Abstract
The COVID-19 (coronavirus disease 2019) pandemic has highlighted the potential role that wastewater-based epidemiology can play in assessing aggregate community health. However, efforts to translate SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) gene copy numbers obtained from wastewater samples into meaningful community health indicators are nascent. In this study, SARS-CoV-2 nucleocapsid (N) genes (N1 and N2) were quantified weekly using reverse transcriptase droplet digital PCR from two municipal wastewater treatment plants for 6 months. Four biomarkers [ammonium, biological oxygen demand (BOD), creatinine and human mitochondrial gene NADH dehydrogenase subunit 5] were quantified and used to normalize SARS-CoV-2 gene copy numbers. These were correlated to daily new case data and 1-, 2- and 3-week cumulative case data. Over the course of the study, the strongest correlations were observed with a 1-day case data lag. However, early measurements were strongly correlated with a 5-day case data lag. This indicates that in the early stages of the pandemic, the wastewater samples may have indicated active COVID-19 cases before clinical indications. Mitochondrial and creatinine normalization methods showed the strongest correlations throughout the study, indicating that human-specific biomarkers were better at normalizing wastewater data than ammonium or BOD. Granger causality tests supported this observation and showed that gene copies in wastewater could be predictive of new cases in a sewershed.
Keywords: mitochondria, ammonia, creatinine, biological oxygen demand (BOD), correlation and causation, sewershed
Normalization of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) concentrations in wastewater with biomarkers allows for comparison with sewershed COVID-19 (coronavirus disease 2019) case data.
Introduction
Coronavirus disease 2019 (COVID-19) is a pandemic that has reignited interest in wastewater-based epidemiology (WBE) as a tool to monitor disease prevalence in a community. COVID-19 is a particularly challenging disease to monitor because both symptomatic and asymptomatic patients shed infectious virus particles even at early incubation stages (Pan et al. 2020, Qian et al. 2020, Rothe et al. 2020). The Centers for Disease Control and Prevention (CDC) estimates that 20–50% of infected persons may be asymptomatic (Centers for Disease Control and Prevention 2020a, Mizumoto et al. 2020). While the availability of clinical diagnostic testing has improved during the pandemic, it remains impractical to test everyone in a community routinely. Evidence continues to mount that COVID-19 infection, long-term morbidity and mortality rates vary by racial and socioeconomic status differences. Poorer and predominantly minority communities are at greater risk of acquiring an infection and, once infected, have higher long-term morbidity and mortality rates (McLaren 2020, Webb Hooper et al. 2020). Explanations for higher infection rates have included limited ability to isolate, higher employment in essential services, threats of eviction or reliance on informal childcare (Quinn et al. 2011). For health outcomes, explanations include comorbid conditions (e.g. obesity and diabetes) (Davis et al. 2017) coupled with limited access to health care (Quinn et al. 2011). In part, higher mortality rates and disproportionate exposure may be linked to reduced testing of marginalized communities, and communities with lower incomes and higher concentrations of racial minorities have less access to general testing (Webb Hooper et al. 2020). Pooled community testing allows public health officials to monitor a community and make informed decisions for the deployment of limited individual testing resources.
While severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for COVID-19, is viewed predominantly as an acute respiratory illness, symptomatic and asymptomatic individuals are known to excrete the virus in their stool (Wölfel et al. 2020). Excreta collected by sanitary distribution system (sewershed) can serve as a central point to assess community health, including the presence of SARS-CoV-2, through the analysis of biological markers. While WBE was originally used to assess biomarkers associated with illicit-drug use, studies have also used WBE as a tool to understand the prevalence of other enteric viruses before the COVID-19 pandemic. This has included tracking poliovirus (Brouwer et al. 2018), hepatitis A (Hellmér et al. 2014), and a variety of enteric and nonenteric viruses (McCall et al. 2020). Recent efforts have detected the SARS-CoV-2 virus in wastewater before any reported cases (Randazzo et al. 2020), and the virus loading was normalized based on generic population measurements (Gonzalez et al. 2020).
However, a biomarker that indicates virus concentrations in wastewater alone does not directly correlate with the number of individuals contributing to a sewershed. Further, the use of census-level community data to normalize the load may not provide the resolution required to understand highly dynamic individual contributions to wastewater. Wastewater treatment plants typically have a distinct diurnal influent flow pattern. Also, when it rains, influent flows can increase by 3–10 times the normal flow rate. Because of these flow patterns, it is unclear whether the best sampling practice is to collect 24-h composite samples or a grab sample at peak flows. For either sample, the specific population size contributing fecal load to the system at a given time is unknown. Chemical analysis of human-specific biomarkers in urban wastewater can be used to estimate the population excreting fecal matter into a wastewater sample (Gracia-Lor et al. 2017). The accurate estimation of population size is necessary to normalize epidemiological data to the per capita level and allows temporal and spatial comparisons to be made (Van Nuijs et al. 2011). Biological oxygen demand (BOD) and ammonium are routinely collected at wastewater facilities, and the biomarkers have been used to estimate population size (Gracia-Lor et al. 2017). However, this estimation is made difficult by industrial wastewater flows that can dilute or contribute to generic biomarkers (i.e. BOD) in wastewater. The ideal biomarker should have a low variance in per capita daily excretion, and the excretion should not vary with season, weather and geographic location. In addition, there should be an understanding of how the excretion of biomarkers may vary diurnally with circadian rhythms. While several compounds linked to human metabolism have been proposed as endogenous biomarkers (creatinine, coprostanol, 1-aminopropan-2-one, 5-hydroxyindoleacetic acid and ammonium) (Gracia-Lor et al. 2017), these have not been widely tested for normalizing viral concentrations to community prevalence.
We present a study that quantified the SARS-CoV-2 virus in two sewersheds over a 24-week period. Along with the SARS-CoV-2 concentration data, the wastewater was assessed for ubiquitous biomarkers as a means to normalize the wastewater strength to account for variations in population contributions to the sewersheds and the effects of infiltration and inflow during wet-weather events. These biomarkers include BOD, ammonium, human mitochondrial DNA and creatinine. The selected biomarkers are routinely collected and available (BOD and ammonium), have a similar half-life to SARS-CoV-2 (creatinine) or are excreted in fecal matter (mitochondrial DNA). Additionally, the biomarkers have reported person equivalents [BOD: 74.7 g day–1 person–1 (Yonker et al. 2012); ammonium: 24.2 g day–1 person–1 (Rudman et al. 1973); creatinine: 2.65 g day–1 person–1 (Brewer et al. 2012); and mitochondrial DNA: 1.4 × 109 gene copies (gc) day–1 person–1 (Caldwell et al. 2007)] to facilitate population size estimates. The creatinine and mitochondrial DNA biomarkers showed strong correlations and causation with community case data. This is in comparison to biomarkers that can be influenced by industrial or agricultural activities (e.g. BOD and ammonium). These methods provide an opportunity to normalize wastewater SARS-CoV-2 concentrations, which could facilitate comparisons across communities.
Methods
Lawrence, KS, wastewater treatment facilities and catchment
The City of Lawrence has a population of ∼96 369 and is home to a major university campus, the University of Kansas. The University of Kansas has a student population of 27 690. Two wastewater treatment facilities serve Lawrence: the Kansas Wastewater Treatment Plant (KAWWWTP) and the Wakarusa Wastewater Treatment Plant (WAKWWTP). Lawrence has two sewer catchments (sewersheds; Fig. 1). The Kansas sewershed (KAWSS) discharges solely to the KAWWWTP. The Wakarusa sewershed (WAKSS) discharges to both the KAWWWTP and WAKWWTP. WAKWWTP is operated as a three-stage Bardenpho with a base daily flow rate of 2 million gallons per day (MGD). The KAWWWTP treats all other wastewater from Lawrence, with a maximum of 25 MGD through the biological treatment with an additional 40 MGD through Actiflo treatment (Veolia, Paris, France). Flow split from the WAKSS was used to account for the fluctuating flows that went to the KAWWWTP from each sewershed and to connect the biomarker concentration data to case data. Total daily values of SARS-CoV-2 genes (viral load) and the mass of each biomarker were proportioned to each sewershed to facilitate comparison with the sewershed case data.
Figure 1.
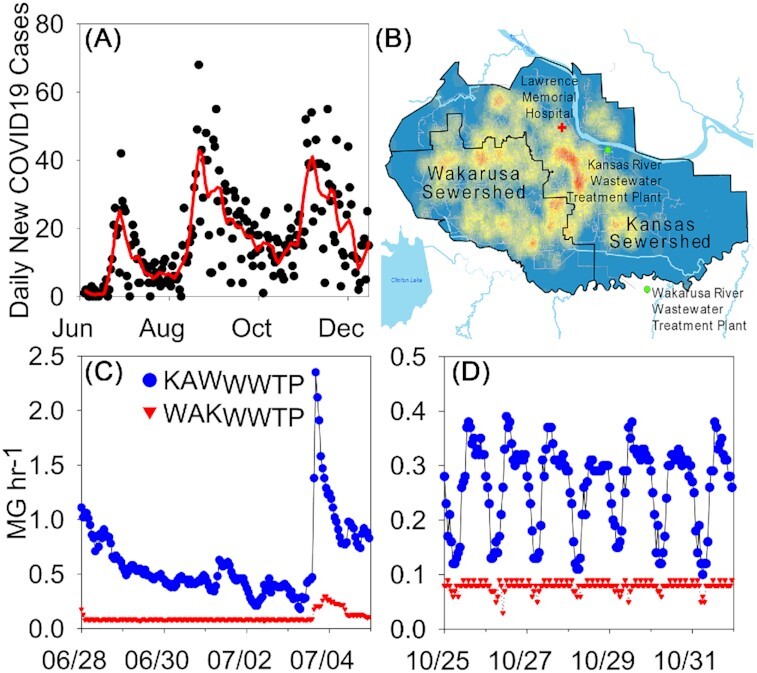
(A) New daily case data with Loess smoothing curve. (B) Map outline of the Lawrence sewersheds, Wakarusa and Kansas, with case densities over the 6-month study. The map highlights the KAWWWTP, the WAKWWTP and the Lawrence Memorial Hospital. (C)Flow data (MG h−1) for the two wastewater treatment plants at a high-flow event. (D) Flow data (MG h−1) for the two wastewater treatment plants demonstrating diurnal flow pattern.
Mass balance to compare wastewater values to case data
To facilitate a direct correlation of the wastewater values to the case data in each sewershed, a mass balance approach was used to allocate biomarkers to each sewershed. Flow meter data were used that allocated the WAKSS flow to the KAWWWTP and WAKWWTP. This fraction of the flow split (x) was used to allocate the KAWSS flow rate (Q) that contributed to the KAWWWTP and WAKWWTP. The allocated flow and the concentration data (C) from each WWTP was used to determine the mass or gene copies (M) of each biomarker present in the sewershed.
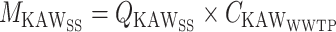 |
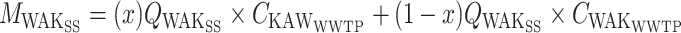 |
Case data
The Lawrence Douglas County Health Department collected case data and provided deidentified data for this study. Case numbers excluded multiple positive tests from the same individual, and the deidentified data were binned into the two sewersheds based on patient residence.
Wastewater collection
One liter composite samples were taken after grit screening and before primary sedimentation. For the KAWWWTP, composite samples were time weighted from 4 June 2020 to 14 August 2020. Subsequent composite samples were taken on a flow weighted basis until 8 December 2020. For the WAKWWTP, samples were collected over the same time range with a transition for sampling method occurring on 27 July 2020. Samples were transported to the University of Kansas Environmental Laboratories on ice in coolers within 6 h. Samples were processed upon receipt with molecular analysis occurring within 24 h. Approximately 100 mL was stored at −80°C for subsequent creatinine and mitochondrial DNA biomarker analysis.
SARS-CoV-2 concentration and RNA extraction
A modified polyethylene glycol (PEG)-8000 precipitation protocol was used to concentrate viral particles from wastewater (Ahmed et al. 2020). Fifty milliliters of wastewater was centrifuged in conical tubes at 1000 × g for 5 min at 4°C to remove large particulate matter. The supernatant (37.5 mL) was transferred to a clean 50-mL conical tube, and 12.5 mL of 48% PEG and 0.12 g bovine serum albumin (BSA) were added to achieve 12% PEG and 0.3% BSA in the final matrix. The sample was centrifuged at 12 000 × g for 2 h at 4°C. The supernatant was aspirated. RNA was extracted from the pellet using the QIAamp Viral RNA Mini Kit manually or with the QIAcube Connect following the manufacturer's protocol. Carrier RNA provided in the kit was utilized in concentrations recommended by the manufacturer to improve recovery. RNA was eluted with 60 µL of nuclease-free water or buffer AVE (RNase-free water with 0.04% NaN3). Two virus surrogates were used to assess the efficiency of the concentration and extraction methods: murine hepatitis virus (MHV) A59 (ATCC VR-764) (Fehr et al. 2015) and HIV-1-coPuro (PURO) (Lucas et al. 2010). Surrogates were spiked into wastewater samples before the first centrifugation, and recovery was calculated using quantitative PCR (qPCR).
SARS-CoV-2 gene and surrogate quantification
The virus was quantified via reverse transcription droplet digital PCR (RT-ddPCR) using CDC RUO nCoV_N1 and nCoV_N2 primers and probe assays (Centers for Disease Control and Prevention 2020b) that target two regions (N1 and N2) on the nucleocapsid (N) gene of the SARS-CoV-2 virus. The RT-ddPCR assay was performed on a QX200 Digital Droplet PCR (ddPCR) system (Bio-Rad Laboratories, Hercules, California) and results were accepted if there were >10 000 partitions (Ciesielski et al. 2021). Quantification of both nCoV_N1 and nCoV_N2 for each sample was performed in a single multiplexed reaction by labeling the 5′ end of the nCoV_N1 probe with FAM and the 5′ end of the nCoV_N2 probe with HEX (Centers for Disease Control and Prevention 2020b). The reaction mix was prepared using One-Step RT-ddPCR Advanced Kit for Probes (Bio-Rad Laboratories) following the manufacturer's protocol. Each sample contained 16.5 µL of reaction mix and 5.5 µL of template RNA prior to droplet generation. Droplet generation for each sample was performed with 20 µL of sample mix and 70 µL of droplet generation oil. Upon successful generation of droplets, the reaction plate containing all the samples was sealed using a PX1 plate sealer (Bio-Rad Laboratories). The sealed plate was then immediately thermal cycled on a C1000 touch thermal cycler (Bio-Rad Laboratories) with a deep well reaction module under the following conditions: one cycle at 50°C for 60 min; one cycle at 95°C for 10 min; 50 cycles of 94°C for 30 s and 55°C for 1 min; one cycle at 98°C for 10 min; and one cycle at 4°C for 30 min. A list of all primers and probes used in ddPCR and qPCR experiments is available (Table S.1, Supporting Information). The droplets were read using the QX200 droplet reader and QuantaSoft software version 1.7 (Bio-Rad Laboratories). Final quantification was performed using QuantaSoft Analysis Pro software version 1.0.596 (Bio-Rad Laboratories).
Biomarker analysis
Four biomarkers were measured to normalize the strength of wastewater to population equivalents including ammonium, BOD5, creatinine and mitochondrial DNA. Ammonium was measured using a TNT 832 Ammonia Nitrogen kit (Hach). BOD5 measurements followed standard methods. On the day of sample collection, ammonium measurements were taken and BOD tests were started. Creatinine was analyzed using a creatinine colorimetric detection assay (Enzo Life Sciences, Farmingdale, NY) (Burgard et al. 2013). To determine mitochondrial DNA content, 25 mL of wastewater was filtered using a Pall 0.2-µm (membrane type) filter (Pall Corporation, Port Washington, NY). The DNA was extracted from the filter using a DNeasy PowerWater Kit (Qiagen, Germantown, MD) with a final elution volume of 100 µL. The DNA was quantified using broad-spectrum double-stranded DNA Qubit assay. The mitochondrial DNA was quantified using qPCR with forward primer mtH-ND5-F, reverse primer mtH-ND5-R and probe mtH-ND5-P (Table S.1, Supporting Information) (Caldwell et al. 2007). The reaction mix was prepared using SsoAdvanced Universal Probes Supermix (Bio-Rad Laboratories) using the manufacturer's protocol. Each qPCR reaction of 20 µL comprised of 15 µL reaction mix and 5 µL template DNA. Quantification was performed on a CFX Connect Real-Time PCR Detection System (Bio-Rad Laboratories) under the following conditions: one cycle at 95°C for 2 min and 40 cycles of 95°C for 10 s, 60°C for 30 s and 72°C for 30 s. Standards were created using gBlock of the mitochondrial DNA sequence (Table S.2, Supporting Information).
Statistical analysis
Statistical analysis was performed in SigmaPlot 14 and R. Correlations were established using Spearman's rank correlation coefficient test, and significance is indicated when P < 0.05. The causality was examined using the Granger causality test, and significance is indicated when P < 0.10. To quantify the magnitude of causality, we also computed an effect size (partial η2) for each Granger causality test. The values of 0.01, 0.06 and 0.14 represent a small, medium and large effect, respectively (Richardson 2011, Cohen 2013).
Results
Cases in Lawrence, KS
In mid-March 2020, Douglas County experienced its first COVID-19 case. Positive cases remained relatively low prior to the start of this study (<6 new cases each day with a cumulative total of 72 cases by 3 June 2020). Over the ∼6-month period, the intensity of new cases varied geographically (Fig. 1A). During the course of this study, three large increases in the number of new cases were observed in the Loess smoothing (Fig. 1B). The first peak corresponds closely with 29th June and a daily case number of 42. The second peak occurred on 21st August, which corresponds closely with the return of students for the Fall semester and a high frequency testing campaign implemented by the University of Kansas. The last peak occurred on 9th November, with a total daily case number of 46.
Wastewater characteristics and biomarker concentrations
In order to capture the number of individuals contributing to the wastewater treatment plant, standard wastewater characteristic parameters were determined in addition to biomarkers that are indicative of population equivalence. BOD (83–326 mg L–1), ammonium (8.7–41.3 mg N L–1), creatinine (0.24–1.14 mg L–1) and mitochondria (5.6 × 107–1.4 × 108 gc L–1) were determined (Fig. S.1, Supporting Information). These values along with flow rates from each sewershed were used to determine the total gene copies or mass of biomarker that each sewershed contributed to the wastewater treatment plant. The mass balance calculations were required to facilitate a comparison with the case data in each sewershed.
Gene quantification with surrogate recoveries
RT-ddPCR N1 and N2 gene quantification had a method detection limit of 1280 and 1050 gene copies (L wastewater)–1, respectively. These detection limits corresponded to 4.9 gcN1 reaction–1 and 4.0 gcN2 reaction–1, which align closely with previously reported detection limits of 2.9 gcN1 reaction–1 and 4.6 gcN2 reaction–1 (Ahmed et al. 2022). Viral surrogates were used to assess the recovery from the PEG concentration. From 4 June to 28 July, the PURO virus-like plasmid was used and had a 10.8 ± 5.2% recovery for samples from the KAWWWTP and 9.5 ± 6.4% for the WAKWWTP. During 29 July and 8 December, MHV was the viral surrogate with recoveries of 2.8 ± 3.1% for KAWWWTP and 1.9 ± 3.0% for the WAKWWTP. Recoveries were strongly influenced by the presence of biosolids, and total MHV recoveries increased to 34% when accounting for the liquid and biosolid phases.
Biomarker normalized viral loads and correlation with case data
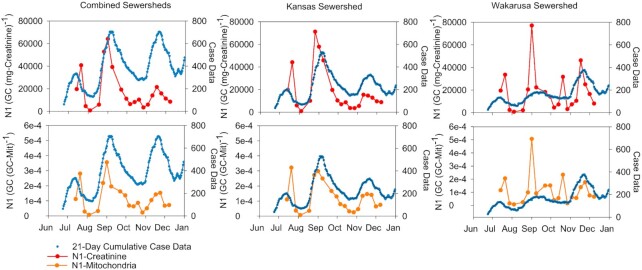
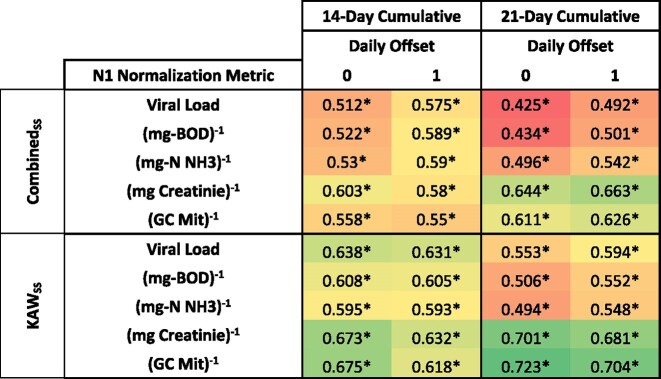
The SARS-CoV-2 N1 and N2 gene copies were normalized to total facility volume, BOD, ammonium, creatinine and mitochondria (Fig. 2). To elucidate if wastewater data was correlated with case data, Spearman's correlation analysis was used. In the first analysis, 10 normalized viral biomarkers (five biomarkers and two viral gene quantification) were compared with different case data scenarios over the entire span of data collection for the combined sewersheds. N1 biomarkers normalized to creatinine and mitochondrial gene copies showed the strongest correlation with 21-day cumulative case counts with a ρ value of 0.644 and 0.611, respectively (Table 1, additional biomarker correlations available in Tables S.3–5, Supporting Information). A slight increase in correlation strength is observed from wastewater concentration data compared with a 1-day lag in case data. This could indicate over the 6-month span that WBE may have provided a 1-day advanced notice of an increase in COVID-19 cases. While significant, positive correlations were observed with case data after a 1-day lag in case data, the strength of the correlation decreased with an increase in the case data lag. N2 normalized values showed no statistically significant correlation with any of the case data comparisons.
Figure 2.
SARS-CoV-2 N1 gene copies normalized to creatinine (top row) and mitochondria (Mit) (bottom row) along with case data. The data are presented for the combined sewersheds (first column), the KAWSS (middle column) and the WAKSS (last column).
Table 1.
Select Spearman's rho values highlighting the positive correlations between SARS-CoV-2 viral load normalized by human-specific biomarkers and case data. Significant correlations (P < 0.05) are indicated by an asterisk. Coloring was used to highlight contrast in correlation coefficient (red, low; green, high) and high and low values in this table were used for the bounds.
|
This study represents a significant geography and time span (6 months) of testing. Accordingly, it is critical to examine these two factors for their independent impact on the strength of the correlation. Geographically, Lawrence is divided into two sewersheds, and the correlation of sewershed case data and biomarkers yielded different correlation results. The KAWSS had the strongest correlation values over the course of the study with 21-day cumulative N1 gene copies normalized to mitochondrial gene copies (ρ = 0.723, Table 1). This was followed by the 21-day case comparison with 1 day of lag (ρ = 0.704). The WAKSS consistently had weaker correlations over the course of the study with the strongest correlation between the N1 gene copies normalized to ammonium (ρ = 0.52). Nonetheless, these correlations were statistically significant. Select N2 normalization metrics had statistically significant correlations for the KAWSS [N2 viral load with 14-day cumulative case data with no lag and 1-day lag (Table S.3, Supporting Information)], but no other statistically significant correlations were observed for normalized N2 values with case data. The general trend of decreasing correlation strengths with an increase in case data lag was observed in each of the separate sewersheds. N2 normalized values showed no statistically significant correlation with any of the case data comparisons for the WAKSS.
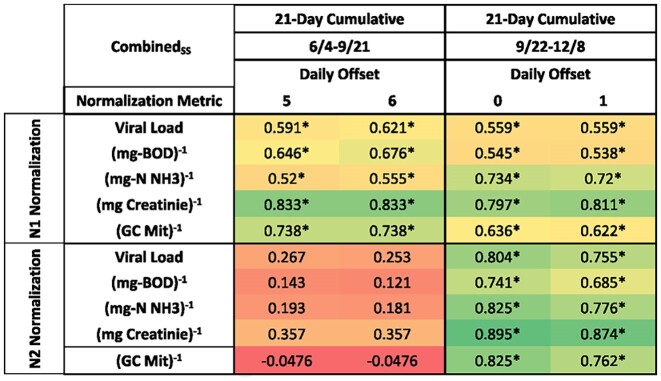
To examine the variability of the data over the course of the study, normalized biomarkers and case data were compared in ∼3-month blocks for the combined sewersheds. Earlier in the pandemic (4th June–21st September), sewershed values were highly correlated with 21-day cumulative values with a 5-day lag (Table 2). Across the five normalization metrics, creatinine and mitochondria showed the highest correlation with a ρ value of 0.833 and 0.738, respectively, for both 5- and 6-day lags.
Table 2.
Select Spearman's rho values highlighting the positive correlations between SARS-CoV-2 viral load normalized by human-specific biomarkers and case data in the combined sewershed for data collected from 4 June 2020 to 21 September 2020 and 22 September 2020 to 8 December 2020. Significant correlations (P < 0.05) are indicated by an asterisk. Coloring was used to highlight contrast in correlation coefficient (red, low; green, high) and high and low values in this table were used for the bounds.
|
Over the second time period (22 September–8 December), wastewater values were strongly correlated with the 21-day cumulative case data with no lag or a 1-day lag. For N1 normalization, creatinine normalization was strongest with 21-day cumulative case data with a 1-day lag (ρ = 0.811). Interestingly, the N2 normalized values were strongly correlated with the creatinine normalized and the 21-day cumulative case data with no lag (ρ = 0.895).
Causal relationships
Correlations do not indicate causation. As such, Granger causality was used to determine links between indicators and case data. Overlaying the correlation and causal data for N1, significant correlations and predictive causal relationships are found between creatinine normalized values with the 1-day lag, 21-day cumulative cases; and 4-day lag, 21-day cumulative cases (Fig. 3). The N1 normalized to gene copies of mitochondria also had significant correlations and predictive causal relationships with 1-day lag, 21-day cumulative cases. The partial eta squared for these causal relationships was 0.37, indicating a large effect size.
Figure 3.
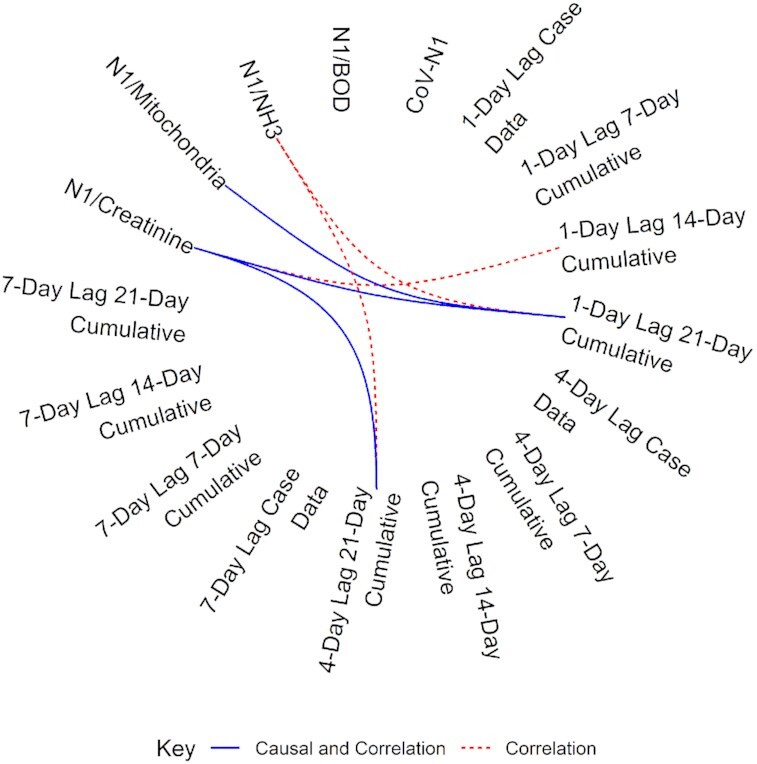
Chord diagram of the correlation and causal relationship between normalized SARS-CoV-2 values and case data. Dashed red lines indicate a positive, significant (P < 0.05) correlation between values. The solid blue lines indicate a similar positive correlation but also a predictive causal relationship between normalized SARS-CoV-2 values and case data (P < 0.10).
Discussion
This work demonstrates the need to normalize SARS-CoV-2 viral load to account for variations in individuals contributing to a sewershed as well as changing sewershed conditions that affect the quantification process. Over the 6-month study period, mitochondrial and creatinine biomarkers yielded the strongest correlations with case data. The amplification of N1 and N2 genes has been recommended to capture SARS-CoV-2 (Centers for Disease Control and Prevention 2020b). In this study, N1 gene and the normalized values had better correlations with case data throughout the study, which aligns well with other studies (Lu et al. 2020, Vogels et al. 2020, D'Aoust et al. 2021). Interestingly, over the second time series, N2 showed strong correlations with case data and mimics findings of a similar study in Southeastern Virginia (Gonzalez et al. 2020). Recent work has also demonstrated that while the PEG method yields some of the highest and consistent recoveries among the different concentration methods, the inclusion of a pre-centrifugation step to remove solids, as was done in this study, increases the uncertainty of the quantification (Pecson et al. 2021).
Initial wastewater monitoring showed an increase in the number of SARS-CoV-2 gene copies before a corresponding increase in case data. This is observed in the wastewater SARS-CoV-2 gene concentration for testing from June to September with strong correlations of wastewater testing 5–6 days before the case data. This observation has also been noted in other studies (e.g. Peccia et al. 2020). In the later testing period (September–December), the correlation coefficients were strongest with no or 1-day case data lag. This observation is likely influenced by the availability of COVID-19 clinical testing (Bibby et al. 2021). Within the Lawrence community, the University of Kansas adopted high-frequency and rapid turnaround testing campaign of 22 563 individuals in late August. This testing campaign added 546 positive cases to the county data within 2 weeks. These tests were broadly available and included individuals with no symptoms or COVID-19 risk factors.
The choice of normalizing biomarkers used in this study was heavily influenced by their ubiquity (i.e. all individuals excrete creatinine and human mitochondrial DNA) (Gracia-Lor et al. 2017). Nonetheless, many other biomarkers can provide normalization to human equivalence. For example, pepper mild mottle virus (PMMoV) is one such candidate (D'Aoust et al. 2021). This virus was found to be the most abundant virus in human stool with relatively low occurrence in animal feces (Rosario et al. 2009, Hamza et al. 2011). The shed rate in human feces is 105–106 copies (g dry weight)–1 (Zhang et al. 2005), and a recent work has normalized SARS-CoV-2 viral copies with PMMoV that were strongly correlated with case data (D'Aoust et al. 2021). However, while several studies detected the presence of PMMoV, other studies have suggested that the presence of PMMoV is diet dependent (Colson et al. 2010). Further, PMMoV may not sufficiently capture the contribution by sick individuals as hospitalized individuals had low shed rates of PMMoV (Colson et al. 2010), a critical concern when assessing the community spread of a disease during a pandemic.
Efforts to normalize the biomarkers to population equivalence increase the uncertainty of the values. BOD, ammonium, mitochondria and creatinine have reported metrics by which to convert to population equivalence. Over the course of this study, the population contributing to the combined sewershed varied from 52 050 to 132 000 based on BOD measurements. This closely aligns with the reported census values and takes into account large population fluxes associated with the University of Kansas. Excretion values are available in the literature for the biomarkers ammonium (Rudman et al. 1973) and creatinine (Smith-Palmer 2002, Barr et al. 2005, Arndt 2009, Brewer et al. 2012). These values indicate that 16 600–48 300 individuals are contributing to the wastewater based on ammonium measurements and 3350–17 314 individuals are contributing to the wastewater based on creatinine measurements. While our mitochondria concentrations in the wastewater match closely with previously reported values (Caldwell and Levine 2009), calculations overestimate the population contributing to the wastewater (estimated range of 1 400 000–3 000 000 individuals) (Caldwell et al. 2007), indicating that further research is required for mitochondrial DNA biomarkers to make stronger connections between individual fecal contribution and wastewater loads.
However, despite the variations in the population equivalence for the biomarkers, there is a clear need to offer a normalizing metric. For this study, viral loads were not well correlated with case data and could be due to limitations of SARS-CoV-2 quantification. However, the values normalized to creatinine and mitochondria were correlated strongly, indicating that the normalizing biomarkers could account for varying conditions that affected SARS-CoV-2 quantification. Two possible phenomena may be responsible for these variations: (i) decay rates and (ii) partitioning. Creatinine has been shown to have first-order decay in gravity-based sewers, especially in the presence of biofilms, with a half-life of 1.03 days (Thai et al. 2014). This half-life compares closely with the first-order decay of SARS-CoV-2 RNA in wastewater conditions, with a half-life of 0.99 day (Bivins et al. 2020). While these half-lives compare favorably, creatinine and SARS-CoV-2 are dissimilar with respect to solubility. Creatinine is a very soluble chemical excreted through the urinary system. In contrast, the SARS-CoV-2 is associated with fecal excretion (Crank et al. 2022), and preliminary evidence suggests that the viral particles partition into solids (Graham et al. 2021, Pecson et al. 2021). On dates when partitioning may affect the quantification of SARS-CoV-2 viruses, the mitochondrial DNA biomarkers may be a more suitable normalization method. Both the SARS-CoV-2 viral particles and mitochondrial biomarkers are excreted in feces. However, the stability of DNA in the environment can be high, with half-lives on the order of days (Dejean et al. 2011).
Conclusion
As WBE work continues, selection of normalizing biomarkers to indicate variation in population and sewershed conditions is critical. This work presents normalization methods to account for the varying strength of wastewater and, specifically, methods to offer preliminary estimates of the number of individuals contributing to a sewershed. The quantification of human-specific biomarkers to normalize SARS-CoV-2 concentrations is an important contribution to realize WBE. Human-specific biomarkers, including creatinine and mitochondrial gene copies, provided stronger correlations for normalized SARS-CoV-2 gene copy values with case data compared with standard BOD and ammonium quantification values.
Acknowledgments
The authors would like to thank Anthony Fehr (University of Kansas) for providing MHV; Mark Johnson (University of Missouri) for providing PURO; the City of Lawrence, KS, for collecting wastewater samples and reporting of biological wastewater markers; the Douglas County Health Department for providing clinical case data; and Xan Wedel for developing the case data map.
Funding
The authors acknowledge funding from the Kansas Department of Health and Environment (JMH and BSMS).
Data availability
The data underlying this article are available in Mendeley Data at (DOI: 10.17632/bfn2hg3dzz.1). (Hutchison 2022).
Supplementary Material
Contributor Information
Justin M Hutchison, Civil, Environmental, and Architectural Engineering, University of Kansas, 1530 W 15th St, Lawrence, KS 66049, USA.
Zhengxi Li, Civil, Environmental, and Architectural Engineering, University of Kansas, 1530 W 15th St, Lawrence, KS 66049, USA.
Chi-Ning Chang, Life Span Institute, University of Kansas, 1000 Sunnyside Ave, Lawrence, KS 66045, USA.
Yasawantha Hiripitiyage, Civil, Environmental, and Architectural Engineering, University of Kansas, 1530 W 15th St, Lawrence, KS 66049, USA.
Megan Wittman, Civil, Environmental, and Architectural Engineering, University of Kansas, 1530 W 15th St, Lawrence, KS 66049, USA.
Belinda S M Sturm, Civil, Environmental, and Architectural Engineering, University of Kansas, 1530 W 15th St, Lawrence, KS 66049, USA.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Ahmed W, Angel N, Edson Jet al. . First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020;728:138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Smith WJ, Metcalfe Set al. . Comparison of RT-qPCR and RT-dPCR platforms for the trace detection of SARS-CoV-2 RNA in wastewater. ACS EST Water. 2022. DOI: 0.1021/acsestwater.1c00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt T. Urine-creatinine concentration as a marker of urine dilution: reflections using a cohort of 45,000 samples. Forensic Sci Int. 2009;186:48–51. [DOI] [PubMed] [Google Scholar]
- Barr DB, Wilder LC, Caudill SPet al. . Urinary creatinine concentrations in the US population: implications for urinary biologic monitoring measurements. Environ Health Perspect. 2005;113:192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K, Bivins A, Wu Zet al. . Making waves: plausible lead time for wastewater based epidemiology as an early warning system for COVID-19. Water Res. 2021;202:117438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A, Greaves J, Fischer Ret al. . Persistence of SARS-CoV-2 in water and wastewater. Environ Sci Technol Lett. 2020;7:937–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer AJ, Ort C, Banta-Green CJet al. . Normalized diurnal and between-day trends in illicit and legal drug loads that account for changes in population. Environ Sci Technol. 2012;46:8305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer AF, Eisenberg JNS, Pomeroy CDet al. . Epidemiology of the silent polio outbreak in Rahat, Israel, based on modeling of environmental surveillance data. Proc Natl Acad Sci USA. 2018;115:E10625–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgard DA, Fuller R, Becker Bet al. . Potential trends in attention deficit hyperactivity disorder (ADHD) drug use on a college campus: wastewater analysis of amphetamine and ritalinic acid. Sci Total Environ. 2013;450–1:242–9. [DOI] [PubMed] [Google Scholar]
- Caldwell JM, Levine JF. Domestic wastewater influent profiling using mitochondrial real-time PCR for source tracking animal contamination. J Microbiol Methods. 2009;77:17–22. [DOI] [PubMed] [Google Scholar]
- Caldwell JM, Raley ME, Levine JF. Mitochondrial multiplex real-time PCR as a source tracking method in fecal-contaminated effluents. Environ Sci Technol. 2007;41:3277–83. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . COVID-19 Pandemic Planning Scenarios. 2020a. https://www.cdc.gov/coronavirus/2019-ncov/hcp/planning-scenarios.html(10 April 2022, date last accessed).
- Centers for Disease Control and Prevention . Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Primer and Probe Information. 2020b. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html(22 March 2020, date last accessed). [Google Scholar]
- Ciesielski M, Blackwood D, Clerkin Tet al. . Assessing sensitivity and reproducibility of RT-ddPCR and RT-qPCR for the quantification of SARS-CoV-2 in wastewater. J Virol Methods. 2021;297:114230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Statistical Power Analysis for the Behavioral Sciences. Burlington, Canada: Academic Press, 2013. [Google Scholar]
- Colson P, Richet H, Desnues Cet al. . Pepper mild mottle virus, a plant virus associated with specific immune responses, fever, abdominal pains, and pruritus in humans. PLoS One. 2010;5:e10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crank K, Chen W, Bivins Aet al. . Contribution of SARS-CoV-2 RNA shedding routes to RNA loads in wastewater. Sci Total Environ. 2022;806:150376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust PM, Graber TE, Mercier Eet al. . Catching a resurgence: increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci Total Environ. 2021;770:145319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J, Penha J, Mbowe Oet al. . Prevalence of single and multiple leading causes of death by race/ethnicity among US adults aged 60 to 79 years. Prev Chronic Dis. 2017;14:E101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejean T, Valentini A, Duparc Aet al. . Persistence of environmental DNA in freshwater ecosystems. PLoS One. 2011;6:e23398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr AR, Athmer J, Channappanavar Ret al. . The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J Virol. 2015;89:1523–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R, Curtis K, Bivins Aet al. . COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186:116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracia-Lor E, Castiglioni S, Bade Ret al. . Measuring biomarkers in wastewater as a new source of epidemiological information: current state and future perspectives. Environ Int. 2017;99:131–50. [DOI] [PubMed] [Google Scholar]
- Graham KE, Loeb SK, Wolfe MKet al. . SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ Sci Technol. 2021;55:488–98. [DOI] [PubMed] [Google Scholar]
- Hamza IA, Jurzik L, Überla Ket al. . Evaluation of pepper mild mottle virus, human picobirnavirus and Torque teno virus as indicators of fecal contamination in river water. Water Res. 2011;45:1358–68. [DOI] [PubMed] [Google Scholar]
- Hellmér M, Paxéus N, Magnius Let al. . Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl Environ Microbiol. 2014;80:6771–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison J. 2022_02_28_FEMS_SARS_COV2”, Mendeley Data, V1, 2022. DOI: 10.17632/bfn2hg3dzz.1.
- Lu X, Wang L, Sakthivel SKet al. . US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26:1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas TM, Lyddon TD, Cannon PMet al. . Pseudotyping incompatibility between HIV-1 and gibbon ape leukemia virus Env is modulated by Vpu. J Virol. 2010;84:2666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall C, Wu H, Miyani Bet al. . Identification of multiple potential viral diseases in a large urban center using wastewater surveillance. Water Res. 2020;184:116160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren J. Racial Disparity in COVID-19 Deaths: Seeking Economic Roots with Census Data. National Bureau of Economic Research, 2020. [Google Scholar]
- Mizumoto K, Kagaya K, Zarebski Aet al. . Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill. 2020;25:2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Chen D, Xia Yet al. . Asymptomatic cases in a family cluster with SARS-CoV-2 infection. Lancet Infect Dis. 2020;20:410–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J, Zulli A, Brackney DEet al. . SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. medRxiv. 2020;12. DOI: 10.1101/2020.05.19.20105999. [Google Scholar]
- Pecson BM, Darby E, Haas CNet al. . Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the US. Environ Sci: Water Res Technol. 2021;7:504–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian G, Yang N, Ma AHYet al. . COVID-19 transmission within a family cluster by presymptomatic carriers in China. Clin Infect Dis. 2020;71:861–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn SC, Kumar S, Freimuth VSet al. . Racial disparities in exposure, susceptibility, and access to health care in the US H1N1 influenza pandemic. Am J Public Health. 2011;101:285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W, Truchado P, Cuevas-Ferrando Eet al. . SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JT. Eta squared and partial eta squared as measures of effect size in educational research. Educ Res Rev. 2011;6:135–47. [Google Scholar]
- Rosario K, Symonds EM, Sinigalliano Cet al. . Pepper mild mottle virus as an indicator of fecal pollution. Appl Environ Microbiol. 2009;75:7261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C, Schunk M, Sothmann Pet al. . Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudman D, DiFulco TJ, Galambos JTet al. . Maximal rates of excretion and synthesis of urea in normal and cirrhotic subjects. J Clin Invest. 1973;52:2241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Palmer T. Separation methods applicable to urinary creatine and creatinine. J Chromatogr B. 2002;781:93–106. [DOI] [PubMed] [Google Scholar]
- Thai PK, O'Brien J, Jiang Get al. . Degradability of creatinine under sewer conditions affects its potential to be used as biomarker in sewage epidemiology. Water Res. 2014;55:272–9. [DOI] [PubMed] [Google Scholar]
- Van Nuijs AL, Mougel J-F, Tarcomnicu Iet al. . Sewage epidemiology: a real-time approach to estimate the consumption of illicit drugs in Brussels, Belgium. Environ Int. 2011;37:612–21. [DOI] [PubMed] [Google Scholar]
- Vogels CB, Brito AF, Wyllie ALet al. . Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer–probe sets. Nat Microbiol. 2020;5:1299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb Hooper M, Napoles AM, Perez-Stable EJ. COVID-19 and racial/ethnic disparities. JAMA. 2020;323:2466–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R, Corman VM, Guggemos Wet al. . Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–9. [DOI] [PubMed] [Google Scholar]
- Yonker SA, Keller JJ, Mitchell PE. Executive Summary Wastewater Facilities Master Plan. Kansas City, MO, 2012. [Google Scholar]
- Zhang T, Breitbart M, Lee WHet al. . RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2005;4:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in Mendeley Data at (DOI: 10.17632/bfn2hg3dzz.1). (Hutchison 2022).