In this study, Lyu et al. investigated the lineage specificity of KLF5, a transcription factor broadly involved in development and regeneration, and report that KLF5 specifically marks the epidermis and that its deletion leads to skin barrier dysfunction in vivo. Their findings establish KLF5 as a disease-relevant transcription factor governing sphingolipid metabolism and barrier function in the skin, likely representing a long-sought secretory lineage-defining factor across tissue types.
Keywords: stem cells, transcription factors, skin epidermis, barrier, secretory, sphingolipid metabolism
Abstract
Stem cells are fundamental units of tissue remodeling whose functions are dictated by lineage-specific transcription factors. Home to epidermal stem cells and their upward-stratifying progenies, skin relies on its secretory functions to form the outermost protective barrier, of which a transcriptional orchestrator has been elusive. KLF5 is a Krüppel-like transcription factor broadly involved in development and regeneration whose lineage specificity, if any, remains unclear. Here we report KLF5 specifically marks the epidermis, and its deletion leads to skin barrier dysfunction in vivo. Lipid envelopes and secretory lamellar bodies are defective in KLF5-deficient skin, accompanied by preferential loss of complex sphingolipids. KLF5 binds to and transcriptionally regulates genes encoding rate-limiting sphingolipid metabolism enzymes. Remarkably, skin barrier defects elicited by KLF5 ablation can be rescued by dietary interventions. Finally, we found that KLF5 is widely suppressed in human diseases with disrupted epidermal secretion, and its regulation of sphingolipid metabolism is conserved in human skin. Altogether, we established KLF5 as a disease-relevant transcription factor governing sphingolipid metabolism and barrier function in the skin, likely representing a long-sought secretory lineage-defining factor across tissue types.
Tissue stem cells undergo long-term self-renewal and multilineage differentiation, serving as fundamental units for tissue remodeling (Joseph and Morrison 2005; Chandel et al. 2016; Ge and Fuchs 2018). Among the key molecular machineries governing stem cell functions are lineage-specific transcription factors (TFs), a group of evolutionarily conserved transcriptional regulators well known for their ability to dictate cell fates (Jaenisch and Young 2008; Sánchez Alvarado and Yamanaka 2014; Iwafuchi-Doi and Zaret 2016). It remains a major challenge, however, to define the tissue-specific roles of lineage TFs and the in vivo contexts under which they exert physiological and pathological functions.
Mammalian skin is an excellent model to address these outstanding questions. As the body's largest organ, skin serves as a physical barrier to provide protection and mediate organismal communications. Epidermal stem cells residing in the basal layer undergo self-renewal and fuel the upward flux of progenies to orchestrate stratification (Rheinwald and Green 1975; Watt 2001; Fuchs 2008; Frye and Benitah 2012; Lopez-Pajares et al. 2013; Perdigoto et al. 2014). These stem cell descendants sequentially form the suprabasal spinous, secretory granular, and protective cornified layers, which culminate in a tight seal that enables the adaptation to terrestrial life (Wertz and Downing 1982; Eckert 1989). Mammalian epidermis differs from other epithelial tissues in its restricted water permeability, a unique property that finds its resemblance in plants and yeast cell walls (Nemes and Steinert 1999; Candi et al. 2005; Proksch et al. 2008; Feingold and Elias 2014). A masterpiece in the making, skin function is heavily dependent on specialized molecular species, including complex lipids and secretory machineries (Candi et al. 2005; Uchida and Holleran 2008; Breiden and Sandhoff 2014; Rabionet et al. 2014; Kihara 2016), the disruption of which leads to a wide spectrum of human cutaneous disorders (Bouwstra and Ponec 2006; Elias et al. 2008; Traupe et al. 2014). Despite epidermal lipid secretion being long appreciated as an intricate multistep process, a major transcriptional orchestrator of this process has yet to be identified. In fact, broadly speaking, whether there is a master secretory lineage factor remains a tantalizing hypothesis, particularly when it relates to diseases.
KLF5 belongs to a family of highly conserved Krüppel-like zinc finger TFs that are essential across the spectrum of metazoan animals (Turner and Crossley 1999; Presnell et al. 2015). KLF5 is important for the development of multiple mammalian organs through its regulation of cell proliferation and regeneration (Nagai et al. 2005; McConnell and Yang 2010) whose lineage specificity, if any, remains elusive. In the skin, KLF5 is highly expressed in the developing and adult epithelia (Conkright et al. 1999; Ohnishi et al. 2000), whose functions and targets therein remain unclear.
Here we found that unlike other KLF family members, KLF5 expression is highly enriched in the epidermis, where it serves as a major epidermal lineage factor. Deletion of KLF5 specifically in the epidermis leads to profound skin defects resulting in impaired survival, accompanied by disrupted lipid envelop ultrastructures and selective reduction of high-complexity sphingolipids. Mechanistically, KLF5 binds to and transcriptionally regulates genes encoding key enzymes that catalyze the biosynthesis, processing, and secretion of epidermal sphingolipids, a preponderance of which are mutated in human diseases. Of significance, skin deficiencies caused by the lack of KLF5 can be rescued by transient lipid-rich dietary interventions, suggesting KLF5's dominant role is to control epidermal lipids. Finally, we showed that KLF5 expression is widely suppressed in human cutaneous disorders manifesting epidermal secretory defects and that KLF5's targets in sphingolipid metabolism are conserved in human skin. Altogether, our work establishes KLF5 as an epidermal transcription factor that governs sphingolipid metabolism and barrier function in the skin. KLF5 likely represents a critical transcription factor relevant to the secretory lineage functions and is involved in human cutaneous diseases.
Results
KLF5 serves as a master epidermal transcription factor
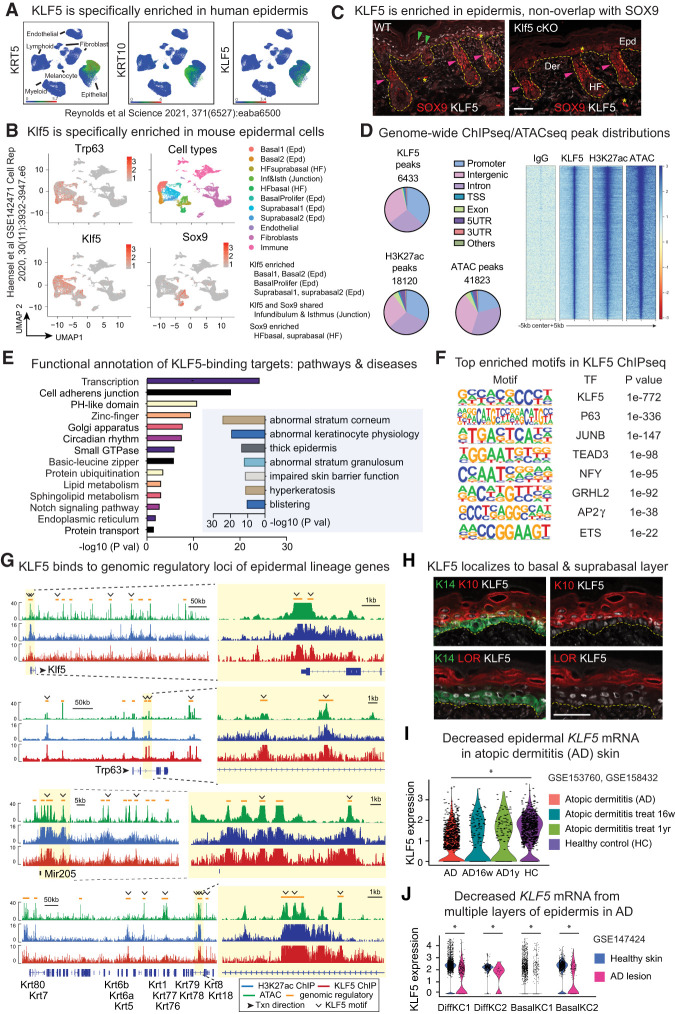
KLF5 is broadly expressed in many cell types (Nagai et al. 2005; McConnell and Yang 2010) and has not been specifically associated with a particular lineage. In the skin, we noticed KLF5 is enriched in the epidermis in both human (Fig. 1A; Reynolds et al. 2021) and mouse (Fig. 1B; Ge et al. 2017; Haensel et al. 2020) samples. In particular, KLF5 specifically labels epidermal basal and suprabasal cells and is excluded from hair follicle stem cells marked by SOX9 (Fig. 1B,C), consistent with previous observations (Ge et al. 2017). We thus set out to examine the possibility that KLF5 serves as an epidermal lineage transcription factor (TF) in the skin.
Figure 1.
KLF5 serves as a lineage-specific epidermal transcription factor. (A) Publicly available healthy human skin scRNA-seq data plotted using UMAP (uniform manifold approximation and projection) depicting the specific expression of KLF5 transcripts in KRT+ or KRT10+ epidermal cells. (B) UMAP of publicly available murine skin scRNA-seq data depicting expression of Klf5 is enriched in Trp63+ epithelial cells, and more specifically epidermal (Epd) populations, while being depleted in hair follicle (HF) cells marked by Sox9. Both Klf5 and Sox9 are expressed in the junction zone (infundibulum and isthmus). (C) Immunofluorescence (IF) shows that KLF5 is specifically expressed in the epidermis (green arrows), nonoverlapping with SOX9, which labels hair follicle stem cells (pink arrows). Klf5 cKO (K14Cre-driven epidermal conditional knockout of Klf5, described later) demonstrates KLF5 antibody specificity. Yellow dashed lines mark the epidermal–dermal border. The asterisk denotes occasional unspecific binding of SOX9 antibody in the epidermal and dermal regions. (Epd) Epidermis, (HF) hair follicle, (Der) dermis. Scale bar, 50 µm. Images are representative and from at least five biologically independent replicates. (D) KLF5 ChIP-seq, H3K27 acetylation ChIP-seq, and ATAC-seq were performed on FACS-purified E17.5 embryonic epidermal cells. Pie charts show genome-wide distributions of KLF5 ChIP-seq, H3K27ac ChIP-seq, and ATAC-seq peaks and the total numbers of peaks called in each case. The majority of peaks are found in the intergenic and intronic regions aside from promoters, corresponding to genomic regulatory elements. (TSS) Transcription start site, (UTR) untranslated region. Heat map of IgG ChIP-seq, KLF5 ChIP-seq, H3K27ac ChIP-seq, and ATAC-seq peaks centered at summit, showing KLF5 is enriched in open chromatin regions (marked by ATAC-seq) and transcriptionally active regions (marked by H3K27ac ChIP-seq). (E) DAVID (left) and GREAT (right) pathway analysis of genes associated with KLF5 ChIP-seq peaks, showing KLF5's close association with epidermal development and skin diseases. Note that GREAT assigns peaks to closest genes and hence is suboptimal in capturing distal enhancer elements. (F) HOMER motif analysis detected the top enriched motifs in KLF5 ChIP-seq peaks, suggesting autoregulatory regulation between KLF5 and other master epidermal TFs, including P63, AP-1 (JUNB), TEAD3, GRHL2, AP2g, ETS, and NFY. NFY has been associated with the pathogenesis of cutaneous squamous carcinomas (Chitsazzadeh et al. 2016). (G) ATAC-seq (green), H3K27ac ChIP-seq (blue), and KLF5 ChIP-seq (red) tracks showing KLF5-associated putative regulatory elements (denoted as orange lines above the tracks) of selected epidermal genes including Klf5 itself, transcription factor Trp63, microRNA miR-205, and keratin cluster. Yellow-shaded zoomed-in regions highlight peaks that contain the KLF5 motifs, represented as open arrowheads. Gene annotation tracks (major isoform represented) are shown below, with taller boxes indicating coding sequence exons, shorter boxes indicating untranslated region exons, and lines indicating introns. Arrows next to gene names indicate direction of transcription (txn), shown for individual gene tracks. (H) IF images show KLF5 is specifically expressed in the basal (overlaps with KERATIN 14 [K14]) and suprabasal (overlaps with KERATIN 10 [K10]) but not granular (nonoverlap with LORICRIN [LOR]) layers. Yellow dashed lines mark the epidermal–dermal border. Scale bar, 50 µm. Images are representative and from at least five biologically independent replicates. (I) scRNA-seq for atopic dermatitis patient skin treated with IL-4Rα blocker dupilumab for 16 wk or 1 yr and healthy control showing KLF5 levels are declined in barrier-deficient skin samples. (J) scRNA-seq for skin samples from atopic dermatitis (AD) patients and healthy donor showing KLF5 levels are declined in AD lesions in both differentiated (Diff KC1/2) and basal (basal KC1/2) keratinocytes. (I,J) Unpaired t-test. (*) P < 0.05.
To assess the genome-wide targets of KLF5, we performed chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) of in vivo epidermal cells purified by fluorescence-activated cell sorting (FACS). In addition, we performed H3K27ac ChIP-seq and assay for transposase-accessible chromatin with sequencing (ATAC-seq) to gauge for transcriptionally active and accessible chromatin regions, respectively, with IgG ChIP-seq as negative control. On a genome-wide level, KLF5 ChIP-seq peaks mirror those of ATAC-seq and H3K27ac ChIP-seq (Fig. 1D), indicating that KLF5 regulates transcription at these genomic regulatory regions. Interestingly, pathway analysis via Database for Annotation, Visualization, and Integrated Discovery (DAVID) and genomic region enrichment of annotations tool (GREAT) pointed to squamous development and epidermal diseases associated with KLF5 (Fig. 1E), suggesting its link to cutaneous pathology (discussed later).
Notably, our chromatin profiling results revealed an intimate relationship between KLF5 and several well-established epidermal lineage TFs, evidenced as follows: While the KLF5 motif is the top scored one in KLF5 ChIP-seq peaks as expected, motifs of P63, AP-1, TEAD, GRHL, AP2g, and ETS, known as important epidermal TFs (Guan et al. 2021), also emerged as highly enriched (Fig. 1F). KLF5 directly binds to the genomic regulatory regions of these core TFs, including itself (Fig. 1G; Supplemental Fig. S1A). This group of TFs likely converges on the regulation of epidermal stem cell identity, reminiscent of an analogous yet distinct transcriptional network formed in hair follicle stem cells (Adam et al. 2015). KLF5 additionally binds to many well-established epidermal lineage genes involved in cell adhesions and junctions, keratins, cornified envelopes, and epithelial microRNAs (Fig. 1G; Supplemental Fig. S1A), collectively known to be targets of epidermal TFs (Guan et al. 2021). About 32% of annotated KLF5 peaks contain KLF5 motifs (Fig. 1G; Supplemental Fig. S1B; Supplemental Data S1). On the other hand, KLF5 does not bind to hair follicle stem cell genes, which are otherwise controlled by the corresponding lineage-specific TFs of hair follicle stem cells (Supplemental Fig. S1C; Adam et al. 2015).
We next examined KLF5 expression relative to epidermal differentiation markers. KLF5 colocalizes strongly with K5 basal and K10 suprabasal layers, but not with LOR- and FLG-marked granular layers (Fig. 1H; Supplemental Fig. S1D), suggesting murine KLF5 is associated with epidermal stem cells and their immediate progenies. Of significance, in human atopic dermatitis (AD) skin, epidermal KLF5 level is decreased compared with healthy control skin, and its decline correlates with disease severity (Fig. 1I), supporting KLF5's association with human skin barrier function. AD-associated KLF5 down-regulation is observed from both the basal and differentiated layers of epidermis (Fig. 1J; Supplemental Fig. S1E). These results together suggested that KLF5 serves as a major epidermal lineage TF.
Epidermal loss of KLF5 leads to skin defects in vivo
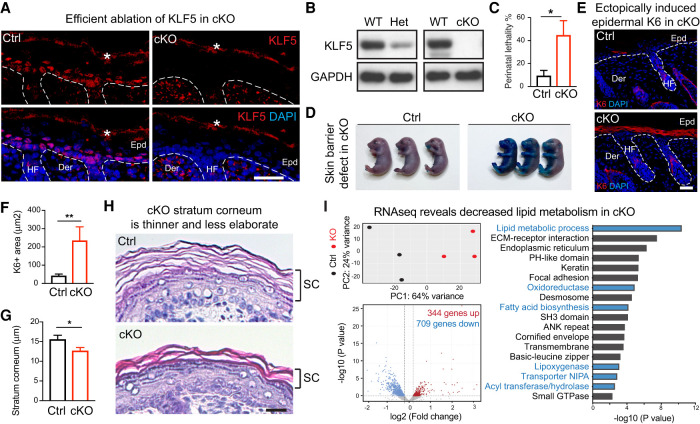
Prompted by its specificity to the epidermis and association with epidermal TFs, we next analyzed KLF5's function in vivo. To this end, we crossed mice harboring the conditional allele Klf5fl/fl (Wan et al. 2008) with those carrying K14Cre (Dassule et al. 2000), which specifically and efficiently abrogated KLF5 expression in the developing epidermis (Figs. 1C, 2A) and isolated keratinocytes (Fig. 2B). Mice with homozygous knockout of Klf5, referred to here as cKO mice, were born at a Mendelian ratio with their heterozygous and wild-type littermates. However, whereas heterozygous and wild-type neonates were indistinguishable (Supplemental Fig. S2A), cKO pups often died shortly after birth (Fig. 2C). Mice harboring heterozygous deletion of Klf5 (Het) were used as controls (Ctrls).
Figure 2.
KLF5 is essential for epidermal function in vivo. All images are representative and from at least five biologically independent replicates. White dashed lines on IF images denote the epidermal–dermal border. (Epd) Epidermis, (HF) hair follicle, (Der) dermis, (WT) wild-type mice, (Het) heterozygous loss mice, (cKO) homozygous loss mice, (Ctrl) control mice (heterozygous loss). (A) IF demonstrates that KLF5 protein is selectively lost in cKO epidermis compared with Ctrl at postnatal day 0. The asterisk denotes background signals at the stratum corneum layers. Scale bar, 50 µm. (B) Western blot analysis shows KLF5 protein is decreased in epidermal cells of heterozygous loss animals (Het) and completely lost in those of cKO compared with WT (wild type). GAPDH served as loading control. (C) Increased perinatal lethality in cKO compared with Ctrl (n = 11 for each group). (D) Toluidine Blue O dye exclusion assay shows cKO failed to exclude dye at embryonic day 18.5, while those from the Ctrl group formed an intact barrier and were able to exclude dye completely. (E,F) IF of KERATIN 6 (K6) and quantifications reveal that K6 protein is strongly induced in cKO epidermis (Ctrl: n = 23; cKO: n = 25). Note the normal expression of K6 in the hair follicle inner root sheath lineage in both Ctrl and cKO sections. Scale bar, 50 µm. (G,H) Hematoxylin and eosin stainings of postnatal day 0 skin sections show notable thinning of the stratum corneum in cKO compared with its elaborate basket weave appearance in Ctrl (Ctrl: n = 18; cKO: n = 25). Scale bar, 60 µm. Dashed line frames show the regions magnified. Scale bar, 20 µm. (C,F,G) Unpaired t-test. (*) P < 0.05, (**) P < 0.01. (I) Bulk RNA-seq was performed on Ctrl and cKO epidermis. Dispase was used to separate epidermis from dermis, dermis was discarded, and epidermis was trypsinized to obtained single-cell resuspensions, which were subjected to FACS sorting that excluded DAPI (dead) and CD45 (immune), and enriched for GFP (epithelial lineage marked by K14Cre;R26YFPfl/fl) and Integrin α6+ basal and suprabasal epithelial cells. (Top left) Principal component analysis showing three Ctrl and three KO samples. (Bottom left) Volcano plot showing differentially expressed genes (709 down-regulated and 344 up-regulated) in cKO compared with the Ctrl (heterozygous) group using P < 0.05 and absolute log2 fold change of >0.2 as the cutoff. (Right) Gene ontology analysis using Database for Annotation, Visualization, and Integrated Discovery (DAVID) reveals that both lipid metabolic process and epidermal development pathways are deregulated in cKO epidermis.
Skin defect-associated perinatal lethality is often suggestive of deficiency in the physical barrier (Nemes and Steinert 1999). Indeed, cKO skin showed immature epidermal formation, reflected by Toluidine Blue O dye penetration at embryonic day 17.5, a stage when their control littermates had already formed intact epidermis (Fig. 2D). cKO neonates also lost weight faster than controls (Supplemental Fig. S2A,B), consistent with their compromised skin function and inability to retain body fluid.
The mammalian skin function is fulfilled by a stepwise differentiation and stratification of its resident epidermal stem cells (Watt 2001; Fuchs 2008). Curiously, we did not observe significant changes in the expression of epidermal stratification genes (Supplemental Fig. S2C,D), and there were no notable differences in proliferation or apoptosis markers in the skin epidermis (Supplemental Fig. S2E–H). Nevertheless, we noted an aberrant induction in the suprabasal layer of keratin 6 (K6) expression in the cKO epidermis (Fig. 2E,F) that is known to be associated with skin barrier stress (Weiss et al. 1984; Stoler et al. 1988). Upon closer inspection of skin histology, we noticed substantially thinner stratum corneum in the cKO epidermis (Fig. 2G,H). Compared with the well-organized classic basket weave structure in controls, the stratum corneum in the cKO mice appeared compressed and less elaborate (Fig. 2H), suggesting that their skin defect could be attributable to the impaired formation of the otherwise protective outermost epidermal layers.
Meanwhile, hair follicle morphogenesis in the Klf5 cKO mice remained intact, as evidenced by the expression of several markers of hair follicle specification and differentiation at this stage (Supplemental Fig. S2I), consistent with the notion that KLF5 is down-regulated in the hair follicle stem cells (Fig. 1C; Supplemental Fig. S1C; Ge et al. 2017). These results together suggest that KLF5 is essential for epidermal function in vivo.
Epidermal sphingolipid metabolism genes are transcriptionally dependent on KLF5
To gain molecular insights into Klf5 loss-induced skin defects, we purified epidermal cells from control and cKO embryos at embryonic day 17.5 by FACS and performed RNA-seq (Supplemental Data S2). Using P < 0.05 and absolute log2 fold change of >0.2 as the cutoff, we found that 1053 genes were differentially expressed in the cKO compared with controls, and among these genes, about two-thirds were down-regulated upon Klf5 ablation (Fig. 2I). Using DAVID analysis, several pathways emerged as being dependent on KLF5; the top pathways included epidermal stratification (extracellular matrix [ECM]–receptor interaction, keratin, focal adhesion, desmosome, and cornified envelope) and the lipid metabolic process (oxidoreductase, fatty acid biosynthesis, transporter, lipoxygenase, and acyl transferase/hydrolase) (Fig. 2I). These results suggest that KLF5 is a crucial regulator of epidermal function.
A priori, defects in epidermal stratification genes such as those encoding keratins, desmosomes, cell adhesions, and ECMs are well-known causal factors leading to epidermal defects (Fuchs et al. 1994; Ihrie et al. 2005; Carulli et al. 2013; Green et al. 2020). Since many KLF5 target genes are critically involved in epidermal stratification, we next asked whether genetically ablating this group of KLF5 targets would recapitulate the KLF5 loss-of-function phenotype. To this end, we examined two candidates that appeared to be regulated by KLF5 in the skin (Supplemental Fig. S2J,K) and have well-established pathological roles: S100a9, a calcium-responsive peptide critically involved in psoriasis (Schonthaler et al. 2013; Mellor et al. 2022; Monteiro et al. 2022), and Col1a1, a type I collagen component of the extracellular matrix involved in fibrosis (Zeisberg and Kalluri 2013; Chen et al. 2022). However, deletion of either S100a9 or Col1a1 using their respective floxed alleles combined with K14Cre (Supplemental Fig. S2J,K) did not elicit any measurable changes during skin development (data not shown), disfavoring them being mediators of KLF5's function in the developing epidermis.
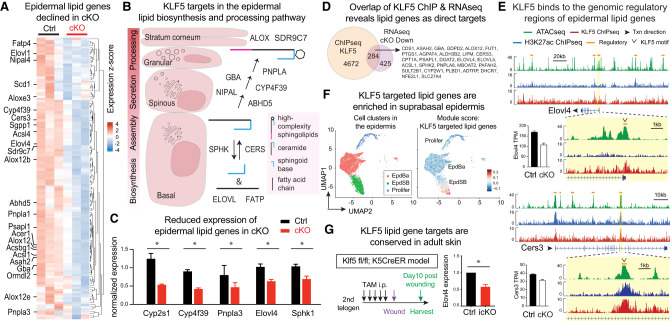
We next searched for alternative KLF5 targets that may regulate skin barrier function. As mentioned previously, several lipid metabolic processes are among the top enriched altered pathways in KLF5-deficient epidermis (Fig. 2I). Analyzing the differentially expressed genes that scored within the lipid metabolism categories (Supplemental Data S2), we noticed that many KLF5-dependent genes were specifically involved in epidermal sphingolipid biosynthesis, processing, and secretion (Fig. 3A,B). This includes the ceramide synthase Cers3, fatty acid elongase Elovl4, fatty acid transporter/acyl-CoA synthetase Fatp4, transacylase Pnpla1, esterase/lipase Abhd5, hydroxylase/oxidoreductase Cyp4f39, lipoxygenase Alox12b, and Aloxe3, among others (Fig. 3A,B). Of significance, many of these putative KLF5 targets are mutated in human skin disorders (Bouwstra and Ponec 2006; Elias et al. 2008; Traupe et al. 2014) and are known to elicit epidermal defects when deleted in mice (Radner et al. 2010; Breiden and Sandhoff 2014; Kihara 2016), phenocopying the KLF5 deficiency that we observed. Using qPCR, we validated several genes whose expression is reduced upon Klf5 ablation in vivo (Fig. 3C). These results provided strong genetic and molecular links between KLF5 and epidermal sphingolipid metabolism in the context of skin barrier formation.
Figure 3.
KLF5 transcriptionally regulates epidermal sphingolipid genes by binding to their genomic regulatory regions. (A) Heat map of differentially expressed genes within the scored lipid metabolic pathways (oxidoreductase, fatty acid biosynthesis, transporter, lipoxygenase, and acyl transferase/hydrolase) highlighting that many genes in epidermal lipid biosynthesis and processing are decreased in cKO compared with Ctrl. (B) Schematic overview of complex sphingolipid biosynthesis and processing in the epidermis. De novo ceramide synthesis occurs in the endoplasmic reticulum of the basal and suprabasal spinous epidermal layers involving ceramide synthase CERS. The inverse process ceramide degradation involves sphingosine kinase SPHK. Preparation of fatty acid substrates involves elongase ELOVL and transporter FATP, among others. Specific to the stratifying epidermis, ceramides are further modified by GBA, NIPAL, PNPLA, AYP4F39, and ABHD5; packaged into lamellar bodies in the granular layer to form high-complexity sphingolipids, including acylceramides; and then secreted into the stratum corneum for postsecretion processing (ALOX and SDR9C7) and ultimately incorporation into the corneocyte lipid envelope and intercellular lipid lamellae. Many more enzymes are involved in the multistep process, and only a few examples are shown here. (C) qPCR validation of epidermal lipid biogenesis and processing genes down-regulated in cKO epidermis compared with Ctrl. n = 3 in each group. Paired t-tests were performed. Data are mean ± SEM. (*) P < 0.05. (D) Venn diagram overlapping KLF5 ChIP-seq peaks (KLF5-bound genes) with Klf5 cKO down-regulated genes as potential KLF5 targets. A list of lipid metabolism genes within the overlap are shown. (E) ATAC-seq (green), H3K27ac ChIP-seq (blue), and KLF5 ChIP-seq (red) tracks showing KLF5-associated putative regulatory elements (denoted as orange lines above the tracks) of selected complex sphingolipid and acylceramide biosynthesis and processing. Bars next to the tracks are RNA-seq transcripts per million reads (TPM) values from Ctrl (black bars) and cKO (white bars) epidermis. (F) scRNA-seq analysis of epidermal populations using the AddModuleScore function of Seurat to denote the high expression of KLF5 targeted sphingolipid metabolism genes (gene list from Fig. 3A; Supplemental Data S2) in suprabasal epidermis. (G) Klf5fl/fl; K5CreER (icKO) and Klf5fl/+; K5CreER (Ctrl) 2-mo-old adult mice were subjected to daily i.p. tamoxifen injection (7.2 uL of 2% tamoxifen per gram of body weight) for 5 d, followed by a 5-mm punch wound (with splint to avoid dermal contraction). Wounded skin regions were harvested 10 d after wounding, and total RNA was extracted for qPCR analysis. N = 3 technical replicates. Paired t-tests were performed. (*) P < 0.05.
Meanwhile, we noted previously that sphingolipid metabolism was scored as a significantly enriched pathway associated with KLF5 binding based on its ChIP-seq (Fig. 1D), suggesting that KLF5 may directly control the expression of epidermal sphingolipid enzymes by binding to their genomic regulatory regions. Indeed, intersecting KLF5 ChIP peaks with differentially expressed genes in Klf5 cKO versus control revealed substantial overlap that 40% of down-regulated genes are bound by KLF5 (Fig. 3D; Supplemental Fig. S3A), suggesting these genes are direct targets of KLF5. For example, two epidermal lipid-processing genes, Cers3 and Elovl4, are bound by KLF5 at their promoters and enhancers, aligning with transcriptionally active and accessible chromatin, while being down-regulated in the Klf5 cKO skin (Fig. 3E; Supplemental Fig. S3B). Further supporting this notion, scRNA-seq analysis of the skin epidermis (Ge et al. 2020) showed notable coexpression of Klf5 and its target lipid genes in the suprabasal epidermis (Fig. 3F; Supplemental Fig. S3C). Moreover, when KLF5 was inducibly ablated in the adult skin using Klf5fl/fl crossed to K5CreER (Van Keymeulen et al. 2011), we found one of its target genes, Elovl4, also decreased (Fig. 3G). These results together suggest that KLF5 directly targets the transcription of multiple sphingolipid metabolism enzymes to govern skin barrier function.
Ultrastructural analysis reveals lipid secretory defects in KLF5-deficient skin
Serving as a water-impermeable seal, the skin stratum corneum is organized into a “brick and mortar” structure composed of flattened dead corneocytes filled with heavily cross-linked keratin filaments (brick) and intercorneocyte spaces sealed with specialized lipids (mortar) (Nemes and Steinert 1999). Our transcriptional observations so far have raised a distinct possibility that KLF5 controls proper epidermal lipid processing and incorporation into the stratum corneum, a major secretory function of the skin. We therefore sought to examine the consequences of epidermal Klf5 deletion at the ultrastructural level via transmission electron microscopy.
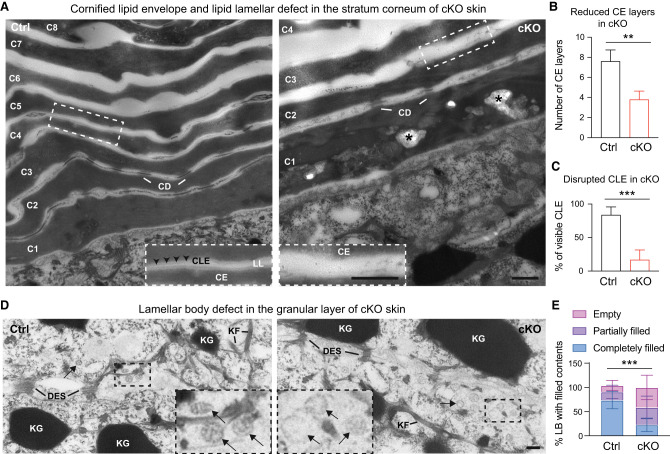
Consistent with immunofluorescent and histological analysis (Fig. 2), the overall organization of the four major layers of the skin epidermis appeared comparable between cKO and Ctrl skin (Supplemental Fig. S4A). Several classic epidermal junctional structures, including hemidesmosomes and desmosomes, also remained intact in the cKO skin (Supplemental Fig. S4B,C), suggesting that KLF5's impact on epidermis is unlikely to be mediated by junctional and adhesion proteins. In contrast, striking differences were noted in the envelope layers. In the control group, the stratum corneum consisted of multiple layers of electron-dense cornified envelope (CE) composed of flattened corneocytes (Fig. 4A, left, C1–C8), wrapped by cornified lipid envelope (CLE) appearing as a lucent band on the external surface of corneocytes, neatly spaced with intercellular lipid lamellae (LL) (Fig. 4A, left, inset). In contrast, in the cKO skin, the CE exhibited significantly fewer and disorganized layers (Fig. 4A, right, C1–C4), and CLE was no longer discernible (Fig. 4A, right, inset), suggesting that loss of KLF5 led to disrupted CLE and intercellular LL in the cornified layer (Fig. 4B,C). Curiously, the CE in the cKO skin not only exhibited fewer layers (Fig. 4B,C), but was also frequently disrupted by unusual lipid aggregates (Fig. 4A, right, asterisk; Supplemental Fig. S4D,E). Case studies in human patients with specific cutaneous lipid secretion defects have documented analogously unusual lipid clusters and undegraded organelles in their stratum corneum due to mutations in lipid-processing enzymes (Arnold et al. 1988; Niemi et al. 1993). This observation is remarkably recapitulated in our genetic model of Klf5 cKO resulting in similarly disrupted lipid structures and secretory deficits in the epidermis, indicating a close association of KLF5 with human skin disease.
Figure 4.
Ultrastructural analysis reveals KLF5 is required for cornified envelope formation and lipid secretory functions. (A) TEM at 25,000× magnification shows fewer and largely disorganized cornified envelope (CE) layers from cKO epidermis (right, C1–C4 cornified layers) compared with Ctrl (left, C1–C8 cornified layers). Dashed line insets show magnified regions from Ctrl and cKO samples and demonstrate in the Ctrl sample well-organized electron-dense CE, attached by a lucent band of cornified lipid envelope (CLE), and further sandwiched by translucent intercellular lipid lamellae (LL). CLE and LL are significantly disrupted in cKO. Asterisks denote abnormal lipid aggregates found in the cKO stratum corneum, reminiscent of case reports from skin congenital disorder patients with barrier defects. (CD) Corneodesmosome, (Der) dermis. Scale bar, 500 nm. (B,C) CE (B) and CLE (C) are quantified. (D) TEM at 25,000× magnification shows the granular layer harboring keratohyalin granules (KG), keratin filaments (KF), and desmosomes (DES). Lamellar bodies (black arrows) show lipid stacks in the Ctrl group but appear largely vacant from cKO. Scale bar, 100 nm. (E) Lamellar bodies are quantified. Unpaired t-tests were performed for B, C, and E. Data are mean ± SEM. (**) P < 0.01, (***) P < 0.001.
Next, we analyzed lamellar bodies, which are lipid granules that reside in the secretory granular layer and contain specialized epidermal lipids serving as the secretory precursors of CLE and intercellular LL (Matoltsy and Parakkal 1965; Squier 1973; Elias and Friend 1975; Lavker 1976). As expected, compared with the control group (Fig. 4D, left, inset), lamellar bodies are incompletely filled and often hollow in the cKO skin (Fig. 4D [right, inset], E). Thus, ultrastructural analysis supported our hypothesis that the loss of KLF5 elicits skin defects due to compromised epidermal lipid processing and secretion. This leads to underfilled lamellar bodies, aberrant CLE, and disrupted intercellular LL, all of which underscore secretory dysfunction in the cKO epidermis.
KLF5 specifically targets high-complexity sphingolipid species in the epidermis
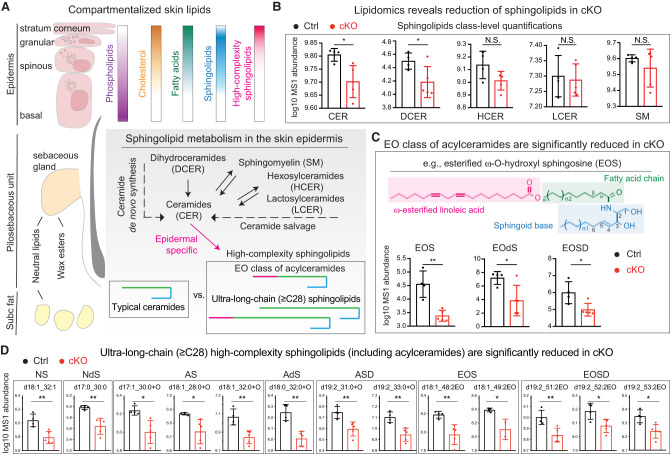
Multiple lipid species are abundantly distributed throughout the mammalian skin in a spatially organized manner, serving diverse functions (Fig. 5A; Long 1970; Gray and Yardley 1975). To further pinpoint the lipid metabolites targeted by KLF5, we performed lipidomics analysis using the neonatal Ctrl and cKO skin. Among >2200 lipid species detected (Supplemental Data S3; Supplemental Fig. S5A), class-level analysis revealed a notable and selective decline in the ceramide (CER) and dihydroceramide (DCER) classes along the ceramide de novo synthesis pathway, but not much in the hexosylceramides (HCER), lactosylceramides (LCER), or sphingomyelins (SM) of the salvage pathway (Fig. 5A,B). On the other hand, no significant alterations were observed in neutral lipids and few changes were detected in phospholipids (Supplemental Fig. S5B). This is consistent with our earlier observations that the overall epidermal organizations remained intact with no detectable changes in proliferation or apoptosis in the cKO skin (Supplemental Fig. S2).
Figure 5.
KLF5 regulates high-complexity sphingolipids and acylceramides in the epidermis. (A) Schematic overview of lipid distribution across different compartments of skin. Phospholipids are abundant in the epidermal basal, spinous, and granular layers and become undetectable in the stratum corneum, whereas cholesterol and fatty acids accumulate continuously with differentiation. On the other hand, neutral lipids are concentrated in the sebaceous glands of the pilosebaceous units and subcutaneous (subc) adipocytes. Wax esters are secreted from the sebaceous glands. Ceramides are the main components of epidermis. CER could be derived from either de novo synthesis from DCER or the salvage pathway from SM, HCER, or LCER. Unique to the epidermis, CERs are further processed into specialized high-complexity sphingolipids, such as the esterified ω-O-hydroxy (EO) class of acylceramides and ultralong chain (≥C28) ceramides. (B–D) Lipidomics profiling was performed on skin samples of postnatal day 0 heterozygous controls (Ctrl, black bar; n = 4) or Klf5 conditional knockout (cKO, red bar; n = 5) mice. Log10 transformed mass spectrometry (MS1) abundance of detected lipid species is plotted on the Y-axis. Note that the Y-axis scales are adjusted according to lipid species abundance for easier visualization. Data are mean ± SD. (*) P < 0.05, (**) P < 0.01, (N.S.) not significant. (B) Lipid class-level analysis reveals selective reduction of CER and DCER but not the HCER, LCER, or sphingomyelin SM. (C) Within further-refined ceramide groups, the largest changes occurred in the EO ceramides. An example of esterified ω-O-hydroxy sphingosine (EOS) is shown. (D) Widespread reduction of ultralong chain (≥C28) ceramides and acylceramides in cKO compared with Ctrl from lipidomics profiling. (N) Nonhydroxy, (A) α-hydroxy (could also be β-hydroxy [B] or ω-hydroxy [O], as the exact position of the −OH group could not be differentiated based on spectrum data), (EO) ω-O-hydroxy, (S) sphingosine, (dS) dihydrosphingosine, (SD) sphingadiene. Not shown here are phytosphingosine and 6-hydroxysphingosine which were not robustly detected.
A major fate for ceramides in the skin is to further mature into epidermal-specific high-complexity sphingolipids to be secreted into the intercellular space in the protective layers (Fig. 5A). Epidermal complex sphingolipids harbor several flavors (Supplemental Fig. S5C; Holleran et al. 2006; Breiden and Sandhoff 2014; Rabionet et al. 2014; Kihara 2016). The most striking lipid loss in the cKO skin occurred in the class of esterified ω-hydroxyceramides, often referred to as acylceramides (Fig. 5C). One such example is shown: esterified ω-O-hydroxyl sphingosine (EOS). Uniquely found in plants, yeast, and mammalian skin, acylceramides are specialized sphingolipids essential to the formation of the stratum corneum (Wertz and Downing 1982; Uchida and Holleran 2008). This finding is particularly interesting in light of the transcriptional deregulation that we observed in multiple genes of Klf5 cKO epidermis, including Pnpla1, Abhd5, Fatp4, and Alox (Fig. 3), all of which encode specialized lipid enzymes responsible for acylceramide processing and secretion (Epp et al. 2007; Radner et al. 2010; Kihara 2016).
Moreover, we also noted in our lipidomics profiling a widespread and preferential reduction of ≥C28 ultralong chain ceramides in cKO skin (Fig. 5D), another unique group of the epidermal lipid secretome (Kihara 2016). In contrast, the majority of typical long chain and very long chain ceramides (<C28) remained unaffected (Supplemental Fig. S5D). The selective reduction of ≥C28 ceramides aligns well with the fact that their rate-limiting enzymes are targeted by KLF5, especially Cers3 and Elovl4 (Fig. 3), which exhibit unique activity toward these epidermal-specific ultralong chain ceramides (Breiden and Sandhoff 2014; Kihara 2016). These results together pointed toward a fundamental role of KLF5 in governing complex sphingolipid metabolism, particularly the critical steps of acylceramides and ultralong chain ceramide maturation and secretion in the skin.
Epidermal deficiency from KLF5 deletion can be mitigated by transient lipid-rich dietary interventions
Our evidence so far strongly suggest that KLF5 as an epidermal lineage TF is steered toward regulating complex sphingolipid biosynthesis, processing, and secretion. If this were true, one would predict that supplementing key lipid substrates and boosting residual enzyme activities might compensate for KLF5 loss and ameliorate skin defects in vivo.
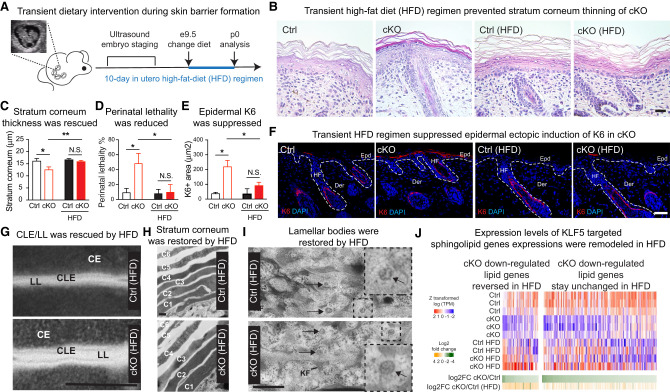
We set out to test this idea by temporarily switching the pregnant female mice to a lipid-rich diet starting at embryonic day 9.5 (E9.5), when epidermal progenitors begin to stratify. We examined the skin phenotype of their newborn pups at postnatal day 0 (P0) (Fig. 6A) and found that our dietary intervention regimen was able to significantly rescue the stratum corneum maturation in the cKO pups to a level comparable with that of Ctrl groups (Fig. 6B,C). Dietary supplement also ameliorated the perinatal lethality phenotype (Fig. 6D) and suppressed the ectopic induction of stress-associated K6 expression (Fig. 6E,F) in cKO pups. Of importance, it largely restored skin integrity at the ultrastructural level, including CLE, intercellular LL, CE, and normalized lamellar body contents (Fig. 6G–I). These results demonstrate that the major function of KLF5 in the skin is to govern sphingolipid metabolism and epidermal secretion.
Figure 6.
Epidermal deficiency from KLF5 ablation can be rescued by transient lipid-rich dietary interventions. (A) Schematics show transient dietary intervention regimen. A pregnant dam was subjected to embryonic staging via ultrasound every 2–3 d once breeding started and until E9.5 was identified, at which point the female was switched to a high-fat diet. Ten days later, pups were born and analyzed. (B,C) Hematoxylin and eosin staining and quantification of postnatal day 0 skin sections from high-fat diet (HFD) demonstrating that there were no longer significant (N.S.) differences between Ctrl and cKO mice with regard to the stratum corneum thickness (Ctrl: n = 12; cKO: n = 20). Scale bar, 60 μm. (D) There was no longer a significant difference observed in perinatal lethality of HFD-fed Ctrl and cKO mice (Ctrl: n = 15; cKO: n = 12). (E,F) IF of K6 and quantifications reveal no significant difference between HFD-fed Ctrl and cKO mice (Ctrl: n = 8; cKO: n = 6). Note the normal expression of K6 in the hair follicle (HF) companion lineage in both Ctrl and cKO sections. (Epd) Epidermis, (Der) dermis. Scale bar, 50 μm. For C–E, unpaired t-test was used. (*) P < 0.05, (**) P < 0.01. (G–I) Transmission electron microscopy (TEM) revealed restored cornified lipid envelope (CLE) and intercellular lipid lamellae (LL) bridging the cornified envelope (CE) (G), rescued cornified envelope organization (H), and largely normalized granular layer lamellar bodies (I) from HFD-fed cKO mice compared with Ctrl. Shown are uncropped images from 25,000× acquisition, and the epidermal angle observed in cKO was due to curvature during sample preparations. (CD) Corneodesmosome, (KG) keratohyalin granules, (KF) keratin filaments. Scale bars: G,H, 500 nm; I, 100 nm. (J) Heat map of RNA-seq data showing that expressions of some sphingolipid metabolism genes are reversed in dietary-rescued skin, while others remain unchanged.
We additionally tested alternative dietary enrichment regimens in which we administrated, via oral gavage, several doses of corn oil to pregnant females starting at E9.5 while maintaining them under a normal diet (Supplemental Fig. S6A). Corn oil is enriched with linoleic acid, which is a critical moiety of acylceramides (Wertz and Downing 1982), a major class of lipid species that we found to be significantly reduced in cKO epidermis. Notably, linoleic acid supplementation in the form of corn oil improved the survival of neonates (Supplemental Fig. S6A), suggesting that dietary linoleic acid could potentially bypass the KLF5 deficiency and normalize acylceramide synthesis activities to restore the barrier. Notably, when attempting to rescue Klf5 deficiency by overexpressing either Cers3 or Elovl4, we did not observe any phenotypic changes in our cultured primary keratinocytes derived from Klf5 cKO skin (Supplemental Fig. S6B,C), suggesting that multiple targets might be responsible for mediating KLF5 function.
To explore the molecular underpinning of diet-mediated phenotypic rescue in vivo, we subjected the cKO and rescued epidermis to RNA-seq. Interestingly, among the lipid genes that are down-regulated in cKO, we observed reversed expression for a subset of them, while others remained unchanged (Fig. 6J; Supplemental Data S4). We speculate that supplementing lipid substrates potentially provide crucial intermediates or enhance residual enzymatic activities to normalize skin function in vivo, while certain lipid gene expression might be responsive to dietary stimuli or modulated by compensatory mechanisms (see the Discussion).
The role of KLF5 in governing sphingolipid metabolism is conserved in human skin
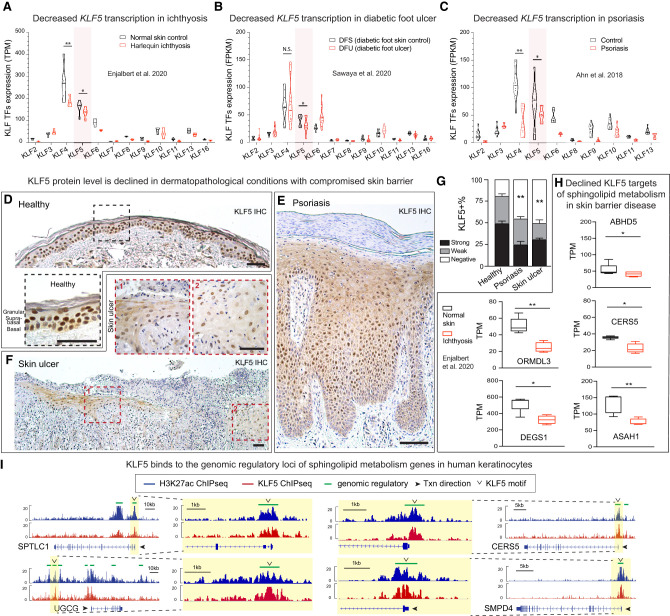
Thus far, several of our observations linked KLF5 to human skin diseases, including pathway-level analysis revealing KLF5 targets associated with epidermal barrier diseases (Fig. 1) and Klf5 cKO phenocopying human skin lipid secretion deficiencies (Fig. 4). To directly examine KLF5's involvement in human skin pathology, we mined publicly available data sets of dermatological diseases in which epidermal secretory dysfunctions have been implicated (see the Supplemental Material). We found that KLF5 is significantly suppressed in ichthyosis (Fig. 7A), a group of cornification disorders that are characterized by universal scaling in the skin (Traupe et al. 2014). KLF5 is also down-regulated in diabetic foot ulcers (Fig. 7B), manifesting nonhealing wounds whose epidermal barrier could not form (Brem and Tomic-Canic 2007). Additionally, KLF5 is reduced in cutaneous inflammatory conditions including psoriasis (Fig. 7C), a disease with a strong component in epidermal barrier pathology and lipid dysregulation (Imokawa et al. 1991; Motta et al. 1994; Bouwstra and Ponec 2006). Interestingly, the decrease of KLF5 in several skin pathologies mirrors that of KLF4, a well-established family member that also governs the skin barrier function (Segre et al. 1999; Szigety et al. 2020). Nevertheless, what distinguishes KLF5 from KLF4, or any other KLF family members, is that KLF5 is uniquely enriched in the epidermis (Supplemental Fig. S7A), consistent with the notion that KLF5 serves as a lineage-specific TF in the skin.
Figure 7.
KLF5 is suppressed in human skin diseases manifesting secretory defects. (A–C) Publicly available data sets were analyzed to show the expression of Krüppel-like factor 5 (KLF5) and other KLF family members: bulk RNA-seq for normal skin and Harlequin ichthyosis patient samples (A), diabetic foot skin control and diabetic foot ulcer (B), and normal skin and psoriasis samples (C). Unpaired t-test was performed. (*) P < 0.05, (**) P < 0.01, (N.S.) not significant. (TPM) Transcripts per million, (FPKM) fragments per kilobase million. (D–F) IHC of KLF5 in human biopsies demonstrates that KLF5 is highly enriched in human normal skin epidermis (D) and is suppressed in psoriasis (E) and skin ulcer (F). Dashed line frames show the corresponding magnified regions. Scale bars, 50 μm. (G) Quantifications of KLF5 expression (strong, weak, or negative) based on immunohistochemistry (IHC) signals from D–F. Healthy: n = 3; psoriasis: n = 6; ulcer: n = 1. One-way ANOVA with multiple comparisons was performed. Data are mean ± SEM. (**) P < 0.01. (H) A publicly available GEO data set (GSE131903) was used to plot RNA-seq TPM data showing decreased expression in ichthyosis sample compared with normal skin. Unpaired t-test was used. (*) P < 0.05, (**) P < 0.01. (I) H3K27ac ChIP-seq (blue) and KLF5 ChIP-seq (red) tracks showing KLF5-associated putative regulatory elements (denoted as green lines above the tracks) of selected sphingolipid metabolism genes in cultured human keratinocytes. Open arrowheads denote KLF5 motifs under KLF5 peaks.
To validate these transcriptional changes, we went on to examine KLF5 protein levels in available human skin biopsies using immunohistochemistry. In healthy skin, KLF5 is highly expressed in the epidermal compartments, including the basal, suprabasal, and granular layers (Fig. 7D). Consistent with the transcriptional data, we found that nuclear KLF5 signals are significantly reduced in psoriatic lesions and ulcerous skin wounds (Fig. 7E,G). These results indicated that KLF5 is prevalently suppressed in cutaneous pathologies exhibiting epidermal defects. Consistently, KLF5 targets involved in sphingolipid metabolism are also decreased in skin diseases such as ichthyosis (Fig. 7H). Finally, we used ChIP-seq to map KLF5-binding targets in human keratinocytes, along with H3K27ac ChIP-seq, which marks transcriptionally active regions. Indeed, as in our observations in mice, KLF5 directly binds to the genomic regulatory regions of genes involved in sphingolipid metabolism in human skin (Fig. 7I; Supplemental Fig. S7B). Taken together, our results suggest that KLF5's function in sphingolipid metabolism is conserved in human skin and is potentially involved in epidermal secretion defects in human cutaneous diseases.
Discussion
KLF5 is a disease-relevant epidermal lineage factor that governs skin sphingolipid metabolism
Widely expressed in multiple cell types, KLF5 is known as a guardian of development and regeneration in many tissues (Nagai et al. 2005; McConnell and Yang 2010), but its lineage specificity, if any, has remained elusive. We found that in the skin, KLF5 marks the epidermis, and its genetic ablation resulted in skin defects with pronounced disruption of lipid envelopes, lamellar bodies, and preferential loss of high-complexity sphingolipids, suggesting its function is tailored to the epidermal lipid biosynthesis and secretory machineries. Drawing analogy to PPARs in fatty acid metabolism (Mangelsdorf et al. 1995) and SREBPs in cholesterol metabolism (Goldstein and Brown 2015), KLF5's role in sphingolipid metabolism stands out. This is further supported by the fact that transient dietary intervention is sufficient to normalize skin function in KLF5-deficient animals, a beneficial effect likely achieved by supplying critical lipid intermediates and increasing residual enzyme activities. The remodeling of lipid gene transcriptional response upon dietary interventions and evoking of other lipid metabolism transcriptional regulators may additionally contribute to the phenotypic rescue. Combined with the observations that KLF5 is widely suppressed in cutaneous diseases, our current work suggests that KLF5 might be a long-sought transcriptional orchestrator of the secretory lineage function via governing sphingolipid metabolism.
Disruption of sphingolipid metabolism underlies several common skin pathologies, including atopic dermatitis, psoriasis, and aging (Imokawa et al. 1991; Motta et al. 1994; Bouwstra and Ponec 2006). This idea is further supported by a recent study showing deregulations of sphingolipids and ceramide metabolism enzymes in atopic dermatitis and psoriasis (Reynolds et al. 2021). Interestingly enough, in Klf5 cKO mice, we noted lipid aggregates due to defective secretion, reminiscent of the diagnostic lipid remnants and undegraded organelles seen in certain ichthyosis patients harboring mutations in acylceramide-processing enzymes (Arnold et al. 1988; Niemi et al. 1993), providing further disease relevance of our models. KLF5 is widely suppressed in human skin pathologies, including ichthyosis, chronic nonhealing wounds, atopic dermatitis, and psoriasis (Figs. 1H, 7), all of which exhibit compromised epidermal secretion. We thus speculate that transient lipid intervention strategies aiming to replenish critical intermediates and enhance epidermal lipid enzyme activities could be beneficial in the context of these dermatological diseases.
Lipid metabolism is tightly associated with epidermal differentiation (Nishizuka 1984; Vietri Rudan et al. 2020). While transcriptional changes in sphingolipid metabolism could often be secondary due to overtly disrupted differentiation programs (Ting et al. 2005; Oberbeck et al. 2019; Szigety et al. 2020), this does not seem to be the case in Klf5 cKO mice, whose differentiation remained largely intact (Supplemental Fig. S2). Meanwhile, epidermal function depends on many other machineries besides lipid metabolism; for example, cell adhesions and junctions (Fuchs et al. 1994; Carulli et al. 2013; Green et al. 2020), whose disruptions are frequently observed in genetic models manifesting epidermal defects (Segre et al. 1999; Ihrie et al. 2005; Ezhkova et al. 2009; LeBoeuf et al. 2010; Sen et al. 2012; Fan et al. 2018; Oberbeck et al. 2019; Szigety et al. 2020; Cohen et al. 2021). We found that many of the epidermal genes are in fact bound by KLF5 (Fig. 1). Nevertheless, epidermal junctions such as desmosomes and hemidesmosomes remained intact in the absence of KLF5 (Supplemental Fig. S4), suggesting possible compensations from other KLF family members (Garrett-Sinha et al. 1996; Segre et al. 1999; Nakamura et al. 2004), a known phenomenon of KLF TFs (Jiang et al. 2008; Moore et al. 2009). Indeed, KLF4 is essential for skin barrier function (Segre et al. 1999; Szigety et al. 2020) and widely impacts lipid metabolism genes (Szigety et al. 2020). KLF5’s transcriptional decline parallels that of KLF4 in human diseases, raising a tantalizing possibility that KLF4 might also regulate skin secretory functions via sphingolipid metabolism, similar to what we have established here for KLF5. In this regard, it is interesting to note that murine KLF5 is enriched in basal and suprabasal cells but impacts the function of granular secretory cells and cornified envelopes nonautonomously. In contrast, KLF4 is enriched in the suprabasal and granular layers (Szigety et al. 2020). In human skin, KLF5 becomes more broadly expressed across all layers, where its function in sphingolipid metabolism is likely conserved.
Finally, additional TFs in epidermal lipid metabolism regulation (Wang et al. 2013; Li et al. 2017; Oberbeck et al. 2019) may also serve as fail-safe mechanisms and join KLF5 to ensure barrier function and organismal survival. In this regard, we noted that in contrast to embryos, dyes no longer penetrated the cKO skin at P0 stage (Supplemental Fig. S2B) despite clear barrier dysfunction detected at the microscopic level by TEM and lipidomics. This suggests that the perinatal lethality that we observed could be attributed to additional factors beyond compromised skin barrier, since K14Cre is active in many squamous epithelia besides skin. Regardless, our evidence collectively suggests that KLF5 plays an essential nonredundant role in sphingolipid metabolism regulation and barrier function, including in adult skin.
A versatile, context-dependent transcription factor, KLF5 functions in the epidermal secretory pathway in the skin
In the skin, epidermal cells in the granular layer are specialized secretory cells that package epidermal lipids into lamellar bodies whose contents are subsequently secreted into the intercorneocyte spaces of the stratum corneum, a fascinating molecular assembly critical to the epidermal function (Matoltsy and Parakkal 1965; Squier 1973; Elias and Friend 1975; Lavker 1976). KLF5 sits at the top of the hierarchy by targeting key rate-limiting enzymes such as CERS3 (Hillmer et al. 2005; Mizutani et al. 2008; Jennemann et al. 2012; Eckl et al. 2013; Radner et al. 2013) and ELOVL4 (McMahon et al. 2007; Vasireddy et al. 2007; Aldahmesh et al. 2011) that exhibit exceptional enzymatic activity toward ultralong chain ceramides (Kihara 2016). KLF5 also regulates acylceramide modification enzymes, including PNPLA1 (Grall et al. 2012; Grond et al. 2017; Hirabayashi et al. 2017; Ohno et al. 2017), ABHD5 (Lefèvre et al. 2001; Radner et al. 2010), FATP4 (Stahl et al. 1999; Herrmann et al. 2001, 2003; Moulson et al. 2003; Hall et al. 2005; Lin et al. 2019; Yamamoto et al. 2020), ALOX12B, and ALOXE3 (Yu et al. 2003; Epp et al. 2007; Krieg et al. 2013), among others, all of which incur causal mutations in human congenital diseases and cutaneous disorders (Holleran et al. 1994; Lefèvre et al. 2001, 2004; Jobard et al. 2002; Akiyama et al. 2005; Klar et al. 2009; Grall et al. 2012). The essential role of KLF5 in skin lipid synthesis and secretion evokes a strong resemblance to its involvement in the production of surfactant lipids in the developing lung (Wan et al. 2008) and goblet cell function in the conjunctiva (Kenchegowda et al. 2011) and the gut (Bell et al. 2013; Nandan et al. 2014). A seamless demonstration of nature's resourcefulness, the ability of KLF5 to regulate multiple enzymes for epidermal lipid production coupled with molecular machineries for lipid assembly and secretion ensures the highly efficient maturation of the epidermis. Thus, we propose that KLF5 is an underappreciated prototype TF coupled to the secretory lineages at large across many tissue types.
Notably, KLF5's regulation of sphingolipid metabolism in the skin is distinct from its reported functions in other aspects of lipid metabolism, such as lipid oxidation and energy expenditure in cardiac and skeletal muscles (Oishi et al. 2005, 2008) and lipid storage in adipocytes (Drosatos et al. 2016). It would be interesting to test whether other lipid metabolism regulatory functions of KLF5 are also directly associated with secretory activities in the future. Such cell type-tailored functionality further highlights the importance of understanding context dependency for tissue-specific transcription factors. A versatile lineage TF across various cell types, KLF5 targets are shaped by the physiological demand of its residing tissues. Its strong association with secretory lineage and broader impact on metabolic diseases across multiple cell types merit future investigations.
KLF5 cooperates with other lineage factors in a context-dependent manner
Transcriptional regulators such as KLF5 execute context-dependent functions at the chromatin level, often in combination with other tissue-specific TFs. In this regard, it is interesting to consider enriched motifs in the genomic vicinity of KLF5-binding regions, including many known epidermal TFs such as P63, GRHL, AP-1, AP2, TEAD, and ETS (Fig. 2; Mills et al. 1999; Yang et al. 1999, 2015; Waikel et al. 2001; Ting et al. 2005; Zenz et al. 2005; Wang et al. 2008; Zhang et al. 2011; Ge et al. 2017; Fan et al. 2018). One intriguing possibility is that KLF5 functionally interacts with these TFs during epidermal development. Nevertheless, these TFs do not appear to be differentially expressed (data not shown) between control and cKO samples, suggesting that compensation or a regulatory hierarchy exists in this epidermal TF network. This also supports the idea that sphingolipid metabolism dysregulation in Klf5 cKO is unlikely to be a secondary effect from the changes of other epidermal TFs. In addition, the GREAT assigns regulatory elements based on proximity, which is at odds with many developmental genes, which often sit in gene deserts and loop to distal enhancers many kilobases away. Therefore, alternative approaches such as chromatin capture-based assays are warranted for better capturing of the regulatory network by master transcription factors like KLF5.
Outside the epidermis, KLF5 reappears in the inner root sheath of hair follicles (Sur et al. 2002), suggesting it may also play a role in hair differentiation, likely in conjunction with GATA3 (Kaufman et al. 2003). Under stress, KLF5 together with SOX9 regulates stem cell lineage infidelity during wound repair and squamous cancer (Ge et al. 2017), whereas overexpression of KLF5 leads to ectodermal dysplasia (Sur et al. 2006). Moreover, context-dependent functions of KLF5, as such, are likely applicable beyond skin. For example, KLF5 is essential for early development (Lin et al. 2010) and embryonic stem cell self-renewal (Ema et al. 2008; Jiang et al. 2008), where it functionally cooperates with pluripotency TFs (Takahashi and Yamanaka 2006). KLF5 also works with MyoD to promote skeletal myogenesis and myoblast differentiation (Hayashi et al. 2016) and with RARα to stimulate angiogenesis and cardiac hypertrophy (Shindo et al. 2002).
Finally, KLF5 is heavily involved in human malignancies (Dong and Chen 2009; McConnell and Yang 2010). In the stratified epithelia, KLF5 regulates squamous cell carcinomas of the esophagus (Tetreault et al. 2013; Jiang et al. 2020) and head and neck (Zhang et al. 2018), likely cooperating with squamous drivers such as P63 and SOX2 (Guan et al. 2019). Given our previous finding that KLF5 cooperates with both SOX9 and ETS2 TFs to regulate stem cell lineage infidelity in skin wounding and squamous cancers (Ge et al. 2017; Ge and Fuchs 2018), our current work suggests that the KLF5 targeted sphingolipid metabolism and secretory pathway might be involved in stem cell function during wound repair and tumorigenesis.
Materials and methods
Key resources
A list of key reagents, constructs, and resources is in Supplemental Data S5.
Experimental model and subject details
Mice
Klf5tm1Jaw/J (stock no. 029787), Tg(KRT14-cre)1Amc/J (stock no. 004782), Krt5tm1.1(cre/ERT2)Blh/J (stock no. 029155), and B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J (stock no. 006148) were obtained from Jackson Laboratory. Col1a1fl/fl was a gift from R. Kalluri, S100a9fl/fl was a gift from S. Grivennikov. All animal studies were performed in compliance with the protocol approved by The University of Texas MD Anderson Cancer Center Institutional Animal Care and Use Committee. Mice were maintained on a mixed C57BL/6J background in a specific pathogen-free barrier facility on a standard light–dark cycle, housed in groups, and monitored for health status every other day. All mice were fed with a standard chow unless otherwise specified. Mice of both sexes were analyzed, and no sex-specific differences were observed. For timed mating experiments, embryonic ages were determined by ultrasound imaging (Fujifilm VisualSonics Vevo 3100). For high-fat diet (HFD) rescue experiments, pregnant females were fed with TestDiet 58Y1 diet containing 4.7% linoleic acid from embryonic day 9.5 until delivery day.
Human samples
Human skin ulcer tissue (SKD241) was purchased from US Biomax, Inc. Details of samples on the array are available on the Biomax website (http://old.biomax.us/tissue-arrays/Skin/SKD241). Additional human skin tissue sections were obtained from the archive at the Department of Pathology, MD Anderson Cancer Center. All human sample studies were in compliance with regulations of the Institutional Review Board and in accordance with requirements in the Human Research Protection Program.
Methods
Histology and immunohistochemistry
Skin samples were fixed in 4% paraformaldehyde (PFA) for 24 h and made into paraffin blocks. Sections were cut at 5 μm thick, deparaffinized with xylene washes twice, and then rehydrated by sequential washes with 100%, 90%, 70%, and 50% ethanol and double-distilled H2O.
Hematoxylin and eosin staining was performed according to the manufacturer's recommendations on the hematoxylin and eosin staining protocol (Newcomer Supply 12013 and 1072).
For immunochemistry, antigens were unmasked by boiling slides in 1× basic antigen retrieval buffer (R&D Systems CTS013) for 30 min. Following antigen retrieval, sections were incubated with 3% H2O2 for 15 min at room temperature and then blocked with blocking buffer (2% gelatin, 5% normal donkey serum, 1% bovine serum albumin, 0.3% Triton X-100 in phosphate-buffered saline [PBS]). The sections were stained with anti-KLF5 (rabbit; 1:500; Abcam) overnight at 4°C. The next day, samples were washed and stained with horseradish peroxidase (HRP)-conjugated donkey antirabbit IgG. Color development was performed using DAB (Thermo Fisher Scientific 34002) at room temperature until color developed (1–5 min) upon inspection under the bright-field microscope.
Immunofluorescence
Skin samples were prefixed in 4% PFA in PBS for 5 min at room temperature and washed extensively in PBS overnight. Samples were embedded in OCT compound (Thermo Fisher Scientific 4585) the next day and cryosectioned at a thickness of 10 μm. Slides were blocked with blocking buffer (2% gelatin, 5% normal donkey serum, 1% bovine serum albumin, 0.3% Triton X-100 in PBS) for 1 h at room temperature and then incubated with primary antibody overnight at 4°C. The following day, slides were washed in 0.3% Triton X-100 in PBS and incubated with the corresponding secondary antibodies and 0.2 μg/mL DAPI. The sections were washed in 0.3% Triton X-100 in PBS and mounted. Imaging was performed with either a Leica DM1000 LED microscope or a Zeiss LSM 800 microscope.
The following primary antibodies and dilutions were used: KLF5 (goat; 1:100; R&D Systems), K6 (rabbit; 1:1000; BioLegend), K10 (rabbit; 1:1000; BioLegend), LOR (rabbit; 1:1000; BioLegend), FLG (rabbit; 1:100; BioLegend), Ki67 (rabbit; 1:500; Thermo Fisher Scientific), cleaved caspase-3 (rabbit; 1:200; Cell Signaling Technology), LRIG1 (goat; 1:200; R&D Systems), SOX9 (rabbit; 1:200; Abcam), P-cadherin (goat; 1:200; R&D Systems), GATA3 (rat; 1:100; Thermo Fisher Scientific), and GFP (chicken; 1:2000; Abcam). Secondary antibodies used included Rhodamine Red-X (RRX)-conjugated donkey antirabbit IgG, RRX-conjugated donkey antirat IgG (1:300; Jackson ImmunoResearch), Alexa Fluor 647-conjugated donkey antigoat IgG, Alexa Fluor 647-conjugated donkey antirat IgG (1:500; Jackson ImmunoResearch), and Alexa Fluor 488-conjugated donkey antichicken IgG (1:500; Jackson ImmunoResearch). Images were processed using ImageJ and Adobe Photoshop 2021.
Isolation of epidermal cells
Following euthanasia, back skin was removed and floated dermis side down on 2.5 U/mL Dispase (Gibco) in PBS overnight at 4°C. Epidermis was peeled from dermis using fine forceps. Isolated epidermis was then incubated with 0.25% trypsin-EDTA (Gibco) for 10 min at 37°C, neutralized by cold PBS containing 2% calcium-chelated fetal bovine serum [FBS(−)], and filtered through a 40-μm cell strainer. Cells were spun down at 300g for 10 min at 4°C and washed once with cold PBS containing 2% FBS(–).
Fluorescence-activated cell sorting (FACS)
Freshly isolated epidermal cells from postnatal day 0 pups from either Klf5fl/fl; K14Cre cKO or Klf5fl/+; K14Cre heterozygous controls were incubated with conjugated antibodies CD49F_PE (rat; 1:250; Thermo Fisher Scientific) and CD45_APC-eFluor780 (rat; 1:1000; Thermo Fisher Scientific) along with 0.2 μg/mL DAPI for 20 min on ice, washed once, and resuspended in cold PBS containing 2% FBS(−). GFP+ was derived from the R26YFPfl/fl allele. Sorting was performed using a 70-μm nozzle on a BD FACSAria equipped with FACSDiva software (BD Biosciences), gating on DAPI−CD45−GFP+ITGA6+ to isolate basal and suprabasal epidermal cells. Postsorting analysis was performed using FlowJo.
Transmission electron microscopy
Fresh postnatal day 0 skin samples were fixed in 2% glutaraldehyde, 4% PFA, and 2 mM CaCl2 in 0.05 M sodium cacodylate buffer (pH 7.2) for 1 h at room temperature and then stored in 4°C until sample submission. Fixed samples were submitted to the High-Resolution Electron Microscopy Facility at MD Anderson Cancer Center for TEM processing. Briefly, submitted samples were postfixed with 0.2% ruthenium tetroxide at room temperature and processed for Epon embedding. Ultrathin sections (60–70 nm) were counterstained with uranyl acetate and lead citrate. Digital images were obtained using the AMT imaging system (Advanced Microscopy Techniques Corp.).
Lipidomics analysis
Fresh postnatal day 0 skin tissues were dissected, snap-frozen in liquid nitrogen, and stored at −80°C until sample submission. Samples were submitted on dry ice to the Metabolomics Core at MD Anderson Cancer Center for lipidomics profiling.
For lipid extraction, murine skin tissue snap-frozen in liquid nitrogen was powderized using an in-house-built tissue smasher. Samples were subsequently extracted using ice-cold isopropanol containing 1% (v/v) 10 mM butylated hydroxytoluene and 2% (v/v) Avanti Splash Lipidomix mass spectrometry standard (330707), both in methanol. A solvent/tissue ratio of 10 µL/mg was used. Samples were homogenized using a liquid nitrogen-cooled Precellys Evolution bead mill homogenizer, vortexed for 10 min at room temperature, and then centrifuged at 17,000g for 20 min at 4°C. Supernatants were transferred to glass autosampler vials for immediate analysis.
For reverse-phase liquid chromatography, the injection volume was 10 µL. Mobile phase A was 60:40 acetonitrile:water containing 0.1% formic acid and 10 mM ammonium acetate. Mobile phase B (MPB) was 90:9:1 isopropanol:acetonitrile:water containing 0.1% formic acid and 10 mM ammonium acetate. The chromatographic method included a Thermo Fisher Scientific Accucore C30 column (2.6 µm, 150 × 2.1 mm) maintained at 40°C, autosampler tray chilling at 15°C, a mobile phase flow rate of 0.200 mL/min, and a gradient elution program as follows: 0–3 min, 30% MPB; 3–13 min, 30%–43% MPB; 13–13.1 min, 43%–50% MPB; 13.1–33 min, 50%–70% MPB; 33–48 min, 70%–99% MPB; 48–55 min, 99% MPB; 55–55.1 min, 99%–30% MPB; and 55.1–60 min, 30% MPB.
For mass spectrometry, a Thermo Fisher Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer with heated electrospray ionization source was operated in data-dependent acquisition mode, in both positive and negative ionization modes, with scan ranges of 150–677 m/z and 675–1500 m/z. Orbitrap resolutions of 120,000 and 30,000 (FWHM) were used for MS1 and MS2–3 acquisitions, respectively, and spray voltages of 3600 V and −2900 V were used for positive and negative ionization modes, respectively. Vaporizer and ion transfer tube temperatures were set at 275°C and 300°C, respectively. The sheath, auxiliary, and sweep gas pressures were 35, 7, and 0 (arbitrary units), respectively. For MS2 and MS3 fragmentation, a hybridized HCD/CID approach was used. Each sample was analyzed using four 10-µL injections, making use of the two aforementioned scan ranges, in both ionization modes. Data were analyzed using Thermo Fisher Scientific LipidSearch software (version 4.2.27) and R scripts written in house.
Barrier function assay
The Toluidine Blue O dye exclusion assay was carried out as previously described (Laurin et al. 2019). In brief, the pregnant female was euthanized at embryonic day (E) 18.5, and fresh E18.5 embryos were isolated; passed through chilled methanol at gradients of 25%, 50%, 75%, and 100%; and immersed in 0.1% Toluidine Blue O solution (Sigma-Aldrich T3260) for 1 min on ice. Embryos were then destained in PBS to reveal dye penetration or exclusion outcomes. Similar procedures were performed on P0 newborns.
Western blot
Cultured keratinocytes from postnatal day 0 pups (Klf5fl/fl or Klf5fl/+) were infected in culture with lentiviral Cre by spinning at 1100g for 30 min at 30°C. Cells were lysed in RIPA buffer (Millipore 20-188) supplemented with protease inhibitor (Roche 04693159001). After centrifugation at 15,000g for 10 min at 4°C, the protein content from the supernatant was quantified using Bradford assay (Bio-Rad 5000205). Equal amounts of protein were loaded and separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, blotted onto polyvinylidene fluoride membranes, and incubated overnight with primary antibodies directed against KLF5 (goat; 1:500; R&D Systems) and GAPDH (rabbit; 1:2000; Cell Signaling Technology). Detection of HRP-conjugated secondary antibodies was performed with Clarity Western ECL substrate (Bio-Rad 1705060), and images were developed on X-ray films. Quantification of Western blot band intensities was performed by densitometry using ImageJ software.
ChIP-seq
ChIP-seq libraries were made from freshly FACS-sorted epidermal cells as described above. Library preparations and analyses were performed as reported previously (Schmidl et al. 2015).
Briefly, 10 million FACS-sorted cells were cross-linked by addition of fresh PFA solution at 1% for 10 min at room temperature, quenched with fresh glycine at 0.125 M for 5 min, and rinsed twice with phospho-buffered saline (PBS) prior to snap-freezing in liquid nitrogen and storing at –80°C. Before ChIP, cells were resuspended, lysed, and sonicated in 0.25% SDS on a Bioruptor Sonicator (Diagenode UCD-200) to solubilize and shear cross-linked DNAs. For sonication, a 10-cycle regimen of a 30-sec sonication followed by a 30-sec rest was used. The resulting whole-cell extract was incubated overnight at 4°C with primary antibody (2.5 μg of antibody per 1 mg of total protein: anti-KLF5 [Abcam ab137676] and anti-H3K27ac [Abcam ab4729]). Ten microliters of Dynal Protein G magnetic beads (Invitrogen 10017D) was added to each sample the following day and incubated for 2–4 h at 4°C, followed by washes with low-salt, high-salt, LiCl, and Tris-EDTA buffer by passing through a magnetic stand six times each. Bound complexes were treated with 5 µL of Tn5 transposase (Illumina 20034198) for 5 min at 37°C and eluted, and cross-linking was reversed by 6–10 h of incubation at 65°C, cleaned up with MinElute PCR purification kit (Qiagen 28004), and PCR-amplified for 10–12 cycles. Libraries were purified by Beckman Coulter AMPureXP beads (Fisher NC9959336) double-sided selection and submitted to Novogene for library quality control (QC). Libraries that passed QC were then pooled and sequenced using PE150 on a HiSeq4000 lane with ∼25 million to 30 million reads output per sample, trimmed to obtain SE50 reads, and demultiplexed.
ChIP-seq alignment, peak calling, and analysis
Raw sequencing 50-bp single-end reads were aligned to the Mus musculus reference genome (version mm10 from Genome Reference Consortium GRCm38) or the Homo sapiens reference genome (version hg19 from Genome Reference Consortium GRCh37) for mouse and human samples, respectively, using the Bowtie package (https://sourceforge.net/projects/bowtie-bio/files/bowtie/1.0.1) with parameters -n 1 -m 1 –best –strata. Unique reads mapped to a single genomic location (allowing default mismatches) were kept for peak identification using the model-based analysis of ChIP-seq 2 (MACS2) (Zhang et al. 2008) pipeline with the parameter –nomodel. Peak BAM files were converted to TDF files and visualized using the Integrative Genomics Viewer (IGV; http://software.broadinstitute.org/software/igv).
Peak BED files were subjected to peak annotation and motif analysis using the HOMER v4.11 annotatePeaks.pl and findMotifs.pl packages (http://homer.ucsd.edu/homer/motif) with default settings. The gene list obtained from peak annotation was then used to perform pathway analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov) and genomic regions enrichment of annotations tool (GREAT) v4.0.4 (http://great.stanford.edu/public/html) with default parameters. Peak BAM files were converted to TDF files and visualized using the Integrative Genomics Viewer (IGV; http://software.broadinstitute.org/software/igv).
ATAC-seq
ATAC-seq libraries were made from freshly FACS-sorted epidermal cells as described above. Library preparations and analyses were performed as reported previously (Buenrostro et al. 2013).
Briefly, 100,000 freshly FACS-sorted cells were subjected to tagmentation reactions with 10 µL of Tn5 transposase (Illumina 20034198), cleaned up with MinElute PCR purification kit (Qiagen 28004), and PCR-amplified for eight to 12 cycles. Libraries were purified by Beckman Coulter AMPureXP beads (Fisher NC9959336) and run on a D1000 High-Sensitivity Tape Station (Agilent 5067-5584) for quality control (QC) prior to sequencing. Libraries that passed QC were pooled and submitted to Novogene, sequenced using paired-end PE150 on a HiSeq4000 lane with ∼50 million reads output per sample, trimmed to obtain SE50 reads, and demultiplexed.
ATAC-seq alignment, peak calling, and analysis
Raw sequencing 50-bp single-end reads were aligned to the genomic builder for Mus musculus (version mm10 from Genome Reference Consortium GRCm38) as previously described (Buenrostro et al. 2013). Briefly, mapping was performed using the Bowtie package (https://sourceforge.net/projects/bowtie-bio/files/bowtie2/2.2.3). Unique reads mapped to a single genomic location (allowing default mismatches) were kept for peak identification using the model-based analysis of ChIP-seq 2 (MACS2) (Zhang et al. 2008) pipeline with the parameter –nomodel. Peak BAM files were converted to TDF files and visualized using the Integrative Genomics Viewer (IGV; http://software.broadinstitute.org/software/igv).
ChIP-seq and ATAC-seq heat map analysis
The peak intensity heat map was generated by extracting the ChIP-seq/ATAC-seq intensities within the 5-kb upstream and 5-kb downstream window of the peak summit. Each row in the heat map represents one ATAC-seq/ChIP-seq peak. The heat map was generated using the Deeptools package using KLF5 ChIP-seq peaks as reference.
RNA-seq and quantitative PCR
FACS-sorted epidermal cells were isolated as described above and lysed with TRI reagent LS (Sigma T3934). Total RNA was isolated with the Direct-zol RNA miniprep kit (Zymo Research R2051).
For RNA-seq, RNAs were submitted to Novogene for quality control and polyA-enriched eukaryotic mRNA-seq library preparation. Libraries were sequenced using PE150 on a NovaSeq lane with ∼20 million reads output per sample.
For quantitative PCR (qPCR), DNA oligo primers were synthesized from Sigma. One microgram of total RNAs was used to generate complementary DNAs (cDNAs) using the SuperScript VILO cDNA synthesis kit (Invitrogen 11754050). cDNA was diluted and used as templates for real-time PCR using Applied Biosystems PowerUp SYBR Green master mix (Thermo Fisher A25743). qPCR was performed on the QuantStudio 7 Flex equipped with QuantStudio real-time PCR software v1.3 (Applied Biosystems and Thermo Fisher) or 7500 real-time PCR system equipped with 7500 software v2.3 (Applied Biosystems and Thermo Fisher). Data were normalized to GAPDH expression and are reported as ΔΔCt values compared with control sample. The qPCR primer sequences used were Cyp2s1 (fw: CTGAGGAAATTCACCCTGCTC; rv: CAAGGGAACAGACGACATTAGAG), Cyp4f39 (fw: TGCACCCAAGGATGAGTTTTT; rv: CAGATGTCGCCGCCATTTAG), Pnpla3 (fw: TCACCTTCGTGTGCAGTCTC; rv: CCTGGAGCCCGTCTCTGAT), Elovl4 (fw: AAGCACGCTCTATCTCCTGTT; rv: CTGCGTTGTATGATCCCATGAA), and Sphk1 (fw: GCTTCTGTGAACCACTATGCTGG; rv: ACTGAGCACAGAATAGAGCCGC).
RNA-seq alignment and differential expression analysis
Raw sequencing 150-bp paired-end reads were aligned to the Mus musculus reference genome (version mm10 from Genome Reference Consortium GRCm38) using a STAR v2.6.0 aligner. Aligned reads were quantified against the reference annotation GRCm38 to obtain FPKM (fragments per million reads) and raw counts using HTSeq (python/3.6/anaconda3/bin/htseq-count with options -m union -a 10 -s no), respectively. TPM (transcripts per million reads) was obtained by using reference annotation mm10.refgenes and running rsem-calculate-expression (1.2.3) with options --bowtie2 (2.2.3).
Differential expression analysis was performed on normalized raw counts using the edgeR package in R (http://bioconductor.org/packages/release/bioc/html/edgeR.html). Genes with absolute log2 fold change of >0.2 and adjusted P-value of <0.05 were considered to be significantly differentially expressed and were plotted on volcano plot using R. A differentially expressed gene list was then used to perform pathway analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov) using default parameters. Differentially expressed genes within selected pathways were used for hierarchical clustering and plotted on heat maps using R.
Statistics analysis and reproducibility
Statistical analyses were performed using Graphpad Prism 8.0 software. All experiments shown were repeated at least three times, and representative data are shown. Unpaired Student's t-test was used to compare two groups, and one-way analysis of variance (ANOVA) test was used for multiple comparisons of more than two groups. P-value of <0.05 was considered statistically significant and is reported in the figures and figure legends. Randomization and experimenter blinding were not performed given the lack of ambiguity in the phenotypes observed.
Data availability
Data generated in this study
Raw and analyzed data reported here are available via NCBI GEO under the accession number GSE168600 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE168600).
Mining of published GEO and related data sets
The following GEO data sets were downloaded: GSE131903, GSE117405, GSE134431, GSE147424, GSE153760, GSE158432, and GSE142471. For RNA-seq GSE131903, raw FastQ files were downloaded and processed using RNA-seq alignment pipeline as described above and TPM was plotted for KLF family members expressed (TPM > 1). For RNA-seq GSE117405 and GSE134431, processed data were downloaded and FPKM was plotted for KLF family members expressed (FPKM > 1). For scRNA-seq GSE147424, GSE153760, GSE158432, and GSE142471, raw gene, barcode, and matrix files were downloaded and analyzed using Seurat package (https://satijalab.org/seurat), and the corresponding cells with KLF5 expression were plotted. For scRNA-seq from Reynolds et al. (2021), plots were generated using web portal https://developmentcellatlas.ncl.ac.uk/datasets/hca_skin_portal.
Supplementary Material
Acknowledgments
We thank Y. You for technical assistance; K. Dunner Jr. for TEM analysis; R. Kalluri and S. Grivennikov for mouse line sharing; R. DePinho, P. Deng, R. Kalluri, M.L. Kirtley, F. Giancotti, and J. Hu for sharing reagents; M. Laurin, S. Ellis, R.C. Adam, C.P. Lu, J. Hu, S. Shalapour, E. Wagner, and M.J. Gacha Garay for scientific discussions; A.K. Singh, K. Rai, and Yiwen Chen for bioinformatics discussions; S.J. Bronson (Research Medical Library) for scientific editing; E. Pettus and the team at the Department of Cancer Biology for administrative support; the Research Animal Support Facility (director V. Jensen) for care of mice in accordance with National Institutes of Health guidelines (Association for Assessment and Accreditation of Laboratory Animal Care International accredited); the Genetically Engineered Mouse Facility (director J. Parker-Thornburg) for mouse recovery and cryopreservation; the High-Resolution Electron Microscopy Facility (director R. Langley) for transmission electron microscopy analysis; the Advanced Cytometry and Sorting Facility at South Campus (director K. Clise-Dwyer) for FACS; the Advanced Technology Genomics Core (director V. Huff) for sequencing; the Advanced Microscopy Core Facility (director T. Zal) for imaging; the High-Performance Computing Center cluster for computational and bioinformatics support; and B. Czako for their work as lipidomics experiment consultant. K.M.M. and Y.C. were supported by Ergon Foundation Post-Doctoral Trainee Fellowships and startup funds to the Kalluri Laboratory. The Metabolomics Core Facility (director P.L.L.) is supported by Cancer Prevention and Research Institute of Texas (CPRIT) grant RP130397 and National Institutes of Health (NIH) grants S10OD012304-01 and P30CA016672. S.V.K. was supported by a University of Texas Health Innovation for Cancer Prevention Research Training Program Predoctoral Fellowship (CPRIT grant RP160015). Y. Ge is a CPRIT Scholar of Cancer Research. This work was supported by grants from the NIH (1K01AR072132), CPRIT (FP00006955), University of Texas Rising STARs program, Cancer Center Support grant new faculty award, Andrew Sabin Family Award, and MD Anderson Cancer Center startup funding awarded to Y. Ge.
Author contributions: Y.L. , Y. Guan, and Y. Ge designed the experiments and wrote the manuscript. L.D. contributed to immunofluorescent staining and bioinformatics analysis. E.H. contributed to skin analysis and barrier assays. Y.J.Y. assisted in genotyping and immunoblot. D.Z. provided technical support in next-generation sequencing. L.J.V., S.V.K., and P.L.L. performed lipidomics analysis. Y.C., K.M.M., and S.G. shared mouse lines. K.J., V.L., K.H.K., V.M., J.D., X.S., J.Z., Y.X., J.W., and K.C. assisted in bioinformatics pipeline optimization and data analysis on RNA-seq, ChIP-seq, and ATAC-seq. P.N. provided histological samples and pathological analysis. Y. Guan performed histological analysis (Figs. 2A,E,H, 6B,F, 7D–F; Supplemental Fig. S2) and assisted with quantifications and figure preparations. Y.L. performed the rest of the experiments. All authors provided intellectual input and vetted and approved the final manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Aricle and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.349662.122.
Competing interest statement
The authors declare no competing interests.
References
- Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, Polak L, Kadaja M, Asare A, Zheng D, et al. 2015. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521: 366–370. 10.1038/nature14289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn R, Yan D, Chang H-W, Lee K, Bhattarai S, Huang Z-M, Nakamura M, Singh R, Afifi L, Taravati K, et al. 2018. RNA-seq and flow-cytometry of conventional, scalp, and palmoplantar psoriasis reveal shared and distinct molecular pathways. Sci Rep 8: 11368. 10.1038/s41598-018-29472-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama M, Sugiyama-Nakagiri Y, Sakai K, McMillan JR, Goto M, Arita K, Tsuji-Abe Y, Tabata N, Matsuoka K, Sasaki R, et al. 2005. Mutations in lipid transporter ABCA12 in harlequin ichthyosis and functional recovery by corrective gene transfer. J Clin Invest 115: 1777–1784. 10.1172/JCI24834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldahmesh MA, Mohamed JY, Alkuraya HS, Verma IC, Puri RD, Alaiya AA, Rizzo WB, Alkuraya FS. 2011. Recessive mutations in ELOVL4 cause ichthyosis, intellectual disability, and spastic quadriplegia. Am J Hum Genet 89: 745–750. 10.1016/j.ajhg.2011.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ML, Anton-Lamprecht I, Melz-Rothfuss B, Hartschuh W. 1988. Ichthyosis congenita type III. Clinical and ultrastructural characteristics and distinction within the heterogeneous ichthyosis congenita group. Arch Dermatol Res 280: 268–278. 10.1007/BF00440599 [DOI] [PubMed] [Google Scholar]
- Bell SM, Zhang L, Xu Y, Besnard V, Wert SE, Shroyer N, Whitsett JA. 2013. Kruppel-like factor 5 controls villus formation and initiation of cytodifferentiation in the embryonic intestinal epithelium. Dev Biol 375: 128–139. 10.1016/j.ydbio.2012.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwstra JA, Ponec M. 2006. The skin barrier in healthy and diseased state. Biochim Biophys Acta 1758: 2080–2095. 10.1016/j.bbamem.2006.06.021 [DOI] [PubMed] [Google Scholar]
- Breiden B, Sandhoff K. 2014. The role of sphingolipid metabolism in cutaneous permeability barrier formation. Biochim Biophys Acta 1841: 441–452. 10.1016/j.bbalip.2013.08.010 [DOI] [PubMed] [Google Scholar]
- Brem H, Tomic-Canic M. 2007. Cellular and molecular basis of wound healing in diabetes. J Clin Invest 117: 1219–1222. 10.1172/JCI32169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G. 2005. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol 6: 328–340. 10.1038/nrm1619 [DOI] [PubMed] [Google Scholar]
- Carulli S, Contin R, De Rosa L, Pellegrini G, De Luca M. 2013. The long and winding road that leads to a cure for epidermolysis bullosa. Regen Med 8: 467–481. 10.2217/rme.13.33 [DOI] [PubMed] [Google Scholar]
- Chandel NS, Jasper H, Ho TT, Passegué E. 2016. Metabolic regulation of stem cell function in tissue homeostasis and organismal ageing. Nat Cell Biol 18: 823–832. 10.1038/ncb3385 [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang S, Tavormina J, Tampe D, Zeisberg M, Wang H, Mahadevan K, Wu C-J, Sugimoto H, Chang C-C, et al. 2022. Oncogenic collagen I homotrimers from cancer cells bind to α3β1 integrin and impact tumor microbiome and immunity to promote pancreatic cancer. Cancer Cell 40: 818–834.e9. 10.1016/j.ccell.2022.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitsazzadeh V, Coarfa C, Drummond JA, Nguyen T, Joseph A, Chilukuri S, Charpiot E, Adelmann CH, Ching G, Nguyen TN, et al. 2016. Cross-species identification of genomic drivers of squamous cell carcinoma development across preneoplastic intermediates. Nat Commun 7: 12601. 10.1038/ncomms12601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen I, Bar C, Liu H, Valdes VJ, Zhao D, Galbo PM Jr, Silva JM, Koseki H, Zheng D, Ezhkova E. 2021. Polycomb complexes redundantly maintain epidermal stem cell identity during development. Genes Dev 35: 354–366. 10.1101/gad.345363.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conkright MD, Wani MA, Anderson KP, Lingrel JB. 1999. A gene encoding an intestinal-enriched member of the Kruppel-like factor family expressed in intestinal epithelial cells. Nucleic Acids Res 27: 1263–1270. 10.1093/nar/27.5.1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. 2000. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 127: 4775–4785. 10.1242/dev.127.22.4775 [DOI] [PubMed] [Google Scholar]
- Dong JT, Chen C. 2009. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell Mol Life Sci 66: 2691–2706. 10.1007/s00018-009-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosatos K, Pollak NM, Pol CJ, Ntziachristos P, Willecke F, Valenti MC, Trent CM, Hu Y, Guo S, Aifantis I, et al. 2016. Cardiac myocyte KLF5 regulates ppara expression and cardiac function. Circ Res 118: 241–253. 10.1161/CIRCRESAHA.115.306383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert RL. 1989. Structure, function, and differentiation of the keratinocyte. Physiol Rev 69: 1316–1346. 10.1152/physrev.1989.69.4.1316 [DOI] [PubMed] [Google Scholar]
- Eckl KM, Tidhar R, Thiele H, Oji V, Hausser I, Brodesser S, Preil ML, Önal-Akan A, Stock F, Müller D, et al. 2013. Impaired epidermal ceramide synthesis causes autosomal recessive congenital ichthyosis and reveals the importance of ceramide acyl chain length. J Invest Dermatol 133: 2202–2211. 10.1038/jid.2013.153 [DOI] [PubMed] [Google Scholar]
- Elias PM, Friend DS. 1975. The permeability barrier in mammalian epidermis. J Cell Biol 65: 180–191. 10.1083/jcb.65.1.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias PM, Williams ML, Holleran WM, Jiang YJ, Schmuth M. 2008. Pathogenesis of permeability barrier abnormalities in the ichthyoses: inherited disorders of lipid metabolism. J Lipid Res 49: 697–714. 10.1194/jlr.R800002-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M, Mori D, Niwa H, Hasegawa Y, Yamanaka Y, Hitoshi S, Mimura J, Kawabe Y, Hosoya T, Morita M, et al. 2008. Krüppel-like factor 5 is essential for blastocyst development and the normal self-renewal of mouse ESCs. Cell Stem Cell 3: 555–567. 10.1016/j.stem.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Enjalbert F, Dewan P, Caley MP, Jones EM, Morse MA, Kelsell DP, Enright AJ, O'Toole EA. 2020. 3D model of harlequin ichthyosis reveals inflammatory therapeutic targets. J Clin Invest 130: 4798–4810. 10.1172/JCI132987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp N, Fürstenberger G, Müller K, de Juanes S, Leitges M, Hausser I, Thieme F, Liebisch G, Schmitz G, Krieg P. 2007. 12R-lipoxygenase deficiency disrupts epidermal barrier function. J Cell Biol 177: 173–182. 10.1083/jcb.200612116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. 2009. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136: 1122–1135. 10.1016/j.cell.2008.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Wang D, Burgmaier JE, Teng Y, Romano RA, Sinha S, Yi R. 2018. Single cell and open chromatin analysis reveals molecular origin of epidermal cells of the skin. Dev Cell 47: 21–37.e5. 10.1016/j.devcel.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingold KR, Elias PM. 2014. Role of lipids in the formation and maintenance of the cutaneous permeability barrier. Biochim Biophys Acta 1841: 280–294. 10.1016/j.bbalip.2013.11.007 [DOI] [PubMed] [Google Scholar]
- Frye M, Benitah SA. 2012. Chromatin regulators in mammalian epidermis. Semin Cell Dev Biol 23: 897–905. 10.1016/j.semcdb.2012.08.009 [DOI] [PubMed] [Google Scholar]
- Fuchs E. 2008. Skin stem cells: rising to the surface. J Cell Biol 180: 273–284. 10.1083/jcb.200708185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Coulombe P, Cheng J, Chan YM, Hutton E, Syder A, Degenstein L, Yu QC, Letai A, Vassar R. 1994. Genetic bases of epidermolysis bullosa simplex and epidermolytic hyperkeratosis. J Invest Dermatol 103: 25S–S30. 10.1038/jid.1994.6 [DOI] [PubMed] [Google Scholar]
- Garrett-Sinha LA, Eberspaecher H, Seldin MF, de Crombrugghe B. 1996. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem 271: 31384–31390. 10.1074/jbc.271.49.31384 [DOI] [PubMed] [Google Scholar]
- Ge Y, Fuchs E. 2018. Stretching the limits: from homeostasis to stem cell plasticity in wound healing and cancer. Nat Rev Genet 19: 311–325. 10.1038/nrg.2018.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Gomez NC, Adam RC, Nikolova M, Yang H, Verma A, Lu CP, Polak L, Yuan S, Elemento O, et al. 2017. Stem cell lineage infidelity drives wound repair and cancer. Cell 169: 636–650.e14. 10.1016/j.cell.2017.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Miao Y, Gur-Cohen S, Gomez N, Yang H, Nikolova M, Polak L, Hu Y, Verma A, Elemento O, et al. 2020. The aging skin microenvironment dictates stem cell behavior. Proc Natl Acad Sci 117: 5339–5350. 10.1073/pnas.1901720117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. 2015. A century of cholesterol and coronaries: from plaques to genes to statins. Cell 161: 161–172. 10.1016/j.cell.2015.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grall A, Guaguère E, Planchais S, Grond S, Bourrat E, Hausser I, Hitte C, Le Gallo M, Derbois C, Kim GJ, et al. 2012. PNPLA1 mutations cause autosomal recessive congenital ichthyosis in golden retriever dogs and humans. Nat Genet 44: 140–147. 10.1038/ng.1056 [DOI] [PubMed] [Google Scholar]
- Gray GM, Yardley HJ. 1975. Different populations of pig epidermal cells: isolation and lipid composition. J Lipid Res 16: 441–447. 10.1016/S0022-2275(20)34494-1 [DOI] [PubMed] [Google Scholar]
- Green KJ, Roth-Carter Q, Niessen CM, Nichols SA. 2020. Tracing the evolutionary origin of desmosomes. Curr Biol 30: R535–R543. 10.1016/j.cub.2020.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grond S, Eichmann TO, Dubrac S, Kolb D, Schmuth M, Fischer J, Crumrine D, Elias PM, Haemmerle G, Zechner R, et al. 2017. PNPLA1 deficiency in mice and humans leads to a defect in the synthesis of omega-O-acylceramides. J Invest Dermatol 137: 394–402. 10.1016/j.jid.2016.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Wang G, Fails D, Nagarajan P, Ge Y. 2019. Unraveling cancer lineage drivers in squamous cell carcinomas. Pharmacol Ther 206: 107448. 10.1016/j.pharmthera.2019.107448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Yang YJ, Nagarajan P, Ge Y. 2021. Transcriptional and signalling regulation of skin epithelial stem cells in homeostasis, wounds and cancer. Exp Dermatol 30: 529–545. 10.1111/exd.14247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haensel D, Jin S, Sun P, Cinco R, Dragan M, Nguyen Q, Cang Z, Gong Y, Vu R, MacLean AL, et al. 2020. Defining epidermal basal cell states during skin homeostasis and wound healing using single-cell transcriptomics. Cell Rep 30: 3932–3947.e6. 10.1016/j.celrep.2020.02.091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AM, Wiczer BM, Herrmann T, Stremmel W, Bernlohr DA. 2005. Enzymatic properties of purified murine fatty acid transport protein 4 and analysis of acyl-CoA synthetase activities in tissues from FATP4 null mice. J Biol Chem 280: 11948–11954. 10.1074/jbc.M412629200 [DOI] [PubMed] [Google Scholar]
- Hayashi S, Manabe I, Suzuki Y, Relaix F, Oishi Y. 2016. Klf5 regulates muscle differentiation by directly targeting muscle-specific genes in cooperation with MyoD in mice. Elife 5: e17462. 10.7554/eLife.17462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann T, Buchkremer F, Gosch I, Hall AM, Bernlohr DA, Stremmel W. 2001. Mouse fatty acid transport protein 4 (FATP4): characterization of the gene and functional assessment as a very long chain acyl-CoA synthetase. Gene 270: 31–40. 10.1016/S0378-1119(01)00489-9 [DOI] [PubMed] [Google Scholar]
- Herrmann T, van der Hoeven F, Gröne HJ, Stewart AF, Langbein L, Kaiser I, Liebisch G, Gosch I, Buchkremer F, Drobnik W, et al. 2003. Mice with targeted disruption of the fatty acid transport protein 4 (Fatp 4, Slc27a4) gene show features of lethal restrictive dermopathy. J Cell Biol 161: 1105–1115. 10.1083/jcb.200207080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmer AM, Hanneken S, Ritzmann S, Becker T, Freudenberg J, Brockschmidt FF, Flaquer A, Freudenberg-Hua Y, Jamra RA, Metzen C, et al. 2005. Genetic variation in the human androgen receptor gene is the major determinant of common early-onset androgenetic alopecia. Am J Hum Genet 77: 140–148. 10.1086/431425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi T, Anjo T, Kaneko A, Senoo Y, Shibata A, Takama H, Yokoyama K, Nishito Y, Ono T, Taya C, et al. 2017. PNPLA1 has a crucial role in skin barrier function by directing acylceramide biosynthesis. Nat Commun 8: 14609. 10.1038/ncomms14609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran WM, Ginns EI, Menon GK, Grundmann JU, Fartasch M, McKinney CE, Elias PM, Sidransky E. 1994. Consequences of β-glucocerebrosidase deficiency in epidermis. Ultrastructure and permeability barrier alterations in Gaucher disease. J Clin Invest 93: 1756–1764. 10.1172/JCI117160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran WM, Takagi Y, Uchida Y. 2006. Epidermal sphingolipids: metabolism, function, and roles in skin disorders. FEBS Lett 580: 5456–5466. 10.1016/j.febslet.2006.08.039 [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Marques MR, Nguyen BT, Horner JS, Papazoglu C, Bronson RT, Mills AA, Attardi LD. 2005. Perp is a p63-regulated gene essential for epithelial integrity. Cell 120: 843–856. 10.1016/j.cell.2005.01.008 [DOI] [PubMed] [Google Scholar]
- Imokawa G, Abe A, Jin K, Higaki Y, Kawashima M, Hidano A. 1991. Decreased level of ceramides in stratum corneum of atopic dermatitis: an etiologic factor in atopic dry skin? J Invest Dermatol 96: 523–526. 10.1111/1523-1747.ep12470233 [DOI] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KS. 2016. Cell fate control by pioneer transcription factors. Development 143: 1833–1837. 10.1242/dev.133900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Young R. 2008. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132: 567–582. 10.1016/j.cell.2008.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennemann R, Rabionet M, Gorgas K, Epstein S, Dalpke A, Rothermel U, Bayerle A, van der Hoeven F, Imgrund S, Kirsch J, et al. 2012. Loss of ceramide synthase 3 causes lethal skin barrier disruption. Hum Mol Genet 21: 586–608. 10.1093/hmg/ddr494 [DOI] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. 2008. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol 10: 353–360. 10.1038/ncb1698 [DOI] [PubMed] [Google Scholar]
- Jiang YY, Jiang Y, Li CQ, Zhang Y, Dakle P, Kaur H, Deng JW, Yu-Tong Lin R, Han L, Xie JJ, et al. 2020. TP63, SOX2, and KLF5 establish a core regulatory circuitry that controls epigenetic and transcription patterns in esophageal squamous cell carcinoma cell lines. Gastroenterology 159: 1311–1327.e19. 10.1053/j.gastro.2020.06.050 [DOI] [PubMed] [Google Scholar]
- Jobard F, Lefèvre C, Karaduman A, Blanchet-Bardon C, Emre S, Weissenbach J, Ozgüc M, Lathrop M, Prud'homme JF, Fischer J. 2002. Lipoxygenase-3 (ALOXE3) and 12(R)-lipoxygenase (ALOX12B) are mutated in non-bullous congenital ichthyosiform erythroderma (NCIE) linked to chromosome 17p13.1. Hum Mol Genet 11: 107–113. 10.1093/hmg/11.1.107 [DOI] [PubMed] [Google Scholar]
- Joseph NM, Morrison SJ. 2005. Toward an understanding of the physiological function of Mammalian stem cells. Dev Cell 9: 173–183. 10.1016/j.devcel.2005.07.001 [DOI] [PubMed] [Google Scholar]
- Kaufman CK, Zhou P, Pasolli HA, Rendl M, Bolotin D, Lim KC, Dai X, Alegre ML, Fuchs E. 2003. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes Dev 17: 2108–2122. 10.1101/gad.1115203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenchegowda D, Swamynathan S, Gupta D, Wan H, Whitsett J, Swamynathan SK. 2011. Conditional disruption of mouse Klf5 results in defective eyelids with malformed meibomian glands, abnormal cornea and loss of conjunctival goblet cells. Dev Biol 356: 5–18. 10.1016/j.ydbio.2011.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A. 2016. Synthesis and degradation pathways, functions, and pathology of ceramides and epidermal acylceramides. Prog Lipid Res 63: 50–69. 10.1016/j.plipres.2016.04.001 [DOI] [PubMed] [Google Scholar]
- Klar J, Schweiger M, Zimmerman R, Zechner R, Li H, Törmä H, Vahlquist A, Bouadjar B, Dahl N, Fischer J. 2009. Mutations in the fatty acid transport protein 4 gene cause the ichthyosis prematurity syndrome. Am J Hum Genet 85: 248–253. 10.1016/j.ajhg.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P, Rosenberger S, de Juanes S, Latzko S, Hou J, Dick A, Kloz U, van der Hoeven F, Hausser I, Esposito I, et al. 2013. Aloxe3 knockout mice reveal a function of epidermal lipoxygenase-3 as hepoxilin synthase and its pivotal role in barrier formation. J Invest Dermatol 133: 172–180. 10.1038/jid.2012.250 [DOI] [PubMed] [Google Scholar]
- Laurin M, Gomez NC, Levorse J, Sendoel A, Sribour M, Fuchs E. 2019. An RNAi screen unravels the complexities of Rho GTPase networks in skin morphogenesis. Elife 8: e50226. 10.7554/eLife.50226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavker RM. 1976. Membrane coating granules: the fate of the discharged lamellae. J Ultrastruct Res 55: 79–86. 10.1016/S0022-5320(76)80083-4 [DOI] [PubMed] [Google Scholar]
- LeBoeuf M, Terrell A, Trivedi S, Sinha S, Epstein JA, Olson EN, Morrisey EE, Millar SE. 2010. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev Cell 19: 807–818. 10.1016/j.devcel.2010.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèvre C, Jobard F, Caux F, Bouadjar B, Karaduman A, Heilig R, Lakhdar H, Wollenberg A, Verret JL, Weissenbach J, et al. 2001. Mutations in CGI-58, the gene encoding a new protein of the esterase/lipase/thioesterase subfamily, in Chanarin–Dorfman syndrome. Am J Hum Genet 69: 1002–1012. 10.1086/324121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèvre C, Bouadjar B, Karaduman A, Jobard F, Saker S, Özguc M, Lathrop M, Prud'homme JF, Fischer J. 2004. Mutations in ichthyin a new gene on chromosome 5q33 in a new form of autosomal recessive congenital ichthyosis. Hum Mol Genet 13: 2473–2482. 10.1093/hmg/ddh263 [DOI] [PubMed] [Google Scholar]
- Li S, Teegarden A, Bauer EM, Choi J, Messaddeq N, Hendrix DA, Ganguli-Indra G, Leid M, Indra AK. 2017. Transcription factor CTIP1/ BCL11A regulates epidermal differentiation and lipid metabolism during skin development. Sci Rep 7: 13427. 10.1038/s41598-017-13347-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Wani MA, Whitsett JA, Wells JM. 2010. Klf5 regulates lineage formation in the pre-implantation mouse embryo. Development 137: 3953–3963. 10.1242/dev.054775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MH, Hsu FF, Crumrine D, Meyer J, Elias PM, Miner JH. 2019. Fatty acid transport protein 4 is required for incorporation of saturated ultralong-chain fatty acids into epidermal ceramides and monoacylglycerols. Sci Rep 9: 13254. 10.1038/s41598-019-49684-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long VJ. 1970. Variations in lipid composition at different depths in the cow snout epidermis. J Invest Dermatol 55: 269–273. 10.1111/1523-1747.ep12259974 [DOI] [PubMed] [Google Scholar]
- Lopez-Pajares V, Yan K, Zarnegar BJ, Jameson KL, Khavari PA. 2013. Genetic pathways in disorders of epidermal differentiation. Trends Genet 29: 31–40. 10.1016/j.tig.2012.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83: 835–839. 10.1016/0092-8674(95)90199-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoltsy AG, Parakkal PF. 1965. Membrane-coating granules of keratinizing epithelia. J Cell Biol 24: 297–307. 10.1083/jcb.24.2.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Yang VW. 2010. Mammalian Krüppel-like factors in health and diseases. Physiol Rev 90: 1337–1381. 10.1152/physrev.00058.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A, Butovich IA, Mata NL, Klein M, Ritter R III, Richardson J, Birch DG, Edwards AO, Kedzierski W. 2007. Retinal pathology and skin barrier defect in mice carrying a Stargardt disease-3 mutation in elongase of very long chain fatty acids-4. Mol Vis 13: 258–272. [PMC free article] [PubMed] [Google Scholar]
- Mellor LF, Gago-Lopez N, Bakiri L, Schmidt FN, Busse B, Rauber S, Jimenez M, Megías D, Oterino-Soto S, Sanchez-Prieto R, et al. 2022. Keratinocyte-derived S100A9 modulates neutrophil infiltration and affects psoriasis-like skin and joint disease. Ann Rheum Dis (in press). 10.1136/annrheumdis-2022-222229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398: 708–713. 10.1038/19531 [DOI] [PubMed] [Google Scholar]
- Mizutani Y, Kihara A, Chiba H, Tojo H, Igarashi Y. 2008. 2-Hydroxy-ceramide synthesis by ceramide synthase family: enzymatic basis for the preference of FA chain length. J Lipid Res 49: 2356–2364. 10.1194/jlr.M800158-JLR200 [DOI] [PubMed] [Google Scholar]
- Monteiro C, Miarka L, Perea-García M, Priego N, García-Gómez P, Álvaro-Espinosa L, de Pablos-Aragoneses A, Yebra N, Retana D, Baena P, et al. 2022. Stratification of radiosensitive brain metastases based on an actionable S100A9/RAGE resistance mechanism. Nat Med 28: 752–765. 10.1038/s41591-022-01749-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, Goldberg JL. 2009. KLF family members regulate intrinsic axon regeneration ability. Science 326: 298–301. 10.1126/science.1175737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta S, Monti M, Sesana S, Mellesi L, Ghidoni R, Caputo R. 1994. Abnormality of water barrier function in psoriasis. Role of ceramide fractions. Arch Dermatol 130: 452–456. 10.1001/archderm.1994.01690040056007 [DOI] [PubMed] [Google Scholar]
- Moulson CL, Martin DR, Lugus JJ, Schaffer JE, Lind AC, Miner JH. 2003. Cloning of wrinkle-free, a previously uncharacterized mouse mutation, reveals crucial roles for fatty acid transport protein 4 in skin and hair development. Proc Natl Acad Sci 100: 5274–5279. 10.1073/pnas.0431186100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai R, Suzuki T, Aizawa K, Shindo T, Manabe I. 2005. Significance of the transcription factor KLF5 in cardiovascular remodeling. J Thromb Haemost 3: 1569–1576. 10.1111/j.1538-7836.2005.01366.x [DOI] [PubMed] [Google Scholar]
- Nakamura T, Unda F, de-Vega S, Vilaxa A, Fukumoto S, Yamada KM, Yamada Y. 2004. The Krüppel-like factor epiprofin is expressed by epithelium of developing teeth, hair follicles, and limb buds and promotes cell proliferation. J Biol Chem 279: 626–634. 10.1074/jbc.M307502200 [DOI] [PubMed] [Google Scholar]
- Nandan MO, Ghaleb AM, Liu Y, Bialkowska AB, McConnell BB, Shroyer KR, Robine S, Yang VW. 2014. Inducible intestine-specific deletion of Krüppel-like factor 5 is characterized by a regenerative response in adult mouse colon. Dev Biol 387: 191–202. 10.1016/j.ydbio.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemes Z, Steinert PM. 1999. Bricks and mortar of the epidermal barrier. Exp Mol Med 31: 5–19. 10.1038/emm.1999.2 [DOI] [PubMed] [Google Scholar]
- Niemi KM, Kuokkanen K, Kanerva L, Ignatius J. 1993. Recessive ichthyosis congenita type IV. Am J Dermatopathol 15: 224–228. 10.1097/00000372-199306000-00005 [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. 1984. The role of protein kinase C in cell surface signal transduction and tumour promotion. Nature 308: 693–698. 10.1038/308693a0 [DOI] [PubMed] [Google Scholar]
- Oberbeck N, Pham VC, Webster JD, Reja R, Huang CS, Zhang Y, Roose-Girma M, Warming S, Li Q, Birnberg A, et al. 2019. The RIPK4–IRF6 signalling axis safeguards epidermal differentiation and barrier function. Nature 574: 249–253. 10.1038/s41586-019-1615-3 [DOI] [PubMed] [Google Scholar]
- Ohnishi S, Laub F, Matsumoto N, Asaka M, Ramirez F, Yoshida T, Terada M. 2000. Developmental expression of the mouse gene coding for the Krüppel-like transcription factor KLF5. Dev Dyn 217: 421–429. [DOI] [PubMed] [Google Scholar]
- Ohno Y, Kamiyama N, Nakamichi S, Kihara A. 2017. PNPLA1 is a transacylase essential for the generation of the skin barrier lipid ω-O-acylceramide. Nat Commun 8: 14610. 10.1038/ncomms14610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K, Nishimura G, Maemura K, Yamauchi T, Kubota N, et al. 2005. Krüppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab 1: 27–39. 10.1016/j.cmet.2004.11.005 [DOI] [PubMed] [Google Scholar]
- Oishi Y, Manabe I, Tobe K, Ohsugi M, Kubota T, Fujiu K, Maemura K, Kubota N, Kadowaki T, Nagai R. 2008. SUMOylation of Krüppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-δ. Nat Med 14: 656–666. 10.1038/nm1756 [DOI] [PubMed] [Google Scholar]
- Perdigoto CN, Valdes VJ, Bardot ES, Ezhkova E. 2014. Epigenetic regulation of epidermal differentiation. Cold Spring Harb Perspect Med 4: a015263. 10.1101/cshperspect.a015263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnell JS, Schnitzler CE, Browne WE. 2015. KLF/SP transcription factor family evolution: expansion, diversification, and innovation in eukaryotes. Genome Biol Evol 7: 2289–2309. 10.1093/gbe/evv141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proksch E, Brandner JM, Jensen JM. 2008. The skin: an indispensable barrier. Exp Dermatol 17: 1063–1072. 10.1111/j.1600-0625.2008.00786.x [DOI] [PubMed] [Google Scholar]
- Rabionet M, Gorgas K, Sandhoff R. 2014. Ceramide synthesis in the epidermis. Biochim Biophys Acta 1841: 422–434. 10.1016/j.bbalip.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Radner FP, Streith IE, Schoiswohl G, Schweiger M, Kumari M, Eichmann TO, Rechberger G, Koefeler HC, Eder S, Schauer S, et al. 2010. Growth retardation, impaired triacylglycerol catabolism, hepatic steatosis, and lethal skin barrier defect in mice lacking comparative gene identification-58 (CGI-58). J Biol Chem 285: 7300–7311. 10.1074/jbc.M109.081877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radner FP, Marrakchi S, Kirchmeier P, Kim GJ, Ribierre F, Kamoun B, Abid L, Leipoldt M, Turki H, Schempp W, et al. 2013. Mutations in CERS3 cause autosomal recessive congenital ichthyosis in humans. PLoS Genet 9: e1003536. 10.1371/journal.pgen.1003536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds G, Vegh P, Fletcher J, Poyner EFM, Stephenson E, Goh I, Botting RA, Huang N, Olabi B, Dubois A, et al. 2021. Developmental cell programs are co-opted in inflammatory skin disease. Science 371: eaa6500. 10.1126/science.aba6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinwald JG, Green H. 1975. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell 6: 331–343. 10.1016/S0092-8674(75)80001-8 [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Yamanaka S. 2014. Rethinking differentiation: stem cells, regeneration, and plasticity. Cell 157: 110–119. 10.1016/j.cell.2014.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya AP, Stone RC, Brooks SR, Pastar I, Jozic I, Hasneen K, O'Neill K, Mehdizadeh S, Head CR, Strbo N, et al. 2020. Deregulated immune cell recruitment orchestrated by FOXM1 impairs human diabetic wound healing. Nat Commun 11: 4678. 10.1038/s41467-020-18276-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl C, Rendeiro AF, Sheffield NC, Bock C. 2015. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods 12: 963–965. 10.1038/nmeth.3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonthaler HB, Guinea-Viniegra J, Wculek SK, Ruppen I, Ximénez-Embún P, Guío-Carrión A, Navarro R, Hogg N, Ashman K, Wagner EF. 2013. S100A8-S100A9 protein complex mediates psoriasis by regulating the expression of complement factor C3. Immunity 39: 1171–1181. 10.1016/j.immuni.2013.11.011 [DOI] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. 1999. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet 22: 356–360. 10.1038/11926 [DOI] [PubMed] [Google Scholar]
- Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, Johnston D, Siprashvili Z, Khavari PA. 2012. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev Cell 21: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo T, Manabe I, Fukushima Y, Tobe K, Aizawa K, Miyamoto S, Kawai-Kowase K, Moriyama N, Imai Y, Kawakami H, et al. 2002. Krüppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med 8: 856–863. 10.1038/nm738 [DOI] [PubMed] [Google Scholar]
- Squier CA. 1973. The permeability of keratinized and nonkeratinized oral epithelium to horseradish peroxidase. J Ultrastruct Res 43: 160–177. 10.1016/S0022-5320(73)90076-2 [DOI] [PubMed] [Google Scholar]
- Stahl A, Hirsch DJ, Gimeno RE, Punreddy S, Ge P, Watson N, Patel S, Kotler M, Raimondi A, Tartaglia LA, et al. 1999. Identification of the major intestinal fatty acid transport protein. Mol Cell 4: 299–308. 10.1016/S1097-2765(00)80332-9 [DOI] [PubMed] [Google Scholar]
- Stoler A, Kopan R, Duvic M, Fuchs E. 1988. Use of monospecific antisera and cRNA probes to localize the major changes in keratin expression during normal and abnormal epidermal differentiation. J Cell Biol 107: 427–446. 10.1083/jcb.107.2.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur I, Undén AB, Toftgård R. 2002. Human Krüppel-like factor5/KLF5: synergy with NF-κB/Rel factors and expression in human skin and hair follicles. Eur J Cell Biol 81: 323–334. 10.1078/0171-9335-00257 [DOI] [PubMed] [Google Scholar]
- Sur I, Rozell B, Jaks V, Bergström A, Toftgård R. 2006. Epidermal and craniofacial defects in mice overexpressing Klf5 in the basal layer of the epidermis. J Cell Sci 119: 3593–3601. 10.1242/jcs.03070 [DOI] [PubMed] [Google Scholar]
- Szigety KM, Liu F, Yuan CY, Moran DJ, Horrell J, Gochnauer HR, Cohen RN, Katz JP, Kaestner KH, Seykora JT, et al. 2020. HDAC3 ensures stepwise epidermal stratification via NCoR/SMRT-reliant mechanisms independent of its histone deacetylase activity. Genes Dev 34: 973–988. 10.1101/gad.333674.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Tetreault MP, Yang Y, Katz JP. 2013. Krüppel-like factors in cancer. Nat Rev Cancer 13: 701–713. 10.1038/nrc3582 [DOI] [PubMed] [Google Scholar]
- Ting SB, Caddy J, Hislop N, Wilanowski T, Auden A, Zhao LL, Ellis S, Kaur P, Uchida Y, Holleran WM, et al. 2005. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science 308: 411–413. 10.1126/science.1107511 [DOI] [PubMed] [Google Scholar]
- Traupe H, Fischer J, Oji V. 2014. Nonsyndromic types of ichthyoses - an update. J Dtsch Dermatol Ges 12: 109–121. [DOI] [PubMed] [Google Scholar]
- Turner J, Crossley M. 1999. Mammalian Krüppel-like transcription factors: more than just a pretty finger. Trends Biochem Sci 24: 236–240. 10.1016/S0968-0004(99)01406-1 [DOI] [PubMed] [Google Scholar]
- Uchida Y, Holleran WM. 2008. Omega-O-acylceramide, a lipid essential for mammalian survival. J Dermatol Sci 51: 77–87. 10.1016/j.jdermsci.2008.01.002 [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C. 2011. Distinct stem cells contribute to mammary gland development and maintenance. Nature 479: 189–193. 10.1038/nature10573 [DOI] [PubMed] [Google Scholar]
- Vasireddy V, Uchida Y, Salem N Jr, Kim SY, Mandal MN, Reddy GB, Bodepudi R, Alderson NL, Brown JC, Hama H, et al. 2007. Loss of functional ELOVL4 depletes very long-chain fatty acids (≥C28) and the unique ω-O-acylceramides in skin leading to neonatal death. Hum Mol Genet 16: 471–482. 10.1093/hmg/ddl480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M, Mishra A, Klose C, Eggert US, Watt FM. 2020. Human epidermal stem cell differentiation is modulated by specific lipid subspecies. Proc Natl Acad Sci 117: 22173–22182. 10.1073/pnas.2011310117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waikel RL, Kawachi Y, Waikel PA, Wang XJ, Roop DR. 2001. Deregulated expression of c-Myc depletes epidermal stem cells. Nat Genet 28: 165–168. 10.1038/88889 [DOI] [PubMed] [Google Scholar]
- Wan H, Luo F, Wert SE, Zhang L, Xu Y, Ikegami M, Maeda Y, Bell SM, Whitsett JA. 2008. Kruppel-like factor 5 is required for perinatal lung morphogenesis and function. Development 135: 2563–2572. 10.1242/dev.021964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Pasolli HA, Williams T, Fuchs E. 2008. AP-2 factors act in concert with Notch to orchestrate terminal differentiation in skin epidermis. J Cell Biol 183: 37–48. 10.1083/jcb.200804030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kirkwood JS, Taylor AW, Stevens JF, Leid M, Ganguli-Indra G, Indra AK. 2013. Transcription factor Ctip2 controls epidermal lipid metabolism and regulates expression of genes involved in sphingolipid biosynthesis during skin development. J Invest Dermatol 133: 668–676. 10.1038/jid.2012.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM. 2001. Stem cell fate and patterning in mammalian epidermis. Curr Opin Genet Dev 11: 410–417. 10.1016/S0959-437X(00)00211-2 [DOI] [PubMed] [Google Scholar]
- Weiss RA, Eichner R, Sun TT. 1984. Monoclonal antibody analysis of keratin expression in epidermal diseases: a 48- and 56-kdalton keratin as molecular markers for hyperproliferative keratinocytes. J Cell Biol 98: 1397–1406. 10.1083/jcb.98.4.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz PW, Downing DT. 1982. Glycolipids in mammalian epidermis: structure and function in the water barrier. Science 217: 1261–1262. 10.1126/science.7112128 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Hattori M, Chamulitrat W, Ohno Y, Kihara A. 2020. Skin permeability barrier formation by the ichthyosis-causative gene FATP4 through formation of the barrier lipid ω-O-acylceramide. Proc Natl Acad Sci 117: 2914–2922. 10.1073/pnas.1917525117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398: 714–718. 10.1038/19539 [DOI] [PubMed] [Google Scholar]
- Yang H, Schramek D, Adam RC, Keyes BE, Wang P, Zheng D, Fuchs E. 2015. ETS family transcriptional regulators drive chromatin dynamics and malignancy in squamous cell carcinomas. Elife 4: e10870. 10.7554/eLife.10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Schneider C, Boeglin WE, Marnett LJ, Brash AR. 2003. The lipoxygenase gene ALOXE3 implicated in skin differentiation encodes a hydroperoxide isomerase. Proc Natl Acad Sci 100: 9162–9167. 10.1073/pnas.1633612100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisberg M, Kalluri R. 2013. Cellular mechanisms of tissue fibrosis. 1. Common and organ-specific mechanisms associated with tissue fibrosis. Am J Physiol Cell Physiol 304: C216–C225. 10.1152/ajpcell.00328.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenz R, Eferl R, Kenner L, Florin L, Hummerich L, Mehic D, Scheuch H, Angel P, Tschachler E, Wagner EF. 2005. Psoriasis-like skin disease and arthritis caused by inducible epidermal deletion of Jun proteins. Nature 437: 369–375. 10.1038/nature03963 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Pasolli HA, Fuchs E. 2011. Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proc Natl Acad Sci 108: 2270–2275. 10.1073/pnas.1019603108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Choi PS, Francis JM, Gao GF, Campbell JD, Ramachandran A, Mitsuishi Y, Ha G, Shih J, Vazquez F, et al. 2018. Somatic superenhancer duplications and hotspot mutations lead to oncogenic activation of the KLF5 transcription factor. Cancer Discov 8: 108–125. 10.1158/2159-8290.CD-17-0532 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated in this study
Raw and analyzed data reported here are available via NCBI GEO under the accession number GSE168600 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE168600).
Mining of published GEO and related data sets
The following GEO data sets were downloaded: GSE131903, GSE117405, GSE134431, GSE147424, GSE153760, GSE158432, and GSE142471. For RNA-seq GSE131903, raw FastQ files were downloaded and processed using RNA-seq alignment pipeline as described above and TPM was plotted for KLF family members expressed (TPM > 1). For RNA-seq GSE117405 and GSE134431, processed data were downloaded and FPKM was plotted for KLF family members expressed (FPKM > 1). For scRNA-seq GSE147424, GSE153760, GSE158432, and GSE142471, raw gene, barcode, and matrix files were downloaded and analyzed using Seurat package (https://satijalab.org/seurat), and the corresponding cells with KLF5 expression were plotted. For scRNA-seq from Reynolds et al. (2021), plots were generated using web portal https://developmentcellatlas.ncl.ac.uk/datasets/hca_skin_portal.