Abstract
A new subunit, YabF, for the KefC K+ efflux system in Escherichia coli has been identified. The subunit is required for maximum activity of KefC. Deletion of yabF reduces KefC activity 10-fold, and supply of YabF in trans restores activity. IS2 and IS10R insertions in yabF can be isolated as suppressors of KefC activity consequent upon the V427A and D264A KefC mutations.
Glutathione (GSH)-gated potassium efflux systems are found in a range of gram-negative bacteria (5, 6). These systems have been most extensively studied for Escherichia coli, where there are two systems, KefC and KefB, that are closely related in their sequence and regulation (6). The efflux systems are maintained in a closed state by GSH or by its nonsulfydryl analogue, ophthalmic acid (10, 17). KefC and KefB are activated by adducts formed by reaction of GSH with electrophilic compounds, such as N-ethylmaleimide (NEM), methylglyoxal, and chlorodinitrobenzene (10). The systems differ in their response to methylglyoxal, with only KefB being strongly activated by this electrophile (12). In addition to regulation by specific ligands, the proteins share many features with eukaryotic channels, and calculated rates of K+ efflux are consistent with channel-like activity (6). Each efflux system was originally identified as the product of a single structural gene, kefC and kefB, for the KefC and KefB systems, respectively (5, 6). In this study, we report that each system has in addition a separate and specific ancillary protein that is required for full activity.
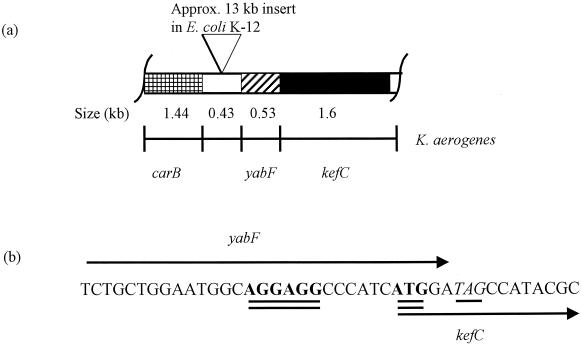
The yabF gene was discovered during the cloning of the kefC locus from Klebsiella aerogenes. Plasmid pASRB1 was derived from a plasmid carrying the K. aerogenes folA gene, which in E. coli lies immediately clockwise after the kefC gene (3). This plasmid was expected to carry the K. aerogenes kefC gene since it restored NEM-elicited K+ efflux to E. coli strain MJF276 (KefB− KefC−) (26) and it complements the E. coli KefCD264A mutation (9). The sequence of the 4.4-kb EcoRI-BamHI insert on pASRB1 was determined on both strands (26) (EMBL submission AJ242913) and was found to carry the 3′ end of the carB gene, kefC, and the open reading frame (orf) yabF (Fig. 1). On the E. coli chromosome, kefC and carB lie 13.7 kb apart and are separated by a number of orfs (4, 23), but the only one conserved at this position between Klebsiella and E. coli is yabF. PCR analysis using K. aerogenes cells as the source of DNA and a forward and reverse primer specific for carB and kefC, respectively, yielded a product of 2.4 kb, the size predicted from the sequence of the insert in pARSB1 (Fig. 1a) (data not shown). Similarly, Southern blots of K. aerogenes DNA digested with EcoRI and BamHI using probes specific to carB and kefC yielded identical 4-kb bands as predicted by the restriction map of pARSB1 (Fig. 1a) (26). Thus, in the K. aerogenes genome carB and kefC are separated only by the orf yabF. The significance of this observation was enhanced by the observation that the yabF orf overlaps that of kefC by 8 bases in both K. aerogenes and E. coli, which suggested that the two genes might be related by function (Fig. 1b). A similar orf, yheR, was found 5′ to kefB and overlaps the 5′ end of the kefB gene of E. coli by 1 bp (20).
FIG. 1.
Diagram showing the organization of the carB and kefC region in K. aerogenes and E. coli (a) and the overlap between yabF and kefC in E. coli (b). (a) The bar indicates the 4.2-kb fragment cloned and sequenced from pASRB1, showing the position of the 3′ end of carB (hatched), the carB-yabF intergenic region (open), yabF (diagonally striped), and kefC (filled). The inverted triangle above the block indicates the approximate position of the 13-kb insert found in the E. coli genome sequence (23). (b) DNA sequence of the E. coli yabF-kefC junction. The arrows indicate the position of the 3′ end of yabF (TAG stop codon underlined) and the 5′ end of kefC (putative Shine-Dalgarno sequence and start codon double underlined).
The yabF orf would encode a soluble 20.2-kDa protein (176 amino acids in E. coli) that exhibits considerable similarity to quinone oxidoreductases and to proteins involved in drug sensitivity (MdaB) (7) (Table 1). K. aerogenes YabF is only 83% identical to the E. coli protein (Table 1), whereas the KefC sequence retains 88% identity to E. coli KefC (data not shown). During the course of this study, the yabF gene of Frag5, the parent strain used in our studies, was found to have three single-base changes from the reported K-12 sequence (P31577), which cause two amino acid changes (N79D and G123V) and a silent alteration (G89). The identical sequence was found in pkC11, which we created in the initial cloning of the yabF-kefC genes (19), and which derives from the Clarke-Carbon cosmid series (8). These base changes were previously noted as conflicts (P31577) with the original K-12 sequence, but from our data it is likely that these differences are common to many E. coli K-12 strains.
TABLE 1.
Sequences with greatest overall similarity to YabF
| Gene sequence accession no. | Organism | Protein and/or function | BlastP scorea | Similarityb
|
Lengthd | |
|---|---|---|---|---|---|---|
| %I | %S | |||||
| AJ242913 | K. aerogenes | YabF; regulator of KefC | 1e−90 | 83 | 92 | 176/176 |
| Z99117 | Bacillus subtilis | YrkL; putative NAD(P)H oxidoreductase | 2e−30 | 39 | 56 | 170/174 |
| Z93767 | Bacillus subtilis | GS14; putative NAD(P)H oxidoreductase | 6e−29 | 38 | 54 | 163/175 |
| U18997 | E. coli | YheR; putative NAD(P)H oxidoreductase | 2e−26 | 40 | 55 | 163/184 |
| Z99106 | Bacillus subtilis | YdeQ; putative NAD(P)H oxidoreductase | 2e−26 | 38 | 54 | 164/197 |
| J02888 | Homo sapiens | Human quinone oxidoreductase (QR2) | 3e−13 | 33 | 48 | 149/231 |
| P15559 | Homo sapiens | Human quinone oxidoreductase (QR1) | 8e−12 | 34 | 56 | 103/274 |
| T23934 | Streptomyces coelicolor | Putative NAD(P)H oxidoreductase | 2e−7 | 37 | 58 | 65/246 |
| U32829 | Haemophilus influenzae | Putative NAD(P)H oxidoreductase | 4e−6 | 29 | 43 | 134/202 |
| D90728 | E. coli | Putative NAD(P)H oxidoreductase | 7e−6 | 31 | 46 | 105/196 |
| C64084 | Haemophilus influenzae | MdaB; modulator of drug activityc | 1e−5 | 23 | 37 | 178/208 |
The BlastP (2.0.12) program (1) at the National Center for Biotechnology Information was used to compare the E. coli YabF protein sequence with available completed genomes. A number of related sequences have been found in the incomplete genome sequences also but have not been included in this analysis. None of the bacterial putative quinone oxidoreductases has been characterized.
%I, percentage of identity; %S, percentage of similarity.
A number of gene products that are related by sequence to the E. coli MdaB (modulator of drug resistance) (7) were found to be related to the YabF protein with similar BlastP scores but have been omitted for clarity. However, the E. coli MdaB protein exhibited a score of 0.005, which has a very low significance.
First number, length of protein (amino acids) over which similarity is significant; second number, length of protein.
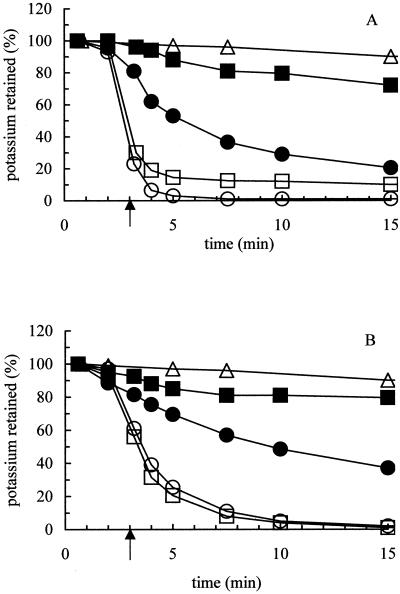
To investigate the function of the YabF and YheR proteins, deletion mutants were constructed that removed yabF and yheR and the adjacent kefB and kefC genes. Regions flanking the yabF-kefC and yheR-kefB genes were amplified by PCR and cloned into pHG165 (2, 24). A restriction site was created between the flanking regions during the PCR amplification, as described previously (25), and the kanamycin resistance cassette of pUC4K (Pharmacia) was then inserted at this restriction site. Integration of the cloned kanamycin cassette DNA, via the flanking regions, was accomplished by transformation of strain JC7623 (15) as described previously (25). Strains MJF362 (Frag5, ΔyabF-kefC) and MJF369 (MJF274, ΔyheR-kefB) were created by P1 transduction of Frag5 and MJF274, respectively, to Kanr (Table 2). From these and related strains carrying either kefB::Tn10 or kefC::Tn10 (5), a series of strains was created that possessed different combinations of YabF, YheR, KefC, and KefB (Table 2). The strains exhibited no significant growth phenotype. Using these strains, NEM-elicited KefC activity was investigated using two plasmids: pkC952 (YabF− KefC+) (19), which expresses KefC but does not carry a complete yabF gene due to deletion of the first 389 bp of the yabF sequence at the 5′ end, and pkC11 (YabF+ KefC+) (9). The KefC activity derived from pkC952 was always less than that obtained with pkC11, as noted previously (9) (see below). Strain MJF276 (YabF+ KefC− YheR+ KefB−), which retains a functional chromosomal yabF gene (12), exhibited high levels of KefC activity when transformed with either pkC952 or pkC11 (Fig. 2A). In contrast, MJF366 (YabF− KefC− YheR+ KefB−) exhibits only low activity when transformed with pkC592, while full efflux activity was seen with pkC11 (Fig. 2A). The residual K+ efflux seen with MJF366/pkC952 was slightly greater than that seen with MJF276, which lacks KefB and KefC activity, and this suggests that the KefC system retains some activity in the absence of YabF (Fig. 2A). Identical data were obtained with strain MJF374 (YheR− KefB− YabF− KefC−), which lacks both YabF and YheR, transformed with pkC952 and pkC11, and this suggests that the YheR protein could not substitute for YabF in activating KefC (data not shown). Thus, YabF is required for NEM-elicited KefC activity. Similar data obtained with plasmid pΔYheR (YheR− KefB+) (20) support the conclusion that YheR is required for KefB activity (Fig. 2B). KefB activity was evident only when YheR either was present on the cloned fragment or was supplied from the chromosome (Fig. 2B). These data suggest that YabF and YheR proteins are required for KefC and KefB activity, respectively, and that this requirement can be met in trans.
TABLE 2.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or description | Reference |
|---|---|---|
| Strains | ||
| Frag5 | F− ΔkdpABC5 thi rha lacZ | 11 |
| JC7623 | F−thr-1 leuB6 hisG4 argE3 thi-1 recB21 recC22 sbcB15 ara-14 tsx-33 supE44 galK2 rfbD1 zyl-5 mtl-1 rpsL31 lacY1 Δ(gpt-proA)62 kdpK51 | 15 |
| MJF104 | Frag5 kefC104 (KefCD264A) kefB::Tn10 | 18 |
| MJF104#29 | MJF104 [IS10R 31 bp upstream of yabF] | This study |
| MJF104#31 | MJF104 [IS10R 31 bp upstream of yabF] | This study |
| MJF116 | Frag5 kefC116 (KefCV427A) kefB::Tn10 | 18 |
| MJF116#22 | MJF116 [IS2 19 bp upstream of yabF] | This study |
| MJF116#5 | MJF116 [yabF::IS10R; 434 bp 3′ to yabF ATG] | This study |
| MJF116#34 | MJF116 [yabF::IS10R; 434 bp 3′ to yabF ATG] | This study |
| MJF116#52 | MJF116 [yabF::IS10R; 436 bp 3′ to yabF ATG] | This study |
| MJF274 | F− ΔkdpABC5 thi rha lacI lacZ trkD1 | 10 |
| MJF276 | MJF274 kefB157 kefC::Tn10 | 21 |
| MJF335 | MJF276 gshA::Tn10(Kan) | 18 |
| MJF362 | Frag5 Δ[yabF-kebfzztabftr;bC] | 26 |
| MJF366 | MJF274 kefB::Tn10 Δ[yabF-kefC] | 20 |
| MJF369 | MJF274 Δ[yheR-kefB] | 20 |
| MJF374 | MJF276 Δ[yabF-kefC] Δ[yheR-kefB] | 20 |
| MJF526 | MJF116#52 gshA::Tn10(Kan) | This study |
| MJF527 | MJF116#22 gshA::Tn10(Kan) | This study |
| MJF528 | MJF104#29 gshA::Tn10(Kan) | This study |
| MJF532 | MJF116 gshA::Tn10(Kan) | This study |
| Plasmids | ||
| pHG165 | Cloning vector; a pBR322 copy number derivative of pUC8 | 24 |
| pASRB1 | pBR322 carrying yabF and kefC from K. aerogenes strain W70 | 3 |
| pkC11 | pHG165 carrying yabF and kefC | 19 |
| pkC952 | pkC11 minus promoter region and first 389 bp of yabF | 19 |
| pCWyabF | XhoI-EcoRV fragment from pKC11; yabF and initial 471 bp of kefC | 26 |
| pKefB | pHG165 carrying yheR and kefB | 20 |
| pΔYheR | pKefB with promoter region and first 188 bp of yheR deleted | 20 |
| pGEXBG4 | pGEX2TK carrying in-frame fusion of GSH S-transferase–YabF | 14 |
FIG. 2.
YabF and YheR are required in trans for the activity of KefC and KefB. Potassium efflux was measured in mutant strains transformed with the appropriate plasmids to allow the contribution of YabF and YheR to be analyzed. Potassium efflux was measured according to methods in previous publications (21), and each experiment has been repeated at least three times. The data shown are representative. The arrow indicates the time of addition of 0.5 mM NEM. (A) KefC activity. Symbols: ▵, strain MJF276 (YheR+ KefB− YabF+ KefC−); ●, MJF276/pkC952; ○, MJF276/pkC11; ■, MJF366/pkC952; □, MJF366/pkC11. (B) KefB activity. Symbols: ▵, strain MJF276; ●, MJF276/pΔYheR; ○, MJF276/pKefB; ■, MJF370/pΔYheR; □, MJF370/pKefB. Strain MJF276 expresses YabF and YheR, MJF366 expresses only YheR, and MJF370 expresses only YabF. Plasmids pkC952 (YabF− KefC+), pkC11 (YabF+ KefC+), pΔYheR (YheR− KefB+), and pKefB (YheR+ KefB+) are described in Table 2. Data obtained with MJF374 (YabF− KefC− YheR− KefB−) transformed with the above plasmids were identical to the data obtained with either MJF366 (A) or MJF370 (B) (data not shown).
Missense mutations in kefC that lead to enhanced spontaneous activity were previously isolated by their failure to grow on medium containing 0.1 mM K+ (K0.1 medium) (5, 11, 17). We have previously shown that kefC null mutants are readily isolated as colonies that grow on K0.1 medium (5, 11). These isolates grow normally on medium containing either 10 mM (K10) or 115 mM (K115) K+, and thus buffers and media with these concentrations were used to allow normal growth and retention of the K+ pool. Independent suppressor mutations were sought that restored growth on K0.1 medium but which retained KefC activity as detected by NEM-elicited K+ efflux. Strains carrying suppressor mutations were isolated from strains MJF104 and MJF116, which carry the KefCD264A and KefCV427A mutations, respectively (18). Overnight cultures of MJF104 or MJF116 (grown at 37°C in K120 minimal medium and glucose [0.2% (wt/vol)] as the carbon source) were washed sequentially in K10 buffer and K0 buffer and then serially diluted in K0 buffer. Dilutions were plated on control, high-potassium (K120) medium, and selective, low-potassium (K0.1) minimal medium (11). Colonies were visible after 48 h on the low-potassium plates, and a single suppressor colony was chosen from each original overnight culture. The frequency of isolation of suppressors of strains MJF116 and MJF104 was approximately 3 × 10−5 and 4 × 10−6, respectively. Six mutant strains exhibited the unmodified kefC gene sequence of the mutant strains used for suppressor isolation (i.e., they retained either the V427A or the D264A mutation). However, PCR analysis of the yabF gene and its flanking regions revealed that the normal ∼950-bp fragment found in the parent (Frag5), MJF104 (KefCD264A), and MJF116 (KefCV427A) was enlarged to approximately 2 to 3 kb. Sequence analysis on both strands revealed that this region of each suppressor strain contained an insertion of either IS10R or IS2 (Table 2). The MJF104 suppressors MJF104#29 and MJF104#31 had IS10R insertions 5′ to the yabF gene approximately 31 bp 5′ to the translation start site. Although both IS10R insertions are in the same orientation, the strains were isolated from independent cultures. MJF116#22 was found to contain an IS2 insertion 5′ to the yabF gene at approximately 19 bp 5′ to the translation start site, which may disrupt the promoter structure. Three MJF116 derivatives, MJF116#52, MJF116#5, and MJF116#34, carried an IS10R element inserted within the coding sequence of yabF (Table 2). The insertion in MJF116#5 and MJF116#34 was at the same position (434 bp into yabF), but the IS10R insertions lie in the opposite orientation (data not shown). Strain MJF116#52 carries an IS10R at bp 436 of yabF (data not shown). Thus, it is likely that the suppression in these strains arose either from inactivation of YabF or from reduced expression of yabF due to disruption of the promoter. Southern hybridization was performed on the suppressor strains and confirmed that the IS10R and IS2 mutations arose by a single duplicative transposition from an insertion sequence (IS) located elsewhere in the genome (data not shown).
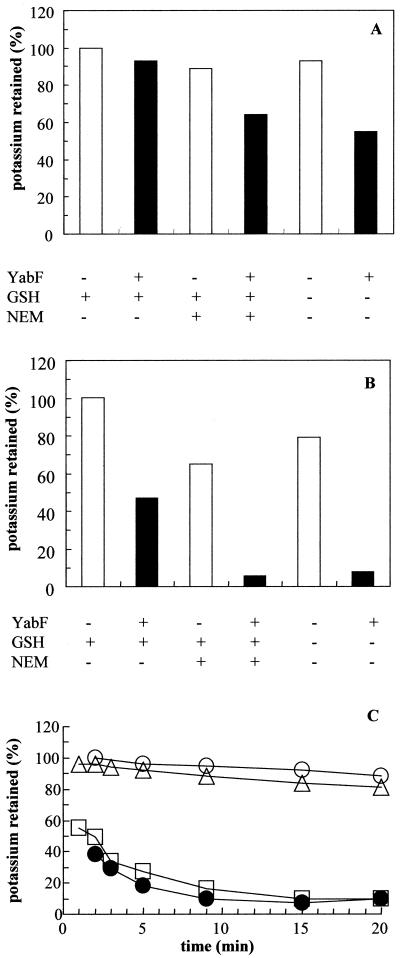
The activity of the KefC system was analyzed in more detail for strains MJF116#52 (KefCV427A, YabF::IS10R) and MJF116#22 (KefCV427A, IS2 insertion 5′ to yabF). These strains differed in their response to the introduction of YabF in trans. Strain MJF116#22 showed only marginal increases in KefC activity when pCWYabF was introduced irrespective of whether GSH was present (Fig. 3A). Similar data were obtained with suppressors of MJF104 both of which had IS insertions 5′ to the yabF gene (data not shown). These data are consistent with suppression arising from diminished expression of both YabF and KefC due to the IS insertion 5′ to the yabF gene. MJF116#52 (KefCV427A, YabF::IS10R) exhibited low rates of both spontaneous (i.e., in the absence of an added electrophile) and NEM-elicited efflux compared with the parent strain MJF116 (KefCV427A) (Fig. 3B). Efflux could be restored by expression of YabF in trans using plasmid pCWYabF (Table 2), which carries the E. coli yabF gene (Fig. 3B), or by pGEXBG4, which expresses a GSH S-transferase–YabF protein fusion (data not shown) (14). Thus, the YabF protein expressed in trans can complement the YabF deficiency of this suppressor strain. GSH-deficient mutants exhibit high rates of spontaneous K+ loss due to deregulation of KefC activity (10, 17, 18), and in most missense mutants of KefC there is synergy between the absence of GSH and the altered KefC protein leading to very rapid K+ efflux (18). In contrast, K+ efflux was negligible in the equivalent GSH-deficient derivative of strain MJF116#52 (KefCV427A, YabF::IS10R), and introduction of pCWYabF restored K+ efflux (Fig. 3C). Thus, YabF is required for activity of KefC even in the absence of GSH.
FIG. 3.
Analysis of KefCV427A suppressor mutants. Potassium efflux was determined as described previously (10, 12). (A) Strains MJF116#22, (KefCV427A, IS2 5′ to yabF) and MJF527 (MJF116#22, GshA−) were transformed with pCWYabF (YabF+) to create strains that possess different combinations of YabF and GSH biosynthesis (Table 2). The strains were incubated in potassium-free medium (18), and either spontaneous K+ efflux (no NEM) or NEM-elicited K+ efflux (NEM added 3 min after resuspension in K+-free medium) was measured. Open columns, YabF− strains; filled columns, YabF+ strains. The percentage of K+ retained 15 min after resuspension in K+-free medium is shown. Low figures indicate high KefC activity. One hundred percent is set for MJF527 immediately after resuspension in K+-free medium. (B) Details are as for panel A, substituting strains MJF116#52 and MJF526 (MJF116#52, GshA−) for strains MJF116#22 and MJF527, respectively. Similar data were obtained with MJF116#5 and MJF116#34 and their GshA− derivatives (data not shown). (C) Potassium efflux from GSH-deficient strains: MJF335 (MJF276, GshA−) (▵), MJF532 (MJF116, GshA−) (□), MJF526 (MJF116#52, GshA−) (○), and MJF526/pCWYabF (●). Spontaneous K+ efflux was measured as described previously (18), and the experiments have been repeated at least three times. The data shown are representative.
From these data, we conclude that YabF is an intrinsic component of the KefC potassium efflux system. A similar protein, YheR, was found for the activity of the KefB system. The ancillary subunits are specific to the individual efflux system since strains possessing YheR but lacking YabF did not display significant KefC activity (MJF366/pkC952 [Fig. 2A]) and vice versa (data not shown). YabF and YheR exhibit only 55% similarity, which may explain the apparent lack of cross activation of the efflux systems. The yabF and yheR orfs and the genes for the integral membrane components (kefC and kefB, respectively) of the efflux systems overlap in sequence and may form operons in E. coli. However, it is likely that the kefC gene can be expressed independently of yabF, since firstly pkC952 expresses KefC, albeit at an approximately 30-fold-lower rate than that of the normal yabF-kefC construct found in pkC11 (9, 12). Secondly, the yabF::IS suppressor mutants of the V427A and D264A mutants of KefC still retain expression of the KefC system, and complementation in trans by the cloned yabF gene restores full KefC activity (Fig. 3B and C). The overlap between the yabF and kefC genes may allow for translational coupling between the two orfs. There is strong secondary structure predicted for the yabF-kefC junction (and also for yheR-kefB), which places the ribosome binding site for the membrane protein in a stem-loop. This may be sufficient to explain the low level of expression from pkC952 (9, 12), since this plasmid does not carry the translation initiation signals for yabF but retains the 3′ sequence that is involved in potential stem-loop formation.
The regulation of KefC and KefB by GSH and GSH metabolites is well established (10, 17, 18). Regulation of K+ efflux via KefC or KefB by GSH is unlikely to be directly mediated by the ancillary subunits since strains lacking YabF do not exhibit high rates of spontaneous potassium loss, which is the phenotype expected for loss of control by GSH (10, 17). Indeed, even in the absence of YabF and YheR, the KefC and KefB efflux systems retain both negative regulation by GSH and activation via GSH adducts (Fig. 2 and 3). The data presented here show that the newly identified subunit is required for maximum activity of the system and therefore provides an additional level of complexity of the efflux systems. The YabF protein shows strong sequence similarity to human quinone oxidoreductases QR1 and QR2 (13). YabF is a shorter protein than either QR1 or QR2, which arises from truncation of the N and C termini (data not shown). The greatest sequence conservation lies around two regions associated with flavin binding, but YabF retains only 4 of the 15 residues that are implicated in binding the flavin. Due to truncation of the C terminus, the YabF protein lacks the NAD(P)H binding site that is present in QR1 (13). It seems unlikely, therefore, that the YabF protein (and by inference YheR, which similarly lacks conservation of the essential residues) has quinone oxidoreductase activity. It seems probable that these proteins have evolved from their role as oxidoreductases to be modulators of KefC (and KefB) activity in a manner similar to that of β subunits of mammalian Shaker channels (16). Therefore, by analogy with the Shaker family of K+ channels (16, 22), we suggest that the KefB and KefC efflux systems each comprise two structural components, KefC with YabF and KefB with YheR, both of which are required to give the functional characteristics of the efflux system. Given their unique role in the activity of KefC and KefB, we now propose that YabF and YheR should be termed KefF and KefG, respectively.
Acknowledgments
S.M., L.S.N., and C.M.W. contributed equally to the work.
This work was supported by a Wellcome Trust Programme grant (040174), a Wellcome Trust Prize studentship to C.M.W., BBSRC studentships to B.C.F. and L.S.N., and a Research Leave Fellowship to I.R.B.
We thank Clare Barson, Joanne Salmon, and Fiona Galbraith for their contributions to this project.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programmes. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Sideman J G, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1987. [Google Scholar]
- 3.Azakami H, Sugino H, Murooka Y. Cloning and nucleotide sequence of a negative regulator for Klebsiella aerogenes arylsulfatase synthesis and identification of the gene folA. J Bacteriol. 1992;174:2344–2351. doi: 10.1128/jb.174.7.2344-2351.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlyn M K B. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol Mol Biol Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booth I R, Epstein W, Giffard P M, Rowland G C. Roles of the trkB and trkC gene products of Escherichia coli in K+ transport. Biochimie. 1985;67:83–90. doi: 10.1016/s0300-9084(85)80233-9. [DOI] [PubMed] [Google Scholar]
- 6.Booth I R, Jones M A, McLaggan D, Nicolaev Y, Ness L S, Wood C M, Miller S, Tötemeyer S, Ferguson G P. Bacterial ion channels. In: Konings W N, Kaback H R, Lolkema J S, editors. Handbook of biological physics. Vol. 2. Amsterdam, The Netherlands: Elsevier Science B.V.; 1996. pp. 693–729. [Google Scholar]
- 7.Chatterjee P K, Sternberg N L. A general genetic approach in Escherichia coli for determining the mechanisms of action of tumoricidal agents—application to DMP-840, a tumoricidal agent. Proc Natl Acad Sci USA. 1995;92:8950–8954. doi: 10.1073/pnas.92.19.8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke L, Carbon J. A colony bank containing synthetic ColE1 hybrid plasmids representative of the entire Escherichia coli genome. Cell. 1976;9:91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- 9.Douglas R M, Ritchie G Y, Munro A W, McLaggan D, Booth I R. The potassium efflux system, KefC, in Escherichia coli: genetic evidence for oligomeric structure. Mol Membr Biol. 1994;11:55–61. doi: 10.3109/09687689409161030. [DOI] [PubMed] [Google Scholar]
- 10.Elmore M J, Lamb A J, Ritchie G Y, Booth I R. Activation of potassium efflux from Escherichia coli by glutathione metabolites. Mol Microbiol. 1990;4:405–412. doi: 10.1111/j.1365-2958.1990.tb00607.x. [DOI] [PubMed] [Google Scholar]
- 11.Epstein W, Kim B S. Potassium transport loci in Escherichia coli K-12. J Bacteriol. 1971;108:639–644. doi: 10.1128/jb.108.2.639-644.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson G P, Munro A W, Douglas R M, McLaggan D, Booth I R. Activation of potassium channels during metabolite detoxification in Escherichia coli. Mol Microbiol. 1993;9:1297–1303. doi: 10.1111/j.1365-2958.1993.tb01259.x. [DOI] [PubMed] [Google Scholar]
- 13.Foster C E, Bianchet M A, Talalay P, Zhao Q, Amzel L M. Crystal structure of human quinone reductase type 2, a metalloflavoprotein. Biochemistry. 1999;38:9881–9886. doi: 10.1021/bi990799v. [DOI] [PubMed] [Google Scholar]
- 14.Fox B C. Ph.D. thesis. Aberdeen, United Kingdom: University of Aberdeen; 2000. [Google Scholar]
- 15.Kushner S R, Nagaishi H, Templin A, Clark A J. Genetic recombination in Escherichia coli: the role of exonuclease 1. Proc Natl Acad Sci USA. 1971;68:824–827. doi: 10.1073/pnas.68.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCormack T, McCormack K. Shaker K+ channel β-subunits belong to an NAD(P)H-dependent oxidoreductase superfamily. Cell. 1984;79:1133–1135. doi: 10.1016/0092-8674(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 17.Meury J, Kepes A. Glutathione and the gated potassium channels of Escherichia coli. EMBO J. 1982;1:339–343. doi: 10.1002/j.1460-2075.1982.tb01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller S, Douglas R M, Carter P, Booth I R. Mutations in the glutathione-gated KefC K+ efflux system of Escherichia coli that cause constitutive activation. J Biol Chem. 1997;272:24942–24947. doi: 10.1074/jbc.272.40.24942. [DOI] [PubMed] [Google Scholar]
- 19.Munro A W, Ritchie G Y, Lamb A J, Douglas R M, Booth I R. The cloning and DNA sequence of the gene for the glutathione-regulated potassium-efflux system KefC of Escherichia coli. Mol Microbiol. 1991;5:607–616. doi: 10.1111/j.1365-2958.1991.tb00731.x. [DOI] [PubMed] [Google Scholar]
- 20.Ness L S. Ph.D. thesis. Aberdeen, United Kingdom: University of Aberdeen; 1996. [Google Scholar]
- 21.Ness L S, Booth I R. Different foci for the regulation of the activity of the KefB and KefC glutathione-gated K+ efflux systems. J Biol Chem. 1999;274:9524–9530. doi: 10.1074/jbc.274.14.9524. [DOI] [PubMed] [Google Scholar]
- 22.Pongs O. Regulation of the activity of voltage-gated potassium channels by β-subunits. Semin Neurosci. 1995;7:137–146. [Google Scholar]
- 23.Rudd K E. Linkage map of Escherichia coli K-12, edition 10: the physical map. Microbiol Mol Biol Rev. 1998;62:985–1019. doi: 10.1128/mmbr.62.3.985-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart G S A B, Lubinsky-Mink S, Jackson C G, Cassel A, Kuhn J. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid. 1986;15:172–181. doi: 10.1016/0147-619x(86)90035-1. [DOI] [PubMed] [Google Scholar]
- 25.Tötemeyer S, Booth N A, Nichols W W, Dunbar B, Booth I R. From famine to feast: the role of methylglyoxal production in Escherichia coli. Mol Microbiol. 1998;27:553–562. doi: 10.1046/j.1365-2958.1998.00700.x. [DOI] [PubMed] [Google Scholar]
- 26.Wood C M. Ph.D. thesis. Aberdeen, United Kingdom: University of Aberdeen; 1996. [Google Scholar]