Abstract
The sequence of the nonredundant region of the Salmonella enterica serovar Typhimurium temperate, serotype-converting bacteriophage P22 has been completed. The genome is 41,724 bp with an overall moles percent GC content of 47.1%. Numerous examples of potential integration host factor and C1-binding sites were identified in the sequence. In addition, five potential rho-independent terminators were discovered. Sixty-five genes were identified and annotated. While many of these had been described previously, we have added several new ones, including the genes involved in serotype conversion and late control. Two of the serotype conversion gene products show considerable sequence relatedness to GtrA and -B from Shigella phages SfII, SfV, and SfX. We have cloned the serotype-converting cassette (gtrABC) and demonstrated that it results in Salmonella serovar Typhimurium LT2 cells which express antigen O1. Many of the putative proteins show sequence relatedness to proteins from a great variety of other phages, supporting the hypothesis that this phage has evolved through the recombinational exchange of genetic information with other viruses.
In 1952, Zinder and Lederberg demonstrated the transfer (generalized transduction) of genetic material between Salmonella enterica serovar Typhimurium (here referred to as serovar Typhimurium) mutants involving a phage intermediary (80). The temperate phage vector, originally called PLT 22, is now commonly referred to as P22 and has continued to be the virus of choice for investigating the genetics of this bacterium.
Morphologically P22 is a member of the virus family Podoviridae, which encompasses viruses with short, noncontractile tails (1). P22 binds to the lipopolysaccharide (LPS) O side chains of serovar Typhimurium or to Escherichia coli strains expressing the serovar Typhimurium rfb cluster (46) via the virion tailspike proteins (65). The latter possess endorhamnosidase activity, which digests the O antigen, permitting passage of the phage through the LPS barrier to the surface of the outer membrane, where tight binding occurs. The linear double-stranded viral DNA enters the host cell, and circularization occurs, mediated by the phage-encoded protein Erf and the host proteins RecA and gyrase (50, 67). The resulting covalent closed and superhelical molecule is the substrate for integration into the host chromosome. The phage integration site, attP22 or ataA, maps within thrW, a gene for threonyl tRNA2 (53), with site-specific recombination being catalyzed, as it is with coliphage λ, by integration host factor (IHF) and integrase (Int) (40, 62). Upon induction, specialized transducing particles may arise, carrying genes adjacent to the att site, as well as a generalized transducing particle carrying only host DNA.
In the lysogenic state, P22 expresses three different systems that may interfere with superinfection by homologous phages. These are immunity conferred by the prophage repressor (c2), superinfection exclusion mediated by the sieA and sieB genes, and serotype conversion. The presence of the C2 protein represses the replication of homoimmune phage genomes, while the sie genes appear to function in preventing phage DNA injection (29, 49). Lysogenization by P22 also results in the addition of an α-linked glucosyl residue to the 6 position of galactose moieties in the LPS O-antigenic tetrameric repeat. This results in a change in serotype from 4,[5],12 to 1,4,[5],12 and prevents the binding of P22 and other serovar Typhimurium phages, a phenomenon known as lysogenic conversion (35, 51). Mutational analysis by Young and his associates determined that gene a1 was responsible for the expression of antigen 1 and showed its relative position in the P22 genome, adjacent to the phage attachment site (attP) (78). Preliminary evidence suggests that this region is highly homologous to the conversion-att-int region from Shigella flexneri bacteriophage SfV (32).
Early transcription events mimic those observed in coliphage λ-infected cells. Transcription is initiated from two promoters, PL and PR, that flank the repressor (c2) gene. The early proteins are 24, a λ N homologue which functions as a transcriptional antiterminator, and Cro, which functions to inhibit transcription from PRM and generally down-regulate transcription from PL and PR, thereby favoring lytic development. Another early transcript is initiated from Pant in the unique immI region, giving rise to an antirepressor, Ant, which functions to inhibit c2 repressor function. Late gene expression is regulated, as it is in coliphage λ, in an antitermination-dependent mechanism involving gp23, a Q homologue (48). The late genes include a holin (gp13), a lysozyme homologue (gp19), and the genes involved in morphogenesis. The last have been extensively studied (45), revealing that, unlike the situation with λ phage morphogenesis, a unique scaffolding protein (gp8) is involved in the formation of a morphogenic core together with portal protein (gp1) and pilot proteins (gp16, -20, and -7). The virus surface is composed almost exclusively of a single protein (gp5). The scaffold is reutilized in subsequent rounds of capsid assembly. In contrast, λ uses the product of a gene, Nu3, to play a transient role as a core or scaffold protein, which is subsequently cleaved, and the λ protein coat is composed of two main proteins, gpD and gpE.
In lytic development, DNA replication is initiated from an origin (Ori) located within gene 18 (7) in a region which shows superficial similarity to that of coliphage λ (gpO-gpP) with the exception that P22 contains a primase (gp18) and a helicase (gp12) (33). Replication requires additional host (55) and viral (75) proteins, leading to the formation of concatemeric molecules (15), perhaps as a result of rolling-circle replication (48). Another aspect distinguishing P22 from λ is that DNA packaging in P22 proceeds from a unique site (pac) located within gene 3 on the concatemeric substrate, resulting in the head-full packaging of a limited series of terminally redundant, circularly permuted genomes. P22 packages about 43.4 kb of DNA (11) that has terminal 1.7-kb direct repeats (48) and is 5 to 8% circularly permuted. More recent molecular studies by Španová indicated that the terminal redundancy is 0.9 kb (2.2%) (64). In the case of coliphage lambda, concatemeric DNA is cut by terminase at specific sites and packaged. The latter results in unique ends with cohesive extended 5′ termini rather than the blunt-ended, terminally repetitious molecules observed with P22.
Many studies have suggested that P22, in spite of its morphology, is a member of the lambdoid family. The layout of its genes is very similar to that of other lambdoid phages, viable λ-P22 hybrids have been formed in vivo, and of the 36 known P22 genes, 23 are believed to have λ analogues (44). These facts confirm the observation of Casjens et al. (19) that “an important feature of the lambdoid phage is that its structure and function are more highly conserved than are actual gene sequences” (48). Another way of looking at this group of phages is that they are a mosaic built up of modules or cassettes (47), and while conserved patterns which suggest familial relationships exist, the overall picture suggests that considerable intervirus or virus-host recombination has occurred, often between viruses infecting distant bacterial groups (26).
Largely because of the morphological difference between λ and P22, the latter has been proposed recently as the type virus for a new genus which includes phages L (14), ES18 (56), LP7 (37), ɛ34 (34), and APSE-1 (72). Schicklmaier and Schmieger used complementation and hybridization to demonstrate sequence similarity between ES18 and P22. Limited DNA sequence data from the att-int region to gene 15 has confirmed this (56). Yet even these phages are morphologically different, and the genome size of ES18 is 46.15 kb as opposed to the value of 41.8 kb for P22. This illustrates the problem of establishing relationships based upon limited data.
P22 has been extensively studied, with current emphasis on capsid morphogenesis (45, 69–71), tailspike protein-ligand interactions (59, 65), elucidation of regulatory circuits (23, 54), and transductional analysis (12, 43, 46). P22-Mu hybrid phages have been constructed carrying Mu termini and an internal fragment containing the P22 pac site (57, 77). These insert randomly into the serovar Typhimurium chromosome, package DNA adjacent to the integration site, and have proved extremely useful in chromosomal mapping. In spite of its historical and current importance, the complete genome sequence of P22 has not been reported; rather, a large number of partial sequences are to be found in GenBank. In this report, we have taken those sequences, aligned them, and extended the sequence. The similarity between P22 proteins and those of other bacteriophages was investigated in order to suggest phylogenetic relationships between different phages.
MATERIALS AND METHODS
Bacteria, bacteriophage, and plasmid vector.
The LT2 wild-type strain of serovar Typhimurium was obtained from N. L. Martin (Queen's University, Kingston, Ontario, Canada). TOP10 cells [genotype, F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG] (Invitrogen) were used for the recombinant DNA techniques. Bacteriophage P22 was obtained from H.-W. Ackermann (Laval University, Québec, Canada).
Media.
Bacteria were grown in Luria-Bertani broth (LB; Difco Laboratories) or on LBA plates (LB with 1.5% [wt/vol] agar). For phage titrations, 3-ml overlays were prepared using LB containing 0.6% (wt/vol) agar. The titers of the phage preparations were determined using the agar overlay technique of Adams (2).
Purification of P22.
A culture of serovar Typhimurium LT2 was grown at 37°C overnight, and 5-ml samples were inoculated into four 2-liter flasks, each containing 500 ml of LB. The flasks were incubated at 37°C with shaking at 180 rpm, and the optical density at 650 nm was periodically monitored. When the optical density reached 0.25, P22 was added to a multiplicity of infection of 5. Following 6.5 h of incubation, 20 ml of chloroform was added to each flask. The phage were separated from the cell debris by centrifuging the flasks at 10,000 × g for 10 min at 4°C, and the clarified lysate was retained. The phage were precipitated using 10% (wt/vol) polyethylene glycol (76), harvested by centrifugation at 10,000 × g for 15 min at 4°C, and resuspended in 20 ml of SM buffer [5.8 g of NaCl, 2 g of MgSO4 · 7H2O, 50 ml of 1 M Tris · Cl (pH 7.5), 5 ml of 2% gelatin solution per liter] with 10% (vol/vol) Triton X-100 (52). Solid CsCl was added to the crude resuspended phage to a concentration of 0.5 g/ml, and the mixture was layered on a CsCl step gradient prepared as described by Sambrook et al. (52). The tubes were centrifuged at 60,000 × g at 4°C for 2 h in the Beckman L8-70 ultracentrifuge with a SW28.1 rotor. The phage were further purified using a CsCl equilibrium gradient. The material from the CsCl step gradient was added to a type 75T Beckman Quick-Seal centrifuge tube, and the volume was topped off with a CsCl solution with a density of 1.5 g/ml. The tube was sealed and centrifuged using a Type 75Ti rotor at 104,000 × g at 4°C for 24 h. The phage were then removed from the tube with a syringe.
To remove the CsCl from the purified phage suspension, the latter was added to a 10K Slide-A-Lyzer dialysis cassette (Pierce) and dialyzed against multiple changes of 50 mM Tris-HCl, pH 8, at 4°C.
Isolation of P22 DNA.
The following were added to the dialyzed phage stock: EDTA to a final concentration of 20 mM, proteinase K (Boehringer Mannheim) to 50 μg/ml, and, sodium dodecyl sulfate to 0.5% (wt/vol). The mixture was incubated at 53°C for 1 h. The lysate was deproteinized by shaking it with phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol/vol]; Fisher Scientific) followed by centrifugation at 16,000 × g for 25 min. The aqueous layer was extracted once more with phenol-chloroform-isoamyl alcohol and then again with chloroform. The final aqueous layer was dialyzed as previously outlined.
The concentration and purity of the isolated DNA was analyzed using the Beckman DU-600 spectrophotometer at wavelengths of 260, 280, and 320 nm, based upon the assumption that 1 A260 = 50 μg of DNA/ml (52). The purified DNA was stored at 4°C.
DNA sequencing.
The DNA primers used for sequencing were designed by examining the regions near the end of the contiguous sequence or near the conflict in the sequence. Potential oligonucleotide primers were analyzed for melting temperature and secondary structures using Net Primer (Premier Biosoft International), and were synthesized by Cortec DNA Service Laboratories. Fluorescent dye dideoxy chain-terminating DNA sequencing was carried out at Cortec using an Applied Biosystems 373XL automated sequencer. Primer walking, using amplification conditions optimized for sequencing lambda clones, was used to determine the sequence directly from the P22 genomic DNA.
Sequence assembly and analysis.
The Applied Biosystems sequence data was collected, stripped of poor-quality data, and assembled into contigs using Seqman II (DNASTAR Inc.). Open reading frames (ORFs) were analyzed using ORF Finder at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and WebGeneMark.HMM (42) (http://genemark.biology.gatech.edu/GeneMark/whmm.cgi). In addition, the Find ORF feature of SeqEdit (DNASTAR) was employed to manually scan the sequence for potential genes. A compendium of online tools (http://www.queensu.ca/micr/faculty/kropinski/online.html) was employed in the analysis of the putative genes. Proteins translated at ORF Finder or “translate tool” (http://www.expasy.ch/tools/dna.html) were scanned for homologues by using BLASTP (5, 6) against the nonredundant GenBank protein database (http://www.ncbi.nlm.nih.gov/blast/blast.cgi). Their molecular masses and isoelectric points were determined online at ProtParam tools (http://www.expasy.ch/tools/protparam.html). Where homologues were identified, the sequences were compared using Clustal W (68) at the European Molecular Biology Laboratory-European Bioinformatics Institute (http://www.ebi.ac.uk/clustalw/). In addition, ALIGN at Genestream (Institute de Génétique Humaine) at its website (http://www2.igh.cnrs.fr/bin/align-guess.cgi) was employed to compare two sequences. Proteins were also scanned against the Prosite (9, 30) and Protein Families (Pfam) (10) databases for conserved motifs at the Swiss Institute for Experimental Cancer Research ProfileScan server (http://www.ch.embnet.org/software/PFSCAN_form.html). To predict transmembrane proteins, two online programs were employed, TMPred (31) at the European Molecular Biology network—Swiss node (http://www.ch.embnet.org/software/TMPRED form.html) and TMHMM (63) at the Center for Biological Sequence Analysis at The Technical University of Denmark (http://www.cbs.dtu.dk/services/TMHMM-1.0/).
For basic analysis of the DNA sequence, including restriction sites and motifs, DNAMAN (Lynnon BioSoft, Vaudreuil, Canada) and Omiga from Oxford Molecular Group (Campbell, Calif.) were employed. The DNA sequence was scanned for putative tRNA species by using tRNAscan-SE (21, 41) at its website (http://www.genetics.wustl.edu/eddy/ tRNAscan-SE/) and FAStRNA (22) (http://bioweb.pasteur.fr/seqanal/interfaces/fastrna.html). Potential IHF-binding sites were assessed using MacTargsearch at SEQSCAN (http://www.bmb.psu.edu/seqscan/seqform1.htm), while rho-independent transcription terminators were analyzed at Bionavigator (http://www.bionavigator.com) using the Genetics Computer Group Terminator program (16, 17).
For comparison with unpublished Salmonella genomic sequences, WU_BLAST 2.0 (http://blast.wustl.edu/) was employed at the Genome Sequencing Center, Washington University School of Medicine (St. Louis, Mo.). For comparison with the sequence of lambda, DNA BLAST 2 (66) was used at the National Center for Biotechnology Information.
Cloning and serotype conversion.
Two PCR primers (CCAAACCACTTAGCAATCAGC and AGCGCTAATTAAACCTAACAACTATGG) were designed to flank the gtrABC cassette. These were used together with Taq DNA polymerase to amplify the gtrABC genes and upstream sequence. The amplicon was ligated into the pCRII-TOPO vector and transformed into the TOP10 chemically competent cells (Invitrogen). The cells were then recovered in SOC medium (52), and after 1 h of incubation at 37°C, aliquots were plated onto LBA plates containing ampicillin (100 μg/ml) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (40 μg/ml). Plasmid DNA was isolated using the alkaline lysis technique (52) and electroporated into serovar Typhimurium LT2. Ampicillin-resistant clones were tested for the ability to agglutinate in anti-O1,2,12 serum (Difco Laboratories).
Nucleotide sequence accession number.
The nucleotide sequence described in this manuscript has been deposited with GenBank and has been assigned accession no. AF217253.
RESULTS AND DISCUSSION
Sequence assembly and analysis.
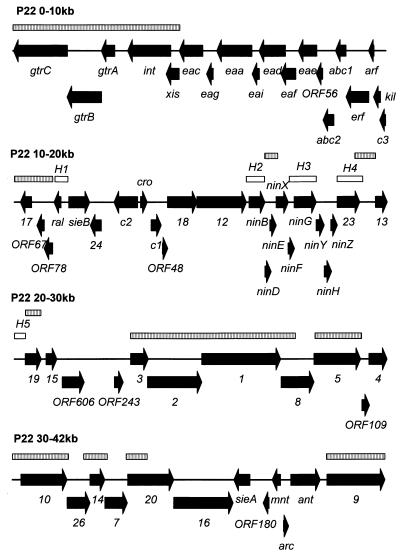
Twenty-four P22 sequences were retrieved from GenBank and assembled, using SeqMan, into four contigs ranging from 0.6 to 24.6 kb. These amalgamated sequences revealed 17 discrepancies. Using primer walking, we have corrected these errors, linked the contigs, and extended the assembly to the ends of the unique sequence. Sequencing from the integrase (int) gene leftward resulted in sequence that was identical to sequence derived from sequencing rightward from gene 9 (tailspike protein) (Fig. 1). This region extends to over 800 bp, the limit of our sequencing reactions, and corresponds to the terminally redundant ends of the genome. To circumvent the problems in presenting the circularly permuted, terminally redundant genome, the map (Fig. 1) was opened adjacent to a 15-bp stem-loop structure (AATAAAAATGGGTGTaaACACCCATTTTTATT [bases in the loop are shown in lowercase]) with a calculated ΔG of −17.1 kcal/mol located downstream of the tailspike protein gene. The unique genomic sequence is 41.7-kb, which is remarkably similar to the value of 41.6 kb calculated by Chisholm and colleagues on the basis of restriction endonuclease digestion (20). The DNA has an overall moles percent GC content of 47.1, which is somewhat less than the published value of 50 (38).
FIG. 1.
ORFs and sequence similarity regions in P22 DNA. The ORFs for which no genetic designation had been made previously are labeled based upon the number of amino acid residues (e.g., ORF-80 would encode a protein with 80 amino acids). The striped and open boxes correspond to regions which have nucleic acid sequence similar to S. paratyphi A or coliphage λ DNA, respectively.
No tRNA genes were discovered with tRNAscan-SE or FAStRNA. Large numbers of IHF-binding sites were discovered with the online program MacTargsearch (24). Since the P22 genome is relatively AT rich, the relevance of many of these is open to question, and the list presented in Table 1 is restricted to those with MacTargsearch scores of ≥50. Lambda protein CII is an activator, stimulating transcription by binding to the face of the DNA opposite that to which the RNA polymerase binds in three promoters: PRE (promoter for repressor establishment), PI (integrase promoter), and PaQ (anti-Q promoter). The P22 homologue, C1 protein, also stimulates transcription from PRE and Pa23 (the P22 Q homologue) (27). P22 C1-binding sites, have the recognition motif TTGC(N6)TTGY (27), while its homologue, lambda CII protein, recognizes TTGC(N6)TTGC (28). A search for TTGC(N6)TTGY identified 11 sites in P22 DNA (Table 1). The site upstream of the integrase (int) gene may represent PaI, a C1-activated anti-integrase promoter (27). Similarly, the two sites immediately downstream from the int gene may also function in the regulation of integrase expression. The significance of the other potential C1-binding sites, primarily located within the morphogenesis genes, is unknown.
TABLE 1.
Potential IHF- and C1-binding sites and rho-independent terminators in P22 DNAa
| Location | Similarity score | Sequence | Potential function |
|---|---|---|---|
| Potential IHF-binding sites | |||
| 427–453 | 57.4 | TTCTTGATATTAACTGTTTATCTTCAA | |
| 2855–2829 | 50.1 | AGATAAAAACTATCAAATTATACATTA | |
| 3032–3006 | 52.4 | CCTTTTAAGTCAACAACATACCACGTC | |
| [3087–3113] | 61.7 | CCAGTTAAATCAAATACTTACGTATTA | |
| [11584–11558] | 53.3 | GCATATGAATCAACTGTTTAAGTGTCA | |
| 12711–12685 | 49.8 | AAGTAACGATAAAATATTTAAGTTTTC | |
| 19623–19597 | 53.5 | CAACTTTATTCAAAAAGTCAATATCAT | |
| 21186–21212 | 54.2 | CACTGAAATTTAACAAGTGACTTTCAG | |
| [21579–21605] | 52.6 | CCGAAAAAATCAATAACTTAGGGATTT | |
| 37218–37244 | 51.4 | TAGAAAAAAACAACCACGCAATCTGCA | |
| Lambda cro/cII | 61.5 | TGCATACATTCAATCAATTGTTATCTA | |
| Potential C1-binding sites | |||
| 2946–2959 | TTGCATCGGTTTGC | ||
| 2956–2969 | TTGCAAGGCTTTGC | ||
| 4647–4660 | TTGCGGGTGCTTGC | PaI | |
| 13809–13796 | TTGCGAGTGCTTGT | PRE | |
| 18448–18435 | TTGCCTAACCTTGC | ||
| 19237–19224 | TTGCGAGCACTTGC | ||
| 24346–24333 | TTGCCCGTATTTGT | ||
| 26998–26985 | TTGCCGGGTCTTGT | ||
| 27542–27529 | TTGCCGGGTCTTGT | ||
| 34184–34171 | TTGCTGCGGATTGT | ||
| 40808–40795 | TTGCGAGAGGTTGT | ||
| Rho-independent terminators | |||
| 2871–2900 | ATTGATCGTTGTTACCGATCAATTTTTATT | ||
| 5152–5179 | ACCGCCATCAGGCGGCTTGGTGTTCTTT | ||
| [5167–5144] | GCCGCCTGATGGCGGTTTTTTATT | ||
| 10157–10188 | AGCCGCACTCAGGCGGCTGTCGTTTCTTCTTT | ||
| [10174–10148] | AGCCGCCTGAGTGCGGCTTTTTCATAT | ||
| [10519–10490] | TTGCCGCTCTATATGGGCGGCATTCTTTTT | ||
| [18516–18488] | CTCGCTTTCACAGCGGCTTTCTCTTCGTT | ||
| 21218–21246 | TGCCTCGCAGATGCGGGGCGTTTTTGTAT | ||
| 22161–22187 | AGCCGCTTACTTAGCGGCTTGACGTTT | ||
| [22179–22154] | AGCCGCTAAGTAAGCGGCTTTTTTAT | ||
| 37691–37723 | AGCCGGAGTGACCCGGCTTGATTATTACTTTTT | ||
| [37708–37682] | AGCCGGGTCACTCCGGCTTTTTGATAT | ||
| 39487–39512 | ACCCAGCTTCGGCTGGGTTTTTTTAT | ||
| 39634–39660 | ACCGTAGCCATGCTGCGGCAATTCCTT | ||
| [39652–39623] | GCCGCAGCATGGCTACGGTGAATTTTTTGT | ||
| 40243–40265 | TCCCGCATTGCCGGGGTTTTTAT |
In the case of IHF-binding sites, we have included the similarity scores calculated by MacTargsearch (24). The bases which are identical to the consensus sequence are highlighted in boldface. For comparison, we have included a characterized IHF-binding site in coliphage λ. Putative C1-binding sites were determined by scanning the sequence for the consensus sequence (TTGCN6TTGY), while rho-independent terminators were determined using the Genetics Computer Group terminator program. Only those sites for which there was evidence of a stem-loop structure (boldface and underlined) followed by a region rich in thymine residues were included. The sites in brackets are those associated with the complementary strand.
Using the search algorithm of Brendel and colleagues (16, 17), we were able to identify 16 potential rho-independent terminators (Table 1). Interestingly, almost all of these lie in intergenic regions. Other stem-loop structures were identified immediately downstream of gene 16 (16453 to 16476; AGGCCTGCtggtaatcGCAGGCCT; −10.9 kcal/mol), gene ORF202 (22160 to 22180; AAGCCGCTtacttAGCGGCTT; −9.3 kcal/mol), and ant (39560 to 39582; GACCTACAAaaaaaTTGTAGGTC; −9.0 kcal/mol) (bases in loops are shown in lowercase; bases shown by capital letters form hydrogen bonds). If the last functions as a transcriptional terminator, then gene 9 must possess its own promoter. This has been proposed (13, 58).
Sequence similarity between P22 and other phage and Salmonella DNA.
P22 DNA shares 13.5% sequence similarity with that of phage lambda DNA as shown by hybridization experiments (61). Using the BLAST2 algorithm of Tatusova and Madden, we identified five >300-bp regions in lambda DNA which shared >85% sequence identity with the P22 sequence (66). These are indicated in Fig. 1. The regions of greatest DNA sequence similarity correspond to genes ninB, ninG, and 23. This is also verified at the amino acid level.
Wu_BLAST analysis against the incomplete Salmonella genomes at the Washington University School of Medicine Genome Sequencing Center indicated strong regions of sequence identity (>90% identity), particularly to Salmonella paratyphi A in the regions shown in Fig. 1. The strongly conserved regions include those associated with integration and O antigen conversion and with morphogenesis. These results suggest that S. paratyphi A probably harbors a prophage that is quite similar to P22. Homology with the genomic sequence of serovar Typhimurium and Salmonella typhi is restricted to a region at the left end of the molecule (from 1.5 to 2.8 kb). This will require reassessment when these genomes are completely sequenced.
P22 ORF analysis.
Many of the P22 ORFs were previously identified (48). We reanalyzed the sequence data, updating the positions of the previously identified ORFs and making limited corrections. For example, ORF67 was previously referred to as ORF87 due to a mistake in the DNA sequence. The sequence was reanalyzed using more modern algorithms, revealing a total of 65 ORFs, 29 (45%) of which showed no similarity to those of any other protein in the GenBank protein database. We were very cautious in defining what constituted an ORF, relying on the work of previous workers or the presence of a clearly recognizable ribosome-binding site. This manuscript will concentrate on those genes that have been identified by the current authors. A complete list of P22 ORFs is displayed in Table 2.
TABLE 2.
Characterization of the genes of phage P22
| Gene | Froma | Toa | Strand | Mass (kDa) | Function | Related phage sequencesb | BlastP e value | % Identity |
|---|---|---|---|---|---|---|---|---|
| gtrC | 17 | 1474 | − | 55.2 | O-antigen conversion; glucosyl transferase | |||
| gtrB | 1464 | 2396 | − | 35.1 | O-antigen conversion; bactoprenol glucosyl transferase | AAB72133; SfV orf5 | e-139 | 87 |
| AAC39272; SfII bgt | e-139 | 87 | ||||||
| AF056939; SfX gtrB | e-138 | 85 | ||||||
| gtrA | 2393 | 2755 | − | 13.5 | O-antigen conversion; translocase (flippase) | AF056939; SfX gtrA | 3e-44 | 78 |
| AAB72134; SfV orf6 | 7e-43 | 77 | ||||||
| AAC39271; SfII orf2 | 2e-42 | 77 | ||||||
| int | 3104 | 4267 | − | 44.8 | Integrase | AAB72135; SfV int | 0 | 89 |
| INTD_ECOLI, DLP12 int | e-162 | 71 | ||||||
| F157835_38; APSE-1 int | e-149 | 63 | ||||||
| xis | 4144 | 4494 | − | 12.8 | Excisionase | AAB72136; SfV xis | 9e-52 | 69 |
| eac | 4497 | 5132 | − | 23.9 | Unknown | |||
| eag | 5233 | 5412 | − | 6.6 | Unknown | |||
| eaa | 5509 | 6462 | − | 35.7 | Unknown | BAA84359; VT2-Sa orf76 | 2e-22 | 26 |
| F125520_78; 933W L0140 | 2e-22 | 28 | ||||||
| eai | 6466 | 6660 | − | 7.0 | Unknown | |||
| ead | 6657 | 7367 | − | 26.8 | Unknown | VE22_LAMBD; λ ea22 | 1e-13 | 31 |
| eaf | 7247 | 7642 | − | 15.2 | Unknown | |||
| eae | 7715 | 8212 | − | 18.1 | Unknown | |||
| ORF-56 | 8200 | 8370 | − | 6.6 | Unknown | |||
| abc2 | 8381 | 8674 | − | 11.6 | Anti-RecBCD protein | |||
| abc1 | 8721 | 9005 | − | 10.9 | Anti-RecBCD protein | |||
| erf | 9005 | 9622 | − | 23.0 | Recombination protein | AAD04639; H-19B erf | 4e-79 | 74 |
| AAA92165; c2 e15 | 6e-17 | 32 | ||||||
| arf | 9619 | 9762 | − | 5.5 | Recombination protein | |||
| kil | 9752 | 9940 | − | 6.9 | Unknown | |||
| c3 | 9921 | 10079 | − | 5.7 | Regulatory protein | RPC3_LAMBD; λ cIII | 4e-8 | 59 |
| F125520_17; 933W cIII | 3e-7 | 57 | ||||||
| 17 | 10165 | 10476 | − | 12.2 | Superinfection exclusion | AAD04642; H-19B gp17 | 6e-50 | 84 |
| ORF-67 | 10624 | 10827 | − | 7.8 | Unknown | |||
| ORF-78 | 10827 | 11063 | − | 8.6 | Unknown | |||
| ral | 11100 | 11294 | − | 7.4 | Antirestriction protein | VRAL_LAMBD; λ ral | 9e-22 | 73 |
| VRAL_BPPH3; φ21 ral | 9e-22 | 62 | ||||||
| sieB | 11509 | 12087 | + | 22.4 | Superinfection exclusion | |||
| 24 | 12108 | 12410 | − | 11.0 | Antitermination | CAA63998; L 24 | 5e-35 | 73 |
| CAA60872; ES18 24 | 2e-31 | 64 | ||||||
| c2 | 12764 | 13414 | − | 24.1 | Repressor | CAA63999; L c2 | 9e-57 | 53 |
| S32822; 434 cI | 1e-51 | 49 | ||||||
| RPC1_LAMBD; λ cI | 1e-27 | 35 | ||||||
| BAA84306; VT2-Sa cI | 8e-26 | 33 | ||||||
| cro | 13495 | 13680 | + | 6.8 | Antirepressor | CAB39982; 21 cro | 3e-28 | 98 |
| c1 | 13787 | 14065 | + | 10.2 | Transcriptional activator | RPC2_BP434; 434 cII | 5e-19 | 48 |
| RPC2_LAMBD; λ cII | 6e-19 | 48 | ||||||
| ORF-48 | 14100 | 14246 | + | 5.8 | Unknown | S42399; HKO22 | 4e-17 | 85 |
| 18 | 14239 | 15054 | + | 30.6 | DNA replication | CAA60876; ES18 gp18 | 6e-24 | 29 |
| AAD04647; H-19B gpO | 4e-22 | 29 | ||||||
| AF125520_28; 933W gpO | 6e-22 | 28 | ||||||
| 12 | 15051 | 16427 | + | 50.1 | DNA replication (helicase) | CAA09719; P1 ban | 2e-55 | 32 |
| BAA84310; VT2-Sa P | 2e-48 | 30 | ||||||
| S43527; HKO22 P | 1e-40 | 27 | ||||||
| ninB | 16501 | 16938 | + | 16.4 | Unknown | CAB39988; 21 ninB | 4e-77 | 97 |
| Y146_LAMBD; λ orf146 | 1e-76 | 96 | ||||||
| ninD | 16935 | 17108 | + | 7.0 | Unknown | Y57_LAMBD; λ orf57 | 7e-23 | 73 |
| ninE | 17075 | 17251 | + | 72.0 | Unknown | BAA84316; VT2-Sa orf33 | 2e-29 | 95 |
| CAB39989; 21 ninE | 9e-29 | 93 | ||||||
| AAD04650; H-19B orf58 | 1e-28 | 97 | ||||||
| NINE_LAMBD; λ ninE | 8e-28 | 92 | ||||||
| ninX | 17248 | 17586 | + | 12.5 | Unknown | |||
| ninF | 17579 | 17755 | + | 6.4 | Unknown | AAD04651; H-19B nin orf-58-B | 4e-26 | 95 |
| Y56_LAMBD; λ orf56 | 5e-21 | 79 | ||||||
| CAB39990; 21 ninF | 2e-20 | 79 | ||||||
| ninG | 17748 | 18359 | + | 24 | Unknown | CAB39991; 21 ninG | e-112 | 94 |
| Y204_LAMBD; λ orf204 | e-112 | 94 | ||||||
| CAB39297; 933W orf15 | e-105 | 89 | ||||||
| AAD04653; H-19B nin orf-204 | e-101 | 87 | ||||||
| ninY | 18356 | 18580 | + | 8.6 | Unknown | |||
| ninH | 18577 | 18780 | + | 7.9 | Unknown | Q1BP0L; λ nin | 8e-19 | 61 |
| CAB39298; 933W orf16 | 2e-18 | 64 | ||||||
| AAD04654; H-19B nin orf-59 | 2e-18 | 61 | ||||||
| ninZ | 18761 | 18940 | + | 6.9 | Unknown | |||
| 23 | 18937 | 19560 | + | 22.3 | Antitermination | CAA09704; PS34 gp23 | e-112 | 95 |
| REGQ_LAMBD; λ Q | e-112 | 95 | ||||||
| S28977; HKO22 Q | e-112 | 95 | ||||||
| 13 | 19995 | 20321 | + | 11.7 | Lysis (holin) | VLYS_LAMBD; λ S | 2e-47 | 89 |
| 19 | 20302 | 20742 | + | 16.1 | Lysozyme | CAA47617.1; ES18 gp19 | 2e-75 | 97 |
| AF157835_13; APSE-1 P13 | 4e-26 | 43 | ||||||
| 15 | 20877 | 21176 | + | 11.0 | Endopeptidase (Rz homologue) | CAA09707; PS34 gp15 | 4e-49 | 66 |
| CAA09702; PS3 gp15 | 7e-46 | 60 | ||||||
| BAA84330.1; V2T-Sa Rz | 3e-22 | 35 | ||||||
| AF125520_47; 933W Rz | 5e-21 | 33 | ||||||
| ORF-201 | 21326 | 21931 | + | 22.7 | Unknown | |||
| ORF-80 | 22765 | 23007 | + | 9.0 | Unknown | |||
| 3 | 23212 | 23700 | + | 18.6 | Terminase (small subunit) | CAA09708; PS34 gp3 | 2e-90 | 96 |
| CAA09703; PS3 gp3 | 5e-89 | 95 | ||||||
| TERM_BPLP7; LP7 gp3 | 2e-88 | 94 | ||||||
| 2 | 23678 | 25177 | + | 57.6 | Terminase (large subunit) | VG2_BPLP7; LP7 gp2 | 0 | 75 |
| 1 | 25177 | 27354 | + | 82.7 | Portal protein | AF157835_19; APSE-1 P19 | 1e-96 | 32 |
| 8 | 27368 | 28279 | + | 33.6 | Scaffolding protein | |||
| 5 | 28279 | 29571 | + | 46.8 | Coat protein | |||
| ORF-69 | 29610 | 29819 | + | 7.7 | Unknown (Y7K7_BPP22) | |||
| 4 | 29803 | 30303 | + | 18.0 | DNA stabilization protein | |||
| 10 | 30263 | 31681 | + | 52.5 | Packaged DNA stabilization protein | AF157835_28; APSE-1 P28 | e-174 | 60 |
| 26 | 31685 | 32386 | + | 24.7 | Packaged DNA stabilization protein | |||
| 14 | 32386 | 32841 | + | 17.2 | Unknown | |||
| 7 | 32844 | 33533 | + | 23.4 | DNA transfer protein | AF157835_32; APSE-1 P32 | 1e-61 | 53 |
| 20 | 33544 | 34959 | + | 50.1 | DNA transfer protein | |||
| 16 | 34959 | 36788 | + | 64.4 | DNA transfer protein | AF157835_35; APSE-1 P35 | 0.0 | 64 |
| sieA | 36811 | 37305 | − | 18.7 | Superinfection exclusion | |||
| ORF-59 | 37715 | 37894 | − | 6.4 | Unknown | |||
| mnt | 37994 | 38245 | − | 9.7 | Regulatory protein | |||
| arc | 38336 | 38497 | + | 6.2 | Repressor | |||
| ant | 38566 | 39468 | + | 34.6 | Antirepressor | BAA84318.1; V2T-Sa ant | 9e-46 | 39 |
| AF125520_46; 933W ant | 2e-27 | 27 | ||||||
| 9 | 39679 | 41682 | + | 71.9 | Tailspike protein (endorhamnosidase) | AF128887_1; Sf6 | 9e-44 | 28 |
| AF157835_36; APSE-1 P36 | 3e-33 | 20 |
Nucleotide coordinates corresponding to the first nucleotide of the initiation codon and the last nucleotide of the termination codon.
Where similar sequences exist, the GenBank accession number is followed by the phage name and the gene designation.
The codon utilization of the ORFs of phage P22 and its host are shown in Table 3. For those amino acids that have only two possible codons, phenylalanine, tyrosine, lysine, histidine, glutamine, asparagine, aspartic acid, glutamic acid, and cysteine, only in the last case was there a significant trend away from quasiequal utilization of U/A or C/G in the wobble position. A comparison of codon usage in a highly expressed protein (coat protein; gp5) in comparison to two poorly expressed proteins (Int and C2) revealed some interesting differences. The following codons were not utilized in gp5 (AUA [Ile], CAC [His], and AGA and AGG [Arg]), while CCA (Pro) and GGA (Gly) were underrepresented in the coding region for this protein compared with Int and C2. It has been noted that the λ integrase has a higher proportion of the rare arginine codons, AGA and AGG, and that this influences expression of the gene (79). Of greater interest is the observation that for many amino acids (Phe, Tyr, His, Lys, Leu, and Asp), all of the codons used in P22 differ by ±20% from those in the host bacterium. This would be expected to have a global influence on translation and is also different from the results that we observed with Pseudomonas aeruginosa phage D3 (60).
TABLE 3.
Codon usage of phage P22 ORFs (in boldface) and those of its host, S. enterica
| U | C | A | G | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U | F | 45 | 72 | S | 9 | 19 | Y | 52 | 73 | C | 39 | 57 | U |
| F | 55 | 28 | S | 11 | 14 | Y | 48 | 27 | C | 61 | 43 | C | |
| L | 22 | 25 | S | 21 | 17 | Stop | 31 | 63 | Stop | 46 | 30 | A | |
| L | 27 | 15 | S | 19 | 14 | Stop | 24 | 7 | W | 100 | 100 | G | |
| C | L | 6 | 13 | P | 12 | 22 | H | 56 | 71 | R | 8 | 34 | U |
| L | 9 | 8 | P | 16 | 15 | H | 44 | 29 | R | 15 | 32 | C | |
| L | 12 | 6 | P | 36 | 18 | Q | 46 | 35 | R | 12 | 8 | A | |
| L | 24 | 34 | P | 37 | 45 | Q | 54 | 65 | R | 15 | 12 | G | |
| A | I | 30 | 52 | T | 15 | 21 | N | 51 | 63 | S | 15 | 17 | U |
| I | 32 | 27 | T | 21 | 36 | N | 49 | 37 | S | 25 | 19 | C | |
| I | 38 | 22 | T | 29 | 18 | K | 47 | 75 | R | 23 | 9 | A | |
| M | 100 | 100 | T | 34 | 24 | K | 53 | 25 | R | 27 | 5 | G | |
| G | V | 17 | 28 | A | 16 | 19 | D | 53 | 72 | G | 17 | 31 | U |
| V | 19 | 19 | A | 16 | 24 | D | 47 | 28 | G | 28 | 38 | C | |
| V | 27 | 22 | A | 33 | 19 | E | 57 | 61 | G | 27 | 14 | A | |
| V | 37 | 32 | A | 35 | 38 | E | 43 | 39 | G | 27 | 17 | G | |
The P22 ORFs were analyzed using DNAMAN, while data on 223 S. enterica coding regions in the Codon Usage Database (http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=Salmonella+enterica+[gbbct]) were used for comparison. The left-hand column contains the first nucleotides of the codons. The second nucleotides of the codons are listed across the top, while the third nucleotides are listed in the right-hand column. The numbers represent percentages. The amino acids or functions are given on the left.
Among the new genes discovered are those involved in serotype conversion. Serovar Typhimurium belongs to Salmonella serogroup B, which is characterized by possessing an O antigen repeating unit of a d-mannose-α1→2-l-rhamnose-α1→3-d-galactose trimeric repeat in which the mannosyl residue is substituted (α1→3) with the 3,6-dideoxy hexose abequose (74). This tetrasaccharide is equivalent to O antigen 4. Lysogenization of cells by P22 results in the appearance of O antigen 1, corresponding to the modification of this repeat through the addition of α1→6 glucosyl residues on the galactosyl residues (74). We have identified the three genes involved in the conversion event, which are arranged in the same fashion as many other phage O antigen conversion modules (4).
(i) GtrA.
This 363-bp ORF (45.4 mol% GC) encodes a 13.5-kDa protein with a calculated pI of 9.4. As predicted by TMHMM analysis, this small protein has four transmembrane domains, with the amino and carboxy termini of the protein arrangement on the cytoplasmic side of the inner membrane. The protein shows strong sequence homology to the serotype conversion proteins from the S. flexneri phages SfV, SfII, and SfX, which, like P22, carry out glucosylation of the O antigen. Morphologically, these phages belong to three different virus families: Podoviridae (SfV), Myoviridae (SfII), and Inoviridae (SfX). GtrA also showed homology to the products of defective prophages in S. flexneri (orf1, 77% identity) (3) and E. coli (hypothetical gene b2350, 79% identity). In addition, we have been able to identify homologues in the incomplete Salmonella genomes. P22 GtrA shares 93% sequence identity with proteins in S. typhi, serovar Typhimurium LT2, and S. paratyphi A. Guan et al. have proposed that this highly conserved group of proteins function as flippases, translocating glucosylated undecaprenyl phosphate from the cytoplasmic face to the periplasmic face of the inner membrane in gram-negative cells (25).
(ii) GtrB.
This 933-bp ORF (41.8 mol% GC) encodes a protein with a mass of 35,130 Da and a pI of 8.8. TMHMM revealed two transmembrane domains in the latter two-thirds of the protein, suggesting that both the amino and carboxy termini are cytoplasmic. This protein also was found to exhibit considerable sequence similarity to proteins from Shigella phages SfII, SfV, and SfX (Table 2). In addition, it has 86% sequence identity to a hypothetical 34.6-kDa protein (YFDH_ECOLI; GenBank accession no. P77293) associated with a defective prophage in the E. coli genome. The latter protein is defined as a dolichol-phosphate mannosyl transferase (EC 2.4.1.83; also known as dolichol-phosphate mannose synthase). In S. flexneri, two proteins, GenBank accession no. AAF09026.1 and AAC39272.1, are 87% identical to P22 GtrB. MEME/MAST analyses (8) revealed additional homologues to putative sugar transferases from Synechocystis (P74505), Bacillus (YKCC_BACSU, YKNOT_BACSU), and Streptomyces (CAA20162). Based upon the analysis of the phage SfX conversion genes, GtrB is probably a bactoprenol glucosyl transferase, catalyzing the transfer of glucose from an activated nucleotide intermediate to bactoprenol phosphate (4, 25).
(iii) GtrC.
This 1,458-bp ORF begins with a GTG and is preceded by a TAAGG sequence 9 bp upstream which resembles the consensus for a ribosome-binding site (TAAGGAGGT). The gene would encode a protein of 485 amino acids with a mass and pI of 55,233 Da and 8.7, respectively. Interestingly, the guanine-cytosine content, 31.9 mol%, is considerably less than that of the bulk DNA. BLASTP analysis failed to reveal any related sequences in the protein databases. Examination of the unpublished Salmonella genome sequences showed that, in each case, the gtrAB cassette was followed by a third gene encoding a large protein with multiple transmembrane domains. The product of the third gene in S. paratyphi A was shown to be a GtrC homologue exhibiting 98% sequence identity with the phage P22 protein. This suggests that either this bacterium harbors a phage closely related to P22, as suggested by the DNA similarity data, or that the gtrABC cassette originated from a defective prophage in S. paratyphi. It is worth noting that the base composition of the third gene in the conversion cassettes identified in Shigella and E. coli are all relatively low in GC content (4).
Searches for conserved motifs using Prosite (9, 30) revealed nothing, while Protein Families (10) at the Swiss Institute for Experimental Cancer Research ProfileScan server showed several conserved motifs, which were identified as PF00324 (amino acid permease), PF00344 (eubacterial secY protein), PF00662 (NADH-ubiquinone oxidoreductase), and PF00950 (ABC 3 transport family). An analysis for transmembrane proteins using TMHMM (63) revealed the presence of 11 potential membrane-spanning domains, with the carboxy terminus of the protein probably found in the periplasmic region of the cell. A similarly sized protein with 11 transmembrane domains has been proposed for the putative fucosamine acetylase encoded by P. aeruginosa phage D3, another serotype-converting phage (39; A. M. Kropinski, unpublished results). In addition, it shows superficial structural similarity to the glucosyl transferase gene (bgt) of S. flexneri phage SfX (73) and the chromosomal gtrI gene of this bacterium (3). Since SfX bgt and Shigella gtrI can result in serotype conversion we believe that this gene probably encodes the glucosyl transferase directly involved in serotype conversion while gtrA and gtrB are accessory genes. It would be expected that the specificity of the conversion would lie with this protein, since it must recognize different receptor molecules.
(iv) 23.
The antitermination protein involved in controlling late transcription is defined by gene 23. This protein is highly homologous to a group of proteins from phage including lambda, HK022, and PS34, suggesting similar types of regulation of late-gene expression in these morphologically different viruses.
Cloning putative conversion genes.
Using Martin Reese's Promoter Prediction by Neural Network program (http://www.fruitfly.org/seq_tools/promoter.html), a sequence (TTGATCGGTAACAACGATCAATTAACATGCATTA [promoter −35 and −10 consensus sequences shown in boldface and underlined]) with similarity to sigma70 promoters in E. coli was found 138 bases upstream of the gtrA gene. Using PCR, we amplified the gtrABC genes plus the putative promoter and ligated the amplicon into the pCRII-TOPO vector, which was subsequently transformed into E. coli TOP10 cells. Ampicillin-resistant clones were isolated, and the orientation of the insert relative to the lac promoter was determined by restriction endonuclease digestion. DNA from clones representing both orientations of the gtrABC cluster were electroporated into serovar Typhimurium LT2, and recombinant clones were tested for agglutination with anti-serogroup A sera. All agglutinated, confirming that (i) the amplified segment contained its own promoter and (ii) it encoded the genes necessary for complete seroconversion.
Evolutionary considerations.
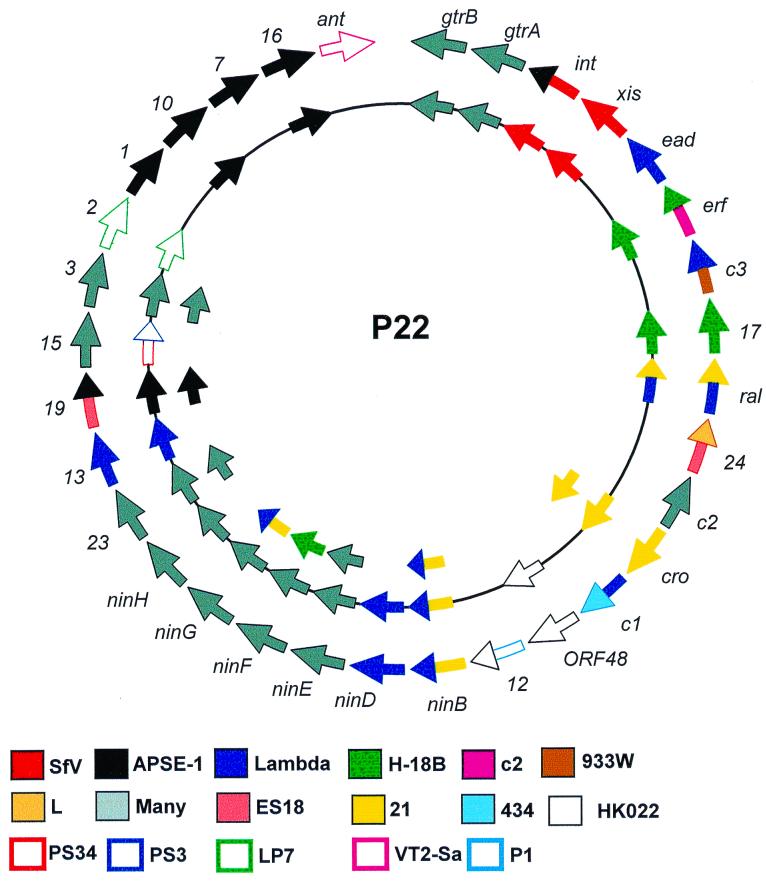
The phylogeny of phages has been discussed in two excellent reviews by Campbell (18) and Casjens et al. (19). Relationships have been hypothesized based on similar morphology, conservation of gene arrangement, ability to recombine, cross hybridization patterns, and sequence identity. Phage evolution can be thought of in terms of the recombinational exchange of gene modules or cassettes (19), and examination of the P22 protein data (Table 2 and Fig. 2) shows clear evidence that this type of evolutionary pathway occurred during P22 evolution. The acquisition of the xis-int-gtrA-gtrB cassette is such a case, where S. flexneri phage SfV also shares this module and similar morphology.
FIG. 2.
Diagrammatic representation of the potential phylogenetic significance of similar proteins in P22 and other phages. The outermost level indicates those proteins that share >30% sequence identity. The genes on the map line correspond to proteins which share ≥60% identity, while the inner arrows refer to proteins which share ≥90% sequence identity. A single color is employed where the P22 protein has only a single homologue. In cases where similarity is to two proteins, the arrowhead and shaft are differently colored. Where homology is shared with ≥3 different phage proteins, the gene is represented in grey.
Roger Hendrix and coworkers have stated that while conserved patterns which indicate familial relationships exist, the overall picture suggests that considerable intervirus or virus-host recombination has occurred, often between distant bacterial groups (26). Their proposition was that all double-stranded-DNA phage genomes are “mosaics with access, by horizontal exchange, to a large common genetic pool but in which access to the gene pool is not uniform for all phages.” This could occur following the superinfection of a common host cell by two different phages or through recombination between superinfecting and resident prophage genomes. Those phages that had the ability to infect different bacterial hosts could then pass on the new genomic segments, ultimately resulting in unrelated bacteriophages possessing homologous genes. The sequence of P22 is yet another example of the extent to which this theory of viral evolution is supported by the sequence data. A significant degree of protein similarity has been found between P22 proteins and the products of genes from members of the families Podoviridae (APSE-1, L, 933W, H-19B, LP-7, PS3, PS34, ES18, and SfV), Siphoviridae (λ, 21, c2, HK022, 434, and VT2-Sa), Myoviridae (P1 and SfII), and even Inoviridae (SfX) (Table 3, Fig. 2). The only unifying characteristic is that here the majority of homologues are to proteins of phages infecting gram-negative bacteria. In specific cases, such as genes 17, cro, and 13(S) and morphogenesis genes 10 and 16, a clear relationship to a single virus isolate can be shown, while the nin genes, c2, and gene 3 appear to be related to genes found in a variety of previously characterized phages.
The best evidence for this genetic reassortment is its apparent randomness. While the holin gene (13) is clearly homologous to that in coliphage lambda, the cognate lysozyme gene (19) is not. The nin region shows genes which are clearly lambdoid interspersed with genes, such as ninX and ninY, which are not. Furthermore, while some morphogenesis genes (1, 10, 7, and 16) show sequence similarity to only APSE-1, the intervening genes, including those for scaffold (8) and coat proteins (5), do not. Last, we have the apparent illogic of interspersing the major right and left primary transcripts with sieB and sieA-mnt-arc-ant genes, which are oriented in the opposite direction. In the latter case in particular, the sieA-ant gene cluster separates the bulk of the morphogenesis genes from the tailspike protein. These sequences have recently been termed morons by Juhala and colleagues (36).
In all of the phages examined to date, a considerable percentage of the ORFs do not encode proteins with homologues in the current databases. This makes it imperative that the current databases be stocked with good-quality annotated sequence data, that complete phage genomes be examined, and that conclusions be drawn from complete rather than partial sequence data.
ACKNOWLEDGMENTS
This research was funded by a grant (to A.M.K.) from the Natural Sciences and Engineering Research Council of Canada.
Thanks are extended to Robert Eves, DNA Sequencing Specialist at Cortec DNA Service Laboratories (Queen's University), for his technical assistance. Thanks are also extended to H. Backhaus, P. B. Berget, S. Casjens, C. A. Conlin, B. Dreiseikelmann, N. C. Franklin, R. W. Hendrix, G. Hobom, B. Hofer, M. Kroeger, C. G. Miller, J. B. Petri, A. R. Poteete, D. Rennell, M. M. Susskind, B. Umlauf, P. Youderian, and other members of the P22 community for making this project possible.
REFERENCES
- 1.Ackermann H W. Tailed bacteriophages: the order Caudovirales. Adv Virus Res. 1998;51:135–201. doi: 10.1016/S0065-3527(08)60785-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams M D. Bacteriophages. New York, N.Y: Interscience Publishers, Inc.; 1959. [Google Scholar]
- 3.Adhikari P, Allison G, Whittle B, Verma N K. Serotype 1a O-antigen modification: molecular characterization of the genes involved and their novel organization in the Shigella flexnerichromosome. J Bacteriol. 1999;181:4711–4718. doi: 10.1128/jb.181.15.4711-4718.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allison G E, Verma N K. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 2000;8:17–23. doi: 10.1016/s0966-842x(99)01646-7. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 6.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–4022. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Backhaus H, Petri J B. Sequence analysis of a region from the early right operon in phage P22 including the replication genes 18 and 12. Gene. 1984;32:289–303. doi: 10.1016/0378-1119(84)90004-0. [DOI] [PubMed] [Google Scholar]
- 8.Bailey T L, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 9.Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1992;11:2013–2088. doi: 10.1093/nar/20.suppl.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bateman A, Birney E, Durbin R, Eddy S R, Finn R D, Sonnhammer E L. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benson N R, Goldman B S. Rapid mapping in Salmonella typhimuriumwith Mud-P22 prophages. J Bacteriol. 1992;174:1673–1681. doi: 10.1128/jb.174.5.1673-1681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson N R, Roth J. A Salmonellaphage-P22 mutant defective in abortive transduction. Genetics. 1997;145:17–27. doi: 10.1093/genetics/145.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berget P B, Poteete A R, Sauer R T. Control of phage P22 tail protein expression by transcription termination. J Mol Biol. 1983;164:561–572. doi: 10.1016/0022-2836(83)90050-5. [DOI] [PubMed] [Google Scholar]
- 14.Bezdek M, Amati P. Properties of P22 and a related Salmonella typhimuriumphage. I. General features and host specificity. Virology. 1967;31:272–278. doi: 10.1016/0042-6822(67)90171-7. [DOI] [PubMed] [Google Scholar]
- 15.Botstein D, Levine M. Intermediates in the synthesis of phage P22 DNA. Cold Spring Harbor Symp Quant Biol. 1968;33:659–667. doi: 10.1101/sqb.1968.033.01.075. [DOI] [PubMed] [Google Scholar]
- 16.Brendel V, Hamm G H, Trifonov E N. Terminators of transcription with RNA polymerase from Escherichia coli: what they look like and how to find them. J Biomol Struct Dyn. 1986;3:705–723. doi: 10.1080/07391102.1986.10508457. [DOI] [PubMed] [Google Scholar]
- 17.Brendel V, Trifonov E N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984;12:4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell A. Comparative molecular biology of lambdoid phages. Annu Rev Microbiol. 1994;48:193–222. doi: 10.1146/annurev.mi.48.100194.001205. [DOI] [PubMed] [Google Scholar]
- 19.Casjens S, Hatfull G F, Hendrix R. Evolution of dsDNA tailed bacteriophage genomes. In: Koonin E, editor. Seminars in virology. London, United Kingdom: Academic Press; 1992. pp. 383–397. [Google Scholar]
- 20.Chisholm R L, Deans R J, Jackson E N, Jackson D A, Rutila J E. A physical gene map of the bacteriophage P22 late region: genetic analysis of cloned fragments of P22 DNA. Virology. 1980;102:172–189. doi: 10.1016/0042-6822(80)90079-3. [DOI] [PubMed] [Google Scholar]
- 21.Eddy S R, Durbin R. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Mabrouk N, Lisacek F. Very fast identification of RNA motifs in genomic DNA. Application to tRNA search in the yeast genome. J Mol Biol. 1996;264:46–55. doi: 10.1006/jmbi.1996.0622. [DOI] [PubMed] [Google Scholar]
- 23.Fields D S, He Y, Al-Uzri A Y, Stormo G D. Quantitative specificity of the Mnt repressor. J Mol Biol. 1997;271:178–194. doi: 10.1006/jmbi.1997.1171. [DOI] [PubMed] [Google Scholar]
- 24.Goodrich J A, Schwartz M L, McClure W R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF) Nucleic Acids Res. 1990;18:4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan S, Bastin D A, Verma N K. Functional analysis of the O antigen glucosylation gene cluster of Shigella flexneribacteriophage SfX. Microbiology. 1999;145:1263–1273. doi: 10.1099/13500872-145-5-1263. [DOI] [PubMed] [Google Scholar]
- 26.Hendrix R, Smith M C, Burns R N, Ford M E, Hatfull G F. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho Y S, Pfarr D, Strickler J, Rosenberg M. Characterization of the transcription activator protein C1 of bacteriophage P22. J Biol Chem. 1992;267:14388–14397. [PubMed] [Google Scholar]
- 28.Ho Y S, Rosenberg M. Characterization of a third, cII-dependent, coordinately activated promoter on phage lambda involved in lysogenic development. J Biol Chem. 1985;260:11838–11844. [PubMed] [Google Scholar]
- 29.Hofer B, Ruge M, Dreiseikelmann B. The superinfection exclusion gene (sieA) of bacteriophage P22: identification and overexpression of the gene and localization of the gene product. J Bacteriol. 1995;177:3080–3086. doi: 10.1128/jb.177.11.3080-3086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofmann K, Bucher P, Falquet L, Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofmann K, Stoffel W. TMbase—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166–175. [Google Scholar]
- 32.Huan P T, Bastin D A, Whittle B L, Lindberg A A, Verma N K. Molecular characterization of the genes involved in O-antigen modification, attachment, integration and excision in Shigella flexneribacteriophage SfV. Gene. 1997;195:217–227. doi: 10.1016/s0378-1119(97)00143-1. [DOI] [PubMed] [Google Scholar]
- 33.Ilyina T V, Gorbalenya A E, Koonin E V. Organization and evolution of bacterial and bacteriophage primase-helicase systems. J Mol Evol. 1992;34:351–357. doi: 10.1007/BF00160243. [DOI] [PubMed] [Google Scholar]
- 34.International Committee on the Taxonomy of Viruses. Virus taxonomy: classification and nomenclature of viruses. 1999. Seventh report of the International Committee on Taxonomy of Viruses. Edited by M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. Carstens, M. K. Estes, S. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. Wickner. Academic Press, New York, N.Y. [Google Scholar]
- 35.Iseki S, Kashiwagi K. Induction of somatic antigen 1 by bacteriophage in Salmonellagroup B. Proc Jpn Acad. 1955;31:558–564. [Google Scholar]
- 36.Juhala R J, Ford M E, Duda R L, Youlton A, Hatfull G F, Hendrix R W. Genetic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 37.Kitamura J, Mise K. A new generalized transducing phage in Salmonella. Jpn J Med Sci Biol. 1970;23:99–102. doi: 10.7883/yoken1952.23.99. [DOI] [PubMed] [Google Scholar]
- 38.Kropinski A M. Bacteriophage DNA: correlation of buoyant density, melting temperature, and the chemically determined base composition. J Virol. 1974;13:753–756. doi: 10.1128/jvi.13.3.753-756.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuzio J, Kropinski A M. O-antigen conversion in Pseudomonas aeruginosaPAO1 by bacteriophage D3. J Bacteriol. 1983;155:203–212. doi: 10.1128/jb.155.1.203-212.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leong J M, Nunes-Duby S, Lesser C F, Youderian P, Susskind M M, Landy A. The phi 80 and P22 attachment sites. Primary structure and interaction with Escherichia coliintegration host factor. J Biol Chem. 1985;260:4468–4477. [PubMed] [Google Scholar]
- 41.Lowe T M, Eddy S R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lukashin A, Borodovsky M. GeneMark.hmm: a new solution for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mann B A, Slauch J M. Transduction of low-copy number plasmids by bacteriophage P22. Genetics. 1997;146:447–456. doi: 10.1093/genetics/146.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1992. [Google Scholar]
- 45.Nakonechny W S, Teschke C M. GroEL and GroES control of substrate flux in the in vivo folding pathway of phage P22 coat protein. J Biol Chem. 1998;273:27236–27244. doi: 10.1074/jbc.273.42.27236. [DOI] [PubMed] [Google Scholar]
- 46.Neal B L, Brown P K, Reeves P R. Use of Salmonella phage P22 for transduction in Escherichia coli. J Bacteriol. 1993;175:7115–7118. doi: 10.1128/jb.175.21.7115-7118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oberto J, Sloan S B, Weisberg R A. A segment of the phage HK022 chromosome is a mosaic of other lambdoid chromosomes. Nucleic Acids Res. 1994;22:354–356. doi: 10.1093/nar/22.3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poteete A R. Bacteriophage P22. In: Calendar R, editor. The bacteriophages. New York, N.Y: Plenum Press; 1988. pp. 647–682. [Google Scholar]
- 49.Ranade K, Poteete A R. Superinfection exclusion (sieB) genes of bacteriophages P22 and lambda. J Bacteriol. 1993;175:4712–4718. doi: 10.1128/jb.175.15.4712-4718.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhoades M, Thomas C A J. The P22 bacteriophage DNA molecule. II. Circular intracellular forms. J Mol Biol. 1968;37:41–61. doi: 10.1016/0022-2836(68)90072-7. [DOI] [PubMed] [Google Scholar]
- 51.Rundell K, Shuster C W. Membrane-associated nucleotide sugar reactions: influence of mutations affecting lipopolysaccharide on the first enzyme of O-antigen synthesis. J Bacteriol. 1975;123:928–936. doi: 10.1128/jb.123.3.928-936.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 53.Sanderson K E, Hessel A, Liu S-L, Rudd K E. The genetic maps of Salmonella typhimurium, edition VIII. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1903–1999. [Google Scholar]
- 54.Schaefer K L, McClure W R. Antisense RNA control of gene expression in bacteriophage P22. I. Structures of sar RNA and its target, ant mRNA. RNA. 1997;3:141–156. [PMC free article] [PubMed] [Google Scholar]
- 55.Schander-Mulfinger U E, Schmieger H. Growth of Salmonella bacteriophage P22 in Escherichia coli dna(Ts) mutants. J Bacteriol. 1980;143:1042–1045. doi: 10.1128/jb.143.2.1042-1045.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schicklmaier P, Schmieger H. Sequence comparison of the genes for immunity, DNA replication, and cell lysis of the P22-related Salmonellaphages ES18 and L. Gene. 1997;195:93–100. doi: 10.1016/s0378-1119(97)00182-0. [DOI] [PubMed] [Google Scholar]
- 57.Schicklmaier P, Wieland T, Schmieger H. Molecular characterization and module composition of P22-related Salmonellaphage genomes. J Biotechnol. 1999;73:185–194. doi: 10.1016/s0168-1656(99)00120-0. [DOI] [PubMed] [Google Scholar]
- 58.Schwarz J J, Berget P B. The isolation and sequence of missense and nonsense mutations in the cloned bacteriophage P22 tailspike protein gene. Genetics. 1989;121:635–649. doi: 10.1093/genetics/121.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seckler R. Folding and function of repetitive structure in the homotrimeric phage P22 tailspike protein. J Struct Biol. 1998;122:216–222. doi: 10.1006/jsbi.1998.3974. [DOI] [PubMed] [Google Scholar]
- 60.Sibbald M J, Kropinski A M. Transfer RNA genes and their significance to codon usage in the Pseudomonas aeruginosalamboid bacteriophage D3. Can J Microbiol. 1999;45:791–796. [PubMed] [Google Scholar]
- 61.Skalka A, Hanson P. Comparison of the distribution of nucleotides and common sequences in deoxyribonucleic acid from selected bacteriophages. J Virol. 1972;9:583–593. doi: 10.1128/jvi.9.4.583-593.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith-Mungo L, Chan I T, Landy A. Structure of the P22 att site. Conservation and divergence in the lambda motif of recombinogenic complexes. J Biol Chem. 1994;269:20798–20805. [PubMed] [Google Scholar]
- 63.Sonnhammer E L L, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. In: Glasgow J, Littlejohn T, Major F, Lathrop R, Sankoff D, Sensen C, editors. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. Menlo Park, Calif: AAAI Press; 1998. pp. 175–182. [PubMed] [Google Scholar]
- 64.Spanova A. Comparison of permuted region lengths in the genomes of related Salmonella typhimuriumphages P22 and L. Folia Microbiol. 1992;37:188–192. doi: 10.1007/BF02933145. [DOI] [PubMed] [Google Scholar]
- 65.Steinbacher S, Miller S, Baxa U, Weintraub A, Seckler R. Interaction of Salmonellaphage P22 with its O-antigen receptor studied by X-ray crystallography. Biol Chem. 1997;378:337–343. doi: 10.1515/bchm.1997.378.3-4.337. [DOI] [PubMed] [Google Scholar]
- 66.Tatusova T A, Madden T L. Blast 2 sequences—a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 67.Thomas C A J, Kelly T J J, Rhoades M. The intracellular forms of T7 and P22 DNA molecules. Cold Spring Harbor Symp Quant Biol. 1968;33:417–424. doi: 10.1101/sqb.1968.033.01.048. [DOI] [PubMed] [Google Scholar]
- 68.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thuman-Commike P A, Greene B, Malinski J A, King J, Chiu W. Role of the scaffolding protein in P22 procapsid size determination suggested by T = 4 and T = 7 procapsid structures. Biophys J. 1998;74:559–568. doi: 10.1016/S0006-3495(98)77814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tuma R, Parker M H, Weigele P, Sampson L, Sun Y, Krishna N R, Casjens S, Thomas G J J, Prevelige P E J. A helical coat protein recognition domain of the bacteriophage P22 scaffolding protein. J Mol Biol. 1998;281:81–94. doi: 10.1006/jmbi.1998.1916. [DOI] [PubMed] [Google Scholar]
- 71.Tuma R, Prevelige P E J, Thomas G J J. Mechanism of capsid maturation in a double-stranded DNA virus. Proc Natl Acad Sci USA. 1998;95:9885–9890. doi: 10.1073/pnas.95.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van der Wilk F, Dullemans A M, Verbeek M, van den Heuvel J F. Isolation and characterization of APSE-1, a bacteriophage infecting the secondary endosymbiont of Acyrthosiphon pisum. Virology. 1999;262:104–113. doi: 10.1006/viro.1999.9902. [DOI] [PubMed] [Google Scholar]
- 73.Verma N K, Verma D J, Huan P T, Lindberg A A. Cloning and sequencing of the glucosyl transferase-encoding gene from converting bacteriophage X (SfX) of Shigella flexneri. Gene. 1999;129:99–101. doi: 10.1016/0378-1119(93)90702-5. [DOI] [PubMed] [Google Scholar]
- 74.Weintraub A, Johnson B N, Stocker B A, Lindberg A A. Structural and immunochemical studies of the lipopolysaccharides of Salmonellastrains with both antigen O4 and antigen O9. J Bacteriol. 1992;174:1916–1922. doi: 10.1128/jb.174.6.1916-1922.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wickner S. DNA-dependent ATPase activity associated with phage P22 gene 12 protein. J Biol Chem. 1984;259:14038–14043. [PubMed] [Google Scholar]
- 76.Yamamoto K R, Alberts B M, Benzinger R, Lawhorne L, Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970;40:734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]
- 77.Youderian P, Sugiono P, Brewer K L, Higgins N P, Elliott T. Packaging specific segments of the Salmonellachromosome with locked-in Mud-P22 prophages. Genetics. 1988;118:581–592. doi: 10.1093/genetics/118.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Young B G, Fukazawa Y, Hartman P. A P22 bacteriophage mutant defective in antigen conversion. Virology. 1964;23:279–283. doi: 10.1016/0042-6822(64)90296-x. [DOI] [PubMed] [Google Scholar]
- 79.Zahn K, Landy A. Modulation of lambda integrase synthesis by rare arginine tRNA. Mol Microbiol. 1996;21:69–76. doi: 10.1046/j.1365-2958.1996.6201335.x. [DOI] [PubMed] [Google Scholar]
- 80.Zinder N D, Lederberg J. Genetic exchange in Salmonella. J Bacteriol. 1952;64:679. doi: 10.1128/jb.64.5.679-699.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]