In this study, Sladky et al. investigated how the PIDDosome–TP53 pathway triggers hepatocyte polyploidization, a tightly controlled process that is initiated at weaning and increases with age. They show that hepatocytes accumulate centrioles during cycles of polyploidization in vivo, and that the presence of excess mature centrioles containing ANKRD26 was required to activate the PIDDosome in polyploid cells.
Keywords: ANKRD26, centrioles, distal appendage, liver, PIDDosome, polyploidy, TP53
Abstract
Hepatocyte polyploidization is a tightly controlled process that is initiated at weaning and increases with age. The proliferation of polyploid hepatocytes in vivo is restricted by the PIDDosome–P53 axis, but how this pathway is triggered remains unclear. Given that increased hepatocyte ploidy protects against malignant transformation, the evolutionary driver that sets the upper limit for hepatocyte ploidy remains unknown. Here we show that hepatocytes accumulate centrioles during cycles of polyploidization in vivo. The presence of excess mature centrioles containing ANKRD26 was required to activate the PIDDosome in polyploid cells. As a result, mice lacking centrioles in the liver or ANKRD26 exhibited increased hepatocyte ploidy. Under normal homeostatic conditions, this increase in liver ploidy did not impact organ function. However, in response to chronic liver injury, blocking centriole-mediated ploidy control leads to a massive increase in hepatocyte polyploidization, severe liver damage, and impaired liver function. These results show that hyperpolyploidization sensitizes the liver to injury, posing a trade-off for the cancer-protective effect of increased hepatocyte ploidy. Our results may have important implications for unscheduled polyploidization that frequently occurs in human patients with chronic liver disease.
The liver is one of the few mammalian tissues that tolerates polyploidy (Donne et al. 2021). Hepatocytes undergo programmed polyploidization that is initiated after weaning and gradually increases with age or in disease states. While diploid hepatocytes proliferate faster in both mice and humans (Wilkinson et al. 2018; Sladky et al. 2020a; Heinke et al. 2022), increasing or decreasing hepatocyte ploidy has no impact on basal liver function (Chen et al. 2012; Pandit et al. 2012; Miettinen et al. 2014; Dewhurst et al. 2020; Lin et al. 2020; Sladky et al. 2020b). In the context of hepatocarcinogenesis, a high degree of liver ploidy has been repeatedly shown to protect against transformation (Kent et al. 2016; Zhang et al. 2018a,b; Sladky et al. 2020a). On the other hand, highly polyploid hepatocytes have been suggested to promote tumorigenesis when undergoing ploidy-reducing cell divisions that generate chromosomally unstable progeny (Duncan et al. 2010; Matsumoto et al. 2020, 2021).
Hepatocyte polyploidization is a tightly regulated process initiated by insulin signaling, miR122, and E2F family members (Celton-Morizur et al. 2009; Chen et al. 2012; Pandit et al. 2012; Hsu et al. 2016). E2F transcription factors control a circuit that both facilitates cytokinesis failure and transiently up-regulates components of the PIDDosome to limit the proliferative capacity of polyploid hepatocytes (Chen et al. 2012; Pandit et al. 2012; Sladky et al. 2020b). The PIDDosome is a multiprotein complex, formed by the death domain-containing proteins PIDD1 and RAIDD, and serves as an activation platform for pro-Caspase-2 (Tinel and Tschopp 2004). Active Caspase-2 proteolytically inactivates the E3 ligase MDM2 to stabilize P53 and arrest the cell cycle (Oliver et al. 2011; Fava et al. 2017). In the liver, the PIDDosome has been shown to restrict polyploidy during development and following regeneration upon partial hepatectomy (Sladky et al. 2020b). How the PIDDosome is activated in polyploid hepatocytes and the physiological benefit of limiting hepatocyte ploidy remain unclear.
Polyploidization leads to the accumulation of both DNA and centrioles. Centrioles are microtubule-based structures that recruit a surrounding pericentriolar material (PCM) to form centrosomes, which organize the interphase microtubule cytoskeleton of most animal cells and form the poles of the mitotic spindle (Nigg and Holland 2018). Centrioles can also be modified to act as basal bodies, which template the formation of cilia and play central roles in cellular signaling, fluid movement, and locomotion (Breslow and Holland 2019). Centriole copy number is strictly controlled in cycling cells through once per cycle duplication. Excessive numbers of centrioles can promote chromosome missegregation that drives tumor development in mice (Coelho et al. 2015; Gönczy 2015; Serçin et al. 2016; Levine et al. 2017). G1 cells contain a younger centriole that lacks appendages and a mature centriole that is decorated with distal appendages that are required for ciliogenesis (Nigg and Holland 2018; Blanco-Ameijeiras et al. 2022). The presence of more than one mature centriole with distal appendages has been shown to trigger the activation of the PIDDosome in cultured cells to signal a P53-dependent block to cell proliferation (Holland et al. 2012; Fava et al. 2017). This signaling depends on the tight clustering of the mature parent centrioles and requires the recruitment of PIDD1 to centrioles by the distal appendage protein ANKRD26 (Burigotto et al. 2021; Evans et al. 2021). Whether centrioles and ANKRD26 act upstream to activate the PIDDosome in tissues in vivo has not been examined.
Mammalian centriole structure and function have been widely studied in cultured cells, but little is known about the cell type-specific roles of centrioles in vivo (Loncarek and Bettencourt-Dias 2018; Breslow and Holland 2019). Here, we investigate the role of centrioles in hepatocytes, a polyploid and nonciliated cell type. We show that centriole signaling is required to restrict the level of hepatocyte polyploidy and plays a critical role in maintaining liver integrity following chronic liver damage.
Results
Polyploid hepatocytes accumulate extra centrioles
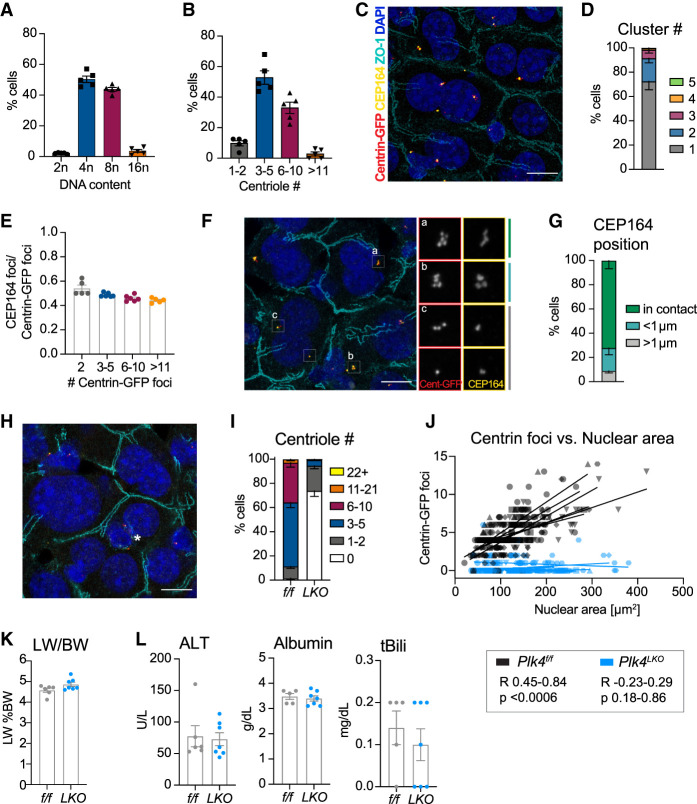
To define whether hepatocytes accumulate centrioles during polyploidization, mouse hepatocytes were isolated from 10-wk-old animals and analyzed for both ploidy and centriole content. More than 90% of hepatocytes had either a 4n or 8n DNA content, and the distribution of centriole numbers tracked closely with these ploidy states (Fig. 1A,B). To rule out artifacts from hepatocyte isolation, we analyzed centrioles in tissue sections from the livers of mice expressing Centrin1-GFP. Although hepatocyte centrioles were frequently present in a single cluster, there was an increase in the number of centriole clusters as centriole number increased (Fig. 1C,D; Supplemental Fig. S1A). Moreover, centriole number scaled with total nuclear area, a proxy for ploidy (Fig. 1J). The fraction of mature parent centrioles marked by the distal appendage protein CEP164 was consistent at ∼50%, which is the expected ratio of mature to immature centrioles. This was independent of the total number of centrioles or animal age (Fig. 1E,F; Supplemental Fig. S1B). Within each centriole cluster, CEP164-positive centrioles were usually in direct contact (63.6%) or <1 µm apart (26.3%) (Fig. 1F,G). Thus, polyploid hepatocytes fulfill the two known requirements for centriole-dependent PIDDosome activation: (1) two or more mature parent centrioles with distal appendages, and (2) centriole clustering with distal appendages in direct contact (Fava et al. 2017; Burigotto et al. 2021; Evans et al. 2021).
Figure 1.
Hepatocytes accumulate centrioles during polyploidization. (A) Flow cytometry analysis of hepatocyte ploidy distribution in Centrin-GFPtg/tg mice. (B) Centriole number in primary hepatocytes from A. N = 5 mice, n = 40–130 cells per mouse, age 10 wk. (C) Representative liver section from Plk4f/f; Centrin-GFPtg/tg mice. Scale bar, 10 µm. (D) Number of centriole clusters per cell quantified in liver sections. N = 6 mice, n = 40–65 cells per mouse. (E) The ratio of CEP164 foci over Centrin-GFP foci analyzed in the same liver sections as in D. (F) Representative image of centriole clusters showing examples of the orientation of distal appendages. Scale bar, 10 µm. (G) Quantification of the distance between the two closest positioned CEP164-stained appendages in a centriole cluster. (H) Representative liver section from a Plk4LKO; Centrin-GFPtg/tg mouse. Asterisk marks a nonhepatocyte cell containing centrioles. Scale bar, 10 µm. (I) Centriole number quantified by counting Centrin-GFP foci in sections from Plk4f/f; Centrin-GFPtg/tg (f/f) and Plk4LKO; Centrin-GFPtg/tg (LKO) livers. N = 6 mice, n = 31–65 cells per mouse. (J) Graph showing the correlation between the number of Centrin-GFP foci and nuclear area in Plk4f/f; Centrin-GFPtg/tg mice (Pearson R = 0.45–0.84, P < 0.0006) and Plk4LKO; Centrin-GFPtg/tg mice (Pearson R = −0.23–0.29, P = 0.18–0.86). Lines represent a linear regression calculated for each mouse. (K,L) Plk4f/f; Centrin-GFPtg/tg (f/f, n = 6) and Plk4LKO; Centrin-GFPtg/tg (LKO, n = 7) mice were analyzed for the liver to body weight ratio (LW/BW) (K) and serum concentrations of alanine transaminase (ALT), albumin, and total bilirubin (tBili) (L). All data are represented as mean ± SEM. Unpaired two-tailed Student's t-test was performed to determine statistical significance in K and L. Only significant results are indicated.
We next sought to determine the role of centrioles in the liver, as hepatocytes lack primary cilia and staining with α-Tubulin showed that centrioles did not function as microtubule-organizing centers in isolated hepatocytes (Supplemental Fig. S1C). To do this, we inactivated PLK4, the master regulator of centriole biogenesis, by combining a conditional Plk4 allele (Plk4f/f) with the liver-specific Alb-Cre (referred to here as Plk4LKO). Plk4LKO mice had a dramatic reduction in centriole number, with ∼70% of hepatocytes lacking centrioles altogether (Fig. 1H,I). Moreover, Plk4LKO hepatocytes lost the scaling of centriole number with nuclear size (Fig. 1J). Strikingly, the loss of centrioles had no impact on liver size, zonation, or architecture (Fig. 1K; Supplemental Fig S1D–F). Serum parameters for liver integrity (ALT and AST) or function (albumin, total bilirubin, cholesterol, and triglycerides) were also unaffected by the depletion of centrioles (Fig. 1L; Supplemental Fig. S1G). Together, these data show that although hepatocytes accumulate centrioles during polyploidization, centriole depletion has no short-term impact on liver function.
Centrioles restrict hepatocyte polyploidization
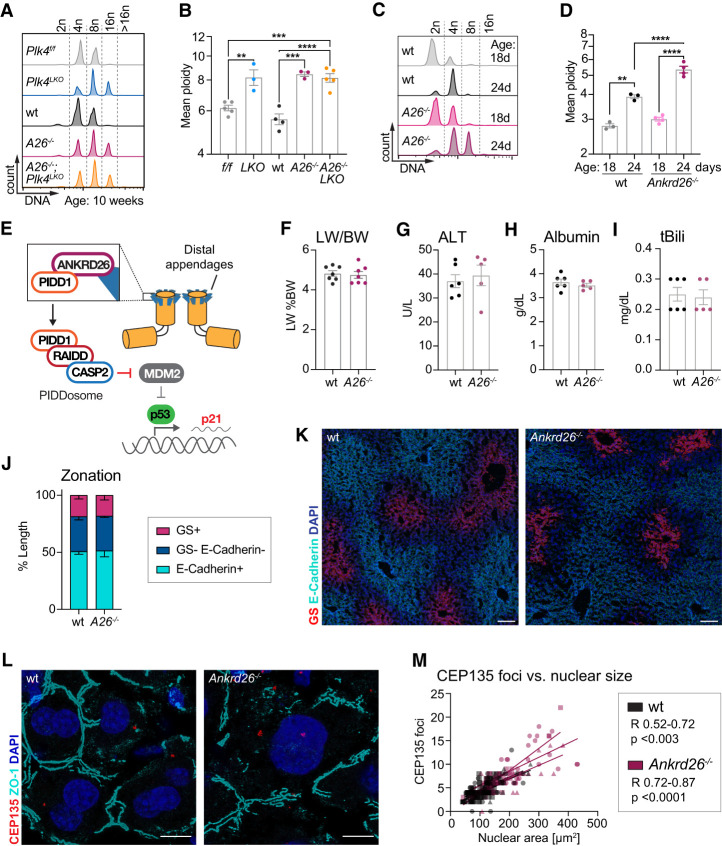
Immunofluorescence imaging and histology indicated that Plk4LKO hepatocytes were larger than in control mice (Fig. 1H; Supplemental Fig. S1F). To determine the effect of centriole loss on hepatocyte ploidy, we analyzed the ploidy distribution of isolated hepatocytes from 10-wk-old Plk4f/f and Plk4LKO mice. Loss of centrioles resulted in increased ploidy, highlighting a critical role of centrioles in restricting polyploidization in the liver (Fig 2A,B; Supplemental Fig. S3A).
Figure 2.
Centrioles and ANKRD26 limit hepatocyte polyploidy without affecting liver function and integrity. (A) Representative histograms show the DNA distribution in primary hepatocytes from 10-wk-old mice analyzed by flow cytometry. (B) Quantification of the mean ploidy as measured in A. Plk4f/f; Centrin-GFPtg/tg (f/f, N = 5), Plk4LKO; Centrin-GFPtg/tg (LKO, N = 3), and C57BL/6J (wt, N = 4) mice served as congenic controls for Ankrd26−/− (A26−/−, N = 3) and Ankrd26−/−; Plk4LKO (A26−/−; LKO, N = 5) animals. For statistical analysis, knockouts were compared with their relevant controls and with each other. (C,D) Representative histograms (C) and quantification of the mean ploidy (D) of primary wt and Ankrd26−/− hepatocytes. Analysis was performed before weaning (18 d) and after weaning (24 d) when polyploidization is induced. For wt 18 d, N = 3; 24 d, N = 3. For Ankrd26−/− 18 d, N = 4; 24 d, N = 3. (E) The presence of clustered excess mature centrioles triggers a proliferative arrest. The distal appendage protein ANKRD26 recruits PIDD1 to the centriole, which triggers PIDDosome formation and pro-Caspase 2 activation. Activated Caspase-2 cleaves MDM2 to stabilize P53, resulting in a p21-dependent cell cycle arrest. (F–I) Liver to body weight ratio (LW/BW) (F) and serum concentrations of alanine transaminase (ALT) (G), albumin (H), and total bilirubin (tBili) (I) measured for wt (N = 6) and Ankrd26−/− (N = 7) mice. (J,K) Quantification of liver zonation on wt and Ankrd26−/− liver sections immunostained for the pericentral marker glutamine synthetase (GS, red) and the periportal marker E-Cadherin (cyan), as described in Supplemental Figure S1D. n = 15–33 liver lobules analyzed in N = 3 mice per genotype. Scale bar, 100 µm. (L) Liver sections from the indicated genotypes immunostained for the centriole component CEP135 (red) and ZO-1 (cyan) to mark cell boundaries. Scale bar, 10 µm. (M) Graph showing the correlation between the number of CEP135 foci and nuclear area in wt (Pearson R = 0.52–0.72, P < 0.003) and Ankrd26−/− (Pearson R = 0.72–0.87, P > 0.0001) livers. Lines show a linear regression performed for each mouse. N = 3 mice, n = 20–60 cells per mouse. All data are shown as mean ± SEM. Statistical significance was assessed using an unpaired two-tailed Student's t-test (F–I), one-way-ANOVA with Sidak's multiple comparisons test (B,D), or two-way-ANOVA with Sidak's multiple comparisons test (J). Only significant results are indicated. (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001.
To specifically investigate the role of centriole distal appendages in controlling hepatocyte polyploidization, we generated an Ankrd26−/− mouse lacking exons 23–30, which contain the region of ANKRD26 required to bind to PIDD1 and promote PIDDosome activation in cells with extra centrioles (Supplemental Fig. S2A; Burigotto et al. 2021; Evans et al. 2021). Deletion of exons 23–30 produced a frameshifted Ankrd26 mRNA that underwent premature termination at amino acid 703 (Supplemental Fig. S2B,C). Ankrd26−/− mouse embryonic fibroblasts failed to cleave and activate pro-Caspase-2 following cytokinesis failure, confirming that the Ankrd26−/− allele cannot support PIDDosome activation to restrict further polyploidization (Supplemental Fig. S2D–F). Ankrd26−/− mice gained weight faster than their heterozygous and wild-type (wt) littermates (Supplemental Fig. S2G). This is consistent with the phenotype reported in an Ankrd26 gene trap mouse model that had impaired ciliary signaling in POMC neurons, leading to hyperphagia and obesity (Bera et al. 2008). To minimize confounding effects from differences in body weight, we used mice up to a maximum of 14 wk of age in our experiments and monitored body weight and serum cholesterol and triglyceride concentrations (Supplemental Figs. S2I,J, S4D,E, S6J).
We analyzed the ploidy distribution of isolated hepatocytes from 10-wk-old wt and Ankrd26−/− mice. Loss of ANKRD26 resulted in an increase in hepatocyte ploidy that was very similar to that observed in Plk4LKO animals (Fig. 2A,B; Supplemental Fig. S3A). This ploidy increase in Ankrd26−/− and Plk4LKO mice began after weaning-induced tetraploidization (24 d), mirroring the previously reported phenotype in mice deficient for PIDDosome components or P53 (Fig. 2C,D; Supplemental Fig. S3B; Sladky et al. 2020b). Like Plk4LKO mice, the increased ploidy in Ankrd26−/− livers had no effect on organ size, zonation, or serum parameters that reflect liver function and integrity (Fig. 2F–K; Supplemental Fig. S2H–J). Double knockout of ANKRD26 and PLK4 (Ankrd26−/−; Plk4LKO) increased the degree of polyploidy to an extent similar to that of loss of ANKRD26 or PLK4 alone, confirming that centrioles and ANKRD26 function in the same pathway to restrict ploidy (Fig. 2A,B).
Staining for the core centriole protein CEP135 revealed that centriole number scaled with nuclear size even in highly polyploid Ankrd26−/− hepatocytes (Fig. 2L,M). As in control animals, centriole clustering remained robust but decreased with centriole number (Supplemental Fig. S3C,D). In summary, these data show that centrioles and ANKRD26 restrict the proliferation of polyploid hepatocytes. Strikingly, loss of this pathway did not affect the function or integrity of the liver under normal homeostatic conditions.
ANKRD26 limits liver injury in response to chronic liver damage
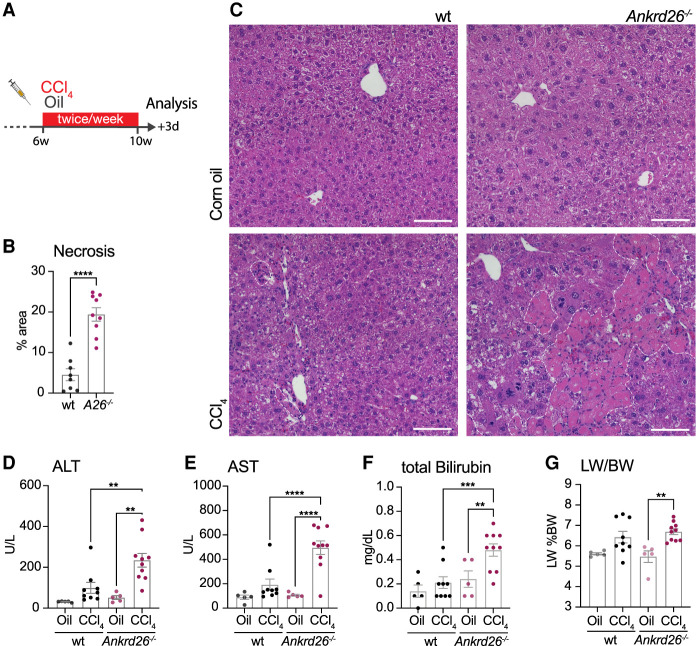
Next, we tested the relevance of centriole-mediated ploidy control in disease conditions. Previous work has shown that PIDDosome- or P53-deficient livers showed a marked increase in ploidy after recovery from a partial hepatectomy but displayed no difference in the overall regenerative capacity of the liver (Sladky et al. 2020b). In contrast to an acute form of liver injury, which induces a limited number of cycles of proliferation, chronic liver disease is characterized by repeated cycles of cell death and compensatory proliferation (Michalopoulos and Bhushan 2021). To model chronic liver injury, wt and Ankrd26−/− mice were injected with CCl4 or corn oil twice per week for 4 wk and analyzed 3 d after the last injection (Fig. 3A). Cyp2E1, which is critical for CCl4 breakdown and toxicity, was expressed equally in wt and Ankrd26−/− livers (Supplemental Fig. S4A). While wt mice experienced a modest injury following CCl4 exposure, Ankrd26−/− animals exhibited massive liver necrosis (Fig. 3B,C). The high degree of liver damage was also reflected by strongly increased ALT and AST serum concentrations and impaired liver function, as marked by elevated total bilirubin concentration (Fig. 3D–F). Serum albumin levels remained unchanged (Supplemental Fig. S4B). In accordance with the severe liver damage, mice treated with CCl4 gained less weight over the course of the treatment than corn oil-treated controls. However, after a recovery period of 4 wk, both genotypes reached the same body weight as age-matched untreated animals (Supplemental Fig. S4C). Chronic CCl4 treatment increased the liver to body weight ratio in Ankrd26−/− mice, and there was a similar trend in wt animals (Fig. 3G). Although Ankrd26−/− mice were heavier than wt mice of the same age, these mice were not obese, and serum triglyceride and cholesterol concentrations were within a normal range (Supplemental Fig. S4C–E; Ullman-Culleré and Foltz 1999).
Figure 3.
ANKRD26 protects against liver damage from chronic CCl4 treatment. (A) Carbon tetrachloride (CCl4) treatment scheme: Six-week-old male mice were injected twice per week with 5 mL/kg CCl4 for 4 wk and analyzed 3 d after the last injection. (B) Liver injury was assessed by measuring the necrotic area in H&E-stained paraffin liver sections from wt (N = 8) and Ankrd26−/− (A26−/−, N = 9) mice. (C) Representative H&E-stained liver sections from CCl4 or corn oil-treated wt and Ankrd26−/− mice. White dashed lines outline necrotic regions. (D–F) Serum concentrations of alanine transaminase (ALT) (D), aspartate transaminase (AST) (E), and total bilirubin (F) measured in corn oil-treated (Oil) or CCl4-treated wt and Ankrd26−/− mice. (G) Liver to body weight ratio (LW/BW) of the same animals. (D–G) For wt control, N = 5. For CCl4, N = 9. For Ankrd26−/− control, N = 5. For CCl4, N = 10. Data are shown as mean ± SEM. Statistical significance was assessed using an unpaired two-tailed Student's t-test (B) or one-way-ANOVA with Sidak's multiple comparisons test (D–G). Only significant results are indicated. (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001.
To further test whether the increased CCl4 sensitivity in Ankrd26−/− mice is due to increased liver ploidy, we induced chronic liver injury in Plk4LKO and control Plk4f/f mice with CCl4 (Fig. 3A; Supplemental Fig. S4F–N). Of note, while Ankrd26−/− mice were on a congenic C57BL/6J background, Plk4f/f and Plk4LKO mice were maintained on a mixed genetic background, which is known to affect the response and susceptibility to CCl4-induced damage (Shi et al. 1997; Walkin et al. 2013; Scholten et al. 2015). Indeed, Plk4f/f and Plk4LKO mouse lines showed an overall increased sensitivity to CCl4, which is in line with the twofold higher Cyp2E1 mRNA levels compared with C57BL6/J and Ankrd26−/− mice (Fig. 3B; Supplemental Fig. S4A,F,H). Increases in relative liver weight were more pronounced in CCl4-treated Plk4LKO mice compared with Plk4f/f littermates (Supplemental Fig. S4I), while serum concentrations of ALT, AST, and total bilirubin were increased similarly in Plk4f/f and Plk4LKO animals (Supplemental Fig. S4J–L). Similar to global ANKRD26 knockout, liver-specific loss of centrioles increased necrosis following chronic exposure to CCl4 (Supplemental Fig. S4F,G). Furthermore, CCl4-treated Plk4LKO animals had reduced serum albumin concentrations, indicating impaired liver function (Supplemental Fig. S4M). Together, these data show that centriole and ANKRD26-dependent signaling limits the severity of CCl4-induced chronic liver injury.
ANKRD26-mediated ploidy control restricts hepatocyte hyperpolyploidization during chronic liver injury
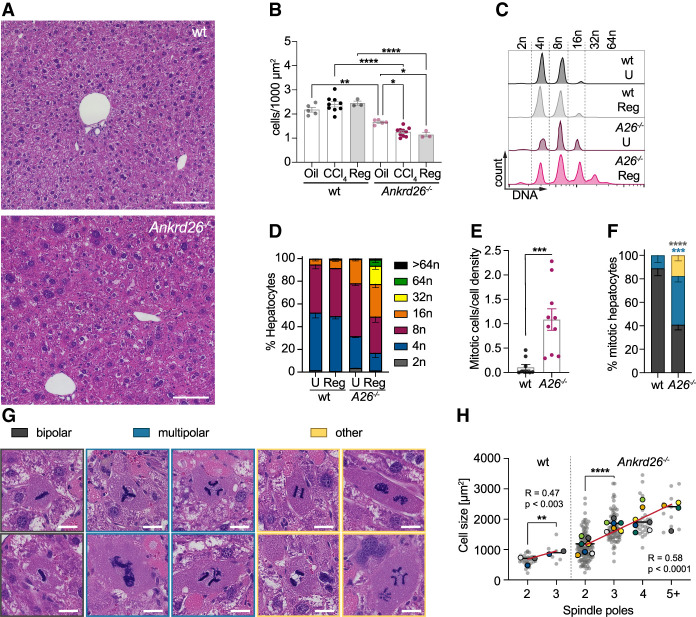
In addition to necrosis, we noticed that Ankrd26−/− hepatocytes appeared larger following chronic CCl4 treatment. We quantified the number of hepatocytes per field of view (cell density) as an inverse readout for cell size. While there was no change in the density of hepatocytes in corn oil-treated animals or CCl4-treated wt and Plk4f/f mice, Ankrd26−/− and Plk4LKO animals exhibited reduced cell density 3 d after the last CCl4 injection (Fig. 4A,B; Supplemental Fig. S5A). This level of cell density was maintained after a recovery period of 28 d (Fig. 4A,B). The comparable number of proliferating Ki67+ hepatocytes and infiltrating leukocytes in untreated and regenerated wt and Ankrd26−/− livers indicated that regeneration was complete at 4 wk (Supplemental Fig. S5B,C,F,G). Liver zonation was modestly altered with a reduced midzone in both regenerated wt and Ankrd26−/− livers (Supplemental Fig. S5D,E). To directly assess hepatocyte ploidy, we measured the DNA content in isolated hepatocytes from wt and Ankrd26−/− mice that had recovered from chronic CCl4 treatment. The degree of polyploidy in recovered wt livers was unchanged compared with age-matched untreated controls. In contrast, loss of ANKRD26 led to an increase in liver ploidy that was exacerbated by chronic CCl4 treatment, resulting in massive hyperpolyploidization with ∼5% of hepatocytes ≥64n (Fig. 4C,D; Supplemental Fig. S5H). Remarkably, despite the high polyploidy in recovered Ankrd26−/− livers, these animals showed no signs of impaired liver function or integrity as measured by serum concentrations of ALT, AST, or total bilirubin (Supplemental Fig. S6G–I).
Figure 4.
ANKRD26 restricts hepatocyte polyploidy during acute and chronic CCl4 exposure. (A) H&E-stained liver sections from regenerated wt and Ankrd26−/− mice 28 d after the last CCl4 injection. Scale bar, 100 µm. (B) Cell density was assessed by hepatocyte count on H&E-stained liver sections. Mice were sacrificed 3 d (CCl4) or 28 d (Reg) after 4 wk of chronic CCl4 exposure, or 3 d after chronic corn oil treatment (Oil). For wt, oil, N = 5; CCl4, N = 9; Reg, N = 3 mice. For Ankrd26−/−, oil, N = 5; CCl4, N = 10; Reg, N = 3 mice. (C) Flow cytometry analysis of the DNA content of hepatocytes isolated from mice of the indicated genotypes after 28 d of regeneration from chronic CCl4 treatment (Reg) or untreated age-matched controls (U). (D) Quantification of the ploidy distribution described in C for wt and Ankrd26−/− (A26−/−). N = 3 mice per condition and genotype. (E) The number of mitotic hepatocytes analyzed 3 d after 4 wk of chronic CCl4 treatment. Mitotic hepatocytes were counted per area and normalized to the cell density of the respective sample. For wt, N = 9. For Ankrd26−/−, N = 10. (F) Mitotic hepatocytes quantified in E were grouped based on mitosis morphology, with examples shown in G. (Gray) Bipolar, (blue) multipolar, (yellow) other. “Other” includes hepatocytes with multiple spindles per cell, lagging chromosomes, and/or DNA bridges. Scale bar, 10 µm. (H) Graph showing the correlation between the size of mitotic hepatocytes and the number of spindle poles for each genotype. For wt, Pearson R = 0.47, P < 0.003. For Ankrd26−/−, Pearson R = 0.58, P < 0.0001. Gray dots show individual cells, and colored dots represent the average of different animals with the means shown as a line. The red line shows a linear regression. Statistical analysis was performed on all cells. For wt, N = 9 mice, n = 39 cells. For Ankrd26−/−, N = 10, n = 177. Data in B–F are represented as mean ± SEM. Statistical significance was assessed using an unpaired two-tailed Student's t-test (E,H, wt), one-way-ANOVA (B,H, Ankrd26−/−), or two-way-ANOVA (F) with Sidak's multiple comparisons test. Only significant results are indicated. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001.
To determine how ANKRD26 deficiency leads to hyperpolyploidization following chronic CCl4 treatment, we examined histological sections. Ankrd26−/− and Plk4LKO livers had an increased number of mitotic figures at 3 d after the last CCl4 treatment (Fig. 4E; Supplemental Fig. S5I). This is in accordance with the higher degree of liver injury and indicates ongoing regeneration. Interestingly, ∼60% of mitoses in Ankrd26−/− hepatocytes showed abnormalities, including multipolar spindles, multiple spindles in a single cell, lagging chromosomes, and DNA bridges (Fig. 4F,G). The number of spindle poles increased in larger hepatocytes of both genotypes, suggesting that centriole clustering efficiency decreases with increased mitotic cell size (Fig. 4H). This explains the prevalence of large hepatocytes with more than two or multilobed nuclei observed in CCl4-treated Ankrd26−/− livers (Fig. 4A,G; Supplemental Fig. S5J). Therefore, the elevated centriole number and DNA content in highly polyploid hepatocytes increases the risk for mitotic errors and thus further polyploidization.
Hyperpolyploidy sensitizes the liver to injury
Ankrd26−/− livers have normal morphology and function and thus represent a valuable model to study the physiological consequence of increases in hepatocyte ploidy. To examine the effect of ploidy on the susceptibility to CCl4-induced cell death, isolated hepatocytes were treated with CCl4 in vitro and cell death was monitored by live imaging. A cell-permeable DNA dye was used to measure nuclear intensity as a proxy for hepatocyte ploidy (Supplemental Fig. S6A). To separate the effects of increased ploidy from ANKRD26 function per se, we examined 35-wk-old wt mice with a ploidy distribution similar to that of 14-wk-old Ankrd26−/− animals (Supplemental Fig. S6D–F). Higher ploidy states did not render hepatocytes more sensitive to CCl4-induced cell death, suggesting that this is unlikely to cause the increased injury in Ankrd26−/− livers (Supplemental Fig. S6B,C).
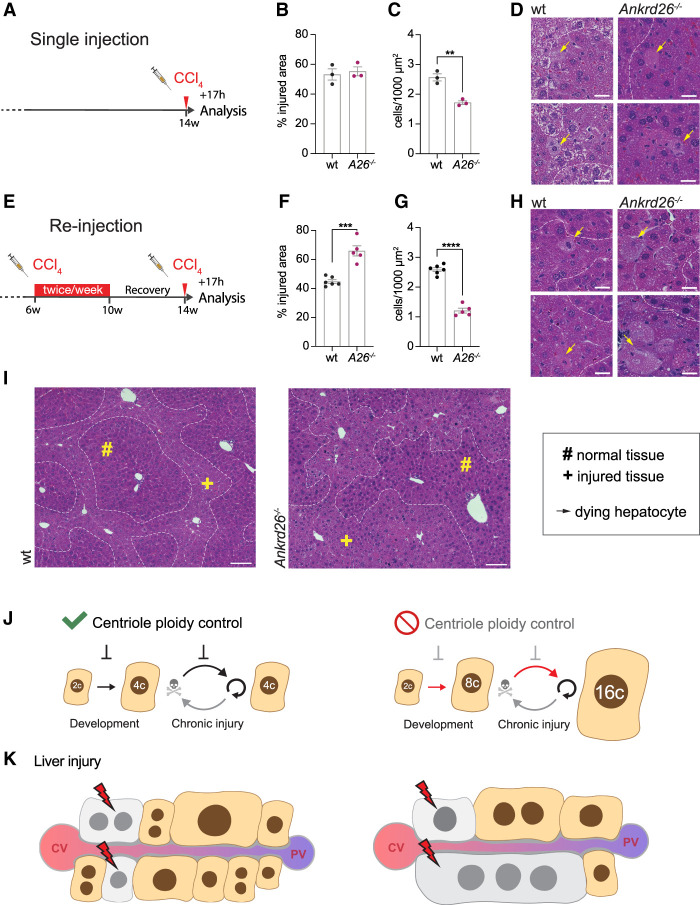
To assess the effect of polyploidy on the susceptibility to CCl4 injury in vivo, we analyzed the acute effect of CCl4 in wt and Ankrd26−/− hepatocytes of different ploidy degrees. First, 14-wk-old mice were injected with a single dose of CCl4 and analyzed 17 h later (Fig. 5A). Despite the overall higher ploidy levels in Ankrd26−/− hepatocytes, there was no difference in the injured area in wt and Ankrd26−/− livers (Figs. 4B–D, 5B–D), although serum ALT and AST concentrations were more highly elevated in Ankrd26−/− mice after a single CCl4 injection (Supplemental Fig. S6G). To generate hyperpolyploid hepatocytes, we performed 4 wk of chronic CCl4 treatment in wt and Ankrd26−/− mice and allowed 4 wk for the livers to fully recover. Reinjection with a single CCl4 dose resulted in significantly higher liver injury in Ankrd26−/− animals (Fig. 5E,F). Ankrd26−/− hepatocytes in the affected region were, on average, more than twice the size of wt hepatocytes (Fig. 5G–I). Thus, the death of the same number of hepatocytes generates a substantially larger damaged area in Ankrd26−/− livers. Serum ALT, AST, and total bilirubin were elevated in reinjected Ankrd26−/− animals, indicating compromised liver function (Supplemental Fig. S6H,I).
Figure 5.
Massive ploidy increases sensitize the liver to injury. (A) Treatment scheme for single injection: Fourteen-week-old male mice were injected with a single dose of 5 mL/kg CCl4 and analyzed after 17 h. (B,C) The injured area (B) and cell density in this region (C) were quantified in H&E-stained wt and Ankrd26−/− (A26−/−) liver sections. N = 3 mice per genotype. (D) Representative close-ups of H&E-stained liver sections show healthy and affected liver tissue separated by dashed lines. Yellow arrows mark dying hepatocytes. Scale bar, 10 µm. (E) Treatment scheme reinjection: Six-week-old male mice were injected biweekly with 5 mL/kg CCl4 for 4 wk. Following 28 d of recovery, the mice were reinjected with a single dose of 5 mL/kg CCl4 and the livers were harvested after 17 h. (F,G) Quantification of the affected area (F) and the cell density within the injured region (G) performed on H&E-stained liver sections also shown in H and I. For wt, N = 6. For Ankrd26−/−, N = 5. (H) Close-ups of wt and Ankrd26−/− livers. Dashed lines separate normal and injured tissue, and yellow arrows indicate dying hepatocytes. Scale bar, 10 µm. (I) Tissue overview of wt and Ankrd26−/− livers treated as shown in E. Scale bar, 100 µm. (#) Intact tissue, (+) affected tissue. (J,K) Model summarizing how centriole-directed ploidy control maintains a low degree of polyploidy during development and chronic injury cycles. The absence of this pathway results in hyperpolyploidy over multiple cycles of compensatory proliferation. (K) Additional injury targeting the same number of cells impacts the hyperpolyploid liver to a greater extent. (CV) Central vein, (PV) portal vein. All data are shown as mean ± S.E.M and statistical significance was determined using an unpaired two-tailed Student's t-test. Only significant results are indicated. (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001.
Taken together, these results suggest that low- and high-ploidy hepatocytes have a similar intrinsic sensitivity to CCl4-induced cell death. However, hyperpolyploid hepatocytes are larger, and thus their death results in the loss of a larger fraction of functional parenchyma (Fig. 5J,K). This reveals that centriole-mediated ploidy control has a critical role in limiting hepatocyte hyperpolyploidization and improving liver robustness.
Discussion
Here, we show that centrioles accumulate during hepatocyte polyploidization but do not function in their canonical roles as organizers of the centrosome or cilium. Instead, in hepatocytes, centrioles serve as signaling platforms that control polyploidization and ensure organ robustness in response to chronic liver damage.
Despite a clear role in limiting hepatocyte polyploidization, the trigger for PIDDosome activation in the liver was unknown. Here, we demonstrate that the centriole distal appendage protein ANKRD26 is essential to limit polyploidy both in juvenile development and during regeneration from CCl4-driven chronic liver injury. We show that polyploid hepatocytes fulfill all the known requirements for ANKRD26-PIDDosome activation, including the presence of two or more centrioles with distal appendages and centriole clustering with distal appendages in direct contact (Fig. 1A–G) (Fava et al. 2017; Burigotto et al. 2021; Evans et al. 2021). In addition, loss of centrioles (Plk4LKO), ANKRD26 deficiency, or both (Ankrd26−/−; Plk4LKO) phenocopy the increased polyploidy observed in mice lacking PIDDosome components, P53, or P21 (Sheahan et al. 2004; Kurinna et al. 2013; Fava et al. 2017; Sladky et al. 2020b). This strongly suggests that centriole-directed PIDDosome activation restricts hepatocyte polyploidization. Further supporting these findings, a recent study showed that P53-deficient mice generate giant hepatocytes following chronic CCl4 treatment, resembling our findings in chronically injured centriole-depleted (Plk4LKO) or ANKRD26 knockout livers (Figs. 3–5; Supplemental Figs. S4, S5; Humpton et al. 2022).
Like ANKRD26 knockout mice, liver-specific centriole depletion increased the degree of liver injury and impaired liver function following chronic CCl4 treatment (Supplemental Fig. S4). This suggests that the increased liver damage upon CCl4 exposure is due to the loss of ANKRD26-based centriole signaling in the liver. The only known function of ANKRD26 outside of PIDDosome activation is its role in ciliary gating (Bera et al. 2008; Yan et al. 2020). Since hepatocytes do not produce cilia, loss of ANKRD26 offers a convenient means to study the effects of liver hyperpolyploidy without impacting the cell cycle machinery or E2F transcriptional circuits (Chen et al. 2012; Dewhurst et al. 2020; Lin et al. 2020). In addition, Ankrd26−/− mice possess normal centriole numbers, and additional functional roles of the PIDDosome in the liver remain intact (Kim et al. 2018, 2021).
Our results show that the “unleashed” cycling of polyploid Ankrd26−/− hepatocytes results in self-propagating polyploidy through the formation of unresolved multipolar mitotic spindles. Similar observations were made in polyploid Drosophila neuroblasts and human cancer cells, where the high DNA content forms a physical barrier to the coalescence of multiple poles into a pseudobipolar spindle for proper chromosome segregation (Goupil et al. 2020). Consequently, these cells undergo mitotic slippage or fail to complete cytokinesis, resulting in further polyploidization. It has been proposed that the in vivo tissue context facilitates centrosome clustering in the liver, and consequently, most multipolar spindles coalesce into a pseudobipolar spindle before chromosome segregation in anaphase (Knouse et al. 2018). Although our results on chronically injured wt livers support these findings (Fig. 4H), ANKRD26-deficient hyperpolyploid hepatocytes showed frequent multipolar mitotic spindles and hepatocytes with three or more nuclei (Fig. 4F–H; Supplemental Figs. S4G, S5J). This suggests that there is a specific centriole and ploidy threshold beyond which centriole clustering is inefficient and successful cell division becomes unlikely (Supplemental Figs. S1A, S3C,D). In patient-derived liver organoids, CRISPR–Cas9-mediated knockout of P53 resulted in a high frequency of multipolar mitotic spindles and increased polyploidy, corroborating the importance of the centriole–P53 axis in humans (Artegiani et al. 2020).
The physiological and pathological consequences of hepatocyte polyploidy are not fully understood. E2f7/8−/− livers that are largely diploid develop spontaneous liver cancer at a young age and are less proficient in adaption to tyrosinemia (Kent et al. 2016; Wilkinson et al. 2019). In contrast, increased polyploidy was shown to confer a cancer-protective effect without impairing liver function (Zhang et al. 2018a; Lin et al. 2020). In line with these studies, Plk4LKO mice and ANKRD26-deficient animals with increased ploidy exhibit normal liver function (Figs. 1L, 2H,I). This raises the question of why the extent of hepatocyte ploidy is limited if it offers a tumor protective function. Our data show that centriole–ANKRD26 signaling dampens hepatocyte polyploidization to ensure stable, modest liver polyploidy in normal conditions and in response to chronic injury. Loss of centriole signaling exacerbated hepatic polyploidy following chronic liver damage. The resultant hyperpolyploid liver was more vulnerable to CCl4-induced damage, as the loss of an equal number of cells generates more damage when the cells are larger (Fig. 5J,K). This effect can be exacerbated by injury-induced inflammation, which further promotes hepatocyte cell death (Huby 2021; Tiegs 2022). However, hyperpolyploid Ankrd26−/− livers did not show signs of increased inflammation (Supplemental Fig. S5F,G), as has been reported when ploidy was increased by liver-specific knockout of CDK1 (Dewhurst et al. 2020). Together, our findings show that a liver comprised of larger, hyperpolyploid cells is more vulnerable to liver damage, posing a trade-off to the protection that polyploidy offers from malignant transformation (Kent et al. 2016; Zhang et al. 2018a,b; Sladky et al. 2020a).
A previous study used a transient knockdown of Anilin to generate livers with increased ploidy but did not observe differences in CCl4-induced injury (Zhang et al. 2018b; Lin et al. 2020). While Anilin depletion increases baseline polyploidy in the liver, it leaves centriole-mediated ploidy control intact, thus preventing hepatocyte hyperpolyploidization. Our data suggest that the severity of liver injury increases with the degree of polyploidy, and this is most apparent in a hyperpolyploid liver with an average DNA content of ≥12n–16n per hepatocyte.
Our findings may have important implications for patients suffering from ploidy-modulating chronic liver disease. This includes two of the main etiologies underlying hepatocellular carcinoma, nonalcoholic fatty liver disease (NAFLD), and hepatitis B virus infection, where ploidy has been shown to increase during disease progression (Toyoda et al. 2005; Gentric et al. 2015; Younossi et al. 2018; Paik et al. 2020; Donne et al. 2021; Sladky et al. 2021). Although chronic liver diseases can be managed clinically (Powell et al. 2021), increases in polyploidy sustained from chronic injury might sensitize the organ to future insults. With the global burden of chronic liver disease rising, it will be important to define the effect of increased polyploidy on organ robustness in patients.
Materials and methods
Mice
Plk4f/f and Ankrd26−/− mice were generated using CRISPR/Cas9. A mix of 30 ng/µL Cas9 protein (PNABio), 0.6 μM tracrRNA (Dharmacon), 0.6 μM crRNA (IDT), and 10 ng/µL ssDNA oligonucleotide (IDT) was diluted in RNase-free injection buffer (10 mM Tris-HCl at pH 7.4, 0.25 mM EDTA) and used for pronuclear injection of one-cell embryos. These embryos were transplanted into pseudopregnant ICR females. Injections and transplantation were performed by the Johns Hopkins University Transgenic Core. sgRNAs were chosen based on the location of the available PAM sites and a minimal number of predicted off-target sites (http://crispor.tefor.net).
To generate Ankrd26−/− mice, two sgRNAs and a DNA repair template were coinjected into C57Bl/6J embryos to delete exons 23–30. This led to a frameshift terminating the protein at amino acid 703. The following sgRNAs and repair template were used: 5′ sgRNA (5′-GCCACACATCCAGGGTCGAG-3′) and 3′ sgRNA (5′-GAAAGCTGTGGTATTCACGC-3′). DNA repair template: 5′-ATGTATGGTGATGTATGCCACTTAGCTCAGCATTGCCCCTCGGATCCCGCAGGGGACCTGGCAGACCCGCCTCTTCCCTGTGTTGCTG-3′.
Primers used for genotyping and sequencing to characterize the offspring were as follows: Ankrd26 wt fw (5′-CACCCACACACACACTGGAT-3′), Ankrd26 wt rev (5′-CACCCACACACACACTGGAT-3′), Ankrd26 KO fw (5′-AAAGTGCCTCCTTCTGCTTG-3′), and Ankrd26 KO rev (5′-CCCCAAAATCTCAGGAATGA-3′).
Pathological assessment by a board-certified veterinary pathologist (Johns Hopkins University Phenotyping and Pathology Core) revealed no obvious abnormalities, aside from the obesity at older age, previously reported for Ankrd26 gene trap animals (Bera et al. 2008). Ankrd26−/− mice were maintained on a C57BL/6J background.
To generate Plk4f/f mice, B6SJL/F2 embryos were coinjected with two sgRNAs and a ssDNA repair template spanning exon 5. The repair template introduced loxP sites into introns 4 and 5 of Plk4 and had 72- or 100-bp homology arms. Primers were as follows: Plk4 sgRNA1 (5′-AAGCTAGGACTTTAATACTC-3′) and Plk4 sgRNA2 (5′-CTGCATGTAGAGGGAAGCTG-3′). Primers used for genotyping and allele characterization were as follows: 976 Plk4_F (5′-TCTTGAGGGGAATTAGATAGCA-3′), 979 Plk4_R (loxP) (5′-CTCACTCAGCCCCAGATAAC-3′), and 981 Plk4_R (5′-TGCAATATGCCATGAGAATGA-3′).
Plk4f/f and Ankrd26−/− mice are available from the Jackson Laboratory (Plk4f/f strain 037549, RRID:IMSR_JAX:037549 and Ankrd26−/− strain 037529, RRID:IMSR_JAX:037529).
Plk4LKO mice were generated by crossing Plk4f/f animals with EGFP-Centrin1 mice (the Jackson Laboratory strain 029363; RRID:IMSR_JAX:029363) and albumin-Cre mice (the Jackson Laboratory strain 003574; RRID:IMSR_JAX:003574) (Postic et al. 1999; Hirai et al. 2016). EGFP-Centrin1 mice were previously crossed to a deleter strain to achieve constitutive transgene expression. Plk4f/f; EGFP-Centrin1tg/tg; AlbCretg mice were maintained on a mixed SJL/C57BL/6J background. AlbCre-negative littermate controls were used for all experiments with this mouse line. Ankrd26−/−; Plk4f/f; AlbCretg mice were kept on a mixed SJL/C57BL/6J background. Both Plk4f/f; EGFP-Centrin1tg/tg and C57BL/6J were included to serve as controls for this line.
Mice were housed under standard conditions in an AAALAC-accredited facility. All animal experiments were approved by the Johns Hopkins University Institute Animal Care and Use Committee (MO21M300). For experiments involving CCl4 treatment, only male animals were used. In all other experiments, age-matched mice were used in a sex-balanced manner.
CCl4 treatment
To induce liver damage, mice were intraperitoneally injected with CCl4 (carbon tetrachloride; Sigma-Aldrich 270652) diluted 1:4 (v/v) in corn oil (Sigma-Aldrich C8267) at a dose of 5 mL/kg. For chronic liver injury, 6-wk-old mice were injected twice per week for 4 wk and analyzed 3 or 28 d later. Corn oil injections served as a control. Mice injected with a single dose of 5 mL/kg CCl4 were harvested after 17 h to assess the acute injury phase. All animals were closely monitored during the treatment period. Only male mice were used for experiments with CCl4.
Generation and culture of mouse embryonic fibroblasts
Embryos were isolated at E12.5–E14.5 and the embryo bodies were digested overnight at 4°C in 0.05% Trypsin-EDTA (Gibco 25300062). Following a 5-min incubation at 37°C, MEF cells were dissociated by pipetting. MEFs were cultured in DMEM (Corning 10-017-CV) containing 10% FBS (Corning 35-010-CV), penicillin, streptomycin (Gibco 10378016), and 0.1 mM β-mercaptoethanol at 37°C in 5% CO2 and 3% O2 atmosphere. To induce cytokinesis failure, MEFs were treated with DMSO (control) or 10 µg/mL Cytochalasin B for 48 h.
Serum parameters
All serum parameters were measured by the Johns Hopkins University Phenotyping and Pathology Core using a DIASYS respons910 automated clinical chemistry analyzer.
Histology
Histological assessment was performed on 4-µm H&E-stained sections of paraffin-embedded liver tissue. The sections were imaged using a Zeiss Axiovert.Z1 slide scanner (Zeiss).
Hepatocyte isolation
Primary mouse hepatocytes were isolated by a retrograde two-step collagen perfusion as described previously (Grompe et al. 1992; Sladky et al. 2020b). Briefly, the animals were anesthetized (avertin, i.p.), and the vena cava was canulated. Following an incision in the portal vein, the vena cava was constricted below the heart for perfusion of the liver at 37°C with 70 mL of EGTA buffer (0.14 M NaCl, 6.7 mM KCl, 10 mM HEPES, 0.1 mM EGTA at pH 7.4) and 40 mL of liberase TL solution (66.7 mM NaCl, 6.7 mM KCl, 4.7 mM CaCl2*2H2O, 10 mM HEPES, at pH 7.45, 25 mg/mL Liberase TL [Roche 05401020001]). Hepatocytes were released by disrupting the liver in DMEM (Corning 10-017-CV) containing 10% FCS, 2 mM L-glutamine, penicillin, and streptomycin (Gibco 10378016). Viable hepatocytes were enriched by low-speed centrifugations (three times at 30g for 3 min) in the same medium. After assessing purity and quality by light microscopy, hepatocytes were fixed in 70% ice-cold ethanol for flow cytometry. Additionally, the cells were allowed to adhere for 3.5 h onto collagen-coated coverslips or tissue culture dishes (0.36 mg/mL rat tail collagen; Corning 3542360).
Flow cytometry
Ethanol-fixed cells were washed twice in 1× PBS and stained with 40 µL/mL propidium iodide in the presence of RNase A (Sigma-Aldrich R6148) in PBS for 15 min at room temperature. The DNA content was measured on a BD FACSCalibur flow cytometer (BD Biosystems) and analyzed using FlowJo (v10.7.2, BD Biosystems).
Hepatocyte live imaging
Freshly isolated hepatocytes were seeded in collagen-coated 12-well plates in DMEM supplemented with 10% FCS, 2 mM L-glutamine, penicillin, and streptomycin. After 3.5 h, when hepatocytes were adherent, the medium was replaced with William's E medium (Gibco 12551032) containing 2 mM L-glutamine, penicillin, and streptomycin. Cells were treated with 4 mM CCl4 (diluted in DMSO) or DMSO as control, and DNA was stained using the cell-permeable dye NucSpot Live 488 (Biontium 40081). The cells were imaged every 1.5 h for 24 h using a CellCyte X live-imaging system (Cytena, 10× objective, 0.345 µm/pixel, HeartOS v5.0.0 software) in a tissue culture incubator at 37°C with 5% CO2.
Immunofluorescence
Fresh tissue was embedded and frozen in TissueTek O.C.T. compound (Sakura Finetek). Twenty-micrometer sections were cut on a Leica CM1950 cryostat and collected on Superfrost Plus microscope slides (Thermo Fisher Scientific). Hepatocytes on collagen-coated coverslips or sectioned tissues were fixed for 4 min each in 1.5% paraformaldehyde (Electron Microscopy Sciences 15714) and in −20°C cold methanol. The samples were blocked in 2.5% FBS, 200 mM glycine, and 0.1% Triton X-100 in PBS for 1 h and incubated with the respective primary antibodies in the same buffer. After washing three times with PBS containing 0.5% TritonX-100, the samples were incubated with the secondary antibodies (Invitrogen) and DAPI. Samples were washed three times and mounted in Prolong Gold antifade (Life Technologies P36930). Primary antibodies used were CEP164 (rabbit polyclonal; 1:500; EMD Millipore Corp. ABE2621), ZO-1 (rat monoclonal antibody; 1:500; eBioscience 14-9776-82), α-Tubulin (rat monoclonal antibody [YL1/2]; 1:1000; Invitrogen MA1-80017), CEP135 (rabbit polyclonal; 1:500; Alexa555-conjugated; homemade), CD45.2 (mouse monoclonal, 106 [RUO]; 1:1000; FITC-conjugated; BD Biosciences 561874), glutamine synthetase (rabbit polyclonal; 1:1000; Abcam ab49873), and E-Cadherin (goat polyclonal; 1:500; R&D Systems AF7480).
Images of tissue sections were acquired on an SP8 confocal microscope (Leica Microsystems) using a Leica 40× 1.30 NA oil objective at 0.2-μm z-sections. For imaging of liver zonation and CD45+ cells, a Leica 20× 0.75 NA oil objective at 1-µm z-sections was used. Adherent hepatocytes were imaged using a DeltaVision Elite system (GE Healthcare) with an Olympus 60× 1.42 NA oil objective at 0.2-µm z-sections.
Image analysis
All imaging analysis was performed blinded using ImageJ (v2.1.0/1.53c, National Institutes of Health, http://imagej.net).
Histology
Cell density was measured by counting the number of hepatocytes per area. Each region selected for counting spanned all three liver zones. Three to five regions with 200–250 cells each were counted per mouse. In the CCl4 condition in Figure 4B, cell density analysis was performed excluding necrotic regions. For Figure 5, B and E, cell density was assessed in the injured area only. Necrosis or injury was analyzed relative to the total area, excluding blood vessels. For every mouse, mitotic figures were analyzed in 18 frames of 250,000 µm2 that covered all liver zones.
Immunofluorescence
For tissue sections, immunofluorescence images were lightning-processed using LAS X software (Leica, v3.5.6.21594). Centriole marker quantification was performed on unprojected z-stacks (16 bit). Centrioles were considered to be in one cluster if the largest distance between them was <2.5 µm. Nuclear area was measured in the image showing the largest cross-section. Hepatocytes were included in the analysis only if the top and bottom of the cell lay within the z-stack. Liver zonation was assessed on lightning-processed, maximum intensity-projected images. In a liver lobule, the GS+, GS−; E-Cadherin−, and E-Cadherin+ fractions between the central vein and portal vein were measured, as shown in Supplemental Figure S1D. Centriole number in isolated hepatocytes was assessed on deconvolved 2D maximum intensity projections (16 bit).
Live imaging analysis
DNA content was measured by subtracting the background signal from nuclear intensity at time point 0 h. Specifically, signal intensity was determined by drawing a circular region of interest (ROIS) around all nuclei of a cell, and a larger circular ROI (ROIL) around the ROIS. The signal in ROIS was calculated using the formula IS − [(IL − IS)/(AL − AS) × AS], where A is area and I is integrated intensity. To correct for variations in DNA dye distribution, the nuclear intensity was normalized to the median value of each frame. Cell death was analyzed using phase-contrast images and defined as membrane blebbing or detachment. Only cells that were healthy and fully attached at 0 h were included in the analysis.
Protein extraction and immunoblotting
Cell pellets were homogenized in RIPA lysis buffer (150 mM NaCl, 50 mM Tris, 1% NP-40, 0.5% sodium deoxycholate 1% SDS, one tablet EDTA-free protease inhibitors), and protein content was determined using a Pierce BCA protein assay (Thermo Fisher Scientific 23227). Protein (50–100 µg) was separated by SDS-PAGE and transferred to a 0.45-µm nitrocellulose membrane (Santa Cruz Biotechnology sc-3724) using a wet transfer system (Bio-Rad). The membranes were probed for CASP2 (rat monoclonal [11B4]; 1:500; EMD Millipore MAB3507) and HSP90 (mouse monoclonal; 1:2000; Santa Cruz Biotechnology sc-13119), and HRP-conjugated secondary antibodies were used for detection (antirat IgG; 1:5000; Cell Signaling Technologies 7077; and antimouse IgG; 1:5000; Cell Signaling Technologies 7076).
RNA extraction and quantitative real time PCR
Total RNA was extracted from homogenized liver tissue using TRIzol reagent (Thermo Fisher Scientific 15596026). SuperScript IV reverse transcriptase (Thermo Fisher Scientific 18090050) was used to generate cDNA with oligo-dT primers. Quantitative real-time PCR was performed in technical triplicates using Ssoadvanced Universal SYBR (Bio-Rad 1725271) on a QuantStudio 6 Flex real-time PCR system (Thermo Fisher Scientific) and analyzed with the QuantStudio real-time PCR software.
Relative expression levels were calculated using the 2−ΔΔCt method normalized to GAPDH. The following primers were used to characterize the Ankrd26 mRNA in Supplemental Figure S2B: fragment i (Ankrd26_Nt-F: 5′-CCTTGGAAAAAGTGGCGTGA-3′ and Ankrd26_Nt-R: 5′-ATCACATTTGGATCGGCCCC-3′), fragment ii (Ankrd26_qPCR-F: 5′-TTCCGAGAGCATTTTGTGTG-3′ and Ankrd26_wt-R: 5′-CCATCCGACCTTTTCATGTT-3′), fragment iii (Ankrd26_del-F: 5′-GTACGAGAACGAGAAAGCCGA-3′ and Ankrd26_del-R: 5′-GTGTACAGCCTCTCTGTGTTGA-3′), fragment iv (Ankrd26_qPCR-F: 5′-TTCCGAGAGCATTTTGTGTG-3′ and Ankrd26_KO-R: 5′-TGGGATATCCATTTCAAGGTG-3′), and fragment v (Ankrd26_Ct-F: 5′-AAAGCAATTGGAGCAGGAGGT-3′ and Ankrd26_Ct-R: 5′-TCTGACTTCTAACCGAAGCGT-3′). The following primers were used to determine the expression of GAPDH and Cyp2E1 mRNAs: mGADPH Fwd: 5′-AATGTGTCCGTCGTGGATCTGA-3′, mGADPH Rev: 5′-TGGGAGTTGCTGTTGAAGTCG-3′, Cyp2E1 Fwd: 5′-TTTCTGCAGGAAAGCGCG-3′, and Cyp2E1 Rev: 5′-CTGCCAAAGCCAATTGTAACAG-3′ (Wilkinson et al. 2018). Fragment iv was PCR-amplified, purified, and characterized by Sanger sequencing using the same primers (Supplemental Fig. S2C).
Statistical analysis
Statistical analysis was performed using GraphPad Prism (v9.0.0, GraphPad software, LLC). Two-tailed, unpaired Student's t-test was used for comparison of two groups. One-way ANOVA or two-way ANOVA with Sidak's multiple comparisons test was used for three or more groups. For correlations, Pearson's correlation coefficients were calculated. The significance levels and tests performed are stated in the figure legends. Only statistically significant results are indicated in the graphs. “N” represents the number of animals and “n” refers to the number of cells analyzed per mouse. Each mouse was considered as a biological replicate.
Data availability
Requests for data and materials should be addressed to A.J.H. (aholland@jhmi.edu).
Supplementary Material
Acknowledgments
We thank Robert Anders for advice and critical suggestions, and Andreas Villunger and Oliver Schmidt for fruitful discussions. Figures were in part created using Biorender.com. A.J.H. acknowledges funding by the National Institutes of Health (R01 GM133897, R01 GM114119, and R01 CA266199). V.C.S received an EMBO postdoctoral fellowship (ALTF 1194-2020).
Author contributions: V.C.S., and A.J.H conceived the study. V.C.S., H.A., D.T.-G., L.T.E., C.G.D., M.A.S., and G.M.L. acquired the data. V.C.S., and T.L. analyzed the data. A.J.H., and V.C.S. acquired the funding. AJ.H. was the project administrator. A.J.H. supervised the study V.C.S., and A.J.H. wrote the original draft of the manuscript. A.J.H., V.C.S., T.L., M.A.S., and G.M.L. reviewed and edited the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.349727.122.
Competing interest statement
The authors declare no competing interests.
References
- Artegiani B, Hendriks D, Beumer J, Kok R, Zheng X, Joore I, Chuva de Sousa Lopes S, van Zon J, Tans S, Clevers H. 2020. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR–Cas9 precision genome editing. Nat Cell Biol 22: 321–331. 10.1038/s41556-020-0472-5 [DOI] [PubMed] [Google Scholar]
- Bera TK, Liu XF, Yamada M, Gavrilova O, Mezey E, Tessarollo L, Anver M, Hahn Y, Lee B, Pastan I. 2008. A model for obesity and gigantism due to disruption of the Ankrd26 gene. Proc Natl Acad Sci 105: 270–275. 10.1073/pnas.0710978105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Ameijeiras J, Lozano-Fernández P, Martí E. 2022. Centrosome maturation – in tune with the cell cycle. J Cell Sci 135: jcs259395. 10.1242/jcs.259395 [DOI] [PubMed] [Google Scholar]
- Breslow DK, Holland AJ. 2019. Mechanism and regulation of centriole and cilium biogenesis. Annu Rev Biochem 88: 691–724. 10.1146/annurev-biochem-013118-111153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burigotto M, Mattivi A, Migliorati D, Magnani G, Valentini C, Roccuzzo M, Offterdinger M, Pizzato M, Schmidt A, Villunger A, et al. 2021. Centriolar distal appendages activate the centrosome-PIDDosome-p53 signalling axis via ANKRD26. EMBO J 40: e104844. 10.15252/embj.2020104844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celton-Morizur S, Merlen G, Couton D, Margall-Ducos G, Desdouets C. 2009. The insulin/Akt pathway controls a specific cell division program that leads to generation of binucleated tetraploid liver cells in rodents. J Clin Invest 119: 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HZ, Ouseph MM, Li J, Pécot T, Chokshi V, Kent L, Bae S, Byrne M, Duran C, Comstock G, et al. 2012. Canonical and atypical E2Fs regulate the mammalian endocycle. Nat Cell Biol 14: 1192–1202. 10.1038/ncb2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho PA, Bury L, Shahbazi MN, Liakath-Ali K, Tate PH, Wormald S, Hindley CJ, Huch M, Archer J, Skarnes WC, et al. 2015. Over-expression of Plk4 induces centrosome amplification, loss of primary cilia and associated tissue hyperplasia in the mouse. Open Biol 5: 150209. 10.1098/rsob.150209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhurst MR, Ow JR, Zafer G, van Hul NKM, Wollmann H, Bisteau X, Brough D, Choi H, Kaldis P. 2020. Loss of hepatocyte cell division leads to liver inflammation and fibrosis. PLoS Genet 16: e1009084. 10.1371/journal.pgen.1009084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donne R, Sangouard F, Celton-Morizur S, Desdouets C. 2021. Hepatocyte polyploidy: driver or gatekeeper of chronic liver diseases. Cancers (Basel) 13: 5151. 10.3390/cancers13205151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, Grompe M. 2010. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467: 707–710. 10.1038/nature09414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans LT, Anglen T, Scott P, Lukasik K, Loncarek J, Holland AJ. 2021. ANKRD26 recruits PIDD1 to centriolar distal appendages to activate the PIDDosome following centrosome amplification. EMBO J 40: e105106. 10.15252/embj.2020105106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava LL, Schuler F, Sladky V, Haschka MD, Soratroi C, Eiterer L, Demetz E, Weiss G, Geley S, Nigg EA, et al. 2017. The PIDDosome activates p53 in response to supernumerary centrosomes. Genes Dev 31: 34–45. 10.1101/gad.289728.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentric G, Maillet V, Paradis V, Couton D, Hermitte AL, Panasyuk G, Fromenty B, Celton-morizur S, Desdouets C. 2015. Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease. J Clin Invest 125: 981–992. 10.1172/JCI73957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. 2015. Centrosomes and cancer: revisiting a long-standing relationship. Nat Rev Cancer 15: 639–652. 10.1038/nrc3995 [DOI] [PubMed] [Google Scholar]
- Goupil A, Nano M, Letort G, Gemble S, Edwards F, Goundiam O, Gogendeau D, Pennetier C, Basto R. 2020. Chromosomes function as a barrier to mitotic spindle bipolarity in polyploid cells. J Cell Biol 219. 10.1083/jcb.201908006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grompe M, Jones SN, Loulseged H, Caskey CT. 1992. Retroviral-mediated gene transfer of human ornithine transcarbamylase into primary hepatocytes of spf and spf-ash mice. Hum Gene Ther 3: 35–44. 10.1089/hum.1992.3.1-35 [DOI] [PubMed] [Google Scholar]
- Heinke P, Rost F, Rode J, Trus P, Simonova I, Lázár E, Feddema J, Welsch T, Alkass K, Salehpour M, et al. 2022. Diploid hepatocytes drive physiological liver renewal in adult humans. Cell Syst 13: 499–507.e12. 10.1016/j.cels.2022.05.001 [DOI] [PubMed] [Google Scholar]
- Hirai M, Chen J, Evans SM. 2016. Generation and characterization of a tissue-specific centrosome indicator mouse line. Genesis 54: 286–296. 10.1002/dvg.22937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Zhu Q, Bauer M, Verma IM, Nigg EA, Cleveland DW. 2012. The autoregulated instability of polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev 26: 2684–2689. 10.1101/gad.207027.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SH, Delgado ER, Otero PA, Teng KY, Kutay H, Meehan KM, Moroney JB, Monga JK, Hand NJ, Friedman JR, et al. 2016. MicroRNA-122 regulates polyploidization in the murine liver. Hepatology 64: 599–615. 10.1002/hep.28573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humpton TJ, Hall H, Kiourtis C, Nixon C, Clark W, Hedley A, Shaw R, Bird TG, Blyth K, Vousden KH. 2022. p53-mediated redox control promotes liver regeneration and maintains liver function in response to CCl4. Cell Death Differ 29: 514–526. 10.1038/s41418-021-00871-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent LN, Rakijas JB, Pandit SK, Westendorp B, Chen H-Z, Huntington JT, Tang X, Bae S, Srivastava A, Senapati S, et al. 2016. E2f8 mediates tumor suppression in postnatal liver development. J Clin Invest 126: 2955–2969. 10.1172/JCI85506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Garcia-Carbonell R, Yamachika S, Zhao P, Dhar D, Loomba R, Kaufman RJ, Saltiel AR, Karin M. 2018. ER stress drives lipogenesis and steatohepatitis via caspase-2 activation of S1P. Cell 175: 133–145.e15. 10.1016/j.cell.2018.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Wang L, Sladky VC, Oh TG, Liu J, Trinh K, Eichin F, Downes M, Hosseini M, Jacotot ED, et al. 2021. PIDDosome-SCAP crosstalk controls diet dependent transition from simple steatosis to steatohepatitis. SSRN Electron J 10.2139/ssrn.4029141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouse KA, Lopez KE, Bachofner M, Amon A. 2018. Chromosome segregation fidelity in epithelia requires tissue architecture. Cell 175: 200–211.e13. 10.1016/j.cell.2018.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurinna S, Stratton SA, Coban Z, Schumacher JM, Grompe M, Duncan AW, Barton MC. 2013. P53 regulates a mitotic transcription program and determines ploidy in normal mouse liver. Hepatology 57: 2004–2013. 10.1002/hep.26233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MS, Bakker B, Boeckx B, Moyett J, Lu J, Vitre B, Spierings DC, Lansdorp PM, Cleveland DW, Lambrechts D, et al. 2017. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev Cell 40: 313–322.e5. 10.1016/j.devcel.2016.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YH, Zhang S, Zhu M, Lu T, Chen K, Wen Z, Wang S, Xiao G, Luo D, Jia Y, et al. 2020. Mice with increased numbers of polyploid hepatocytes maintain regenerative capacity but develop fewer hepatocellular carcinomas following chronic liver injury. Gastroenterology 158: 1698–1712.e14. 10.1053/j.gastro.2020.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Bettencourt-Dias M. 2018. Building the right centriole for each cell type. J Cell Biol 217: 823–835. 10.1083/jcb.201704093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Wakefield L, Tarlow BD, Grompe M. 2020. In vivo lineage tracing of polyploid hepatocytes reveals extensive proliferation during liver regeneration. Cell Stem Cell 26: 34–47.e3. 10.1016/j.stem.2019.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Wakefield L, Peters A, Peto M, Spellman P, Grompe M. 2021. Proliferative polyploid cells give rise to tumors via ploidy reduction. Nat Commun 12: 646. 10.1038/s41467-021-20916-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK, Bhushan B. 2021. Liver regeneration: biological and pathological mechanisms and implications. Nat Rev Gastroenterol Hepatol 18: 40–55. 10.1038/s41575-020-0342-4 [DOI] [PubMed] [Google Scholar]
- Miettinen TP, Pessa HKJ, Caldez MJ, Fuhrer T, Diril MK, Sauer U, Kaldis P, Björklund M. 2014. Identification of transcriptional and metabolic programs related to mammalian cell size. Curr Biol 24: 598–608. 10.1016/j.cub.2014.01.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Holland AJ. 2018. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat Rev Mol Cell Biol 19: 297–312. 10.1038/nrm.2017.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver TG, Meylan E, Chang GP, Xue W, Burke JR, Humpton TJ, Hubbard D, Bhutkar A, Jacks T. 2011. Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol Cell 43: 57–71. 10.1016/j.molcel.2011.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JM, Golabi P, Younossi Y, Mishra A, Younossi ZM. 2020. Changes in the global burden of chronic liver diseases from 2012 to 2017: the growing impact of NAFLD. Hepatology 72: 1605–1616. 10.1002/hep.31173 [DOI] [PubMed] [Google Scholar]
- Pandit SK, Westendorp B, Nantasanti S, Van Liere E, Tooten PCJ, Cornelissen PWA, Toussaint MJM, Lamers WH, De Bruin A. 2012. E2f8 is essential for polyploidization in mammalian cells. Nat Cell Biol 14: 1181–1191. 10.1038/ncb2585 [DOI] [PubMed] [Google Scholar]
- Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, Shelton KD, Lindner J, Cherrington AD, Magnuson MA. 1999. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using Cre recombinase. J Biol Chem 274: 305–315. 10.1074/jbc.274.1.305 [DOI] [PubMed] [Google Scholar]
- Powell EE, Wong VWS, Rinella M. 2021. Non-alcoholic fatty liver disease. Lancet 397: 2212–2224. 10.1016/S0140-6736(20)32511-3 [DOI] [PubMed] [Google Scholar]
- Scholten D, Trebicka J, Liedtke C, Weiskirchen R. 2015. The carbon tetrachloride model in mice. Lab Anim 49: 4–11. 10.1177/0023677215571192 [DOI] [PubMed] [Google Scholar]
- Serçin Ö, Larsimont JC, Karambelas AE, Marthiens V, Moers V, Boeckx B, Le Mercier M, Lambrechts D, Basto R, Blanpain C. 2016. Transient PLK4 overexpression accelerates tumorigenesis in p53-deficient epidermis. Nat Cell Biol 18: 100–110. 10.1038/ncb3270 [DOI] [PubMed] [Google Scholar]
- Sheahan S, Bellamy CO, Treanor L, Harrison DJ, Prost S. 2004. Additive effect of p53, p21 and Rb deletion in triple knockout primary hepatocytes. Oncogene 23: 1489–1497. 10.1038/sj.onc.1207280 [DOI] [PubMed] [Google Scholar]
- Shi Z, Wakil AE, Rockey DC. 1997. Strain-specific differences in mouse hepatic wound healing are mediated by divergent T helper cytokine responses. Proc Natl Acad Sci 94: 10663–10668. 10.1073/pnas.94.20.10663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladky VC, Knapp K, Szabo TG, Braun VZ, Bongiovanni L, Bos H, Spierings DC, Westendorp B, Curinha A, Stojakovic T, et al. 2020a. PIDDosome-induced p53-dependent ploidy restriction facilitates hepatocarcinogenesis. EMBO Rep 21: e50893. 10.15252/embr.202050893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladky VC, Knapp K, Soratroi C, Heppke J, Eichin F, Rocamora-Reverte L, Szabo TG, Bongiovanni L, Westendorp B, Moreno E, et al. 2020b. E2F-family members engage the PIDDosome to limit hepatocyte ploidy in liver development and regeneration. Dev Cell 52: 335–349.e7. 10.1016/j.devcel.2019.12.016 [DOI] [PubMed] [Google Scholar]
- Sladky VC, Eichin F, Reiberger T, Villunger A. 2021. Polyploidy control in hepatic health and disease. J Hepatol 75: 1177–1191. 10.1016/j.jhep.2021.06.030 [DOI] [PubMed] [Google Scholar]
- Tinel A, Tschopp J. 2004. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304: 843–846. 10.1126/science.1095432 [DOI] [PubMed] [Google Scholar]
- Toyoda H, Bregerie O, Vallet A, Nalpas B, Pivert G, Brechot C, Desdouets C. 2005. Changes to hepatocyte ploidy and binuclearity profiles during human chronic viral hepatitis. Gut 54: 297–302. 10.1136/gut.2004.043893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman-Culleré MH, Foltz CJ. 1999. Body condition scoring: a rapid and accurate method for assessing health status in mice. Lab Anim Sci 49: 319–323. [PubMed] [Google Scholar]
- Walkin L, Herrick SE, Summers A, Brenchley PE, Hoff CM, Korstanje R, Margetts PJ. 2013. The role of mouse strain differences in the susceptibility to fibrosis: a systematic review. Fibrogenes Tissue Repair 6: 1–12. 10.1186/1755-1536-6-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson PD, Delgado ER, Alencastro F, Leek MP, Roy N, Weirich MP, Stahl EC, Otero PA, Chen MI, Brown WK, et al. 2018. The polyploid state restricts hepatocyte proliferation and liver regeneration. Hepatology 69: 1242–1258. 10.1002/hep.30286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson PD, Alencastro F, Delgado ER, Leek MP, Weirich MP, Otero PA, Roy N, Brown WK, Oertel M, Duncan AW. 2019. Polyploid hepatocytes facilitate adaptation and regeneration to chronic liver injury. Am J Pathol 189: 1241–1255. 10.1016/j.ajpath.2019.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Chen C, Chen H, Hong H, Huang Y, Ling K, Hu J, Wei Q. 2020. TALPID3 and ANKRD26 selectively orchestrate FBF1 localization and cilia gating. Nat Commun 11: 1–14. 10.1038/s41467-020-16042-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, Bugianesi E. 2018. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 15: 11–20. 10.1038/nrgastro.2017.109 [DOI] [PubMed] [Google Scholar]
- Zhang S, Zhou K, Luo X, Li L, Tu HC, Sehgal A, Nguyen LH, Zhang Y, Gopal P, Tarlow BD, et al. 2018a. The polyploid state plays a tumor-suppressive role in the liver. Dev Cell 44: 447–459.e5. 10.1016/j.devcel.2018.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Nguyen LH, Zhou K, Tu H-C, Sehgal A, Nassour I, Li L, Gopal P, Goodman J, Singal AG, et al. 2018b. Knockdown of anillin actin binding protein blocks cytokinesis in hepatocytes and reduces liver tumor development in mice without affecting regeneration. Gastroenterology 154: 1421–1434. 10.1053/j.gastro.2017.12.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Requests for data and materials should be addressed to A.J.H. (aholland@jhmi.edu).