SUMMARY
The corneal epithelium is renowned for high regenerative potential, which is dependent on the coordinated function of its diverse progenitor subpopulations. However, the molecular pathways governing corneal epithelial progenitor differentiation are incompletely understood. Here, we identify a highly proliferative limbal epithelial progenitor subpopulation characterized by expression of basal cell adhesion molecule (BCAM) that is capable of holocone formation and corneal epithelial sheet generation. BCAM-positive cells can be found among ABCB5-positive limbal stem cells (LSCs) as well as among ABCB5-negative limbal epithelial cell populations. Mechanistically, we show that BCAM is functionally required for cellular migration and differentiation and that its expression is regulated by the transcription factor p63. In aggregate, our study identifies limbal BCAM expression as a marker of highly proliferative corneal epithelial progenitor cells and defines the role of BCAM as a critical molecular mediator of corneal epithelial differentiation.
Graphical abstract
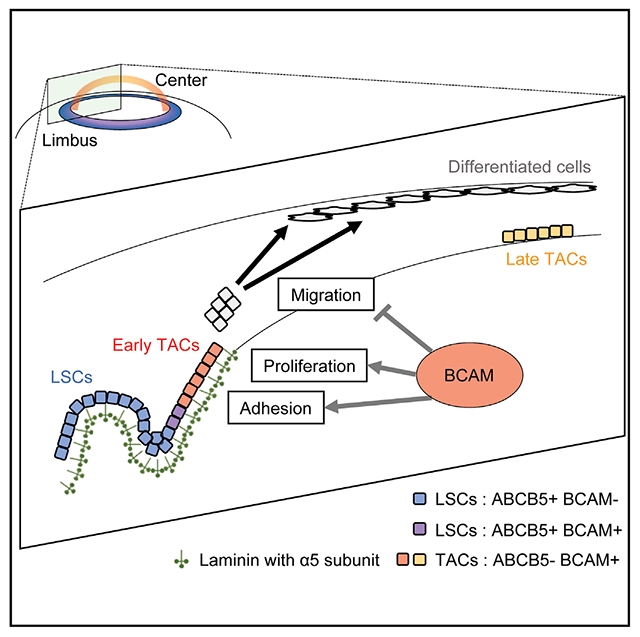
In brief
Using scRNA sequencing of ABCB5-positive human limbal stem cells, Sasamoto et al. identify a BCAM-positive highly proliferative limbal epithelial progenitor subpopulation that is capable of holocone formation and corneal epithelial sheet generation. BCAM regulated by the stem cell transcription factor p63 is functionally required for corneal cell migration and differentiation.
INTRODUCTION
The corneal epithelium is renowned for its high regenerative potential, which is dependent on the coordinated function of its progenitor subpopulations. Corneal epithelial stem cells are located in the cornea’s outermost structure, termed the limbus (Davanger and Evensen, 1971; Kenyon and Tseng, 1989; Schermer et al., 1986), and are also known as limbal stem cells (LSCs). LSCs give rise to corneal transit-amplifying cells (TACs), which can migrate along the basement membrane from the limbus toward the central cornea and generate differentiated corneal epithelial cells (Beebe and Masters, 1996; Castro-Munozledo and Gomez-Flores, 2011; Park et al., 2019; Thoft and Friend, 1983; Yoon et al., 2014). Limbal TACs are considered to be “early” or immature due to their relatively undifferentiated phenotype, as opposed to “late” or mature central cornea TACs, which begin expressing corneal differentiation markers such as KRT12 (Kaplan et al., 2019; Lehrer et al., 1998). Recently, single-cell RNA sequencing (scRNA-seq) analyses of mixed corneal populations were performed and have begun to shed more light on corneal epithelial heterogeneity, including the LSC niche (Catala et al., 2021; Collin et al., 2021; Kaplan et al., 2019; Li et al., 2021).
ATP-binding cassette (ABC) superfamily member ABCB5 represents a molecular marker for LSCs (Jongkhajornpong et al., 2016; Ksander et al., 2014; Kureshi et al., 2015; Mathan et al., 2016; Norrick et al., 2021; Parfitt et al., 2015; Shaharuddin et al., 2016, 2017), and prospectively isolated ABCB5-positive LSCs, but not ABCB5-negative limbal epithelial cells, have been shown to possess a capacity of long-term (>1 year) corneal epithelial restoration in the setting of experimental LSC deficiency (LSCD) (Ksander et al., 2014). In these studies, ABCB5 was found to be preferentially expressed on label-retaining, quiescent LSCs that were depleted in Abcb5 knockout mice due to enhanced proliferation and apoptosis (Ksander et al., 2014). Moreover, ABCB5-positive cell frequency was shown to be significantly reduced in patients with clinical LSCD. While these studies unequivocally demonstrated that LSCs were contained within the ABCB5-positive cell fraction, the molecular mechanisms governing their differentiation have remained largely unknown.
The centripetal migration of corneal progenitors along the basement membrane is essential for corneal epithelial differentiation (Thoft and Friend, 1983). The rate of this migration is tightly controlled by direct interactions between adhesion molecules and laminins (Sekiguchi and Yamada, 2018). Basal cell adhesion molecule (BCAM) is a transmembrane glycoprotein that specifically binds to laminins containing the α5 chain (Kikkawa et al., 2002; Moulson et al., 2001; Parsons et al., 2001a). Laminin α5 preferentially localizes to the basement membrane of the limbal epithelium and is essential for stem cell self-renewal (Domogatskaya et al., 2008; Polisetti et al., 2017). While BCAM has additionally been found to be expressed by immortalized corneal epithelial cells (Hasenson et al., 2005), a function of BCAM in progenitor differentiation and corneal epithelial formation has not been described to date.
Here, we show that limbal BCAM expression identifies a highly proliferative limbal epithelial progenitor subpopulation that is capable of holocone formation and corneal epithelial sheet generation. BCAM-positive cells can be found among ABCB5-positive LSCs as well as among ABCB5-negative limbal epithelial cell populations. Mechanistically, we demonstrate that BCAM is functionally required for cellular migration and differentiation and that its expression is regulated by the transcription factor p63. In aggregate, our study identifies limbal BCAM expression as a marker of highly proliferative corneal epithelial progenitor cells and defines the role of BCAM as a critical molecular mediator of corneal epithelial differentiation.
RESULTS
Limbal BCAM expression marks corneal epithelial progenitors
Based on the reports of BCAM expression in immortalized corneal epithelial cell lines (Hasenson et al., 2005) and BCAM function as a laminin α5 receptor (Kikkawa et al., 2002; Moulson et al., 2001; Parsons et al., 2001a), we hypothesized that limbal BCAM-positive cells might represent a progenitor population in the cornea (Figures 1A and 1B). Flow cytometry analysis revealed that BCAM was expressed by 11.4% ± 6.6% of freshly isolated imbal epithelial cells (n = 16) (Figure S1A). 20.18% ± 3.40% of ABCB5-positive LSCs and 20.59% ± 13.46% of ABCB5-negative limbal epithelial cells co-expressed BCAM when analyzed by dual-color flow cytometry in additional samples (n = 3) (Figures 1C and S1B). In vitro culture expansion resulted in significantly increased proportions of BCAM-positive cells. Specifically, in in-vitro-expanded limbal epithelial cultures, 88.86% ± 3.77% of ABCB5-positive LSCs and 90.46% ± 5.31% ABCB5-negative limbal epithelial cells were BCAM positive (n = 5) (Figure 1D). Side-by-side comparison revealed high proliferative potential of both BCAM-positive/ABCB5-positive LSCs and BCAM-positive/ABCB5-negative limbal epithelial subpopulations compared with both BCAM-negative/ABCB5-positive LSCs and BCAM-negative/ABCB5-negative limbal epithelial cells (Figure 1E). Purified BCAM-positive/ABCB5-positive LSCs exhibited both stem cell-intrinsic self-renewal capacity, as evidenced by 3.33% ± 2.28% positivity for this phenotype in 9.37 ± 6.56-fold cell number-expanded 17-day cultures (n = 3), as well as stem cell-characteristic differentiation capacity, through giving rise, de novo, to a majority of BCAM-positive/ABCB5-negative cells in such cultures (93.28% ± 2.07% of cells, n = 3) (Figure 1F). Notably, a small number of BCAM-negative/ABCB5-positive cells (0.45% ± 0.07%, n = 3) (Figure 1F) could also be detected in these cultures. These findings suggest that limbal BCAM-positive cells represent a corneal progenitor population with high proliferative potential.
Figure 1. BCAM marks limbal corneal epithelial progenitors.
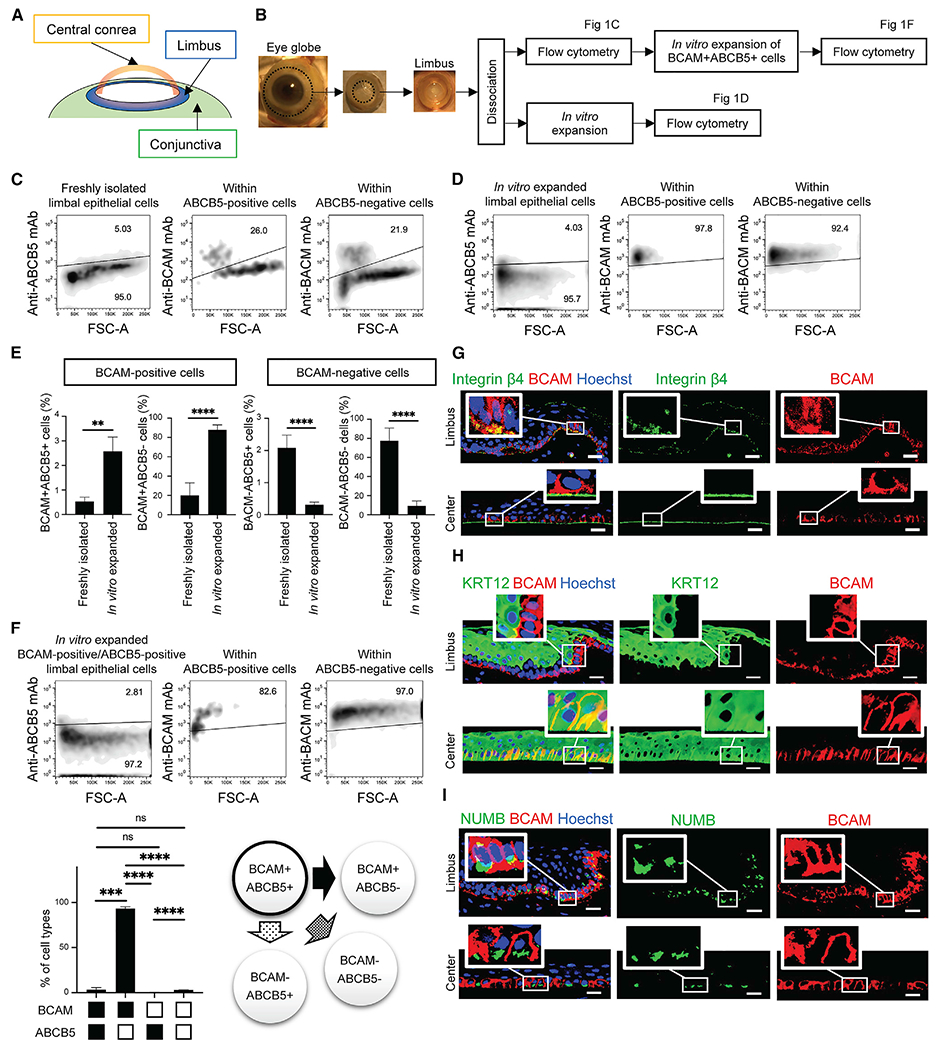
(A) Schematic illustration of the ocular surface anatomy.
(B) Schematic illustration of the experimental design in (C)–(F).
(C) Representative flow cytometric analyses of ABCB5 and BCAM expression in freshly isolated limbal epithelial cells. FSC, forward scatter; A, area. n = 3.
(D) Representative flow cytometric analyses of ABCB5 and BCAM expression in in-vitro-expanded limbal epithelial cell cultures. FSC, forward scatter; A, area. n = 5.
(E) Bar graphs represent the relative percentages of ABCB5-positive/BCAM-positive, ABCB5-positive/BCAM-negative, ABCB5-negative/BCAM-positive, and ABCB5-negative/BCAM-negative subpopulations in freshly isolated limbal epithelial cells and in-vitro-expanded limbal epithelial cultures (mean ± SD; **p < 0.01, ****p < 0.0001). Data were analyzed using a t test.
(F) Top, representative flow cytometric analyses of ABCB5 and BCAM expression in in-vitro-expanded ABCB5-positive/BCAM-positive limbal epithelial cells. Bottom left, bar graph illustrates the relative percentages of ABCB5-positive/BCAM-negative, ABCB5-positive/BCAM-positive, ABCB5-negative/BCAM-positive, and ABCB5-negative/BCAM-positive subpopulations (mean ± SD; n = 3; ***p < 0.001, ****p < 0.0001). Data were analyzed using a Tukey’s multiple comparisons test. Bottom right, schematic illustration of the ABCB5-positive/BCAM-positive cell fate. FSC, forward scatter; A, area.
(G) Representative immunostaining analyses of integrin β4 (green) and BCAM (red) expression in the limbus and central cornea. Nuclei stained with Hoechst 33342 (blue), n = 3. Scale bar, 20 μm.
(H) Representative immunostaining analyses of KRT12 (green) and BCAM (red) co-expression in the limbus and central cornea. Nuclei stained with Hoechst 33342 (blue), n = 3. Scale bar, 20 μm.
(I) Representative immunostaining analyses of NUMB (green) and BCAM (red) co-expression in the limbus and central cornea. Nuclei stained with Hoechst 33342 (blue), n = 3. Scale bar, 20 μm.
Immunofluorescent analyses of human cornea revealed that BCAM was expressed only in cells forming the basal layer of the limbus and the central cornea, i.e., known locations of stem cells and corneal TACs (Figure 1G) (Lehrer et al., 1998). BCAM-positive epithelial cells in the basal layer co-expressed integrin β4, a TAC marker (Hayashi et al., 2016) (Figure 1G). Limbal BCAM-positive cells did not express the corneal differentiation marker KRT12 (Figure 1H), indicating an early TAC phenotype (Lehrer et al., 1998). In contrast, central corneal BCAM-positive cells were KRT12 positive (Figure 1H), suggesting a late TAC phenotype (Lehrer et al., 1998). Additionally, fluorescence-activated cell sorted (FACS) BCAM-positive limbal epithelial cells exhibited higher levels of BCAM mRNA and lower levels of KRT12 mRNA expression compared with BCAM-negative cells (fold change BCAM-positive versus -negative cells: BCAM: 4.14 ± 3.90, p = 0.0078; KRT12: 0.42 ± 0.32, p = 0.0078, n = 8) (Figure S1C). No significant difference in BCAM-positive cell frequency was observed between the limbus and the central cornea (Figure S1D). Moreover, NUMB, a segregating determinant of asymmetric cell division (Knoblich, 2008), was expressed by a subset of BCAM-positive cells along the entire corneal epithelium (Figure 1I), suggesting that BCAM-positive progenitor cells both in the limbus and the central cornea can undergo asymmetric cell division, giving rise to more differentiated, BCAM-negative cells.
scRNA-seq of ABCB5-positive LSCs reveals a distinct BCAM-expressing subpopulation
To further examine the BCAM-positive cell subpopulation that co-expressed ABCB5, we performed targeted scRNA-seq of ABCB5-positive LSCs prospectively isolated from the limbus of human donors, as described previously (Figures 1B, 2A, and S2A) (Ksander et al., 2014; Sasamoto et al., 2020). Donor characteristics (n = 3) are described in Table S1. After batch correction with Harmony (Figure S2B), Seurat clustering identified a BCAM-low cluster 1 and a BCAM-high cluster 2 within ABCB5-positive LSCs (Figure 2B). While both clusters expressed the eye transcription factor PAX6 and LSC markers KRT15 and TP63 (Figure 2C), cluster 2 was also distinguished by the presence of cells with higher levels of MYC mRNAs (Figures 2D and S2C), which is known to play a role in stem cell to TAC progression (Arnold and Watt, 2001). Also observed in cluster 2 were reduced p15 and enhanced Cyclin D2 expression associated with the G1- to S-phase transition (Bretones et al., 2015) (Figure 2D). The MYChighP15lowCyclinD2high molecular phenotype of BCAM-positive/ABCB5-positive LSCs is consistent with their high proliferative potential (Figure 1E).
Figure 2. scRNA-seq analysis of ABCB5-positive LSCs identifies stem cell heterogeneity.
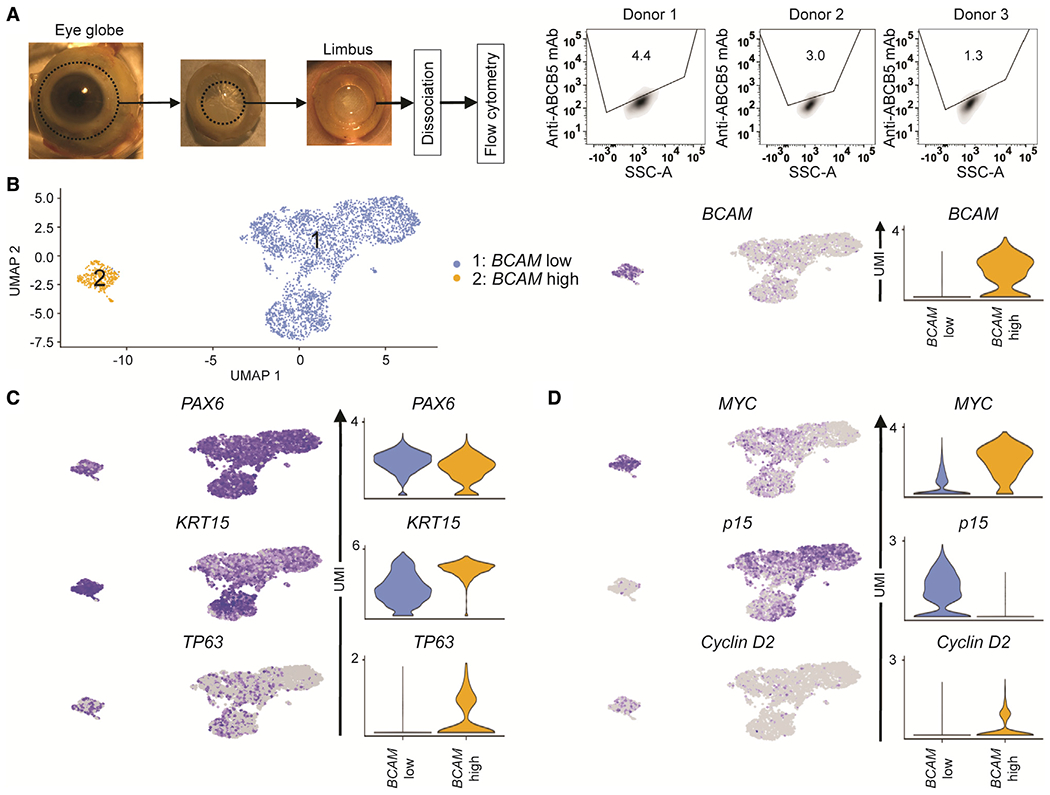
(A) Left, illustration of ABCB5-positive LSC isolation from human limbus. Right, flow cytometric analyses of ABCB5 expression in human donor limbal tissues used for scRNA-seq analyses (n = 3).
(B) Uniform manifold approximation and projection (UMAP) plot of 3,417 limbal epithelial cells isolated from three human donors (left). Feature plot (middle) and violin plot (right) depicting the expression of BCAM.
(C) Feature plots (left) and violin plots (right) depicting the expression of the eye transcription factor PAX6 and the LSC markers KRT15 and TP63.
(D) Feature plots (left) and violin plots (right) illustrating expression of MYC, p15, and Cyclin D2.
Limbal BCAM-positive cells possess holoclone-forming capacity
To test self-renewal potential, we performed comparative colony-forming assays and holoclone assays of in-vitro-expanded BCAM-positive and BCAM-negative cells. We found that compared with limbal BCAM-negative cells, BCAM-positive limbal epithelial cells had higher colony-forming efficiency (CFE) (BCAM-positive versus -negative cells: 14.45% ± 4.64% versus 2.25% ± 1.79%, p < 0.0001, n = 12) (Figure 3A), and generated a significantly higher number of holoclones (5.06% ± 2.65%, p = 0.0028, n = 7) (Figures 3B and S3A). Central-cornea-derived BCAM-positive cells also possessed higher CFE than central-cornea-derived BCAM-negative cells (BCAM-positive versus-negative cells: 3.97% ± 4.52% versus 0.71% ± 1.11%, p = 0.0479, n = 7) (Figure 3C), which, however, was significantly lower than the CFE of BCAM-positive limbal cells (limbus versus central cornea: 15.09% ± 5.41% versus 3.97% ± 4.52%, p = 0.0081, n = 7). Compared with limbal BCAM-positive cells, central cornea BCAM-positive cells also formed 31.7% ± 33.3% smaller colonies (p = 0.0453, n = 7) (Figure S3B) and exhibited a 91.9% ± 13.0% lower proliferation rate (p < 0.0001, n = 5) (Figure S3C). While the holoclone-forming capacity of limbal BCAM-positive cells is indicative of the cell-intrinsic self-renewal capacity, the high CFE is consistent with their proliferative phenotype.
Figure 3. Corneal differentiation potential of limbal BCAM-positive cells.
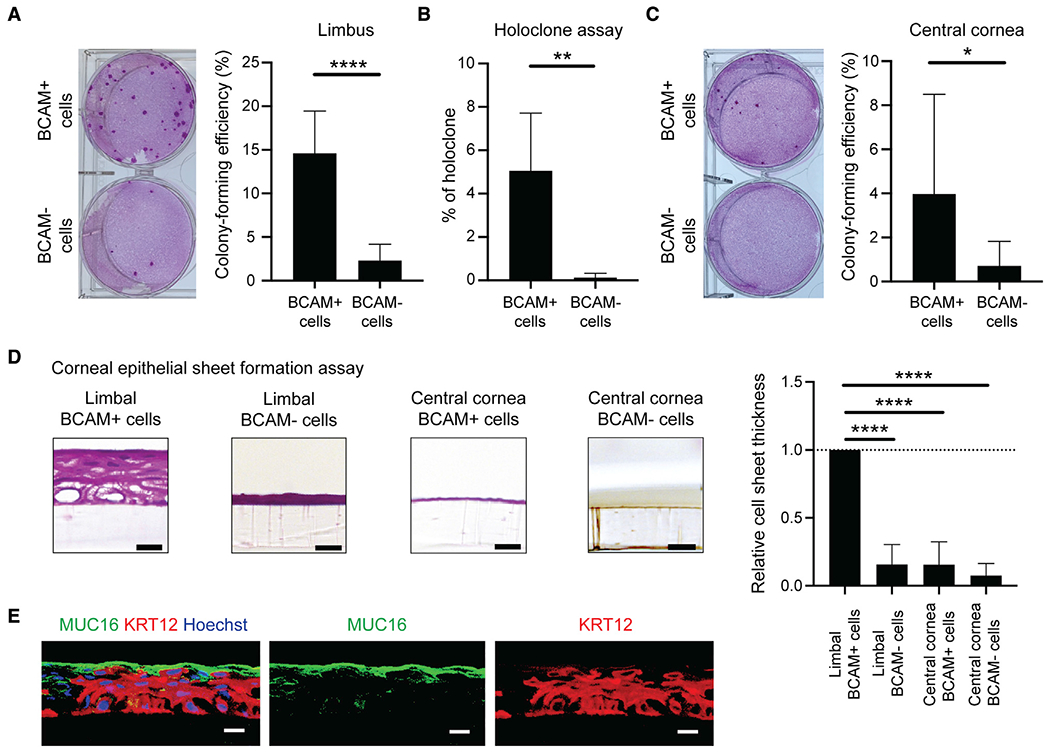
(A) Left, representative macroscopic images of the cell colonies generated by limbal BCAM-positive and BCAM-negative cells. Individual colonies are stained with Rhodamine B (pink). Right, bar graph depicts comparative analyses of colony-forming efficiency (mean ± SD; n= 12; ****p < 0.0001). Data were analyzed using a paired t test.
(B) Bar graph illustrates comparative analyses of limbal BCAM-positive and -negative cell holoclone-forming efficiency (mean ± SD; n = 7; **p < 0.01). Data were analyzed using a paired t test.
(C) Left, representative macroscopic images of the cell colonies generated by central corneal BCAM-positive and -negative cells. Right, bar graph depicts comparative analyses of colony-forming efficiency (mean ± SD; n = 7; *p < 0.05). Data were analyzed using a paired t test.
(D) Left, representative H&E-stained images of the cell sheets generated by BCAM-positive and -negative cells isolated from the limbus and central cornea. Right, bar graph depicts the comparative thickness analyses of the cell sheets generated by BCAM-positive and -negative cells isolated from the limbus and central cornea (mean ± SD; n = 7; ****p < 0.0001). Data were analyzed using a Tukey’s multiple comparisons test.
(E) Representative immunostaining analyses of MUC16 (green) and KRT12 (red) expression in the cell sheet derived from limbal BCAM-positive cells. Nuclei stained with Hoechst 33342 (blue), n = 4. Scale bar, 20 mm.
Stratified corneal epithelial formation by limbal BCAM-positive cells
To determine the role of BCAM in the formation of stratified corneal epithelium, we cultured BCAM-positive or -negative cells isolated from the limbus and the central cornea in corneal maturation medium in the presence of a 3T3-J2 feeder layer, as previously described (Chen et al., 2017; Hayashi et al., 2016, 2017). At day 20 of culture, histological examination revealed that only limbal BCAM-positive cells gave rise to multilayered epithelial sheets (Figure 3D) that expressed the markers of mature corneal epithelium, MUC16 and KRT12 (Figure 3E). Additionally, quantitative analyses showed significantly lower thicknesses of epithelial sheets that arose from limbal BCAM-negative cells (84.3% ± 14.7%, p < 0.0001), central corneal BCAM-positive cells (84.4 %± 16.9%, p < 0.0001), or central corneal BCAM-negative cells (92.5% ± 8.9%, p < 0.0001) (Figure 3D) compared with those epithelial sheets generated by limbal BCAM-positive cells.
Laminin α5 is secreted by limbal BCAM-positive cells and deposited in the vicinity of BCAM-positive limbal progenitors
Based on the function of laminin α5 as a BCAM ligand (Parsons et al., 2001b) and the reported contribution of laminin α5 to corneal epithelial sheet formation (Shibata et al., 2018), we hypothesized that the unique ability of limbal BCAM-positive cells to efficiently generate stratified epithelium might be explained by BCAM/laminin α5 interactions. scRNA-seq showed that LAMA5 was expressed by both clusters 1 and 2 (Figure 4A). Immunostaining of human corneas showed preferential laminin α5 expression in the limbus, as also observed by Polisetti et al. (2017) (Figure 4B). Furthermore, western blot analyses showed laminin α5 protein production by in-vitro-expanded limbal epithelial cell cultures (Figure 4C). Since only BCAM-positive cells isolated from the limbus and not from the central cornea were capable of generating a stratified epithelial sheet, these results suggested that laminin α5 expressed by limbal BCAM-positive cells drives corneal differentiation through modulation of BCAM signaling.
Figure 4. ABCB5-positive LSCs express the BCAM ligand laminin α5.
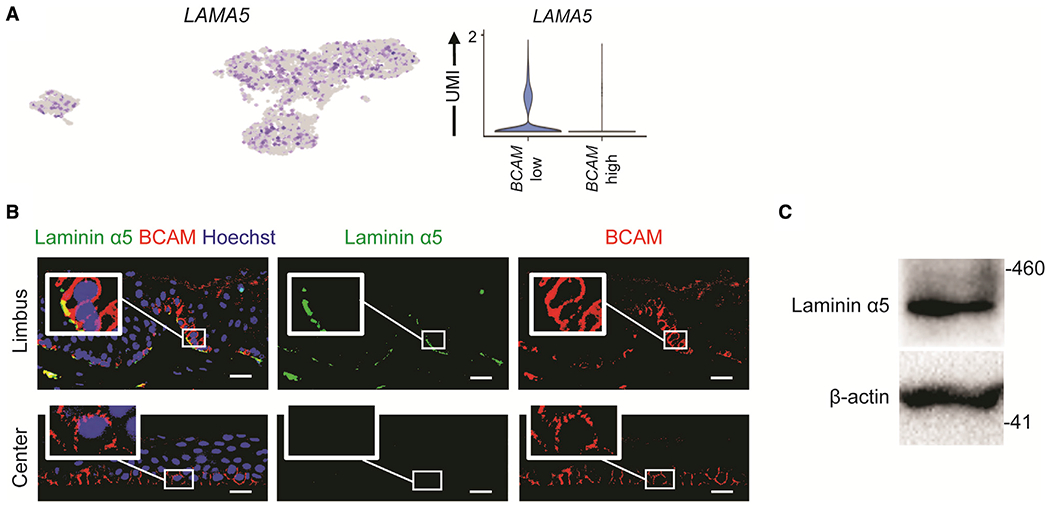
(A) Left, a feature plot, and right, a violin plot depicting LAMA5 expression in each cluster.
(B) Representative immunostaining analyses of laminin α5 (green) and BCAM (red) expression in the limbus and central cornea. Nuclei stained with Hoechst 33342 (blue). n = 3. Scale bar: 20 μm.
(C) Western blot analyses of laminin α5 expression in cultured limbal epithelial cells.
BCAM maintains corneal epithelial stratification through regulation of cell adhesion and migration
To test whether BCAM was functionally required for corneal epithelial stratification through differentiation, we performed BCAM knockdown in limbal epithelial cell cultures using two distinct siRNAs, designated as BCAM KD#1 and BCAM KD#2 (Figures S4A and S4B). We found that BCAM knockdown led to inhibited differentiation as evidenced by significant reduction of corneal epithelial sheet thickness (BCAM KD#1: 35.3% ± 23.1%, p = 0.0120; BCAM KD#2: 55.5% ± 23.3%, p = 0.0013, n = 7) (Figure 5A). As a potential mechanism for deficient differentiation-dependent epithelial sheet formation, we found that downregulation of BCAM expression also resulted in reduction of cell adhesion and proliferation, as evidenced by a significantly attenuated CFE (control versus BCAM KD#1: 31.35% ± 15.92% versus 16.61% ± 8.02%, p = 0.0055; control versus BCAM KD#2: 31.35% ± 15.92% versus 3.99% ± 5.13%, p = 0.0013, n = 8) (Figure 5B) and significant induction of TAC migration, as assessed in a wound-closure migration assay (F (2.988, 17.93) = 3.223, p = 0.0475; n = 7) (Figure 5C).
Figure 5. BCAM maintains corneal epithelial stratification through regulation of cell adhesion and migration.
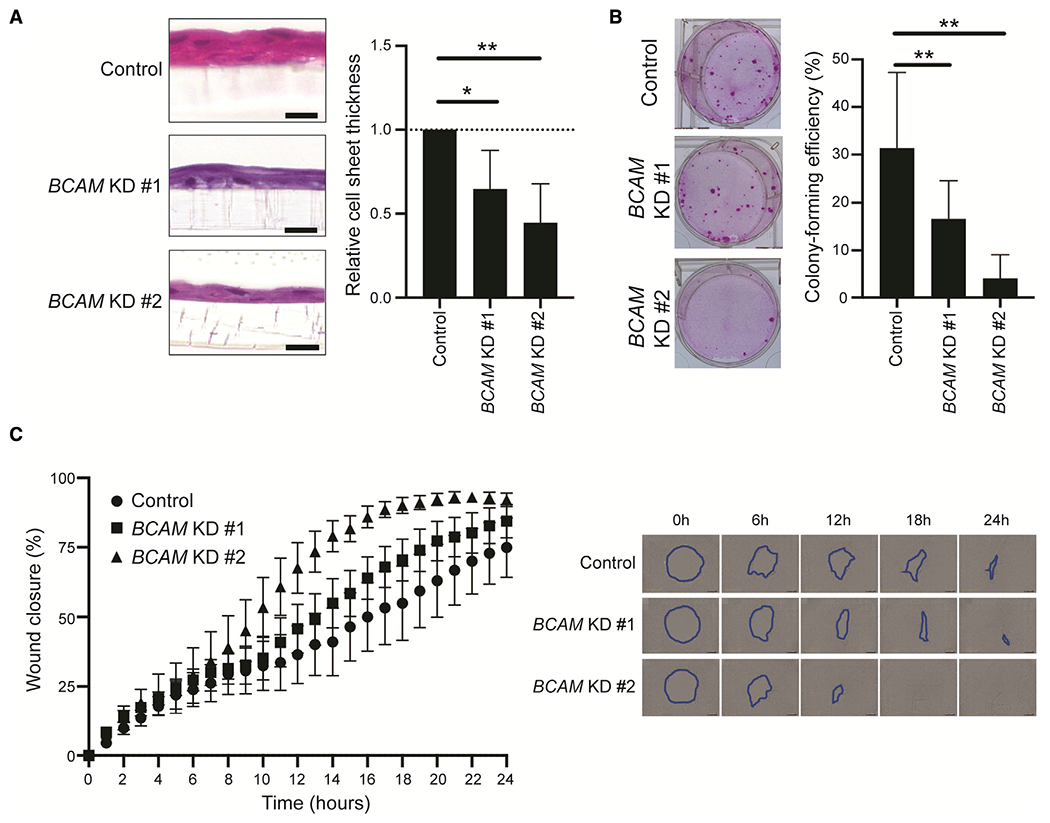
(A) Left, representative H&E-stained images of the cell sheets generated by limbal epithelial cells subjected to siRNA-induced BCAM blockade. Right, bar graph depicts the comparative thickness analyses of the cell sheets generated by control and BCAM siRNA-treated cells (mean ± SD; n = 7; *p < 0.05, **p < 0.01). Data were analyzed using a Dunnett’s multiple comparisons test. KD, knockdown.
(B) Left, representative macroscopic images of the cell colonies generated by control and BCAM siRNA-treated cells. Right, bar graph depicts comparative analyses of colony-forming efficiency (mean ± SD; n = 8; **p < 0.01). Data were analyzed using a Dunnett’s multiple comparisons test.
(C) Left, line graph represents the quantitative analyses of wound closure in control and BCAM siRNA-treated cells (mean ± SEM; n = 7; p = 0.0475). Data were analyzed using a two-way ANOVA. A representative phase-contrast image is shown on the right.
BCAM is a downstream target of the LSC transcription factor p63
Based on the well-established expression of the ectodermal lineage transcription factor p63 in limbal basal epithelial cells (Pellegrini et al., 2001) and on TP63 mRNA expression detected in the BCAMhigh cluster detected by scRNA-seq, we hypothesized that p63 might be involved in the induction of BCAM. Immunostaining of human corneas revealed high p63 nuclear expression in BCAM-positive limbal epithelial cells (Figure 6A). To further dissect the role of p63 in regulating BCAM, we first performed TP63 knockdown in limbal epithelial cell cultures using two distinct small interfering RNAs (siRNAs), designated as TP63 KD#1 and TP63 KD# 2 (Figures S5A and S5B). We found that TP63 knockdown led to significant reduction of BCAM mRNA (TP63 KD#1: 56.6% ± 14.7%, p < 0.0001; TP63 KD# 2: 67.7% ± 12.4%, p < 0.0001, n = 8) and protein (TP63 KD#1: 37.1% ± 10.2%, p = 0.0006; TP63 KD# 2: 55.1% ± 14.5%, p = 0.0005; n = 6) expression levels (Figures 6B, 6C, and S5C).
Figure 6. BCAM is a downstream target of the transcription factor p63.
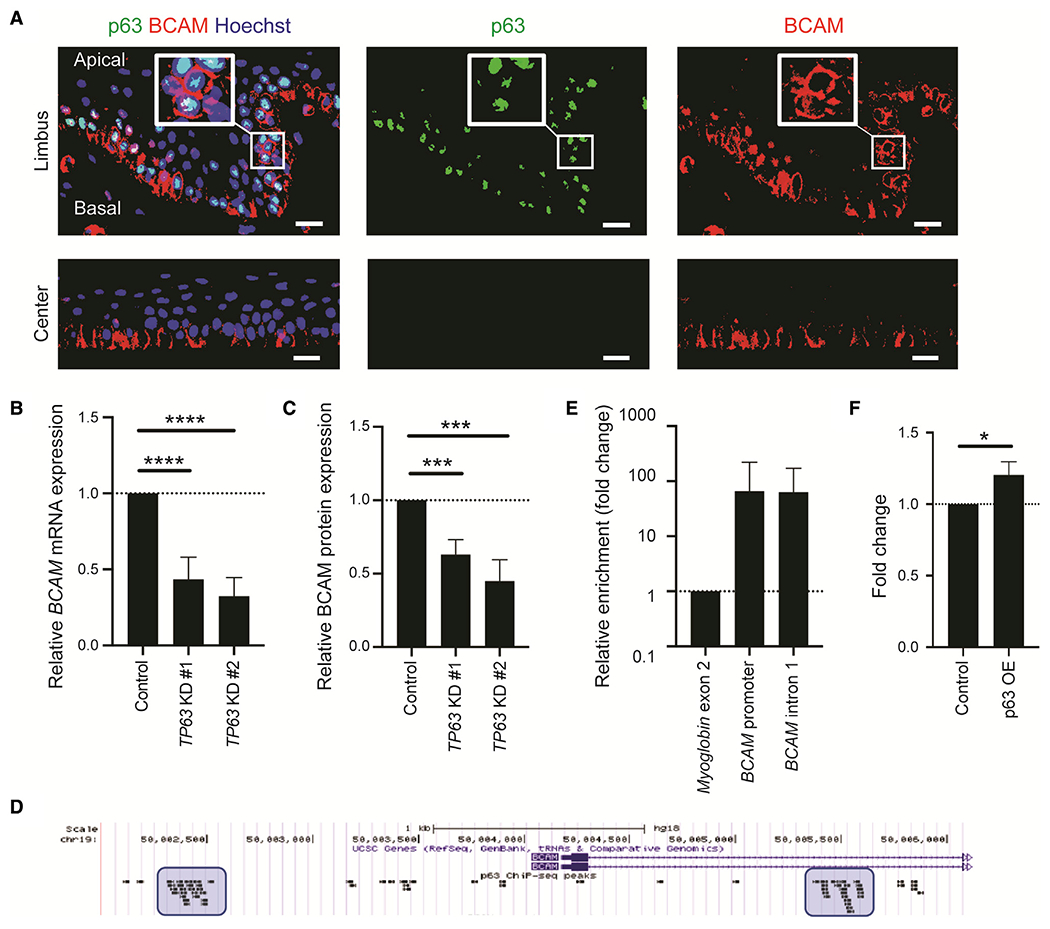
(A) Representative immunostaining analysis of p63 (green) and BCAM (red) expression in the limbus and central cornea. Nuclei stained with Hoechst 33342 (blue). n = 3. Scale bar: 20 μm.
(B) Bar graph depicts BCAM expression in control and TP63 siRNA-treated limbal epithelial cells (mean ± SD; n = 8; ****p < 0.0001). Data were analyzed using a Dunnett’s multiple comparisons test. KD, knockdown.
(C) Bar graph illustrates relative percentage of BCAM-positive cells in control and TP63 siRNA-treated limbal epithelial cultures analyzed by flow cytometry (mean ± SD; n = 6; ***p < 0.001). Data were analyzed using a Dunnett’s multiple comparisons test.
(D) The graph illustrates p63 binding sites within 2-kb up- and downstream of the BCAM transcription start site (TSS) of BCAM identified by p63 ChIP-seq in human skin (Kouwenhoven et al., 2010). P63 ChIP-seq data were downloaded from GEO: GSE17611 and visualized using the UCSC genome browser.
(E) Bar graph depicts enrichment of the BCAM promoter/enhancer and the BCAM first intron sequences in anti-p63 antibody immunoprecipitates compared with the isotype control immunoprecipitates as determined by ChIP-qPCR analyses. Myoglobin exon 2 sequence was used as a negative control (mean ± SD; n = 6).
(F) Bar graph represents the fold change in luciferase activity in ΔNp63α-overexpressing and control 293T cells co-transfected with luciferase reporter vector containing the promoter region 2-kb upstream of BCAM TSS (mean ± SD; n = 4; *p < 0.05). Data were analyzed using a paired t test. OE, overexpression.
Next, we examined whether BCAM represents a transcriptional target of p63. Previously published studies had revealed the existence of two clusters of p63 binding peaks located 2-kb upstream and downstream of the BCAM transcription start site (TSS): one at the promoter/enhancer region, and another at the first intron of the BCAM gene in human keratinocytes (Kouwenhoven et al., 2010) (Figure 6D). To test whether p63 provides upstream control of BCAM transcription, we first performed a p63 chromatin immunoprecipitation (ChIP) assay examining these specific binding sites. We found 65.6 ± 155.1-fold enrichment of the BCAM promoter and 63.4 ± 108.3-fold enrichment of intronic genomic DNA in p63 antibody immunoprecipitates compared with isotype control immunoprecipitates by qRT-PCR (Figure 6E). Next, using a luciferase reporter assay, we tested whether overexpression of the LSC-specific TP63 isoform ΔNp63α in 293T cells leads to activation of BCAM promoter activity. Cells were co-transfected with a ΔNp63α vector containing a full ΔNp63α open reading frame (ORF) or an empty control vector and a luciferase reporter vector that contained the 2-kb promoter region upstream of BCAM TSS (Figure S5D). Compared with empty vector controls, cells transfected with the ΔNp63α vector showed 20.4% ± 9.3% induction of the luciferase signal (p = 0.0220, n = 4) (Figure 6F), indicating that ΔNp63α provides upstream control of BCAM promoter activity.
DISCUSSION
In the current study, we identify a BCAM-positive limbal cell population capable of holoclone formation and corneal epithelial differentiation. Using scRNA-seq, flow cytometry, and in situ immunofluorescence analyses, we show that while limbal BCAM-positive cells are considerably different from ABCB5-positive LSCs, they also possess high self-renewal and proliferative potential and are uniquely capable of corneal epithelial sheet formation.
BCAM-positive cells are present among ABCB5-positive LSCs and ABCB5-negative limbal epithelial cells. Even though they constitute only a minority subpopulation in freshly isolated limbal cell suspensions, in in-vitro-expanded limbal epithelial cultures, BCAM is expressed by the majority of cells. Side-by-side comparison reveals high proliferative potential of BCAM-expressing cells among ABCB5-positive LSCs and ABCB5-negative limbal epithelial cells compared with BCAM-negative cell populations and demonstrates that in-vitro-expanded limbal epithelial cultures contain mostly TACs with a significant depletion of quiescent LSCs.
Furthermore, our study uncovers functional TAC heterogeneity, showing that corneal epithelial differentiation is sustained by BCAM-positive cells located in the limbus. Specifically, siRNA-induced BCAM depletion attenuated CFE and differentiation-dependent epithelial sheet formation in our experiments, indicating that BCAM is functionally required for TAC self-renewal, adhesion, proliferation, and migration. Detected expression of NUMB in BCAM-positive basal corneal epithelial cells and the lack of BCAM expression in the suprabasal epithelial layer hereby also suggest that BCAM-positive basal cells divide asymmetrically and produce BCAM-negative daughter cells. Thus, loss of BCAM expression by TACs might be required for their apical migration and terminal differentiation.
Laminin α5, a ligand of BCAM, was detected by immunostaining specifically in the limbal basement membrane in close proximity to BCAM-expressing early TACs. Given the essential role of laminin α5 in stem cell renewal (Domogatskaya et al., 2008), our results suggest that the superior cell CFE of limbal BCAM-positive cells is dependent not only on BCAM alone as demonstrated but also potentially on specific BCAM/laminin α5 interactions. Such interactions might explain the exclusive ability of limbal BCAM-positive cells compared with central-cornea-expressed BCAM-positive cells to generate a stratified corneal epithelial sheet, an additional possibility of investigation for which our findings provide a rationale. Our findings of production of laminin α5 by cultured limbal epithelial cells also suggest that BCAM-positive cells might contribute to basement membrane formation within the limbal niche.
Importantly, our results also reveal for the first time that the transcription factor p63 (Gonzalez et al., 2018; Pellegrini et al., 2001) can induce BCAM expression in limbal epithelial cells through upstream control of the BCAM promoter/enhancer. This mechanism resembles that of other BCAM-expressing epithelial tissues such as, for example, the skin, where p63 binding to the BCAM promoter was first identified (Kouwenhoven et al., 2010). Additional transcription factors expressed by ABCB5-positive LSCs, including MYC and ID4, might also be involved in the induction of BCAM expression in ABCB5-positive LSCs, but these possibilities require further investigation.
In aggregate, our study demonstrates that limbal BCAM expression characterizes a progenitor population capable of holoclone formation and corneal epithelial sheet generation and identifies a critical role of BCAM as a regulator of TAC differentiation into mature corneal epithelium. Moreover, our results will now allow prospective isolation of both ABCB5-positive LSCs and BCAM-positive TACs for further study and therapeutic modulation.
Limitations of the study
While this study establishes a unique capacity of BCAM-positive limbal cells for holoclone formation, proliferation, and differentiation-dependent corneal epithelial sheet formation, we have not yet examined their potential contribution to long-term corneal maintenance following transplantation. Additionally, BCAM promoter mutational studies would be required to further support the notion that BCAM is a direct transcriptional target of p63.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Natasha Y. Frank (nyfrank@bwh.harvard.edu).
Materials availability
BCAM reporter vector generated in this study have been deposited to Addgene # 188391.
Data and code availability
The scRNA-seq data were deposited to the Gene Expression Omnibus (GEO): GSE156524.
The R code used for the single-cell analysis is available on Github at https://github.com/cataalee/ABCB5-limbus/blob/main/ABCB5_limbus.R (https://doi.org/10.5281/zenodo.6822009).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human cell source
Cadaveric human donor whole eye globes and corneas derived from consented donors according to Institutional Review Board (IRB)-approved protocols were obtained from the Saving Sight (Kansas City, MO) and CorneaGen (Seattle, WA) eye banks. The donor characteristics are described in Tables S1 and S2. Limbal and central corneal epithelial cells were harvested as reported previously (Sasamoto et al., 2020). Briefly, the whole corneas were dissected from the eye globes and the central corneas were separated from the limbus using an 8 mm disposable biopsy punch (Integra LifeSciences, Plainsboro, NJ) and the corneal endothelium was removed mechanically. Limbal and central corneal epithelial cells were collected after 1-h incubation with PluriSTEM Dispase II Solution (MilliporeSigma, Burlington, MA) at 37°C and dissociated using TrypLE Express Enzyme (Thermo Fisher Scientific, Waltham, MA) at 37°C for 30 min. Cell cultures were maintained in DMEM/F12 medium (Thermo Fisher Scientific) supplemented with 10 ng/mL keratinocyte growth factor (KGF) (PeproTech, Rocky Hill, NJ), 10 μM Y-27632 (Tocris Bioscience, Bristol, UK) and B-27 Supplement (Thermo Fisher Scientific) (Miyashita et al., 2013).
293T cells (Clontech Laboratories, Mountain View, CA) were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific).
Rodent cell source
3T3-J2 cell line (Kerafast, Boston, MA) was maintained in DMEM (Thermo Fisher Scientific) supplemented with 10% calf serum (GE Healthcare Life Sciences, Marlborough, MA).
METHOD DETAILS
Flow cytometry
ABCB5-positive LSCs were isolated as described previously (Sasamoto et al., 2020). Briefly, dissociated limbal epithelial cells were resuspended with phosphate-buffered saline (PBS) (GE Healthcare Life Sciences) supplemented with 2% FBS and incubated for 30 min on ice with 2.5μg/ml mouse anti-ABCB5 monoclonal antibody (mAb) clone 3C2-1D12 (Frank et al., 2003; Ksander et al., 2014) conjugated with Alexa Fluor 647 (Thermo Fisher Scientific) and 0.125μg/ml PE-conjugated anti-CD45 mAb (clone 2D1, BioLegend, San Diego, CA). Cells were washed twice with PBS supplemented with 2% FBS and incubated with 30nM SYTOX Green Dead Cell Stain (Thermo Fisher Scientific) to exclude nonviable cells. For BCAM-positive cell isolation, the cells were incubated with 4μg/ml VioBright FITC-conjugated anti-BCAM mAb (Miltenyi Biotec, Bergisch Gladbach, Germany) and Propidium Iodide Staining Solution (BD Biosciences, San Jose, CA). For co-labeling of ABCB5 and BCAM in freshly isolated limbal epithelial cells, the cells were incubated with 2.5μg/ml mouse anti-ABCB5 monoclonal antibody (mAb) (clone 3C2-1D12) conjugated with Alexa Fluor 647, 0.5μg/ml Brilliant Violet 421-conjugated CD45 mAb (clone 2D1, BioLegend), 4μg/ml VioBright FITC-conjugated anti-BCAM mAb (Miltenyi Biotec) and Propidium Iodide Staining Solution. For analysis of ABCB5 and BCAM expression in in vitro-cultured limbal epithelial cells, the cells were incubated with 2.5μg/ml mouse anti-ABCB5 mAb (clone 3C2-1D12) conjugated with Alexa Fluor 647, 4μg/ml VioBright FITC-conjugated anti-BCAM mAb (Miltenyi Biotec) and GloCell Fixable Viability Dye Violet 450 (STEMCELL Technologies, Vancouver, Canada).
Cell sorting was performed using FACSAria II cell sorter (BD Biosciences) using a 100 μm nozzle. Cell analysis was performed using FACSCalibur (BD Biosciences) and FACSCelesta (BD Biosciences). Data were analyzed using BD FACSDiva v8.0.1 and FlowJo v10.6.1. Median cell size was determined by the forward scatter (FSC).
Immunofluorescence staining
Whole globes were fixed with 10% neutral buffered formalin (Fisher Scientific, Pittsburgh, PA) at 4°C overnight and transferred into 70% ethanol until the paraffin-embedding. The tissue sections were deparaffinized and subjected to antigen retrieval prior to antibody incubation. For the fresh frozen tissue analyses, cornea-containing areas of the whole globes were cryopreserved with the TissueTek® O.C.T Compound (Sakura, Tokyo, Japan). Stratified corneal epithelial cell sheets were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 10 min at room temperature and cryopreserved with the TissueTek® O.C.T compound. The tissue sections were permeabilized and blocked with a buffer containing 5 % normal donkey serum (Jackson ImmunoResearch Laboratories, West Grove, PA) and 0.3% Triton X-100 (MilliporeSigma) for 30 min at room temperature, and incubated with primary antibodies in the blocking buffer overnight at 4°C. The following primary antibodies were used: rabbit anti-BCAM polyclonal antibody (pAb) (1:100, NOVUS Biologicals, Centennial, CO), mouse anti-Integrin β4 mAb (1:200, NOVUS Biologicals), mouse anti-KRT12 mAb (1:100, Santa Cruz Biotechnology), rabbit anti-KRT12 mAb (1:400, Abcam, Cambridge, UK), mouse anti-NUMB mAb (1:100, Satna Cruz Biotechnology), mouse anti-MUC16 mAb (1:400, Abcam), mouse anti-LAMA5 mAb (1:200, Atlas Antibodies), and mouse anti-p63 mAb (1:100, Abcam). After washing with Tris-buffered saline (TBS) (Boston BioProducts, Ashland, MA), the slides were incubated with Alexa Fluor 488-conjugated mouse secondary antibody (Abcam) and Alexa Fluor 568-conjugated rabbit secondary antibody (Abcam) for 1 h at room temperature. Slides were stained with Hoechst 33342 (Thermo Fisher Scientific) for 10 min at room temperature and washed with TBS before sealing the slides with ProLong Gold Antifade Mountant (Thermo Fisher Scientific). Images were taken by C2+ confocal microscope (Nikon, Tokyo, Japan) and analyzed by NIS-Elements AR v4.30.01 (Nikon).
RNA extraction, reverse transcription and quantitative PCR (qPCR)
Total RNA was extracted using RNeasy Plus Mini Kit (QIAGEN, Hilden, Germany). After the removal of contaminating genomic DNA by DNA-free™ DNA Removal Kit (Thermo Fisher Scientific), cDNA was synthesized by High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). qPCR was performed with TaqMan™ Gene Expression Assay probes (Thermo Fisher Scientific) and TaqMan™ Fast Universal PCR Master Mix (Thermo Fisher Scientific). TaqMan™ probes used in the current study were: GAPDH (Hs99999905_m1), BCAM (Hs00170663_m1), KRT12 (Hs00165015_m1) and TP63 (Hs00978339_m1). The cycling conditions were 95°C for 20 s and 50 cycles of [95°C/1 s; 60°C/20 s] with StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific). ΔΔCt was calculated using GAPDH as a reference gene.
Single-cell RNA-sequencing
ABCB5-positive LSCs purified by flow cytometry were resuspended in 0.4% bovine serum albumin (BSA) (MilliporeSigma) in PBS at a concentration of 1,000 cells/μl. 7,000 cells were loaded onto a single lane (Chromium chip, 10X Genomics, Pleasanton, CA) followed by encapsulation in lipid droplets (Single Cell 3′ kit v3,10X Genomics), cDNA creation, and library generation as per manufacturer’s protocol. cDNA libraries were sequenced to an average of 61,237 reads per cell using an Illumina Nextseq 500 (Illumina, San Diego, CA). The reads were pre-processed with Cell Ranger v2.1.0 (10X Genomics), which demultiplexed cells from different samples and quantified transcript counts per putative cell (3,768 cells total). Quantification was performed using the STAR aligner (Dobin et al., 2013) against the hg19 transcriptome. Cell Ranger aggr was used to combine the three libraries and to normalize the results based on the mapped sequencing depth. Downstream analysis was performed in the R programming environment. The Seurat (v3.2.3) (Butler et al., 2018) object was created using recommended settings (min.cells = 3, min.features = 200). Cells expressing <400 or >4000 genes were filtered out to exclude noncell and cell aggregates. Cells expressing >10% mitochondrial genes were filtered out to exclude dead or dying cells. Cells expressing >2 MLANA or TYRP1 and >2 KERA or LUM genes, indicative of melanocyte and stromal contamination, respectively, were removed, leaving a total of 3,417 cells for downstream analysis. Harmony (v1.0) was implemented to adjust for individual donor batch effects and required 10 iterations to reach convergence (Figure S2B). After log-normalization and data scaling, principal component analysis (PCA) was performed. After visualization of the PC loadings, JackStraw calculations, and elbow plotting, the first 20 principal components were used for clustering analysis. The results of the clustering were visualized using Uniform Manifold Approximation and Projection (UMAP). Markers for the cells in Cluster 2 compared to Cluster 1 were identified using FindAllMarkers (min.pct = 0.25, logfc.threshold = 0.25). Clusters were assigned using these markers.
Colony-forming assay
Colony-forming assay (CFA) was performed as previously reported (Sasamoto et al., 2020). Briefly, limbal epithelial cells were seeded on the mitomycin C (MMC) (MilliporeSigma)-treated 3T3-J2 feeder cell layer at 500 cells per well on 6-well plates. The cells were cultured in keratinocyte culture medium (KCM) supplemented with 10 ng/mL KGF and 10 μM Y-27632. KCM was composed of DMEM without glutamine and Ham’s F-12 Nutrient Mix (Thermo Fisher Scientific) combined at 3:1 ratio, supplemented with 10% FBS, 0.4μg/ml hydrocortisone hydrogen succinate (MilliporeSigma), 2nM 3,3′,5-triiodo-l-thyronine sodium salt (MilliporeSigma), 1 nM cholera toxin (List Biological Laboratories, Campbell, CA), 2.25μg/ml bovine transferrin HOLO form (Thermo Fisher Scientific), 2mM L-glutamine (Thermo Fisher Scientific), 0.5% (vol/vol) insulin transferrin selenium solution (Thermo Fisher Scientific), and 1% (vol/vol) penicillin-streptomycin solution (GE Healthcare Life Sciences). After the 10-day culture, the colonies were fixed with 10% neutral buffered formalin and stained with Rhodamine B (MilliporeSigma). The colony-forming efficiency was calculated as a ratio of the colony numbers per well to the seeded cell numbers and shown as a percentage. The colony size was calculated using ImageXpress Micro Confocal High-Content Imaging System (Molecular Devices, San Jose, CA). For the holoclone assay, each colony was picked up, dissociated using TrypLE Express, and seeded on the MMC-treated 3T3-J2 cells in the 6-well plates. After the 10-day culture, the colonies were fixed with 10% neutral buffered formalin and stained with Rhodamine B. The holoclone formation was evaluated as previously described (Barrandon and Green, 1987).
Cell sheet formation assay
FACS sorted cells were seeded on iMatrix-511 (Nacalai Tesque, Kyoto, Japan)-coated 24 well inserts (Corning, Corning, NY) at 500 cells per well and cultured in DMEM/F12 medium supplemented with 10 ng/mL KGF and 10 μM Y-27632 for 10 days. At day 10, the inserts were transferred onto 3T3-J2 cell-seeded 24 well plates and grown in KCM supplemented with 10 ng/mL KGF and 10 μM Y-27632. The cells were airlifted at day 15 in order to induce the cell sheet differentiation and the sheets were harvested at day 20.
Hematoxylin and eosin (HE) staining and sheet thickness quantification
Fresh frozen corneal epithelial sheet sections were stained using Hematoxylin and Eosin Stain Kit (Vector Laboratories, Burlingame, CA), and images were obtained with ECLIPSE 80i microscope (Nikon). Epithelial sheet thickness was calculated from the cross-sectional images using NIS-Elements AR v4.13.05 software.
Western blot analyses
Cultured cells were dissolved in RIPA buffer (Cell Signaling Technology, Danvers, MA) supplemented with cOmplete™ Protease Inhibitor Cocktail (MilliporeSigma). The samples were incubated for 30 min on ice, and debris were removed by centrifugation. Subsequently, the protein concentration was measured using Bio-Rad Protein Assay (Bio-Rad, Hercules, CA). The lysates were mixed with SDS-sample buffer (Boston BioProducts) and 2-mercaptoethanol (MilliporeSigma) and denatured for 10 min at 95°C. The proteins were separated using SDS-PAGE gel electrophoresis and transferred on the PVDF blotting membranes (GE Healthcare Life Sciences). The membranes were subsequently treated with a blocking buffer containing 5% blotting-grade blocker (Bio-Rad) for 1 h at room temperature and incubated with primary antibodies diluted in the blocking buffer overnight at 4°C. Primary antibodies used in the current study were: rabbit anti-β-actin pAb (1:1000, Cell Signaling Technology), rabbit anti-laminin α5 pAb (1:500, GeneTex), and rabbit anti-p63 mAb (1:1000, Abcam). After washing with TBS with Tween 20 (MilliporeSigma) (TBS-T), the membranes were incubated with HRP-conjugated rabbit secondary antibody (Cell Signaling Technology) for 1 h at room temperature. Following a subsequent wash with TBS-T, the protein signal was developed by Western Lightning Plus-ECL (PerkinElmer, Waltham, MA), and images were taken by ChemiDoc MP Imaging System (Bio-Rad). Protein expression levels were calculated using Image Lab software v5.2.1 (Bio-Rad) and normalized to the expression of β-actin.
RNA interference
RNA interference was performed as previously described (Fujimoto et al., 2019). Briefly, cells were transfected with siRNA using Lipofectamine™ RNAiMAX Transfection Reagent (Thermo Fisher Scientific) immediately after the cell seeding. The following Silencer™ Select siRNAs (Thermo Fisher Scientific) were used: Silencer™ Select Negative Control No.1 siRNA, BCAM siRNAs (s8336 and s8337) and TP63 siRNAs (s531582 and s531583). For the knockdown experiment during the cell sheet formation, the BCAM siRNAs were applied to the seeded cells cultured on the iMatrix-511-coated 24 well inserts in DMEM/F12 medium supplemented with 10 ng/mL KGF and 10 μM Y-27632.
Migration assay
Migration assay was performed using OrisTM Cell Migration Assay Kit (Platypus Technologies, Madison, WI, USA) as per manufacturer’s instructions. Cells were seeded at 1 × 105 cells per well in the 96-well plates. The wells contained 2 mm diameter stoppers inserted in the central area of each well to prevent cell growth. The cells were transfected with BCAM siRNAs immediately after seeding, and the stoppers were removed two days after the transfection when the surrounding cells became confluent. Cell migration was monitored by IncuCyte® Live-Cell Analysis Systems (Essen BioScience, Ann Arbor, MI) for 24 h and calculated as a percentage of wound closure over time.
Chromatin immunoprecipitation (ChIP)-qPCR
The p63-ChIP assay was performed using EZ-Magna ChIP™ A/G Chromatin Immunoprecipitation Kit (MilliporeSigma) as per manufacturer′s instructions. Briefly, after cross-linking protein and DNA with 1% formaldehyde (MilliporeSigma), the DNA sonication was performed using COVARIS S220 (COVARIS, Woburn, MA). The samples were incubated with mouse anti-p63 mAb (Abcam) or mouse isotype control mAb (BioLegend) overnight at 4°C. The DNA was isolated using a DNA extraction reagents from EZ-Magna ChIP™ A/G Chromatin Immunoprecipitation Kit (MilliporeSigma). qPCR analyses were performed using PowerSYBR Green PCR Master Mix (Thermo Fisher Scientific) with the following primers: 5′-TATCCTGGGATGCTCCAGTC-3′ and 5′-TGCTCAGGGAAGGTTTGTTT-3′ for the BCAM promoter region, 5′-TGTCCCTCCCACATTCCTGA-3′ and 5′-TTAGGCGGGATGGATGTGTG-3′ for the BCAM intron 1 region, and 5′-AAGTTTGACAAGTTCAAGCACCTG-3′ and 5′-TGGCACCATGCTTCTTTAAGTC-3′ the Myoglobin exon 2 region, which was used as a negative control. The cycling conditions were 95°C for 10 min and 50 cycles of (95°C /15 s; 60°C /60 s) using StepOnePlus™ Real-Time PCR System. Fold enrichment of DNA in anti-p63 immunoprecipitates was calculated against DNA immunoprecipitated with mouse isotype control mAb.
Luciferase reporter assay
The BCAM luciferase reporter assay was performed using ONE-Glo™ + Tox Luciferase Reporter and Cell Viability Assay kit (Promega, Madison, WI, USA). 293T cells were seeded at 4.0 × 104 cells per well in 96-well plates. After 24-h culture, cells were co-transfected with BCAM promoter-pRB-Puro-TurboRFPmyc-T2A-Luciferase reporter vector and either with ΔNp63α-FLAG pcDNA3.1/hygro plasmid or no insert control pcDNA3.1/hygro plasmid using the Lipofectamine™ 3000 Transfection Reagent (Thermo Fisher Scientific). The BCAM promoter-pRB-Puro-TurboRFPmyc-T2A-Luciferase reporter vector was constructed by cloning the BCAM promoter region located 2kb upstream of the BCAM first exon (Figure S5D) into a pRB-Puro-TurboRFPmyc-T2A-Luciferase reporter vector. ΔNp63α-FLAG cDNA cloned into pcDNA3.1/hygro plasmid was gifted by David Sidransky (Addgene plasmid #26979 ; http://n2t.net/addgene:26979 ; RRID:Addgene_26979) (Addgene, Watertown, MA, USA). The empty pcDNA3.1/hygro vector without the insert was used as a negative control. The cells were harvested after 48 h incubation. Cell viability and firefly luciferase activity were measured in the same assay well using ONE-Glo™ + Tox Luciferase Reporter and Cell Viability Assay kit. Luciferase activity was corrected for the cell viability.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Two-sided tests were used in the statistical analyses. Appropriate statistical tests were used for all data sets depicted in the figures, with data meeting the assumptions of the tests. Variations within each group were estimated and similar between statistically compared groups. The data are presented as the mean ± standard deviation (SD) or mean ± standard error (SEM) of three or more independent experiments. The relevant statistical tests are described in the figure legends. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-ABCB5 (clone 3C2-1D12) | Frank lab | PMID: 25030174 |
| Mouse monoclonal anti-CD45-PE | BioLegend | 368510 Cat# 368510; RRID:AB_2566370 |
| Mouse monoclonal anti-CD45- Brilliant Violet 421 | BioLegend | 368522 Cat# 368522; RRID:AB_2687375 |
| Human monoclonal anti-BCAM-VioBright FITC | Miltenyi Biotec | 130-104-839 Cat# 130-104-839; RRID:AB_2656519 |
| Rabbit polyclonal anti-BCAM | NOVUS Biologicals | NBP2-31994 Cat# NBP2-31994: RRID:AB_2922815 |
| Mouse monoclonal anti- Integrin β4 | NOVUS Biologicals | NB100-65599 Cat# NB100-65599; RRID:AB_959522 |
| Mouse monoclonal anti-KRT12 | Santa Cruz Biotechnology | sc-515882 Cat# sc-515882; RRID:AB_2922816 |
| Rabbit monoclonal anti-KRT12 | Abcam | ab185627 Cat# ab185627; RRID:AB_2889825 |
| Mouse monoclonal anti-NUMB | Santa Cruz Biotechnology | sc-136554 Cat# sc-136554; RRID:AB_10611794 |
| Mouse monoclonal anti-MUC16 | Abcam | ab1107 Cat# ab1107; RRID:AB_297721 |
| Mouse monoclonal anti-laminin α5 | Atlas Antibodies | AMAb91124 Car# AMAb91124; RRID:AB_2665809 |
| Rabbit polyclonal anti-laminin α5 | GeneTex | GTX55867 Cat# GTX55867; RRID:AB_2887940 |
| Mouse monoclonal anti-p63 | Abcam | ab735 Cat# ab735; RRID:AB_305870 |
| Rabbit monoclonal anti-p63 | Abcam | ab124762 Cat# ab124762; RRID:AB_10971840 |
| Rabbit polyclonal anti-β-actin | Cell Signaling Technology | 8457L Cat# 8457; RRID:AB_10950489 |
| Donkey anti-Mouse IgG- Alexa Fluor 488 | Thermo Fisher Scientific | A21202 Cat# A21202; RRID:AB_141607 |
| Donkey anti-Rabbit IgG- Alexa Fluor 568 | Thermo Fisher Scientific | A10042 Cat# A10042; RRID:AB_2534017 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling Technology | 7076S Cat# 7076; RRID:AB_330924 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | 7074S Cat# 7074; RRID:AB_2099233 |
| Bacterial and virus strains | ||
| deltaNp63alpha-FLAG | Addgene | 26979 |
| BCAM promoter-pRB-Puro-TurboRFPmyc-T2A-Luciferase | This manuscript | 188391 |
| Biological samples | ||
| Human whole globes and corneas | Saving Sight (Kansas City, MO) andN/A CorneaGen (Seattle, WA) eye banks | |
| Chemicals, peptides, and recombinant proteins | ||
| PluriSTEM Dispase II Solution | MilliporeSigma | SCM133 |
| TrypLE™ Express Enzyme | Thermo Fisher Scientific | 12605036 |
| DMEM/F-12 | Thermo Fisher Scientific | 11320033 |
| Recombinant Human KGF (FGF-7) | PeproTech | 100-19 |
| Y-27632 dihydrochloride | TOCRIS | 1254 |
| B-27 Supplement (50X), serum free | Thermo Fisher Scientific | 17504001 |
| DMEM, high glucose, pyruvate | Thermo Fisher Scientific | 11995-065 |
| Fetal Bovine Serum (FBS) | Thermo Fisher Scientific | 10438026 |
| HyClone Phosphate Buffered Saline solution | GE Healthcare | SH30256.01 |
| SYTOX Green Dead Cell Stain | Thermo Fisher Scientific | S34860 |
| Propidium Iodide Staining Solution | BD Biosciences | 556463 |
| GloCell™ Fixable Viability Dye Violet 450 | STEMCELL Technologies | 75009 |
| Bovine serum albumin (BSA) | MilliporeSigma | A7030 |
| 10% neutral buffered formalin | Fisher Scientific | SF100-4 |
| TissueTek® O.C.T Compound | Sakura | 4583 |
| 16% paraformaldehyde | Electron Microscopy Sciences | 15710 |
| Normal donkey serum | Jackson Immuno Research Laboratories | 017-000-121 |
| Triton™ X-100 | MilliporeSigma | X-100 |
| Tris-Buffered Saline (TBS) | Boston BioProducts | BM301 |
| Hoechst 33342 | Thermo Fisher Scientific | H3570 |
| ProLong Gold Antifade Mountant | Thermo Fisger Scientific | P36934 |
| TaqMan™ Fast Universal PCR Master Mix | Thermo Fisher Scientific | 4366072 |
| Mitomycin C from Streptomyces caespitosus | MilliporeSigma | M4287 |
| DMEM, high glucose, pyruvate, no glutamine | Thermo Fisher Scientific | 10313021 |
| Ham’s F-12 Nutrient Mix | Thermo Fisher Scientific | 11765054 |
| Hydrocortisone hydrogen succinate | MilliporeSigma | BP187 |
| 3,3,5-Triiodo-L-thyronine sodium salt | MilliporeSigma | T2752 |
| Transferrin, Bovine (Holo form), lyophilized | Thermo Fisher Scientific | 11107047 |
| L-glutamine | Thermo Fisher Scientific | 25030081 |
| Insulin-Transferrin-Selenium (ITS-G) (100X) | Thermo Fisher Scientific | 41400045 |
| Cholera Toxin (AZIDE-FREE) from Vibrio cholerae | List Biological Laboratories | 100B |
| HyClone Penicillin-Streptomycin 100X solution | GE Healthcare | SV30010 |
| Rhodamine B | MilliporeSigma | R6626 |
| iMatrix-511 | Nacalai Tesque | 892012 |
| RIPA buffer | Cell Signaling Technology | 9806S |
| cOmplete™ Protease Inhibitor Cocktail | MilliporeSigma | 11873580001 |
| SDS-sample buffer | Boston BioProducts | BP111NR |
| 2-mercaptoethanol | MilliporeSigma | M3148 |
| PVDF Blotting Membrane | GE healthcare | 10600023 |
| Blotting-Grade Blocker | Bio-Rad | 1706404 |
| Tween 20 | MilliporeSigma | P1379 |
| Western Lightning Plus-ECL | PerkinElmer | NEL104001EA |
| Lipofectamine™ RNAiMAX Transfection Reagent | Thermo Fisher Scientific | 13778075 |
| Lipofectamine™ 3000 Transfection Reagent | Thermo Fisher Scientific | L3000008 |
| PowerSYBR Green PCR Master Mix | Thermo Fisher Scientific | 4367659 |
| Critical commercial assays | ||
| Alexa Fluor™ 647 Antibody Labeling Kit | Thermo Fisher Scientific | A20186 |
| Chromium Single Cell B Chip Kit | 10X Genomics | PN-1000073 |
| Chromium Single Cell 3’ GEM, Library & Gel Bead Kit v310X Genomics | PN-1000075 | |
| RNeasy plus Mini Kit | QIAGEN | 74136 |
| DNA-free™ DNA Removal Kit | Thermo Fisher Scientific | AM1906 |
| High-Capacity cDNA Reverse Transcription Kit | Thermo Fisher Scientific | 4368814 |
| Hematoxylin and Eosin Stain Kit | Vector Laboratories | H-3502 |
| OrisTM Cell Migration Assay Kit | Platypus Technologies | CMA1.101 |
| EZ-Magna ChIP™ A/G Chromatin Immunoprecipitation Kit | MilliporeSigma | 17-10086 |
| ONE-Glo™ + Tox Luciferase Reporter and Cell Viability Assay kit | Promega | E7110 |
| Deposited data | ||
| Single-cell RNA sequencing data | This manuscript | GEO: GSE156524 |
| p63 ChIP-seq data | Kouwenhoven et al. (2010) | GEO: GSE17611 |
| Experimental models: Cell lines | ||
| 3T3-J2 Cell Line | Kerafast | EF3003 |
| 293T cell line | Clontech | 632180 |
| Oligonucleotides | ||
| TaqMan probes | Thermo Fisher Scientific | 4331182 |
| Silencer™ Select siRNAs | Thermo Fisher Scientific | 4427037 |
| Silencer™ Select Negative Control No. 1 siRNA | Thermo Fisher Scientific | 4390843 |
| Software and algorithms | ||
| GraphPad Prism | GraphPad Software | 8 |
| BD FACSDiva | BD Biosciences | 8.0.1 |
| FlowJo | BD Biosciences | 10.6.1 |
| Cell Ranger | 10X Genomics | 2.1 |
| STAR aligner 2.5.1b | Dobin et al., (2013) | https://github.com/alexdobin/STAR |
| R version 3.6.3 | R Foundation | https://www.r-project.org/foundation/ |
| RStudio Desktop 1.1.463 | RStudio | https://www.rstudio.com/products/rstudio-desktop/ |
| Seurat 3.2.3 | Satija et al., (2015) | https://satijalab.org/seurat/ |
| Harmony | 1.0 | |
| R code for single-cell RNA sequencing data | Github | https://github.com/cataalee/ABCB5-limbus/blob/main/ABCB5_limbus.R |
| NIS-elements AR | Nikon | 4.30.01 |
| Image Lab | Bio-Rad | 5.2.1 |
| Adobe Illustrator | Adobe | 24.2.1 |
Highlights.
BCAM-positive limbal cells are capable of holoclone formation and corneal differentiation
The limbus-specific BCAM ligand laminin α5 is required for stem cell self-renewal
Gene knockdown of BCAM inhibits corneal epithelial sheet formation
Subsets of ABCB5-positive LSCs and ABCB5-negative limbal epithelial cells express BCAM
ACKNOWLEDGMENTS
We would like to thank the patients for their generous tissue donations that enabled this research. This work was supported by NIH/NEI grants 1K99EY031741 to Y.S. and 1R01EY025794 and R24EY028767 to N.Y.F., B.R.K., and M.H.F.; NIH/NEI Schepens Core grant P30EY003790 to B.R.K.; NIH/NHLBI grant 1R01HL161087 to G.F.M., M.H.F., and N.Y.F.; NIH/NIBIB grant 2T32EB016652-06 to C.A.A.L.; NIH/NIDDK R01-DK076683 to F.H.; Alcon Young Investigator Grant and Japan Eye Bank Association Overseas Award to Y.S.; and VA R&D Merit Review Award 1I01RX000989 and a Harvard Stem Cell Institute seed grant award to N.Y.F. F.B. was supported by a fellowship grant (404527522) from the German Research Foundation (DFG). We thank the BRI Single Cell Genomics Core, BWH Pathology Core, DF/HCC Core, DFCI Flow Cytometry Core, BWH Flow Cytometry Core, BCH IDDRC Molecular Genetics Core Facility, and BCH Assay Development & Screening Facility for their technical assistance.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111166.
DECLARATION OF INTERESTS
M.H.F., B.R.K., and N.Y.F. are inventors or co-inventors of US and international patents assigned to Brigham and Women’s Hospital, Boston Children’s Hospital, the Massachusetts Eye and Ear Infirmary, and/or the VA Boston Healthcare System, Boston, MA, licensed to Ticeba GmbH (Heidelberg, Germany) and Rheacell GmbH & Co. KG (Heidelberg, Germany). M.H.F. serves as a scientific advisor to Ticeba GmbH and Rheacell GmbH & Co. KG.
REFERENCES
- Arnold I, and Watt FM (2001). c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr. Biol 11, 558–568. 10.1016/s0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- Barrandon Y, and Green H (1987). Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl. Acad. Sci. USA 84, 2302–2306. 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe DC, and Masters BR (1996). Cell lineage and the differentiation of corneal epithelial cells. Invest. Ophthalmol. Vis. Sci 37, 1815–1825. [PubMed] [Google Scholar]
- Bretones G, Delgado MD, and León J (2015). Myc and cell cycle control. Biochim. Biophys. Acta 1849, 506–516. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Murñozledo F, and Gómez-Flores E (2011). Challenges to the study of asymmetric cell division in corneal and limbal epithelia. Exp. Eye Res 92, 4–9. 10.1016/j.exer.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Catala P, Groen N, Dehnen JA, Soares E, van Velthoven AJ, Nuijts RM, Dickman MM, and LaPointe VL (2021). A single cell transcriptome atlas reveals the heterogeneity of the healthy human cornea and identifies novel markers of the corneal limbus and stroma. Preprint at bioRxiv. 10.1101/2021.07.07.451489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lan J, Liu D, Backman LJ, Zhang W, Zhou Q, and Danielson P (2017). Ascorbic acid promotes the stemness of corneal epithelial stem/progenitor cells and accelerates epithelial wound healing in the cornea. Stem Cells Transl. Med 6, 1356–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin J, Queen R, Zerti D, Bojic S, Dorgau B, Moyse N, Molina MM, Yang C, Dey S, and Reynolds G (2021). A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells. Ocul. Surf 21, 279–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanger M, and Evensen A (1971). Role of the pericorneal papillary structure in renewal of corneal epithelium. Nature 229, 560–561. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domogatskaya A, Rodin S, Boutaud A, and Tryggvason K (2008). Laminin-511 but not-332, −111, or-411 enables mouse embryonic stem cell self-renewal in vitro. Stem cells 26, 2800–2809. [DOI] [PubMed] [Google Scholar]
- Frank NY, Pendse SS, Lapchak PH, Margaryan A, Shlain D, Doeing C, Sayegh MH, and Frank MH (2003). Regulation of progenitor cell fusion by ABCB5 P-glycoprotein, a novel human ATP-binding cassette transporter. J. Biol. Chem 278, 47156–47165. [DOI] [PubMed] [Google Scholar]
- Fujimoto S, Hayashi R, Hara S, Sasamoto Y, Harrington J, Tsujikawa M, and Nishida K (2019). KLF4 prevents epithelial to mesenchymal transition in human corneal epithelial cells via endogenous TGF-beta2 suppression. Regen. Ther 11, 249–257. 10.1016/j.reth.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez G, Sasamoto Y, Ksander BR, Frank MH, and Frank NY (2018). Limbal stem cells: identity, developmental origin, and therapeutic potential. Wiley Interdiscip. Rev. Dev. Biol 7. 10.1002/wdev.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenson S, Määttä M, Rousselle P, Kikkawa Y, Miner JH, Tervo T, and Virtanen I (2005). The immortalized human corneal epithelial cells adhere to laminin-10 by using Lutheran glycoproteins and integrin alpha3beta1. Exp. Eye Res 81, 415–421. 10.1016/j.exer.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Hayashi R, Ishikawa Y, Sasamoto Y, Katori R, Nomura N, Ichikawa T, Araki S, Soma T, Kawasaki S, Sekiguchi K, et al. (2016). Co-ordinated ocular development from human iPS cells and recovery of corneal function. Nature 531, 376–380. 10.1038/nature17000. [DOI] [PubMed] [Google Scholar]
- Hayashi R, Ishikawa Y, Katori R, Sasamoto Y, Taniwaki Y, Takayanagi H, Tsujikawa M, Sekiguchi K, Quantock AJ, and Nishida K (2017). Coordinated generation of multiple ocular-like cell lineages and fabrication of functional corneal epithelial cell sheets from human iPS cells. Nat. Protoc 12, 683–696. [DOI] [PubMed] [Google Scholar]
- Jongkhajornpong P, Nakamura T, Sotozono C, Nagata M, Inatomi T, and Kinoshita S (2016). Elevated expression of ABCB5 in ocular surface squamous neoplasia. Sci. Rep 6, 20541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Wang J, Wray B, Patel P, Yang W, Peng H, and Lavker RM (2019). Single-cell RNA transcriptome helps define the limbal/corneal epithelial stem/early transit amplifying cells and how autophagy affects this population. Invest. Ophthalmol. Vis. Sci 60, 3570–3583. 10.1167/iovs.19-27656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon KR, and Tseng SC (1989). Limbal autograft transplantation for ocular surface disorders. Ophthalmology 96, 709–722. [DOI] [PubMed] [Google Scholar]
- Kikkawa Y, Moulson CL, Virtanen I, and Miner JH (2002). Identification of the binding site for the Lutheran blood group glycoprotein on laminin α5 through expression of chimeric laminin chains in vivo. J. Biol. Chem 277, 44864–44869. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. (2008). Mechanisms of asymmetric stem cell division. Cell 132, 583–597. [DOI] [PubMed] [Google Scholar]
- Kouwenhoven EN, van Heeringen SJ, Tena JJ, Oti M, Dutilh BE, Alonso ME, de la Calle-Mustienes E, Smeenk L, Rinne T, Parsaulian L, et al. (2010). Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 6, e1001065. 10.1371/journal.pgen.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksander BR, Kolovou PE, Wilson BJ, Saab KR, Guo Q, Ma J, McGuire SP, Gregory MS, Vincent WJB, Perez VL, et al. (2014). ABCB5 is a limbal stem cell gene required for corneal development and repair. Nature 511, 353–357. 10.1038/nature13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kureshi AK, Dziasko M, Funderburgh JL, and Daniels JT (2015). Human corneal stromal stem cells support limbal epithelial cells cultured on RAFT tissue equivalents. Sci. Rep 5, 16186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer MS, Sun TT, and Lavker RM (1998). Strategies of epithelial repair: modulation of stem cell and transit amplifying cell proliferation. J. Cell Sci 111, 2867–2875. [DOI] [PubMed] [Google Scholar]
- Li J-M, Kim S, Zhang Y, Bian F, Hu J, Lu R, Pflugfelder SC, Chen R, and Li D-Q (2021). Single-cell transcriptomics identifies a unique entity and signature markers of transit-amplifying cells in human corneal limbus. Invest. Ophthalmol. Vis. Sci 62, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathan JJ, Ismail S, McGhee JJ, McGhee CNJ, and Sherwin T (2016). Sphere-forming cells from peripheral cornea demonstrate the ability to repopulate the ocular surface. Stem Cell Res. Ther 7, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita H,Yokoo S,Yoshida S, Kawakita T, Yamagami S,Tsubota K, and Shimmura S (2013). Long-term maintenance of limbal epithelial progenitor cells using rho kinase inhibitor and keratinocyte growth factor. Stem Cells Transl. Med 2, 758–765. 10.5966/sctm.2012-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulson CL, Li C, and Miner JH (2001). Localization of Lutheran, a novel laminin receptor, in normal, knockout, and transgenic mice suggests an interaction with laminin α5 in vivo. Dev. Dyn 222, 101–114. [DOI] [PubMed] [Google Scholar]
- Norrick A, Esterlechner J, Niebergall-Roth E, Dehio U, Sadeghi S, Schröder HM, Ballikaya S, Stemler N, Ganss C, Dieter K, et al. (2021). Process development and safety evaluation of ABCB5+ limbal stem cells as advanced-therapy medicinal product to treat limbal stem cell deficiency. Stem Cell Res. Ther 12, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfitt GJ, Kavianpour B, Wu KL, Xie Y, Brown DJ, and Jester JV (2015). Immunofluorescence tomography of mouse ocular surface epithelial stem cells and their niche microenvironment. Invest. Ophthalmol. Vis. Sci 56, 7338–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Richardson A, Pandzic E, Lobo EP, Lyons JG, and Di Girolamo N (2019). Peripheral (not central) corneal epithelia contribute to the closure of an annular debridement injury. Proc. Natl. Acad. Sci. USA 116, 26633–26643. 10.1073/pnas.1912260116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons SF, Lee G, Spring FA, Willig T-N, Peters LL, Gimm JA, Tanner MJ, Mohandas N, Anstee DJ, and Chasis JA (2001a). Lutheran blood group glycoprotein and its newly characterized mouse homologue specifically bind α5 chain-containing human laminin with high affinity. Blood 97, 312–320. [DOI] [PubMed] [Google Scholar]
- Parsons SF, Lee G, Spring FA, Willig TN, Peters LL, Gimm JA, Tanner MJ, Mohandas N, Anstee DJ, and Chasis JA (2001b). Lutheran blood group glycoprotein and its newly characterized mouse homologue specifically bind alpha5 chain-containing human laminin with high affinity. Blood 97, 312–320. 10.1182/blood.v97.1.312. [DOI] [PubMed] [Google Scholar]
- Pellegrini G, Dellambra E, Golisano O, Martinelli E, Fantozzi I, Bondanza S, Ponzin D, McKeon F, and De Luca M (2001). p63 identifies keratinocyte stem cells. Proc. Natl. Acad. Sci. USA 98, 3156–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polisetti N, Sorokin L, Okumura N, Koizumi N, Kinoshita S, Kruse FE, and Schlötzer-Schrehardt U (2017). Laminin-511 and-521-based matrices for efficient ex vivo-expansion of human limbal epithelial progenitor cells. Sci. Rep 7, 5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasamoto Y, Sasamoto N, Tran J, Mishra A, Ksander BR, Frank MH, and Frank NY (2020). Investigation of factors associated with ABCB5-positive limbal stem cell isolation yields from human donors. Ocul. Surf 18, 114–120. 10.1016/j.jtos.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell J, Gennert D, et al. (2015). Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33, 495–502. 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermer A, Galvin S, and Sun TT (1986). Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J. Cell Biol 103, 49–62. 10.1083/jcb.103.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi R, and Yamada KM (2018). Basement membranes in development and disease. In Current Topics in Developmental Biology (Elsevier), pp. 143–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaharuddin B, Osei-Bempong C, Ahmad S, Rooney P, Ali S, Oldershaw R, and Meeson A (2016). Human limbal mesenchymal stem cells express ABCB5 and can grow on amniotic membrane. Regen. Med 11, 273–286. [DOI] [PubMed] [Google Scholar]
- Shaharuddin B, Ahmad S, Md Latar N, Ali S, and Meeson A (2017). A human corneal epithelial cell line model for limbal stem cell biology and limbal immunobiology. Stem Cells Transl. Med 6, 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Hayashi R, Okubo T, Kudo Y, Katayama T, Ishikawa Y, Toga J, Yagi E, Honma Y, Quantock AJ, et al. (2018). Selective laminin-directed differentiation of human induced pluripotent stem cells into distinct ocular lineages. Cell Rep. 25, 1668–1679.e5. 10.1016/j.celrep.2018.10.032. [DOI] [PubMed] [Google Scholar]
- Thoft RA, and Friend J (1983). The X, Y, Z hypothesis of corneal epithelial maintenance. Invest. Ophthalmol. Vis. Sci 24, 1442–1443. [PubMed] [Google Scholar]
- Yoon JJ, Ismail S, and Sherwin T (2014). Limbal stem cells: central concepts of corneal epithelial homeostasis. World J. Stem Cells 6, 391–403. 10.4252/wjsc.v6.i4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data were deposited to the Gene Expression Omnibus (GEO): GSE156524.
The R code used for the single-cell analysis is available on Github at https://github.com/cataalee/ABCB5-limbus/blob/main/ABCB5_limbus.R (https://doi.org/10.5281/zenodo.6822009).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


