Abstract
The cbsA gene of Lactobacillus crispatus strain JCM 5810, encoding a protein that mediates adhesiveness to collagens, was characterized and expressed in Escherichia coli. The cbsA open reading frame encoded a signal sequence of 30 amino acids and a mature polypeptide of 410 amino acids with typical features of a bacterial S-layer protein. The cbsA gene product was expressed as a His tag fusion protein, purified by affinity chromatography, and shown to bind solubilized as well as immobilized type I and IV collagens. Three other Lactobacillus S-layer proteins, SlpA, CbsB, and SlpnB, bound collagens only weakly, and sequence comparisons of CbsA with these S-layer proteins were used to select sites in cbsA where deletions and mutations were introduced. In addition, hybrid S-layer proteins that contained the N or the C terminus from CbsA, SlpA, or SlpnB as well as N- and C-terminally truncated peptides from CbsA were constructed by gene fusion. Analysis of these molecules revealed the major collagen-binding region within the N-terminal 287 residues and a weaker type I collagen-binding region in the C terminus of the CbsA molecule. The mutated or hybrid CbsA molecules and peptides that failed to polymerize into a periodic S-layer did not bind collagens, suggesting that the crystal structure with a regular array is optimal for expression of collagen binding by CbsA. Strain JCM 5810 was found to contain another S-layer gene termed cbsB that was 44% identical in sequence to cbsA. RNA analysis showed that cbsA, but not cbsB, was transcribed under laboratory conditions. S-layer-protein-expressing cells of strain JCM 5810 adhered to collagen-containing regions in the chicken colon, suggesting that CbsA-mediated collagen binding represents a true tissue adherence property of L. crispatus.
Species of Lactobacillus are major members of the indigenous bacterial flora in the gastrointestinal and genital tracts of humans and animals. They represent a major bacterial type within the mammalian body and reach a density of 1010 bacteria/g in human feces (27). The molecular mechanisms of Lactobacillus colonization and adhesion within the intestinal tract have remained poorly characterized. This is in contrast to our detailed knowledge on the various adhesin-receptor interactions displayed by pathogenic bacteria (reviewed in references 17, 30, and 47). Bacterial colonization of and adhesion to tissue surfaces has been proposed to be important for the establishment of a stable normal flora in the mammalian intestine (13, 36). Evidence for this hypothesis, however, is limited, in part due to our poor knowledge of the adhesive surface components expressed by lactobacilli. Considering the large number of lactobacilli in the bodies of humans and other animals and the ecological, biotechnological, and health-associated importance of these bacteria, knowledge of their colonization mechanisms could be very important. At present, two proteinaceous adhesins on lactic acid bacteria have been described, a solute-binding component of the bacterial ATP-binding cassette (ABC) transporter system (34) and an S-layer protein (36, 43), which both bind to collagens.
S-layers are paracrystalline surface protein arrays commonly expressed by species of the domains Bacteria and Archaea (reviewed in references 3 and 38). Most S-layers are composed of a single protein species which greatly varies in size in different bacterial genera. The primary sequences of the S-layer proteins exhibit little similarity, but their amino acid compositions are similar. The S-layer protein is the major single protein species and represents 10 to 20% of the total cellular protein of the bacterial cell (reviewed in reference 6). The S-layer proteins are transported over the membrane and assembled into a two-dimensional layer on the bacterial surface. Diverse functions have been attributed to the S-layers of individual bacterial species, including mediation of bacterial attachment to host tissues (11, 36, 43). This has been well characterized for the S-layer of the fish pathogen Aeromonas salmonicida, where the S-layer binds to proteins of the extracellular matrix and potentiates the establishment of systemic infection in fish (reviewed in reference 28).
S-layers commonly occur in Lactobacillus isolates belonging to DNA homology groups A1 to A4 but are absent from isolates belonging to homology groups B1 and B2 (24). The primary structures of only a few lactobacillus S-layer proteins are known (4, 5, 9, 45). The predicted S-layer proteins are 43 to 46 kDa in size and exhibit conserved C-terminal amino acid sequences but variable N-terminal and central parts of the proteins. Recent work has indicated that strains of lactobacilli harbor multiple genes for S-layer homologs whose expression is subjected to phase variation. Some of the genes are silent and lack promoters but can be translocated to an expression site via an inversion of a chromosomal segment (8). The functions of lactobacillus S-layers are largely unknown, but adhesive characteristics have been suggested in two cases (36, 43). The lactobacillus S-layers probably play an important role in maintenance of cellular functions, since no specific knockout mutants for lactobacilli of the Acidophilus group have been reported.
Various human pathogenic bacteria exhibit specific adhesiveness to the mammalian extracellular matrix proteins collagens (reviewed in references 30 and 47). Adherence to collagens is generally thought to promote bacterial colonization at damaged tissue sites, such as wounds, and is essential in enterobacterial invasion from the intestine into the circulation in orally infected mice (33, 40). The majority of Lactobacillus isolates of human or animal origin express adhesiveness to collagens (1, 26). The biological function(s) of collagen binding by lactobacilli has remained poorly characterized, but adherence may play a role in bacterial colonization of tooth surfaces (26) and in pathogenesis of infective endocarditis by lactobacilli (14). We have described adhesiveness of strain JCM 5810 of Lactobacillus crispatus to human subintestinal extracellular matrix, to mouse basement membrane, and to human collagen types (43). The adhesive surface structure was preliminarily characterized as an S-layer protein, which we have named CbsA (for “collagen-binding S-layer protein A”). We report here the expression of the cbsA gene and the characterization of collagen-binding regions within the CbsA molecule.
MATERIALS AND METHODS
Bacterial strains.
The S-layer-expressing strains L. crispatus JCM 5810 and L. acidophilus ATCC 4356 (JCM 1132) originated from the Japan Collection of Microorganisms, Wako, Japan, and their adhesive characteristics have been described previously (43). The primary structure of the S-layer protein SlpA of L. acidophilus ATCC 4356 has been described previously (4); the strain also has a silent S-layer protein gene (5). The strains representing DNA homology groups A1, A2, A3, A4, B1, and B2 of the Acidophilus group (18), originated from the Japan Collection of Microorganisms and were tested for the presence of the cbsA gene homolog. The bacteria were cultivated in static MRS broth as previously described (43). Escherichia coli strain XL1 Blue MRF′ (Stratagene, La Jolla, Calif.) was used as a host for the cloning of cbsA, and E. coli strain M15(pREP4) (Qiagen GmbH, Hilden, Germany) was used for expression of the S-layer proteins. They were cultivated at 37°C in Luria broth containing ampicillin (100 μg/ml), tetracycline (12.5 μg/ml), or kanamycin (25 μg/ml) when appropriate.
Isolation of chromosomal DNA.
To achieve efficient cell lysis, the procedure to isolate chromosomal DNA from L. crispatus JCM 5810 was modified from the procedures of Vidgrén et al. (45) and Boot et al. (4). Bacteria were grown in 100 ml of MRS medium to an optical density at 695 nm of 1.0, collected, and suspended in 3 ml of 6 M guanidine hydrochloride solution. After incubation for 20 min at 20°C, the cells were washed with 20 mM Tris buffer (pH 8.2) and resuspended in 2.5 ml of the buffer. Then 5 ml of 24% polyethylene glycol 20000 (Pharmacia Biotech, Uppsala, Sweden) and 2.5 ml of lysozyme solution (4 mg/ml; Boehringer, Mannheim, Germany) were added, and the mixture was incubated at 37°C for 60 min. A 5-ml volume of 0.2 M EDTA was then added, and the cells were collected by centrifugation (3,000 × g at 4°C for 15 min). The cells were resuspended in 10 ml of Tris buffer containing 50 μl of mutanolysin solution (15,000 U/ml; Sigma Chemical Co., St. Louis, Mo.). After incubation for 1 h at 37°C, the cells were lysed by adding 1.5 ml of 9% Sarkosyl (Sigma Chemical Co.) and 3 ml of 5 M NaCl solution. Finally, 2.9 ml of 5 M sodium perchlorate was mixed with the cell lysate, and the suspension was extracted with chloroform-isoamyl alcohol (24:1). After ethanol precipitation, chromosomal DNA was collected with a glass rod and dissolved in water.
Cloning, sequencing, and PCR mutagenesis of cbsA, and dot blot hybridization.
Standard recombinant DNA techniques were used (35). Enzymes were obtained from Promega (Madison, Wis.) or New England Biolabs (Beverly, Mass.). Chromosomal DNA from L. crispatus JCM 5810 was digested with Sau3A, separated on a 0.8% agarose gel, and transferred to a Hybond-N nylon membrane (Amersham, Little Chalfont, United Kingdom) by capillary blotting. Two degenerate oligonucleotide mixes, 5′-ATGAA(C/T)AT(A/C/T)GA(C/T)GTIGA(C/T)GC-3′ and 5′-TA(C/T)AA(C/T)TCIGCIACIGTIGCIATG-3′ complementary to amino acid sequences MNIDVD and YNSATVAM that were obtained by sequencing of CbsA peptides (see below), were used as probes in Southern hybridization to localize cbsA in chromosomal DNA fragments. Oligonucleotides were labeled by the enhanced chemiluminescesce 3′-oligolabeling and detection system (Amersham). The identified fragment was purified from the agarose gel by phenol extraction and ligated to BamHI-digested pBluescriptII KS(+) vector (Stratagene) to obtain plasmid pJS1, which was transformed to E. coli XL1 Blue MRF′ by electroporation. Transformants were screened by colony hybridization using as probes the above-mentioned oligonucleotides end-labeled with [γ-32P]dATP (Amersham). The 3′ region of cbsA not present in pJS1 was amplified for sequencing by the inverse PCR technique. Chromosomal DNA was digested with the restriction enzyme HaeIII, the resulting fragments were circularized by ligation, and PCR was performed using the primers 5′-CTCGTTACAACTCAGCAAC-3′ and 5′-GCTTAACATTAACAGTAGCACCG-3′ obtained from the cbsA sequence. A fragment of ca. 800 bp was obtained and cloned into pBluescript II KS(+) for sequencing, to give plasmid pJS2. DNA sequencing was performed by the dideoxy chain termination method using the T7 sequencing kit (Pharmacia, Uppsala, Sweden) with [α-35S]dATP (Amersham). Universal and reverse primers of the pBluescript vector and internal primers from the cbsA sequence were used.
The hybrid CbsA-SlpA and CbsA-SlpnB proteins as well as deletions and point mutations in cbsA were created by a recombinant PCR amplification procedure (16), where the switch or the mutation site was incorporated in the internal primer pair. The outside primer pair was the same as used in amplifying His-tagged cbsA or slpA (see below). The internal primer sequences were designed on the basis of the cbsA, slpA, and slpnB sequences and the desired mutation. Two pairs of hybrid S-layer proteins were created by fusing the 3′ region of cbsA into the 5′ region of slpA and slpnB and vice versa. Cloning of the recombinant PCR fragments into pQE-30, their expression, and purification of the His-tagged hybrid proteins were as described for CbsA. The nucleotide sequence at each substitution or mutation site was determined before testing of the mutated proteins.
The HpaI DNA fragment (nucleotides 226 to 1044) of cbsA purified from an agarose gel was used to detect homologous sequences in the DNA from 11 other strains of L. acidophilus representing DNA homology groups A1 to A4, B1 and B2. The probe fragment was labeled by the enhanced chemiluminiscence direct labeling and detection system, hybridization was performed at 42°C for 16 h, and the washes were done at 42°C in a low-stringency buffer (6 M urea, 0.4% sodium dodecyl sulfate [SDS], 0.5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) twice for 20 min each and then at 20°C in 2× SSC twice for 5 min each. A 2-μg portion of DNA was analyzed in each dot blot hybridization.
DNA and protein sequence analysis and homology searches were done using the FASTA package programs Align (algorithm of Lipman and Pearson) for global and Lalign for local alignment (available at http://molbiol.soton.ac.uk/compute/align.html and http://www.ch.embnet.org/software/LALIGN_form.html). Multiple-sequence alignments were done by using Clustal W version 1.7 (http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html). Homology searches in the databases were done by using Blast (http://www.ncbi.nlm.nih.gov/BLAST) and the MEME motif discovery tool (http://www.sdsc.edu/MEME) (2).
Cloning of cbsB.
Chromosomal DNA from L. crispatus JCM 5810 was digested with XhoI, which cuts the cbsA gene at position 950, and used as a template in PCR. The primers were 5′-GTTAGCGCTGCTGCTGCTGC-3′ and 5′-TTAATTAAAAGTTTGAAGCCTTTAC-3′ with terminal extensions containing BamHI and EcoRI-EcoRV sites, respectively. The 1.4-kb DNA fragment obtained by PCR was digested with XhoI to avoid any contamination with cbsA and cloned into BamHI- and EcoRI-digested pUC19 to yield plasmid pUKB36. The nucleotide sequence of this fragment was determined for both strands.
RNA analysis.
Total RNA was extracted by the methods of Chomczynski and Sacchi (10) from exponentially growing cells of L. crispatus JCM 5810 and separated in 1.5% (wt/vol) agarose gel in 1× MOPS buffer (0.02 M morpholinepropanesulfonic acid [MOPS], 5 mM sodium acetate, 1 mM EDTA [pH 7]). The gel was soaked twice for 20 min in 10× SSC buffer and transferred to Hybond filter (Amersham). RNA was cross-linked to the filter by UV light. Probes complementary to the coding region of cbsA (5′-CTGTTGTTTGCACTTGAAACGGCG-3′) and cbsB (5′-AGTAGTAGTTGCAGTAGAGTCAGC-3′) were labeled with [γ-32P]ATP by a standard T4 polymerase kinase reaction. Prehybridization, hybridization, and washings were performed in Rapid-hyb hybridization buffer (Amersham) as specified by the manufacturer.
Expression and purification of the S-layer proteins.
For expression, the DNA regions of cbsA, cbsB, slpnB, and slpA encoding the mature S-layer proteins, were PCR amplified and cloned into the pQE-30 expression vector (Qiagen GmbH). The primers were designed on the basis of the nucleotide sequences of cbsA, cbsB, slpnB, and slpA and contained in the forward primer a 5′ BamHI restriction site and in the reverse primer a SacI restriction site after the stop codon. The S-layer His-tagged fusion proteins were purified from recombinant E. coli induced with 1 mM isopropyl-β-d-thiogalactoside by lysing the cells and dissolving the cytoplasmic precipitate in 6 M guanidine hydrochloride. The S-layer His-tagged fusions were purified from the cell lysate by Ni-nitrilotriacetate affinity chromatography in 0.1 M sodium phosphate–0.01 M Tris-HCl (pH 8.0) buffer containing 6 M guanidine hydrochloride and by elution in a stepwise pH gradient in 8 M urea buffer as described in the manufacturer's instructions (Qiagen GmbH). Fractions with the S-layer proteins were extensively dialyzed against phosphate-buffered saline (PBS) at 4°C, and the self-assembled S-layer proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (20) before use in adherence tests. S-layer proteins from the wild-type strains were extracted by guanidine hydrochloride as detailed previously (23, 24).
Adherence assays.
For binding tests with solubilized collagens, human type I and IV collagens (100 μg; Sigma) were labeled with carrier-free Na125I (Amersham) by the Iodogen method (22); the activity obtained ranged between 5 × 105 and 10 × 105 cpm/μg in different lots. For blotting, 4 μg of the S-layer protein was applied by gravity flow to a nitrocellulose membrane in a Bio-Dot microfiltration apparatus (Bio-Rad Laboratories, Richmond, Calif.). The quantity of the S-layer proteins was ascertained from Coomassie blue-stained SDS-PAGE gels (20) using the Tina (version 2.0) image analysis program (Raytest Isotopenmessgeräte Gmbh, Straubenhardt, Germany). Binding of 125I-labeled collagen (500 ng/ml) to the S-layer variants and the inhibition assays with solubilized, unlabeled type I, IV or V collagen (Sigma) or fibronectin (Collaborative Biomedical Products, Bedford, Mass.), were performed as detailed elsewhere (43). The inhibitory proteins were tested at 50 μg/ml.
Binding of S-layer proteins to immobilized type I and IV collagens, laminin, fibronectin, and bovine serum albumin (BSA) was tested by enzyme-linked immunosorbent assay (ELISA) technology using the His6 monoclonal antibody (Clontech Inc., Palo Alto, Calif.) and alkaline phosphatase-tagged anti-mouse immunoglobulins as secondary antibodies essentially as specified by the manufacturer. To minimize nonspecific binding, the S-layer proteins were kept in 0.05% (wt/vol) octyl glucopyranoside in PBS overnight at 4°C and then diluted into PBS containing 0.05% (vol/vol) Triton X-100 and 0.05% (wt/vol) BSA; this buffer was also used throughout the binding assays. Detection was performed with 1 mg of disodium p-nitrophenyl phosphate per ml in diethanolamine-MgCl2 buffer.
Double staining of frozen sections of chicken colon with fluorescein isothiocyanate (FITC)-tagged bacteria and the monoclonal antibody P3X63Ag8.653 and tetramethylrhodamine isothiocyanate (TRITC)-conjugated anti-mouse immunoglobulins was performed essentially as described earlier (19). The monoclonal antibody is against type III collagen from chicken and was developed by Richard Mayne; it was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, University of Iowa, Iowa City, Iowa. We used FITC-labeled bacteria at concentrations of 109 and 108 cells/ml, and incubation on tissue sections was carried out for 1 h at room temperature. The tissue sections with adherent FITC-labeled bacteria and with TRITC-labeled tissue marker were examined in a Bx50 (Olympus Optical Co., Tokyo, Japan) microscope equipped with filters for FITC and TRITC, and the images were digitally recorded using the Image-Pro Plus version 4.0 program (Media Cybernetics, Silver Spring, Mo.).
Aggregation of JCM 5810 cells by His-CbsA proteins was analyzed by incubating 2 × 109 bacteria under gentle agitation with 10 μg of the His-CbsA protein or peptide for 1 h at room temperature in a total volume of 300 μl of PBS. The bacterial suspensions were then stained with methylene blue and examined by light microscopy.
Transmission electron microscopy.
For negative staining, a small amount of bacterial suspension or purified protein in PBS or water was transferred onto Pioloform and carbon-coated 150-mesh nickel grids. The samples were stained with 1% (wt/vol) phosphotungstate (pH 7.3), and the electron micrographs were obtained with a JEOL 1200-EX transmission electron microscope operating at 60 kV.
Protein digestion, peptide separation, and mass spectrometry.
The S-layer protein was extracted from the bacterial cell surface with 2 M guanidine hydrochloride (15), and the polypeptides were separated in a 12% polyacrylamide gel in the presence of SDS. After being stained with Coomassie brilliant blue, the S-layer was in-gel digested with Lysylendopeptidase C (Wako GmbH, Neuss, Germany) as described previously (29). The resulting peptides were separated in a SMARTSystem (Pharmacia Biotech) by reversed-phase chromatography on a 1- by 150 mm Vydac C8 column (MIC-15-05-C8P3; LC-Packings, Amsterdam, The Netherlands) using a flow rate of 40 μl/min and detection at 214 nm. A linear gradient of acetonitrile (0 to 40% in 120 min) in 0.1% trifluoroacetic acid was used for elution. Peptides were collected automatically. The molecular masses of the collected peptides were determined by matrix-assisted laser desorption-ionization–time-of-flight (MALDI-TOF) mass spectrometry in the delayed-extraction mode with a Biflex mass spectrometer (Bruker-Franzen Analytik, Bremen, Germany), using a 337 nitrogen laser. A thin-layer matrix preparation with saturated α-cyano-4-hydroxycinnamic acid in acetone was used (46). An aliquot (0.5 μl) of matrix was deposited on a stainless steel target plate and allowed to dry, after which 0.5 μl of the sample was added on top of the matrix spot.
Protein and peptide sequencing.
Protein and peptide sequencing was performed using a Procise 494A sequencer (Applied Biosystems, Perkin-Elmer, Foster City, Calif.). Selected peptides from the reverse-phase chromatography were directly applied to the BioBrene (Applied Biosystems)-treated fiberglass disks and subjected to Edman degradation. For N-terminal sequence analysis, the S-layer peptide was separated by SDS-PAGE (12% polyacrylamide) followed by electroblotting (25) onto a ProBlott polyvinylidene difluoride membrane (Applied Biosystems) and visualized by Coomassie brilliant blue staining. The stained protein band was cut out and loaded into the sequencer.
Nucleotide sequence accession numbers.
The nucleotide sequence of cbsA has been deposited in GenBank under accession number AF001313, cbsB has been deposited under accession number AF079365, slpNA has been deposited under accession number AF253043, and slpNB has been deposited under accession number AF253044.
RESULTS
Cloning and sequence analysis of the CbsA-encoding DNA fragment.
The S-layer was extracted from the L. crispatus JCM 5810 cell surface, and the 43-kDa CbsA was separated by SDS-PAGE and subjected to N-terminal and internal peptide sequence analysis. The obtained 14-mer N-terminal sequence was DAVSSANNSNLGNN. Degenerate oligonucleotide probes designed on the basis of two of the four internal amino acid sequences obtained by peptide sequencing were used in Southern hybridization to locate cbsA on genomic DNA of L. crispatus JCM 5810. The DNA region was cloned, and both strands of 1,584-bp DNA fragment containing cbsA were sequenced (GenBank accession number AF001313). Sequence analysis of cbsA revealed an open reading frame (ORF) encoding a polypeptide of 440 amino acids that contained the N-terminal sequence as well as the four peptide sequences obtained from the CbsA protein (data not shown).
The ORF encodes a signal sequence of 30 amino acids, and the calculated molecular weight of the mature form was 43,910. The calculated mass of 16 peptides in region 1 to 388 of ORF matched the mass of CbsA peptides determined by mass spectrometry (details not shown). The cbsA sequence contains a putative ribosome-binding site (37), AGGAGG, 10 bases upstream of the translational initiation codon ATG. The predicted amino acid sequence (Fig. 1) has typical features of an S-layer protein (3, 4, 38); it has a high content of hydrophobic amino acid residues (37%) as well as of serine (6%) and threonine (13%), and it lacks cysteine residues. In Fig. 1, the predicted amino acid sequence of CbsA is compared to that of seven other Lactobacillus S-layer proteins. In individual sequence alignments, CbsA exhibited an identity of 48 to 74% with the S-layer protein sequences used for Fig. 1. The MEME program for protein motif discovery and the Clustal W multialignment program revealed considerable sequence identity and similarity in the eight S-layer proteins, in particular at the C-terminal regions, whereas the N-terminal regions were more variable. Of the eight S-layer proteins included in Fig. 1, information on collagen binding was available for CbsA of L. crispatus JCM 5810 and SlpA of L. acidophilus ATCC 4356 (43), CbsB and SlpnB were tested in this study (see below), and the binding activities of SlpH1, M247, SlpnA, and SlpB remain unknown. Of the individual S-layer proteins, CbsA exhibited highest sequence identity (74%) to SlpnB, which did not bind collagens (see below). SlpnB is a nonexpressed gene of L. crispatus LGM 12003 (B. Martínez and P. Pouwels, unpublished data).
FIG. 1.
Predicted amino acid sequence of the mature CbsA protein of L. crispatus JCM 5810. Below the CbsA sequence are shown conserved regions in eight lactobacillus S-layer sequences identified by the MEME program for protein motif detection. The other S-layer proteins were SlpA (Genbank accession number X89375) (5), SlpH1 (X91199) (9), CbsB (AF079365), M247 (AJ007839), SlpB (X89376; 6), SlpnA (AF253043) (Martinez and Pouwels, unpublished), and SlpnB (AF253044) (Martinez and Pouwels, unpublished). The amino acid substitutions between CbsA and Slpa or SlpnB, as well as the deletion sites, are indicated in boldface type above the CbsA sequence. The amino acid positions where the switches in the hybrid CbsA-SlpA and CbsA-SlpnB molecules were constructed are also shown.
Identification, cloning and characterization of cbsB.
Many S-layer-containing Lactobacillus isolates in the Acidophilus group possess a silent as well as an expressed copy of an S-layer gene (7, 8), which have marked homology at the 5′ and 3′ ends. To analyze whether L. crispatus JCM 5810 also has two S-layer genes, we PCR amplified DNA fragments from XhoI-digested chromosomal DNA of JCM 5810 by using as primers nucleotides of the signal sequence as well as the 3′ end of cbsA, since these regions are highly conserved in the S-protein genes characterized in the Acidophilus group. XhoI cuts cbsA at nucleotide 950. The amplified 1.4-kb DNA fragment was cut with XhoI to remove any contaminating cbsA and cloned into pUC19 and sequenced. The sequence does not contain an XhoI site. The translated ORF of this DNA, which we termed cbsB, encoded a predicted protein of 453 amino acids (data not shown). The first 23 N-terminal amino acids are identical to those in the signal sequence of CbsA, the predicted mature CbsB of 429 amino acids is 43% identical to CbsA in sequence and showed the typical features of an S-layer protein.
Expression of cbsA and cbsB in E. coli and L. crispatus.
To confirm that CbsA indeed is a collagen-binding S-layer protein on the surface of L. crispatus and to analyze the binding properties of CbsB, SlpA, and SlpnB, the proteins were expressed and purified from E. coli as His-tagged fusion proteins. The DNA fragment encoding the mature CbsA (residues 1 to 410) was cloned into the pQE-30 expression vector and expressed as an N-terminal His-tagged fusion protein in E. coli M15(pREP4). The His-CbsA protein was purified by affinity chromatography in the presence of 8 M urea, and the resulting protein precipitated during subsequent dialysis against PBS. The results of the SDS-PAGE analysis of the CbsA isolated from L. crispatus cells as well as of His-CbsA are shown in Fig. 2. In electron microscopy, the proteins exhibited a similar crystalline structure with sheet morphology (Fig. 3). By similar procedures, we isolated SlpA from L. acidophilus ATCC 4356 cells and as a His-tagged fusion protein from recombinant E. coli, as well as His-CbsB (Fig. 2) and His-SlpnB from E. coli (data not shown).
FIG. 2.
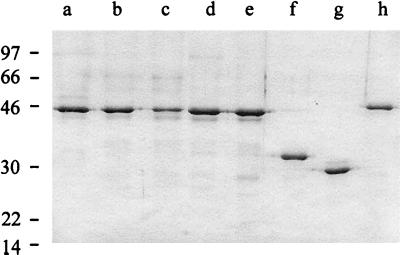
SDS-PAGE analysis of selected S-layer proteins. Lanes: a, CbsA extracted from L. crispatus JCM 5810 cells; b, His-CbsA from recombinant E. coli; c, the hybrid S-layer protein CbsA1–287/SlpA290–413; d, His-CbsA D130N; e, His-CbsA Δ91–96; f, His-CbsA1–287; g, His-CbsA42–287; h, His-CbsB. The migration distances of molecular mass marker peptides are indicated on the left in kilodaltons.
FIG. 3.
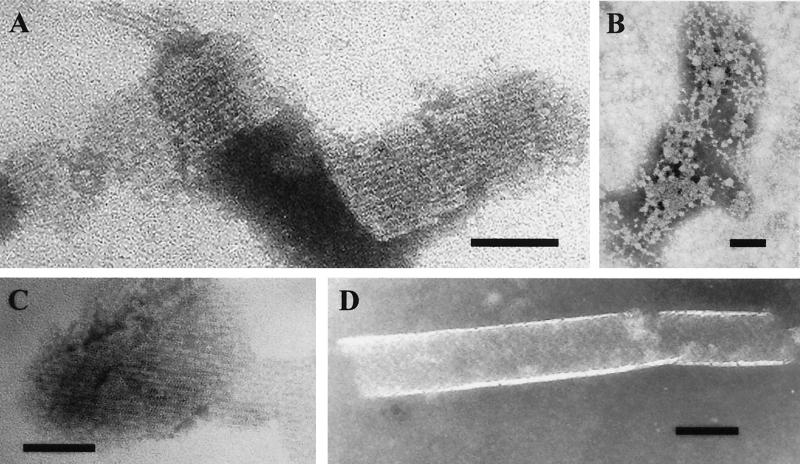
Transmission electron microscopy of the CbsA preparations. The S-layer proteins are His-CbsA from recombinant E. coli (A), the polypeptide His-CbsA1–250 (B), the polypeptide His-CbsA1–287 (C), and the CbsA protein with the substitution KSDV257TANN (D). Bar, 100 nm.
The amino acid sequence data described above suggested that CbsA, but not CbsB, is expressed on the surface of L. crispatus JCM 5810 cells. To confirm this, total RNA from logarithmically growing JCM 5810 cells was extracted and the cbsA and cbsB transcript levels were evaluated by Northern blot analysis. The probes were designed from the coding sequence corresponding to the N terminus of CbsA or CbsB, and their similar labelling was confirmed by Southern hybridization using digested chromosomal DNA as the template (data not shown). In Northern blotting, the cbsA-specific probe detected a transcript with an apparent size of 1.5 kb, which is close to the predicted size of a monocistronic cbsA transcript, whereas no signal was detected with the cbsB probe (data not shown).
Binding of solubilized collagens to the S-layer proteins.
We next tested the binding of 125I-labeled type IV and I collagens to CbsA and His-CbsA immobilized on a nitrocellulose membrane. We also assessed the binding to SlpA, His-SlpA, His-CbsB, His-SlpnB, and BSA (as a control). The type IV and I collagens efficiently bound to His-CbsA as well as to CbsA, whereas no binding to the other Lactobacillus S-layer proteins was observed (Table 1). Binding of 125I-labeled type I and IV collagens to His-CbsA as well as to CbsA was effectively inhibited (by >99%) by a 100-fold excess of solubilized, unlabeled type I or IV collagens; unlabeled type V collagen under the same conditions inhibited binding by 94%, and fibronectin inhibited it by <1%. The inhibition patterns were highly similar to the one we previously detected for binding of 125I-type IV collagen to JCM 5810 cells (43), and we concluded that the collagen binding observed with His-CbsA and CbsA was specific.
TABLE 1.
Binding of 125I-labeled type I and IV collagen to the S-layer proteins and formation of a crystalline layer by the proteins
| Test protein | Binding (%)a of
|
Formation of crystalline layerb | |||
|---|---|---|---|---|---|
|
125I-collagen type I
|
125I-collagen type IV
|
||||
| Mean | Range | Mean | Range | ||
| Native or His-tag fusion S-layer proteinc | |||||
| CbsA | 120 | 81–145 | 122 | 66–162 | + |
| His-CbsA | 100 | 100 | + | ||
| SlpA | <1 | 0–1 | <1 | 0–1 | + |
| His-SlpA | <1 | 0–1 | <1 | 0–1 | + |
| His-CbsB | <1 | 0–1 | <1 | 0–1 | + |
| His-SlpnB | 5 | 4–6 | 2 | 0–4 | + |
| Hybrid His-tag fusion S-layer protein | |||||
| His-CbsA1–212/SlpA208–413 | <1 | 0–1 | 1 | 0–2 | − |
| His-SlpA1–207/CbsA213–410 | <1 | 0–1 | <1 | 0–1 | − |
| His-CbsA1–287/SlpA290–413 | 61 | 22–111 | 72 | 40–92 | + |
| His-SlpA1–289/CbsA288–410 | <1 | 0–1 | <1 | 0–1 | + |
| His-CbsA1–28/SlpnB20–409 | <1 | 0–1 | <1 | 0–1 | + |
| His-SlpnB1–19/CbsA29–410 | 105 | 87–153 | 97 | 74–117 | + |
| His-CbsA1–81/SlpnB73–409 | <1 | 0 | <1 | 0–1 | − |
| His-SlpnB1–72/CbsA82–410 | 78 | 27–131 | 61 | 44–86 | + |
| His-CbsA1–194/SlpnB187–409 | <1 | 0–1 | <1 | 0–1 | + |
| His-SlpnB1–186/CbsA195–410 | <1 | 0–1 | <1 | 0–1 | + |
| His-CbsA1–212/SlpnB205–409 | <1 | 0–1 | <1 | 0–1 | + |
| His-SlpnB1–204/CbsA213–410 | <1 | 0–1 | <1 | 0–1 | + |
| His-CbsA1–250/SlpnB250–409 | <1 | 0–1 | <1 | 0–1 | − |
| His-SlpnB1–249/CbsA251–410 | <1 | 0–1 | <1 | 0–1 | − |
| His-CbsA1–287/SlpnB287–409 | 54 | 39–96 | 73 | 47–98 | + |
| His-SlpnB1–286/CbsA288–410 | 2 | 0–3 | 4 | 4–5 | + |
| Polypeptide | |||||
| His-CbsA1–212 | 2 | 0–2 | <1 | 0–1 | − |
| His-CbsA1–250 | 1 | 0–2 | 1 | 1–2 | − |
| His-CbsA1–287 | 77 | 65–83 | 73 | 27–77 | + |
| His-CbsA42–287 | 4 | 2–5 | <1 | 0–1 | − |
| His-CbsA288–410 | 2 | 2–3 | <1 | 0–1 | − |
| Mutated CbsA protein | |||||
| His-CbsA Δ22–26 | 2 | 0–2 | 8 | 5–15 | + |
| His-CbsA Δ91–96 | 22 | 20–24 | 27 | 19–33 | + |
| His-CbsA NNN14INL | 88 | 50–131 | 74 | 32–100 | + |
| His-CbsA F19S | 83 | 53–109 | 80 | 52–109 | + |
| His-CbsA D130N | 56 | 40–68 | 64 | 39–78 | + |
| His-CbsA N226A | 33 | 24–44 | 34 | 22–40 | + |
| His-CbsA KSDV257TANN | <1 | 0–1 | <1 | 0–1 | + |
| His-CbsA K257T | 91 | 80–102 | 92 | 83–99 | + |
| His-CbsA S258A | 10 | 6–13 | 9 | 4–13 | + |
| His-CbsA D259N | 77 | 63–87 | 87 | 80–98 | + |
| His-CbsA V260N | <1 | 0–1 | 1 | 0–1 | + |
| His-CbsA TA264SK | 52 | 44–64 | 49 | 36–68 | + |
| His-CbsA P268A | 51 | 34–66 | 44 | 36–54 | + |
| Control | |||||
| BSA | <1 | 0 | |||
Results are mean values from at least six independent assays; the range gives the lowest and the highest value obtained in the assays. The binding value shown by His-CbsA was assigned as 100% in each individual test.
Formation of the S-layer was determined by transmission electron microscopy of the purified proteins negatively stained with phosphotungstate acid.
“Native” denotes to the S-layer extracted from Lactobacillus cells. His-tag fusion proteins were isolated from E. coli cells.
Binding properties of His-CbsA/SlpA and His-CbsA/SlpnB hybrid molecules.
The sequence comparison (Fig. 1) indicated that most differences between CbsA and the other S-layer proteins reside in the N-terminal two-thirds of the mature proteins. To test whether the collagen-binding activity in CbsA also is located in that region, we constructed His-tagged hybrid S-layer molecules where the N or C terminus was exchanged between CbsA and SlpA or SlpnB. We initiated the hybrid constructions with SlpA because its sequence was available. During the work, however, sequencing of slpnB revealed that SlpnB has higher identity to CbsA, and we therefore also made CbsA-SlpnB hybrid molecules. The hybrid molecules are listed in Table 1; the numbering refers to the mature form of CbsA in Fig. 1 and the corresponding sites in SlpA or SlpnB. Binding of collagens to the hybrid molecules is also given in Table 1. With the four CbsA-SlpA hybrids, collagen binding was observed with the hybrid His-CbsA1–287/SlpA290–413 that contained the N-terminal two-thirds of CbsA fused to the C terminus of SlpA, whereas the counterhybrid with the C terminus from CbsA as well as the hybrids created at site 212 of CbsA did not show binding. As an example, the SDS-PAGE analysis of the His-CbsA1–287/SlpA290–413 preparation is shown in Fig. 2.
Similarly, the hybrid His-CbsA1–287/SlpnB287–409 efficiently bound collagens, whereas the corresponding hybrid SlpnB1–286/CbsA288–410 was inactive in collagen binding. The hybrids His-SlpnB1–19/CbsA29–410 and His-SlpnB1–72/CbsA82–410 bound collagens, indicating that the extreme N terminus can be substituted between CbsA and SlpnB without loss of binding. However, the hybrids with 194, 212, or 250 N-terminal amino acids of CbsA fused to the C terminus of SlpnB did not support adherence.
Binding of solubilized collagens to His-CbsA polypeptides and His-CbsA mutants.
To further characterize the region in CbsA that binds to collagen, we constructed five His-tagged polypeptides, His-CbsA1–287, His-CbsA1–250, His-CbsA1–212, His-CbsA42–287 (Fig. 2), and His-CbsA288–410, and tested their capacity to bind solubilized type I and IV collagens. His-CbsA1–287 bound radiolabeled collagens with an efficiency (ca. 70%) similar to that shown by the collagen-binding hybrid S-layer proteins, whereas polypeptides 1 to 212 and 1 to 250 showed a low level of binding (Table 1). The C-terminal peptide His-CbsA288–410 exhibited low-level (2%) binding to type I collagen but no detectable binding to solubilized type IV collagen.
We also constructed mutations in His-CbsA at sites where the CbsA sequence differs from that of SlpA or SlpnB; these were deletions (Δ22–26 and Δ91–96) or mutations replacing residues in CbsA with the corresponding ones in SlpA or SlpnB. Figure 2 shows an SDS-PAGE analysis of selected proteins; the binding of radiolabeled collagen to the constructs is given in Table 1. The two deletions of 5 or 6 amino acids in the N-terminal region decreased the binding by 90 or 70%, respectively, while the substitutions D130N, N226A, TA264SK, and P268A caused reduction of binding by 40 to 70% and the substitutions NNN141INL and F19S had less effect on the binding. A dramatic loss of binding activity was seen with the substitution mutant KSDV257TANN. Analysis of the single amino acid substitutions at this site revealed that the substitutions S258A and V260N had a greatly reduced binding activity whereas the substitutions K257T and D259N had less effect.
Binding of S-layer proteins to immobilized collagens.
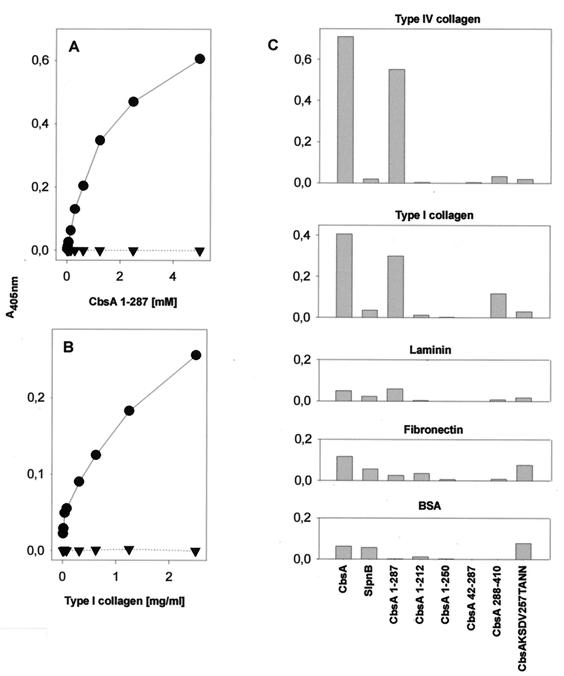
In tissues, collagens occur in insoluble, immobilized forms, and a number of bacteria and bacterial adhesins interact differentially with the immobilized and the soluble forms of collagens (reviewed in reference 47). We therefore tested the binding of selected S-layer proteins to immobilized type I and IV collagens. The binding was measured by ELISA technology using monoclonal anti-His-tag antibodies and secondary antibodies; the S-layer proteins or peptides used in the study were treated with 0.05% octyl glucopyranoside and then diluted in 0.05% Triton X-100 buffer to minimize protein aggregation and nonspecific binding. The peptide His-CbsA1–287 efficiently bound to immobilized type I collagen, and the binding was dependent on the amount of the CbsA1–287 peptide added to the mixture (Fig. 4A), as well as on the amount of type I collagen used for coating (Fig. 4B), and was therefore considered specific. The His-CbsA1–287 peptide bound to type IV and I collagens but not to laminin, fibronectin, or BSA (Fig. 4C). The complete S-layer protein His-CbsA also bound to collagens but not significantly to laminin, fibronectin, or BSA; its binding to collagens was slightly more efficient than that shown by His-CbsA1–287. The S-layer protein SlpnB and the mutated protein His-CbsA KSDV257TANN showed poor binding to the test proteins, as did the His-CbsA1–212, His-CbsA1–250, and His-CbsA42–287 peptides. The C-terminal peptide His-CbsA288–410, however, bound to type I collagen, at a level of 35% of that shown by His-CbsA1–287, but did not show detectable binding to type IV collagen, laminin, fibronectin, or BSA (Fig. 4).
FIG. 4.
Binding of His-tagged S-layer proteins to extracellular matrix proteins immobilized on plastic was determined by ELISA technology with monoclonal anti-His-tag antibodies and secondary antibodies. (A) Binding in increasing concentrations of the His-CbsA1–287 polypeptide to type I collagen (●) and BSA (▾). (B) Binding of the polypeptide (1.2 mM) to wells coated with increasing concentrations of type I collagen (●) or BSA (▾). (C) Binding of S-layer proteins to type IV and I collagens, laminin, fibronectin, and BSA immobilized on plastic. A405nm, absorbance at 405 nm.
The peptide His-CbsA1–287 migrated in 0.05% octyl glucopyranoside buffer as polymers of >200,000 Da in apparent molecular mass as estimated by gel filtration (details not shown), indicating that the detergent did not solubilize the S-layer into subunits.
Adherence of bacteria to chicken tissue.
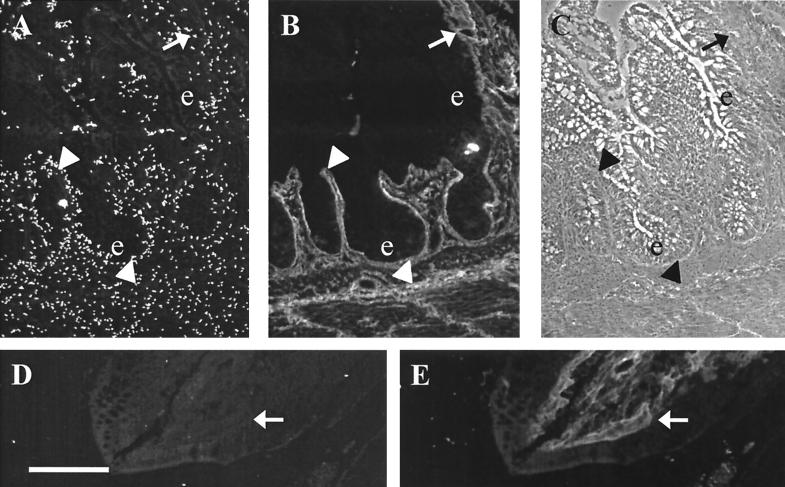
Isolates of L. crispatus are recovered from chicken and human intestine or feces (27), and strain JCM 5810 had been isolated from chicken feces. The binding experiments described above were performed with purified human collagens since they were commercially available, leaving open the question whether CbsA also recognizes chicken connective tissue. We approached this problem by analyzing the binding sites for CbsA on frozen sections of chicken colon, using double staining of the tissue sections with FITC-tagged bacteria and TRITC-tagged monoclonal antibody against chicken type III collagen, by methods we had developed earlier with pathogenic bacteria. The JCM 5810 cells showed no epithelial adherence in the colon but adhered to connective tissue sites stained by the antibody. Bacteria were also detected at basolaterial aspects of epithelial cells (Fig. 5). Removal of the S-layer from JCM 5810 cell surface by guanidine hydrochloride extraction (43) nearly completely abolished bacterial adherence to chicken tissue (Fig. 5D and E).
FIG. 5.
Adherence of FITC-tagged cells of L. crispatus JCM 5810 to frozen section of chicken colon detected by fluorescence microscopy. (A) FITC-tagged bacteria adherent on the colon tissue. (B) The same microscopic field for staining with the TRITC-labeled monoclonal antibody against chicken type III collagen. (C) The same field by light microscopy. (D) Adhesion of FITC-labeled JCM 5810 cells after removal of the S-layer by guanidine hydrochloride extraction. (E) The same microscopic field for anti-type III collagen staining. The bacteria were tested at 108 cells/ml. Arrows indicate connective-tissue regions rich in type III collagen, arrowheads indicate basolateral regions of epithelial cells, and e indicates epithelial cells. Bar, 100 μm. Note the adherence of JCM 5810 cells to type III collagen-rich areas and to basolateral aspects of the epithelial cells as well as the poor adhesion by JCM 5810 cells lacking the S-layer.
Formation of a regular S layer by hybrid and mutated molecules.
We next used transmission electron microscopy to analyze the ability of each molecule to form a regular S-layer structure. Selected examples are shown in Fig. 3. In Table 1 we have listed the formation of a crystalline layer with a periodic, regular morphology as observed by negative-stain transmission electron microscopy. The His-CbsA molecules formed crystalline sheet structure (Fig. 3A). Similar structures were observed with the CbsA molecules extracted from the L. crispatus cell surface (data not shown) and with the CbsA1–287 peptide (Fig. 3C). The shorter N-terminal peptides CbsA1–250 (Fig. 3B), CbsA1–212, and CbsA42–287 (data not shown) were aggregated but did not form a regular layer structure. Nine of the CbsA hybrids or mutants constructed in this study failed to polymerize into a crystalline layer; none of these molecules exhibited collagen binding (Table 1). The His-CbsA KSDV257TANN substitution mutant, which had completely lost the capability of collagen binding, formed a structure of cylinder-like morphology 90 nm in diameter (Fig. 3D). Similar cylinder morphology was detected with the substitution mutant V260N, whereas the substitution mutants K257T, D258A, and D259N exhibited sheet-like crystalline layers (data not shown). It was obvious that this morphological difference was not an all-or-nothing property, since minor amounts of cylinder-like structures were observed with His-CbsA and His-CbsA1–287 as well. However, only cylinder-like structures were observed with the His-CbsA KSDV257TANN and His-CbsA V260N mutants.
Binding of the S-layer proteins to the JCM 5810 cells.
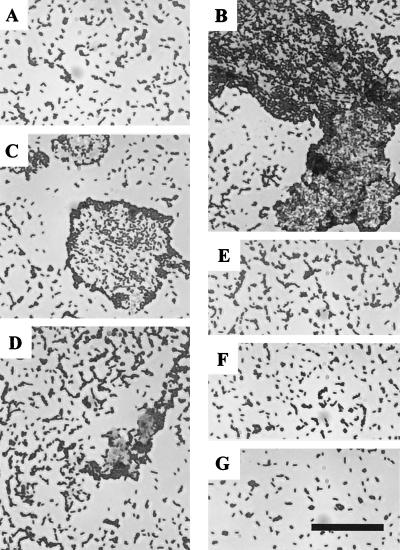
The ELISA experiments in Fig. 4A and B indicated that the binding of His-CbsA1–287 to collagen did not become saturated. Binding analysis by the Biacore technology indicated the same phenomenon, and no inhibition by His-CbsA1–287 of the binding of radiolabeled collagens to JCM 5810 cells was observed (data not shown). These findings suggested that the S-layer proteins bound to each other as well as to the JCM 5810 cell surface. We analyzed this by incubating 2 × 109 JCM 5810 cells with 10 μg of the S-layer protein for 1 h and visualizing the cell suspensions by light microscopy after staining them with methylene blue. The JCM 5810 cell population without added His-CbsA was not significantly aggregated, whereas addition of the His-CbsA or the His-CbsA1–287 proteins caused the formation of massive aggregates with bacterial cells embedded in the S-layer protein (Fig. 6). Such visible aggregate formation was not seen with His-CbsA1–212, His-CbsΔ1–250, or His-CbsA42–287, whereas small aggregates were detected in the presence of His-CbsA288–410 (Fig. 6). Small precipitates were detected by light microscopy in the His-CbsA protein preparations without added cells; these were morphologically different from bacterial aggregates (data not shown).
FIG. 6.
Aggregation of L. crispatus cells in the presence of S-layer proteins. Bacteria (2 × 109) were incubated for 1 h with the His-CbsA proteins (10 μg) in PBS, stained with methylene blue, and then examined by light microscopy. (A) JCM 5810 cells without added His-CbsA protein. (B to G) Cells in the presence of His-CbsA (B), His-CbsA1–287 (C), His-CbsA288–410 (D), His-CbsA1–250 (E), His-CbsA1–212 (F), and His-CbsA42–287 (G). Bar, 10 μm.
Presence of cbsA homologs in lactobacilli and other bacteria.
We used the 819-bp HpaI fragment of cbsA as a probe to detect homologous DNA fragments in DNA from 11 lactobacillus isolates representing DNA homology groups A1 to A4, B1 and B2 of the Acidophilus group (two isolates in each group). Five of these isolates bound collagens (data not shown). Strain JCM 5810 is of the homology group A2. The probe fragment encodes amino acids −2 to +271 (where +1 is the first residue in the mature CbsA), i.e., a region which is not conserved in the S-layer proteins (Fig. 1) but is involved in collagen-binding. None of the test strains gave a signal with the probe, which strongly reacted with DNA from the homologous strain (data not shown).
The collagen-binding region has been characterized in three bacterial surface proteins, the S-layer-like protein YadA of Yersinia enterocolitica (48) and the collagen-binding adhesins CNA of Staphylococcus aureus (30) and Ace of Enterococcus faecalis (32). The predicted sequence of CbsA shows only 13 to 16% identity to these collagen-binding protein regions. The highest local identity score was 24% in an 81-amino-acid overlap of residues 92 to 172 in CbsA and 88 to 166 in CNA and 22% in a 73-amino-acid overlap of residues 211 to 282 in CbsA and 35 to 103 in CNA. We found 17 to 20% sequence identity of CbsA to the collagen- and laminin-binding S-layer protein of Aeromonas salmonicida (28), the collagen-binding ABC transporter protein of Lactobacillus reuteri (34), and the type V collagen-binding fimbrial adhesin MrkD of Klebsiella pneumoniae (41).
DISCUSSION
The ability to bind to collagens is expressed by 70% of Lactobacillus isolates (1, 14, 26), and it appears that lactobacilli express multiple adhesin types interacting with these abundant tissue proteins. Roos et al. (34) recently described Cnb, a 29-kDa protein of L. reuteri with an affinity to solubilized type I collagen. Cnb is a putative ABC transporter protein and has no significant sequence homology to CbsA. Our results confirm the function of the S-layer protein CbsA of L. crispatus as a collagen-binding protein and show that adhesion to collagens is not expressed by all S-layer proteins of Lactobacillus isolates in the Acidophilus group. Despite its common occurrence, the biological functions of collagen binding by lactobacilli remain open. Our finding that the S-layer-protein-expressing cells of JCM 5810 adhered to collagen-rich regions in the colon tissue of chicken, the natural host for JCM 5810, supports the notion that collagen binding represents a true tissue-binding property of L. crispatus. Adhesiveness to collagens has been proposed to enhance Lactobacillus colonization at tooth surfaces and to promote the spread of lactobacilli from the oral cavity to cause infective endocarditis (14, 26). However, lactobacilli are most numerous in the intestine, and the role of collagen binding in colonization by intestinal lactobacilli remains to be determined.
The ability of CbsA to bind collagens was reduced by ca. 20% by the N-terminal fusion to the 6-mer His-tag peptide, as indicated by the collagen-binding efficiency of CbsA from lactobacilli and His-CbsA from recombinant E. coli. Both CbsA and His-CbsA were solubilized by guanidine hydrochloride during their isolation and, when dialyzed into a physiological buffer, formed morphologically highly similar S-layer-like structures, which also resembled those seen by electron microscopy on the JCM 5810 cell surface. Refolding into a morphologically or functionally correct form in a detergent- or chaotropic agent-free medium has also been observed with other lactobacillus S-layer proteins (21, 23), as well as with the collagen-binding S-layer-like protein YadA of Y. enterocolitica (33). Binding of radiolabeled collagen by His-CbsA from E. coli and by CbsA from L. crispatus and on the other hand by JCM 5810 cells (43) was similarly inhibited by unlabeled extracellular matrix proteins. These findings indicate that the N-terminal fusion and expression in a heterologous host species did not significantly change the collagen-binding specificity or polymerization of CbsA into a crystalline layer.
It appears that CbsA contains two peptide regions binding to collagens, the major one at 1 to 287 reacting with type I and IV collagens, and another, weaker type I collagen-binding site within the C-terminal region. The His-CbsA288–410 peptide bound to immobilized type I collagen but not to type IV collagen, and its binding to solubilized type I collagen was barely detectable. It could be that the peptide recognizes the immobilized form of collagen or, more probably, due to low affinity it needs a high receptor density in the solid-phase assay. The C-terminal regions in Lactobacillus S-layer proteins are conserved, but the binding levels of His-SlpnB, His-CbsB, and His-SlpA to immobilized or solubilized collagens are very low. The C-terminal His-CbsA288–401 peptide did not form an S-layer, and it remains open whether this low binding activity represents a cryptic binding site exposed in the peptide or whether it is indeed another collagen-binding site within the CbsA S-protein.
It appears that the major collagen-binding region within CbsA is large and that a crystalline structure with sheet morphology is important for optimal binding. This is suggested by the behavior of the His-CbsA peptides and mutated proteins as well as the His-CbsA/SlpA and His-CbsA/SlpnB hybrid molecules; i.e., those that did not form an S-layer also failed to bind collagens. This is different to what has been found with the laminin- and fibronectin-binding S-layer protein A of A. salmonicida, where a soluble 38-kDa N-terminal peptide from the 51-kDa A-layer protein was found to express adhesiveness but did not assemble into a tetragonal array (11, 42). Similarly, the 302-amino-acid collagen-binding region in the S-layer-like protein YadA of Y. enterocolitica has been functionally expressed as an internal peptide fusion in flagella of E. coli (48). Collagen binding and crystalline-layer formation were exhibited by the His-CbsA1–287 peptide but not by His-CbsA1–212, His-CbsA1–250, or His-CbsA42–287, indicating that the region from 1 to 287 in CbsA contains information for both processes. It is interesting that the substitution mutants KSDV257TANN and V260N failed to adhere to collagens and predominantly exhibited a crystalline layer with a cylinder-like morphology. This finding also indicates the importance of the sheet-like crystalline structure in correct expression of the CbsA residues interacting with collagens. The assembled structures of CbsA and mutated proteins must be analyzed in more detail. Our ongoing work by Fourier analyses of the electron microscopic images has suggested that the CbsA crystalline layers have oblique structure and that the KSDV256TANN substitution mutant indeed forms a crystal structure different from that of CbsA.
Binding analyses of the His-CbsA peptides and mutants indicate that the major collagen-binding region is within the 287 N-terminal residues of CbsA; however, the role of the extreme N terminus of CbsA in collagen binding remains uncertain. The His-CbsA-derived peptide from 1 to 287 exhibited adhesiveness and formed an S-layer, whereas the peptide from 42 to 287 failed to adhere and to polymerize into an S-layer. The hybrid S-layer proteins His-SlpnB1–19/CbsA29–410 and His-SlpnB1–72/CbsA82–410 exhibited efficient collagen binding, indicating that the N termini could be exchanged between CbsA and SlpnB without a loss of collagen binding. This can be explained either by complementation of residues in CbsA important for binding by the corresponding residues in SlpnB or by a lack of a role of CbsA residues 1 to 72 in the binding. The former hypothesis is supported by the low binding efficiency of the deletion derivative His-CbsAΔ22–26, and the latter hypothesis is supported by the adhesiveness exhibited by the substitution mutant proteins NNN14INL and F19S.
CbsA resembles the other well-characterized collagen-binding bacterial proteins, CNA of S. aureus (39), Ace of E. faecalis (32), and the S-layer-like protein YadA of Y. enterocolitica (40, 48) in that the collagen-binding region is large. In YadA, the binding region is ca. 300 amino acids and probably discontinuous. The crystal structure of the 168-amino-acid collagen-binding domain CBD of CNA revealed that the binding site is located along a groove on a concave β-sheet with a structure complementary to the helical structure of collagen. A similar structural motif was recently identified in Ace (32). Secondary-structure predictions for CbsA gave a high content (37%) of β-sheet, as also found in CBD and Ace. However, we found only weak identity of CbsA to the other bacterial collagen-binding proteins, and the highest local identity (22 to 24%) was detected in amino acid overlaps at 92 to 172 in CbsA and 88 to 166 in CNA, as well as 211 to 282 in CbsA and 35 to 103 in CNA. These sequences in CbsA are within the binding region but in CNA correspond to sites that are either in the vicinity of or partially within the region important for collagen binding. Furthermore, some of the residues in CBD found to interact with collagen do not have homologs within the CbsA collagen-binding regions. It therefore seems likely that the structural motifs detected in CNA and Ace are not directly applicable to collagen binding by the rigid CbsA S-layer. We found that replacement of residues D130, N226, S258, V260, TA264, and P268A, which are located within the collagen-binding region in CbsA, decreased the binding without affecting the formation of a crystalline layer, suggesting that these residues are involved in the adherence by CbsA.
Very little is presently known about the incorporation of S-layer subunits into a growing S-layer (for a recent review, see reference 12). Our results suggest that the region from 250 to 287 in CbsA is important for the assembly of subunits into a crystalline layer. First, the peptide from 1 to 287 was the only His-CbsA peptide that polymerized into an S-layer-like structure and aggregated the JCM 5810 cells; the latter probably resulted from binding of the peptide to S-layer covering the JCM 5810 cell surface. On the other hand, the peptide from 42 to 287 failed to form a crystalline layer and to aggregate JCM 5810 cells, indicating a role for the extreme N terminus of CbsA in the assembly. Second, the CbsA-SlpnB hybrids created at residue 250 in CbsA failed to form a crystalline layer whereas the other CbsA-SlpnB hybrids polymerized into a periodic structure. The substitution mutant proteins His-CbsA KSDV257TANN and His-CbsA V260N formed a crystalline structure with different morphology from that of His-CbsA. A search for protein motifs in lactobacillus S-layer proteins indicated that this region is conserved in Lactobacillus S-layer proteins, which also contain shorter conserved regions in the N terminus. Such conserved blocks might be important for correct assembly of the rigid S-layer structure. Our results suggest that the extreme C terminus of CbsA plays no significant role in S-layer assembly. This conserved region could be involved in the secretion of the S-layer subunit onto the Lactobacillus cell surface. It is interesting that the His-CbsA-SlpA hybrids and the His-CbsA-SlpnB hybrids created at site 212 of CbsA behaved differently: the His-CbsA-SlpA hybrids failed to polymerize into a crystalline layer, whereas the His-CbsA-SlpnB hybrids formed a periodic polymer. This suggests that the structural constraints in the S-layer assembly by SlpA and SlpnB are differentially compatible with the CbsA terminal regions, which may reflect the sequence differences at the N-terminal half of these S-proteins.
As noted for many Lactobacillus isolates (7), strain JCM 5810 of L. crispatus has two homologous S-layer genes, cbsA and cbsB. We found no evidence for the expression of CbsB under the growth conditions we used, nor was a cbsB-specific RNA transcript detected, suggesting that cbsB is a silent gene. His-CbsB from E. coli polymerized into a crystalline layer, but we could not detect any collagen binding by His-CbsB.
S-layers are expressed by isolates belonging to homology groups A1 to A4 of the Acidophilus group of Lactobacillus. When using DNA hybridization, we detected a cbsA homolog only in isolate JCM 5810, which is a member of homology group A2. Furthermore, we have not detected collagen binding by S-layer proteins from 12 additional isolates of L. crispatus (S. Tankka, and J. Sillanpää, unpublished data). These findings suggest that CbsA is not common in these bacteria and also are in agreement with the observations that Lactobacillus S-layers exhibit marked sequence variability, even among isolates representing the same Acidophilus homology group (7). We are currently analyzing how this sequence variability is correlated with adhesive functions of Lactobacillus S-layer proteins.
ACKNOWLEDGMENTS
This study was supported by the Academy of Finland (project 40836, 44600, 44168, and 164916) and by the University of Helsinki.
We thank Nina Rautonen and Hannele Kettunen, Danisco Ltd., Kantvik, Finland, for help in obtaining the intestinal tissue samples. Electron Microscopy was performed at the Section of Electron Microscopy, Institute of Biotechnology, University of Helsinki.
REFERENCES
- 1.Aleljung P, Paulsson M, Emödy L, Andersson M, Naidu A S, Wadström T. Collagen binding by lactobacilli. Curr Microbiol. 1991;23:33–38. [Google Scholar]
- 2.Bailey T, Elkan C. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. Menlo Park, Calif: AAAI Press; 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers; pp. 28–36. [PubMed] [Google Scholar]
- 3.Beveridge T J. Bacterial S-layers. Curr Opin Struct Biol. 1994;4:204–212. [Google Scholar]
- 4.Boot H J, Kolen C P A M, van Noort J M, Pouwels P H. S-layer protein of Lactobacillus acidophilus ATCC 4356: purification, expression in Escherichia coli and nucleotide sequence of the corresponding gene. J Bacteriol. 1993;175:6089–6096. doi: 10.1128/jb.175.19.6089-6096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boot H J, Kolen C P A M, Pouwels P H. Identification, cloning, and nucleotide sequence of a silent S-layer protein gene of Lactobacillus acidophilus ATCC 4356 which has extensive similarity with the S-layer protein gene of this species. J Bacteriol. 1995;177:7222–7230. doi: 10.1128/jb.177.24.7222-7230.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boot H J, Pouwels P H. Expression, secretion and antigenic variation of bacterial S-layer proteins. Mol Microbiol. 1996;21:1117–1123. doi: 10.1046/j.1365-2958.1996.711442.x. [DOI] [PubMed] [Google Scholar]
- 7.Boot H J, Kolen C P A M, Pot B, Kersters K, Pouwels P H. The presence of two S-layer-protein-encoding genes is conserved among species related to Lactobacillus acidophilus. Microbiology. 1996;142:2375–2384. doi: 10.1099/00221287-142-9-2375. [DOI] [PubMed] [Google Scholar]
- 8.Boot H J, Kolen C P A M, Pouwels P H. Interchange of the active and silent S-layer protein genes of Lactobacillus acidophilus by inversion of the chromosomal slp segment. Mol Microbiol. 1996;21:799–809. doi: 10.1046/j.1365-2958.1996.401406.x. [DOI] [PubMed] [Google Scholar]
- 9.Callegari M L, Riboli B, Sanders J W, Cocconcelli P S, Kok J, Venema G, Morelli L. The S-layer gene of Lactobacillus helveticus CNRZ 892: cloning, sequence and heterologous expression. Microbiology. 1998;144:719–726. doi: 10.1099/00221287-144-3-719. [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski P, Sacchi N. Single-step method for RNA isolation by acid guanidium thiocyanate-phenolchloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.Doig P, Emödy L, Trust T J. Binding of laminin and fibronectin by the trypsin-resistant major structural domain of the crystalline virulence surface array protein of Aeromonas salmonicida. J Biol Chem. 1992;267:43–49. [PubMed] [Google Scholar]
- 12.Fernández L A, Berenguer J. Secretion and assembly of regular surface structures in Gram-negative bacteria. FEMS Microbiol Rev. 2000;24:21–44. doi: 10.1111/j.1574-6976.2000.tb00531.x. [DOI] [PubMed] [Google Scholar]
- 13.Fuller R. Ecological studies on the Lactobacillus flora associated with the crop epithelium of the fowl. J Appl Bacteriol. 1973;36:131–139. [Google Scholar]
- 14.Harty D W S, Patrikakis M, Know K W. The identification of Lactobacillus strains isolated from patients with infective endocarditis and comparison of their surface-associated properties with those of other strains of the same species. Microb Ecol Health Dis. 1993;6:191–201. [Google Scholar]
- 15.Hellman U, Wernstedt C, Gonez J, Heldin C-H. Improvement of an “in-gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal Biochem. 1995;224:451–455. doi: 10.1006/abio.1995.1070. [DOI] [PubMed] [Google Scholar]
- 16.Higuchi R, Krummel B, Saiki R K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hultgren S J, Normark S, Abraham S N. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu Rev Microbiol. 1991;45:383–415. doi: 10.1146/annurev.mi.45.100191.002123. [DOI] [PubMed] [Google Scholar]
- 18.Johnsson J L, Phelps C F, Cummins C S, London J, Gasser F. Taxonomy of the Lactobacillus acidophilus group. Int J Syst Bacteriol. 1980;30:53–68. [Google Scholar]
- 19.Korhonen T K, Parkkinen J, Hacker J, Finne J, Pere A, Holthöfer H. Binding of Escherichia coli S fimbriae to human kidney epithelium. Infect Immun. 1984;54:322–327. doi: 10.1128/iai.54.2.322-327.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lortal S. Crystalline surface-layers of the genus Lactobacillus. In: Beveridge T J, Brawerman G, editors. Advances in paracrystalline bacterial surface layers. New York, N.Y: Plenum Press; 1990. pp. 57–65. [Google Scholar]
- 22.Markwell M A K, Fox C F. Surface-specific iodination of membrane proteins of viruses and eukaryotic cells using 1,3,4,6-tetracholoro-3α,6α-diphenylglycouracil. Biochemistry. 1978;17:4807–4817. doi: 10.1021/bi00615a031. [DOI] [PubMed] [Google Scholar]
- 23.Masuda K, Kawata T. Reassembly of the regularly arranged subunits in the cell wall of Lactobacillus brevis and their reattachment to cell wall. Microbiol Immunol. 1980;24:299–308. doi: 10.1111/j.1348-0421.1980.tb02833.x. [DOI] [PubMed] [Google Scholar]
- 24.Masuda K, Kawata T. Distribution and chemical characterization of regular arrays in the cell wall of strains of the genus Lactobacillus. FEMS Microbiol Lett. 1983;20:145–150. [Google Scholar]
- 25.Matsudaria P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 26.McGrady J A, Butcher W G, Beighton D, Switalski L M. Specific and charge interactions mediate collagen recognition by oral lactobacilli. J Dent Res. 1995;74:649–657. doi: 10.1177/00220345950740020501. [DOI] [PubMed] [Google Scholar]
- 27.Mitsuoka T. The human gastrointestinal tract. In: Wood B J B, editor. The lactic acid bacteria. 1. The lactic acid bacteria in health and disease. New York, N.Y: Elsevier Applied Science; 1992. pp. 69–114. [Google Scholar]
- 28.Noonan B, Trust T J. The synthesis, secretion and role in virulence of the paracrystalline surface protein layers of Aeromonas salmonicida and A. hydrophila. FEMS Microbiol Lett. 1997;154:1–7. doi: 10.1111/j.1574-6968.1997.tb12616.x. [DOI] [PubMed] [Google Scholar]
- 29.Nyman T A, Tölö H, Parkkinen J, Kalkkinen N. Identification of nine interferon-α subtypes produced by Sendai virus-induced human peripheral blood leucocytes. Biochem J. 1998;329:295–302. doi: 10.1042/bj3290295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patti J M, Allen B L, McGavin M J, Höök M. MSCRAMM-mediated adherence of microorganisms to host tissue. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 31.Patti J M, House-Pompeo K, Boles J O, Garza N, Gurusiddappa S, Hook M. Critical residues in the ligand binding site of the Staphylococcus aureus collagen-binding adhesin (MSCRAMM) J Biol Chem. 1995;270:12005–12011. doi: 10.1074/jbc.270.20.12005. [DOI] [PubMed] [Google Scholar]
- 32.Rich R L, Kreikemeyer B, Owens R T, LaBrenz S, Narayana S V L, Weinstock G M, Murray B E, Höök M. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J Biol Chem. 1999;274:26939–26945. doi: 10.1074/jbc.274.38.26939. [DOI] [PubMed] [Google Scholar]
- 33.Roggenkamp A, Neuberger H-R, Flugel A, Schmoll T, Heeseman J. Substitution of two histidine residues in YadA protein of Yersinia enterocolitica abrogates collagen binding, cell-adherence and mouse virulence. Mol Microbiol. 1995;16:1207–1219. doi: 10.1111/j.1365-2958.1995.tb02343.x. [DOI] [PubMed] [Google Scholar]
- 34.Roos S, Aleljung P, Robert N, Lee B, Wadström T, Lindberg M, Jonsson H. A collagen binding protein from Lactobacillus reuteri is part of an ABC transporter system. FEMS Microbiol Lett. 1996;144:33–38. doi: 10.1111/j.1574-6968.1996.tb08505.x. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Schneitz C, Nuotio L, Lounatmaa K. Adhesion of Lactobacillus acidophilus to avian intestinal epithelial cells mediated by the crystalline bacterial cell surface layer (S-layer) J Appl Bacteriol. 1993;74:290–294. doi: 10.1111/j.1365-2672.1993.tb03028.x. [DOI] [PubMed] [Google Scholar]
- 37.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sleytr U B, Beveridge T J. Bacterial S-layers. Trends Microbiol. 1999;7:253–260. doi: 10.1016/s0966-842x(99)01513-9. [DOI] [PubMed] [Google Scholar]
- 39.Symersky J, Patti J M, Carson M, House-Pompeo K, Teale M, Moore D, Jin L, Schneider A, DeLucas L J, Höök M, Narayana S V L. Structure of the collagen-binding domain from a Staphylococcus aureus adhesin. Nat Struct Biol. 1997;4:833–838. doi: 10.1038/nsb1097-833. [DOI] [PubMed] [Google Scholar]
- 40.Tamm A, Tarkkanen A-M, Korhonen T K, Kuusela P, Toivanen P, Skurnik M. Hydrophobic domains affect the collagen-binding specificity and surface polymerization as well as virulence potential of the YadA protein of Yersinia enterocolitica. Mol Microbiol. 1993;10:995–1011. doi: 10.1111/j.1365-2958.1993.tb00971.x. [DOI] [PubMed] [Google Scholar]
- 41.Tarkkanen A-M, Allen B L, Westerlund B, Holthöfer H, Kuusela P, Risteli L, Clegg S, Korhonen T K. Type V collagen as the target for type-3 fimbriae, enterobacterial adherence organelles. Mol Microbiol. 1990;4:1353–1361. doi: 10.1111/j.1365-2958.1990.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 42.Thomas S, Austin J W, McCubbin W D, Kay C M, Trust T. Roles of structural domains in the morphology and surface anchoring of the tetragonal paracrystalline array of Aeromonas hydrophila. J Mol Biol. 1992;228:652–661. doi: 10.1016/0022-2836(92)90847-d. [DOI] [PubMed] [Google Scholar]
- 43.Toba T, Virkola R, Westerlund B, Björkman Y, Sillanpää J, Vartio T, Kalkkinen N, Korhonen T K. A collagen-binding S-layer protein in Lactobacillus crispatus. Appl Environ Microbiol. 1995;61:2467–2471. doi: 10.1128/aem.61.7.2467-2471.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner M S, Timms P, Hafner L M, Giffard P M. Identification and characterization of a basic cell surface-located protein from Lactobacillus fermentum BR11. J Bacteriol. 1997;179:3310–3316. doi: 10.1128/jb.179.10.3310-3316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vidgrén G, Palva I, Pakkanen R, Lounatmaa K, Palva A. S-layer protein gene of Lactobacillus brevis: cloning by polymerase chain reaction and determination of the nucleotide sequence. J Bacteriol. 1992;174:7419–7427. doi: 10.1128/jb.174.22.7419-7427.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vorm O, Roepstorff P, Mann M. Improved resolution and very high sensitivity in MALDI TOF of matrix surfaces made by fast evaporation. Anal Chem. 1994;66:3281–3287. [Google Scholar]
- 47.Westerlund B, Korhonen T K. Bacterial proteins binding to the mammalian extracellular matrix. Mol Microbiol. 1993;9:687–694. doi: 10.1111/j.1365-2958.1993.tb01729.x. [DOI] [PubMed] [Google Scholar]
- 48.Westerlund-Wikström B, Tanskanen J, Virkola R, Hacker J, Lindberg M, Skurnik M, Korhonen T K. Functional expression of adhesive peptides as fusions to Escherichia coli flagellin. Protein Eng. 1997;11:1319–1326. doi: 10.1093/protein/10.11.1319. [DOI] [PubMed] [Google Scholar]