Abstract
Pasteurella multocida is a mucosal pathogen that colonizes the respiratory system of susceptible hosts. Most isolates of P. multocida produce sialidase activity, which may contribute to colonization of the respiratory tract or the production of lesions in an active infection. We have cloned and sequenced a sialidase gene, nanH, from a fowl cholera isolate of P. multocida. Sequence analysis of NanH revealed that it exhibited significant amino acid sequence homology with many microbial sialidases. Insertional inactivation of nanH resulted in a mutant strain that was not deficient in sialidase production. However, this mutant exhibited reduced enzyme activity and growth rate on 2-3′ sialyl lactose compared to the wild type. Subsequently, we demonstrated the presence of two sialidases by cloning another sialidase gene that differed from nanH in DNA sequence and substrate specificity. NanB demonstrated activity on both 2-3′ and 2-6′ sialyl lactose, while NanH demonstrated activity only on 2-3′ sialyl lactose. Neither enzyme liberated sialic acid from colominic acid (2-8′ sialyl lactose). Recombinant E. coli containing the sialidase genes were able to utilize several sialoconjugants when they were provided as sole carbon sources in minimal medium. These data suggest that sialidases have a nutritional function and may contribute to the ability of P. multocida to colonize and persist on vertebrate mucosal surfaces.
Pasteurella multocida is a gram-negative coccobacillus of the family Pasteurellaceae and is a normal inhabitant of the upper respiratory system of many animals (24). The organism has a broad host range and is commonly a secondary pathogen in upper respiratory infections. Serotype D virulent isolates are toxigenic, but all serotypes produce capsules which confer serum resistance and resistance to phagocytosis (42). However, it is unusual to isolate a P. multocida strain that does not produce sialidase activity (40). Sialidases (neuraminidases; EC 3.2.1.18) are enzymes that liberate sialic acid from sialylconjugated glycoproteins, glycolipids, or colominic acids by cleaving alpha-ketosidic linkages. It is hypothesized that sialidase contributes to the virulence of some pathogenic organisms, especially those that inhabit and invade mucosal surfaces (7). Drzeniek (14) found sialidase activity in bacterial isolates that belong to the orders Pseudomonadales and Eubacteriales, and sialidases have been cloned from Clostridium species (35, 36, 37), Vibrio cholerae (48), Streptococcus pneumoniae (4, 5), Micromonospora viridifaciens (38), and Salmonella enterica serotype Typhimurium (21). Many of these bacterial sialidases have about 20% similarity at the amino acid level (21).
Sialidases have been implicated as bacterial virulence factors (7, 34). It has been shown that a sialidase-deficient mutant of S. pneumoniae was less able to colonize and persist on mucosal surfaces than the wild type (46). In addition, a Bacteroides fragilis sialidase-deficient mutant was attenuated in the rat abscess model (18). The role of sialidase in virulence and nutrition of bacteria is implied further by recent reports of possession of more than one sialidase by a number of virulent bacteria, e.g., some clostridial species (35) and S. pneumoniae (4).
The bacterial sialidases are divided into two groups based on size (49). The Salmonella sialidase and one of the Clostridium perfringens sialidases are approximately 40 kDa in size and are considered to be members of the “small” sialidase family, while the sialidases of Clostridium tertium and V. cholerae are greater than 80 kDa. The larger enzymes generally have multiple protein domains in addition to the sialidase domain (8, 17, 49). There are ambiguities in the literature regarding the size of the sialidases from P. multocida. Drzeniek et al. (13) isolated an enzyme estimated to be 250 kDa. White et al. (50) and Straus et al. (45) purified sialidases from multiple serotypes of P. multocida and estimated their sizes to be in excess of 250 kDa. However, Ifeanyi and Bailie (22) isolated a 36-kDa protein which possessed sialidase activity. These reports suggest the possibility of multiple sialidase enzymes in P. multocida. In this report we describe the cloning and characterization of the genes encoding two sialidases of P. multocida.
MATERIALS AND METHODS
Strains and culture conditions.
P. multocida isolates 86-1913, P1059, and X-73 have been characterized previously (19, 25). A spontaneously occurring nalidixic acid-resistant derivative of P. multocida P-1059, isolated by plating on brain heart infusion agar (Difco Laboratories, Detroit, Mich.) containing 50 μg of nalidixic acid per ml, was used as the wild-type parent for mutant construction. Escherichia coli XL1-Blue was used to initially screen the genomic library for sialidase-producing recombinants. E. coli DH5α was used as the host strain for subcloning and expression of sialidase constructs. Bacterial strains and isolates were grown in brain heart infusion broth or agar (Difco Laboratories) at 37°C. The media were supplemented with ampicillin (100 μg/ml) and/or nalidixic acid (60 μg/ml) and/or kanamycin (50 μg/ml) for the selection of recombinants. However, for maximum production of sialidase, cells were grown in RPMI 1640 containing 25 mM HEPES and 2 mM N-acetylmannosamine (13). Unless otherwise indicated, chemicals were obtained from Sigma Chemical Co. (St. Louis, Mo.).
Cloning of sialidase genes.
Genomic DNA from P. multocida isolate 86-1913 was extracted from cell suspensions by the cetyltrimethylammonium bromide method described by Ausubel et al. (3). A genomic library was prepared in λZAP-EXPRESS according to the manufacturer's directions (Stratagene, Chatsworth, Calif.). Recombinant plaques were screened in E. coli XL1-Blue for sialidase activity using 2′-(4-methylumbelliferyl)α-d-N-acetylneuraminic acid (4MU-Neu5Ac) in the filter paper test method described by Moncla and Braham (28). A cosmid library was also created in the broad-host-range vector pJRD215 by standard methods (3, 11). Recombinant sialidase-positive clones were screened in DH5α as described above.
DNA sequence analysis.
DNA sequencing was performed by dideoxy-termination in an ABI automated sequencer at the Molecular Genetics Instrumentation Facility at The University of Georgia by using primer walking. DNA sequences were compared to other sequences in GenBank using the online BLAST search engine at the National Center for Biotechnology Information (http: //www.ncbi.nlm.nih.gov/). DNA sequence analysis was performed with Gene Runner 3.04 (Hastings Software, Hastings, N.Y.) or Vector NTI (Informax, North Bethesda, Md.).
Construction of a sialidase-deficient mutant.
A sialidase-deficient mutant was produced by insertional inactivation by a single crossover event. A 512-bp internal portion of the sialidase gene was amplified by PCR using the primers F1 (5′-GCTTTGATGGCAGTTTATATGTG-3′) and R2 (5′-TGAAGGAGCCGCTGTAGTCG-3′) with denaturation for 1 min at 94°C, renaturation for 1 min at 55°C, and primer extension for 1 min at 72°C in a 30-cycle program using the Amplitron II thermocycler (Fisher Scientific, Pittsburgh, Pa.). The reaction mixture contained 2 mM MgCl2, 50 mM Tris (pH 7.4), 0.1 mM primer, 0.2 mM nucleotides, and 1 U of Taq polymerase per 20 μl. The identity of the PCR product was confirmed by visual analysis of fragment size using agarose gel electrophoresis and by DNA sequencing. Nucleotides, polymerase, and buffer were purchased from Boehringer Mannheim (Indianapolis, Ind.). The 500-bp amplicon was cloned into pCRII (Invitrogen, San Diego, Calif.), and then the fragment was subcloned into the XbaI/SacI sites of pGPKan, producing pMZ1. The suicide vector pGPKan, a derivative of pGP704, was constructed by removing the PstI fragment containing the ampicillin resistance gene and replacing it with the 1-kb PstI fragment of pUC-4k containing a kanamycin resistance gene. pMZ1 was transformed into E. coli SM10 (RecA::RP4-2Tc::Mu::) by electroporation (41). pMZ1 was transferred to P. multocida P-1059 by conjugation (26). Kanamycin-resistant transconjugants were selected, and nanH was confirmed to be insertionally inactivated by DNA-DNA hybridization.
DNA-DNA hybridization.
Probes specific for each sialidase gene were produced by restriction enzyme digestion of relevant clones. The DNA fragment was gel isolated, labeled with digoxigenin by nick translation according to the manufacturer's protocol (Boehringer Mannheim), and added to 50 ml of hybridization buffer (750 mM sodium chloride, 75 mM sodium citrate, 0.1% N-lauryl sarcosine, 0.02% sodium dodecyl sulfate [SDS], 1% blocking reagent [Boehringer Mannheim] [pH 7.0]). Genomic DNA from P. multocida isolates was extracted from cell suspensions by the cetyltrimethylammonium bromide method (3). Genomic DNA was digested with HindIII and separated on a 0.7% agarose gel. DNA was transferred and hybridized using the protocol for Southern blotting on nylon membranes according to Current Protocols in Molecular Biology (3). Washes were performed with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% SDS at 68°C.
Sialidase purification.
Isolation of the sialidase was performed by detergent (n-octyl glucoside) extraction from whole cells, and the enzyme was purified by a combination of hydrophobic and size exclusion chromatography. Unless otherwise indicated, all incubations, centrifugations, and washes were performed at 4°C.
E. coli DH5α cells harboring pNEU101 were grown with shaking (150 rpm) in 1.6 liters of brain heart infusion broth containing 100 μg of ampicillin per ml for 16 h at 37°C. Cells were pelleted by centrifugation (3,500 × g) for 15 min. The pellet was washed in 500 ml of phosphate-buffered saline (pH 7). The cells were then washed with a buffer containing 50 mM Tris HCl, 1 mM EDTA, and 1% Triton X-100 (pH 7.5). Cells were resuspended in 20 ml of extraction buffer (50 mM Tris HCl, 2% n-octyl glucoside [Fisher Scientific] [pH 7.5]) containing 1 mM EDTA, phenylmethylsulfonyl fluoride (PMSF; 100 μg/ml), and pepstatin (1 μg/ml) and mixed thoroughly by low-speed vortexing for 15 min at room temperature. Cells were pelleted by centrifugation at 10,000 × g for 15 min. This step was repeated twice with 15 ml of extraction buffer. The supernatants containing the extracted sialidase and were pooled and filtered through a 0.2-μm-pore-size filter (Whatman, Hillsboro, Oreg.).
Saturated ammonium sulfate was added slowly at room temperature to the filtered extract to 40% saturation. The mixture was allowed to settle for an hour at room temperature, and then the precipitate was removed by centrifugation at 5,000 × g for 10 min. The supernatant was collected, the ammonium sulfate concentration was increased to 60% saturation, and the mixture was allowed to settle and collected as described above. The sediment, containing the sialidase activity, was washed once with 60% ammonium sulfate solution and then was dissolved in 20 ml of solubilization buffer (50 mM Tris HCl, 1% Triton X-100, 5 mM EDTA, 100 μg of PMSF per ml, 1 μg of pepstatin per ml [pH 7.5]). The suspension was dialyzed against 20 mM sodium citrate buffer at pH 5.1. EDTA was added to the dialysate to 5 mM, and then the mixture was centrifuged at 10,000 × g for 10 min. The supernatant was filtered through a 0.2-μm-pore-size filter, and the filtrate was loaded at a rate of 10 cm/h onto a carboxymethyl (CM) Sepharose column (1.5 by 1.5 cm) preequilibrated with CM loading buffer (20 mM sodium citrate [pH 5.1], 1% Triton X-100, 5 mM EDTA). The column was washed with 10 bed volumes of the loading buffer. Bound sialidase was eluted at a rate of 5 cm/h with a combined increasing gradient of pH and ionic strength, with the CM loading buffer as the starting buffer and 100 mM sodium citrate–1% Triton X-100–5 mM EDTA (pH 5.9) as the ending buffer. Ten column volumes of each buffer was used, 2-ml fractions were collected, and those containing sialidase activity were identified by a filter paper spot assay (28).
The CM-eluted fractions containing sialidase activity were pooled and loaded directly onto a p-aminophenyl oxamic acid column (0.5 by 1.5 cm) preequilibrated with loading buffer (50 mM sodium citrate, 1% Triton X-100, 5 mM EDTA [pH 5.5]). The column was washed with 10 bed volumes of loading buffer and then with 3 volumes of wash buffer (50 mM sodium citrate buffer, 2% n-octyl glucoside, 1 mM EDTA [pH 5.5]). Sialidase was eluted with a second buffer (50 mM Tris HCl, 2% n-octyl glucoside, 1 mM EDTA, 1 M NaCl [pH 7.5]).
A 1.5- by 50-cm Sephacryl-200 HR column (Amersham-Pharmacia, Uppsala, Sweden) was packed according to the manufacturer's protocol. The column was equilibrated with an n-octyl glucoside buffer (50 mM Tris HCl [pH 7.5], 2% n-octyl glucoside, 1 mM EDTA, 0.5 M NaCl). About 1.5 ml of the oxamic acid affinity column eluate was applied to the column and was run at flow rate of 8 ml/h. Two-milliliter fractions were collected and screened for sialidase activity by the filter paper spot test. The sialidase active fractions were pooled and passed through a G-25 (Amersham-Pharmacia) desalting column (3 by 25 cm) preequilibrated with Triton buffer (20 mM Tris HCl, 1% Triton X-100, 5 mM EDTA [pH 8.5]). The eluted sialidase active volume was passed through a DEAE Sepharose anion-exchange column (0.5 by 1.5 cm) preequilibrated with Triton buffer. The sialidase passed unbound.
Sialidase assays.
Sonicated bacterial cell suspensions were used for comparing sialidase activities of various P. multocida isolates and strains. Bacterial cells were grown in 20 ml of broth culture. The E. coli cells were sedimented by centrifuging at 3,000 × g for 10 min. The P. multocida cells were treated with hyaluronidase (4 U/ml) for an hour at 37°C in a shaking incubator and then harvested by centrifugation (4,000 × g for 15 min at 4°C). The cells were suspended in 10 ml of 50 mM Tris HCl buffer (pH 7.5), harvested a second time, and then resuspended in 1 ml of lysis buffer (10 mM Tris HCl, 1 mM EDTA, 0.1 M NaCl [pH 8.0]). PMSF (1 mg/ml), pepstatin A (100 mg/ml), and lysozyme (200 mg/ml) were added, and the mixtures were incubated on ice for 30 min. The cells were lysed by sonication using a Branson 450 sonifier (Branson Ultrasonics, Danbury, Conn.) set at output level 2, cycle 20%, for 5 min. The lysate suspensions were used as crude sialidase preparations for the enzyme assays.
Various strains and clones were screened for sialidase activity by using 4MU-Neu5Ac in a filter paper spot test as described by Moncla and Braham (28). Quantitative assays were done fluorometrically and spectrophotometrically on positive strains or clones. The fluorometric assay was done by the method of Potier et al. using the 4MU-Neu5Ac substrate (29, 32). The reaction mixture contained 0.1 mM 4MU-Neu5Ac and the enzyme preparation in reaction buffer (0.05 M sodium phosphate [pH 6.8] with 0.1 mM NaCl and 0.05% bovine serum albumin) in a total volume of 0.2 ml. The relative amount of the enzyme was such that less than 20% of the substrate was hydrolyzed at the end of the reaction in order to produce a linear response. The reaction mixtures were incubated at 37°C for 3 min, and then the reaction was stopped by adding 1.8 ml of 0.5 M Na2CO3. Relative fluorescence was measured with a TKO 100 minifluorometer (Hoefer, San Francisco, Calif.) at a fixed excitation wavelength of 365 nm and emission at 458 nm, according to the manufacturer's protocol. One unit of the enzyme activity was defined as the amount of activity that released 1 mmol of 4-methylumbelliferone per min under the above-described reaction conditions.
The specificities of the sialidase preparations were determined spectrophotometrically by using 2-3′ or 2-6′ sialyl lactose as the substrate. The reaction mixtures contained 1.0 mM substrate with 2.25 × 10−3 U of sialidase preparation in reaction buffer (0.05 M NaPO4, 0.1 mM NaCl, 0.05% bovine serum albumin [pH 6.8]), in a total volume of 0.1 ml. Reaction mixtures were incubated at 37°C for 10 min, and reactions were stopped by the addition of 50 μl of 0.125 N H2SO4. Sialic acid liberated during the incubation was measured using the method of Aminoff (1).
Effect of pH on sialidase activity.
Crude enzyme preparations from P. multocida isolates and the sialidase-producing clones, as well as purified NanH, were tested at various pHs. For various pH ranges the following buffer systems were used at a concentration of 100 mM: citrate-phosphate buffer (pH 3 to 7.1), Tris HCl buffer (pH 7.6 to 9), and glycine-NaOH buffer (pH 9.5 to 10.5). The activities of the sialidases were assayed fluorometrically using 4MU-Neu5Ac as the substrate as described above.
Utilization of sialylconjugants.
Sialic acid utilization studies for P. multocida were performed with minimal media (23) in a microtiter plate containing 0.1% mucin, fetuin, and 2-3′ or 2-6′ sialyl lactose. Changes in optical density at 630 nm were detected using a microplate reader (Bio-Tek Instrument, Inc., Winooski, Vt.). The ability of E. coli DH5α containing either the sialidase-producing clones or the vector plasmids to utilize sialylconjugants was studied in minimal media, using various N-acetylneuraminic acid-bound substrates as the sole source of carbon. M-9 salt solution (39) containing 4 μg of thiamine per ml was used as the base medium, to which sialylconjugants were added so that the bound N-acetylneuraminic acid concentration was 0.1%. The sialylconjugants used included 2-3′ or 2-6′ sialyl lactose, colominic acid, α-acid glycoprotein (orosomucoid), and fetuin. Glucose was used as the positive control at a concentration of 0.1%. Approximately 105 bacterial cells, grown to mid-log phase in M-9–glucose medium, were inoculated into 100 μl of the minimal media in a 96-well plate. Plates were incubated at 37°C in a shaker incubator. Changes in optical density at 630 nm were measured using a microplate reader.
Nucleotide sequence accession numbers.
The DNA sequences of nanH and nanB were deposited in GenBank under the accession numbers AF274869 and AF274868, respectively.
RESULTS
Cloning and sequence analysis of the sialidase gene, nanH, from P. multocida.
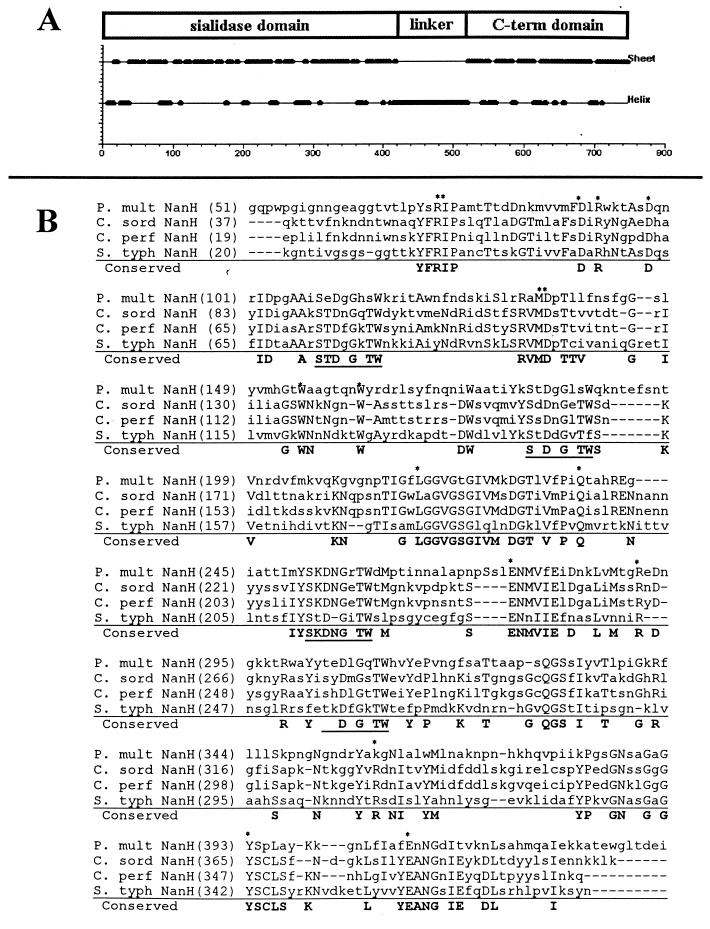
A sialidase-producing plaque was identified from the lambda library. The construct contained a 3.2-kb insert which was subcloned into pMMB67HE, using the KpnI and SacI sites in the multicloning site of both vectors (15). This construct, pNeu101, was used to transform E. coli DH5α to sialidase production. The insert was sequenced and contained a 2,180-bp open reading frame, nanH, estimated to produce an 80-kDa protein. The predicted N-terminal amino acid sequence of NanH contained a 400-amino-acid (aa) region that shared approximately 50% similarity to the S. enterica serotype Typhimurium and the small clostridial sialidases (Fig. 1). Sequence analysis indicated the presence of a hydrophobic N-terminal signal sequence, suggesting that the protein is transported through the inner membrane (data not shown). Four aspartate boxes (S-X-D-X-G-X-TW) were identified, as well as a FRIP motif that was consistent with the expected motifs for bacterial sialidases. However, the nonsialidase domain(s) shared little similarity with known proteins.
FIG. 1.
Amino acid sequence alignment of P. multocida NanH and related bacterial sialidases. (A) Putative domains of NanH. Protein secondary structure was predicted using the algorithm of Garnier et al. (16). The N-terminal 400 aa of NanH are primarily β sheet, which is consistent with the structure of sialidases. (B) Conservation of specific residues (boldface) between P. multocida (P. mult) NanH and sialidases from Clostridium sordelli (C. sord), C. perfringens (C. perf) (small sialidase), and Salmonella serotype Typhimurium (S. typh). Residues believed to be located in the enzyme active site are marked with asterisks; aspartate boxes are underlined (9, 10). The alignment was generated using the AlignX program of VectorNTI (Informax). The linker and C-terminal domains do not exhibit significant homology to other known proteins.
Purification of NanH sialidase.
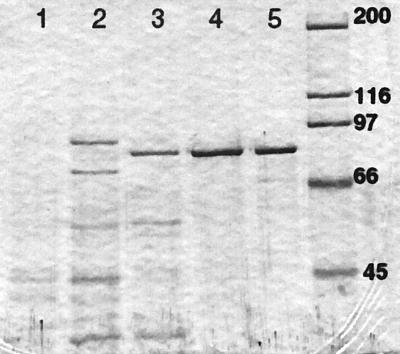
Solubilization and purification of the sialidase required detergent throughout all steps, suggesting that the protein was membrane associated. The purification procedure using E. coli harboring nanH yielded 6% recovery with 950-fold purification. The degree of purification at different steps of the procedure is shown in Table 1 and Fig. 2. Freshly purified NanH migrated as a single band at 80 ± 0.5 kDa, in agreement with the DNA sequence prediction. Samples stored in the refrigerator for 3 or more days showed a breakdown product at 35.5 ± 0.5 kDa. In addition, the sialidase failed to enter the separating gel in nondenaturing polyacrylamide electrophoresis, but samples prepared in denaturing buffer containing 0.2% SDS (without boiling) showed that the sialidase entered the gel. However, the activity was associated with a relatively high molecular mass, approximately 200 kDa. These findings suggest a high isoelectric point and/or a strong tendency of the enzyme to form multimers.
TABLE 1.
NanH purification from recombinant E. coli
| Purification step | Total activity (U)a | Sp actb | Purification factorc | % Recoveryd |
|---|---|---|---|---|
| Whole-cell lysate | 1,734 | 0.002 | 1 | 100 |
| Detergent extract | 1,436 | 0.005 | 2.5 | 83 |
| Ammonium sulfate precipitate | 816 | 0.012 | 6.0 | 47 |
| CM Sepharose | 363 | 0.248 | 124 | 21 |
| p-Aminophenyl oxamate | 267 | 0.637 | 319 | 15 |
| Sephacryl 200HR | 141 | 1.614 | 807 | 8 |
| DEAE Sepharose | 101 | 1.900 | 950 | 6 |
One unit of activity is defined as the amount of sialidase that releases 1 μmol of sialic acid per min.
Units of sialidase activity per microgram of protein.
Calculated by dividing the specific activity of each preparation by the specific activity of the whole-cell lysate.
Determined by the following formula: (total units of activity of each preparation)/(total units of the whole cell lysate) × 100.
FIG. 2.
NanH purification demonstrated by SDS-polyacrylamide gel electrophoresis separation of proteins at various stages of the purification procedure. Samples were separated on an 8% gel and stained with Coomassie brilliant blue R-250. Approximately equal amounts of sialidase activity (units) were loaded into each lane. Lane 1, n-octyl glucoside (2%) extraction from whole cells; lane 2, ammonium sulfate precipitation (40 to 60%) of the detergent extraction; lane 3, CM-Sepharose sialidase-positive fraction, eluted with a NaCl gradient; lane 4, sialidase-positive fraction after separation on Sephacryl 200-HR; lane 5, DEAE Sepharose sialidase-positive fraction; lane 6, molecular weight markers (molecular weights are on the right, in thousands).
Substrate specificity of NanH.
Specific hydrolysis kinetics of purified NanH was measured using 4MU-Neu5Ac as the substrate. The Km was determined to be 63.11 ± 1.05 μM, the Vmax was 4.17 ± 0.04 μmol/liter/min, and the specific activity was 1.9 ± 0.02 μmol/μg/min. NanH demonstrated a significant kinetic preference for α2-3′ sialyl linkages over α2-6′ linkages when tested for activity on naturally occurring oligosaccharide substrates (Table 2). Almost no sialic acid was released from colominic acid, which is composed of 2-8′-linked sialyl lactose. The release of sialic acid from complex conjugants that contain few 2-3′ sialyl linkages, such as ganglioside GM1, was significantly lower than those containing a higher percentage of 2-3′ linkages, such as acid glycoprotein or fetuin. NanH demonstrated hydrolytic activity on mucin, serum proteins (glycoprotein, fetuin, and apotransferrin), and gangliosides (GD1a and GT1b).
TABLE 2.
Substrate specificity of purified recombinant NanH for sialyl conjugants
| Substrate | Rate of hydrolysisa | Relative rateb |
|---|---|---|
| Sialyl α2-3′ lactose | 517 ± 29 | 100 |
| Sialyl α2-6′ lactose | 12 ± 1 | 2 |
| Bovine submaxillary mucin | 122 ± 6 | 23 |
| α-Acid glycoprotein | 360 ± 25 | 70 |
| Apo-transferrin | 115 ± 11 | 22 |
| Fetuin | 343 ± 33 | 66 |
| Colominic acid | 5 ± 1.6 | 1 |
| Ganglioside GM1 | 1 ± 1 | 0.2 |
| Ganglioside GD1a | 141 ± 1 | 27 |
| Ganglioside GT1b | 70 ± 9 | 13 |
Micromoles of sialic acid released per milligram of purified sialidase per minute at 37°C.
Normalized with respect to the value for α2-3′ sialyl lactose.
NanH mutant characteristics.
The NanH-deficient P. multocida mutant was constructed by insertional inactivation of the nanH gene in isolate P1059. The nanH mutation was confirmed by Southern blotting (data not shown), which showed a gel shift corresponding to the size of pMZ1 inserted into the nanH-containing genomic fragment. A sialidase assay using the filter paper spot test demonstrated that the mutant produced activity. However, a quantitative assay showed that the mutant produced only 4% of wild-type activity, suggesting that NanH was defective but the strain harbored another sialidase gene. In addition, whole-cell lysate of wild-type cultures demonstrated a 1.8-fold preference for the 2-3′ linkage, while whole-cell lysate of the mutant showed a 2.1-fold kinetic preference for 2-6′ sialyl lactose, again suggesting that the strain was NanH deficient but expressed an additional sialidase (Table 3). Purified NanH and whole-cell lysate of E. coli harboring nanH characteristically exhibited a 25-fold kinetic preference for 2-3′ sialyl lactose over 2-6′ sialyl lactose.
TABLE 3.
Specific enzymatic hydrolysis of 2-3′ and 2-6′ sialyl lactose by sialidase preparations from P. multocida isolates and clones
| Sialidase sourcea | Sialic acid release from sialyl lactoseb
|
Ratio of 2-3′ to 2-6′ activity | |
|---|---|---|---|
| 2-3′ linked | 2-6′ linked | ||
| NanH (pNEU101) | 2.5 | 0.1 | 25.0 |
| NanB (pAH502) | 1.8 | 6.4 | 0.3 |
| P. multocida 86-1913 | 1.3 | 0.2 | 6.5 |
| P. multocida X-73 | 1.5 | 2.4 | 0.6 |
| P. multocida P1059 | 1.4 | 0.8 | 1.8 |
| P. multocida P1059 NanH mutant | 0.9 | 1.2 | 0.8 |
Sonicated bacterial cell lysates were used as crude sialidase preparations for the enzyme assays. Sialidase-encoding constructs pNeu101 and pAH502 were expressed in E. coli DH5α. E. coli cells harboring only vector did not produce any detectable sialidase activity.
Results are expressed as micromoles of sialic acid (Neu5Ac) released per unit of enzyme per minute. A total of 2 × 10−3 U of sialidase was incubated with 1 mM substrate for 10 min. One unit of sialidase activity was defined as the amount that releases 1 μmol of 4-methylumbelliferone from 4MU-Neu5Ac per min.
The contribution of NanH to nutrient acquisition was investigated by growth studies using the sialoconjugants fetuin, mucin, and sialyl lactose. The NanH-deficient mutant exhibited reduced growth compared to the wild type when 2-3′ sialyl lactose was provided as the sole carbon source (data not shown). The mutant was indistinguishable from the wild type in growth on mucin, fetuin, or 2-6′ sialyl lactose, showing an increase in turbidity over 24 h. The ability of the mutant to grow on the 2-6′ sialyl lactose also suggested the presence of an additional sialidase as well as the ability of P. multocida to utilize sialyl conjugants as carbon sources.
Cloning and sequence analysis of the sialidase gene, nanB, from P. multocida.
Since the NanH-deficient mutant was not deficient in sialidase activity, we sought to identify a second sialidase gene. A sialidase-producing clone, pAH502, was isolated from the cosmid library. A 7.2-kb HindIII/EcoRI fragment of the cosmid insert was subcloned into pUC18 (pMZ506) and sequenced. The derived amino acid sequence of a 3,210-bp open reading frame contained an N-terminal signal sequence and exhibited homology to sialidase proteins (Fig. 3). Since the DNA sequence showed no significant homology with the nanH gene, this sialidase gene was designated nanB; NanB is predicted to be 120 kDa. The sialidase domain resided within the N-terminal 510 aa, where the predicted amino acid sequence exhibited approximately 50% homology to S. pneumoniae NanA and the large clostridial sialidase proteins but only 20% homology to P. multocida NanH. NanB contained the expected sialidase motifs, i.e., a FRIP motif and four aspartate boxes. This group of sialidases, however, appeared not to contain the conserved tryptophan residues expected to occupy the hydrophobic pocket of the active enzyme (9, 10). In the small sialidases, including P. multocida NanH, these residues were found within a 60- to 70-aa region between the first two aspartate boxes (Fig. 1). This region was variable in the NanB-related group and contained approximately 120 aa residues; a conserved tryptophan residue occurred after the second aspartate box, suggesting that the active site may be different in this group (Fig. 3).
FIG. 3.
Amino acid sequence alignment of P. multocida NanB and related bacterial sialidases. (A) Putative domains of NanB. Protein secondary structure was predicted using the algorithm of Garnier et al. (16). (B) Conservation of specific residues (boldface) between P. multocida NanB (P. mult) and four other sialidases: S. pneumoniae (S. pneu) NanA, C. perfringens (C. perf) large sialidase NanI, Clostridium septicum (C. sept) sialidase, and C. tertium (C. tert) sialidase. Residues believed to be located in the enzyme active site are marked with asterisks; aspartate boxes are underlined (9, 10). The alignment was generated using the AlignX program of VectorNTI (Informax).
The C-terminal portion of NanB demonstrates homology to the family of autotransporter proteins, such as the temperature-sensitive hemagglutinin of avian E. coli (12), Haemophilus influenzae adhesion-penetration protein (44) and immunoglobulin A (IgA) protease (33), and Helicobacter pylori vacuolating cytotoxin (2) (Fig. 4). This group of proteins is believed to transport themselves across the outer membrane after spontaneous insertion of the C terminus in the membrane to form a β-barrel configuration (20, 27). The autotransporter domains consisted of an even number of amphipathic β-sheets that were proposed to form 14 transmembrane spanning regions making up the predicted autotransporter pore.
FIG. 4.
Amino acid sequence alignment of the C-terminal domain of P. multocida NanB (P. mul) and related autotransporter domains from H. influenzae (H. inf), Helicobacter pylori (H. pyl), and E. coli (E. col). Specific conserved residues identified by Loveless and Saier (27) among the putative autotransporter proteins are shown in boldface. Regions predicted by Loveless and Saier to contain transmembrane β-strands are marked with numbered lines. The alignment was generated using the AlignX program of VectorNTI (Informax).
Detection of the nanH and nanB genes in P. multocida.
In order to confirm the existence of multiple sialidase genes within isolates, DNA-DNA hybridization was performed. The nanH probe was produced by excising the 3.2-kb SacI/KpnI insert from pNeu101, and the nanB probe was produced by excising a 2-kb HindIII/EcoRI fragment, containing a portion of nanB, from pMZ506. The nanH and nanB fragments were used to probe genomic DNA isolated from P. multocida isolates X-73 (serotype 1) and 86-1913 (serotype 3,4). The nanH probe hybridized with genomic DNA isolated from 86-1913 but not with the nanB-encoding cosmid or chromosomal DNA from isolate X-73 (data not shown). The nanB probe hybridized with genomic DNA from both isolates as well as with the nanB-encoding cosmid but not with the nanH clone, confirming the presence of two genetically distinct enzymes in isolate 86-1913.
pH activity range of the sialidases.
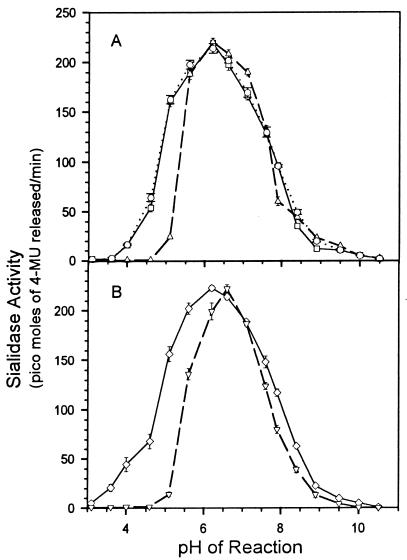
Figure 5 shows the activity of the sialidases at different pHs. The pH optimum of the two cloned sialidases varied between 6.2 and 6.8, depending on the buffer system (Fig. 5A). In the citrate-phosphate buffer the optimum was in a lower range than that in the phosphate or acetate buffer. Although no significant variation in the pH optima of the two cloned enzymes was observed, NanH had significant activity at pH 4 to 5. Both enzymes produced activity up to pH 9.
FIG. 5.
Effect of pH on sialidase activity, determined by using the fluorescent substrate 4MU-Neu5Ac. (A) Crude recombinant sialidase preparations from E. coli DH5α (□, pNEU101 [nanH]; ▵, pAH502 [nanB]) and purified recombinant NanH (○). (B) Activity of two wild-type P. multocida isolates, 86-1913 (◊) and X-73 (▿). The rates of release of 4-methylumbelliferone (4-MU) from the substrate were measured fluorometrically. The buffers used were citrate-phosphate (pH 3.1 to 7.1), Tris-HCl (pH 7.6 to 8.9), and glycine-NaOH (pH 9.5 to 10.5). Equal amounts of activity were used for each assay. Each reaction was done in triplicate, results are plotted as means ± standard deviations.
The pH activity curves of the enzyme preparation from the P. multocida isolates showed similar results (Fig. 5B). However, the activity curve of isolate 86-1913 was broader than that of X-73, suggesting that the combined action of the two enzymes covers a broader pH range than either one alone.
Substrate specificity of the sialidases.
Bacterial cell lysates were assayed for enzymatic specificity using 2-3′ and 2-6′ sialyl lactose as the substrates (Table 3). There were distinct differences between the substrate-specific activity between NanH and NanB. NanH released sialic acid from 2-3′ sialyl lactose much more rapidly than from the 2-6′ substrate (ratio = 25). In contrast, NanB released sialic acid from 2-3′ sialyl lactose at a slightly lower rate than from 2-6′-linked substrate (ratio = 0.3). These differences were reflected in the specificities of the preparations from wild-type P. multocida. Isolate 86-1913, which contains both nan genes, exhibited higher activity on 2-3′ sialyl lactose (ratio = 6), while X-73, which lacks a conserved copy of nanH, exhibited similar activity on both substrates (ratio = 0.6). Isolate P1059, which also contained both genes, appeared to be intermediate between the two (ratio = 1.8). Abolishing NanH activity by a mutation in P1059 shifted the ratio toward 2-6′ activity (ratio = 0.8).
Contribution of sialidases to utilization of sialyl conjugants.
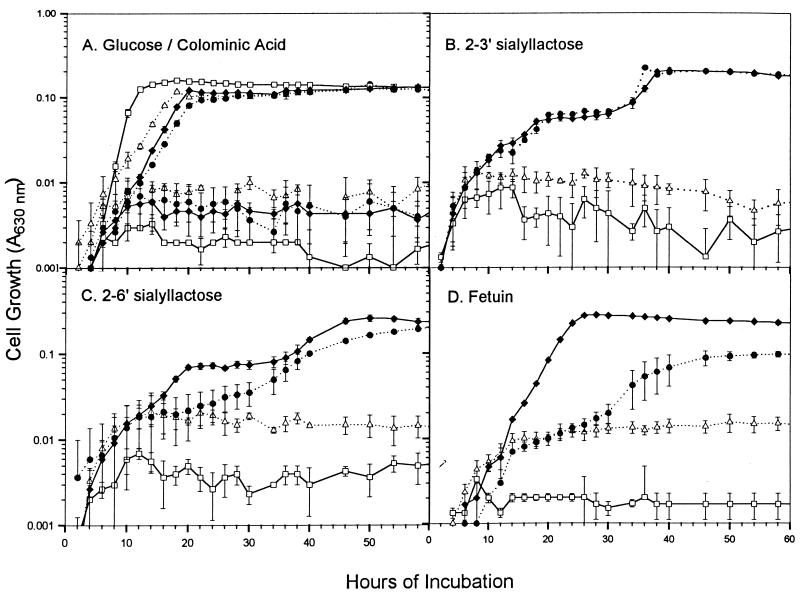
Representative growth curves of the sialidase-producing clones in E. coli DH5α are shown in Fig. 6. All the strains were capable of growing in glucose, but none of them utilized colominic acid, a 2-8′dimer of sialic acid. No vector-containing controls grew in any of the media containing bound sialic acid as the sole source of carbon. In contrast, E. coli harboring either of the sialidase-producing clones grew in the minimal media containing 2-3′ sialyl lactose as the sole source of carbon. The clone containing nanB grew more readily in 2-6′ sialyl lactose and in fetuin than the clone containing nanH. The sialidase-producing clones exhibited biphasic growth on sialyl lactose, which may have resulted from slight contamination of the substrates with another carbon source or from the need for E. coli cells to induce the chromosomal enzyme activities needed to metabolize sialic acid (31, 47). Both the sialic acid permease (NanT) and aldolase (NanA) have been shown to be quickly induced by sialic acid (31). However, the aldolase degrades sialic acid to pyruvate and N-acetylmannosamine, which is poorly utilized by E. coli (31). The sialidases would liberate lactose as well as sialic acid from cleavage of sialyl lactose; however, the DH5α E. coli strain used in the growth studies lacks an intact copy of the lactose operon. Biphasic growth was not seen when fetuin was used as the carbon source, suggesting that the sialyl lactose preparations may have contained carbon source contamination.
FIG. 6.
Growth curves of E. coli DH5α cells containing P. multocida sialidase genes. Growth in minimal media containing either 0.1% glucose or colominic acid (2-8′ N-acetylneuraminic acid) as the sole carbon source (A) or in 2-3′ sialyl lactose (B), 2-6′ sialyl lactose (C), or fetuin (D). Each substrate provides 0.1% bound sialic acid. ▵, pMMB67HE in DH5α (vector control); ●, pNEU101 (nanH) in DH5α; □, pJRD215 in DH5α (vector control); ⧫, pAH502 (nanB) in DH5α. Each assay was done in triplicate wells; results are plotted as means ± standard deviations.
DISCUSSION
NanB appears to be a novel member of the autotransporter family of proteins, a newly identified system of protein secretion (20, 27). It is the first sialidase shown to possess the putative channel-forming C-terminal domain believed to be involved in the translocation of the protein from the bacterial outer membrane. While NanH did not demonstrate significant amino acid sequence homology to this group of proteins, it did exhibit similar predicted sequence characteristics. The majority of autotransporter proteins exhibit 14 predicted antiparallel amphipathic β strands, which are proposed to make up the channel-forming domain (20). The C-terminal domain of NanH is predicted to be heavily composed of β sheets that have an amphipathic character. In addition to the β-sheet structure, the putative final membrane-spanning segments of autotransporter proteins share a consensus amino acid motif [(Y/V/I/F/W)-X-(F/W)] where the terminal residue is always phenylalanine or tryptophan preceded by alternating charged (or polar) and aromatic (or hydrophobic) residues (20). This motif occurs several times in the C-terminal 20 aa of NanH, with the sequence N-P-F occurring most terminally.
We found both NanH and NanB to be outer membrane-associated proteins. Both amino acid sequences contained putative signal peptide domains for inner membrane translocation, but with the exception of the autotransporter domains we did not detect any other motifs that mediate outer membrane translocation. Some autotransporter proteins contain protease activity, which may augment their secretion from the cell membrane (20, 27). We did not detect protease motifs or soluble sialidase activity in the supernatants of cultures; however, White et al. (50) and Straus et al. (45) reported detection of secreted sialidases from P. multocida and Pasteurella haemolytica.
Both NanH and NanB demonstrated significant homology with other bacterial sialidases. NanH demonstrated greatest similarity to the small sialidases, revealing the possibility of additional conserved residues in this group. NanB demonstrated homology to the large clostridial sialidases and NanA of S. pneumoniae. While this group contains some of the conserved residues which were identified in the Salmonella serotype Typhimurium sialidase to be involved in substrate binding and hydrolysis (10). Significant differences were observed, especially in the residues believed to occur in the area of the hydrophobic pocket. These differences may contribute to the ability of the enzymes to hydrolyze naturally occurring sialic acid-containing substrates.
Sialic acid is covalently bound to the side chains of mucin by a 2-6′ glycosidic bond; however, the amount and types of sialic acids present in mucin vary by species of animal and system of isolation. For example, 22% of the dry weight of bovine submaxillary mucin is sialic acid, but the vast majority of the sialic acid is N-acetylneuraminic acid, with minor amounts of N-glycolylneuraminic acid (30). Swine submaxillary mucin contains only 1% sialic acid, which is almost exclusively N-glycolylneuraminic acid (30). Bacterial sialidases liberate both forms of sialic acid (7). The serum proteins fetuin and acid glycoprotein contain a mix of 2-3′ and 2-6′ sialic acid linkages (6, 30). The P. multocida sialidases differed in their specificity for these linkages, but expression of both enzymes should enhance the bacterium's ability to liberate sialic acid from a variety of sources. In addition, both enzymes have high activity across a broad pH range, suggesting that both the enzymes are suitable for action in many host environments, including mucosal secretions and serum. Possession of both sialidase enzymes should greatly enhance the metabolic capability of an organism attempting to colonize different species of animals or grow in different tissues. P. multocida is particularly good at both.
The growth of the sialidase clones in minimal media containing various forms of bound sialic acid corresponded very well with what could be predicted from the specificity assay. Both the nanH- and nanB-harboring E. coli strains grew well in the medium containing 2-3′ sialyl lactose. In contrast, the nanH-containing strain was attenuated in growth in 2-6′ sialyl lactose, corroborating the finding that NanH has poor activity on 2-6′ linkages. The nanH-harboring strain was also less able than expected to utilize fetuin, which contains 40% 2-3′- and 60% 2-6′-linked sialic acid (43), suggesting that NanB can liberate some forms of bound sialic acid which are not cleavable by NanH. Since fetuin contains both N- and O-linked oligosaccharides (43, 51), it is possible that NanB is able to cleave both those linkages. However, possession of both sialidases gives the organism remarkable versatility in its access to sialic acid as a carbon source.
The demonstration of two distinct sialidases in P. multocida also clarifies the controversy on the nature of sialidases from P. multocida. Molecular masses reported by various authors ranged from 36 kDa (22) to 250 kDa (13) to 500 kDa (45). We think these differences in assessments were due to the presence of more than one sialidase and possibly also to incomplete solubilization of the membrane-bound protein. Studies are under way to determine the prevalence of these sialidase genes among the virulent P. multocida isolates and to further characterize the genes and their products.
ACKNOWLEDGMENTS
This work was supported by the U.S. Poultry and Egg Association, the Veterinary Medical Experiment Station, and a National Institute of Health minority supplement (Margie Lee).
We thank Frank Gherardini, John Maurer, and Duncan Krause for their help in the protein studies and Eric Vimr for guidance and tutelage in approaches to characterizing sialidases.
Footnotes
In memory of Rick Rimler, who knew so much about P. multocida.
REFERENCES
- 1.Aminoff D. Methods for the quantitative estimation of N-acetylneuraminic acid and their application to hydrolysates of sialomucoids. Biochem J. 1961;81:384–392. doi: 10.1042/bj0810384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atherton J C, Cao P, Peek R M, Jr, Tummuru M K, Blaser M J. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Boston, Mass: Greene Publishing Associates, Inc., and John Wiley & Sons Inc.; 1993. [Google Scholar]
- 4.Berry A M, Lock R A, Paton J C. Cloning and characterization of nanB, a second Streptococcus pneumoniae neuraminidase gene, and purification of the NanB enzyme from recombinant Escherichia coli. J Bacteriol. 1996;178:4854–4860. doi: 10.1128/jb.178.16.4854-4860.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camara M, Boulnois G J, Andrew P W, Mitchell T J. A neuraminidase from Streptococcus pneumoniae has the features of a surface protein. Infect Immun. 1994;62:3688–3695. doi: 10.1128/iai.62.9.3688-3695.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cointe D, Leroy Y, Chirat F. Determination of the sialylation level and of the ratio alpha-(2→3)/alpha-(2→6) sialyl linkages of N-glycans by methylation and GC/MS analysis. Carbohydr Res. 1998;311:51–59. doi: 10.1016/s0008-6215(98)00196-7. [DOI] [PubMed] [Google Scholar]
- 7.Corfield T. Bacterial sialidases—roles in pathogenicity and nutrition. Glycobiology. 1992;2:509–521. doi: 10.1093/glycob/2.6.509. [DOI] [PubMed] [Google Scholar]
- 8.Crennell S, Garman E, Laver G, Vimr E, Taylor G. Crystal structure of Vibrio cholerae neuraminidase reveals dual lectin-like domains in addition to the catalytic domain. Structure. 1994;2:535–544. doi: 10.1016/s0969-2126(00)00053-8. [DOI] [PubMed] [Google Scholar]
- 9.Crennell S J, Garman E F, Laver W G, Vimr E R, Taylor G L. Crystal structure of a bacterial sialidase (from Salmonella typhimurium LT2) shows the same fold as an influenza virus neuraminidase. Proc Natl Acad Sci USA. 1993;90:9852–9856. doi: 10.1073/pnas.90.21.9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crennell S J, Garman E F, Philippon C, Vasella A, Laver W G, Vimr E R, Taylor G L. The structures of Salmonella typhimurium LT2 neuraminidase and its complexes with three inhibitors at high resolution. J Mol Biol. 1996;259:264–280. doi: 10.1006/jmbi.1996.0318. [DOI] [PubMed] [Google Scholar]
- 11.Davison J, Heusterspreute M, Chevalier N, Ha-Thi V, Brunel R. Vectors with restriction site banks—pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene. 1987;51:275–280. doi: 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- 12.Dozois C M, Dho-Moulin M, Bree A, Fairbrother J M, Desautels C, Curtiss R., III Relationship between Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of Tsh genetic region. Infect Immun. 2000;68:4145–4154. doi: 10.1128/iai.68.7.4145-4154.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drzeniek R, Scharmann W, Balke E. Neuraminidase and N-acetylneuraminate pyruvate-lyase of Pasteurella multocida. J Gen Microbiol. 1972;27:357–368. doi: 10.1099/00221287-72-2-357. [DOI] [PubMed] [Google Scholar]
- 14.Drzeniek R. Viral and bacterial neuraminidases. Curr Top Microbiol Immunol. 1972;59:35–75. doi: 10.1007/978-3-642-65444-2_2. [DOI] [PubMed] [Google Scholar]
- 15.Furste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 16.Garnier J, Osguthorpe D F, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 17.Gaskell A, Crennell S, Taylor G. The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll. Structure. 1995;3:1197–1205. doi: 10.1016/s0969-2126(01)00255-6. [DOI] [PubMed] [Google Scholar]
- 18.Godoy V G, Dallas M M, Russo T A, Malamy M H. A role for Bacteroides fragilis neuraminidase in bacterial growth in two model systems. Infect Immun. 1993;61:4415–4426. doi: 10.1128/iai.61.10.4415-4426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heddleston K L, Rebers P A. Fowl cholera: cross-immunity induced in turkey with formalin-killed in-vivo-propagated Pasteurella multocida. Avian Dis. 1972;16:578–586. [PubMed] [Google Scholar]
- 20.Henderson I R, Navarro-Garcia F, Natarro J P. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 1998;6:370–378. doi: 10.1016/s0966-842x(98)01318-3. [DOI] [PubMed] [Google Scholar]
- 21.Hoyer L L, Hamilton A C, Steenbergen S M, Vimr E R. Cloning, sequencing and distribution of the Salmonella typhimurium LT2 sialidase gene, nanH, provides evidence for interspecies gene transfer. Mol Microbiol. 1992;6:873–884. doi: 10.1111/j.1365-2958.1992.tb01538.x. [DOI] [PubMed] [Google Scholar]
- 22.Ifeanyi F G, Bailie W E. Passive protection of mice with antiserum to neuraminidase from Pasteurella multocida serotype A:3. Vet Res Commun. 1992;16:97–105. doi: 10.1007/BF01839006. [DOI] [PubMed] [Google Scholar]
- 23.Jablonski P E, Jaworski M, Hovde C J. A minimal medium for growth of Pasteurella multocida. FEMS Microbiol Lett. 1996;140:165–169. doi: 10.1111/j.1574-6968.1996.tb08331.x. [DOI] [PubMed] [Google Scholar]
- 24.Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams and Wilkins; 1984. p. 550. [Google Scholar]
- 25.Lee M D, Brown J, Wooley R E, Glisson J R. The relationship of pathogenicity to the growth of 3,4 Pasteurella multocida isolates in normal turkey plasma. Avian Dis. 1988;32:509–512. [PubMed] [Google Scholar]
- 26.Lee M D, Henk A. Tn10 insertional mutagenesis in Pasteurella multocida. Vet Microbiol. 1996;50:143–148. doi: 10.1016/0378-1135(96)00007-7. [DOI] [PubMed] [Google Scholar]
- 27.Loveless B J, Saier M H. A novel family of channel-forming, autotransporting, bacterial virulence factors. Mol Membr Biol. 1997;14:113–123. doi: 10.3109/09687689709048171. [DOI] [PubMed] [Google Scholar]
- 28.Moncla B J, Braham P. Detection of sialidase (neuraminidase) activity in Actinomyces species by using 2′-(4-methylumbelliferyl)α-d-N-acetylneuraminic acid in a filter paper spot test. J Clin Microbiol. 1989;27:182–184. doi: 10.1128/jcm.27.1.182-184.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers R W, Lee R, Lee T Y C, Thomas G H, Reynolds L W, Uchida Y. The synthesis of 4-methylumbelliferyl-α-ketoside of N-acetylneuraminic acid and its use in a fluorometric assay for neuraminidase. Anal Biochem. 1980;101:166–174. doi: 10.1016/0003-2697(80)90056-1. [DOI] [PubMed] [Google Scholar]
- 30.Pigman W, Gottschalk A. Submaxillary gland glycoproteins. In: Gottschalk A, editor. Glycoproteins. Their composition, structure, and function. Amsterdam, The Netherlands: Elsevier; 1966. pp. 434–445. [Google Scholar]
- 31.Plumbridge J, Vimr E. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J Bacteriol. 1999;181:47–54. doi: 10.1128/jb.181.1.47-54.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potier M, Mameli L, Belisle M, Dallaire L, Melancon S B. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-d-N-acetylneuraminate) substrate. Anal Biochem. 1979;94:287–296. doi: 10.1016/0003-2697(79)90362-2. [DOI] [PubMed] [Google Scholar]
- 33.Poulsen K, Brandt J, Hjorth J P, Thogersen H C, Kilian M. Cloning and sequencing of the immunoglobulin A1 protease gene (iga) of Haemophilus influenzae serotype B. Infect Immun. 1989;57:3097–3105. doi: 10.1128/iai.57.10.3097-3105.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roggentin P, Schauer R, Hoyer L L, Vimr E R. The sialidase superfamily and its spread by horizontal gene transfer. Mol Microbiol. 1993;9:915–921. doi: 10.1111/j.1365-2958.1993.tb01221.x. [DOI] [PubMed] [Google Scholar]
- 35.Roggentin P, Kleineidam R G, Schauer R. Diversity in the properties of two sialidase isoenzymes produced by Clostridium perfringens spp. Biol Chem Hoppe-Seyler. 1995;376:569–575. doi: 10.1515/bchm3.1995.376.9.569. [DOI] [PubMed] [Google Scholar]
- 36.Rothe B, Roggentin P, Frank R, Blocker H, Schauer R. Cloning, sequencing and expression of a sialidase gene from Clostridium sordellii G12. J Gen Microbiol. 1989;135:3087–3096. doi: 10.1099/00221287-135-11-3087. [DOI] [PubMed] [Google Scholar]
- 37.Rothe B, Roggentin P, Schauer R. The sialidase gene from Clostridium septicum: cloning, sequencing, expression in Escherichia coli and identification of conserved sequences in sialidases and other proteins. Mol Gen Genet. 1991;226:190–197. doi: 10.1007/BF00273603. [DOI] [PubMed] [Google Scholar]
- 38.Sakurada K, Ohta T, Hasegawa M. Cloning, expression, and characterization of the Micromonospora viridifaciens neuraminidase gene in Streptomyces lividans. J Bacteriol. 1992;174:6896–6903. doi: 10.1128/jb.174.21.6896-6903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Scharmann W, Drzeniek R, Blobel H. Neuraminidase of Pasteurella multocida. Infect Immun. 1970;1:319–320. doi: 10.1128/iai.1.3.319-320.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon R, Piefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 42.Snipes K P, Ghazikhanian B Y, Hirsh D C. Fate of Pasteurella multocida in the blood vascular system of turkeys following intravenous inoculation: comparison of an encapsulated, virulent strain with its avirulent, acapsular variant. Avian Dis. 1987;31:254–259. [PubMed] [Google Scholar]
- 43.Spiro R G, Bhoyroo V D. Structure of the O-glycosidically linked carbohydrate units of fetuin. J Biol Chem. 1974;249:5704–5717. [PubMed] [Google Scholar]
- 44.St. Geme J W, III, de la Morena M L, Falkow S. A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol Microbiol. 1994;2:217–233. doi: 10.1111/j.1365-2958.1994.tb01283.x. [DOI] [PubMed] [Google Scholar]
- 45.Straus D C, Jolley W L, Purdy C W. Characterization of neuraminidases produced by various serotypes of Pasteurella multocida. Infect Immun. 1996;64:1446–1449. doi: 10.1128/iai.64.4.1446-1449.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tong H H, Blue L E, James M A, DeMaria T F. Evaluation of the virulence of a Streptococcus pneumoniae neuraminidase-deficient mutant in nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect Immun. 2000;68:921–924. doi: 10.1128/iai.68.2.921-924.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vimr E R, Troy F A. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J Bacteriol. 1985;164:845–853. doi: 10.1128/jb.164.2.845-853.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vimr E R, Lawrisuk L, Galen J, Kaper J B. Cloning and expression of the Vibrio cholerae neuraminidase gene nanH in Escherichia coli. J Bacteriol. 1988;170:1495–1504. doi: 10.1128/jb.170.4.1495-1504.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vimr E R. Microbial sialidases: does bigger always mean better? Trends Microbiol. 1994;2:271–277. doi: 10.1016/0966-842x(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 50.White D J, Jolley W L, Purdy C W, Straus D C. Extracellular neuraminidase production by a Pasteurella multocida A:3 strain associated with bovine pneumonia. Infect Immun. 1995;63:1703–1709. doi: 10.1128/iai.63.5.1703-1709.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yet M-G, Chin C C Q, Wold F. The covalent structure of individual N-linked glycopeptides from ovomucoid and asialofetuin. J Biol Chem. 1988;263:111–117. [PubMed] [Google Scholar]