In this Perspective, Tartakoff et al. discuss the structured versus condensate-based duality as being of central importance for understanding ribosome biogenesis. In addition, they emphasize that experimental probes of the nucleolus must be functional and be expressed at physiological levels if they are to report on the normal biology of subunit maturation.
Keywords: condensates, nucleolus, ribosome genesis
Abstract
The nucleolus is best known for housing the highly ordered assembly line that produces ribosomal subunits. The >100 ribosome assembly factors in the nucleolus are thought to cycle between two states: an operative state (when integrated into subunit assembly intermediates) and a latent state (upon release from intermediates). Although it has become commonplace to refer to the nucleolus as “being a multilayered condensate,” and this may be accurate for latent factors, there is little reason to think that such assertions pertain to the operative state of assembly factors.
The nucleolus is embedded in the nucleoplasm, where it constitutes a compositionally distinct territory. Along with experimental studies of fluorescent proteins that localize to the nucleolus, this partitioning of the nuclear volume has led to repeated assertions that the nucleolus should be considered to be a condensate.
Biomolecular condensates (or membraneless organelles within cells) are composed of multivalent proteins, along with RNA species. These objects tend to be spherical and isotropic and can participate in homotypic fusion. As we discuss below, it remains an open question whether or how condensates participate in ribosome genesis.
In an alternative view, the functional essence of the nucleolus is not that of a condensate but is highly structured. This view dates back to the electron micrographic studies of rDNA repeats (“Miller chromatin spreads”) that showed characteristic “Christmas tree” arrangements of nascent rRNPs that progressively lengthen, form terminal knobs, and undergo cleavage (Miller and Beatty 1969; Osheim et al. 2009). The closely packed “cyclopean” structures of corresponding massive subunit precursors have more recently been imaged by cryo-EM (Baßler and Hurt 2019; Klinge and Woolford 2019; Black and Johnson 2021).
Are these two views compatible?
The generic term “condensate” indicates a separation of territories (often referred to as phases) without implying any underlying mechanism. It is therefore far from specific, especially when used to describe elements within living cells. In contrast, in vitro condensates formed from recombinant proteins have been better characterized. In the best-studied examples, their coherence depends on many low-affinity interactions among their multivalent proteins, often dependent on “intrinsically disordered regions” (IDRs). If more than one “type” of in vitro condensate coexists, they can exhibit liquid–liquid phase separation (Oldfield and Dunker 2014; Wang et al. 2016; Protter et al. 2018; Mao et al. 2019; Riback et al. 2020; Stenström et al. 2020; Lafontaine et al. 2021).
During early stages of their maturation, nascent small and large ribosomal subunit (SSU and LSU, respectively) precursors are tethered to the rDNA axis that structures their maturation, thereby reducing the dimensionality of assembly. In this process, what role(s) might one or more type of condensates play? Here we discuss the structured versus condensate-based duality as being of central importance for understanding ribosome biogenesis. In addition, we emphasize that experimental probes of the nucleolus must be functional and be expressed at physiological levels if they are to report on the normal biology of subunit maturation.
Known subcompartments of the nucleolus
In many types of higher eukaryotic cells, one can detect three classical subcompartments within the nucleolus. In favorable circumstances, they can be distinguished ultrastructurally and by localizing selected proteins, some of which are known to be ribosome assembly factors (AFs). The compartments are the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC). Analysis of the significance and composition of these compartments is incomplete and is complicated by the realization that the localizations of many AFs are dynamic and can shift according to whether ribosomal subunits are being produced (Phair and Misteli 2000; Chen and Huang 2001; Ide et al. 2020; Lafontaine et al. 2021; Tartakoff et al. 2021). In fact, many AFs that are loaded onto immature subunits in the nucleolus travel with them to the nucleoplasm or the cytoplasm and then recycle. Others reside primarily in the nucleoplasm or cytoplasm (Hernandez-Verdun et al. 2010; Panse and Johnson 2010; Zisser et al. 2018; Baßler and Hurt 2019).
rDNA and rDNA transcription have been localized to the FC (or FC/DFC interface). Nascent rRNP intermediates then extend from the DFC into the GC, where endonucleolytic cleavage along with extensive remodeling are thought to allow immature subunits to be released (Scheer and Hock 1999; Raška et al. 2006; Pederson 2011). The existence of these subcompartments has given rise to the suggestion that each one may be a distinct condensate (Feric et al. 2016; Lafontaine et al. 2021).
The significance of subcompartments has been further investigated in Saccharomyces cerevisiae, where an underlying tripartite organization is also detected. The physical properties of yeast (and human) nucleolar AFs are notably diverse (size: 10–280 kDa, predicted disorder: <10%–80%, isoelectric point: pH 4–11). The average titer of each AF (∼0.5 mg/mL in the nucleolus) is stoichiometrically comparable with that of nascent rRNA (Tartakoff et al. 2021; Lin et al. 2022).
In yeast, the nucleolus can be seen to have a “coaxial” structure, with the rDNA axis and its most closely associated proteins being surrounded by two layers of AFs and corresponding rRNA segments. The inner layer includes proteins that contribute to both types of subunit. It is conspicuously enriched in AFs that assemble the SSU. The outer layer, in contrast, is dedicated to production of the LSU. When subunit production is halted, many AFs that otherwise would localize to the inner layer relocate to the outer layer/volume. Like the GC, it extends to the surface of the nucleolus (Tartakoff et al. 2021).
As diagrammed in Figure 1, elongation of rRNA is therefore thought to entail the sequential recruitment of specific AFs from this surrounding reservoir of latent AFs, bringing them to the nascent rRNA (Lin et al. 2022). Assuming that these AFs are most stable in the outer layer, this recruitment could build potential energy into the system, thereby driving subsequent vectorial transport in a thermodynamically downhill direction as rRNP intermediates move centrifugally away from the rDNA axis (Tartakoff et al. 2021). The size of the latent pool presumably depends on the rate of subunit assembly. In biophysical parlance, the latent reservoir may be described as an “active emulsion” (Weber et al. 2019).
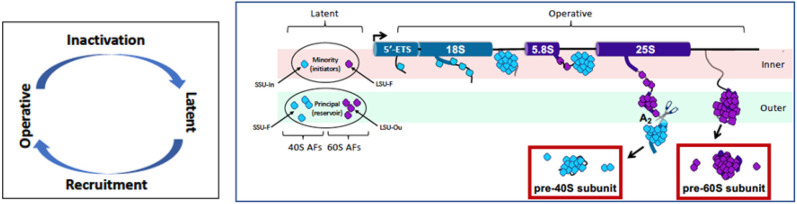
Figure 1.
The left panel illustrates the cyclic behavior of ribosome assembly factors, which alternate between having been recruited to immature subunits (operative state) and having been released from them (latent state). The right panel summarizes the processing of nascent subunits. The rDNA axis is at the top designates the segments that code for distinct domains of rRNA. When latent, almost all assembly factors are broadly distributed, occupying the outer compartment, as indicated at the left. Upon initiation of transcription, these factors are recruited to specific binding sites of nascent rRNPs, progressively forming particulate intermediates that extend from the inner into the outer layer and are ultimately released after endonucleolytic cleavage. The inner layer seems roughly equivalent to the DFC, while the outer layer corresponds to the GC. For a more detailed description, see Tartakoff et al. (2021) and Lin et al. (2022).
The clarity of visualization of the coaxial layers in yeast is made possible by (1) in vivo “linearization” of rDNA upon cell cycle arrest (in contrast to its normal tortuous path throughout the nucleolus) and (2) the observation that nascent rRNA in yeast is cleaved in the ITS1 region that lies between the segments that are destined for the SSU and LSU (Osheim et al. 2004). This conveniently removes nearly mature SSU precursors once they have been assembled, thereby making it possible to visualize the underlying structure.
The biological importance of condensates
In vitro condensates resemble organelles that lack a surrounding membrane (Matera et al. 2009; Lin et al. 2015; Banani et al. 2017; McSwiggen et al. 2019; Chen and Mayr 2022; Sharp et al. 2022). In vivo condensates might enable pathways to function more efficiently for the following reasons: (1) They shield internal components from the surrounding milieu. (2) They promote the thermodynamically downhill vectorial transfer of cargoes to contiguous compartments (e.g., newly assembled subunit precursors, as described above). (3) They concentrate relevant reactants within their interior (e.g., Zhao et al. 2015).
In order to probe the interior of the nucleolus, investigators have studied fluorescent proteins that localize to the nucleolus after microinjection into cells and upon expression in living cells (e.g., Brangwynne et al. 2011; Feric et al. 2016; Pillet et al. 2017; Yao et al. 2019; Riback et al. 2020; Lafontaine et al. 2021). Interpretation of these experiments is based on the assumption that the exogenous tracers exhibit behavior that is characteristic of endogenous nucleolar proteins. It has often been taken for granted that they localize to condensates.
Limitations of experiments using fluorescent reporters
Our concerns with such experiments are largely distinct from others who have scrutinized the near universality of condensate biology (Alberti et al. 2019; McSwiggen et al. 2019; Narlikar et al. 2021). There are several issues:
Issue 1: Observations made using fluorescent tracers that are expressed at supraphysiologic levels—or lack functional activity—seem likely to report (at best) on the condition of latent AFs, rather than those that are directly engaged in subunit maturation. In fact, none of the tracers that have been (over)expressed have been proven to retain function, including derivatives of the GC and DFC markers B23/NPM and fibrillarin/FBL/NOP1, respectively. Among the tracers that have been used are engineered reporters that have little resemblance to nucleolar AFs (e.g., Emmott and Hiscox 2009; Scott et al. 2011; Martin et al. 2015; Bracha et al. 2018; Zhu et al. 2019; Riback et al. 2020).
Issue 2: If tracers reach a sufficiently high level within the nucleolus, they could initiate local condensate formation. In this event, they may recapitulate in vitro condensate formation behavior at intracellular sites without being representative of the condition of endogenous proteins engaged in ribosome biogenesis. Considering the elaborate biophysical studies that have depended on the use of fluorescent tracers, this concern is especially important.
Issue 3: The key cell type used for critical early studies (late-stage Xenopus oocytes) produces few if any ribosomes (Brown and Littna 1964; Feric et al. 2016). Therefore, the observations made, which led to formulation of the multilayer condensate model, are unlikely to pertain to the operative form of tracers.
Issue 4: There is no reason to expect that studies focused on a single stage of subunit assembly (e.g., the GC markers mentioned above) report on events that occur during earlier or later stages of assembly.
For these several reasons, tracers that concentrate in the nucleolus may or may not report on subunit assembly per se. Given present gene replacement options, greater clarity could surely be achieved in some cases by using tracers that are expressed at normal levels and are known to be functional in genetic complementation assays.
The dual nature of the nucleolus
As a minimal hypothesis, the nucleus can be modeled as including both a nucleosome-filled volume (chromatin) and an “excluded” nucleolar domain, each of which has self-coherent properties (Hult et al. 2017; Maeshima et al. 2021; Lin et al. 2022). Many nucleolar AFs, like other proteins, include sequences that are predicted to be disordered (Stenström et al. 2020; Lin et al. 2022). However, it is not evident that any resulting interactions are directly relevant to subunit maturation. Moreover, multiple AFs have other types of motifs that can mediate protein–protein binding (WD repeats, HEAT/ARM repeats, complementary charge characteristics, etc.) (Woolford and Baserga 2013; Baßler et al. 2017; Vincent et al. 2018; Lin et al. 2022). Thus, although latent AFs may form condensates, there is little evidence that operative forms should be considered to be part of condensates. Asserting that the nucleolus is a condensate, rather than proving that this is the case, inverts the normal deductive process.
Future studies of the nucleolus
The critical biological roles of the nucleolus and its high complexity have intrigued investigators for decades. Moreover, the nucleolus provides an exaggerated and accessible model of transcription that is relevant to understanding the production and processing of other varieties of RNA. Investigation of the nucleolus has been energized by suggestions that it is fundamentally a multilayered condensate. The facile use of the term “condensate,” unfortunately, has often been used without explicit justification and without attempting to integrate much of the biochemical and cell-biological knowledge that is available. A central unresolved question is whether the many proteins that concentrate in the nucleolus owe their absence from the rest of the nucleoplasm primarily to some shared characteristic that gives them mutual coherence or, rather, that chromatin has some property that restricts intermixing.
Acknowledgments
Our work is supported by National Institutes of Health grant R01GM089872.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.349748.122.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Alberti S, Gladfelter A, Mittag T. 2019. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176: 419–434. 10.1016/j.cell.2018.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baßler J, Hurt E. 2019. Eukaryotic ribosome assembly. Annu Rev Biochem 88: 281–306. 10.1146/annurev-biochem-013118-110817 [DOI] [PubMed] [Google Scholar]
- Baßler J, Ahmed YL, Kallas M, Kornprobst M, Calviño FR, Gnädig M, Thoms M, Stier G, Ismail S, Kharde S, et al. 2017. Interaction network of the ribosome assembly machinery from a eukaryotic thermophile. Protein Sci 26: 327–342. 10.1002/pro.3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JJ, Johnson AW. 2021. Genetics animates structure: leveraging genetic interactions to study the dynamics of ribosome biogenesis. Curr Genet 67: 729–738. 10.1007/s00294-021-01187-y [DOI] [PubMed] [Google Scholar]
- Bracha D, Walls MT, Wei MT, Zhu L, Kurian M, Avalos JL, Toettcher JE, Brangwynne CP. 2018. Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell 175: 1467–1480.e13. 10.1016/j.cell.2018.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA. 2011. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci 108: 4334–4339. 10.1073/pnas.1017150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Littna E. 1964. Variations in the synthesis of stable RNA's during oogenesis and development of Xenopus laevis. J Mol Biol 8: 688–695, IN10. 10.1016/S0022-2836(64)80117-0 [DOI] [PubMed] [Google Scholar]
- Chen D, Huang S. 2001. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J Cell Biol 153: 169–176. 10.1083/jcb.153.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Mayr C. 2022. A working model for condensate RNA-binding proteins as matchmakers for protein complex assembly. RNA 28: 76–87. 10.1261/rna.078995.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmott E, Hiscox JA. 2009. Nucleolar targeting: the hub of the matter. EMBO Rep 10: 231–238. 10.1038/embor.2009.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. 2016. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165: 1686–1697. 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D, Roussel P, Thiry M, Sirri V, Lafontaine DL. 2010. The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip Rev RNA 1: 415–431. 10.1002/wrna.39 [DOI] [PubMed] [Google Scholar]
- Hult C, Adalsteinsson D, Vasquez PA, Lawrimore J, Bennett M, York A, Cook D, Yeh E, Forest MG, Bloom K. 2017. Enrichment of dynamic chromosomal crosslinks drive phase separation of the nucleolus. Nucleic Acids Res 45: 11159–11173. 10.1093/nar/gkx741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide S, Imai R, Ochi H, Maeshima K. 2020. Transcriptional suppression of ribosomal DNA with phase separation. Sci Adv 6: eabb5953. 10.1126/sciadv.abb5953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S, Woolford JL Jr. 2019. Ribosome assembly coming into focus. Nat Rev Mol Cell Biol 20: 116–131. 10.1038/s41580-018-0078-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. 2021. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol 22: 165–182. 10.1038/s41580-020-0272-6 [DOI] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R. 2015. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell 60: 208–219. 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Rajan S, Lemberg S, Altawil M, Anderson K, Bryant R, Cappeta S, Chin B, Hamdan I, Hamer A, et al. 2022. Production of nascent ribosome precursors within the nucleolar microenvironment of Saccharomyces cerevisiae. Genetics 221: iyac070. 10.1093/genetics/iyac070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K, Iida S, Tamura S. 2021. Physical nature of chromatin in the nucleus. Cold Spring Harb Perspect Biol 13: a040675. 10.1101/cshperspect.a040675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S, Kuldinow D, Haataja MP, Košmrlj A. 2019. Phase behavior and morphology of multicomponent liquid mixtures. Soft Matter 15: 1297–1311. 10.1039/C8SM02045K [DOI] [PubMed] [Google Scholar]
- Martin RM, Ter-Avetisyan G, Herce HD, Ludwig AK, Lättig-Tünnemann G, Cardoso MC. 2015. Principles of protein targeting to the nucleolus. Nucleus 6: 314–325. 10.1080/19491034.2015.1079680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. 2009. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell 17: 639–647. 10.1016/j.devcel.2009.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggen DT, Mir M, Darzacq X, Tjian R. 2019. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev 33: 1619–1634. 10.1101/gad.331520.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OL Jr, Beatty BR. 1969. Visualization of nucleolar genes. Science 164: 955–957. 10.1126/science.164.3882.955 [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Myong S, Larson D, Maeshima K, Francis N, Rippe K, Sabari B, Strader L, Tjian R. 2021. Is transcriptional regulation just going through a phase? Mol Cell 81: 1579–1585. 10.1016/j.molcel.2021.03.046 [DOI] [PubMed] [Google Scholar]
- Oldfield CJ, Dunker AK. 2014. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem 83: 553–584. 10.1146/annurev-biochem-072711-164947 [DOI] [PubMed] [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell 16: 943–954. 10.1016/j.molcel.2004.11.031 [DOI] [PubMed] [Google Scholar]
- Osheim YN, French SL, Sikes ML, Beyer AL. 2009. Electron microscope visualization of RNA transcription and processing in Saccharomyces cerevisiae by Miller chromatin spreading. Methods Mol Biol 464: 55–69. 10.1007/978-1-60327-461-6_4 [DOI] [PubMed] [Google Scholar]
- Panse VG, Johnson AW. 2010. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci 35: 260–266. 10.1016/j.tibs.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. 2011. The nucleolus. Cold Spring Harb Perspect Biol 3: a000638. 10.1101/cshperspect.a000638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Misteli T. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404: 604–609. 10.1038/35007077 [DOI] [PubMed] [Google Scholar]
- Pillet B, Mitterer V, Kressler D, Pertschy B. 2017. Hold on to your friends: dedicated chaperones of ribosomal proteins: dedicated chaperones mediate the safe transfer of ribosomal proteins to their site of pre-ribosome incorporation. Bioessays 39: 1–12. 10.1002/bies.201600153 [DOI] [PubMed] [Google Scholar]
- Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, Rosen MK, Parker R. 2018. Intrinsically disordered regions can contribute promiscuous interactions to RNP granule assembly. Cell Rep 22: 1401–1412. 10.1016/j.celrep.2018.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raška I, Shaw PJ, Cmarko D. 2006. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol 18: 325–334. 10.1016/j.ceb.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, Wei MT, Kriwacki RW, Brangwynne CP. 2020. Composition-dependent thermodynamics of intracellular phase separation. Nature 581: 209–214. 10.1038/s41586-020-2256-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Hock R. 1999. Structure and function of the nucleolus. Curr Opin Cell Biol 11: 385–390. 10.1016/S0955-0674(99)80054-4 [DOI] [PubMed] [Google Scholar]
- Scott MS, Troshin PV, Barton GJ. 2011. Nod: a nucleolar localization sequence detector for eukaryotic and viral proteins. BMC Bioinformatics 12: 317. 10.1186/1471-2105-12-317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA, Chakraborty AK, Henninger JE, Young RA. 2022. RNA in formation and regulation of transcriptional condensates. RNA 28: 52–57. 10.1261/rna.078997.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenström L, Mahdessian D, Gnann C, Cesnik AJ, Ouyang W, Leonetti MD, Uhlén M, Cuylen-Haering S, Thul PJ, Lundberg E. 2020. Mapping the nucleolar proteome reveals a spatiotemporal organization related to intrinsic protein disorder. Mol Syst Biol 16: e9469. 10.15252/msb.20209469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartakoff AM, Chen L, Raghavachari S, Gitiforooz D, Dhinakaran A, Ni CL, Pasadyn C, Mahabeleshwar GH, Pasadyn V, Woolford JL Jr. 2021. The nucleolus as a polarized coaxial cable in which the rDNA axis is surrounded by dynamic subunit-specific phases. Curr Biol 31: 2507–2519.e4. 10.1016/j.cub.2021.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent NG, Charette JM, Baserga SJ. 2018. The SSU processome interactome in Saccharomyces cerevisiae reveals novel protein subcomplexes. RNA 24: 77–89. 10.1261/rna.062927.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Uversky VN, Kurgan L. 2016. Disordered nucleiome: abundance of intrinsic disorder in the DNA- and RNA-binding proteins in 1121 species from eukaryota, bacteria and archaea. Proteomics 16: 1486–1498. 10.1002/pmic.201500177 [DOI] [PubMed] [Google Scholar]
- Weber CA, Zwicker D, Jülicher F, Lee CF. 2019. Physics of active emulsions. Rep Prog Phys 82: 064601. 10.1088/1361-6633/ab052b [DOI] [PubMed] [Google Scholar]
- Woolford JL Jr, Baserga SJ. 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681. 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao RW, Xu G, Wang Y, Shan L, Luan PF, Wang Y, Wu M, Yang LZ, Xing YH, Yang L, et al. 2019. Nascent pre-rRNA sorting via phase separation drives the assembly of dense fibrillar components in the human nucleolus. Mol Cell 76: 767–783.e11. 10.1016/j.molcel.2019.08.014 [DOI] [PubMed] [Google Scholar]
- Zhao H, Chiaro CR, Zhang L, Smith PB, Chan CY, Pedley AM, Pugh RJ, French JB, Patterson AD, Benkovic SJ. 2015. Quantitative analysis of purine nucleotides indicates that purinosomes increase de novo purine biosynthesis. J Biol Chem 290: 6705–6713. 10.1074/jbc.M114.628701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Richardson TM, Wacheul L, Wei MT, Feric M, Whitney G, Lafontaine DLJ, Brangwynne CP. 2019. Controlling the material properties and rRNA processing function of the nucleolus using light. Proc Natl Acad Sci 116: 17330–17335. 10.1073/pnas.1903870116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisser G, Ohmayer U, Mauerhofer C, Mitterer V, Klein I, Rechberger GN, Wolinski H, Prattes M, Pertschy B, Milkereit P, et al. 2018. Viewing pre-60S maturation at a minute's timescale. Nucleic Acids Res 46: 3140–3151. 10.1093/nar/gkx1293 [DOI] [PMC free article] [PubMed] [Google Scholar]