Abstract
Cell division cycle-associated 5 (CDCA5) plays a critical role in the progression of various human cancers by regulating cell cycle-related proteins; however, the function of CDCA5 in breast cancer (BC) is poorly understood. The aim of the present study was to investigate the expression level of CDCA5 in BC and its effect on BC progression. CDCA5 was found to be highly expressed in patients with BC, as well as in BC cell lines. It was also found that a high CDCA5 expression in BC was significantly associated with a shorter survival rate. In addition, the expression level of CDCA5 was significantly increased in stem cells derived from suspension-cultured BC cells, as compared to adherent-cultured cells. CDCA5 knockdown in MCF7 and SKBR3 cells significantly reduced cell proliferation, migration and clone formation. At the same time, the stemness capacity of BC cells, determined by analyzing cancer stem cell marker expression and mammosphere formation, was also markedly diminished following the knockdown of CDCA5. In addition, in vivo experiments demonstrated that CDCA5 knockdown in MCF7 cells markedly reduced tumor growth. On the whole, the present study demonstrates that CDCA5 may be used as a prognostic biomarker and therapeutic target for BC.
Keywords: cell division cycle-associated 5, breast cancer stem cells, proliferation, migration, mammosphere formation
Introduction
Breast cancer (BC) is the most common cancer type affecting women worldwide (1). Despite the benefits arising from expanding access to high-quality prevention methods and early detection, as well as the rapid development of treatment modalities, such as surgical resection, endocrine therapy, chemotherapy and immunotherapy, the mortality rate associated with BC remains relatively high and a further decrease is warranted (2,3). Although certain molecular markers have been extensively characterized, including immunohistochemical markers (i.e., estrogen receptor, progesterone receptor, human epidermal growth factor receptor 2 and Ki-67), genomic markers [i.e., breast invasive carcinoma (BRCA)1, BRCA2 and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α] and immunomarkers (i.e., programmed death-ligand 1), new biomarkers and new biomarker combinations still need to be developed, due to the high tumor heterogeneity of BC.
Cancer stem cells (CSCs) retain the properties of self-renewal, differentiation and drug-resistance, all of which are considered to lead to a poor therapeutic response in BC, as well as tumor recurrence and metastasis (4–6). CSCs have been identified in various types of cancer, including BC. The most common markers of BC stem cells (BCSCs) are CD44+CD24− and aldehyde dehydrogenase (ALDH)+ (7,8). Although a number of studies have identified that targeting CSCs may be an effective approach to anticancer treatment, more CSC-related markers and mechanisms need to be elucidated.
Human cell division cycle-associated 5 (CDCA5; also known as sororin or p35), which is located at 11q13.1, is a critical regulator of sister chromatid in mitosis, stabilizing the association between cohesion complex and chromatin, and is involved in mitotic cell cycle and double-strand break repair (9,10). In addition, CDCA5 is necessary for the stable cohesion of chromatids during the S and G2/M phases and is then degraded through anaphase-promoting complex-dependent ubiquitination during the G0/G1 phase (11–13). Diseases associated with CDCA5 include Cornelia de Lange syndrome (14) and Robert-Sc phocomelia syndrome (15). A significantly increased CDCA5 expression has been reported in various human tumor tissues, including lung cancer (16), urothelial carcinoma (17), oral squamous cell carcinoma (18), gastric cancer (19,20) and hepatocellular carcinoma (21) tissues. Hence, CDCA5 may be a poor prognostic biomarker for patients with these types of cancer. However, the clinical significance and biological function of CDCA5 in BC are not yet fully understood.
In the present study, a link was identified between aberrant CDCA5 expression and a poor prognosis of patients with BC by analyzing The Cancer Genome Atlas (TCGA). Furthermore, it was found that BC cells and BCSCs exhibited a higher CDCA5 expression than normal breast and adherent cells, respectively. Finally, CDCA5 knockdown in BC cells inhibited cell proliferation, migration and CSC activity. These findings thus suggest that CDCA5 may be a novel prognostic biomarker for BC.
Materials and methods
Bioinformatics analysis
UALCAN (22) is a comprehensive, user-friendly and interactive web resource for analyzing OMICS data from the TCGA, MET500 and CPTAC databases. CDCA5 mRNA expression was investigated in various tumor samples (including BC samples) through a pan-cancer view of TCGA cancers (using tumor and normal samples) and the expression of CDCA5 was investigated, particularly in BRCA, based on sample types, individual cancer stages and TP53 mutation status.
The Kaplan-Meier Plotter (23) is an online tool that was used herein to perform a meta-analysis discovery and validation of survival biomarkers, including >5,400 genes from the Gene Expression Omnibus (GEO), European Genome-phenome Archive (EGA) and TCGA databases, covering breast (n=7,830), ovarian (n=2,190), lung (n=3,452) and gastric (n=1,440) cancer. CDCA5 (Affymetrix ID: 224753_at) was analyzed by selecting all probe sets per gene in probe set options and a median split in the patient options. The overall survival (OS), disease-free survival (DFS) and distant metastasis-free survival (DMFS) of patients with BC were analyzed by the use of the log-rank test.
LinkedOmics (24) is a publicly available portal that includes three analytical modules: LinkFinder, LinkInterpreter and LinkCompare. The LinkInterpreter module performs enrichment analysis was based on Gene Ontology (GO), biological pathways and network modules, among other functional categories. CDCA5 was analyzed using the Pearson's correlation coefficient with RNAseq data on the TCGA_BRCA sample cohort for GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis.
StarBase (25) is an open-source platform that systematically identifies the RNA-RNA and protein-RNA interaction networks from 108 CLIP-Seq (PAR-CLIP, HITS-CLIP, iCLIP and CLASH) datasets generated by 37 independent studies. The correlation between CDCA5 and stemness transcription factors SRY-box transcription factor 2 (SOX2), POU class 5 homeobox 1 (POU5F1, also known as OCT4) and c-MYC in BRCA was analyzed.
Cells and cell culture
The human normal mammary epithelial cell line, MCF10A (cat no. CL-0525), was purchased from Wuhan Procell Company and cultured in MCF10A special medium (Procell Co., Ltd.). The human BC cell lines, SKBR3 (cat no. 1101HUM-PUMC000085), MCF7 (cat no: 1101HUM-PUMC000013), MDA-MB-231 (cat no. 1101HUM-PUMC000014) and Hs578T (cat no. 1101HUM-PUMC000670) and MDA-MB-468 (cat no. 1101HUM-PUMC000249), were purchased from the National Infrastructure of Cell line Resource (NICR). The SKBR3, MCF7, MDA-MB-231 and Hs578T cells were cultured in DMEM (Meilun Biotech Co., Ltd.) and the MDA-MB-468 cells were cultured in RPMI-1640 (Meilun Biotech Co., Ltd.) medium supplemented with 10% fetal bovine serum (HyClone; Cytiva), 100 U/ml penicillin and 100 µg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.) in a 5% CO2 incubator at 37°C.
Lentivivrus infection
shRNA vectors for sh-CDCA5 and a control vector were purchased from Tsingke Biotechnology Co. Ltd. The shRNA sequences were as follows: sh-CDCA5 forward, 5′-CCGGGGACGCCAGAGACTTGGAAATCTCGAGATTTCCAAGTCTCTGGCGTCCTTTTTG-3′ and reverse, 5′-AATTCAAAAAGGACGCCAGAGACTTGGAAATCTCGAGATTTCCAAGTCTCTGGCGTCC-3′. The second-generation three-plasmid lentivirus system was used. When the 293T (Procell Co., Ltd.) cells were in the logarithmic growth phase, a total of 10 µg of the target plasmid and the helper plasmid in a ratio of 5:3:2 were incubated with PEI transfection reagent at 37°C for 30 min. After 72 h, the cell supernatant was collected, cell debris was removed, the virus was collected by centrifugation with PEG8000, incubated with target cells MCF7 and SKBR3 for 4 h, the medium was changed, and puromycin was started after 48 h. Screening, continuous drug screening for 2 weeks, to obtain stable cell lines.
The SKBR3 and MCF7 BC cells were seeded in 6-well plates (5×104 cells/well) at 60% confluency and then infected with the sh-CDCA5 lentivirus. After 48 h, the stably transfected cells were established using selection with 2 µg/ml puromycin for 2 weeks.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from the BC cell lines using the Ultrapure RNA kit (CWBio), according to the manufacturer's instructions. Reverse transcription was conducted using HiScript® II Q RT SuperMix for qPCR (Vazyme Biotech Co., Ltd.) at 50°C for 15 min and 85°C for 5 sec. The mRNA levels were determined using Hieff® qPCR SYBR®-Green Master Mix (Yeasen Biotechnology Co., Ltd.) on a Bio-Rad CFX Maestro (Bio-Rad Laboratories, Inc.) with the following PCR cycling conditions: 95°C 5 min; 95°C 15 sec; 55°C 15 sec; and 72°C 30 sec for 30 cycles. GAPDH served as the quantitative control to normalize the mRNA expression levels of the target genes. The primer sequences for the detected genes are listed in the Table I.
Table I.
Gene specific primers used for reverse transcription-quantitative PCR.
| Gene | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| GAPDH | TTAAAAGCAGCCCTGGTGAC | CTCTGCTCCTCCTGTTCGAC |
| SOX2 | GCCGAGTGGAAACTTTTGTCG | GGCAGCGTGTACTTATCCTTCT |
| C-MYC | GGCTCCTGGCAAAAGGTCA | CTGCGTAGTTGTGCTGATGT |
| OCT4 | TTTTGGTACCCCAGGCTATGGGAG | GTTTGAATGCATGGGAGAGCCCAG |
| CDCA5 | GAGGTCCCAGCGGAAATCAG | TCTTTAAGACGATGGGCTTTCTG |
Western blot analysis
Protein samples were prepared using RIPA lysis buffer (Meilun Biotech Co., Ltd.) containing protease inhibitors on ice for 15 min, and were then centrifuged at 12,000 × g for 15 min at 4°C. Protein concentrations were measured using the BCA Protein Assay kit (Beyotime Institute of Biotechnology). Equal amounts of protein were separated by 10% SDS-PAGE, and transferred to PVDF membranes (MilliporeSigma). They were then incubated with TRIS-buffered saline (TBST) containing 5% skim milk at 37°C for 2 h, followed by incubation with primary antibodies targeting CDCA5 rabbit mAb (dilution, 1:2,000; cat no. A4044; ABclonal Science, Inc.), OCT4 rabbit mAb (dilution, 1:2,000; cat no. T55781; Abmart Inc.), c-MYC rabbit mAb (dilution, 1:2,000; cat no. T55150; Abmart Inc.), SOX2 rabbit mAb (dilution, 1:2,000; cat no. T55268; Abmart Inc.) and GAPDH rabbit mAb (dilution, 1:2,000; cat no. A19056; ABclonal Science, Inc.) overnight at 4°C. Following three 5-min washes in TBST, membranes were incubated with HRP goat anti-rabbit secondary antibodies (dilution, 1:2,000; cat no. AS014; ABclonal Science, Inc.) at room temperature for 1.5 h. Chemiluminescent signals were detected using enhanced chemiluminescence detection reagents (Meilun Biotech Co., Ltd.).
Cell proliferation assay
The MCF7 and SKBR3 cells were seeded in 96-well plates at 2,000 cells/well, each well containing 100 µl culture medium. A Cell Counting Kit-8 assay (Beyotime Institute of Biotechnology) was performed at 24, 48, 72 and 96 h, according to the manufacturer's instructions. The resulting solution was quantified using a microplate reader (FC; Thermo Fisher Scientific, Inc.). The light absorption value at 450 nm was detected, and the cell growth curve was plotted with time as the abscissa and absorbance as the ordinate.
Colony formation assay
Following selection with puromycin, the stably transfected BC cells subjected to CDCA5 knockdown and the control cells were seed in 6-well plates (1×103 cells/well) and kept in culture for 14 days to yield cell colonies. Following fixation with 4% paraformaldehyde for 15 min, the colonies were stained at room temperature with 0.1% crystal violet solution (Beyotime Institute of Biotechnology) for 10 min. Colonies were photographed and counted.
Cell migration assay
BC cell migration was examined using a Transwell assay with 8-µm well chambers (Corning, Inc.). The treated BC cells were harvested and resuspended in medium to a final concentration of 1×104 cells/ml. Subsequently, 0.2 ml BC cell suspension was added to the upper chamber, and 0.5 ml medium containing 20% FBS was added to the lower chamber. Following 24 h of incubation at 37°C, the migrated BC cells were stained with 0.1% crystal violet at room temperature for 10 min (Beyotime Institute of Biotechnology) and quantified under a microscope (Olympus/IX83, Japan).
Mammosphere formation assay
The BCSCs were cultured as previously described (26–28). In brief, the CSCs were enriched in ultra-low adsorption 6-well plates (Corning, Inc.) with SKBR3 and MCF7 cells using serum-free DMEM/F12 medium supplemented with epidermal growth factor (20 ng/ml, T&L Biological Technology), basic fibroblast growth factor (10 ng/ml, T&L Biological Technology) and 2% B27 (Gibco; Thermo Fisher Scientific, Inc.) for 14 days. The numbers and volume of CSCs were calculated.
Xenograft model in vivo
A total of 12 3-week-old female nude mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. for xenotransplantation experiments. The mice were housed in plastic cages (three mice/cage) under controlled conditions of light (12-h light/dark cycle), temperature (23±2°C) and humidity (55%), and were provided with free access to food and water. These stable cells were trypsinized and suspended in PBS. A total volume of 0.12 ml PBS containing 1×106 MCF7 cells was subcutaneously injected into the right flanks of the mice. The 12 mice were divided into two groups (6 mice per group) as follows: One group was injected with the MCF7-shNC cell line, and the other group was injected with the MCF7-shCDCA5 cell line. All mice that reached the study endpoint were euthanized by cervical dislocation under 2–3% isoflurane anesthesia. The humane endpoints were deemed as the time when the xenograft tumor diameter was >20 mm, the xenograft tumor reached >20% of the animal body weight, a body weight loss of >20% occurred due to tumor growth, and signs of immobility, inability to eat, ulceration, infection or necrosis were observed. Death was verified by the observation of pupil dilation, as well as the cessation of breath and heartbeat. The animal research protocol was reviewed and approved by the Animal Protection Committee of Wuhan University of Science and Technology (Wuhan, China).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5.0 (GraphPad, Inc.). The results are presented as the mean ± SD from at least three independent experiments. Single comparisons between two groups were performed using the unpaired Student's t-test. Comparisons between three or more groups were determined using ANOVA followed by Tukey's post hoc test. Kaplan-Meier survival analysis was used to assess cumulative survival probability by the use of the log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of CDCA5 in different types of cancer
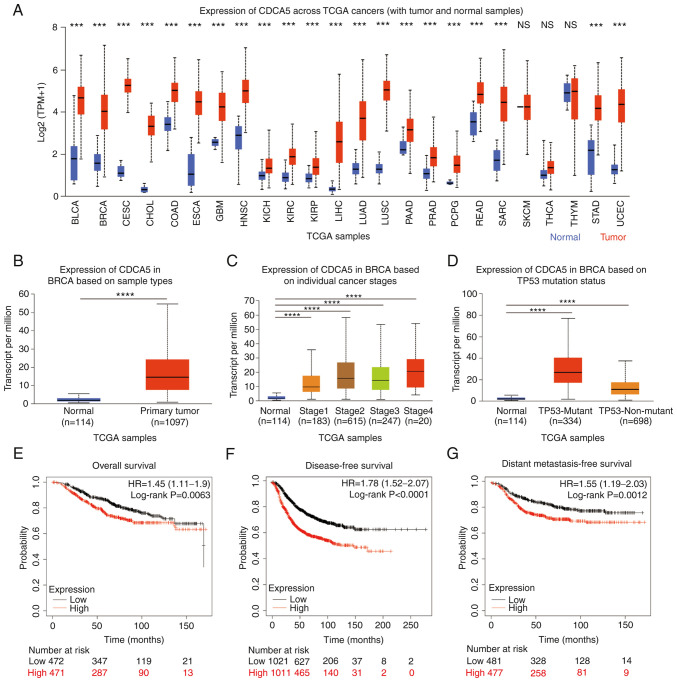
The mRNA expression levels of CDCA5 were examined in different types of cancer from the UALCAN database (Fig. 1A). The results revealed that, in the majority of cancer types, the expression of CDCA5 was significantly increased, including bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), esophageal carcinoma (ESCA), liver hepatocellular carcinoma (LIHC), lung squamous cell carcinoma (LUSC), sarcoma (SARC) and uterine corpus endometrial carcinoma (UCEC). There were only a few cancer types where CDCA5 was not clearly increased, such as thymoma (THYM; normal median value of 4.91 vs. tumor median value of 4.969) and skin cutaneous melanoma (SKCM) with the same expression level of CDCA5. TCGA pan-cancer abbreviations and noun comparisons are presented in Table SI.
Figure 1.
High CDCA5 expression across TCGA cancers and invasive breast cancer. (A) CDCA5 mRNA expression in various types of cancer compared with normal tissues from the UALCAN database. ***P<0.001. Expression of CDCA5 in BRCA based on (B) sample types, (C) individual cancer stages and (D) TP53 mutation status from TCGA samples. (E) Overall, (F) disease-free and (G) distant metastasis-free survival curves from the Kaplan-Meier Plotter, and the evaluation of the impact of the low (black line) and high (red line) CDCA5 expression. ****P<0.0001. CDCA5, cell division cycle-associated 5; TCGA, The Cancer Genome Atlas; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, Colon adenocarcinoma; ESCA, Esophageal carcinoma; GBM, glioblastoma; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PAAD, pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; READ, rectal adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; THCA, thyroid carcinoma; STAD, stomach adenocarcinoma; UCEC, uterine corpus endometrial carcinoma.
Expression of CDCA5 in invasive BC
Similarly, the expression of CDCA5 in BC and normal tissues was analyzed using the UALCAN database. CDCA5 expression was high in 1,097 BC primary tumor tissues compared with 114 normal tissues (P<0.0001; Fig. 1B). The expression of CDCA5 was associated with various clinical features, such as individual cancer stages (normal vs. stage 1, P<0.0001; stage 2, P<0.0001; stage 3, P<0.0001; and stage 4, P<0.0001; Fig. 1C), TP53 mutation status (normal vs. TP53 mutant, P<0.0001; and TP53-non-mutant, P<0.0001, Fig. 1D).
Prognostic value of CDCA5 in patients with BC
Based on the upregulated expression of CDCA5 in BC, its prognostic value in patients with BC was analyzed using the Kaplan-Meier Plotter. Compared with the low CDCA5 expression group, the high CDCA5 expression group exhibited a worse OS (P=0.0063, HR=1.45; Fig. 1E), DFS (P<0.0001, HR=1.78; Fig. 1F) and DMFS (P=0.0012, HR=1.55; Fig. 1G).
CDCA5 co-expressed genes and functional enrichment analysis
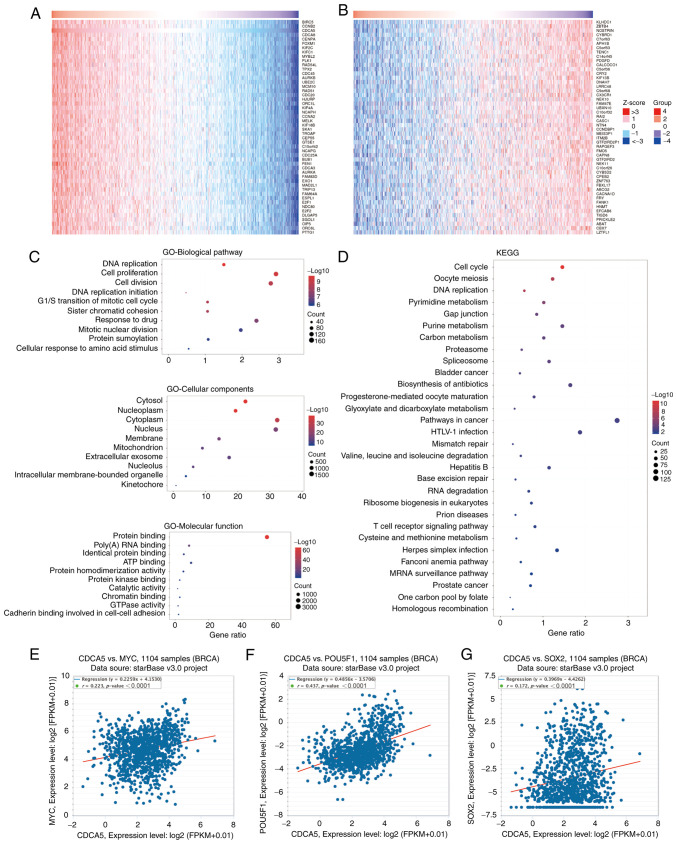
Using the LinkedOmics database, the genes co-expressed with CDCA5 were identified, and the top 50 positively (Fig. 2A) and negatively (Fig. 2B) associated genes are presented in the heat maps (Fig. 2A and B). Further information about the top 50 positively and negatively associated genes is presented in Tables SII and SIII. In addition, GO (Fig. 2C) and KEGG (Fig. 2D) functional enrichment analysis was performed on the co-expressed genes. GO biological pathway enrichment analysis (Table SIV) indicated that these genes were involved in a variety of processes, including DNA replication (P<0.0001), cell proliferation (P<0.0001) and the G1/S transition of mitotic cell cycle (P<0.0001). GO cellular component (Table SV) indicated various cellular structures, including intracellular membrane-bounded organelles (P<0.0001), extracellular exosomes (P<0.0001) and mitochondria (P<0.0001). On the other hand, GO molecular function (Table SVI) indicated that these genes were involved in protein binding (P<0.0001), poly(A) RNA binding (P<0.0001) and ATP binding (P<0.0001). Moreover, KEGG functional enrichment analysis (Table SVII) revealed that these genes were closely linked to pathways, such as T-cell receptor signaling, Fanconi anemia and MRNA surveillance pathways. In addition, CDCA5 exhibited a strong positive correlation with the stemness transcription factors, c-MYC (Fig. 2E), OCT4/POU5F1 (Fig. 2F) and SOX2 (Fig. 2G).
Figure 2.
CDCA5 co-expressed genes and functional enrichment analysis. Heatmap of CDCA5 co-expressed genes in BC using LinkedOmics. (A) The top 50 positively and (B) 50 negatively correlated genes are displayed. (C) GO enrichment analysis (top 10 terms of each subtype: biological pathway, cellular component and molecular function) of genes co-expressed with CDCA5. (D) KEGG enrichment analysis (top 30 terms) of genes co-expressed with CDCA5. Correlation analysis between CDCA5 and (E) c-MYC, (F) POU5F1/OCT4 and (G) SOX2. CDCA5, cell division cycle-associated 5; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BRCA, breast invasive carcinoma; POU5F1, POU5F1 (also known as OCT4).
mRNA and protein expression of CDCA5 in BC cells and CSCs
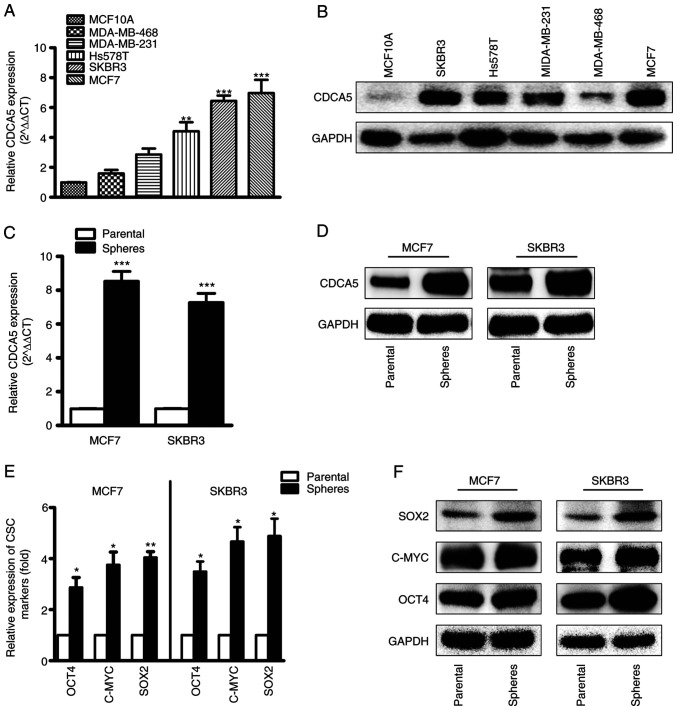
CDCA5 expression was high in the MDA-MB-231, MDA-MB-468, Hs578T, MCF7 and SKBR3 BC cell lines, and low in the MCF10A normal breast cell lines. The mRNA (Fig. 3A) and protein (Fig. 3B) levels of CDCA5 were determined using RT-qPCR and western blot analysis. By comparison, the SKBR3 and MCF7 cell lines exhibited higher CDCA5 expression levels compared to the other cell lines; thus, these two cell lines were selected for CDCA5 knockdown experiments. Furthermore, CDCA5 expression was significantly upregulated in mammospheres compared with CDCA5 adherent cells (Fig. 3C and D). To explore whether or not the mammospheres were CSCs, the levels of CSC markers, including SOX2, c-MYC and OCT4 were determined (Fig. 3E and F). The increased levels of the CSC markers indicated that the mammospheres contained a large number of CSCs.
Figure 3.
Expression levels of CDCA5 in BC cell lines and CSCs. (A) The mRNA and (B) protein levels of CDCA5 in normal and tumor breast cell lines were determined using reverse transcription-quantitative PCR and western blot analysis. (C) The mRNA and (D) protein levels of CDCA5 in mammospheres and tumor-attached BC cells were determined. (E) The mRNA and (F) protein levels of CSC markers in mammospheres and tumor-attached BC cells were determined. Three independent experiments were performed, and GAPDH was used as an internal control. *P<0.05, **P<0.01 and ***P<0.001, vs. MCF10A cells or parental control. CDCA5, cell division cycle-associated 5; BC, breast cancer.
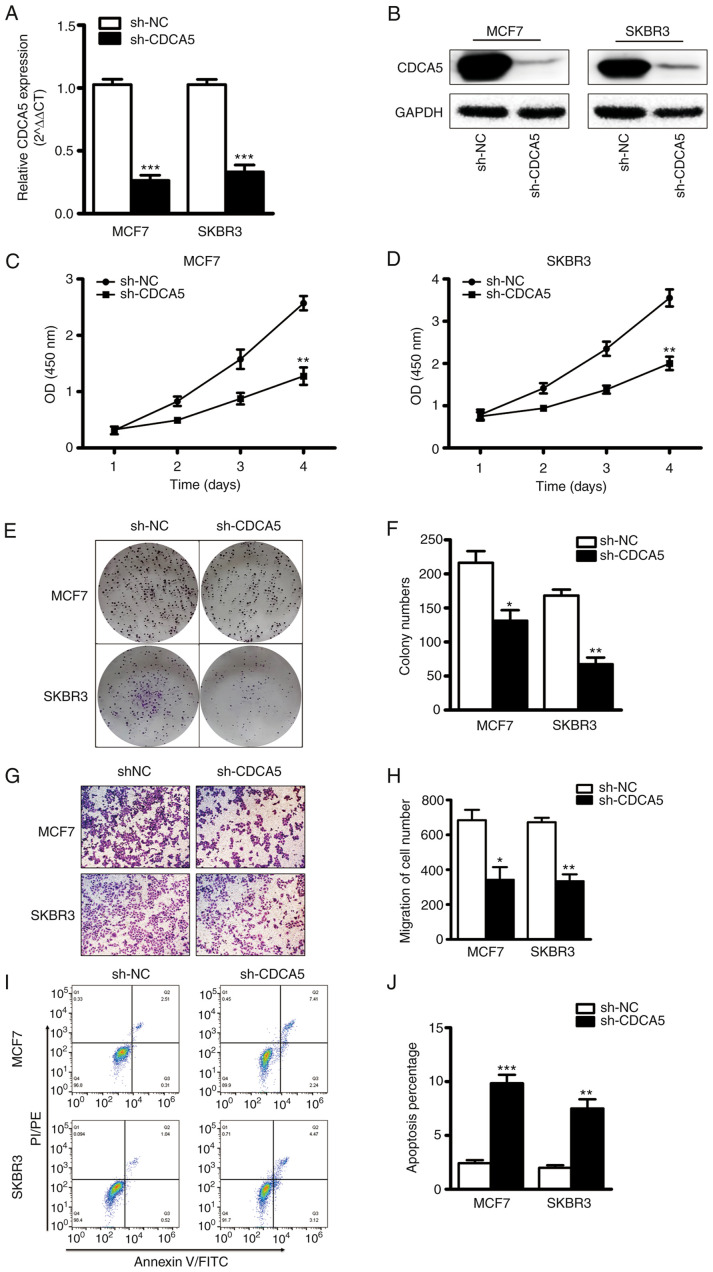
CDCA5 knockdown inhibits BC cell proliferation and migration
The effect of CDCA5 on BC cell function was explored in vitro. The MCF7 and SKBR3 cells were subjected CDCA5 knockdown and thus two stable cell lines with CDCA5 knockdown were established (Fig. 4A and B). The results of CCK-8 assay revealed that the proliferative capacity of the MCF7 and SKBR3 cells was significantly decreased following CDCA5 knockdown (Fig. 4C and D). Furthermore, colony formation assays revealed that the clone formation ability of the MCF7 and SKBR3 cells was significantly reduced following CDCA5 knockdown (Fig. 4E). The statistical analysis results of the number of colonies revealed a significant difference between the two groups (Fig. 4F). The experimental results also demonstrated that when CDCA5 was knocked down in the SKBR3 or MCF7 BC cells, their migratory ability was significantly decreased (Fig. 4G). The statistical analysis results of the number of colonies revealed a significant difference between the two groups (Fig. 4H). In addition, as shown in Fig. 4I and J, CDCA5 knockdown significantly promoted the apoptosis of the MCF7 and SKBR3 cells. These data thus suggested that the proliferative and migratory capacity of BC cells was reduced following CDCA5 knockdown.
Figure 4.
CDCA5 knockdown inhibits BC cell proliferation and migration. (A) mRNA and (B) protein levels of CDCA5 in MCF7 and SKBR3 cells following CDCA5 knockdown. GAPDH was used as an internal control. CCK-8 assay was used to detect (C) MCF7 and (D) SKBR3 cell growth following CDCA5 knockdown. The absorbance value at a wavelength of 450 nm was detected. (E) Cell colony formation assay was used to detect BC cell growth following CDCA5 knockdown. (F) The number of colonies from G was counted. (G) The Transwell invasion assay detected the migration of the BC cells following CDCA5 knockdown. (H) The number of migrated cells in E was counted and plotted on a graph. (I and J) Apoptosis assay was detected by Annexin V/PI using flow cytometry. *P<0.05, **P<0.01 and ***P<0.001, vs. sh-NC. CDCA5, cell division cycle-associated 5; BC, breast cancer.
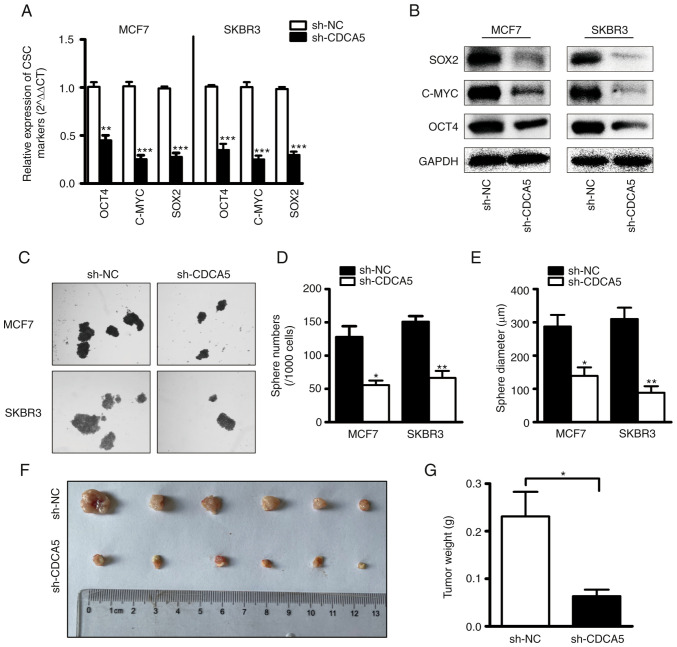
CDCA5 knockdown suppresses BC cell stemness
To explore the role of CDCA5 in the regulation of BCSCs, BC cells subjected to CDCA5 knockdown and their corresponding control cells were utilized. CSC-related genes (SOX2, OCT4 and c-MYC) have frequently been used to recognize CSCs in clinical tissues and several cancer cell lines (29–31). The mRNA (Fig. 5A) and protein (Fig. 5B) levels of SOX2, OCT4 and c-MYC were examined using RT-qPCR and western blot analysis. Compared with the control cells, transfection with sh-CDCA5 led to a markedly decreased expression of the stemness-related genes. These findings indicated that CDCA induced alterations in the levels of stem cell markers. Furthermore, the capacity of BC cells in which CDCA5 was knocked down to form tumor spheroids was examined. As shown in Fig. 5C, the cells transfected with sh-CDCA5 formed less spheres than the vector control cells, as was expected. The statistical analysis results of the number and diameter of spheroids revealed a significant difference between the two groups (Fig. 5D and E). It was thus concluded that CDCA5 promotes BC cell stemness.
Figure 5.
CDCA5 knockdown inhibits BC cellular stemness and growth in vivo. (A) mRNA and (B) protein levels of cancer stem cell markers in sh-CDCA5 and sh-NC BC cells were determined using reverse transcription-quantitative PCR and western blot analysis. (C) Representative images of mammospheres in sh-CDCA5 and sh-NC BC cells. The (D) number and (E) diameter of mammospheres from panel C was counted. CDCA5 knockdown inhibits BC tumor growth in vivo. (F) Volumes of MCF7 subcutaneous xenograft tumors were compared between the sh-CDCA5 and sh-NC groups (n=6). (G) Tumor weights were compared between the sh-CDCA5 and sh-NC groups (n=6). *P<0.05, **P<0.01 and ***P<0.001, vs. sh-NC. CDCA5, cell division cycle-associated 5; BC, breast cancer.
CDCA5 knockdown suppresses tumor growth in vivo
To investigate the effects of CDCA5 expression on BC growth, a mouse xenograft model was established. Since the protein expression of CDCA5 was higher in the MCF7 cell line than in the SKBR3 cells, the MCF7 cells were selected for use in vivo experiments. Another reason for this selection was that the MCF7 cells are more capable of forming tumors in vivo. A total of 12 nude female mice were equally divided into two groups, and sh-NC and sh-CDCA5 cells were subcutaneously injected. The detected ratio of tumor growth to body weight was higher in the sh-NC group than in the sh-CDCA5 group (Fig. 5F and G). This indicated that the growth of MCF7 cells in vivo was inhibited following CDCA5 knockdown.
Discussion
The mammalian cell cycle is induced by the sequential activation or inactivation of proteins that regulate various phases of the cell cycle (32). The loss of normal cell cycle due to the dysregulation of several cell cycle-related genes can lead to cancer development (33). CDCA5 is a key regulator of DNA repair and chromosome segregation, and belongs to a family of cell division cycle-related proteins. The primary role of CDCA5 is to promote sister chromatid association and ensure accurate sister chromatid segregation to maintain genome integrity (34). CDCA5 is aberrantly expressed in various types of cancer, such as hepatocellular carcinoma (35–37), bladder cancer (38), prostate cancer (39) and renal clear cell carcinoma (40), rendering it a potentially important prognostic marker and potential therapeutic target for cancer patients. However, only a limited number of studies (41–43) to date have investigated the expression and function of CDCA5 in BRCA.
In the present study, a significant upregulation of CDCA5 expression was observed in BC cells. Moreover, patients with a high expression of CDCA5 were found to have a worse survival. The GO and KEGG function enrichment analysis revealed that CDCA5 was a critical regulator of the cell cycle and DNA repair. Furthermore, the knockdown of CDCA5 significantly inhibited cell growth and tumorigenesis in vitro and in vivo. Therefore, consistent with the findings of previous research (43), the results of the present study support an oncogenic role for CDCA5 in BC progression and suggest the possibility of the use of CDCA5 as a therapeutic target for breast cancer.
The CSC theory highlights that CSCs, a minor population of tumor cells, also referred to as tumor-initiating cells, harbor the properties of self-renewal, differentiation and drug-resistance, which contribute to metastasis and relapse (44,45). The present study further explored the association of CDCA5 with CSCs to examine whether CDCA5 is involved in the regulation of CSCs. As was predicted, CDCA5 was significantly upregulated in BCSCs, accompanied by the significant upregulation of cancer stem cell transcription factors, including SOX2, OCT4, c-MYC. CDCA5 knockdown significantly inhibited the expression of CSC-related transcription factors, and significantly inhibited mammosphere formation, such as the size and number of tumor spheroids. Of course, the present study has limitations compared to other studies on CDCA5. For example, the present study did not further explore and examine the in-depth mechanisms underlying the regulation of BCSCs by CDCA5, but only observed that CDCA5 regulated BCSCs. In addition, it is unknown whether CDCA5 also has the ability to regulate CSCs in other types of cancer. In the tumor microenvironment, the association between CDCA5 and immune infiltration is also worthy of further study.
In conclusion, the present study indicated that CDCA5 plays a critical role in BC progression by promoting BC cell proliferation, migration and cellular stemness. Therefore, CDCA5 may serve as a novel prognostic biomarker and therapeutic target for patients with BC.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Cao Yuan at the Analytical and Testing Center of Wuhan University of Science and Technology (Wuhan, China) for providing assistance with the flow cytometric analysis.
Funding Statement
The present study was supported by the National Natural Science Foundation of China (grant nos. 31501149, 31770815 and 31570764), the Wuhan Health and Family Planning Scientific Research Project (grant no. WX21Q49), the Hubei Natural Science Foundation (grant nos. 2017CFB537, 2019CFB398, 2019CFB368 and 2021CFB230), the Educational Commission of Hubei (grant no. B2020001), the Hubei Province Health and Family Planning Scientific Research Project (grant nos. WJ2021Q051, WJ2019M255 and ZY2021Q005), the Frontier Project of Applied Basic Research in Wuhan (grant no. 2020020601012250) and the Hubei Province Technology Innovation Special Major Project (Project no. 2019ACA168).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
XHL and TCZ participated in the research design. HH, YX, XYZ, YD, FJW and YH performed the experiments and analyzed the data, and HH was the major contributor to the writing of the manuscript. HH and YX confirm the authenticity of all the raw data. All the authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The animal research protocol was reviewed and approved by the Animal Protection Committee of Wuhan University of Science and Technology (Wuhan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Loibl S, Poortmans P, Morrow M, Denkert C, Curigliano G. Breast cancer. Lancet. 2021;397:1750–1769. doi: 10.1016/S0140-6736(20)32381-3. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, Jemal A, Siegel RL. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69:438–451. doi: 10.3322/caac.21583. [DOI] [PubMed] [Google Scholar]
- 3.Ginsburg O, Bray F, Coleman MP, Vanderpuye V, Eniu A, Kotha SR, Sarker M, Huong TT, Allemani C, Dvaladze A, et al. The global burden of women's cancers: A grand challenge in global health. Lancet. 2017;389:847–860. doi: 10.1016/S0140-6736(16)31392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 6.Peng F, Li TT, Wang KL, Xiao GQ, Wang JH, Zhao HD, Kang ZJ, Fan WJ, Zhu LL, Li M, et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017;8:e2569. doi: 10.1038/cddis.2016.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charafe-Jauffret E, Ginestier C, Bertucci F, Cabaud O, Wicinski J, Finetti P, Josselin E, Adelaide J, Nguyen TT, Monville F, et al. ALDH1-positive cancer stem cells predict engraftment of primary breast tumors and are governed by a common stem cell program. Cancer Res. 2013;73:7290–7300. doi: 10.1158/1538-7445.AM2013-244. [DOI] [PubMed] [Google Scholar]
- 9.Rankin S, Ayad NG, Kirschner MW. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol Cell. 2005;18:185–200. doi: 10.1016/j.molcel.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 10.Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011;12:449–462. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitz J, Watrin E, Lénárt P, Mechtler K, Peters JM. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr Biol. 2007;17:630–636. doi: 10.1016/j.cub.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 12.Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, Peters JM. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 2010;143:737–749. doi: 10.1016/j.cell.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 13.Watrin E, Demidova M, Watrin T, Hu Z, Prigent C. Sororin pre-mRNA splicing is required for proper sister chromatid cohesion in human cells. EMBO Rep. 2014;15:948–955. doi: 10.15252/embr.201438640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyle MI, Jespersgaard C, Nazaryan L, Ravn K, Brøndum-Nielsen K, Bisgaard AM, Tümer Z. Deletion of 11q12.3-11q13.1 in a patient with intellectual disability and childhood facial features resembling Cornelia de Lange syndrome. Gene. 2015;572:130–134. doi: 10.1016/j.gene.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 15.Safran M, Rosen N, Twik M, BarShir R, Stein TI, Dahary D, Fishilevich S, Lancet D. Practical Guide Life Sci Databases; 2021. The GeneCards Suite; pp. 27–56. [Google Scholar]
- 16.Nguyen MH, Koinuma J, Ueda K, Ito T, Tsuchiya E, Nakamura Y, Daigo Y. Phosphorylation and activation of cell division cycle associated 5 by mitogen-activated protein kinase play a crucial role in human lung carcinogenesis. Cancer Res. 2010;70:5337–5347. doi: 10.1158/0008-5472.CAN-09-4372. [DOI] [PubMed] [Google Scholar]
- 17.Chang IW, Lin VC, He HL, Hsu CT, Li CC, Wu WJ, Huang CN, Wu TF, Li CF. CDCA5 overexpression is an indicator of poor prognosis in patients with urothelial carcinomas of the upper urinary tract and urinary bladder. Am J Transl Res. 2015;7:710–722. [PMC free article] [PubMed] [Google Scholar]
- 18.Tokuzen N, Nakashiro K, Tanaka H, Iwamoto K, Hamakawa H. Therapeutic potential of targeting cell division cycle associated 5 for oral squamous cell carcinoma. Oncotarget. 2016;7:2343–2353. doi: 10.18632/oncotarget.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Shen M, Zhou G. Upregulation of CDCA5 promotes gastric cancer malignant progression via influencing cyclin E1. Biochem Biophys Res Commun. 2018;496:482–489. doi: 10.1016/j.bbrc.2018.04.160. [DOI] [PubMed] [Google Scholar]
- 20.Chen T, Huang Z, Tian Y, Wang H, Ouyang P, Chen H, Wu L, Lin B, He R. Role of triosephosphate isomerase and downstream functional genes on gastric cancer. Oncol Rep. 2017;38:1822–1832. doi: 10.3892/or.2017.5846. [DOI] [PubMed] [Google Scholar]
- 21.Shen Z, Yu X, Zheng Y, Lai X, Li J, Hong Y, Zhang H, Chen C, Su Z, Guo R. CDCA5 regulates proliferation in hepatocellular carcinoma and has potential as a negative prognostic marker. Onco Targets Ther. 2018;11:891–901. doi: 10.2147/OTT.S154754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Győrffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol J. 2021;19:4101–4109. doi: 10.1016/j.csbj.2021.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saadin K, White IM. Breast cancer stem cell enrichment and isolation by mammosphere culture and its potential diagnostic applications. Expert Rev Mol Diagn. 2013;13:49–60. doi: 10.1586/erm.12.117. [DOI] [PubMed] [Google Scholar]
- 28.Qin Y, Hou Y, Liu S, Zhu P, Wan X, Zhao M, Peng M, Zeng H, Li Q, Jin T, et al. A Novel long non-coding RNA lnc030 maintains breast cancer stem cell stemness by stabilizing SQLE mRNA and increasing cholesterol synthesis. Adv Sci (Weinh) 2021;8:2002232. doi: 10.1002/advs.202002232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ai L, Mu S, Sun C, Fan F, Yan H, Qin Y, Cui G, Wang Y, Guo T, Mei H, et al. Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing piRNA-823 expression and DNMT3B activation. Mol Cancer. 2019;18:88. doi: 10.1186/s12943-019-1011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang T, Guo C, Xia T, Zhang R, Zen K, Pan Y, Jin L. LncCCAT1 promotes breast cancer stem cell function through activating WNT/β-catenin signaling. Theranostics. 2019;9:7384–7402. doi: 10.7150/thno.37892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma XL, Hu B, Tang WG, Xie SH, Ren N, Guo L, Lu RQ. CD73 sustained cancer-stem-cell traits by promoting SOX9 expression and stability in hepatocellular carcinoma. J Hematol Oncol. 2020;13:11. doi: 10.1186/s13045-020-0845-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dominguez-Brauer C, Thu KL, Mason JM, Blaser H, Bray MR, Mak TW. Targeting mitosis in cancer: Emerging strategies. Mol Cell. 2015;60:524–536. doi: 10.1016/j.molcel.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 33.López-Lázaro M. The stem cell division theory of cancer. Crit Rev Oncol Hematol. 2018;123:95–113. doi: 10.1016/j.critrevonc.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 34.Zhang N, Pati D. Sororin is a master regulator of sister chromatid cohesion and separation. Cell Cycle. 2012;11:2073–2083. doi: 10.4161/cc.20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H, Chen J, Zhao L, Song W, Xuan Z, Chen J, Li Z, Song G, Hong L, Song P, Zheng S. CDCA5, transcribed by E2F1, promotes oncogenesis by enhancing cell proliferation and inhibiting apoptosis via the AKT pathway in hepatocellular carcinoma. J Cancer. 2019;10:1846–1854. doi: 10.7150/jca.28809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian Y, Wu J, Chagas C, Du Y, Lyu H, He Y, Qi S, Peng Y, Hu J. CDCA5 overexpression is an Indicator of poor prognosis in patients with hepatocellular carcinoma (HCC) BMC Cancer. 2018;18:1187. doi: 10.1186/s12885-018-5072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Xia C, Pu M, Dai B, Yang X, Shang R, Yang Z, Zhang R, Tao K, Dou K. Silencing of CDCA5 inhibits cancer progression and serves as a prognostic biomarker for hepatocellular carcinoma. Oncol Rep. 2018;40:1875–1884. doi: 10.3892/or.2018.6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu G, Xu Z, Chen X, Pan H, Wang Y, Jin B. CDCA5 functions as a tumor promoter in bladder cancer by dysregulating mitochondria-mediated apoptosis, cell cycle regulation and PI3k/AKT/mTOR pathway activation. J Cancer. 2020;11:2408–2420. doi: 10.7150/jca.35372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji J, Shen T, Li Y, Liu Y, Shang Z, Niu Y. CDCA5 promotes the progression of prostate cancer by affecting the ERK signalling pathway. Oncol Rep. 2021;45:921–932. doi: 10.3892/or.2021.7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang X, Huang Y, Lv Z, Wang T, Feng H, Wang H, Du S, Wu S, Shen D, Wang C, et al. Loss of cell division cycle-associated 5 promotes cell apoptosis by activating DNA damage response in clear cell renal cell carcinoma. Int J Oncol. 2022;61:87. doi: 10.3892/ijo.2022.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phan NN, Wang CY, Li KL, Chen CF, Chiao CC, Yu HG, Huang PL, Lin YC. Distinct expression of CDCA3, CDCA5, and CDCA8 leads to shorter relapse free survival in breast cancer patient. Oncotarget. 2018;9:6977–6992. doi: 10.18632/oncotarget.24059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin X, Wang D, Lei M, Guo Y, Cui Y, Chen F, Sun W, Chen X. TPI1 activates the PI3K/AKT/mTOR signaling pathway to induce breast cancer progression by stabilizing CDCA5. J Transl Med. 2022;20:191. doi: 10.1186/s12967-022-03370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Yao J, Zhu Y, Zhao X, Lv J, Sun F. Knockdown of CDCA5 suppresses malignant progression of breast cancer cells by regulating PDS5A. Mol Med Rep. 2022;25:209. doi: 10.3892/mmr.2022.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerger A, Zhang W, Yang D, Bohanes P, Ning Y, Winder T, LaBonte MJ, Wilson PM, Benhaim L, Paez D, et al. Common cancer stem cell gene variants predict colon cancer recurrence. Clin Cancer Res. 2011;17:6934–6943. doi: 10.1158/1078-0432.CCR-11-1180. [DOI] [PubMed] [Google Scholar]
- 45.Fabrizi E, di Martino S, Pelacchi F, Ricci-Vitiani L. Therapeutic implications of colon cancer stem cells. World J Gastroenterol. 2010;16:3871–3877. doi: 10.3748/wjg.v16.i31.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.