Abstract
The methyl-accepting chemotaxis proteins (MCPs) are concentrated at the cell poles in an evolutionarily diverse panel of bacteria and an archeon. In elongated cells, the MCPs are located both at the poles and at regions along the length of the cells. Together, these results suggest that MCP location is evolutionarily conserved.
Prokaryotic processes, such as cell division and chemotaxis, often depend on the asymmetric or nonuniform distribution of proteins to subcellular locations (for a recent review on this subject, see reference 31). The methyl-accepting chemotaxis proteins (MCPs) are components of the chemotactic response system in Bacteria and Archaea (1, 12, 16, 35). The subcellular location of MCPs has been determined for four prokaryotes: Caulobacter crescentus (2), Escherichia coli (23, 24), Bacillus subtilis (17), and Rhodobacter sphaeroides (14). In these species, the MCPs are concentrated primarily at the cell poles. The location of the MCPs may be involved in the regulation of E. coli chemotaxis (4, 8, 20), as MCP clustering has been implicated in regulating ligand binding and signaling in this bacterium (7, 10, 19, 21, 22, 34). An understanding of MCP localization in a wide range of other bacteria and archaea could illuminate the relationship between MCP location and regulation of chemotaxis.
Previous microscopy studies that have explored the subcellular location of MCPs have utilized anti-MCP antibodies raised against the MCP of the organism being studied (2, 17, 23, 24). Hazelbauer and coworkers have reported that antibodies raised against Trg, an E. coli MCP, can be used in Western blottings to identify proteins antigenically related to the E. coli MCPs from a number of different organisms (1, 26, 27). These results suggest that anti-Trg antibodies could be used in fluorescence microscopy experiments as convenient reagents to visualize the subcellular location of MCPs in a variety of organisms.
To explore the conservation of MCP location, we selected a representative panel of bacterial species and an archaeon. The species were chosen to represent chemotactically active strains that, relative to E. coli, are either evolutionarily (5, 18) similar (e.g., Vibrio furnissii [36]), divergent (e.g., Spirochaeta aurantia [11]), or highly divergent (e.g., Halobacterium salinarium [29, 33]).
Cells were grown in the following liquid media supplemented with 10 mM d-galactose: E. coli, Luria broth (LB) (1% tryptone, 0.5% yeast extract, 0.5% NaCl); V. furnissii, high-salt LB (1% tryptone, 0.5% yeast extract, 2% NaCl); S. aurantia, GTY (10 mM d-glucose, 4% tryptone, 2% yeast extract, 10 mM phosphate buffer [pH 7.0]); H. salinarium, complex medium (27 mM KCl, 166 mM MgSO4, 10 mM sodium citrate, 4.3 M NaCl, 1% peptone, 2 mM CaCl2). We used anti-Trg antibodies to determine the location of MCPs by fluorescence microscopy (23, 24).
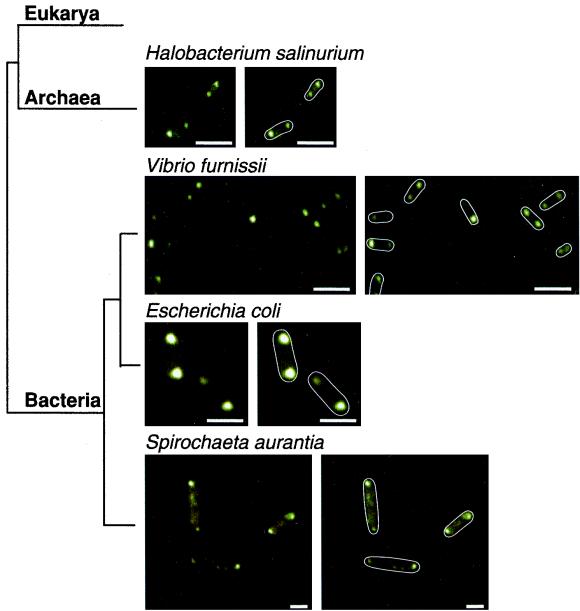
As has been seen in investigations of C. crescentus (2), E. coli (10, 23, 24), B. subtilis (16), and R. sphaeroides (14), fluorescence is observed primarily at the poles of both V. furnissii and S. aurantia (Fig. 1), which suggests that the MCPs are localized to the cell poles in these bacteria. Similar results were obtained with an anti-Tsr antibody (data not shown). Interestingly, a polarly localized fluorescence pattern also is observed with the archaeal species H. salinarium (Fig. 1), despite the divergence between this organism and E. coli (5, 18). Thus, in each of the organisms studied, proteins that are antigenically related to the E. coli MCPs are localized. The predominant sites of concentration are the cell poles.
FIG. 1.
Schematic representation of relative evolutionary relationships (5, 18) and fluorescence patterns for cells treated with anti-Trg antibody (1:400) followed by a fluorescein-labeled anti-rabbit immunoglobulin G secondary antibody (1:200). Duplicate figures on the right display outlines of the cells. The result with E. coli AW405 confirms our previous results (23). Images such as those presented were obtained in >70% of the cells observed in at least three independent experiments. Bars, 2 μm. Branch lengths are not based on phylogenetic distances.
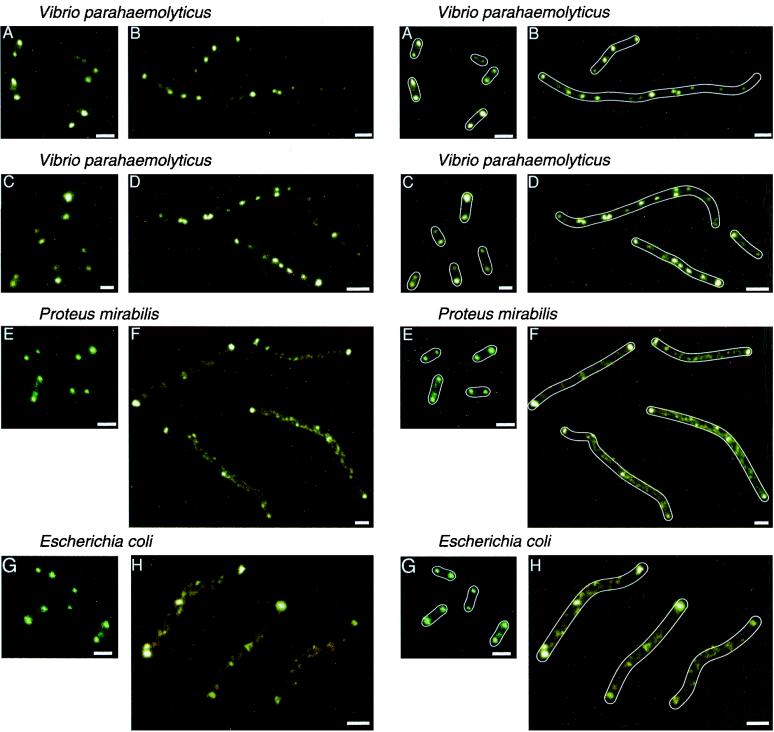
We next investigated the location of MCPs in elongated cells. Bacterial cell elongation occurs after treatment with β-lactam antibiotics, such as cephalexin (13, 24, 28, 30), from mutations in the cell division gene lonS (32), or upon differentiation to swarmer cells (3, 6, 9, 15, 25). We have previously reported that the MCPs of cephalexin-treated E. coli cells are localized not only to the cell poles but also at intervals along the length of the elongated cell (24). Treatment of E. coli with cephalexin, therefore, results in deviation from the primarily polar MCP localization observed in the normal-sized cells. To investigate the MCP pattern in elongated cells generated by means other than cephalexin treatment, we conducted fluorescence microscopy experiments in a lonS mutant of Vibrio parahaemolyticus LM1017 (32) and swarmer cells of V. parahaemolyticus BB22, Proteus mirabilis BB2000, and E. coli ATCC 25922.
The MCPs of the normal-sized cells from which the elongated cells were generated were located at the cell poles in each case (Fig. 2A, C, E, and G). The fluorescence patterns of V. parahaemolyticus LM4420, an elongated mutant deficient in LonS (32), revealed that the MCPs are localized not only at the poles but also at intervals along the cell (Fig. 2B). Similar results were observed in the wild-type naturally occurring swarmer cells of V. parahaemolyticus; the MCPs are concentrated both at the cell poles and at intervals (Fig. 2D). In contrast, the fluorescence intensity at the poles of swarmer cells of P. mirabilis (Fig. 2F) and E. coli (Fig. 2H) is more pronounced than it is at intervals along the cells. This suggests that the concentration of MCPs at the poles is higher than that along the length of these cells. This pattern of MCP localization may be related to the observation that E. coli swarmer cells run essentially all the time (R. Harshey, unpublished data); the probability that the transient phosphorylated CheY (which causes tumbling) can migrate from the site where it is generated (MCPs) to the site where it acts (the flagella) is low. In contrast, E. coli cephalexin-produced filaments have MCP clusters throughout, and both running and stopping (the equivalent of tumbling in filamentous bacteria) are observed commonly (24).
FIG. 2.
Comparison of the MCP fluorescence patterns in normal-sized and elongated cells. (A) Normal-sized V. parahaemolyticus LM1017. (B) V. parahaemolyticus lonS mutant LM4420 derived from LM1017. (C) Normal-sized V. parahaemolyticus BB22. (D) Elongated swarmer cells of V. parahaemolyticus BB22 taken from 2% agar high-salt LB plates at 30°C. (E) Normal-sized P. mirabilis. (F) P. mirabilis swarmer cells from 1.5% agar LB plates at 37°C. (G) Normal-sized E. coli ATCC 25922. (H) E. coli ATCC 25922 swarmer cells from 0.55% agar LB plates supplemented with 0.5% glucose at 30°C. Duplicate figures on the right display drawn outlines of the cells. Images such as those presented were obtained in >90% of the cells observed in three independent experiments. Bars, 2 μm.
The regulation of E. coli chemotaxis may be modulated by proximity of the MCPs (4, 8, 20). Here we report that the MCPs are polarly localized in a panel of bacteria that are evolutionarily similar (Vibrio sp. and P. mirabilis) and divergent (S. aurantia) from E. coli. The MCPs also are localized at the poles in a more distantly related archaeon (H. salinarium). Additionally, the MCPs of all of the elongated cells that we studied are located at the poles and at intervals along the cell, although the distribution of MCPs varies (Fig. 2). Moreover, in both normal-sized and elongated cells, the MCPs typically are localized and not distributed randomly. These data suggest that, as with C. crescentus, E. coli, B. subtilis, and R. sphaeroides chemotaxis, MCP clustering may be involved, here also, in the regulation of chemotaxis. Investigations of MCP localization in organisms such as E. coli, therefore, may lead to a greater understanding of the regulation of chemotactic responses in other bacteria and in archaea.
Acknowledgments
We are grateful to Peter Greenberg for helpful comments. For supplying antibodies, we thank Gerald Hazelbauer (anti-Trg) and Sandy Parkinson (anti-Tsr). Bacterial strains originated from Robert Belas (P. mirabilis BB2000 and V. parahaemolyticus BB22), P. Greenberg (S. aurantia JI), Rasika Harshey (E. coli ATCC 25922), Linda McCarter (V. parahaemolyticus LM1017 and LM4420), Saul Roseman (V. furnissii SR1514), and John Spudich (H. salinarium S9). Sebastian Bednarek provided access to fluorescence microscopy equipment.
This research was supported in part by grants from the NSF (IBN-9807789 to J.A.) and the NIH (GM52214 to J.A.; GM-55984 to L.L.K.). J.E.G. thanks the NIH Biotechnology Training Program for a predoctoral fellowship (T32GM08349). A.C.L. acknowledges the NSF for support.
REFERENCES
- 1.Alam M, Hazelbauer G L. Structural features of methyl-accepting taxis proteins conserved between archaebacteria and eubacteria revealed by antigenic cross-reaction. J Bacteriol. 1991;173:5837–5842. doi: 10.1128/jb.173.18.5837-5842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alley M R K, Maddock J R, Shapiro L. Polar localization of a bacterial chemoreceptor. Genes Dev. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- 3.Belas R, Erskine D, Flaherty D. Proteus mirabilis mutants defective in swarmer cell differentiation and multicellular behavior. J Bacteriol. 1991;173:6279–6288. doi: 10.1128/jb.173.19.6279-6288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bray D, Levin M D, Morton-Firth C J. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 5.Brown J R, Doolittle W F. Archaea and the prokaryotic-to-eukaryotic transition. Microbiol Mol Biol Rev. 1997;61:456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkart M, Toguchi A, Harshey R M. The chemotaxis system, but not chemotaxis, is essential for swarming motility in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2568–2573. doi: 10.1073/pnas.95.5.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cochran A G, Kim P S. Imitation of Escherichia coli aspartate receptor signaling in engineered dimers of the cytoplasmic domain. Science. 1996;271:1113–1116. doi: 10.1126/science.271.5252.1113. [DOI] [PubMed] [Google Scholar]
- 8.Duke T A J, Bray D. Heightened sensitivity of a lattice of membrane receptors. Proc Natl Acad Sci USA. 1999;96:10104–10108. doi: 10.1073/pnas.96.18.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser G M, Hughes C. Swarming motility. Curr Opin Microbiol. 1999;2:630–635. doi: 10.1016/s1369-5274(99)00033-8. [DOI] [PubMed] [Google Scholar]
- 10.Gestwicki J E, Strong L E, Kiessling L L. Tuning chemotactic responses with synthetic multivalent ligands. Chem Biol. 2000;7:583–591. doi: 10.1016/s1074-5521(00)00002-8. [DOI] [PubMed] [Google Scholar]
- 11.Goulbourne E A, Jr, Greenberg E P. Chemotaxis of Spirochaeta aurantia: involvement of membrane potential in chemosensory signal transduction. J Bacteriol. 1981;148:837–844. doi: 10.1128/jb.148.3.837-844.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grebe T W, Stock J. Bacterial chemotaxis: the five sensors of a bacterium. Curr Biol. 1998;8:R154–R157. doi: 10.1016/s0960-9822(98)00098-0. [DOI] [PubMed] [Google Scholar]
- 13.Greenwood D, O'Grady F. Comparison of the response of Escherichia coli and Proteus mirabilis to seven β-lactam antibiotics. J Infect Dis. 1973;128:211–222. doi: 10.1093/infdis/128.2.211. [DOI] [PubMed] [Google Scholar]
- 14.Harrison D M, Skidmore J, Armitage J P, Maddock J R. Localization and environmental regulation of MCP-like proteins in Rhodobacter sphaeroides. Mol Microbiol. 1999;31:885–892. doi: 10.1046/j.1365-2958.1999.01226.x. [DOI] [PubMed] [Google Scholar]
- 15.Harshey R M. Bees aren't the only ones: swarming in Gram-negative bacteria. Mol Microbiol. 1994;13:389–394. doi: 10.1111/j.1365-2958.1994.tb00433.x. [DOI] [PubMed] [Google Scholar]
- 16.Kathariou S, Greenberg E P. Chemoattractants elicit methylation of specific polypeptides in Spirochaeta aurantia. J Bacteriol. 1983;156:95–100. doi: 10.1128/jb.156.1.95-100.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirby J R, Niewold T B, Maloy S, Ordal G W. CheB is required for behavioral responses to negative stimuli during chemotaxis in Bacillus subtilis. Mol Microbiol. 2000;35:44–57. doi: 10.1046/j.1365-2958.2000.01676.x. [DOI] [PubMed] [Google Scholar]
- 18.Koonin E V, Galperin M Y. Prokaryotic genomes: the emerging paradigm of genome-based microbiology. Curr Opin Genet Dev. 1997;7:757–763. doi: 10.1016/s0959-437x(97)80037-8. [DOI] [PubMed] [Google Scholar]
- 19.Le Moual H, Quang T, Koshland D E., Jr Methylation of the Escherichia coli chemotaxis receptors: intra- and interdimer mechanisms. Biochemistry. 1997;36:13441–13448. doi: 10.1021/bi9713207. [DOI] [PubMed] [Google Scholar]
- 20.Levit M N, Liu Y, Stock J B. Stimulus response coupling in bacterial chemotaxis: receptor dimers in signaling arrays. Mol Microbiol. 1998;30:459–466. doi: 10.1046/j.1365-2958.1998.01066.x. [DOI] [PubMed] [Google Scholar]
- 21.Li G, Weis R M. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Levit M, Lurz R, Surette M G, Stock J B. Receptor-mediated protein kinase activation and the mechanism of transmembrane signaling in bacterial chemotaxis. EMBO J. 1997;16:7231–7240. doi: 10.1093/emboj/16.24.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maddock J R, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 24.Maki N, Gestwicki J E, Lake E M, Kiessling L L, Adler J. Motility and chemotaxis in filamentous cells of Escherichia coli. J Bacteriol. 2000;182:4337–4342. doi: 10.1128/jb.182.15.4337-4342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarter L, Silverman M. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol Microbiol. 1990;4:1057–1062. doi: 10.1111/j.1365-2958.1990.tb00678.x. [DOI] [PubMed] [Google Scholar]
- 26.Morgan D G, Baumgartner J W, Hazelbauer G L. Proteins antigenically related to methyl-accepting chemotaxis proteins of Escherichia coli detected in a wide range of bacterial species. J Bacteriol. 1993;175:133–140. doi: 10.1128/jb.175.1.133-140.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowlin D M, Nettleton D O, Ordal G W, Hazelbauer G L. Chemotactic transducer proteins of Escherichia coli exhibit homology with methyl-accepting proteins from distantly related bacteria. J Bacteriol. 1985;163:262–266. doi: 10.1128/jb.163.1.262-266.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rolinson G N. Effect of β-lactam antibiotics on bacterial cell growth rate. J Gen Microbiol. 1980;120:317–323. doi: 10.1099/00221287-120-2-317. [DOI] [PubMed] [Google Scholar]
- 29.Rudolph J, Oesterhelt D. Deletion analysis of the che operon in the archaeon Halobacterium salinarium. J Mol Biol. 1996;258:548–554. doi: 10.1006/jmbi.1996.0267. [DOI] [PubMed] [Google Scholar]
- 30.Segall J E, Ishihara A, Berg H C. Chemotactic signaling in filamentous cells of Escherichia coli. J Bacteriol. 1985;161:51–59. doi: 10.1128/jb.161.1.51-59.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shapiro L, Losick R. Dynamic spatial regulation in the bacterial cell. Cell. 2000;100:89–98. doi: 10.1016/s0092-8674(00)81686-4. [DOI] [PubMed] [Google Scholar]
- 32.Stewart B J, Enos-Berlage J L, McCarter L L. The lonS gene regulates swarmer cell differentiation of Vibrio parahaemolyticus. J Bacteriol. 1997;179:107–114. doi: 10.1128/jb.179.1.107-114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sundberg S A, Alam M, Lebert M, Spudich J L, Oesterhelt D, Hazelbauer G L. Characterization of Halobacterium halobium mutants defective in taxis. J Bacteriol. 1990;172:2328–2335. doi: 10.1128/jb.172.5.2328-2335.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatsuno I, Homma M, Oosawa K, Kawagishi I. Signaling by the Escherichia coli aspartate chemoreceptor Tar with a single cytoplasmic domain per dimer. Science. 1996;274:423–425. doi: 10.1126/science.274.5286.423. [DOI] [PubMed] [Google Scholar]
- 35.Yao V J, Spudich J L. Primary structure of an archeabacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc Natl Acad Sci USA. 1992;89:11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu C, Bassler B L, Roseman S. Chemotaxis of the marine bacterium Vibrio furnissii to sugars: a potential mechanism for initiating the chitin catabolic cascade. J Biol Chem. 1993;268:9405–9409. [PubMed] [Google Scholar]