Abstract
Switching between the pathogenic smooth (1116S) and nonpathogenic rough (1116R) forms of Pseudomonas tolaasii occurs due to the reversible duplication of a 661-bp element within the pheN locus. Disruption of the chromosomal recA locus of 1116S and 1116R produced strains 1116SrecA and 1116RrecA, respectively, which showed typical loss of UV resistance. Switching from the smooth to the rough form was virtually eliminated in the 1116SrecA strain, whereas the extent of switching from the rough to the smooth form was almost identical in 1116R and 1116RrecA. It is concluded that phenotypic switching from 1116S to 1116R is recA dependent whereas that from 1116R to 1116S is recA independent.
Pseudomonas tolaasii is the causal agent of the economically important brown blotch disease of the cultivated mushroom Agaricus bisporus (Lange) Imbach (19). Colonies of the wild-type strain of P. tolaasii (designated 1116S) are opaque, mucoid, pathogenic, and nonfluorescent and produce tolaasin (6). In contrast, colonies of the stable phenotypic variant form of 1116S (designated 1116R) are translucent, nonmucoid, nonpathogenic, and fluorescent and do not produce tolaasin (6). Phenotypic switching in P. tolaasii from 1116S to 1116R is due to a reversible 661-bp duplication in the putative kinase domain of the regulatory locus pheN (8). This DNA duplication causes a frameshift mutation in the predicted pheN open reading frame (ORF), resulting in a truncated pheN′ ORF encoding a 77-kDa protein, which lacks part of the PheN sensor domain (8). Spontaneous switching from 1116R to 1116S occurs by precise deletion of the 661-bp element, restoring full functionality of PheN (8). This reversible mutation within pheN therefore represents one mechanism whereby P. tolaasii can switch its phenotype between pathogenic and nonpathogenic forms.
Studies with Escherichia coli and other bacteria have shown that RecA-dependent DNA recombination is the main mechanism of general homologous recombination (4). RecA-dependent recombination has been demonstrated in antigenic variation in Borrelia hermsii (17), in differential expression of surface layer proteins in Campylobacter fetus (3), in the instability of capsule production in Haemophilus influenzae type b (12), in switching of type IV pilin in Neisseria gonorrhoeae (15), in amplification of toxin genes of Vibrio cholerae (5), and in virulence determination in Yersinia pestis (10). In this paper we show that RecA plays a functional role in DNA rearrangement associated with phenotypic switching in P. tolaasii.
Putative cosmid clones containing the P. tolaasii recA gene were isolated by functional complementation of the recA-deficient E. coli host strain HB101 after en masse mobilization of a P. tolaasii genomic library and selection at 25°C on Pseudomonas agar F (PAF) (21) plates containing 0.02% methyl methanesulfonate (2). Complementation of E. coli DH5α (9) showed that pAPRL1 (Fig. 1) was able to restore RecA function as assessed by methyl methanesulfonate sensitivity. Subcloning of pAPRL1 in pBS (SK+) and subsequent DNA sequence analysis of subclones pAPR1 and pAPR2 revealed the presence of two ORFs of 1,059 and 468 nucleotides (EMBL accession number, recAX, AJ249265), coding for two proteins of 353 and 155 residues with predicted molecular masses of 37.6 and 17.9 kDa, which were designated recA and recX, respectively. This region had >90% homology to the P. fluorescens recA-recX region (1, 2).
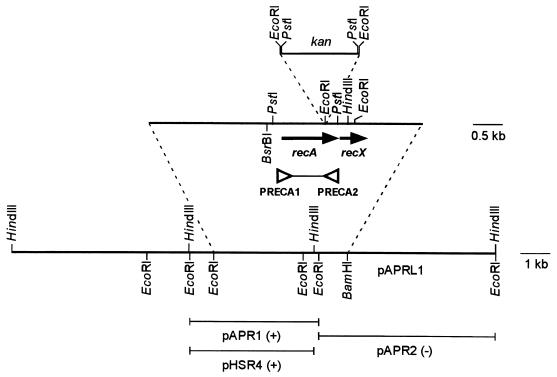
FIG. 1.
Restriction map of pAPRL1 showing the positions of recA and recX and the ability or inability of clones pAPR1 and pAPR2 to restore methyl methanesulfonate resistance (indicated by + and −, respectively) to E. coli HB101 and of pHSR4 to restore UV resistance (indicated by +) to P. tolaasii 1116SrecA. The primers used for recA PCR PRECA1 and PRECA2 are indicated as open triangles. Cloning sites, the kanamycin resistance cassette kan used, and the site of disruption in recA are also shown.
A 1.4-kb BsrBI-HindIII fragment from pAPR1 containing the P. tolaasii recA gene was ligated into SmaI-HindIII-digested pBS (SK+) to generate pHSR1. The 1.3-kb EcoRI fragment containing the kanamycin cassette from pUC4K (Promega) was ligated into the EcoRI site within the recA gene in pHSR1 to yield plasmid pHSR2. The 2.7-kb XbaI-ApaI fragment containing the recA knockout construct was ligated into XbaI-ApaI-digested pKNG101 (13) to give the suicide plasmid construct pHSR3, which was transformed into E. coli CC118 λ pir (11). Marker exchange mutagenesis following triparental mating, between P. tolaasii 1116S or 1116R together with the E. coli CC118 λ pir strain containing pHSR3 and E. coli containing the tra+ helper plasmid pNJ5000 (7), was as described previously (13, 14). Putative recA-disrupted strains, which were kanamycin resistant, streptomycin sensitive, and able to grow in the presence of 10% (wt/vol) sucrose, designated 1116SrecA and 1116RrecA, were selected. The site of recA disruption was confirmed by Southern hybridization of genomic DNA of 1116SrecA and 1116RrecA with an appropriate 1.1-kb recA probe (produced by PCR with primers PRECA1 [5′-AATGGACGACAACAA-3′, recA residues 413 to 427, accession number AJ249265] and PRECA2 [5′-GAACAGCGACGAGTG-3′, recA residues 1514 to 1500, accession number AJ249265] from 25 ng of pAPR1 [Fig. 1]) under optimum PCR conditions. The observed change in band sizes in 1116SrecA and 1116RrecA compared to 1116S and 1116R was consistent with disruption of chromosomal recA with the kanamycin resistance cassette at the EcoRI site (data not shown). The functional disruption of the recA transcript was further confirmed by Northern blotting using total cellular RNA and a recA-recX-specific probe. No recA or recX transcripts were detected in the recA-disrupted strains (1116SrecA and 1116RrecA) of P. tolaasii, whereas a positive signal was observed in Northern blots of wild-type P. tolaasii (total RNA extracted 6 h after induction with 2 μg of ofloaxin per ml [data not shown]).
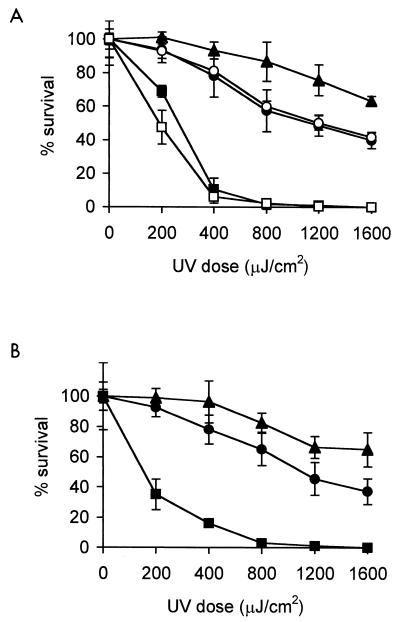
Both 1116SrecA and 1116RrecA were killed by lower doses of UV than were 1116S and 1116R, respectively (Fig. 2), and UV resistance was at least 75% restored at a UV dose of 400 μJ/cm2 by complementation with pHSR4 (a pLAFR3 clone containing a 4.3-kb HindIII fragment of pAPRL1 with a functional copy of recA [Fig. 1]) in both recA-disrupted strains and with pAPRL1 (containing a functional copy of recA/X [Fig. 1]) in 1116SrecA.
FIG. 2.
UV kill curves for wild-type, recA-disrupted, and complemented strains of P. tolaasii 1116S (A) and 1116R (B). ▴, wild type; ■, recA disrupted strain; □, recA disrupted strain complemented with pLAFR3; ●, recA disrupted strain complemented with pHSR4; ○, recA disrupted strain complemented with pAPRL1. Cells were plated on Pseudomonas F plates, exposed to UV light in a UV Stratalinker (Stratagene), and incubated at 25°C in the dark for 2 days. Error bars indicate the standard deviation from triplicate samples.
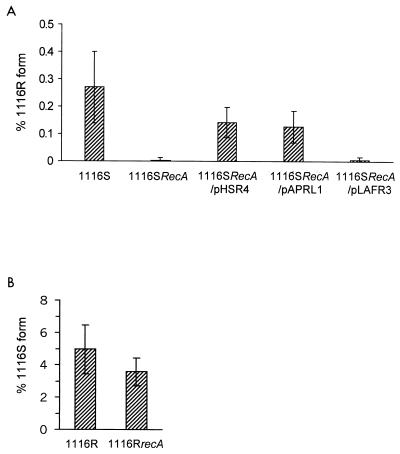
The effect of disruption of the chromosomal recA locus on phenotypic switching was determined for both 1116S and 1116R. The extent of switching from 1116S to 1116R was determined in a shake culture, inoculated with a single 2-day colony of 1116S, after 7 days by measuring the percentage of S and R forms in the resulting population. Since the culture reaches stationary phase after 2 to 3 days, it is not appropriate in this assay to relate the extent of switching to the number of generations of the test organism. Under these conditions, 0.2 to 0.3% of the 1116S population were present as the 1116R phenotype, whereas in 1116SrecA, this was reduced to less than 0.01% and was restored to 0.14% when complemented with pHSR4 (Fig. 3A). Colony PCR analysis (data not shown) of five randomly chosen R forms arising from 1116SrecA, with PHEN1 (5′-GGGCTATTTCACCTGGAT-3′, pheN residues 339 to 357, accession number U25692 [6]) and PHEN2 (5′-GCGGATTTCGTGGCTCAT-3′, pheN residues 1132 to 1114, accession number U25692 [6]) primers which flank the 661-bp duplication site in pheN (8), established that in each case no duplication had occurred in the pheN locus as seen in the 1116S-to-1116R conversion (data not shown). In contrast, the expected 661-bp duplication was observed by PCR analysis of five R forms arising from 1116SrecA/pHSR4 using the same primers (data not shown). When 1116SrecA was complemented with pAPRL1 containing the whole recA/X locus, the extent of switching was restored to 0.13%, which was not significantly different (at the 99% confidence level) from that in 1116SrecA/pHSR4 (Fig. 3A). PCR analysis (as described above) of five randomly chosen colonies of R forms arising from 1116SrecA/pAPRL1 confirmed that in all cases the expected 661-bp duplication had occurred in the pheN locus (data not shown). Finally, introduction of pLAFR3 into 1116SrecA had no effect on the extent of switching observed (Fig. 3A). Although both recA and recX functions are disrupted in 1116SrecA and these functions should both be restored in 1116SrecA/pAPRL1, the ability of pHSR4, which contains only the recA gene (Fig. 1), to restore phenotypic switching establishes that conversion of 1116S to 1116R forms is recA dependent. However in a recA-deficient background, phenotypic switching occurs to a reduced extent and is due to a mechanism other than the duplication event observed in 1116S (8).
FIG. 3.
(A) Extent of phenotypic switching from P. tolaasii 1116S, 1116SrecA, 1116SrecA/pHSR4, 1116SrecA/pAPRL1, and 1116SrecA/pLAFR3 to the corresponding R form. Serial dilutions of a shake culture (7 days at 25°C) in Pseudomonas F broth with appropriate antibodies were plated on PAF agar plates and incubated at 25°C for 2 days, and colonies were scored for phenotype. The extent of switching was calculated as the percentage of R forms in the total number of colonies. Error bars are from 10 replicate determinations in two independent experiments. The mean values for 1116SrecA and 1116SrecA/pLAFR3 are significantly different at the 99% confidence level (using the χ2 test) from the mean values for 1116S, 1116SrecA/pHSR4, and 1116SrecA/pAPRL1. (B) Extent of phenotypic switching from P. tolaasii 1116R and 1116RrecA to the S form in 7-day-old colonies. Serial dilutions of an 18-h culture grown at 25°C were plated out on PAF agar plates with appropriate antibiotics to give 10 to 15 colonies per plate. After incubation for 7 days at 25°C, three individual colonies were randomly selected, removed, and resuspended in 5 ml of sterile deionized water, and serial dilutions were plated out on PAF agar. The resulting colonies were scored for phenotype after a 2-day incubation at 25°C, and the extent of switching was calculated as the percentage of S forms in the total number of colonies. Error bars are from 10 replicate determinations in two independent experiments. The mean values are not significantly different at the 99% confidence level (using the χ2 test).
The extent of switching from 1116R to 1116S was determined by analyzing the percentage of S and R forms in 7-day-old colonies of 1116R on PAF. In this assay the S form appears from day 4 onward as microcolonies within the 1116R colony, and it is not possible to determine the number of generations of the 1116R and 1116S forms. Under these conditions, the extent of switching from the wild-type 1116R to the 1116S form was 4.8%, and there was no significant reduction in the extent of switching in 1116RrecA (Fig. 3B). These data establish that recA is not required for the 1116R-to-1116S reversion. Analysis of five independently isolated S forms from 1116RrecA using the PHEN1 and PHEN2 primers described above confirmed the expected precise deletion of the 661-bp fragment in pheN in all colonies examined, as seen in 1116S forms arising from 1116R (data not shown). It is therefore concluded that under the assay conditions used, transition of the pathogenic form (1116S) to the nonpathogenic form (1116R) is dependent on RecA function but that reversion from 1116R to 1116S is independent of RecA function.
Disruption of recA in 1116S and 1116R also abolished expression of the downstream ORF recX (data not shown). Plasmids pAPRL1 and pHSR4 restored UV resistance and the extent of switching to the 1116R form in 1116SrecA (Fig. 2) to the same extent. This argues against a direct role of recX in pheN duplication-related phenotypic switching from 1116S to 1116R forms in P. tolaasii. Although the precise role of recX in recA-mediated recombination is still under investigation, the ability of recX to reduce the toxicity of recA overexpression has been demonstrated in P. aeruginosa (18), Streptococcus lividans (20), and Mycobacterium smegmatis (16).
The biological significance of the phenotypic switch in P. tolaasii remains to be established. The higher extent of reversion from the avirulent 1116R form to the virulent 1116S form may be important in the epidemiology of brown blotch disease. The failure to find the origin of primary inoculum in mushroom farms may be due to the inadequacy of current diagnostic tests, which are able to detect only the 1116S form (21). Thus, the 1116R form may constitute the primary inoculum, which could revert to the virulent form either spontaneously or as a result of receiving environmental cues.
Acknowledgments
Himanshu Sinha and Arnab Pain contributed equally to the work.
We thank Bin Han and Chris Baldwin for their helpful discussions and Marcus Jarman for his technical assistance.
This work was supported by the Cambridge Commonwealth Trust (H.S. and A.P.) and was performed within the Cambridge Centre for Molecular Recognition under the provisions of licence PHF174B/63/90 issued by the Ministry of Agriculture, Fisheries and Food under the Plant Health (Great Britain) Order 1987.
REFERENCES
- 1.De Mot R, Schoofs G, Vanderleyden J. A putative regulatory gene downstream of recA is conserved in Gram-negative and Gram-positive bacteria. Nucleic Acids Res. 1994;22:1313–1314. doi: 10.1093/nar/22.7.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Mot R, Laeremans T, Schoofs G, Vanderleyden J. Characterisation of the recA gene from Pseudomonas fluorescens OE 28.3 and construction of a recA mutant. J Gen Microbiol. 1993;139:49–57. doi: 10.1099/00221287-139-1-49. [DOI] [PubMed] [Google Scholar]
- 3.Dworkin J, Blaser M J. Molecular mechanisms of Campylobacter fetus surface layer protein expression. Mol Microbiol. 1997;26:433–440. doi: 10.1046/j.1365-2958.1997.6151958.x. [DOI] [PubMed] [Google Scholar]
- 4.Dybvig K. DNA rearrangements, and phenotypic switching in prokaryotes. Mol Microbiol. 1993;10:465–471. doi: 10.1111/j.1365-2958.1993.tb00919.x. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg I, Mekalanos J J. Cloning of the Vibrio cholerae recA gene and construction of a Vibrio cholerae recA mutant. J Bacteriol. 1986;165:715–722. doi: 10.1128/jb.165.3.715-722.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grewal S I S, Han B, Johnstone K. Identification and characterization of a locus which regulates multiple functions in Pseudomonas tolaasii, the cause of brown blotch disease of Agaricus bisporus. J Bacteriol. 1995;177:4658–4668. doi: 10.1128/jb.177.16.4658-4668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grinter N J. A broad-host-range vector transposable to various replicons. Gene. 1983;21:133–143. doi: 10.1016/0378-1119(83)90155-5. [DOI] [PubMed] [Google Scholar]
- 8.Han B, Pain A, Johnstone K. Spontaneous duplication of a 661 bp element within a two component sensor regulator gene causes phenotypic switching in the colonies of Pseudomonas tolaasii, cause of brown blotch disease of mushrooms. Mol Microbiol. 1997;25:211–218. doi: 10.1046/j.1365-2958.1997.4411811.x. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–560. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 10.Hare J M, McDonough K A. High-frequency RecA-dependent and -independent mechanisms of Congo Red binding mutations in Yersinia pestis. J Bacteriol. 1999;181:4896–4904. doi: 10.1128/jb.181.16.4896-4904.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoiseth S K, Moxon E R, Silver R V. Genes involved in Haemophilus influenzae type b capsule expression are part of an 18-kilobase tandem duplication. Proc Natl Acad Sci USA. 1986;83:1106–1110. doi: 10.1073/pnas.83.4.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaniga K, Delor I, Cornelis G R. A wide host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene in Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 14.McGowan S J, Sebaihia M, O'Leary S, Hardie K R, Williams P, Stewart G S A B, Bycroft B W, Salmond G P C. Analysis of the carbapenem gene cluster of Erwinia carotovora: definition of the antibiotic biosynthetic genes and evidence for a novel β-lactam resistance mechanism. Mol Microbiol. 1997;26:545–556. doi: 10.1046/j.1365-2958.1997.6001974.x. [DOI] [PubMed] [Google Scholar]
- 15.Mehr I J, Seifert H S. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol Microbiol. 1998;30:697–710. doi: 10.1046/j.1365-2958.1998.01089.x. [DOI] [PubMed] [Google Scholar]
- 16.Papavinasasundaram K G, Movahedzadeh F, Keer J T, Stoker N G, Colston M J, Davis E O. Mycobacterial recA is cotranscribed with a potential regulatory gene called recX. Mol Microbiol. 1997;24:141–153. doi: 10.1046/j.1365-2958.1997.3441697.x. [DOI] [PubMed] [Google Scholar]
- 17.Restrepo B I, Kitten T, Carter C J, Infante D, Barbour A G. Subtelomeric expression regions of Borrelia hermsii linear plasmids are highly polymorphic. Mol Microbiol. 1992;6:3299–3311. doi: 10.1111/j.1365-2958.1992.tb02198.x. [DOI] [PubMed] [Google Scholar]
- 18.Sano Y. Role of the recA-related gene adjacent to the recA gene in Pseudomonas aeruginosa. J Bacteriol. 1993;175:2451–2454. doi: 10.1128/jb.175.8.2451-2454.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tolaas A G. A bacterial disease of cultivated mushrooms. Phytopathology. 1915;5:51–54. [Google Scholar]
- 20.Vierling S, Weber A, Wohlleben, W. T, Muth G. Transcriptional and mutational analyses of the Streptomyces lividans recX gene and its interference with RecA activity. J Bacteriol. 2000;182:4005–4011. doi: 10.1128/jb.182.14.4005-4011.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong W C, Preece T F. Identification of Pseudomonas tolaasii: the white line in agar and mushroom tissue block rapid pitting tests. J Appl Bacteriol. 1979;47:401–407. [Google Scholar]