Abstract
High fat diet (HFD) consumption alters the synchronized circadian timing system resulting in harmful loss, gain or shift of transcriptional oscillations. The aryl hydrocarbon receptor (AhR) shares structural homology to clock genes, containing both PAS domains and basic helix-loop helix structural motifs, allowing for interaction with components of the primary circadian feedback loop. Activation of AhR alters circadian rhythmicity, primarily through inhibition of Clock/Bmal1-mediated regulation of Per1. AhR-deficient mice are protected from diet-induced metabolic dysfunction, exhibiting enhanced insulin sensitivity and glucose tolerance. This study examined whether AhR haploinsufficiency can also protect against diet-induced alterations in rhythm. After feeding AhR+/+ and AhR+/− mice an HFD (60% fat) for 15 weeks, samples were collected every 4 hours over a 24-hour period. HFD altered the rhythm of serum glucose and the metabolic transcriptome, including hepatic nuclear receptors Rev-erba and PPARy in wild-type c57bl6/j mice. AhR reduction provided protection against diet-induced transcriptional oscillation changes; serum glucose and metabolic gene rhythms were protected from the disruption caused by HFD feeding. These data highlight the critical role of AhR signaling in the regulation of metabolism and provide a potential therapeutic target for diseases characterized by rhythmic desynchrony.
Keywords: Aryl hydrocarbon receptor, circadian disruption, high fat diet
Introduction
Chronic low-level exposure to dioxins is linked to insulin resistance and development of Type 2 Diabetes through activation of the ligand-activated transcription factor, aryl hydrocarbon receptor (AhR) and subsequent upregulation of Phase I and Phase II enzymes (Bock and Kohle 2006; Ko et al., 1996; Mimura and Fujii-Kuriyama 2003). Human exposure to high levels of AhR agonists following industrial accidents and herbicidal warfare during the Vietnam War has provided opportunities to examine the consequences of AhR activation. High dioxin levels increased serum glucose and resulted in greater prevalence of hyperinsulinemia and diabetes (Henriksen et al., 1997; Remillard and Bunce 2002; Silverstone et al., 2012). Thus, ligand-activated AhR alters glucose metabolism, performance on glucose tolerance tests, insulin levels and increases the risk of diabetes mellitus (Henriksen et al., 1997; Remillard and Bunce 2002). However, the mechanisms by which AhR activation produces metabolic dysfunction remain unknown and are the subject of this investigation.
Around 90% of human exposure to dioxins occurs through diet, primarily by consumption of animal fat (USEPA 2004). Although the classical, high affinity AhR agonists found in animal fat are commonly toxic pollutants, AhR also binds “non-classical” ligands found in natural dietary products including indole metabolites from cruciferous plants and flavonoids in fruits and vegetables, leading to physiological activation (Gasiewicz et al., 1996; Nguyen and Bradfield 2008). Additionally, the arachidonic acid metabolite, lipoxin A4, prostaglandins, specifically Prostaglandin G2, and an arachidonic acid metabolite produced in inflammatory disease conditions, 12(R)-hydroxy-5(Z),8(Z),10E, 14(Z)-eicosatetraenoic acid [12(R)-HETE], can act as AhR agonists (Chiaro et al., 2008; Nguyen and Bradfield 2008). Recently, lipid and lipid derivatives, such as oxidized low-density lipoproteins, have been identified as AhR agonists (Moyer et al., 2016; Nguyen and Bradfield 2008). With a wide variety of AhR agonists present within food, or present due to secondary effects of diet, AhR activation is likely common, and the potential metabolic consequences are not understood.
Decreased AhR levels and/or suppressed activation in rodent models protect against metabolic disorders. Mice lacking AhR expression have enhanced insulin sensitivity and improved glucose tolerance (Wang et al., 2011). Additionally, the presence of a low-affinity AhR allele or inhibition of AhR activation protects against diet-induced metabolic dysfunction, as demonstrated by reduced fat mass and preserved hepatic function (Kerley-Hamilton et al., 2012; Moyer et al., 2016). Similarly, AhR−/− mice are resistant to the harmful effects of diet-induced obesity through protection against hepatic steatosis, insulin resistance, and inflammation (Xu et al., 2015).
Metabolic health is preserved by synchronization of the body’s autogenous circadian rhythms with environmental light/dark cycles. Disruption of endogenous circadian rhythms is linked to a higher body mass index, increased triglycerides, glucose dysregulation, obesity and metabolic syndrome (Esquirol et al., 2011). Among other effects, consumption of a high fat diet (HFD) increases fat burden, thereby enhancing accumulation of lipophilic compounds such as AhR agonists, and disrupts circadian regulation of gene expression through decreased binding and/or phase advancement of Circadian Locomotor Output Cycles Kaput (Clock)/Brain Muscle Aryl Hydrocarbon Receptor Nuclear Translocator-Like (Bmal1) to target gene promoters. A large number of metabolites in carbohydrate, lipid, metabolic and xenobiotic pathways are regulated by Clock/Bmal1 binding to their promoter regions (Eckel-Mahan et al., 2012; Eckel-Mahan et al., 2013; Marcheva et al., 2013). Interestingly, the PAS domain protein structure of AhR, which is similar to that of Clock and Bmal1, allows for heterodimer formation and crosstalk between AhR signaling and the molecular circadian clock (Anderson et al., 2013; Gu et al., 2000; McIntosh et al., 2010; Reisz-Porszasz et al., 1994). Similar to HFD, persistent activation of AhR alters circadian rhythmicity, primarily through inhibition of Clock/Bmal1-mediated target genes (Mukai et al., 2008; Xu et al., 2010; Xu et al., 2013). If AhR activation suppresses rhythmic expression of Clock/Bmal1 target genes, then reduction in AhR may enhance expression of these same targets, and possibly counteract the effects of HFD. Therefore, the relationship between AhR and HFD-induced disruption of Clock/Bmal1 activity deserves further attention.
We hypothesized that a reduction in AhR will protect against HFD-induced alterations in metabolic rhythms. To test this hypothesis, rhythmicity was explored in wild-type (WT) and AhR+/− mice fed a HFD (60%) for 15 weeks. HFD disrupted the rhythmic expression of glucose, nuclear receptors and metabolic genes important to fatty acid synthesis and gluconeogenesis in WT mice. AhR haploinsufficiency protected against diet-induced disruption of metabolic rhythms and provided resistance to changes in rhythm amplitude, expression and acrophase. Maintenance of healthy metabolic rhythms contributes to the AhR+/− phenotype and further supports the integrated role of AhR in rhythms and metabolic homeostasis.
Materials and methods
Animals
Animal protocols were approved by Southern Illinois University School of Medicine’s institutional animal care and use committee. Male WT c57bl6/j mice and AhR+/− c57bl6/j mice (Bradfield strain) (Schmidt et al., 1996) were housed 3–4 mice per cage (30 × 19 × 12.5 cm) within light tight chambers with circulating air, temperature maintained at 21°C and humidity of 44%. Mice used for activity monitoring were individually housed. All animals had free access to water and either normal chow diet (LabDiet Rodent 5001) (10% fat) or a HFD (Research Diets-D12451) (60% fat) unless otherwise stated. At 6 weeks of age, mice were entrained to a schedule of 12/12 hour light/dark cycles for 2 weeks at a light intensity of 400 lux, where lights were on from 10 pm to 10 am and lights off from 10 am to 10 pm.
Experimental design
WT and AhR+/− mice were randomly assigned to either control diet or HFD conditions (N = 25/group) (N = 50/genotype). Animals were exposed to the control diet or HFD for 15 weeks. Subsequently, mice (WT, Control) (WT, HFD) (AhR+/−, Control) (AhR+/−, HFD) were sacrificed by cervical dislocation without anesthesia for tissue collection at 6 time points across a 24-hour period (N = 3 mice/time point) (18 mice/group), where sacrifice was performed at Zeitgeber time 0 (ZT0, time of lights on), ZT4, ZT8, ZT12 (lights off), ZT16 and ZT20. To examine behavior, 4 animals/group were randomly assigned for activity monitoring and housed in individual cages for the duration of the study. Three mice per group were randomly assigned to the acute insulin stimulation experiment performed after 15 weeks exposure to feeding schedules.
Activity monitoring
Animals assigned to activity monitoring (4 mice/group) were housed in cages within the light chambers fitted with infra-red activity detectors (Minimitter, Bend, OR). Activity data were collected week 15 of the feeding schedule. Actiview software was used to collect activity data into 10-minute bins. Data were analyzed using Clocklab software (Actimetrics, Evanston, IL). Average activity during lights on and lights off was calculated from actogram counts/minute. Activity onset was determined from actograms.
Body weight, food intake and glucose
Weight (g), food intake (kcal) and blood glucose (mg/dL) were measured weekly at 9 am on Tuesdays during a time period of lights on (ZT11). On Thursday of week 8, mice were fasted for 6 hours starting at 8 am (ZT10-ZT16). Tail vein blood glucose levels (mg/dL) were measured by a True Track Blood Glucose Meter (Niopro Diagnostics Inc., Fort Lauderdale, FL). Weight of white adipose tissue from epididymal fat pads and blood glucose rhythm were measured in mice fed ad libitum during time-course tissue collection.
Acute insulin stimulation
Acute insulin stimulation was performed at the end of week 15 at ZT16 as previously described (Xu et al., 2015). Protein samples from liver were subjected to Western blot analysis to measure Phospho-AKT (Ser473) (1:1000; Cell Signaling) and Phospho-AKT (Thr308) (1:1000; Cell Signaling). Total Akt (1:2000; Cell Signaling) and β-Actin (1:4000; Sigma-Aldrich, St. Louis, MO) were used as internal controls (Xu et al., 2015).
Glucose and insulin tolerance tests
Insulin tolerance tests were performed (18 mice/group) after 12 weeks on the feeding schedule following a 6-hour fasting period from 8 am to 2 pm (ZT10-ZT16). A bolus of insulin (0.75 units/kg diluted in 0.9% NaCl; Sigma) was injected intraperitoneally and blood glucose was measured 15, 30, 60 and 90 minutes following insulin injection. Glucose tolerance tests were performed in the same mice following 13 weeks exposure to the feeding schedule (18 mice/group). Following an overnight fast (16 hours starting at 5 pm) (ZT19-ZT11), basal blood glucose levels were measured. A 10% solution of D-Glucose (1 g/kg body weight) diluted in 0.9% NaCl was injected intraperitoneally followed by blood glucose measurement at 15,30, 60, 90 and 120 minutes post injection.
Hematoxylin & Eosin staining
Sections of adipose, liver and pancreas from 6 mice/group (3 samples from ZT0 and ZT12/group) were collected for histological examination. Formalin-fixed paraffin-embedded tissue was sectioned and stained with hematoxylin and eosin by the Histology Department at Memorial Medical Hospital, Springfield Illinois. Adipose and pancreatic islet diameters were measured using SPOT Advanced Modular Imaging Software for microscopy manufactured by SPOT Imaging Solutions (Sterling Heights, MI). For adipose tissue (N = 6 animals/group) 5 randomly selected areas of each slide (5 slides/mouse) were selected for analysis of adipocyte area using the ImageJ method as described by Parlee et al. (2014).
Insulin ELISA
At sacrifice, whole blood was collected from heart, centrifuged at 367 ×g for 20 minutes to separate plasma, and kept at −80°C. An Ultra Sensitive Mouse Insulin ELISA Kit from Crystal Chem Inc (Downer’s Grove, IL). was used according to the manufacturer’s instructions to measure insulin concentrations from ad libitum, fed mouse serum collected every 4 hours.
Real-Time quantitative polymerase chain reaction
Liver tissue was snap frozen in liquid nitrogen and stored at −80°C. TRI-Reagent (Sigma) was used to extract total RNA from liver tissue according to the manufacturer’s instructions. Complementary DNA was synthesized using Random Primers (Promega, Madison, WI) and Reverse Transcriptase (Promega). Quantitative polymerase chain reaction (qPCR) was performed using Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA) in a StepOnePlus™ Real-Time PCR System (Applied Biosystems). Data were analyzed by StepOne Software v2.1 (Applied Biosystems). Each sample was normalized with β-actin. Each experiment included a negative reverse transcriptase and a negative RNA control. Primer sequences of circadian clock and metabolic genes tested are listed in Table 1.
Table 1.
Primer sequences used in real-time PCR.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Acc | GCCTCCGTCAGCTCAGATAC | ATGTGAAAGGCCAAACCATC |
| Actin | CACCCGCGAGCACAGCTTCTT | TTTGCACATGCCGGAGCCGTT |
| AhR | TTCTTAGGCTCAGCGTCAGCTA | GCAAATCCTGCCAGTCTCTGAT |
| Bmal1 | CTACAAGCCAACATTTCTATCAGATG | GGTCACATCCTACGACAAACAAAA |
| Cry1 | GCCGGCTCTTCCAACGT | TCGTAGATTGGCATCAAGATCCT |
| Cyp1A1 | GTGCATCGGAGAGACCATTG | GGTAGGAGTCATATCCACCTT |
| Cyp1B1 | ACATCCCCAAGAATACGGTC | TAGACAGTTCCTCACCGATG |
| Fasn | GTCACCACAAGCCTGGACCGC | CTGGCCATAGGTGCCGCCTG |
| Pepck | CCACAGCTGCTGCAGAACA | GAAGGGTCGCATGGCAAA |
| Per1 | TCCTCAACCGCTTCAGAGATC | CGGGAACGCTTTGCTTTAGA |
| PPARα | ACTGGTAGTCTGCAAAACCAAA | AGAGCCCCATCTGTCCTCTC |
| PPARγ | TCTGGGAGATTCTCCTGTTGA | GGTGGGCCAGAATGGCATCT |
| Rev-erbα | Qiagen | Qiagen |
| Srebp1c | CCATCGACTACATCCGCTTCTT | ACTTCGTAGGGTCAGGTTCTC |
Statistical analysis
Data were analyzed by two-way ANOVA, Student’s t-test or cosinor analysis where appropriate. For rhythmic data, two-way ANOVAs were performed to examine mean differences among independent variables, group (2 levels) and ZT (6 levels). To further investigate rhythmic data, amplitude, acrophase and phase shift were determined by cosinor analysis performed in RStudio (Version 0.99.484). Cosinor analysis fits rhythmic data with a known period to a smooth line using the least squares regression method. Once the data are fit to a smooth line, acrophase and amplitude can be calculated (Refinetti et al., 2007). Activity data were collected in 10-minute bins and a curve was generated for each animal prior to analysis. Because each data point in gene expression or serum samples represented a different animal, data collected at different ZTs were averaged first and then analyzed by cosinor analysis to generate one set of acrophase and amplitude outputs for each set of data. Sample variance is represented as ±SEM.
Results
AhR+/− mice are protected against HFD-induced metabolic dysfunction
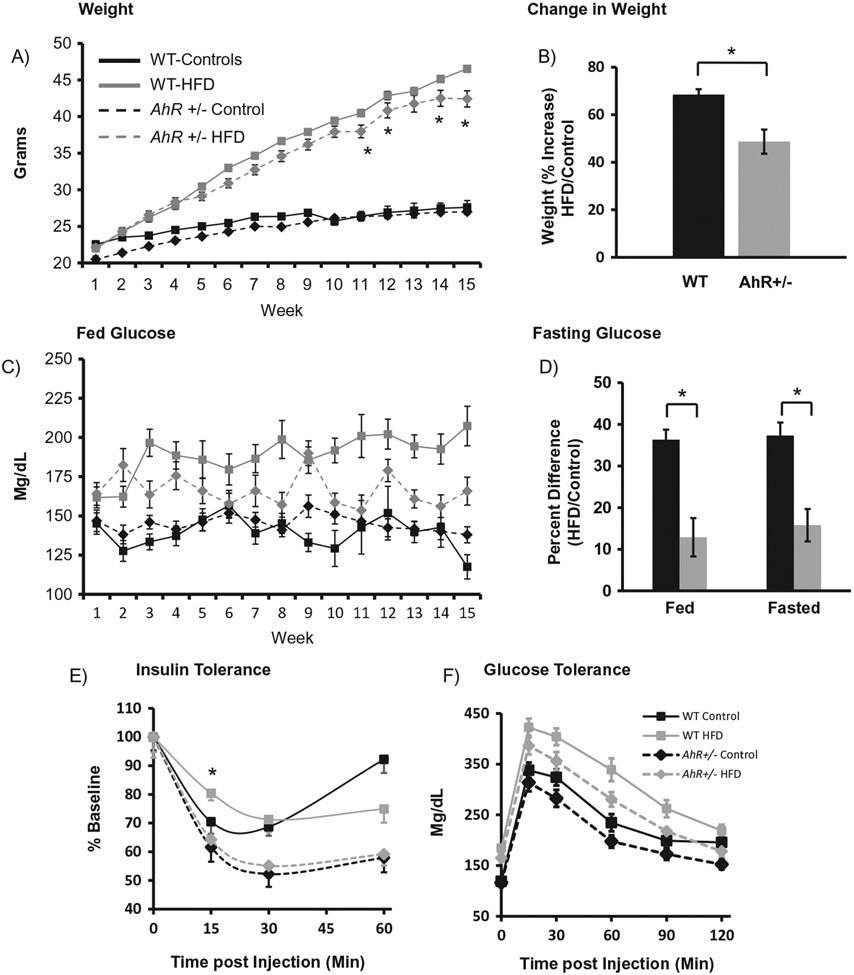
Our previous study demonstrated that AhR haploinsufficiency affords protection against HFD-induced metabolic dysfunction. AhR has also been implicated as a modulator of the molecular circadian clock, especially in peripheral tissues. Specifically, AhR activation can inhibit transcription and rhythmicity of Clock/Bmal1 target genes, leading to dampened rhythms. Because rhythms are important in regulation of metabolic function, this study was undertaken to explore the whether AhR reduction can also prevent HFD-induced changes in metabolic function in peripheral tissues. As we have previously shown, kilocalorie intake was significantly increased by HFD, regardless of genotype, data not shown. HFD significantly increased both weight and fed blood glucose levels in WT and AhR+/− mice; the increase was significantly less in AhR+/− HFD mice during the last few weeks of the experiment (Figure 1A). Expression of weight as percent increase within genotype reveals that WT mice gained more weight on HFD than did AhR+/− mice (Figure 1B). Fed glucose increased in both genotypes (Figure 1C). Expression of fed and fasted glucose as percent difference (HFD/Control) within genotype demonstrated that both fed and fasted glucose were increased more in the WT mice compared to the AhR+/− (Figure 1D). As expected, HFD altered insulin sensitivity in the WT mice, as demonstrated by higher levels of glucose in HFD-fed WT mice 15 minutes after insulin injection (Figure 1E). There were no significant differences between AhR+/− controls and AhR+/− HFD mice (Figure 1e). Improved insulin sensitivity of AhR-deficient mice compared to WT mice during insulin tolerance testing has been previously described (Wang et al., 2011). WT and AhR+/− HFD mice showed increased glucose levels during glucose tolerance testing (Figure 1F, G). Area under the curve calculations for GTT demonstrate that AhR+/− mice have decreased overall glucose load, which is preserved in the AhR+/− mice under HFD conditions (Figure 1G). Acute insulin stimulation confirmed insulin resistance in WT HFD fed mice, reflected by reduced hepatic insulin-stimulated pAKT (Ser 473 and Thr 308)/total AKT levels. AhR+/− HFD mice were protected from reduction in insulin-stimulated pAKT (Ser473) levels, thereby confirming enhanced insulin sensitivity (Figure 1H).
Figure 1.
AhR+/− mice are protected from HFD-induced changes in body weight and metabolic parameters. Body weights of WT and AhR+/− mice were increased by HFD. Two-way ANOVA revealed no differences in weight when animals were on the control diet. However, HFD-fed WT mice were significantly heavier at weeks 11,12,14 and 15; p < 0.05 is indicated by *. There was an interaction, p < 0.05, between genotype and diet (A). WT mice had gained significantly more weight when expressed as percent increase (HFD/Control), p < 0.05 (*, Student’s t-test) (B). Fed glucose was not different between genotypes on the control diet; HFD-fed mice had elevated fed glucose (two-way ANOVA, p < 0.05). There was an interaction between genotype and diet (p < 0.05), indicating that AhR+/− mice on HFD had lower fed glucose than HFD-fed WT mice (C). Fed and fasting glucose were elevated more in WT mice compared to controls. * indicates significance between genotypes in percent change (p < 0.05, Student’s t-test) (D). Insulin tolerance tests were performed at 12 weeks. Mice were fasted for 6 hours followed by intraperitoneal injection of insulin (0.75 units/kg) (E). Glucose tolerance tests were performed at 13 weeks HFD. Mice were fasted overnight (16 hours) followed by intraperitoneal injection of glucose (1 g/kg body weight) (F). Area under the curve for the glucose tolerance test. * p < 0.05 for ANOVA with Tukey’s post hoc analysis (G). Data in A-G are represented as mean ± SEM. N = 17–18. At the end of 15 weeks, mice assigned to acute insulin stimulation were fasted for 6 hours followed by insulin stimulation (0.75 units/kg) via the vena cava. Basal and insulin-stimulated hepatic pAKT/Total Akt levels were compared using Western blotting (H). * p < 0.05 for one-way ANOVA with Tukey’s post hoc test. Data are represented as mean ± SEM. N = 3 per group. Formalin-fixed paraffin-embedded tissue sections of liver and pancreas were stained with Hematoxylin and Eoxin (I, representative data). Hepatic steatosis appeared less severe in AhR+/− HFD mice. Black bars indicate 50 μm. Pancreatic islet diameter (J) expressed as percent change from WT control in WT mice; * indicates p < 0.05 determined by ANOVA with Tukey’s post hoc analysis.
Other HFD-induced changes were also examined. Hepatic steatosis was present in both WT HFD and AhR+/− HFD mice, although steatosis visually appeared less severe in AhR+/− mice (Figure 1I, top row). Pancreatic islet cell diameter was reduced in AhR+/− mice on a normal chow diet compared to WT; HFD increased islet diameter in both genotypes (Figure 1I, bottom row; Figure 1J). Islets in all groups stained positively for insulin, indicating that production of insulin is likely preserved (data not shown).
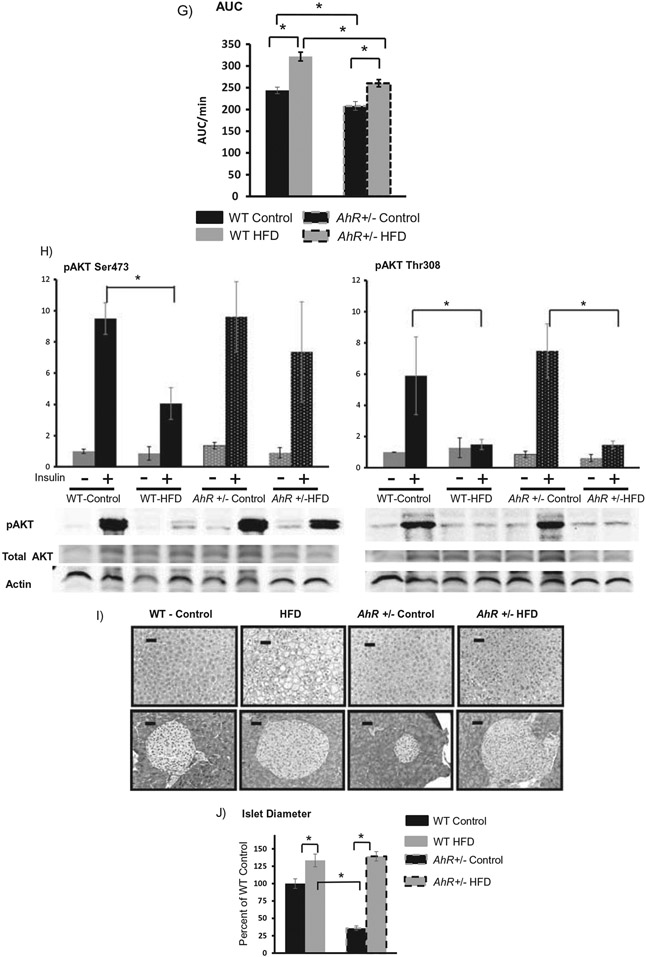
Epididymal fat was examined more extensively. Histology (H&E staining) indicates differences in adipocyte size (Figure 2A). Reduced adipocity could result from smaller adipocytes, decreased adipocyte number or both. AhR+/− mice have a larger proportion of smaller fat cells shown as a leftward shift in the Frequency vs. Adipocyte area plot (Figure 2B), and a significant increase in the frequency of cells whose area was less than 2000 μm2 (Figure 2C). HFD caused a rightward shift in the Frequency vs. Adipocyte area plot for both genotypes (Figure 2B). However, the frequency of adipocytes larger than 2000 μm2 was much greater in the WT mice after HFD (Figure 2B,C). To estimate relative numbers of adipocytes, a correlation was performed between white adipose tissue weight and mean adipocyte volume for each animal. As expected, there was a linear relationship between mean adipocyte diameter and fat pad mass in HFD-fed AhR+/− (R2 = 0.955) and WT (R2 = 0.83). Linear regression analysis of these data indicates that the slope of the regression line is significantly steeper (p < 0.0001) in WT mice (Figure 2d). A flatter slope suggests that AhR+/− mice have an increased number of smaller adipocytes within the fat pad. The smaller, more numerous adipocytes support the enhanced insulin sensitivity in the AhR+/− mice.
Figure 2.
AhR+/− mice have smaller, more numerous adipocytes. Representative formalin fixed, paraffin-embedded sections of epididymal adipose tissue show reduced adipocyte size in AhR+/− mice (A). Frequency distribution reveals that AhR+/− mice have a larger proportion of small adipocytes compared to WT controls (B). HFD produces a rightward shift of the frequency plot in both genotypes. AhR+/− mice have more numerous, smaller adipocytes (<2000 μm2) compared to WT on HFD (C). Data are analyzed by ANOVA with Tukey’s post hoc analysis. *p < 0.05. Linear relationship between epididymal white adipose tissue weight and average adipocyte volume for WT (R2 = 0.83) and AhR+/− (R2 = 0.955) (D). Slopes were statistically different using Pearson’s correlation (p < 0.0001). Data are represented as mean ± SEM. N = 6 per group. Black bars in (A) represent 50 μm.
AhR+/− mice are protected from HFD-induced changes in activity onset
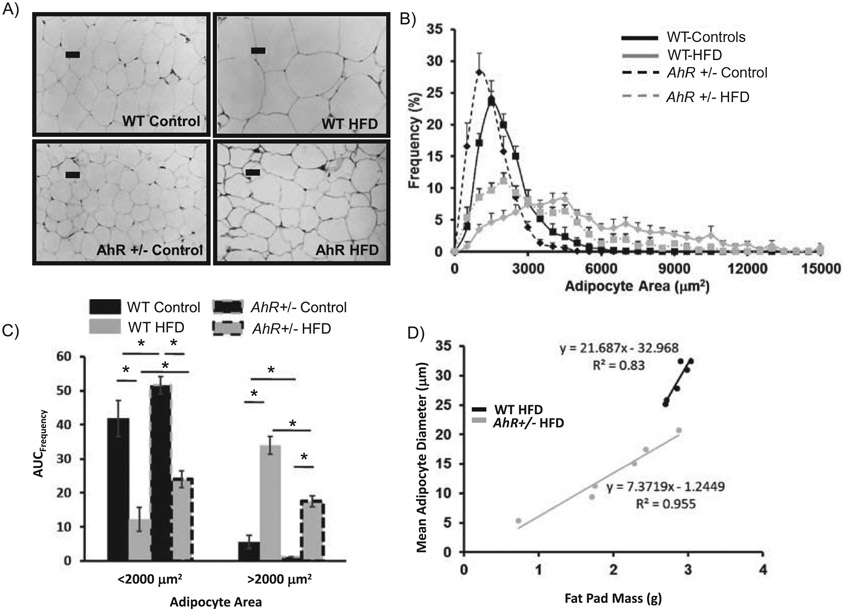
Diurnal activity rhythms were examined during the last week of the study with the expectation that there would be no differences in activity between genotypes. Our previous studies have suggested that AhR has only a modest effect on the central pacemaker, but a more pronounced effect on peripheral rhythms (Mukai et al., 2008). The amplitude of behavioral circadian rhythms is dampened in animals exposed to a HFD. The amplitude of nocturnal activity was reduced in both WT and AhR+/− mice fed a HFD (Figure 3A,B) (Table 2). In WT animals, HFD delayed the onset of activity by more than 30 minutes (Figure 3C). Activity onset was not altered by diet in the AhR+/− mice (Figure 3A-C). Acrophase was not significantly altered by HFD in either genotype (Table 2). Based on previous studies that indicate a greater role for AhR in regulating circadian rhythms in peripheral clocks, the remainder of the rhythm studies focused on the metabolically important liver clock.
Figure 3.
Infrared beam detection was used for behavioral analysis of activity during week 15. Data were analyzed from actogram beam breaks/minute and averaged into hourly activity (A). Total lights on and lights off activity was averaged (B) and activity onset time was determined from actograms (C). Data are represented as mean ± SEM. N = 4 per group. Cosinor analysis was used to determine changes in amplitude, phase. Activity onset was determined from actograms. One-way ANOVA was used to determine differences in lights on/lights off activity in (B); * indicates p < 0.05.
Table 2.
Cosinor analysis of amplitude, acrophase and mesor.
| Amplitude |
Acrophase |
Mesor |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT controls |
p Value | WT- HFD |
p Value | AhR+/− controls |
p Value | AhR+/− HFD |
p Value | WT controls |
WT- HFD |
AhR+/− controls |
AhR+/− HFD |
WT controls |
WT- HFD |
AhR+/− controls |
AhR+/− HFD |
|
| Activity | 2.40 | *<1 × 10−4 | 0.5 | *0.01 | 2.50 | *<1 × 10−4 | 0.90 | *<1 × 10−4 | 14 | 16 | 13 | 13 | 2.71 | 1.03 | 2.81 | 1.54 |
| Glucose | 8.05 | *0.02 | 37.12 | 0.07 | 19.67 | *<1 × 10−4 | 13.53 | *0.05 | 5 | 10 | 11 | 10 | 152.10 | 199.50 | 128.8 | 161.6 |
| Insulin | 0.92 | *<1 × 10−4 | 0.84 | 0.48 | 0.62 | *0.01 | 3.31 | 0.14 | 0 | 19 | 17 | 20 | 3.28 | 6.68 | 0.97 | 6.67 |
| AhR | 0.28 | *1 × 10−3 | 0.40 | *<1 × 10−4 | 0.18 | *<1 × 10−4 | 0.19 | *0.05 | 7 | 6 | 7 | 9 | 0.90 | 0.96 | 0.52 | 0.52 |
| Cyp1A1 | 0.18 | 0.33 | 1.01 | 0.14 | 0.20 | *<1 × 10−4 | 0.21 | *0.02 | 2 | 8 | 7 | 9 | 1.10 | 0.87 | 0.49 | 0.35 |
| Cyp1B1 | 0.25 | 0.12 | 1.15 | *0.05 | 0.14 | *<1 × 10−4 | 0.13 | 0.09 | 14 | 9 | 8 | 10 | 1.12 | 0.89 | 0.20 | 0.23 |
| Per1 | 18.30 | *0.02 | 12.41 | *<1 × 10−4 | 38.80 | *<1 × 10−4 | 49.24 | *0.01 | 11 | 11 | 12 | 11 | 11.99 | 10.04 | 33.53 | 33.99 |
| Bmal1 | 0.36 | *<1 × 10−4 | 0.53 | *<1 × 10−4 | 1.21 | *<1 × 10−4 | 1.30 | *<1 × 10−4 | 23 | 22 | 0 | 22 | 0.36 | 0.54 | 0.90 | 1.05 |
| Cry1 | 0.42 | *<1 × 10−4 | 0.65 | *<1 × 10−4 | 0.81 | *0.02 | 1.91 | *<1 × 10−4 | 18 | 19 | 22 | 20 | 0.69 | 1.03 | 1.20 | 2.12 |
| Rev-erbα | 4.29 | *<1 × 10−4 | 3.25 | *1 × 10−4 | 9.07 | *0.01 | 8.19 | *4 × 10−4 | 7 | 2 | 7 | 8 | 3.29 | 2.87 | 5.66 | 5.57 |
| PPARα | 0.94 | *0.01 | 0.37 | 0.33 | 1.66 | *<1 × 10−4 | 2.06 | *<1 × 10−4 | 11 | 10 | 10 | 11 | 1.59 | 2.25 | 2.23 | 2.80 |
| PPARγ | 0.42 | *0.05 | 2.24 | *0.03 | 0.63 | 0.08 | 2.63 | 0.12 | 17 | 7 | 11 | 12 | 1.25 | 4.63 | 1.36 | 4.85 |
| Srebp1c | 0.25 | 0.19 | 0.51 | *0.05 | 1.47 | *<1 × 10−4 | 2.12 | *0.01 | 12 | 9 | 10 | 11 | 1.11 | 1.96 | 2.04 | 2.71 |
| Acc | 0.29 | 0.14 | 0.67 | 0.08 | 0.67 | *0.02 | 1.00 | *<1 × 10−4 | 9 | 8 | 10 | 12 | 0.95 | 1.79 | 1.33 | 1.71 |
| Fasn | 0.54 | *<1 × 10−4 | 0.86 | *0.01 | 1.68 | 0.07 | 0.65 | *<1 × 10−4 | 12 | 8 | 10 | 12 | 1.37 | 2.15 | 2.90 | 1.75 |
| Pepck | 3.24 | *<1 × 10−4 | 0.70 | 0.07 | 1.02 | *<1 × 10−4 | 1.72 | 0.11 | 12 | 10 | 10 | 12 | 3.63 | 2.60 | 2.14 | 3.43 |
indicates statistical significance as determined by the cosinor analysis.
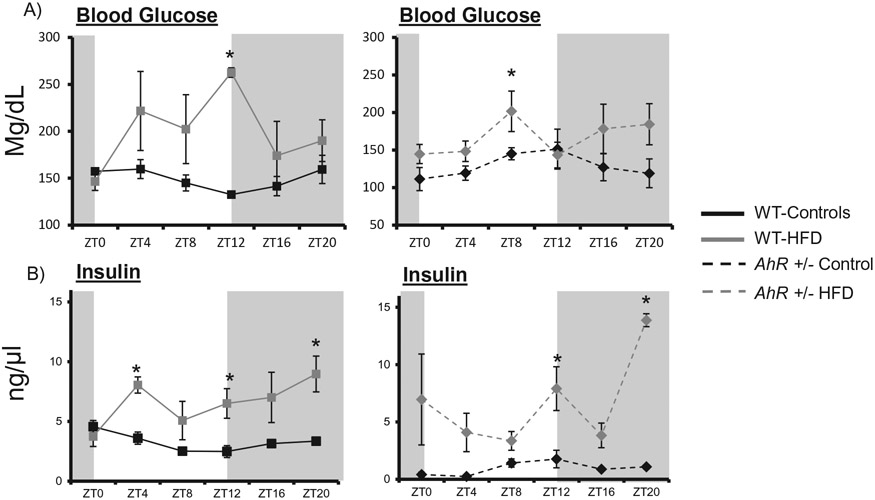
AhR+/− mice are protected from HFD-induced changes in glucose rhythmicity
The major focus of this study was to examine changes in diurnal patterns of metabolic parameters. WT control mice displayed a rhythmic expression of blood glucose that peaked near the beginning of the lights on period (Figure 4A). Acrophase was shifted from ZT5 in WT controls to ZT10 in WT mice fed the HFD (Figure 4A, Table 2). HFD enhanced glucose at ZT8 in AhR+/− mice (Figure 4B), but acrophase was not affected (Table 2). Insulin levels were rhythmic in the WT mice, with a peak early in the day. Rhythm and acrophase were altered in the HFD-fed WT mice (Figure 4A, Table 2). Insulin levels were also enhanced in HFD-fed AhR+/− mice (Figure 4B).
Figure 4.
Insulin is elevated in AhR+/− and WT mice on HFD. Blood glucose samples were obtained from tail vein at each ZT and analyzed by a True Track Blood Glucose Meter in WT and AhR+/mice (A). Two-way ANOVA revealed differences between genotypes (p < 0.05); there was an interaction between genotype and diet (p < 0.05). Cosinor analysis of amplitude and phase is listed in Table 2. Serum insulin was analyzed by ELISA in WT (B). Two-way ANOVA demonstrated that insulin was elevated by HFD (p < 0.01, WT; p < 0.01 AhR+/−). There was no significant interaction between diet and genotype. Data are represented as mean ± SEM. N = 3 per ZT time point. Data are analyzed by two-way ANOVA, *p < 0.05.
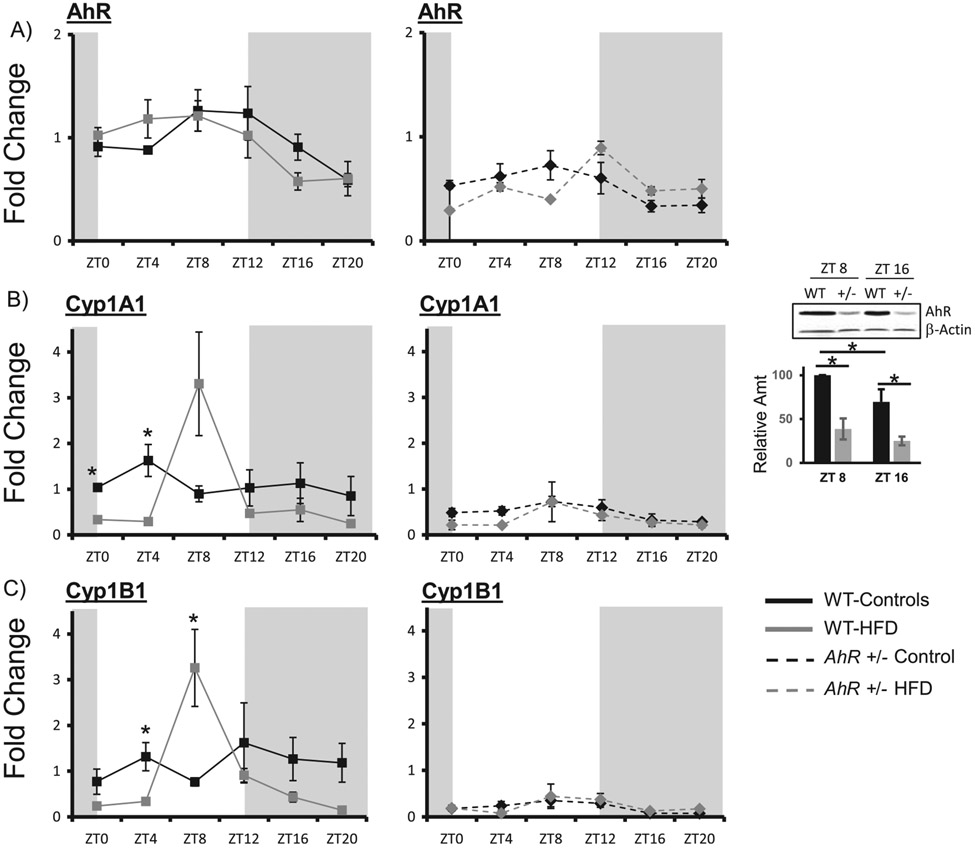
Diet-induced obesity alters the rhythmic expression of clock genes and AhR target genes Cyp1A1 and Cyp1B1 in livers of WT mice
AhR mRNA levels are significantly reduced in AhR+/− mice (Table 2, mesor for AhR). AhR+/− mice do, however, retain a diurnal rhythm in AhR mRNA that is in phase with the WT controls. HFD did not alter phase or amplitude of AhR mRNA (Figure 5A, Table 2). Immunoblot for AhR protein indicates that protein levels are reduced in the AhR+/− mice to less than 50% of WT. WT showed an increase in AhR protein at ZT8, which parallels the AhR mRNA. AhR protein was higher at ZT8 compared to ZT16 in WT controls (Figure 5A, right panel). Proteins levels were not examined in HFD-fed mice. AhR target genes cytochrome P450, family 1, member A1 (Cyp1A1) and cytochrome P450, family 1, subfamily B (Cyp1B1), which correlates with the increased protein, were enhanced at ZT8 in WT mice, suggesting AhR activation. These effects were absent in AhR+/− mice (Figure 5B, C) (Table 2).
Figure 5.
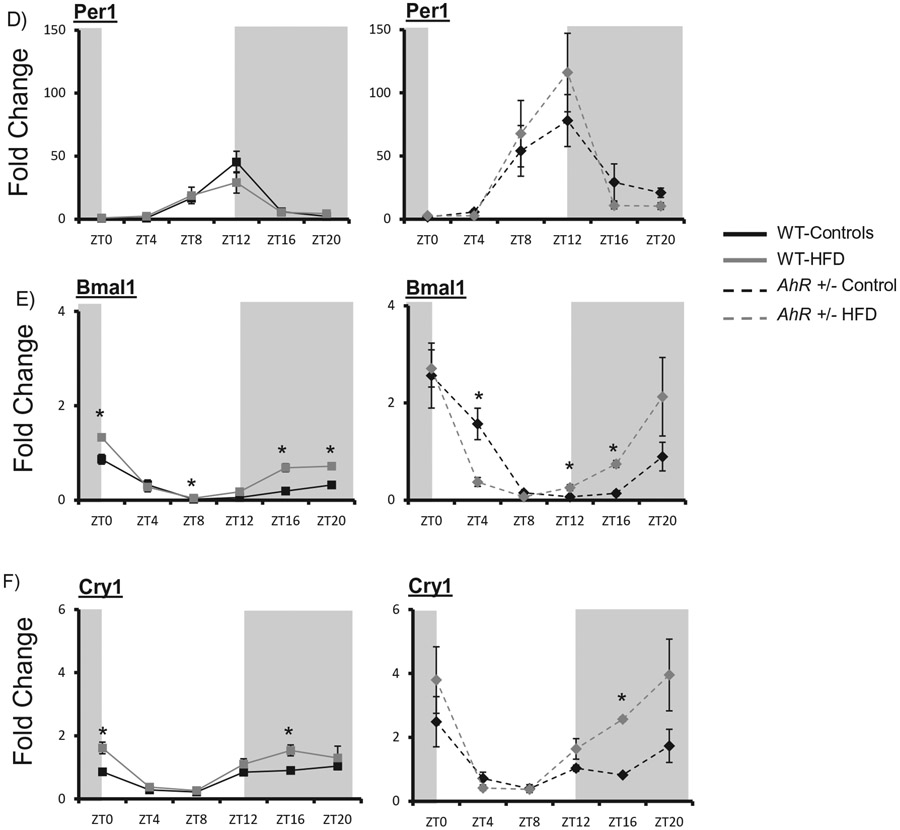
HFD alters AhR target gene expression in WT mice. Liver samples were collected at each ZT followed by RNA extraction. Protein was extracted from samples at two time points and AhR was detected by Western blot. Quantitative PCR was used to measure mRNA levels. Amplitude and phase of oscillation were determined by cosinor analysis and are listed in Table 2. Two-way ANOVA was used to explore differences in mean expression levels. AhR was reduced in AhR+/− mice (p < 0.01) at all times of day. Diet did not change levels of AhR, p = 0.386 and AhR+/− mice (A) p = 0.086. AhR protein was decreased in AhR+/− mice (p < 0.01). There was an interaction between ZT and genotype (p < 0.05). * indicates significance with p < 0.05. AhR target genes Cyp1a1 in WT and AhR+/− mice showed a difference between genotypes (p < 0.01) (B); there was an interaction between diet and genotype (p < 0.05 as indicated by *). Cyp1B1 was decreased in AhR+/− mice (C) p = 0.002. There was an interaction between diet and genotype (p < 0.05 as indicated by *). Per 1 was elevated at ZT 8 and 12 in AhR+/− mice compared to WT. Diet had no effect and there was no interaction between diet and genotype (D). Bmal1 was elevated in in AhR+/− mice at ZT0, 4 and 20 compared to WT mice (* indicates p < 0.05) (E). There was an interaction between diet and genotype (p < 0.05). Cry1 was elevated at ZT0 in AhR+/− mice (F); there was an effect of genotype and diet. * indicates p < 0.05. Data are represented as mean ± SEM. N = 3 per ZT.
Rhythmic expression of hepatic clock genes was generally increased in AhR+/− mice compared to WT on control diets. HFD had little effect on rhythmic expression of hepatic circadian clock genes (Figure 5d-f). In WT control mice, there was a tendency for a decrease in Period 1 (Per1) amplitude (Table 2). The amplitude of the Per1 rhythm increased compared to WT, but not affected by diet in AhR+/− mice (Figure 5D). Bmal1 and Cryptochrome 1 (Cry1) amplitudes were not altered by diet, but were increased in AhR+/− compared to WT (Figure 5E, F).
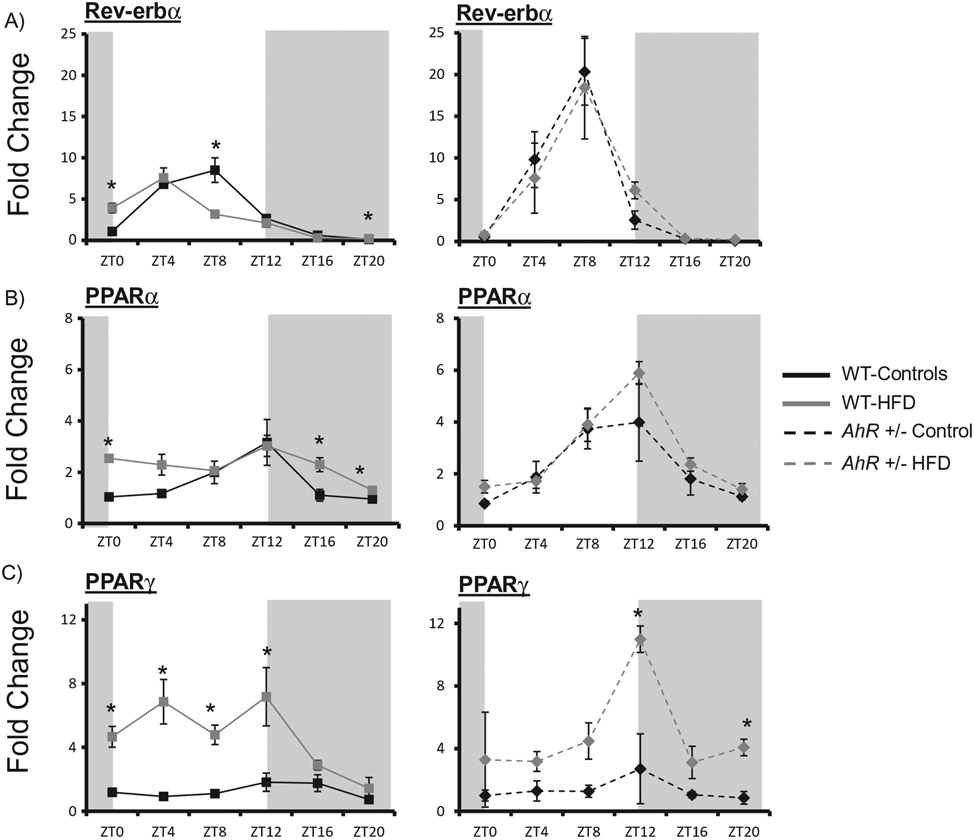
AhR+/− mice are protected against HFD-induced disruption of metabolic gene expression in liver
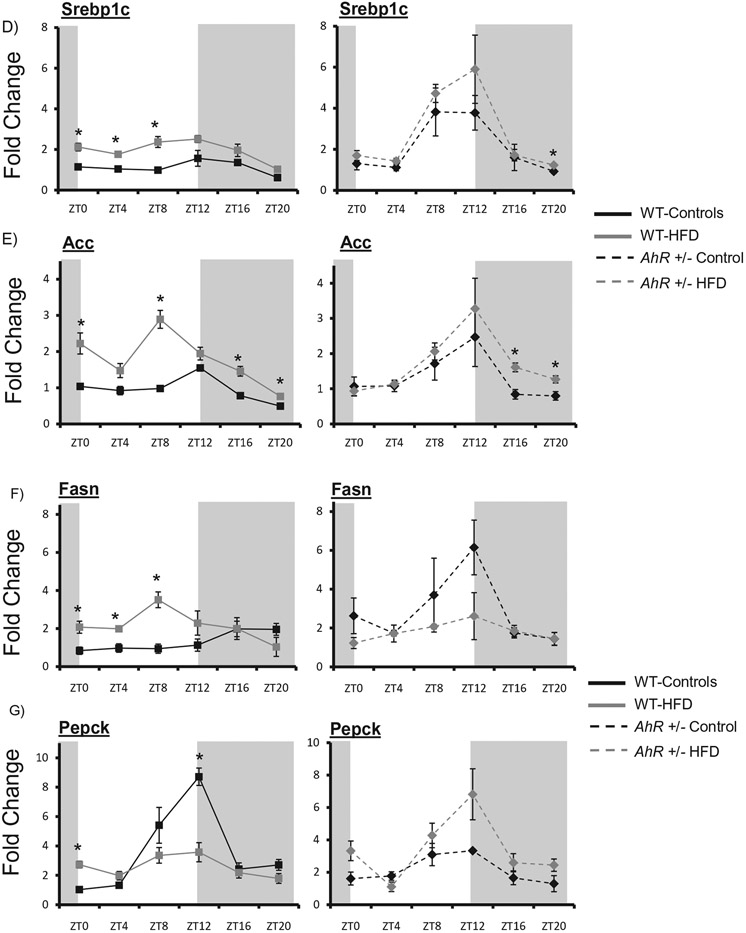
The clock-controlled nuclear receptor subfamily 1, group D, member 1 (Rev-erbα) was phase advanced in response to HFD feeding in WT mice. AhR+/− mice were protected from HFD-induced changes in hepatic Rev-erba expression (Figure 6A) (Table 2). Peroxisome proliferator-activated receptor alpha (PPARα) was enhanced at ZT0, ZT16 and ZT20 in WT HFD fed mice and the rhythm was abolished (Table 2). However, HFD did not significantly alter PPARα expression in AhR+/− mice (Figure 6B) (Table 2). Notably, AhR+/− mice on a control diet displayed increased amplitude rhythms for both Rev-erbα and PPARα. The clock-controlled nuclear receptor Peroxisome proliferator-activated receptor gamma (PPARγ) was enhanced at four time points in WT HFD mice and the rhythm was also abolished. Interestingly, in WT HFD mice PPARγ was enhanced during the lights on period when mice are normally resting. However, increases in PPARγ occurred during the lights off period in AhR+/− mice during their normal active period (Figure 6C).
Figure 6.
AhR+/− mice are protected against HFD-induced changes in clock-controlled nuclear receptors and metabolic gene expression. Liver samples were collected at each ZT followed by RNA extraction. Quantitative PCR was used to measure mRNA levels of nuclear receptors. Amplitude and phase of oscillation was determined by cosinor analysis and are listed in Table 2. Two-way ANOVA was used to explore differences in mean expression levels. Rev-erbα was elevated at ZT8 in AhR+/− mice compared to WT (p < 0.05). There was an interaction between diet and genotype indicating that dietary effects were genotype-specific (A). PPARα was elevated at ZT12 in AhR+/− mice compared to WT (p < 0.05). There was an interaction between diet and genotype indicating that dietary effects were genotype-specific (B). PPARy was not different between WT and AHR+/− mice under control diet conditions (C). HFD elevated PPARy in WT mice; there was an interaction between diet and genotype. Srebp1c was elevated in AhR+/− mice compared to WT at ZTs 8 and 12 (p < 0.05). HFD elevated this gene at ZTs 0, 4 and 8 (p < 0.05 indicated by *); there was an interaction between diet and genotype (D). Acc was elevated at ZT12, but reduced at ZTs 0 and 4 in AhR+/− mice compared to WT mice (E). HFD increased Acc in WT as indicated by * (p < 0.05). There was an interaction between diet and genotype (p < 0.05). Fasn was elevated at ZT12 in AhR+/− mice compared to WT mice (F). HFD elevated Fasn in WT during the day (p < 0.05 as indicated by *). Pepck was reduced in AhR+/− mice at ZTs 8 and 12 compared to WT mice (G). Data are represented as mean ± SEM. N = 3 per ZT. Data are analyzed by two-way ANOVA, *p < 0.05.
Levels of the lipid metabolism genes Sterol regulatory element-binding transcription factor 1c (Srebplc), Acetyl-CoA carboxylase (Acc) and Fatty acid synthase (Fasn) were enhanced in liver of WT HFD mice, occurring predominately throughout the lights on phase, the same period of AhR activation (Figure 6D-F). AhR+/− mice were protected from increases in gene expression of Srebplc, Acc and Fasn during the lights on period (Figure 6D-F) (Table 2). Rhythmic amplitude of the gluconeogenic gene Phosphoenolpyruvate carboxykinase (Pepck) was decreased in WT HFD mice. AhR+/− HFD mice were protected against changes in Pepck expression (Figure 6G) (Table 2). Overall, AhR+/− mice were protected against HFD-induced changes in nuclear receptor and metabolic gene expression (Figure 6).
Discussion
Diet-induced obesity disrupts behavioral and molecular rhythms through altered activity onset, metabolic outputs and gene expression. Intercommunication between the central clock and peripheral circadian network, accomplished by metabolic signal feedback from the periphery to the suprachiasmatic nucleus (SCN), can alter circadian rhythms (Rutter et al., 2002). Metabolic cues, such as circulating glucose levels, are sensed by the ventromedial hypothalamus, which provides direct glutamatergic input to the SCN and indirect, neuropeptide Y input to the SCN via the intergeniculate leaflet (Challet 2010). In this study, HFD (60% fat) reduced nocturnal activity and shifted activity onset in WT animals, in congruence with previous reports (Kohsaka et al., 2007). However, Mendoza et al. did not report a shift in activity onset in WT mice fed a HFD, measured by wheel running. Differences between studies can be explained by variation between wheel running and activity monitoring, telemetry implantation, duration of HFD feeding, and differences in age and diet (Mendoza et al., 2008). Activity patterns in AhR+/− mice were not significantly different than WT on normal diets. However, AhR+/− mice were protected from diet-induced shifting of activity onset. Although rhythms in the SCN and hypothalamus were not explored in this study, they will be the subject of future investigation. Our previous studies indicate that the primary interaction of AhR with the molecular clock occurs in peripheral tissues, suggesting that metabolic feedback to the central oscillator is the most likely explanation for preservation of central clock function in HFD-fed AhR+/− mice. However, direct actions at the SCN cannot be completely discounted.
AhR+/− mice were protected from metabolic dysfunction associated with HFD-induced obesity, in congruence with previous findings (Xu et al., 2015). Reduced hepatic steatosis in AhR+/− mice correlated with protection from HFD-induced increases in PPARγ (Gavrilova et al., 2003). AhR-dependent hepatic steatosis has previously been reported as a consequence of enhanced hepatic fatty acid uptake and storage and inhibition of fatty acid efflux and β-oxidation pathways (Angrish et al., 2012; Lee et al., 2010). Our lab has previously shown that AhR−/− mice have significantly lower fed blood glucose levels compared to WT controls, contributing to their protection from metabolic dysfunction (Xu et al., 2015).
Although both genotypes had an increase in fasting glucose on HFD, glucose was lower in AhR+/− HFD mice. HFD reduced sensitivity to insulin in WT mice; AhR+/− were protected from HFD-induced changes in insulin sensitivity. However, WT and AhR+/− HFD mice displayed higher glucose levels in response to glucose challenge. Blood glucose levels in response to glucose challenge depend on insulin secretion, insulin action and glucose effectiveness (Ayala et al., 2010). Since AhR+/− mice were protected from HFD-induced changes in responsiveness to a bolus of insulin, differences may be a result of altered insulin secretion or glucose effectiveness. A substantial increase in pancreatic islet size in HFD-fed AhR+/− mice may provide a mechanism for increased insulin secretion; AhR+/− mice did show an overall increase in serum insulin at the time of sacrifice. All groups positively stained for insulin within islets (Data not shown). A limitation of the study was that insulin levels were not measured during glucose tolerance testing. If AhR+/− insulin levels were higher than WT insulin levels during glucose tolerance testing, further investigation using hyperglycemic clamps or isolated islets would be needed to confirm enhanced insulin secretion from pancreatic β cells in AhR+/− mice. If insulin levels were similar in AhR+/− and WT mice, enhanced glucose tolerance in AhR+/− mice could be explained by improved insulin action. Future studies should measure insulin levels during glucose tolerance testing and examine clock gene expression within pancreatic islets.
To further investigate changes in insulin resistance, insulin-stimulated pAKT/total AKT levels were measured. WT HFD mice had significantly lower insulin-stimulated hepatic pAKT (Ser 473 and Thr 308)/Total AKT levels, previously reported to be attenuated in db/db obese mice (Shao et al., 2000). However, AhR+/− mice were protected from the HFD-induced reduction of insulin-stimulated pAKT (Ser473)/total AKT. Previously our lab has shown AhR+/− mice protected at pAKT (Ser473 and Thr 308) (Xu et al., 2015). Differences may be due to animal variability, insulin-stimulation technique or duration of HFD feeding at time of sample collection. Although maximal AKT activity is dependent on phosphorylation of both Ser473 and Thr 308, they are regulated by separate kinases (Alessi et al., 1996). Phosphorylation at Ser473 is regulated by mTOR complex 2 (mTORC2) and DNA-dependent protein kinase (DNA-PK) (Feng et al., 2004; Sarbassov et al., 2005). Thr308 phosphorylation is dependent on 3-phohphoinositide-dependent protein kinase 1 (PDK1) (Alessi et al., 1996, 1997). In the current study, insulin-stimulated Thr 308 phosphorylation by PDK1 may have been compromised, resulting in only partial protection of the AKT signaling pathway by Ser473.
The central circadian clock, the SCN, relays autonomic signals to the liver to influence activity and feeding that generates rhythmic concentrations of serum blood glucose (Buijs et al., 2001; Kalsbeek et al., 2008; Yamamoto et al., 1987). Mice fed a HFD (45% fat) for 6 weeks have significantly increased blood glucose levels with a peak expression during the lights on period (Kohsaka et al., 2007). In the current study, glucose levels were increased at ZT12 and cosinor analysis determined that the glucose rhythm was altered in WT mice. HFD-induced changes in glucose levels were less severe in AhR+/− mice. The mesor of glucose indicates that the levels of glucose, although increased in both genotypes on HFD, were lower in the AhR+/− mice. Future studies should investigate circadian expression in the SCN and mediobasal hypothalamus (MBH) to further compare the central clock and metabolic homeostasis in WT and AhR+/− mice.
Previous studies generally indicate a link between AhR and the circadian clock (Anderson et al., 2013; Garrett and Gasiewicz, 2006; Mukai et al., 2008; Xu et al., 2010; Xu et al., 2013). If AhR activation suppresses rhythmic expression of Clock/Bmal1 target genes, then reduction in AhR may enhance expression of these same targets, and possibly counteract the effects of HFD. However, one study observed no significant changes in Per2 luciferase measured in explants following treatment with the AhR agonist 2,3,7,8-tetrachlorodibenzodioxin (TCDD) (Pendergast and Yamazaki 2012). Differences in explant models and specific TCDD-induced effects on Per2 could explain discrepancies between this study and the others. Following AhR activation, the AhR:Arnt heterodimer interacts with AhR response elements to initiate transcription of AhR target genes (Denison et al., 1988; Ko et al., 1996). Canonical AhR target genes Cyp1A1 and Cyp1B1 are xenobiotic metabolizing genes typically used to indicate activity of AhR (Fujii-Kuriyama and Mimura 2005; Ko et al., 1996). Cyp1B1 was significantly enhanced at ZT8 in liver of WT mice suggesting AhR activation. This was unexpected since the 60% HFD obtained from Research diets has not been reported to contain dioxins (www.researchdiets.com/opensource-diets/diet-induced-disease-models/obesity). Dioxins, hydrophobic halogenated aromatic hydrocarbons, are highly lipophilic, accumulate in fat and have a very long half-life so it is possible that low levels of dioxin are contained within the porcine lard in the HFD formula (Michalek et al., 1996; Schecter et al., 2006). However, high dioxin levels within a controlled diet source are unlikely. Previous studies have not reported upregulation of Cyp1 genes following Western diet feeding (Kerley-Hamilton et al., 2012). However, these studies collected samples from a single time point and may have missed the time of AhR activation.
AhR also binds “nonclassical” ligands found in natural products within the diet (Denison et al., 2002; Gasiewicz et al., 1996; Nguyen and Bradfield 2008). Dietary AhR ligands include indole metabolites and flavonoids in addition to lipoxin A4, prostaglandins and arachidonic acid metabolites (Chiaro et al., 2008; Nguyen and Bradfield 2008). Additionally, lipid and lipid derivatives, such as oxidized low-density lipoproteins, have been identified as AhR agonists (Nguyen and Bradfield 2008). In contrast to the sustained AhR activation induced by TCDD, dietary AhR activation is likely more transient as dietary indole derivatives are rapidly converted to inactive metabolites (Bock and Kohle 2006; Mitchell and Elferink 2009; Nguyen and Bradfield 2008). Although changes in AhR target gene expression are likely transient, future studies should determine rhythmic expression of Cyp1A1 and Cyp1B1 protein levels. AhR promiscuity and the wide variety of possible ligands results in many possible targets to explain the HFD-induced activation of AhR.
The clock genes Per1, Bmal1 and Cry1 are integral components of the transcriptional feedback loop that regulate circadian rhythms (Buhr and Takahashi 2013). In general, AhR+/− mice have increased amplitude in clock gene rhythms in the liver. HFD-induced changes in hepatic gene expression were similar between genotypes. Similar to previous reports, HFD weakly shifted the phase or attenuated rhythmic expression of core clock genes (Eckel-Mahan et al., 2013). Rev-erbα, a transcriptional target of Clock/Bmal1, influences gluconeogenesis and links circadian rhythms and metabolism by creating a feedback loop to inhibit Bmal1 (Buhr and Takahashi 2013; Marcheva et al., 2013). The rhythmic mRNA expression of Rev-erbαa is phase advanced in liver during the lights on period in mice fed a HFD (Kohsaka et al., 2007). AhR+/− mice were protected from the HFD-induced shift in Rev-erbα. PPARα, a ligand activated transcription factor and nuclear receptor, augments the link between the circadian clock and metabolism (Mangelsdorf et al., 1995). Identified as a Clock/Bmal1 target gene, PPARα exhibits circadian expression and acts as an essential component to maintain peripheral circadian rhythms and lipid homeostasis (Marcheva et al., 2013). PPARα was significantly enhanced at three time points measured in WT HFD mice. AhR+/− mice were protected from HFD-induced increases in hepatic PPARα.
HFD produces de novo oscillations of gene expression in metabolic pathways by promoting the use of additional signaling pathways and transcription factors such as PPARγ and Sreb to reorganize metabolism in the liver (Eckel-Mahan et al., 2013). PPARγ, an additional clock-controlled target gene, forms a heterodimer with the retinoic X receptor (RXR) and binds to PPAR response elements (PPREs) in the promoter regions of target genes (Eckel-Mahan et al., 2012; Kliewer et al., 1992). Hepatic expression of PPARγ is elevated in animal models of diabetes and insulin resistance where its activation promotes hepatic steatosis (Gavrilova et al., 2003). PPARγ expression was significantly enhanced in WT HFD mice. Interestingly, PPARγ was enhanced during the lights off period in AhR+/− HFD mice, when mice are normally active.
An additional contributor to HFD-induced transcriptional reprogramming, Srebp, and its target genes Acc and Fasn are regulators of lipid metabolism that oscillate in the liver (Brewer et al., 2005). Srebp1c rhythmic expression and binding to Sterol Response Elements (SRE) is influenced not only by diet but also by Rev-erbα, connecting the peripheral circadian clock to fatty acid and triglyceride biosynthesis (Gilardi et al., 2014; Horton et al., 2002; Inoue et al., 2005; Le Martelot et al., 2009). Additionally, insulin increases Srebp1c mRNA and Srebp1c downstream target genes, while glucagon suppresses Srebp1c (Brewer et al., 2005; Horton et al., 2002; Shimomura et al., 1999). HFD enhances Srebp1c, Acc and Fasn gene expression in WT mice (Kohsaka et al., 2007). Increased insulin levels and phase advanced Rev-erbα expression in WT HFD mice likely contribute to enhanced and shifted lipogenic gene expression. AhR+/− mice were protected against HFD-induced changes in Rev-erbα and Rev-erbα target genes Srebp1c, Acc and Fasn (Figure 4A, D-F). Since insulin was enhanced in both WT and AhR+/− groups, regulation of Srebp1c and target genes is more highly dependent on Rev-erbα.
Rhythmic amplitude of the gluconeogenic gene Pepck was decreased in the HFD WT group. Decreased Pepck under HFD conditions parallels AhR activation by TCDD which disrupts hepatic gluconeogenesis (Stahl et al., 1993). AhR+/− mice were protected from HFD-induced changes in Pepck expression. Decreased AhR expression protected mice from HFD-induced changes of the nuclear receptors Rev-erbα, PPARα and PPARγ, the lipogenic genes Srebp1c, Acc and Fasn, and the gluconeogenic gene Pepck. AhR+/− mice were protected against changes in gene expression, amplitude and phase advances and maintained metabolic homeostasis. Because of baseline differences in gene expression under control conditions in WT and AhR+/− mice, HFD-induced changes were compared within genotypes. Potential differences in response to HFD feeding could potentially be affected by baseline levels.
AhR can interrupt Clock/Bmal1 transcriptional activity by forming a heterodimer with Bmal1 and decreasing Clock/Bmal1 interactions (Xu et al., 2010). Decreased expression of AhR may, therefore, increase the efficiency of Clock/Bmal1 binding, thereby promoting transcriptional activity of this complex (Xu et al., 2010). Because HFD feeding alters Clock/Bmal1-mediated transcriptional regulation of metabolic rhythms through acrophase shifts and amplitude changes, we sought to determine the role of AhR in diet-induced disruption of metabolic rhythms. Metabolic effects of AhR reduction in the liver, independent from the circadian clock, must also be considered as an alternative mechanism and studied further. The AhR target genes Cyp1A1 and Cyp1B1 were altered in WT HFD mice, indicating that AhR is activated specifically during the lights-on period. AhR+/− mice were protected from HFD-induced shifts in activity onset, insulin resistance and changes in the acrophase of blood glucose. Additionally, AhR+/− mice were protected against HFD-induced reorganization of transcriptional profiles displaying resistance to alterations in amplitude and phase shifts of nuclear receptors, lipid metabolism, and gluconeogenic genes that contributed to an attenuated phenotype of diet-induced metabolic dysfunction. This study furthers our understanding of the complex role AhR plays in HFD-induced obesity and reveals AhR as a new target of interest for conservation of whole animal metabolic homeostasis.
Funding
This study was funded by NIEHS (ES017774) to S.A.T.
Footnotes
Declaration of interest
The authors declare no conflict of interest in this work.
References
- Alessi D, Andjelkovic M, Caudwell B, et al. (1996). Mechanism of activation of protein kinase B by insulin and IGF-1. Embo J. 15:6541–51. [PMC free article] [PubMed] [Google Scholar]
- Alessi D, James S, Downes C, et al. (1997). Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 7:261–69. [DOI] [PubMed] [Google Scholar]
- Anderson G, Beischlag T, Vinciquerra M, Mazzoccoli G. (2013). The circadian clock circuitry and the AHR signaling pathway in physiology and pathology. Biochem Pharmacol. 85:1405–16. [DOI] [PubMed] [Google Scholar]
- Angrish M, Mets B, Jones A, Zacharewski T. (2012). Dietary fat is a lipid source in 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-elicited hepatic steatosis in C57BL/6 mice. Toxicol Sci. 128:377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala J, Samuel V, Morton G, et al. (2010). Standard operating procedures for describing and performing metabolic tests for glucose homeostasis in mice. Dis Model Mech. 3:525–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock K, Kohle C. (2006). Ah receptor: Dioxin-mediated toxic responses as hints to deregulated physiologic function. Biochem Pharmacol. 72:393–404. [DOI] [PubMed] [Google Scholar]
- Brewer M, Lange D, Baler R, Anzulovich A. (2005). SREBP-1 as a transcriptional integrator of circadian and nutritional cues in the liver. J Biol Rhythms. 20:195–205. [DOI] [PubMed] [Google Scholar]
- Buhr E, Takahashi J. (2013). Molecular components of the mammalian circadian clock. Handb Exp Pharmacol. 217:3–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijs R, Chun S, Niijima A, et al. (2001). Parasympathetic and sympathetic control of the pancreas: A role for the hypothalamic centers that are involved in the regulation of food intake. J Comp Neurol. 431:405–23. [DOI] [PubMed] [Google Scholar]
- Challet E (2010). Interactions between light, mealtime and calorie restriction to control daily timing in mammals. J Comp Physiol B. 180:631–44. [DOI] [PubMed] [Google Scholar]
- Chiaro C, Patel R, Perdew G. (2008). 12(R)-Hydroxy-5(Z),8(Z),10(E),14(Z)-eicosatetraenoic acid [12(R)-HETE], an arachidonic acid derivative, is an activator of the aryl hydrocarbon receptor. Mol Pharmacol. 74:1649–56. [DOI] [PubMed] [Google Scholar]
- Denison M, Fisher J, Whitlock J. (1988). The DNA recognition site for the dioxin-Ah receptor complex. Nucleotide sequence and functional analysis. J Biol Chem. 263:17221–24. [PubMed] [Google Scholar]
- Denison M, Pandini A, Nagy S, et al. (2002). Ligand binding and activation of the Ah receptor. Chem Biol Interact. 141:3–24. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan K, Patel V, de Mateo S, et al. (2013). Reprogramming of the circadian clock by nutritional challenge. Cell. 155:1464–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan K, Patel V, Mohney R, et al. (2012). Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci USA. 109:5541–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquirol Y, Perret B, Ruidavets J, et al. (2011). Shift work and cardiovascular risk factors: New knowledge from the past decade. Arch Cardiovasc Dis. 104:636–68. [DOI] [PubMed] [Google Scholar]
- Feng J, Park J, Cron P, et al. (2004). Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J Biol Chem. 279:41189–96. [DOI] [PubMed] [Google Scholar]
- Fujii-Kuriyama Y, Mimura J. (2005). Molecular mechanisms of AhR functions in the regulation of cytochrome P450 genes. Biochem Biophys Res Commun. 338:311–17. [DOI] [PubMed] [Google Scholar]
- Garrett R, Gasiewicz T. (2006). The aryl hydrocarbon receptor agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin alters the circadian rhythms, quiescence, and expression of clock genes in murine hematopoietic stem and progenitor cells. Mol Pharmacol. 69:2076–83. [DOI] [PubMed] [Google Scholar]
- Gasiewicz T, Kende A, Rucci G, et al. (1996). Analysis of structural requirements for Ah receptor antagonist activity: Ellipticines, flavones, and related compounds. Biochem Pharmacol. 52:1787–803. [DOI] [PubMed] [Google Scholar]
- Gavrilova O, Haluzik M, Matsusue K, et al. (2003). Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J Biol Chem. 278:34268–76. [DOI] [PubMed] [Google Scholar]
- Gilardi F, Migliavacca E, Naldi A, et al. (2014). Genome-wide analysis of SREBP1 activity around the clock reveals its combined dependency on nutrient and circadian signals. PLoS Genet. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Hogenesch J, Bradfield C. (2000). The PAS super-family: Sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol. 40:519–61. [DOI] [PubMed] [Google Scholar]
- Henriksen G, Ketchum N, Michalek J, Swaby J. (1997). Serum dioxin and diabetes mellitus in veterans of Operation Ranch Hand. Epidemiology. 8:252–58. [DOI] [PubMed] [Google Scholar]
- Horton J, Goldstein J, Brown M. (2002). SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 109:1125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue I, Shinoda Y, Ikeda M, et al. (2005). CLOCK/BMAL1 is involved in lipid metabolism via transactivation of the peroxisome prolfierator-activated receptor (PPAR) response element. J Atheroscler Thromb. 12:169–74. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, Foppen E, Schalij C, et al. (2008). Circadian control of the daily plasma gluocse rhythm: An interplay of GABA and glutamate. PLoS One. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerley-Hamilton J, Trask H, Ridley C, et al. (2012). Obesity is mediated by differential aryl hydrocarbon receptor signaling in mice fed a western diet. Environ Health Perspect. 120:1252–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer S, Umesono K, Mangelsdorf D, Evans R. (1992). Retinoid X receptor interacts with nuclear receptors in retinoic acid, thyroid hormone and vitamin D3 signalling. Nature. 355:446–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko H, Okino S, Ma Q, Whitlock J. (1996). Dioxin-induced CYP1A1 transcription in vivo: The aromatic hydrocarbon receptor mediates transactivation, enhancer-promoter communication, and changes in chromatin structure. Mol Cell Biol. 16:430–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka A, Laposky A, Ramsey K, et al. (2007). High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 6:414–21. [DOI] [PubMed] [Google Scholar]
- Lee J, Wada T, Febbraio M, et al. (2010). A novel role for the dioxin receptor in fatty acid metabolism and hepatic steatosis. Gastroenterology. 139:653–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Martelot G, Claudel T, Gatfield D, et al. (2009). REV-ERBα participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf D, Thummel C, Beato M, et al. (1995). The nuclear receptor superfamily: The second decade. Cell. 83:835–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B, Ramsey K, Peek C, et al. (2013). Circadian clocks and metabolism. Handb Exp Pharmacol. 217:127–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh B, Hogenesch J, Bradfield C. (2010). Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu Rev Physiol. 72:625–45. [DOI] [PubMed] [Google Scholar]
- Mendoza J, Pevet P, Challet E. (2008). High-fat feeding alters the clock synchronization to light. J Physiol. 586:5901–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek J, Pirkle J,Caudill S, et al. (1996). Pharmacokinetics of TCDD in veterans of operation ranch hand: 10-year follow-up. J Toxicol Environ Health. 47:209–20. [DOI] [PubMed] [Google Scholar]
- Mitchell K, Elferink C. (2009). Timing is everything: Consequences of transient and sustained AhR activity. Biochem Pharmacol. 77:947–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura J, Fujii-Kuriyama Y. (2003). Functional role of AhR in the expression of toxic effects by TCDD. Biochim Biophys Acta. 1619:263–68. [DOI] [PubMed] [Google Scholar]
- Moyer B, Rojas I, Kerley-Hamilton J, et al. (2016). Inhibition of the aryl hydrocarbon receptor prevents Western diet-induced obesity. Model for AHR activation by kynurenine via oxidized-LDL, TLR2/4, TGFβ, and IDO1. Toxicol Appl Pharmacol. 300:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai M, Lin T, Peterson R, et al. (2008). Behavioral rhythmicity of mice lacking AhR and attenuation of light-induced phase shift by 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Rhythms. 23:200–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L, Bradfield C. (2008). The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol. 21:102–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlee S, Lentz S, Mori H, MacDougald O. (2014). Quantifying size and number of adipocytes in adipose tissue. Methods Enzymol. 537:93–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergast J, Yamazaki S. (2012). The mammalian circadian system is resistant to dioxin. J Biol Rhythms. 27:156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refinetti R, Lissen G, Halberg F. (2007). Procedures for numerical analysis of circadian rhythms. Biol Rhythm Res. 38:275–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisz-Porszasz S, Probst M, Fukunaga B, Hankinson O. (1994). Identification of functional domains of the aryl hydrocarbon receptor nuclear translocator protein (ARNT). Mol Cell Biol. 12:6075–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remillard R, Bunce N. (2002). Linking dioxins to diabetes: Epidemiologiy and biologic plausibility. Environ Health Perspect. 110:853–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J, Reick M, McKnight S. (2002). Metabolism and the control of circadian rhythms. Annu Rev Biochem. 71:307–31. [DOI] [PubMed] [Google Scholar]
- Sarbassov D, Guertin D, Ali S, Sabatini D. (2005). Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 307:1098–101. [DOI] [PubMed] [Google Scholar]
- Schecter A, Birnbaum L, Ryan J, Constable J. (2006). Dioxins: An overview. Environ Res. 101:419–28. [DOI] [PubMed] [Google Scholar]
- Schmidt J, Su G, Reddy J, et al. (1996). Characterization of a murine Ahr null allele: Involvement of the Ah receptor in hepatic growth and development. Proc Natl Acad Sci USA. 93:6731–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Yamashita H, Qiao L, Friedman J. (2000). Decreased Akt kinase activity and insulin resistance in C57BL/KsJ-Leprdb/db mice. J Endocrinol. 167:107–15. [DOI] [PubMed] [Google Scholar]
- Shimomura I, Bashmakov Y, Ikemoto S, et al. (1999). Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci USA. 96:13656–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A, Rosenbaum P, Weinstock R, et al. (2012). Polychlorinated biphenyl (PCB) exposure and diabetes: Results from the Anniston Community Health Survey. Environ Health Prospect. 120:727–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl B, Beer D, Weber L, Rozman K. (1993). Reduction of hepatic phosphoenolpyruvate carboxykinase (PEPCK) activity by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) is due to decreased mRNA levels. Toxicology. 79:81–95. [DOI] [PubMed] [Google Scholar]
- [USEPA] U.S. EPA. (2004). Exposure and human health reassessment of 2,3,7,8-tetrachlorodibenzo-p-dioxin and related compounds. National Academy Sciences (External Review Draft) U.S. Environmental Protection Agency, Washington, DC, EPA/600/P-00/001Cb. [Google Scholar]
- Wang C, Xu C, Krager S, et al. (2011). Aryl hydrocarbon receptor deficiency enhances insulin sensitivity and reduces PPAR-α pathway activity in mice. Environ Health Perspect. 119:1739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Krager S, Liao D, Tischkau S. (2010). Disruption of CLOCK-BMAL1 transcriptional activity is responsible for aryl hydrocarbon receptor-mediated regulation of Period1 gene. Toxicol Sci. 115:98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang C, Krager S, et al. (2013). Aryl hydrocarbon receptor activation attenuates Per1 gene induction and influences circadian clock resetting. Toxicol Sci. 132:368–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang C, Zhang Z, et al. (2015). Aryl hydrocarbon receptor deficiency protects mice from diet-induced adiposity and metabolic disorders through increased energy expenditure. Int J Obes (Lond). 39:1300–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Nagai K, Nakagawa H. (1987). Role of SCN in daily rhythms of plasma glucose, FFA, insulin and glucagon. Chronobiol Int. 4:483–91. [DOI] [PubMed] [Google Scholar]