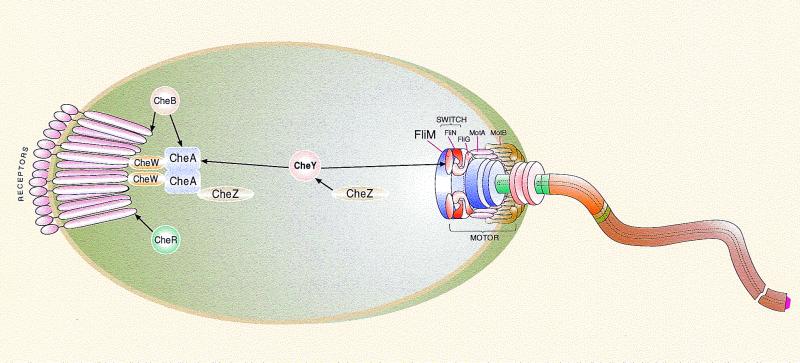
Chemotaxis is a mechanism by which bacteria efficiently and rapidly respond to changes in the chemical composition of their environment, approaching chemically favorable environments and avoiding unfavorable ones. This behavior is achieved by integrating signals received from receptors that sense the environment and modulating the direction of flagellar rotation accordingly (for reviews, see references 39, 43, and 100). Early studies in the modern era, initiated some 4 decades ago (1), uncovered the behavioral response of cells to changes in the chemical composition of their environment and the correlation between flagellar rotation and the swimming mode of the cells. They also identified most of the gene products involved in chemotaxis (for reviews, see references 50 and 56). The mode of signal transduction began to be understood only in the mid-1980s, when the possibilities of electrical signaling and a direct interaction between the receptors and flagella were eliminated (for a review, see reference 41). The possibility of indirect interaction between the receptors and flagella via a protein that is activated by the receptors and inactivated as it diffuses through the cytoplasm was then raised (96). Subsequently, sequential transient phosphorylation of chemotaxis proteins was found to be a key process in signal transduction (for a review, see reference 25). During the last decade, it was established that the signal in bacteria such as Escherichia coli and Salmonella enterica serovar Typhimurium is transduced via protein-protein interactions. These interactions have been extensively studied, contributing greatly to the elucidation of the chemotaxis-signaling cascade.
The chemotactic response in bacteria such as E. coli and Salmonella serovar Typhimurium is accomplished by signal transmission between two supramolecular complexes—the receptor complexes, located mainly at the pole(s) of the cell, and the flagellar-motor complexes (usually 5 to 10 complexes per cell), randomly distributed around the cell and embedded within the cell membrane. A messenger protein, CheY, shuttles back and forth between the complexes and transduces the signal from the receptors to the flagella (Fig. 1). The interaction of this messenger protein with the flagellar-motor supramolecular complex increases the probability of shifting the direction of flagellar rotation from the default direction, counterclockwise (CCW), to clockwise (CW) (for a review see reference 38). The consequence of CW rotation is an abrupt turning motion (tumbling), after which (when the default direction resumes) the cell swims in a new direction. Here we review the protein-protein interactions involved in chemotactic signaling, including interactions within the supramolecular complexes, interactions between the complexes and the messenger protein CheY, and interactions between CheY and the proteins that regulate its signaling state. Interactions involved in the signaling pathway leading to adaptation will also be reviewed. We will mainly focus on functional aspects of the interactions. The reader is referred to references 13, 35, 43, 54, and 81 for more-detailed structural aspects. Because this is a minireview, the reference list is incomplete. Whenever possible, reference is made to reviews or papers that provide access to the original literature.
FIG. 1.
Simplified scheme of protein-protein interactions during chemotactic signal transduction in bacteria. The black arrows represent regulated interactions. The receptor shown is an MCP. The scheme is not drawn to scale.
INTERACTIONS WITHIN THE RECEPTOR SUPRAMOLECULAR COMPLEX
In bacteria such as E. coli and S. enterica serovar Typhimurium, two kinds of receptors monitor the chemical composition of the environment: chemotaxis-specific receptors (named MCPs for methyl-accepting chemotaxis proteins), and dual-function receptors involved in both chemotaxis and transport of the ligand (for reviews, see references 37 and 39). Here we will deal with the chemotaxis-specific receptors. There are five chemotaxis-specific, transmembrane receptors mediating responses to specific attractant and repellent stimuli. Four of these receptors are common to both E. coli and Salmonella serovar Typhimurium: Tar, Tsr, Trg, and Aer. One receptor is unique to each species: Tap in E. coli and Tcp in Salmonella serovar Typhimurium. The abundance of the chemotaxis-specific receptors varies, with Tsr and Tar being highly abundant and Tap, Trg, and Aer being less prevalent (12, 43, 50, 89).
Composition of the receptor supramolecular complex.
The chemotaxis-specific receptors are stable homodimers, connected via a linker protein, CheW, to a histidine kinase, CheA, generating stable ternary complexes (Fig. 1) (for a review, see reference 43). The paradigm for many years was that each ternary complex is built from one receptor dimer, two CheW molecules, and one CheA dimer. Accumulated evidence suggests, however, that the actual structure of the receptor complex is more elaborate. Thus, immunoelectron microscopical studies indicated that, in the presence of CheA and CheW, the receptors form large clusters, located mainly at one (primarily) or both poles of the cell (74). Furthermore, formation of active supramolecular complexes, consisting of about seven receptors, two or four CheW molecules, and one CheA dimer (Fig. 1), was demonstrated in vitro with purified proteins (65). The recently resolved crystal structure of the cytoplasmic domain of the Tsr receptor has revealed that the tails of three dimer receptors come together and form a trimeric structure (54). Considering this structure, Shimizu et al. (97) elegantly examined plastic models of the proteins involved in the receptor supramolecular complexes. The models, generated by three-dimensional technology, predicted that these supramolecular complexes form a two-dimensional hexagonal lattice, built from trigonal units. Each unit is composed of three MCP dimers, three molecules of CheW, and three monomers of CheA, joined to CheA monomers of another unit at their dimerization domain. One might wonder what is the cause of the different MCP-CheA-CheW stoichiometries in the seven-dimer model (65) and the lattice model (97). This could, perhaps, be attributed to the leucine zipper dimerization domain fused, in the experiments that lead to the former model, with the N terminus of the cytoplasmic receptor fragment.
The finding that there are two forms of CheA in the cell—full length (CheAL [L for long]) and short (CheAS)—introduced further complications to elucidating the composition of the receptor supramolecular complex. CheAS is generated by translation of cheA from an alternative start site and is 97 residues shorter than CheAL (55). Under optimal motility conditions, the CheAS/CheAL ratio in the cell is 1:1 (110). However, the relative amounts of CheAS and CheAL within the receptor supramolecular complexes are not known.
While it is believed that the high-order structure of the receptor supramolecular complex has a role in chemotactic signaling (see below), the composition and stoichiometry of the five different MCPs within these receptor supramolecular complexes are not known. If each supramolecular complex contains only seven receptor dimers (65), the supramolecular complexes must differ from each other with respect to their MCP composition. This is because the low-abundance receptors cannot be fully functional (58, 62) and cannot be clustered (71) unless they interact with the high-abundance receptors. Thus, if the MCP composition in all the receptor supramolecular complexes were the same, each complex would be composed of at least 19 or 20 receptor dimers according to the known stoichiometry between the different types of MCPs (10 or 11 Tsr, 6 Tar, and 1 Aer, Tap and Trg [39]). If, on the other hand, the receptor supramolecular complex is indeed arranged as a lattice (97), any MCP combination may be possible. We favor the possibility of a lattice because of the flexibility that it provides to the system.
Transmembrane signal transduction.
A common mechanism for receptor signaling in many signal transduction systems involves receptor dimerization. Since the chemotaxis receptors are stable homodimers and since it was demonstrated that intersubunit cross-linking of engineered cysteine residues does not affect the signaling properties of the receptors, dimerization does not seem to be involved in chemotactic signaling (for reviews, see references 43 and 59). Furthermore, studies of changes in receptor structure, induced by ligand binding, revealed only subtle conformational changes (43, 59, 85). Therefore, an important, yet unsolved question is how the chemotaxis receptors transduce the sensory signal across the membrane.
Two main models for transmembrane signaling have been suggested. According to one model, which is based on the analysis of crystallographic data of the ligand-free and ligand-bound sensing domain of Tar (reference 117 and references cited therein) and is supported by subsequent crystallographic data (31), transmembrane signaling is performed by intersubunit rotation of the receptor monomers within the receptor dimer (117). According to the other model, which is based on a different analysis of the same crystallographic data, on 19F nuclear magnetic resonance (NMR) studies of the sensing domain of Tar, and on electron paramagnetic resonance studies of a spin-labeled Tar receptor, transmembrane signaling is performed by conformational changes within a single subunit of the receptor dimer (reference 85 and references cited therein). This model suggests that the signal is transduced within a receptor monomer across the membrane by a piston-like displacement of one transmembrane segment relative to the other. The ability of a Tar receptor with a single cytoplasmic domain per dimer to transduce chemotactic signals is in line with this model (45, 102). The question of how the subtle conformational changes result in large signals within the cytoplasm may be answered by assuming that the receptor-coupled enzymes can detect small changes in the receptor conformation (85) or that lateral signaling within the supramolecular complex is involved (59, 61). In line with the latter possibility, it was found in vitro that subsaturating concentrations of attractant accelerate formation of active supramolecular complexes (61). This implies that not only existing interactions but also the assembly or disassembly of the complexes may contribute to signal regulation (61). (See references 37 and 41 for elimination of older potential possibilities of transmembrane signaling.)
Intracomplex signal propagation.
Some years ago, Ames and Parkinson isolated tsr mutants locked in either CW or CCW rotation (3). The finding of such mutants raised the possibility that each receptor exists in one of two active signaling states, CW or CCW, and that the receptor output is controlled by modulating the ratio between these states (3, 4). However, without some additional mechanism, such a situation cannot explain how signals from a small fraction of receptors (e.g., signals generated by low-abundance receptors or by low-occupied high-abundance receptors) can be heard on top of much stronger, conflicting signals from nonstimulated receptors.
Such an additional mechanism may be provided by modulation of the interreceptor interactions within the supramolecular complexes. Indirect supportive evidence for changes in the aggregation state at different signaling states of the receptors was provided by Long and Weis (67), who studied locked mutants of the Tar receptor. They found a correlation between the swimming phenotype of tar mutants and the oligomerization properties of the receptor cytoplasmic fragment. Most of the cytoplasmic fragments studied, which were derived from mutants locked in the CCW state, formed oligomers at neutral pH. In contrast, cytoplasmic fragments derived from mutants locked in the CW state or from the wild-type strain did not exhibit any significant oligomer formation. Accordingly, Long and Weis suggested that subunit interactions within the cytoplasmic region are stronger in the attractant-bound form of the receptor than in the attractant-free form. Cooperativity of the kinase activity of CheA with respect to the stimulus concentration (22, 61) is also in line with the possibility of transmembrane signaling mechanism involving receptor aggregates and stimulus-induced changes in the aggregation states of the receptors.
How can such modulation of the interreceptor interactions make signals from a small fraction of receptors be heard on top of conflicting signals from the nonstimulated receptors? Two possibilities have been proposed—a shut off mechanism and an amplification mechanism. According to the first possibility, a change in the packing of the receptors within the cluster, stimulated by occupying a small fraction of the receptors, might act as a shutoff mechanism for other receptors in the cluster, resulting in modulation of the kinase activity according to the signal from the stimulated receptor (Y. Blat, personal communication). According to the other possibility, changes in interreceptor interactions are involved in amplification of the signals from the stimulated receptors. Thus, Bray et al. (27) calculated that a mechanism by which ligand binding changes the activity of a receptor, which then propagates to neighboring receptors in the cluster, can account quantitatively for the high sensitivity and response range of E. coli. Levit et al. (59) suggested that ligand binding changes the packing of the receptors and CheA within the cluster and, consequently, CheA acquires an active or inactive conformation, depending on the signal. These suggestions call for direct experimental evidence.
The information sensed by the receptors is transmitted within the receptor supramolecular complex to regulate the kinase activity of CheA. It has been shown in vitro that the kinase activity is inhibited by attractants (21, 82). (No reports regarding the in vitro effect of repellents on modulation of the kinase activity are available.) It is therefore generally assumed that, in vivo, receptor-occupied attractants and repellents inhibit and activate the kinase, respectively. The signal from the receptors is received by the receptor-binding domain of CheA. Then, from the phosphotransfer domain of CheA, which contains the phosphorylation site His48, it is transmitted to the downstream proteins (CheY and CheB) (43, 100).
INTERACTIONS BETWEEN THE MESSENGER PROTEIN AND THE SUPRAMOLECULAR COMPLEXES
Signals generated by the receptor supramolecular complex are transmitted to the flagellar-motor supramolecular complex by the messenger protein, CheY. This protein—a response regulator in the superfamily of two-component regulatory systems (for a review, see reference 86)—is a small (14-kDa), single-domain molecule which, despite its small size, is multifunctional.
Interaction of CheY with the receptor supramolecular complex.
Once CheA is autophosphorylated, it rapidly transfers the phosphate group to Asp57 of CheY (23). Studies of the crystal structure of the complex between CheY and a fragment, which contains the CheY-binding domain of CheA and which is 75 or 134 residues long (reference 80 or 111, respectively), indicated that the binding triggers conformational changes in CheY, propagating from its CheA-binding surface to its active site (80, 111). Presumably these conformational changes are necessary for CheY phosphorylation. An outcome of the phosphorylation is a reduced affinity of CheY to CheA. Consequently, the phosphorylated form of CheY (CheY∼P) is released from the receptor supramolecular complex (63, 95).
It should be noted that CheY can also be phosphorylated by small phosphodonors such as acetyl phosphate (69). Although acetyl phosphate is present within the cell and its level varies with the growth phase and conditions (78, 87), the rate at which it phosphorylates CheY is much lower than that of CheA-mediated phosphorylation (77). Therefore, while the role of acetyl phosphate-mediated phosphorylation of CheY in vivo is questionable, it is a useful tool for in vitro phosphorylation of CheY. This CheA-independent phosphorylation indicates that CheY, like other response regulators, can catalyze its own phosphorylation.
Interaction of CheY with the flagellar-motor supramolecular complex.
Phosphorylation of CheY not only reduces the affinity of the protein to CheA; it also elevates the affinity of CheY to the protein FliM (79, 112), which is a component of the flagellar-motor supramolecular complex. Consequently, when CheY∼P is released from the receptor supramolecular complex, it diffuses in the cytoplasm and interacts, via FliM, with the flagellar-motor complex. Genetic studies suggested that the N terminus of FliM is involved in CheY binding (104). Biochemical studies localized the CheY-binding domain to the 16 N-terminal residues of FIiM (28). However, the possibility that these residues do not form the whole CheY-binding site cannot be eliminated (76). The end result of this phosphorylation-dependent interaction is an increased probability of flagellar rotation in the CW direction (8). Gradual production of CheY under intracellular phosphorylating conditions (2, 33, 57) or intracellular production of an active CheY mutant protein (92) revealed that this probability increases sigmoidally. Studies of the correlation between CW rotation and the intracellular level of CheY∼P in individual cells demonstrated that the increase in CW rotation is very steep (33), suggesting high cooperativity of CheY∼P binding or of processes within the flagellar-motor supramolecular complex subsequent to the binding.
In a recent study aimed at determining how many FliM molecules within a single flagellar-motor supramolecular complex should be occupied by CheY∼P to generate CW rotation, wild-type FliM (FliMWT) and a mutant FliM protein that is almost locked in CW rotation (FliMCW) were coexpressed in a gutted strain that lacks, among other chemotaxis proteins, CheY (A. Bren and M. Eisenbach, submitted for publication). Surprisingly, a probability of 50% of CW rotation was achieved only when ∼90% of the FliM molecules at the flagellar-motor supramolecular complex were FliMCW molecules. Around this fraction of FliMCW molecules within the complex, the gain of CW rotation with the increased fraction of FliMCW was very steep. This steepness suggests that if FliMCW correctly reflects FliMWT occupied by CheY∼P, the high cooperativity, discussed in the preceding paragraph, is primarily a reflection of cooperativity of a postbinding step.
Regulation of CheY activity.
The phosphorylation level of CheY, which controls its CW rotation-generating activity, is determined by the rates of CheY phosphorylation (by CheA) and dephosphorylation (either spontaneously or, in enhanced manner, by CheZ). As described below, both enzyme-mediated processes are regulated, each by a different mechanism.
(i) Mechanism of CheY activation.
The changes, which occur in CheY upon phosphorylation and prompt its ability to bind to FliM and generate CW rotation, are not fully understood. NMR studies of CheY at a steady-state level of phosphorylation indicated that, upon phosphorylation, CheY undergoes conformational changes that are not restricted to the vicinity of the phosphorylation site but rather propagate along most of the protein (36, 68). The amino acid residues, the electronic environment of which changes in response to phosphorylation, were identified. However, the extent of these conformational changes is not yet clear because there is no direct correlation between the perturbation in a given NMR's chemical shift and the extent of the related conformational change (35). Accurate information about the extent of the conformational changes could be obtained from X-ray crystallography of the phosphorylated and nonphosphorylated forms of CheY. However, due to the short life span of the phosphorylated state (half-life of 20 s [29, 51]), it is very difficult to crystallize CheY∼P. To circumvent this difficulty, two main approaches have been taken. In one approach, analogs of CheY∼P in which the phosphate group was stabilized or replaced by an analogous group were prepared (48, 116). In the other approach, mutant CheY proteins with different functions, including nonactive and phosphorylation-independent active proteins, were isolated. Some analogs and mutant proteins are listed in Table 1. The X-ray structures of several of these proteins were resolved, aiming at finding a correlation between their function and structure (44, 47, 52, 94, 121). Unlike the large NMR shifts observed between the phosphorylated and nonphosphorylated forms of CheY, no major conformational differences which extend beyond the vicinity of the phosphorylation site could be detected between the crystal structures of the various studied forms of CheY (44, 47, 52, 94, 121). The reason for the apparent difference between the X-ray and NMR results is not known. One possibility is that subtle structural changes might result in relatively large perturbations in the NMR's chemical shifts. Another possibility is that the protein might be in a number of dynamic states, most of which might not be represented by its crystal structure. Whatever the extent of the conformational changes that occur in CheY upon phosphorylation, they (rather than the phosphate group per se) appear to be sufficient for activating CheY. This is evident from the observations that the mutant protein CheY13DK/106YW, which is active without phosphorylation (92), binds FliM in vitro (91) and generates CW rotation in vivo (92).
TABLE 1.
Properties of some CheY mutant proteins and analogs
| CheY protein | FliM-binding activity of:
|
CW rotation-generating activity | Orientation of the side chain of residue 106 | |
|---|---|---|---|---|
| CheY | CheY∼P | |||
| WTa | Low (112) | High (112) | Active when phosphorylated (8) | Inward and outward (106) |
| CheY87TI | Like WT (121) | Inapplicableb | Nonactive (6, 121) | Outward (44) |
| CheY13DK | Like WT (94, 113) | Inapplicablec | Active without being phosphorylated (2, 24, 26) | Outward (52) |
| CheY106YW | Like WT (120) | Like WT (120) | Hyperactive when phosphorylated (120) | Inward (121) |
| CheY13DK/106YW | Like CheY∼PWT (47, 91, 94) | Inapplicablec | Active without being phosphorylated (92) | Outward (P. Matsumura, personal communication) |
| CheY87TI/106YW | Like WT (121) | Inapplicableb | Nonactive (121) | Outward (121) |
| CheY95IV | Higher than WT (93, 94) | Higher than WT (93, 94) | Hyperactive when phosphorylated (93, 94) | Outward (94) |
| Phosphono-CheY | Like CheY∼PWT (47, 48) | Inapplicable | Not determined | Inward (47) |
| BeFX·CheY | Like CheY∼PWT (116) | Inapplicable | Not determined | Inward (32) |
WT, wild type.
The mutant protein is not phosphorylated by acetyl phosphate (121).
The mutant protein is not phosphorylated.
A prominent difference between the various studied forms of CheY, as revealed from their X-ray structures, is the orientation of the side chain of Tyr106, located on the face of the molecule. This side chain appears in wild-type CheY as a mixture of inward and outward conformations, whereas in all the other studied mutant proteins and analogs, the side chain is found in only one orientation (Table 1). It was proposed that phosphorylation of Asp57 initiates conversion of Tyr106 from a solvent-exposed orientation to a more internal position, possibly as a consequence of repositioning residues Thr87 (which appears to form a hydrogen bond with Asp57 [32]) and Lys109 (which may form a hydrogen bond with the oxygen atoms of the phosphate group) (47). This notion is supported by the observations that (i) in the analogs phosphono-CheY (47) and BeFX · CheY (32), the side chain of residue 106 is oriented inwardly, and (ii) in the response regulators FixJ (14) and Spo0A (60) (whose structures were resolved in both the phosphorylated and nonphosphorylated forms), the side chains of the residues that are homologous to Tyr106 of CheY appear to be shifted, upon phosphorylation, from a solvent-exposed orientation to an interior position. The finding that the side chain of Tyr106 is oriented outwardly in the complex between CheY and the CheY-binding domain of CheA (80, 111) further suggests that repositioning of Tyr106 might be involved in the release of CheY from CheA and in its subsequent binding to FliM. It thus seems that the rotameric state of Tyr106 may be important for determining the activity of CheY (32, 47, 121). However, there appears to be inconsistency in the Tyr106 orientation in the available structures of CheY, for which reason the situation still seems to be somewhat ambiguous. For example, in the phosphorylation-independent active mutants CheY13DK (52) and CheY13DK/106YW (P. Matsumura, personal communication), the side chain of residue 106 is in a solvent-exposed position rather than inwardly oriented (Table 1). Apparently, more experiments are required to deduce the mechanism of CheY activation and resolve the involvement of Tyr106 in this activation.
Phosphorylation is not the only chemical modification that CheY undergoes and not the only one that activates the protein. CheY also undergoes lysine acetylation (10), primarily at residues 92 and 109 (88). This acetylation increases, to a large extent, the CW rotation-causing activity of CheY both in vitro (in cytoplasm-free envelopes) (10) and in vivo (7, 88). However, unlike CheY phosphorylation, the acetylation does not affect the binding of CheY to FliM, suggesting that it is involved in a post-FliM binding step (88). Although it was recently demonstrated that this chemical modification is involved in chemotaxis (R. Barak and M. Eisenbach, unpublished data), its role is still an open question.
(ii) Mechanism of CheY deactivation.
A major player in the mechanism of CheY deactivation and CW termination is the phosphatase CheZ, which effectively accelerates CheY∼P dephosphorylation (51). (The term phosphatase is used here in the broader sense and does not imply the mechanism of CheZ action.) Biochemical studies revealed that, similar to FliM, CheZ binds CheY in a phosphorylation-dependent manner (15). The CheY-binding domain is located at the C terminus of CheZ (16). Following binding to CheY∼P and a subsequent 50- to 100-ms delay, CheZ is turned on and CheY∼P is dephosphorylated (19). The delay might ensure that the phosphatase activity of CheZ is modulated only after a chemotactic response is established, so that the gain of the response is not reduced. (The response to a negative stimulus is completed within ∼50 ms [53].) The activation of the phosphatase depends, with positive cooperativity, on CheY∼P concentration, and it appears to involve oligomerization of CheZ which, otherwise, is in a dimeric form (17–19, 91). While bound to FliM of the flagellar-motor supramolecular complex, CheY∼P is protected from dephosphorylation by CheZ (29). This suggests that, as part of the mechanism of CW termination, CheZ dephosphorylates free CheY∼P and causes dissociation of CheY∼P from the flagellar-motor complex by shifting the equilibrium between bound and free CheY∼P (29).
Under reducing conditions, CheZ also interacts with CheAS and forms a CheZ-CheAS complex at a ratio of 5:1. The consequence of this interaction is an increase in the phosphatase activity of CheZ (109). In addition, by analyzing cells expressing a functional, full-length CheZ fused with green or yellow fluorescent protein, it was recently found that at least some of the CheZ molecules are localized in clusters at the cell's poles (98; M. Manson, personal communication). This observation suggests that CheZ, like all the other cytoplasmic chemotaxis proteins, can be attached to the receptor supramolecular complex. This attachment might position CheZ in close proximity to CheAS. In view of this observation, it might be tempting to speculate that CheZ-CheAS interaction at the receptor supramolecular complex may play a role in the rapid CheY∼P dephosphorylation that presumably occurs during an attractant response. According to this speculation, CheZ at the receptor supramolecular complex is involved in CCW rotation generation in response to attractants, whereas free CheZ is involved, after a delay, in CW rotation termination following a repellent response. Such a possibility raises a number of intriguing questions, a few of which follow. (i) It was found that mutants which do not express CheAS are unimpaired in their ability to respond to attractants under standard assay conditions (90). If a CheZ-CheAS interaction is indeed involved in CCW rotation generation, how does the absence of CheAS have no effect on the response to attractants? (ii) If CheZ and CheAS are in constant interaction with each other within the receptor supramolecular complex (because the cytoplasm is a reducing environment [109]), how is the activity of CheZ modulated by attractants rather than being constitutively active? (iii) If CheZ and CheAS interact only when a ligand is bound to the receptor supramolecular complex, how is the activation of CheZ sufficiently fast and not delayed by its oligomerization?
(iii) Regulation of the interactions of CheY with its targets.
Even though CheY can bind to three proteins—CheA, CheZ, and FliM, it can bind to only one protein at a time. This is due to the fact that the C-terminal portion of CheY is involved in the binding to all these proteins, forming an overlapping binding surface (references 79, 80, 111, and 122 and references cited therein). Since the affinity of CheA is higher to nonphosphorylated CheY than to CheY∼P whereas that of FliM and CheZ is higher to CheY∼P, it is reasonable that, upon phosphorylation, the conformation of the C-terminal portion of CheY changes from one that recognizes mainly CheA to one that recognizes mainly FliM and CheZ. In line with this notion, the CheY-binding regions of CheZ (16) and FliM (28) were found to share several common features (28, 79). Furthermore, the binding constants of peptides, which contain these regions, to CheY∼P were found to be similar (79). Taken together, these observations suggest that FliM and CheZ compete for CheY∼P and that the inability of CheZ to dephosphorylate FliM-bound CheY∼P (29) may be the result of its inability to bind to CheY∼P when the latter is bound to FliM.
INTERACTIONS WITHIN THE FLAGELLAR-MOTOR SUPRAMOLECULAR COMPLEX
Composition of the complex.
The flagellar-motor supramolecular complex, located at the base of the flagellum, is built, like any other electrical motor, from a rotor and stator. The rotor is built from the proteins FliG, FliF, and probably also FliM and FliN. The stator is built from the proteins MotA and MotB, which form a proton channel anchored to the cell wall by MotB. An inward proton flow through this channel generates the torque for rotation (for a comprehensive review of the flagellar motor, see reference 73). The proteins FliG, FliM, and FliN constitute a gearbox, termed a switch, extending into the cytoplasm (for a review, see reference 72). The switch is the element of the supramolecular complex onto which CheY∼P docks and which determines the direction of flagellar rotation (for a review, see reference 9). Each flagellar-motor supramolecular complex contains ∼27 copies of FliF, ∼35 copies of FliM, ∼35 copies of FliG, ∼100 copies of FliN, and an unknown number of MotA and MotB molecules that form eight force-generating units (references 73, 103, and 118 and references cited therein). In the complex, the switch proteins interact with each other (references 75, 83, 101, 105, and 118 and references therein). FliG, in addition, binds to MotA (46, 101, 119) and thus appears to link the rotor and the stator. It also interacts with FliF and thereby links the switch to the central element of the motor (references 73, 75, and 83 and references therein).
Function of the complex.
The mechanism underlying flagellar rotation appears to involve electrostatic interactions between the rotor and stator (119), but other mechanisms have also been suggested (for a few recent examples, see references 84 and 103). A number of models involving electrostatic interactions have been proposed (for a review, see reference 30; for the more-recent models, see references 42, 107, and 108). According to the most recent one (108), rotation is generated by electrostatic interaction between a proton in the proton channel and alternating tilted rows of fixed positive and negative charges on the rotor. Switching from one direction of rotation to the other involves, according to the model, a change in the angle of tilt. In line with this notion, it was hypothesized on the basis of the crystal structure of the C-terminal domain of FliG (the domain that interacts with MotA) that, at each direction of rotation, different subsets of charged residues of FliG interact with the stator (66).
The models mentioned above provide potential mechanistic modes of switching from one direction of rotation to the other. However, actually nothing is known about signal propagation within the switch itself subsequent to CheY∼P-FliM binding. Kuo and Koshland (57) proposed that CW rotation generation by CheY involves more than one kinetic state. Indeed, it appears that CheY∼P binding to FliM is essential but, yet, insufficient for the generation of CW rotation. This is based on two main lines of evidence. (i) Phosphorylation of CheY in cytoplasm-free envelopes does not result in enhanced CW rotation, unless additional, unidentified cytoplasmic constituents (not chemotaxis proteins) are present (8). (ii) There is a lack of correlation between the ability of some mutant CheY proteins to bind FliM in vitro and to generate CW rotation in vivo (40, 97a). Indeed, this lack of correlation could, in principle, be explained by assuming that there is a difference between the affinities of CheY∼P to FliMCW and FliMCCW and that this difference is not observed when purified FliM is employed. However, both lines of evidence taken together suggest that one or more regulatable post-CheY∼P-FliM binding events should occur for the generation of CW rotation.
INTERACTIONS INVOLVED IN ADAPTATION
Adaptation, namely, restoration of the prestimulus behavior in the presence of the stimulus, is an essential component of every behavioral system, chemotaxis included (56, 99). Adaptation in bacterial chemotaxis is controlled by a feedback mechanism that modulates the methylation level of the MCP receptors. Two enzymes, CheB and CheR, are involved in this mechanism by interacting with the receptor supramolecular complexes and chemically modifying them.
CheR is a methyltransferase, which catalyzes S-adenosylmethionine-dependent methylation of specific glutamate residues (four to six methylatable residues for each MCP) on the cytoplasmic portion of the receptors during adaptation to positive stimuli (for a review, see reference 37). The outcome is an enhancement of CheA autophosphorylation and, thereby, transmission of a CW signal (20, 82).
CheB is a methylesterase that demethylates the receptors during adaptation to negative stimuli (for a review, see reference 37). It also has an amidase activity that catalyzes the conversion of specific glutamine residues of the MCP receptors into glutamate residues. The interaction of CheB with the receptor results in hydrolysis of the methyl ester bond on the side chain of the glutamate residue, and the receptor undergoes demethylation. The outcome of this demethylation is inhibition of CheA autophosphorylation and, thereby, transmission of a CCW signal (20, 82).
Thus, the relative rates of the methylation and demethylation reactions determine the steady-state level of receptor methylation, and this level regulates the kinase activity of CheA. This regulation occurs only after the initial chemotactic response. Recently it was demonstrated that a high methylation level decreases the affinity of the receptor supramolecular complex to attractants (22, 61) (up to 10,000-fold for serine [61]), suggesting that the methylation level regulates ligand binding to receptor supramolecular complexes. This modulation of ligand-binding affinity extends the range of the chemotactic response and suggests that the cells adapt not only by methylation-dependent modulation of the kinase activity but also by decreasing the extent of stimulant binding to the receptor complex. It has also been proposed that the delayed activation of CheZ discussed above might be involved in the adaptation process (19).
CheR-receptor interaction.
CheR is a 32-kDa, two-domain protein (35, 100). The N-terminal domain appears to be involved in MCP recognition. It contains positively charged residues that might complement the negatively charged residues in the methylation region of the MCPs. The actual binding of CheR to the MCPs is, however, carried out by its C-terminal domain. This domain also contains features that are common for S-adenosylmethionine-dependent methyltransferases.
Two separate domains of the receptor are involved in the interaction with CheR: a binding domain onto which CheR docks and a domain that is methylated by CheR. The docking site of CheR on the receptor consists of the last five residues of the latter (114). Intriguingly, this pentapeptide is present only in high-abundance receptors, suggesting that CheR methylates the low-abundance receptors while it is docked onto a high-abundance receptor (114). This suggestion, which was later confirmed experimentally (58, 62), explains the poor methylation of the low-abundance receptors in mutant strains, which lack the high-abundance receptors, and the resulting defective adaptation of these mutants (49, 115).
CheB-receptor interaction.
CheB is a response-regulator protein whose activity, like that of the other chemotactic response regulator CheY, is regulated by CheA-mediated phosphorylation (51, 70). It is a 35-kDa protein, containing two domains: a regulatory domain at the N terminus that undergoes aspartate phosphorylation and an effector domain at the C terminus that possesses amidase and esterase activities (35, 100). The regulatory domain is homologous to the entire length of CheY. When this domain is not phosphorylated, it inhibits the esterase activity of CheB; when phosphorylated, it stimulates this activity (references 5, 34, and 70 and references therein). On the basis of structural and biochemical data, it was proposed that phosphorylation of the regulatory domain results in reorganization of its interface, exposing the active site to the receptor, and simultaneously stimulating the methylesterase activity of CheB (5, 34). The interaction between CheB and the receptors is complex and probably involves several regions on the surface of CheB (35). The docking site of CheB on the receptors is the same C-terminal pentapeptide onto which CheR docks (11). This finding raised the possibility that the relative rates of the methylation and demethylation processes might be influenced by competition between the two enzymes on binding to this site (11).
CheB-CheA interaction.
CheB has two targets of interaction at the receptor supramolecular complex: one is the receptor itself, and the other is CheA. During the latter interaction, CheA phosphorylates CheB with a consequent release of CheB from CheA and its binding to the receptors. CheB and CheY bind, apparently with comparable affinities, to the same domain of CheA, and they therefore compete with each other for binding to CheA (63). Interestingly, even though the N-terminal domain of CheB is homologous to the entire length of CheY, the residues of CheY involved in CheA binding are not conserved in CheB. This suggests that CheY and CheB may bind to overlapping, but yet distinct, sites on CheA (80, 111).
CONCLUSIONS
Protein-protein interactions are the heart of the chemotactic response, which is accomplished, as shown in Fig. 1, by signal transmission between two supramolecular complexes—the receptor complex and the flagellar-motor complex. The signal is transduced by the messenger protein CheY. During the last 3 decades, most, if not all, of the chemotaxis proteins and the interactions between these proteins have been identified. The interactions are summarized in Table 2. For many of the proteins listed in Table 2, a detailed structure is available.
TABLE 2.
Direct interactions involved in chemotaxis
| Protein | Known direct interactions |
|---|---|
| MCP receptor | Intradimer, interdimer, CheW, CheB, CheR, CheAa |
| CheW | MCP, CheA |
| CheAL | Intradimer, CheW, CheY, CheB, CheAS, MCPa |
| CheAS | CheAL, CheW, CheZ |
| CheY | CheA |
| CheY∼P | FliM, CheZ |
| CheZ | CheY∼P, CheAS |
| CheR | MCP |
| CheB | CheA |
| CheB∼P | MCP |
| FliM | CheY∼P, FliG, FliN |
| FliN | FliM, FliG |
| FliG | FliM, FliN, FliF, MotA |
| FliF | FliG |
| MotA | MotB, FliG |
| MotB | MotA |
Based on genetic observations only (64).
Although the signaling cascade in bacterial chemotaxis is one of the best-understood signal transduction systems, many major questions are still open. A few examples follow. (i) How are the sensory signals transduced across the membrane by the chemotaxis receptors? (ii) How do receptors of different abundance generate signals of similar strength? (iii) Is CheAS involved in chemotaxis and, if so, how? (iv) What is the function of the CheZ molecules located at the receptor supramolecular complex? (v) How is CheY activated by acetylation, and what is the role of this way of activation? (vi) How is the signal propagated within the switch subsequent to CheY∼P binding? Structural details of the proteins involved and of the complexes formed between them, together with novel approaches for studying functional aspects of the system, might bring us closer to complete understanding of how signals are heard during bacterial chemotaxis.
ACKNOWLEDGMENTS
M.E. is the incumbent of the Jack and Simon Djanogly Professorial Chair in Biochemistry. Work in our laboratory was supported by the MINERVA Foundation in Germany, by grant 96-00013 from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel, and by grant 111/99 from the Israel Science Foundation.
REFERENCES
- 1.Adler J. Chemotaxis in Escherichia coli. Cold Spring Harbor Symp Quant Biol. 1965;30:289–292. doi: 10.1101/sqb.1965.030.01.030. [DOI] [PubMed] [Google Scholar]
- 2.Alon U, Camarena L, Surette M G, Arcas B A Y, Liu Y, Leibler S, Stock J B. Response regulator output in bacterial chemotaxis. EMBO J. 1998;17:4238–4248. doi: 10.1093/emboj/17.15.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames P, Parkinson J S. Transmembrane signaling by bacterial chemoreceptors: E. coli transducers with locked signal output. Cell. 1988;55:817–826. doi: 10.1016/0092-8674(88)90137-7. [DOI] [PubMed] [Google Scholar]
- 4.Ames P, Parkinson J S. Constitutively signaling fragments of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 1994;176:6340–6348. doi: 10.1128/jb.176.20.6340-6348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anand G S, Goudreau P N, Stock A M. Activation of methylesterase CheB: evidence of a dual role for the regulatory domain. Biochemistry. 1998;37:14038–14047. doi: 10.1021/bi980865d. [DOI] [PubMed] [Google Scholar]
- 6.Appleby J L, Bourret R B. Proposed signal transduction role for conserved CheY residue Thr87, a member of the response regulator active-site quintet. J Bacteriol. 1998;180:3563–3569. doi: 10.1128/jb.180.14.3563-3569.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barak R, Abouhamad W N, Eisenbach M. Both acetate kinase and acetyl coenzyme A synthetase are involved in acetate-stimulated change in the direction of flagellar rotation in Escherichia coli. J Bacteriol. 1998;180:985–988. doi: 10.1128/jb.180.4.985-988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barak R, Eisenbach M. Correlation between phosphorylation of the chemotaxis protein CheY and its activity at the flagellar motor. Biochemistry. 1992;31:1821–1826. doi: 10.1021/bi00121a034. [DOI] [PubMed] [Google Scholar]
- 9.Barak R, Eisenbach M. Regulation of interaction between signaling protein CheY and flagellar motor during bacterial chemotaxis. Curr Top Cell Regul. 1996;34:137–158. doi: 10.1016/s0070-2137(96)80005-7. [DOI] [PubMed] [Google Scholar]
- 10.Barak R, Welch M, Yanovsky A, Oosawa K, Eisenbach M. Acetyladenylate or its derivative acetylates the chemotaxis protein CheY in vitro and increases its activity at the flagellar switch. Biochemistry. 1992;31:10099–10107. doi: 10.1021/bi00156a033. [DOI] [PubMed] [Google Scholar]
- 11.Barnakov A N, Barnakova L A, Hazelbauer G L. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc Natl Acad Sci USA. 1999;96:10667–10672. doi: 10.1073/pnas.96.19.10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bibikov S I, Biran R, Rudd K E, Parkinson J S. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bilwes A M, Alex L A, Crane B R, Simon M I. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 14.Birck C, Mourey L, Gouet P, Fabry B, Schumacher J, Rousseau P, Kahn D, Samama J P. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure Fold Design. 1999;7:1505–1515. doi: 10.1016/s0969-2126(00)88341-0. [DOI] [PubMed] [Google Scholar]
- 15.Blat Y, Eisenbach M. Phosphorylation-dependent binding of the chemotaxis signal molecule CheY to its phosphatase, CheZ. Biochemistry. 1994;33:902–906. doi: 10.1021/bi00170a008. [DOI] [PubMed] [Google Scholar]
- 16.Blat Y, Eisenbach M. Conserved C-terminus of the phosphatase CheZ is a binding domain for the chemotactic response regulator CheY. Biochemistry. 1996;35:5679–5683. doi: 10.1021/bi9530447. [DOI] [PubMed] [Google Scholar]
- 17.Blat Y, Eisenbach M. Mutants with defective phosphatase activity show no phosphorylation-dependent oligomerization of CheZ, the phosphatase of bacterial chemotaxis. J Biol Chem. 1996;271:1232–1236. doi: 10.1074/jbc.271.2.1232. [DOI] [PubMed] [Google Scholar]
- 18.Blat Y, Eisenbach M. Oligomerization of the phosphatase CheZ upon interaction with the phosphorylated form of CheY, the signal protein of bacterial chemotaxis. J Biol Chem. 1996;271:1226–1231. doi: 10.1074/jbc.271.2.1226. [DOI] [PubMed] [Google Scholar]
- 19.Blat Y, Gillespie B, Bren A, Dahlquist F W, Eisenbach M. Regulation of phosphatase activity in bacterial chemotaxis. J Mol Biol. 1998;284:1191–1199. doi: 10.1006/jmbi.1998.2224. [DOI] [PubMed] [Google Scholar]
- 20.Borkovich K A, Alex L A, Simon M I. Attenuation of sensory receptor signaling by covalent modification. Proc Natl Acad Sci USA. 1992;89:6756–6760. doi: 10.1073/pnas.89.15.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borkovich K A, Simon M I. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell. 1990;63:1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- 22.Bornhorst J A, Falke J J. Attractant regulation of the aspartate receptor-kinase complex: limited cooperative interactions between receptors and effects of the receptor modification state. Biochemistry. 2000;39:9486–9493. doi: 10.1021/bi0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourret R B, Borkovich K A, Simon M I. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- 24.Bourret R B, Drake S K, Chervitz S A, Simon M I, Falke J J. Activation of the phosphosignaling protein CheY. II. Analysis of activated mutants by 19F NMR and protein engineering. J Biol Chem. 1993;268:13089–13096. [PMC free article] [PubMed] [Google Scholar]
- 25.Bourret R B, Hess J F, Borkovich K A, Pakula A A, Simon M I. Protein phosphorylation in chemotaxis and two-component regulatory systems of bacteria. J Biol Chem. 1989;264:7085–7088. [PubMed] [Google Scholar]
- 26.Bourret R B, Hess J F, Simon M I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci USA. 1990;87:41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bray D, Levin M D, Morton-Firth C J. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 28.Bren A, Eisenbach M. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J Mol Biol. 1998;278:507–514. doi: 10.1006/jmbi.1998.1730. [DOI] [PubMed] [Google Scholar]
- 29.Bren A, Welch M, Blat Y, Eisenbach M. Signal termination in bacterial chemotaxis: CheZ mediates dephosphorylation of free rather than switch-bound CheY. Proc Natl Acad Sci USA. 1996;93:10090–10093. doi: 10.1073/pnas.93.19.10090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caplan S R, Kara-Ivanov M. The bacterial flagellar motor. Int Rev Cytol. 1993;147:97–164. doi: 10.1016/s0074-7696(08)60767-6. [DOI] [PubMed] [Google Scholar]
- 31.Chi Y I, Yokota H, Kim S H. Apo structure of the ligand-binding domain of aspartate receptor from Escherichia coli and its comparison with ligand-bound or pseudoligand-bound structures. FEBS Lett. 1997;414:327–332. doi: 10.1016/s0014-5793(97)01027-2. [DOI] [PubMed] [Google Scholar]
- 32.Cho H S, Lee S Y, Yan D, Pan X, Parkinson J S, Kustu S, Wemmer D E, Pelton J G. NMR structure of activated CheY. J Mol Biol. 2000;297:543–551. doi: 10.1006/jmbi.2000.3595. [DOI] [PubMed] [Google Scholar]
- 33.Cluzel P, Surette M, Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 34.Djordjevic S, Goudreau P N, Xu Q P, Stock A M, West A H. Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc Natl Acad Sci USA. 1998;95:1381–1386. doi: 10.1073/pnas.95.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Djordjevic S, Stock A M. Structural analysis of bacterial chemotaxis proteins: components of a dynamic signaling system. J Struct Biol. 1998;124:189–200. doi: 10.1006/jsbi.1998.4034. [DOI] [PubMed] [Google Scholar]
- 36.Drake S K, Bourret R B, Luck L A, Simon M I, Falke J J. Activation of the phosphosignaling protein CheY. I. Analysis of the phosphorylated conformation by19F NMR and protein engineering. J Biol Chem. 1993;268:13081–13088. [PMC free article] [PubMed] [Google Scholar]
- 37.Eisenbach M. Signal transduction in bacterial chemotaxis. Mod Cell Biol. 1991;10:137–208. [Google Scholar]
- 38.Eisenbach M. Control of bacterial chemotaxis. Mol Microbiol. 1996;20:903–910. doi: 10.1111/j.1365-2958.1996.tb02531.x. [DOI] [PubMed] [Google Scholar]
- 39.Eisenbach M. Bacterial chemotaxis. In: Robertson S, editor. Embryonic encyclopedia of life sciences. [Online.] Macmillan Publishers, Ltd., London, England. http://www.els.net. 2000. [Google Scholar]
- 40.Eisenbach M, Caplan S R. Bacterial chemotaxis: unsolved mystery of the flagellar switch. Curr Biol. 1998;8:R444–R446. doi: 10.1016/s0960-9822(98)70288-x. [DOI] [PubMed] [Google Scholar]
- 41.Eisenbach M, Margolin Y, Ravid S. Excitatory signaling in bacterial chemotaxis. In: Eisenbach M, Balaban M, editors. Sensing and response in microorganisms. Amsterdam, The Netherlands: Elsevier; 1985. pp. 43–61. [Google Scholar]
- 42.Elston T C, Oster G. Protein turbines I: the bacterial flagellar motor. Biophys J. 1997;73:703–721. doi: 10.1016/S0006-3495(97)78104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Falke J J, Bass R B, Butler S L, Chervitz S A, Danielson M A. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ganguli S, Wang H, Matsumura P, Volz K. Uncoupled phosphorylation and activation in bacterial chemotaxis - the 2.1-Å structure of a threonine to isoleucine mutant at position 87 of CheY. J Biol Chem. 1995;270:17386–17393. [PubMed] [Google Scholar]
- 45.Gardina P J, Manson M D. Attractant signaling by an aspartate chemoreceptor dimer with a single cytoplasmic domain. Science. 1996;274:425–426. doi: 10.1126/science.274.5286.425. [DOI] [PubMed] [Google Scholar]
- 46.Garza A G, Harris-Haller L W, Stoebner R A, Manson M D. Motility protein interactions in the bacterial flagellar motor. Proc Natl Acad Sci USA. 1995;92:1970–1974. doi: 10.1073/pnas.92.6.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halkides C J, McEvoy M M, Casper E, Matsumura P, Volz K, Dahlquist F W. The 1.9 Å resolution crystal structure of phosphono-CheY, an analogue of the active form of the response regulator, CheY. Biochemistry. 2000;39:5280–5286. doi: 10.1021/bi9925524. [DOI] [PubMed] [Google Scholar]
- 48.Halkides C J, Zhu X Y, Phillion D P, Matsumura P, Dahlquist F W. Synthesis and biochemical characterization of an analogue of CheY-phosphate, a signal transduction protein in bacterial chemotaxis. Biochemistry. 1998;37:13674–13680. doi: 10.1021/bi9806293. [DOI] [PubMed] [Google Scholar]
- 49.Hazelbauer G L, Engström P. Parallel pathways for transduction of chemotactic signals in Escherichia coli. Nature. 1980;283:98–100. doi: 10.1038/283098a0. [DOI] [PubMed] [Google Scholar]
- 50.Hazelbauer G L, Harayama S. Sensory transduction in bacterial chemotaxis. Int Rev Cytol. 1983;81:33–70. doi: 10.1016/s0074-7696(08)62334-7. [DOI] [PubMed] [Google Scholar]
- 51.Hess J F, Oosawa K, Kaplan N, Simon M I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 52.Jiang M Y, Bourret R B, Simon M I, Volz K. Uncoupled phosphorylation and activation in bacterial chemotaxis: the 2.3 Å structure of an aspartate to lysine mutant at position 13 of CheY. J Biol Chem. 1997;272:11850–11855. doi: 10.1074/jbc.272.18.11850. [DOI] [PubMed] [Google Scholar]
- 53.Khan S, Castellano F, Spudich J L, McCray J A, Goody R S, Reid G P, Trentham D R. Excitatory signaling in bacteria probed by caged chemoeffectors. Biophys J. 1993;65:2368–2382. doi: 10.1016/S0006-3495(93)81317-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim K K, Yokota H, Kim S-H. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 55.Kofoid E C, Parkinson J S. Tandem translation starts in the cheA locus of Escherichia coli. J Bacteriol. 1991;173:2116–2119. doi: 10.1128/jb.173.6.2116-2119.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koshland D E. Bacterial chemotaxis as a model behavioral system. New York, N.Y: Raven Press; 1980. [Google Scholar]
- 57.Kuo S C, Koshland D E. Multiple kinetic states for the flagellar motor switch. J Bacteriol. 1989;171:6279–6287. doi: 10.1128/jb.171.11.6279-6287.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Le Moual H, Quang T, Koshland D E. Methylation of the Escherichia coli chemotaxis receptors: intra- and interdimer mechanisms. Biochemistry. 1997;36:13441–13448. doi: 10.1021/bi9713207. [DOI] [PubMed] [Google Scholar]
- 59.Levit M N, Liu Y, Stock J B. Stimulus response coupling in bacterial chemotaxis: receptor dimers in signalling arrays. Mol Microbiol. 1998;30:459–466. doi: 10.1046/j.1365-2958.1998.01066.x. [DOI] [PubMed] [Google Scholar]
- 60.Lewis R J, Brannigan J A, Muchova K, Barak I, Wilkinson A J. Phosphorylated aspartate in the structure of a response regulator protein. J Mol Biol. 1999;294:9–15. doi: 10.1006/jmbi.1999.3261. [DOI] [PubMed] [Google Scholar]
- 61.Li G, Weis R M. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 62.Li J Y, Li G Y, Weis R M. The serine chemoreceptor from Escherichia coli is methylated through an inter-dimer process. Biochemistry. 1997;36:11851–11857. doi: 10.1021/bi971510h. [DOI] [PubMed] [Google Scholar]
- 63.Li J Y, Swanson R V, Simon M I, Weis R M. The response regulators CheB and CheY exhibit competitive binding to the kinase CheA. Biochemistry. 1995;34:14626–14636. doi: 10.1021/bi00045a003. [DOI] [PubMed] [Google Scholar]
- 64.Liu J, Parkinson J S. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc Natl Acad Sci USA. 1989;86:8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y, Levit M, Lurz R, Surette M G, Stock J B. Receptor-mediated protein kinase activation and the mechanism of transmembrane signaling in bacterial chemotaxis. EMBO J. 1997;16:7231–7240. doi: 10.1093/emboj/16.24.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lloyd S A, Whitby F G, Blair D F, Hill C P. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature. 1999;400:472–475. doi: 10.1038/22794. [DOI] [PubMed] [Google Scholar]
- 67.Long D G, Weis R M. Oligomerization of the cytoplasmic fragment from the aspartate receptor of Escherichia coli. Biochemistry. 1992;31:9904–9911. doi: 10.1021/bi00156a007. [DOI] [PubMed] [Google Scholar]
- 68.Lowry D F, Roth A F, Rupert P B, Dahlquist F W, Moy F J, Domaille P J, Matsumura P. Signal transduction in chemotaxis—a propagating conformation change upon phosphorylation of CheY. J Biol Chem. 1994;269:26358–26362. [PubMed] [Google Scholar]
- 69.Lukat G S, McCleary W R, Stock A M, Stock J B. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lupas A, Stock J. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J Biol Chem. 1989;264:17337–17342. [PubMed] [Google Scholar]
- 71.Lybarger S R, Maddock J R. Differences in the polar clustering of the high- and low-abundance chemoreceptors of Escherichia coli. Proc Natl Acad Sci USA. 2000;97:8057–8062. doi: 10.1073/pnas.130195397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Macnab R M. Flagellar switch. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 181–199. [Google Scholar]
- 73.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 123–145. [Google Scholar]
- 74.Maddock J R, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 75.Marykwas D L, Schmidt S A, Berg H C. Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J Mol Biol. 1996;256:564–576. doi: 10.1006/jmbi.1996.0109. [DOI] [PubMed] [Google Scholar]
- 76.Mathews M A A, Tang H L, Blair D F. Domain analysis of the FliM protein of Escherichia coli. J Bacteriol. 1998;180:5580–5590. doi: 10.1128/jb.180.21.5580-5590.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mayover T L, Halkides C J, Stewart R C. Kinetic characterization of CheY phosphorylation reactions: comparison of P-CheA and small-molecule phosphodonors. Biochemistry. 1999;38:2259–2271. doi: 10.1021/bi981707p. [DOI] [PubMed] [Google Scholar]
- 78.McCleary W R, Stock J B. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem. 1994;269:31567–31572. [PubMed] [Google Scholar]
- 79.McEvoy M, Bren A, Eisenbach M, Dahlquist F W. Identification of the binding interfaces on CheY for two of its targets, the phosphatase CheZ and the flagellar switch protein FliM. J Mol Biol. 1999;289:1423–1433. doi: 10.1006/jmbi.1999.2830. [DOI] [PubMed] [Google Scholar]
- 80.McEvoy M M, Hausrath A C, Randolph G B, Remington S J, Dahlquist F W. Two binding modes reveal flexibility in kinase/response regulator interactions in the bacterial chemotaxis pathway. Proc Natl Acad Sci USA. 1998;95:7333–7338. doi: 10.1073/pnas.95.13.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mowbray S L, Sandgren M O J. Chemotaxis receptors: a progress report on structure and function. J Struct Biol. 1998;124:257–275. doi: 10.1006/jsbi.1998.4043. [DOI] [PubMed] [Google Scholar]
- 82.Ninfa E G, Stock A, Mowbray S, Stock J. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J Biol Chem. 1991;266:9764–9770. [PubMed] [Google Scholar]
- 83.Oosawa K, Ueno T, Aizawa S. Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J Bacteriol. 1994;176:3683–3691. doi: 10.1128/jb.176.12.3683-3691.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oplatka A. Do the bacterial flagellar motor and ATP synthase operate as water turbines? Biochem Biophys Res Commun. 1998;249:573–578. doi: 10.1006/bbrc.1998.8969. [DOI] [PubMed] [Google Scholar]
- 85.Ottemann K M, Xiao W, Shin Y K, Koshland D E. A piston model for transmembrane signaling of the aspartate receptor. Science. 1999;285:1751–1754. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- 86.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 87.Prüss B M, Wolfe A J. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol Microbiol. 1994;12:973–984. doi: 10.1111/j.1365-2958.1994.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 88.Ramakrishnan R, Schuster M, Bourret R B. Acetylation at Lys-92 enhances signaling by the chemotaxis response regulator protein CheY. Proc Natl Acad Sci USA. 1998;95:4918–4923. doi: 10.1073/pnas.95.9.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rebbapragada A, Johnson M S, Harding G P, Zuccarelli A J, Fletcher H M, Zhulin I B, Taylor B L. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sanatinia H, Kofoid E C, Morrison T B, Parkinson J S. The smaller of two overlapping cheA gene products is not essential for chemotaxis in Escherichia coli. J Bacteriol. 1995;177:2713–2720. doi: 10.1128/jb.177.10.2713-2720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Scharf B E, Fahrner K A, Berg H C. CheZ has no effect on flagellar motors activated by CheY13DK106YW. J Bacteriol. 1998;180:5123–5128. doi: 10.1128/jb.180.19.5123-5128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Scharf B E, Fahrner K A, Turner L, Berg H C. Control of direction of flagellar rotation in bacterial chemotaxis. Proc Natl Acad Sci USA. 1998;95:201–206. doi: 10.1073/pnas.95.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schuster M, Abouhamad W N, Silversmith R E, Bourret R B. Chemotactic response regulator mutant CheY95IV exhibits enhanced binding to the flagellar switch and phosphorylation-dependent constitutive signalling. Mol Microbiol. 1998;27:1065–1075. doi: 10.1046/j.1365-2958.1998.00756.x. . (Erratum, 28:863.) [DOI] [PubMed] [Google Scholar]
- 94.Schuster M, Zhao R, Bourret R B, Collins E J. Correlated switch binding and signaling in bacterial chemotaxis. J Biol Chem. 2000;275:19752–19758. doi: 10.1074/jbc.M909908199. [DOI] [PubMed] [Google Scholar]
- 95.Schuster S C, Swanson R V, Alex L A, Bourret R B, Simon M I. Assembly and function of a quaternary signal transduction complex monitored by surface plasmon resonance. Nature. 1993;365:343–347. doi: 10.1038/365343a0. [DOI] [PubMed] [Google Scholar]
- 96.Segall J E, Ishihara A, Berg H C. Chemotactic signaling in filamentous cells of Escherichia coli. J Bacteriol. 1985;161:51–59. doi: 10.1128/jb.161.1.51-59.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shimizu T S, Le Novère N, Levin M D, Beavil A J, Sutton B J, Bray D. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nature Cell Biol. 2000;2:792–796. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]
- 97a.Silversmith R E, Bourret R B. Throwing the switch in bacterial chemotaxis. Trends Microbiol. 1999;7:16–22. doi: 10.1016/s0966-842x(98)01409-7. [DOI] [PubMed] [Google Scholar]
- 98.Sourjik V, Berg H C. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- 99.Springer M S, Goy M F, Adler J. Protein methylation in behavioral control mechanisms and in signal transduction. Nature. 1979;280:279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- 100.Stock J B, Surette M G. Chemotaxis. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1103–1129. [Google Scholar]
- 101.Tang H, Braun T F, Blair D F. Motility protein complexes in the bacterial flagellar motor. J Mol Biol. 1996;261:209–221. doi: 10.1006/jmbi.1996.0453. [DOI] [PubMed] [Google Scholar]
- 102.Tatsuno I, Homma M, Oosawa K, Kawagishi I. Signaling by the Escherichia coli aspartate chemoreceptor Tar with a single cytoplasmic domain per dimer. Science. 1996;274:423–425. doi: 10.1126/science.274.5286.423. [DOI] [PubMed] [Google Scholar]
- 103.Thomas D R, Morgan D G, DeRosier D J. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc Natl Acad Sci USA. 1999;96:10134–10139. doi: 10.1073/pnas.96.18.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Toker A S, Kihara M, Macnab R M. Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium. J Bacteriol. 1996;178:7069–7079. doi: 10.1128/jb.178.24.7069-7079.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Toker A S, Macnab R M. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J Mol Biol. 1997;273:623–634. doi: 10.1006/jmbi.1997.1335. [DOI] [PubMed] [Google Scholar]
- 106.Volz K, Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7-Å resolution. J Biol Chem. 1991;266:15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- 107.Walz D, Caplan S R. An electrostatic model of the bacterial flagellar motor. Bioelectrochem Bioenerg. 1998;47:19–24. [Google Scholar]
- 108.Walz D, Caplan S R. An electrostatic mechanism closely reproducing observed behavior in the bacterial flagellar motor. Biophys J. 2000;78:626–651. doi: 10.1016/S0006-3495(00)76622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang H, Matsumura P. Characterization of the CheAs/CheZ complex: a specific interaction resulting in enhanced dephosphorylating activity on CheY-phosphate. Mol Microbiol. 1996;19:695–703. doi: 10.1046/j.1365-2958.1996.393934.x. [DOI] [PubMed] [Google Scholar]
- 110.Wang H, Matsumura P. Phosphorylating and dephosphorylating protein complexes in bacterial chemotaxis. J Bacteriol. 1997;179:287–289. doi: 10.1128/jb.179.1.287-289.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Welch M, Chinardet N, Mourey L, Birck C, Samama J-P. Structure of the CheY-binding domain of histidine kinase CheA in complex with CheY. Nat Struct Biol. 1998;5:25–29. doi: 10.1038/nsb0198-25. [DOI] [PubMed] [Google Scholar]
- 112.Welch M, Oosawa K, Aizawa S-I, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Welch M, Oosawa K, Aizawa S-I, Eisenbach M. Effects of phosphorylation, Mg2+, and conformation of the chemotaxis protein CheY on its binding to the flagellar switch protein FliM. Biochemistry. 1994;33:10470–10476. doi: 10.1021/bi00200a031. [DOI] [PubMed] [Google Scholar]
- 114.Wu J G, Li J Y, Li G Y, Long D G, Weis R M. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- 115.Yamamoto K, Macnab R M, Imae Y. Repellent-response functions of the Trg and Tap chemoreceptors of Escherichia coli. J Bacteriol. 1990;172:383–388. doi: 10.1128/jb.172.1.383-388.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yan D, Cho H S, Hastings C A, Igo M M, Lee S-Y, Pelton J G, Stewart V, Wemmer D E, Kustu S. Beryllofluoride mimics phosphorylation of NtrC and other bacterial response regulators. Proc Natl Acad Sci USA. 1999;96:14789–14794. doi: 10.1073/pnas.96.26.14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yeh J I, Biemann H-P, Privé G G, Pandit J, Koshland D E, Kim S-H. High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. J Mol Biol. 1996;262:186–201. doi: 10.1006/jmbi.1996.0507. [DOI] [PubMed] [Google Scholar]
- 118.Zhao R H, Pathak N, Jaffe H, Reese T S, Khan S. FliN is a major structural protein of the c-ring in the Salmonella typhimurium flagellar basal body. J Mol Biol. 1996;261:195–208. doi: 10.1006/jmbi.1996.0452. [DOI] [PubMed] [Google Scholar]
- 119.Zhou J D, Lloyd S A, Blair D F. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci USA. 1998;95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhu X Y, Amsler C D, Volz K, Matsumura P. Tyrosine 106 of CheY plays an important role in chemotaxis signal transduction in Escherichia coli. J Bacteriol. 1996;178:4208–4215. doi: 10.1128/jb.178.14.4208-4215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhu X Y, Rebello J, Matsumura P, Volz K. Crystal structures of CheY mutants Y106W and T87I/Y106W: CheY activation correlates with movement of residue 106. J Biol Chem. 1997;272:5000–5006. doi: 10.1074/jbc.272.8.5000. [DOI] [PubMed] [Google Scholar]
- 122.Zhu X Y, Volz K, Matsumura P. The CheZ-binding surface of CheY overlaps the CheA- and FliM-binding surfaces. J Biol Chem. 1997;272:23758–23764. doi: 10.1074/jbc.272.38.23758. [DOI] [PubMed] [Google Scholar]