Abstract
Porphyromonas gingivalis is a gram-negative, anaerobic coccobacillus that has been implicated as a major etiological agent in the development of chronic periodontitis. In this paper, we report the characterization of a protein, IhtB (iron heme transport; formerly designated Pga30), that is an outer membrane hemin-binding protein potentially involved in iron assimilation by P. gingivalis. IhtB was localized to the cell surface of P. gingivalis by Western blot analysis of a Sarkosyl-insoluble outer membrane preparation and by immunocytochemical staining of whole cells using IhtB peptide-specific antisera. The protein, released from the cell surface, was shown to bind to hemin using hemin-agarose. The growth of heme-limited, but not heme-replete, P. gingivalis cells was inhibited by preincubation with IhtB peptide-specific antisera. The ihtB gene was located between an open reading frame encoding a putative TonB-linked outer membrane receptor and three open reading frames that have sequence similarity to ATP binding cassette transport system operons in other bacteria. Analysis of the deduced amino acid sequence of IhtB showed significant similarity to the Salmonella typhimurium protein CbiK, a cobalt chelatase that is structurally related to the ATP-independent family of ferrochelatases. Molecular modeling indicated that the IhtB amino acid sequence could be threaded onto the CbiK fold with the IhtB structural model containing the active-site residues critical for chelatase activity. These results suggest that IhtB is a peripheral outer membrane chelatase that may remove iron from heme prior to uptake by P. gingivalis.
Porphyromonas gingivalis, a black-pigmented, gram-negative, anaerobic coccobacillus, has been implicated as a major pathogen in the development and progression of chronic periodontitis, an inflammatory disease resulting in the destruction of the supporting tissues of the teeth (19, 24, 45, 46). P. gingivalis has an essential requirement for iron, which it prefers in the form of heme. Heme can be acquired from a range of hemoproteins at low concentrations (<10 μM), including hemoglobin, cytochrome c, haptoglobin-hemoglobin and hemopexin (5, 20, 31). The growth and virulence of P. gingivalis are dependent on heme availability (32) such that growth under conditions of heme excess has been reported to enhance the virulence of the bacterium in a murine model of infection (30), although Genco et al. (13) have suggested that heme limitation results in increased virulence.
Heme and other iron complexes including siderophores are used as a source of iron by a variety of gram-negative bacteria, including Yersinia, Escherichia, and Vibrio spp. (51). Typically, a TonB-linked outer membrane receptor transports heme into the periplasmic space, where it is transported intact into the cell by a multicomponent periplasmic binding protein-dependent ATP binding cassette (ABC) transport system (34).
The heme uptake mechanisms of P. gingivalis have not been well characterized, although a variety of cell envelope heme-binding proteins have been reported (6, 22, 44). P. gingivalis siderophores have not been identified (6). Heme has been shown to bind to the P. gingivalis cell surface and is then transported into the cell by a process that requires energy (14). Both protoporphyrin IX (PPIX) and nonradiolabeled heme compete for radiolabeled heme binding, indicating that at least one outer membrane receptor specific for the PPIX ring is involved in heme binding (14). Smalley et al. (44) have suggested that P. gingivalis has both high- and low-affinity binding sites for heme.
Bramanti and Holt (4) demonstrated that 10 cell surface-associated proteins ranging in size from 26 to 80 kDa were expressed when P. gingivalis was grown under heme-limited conditions. These authors proposed that a 26-kDa protein that is produced by P. gingivalis under conditions of heme limitation has a role in heme binding and uptake (7). Smalley et al. (43) have also identified P. gingivalis heme-repressible proteins and suggested that these proteins may belong to a binding system for heme uptake that is induced by low levels of environmental heme. However, these proposed systems have not been defined.
We have previously purified an antigenic 30-kDa protein, IhtB (iron heme transport; formerly designated Pga30), from chloroform-treated P. gingivalis W50 (17). This protein was identified by its strong reactivity in a Western blot using serum from a control subject who harbored subgingival P. gingivalis but did not display clinical signs of periodontitis. However, IhtB was not recognized by sera from eight patients with moderate to severe periodontitis who also harbored subgingival P. gingivalis (17).
We report here the characterization of IhtB as an outer membrane hemin-binding protein, and we also report the cloning and sequence analysis of the ihtB gene. We propose that IhtB is a peripheral outer membrane chelatase involved in iron transport by P. gingivalis.
MATERIALS AND METHODS
Bacterial strains and maintenance.
P. gingivalis W50 was grown routinely in brain heart infusion broth supplemented with 0.5% (wt/vol) l-cysteine and 1 μg of hemin per ml in an anaerobe chamber (MK3 Anaerobic Workstation; Don Whitely Scientific, Adelaide, South Australia, Australia) with an atmosphere of 10% CO2, 5% H2, and 85% N2 at 37°C. Escherichia coli strain JM109 was grown aerobically in Luria-Bertani broth at 37°C. E. coli clones harboring pUC18 plasmids were grown in Luria-Bertani broth supplemented with ampicillin (100 μg/ml) at 37°C.
Genomic library construction and screening.
The P. gingivalis W50 λ GEM-12 genomic library described previously (42) was screened using synthetic degenerate oligonucleotides corresponding to the amino acid sequence ENKGEAT, derived from the N-terminal sequence of the previously purified IhtB protein (17) using standard techniques (37). Oligonucleotide probes were 5′ end labeled using [γ-32P]ATP and T4 polynucleotide kinase. Approximately 3,000 plaques were screened by lifting onto nylon membranes and hybridized overnight with radiolabeled oligonucleotides in hybridization buffer (90 mM sodium citrate, 150 mM NaCl, 0.25% [wt/vol] sodium dodecyl sulfate [SDS], 1× Denhardt's solution [37], 48°C, 2 h). The filters were washed in 90 mM sodium citrate–0.1% (wt/vol) SDS at 48°C. A positively hybridizing λ clone was selected, and a 4.6-kb BamHI fragment was recovered and ligated into BamHI-digested, bovine alkaline phosphatase-treated pUC18 (Amersham Pharmacia Biotech, Castle Hill, New South Wales, Australia) and transformed into E. coli JM109 by electroporation (37).
DNA sequencing and sequence analysis.
Double-stranded plasmid template DNA was prepared by the procedure of Li and Schweizer (28). The DNA was sequenced in both directions using sequence-derived, synthetic oligonucleotides by the dideoxy-chain termination method (38) with a Sequenase version 2.0 nucleotide sequencing kit used as recommended by the manufacturer (United States Biochemicals, Cleveland, Ohio). Nucleotide and deduced amino acid sequence data were analyzed using program suites accessed by the Australian National Genomic Information Service or by the National Center for Biotechnology Information website at http://www.ncbi.nlm.nih.gov. The preliminary sequence data of the P. gingivalis W83 genome was obtained from The Institute for Genomic Research website at http://www.fasta.genome.ad.jp/.
IhtB conformational modeling.
Using GeneFold (Tripos Inc., St. Louis, Mo.), the deduced amino acid sequence of IhtB was threaded onto the members of a library of nonredundant protein folds derived from structures deposited in the Protein Data Bank (PDB) including the Salmonella typhimurium CbiK fold (PDB code lqgo) and the Bacillus subtilis PPIX ferrochelatase fold (PDB code 1ak1) to identify the fold most likely to be adopted by IhtB. A model of IhtB was constructed based on the CbiK fold, identified by GeneFold, using Sybyl (Tripos). High-energy contacts between amino acyl residues in the model were progressively relieved by simulated annealing at 20 K using the amber force field. A distance-dependent dielectric ɛ = r and a nonbond cutoff distance of 8 Å were used. Simulations were performed for 400 to 500 fs with a time step of 0.1 fs, a nonbonded reset interval and momentum removal at 25-fs intervals, and a thermal bath coupling of 10 fs. The final model was energy minimized to a maximum derivative of 0.05 kcal mol−1 Å−1 using the mmff94s force field. The program what if was used to assess the quality of the model (48).
Northern and Southern analyses.
Northern and Southern blots were prepared as described previously (42). Nylon membranes were hybridized at 48°C in hybridization buffer with the oligonucleotide probe 5′-GATCGGGATAAGCTGCGGC-3′, antisense to the deduced amino acid sequence AAAYPDQ found in IhtB. Membranes were washed to 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at 48°C.
Primer extension analysis.
Total P. gingivalis RNA was isolated as described previously (42). Primer extension was performed using 10 μg of total RNA and the AMV Reverse Transcriptase Primer Extension System in accordance with the manufacturer's (Promega Corporation, Madison, Wis.) instructions.
Production of IhtB peptide-specific antisera.
A synthetic 15-mer peptide corresponding to C-terminal residues 279 to 293 of IhtB was used to raise IhtB peptide-specific antisera. The peptide, including an N-terminal Cys for attachment, with the sequence CIRNIWLKHMKATSAR, was commercially synthesized and conjugated to diphtheria toxoid by Chiron Mimotopes (Melbourne, Victoria, Australia). The peptide-diphtheria toxoid conjugate was then emulsified with Freund's incomplete adjuvant and injected subcutaneously at four sites into the backs of two Dutch rabbits and one New Zealand White rabbit. Immunization was carried out three times at 4-week intervals. Ten days after the final immunization, blood from each rabbit was analyzed by enzyme-linked immunosorbent assay using unconjugated peptide as the absorbed antigen. Three days later, the rabbits were sacrificed and their blood was collected by cardiac puncture. The blood was incubated at 37°C for 1 h to enhance clot formation and then incubated at 4°C overnight to encourage clot retraction. The serum was separated from the clot and stored at −70°C until required.
Cell fractionation.
Outer membrane fractions of P. gingivalis were prepared by the Sarkosyl method (11, 12, 22). In short, P. gingivalis cells were harvested from a 500-ml culture during the late exponential growth phase by centrifugation (5,000 × g, 20 min, 4°C) and suspended in 5 ml of 20 mM Tris-HCl (pH 7.4)–10 mM EDTA–1 mM Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK)–1% sodium lauryl sarcosinate. The cells were disrupted by three passages through a French pressure cell at 109 MPa (SLM-Aminco, Urbana, Ill.). Whole cells were removed by centrifugation (5,000 × g, 20 min, 4°C). The Sarkosyl-insoluble (outer membrane) fraction was separated by high-speed centrifugation (100,000 × g, 30 min). The pellet (outer membrane fraction) was washed three times in 0.5% sodium lauryl sarcosinate and then resuspended in 20 mM Tris-HCl (pH 7.4)–10 mM EDTA–1 mM TLCK and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blot assay after boiling of the outer membrane fraction in SDS sample buffer for 10 min (3).
SDS-PAGE and Western blot analysis.
Proteins to be subjected to Western blot analysis were separated using a discontinuous SDS-PAGE system in accordance with the method of Laemmli (23) with a 12% (wt/vol) polyacrylamide separating gel and a 4% (wt/vol) polyacrylamide stacking gel. Proteins were stained with 0.1% Coomassie brilliant blue R-250 in 45.5% (vol/vol) ethanol–9% (vol/vol) acetic acid. Gels were destained in 25% (vol/vol) ethanol–8% (vol/vol) acetic acid. Prestained standards (Bio-Rad Laboratories, Hercules, Calif.) were included. Following SDS-PAGE, protein transfer was performed in 10% (wt/vol) methanol at a constant voltage of 60 V for 90 min onto a polyvinylidene difluoride membrane (proBlott; PE Biosystems, Scoresby, Victoria, Australia). N-terminal sequencing of transblotted proteins was done as previously described (3). For Western blot analysis, the membrane was blocked with 50% (wt/vol) skim milk powder in TN buffer (25 mM Tris-HCl, 0.5 M NaCl [pH 7.5]) at 25°C for 1 h. The membrane was then incubated with the primary antibody (IhtB peptide-specific antiserum, 1/100) at 4°C overnight. The membrane was washed three times in TN buffer and then incubated at 25°C for 2 h with the second antibody (horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G [IgG] diluted 1/1,000 in TN buffer). The membrane was then washed three times in TN buffer. Binding of the goat anti-rabbit IgG was visualized with 0.5% (wt/vol) 4-chloro-1-naphthol, 16% (vol/vol) methanol, and 0.015% (vol/vol) H2O2 in TN buffer.
Immunocytochemical localization of IhtB on P. gingivalis cells.
Exponentially growing P. gingivalis cells were adsorbed onto Formvar-coated nickel grids by application of 10 μl of a P. gingivalis culture. The grids were then dried onto 2% (wt/vol) Noble agar, blocked by flotation on 1% (wt/vol) casein in phosphate-buffered saline (PBS; 10 mM Na2HPO4-NaH2PO4, 150 mM NaCl [pH 7.2]) for 10 min, and then incubated with the primary antibody overnight at 4°C. The primary antibody was rabbit IhtB peptide-specific antisera or nonspecific rabbit sera diluted 1/20 in casein-PBS. A control of 1% (wt/vol) casein in PBS was also included when the primary antibody was omitted. The grids were washed with casein-PBS for 30 min and then incubated with goat anti-rabbit IgG conjugated with 10-nm colloidal gold (Sigma) diluted 1/20 for 2 h at 25°C. After incubation, the grids were washed with 1% (wt/vol) casein in PBS for 30 min, followed by a PBS wash for 15 min, and then fixed with 2.5% (vol/vol) glutaraldehyde in 0.1 M HEPES-PBS for 2 min. The grids were finally washed with distilled water for 15 min, negatively stained with 2% (vol/vol) ammonium molybdate, and then examined using a Philips CM10 electron microscope operating at 60 kV.
Preparation of cell surface and periplasmic proteins.
P. gingivalis cell surface and periplasmic proteins were released using chloroform treatment as described previously (17). Briefly, exponentially growing P. gingivalis cells were harvested from a 500-ml culture by centrifugation (5,000 × g, 20 min, 4°C). Chloroform (10 ml per liter of original culture) was added to the cell pellet and incubated at 25°C for 15 min with gentle rocking. Buffer A (50 mM NaCl, 20 mM Tris-HCl, 10 mM EDTA [pH 8.0]; 50 ml per liter of original culture) was added, and the suspension was centrifuged (5,000 × g, 20 min, 4°C) to remove whole cells. The upper aqueous phase was removed and further clarified by centrifugation (20,000 × g, 30 min, 4°C). A protein concentration of 4.1 mg/ml was obtained in the final extract.
Hemin-agarose binding.
Hemin-agarose binding was performed essentially as described by Lee (25). Briefly, 200 μl of hemin-agarose (Sigma-Aldrich Pty. Ltd., Castle Hill, New South Wales, Australia) was washed with 100 mM NaCl–25 mM Tris-HCl (pH 7.4). Washing was performed three times by suspension of the agarose in 1 ml of buffer, followed by centrifugation (10,000 × g, 5 min). The P. gingivalis cell surface and periplasmic extract (500 μl) was incubated with the hemin-agarose for 3 h at 37°C with gentle mixing. The sample was centrifuged (10,000 × g, 5 min), and the supernatant was removed. The hemin-agarose was washed three times as described above, and bound proteins were eluted by incubation for 2 min with 2 M guanidine-HCl (100 μl), which was separated from the hemin-agarose beads by centrifugation (10,000 × g, 5 min). A negative control, omitting the hemin-agarose, was included and treated in an identical manner. Proteins in the 2 M guanidine-HCl eluants were analyzed by SDS-PAGE and Western blotting (see above).
Growth studies.
Heme-limited cells of P. gingivalis were produced by three passages in basal medium (1% [wt/vol] Proteose Peptone, 0.5% [wt/vol] Trypticase Peptone, 0.5% [wt/vol] yeast extract, 0.25% [wt/vol] KCl) supplemented with 0.5% (wt/vol) l-cysteine (BM) using a 10% inoculum and incubation for 24 h in an anaerobe chamber at 37°C. BM containing ∼1.0 × 109 heme-limited P. gingivalis cells per ml was divided into 2-ml aliquots and centrifuged (4,000 × g, 10 min, 37°C). Cell pellets were suspended and incubated for 1 h at 37°C in 1 ml of (i) IhtB peptide-specific antisera that had been heat inactivated at 56°C for 30 min, (ii) heat-inactivated normal rabbit sera (NRS), or (iii) BM with hemin (1 μg/ml). The cells were washed (three times) in BM (1 ml) with hemin, followed by suspension in the same medium (300 μl). Each P. gingivalis cell suspension was inoculated into 2.7 ml of BM with hemin, and growth was monitored spectrophotometrically at a wavelength of 650 nm.
Nucleotide sequence accession number.
The nucleotide sequence of the ihtB ORF has been deposited in the GenBank database and assigned accession no. AF195649.
RESULTS
Cloning and sequence analysis of ihtB.
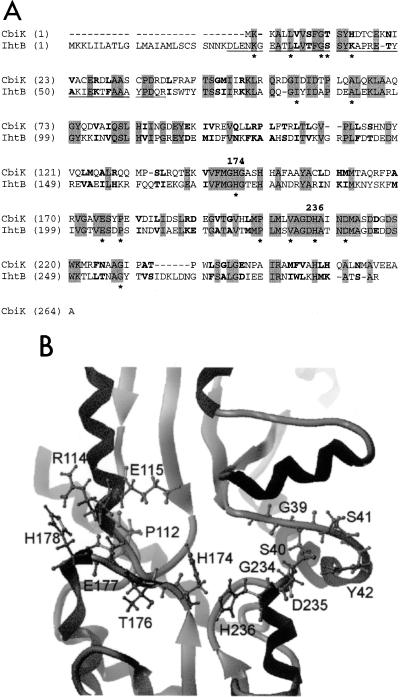
A P. gingivalis genomic library constructed in λ GEM-12 was screened using degenerate oligonucleotides corresponding to the N-terminal amino acid sequence ENKGEAT of the previously purified protein IhtB, formerly designated Pga30 (17). Southern blot analysis revealed a 4.6-kb BamHI fragment that positively hybridized with the oligonucleotides corresponding to the IhtB sequence. The 4.6-kb BamHI fragment was ligated into BamHI-digested pUC18 and transformed into electrocompetent E. coli JM109 cells. Plasmid DNA was recovered from transformed cells, and the insert was sequenced in both directions. The insert DNA nucleotide sequence contained a single complete open reading frame (ORF) that we have designated ihtB (iron heme transport). The ihtB ORF comprises 882 bp. The deduced amino acid sequence of IhtB contained 293 amino acyl residues with a predicted molecular mass of 32.4 kDa. The deduced IhtB sequence was found to contain an additional 24 amino acyl residues N terminal to the sequence identified for the previously purified IhtB protein (see underlining in Fig. 6A) (17). The first 19 residues of this deduced N-terminal sequence is typical of a prokaryotic leader sequence and contains a net positive charge at the N terminus (the start Met followed by two Lys residues), followed by a stretch of hydrophobic residues (residues 4 to 17). This hydrophobic stretch terminates with the sequence 15Ala-Met-Leu-Ser-Cys19, which conforms to the consensus signal peptidase II cleavage site and lipoprotein attachment site (18).
FIG. 6.
(A) Alignment of CbiK and IhtB deduced amino acid sequences generated by GeneFold. Shading indicates identical residues, and bold indicates conservative substitutions. Residues that are conserved across the entire anaerobic, ATP-independent cobalt and ferrochelatase class of proteins (40) are indicated by asterisks. Numbering indicates the position in the IhtB sequence of the His residues (His174 and His236) proposed to be critical for chelatase activity. The N-terminal sequence obtained for the purified N-terminally truncated protein (17) is underlined. (B) Ribbon diagram of the active site of the IhtB conformational model showing the putative heme-binding site constructed using the Sybyl program. Residues believed to be important for substrate binding and active-site conformation are labeled and have side chains displayed.
A putative Shine-Dalgarno sequence (AAGAA) was identified six bases upstream from the suggested start codon (ATG). Primer extension analysis of ihtB showed that the transcription start point is 267 bases upstream from the methionine translation start codon (21). A nearly perfect invert repeat sequence was identified 45 nucleotides downstream from the stop codon, between bases 1391 and 1439 (AGAGCACTtATCGAGAAgaaggacttgccgatTTCTCGATgAGTGCTCT). This was immediately followed by a stretch of T residues, suggesting that this represents a rho factor-independent RNA hairpin transcription terminator (50). The size of the predicted transcript from the transcription start point to the proposed termination site is in agreement with the Northern blot result that showed a transcript of the appropriate size, 1.3 kb (Fig. 1).
FIG. 1.
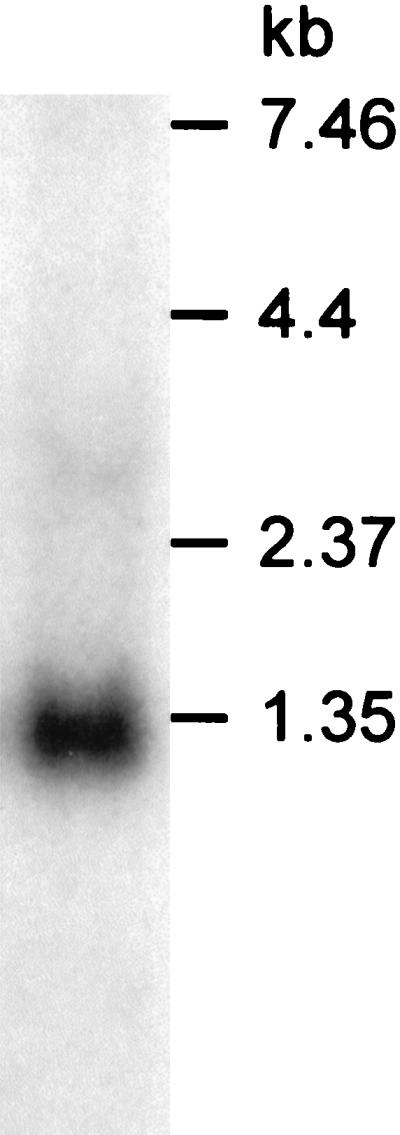
Northern blot analysis of P. gingivalis RNA. Hybridization with an oligonucleotide probe specific for ihtB showed a single transcript of 1.3 kb. The relative positions of RNA molecular size markers are indicated.
When the deduced amino acid sequence of IhtB was compared with sequences in the databases, significant sequence identity was found only with CbiK from S. typhimurium, a member of the ATP-independent ferrochelatase family of enzymes (36). An overall sequence identity of 37.6% (99 of 264 residues) and a sequence similarity of 59.8% (158 of 264 residues) between IhtB and CbiK were found.
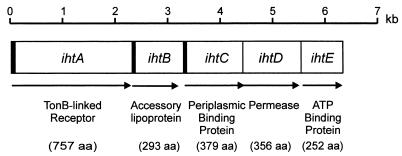
An ORF, which we have designated ihtA, was located immediately upstream of ihtB using The Institute for Genomic Research P. gingivalis W83 preliminary genomic sequence data and the ORF finder program from the National Center for Biotechnology Information (Fig. 2). Sequence analysis revealed that the deduced amino acid sequence encoded by ihtA had identity with TonB-linked outer membrane receptors involved in iron complex and vitamin B12 transport in other gram-negative bacteria, including the ferric receptor of Campylobacter coli with 25% identity (149 of 591 residues) and 41% similarity (248 of 591 residues), the colicin I receptor precursor of E. coli with 25% identity (158 of 617 residues) and 41% similarity (260 of 617 residues), the ferric enterobactin receptor of Bordetella pertussis with 24% identity (175 of 727 residues) and 39% similarity (287 of 727 residues), and the vitamin B12 receptor precursor of S. typhimurium with 22% identity (142 of 618 residues) and 40% similarity (255 of 618 residues) (2, 15, 16, 49). The presence of a putative leader sequence and TonB box III in the N-terminal region of the deduced amino acid sequence of IhtA further supports the proposed function of this protein.
FIG. 2.
The ihtABCDE genetic locus of P. gingivalis that encodes a proposed iron transport system. The putative function and size of the product of each gene are shown. The arrows indicate the relative positions of each of the predicted coding regions, and the shaded areas indicate the presence of leader sequences. aa, amino acids.
Located directly downstream of ihtB were three other predicted ORFs, which we have designated ihtC to -E (Fig. 2). Sequence analysis showed that the deduced IhtE protein contains the Walker A and B and ABC signature sequence motifs that are characteristic of ATP-binding proteins of ABC transport systems (39). IhtE also displayed significant sequence identity (36% over 227 amino acids) to FepC, the ferric enterobactin transport ATP-binding protein of E. coli (41). Sequence analysis of the deduced amino acid sequence of IhtD revealed little identity to other known proteins. However, this hypothetical protein is hydrophobic and contains nine putative membrane-spanning domains, as predicted by TopPred 2. This is consistent with this protein being an inner membrane permease. The deduced amino acid sequence of IhtC showed little identity to other known proteins. However, the presence of a putative leader sequence and a region that has identity to the iron complex-binding motif of gram-negative bacterial periplasmic binding proteins (47) is consistent with this being a periplasmic binding protein of an ABC transport system.
Cellular localization of IhtB.
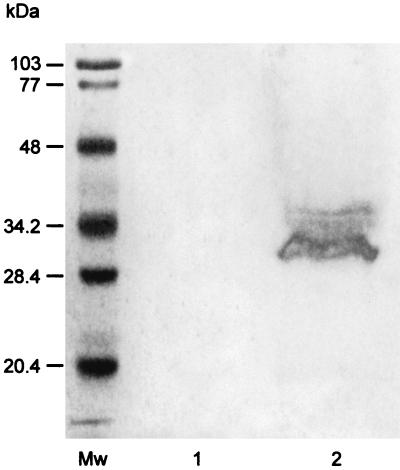
IhtB was localized to the outer membrane of P. gingivalis by subjecting a Sarkosyl-insoluble outer membrane preparation to SDS-PAGE and Western blot analysis using the IhtB peptide-specific antisera. A diffuse band at 32 to 34 kDa, which was N terminally blocked and reactive with the IhtB peptide-specific antisera was detected in the Sarkosyl-insoluble outer membrane fraction that was not seen in the cytoplasmic fraction (Fig. 3). These results confirm the specificity of the peptide-specific antisera for IhtB, and the diffuse nature of the band, as well as the blocked N terminus, is consistent with the proposed N-terminal lipid modification (membrane attachment).
FIG. 3.
Western blot analysis of cell fractions produced by French pressure treatment of P. gingivalis and probed with IhtB peptide-specific antisera. Mw, molecular size markers; lane 1, cytoplasmic fraction; lane 2, Sarkosyl-insoluble outer membrane fraction.
IhtB was further localized to the cell surface of P. gingivalis by transmission electron microscopy of whole cells after immunolabeling with the IhtB peptide-specific antisera and a gold-conjugated secondary antibody. Analysis of 50 grid squares (100 by 100 μm) for both specifically and nonspecifically labeled cells revealed that cells incubated with the IhtB peptide-specific antisera were surface labeled with 25.15 ± 8.76 gold particles/μm2, which was significantly greater labeling (P < 0.001, using Student's t test) than that of cells incubated with the nonspecific rabbit sera, which were surfaced labeled with only 2.90 ± 1.71 gold particles/μm2.
Hemin-agarose binding.
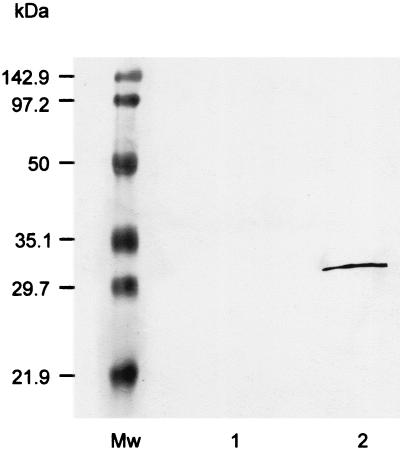
Hemin-binding proteins present in cell surface and periplasmic proteins released by chloroform treatment of P. gingivalis were identified using Western blot analysis of proteins that bound to hemin-agarose. Proteins eluted from the hemin-agarose with 2 M guanidine-HCl were separated using SDS-PAGE and then subjected to Western blot analysis using IhtB peptide-specific antisera. This analysis revealed a single sharp band at 30 kDa (Fig. 4). This protein was identified by N-terminal sequence analysis as the N-terminally truncated IhtB previously identified (17). This band was not seen on omission of the hemin-agarose (Fig. 4) or on the use of unmodified agarose.
FIG. 4.
Western blot analysis of P. gingivalis outer membrane and periplasmic proteins eluted from hemin-agarose with 2 M guanidine-HCl. This Western blot analysis used IhtB peptide-specific antisera. Mw, molecular size markers; lane 1, control omitting the hemin-agarose; lane 2, hemin-agarose eluant.
Growth studies.
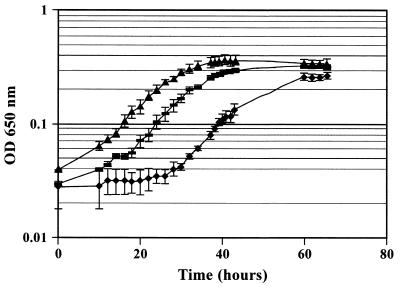
When cells were passaged three times in liquid basal medium containing no added heme, P. gingivalis growth during the third passage was comparable to that seen in the first passage. However, growth in this medium was not sustainable for a fourth passage (data not shown). The heme-depleted P. gingivalis cells from the third passage were harvested by centrifugation and preincubated with heat-inactivated IhtB peptide-specific antisera, heat-inactivated NRS, or growth medium. Growth of the P. gingivalis cells preincubated with heat-inactivated NRS was slightly stimulated; however, growth of cells preincubated with IhtB peptide-specific heat-inactivated antisera was inhibited, as shown by the 20-h increase in lag phase compared with the cells preincubated with media (Fig. 5). Preincubation of heme-replete cells with the IhtB peptide-specific, heat-inactivated antisera, however, did not result in growth inhibition (data not shown).
FIG. 5.
Growth of P. gingivalis in batch culture in basal medium with hemin (1 μg/ml). Heme-depleted cells of P. gingivalis were preincubated for 1 h at 37°C in IhtB peptide-specific antiserum which had been heat inactivated at 56°C for 30 min (⧫), heat-inactivated NRS (▴), or basal medium with hemin (■). Points represent the mean of three independent determinations. OD, optical density.
IhtB conformational model.
Molecular modeling of IhtB demonstrated that the IhtB amino acid sequence could be threaded onto the fold of S. typhimurium CbiK in preference to any other known fold in the PDB. The alignment of the deduced amino acid sequences of CbiK and IhtB generated by GeneFold (Tripos) is presented in Fig. 6A. The Z scores obtained supported the validity of the IhtB conformational model (48). The model fold is bilobal in structure, consisting of two similar domains that each contain a four-stranded parallel β-sheet flanked by α-helices, with the active site located in a deep rectangular cleft between the two domains. Figure 6B shows the active site of the model structure, highlighting residues involved in the formation of the active site and substrate binding. The histidine residues in CbiK that have been identified as being functionally critical for chelatase activity have analogous residues in the IhtB structural model (His174 and His236).
DISCUSSION
Iron has been shown to play an essential role in the growth and virulence of P. gingivalis and is preferred by the bacterium in the form of heme (5). In this study, we have characterized a hemin-binding peripheral outer membrane protein of P. gingivalis (IhtB) that we propose is a chelatase involved in iron removal for uptake.
The deduced amino acid sequence of IhtB (Fig. 6A) exhibited significant similarity to only one other known protein, CbiK from S. typhimurium (37.6% identity). CbiK is an anaerobic cobalt chelatase that inserts cobalt into the tetrapyrrole ring of precorrin and has been proposed to be part of the cobalamin biosynthesis pathway in S. typhimurium (35, 40). CbiK is a single-subunit, ATP-independent enzyme that, on the basis of biochemical characterization and sequence similarity, has been proposed to be a member of a class of enzymes including the PPIX ferrochelatase (HemZ), sirohydrochlorin ferrochelatases (CysG and Met8P), and the other known anaerobic cobalt chelatase (CbiX) (40). Although there is low overall sequence identity between the proteins within the PPIX ferrochelatase-anaerobic cobalt chelatase class, 14 residues are conserved within this class of enzyme. IhtB contains 13 of these 14 residues, and the only nonidentical residue is a conservative Thr→Ser substitution at position 40 in IhtB (Fig. 6A). PPIX ferrochelatases catalyze the insertion of iron into PPIX to produce protoheme and have also been shown to remove Fe2+ from heme when an Fe2+ sink exists (1, 29). CbiK is conformationally similar to the PPIX ferrochelatase of B. subtilis, although there is little sequence identity (1, 40). CbiK has also been shown to function as a ferrochelatase, as well as a cobalt chelatase (35). Both CbiK and the PPIX ferrochelatase are bilobal in structure, consisting of two similar domains that each contain a four-stranded parallel β-sheet flanked by α-helices. The active site of these enzymes is located in a deep rectangular cleft between these two domains (40). Molecular modeling of IhtB demonstrated that the IhtB amino acid sequence could be threaded onto the chelatase fold. The histidine residues in CbiK that have been identified as being functionally critical for chelatase activity have analogous residues in the IhtB structural model (His174 and His236). These analogous histidine residues occupy similar structural locations in the IhtB model, although this was not a constraint applied during model building (Fig. 6B). The chelatase active site is defined by four loops (Fig. 6B). Two of these loops (to the right in Fig. 6B) are largely conserved between CbiK and IhtB, with only the conservative Thr40Ser (IhtB numbering) change. The other two loops display less conservative changes, with Asn112Pro and Asp114Arg substitutions in the upper left loop and Ala176Thr and Ser177Glu substitutions in the lower left loop. These residues are located toward the ends of the cleft and presumably interact with functional groups located on the periphery of the tetrapyrrole group and may thus be responsible for the substrate specificity of the chelatase. Recently, ihtB has been expressed in E. coli and the recombinant protein exhibited chelatase activity (M. Warren, personal communication).
The CbiK protein is part of a cobalamin biosynthesis pathway of S. typhimurium and has been reported to have an intracellular location (35). However, in this study, IhtB has been localized to the cell surface of P. gingivalis. The N-terminal 19 residues of the deduced amino acid sequence of IhtB have the characteristics of a typical prokaryotic leader sequence, which is consistent with an extracellular location for the protein (33). This sequence is not present in CbiK, and it appears that IhtB has an additional 27 N-terminal amino acyl residues compared with CbiK (Fig. 6A).
The amino acid sequence and conformational similarity to CbiK and the presence of the active-site residues of the chelatases suggest that IhtB plays a role in iron removal from heme at the cell surface, where the Iht transport system would act as an iron sink. Hemin-agarose binding of P. gingivalis cell surface and periplasmic proteins, followed by Western blot analysis of bound proteins eluted with 2 M guanidine-HCl using IhtB peptide-specific antisera, confirmed that IhtB is a hemin-binding protein. Presumably, the oxidized form of the iron (Fe3+) in hemin prevented iron removal by the IhtB Fe2+ chelatase activity.
Three P. gingivalis heme-binding outer membrane proteins have been identified by other investigators when cells were grown under conditions of hemin limitation. A 32-kDa protein and a 30-kDa protein were identified by staining of SDS-PAGE gels with the chromogenic substrate tetramethylbenzidine, a method that utilizes the intrinsic peroxidase activity possessed by heme (22, 44). The third known heme-binding outer membrane protein of P. gingivalis is a 26-kDa protein, Omp26, that has been shown to be involved in heme uptake and appears to move across the outer membrane depending upon the heme concentration in the growth medium (6, 7, 8). As an N-terminally truncated IhtB protein was originally purified from P. gingivalis and characterized by SDS-PAGE as a 30-kDa protein (17), it is possible that IhtB, with a predicted molecular mass of 32.4 kDa for the full-length protein, represents the 30- and 32-kDa heme-binding proteins initially identified by Smalley et al. (44).
It is interesting that the N-terminal amino acyl residue of the previously purified N-terminally truncated IhtB (Pga30) protein (17) is immediately preceded by a Lys residue (Lys24) in the deduced IhtB protein sequence (Fig. 6A). P. gingivalis W50 produces a cell surface proteinase, Kgp (formerly designated PrtK), which cleaves specifically at Lys residues (3), and it is therefore likely that the cleavage of IhtB by Kgp enabled release of the protein from the outer membrane upon chloroform treatment (17). The N-terminally truncated protein appeared as a sharp 30-kDa band upon SDS-PAGE (Fig. 4), whereas the membrane-attached form of the protein appeared as a diffuse 32- to 34-kDa band (Fig. 3) which was N-terminally blocked, being consistent with the proposed N-terminal lipid modification of the membrane form of the protein.
Incubation of heme-limited P. gingivalis, but not heme-replete cells, with heat-inactivated IhtB peptide-specific antiserum prior to inoculation into medium containing hemin resulted in a substantially increased lag time compared with incubation in NRS or media (Fig. 5). This result is consistent with the proposal that IhtB is involved in transport of iron into the cell and also provides further evidence for the surface location of IhtB.
Iron and iron complex transport in gram-negative bacteria has been shown to be mediated via TonB-linked outer membrane receptors combined with inner membrane ABC transport systems (9). The ORF located immediately upstream of ihtB encodes a protein (IhtA) whose deduced amino acid sequence contains a TonB box III motif and displays significant sequence similarity to TonB-linked outer membrane receptors involved in iron complex and vitamin B12 transport in other gram-negative bacteria. Three ORFs (ihtCDE) located immediately downstream of ihtB showed similarity to an inner membrane ABC iron complex transport system, and these ORFs have been shown to be cotranscribed as part of an operon using reverse transcription-PCR (N. Slakeski et al., unpublished data). Together, these data suggest that IhtB is part of a transport system involved in iron transport in P. gingivalis. IhtB appears to be a peripheral outer membrane hemin-binding chelatase that potentially acts as an accessory protein for iron removal from heme prior to iron transport through the TonB-linked receptor. IhtA and IhtB may therefore be analogous to the system encoded by the tbpBA operon of Neisseria meningitidis, which is essential for transferrin utilization (26). The tbpA gene encodes an iron-repressible TonB-linked outer membrane receptor, while tbpB encodes a peripheral outer membrane accessory lipoprotein. These proteins act as a two-component receptor system that has been shown to remove iron from transferrin prior to transport into the periplasm (10).
In conclusion, we have characterized an antigenic, peripheral outer membrane hemin-binding protein of P. gingivalis W50 and suggest that this protein is part of an iron transport system that is important for the growth of P. gingivalis.
ACKNOWLEDGMENTS
We gratefully acknowledge the excellent technical assistance of Caroline Moore, Stephen Cleal, Chris Poon, and Peter Riley. Preliminary sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org.
This research was supported by a University of Melbourne Postgraduate Research Scholarship awarded to A. Hendtlass. Sequencing of P. gingivalis was accomplished with support from National Institute of Dental and Craniofacial Research grant DE-12082.
REFERENCES
- 1.Al-Karadaghi S, Hansson M, Nikonov S, Jonsson B, Hederstedt L. Crystal structure of ferrochelatase: the terminal enzyme in heme biosynthesis. Structure. 1997;5:1501–1510. doi: 10.1016/s0969-2126(97)00299-2. [DOI] [PubMed] [Google Scholar]
- 2.Beall B, Sanden G N. A Bordetella pertussis FepA homologue required for utilization of exogenous ferric enterobactin. Microbiology. 1995;141:3193–3205. doi: 10.1099/13500872-141-12-3193. [DOI] [PubMed] [Google Scholar]
- 3.Bhogal P S, Slakeski N, Reynolds E C. A cell-associated protein complex of Porphyromonas gingivalis W50 composed of Arg- and Lys-specific cysteine proteinases and adhesins. Microbiology. 1997;143:2485–2495. doi: 10.1099/00221287-143-7-2485. [DOI] [PubMed] [Google Scholar]
- 4.Bramanti T, Holt S. Iron-regulated outer membrane proteins in the periodontopathogenic bacterium Bacteroides gingivalis. Biochem Biophys Res Commun. 1990;166:1146–1154. doi: 10.1016/0006-291x(90)90986-w. [DOI] [PubMed] [Google Scholar]
- 5.Bramanti T E, Holt S C. Roles of porphyrins and host iron transport proteins in regulation of growth of Porphyromonas gingivalis W50. J Bacteriol. 1991;173:7330–7339. doi: 10.1128/jb.173.22.7330-7339.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bramanti T E, Holt S C. Localization of a Porphyromonas gingivalis 26-kilodalton heat-modifiable, hemin-regulated surface protein which translocates across the outer membrane. J Bacteriol. 1992;174:5827–5839. doi: 10.1128/jb.174.18.5827-5839.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bramanti T E, Holt S C. Hemin uptake in Porphyromonas gingivalis: Omp26 is a hemin-binding surface protein. J Bacteriol. 1993;175:7413–7420. doi: 10.1128/jb.175.22.7413-7420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bramanti T, Holt S. Effects of porphyrins and host iron transport proteins on outer membrane protein expression in Porphyromonas gingivalis: identification of a novel 26 kDa hemin-repressible surface protein. Microb Pathog. 1993;13:61–73. doi: 10.1016/0882-4010(92)90032-j. [DOI] [PubMed] [Google Scholar]
- 9.Braun V. Energy-coupled transport and signal transduction through the Gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol Rev. 1995;16:295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 10.Chen C-Y, Berish S A, Morse S A, Mietzner T A. The ferric iron binding protein of pathogenic Neisseria spp. functions as a periplasmic transport protein in iron acquisition from human transferrin. Mol Microbiol. 1993;10:311–318. doi: 10.1111/j.1365-2958.1993.tb01957.x. [DOI] [PubMed] [Google Scholar]
- 11.Deslauriers M, ni Eidhin D, Lamonde L, Mouton C. SDS-PAGE analysis of protein and lipopolysaccharide of extracellular vesicles and Sarkosyl-insoluble membranes from Bacteroides gingivalis. Oral Microbiol Immunol. 1990;5:1–7. doi: 10.1111/j.1399-302x.1990.tb00217.x. [DOI] [PubMed] [Google Scholar]
- 12.Filip C, Fletcher G, Wulff J L, Earhart C F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genco C. Regulation of hemin and iron transport in Porphyromonas gingivalis. Adv Dent Res. 1995;9:41–47. doi: 10.1177/08959374950090010801. [DOI] [PubMed] [Google Scholar]
- 14.Genco C A, Odusanya B M, Brown G. Binding and accumulation of hemin in Porphyromonas gingivalis are induced by hemin. Infect Immun. 1994;62:2885–2892. doi: 10.1128/iai.62.7.2885-2892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griggs D W, Tharp B B, Konisky J. Cloning and promoter identification of the iron-regulated cir gene of Escherichia coli. J Bacteriol. 1987;169:5343–5352. doi: 10.1128/jb.169.12.5343-5352.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerry P, Perez-Casal J, Yao R, McVeigh A, Trust T J. A genetic locus involved in iron utilization unique to some Campylobacter strains. J Bacteriol. 1997;179:3997–4002. doi: 10.1128/jb.179.12.3997-4002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendtlass, A., S. G. Dashper, and E. C. Reynolds. Purification of an antigenic protein (Pga30) from Porphyromonas gingivalis. Oral Microbiol. Immunol., in press. [DOI] [PubMed]
- 18.Hofmann K, Bucher P, Falquet L, Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holt S C, Ebersole J, Felton J, Brunsvold M, Korman K S. Implantation of Bacteroides gingivalis in non-human primates initiates progression of periodontitis. Science. 1988;239:55–57. doi: 10.1126/science.3336774. [DOI] [PubMed] [Google Scholar]
- 20.Inoshita E, Iwakura K, Amano A, Tamagawa H, Shizukuishi S. Effect of transferrin on the growth of Porphyromonas gingivalis. J Dent Res. 1991;70:1258–1261. doi: 10.1177/00220345910700090501. [DOI] [PubMed] [Google Scholar]
- 21.Jackson C A, Hoffmann B, Slakeski N, Cleal S, Hendtlass A, Reynolds E C. A consensus Porphyromonas gingivalis promoter sequence. FEMS Microbiol Lett. 2000;186:133–138. doi: 10.1111/j.1574-6968.2000.tb09094.x. [DOI] [PubMed] [Google Scholar]
- 22.Kim S, Chu L, Holt S. Isolation and characterisation of a hemin-binding cell envelope protein from Porphyromonas gingivalis. Microb Pathog. 1996;20:65–70. doi: 10.1006/mpat.1996.0043. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lamont R J, Jenkinson H F. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee B C. Isolation of an outer membrane hemin-binding protein of Haemophilus influenzae type B. Infect Immun. 1992;60:810–816. doi: 10.1128/iai.60.3.810-816.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legrain M, Mazarin V, Irwin S W, Bouchon B, Quentin-Millet M J, Jacobs E, Schryvers A B. Cloning and characterization of Neisseria meningitidis genes encoding the transferrin-binding proteins Tbp1 and Tbp2. Gene. 1993;130:73–80. doi: 10.1016/0378-1119(93)90348-7. [DOI] [PubMed] [Google Scholar]
- 27.Lewis J P, Dawson J A, Hannis J C, Muddiman D, Macrina F L. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J Bacteriol. 1999;181:4905–4913. doi: 10.1128/jb.181.16.4905-4913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M, Schweizer H P. Resolution of common DNA sequencing ambiguities of GC-rich DNA templates by terminal deoxynucleotidyl transferase without dGTP analogues. Focus. 1993;15:19–20. [Google Scholar]
- 29.Loeb K. Ferrochelatase activity and protoporphyrin IX utilization in Haemophilus influenzae. J Bacteriol. 1995;177:3613–3615. doi: 10.1128/jb.177.12.3613-3615.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsh P D, McDermid A S, McKee A S, Baskerville A. The effect of growth rate and haemin on the virulence and proteolytic activity of Porphyromonas gingivalis W50. Microbiology. 1994;140:861–865. doi: 10.1099/00221287-140-4-861. [DOI] [PubMed] [Google Scholar]
- 31.Mayrand D, Holt S C. Biology of asaccharolytic black-pigmented Bacteroides species. Microbiol Rev. 1988;52:134–152. doi: 10.1128/mr.52.1.134-152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKee A, McDermid A, Baskerville A, Dowsett B, Elwood D, Marsh P. Effect of hemin on the physiology and virulence of Bacteroides gingivalis W50. Infect Immun. 1986;52:349–355. doi: 10.1128/iai.52.2.349-355.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neidhart F C, Ingram J L, Schaechter M. Physiology of the bacterial cell: a molecular approach. Sunderland, Mass: Sinauer Associates, Inc.; 1990. [Google Scholar]
- 34.Nikaido H, Hall J A. Overview of bacterial ABC transporters. Methods Enzymol. 1998;292:3–20. doi: 10.1016/s0076-6879(98)92003-1. [DOI] [PubMed] [Google Scholar]
- 35.Raux E, Thermes C, Heathcote P, Rambach A, Warren M J. A role for Salmonella typhimurium cbiK in cobalamin (Vitamin B12) and siroheme biosynthesis. J Bacteriol. 1997;179:3202–3212. doi: 10.1128/jb.179.10.3202-3212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth J R, Lawrence J G, Rubenfield M, Kieffer-Higgins S, Church G M. Characterization of cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J Bacteriol. 1993;175:3303–3316. doi: 10.1128/jb.175.11.3303-3316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Sanger F, Nicklen S, Coulsen A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 40.Schubert H L, Raux, E. E, Wilson K S, Warren M J. Common chelatase design in the branched tetrapyrrole pathways of heme and anaerobic cobalamin synthesis. Biochemistry. 1999;38:10660–10669. doi: 10.1021/bi9906773. [DOI] [PubMed] [Google Scholar]
- 41.Shea C M, McIntosh M A. Nucleotide sequence and genetic organization of the ferric enterobactin transport system: homology to other periplasmic binding protein-dependent systems in Escherichia coli. Mol Microbiol. 1991;5:1415–1428. doi: 10.1111/j.1365-2958.1991.tb00788.x. [DOI] [PubMed] [Google Scholar]
- 42.Slakeski N, Cleal S M, Reynolds E C. Characterisation of a Porphyromonas gingivalis gene prtR that encodes an arginine-specific thiol proteinase and multiple adhesins. Biochem Biophys Res. 1996;224:605–610. doi: 10.1006/bbrc.1996.1073. [DOI] [PubMed] [Google Scholar]
- 43.Smalley J W, Birss A J, McKee A S, Marsh P D. Haemin-restriction influences hemin-binding haemagglutination and protease activity of cells and extracellular membrane vesicles of Porphyromonas gingivalis W50. FEMS Microbiol Lett. 1991;69:63–67. doi: 10.1016/0378-1097(91)90647-s. [DOI] [PubMed] [Google Scholar]
- 44.Smalley J W, Birss A J, McKee A S, Marsh P D. Haemin-binding proteins of Porphyromonas gingivalis W50 grown in a chemostat under haemin-limitation. J Gen Microbiol. 1993;139:2145–2150. doi: 10.1099/00221287-139-9-2145. [DOI] [PubMed] [Google Scholar]
- 45.Socransky S S, Haffajee A D, Cugini M A, Smith C, Kent R L. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 46.Socransky S S, Haffajee A D. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992;63:322–331. doi: 10.1902/jop.1992.63.4s.322. [DOI] [PubMed] [Google Scholar]
- 47.Tam R, Saier M H., Jr Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vriend G. WHAT IF: a molecular modelling guide and drug design. J Mol Graph. 1990;58:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 49.Wei B Y, Bradbeer C, Kadner R J. Conserved structural and regulatory regions in the Salmonella typhimurium btuB gene for the outer membrane vitamin B12 transport protein. Res Microbiol. 1992;143:459–466. doi: 10.1016/0923-2508(92)90091-2. [DOI] [PubMed] [Google Scholar]
- 50.Wilson K S, von Hippel P H. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Biochemistry. 1995;92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woolridge K G, Williams P H. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol Rev. 1993;12:325–348. doi: 10.1111/j.1574-6976.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]