Abstract
The transcription factor NNR from Paracoccus denitrificans was expressed in a strain of Escherichia coli carrying a plasmid-borne fusion of the melR promoter to lacZ, with a consensus FNR-binding site 41.5 bp upstream of the transcription start site. This promoter was activated by NNR under anaerobic growth conditions in media containing nitrate, nitrite, or the NO+ donor sodium nitroprusside. Activation by nitrate was abolished by a mutation in the molybdenum cofactor biosynthesis pathway, indicating a requirement for nitrate reductase activity. Activation by nitrate was modulated by the inclusion of reduced hemoglobin in culture media, because of the ability of hemoglobin to sequester nitric oxide and nitrite. The ability of nitrate and nitrite to activate NNR is likely due to the formation of NO (or related species) during nitrate and nitrite respiration. Amino acids potentially involved in NNR activity were replaced by site-directed mutagenesis, and the activities of NNR derivatives were tested in the E. coli reporter system. Substitutions at Cys-103 and Tyr-35 significantly reduced NNR activity but did not abolish the response to reactive nitrogen species. Substitutions at Phe-82 and Tyr-93 severely impaired NNR activity, but the altered proteins retained the ability to repress an FNR-repressible promoter, so these mutations have a “positive control” phenotype. It is suggested that Phe-82 and Tyr-93 identify an activating region of NNR that is involved in an interaction with RNA polymerase. Replacement of Ser-96 with alanine abolished NNR activity, and the protein was undetectable in cell extracts. In contrast, NNR in which Ser-96 was replaced with threonine retained full activity.
Denitrification is the respiratory reduction of nitrate to dinitrogen via the intermediates nitrite, nitric oxide (NO), and nitrous oxide. Since NO is toxic, there is a requirement in denitrifying bacteria to regulate the expression and activity of the enzymes of the pathway in order to maintain a low intracellular concentration of NO. There is increasing evidence to indicate that NO itself acts as a signal molecule that interacts (either directly or indirectly) with transcriptional regulators, which coordinate the expression of the enzymes that make and consume NO. For Paracoccus denitrificans, activation of the nitrite reductase (nirS) and nitric oxide reductase (norCBQDEF) operons by NO requires a transcription factor designated NNR that belongs to the FNR/CRP family (26, 27). An nnr mutant of P. denitrificans has undetectable levels of nitrite reductase activity and reduced levels of NO reductase activity when grown under anaerobic denitrifying conditions (27). The activities of the nirS and norC promoters are substantially reduced in the nnr mutant (28). The nirS and norC promoters are activated by nitrate and nitrite under anaerobic conditions in vivo, though the true signal might be NO, generated by the reduction of nitrate and nitrite. This is supported by the observation that sodium nitroprusside (SNP) (a source of NO+) also efficiently activates the nirS and norC promoters (27). The NNR-regulated nirI gene encodes another protein that is required for nirS expression (21). In a nitrite reductase-deficient mutant, the nirI promoter can be activated by coculturing with an NO reductase-deficient strain that acts as the source of NO (28). Taken together, these observations indicate that for P. denitrificans, NO (or perhaps a chemical species related to NO) acts as a signal to activate expression of the nitrite and NO reductases and that transcriptional activation requires NNR. A similar situation exists for Rhodobacter sphaeroides, for which a transcriptional regulator designated NnrR activates expression of the nitrite and NO reductase genes in response to NO (12, 25). In Pseudomonas aeruginosa, the nirS and norC promoters are activated by another FNR/CRP family member, designated DNR (1), which is closely related to NNR in sequence. DNR is responsive to nitrite in vivo (8), which may reflect the fact that nitrite can be reduced to NO by the nitrite reductase. Nevertheless, there is no direct evidence that DNR is an NO sensor, as are NNR and NnrR. In Pseudomonas stutzeri, there are at least four members of the FNR/CRP family, one of which, DnrD, activates the expression of nitrite reductase and NO reductase (29). DnrD is a close relative of NNR, though it is not known whether it also is responsive to NO. The expression of the genes encoding DNR and DnrD is activated in anaerobically growing cultures (1, 29), whereas NNR is expressed constitutively (28).
The mechanism by which NO activates NNR is not known, nor is it known whether NO interacts directly with the protein or whether there is a signal transduction pathway with additional components. Alignment of the NNR sequence with those of other known (NnrR) and possible (DNR, DnrD) NO sensors provides few clues as to possible signaling mechanisms. The primary structure of NNR and the sequence of its probable binding sites in the promoters that it regulates suggest that NNR has the same DNA binding specificity as the FNR protein of Escherichia coli. This has recently been confirmed by mutagenesis of the NNR binding site in the nirI-nirS intergenic region of P. denitrificans (21) and of the NNR binding site in the norC promoter (Hutchings and Spiro, unpublished data). The common DNA binding specificity raises the possibility that NNR might activate FNR-dependent promoters in E. coli. This paper reports the successful development of a system for studying NNR activity in E. coli and its use to characterize seven NNR proteins with single amino acid substitutions. Several important conclusions can be drawn about the properties of NNR.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
E. coli strain JM83 [ara Δ(lac-proAB) rpsL φ80lacZΔM15] was used for all routine DNA manipulations, and JRG1728 [ΔlacX74 galU galK rpsL Δ(ara-leu) Δ(tyrR-fnr-rac-trg)] (23) was used as the host for the reporter system. To create a mobAB mutant of JRG1728, a P1 lysate grown on TP1000 [araD Δ(argF-lac)U169 rpsL relA flbB ptsF devC rbsR mobAB::kan] (17) was used to transduce JRG1728 to kanamycin resistance. The Mob− phenotype of JRG1728 mobAB::kan was confirmed by showing that unlike its parent, it is unable to accumulate nitrite when grown in media containing nitrate, indicating a total absence of nitrate reductase activity. For β-galactosidase assays, E. coli strains were grown in Lennox (L) broth (tryptone [10 g liter−1], yeast extract [5 g liter−1], NaCl [5 g liter−1]) supplemented with 0.5% glucose and 50 mM nitrate, 2 mM nitrite, or 100 μM sodium nitroprusside, as indicated, or in M9 minimal medium (15) with similar supplements. Aerobic cultures (10 ml in 250-ml flasks) were shaken at 250 rpm; anaerobic cultures were grown in standing bottles filled to the top, in both cases at 37°C. The plasmids used were pRW2A/FF, which contains an FNR-activated lacZ reporter (13), and the glutathione S-transferase fusion vector pGEX-KG (7). To clone the nnr gene, the coding region was amplified using PCR with a 5′ primer (5′-GGCATATGAACGCCCCCCTGCCCG-3′) that introduced an NdeI site around the start codon of the gene and a stop codon into the reading frame of the lacZ alpha peptide. The PCR product was phosphorylated with T4 polynucleotide kinase and ligated into SmaI-digested and dephosphorylated pUC18. A clone, designated pNNR, with the nnr gene in the same orientation as the lac promoter was selected, and the sequence of the insert was confirmed.
PCR mutagenesis.
To incorporate single or double point mutations into the nnr gene, two appropriate complementary primers, both containing the mutation(s), were used in an amplification reaction with plasmid DNA as the template. The template DNA was then removed by treatment with DpnI (which digests only methylated DNA), and the remaining DNA was used to transform JM83. Each reaction mixture contained 25 ng of template DNA, 5 μl of Pwo buffer, 1.5 μl of deoxynucleoside triphosphate mix (50 μM), 4 μM (each) primer, and 0.5 μl of Pwo polymerase (5 U/μl), in a total volume of 50 μl. Reaction conditions were 94°C for 5 min and then 25 cycles of 94°C for 30 s, 67°C for 30 s, and 72°C for 10 min, followed by 72°C for 15 min. The reaction mixtures were transferred to 1.5-ml tubes and treated with 10 U of DpnI for 30 min at 37°C and then 72°C for 30 min. After cooling on ice, each reaction mixture was treated with 2 U of T4 DNA ligase for 1 h and then used to transform competent JM83. Control reaction mixtures contained no primers. Mutant DNAs were sequenced on both strands using an ABI Prism automated sequencer. Other general recombinant DNA techniques were performed as described by Sambrook et al. (20).
Analytical methods.
β-Galactosidase was assayed in duplicate according to the method of Miller (15) on at least three independently grown log-phase cultures. Experiments in which reduced hemoglobin was added to cultures were performed by a modification of the method of Kwiatkowski and Shapleigh (12). Ten-milliliter cultures were grown aerobically to log phase and then transferred to 15-ml bottles sealed with Suba seals. The bottles were shaken for 30 min at 37°C to remove the residual oxygen. Additions of hemoglobin and nitrate were then made as required, and the bottles were incubated without shaking for a further 2.5 h before β-galactosidase was assayed. Absorption spectra were collected from culture supernatants in an Aminco DW2000 dual beam spectrophotometer, with a supernatant from a similar culture grown in the absence of hemoglobin as the reference. Reduced human hemoglobin (Sigma) was prepared in an anaerobic cabinet as a 0.5 mM solution in 50 mM morpholinepropanesulfonic acid (pH 7.5), according to the method of Bazylinski et al. (3). The concentration of nitrate in L broth was measured with a Dionex DX-100 Ion Chromatograph using a Dionex Ionpac AS4A column.
Preparation of anti-NNR antiserum and immunoblotting.
The nnr gene was cloned into the glutathione S-transferase fusion vector pGEX-KG, and the fusion protein was purified on a glutathione affinity column. The column-bound fusion protein was cleaved with thrombin, and NNR was eluted. Full details of the cloning and purification will be published elsewhere. An antiserum against the purified NNR was raised in rabbit by Abcam Limited (Cambridge, United Kingdom). Cultures were grown under the same conditions used for β-galactosidase assays; cells were harvested and disrupted by sonication at 4°C. Protein was assayed (using the bicinchoninic acid assay; Sigma) in the soluble fractions, and 50 μg of protein from each sample was separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. The gel was assembled into a Novablot semi-dry blotter (Pharmacia) with a nitrocellulose membrane and three layers of filter paper soaked in transfer buffer (glycine [2.93 g liter−1], Tris [5.81 g liter−1], SDS [0.38 g liter−1], methanol [20%]), and proteins were transferred at 150 mA for 30 min. The membrane was soaked in blocking solution (10% dried milk–0.3% Tween 20 in phosphate-buffered saline [PBS]) for 1 h and then incubated for 1 h at room temperature with the anti-NNR antiserum diluted 100-fold in blocking solution. The membrane was rinsed (twice for 10 min) in wash buffer (0.3% Tween 20 in PBS) and then incubated for 1 h with anti-rabbit antibody (alkaline phosphatase conjugate) diluted 10,000-fold in blocking solution. The membrane was rinsed twice in wash buffer and then in PBS, and then was incubated in 20-ml reaction buffer (100 mM NaCl, 5 mM MgCl2, 100 mM Tris-HCl [pH 9]), 15 μl of nitroblue tetrazolium (50 mg ml−1), and 60 μl of 5-bromo-4-chloro-3-indolylphosphate (50 mg ml−1) for 10 min.
RESULTS
Correction of the NNR sequence.
Nucleotide sequencing of the nnr gene revealed an error in the previously reported sequence (26); the CG dinucleotide at coordinates 683 to 684 (EMBL accession no. U17435) is GC in the correct sequence. In the predicted protein sequence, this changes the reported sequence RLRNDGV (residues 197 to 203) to RLRKHGV. Reexamination of the original sequence data confirmed the error and this correction.
NNR activates an FNR-dependent promoter in response to reactive nitrogen species.
The nnr gene of P. denitrificans was cloned into pUC18 under the control of the lac promoter to generate a plasmid designated pNNR. This was introduced into an fnr mutant of E. coli containing a second plasmid (pRW2A/FF), which carries a derivative of the melR promoter, from which transcription depends upon the binding of FNR to a consensus FNR-binding site centered at −41.5 with respect to the transcription start site (13). The promoter is fused to lacZ such that any ability of NNR to activate transcription would be manifested as β-galactosidase activity. In aerobically grown cultures, NNR did not activate transcription from the FF-melR promoter (Table 1). Under anaerobic growth conditions, there was a 14-fold stimulation of the promoter by NNR and a further 9-fold stimulation in the presence of 100 μM SNP, a nitrosating agent that is a source of NO+ (this concentration of SNP has a negligible effect on anaerobic growth). The S-nitrosothiols S-nitrosoglutathione and S-nitrosoacetylpenicillamine stimulated NNR-dependent transcription to an extent similar to that stimulated by SNP (data not shown). Thus, NNR responds to reactive nitrogen species, and specifically to SNP, similarly in E. coli and in P. denitrificans (28). This implies that the mechanism by which NNR is activated by NO, whether it is direct or indirect, functions in the heterologous background. The activity of NNR in the E. coli system further implies that it is able to activate transcription catalyzed by E. coli RNA polymerase containing the major sigma factor (ς70), since the synthetic melR promoter is transcribed by ς70-containing RNA polymerase (22). This is, potentially, a significant finding, in light of speculation that in P. denitrificans, NNR might activate RNA polymerase containing an alternative sigma factor (2). In similar experiments, NNR showed no ability to activate a class I promoter, where the FNR binding site is centered at −61.5 with respect to the transcription start site (data not shown).
TABLE 1.
β-Galactosidase activities directed by the FF-melR promoter in the presence of plasmids expressing NNR and FNRa
| Strain (mutation) | Plasmid | β-Galactosidase activityb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Aerobic growth
|
Anaerobic growth
|
||||||||
| LG | LG + nitrate | LG + nitrite | LG + SNP | LG | LG + nitrate | LG + nitrite | LG + SNP | ||
| JRG1728 | pNNR | 30 ± 4 | 41 ± 5 | 45 ± 3 | 74 ± 11 | 433 ± 18 | 1,620 ± 202 | 1,213 ± 165 | 3,965 ± 402 |
| JRG1728 | pGS24 | 507 ± 39 | 511 ± 98 | 566 ± 38 | 610 ± 62 | 3,211 ± 260 | 1,595 ± 186 | 2,745 ± 220 | 2,740 ± 114 |
| JRG1728 | pUC18 | 21 ± 2 | 27 ± 4 | ND | ND | 47 ± 6 | 58 ± 7 | ND | ND |
| JRG1728 (mobAB::kan) | pNNR | ND | ND | ND | ND | 47 ± 3 | 48 ± 22 | 2,480 ± 362 | 1,032 ± 170 |
Activities were measured in duplicate on at least three independently grown cultures; standard errors are shown. Growth of cultures was aerobic or anaerobic, in LG, to which was added 50 mM nitrate, 2 mM nitrite, or 100 μM SNP, as indicated. E. coli JRG1728(pRW2A/FF) and JRG1728(mobAB::kan)(pRW2A/FF) were transformed with plasmids expressing NNR (pNNR), FNR (pGS24), or with the vector (pUC18) as a control. Units of β-galactosidase activity are as defined by Miller (15).
ND, not determined.
It was surprising that NNR-dependent transcription from the FF-melR promoter could be observed under anaerobic growth conditions without additions to the medium or in the presence of either nitrate or nitrite (Table 1). In defined minimal media, a similar response to nitrate, nitrite, and SNP was observed, but there was no activation under anaerobic conditions in the absence of these additions (data not shown). This suggests that activation in rich medium in the absence of additions might be due to the presence of traces of nitrate in L broth. Using ion chromatography, the concentration of nitrate in the L broth used for these experiments was found to be approximately 1.5 mM (data not shown). One possible explanation for the apparent activation of NNR by nitrate and nitrite is that utilization of these electron acceptors in E. coli is accompanied by the accumulation of traces of NO (11). E. coli has three nitrate reductases capable of reducing nitrate to nitrite, all of which are molybdoenzymes (16). A mutation (mobAB::kan) in the molybdenum cofactor biosynthesis pathway (17) was introduced into JRG1728 to generate a strain devoid of nitrate reductase activity. In this strain, there was no NNR-dependent expression in unamended L broth or in the presence of added nitrate (Table 1), indicating that the effect of nitrate in the parent strain requires nitrate reductase activity. However, the ability of nitrite to activate NNR was not affected by the mobAB mutation (Table 1). This excludes the possibility that the activating effect of nitrite is due to the ability of the E. coli membrane-bound nitrate reductase to reduce nitrite to NO (11).
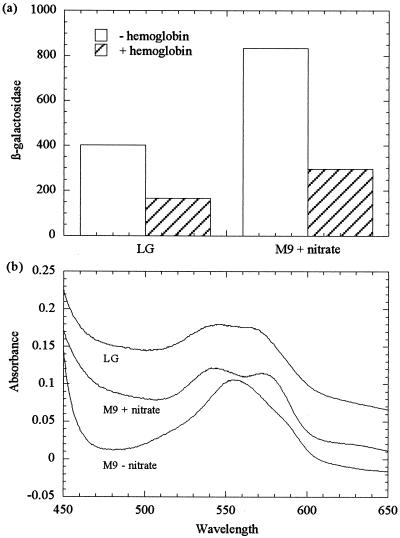
The ability of hemoglobin to trap NO in growing cultures and so to modulate the activity of NO-responsive promoters (6, 12, 28) was used to explore further the nature of the signal(s) activating NNR. The addition of 4 μM hemoglobin to cultures grown anaerobically in minimal medium containing 2 mM nitrate reduced NNR-mediated activation by a factor of 3 (Fig. 1). The spectra of hemoglobin from supernatants of cultures grown in the presence of nitrate showed absorption maxima at approximately 546 and 571 nm (Fig. 1), which is consistent with the formation of a hemoglobin-NO complex (6, 12). Addition of hemoglobin to cultures grown in L broth plus glucose (LG) reduced expression by a factor of 2.5 and resulted in the formation of an NO-hemoglobin complex (Fig. 1), which is further evidence that activation of NNR in L broth is due to the reduction of traces of nitrate. Hemoglobin also reacts with nitrite, though much more slowly than with NO, to produce a rather similar spectrum that also has a broad absorption feature at around 630 nm (6). This feature is just discernible in the spectrum of hemoglobin from the supernatant of a culture grown in minimal medium supplemented with nitrate (Fig. 1), suggesting that hemoglobin might be acting by sequestering nitrite rather than, or as well as, NO. Thus, the data presented herein do not rigorously exclude the possibility that nitrite itself is able to activate NNR. An alternative possibility is that in E. coli, nitrite reduction to ammonia by the multiheme and siroheme nitrite reductases is accompanied by the release of traces of NO, which results in activation of NNR. Hemoglobin had no significant effect on NNR activity in anaerobic cultures growing on 2 mM nitrite (not shown). This may be because the large excess of nitrite reacts with hemoglobin, preventing the formation of the NO-hemoglobin complex, but it is also consistent with the possibility that nitrite itself activates NNR, so that sequestration of NO by hemoglobin in a culture containing 2 mM nitrite has no effect on NNR activity.
FIG. 1.
Effect of hemoglobin on NNR-mediated gene expression. (a) JRG1728 (pRW2A/FF; pNNR) was assayed for β-galactosidase following anaerobic growth on LG (with no added nitrate) or M9 minimal medium supplemented with 2 mM nitrate. Cultures were supplemented with 2 μM (LG) or 4 μM (M9) prereduced hemoglobin, as indicated. (b) Absorption spectra of supernatants from cultures grown in M9 in the presence and absence of nitrate or in L broth plus glucose, as indicated.
Derivatives of the FF-melR promoter have been constructed in which the consensus FNR binding site is changed to a CRP binding site (CC), or to a site bound by neither FNR nor CRP (NN), by two symmetrically related substitutions (13). Neither of these promoters was significantly activated by NNR in the E. coli system (data not shown), confirming that the DNA binding specificity of NNR is the same as, or very similar to, that of FNR.
It has been suggested that the activity of FNR might be sensitive to NO, since the oxygen-labile Fe-S center of FNR is a potential target for NO (30), and it has recently been reported that the CydR (FNR) protein of Azotobacter vinelandii is inactivated by NO in vitro (33). As expected, FNR (expressed from the fnr promoter in pGS24) activated the FF-melR promoter most efficiently under anaerobic growth conditions (Table 1). Activation was not affected by the presence of SNP in growth media (Table 1), suggesting that FNR activity is insensitive to SNP-derived NO+ in vivo, at least under the conditions used in these experiments.
Characterization of altered NNR proteins.
The E. coli-based system provides a facile means to characterize NNR proteins with substitutions in amino acids that may be important for NNR activity. Potential targets for NO modification in NNR include tyrosine residues (by nitrosylation or dityrosine bridge formation) and cysteine residues (by S nitrosylation). Tyr-93 and Tyr-35 of NNR were replaced with phenylalanine, and Cys-103 was replaced with serine. A fourth residue, Phe-82, was altered to both alanine and tyrosine, because Phe-82 is conserved in the NnrR protein of R. sphaeroides, the DNR protein of P. aeruginosa, and the DnrD protein of P. stutzeri but is not conserved in other members of the FNR/CRP family. Serine-96 of NNR was also targeted for mutagenesis, because the above-mentioned relatives of NNR all have either serine or threonine at this position, while other members of the FNR/CRP family do not. The altered genes were introduced into the E. coli reporter system, and β-galactosidase activity was used as a measure of the ability of their products to activate transcription. Both NNR Y35F and NNR C103S showed a reduced ability to activate transcription of the FF-melR promoter but remained significantly responsive to SNP, though less so than the wild-type protein (Table 2). This suggests that in both cases, the residue is not essential for the NO activation of NNR but may play a significant role. On the other hand, replacement of Tyr-93 with phenylalanine abolished the activity of NNR, and replacement of Phe-82 with either alanine or tyrosine substantially reduced activity (Table 2). Replacement of Ser-96 with alanine almost completely abolished the activity of NNR. On the other hand, replacement with threonine had no significant effect on NNR activity in the presence of SNP and significantly increased the basal level of activity seen in aerobic cultures and the intermediate level of activity in anaerobic LG-grown cultures (Table 2). The apparent decrease in the activation of NNR S96T by SNP is a consequence of the increased basal level of activity of this protein. Some of the altered proteins showed different responses to anaerobic growth on LG and on LG amended with SNP. For example, the F82Y and Y93F proteins retained a significant response to anaerobic growth on LG but showed little or no additional response to SNP (Table 2). Similarly, the C103S protein appeared to be specifically impaired in the SNP response. One possible explanation is that the nitrate-derived signal persists throughout growth of the culture, whereas SNP is unstable and likely to be depleted during early stages of growth. The activating effect of SNP on proteins retaining low levels of activity may therefore be less apparent. An alternative possibility is that the mutant proteins retain some responsiveness to nitrite (assuming nitrite itself can activate NNR) but not to SNP.
TABLE 2.
β-Galactosidase activities directed by the FF-melR promoter in the presence of plasmids expressing NNR and its altered derivativesa
| Plasmid (mutation) | β-Galactosidase activity
|
Fold activationb | ||
|---|---|---|---|---|
| Aerobic growth
|
Anaerobic growth
|
|||
| LG | LG | LG + SNP | ||
| pNNR | 30 ± 4 | 433 ± 18 | 3965 ± 402 | 130 |
| pNNR (Y35F) | 24 ± 1 | 107 ± 11 | 766 ± 4 | 32 |
| pNNR (F82A) | 16 ± 1 | 49 ± 7 | 123 ± 45 | 7.7 |
| pNNR (F82Y) | 30 ± 3 | 113 ± 6 | 168 ± 44 | 5.6 |
| pNNR (Y93F) | 29 ± 1 | 129 ± 10 | 37 ± 18 | 1.3 |
| pNNR (S96A) | 18 ± 4 | 52 ± 6 | 69 ± 30 | 3.8 |
| pNNR (S96T) | 139 ± 40 | 1090 ± 47 | 4205 ± 954 | 30 |
| pNNR (C103S) | 16 ± 3 | 451 ± 34 | 1327 ± 248 | 83 |
Activities were measured in duplicate on at least three independently grown cultures; standard errors are shown. Cultures were grown aerobically and anaerobically in LG to which 100 μM SNP was added, as indicated. E. coli JRG1728(pRW2A/FF) was transformed with plasmids expressing NNR (pNNR) and its derivatives expressing altered NNR proteins. Units of β-galactosidase activity are as defined by Miller (15).
The fold activation is the ratio of the activity in cultures grown anaerobically in LG plus SNP to that of cultures grown aerobically in LG.
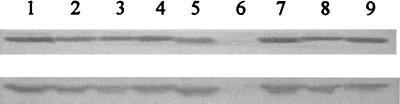
To test whether the consequences of mutagenizing nnr were due to specific effects on the activity of NNR or to changes in the protein's stability or expression level, the abundance of altered proteins in E. coli cell extracts was evaluated by immunoblotting (Fig. 2). Cultures of the same strains used for assays of NNR activity were grown aerobically and anaerobically, and equal amounts of cellular protein were separated by SDS-polyacrylamide gel electrophoresis. Proteins were transferred to a nylon membrane by Western blotting and probed with a polyclonal antiserum raised against purified NNR protein. This revealed that the cellular concentrations of mutant proteins were approximately equal, with the exception of proteins with a substitution at Ser-96. The S96A protein was consistently undetectable in cell extracts grown under a variety of conditions. The same result was demonstrated for three independently isolated mutants and was reproduced when the mutant coding region was subcloned into another vector. Thus, the lack of expression, or instability, of NNR S96A is a direct consequence of the single amino acid substitution. The inactivity of NNR S96A in the in vivo assay (Table 2) can therefore be ascribed to the absence of the protein from the cell. The S96T protein was detectable in cell extracts, to slightly higher concentrations than those of the other proteins (Fig. 2). The increased basal-level activity of this protein (Table 2) may therefore be a simple consequence of its increased abundance in the cell. Taken together, these results point to serine-96 being important for the stability of the NNR protein; an additional functional role for this residue in NNR activity cannot be excluded at this stage.
FIG. 2.
Western blot of NNR and its altered derivatives probed with an anti-NNR antiserum. E. coli JRG1728(pRW2A/FF) transformed with the appropriate NNR-expressing plasmid was grown aerobically (upper panel) and anaerobically (lower panel) in LG plus nitrate. Equal amounts of protein were run on an SDS-polyacrylamide gel, which was then transferred to a nylon membrane, which was probed with an anti-NNR antiserum. Lanes: 1, wild-type NNR; 2, NNR Y35F; 3, NNR F82A; 4, NNR F82Y; 5, NNR Y93F; 6, NNR S96A; 7, NNR S96T; 8, NNR C103S; 9, pure NNR.
NNR represses an FNR-repressible promoter in response to reactive nitrogen species.
A derivative of the gal promoter has been constructed which is subject to simple repression by FNR, binding to a site overlapping the −35 sequence (31). This promoter (FF-galΔ4) was repressed only about 1.4-fold by NNR when cultures were grown anaerobically in the presence of 100 μM SNP. Repression by NNR was more efficient in cultures containing higher concentrations of SNP, but the reagent became rather toxic at these increased concentrations. It was found that the use of nitrate to activate NNR led to a more efficient repression in this assay (Table 3), presumably because nitrate provides a signal that persists throughout the growth period of the culture, whereas SNP might become exhausted, leading to derepression of the promoter. Even under these conditions, NNR is a much poorer repressor of the FF-galΔ4 promoter than is FNR, for reasons that are not clear. Nevertheless, repression of this promoter could be used to test the ability of mutant NNR proteins to bind to DNA in vivo, and these tests were done in media containing nitrate. The S96T protein was a significantly better repressor of the FF-galΔ4 promoter than the wild-type protein (Table 3). This may reflect the slightly greater cellular abundance (Fig. 2) of this protein and its somewhat enhanced activity (Table 2). No significant repression of the NN-galΔ4 derivative was observed with NNR (results not shown), confirming that the repressing effect of NNR involves interaction with the FF site.
TABLE 3.
β-Galactosidase activities directed by the FF-gal Δ4 promoter in the presence of plasmids expressing NNR and its altered derivatives and FNRa
| Plasmid | β-Galactosidase activity
|
Fold repressionb | |
|---|---|---|---|
| Aerobic growth (LG) | Anaerobic growth (LG + nitrate) | ||
| pNNR | 348 ± 18 | 122 ± 2 | 2.9 |
| pGS24 | 231 ± 5 | 7 ± 1 | 33 |
| pNNR (Y35F) | 347 ± 21 | 142 ± 27 | 2.4 |
| pNNR (F82A) | 248 ± 9 | 220 ± 11 | 1.1 |
| pNNR (F82Y) | 266 ± 63 | 122 ± 17 | 2.2 |
| pNNR (Y93F) | 368 ± 26 | 111 ± 7 | 3.3 |
| pNNR (S96T) | 413 ± 67 | 67 ± 11 | 6.2 |
| pNNR (C103S) | 281 ± 6 | 93 ± 9 | 3.0 |
Activities were measured in duplicate on at least three independently grown cultures; standard errors are shown. Cultures were grown anaerobically in LG, to which was added 50 mM nitrate, as indicated. E. coli JRG1728(pRW50/FFgal Δ4) was transformed with plasmids expressing NNR (pNNR), its derivatives expressing altered NNR proteins, as indicated, and FNR (pGS24). Units of β-galactosidase activity are as defined by Miller (15).
The fold repression is the ratio of activities in cultures grown aerobically in LG to the activities in cultures grown anaerobically in LG plus SNP.
The particularly severe effects of the Y93F, F82A, and F82Y mutations (Table 2) may indicate that these residues are specifically required for activation of NNR by NO, have a purely structural role, or are involved in making activating contacts with RNA polymerase. These possibilities were resolved by evaluating the ability of NNR derivatives to repress transcription from the repressible FF-galΔ4 promoter (repression requires DNA binding only and does not involve contacts with RNA polymerase [31]). NNR Y93F repressed FF-galΔ4 at least as efficiently as the wild-type NNR protein (Table 3), implying that NNR Y93F binds to DNA as well as the wild-type protein does. Therefore, Tyr-93 may have a role in making an activating contact with RNA polymerase, rather than in the signal recognition mechanism of NNR. In other words, Y93F may be a positive control mutation that identifies an activating region of the NNR protein that is involved in an interaction with RNA polymerase. The F82Y protein also retained a significant ability to repress the FF-galΔ4 promoter (Table 3), suggesting that Phe-82 may also have a role in interacting with RNA polymerase. However, the F82A protein appeared not to function as a repressor, for reasons that are not clear. The two proteins that retained significant activity in the activation assay (Y35F and C103S) were also functional in the repression assay (Table 3).
DISCUSSION
This work has demonstrated that the NNR protein of P. denitrificans can be activated in E. coli by exposure to SNP in anaerobic cultures or by anaerobic growth on nitrate or nitrite. The reason that NNR can be activated only in anaerobic cultures is not clear, but it may be related to the fact that E. coli deals with reactive nitrogen species in different ways in the presence and absence of oxygen (10). SNP is one of several agents that can induce a nitrosative stress in E. coli, which, among other things, results in the derepression of a flavohemoglobin that plays a role in protection against nitrosating agents and NO-related species (14). The pattern of expression of the flavohemoglobin gene hmp is rather similar to the pattern of NNR-mediated gene expression in E. coli, in that hmp is activated under anaerobic growth conditions by nitrate, nitrite, and SNP (14, 18). It has been suggested that nitrate and nitrite might activate hmp by acting as substrates for the endogenous generation of NO, perhaps through the ability of oxidases to reduce nitrite to NO or through nonenzymatic reduction of nitrite (18). The ability of nitrate and nitrite to activate NNR may be explained in a similar way. On the other hand, the possibility that nitrite itself activates NNR cannot be excluded by the data from experiments performed with E. coli. In a P. denitrificans nitrite reductase mutant, which cannot reduce nitrite to NO, the activities of the NNR-regulated norC promoter and of the NO reductase itself are at near wild-type levels (27, 28). This also implies that either nitrite itself or NO derived from nitrite (in a nitrite reductase-independent reaction) can activate NNR.
Nitrosative stress also results in activation of the transcriptional regulator OxyR, which controls oxidative and nitrosative stress response regulons (9). Nitrosative stress (but not NO itself) causes S nitrosylation and activation of OxyR (9), though it is not clear whether SNP in particular elicits this response. Nitrate respiration in E. coli also causes NO release (10; this study) and nitrosative stress. The activation of NNR by the presence of nitrate in cultures of E. coli requires nitrate reductase activity. Thus, it might be tempting to speculate that the activation of NNR by SNP and by nitrate respiration is a consequence of S nitrosylation of NNR (analogous to the mechanism of activation of OxyR). However, the fact that the C103S mutant of NNR retains a significant response to SNP argues against this idea. NO also activates the SoxR regulatory protein of E. coli by direct nitrosylation of an iron sulfur center (5). Given that NNR has only a single cysteine residue (which is, at least partially, dispensable), this seems to be an unlikely mechanism for the activation of NNR. Besides reaction with thiol groups, formation of metal-NO adducts is the most common mechanism by which proteins can be influenced by NO (24). The sequence of NNR provides no indication that the protein might contain a metal ion, and preliminary characterization of the purified protein has also provided no evidence for the presence of metal ions (unpublished observations). Thus, the mechanism of activation of NNR remains obscure, and the possibility that there is a signal transduction pathway involving additional proteins (conserved in E. coli) cannot be excluded at this stage.
The ability to study NNR activity in E. coli will facilitate the design of experiments aimed at resolving mechanistic questions. A number of residues are herein shown to have important roles in NNR activity. Preliminary indications are that Phe-82 and Tyr-93 may identify an activating region (AR) of NNR involved in an interaction with RNA polymerase. Interestingly, Tyr-93 of NNR aligns closely with Phe-112 of FNR, which has been shown to be part of the activating region, designated AR3, that is involved in transcription activation at class II promoters (19). The failure of NNR to activate a class I promoter suggests that at least in E. coli, NNR lacks a functional AR1 that is required for interaction with RNA polymerase when the activator is bound at −61.5 (19). In this respect, NNR is rather similar to FNR, which has a marked preference for class II promoters (19, 32). Thus, by analogy with the CRP/FNR system (4, 19), it appears as though NNR has a functional AR3 but no AR1. The activity of AR3 in CRP requires Glu-58 (4), which is conserved in NNR.
Tyrosine-35 and cysteine-103 may play important roles in NNR activity but are not the sole determinants of the response to NO, since mutations at these residues do not produce a null phenotype. Serine-96 is clearly an important residue, though properties of mutant proteins with a substitution at this position can be explained solely in terms of protein stability and abundance in the cell. Further characterization of NNR activity in both E. coli and P. denitrificans will shed additional light on the mechanisms of signal perception and transcription activation.
ACKNOWLEDGMENTS
This work was supported by research grants from the Biotechnology and Biological Sciences Research Council.
We are grateful to Jeff Green, Steve Busby, and Tracy Palmer for generous gifts of strains and plasmids and for helpful discussions and to Michael Hill for help with ion chromatography.
REFERENCES
- 1.Arai H, Igarashi Y, Kodama T. Expression of the nir and nor genes for denitrification of Pseudomonas aeruginosa requires a novel CRP/FNR-related transcriptional regulator, DNR, in addition to ANR. FEBS Lett. 1995;371:73–76. doi: 10.1016/0014-5793(95)00885-d. [DOI] [PubMed] [Google Scholar]
- 2.Baker S C, Ferguson S J, Ludwig B, Page M D, Richter O-M H, van Spanning R J M. Molecular genetics of the genus Paracoccus: metabolically versatile bacteria with bioenergetic flexibility. Microbiol Mol Biol Rev. 1998;62:1046–1078. doi: 10.1128/mmbr.62.4.1046-1078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazylinski D A, Goretski J, Hollocher T C. On the reaction of trioxodinitrate (II) with hemoglobin and myoglobin. J Am Chem Soc. 1985;107:7986–7989. [Google Scholar]
- 4.Busby S, Ebright R H. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 5.Ding H, Demple B. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centres in the SoxR transcription activator. Proc Natl Acad Sci USA. 2000;97:5146–5150. doi: 10.1073/pnas.97.10.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goretski J, Hollocher T C. Trapping of nitric oxide produced during denitrification by extracellular hemoglobin. J Biol Chem. 1988;263:2316–2323. [PubMed] [Google Scholar]
- 7.Guan K, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 8.Hasegawa N, Arai H, Igarashi Y. Activation of a consensus FNR-dependent promoter by DNR of Pseudomonas aeruginosa in response to nitrite. FEMS Microbiol Lett. 1998;166:213–217. doi: 10.1111/j.1574-6968.1998.tb13892.x. [DOI] [PubMed] [Google Scholar]
- 9.Hausladen A, Privalle C T, Keng T, DeAngelo J, Stamler J S. Nitrosative stress: activation of the transcription factor OxyR. Cell. 1996;86:719–729. doi: 10.1016/s0092-8674(00)80147-6. [DOI] [PubMed] [Google Scholar]
- 10.Hausladen A, Gow A J, Stamler J S. Nitrosative stress: metabolic pathways involving the flavohemoglobin. Proc Natl Acad Sci USA. 1998;95:14100–14105. doi: 10.1073/pnas.95.24.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji X-B, Hollocher T C. Reduction of nitrite to nitric oxide by enteric bacteria. Biochem Biophys Res Commun. 1988;157:106–108. doi: 10.1016/s0006-291x(88)80018-4. [DOI] [PubMed] [Google Scholar]
- 12.Kwiatkowski A V, Shapleigh J P. Requirement of nitric oxide for induction of genes whose products are involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J Biol Chem. 1996;271:24382–24388. doi: 10.1074/jbc.271.40.24382. [DOI] [PubMed] [Google Scholar]
- 13.Lodge J, Williams R, Bell A, Chan B, Busby S. Comparison of promoter activities in Escherichia coli and Pseudomonas aeruginosa: use of a new broad-host-range promoter-probe plasmid. FEMS Microbiol Lett. 1990;67:221–226. doi: 10.1016/0378-1097(90)90199-z. [DOI] [PubMed] [Google Scholar]
- 14.Membrillo-Hernandez J, Coopamah M D, Anjum M F, Stevanin T M, Kelly A, Hughes M N, Poole R K. The flavohemoglobin of Escherichia coli confers resistance to a nitrosating agent, a “nitric oxide releaser,” and paraquat and is essential for transcriptional responses to oxidative stress. J Biol Chem. 1999;274:748–754. doi: 10.1074/jbc.274.2.748. [DOI] [PubMed] [Google Scholar]
- 15.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 16.Moreno-Vivián C, Cabello P, Martínez-Luque M, Blasco R, Castillo F. Prokaryotic nitrate reduction: molecular properties and functional diversity among bacterial nitrate reductases. J Bacteriol. 1999;181:6573–6584. doi: 10.1128/jb.181.21.6573-6584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer T, Santini C L, Iobbi-Nivol C, Eaves D J, Boxer D H, Giordano G. Involvement of the narJ and mob gene products in distinct steps in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli. Mol Microbiol. 1996;20:875–884. doi: 10.1111/j.1365-2958.1996.tb02525.x. [DOI] [PubMed] [Google Scholar]
- 18.Poole R K, Anjum M F, Membrillo-Hernandez J, Kim S O, Hughes M N, Stewart V. Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J Bacteriol. 1996;178:5487–5492. doi: 10.1128/jb.178.18.5487-5492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ralph E T, Guest J R, Green J. Altering the anaerobic transcription factor FNR confers a hemolytic phenotype on Escherichia coli K12. Proc Natl Acad Sci USA. 1998;95:10449–10452. doi: 10.1073/pnas.95.18.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Saunders N F W, Houben E N G, Koefoed S, de Weert S, Reijnders W N M, Westerhoff H V, de Boer A P N, van Spanning R J M. Transcription regulation of the nir gene cluster encoding nitrite reductase of Paracoccus denitrificans involves NNR and NirI, a novel type of membrane protein. Mol Microbiol. 1999;34:24–36. doi: 10.1046/j.1365-2958.1999.01563.x. [DOI] [PubMed] [Google Scholar]
- 22.Savery N J, Lloyd G S, Kainz M, Gaal T, Ross W, Ebright R H, Gourse R L, Busby S J W. Transcription activation at Class II CRP-dependent promoters: identification of determinants in the C-terminal domain of the RNA polymerase α subunit. EMBO J. 1998;17:3439–3447. doi: 10.1093/emboj/17.12.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spiro S, Guest J R. Inactivation of the FNR protein of Escherichia coli by targeted mutagenesis in the N-terminal region. Mol Microbiol. 1988;2:701–707. doi: 10.1111/j.1365-2958.1988.tb00080.x. [DOI] [PubMed] [Google Scholar]
- 24.Stamler J S. Redox signalling: nitrosylation and related target interactions of nitric oxide. Cell. 1994;78:931–936. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 25.Tosques I E, Shi J, Shapleigh J P. Cloning and characterization of nnrR, whose product is required for the expression of proteins involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J Bacteriol. 1996;178:4958–4964. doi: 10.1128/jb.178.16.4958-4964.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Spanning R J M, De Boer A P N, Reijnders W N M, Spiro S, Westerhoff H V, Stouthamer A H, Van der Oost J. Nitrite and nitric oxide reduction in Paracoccus denitrificans is under the control of NNR, a regulatory protein that belongs to the FNR family of transcriptional activators. FEBS Lett. 1995;360:151–154. doi: 10.1016/0014-5793(95)00091-m. [DOI] [PubMed] [Google Scholar]
- 27.Van Spanning R J M, de Boer A P N, Reijnders W N M, Westerhoff H V, Stouthamer A H, Van Der Oost J. FnrP and NNR of Paracoccus denitrificans are both members of the FNR family of transcriptional activators but have distinct roles in respiratory adaptation in response to oxygen limitation. Mol Microbiol. 1997;23:893–907. doi: 10.1046/j.1365-2958.1997.2801638.x. [DOI] [PubMed] [Google Scholar]
- 28.Van Spanning R J M, Houben E, Reijnders W N M, Spiro S, Westerhoff H V, Saunders N. Nitric oxide is a signal for NNR-mediated transcription activation in Paracoccus denitrificans. J Bacteriol. 1999;181:4129–4132. doi: 10.1128/jb.181.13.4129-4132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vollack K U, Härtig E, Körner H, Zumft W G. Multiple transcription factors of the FNR family in denitrifying Pseudomonas stutzeri: characterization of four fnr-like genes, regulatory responses and cognate metabolic processes. Mol Microbiol. 1999;31:1681–1694. doi: 10.1046/j.1365-2958.1999.01302.x. [DOI] [PubMed] [Google Scholar]
- 30.Watmough N J, Butland G, Cheesman M R, Moir J W B, Richardson D J, Spiro S. Nitric oxide in bacteria: synthesis and consumption. Biochim Biophys Acta. 1999;1411:456–474. doi: 10.1016/s0005-2728(99)00032-8. [DOI] [PubMed] [Google Scholar]
- 31.Williams S M, Wing H J, Busby S J W. Repression of transcription initiation by Escherichia coli FNR protein: repression by FNR can be simple. FEMS Microbiol Lett. 1998;163:203–208. doi: 10.1111/j.1574-6968.1998.tb13046.x. [DOI] [PubMed] [Google Scholar]
- 32.Wing H J, Williams S M, Busby S J W. Spacing requirements for transcription activation by Escherichia coli FNR protein. J Bacteriol. 1995;177:6704–6710. doi: 10.1128/jb.177.23.6704-6710.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu G H, Cruz-Ramos H, Hill S, Green J, Sawers G, Poole R K. Regulation of cytochrome bd expression in the obligate aerobe Azotobacter vinelandii by CydR (Fnr)—sensitivity to oxygen, reactive oxygen species, and nitric oxide. J Biol Chem. 2000;275:4679–4686. doi: 10.1074/jbc.275.7.4679. [DOI] [PubMed] [Google Scholar]