at a minimum, bind to the same chemokines as EVA-R. However, surprisingly we found that the binding spectra of these two chimeras were distinct from those of the parental evasins. CHI-PR exhibited selective but very weak binding to CCL18 (Figs. 3B and 4C). Notably this chemokine also bound to both of the parental evasins (Fig. 3B). CHI-RP recognized three of the same chemokines as EVA-R (CCL3, CCL4, and CCL13) and three additional chemokines and did not bind detectably to CCL14, which is a fairly high-affinity binder to both parental evasins (Kd ∼ 32 nM to EVA-R and ∼1.8 nM to EVA-P). These data further indicate that the N-terminal region and body of these evasins may work cooperatively to recognize cognate chemokines.
Chemokine binding by chimeras of EVA-R and EVA-1
EVA-R and EVA-1 have very similar, narrow-spectrum chemokine recognition profiles. CHI-R1 displayed a similar chemokine recognition profile to EVA-1, except for loss of binding to CCL18 (Fig. 3C). This suggests that the body of EVA-1 may dominate binding to CCL3, CCL4, and CCL14, whereas binding to CCL18 requires some element(s) of the EVA-1 N-terminal region. In contrast, CHI-1R exhibited absolute selectivity to CCL14 (Kd ∼ 60 nM) over all other chemokines (Kd > 500 nM) (Figs. 3C and 4D). This is similar to the above observation that CHI-4R exhibited absolute specificity for CCL8, again supporting a role for cooperative chemokine-binding interactions by the body and N-terminal region of this chimeric evasin.
Chemokine binding by chimeras of EVA-P and EVA-1
Finally, considering that the equivalent regions of EVA-1 and EVA-R did not functionally compensate for each other in chimeras of these two evasins, we postulated that substituting regions of these two evasins into EVA-P would affect chemokine recognition differently. In support of this hypothesis, we found that the chemokine recognition spectrum of CHI-P1 (Fig. 3D) differed substantially from that of CHI-PR (Fig. 3B). This comparison supports an important role for the evasin body in defining chemokine selectivity. The final chimera CHI-1P exhibited the broadest chemokine recognition profile of all the chimeras tested, binding to most of the same chemokines as EVA-P, but with reduced affinity (Fig. 3D).
Discussion
Tick evasins are natural chemokine binders that inhibit receptor signaling and have the potential to be modified for use as clinical anti-inflammatory agents. However, in order to appropriately engineer these tick-derived biomolecules, we need an improved understanding of the factors that influence the affinity and selectivity of evasins for target chemokines. In the current study, we characterized the evasin EVA-R for the first time and analyzed chimeras between EVA-R and three other evasins (EVA-1, EVA-4, and EVA-P) to investigate the role of the evasin N-terminal region in chemokine recognition. Although the results support a key role for the evasin N terminus, they also show that the N terminus works cooperatively with the body of the evasin to achieve chemokine binding selectivity.
Role of the evasin N-terminal region in chemokine recognition
The current data clearly demonstrate that the N-terminal sequence has a major influence on the chemokine selectivity of class A evasins. For example, replacing the N terminus of EVA-P with that of EVA-R yielded a chimera that bound to only 6 of the 14 chemokine ligands of EVA-P (CHI-RP, Fig. 3B). The importance of the N terminus is consistent with several previous reports. Bonvin et al. (22) showed that two residues in the N-terminal region of EVA-4 (Glu16 and Tyr19) were critical for binding to CCL5. Eaton et al. (20) showed that replacing the N terminus and the first β-sheet of EVA-1 with those of EVA-P972 caused a substantial change in chemokine selectivity. More recently, we have shown that truncations and point mutations in the N-terminal region of EVA-P alter the chemokine selectivity profile of this evasin (16).
Importance of other evasin elements
While our results support the importance of the evasin N-terminal region, they also indicate that the N-terminal region alone is not sufficient to define the chemokine selectivity of each evasin, indicating that other regions of the structure contribute to binding affinity and selectivity. For example, EVA-4 and CHI-4R, which have the same N-terminal region, exhibited starkly different selectivity profiles (Fig. 3A). Similarly, EVA-R bound to CCL13 with similar or higher affinity compared to CCL3 and CCL4, whereas CHI-R1, which has the same N-terminal region as EVA-R, failed to bind to CCL13 but retained binding to CCL3 and CCL4 (Fig. 3C).
Until recently, only a single structure of an evasin:chemokine complex had been reported, the structure of EVA-1 bound to CCL3 (18). However, we have recently described the structures of EVA-P bound to each of two wildtype chemokines (CCL7 and CCL17) and a chimeric chemokine (in which the N-terminal and N-loop regions of CCL7 were replaced by those of CCL8), as well as the structure of an EVA-P mutant (F31A) bound to CCL2 (16). These structures clearly support the key role of the evasin N-terminal region but also indicate two other regions that contribute to selective recognition of different chemokines. First, several noncontiguous residues from the first two β-sheets form a hydrophobic pocket that binds to the first residue of the chemokine N-loop, a conserved large hydrophobic amino acid. The residues forming this hydrophobic pocket differ among the four evasins studied here (boxes in Fig. 2A), potentially contributing to their chemokine binding selectivity. Second, the C-terminal tail of the evasin is in close proximity to the N-terminal tail of the chemokine. In most structures, these regions are largely disordered, and the resolved residues show minimal direct contacts. However, the structure of EVA-P bound to the CCL8-CCL7 chimera displays a more extensive and ordered interaction surface involving the N-terminal regions of CCL8 (16). C-terminal truncation mutants provided clear evidence that the EVA-P C-terminal residues contribute to high affinity binding of (and extremely slow dissociation from) CCL8. The C-terminal regions of the four evasins analyzed in the current study differ substantially both in their lengths and amino acid sequences (italics in Fig. 2A), suggesting that the interactions of these regions with the chemokine N termini may influence the chemokine binding affinities and selectivities of these evasins.
Cooperativity between structural elements
Whereas the current results provide evidence that the N-terminal and other regions of the evasins are involved in chemokine recognition, they also suggest that the interactions of these different elements are cooperative rather than simply additive. Perhaps the best evidence for cooperativity comes from the chimera CHI-4R. This chimera is formed from segments of EVA-4 and EVA-R. Both of these parental evasins bind to the chemokines CCL3, CCL4, CCL13, CCL14, and CCL18. The chimera CHI-4R is well-folded and capable of chemokine binding (to CCL8; Fig. 3A). However, it does not bind detectably to any of the five chemokines that bind to both parental evasins (Fig. 3A). Thus, binding to these chemokines is supported by the N-terminal region of EVA-4 in the context of the body of EVA-4 or by the N-terminal region of EVA-R in the context of the body of EVA-R, but it is not supported by the N-terminal region of EVA-4 in the context of the body of EVA-R. This clearly indicates that the different structural regions of the evasin recognize the chemokines in a cooperative manner.
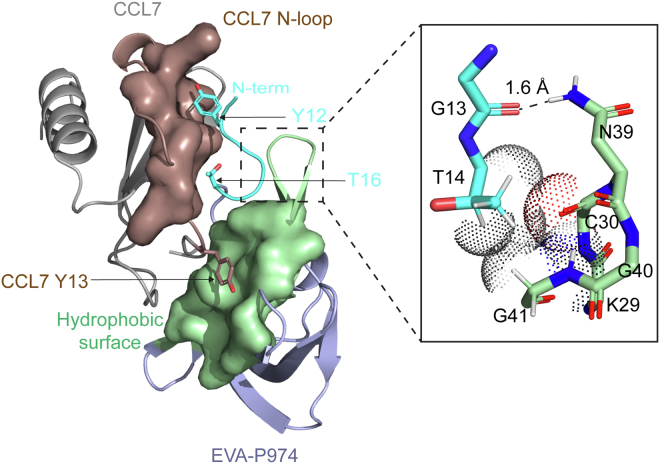
Although we had not anticipated that the different regions of the evasins would exhibit such clear cooperativity, we can rationalize this observation based on the recent structure of EVA-P bound to CCL7 (Fig. 5) (16). The structure showed that the chemokine N-loop interacts with both the body and the flexible N terminus of the evasin. Moreover, point mutations verified that these interactions contribute to the binding energy. Specifically, the first residue of the chemokine N-loop (Tyr13 in CCL7) interacts with a hydrophobic pocket in the body of the evasin (green surface in Fig. 5), and several chemokine N-loop residues, together with residues in the third β-strand, form a well-defined surface (brown surface in Fig. 5) that interacts with evasin N-terminal residues Tyr12 and Thr16. However, the structure also revealed well-defined, direct intramolecular interactions (a hydrogen bond and several van der Waals interactions) between the N-terminal region and the body of EVA-P (Fig. 5, expanded region). These interactions appear to position the N-terminal region optimally to bind to the chemokine N-loop. Thus, exchange of the N terminus between the bodies of two different evasins may disrupt these intramolecular interactions, altering the ability of the N-terminal region to interact optimally with the chemokine and providing a structural rationale for the observed cooperativity between the evasin body and N-terminal region.
Figure 5.
Possible structural basis of chemokine binding cooperativity displayed by the evasin N-terminal region and body. (main image) Structure (PDB code: 7S58) of EVA-P (light blue and cyan ribbons; hydrophobic surface in green) bound to CCL7 (gray ribbons; N-loop surface in brown). Key residues in the EVA-P N terminus for binding to the CCL7 N-loop surface are shown as cyan sticks. CCL7 residue Tyr13, which interacts with the hydrophobic surface of EVA-P is shown as brown sticks. (expanded region) Residues in the EVA-P N-terminal region (cyan sticks) form intramolecular interactions (hydrogen bond and van der Waals interactions) to a hairpin turn connected to the hydrophobic surface of EVA-P, suggesting that interactions of the EVA-P N-terminal region and hydrophobic surface may be coupled to each other.
Multiple class A evasins from R. pulchellus
EVA-R is the third class A evasin from the tick R. pulchellus whose chemokine binding has been characterized (15, 19, 20, 25, 26, 27). We found that EVA-R bound to only six human CC chemokines, whereas other R. pulchellus evasins (EVA-P467 and EVA-P672) have been reported to bind up to 14 human CC chemokines, depending on the position of an affinity tag (15, 20). Curiously, the human chemokines recognized by EVA-R are a subset of those recognized by these other two evasins, which bind to similar sets of chemokines. This raises the question of whether there is some advantage to the tick of producing all three evasins. On the other hand, analysis of transcriptome databases identifies another 17 putative class A evasins (yet to be validated) from R. pulchellus, and it seems likely that some of these evasins bind to different groups of CC chemokines. Interestingly, R. pulchellus, which is endemic to parts of Africa, parasitizes a wide variety of species, including cattle, horses, zebras, rhinoceros, elephants, giraffes, baboons, and humans, as well as some birds (28, 29, 30), and can transmit viral and bacterial pathogens. It is tempting to speculate that by producing a cocktail of salivary evasins that inhibit numerous chemokines, R. pulchellus enhances both its survival on a particular host and its transmission between different host species.
Concluding remarks
Based on an emerging structural understanding of chemokine recognition by class A evasins and comparison of evasin sequences, we anticipated that swapping the flexible N-terminal region between different evasins would yield variants with altered selectivity for target chemokines. Our results verify this prediction but show that the selectivity does not change in an easily predictable manner. In particular, as a consequence of cooperativity between the chemokine interactions of the N-terminal region and other regions of the evasins, swapping the N-terminal regions can give rise to chimeras with novel chemokine selectivity profiles, including, fortuitously, high selectivity for a single chemokine. Thus, while it will be difficult to rationally design evasin variants with desired chemokine selectivity, our results suggest that an efficient approach to discovering such variants may be the screening of combinatorial libraries of evasin chimeras.
Experimental procedures
Protein expression and purification
All parental and chimeric evasins were expressed in Expi293F suspension cells (Thermo Fischer Scientific). The genes encoding parental and chimeric evasins, containing a His6-tag, an Avi-tag (LNDIFEAQKIEWHE) and a (G4S)3 linker on the N terminus, were cloned into the pSecTag2a expression vector using restriction enzymes SfiI and XhoI (New England Biolabs) and transfected into the cells using polyethylenimine (PEI) (Polysciences Inc.). Briefly, Expi 293F cells were grown to a density of 2 × 106 cells/ml in 90 ml of Expi293 Expression Medium (Thermo Fischer Scientific). For transfection, 100 μg DNA and 400 μg PEI were mixed with 9.5 ml of PBS, vortexed briefly, incubated for 20 min at 25 °C, and added dropwise to the cell suspension with gentle swirling. The cells were then grown in a shaking incubator at 125 rpm, 37 °C and 5% CO2 for 85 to 90 h. The cell suspension was centrifuged at 4300 x g for 15 min, and the culture media containing the secreted protein were collected and purified using subsequent chromatographic procedures. The culture media were loaded onto a 5 ml HisTrap FF column equilibrated with 20 mM Tris, 500 mM NaCl, 10% glycerol, pH 8.0. The column was washed with the same buffer containing 20 mM imidazole, and the bound His6-fusion protein was eluted with the same buffer containing 250 mM imidazole. The eluted proteins were concentrated to 1 ml using an Amicon Ultra-15 centrifugal filter (Merck) and purified by size-exclusion chromatography, using Superdex 75 Increase 10/300 Gl column (Cytiva) with buffer containing 10 mM Hepes, 150 mM NaCl, 5% glycerol, pH 7.4, and a flow rate of 0.7 ml/min. The fractions containing the eluted proteins were pooled and used immediately or flash frozen and stored at −80 °C for further use.
Quality control analysis of the purified proteins
The purity and homogeneity of parental evasins and chimeras were analyzed by reversed-phase high-performance liquid chromatography, analytical size-exclusion chromatography, and SDS-PAGE. For reversed-phase high-performance liquid chromatography, 10 μl of purified protein was loaded onto a C4 column equilibrated with water with 0.1% trifluoroacetic acid. The protein was then eluted with gradient flow (1% increment per minute) of acetonitrile with 0.1% trifluoroacetic acid. For analytical size-exclusion chromatography, 10 μl of purified protein was loaded onto a Yarra SEC-2000 300 × 4.6 mm LC column (Phenomenex) and eluted with 0.1 M phosphate saline buffer, pH 6.8, at a flow rate of 0.35 ml/min. The calibration curve was prepared by running protein standards under the same condition. In all cases, the proteins were monitored by observing the absorbance at 280 nm. SDS-PAGE was performed under nonreducing conditions and the gels stained with colloidal Coomassie blue for protein visualization.
Deglycosylation of purified evasin
Mammalian expressed and purified EVA-R was deglycosylated using PNGase F (New England Biolabs) under denaturing conditions. 10 μg of EVA-R was mixed with 10× glycoprotein denaturing buffer and denatured by heating the mixture at 100 °C for 10 min. The denatured protein was then chilled and mixed with 10× GlycoBuffer 2, nonyl phenoxypolyethoxylethanol (NP-40), and PNGase F. The reaction mixture was incubated at 37 °C for 1 h. The deglycosylated protein was assessed by SDS-PAGE and visualized by staining with colloidal Coomassie blue.
Thermal unfolding using nano-DSF
The thermal unfolding of parental and chimeric evasins was performed using nano-DSF (Prometheus NT.48, Nano Temper Technologies). Capillaries were filled with 15 μl of protein sample and exposed to an increasing temperature gradient from 25 °C to 95 °C at a ramp of 1 °C/min. The intrinsic fluorescence was monitored in real time, with an excitation wavelength of 280 nm and emission wavelengths of 330 nm and 350 nm. The ratio of fluorescence emission intensities at 350 nm to 330 nm was normalized and fitted in GraphPad Prism using a sigmoidal function to yield the melting point (Tm). Data shown and Tm values represent the average of three independent replicates.
Site-specific biotinylation of Avi-tagged proteins
Purified parental and chimeric evasins were buffer exchanged to 10 mM Tris (pH 8.0). Site-specific biotinylation of the Avi-tag was performed by incubating evasins with 500 μM D-biotin, 100 mM magnesium acetate, and 100 mM ATP in the presence of 2.5 μg Bir-A ligase (produced in-house) at 37 °C for 1 h in buffer containing 500 mM Bicine, pH 8.3. Biotinylated proteins were further purified from the mixture by size-exclusion chromatography as described above. The fractions containing the biotinylated proteins were pooled for chemokine binding analysis.
Chemokine binding analysis by SPR
Some chemokines (CCL2, CCL7, CCL8, CCL11, CCL16, CCL17, and CCL18) were produced in-house, as described (16), with quality verified using HPLC and MS, whereas others were purchased from PeproTech. The chemokine binding analysis of evasins was performed by SPR using a Biacore T100 instrument (GE Healthcare), Biotin CAPture kit Series S (Cytiva), and running buffer containing 10 mM Hepes, 500 nM NaCl, 0.002% Tween 20, 3 mM EDTA, 1 mg/ml carboxymethyl dextran, pH 7.5. Briefly, a sensor chip was coated with Biotin CAPture reagent (2 μl/min,120 s). Biotinylated evasins (0.1–0.2 μM) in running buffer were immobilized at a flow rate of 10 μl/min to achieve an increase of 100 to 150 response units (RU). For the initial binding screening, 500 nM of each available chemokine (either produced in-house or purchased from PeproTech) in the running buffer was flowed through the evasin-immobilized sensor chip at a flow rate of 30 μl/min for 240 s, followed by 600 s of buffer alone to allow dissociation. For binding kinetics and affinities, the chemokines that exhibited binding on initial screening were then flowed through an evasin-immobilized sensor chip at five increasing concentrations (31.25, 63.5, 125, 250, and 500 nM). The association rate constants, dissociation rate constants, and binding affinities were determined by fitting the obtained sensorgrams with 1:1 binding kinetics using Biacore T100 Evaluation Software. Each experiment was repeated three times independently.
cAMP inhibition assay
Functional inhibition of chemokine-mediated receptor signaling by evasins was carried out using cAMP-based bioluminescence resonance energy transfer (BRET) biosensor assay (31). Briefly, FlpInCHO cells stably expressing chemokine receptor CCR5 were cultured in Dulbecco’s Modified Eagle Medium, supplemented with 5% fetal bovine serum and 1% penicillin/streptomycin, overnight, at 37 °C, in a humidified incubator maintained with 5% CO2. For transient transfection, 5 μg of plasmid containing CAMYEL biosensor was mixed with 500 μl of PBS and vortexed thoroughly. PEI (30 μg) was added, and the mixture was vortexed and incubated for 20 min at 25 °C, then added dropwise into a 10-cm culture dish containing four million adherent cells (32). After 24 h incubation, the transfected cells were detached using PBS-EDTA solution and transferred to a 96-well culture plate (50,000 cells/well), then further incubated for 18 h. Cells were then washed twice and equilibrated with Hank’s Balanced Salt Solution (Gibco) for 10 min at 37 °C, incubated with 5 μM Coelenterazine-H (Nanolight Technology) for 10 min. To obtain chemokine concentration–response curves, cells were activated by different concentrations of either CCL3, CCL4 or CCL8 for 10 min, followed by the addition of 10 μM forskolin (Sigma Aldrich). After a further 10 min, the dual luminescence emissions by RLuc and YFP were measured at a wavelength range of 445 to 505 nm and 505 to 565 nm, respectively, using a BRET one plus module in a PHERAstar FS plate reader (BMG Labtech). BRET ratio was obtained by calculating the ratio of YFP to RLuc emissions. Based on the concentration–response evaluation, cells were then treated with CCL3 (60 nM) or CCL4 (80 nM) or CCL8 (40 nM) either alone or preincubated with different concentrations of evasins, and the BRET ratio was measured as described above. The change in BRET ratio is expressed as a percentage of forskolin-induced cAMP inhibition of the chemokine in the absence of evasin.
Data availability
Experimental raw data are available to be shared upon request to the corresponding authors.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank David Steer (staff of Monash Proteomics & Metabolomics Facility) for assistance with mass spectroscopy and Geoffrey Kong (staff of Monash Macromolecular Crystallisation Facility) for assistance with nano-DSF.
Author contributions
P. A., R. P. B., R. L., R. J. P., D. J., and M. J. S. methodology; P. A., S. R. D., D. J., and R. P. B. investigation; P. A., S. R. D., R. P. B., and M. J. S. formal analysis; P. A., R. P. B., and M. J. S. writing-original draft; P. A., S. R. D., D. J., R. P. B., R. L., R. J. P. and M. J. S. writing-review and editing; P. A., R. L., R. J. P., R. P. B., and M. J. S. conceptualization; P. A. and R. P. B. validation; R. L., R. P. B., and M. J. S. supervision; R. J. P., R. P. B., and M. J. S. funding acquisition; R. P. B. and M. J. S. project administration.
Funding and additional information
This research was supported by National Health and Medical Research Council Project Grants APP1140867 (M. J. S. and R. J. P.) and APP1140874 (M. J. S.) and Investigator Grant APP1174941 (R. J. P.) by a Bridging Postdoctoral Fellowship from Monash University (R. P. B.) and by a Faculty Platform Access Grant from Monash University (R. P. B. and M. J. S.).
Edited by Karen Fleming
Contributor Information
Ram Prasad Bhusal, Email: ram.bhusal@monash.edu.
Martin J. Stone, Email: martin.stone@monash.edu.
Supporting information
References
- 1.Keane M.P., Strieter R.M. Chemokine signaling in inflammation. Crit. Care Med. 2000;28:N13–N26. doi: 10.1097/00003246-200004001-00003. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H., Chen K., Tan Q., Shao Q., Han S., Zhang C., et al. Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5. Nat. Commun. 2021;12:4151. doi: 10.1038/s41467-021-24438-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kufareva I., Salanga C.L., Handel T.M. Chemokine and chemokine receptor structure and interactions: Implications for therapeutic strategies. Immunol. Cell Biol. 2015;93:372–383. doi: 10.1038/icb.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao S., Wu B., Stevens R.C. Advancing chemokine GPCR structure based drug discovery. Structure. 2019;27:405–408. doi: 10.1016/j.str.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 5.González-Motos V., Kropp K.A., Viejo-Borbolla A. Chemokine binding proteins: an immunomodulatory strategy going viral. Cytokine Growth Factor Rev. 2016;30:71–80. doi: 10.1016/j.cytogfr.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharya S., Nuttall P.A. Phylogenetic analysis indicates that evasin-like proteins of ixodid ticks fall into three distinct classes. Front. Cell Infect. Microbiol. 2021;11:769542. doi: 10.3389/fcimb.2021.769542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhusal R.P., Eaton J.R.O., Chowdhury S.T., Power C.A., Proudfoot A.E.I., Stone M.J., et al. Evasins: tick salivary proteins that inhibit mammalian chemokines. Trends Biochem. Sci. 2020;45:108–122. doi: 10.1016/j.tibs.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonvin P., Power C.A., Proudfoot A.E.I. Evasins: therapeutic potential of a new family of chemokine-binding proteins from ticks. Front. Immunol. 2016;7:208. doi: 10.3389/fimmu.2016.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Déruaz M., Bonvin P., Severin I.C., Johnson Z., Krohn S., Power C.A., et al. Evasin-4, a tick-derived chemokine-binding protein with broad selectivity can be modified for use in preclinical disease models. FEBS J. 2013;280:4876–4887. doi: 10.1111/febs.12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montecucco F., Mach F., Lenglet S., Vonlaufen A., Gomes Quinderé A.L., Pelli G., et al. Treatment with Evasin-3 abrogates neutrophil-mediated inflammation in mouse acute pancreatitis. Eur. J. Clin. Invest. 2014;44:940–950. doi: 10.1111/eci.12327. [DOI] [PubMed] [Google Scholar]
- 11.Russo R.C., Alessandri A.L., Garcia C.C., Cordeiro B.F., Pinho V., Cassali G.D., et al. Therapeutic effects of evasin-1, a chemokine binding protein, in bleomycin-induced pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2011;45:72–80. doi: 10.1165/rcmb.2009-0406OC. [DOI] [PubMed] [Google Scholar]
- 12.Braunersreuther V., Montecucco F., Pelli G., Galan K., Proudfoot A.E., Belin A., et al. Treatment with the CC chemokine-binding protein evasin-4 improves post-infarction myocardial injury and survival in mice. Thromb. Haemost. 2013;110:807–825. doi: 10.1160/TH13-04-0297. [DOI] [PubMed] [Google Scholar]
- 13.Frauenschuh A., Power C.A., Déruaz M., Ferreira B.R., Silva J.S., Teixeira M.M., et al. Molecular cloning and characterization of a highly selective chemokine-binding protein from the tick rhipicephalus sanguineus. J. Biol. Chem. 2007;282:27250–27258. doi: 10.1074/jbc.M704706200. [DOI] [PubMed] [Google Scholar]
- 14.Lee A.W., Deruaz M., Lynch C., Davies G., Singh K., Alenazi Y., et al. A knottin scaffold directs the CXC-chemokine–binding specificity of tick evasins. J. Biol. Chem. 2019;294:11199–11212. doi: 10.1074/jbc.RA119.008817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh K., Davies G., Alenazi Y., Eaton J.R.O., Kawamura A., Bhattacharya S. Yeast surface display identifies a family of evasins from ticks with novel polyvalent CC chemokine-binding activities. Sci. Rep. 2017;7:4267. doi: 10.1038/s41598-017-04378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhusal R.P., Aryal P., Devkota S.R., Pokhrel R., Gunzburg M.J., Foster S.R., et al. Structure-guided engineering of tick evasins for targeting chemokines in inflammatory diseases. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2122105119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denisov S.S., Ramírez-Escudero M., Heinzmann A.C.A., Ippel J.H., Dawson P.E., Koenen R.R., et al. Structural characterization of anti-CCL5 activity of the tick salivary protein evasin-4. J. Biol. Chem. 2020;295:14367–14378. doi: 10.1074/jbc.RA120.013891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dias J.M., Losberger C., Déruaz M., Power C.A., Proudfoot A.E.I., Shaw J.P. Structural basis of chemokine sequestration by a tick chemokine binding protein: the crystal structure of the complex between evasin-1 and CCL3. PLoS One. 2010;4 doi: 10.1371/journal.pone.0008514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alenazi Y., Singh K., Davies G., Eaton J.R.O., Elders P., Kawamura A., et al. Genetically engineered two-warhead evasins provide a method to achieve precision targeting of disease-relevant chemokine subsets. Sci. Rep. 2018;8:6333. doi: 10.1038/s41598-018-24568-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eaton J.R.O., Alenazi Y., Singh K., Davies G., Geis-Asteggiante L., Kessler B., et al. The N-terminal domain of a tick evasin is critical for chemokine binding and neutralization and confers specific binding activity to other evasins. J. Biol. Chem. 2018;293:6134–6146. doi: 10.1074/jbc.RA117.000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franck C., Foster S.R., Johansen-Leete J., Chowdhury S., Cielesh M., Bhusal R.P., et al. Semisynthesis of an evasin from tick saliva reveals a critical role of tyrosine sulfation for chemokine binding and inhibition. Proc. Natl. Acad. Sci. U. S. A. 2020;117:12657–12664. doi: 10.1073/pnas.2000605117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonvin P., Dunn S.M., Rousseau F., Dyer D.P., Shaw J., Power C.A., et al. Identification of the pharmacophore of the CC chemokine-binding proteins Evasin-1 and -4 using phage display. J. Biol. Chem. 2014;289:31846–31855. doi: 10.1074/jbc.M114.599233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah N.B., Duncan T.M. Bio-layer interferometry for measuring kinetics of protein-protein interactions and allosteric ligand effects. J. Vis. Exp. 2014;18 doi: 10.3791/51383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Déruaz M., Frauenschuh A., Alessandri A.L., Dias J.M., Coelho F.M., Russo R.C., et al. Ticks produce highly selective chemokine binding proteins with antiinflammatory activity. J. Exp. Med. 2008;205:2019–2031. doi: 10.1084/jem.20072689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayward J., Sanchez J., Perry A., Huang C., Rodriguez Valle M., Canals M., et al. Ticks from diverse genera encode chemokine-inhibitory evasin proteins. J. Biol. Chem. 2017;292:15670–15680. doi: 10.1074/jbc.M117.807255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotál J., Langhansová H., Lieskovská J., Andersen J.F., Francischetti I.M., Chavakis T., et al. Modulation of host immunity by tick saliva. J. Proteomics. 2015;128:58–68. doi: 10.1016/j.jprot.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vieira A.T., Fagundes C.T., Alessandri A.L., Castor M.G., Guabiraba R., Borges V.O., et al. Treatment with a novel chemokine-binding protein or eosinophil lineage-ablation protects mice from experimental colitis. Am. J. Pathol. 2009;175:2382–2391. doi: 10.2353/ajpath.2009.090093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akinyi M.Y., Tung J., Jeneby M., Patel N.B., Altmann J., Alberts S.C. Role of grooming in reducing tick load in wild baboons (Papio cynocephalus) Anim. Behav. 2013;85:559–568. doi: 10.1016/j.anbehav.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wanzala W., Okanga S. Ticks (Acari: ixodidae) associated with wildlife and vegetation of Haller park along the Kenyan coastline. J. Med. Entomol. 2006;43:789–794. doi: 10.1603/0022-2585(2006)43[789:taiaww]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 30.Wanzala W., Ondiaka S.N. Tick-borne lymphadenopathy-like condition in an African woman in Kenya. J. Res. Med. Sci. 2013;18:918–921. [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang L.I., Collins J., Davis R., Lin K.M., DeCamp D., Roach T., et al. Use of a cAMP BRET sensor to characterize a novel regulation of cAMP by the sphingosine 1-phosphate/G13 pathway. J. Biol. Chem. 2007;282:10576–10584. doi: 10.1074/jbc.M609695200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim H.D., Lane J.R., Canals M., Stone M.J. Systematic assessment of chemokine signaling at chemokine receptors CCR4, CCR7 and CCR10. Int. J. Mol. Sci. 2021;22:4232. doi: 10.3390/ijms22084232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Experimental raw data are available to be shared upon request to the corresponding authors.