Abstract
Background and Aims
Stomach cells can be converted to insulin-producing cells by Neurog3, MafA, and Pdx1 overexpression. Enteroendocrine cells can be similarly made to produce insulin by the deletion of FOXO1. Characteristics and functional properties of FOXO1-expressing stomach cells are not known.
Methods
Using mice bearing a FOXO1-GFP knock-in allele and primary cell cultures, we examined the identity of FOXO1-expressing stomach cells and analyzed their features through loss-of-function studies with red-to-green fluorescent reporters.
Results
FOXO1 localizes to a subset of Neurog3 and parietal cells. FOXO1 deletion ex vivo or in vivo using Neurog3-cre or Atp4b-cre increased numbers of parietal cells, generated insulin- and C-peptide-immunoreactive cells, and raised Neurog3 messenger RNA. Gene expression and ChIP-seq experiments identified the cell cycle regulator cyclin E1 (CCNE1) as a FOXO1 target.
Conclusion
FOXO1 is expressed in a subset of stomach cells. Its ablation increases parietal cells and yields insulin-immunoreactive cells, consistent with a role in lineage determination.
Keywords: Enteroendocrine Cells, Diabetes Treatment, Cell Cycle, Parietal Cells, Transcription Factor
Introduction
The gut is the largest endocrine organ and synthesizes hormones that regulate metabolism.1 Identified by their hormone product, endocrine cells primarily reside in gastrointestinal tract-specific patterns to support their physiological function. For example, the appetite-stimulating hormone ghrelin is produced by endocrine cells predominantly located in the stomach,2 whereas incretins GLP-1 and GIP are produced by L- and K-cells, respectively, primarily found in the colon.3 There is a remarkable diversity among intestinal endocrine cells with more 20 unique subtypes of endocrine cells.4 In fact, most enteroendocrine cells can produce more than one hormone.5,6
Endocrine cells are derived from progenitor cells that express the transcription factor Neurogenin3 (Neurog3). Differentiation of endocrine cells from this shared Neurog3 precursor also appears to follow a tract-specific logic.7 Interestingly, there is also transient expression of hormones in endocrine cells during the differentiation process,8 suggesting that cells may have the ability to acquire more than one identity throughout differentiation. The programs that regulate the differentiation of Neurog3-expressing cells remain unclear.
The transcription factor FOXO1 is important in the maintenance of cell identity, including in insulin-secreting pancreatic β-cells.9,10 In the adult gut, FOXO1 is restricted to endocrine progenitors11 and serotonin-producing cells.12 Its ablation in either cell type gives rise to insulin-producing cells in the intestine in a cell nonautonomous manner.11,12 A similar result has been obtained by combined gain-of-function of Pdx1, Neurog3, and MafA,13,14 3 important factors in β-cell differentiation and function.15 Interestingly, using this approach, insulin-producing cells were also found in the stomach at nearly twice the frequency of the intestine.14 As one of the effects of FOXO1 is to repress Neurogenin316 and Pdx1 transcription,17 while promoting MafA activity,18 it is possible that the 2 approaches are mechanistically related. However, the distribution and function of FOXO1 in gastric cells have not been explored.
Little is known about the effect of FOXO ablation on the distribution and function of other specialized cells gut, including endocrine cells. FOXO1 deletion in Neurog3+ cells increases the number of gut Neurog3+ cells in mice11 and in human fetal pancreas.19 It also increases the number of cells positive for Chromogranin A, a general marker of endocrine cells, but not of serotonin- and cholecystokinin (CCK)-producing cells.11 When FOXO1 is inhibited in human intestinal organoids, Neurog3 and chromogranin A messenger RNA (mRNA) also increase.12 However, it remains unclear how FOXO1 ablation alters the distribution of these cell populations.
To understand the role of FOXO1 in enteroendocrine cell specification and their potential involvement in the conversion of stomach cells to insulin-producing cells, we surveyed FOXO1 localization and lineage traced its expression in the stomach.
Results
FOXO1 Localization in the Stomach
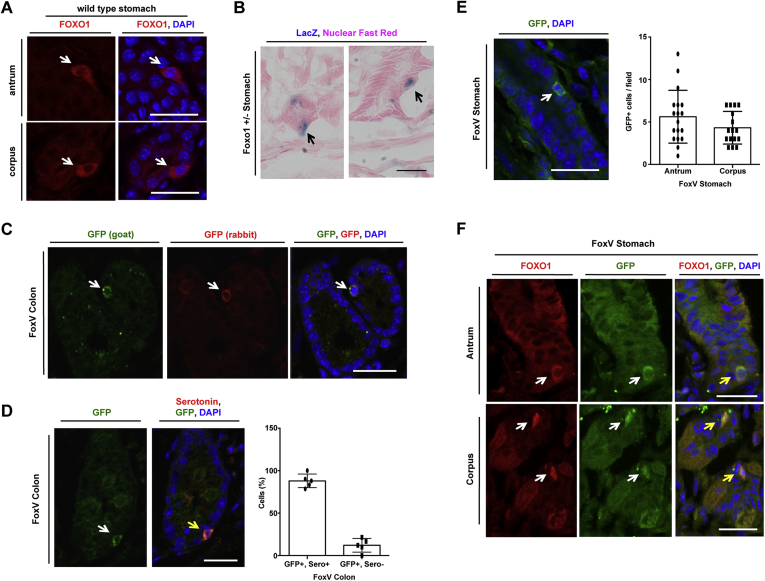
We surveyed FOXO1 localization in the stomach. We found FOXO1-positive cells by immunohistochemistry in wild-type (WT) mice (Figure 1A) and by LacZ immunoreactivity in mice carrying a LacZ reporter knock-in of the Foxo1 locus (Figure 1B).20 Using mice carrying a FOXO1-GFP knock-in allele (FoxV)21 and 2 different GFP antibodies, we confirmed the presence of FoxV-positive cells in the colon (Figure 1C). These cells were serotonin positive, consistent with previous findings of FOXO1-expressing gut cells (Figure 1D).12 We also discovered GFP+ cells in the stomach (Figure 1E). These cells were equally distributed between antrum and corpus in the stomach (Figure 1E) and co-localized with FOXO1 immunohistochemistry (Figure 1F).
Figure 1.
FOXO1+ cells in gastric epithelium. (A) Immunofluorescent staining of wild-type mouse stomach. (B) Stomach of Foxo1+/– mice stained for β-galactosidase (black arrows) counterstained with Nuclear Fast Red. (C) GFP immunofluorescence of FoxV+ cells in the colon of Foxo1-Venus mice (FoxV) with 2 different antibodies. (D) GFP and serotonin immunofluorescence of FoxV colon (left) with quantification (right). (E) GFP immunofluorescence (left) with quantification (right) in the antrum and corpus of FoxV mice. (F) GFP and FOXO1 double immunofluorescence of FoxV stomach (white arrow denotes FoxV cell, yellow arrow denotes co-localization) Blue: DAPI. Scale bar is 25 μm. Data are presented as mean ± standard deviation. N ≥ 5 mice.
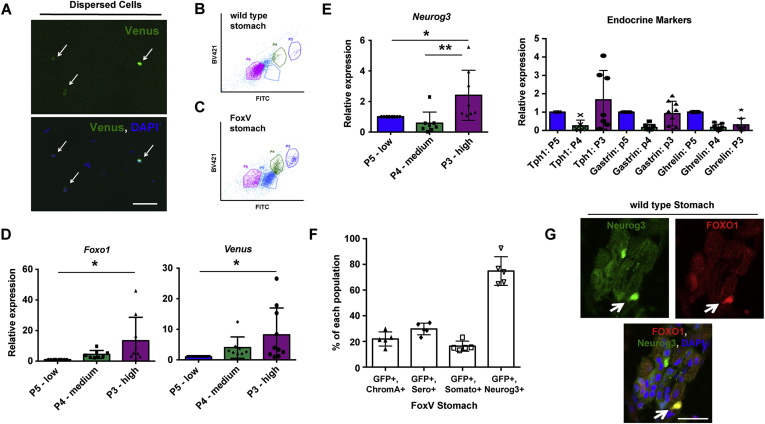
To further characterize these cells, we flow-sorted stomach cells from FoxV and WT mice by fluorescence-activated cell sorting (FACS). We detected 3 populations with differential fluorescence intensity (Figure 2A) in FoxV, but not in control mice: P5 (Venus low), P4 (Venus medium), and P3 (Venus high; Figure 2B and C). P3 cells were enriched for Foxo1, Venus (Figure 2D), and Neurogenin3 (Neurog3) mRNA, the latter which marks endocrine progenitor cells, but not for markers of mature endocrine cells compared with controls (Figure 2E). Accordingly, immunohistochemistry demonstrated that ∼70% of Neurog3+ cells co-localized with GFP, whereas only 20% of cells staining positive for chromogranin A, serotonin, or somatostatin were also GFP+ (Figure 2F). Neurog3+ cells identified using a fluorescent reporter also co-localized with FOXO1 (Figure 2G).
Figure 2.
FoxV+ stomach cells are enriched for the endocrine progenitor marker Neurogenin3 (Neurog3). (A) Cells isolated from Foxo1-Venus (FoxV) mouse after dispersion of stomach glands into single cells. White arrow denotes FoxV+ cell depicted by endogenous fluorescence. DAPI counterstains nuclei. (B, C) FITC dot plot of stomach cells isolated from wild-type (panel B) and FoxV (panel C) mice. (D, E) Gene expression changes for the indicated genes in Venus low (P5), medium (P4), or high (P3) cell populations (depicted in panel C) of stomach cells isolated from FoxV mice using FACS. (F) Quantification of fluorescent staining for GFP co-localization with cell markers: chromogranin A (ChromA), serotonin (Sero), somatostatin (Somato), and neurogenin3 (Neurog3) in FoxV stomach. Venus protein is recognized by GFP antibody. N = 5 mice. (G) Immunofluorescence of Neurog3-GFP stomach tissue stained for FOXO1 (red). Green signal denotes endogenous GFP fluorescence in Neurog3+ cells. ∗P < .05 vs P5 and ∗∗P < .01 vs P5. Gene expression is normalized to β-actin. Scale bar is 50 μm. Data are presented as mean ± standard deviation. Gene expression data shown are ≥8 independent experiments.
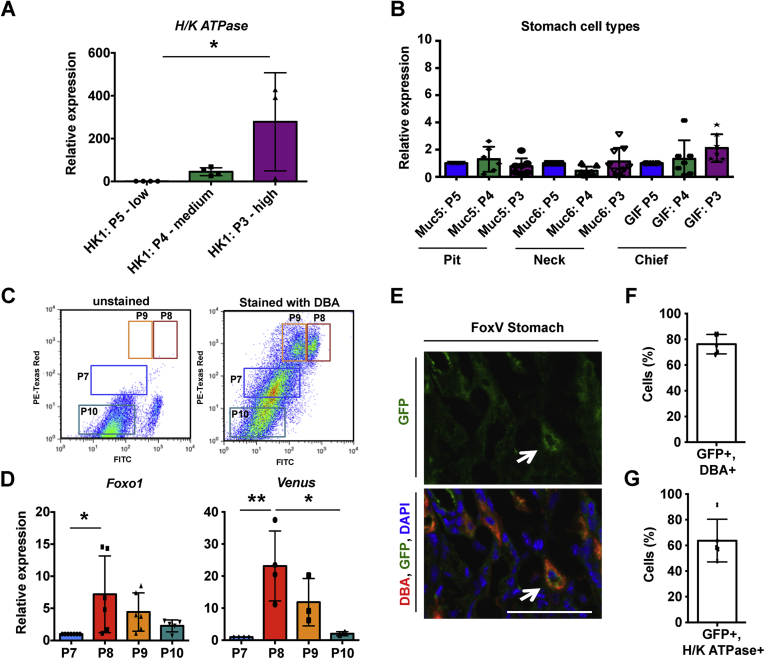
P3 cells were also enriched in H/K ATPase, a marker of acid-secreting parietal cells (Figure 3A and B). To determine whether parietal cells expressed FOXO1, we labeled them with fluorescently conjugated lectin Dolichos biflorus agglutinin (DBA) and subjected them to FACS (Figure 3C). DBA+ cells had elevated levels of Foxo1 and Venus mRNA (Figure 3D). Furthermore, immunofluorescence of stomach sections from FoxV mice demonstrated an overlap of GFP+ cells and DBA or H/K ATPase (Figure 3E–G). These data show that FOXO1-expressing stomach cells are heterogeneous.
Figure 3.
A subset of FOXO1+ parietal cells. (A) H/K ATPase expression in Venus low (P5), medium (P4), or high (P3) cells (Figure 2, panel C) isolated from Foxo1-Venus (FoxV) mice. (B) Expression of gastric cell markers in P5, P4, and P3 sorted FoxV cells. (C) FACS dot plot of stomach cells isolated from FoxV mice and stained with Rhodamine-labeled Dolichos biflorus agglutinin (DBA) to detect parietal cells. (D) Gene expression of sorted cells collected in panel C. (E) GFP and Rhodamine-labeled DBA immunofluorescence in FoxV mice. White arrow denotes FoxV cell. DAPI counterstains nuclei. (F) Quantification of panel E. (G) Quantification of GFP and H/K ATPase (parietal cell marker) co-localization in FoxV tissue. Gene expression is normalized to β-actin. ∗P < .05 and ∗∗P < .01. Scale bar is 50 μm. Data are presented as mean ± standard deviation. Gene expression data shown are ≥4 independent experiments and histological quantification data represent N = 5 mice.
FOXO1 Ablation Generates Insulin-Immunoreactive Cells
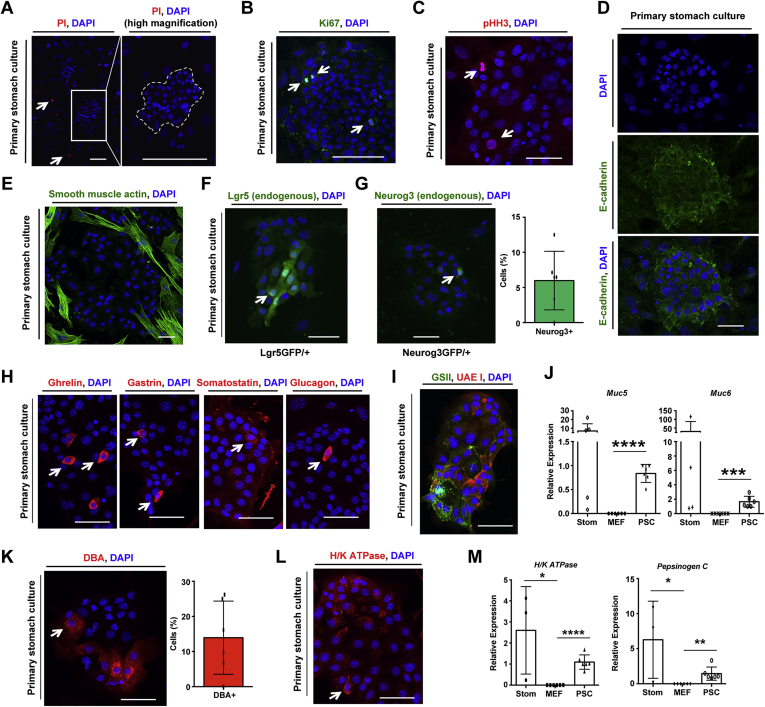
To study the function of FOXO1 in the previously mentioned cell types, we developed a mouse embryonic fibroblast (MEF) co-culture system22, 23, 24 for primary stomach cells. When cultured on growth-arrested MEFs, stomach epithelial cells formed colonies, identified by negative propidium iodide (Figure 4A) and positive Ki67 and phospho-Histone H3 staining (Figure 4B and C). Cultures survive for up to a week, maintaining their epithelial identity, as indicated by positive E-cadherin (Figure 4D) and negative mesenchymal marker smooth muscle actin staining, which is enriched in MEFs (Figure 4E).
Figure 4.
Two-dimensional primary stomach cultures. (A) Propidium iodide (PI, red) staining of primary stomach cells from wild-type mice (outlined by dashed line) cultured on mouse embryonic fibroblasts (MEF). (B) Ki67 and (C) phospho-Histone H3 (pHH3) staining of primary stomach cell cultures (PSC). (D) E-cadherin immunofluorescence of stomach cells. (E) Smooth muscle actin localization to MEF by immunofluorescence of PSC. (F) and (G) Stomach cells isolated from the indicated reporter mice demonstrating the presence of Lgr5+ stem and Neurog3+ endocrine progenitor cells (with quantification) demarcated by endogenous GFP fluorescence. (H) Immunofluorescence of wild-type PSC for indicated endocrine cell markers. (I) Fluorescent staining of wild-type PSC using fluorescently conjugated lectins: Ulex europaeus agglutinin I (UEA I, demarcates mucous pit cells) and Griffonia simplicifolia lectin II (GSII, demarcates mucous neck cells). (J) Gene expression of mucous cell markers in wild-type stomach tissue (Stom), MEF, and PSC. (K, L) Fluorescent staining of wild-type PSC for parietal cell markers using Rhodamine-labeled DBA lectin (panel K, with quantification) and H/K ATPase (panel L). (M) Gene expression for parietal and chief (Pepsinogen C) cell markers. Gene expression is normalized to GAPDH. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. White arrow denotes cell positive for the indicated marker. DAPI: nuclei. Scale bar is 50 μm. Data are presented as mean ± standard deviation. Each dot represents individual mouse or primary stomach culture preparation from N = 6 mice.
The cellular makeup of primary stomach cultures (PSC) was similar to previously described organoid culture systems.25,26 Importantly, PSC had Lgr5+ stem cells and Neurog3+ endocrine progenitor cells (Figure 4F and G). It was also possible to find differentiated stomach endocrine cells (Figure 4H). Specifically, ghrelin- and gastrin-producing cells were prominent in these cultures. It was also possible to find somatostatin+ and glucagon+ cells, although these cells were rare. Markers of mucous pit and neck cells were also detected in PSC (Figure 4I), and this was confirmed by gene expression analysis (Figure 4J). It is interesting to note that PSC also contained markers of parietal cells and chief cells (Figure 4K–M).
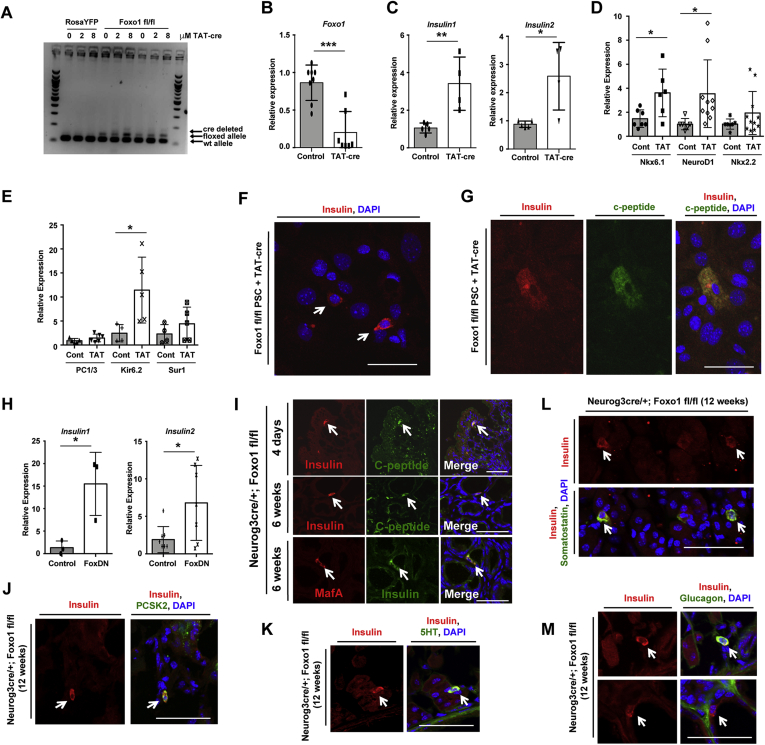
To investigate the effects of FOXO1 ablation, we isolated cells from Foxo1fl/fl mice and induced recombination in PSC by adding TAT-cre to the culture medium. DNA and RNA polymerase chain reaction (PCR) demonstrated locus deletion (Figure 5A) and decreased Foxo1 mRNA (Figure 5B) after treatment of PSC with TAT-cre. Similar to findings in the intestine of Neurog3-cre Foxo1 knockout mice,11 FOXO1 deletion in stomach cultures yielded insulin1 and insulin2 mRNA (Figure 5C). Removal of FOXO1 also increased the expression of β-cell transcription factors Nkx6.1 and NeuroD1 (Figure 5D) and the potassium channel Kir6.2 (Figure 5E). Insulin- and C-peptide immunoreactive cells were exceedingly rare upon ablation of Foxo1 (Figure 5F and G), preventing an effort to quantitate them. We also performed loss-of-function experiments by transducing cells with a dominant-negative FOXO1 mutant and observed increased insulin1 and insulin2 mRNA (Figure 5H).
Figure 5.
Foxo1 ablation. (A) DNA and (B–E) mRNA levels following Foxo1 ablation with TAT-cre in primary stomach cultures (PSC). (F, G) Insulin (red) and C-peptide (green) immunofluorescence of indicated stomach cells treated with TAT-cre. (H) Insulin expression after transduction of PSC with dominant-negative Foxo1. (I) Top and Middle: Immunofluorescence for insulin (red) and C-peptide (green) in mice lacking Foxo1 in Neurogenin3+ cells (Neurog3cre/+; Foxo1 fl/fl) at indicated age. Bottom: Immunostaining for insulin (green) and MafA (red) in Neurog3cre/+; Foxo1 fl/fl mice. (J–M) Stomach tissue from 12-week-old Neurog3cre/+; Foxo1fl/fl mice stained for insulin (red) and other cell markers (green). Gene expression is normalized to the epithelial marker E-cadherin. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. Scale bar is 50 μm. Data are presented as mean ± standard deviation. Each dot represents individual primary stomach culture preparation with N ≥ 3 independent experiments.
To investigate the possibility of generating gastric insulin+ cells in vivo, we ablated FOXO1 in endocrine progenitors using Neurog3-cre transgenic mice (Neurog3cre). Similar to primary cultures, we detected insulin- and C-peptide immunoreactive cells in the gastric epithelium (Figure 5I). These insulin-positive cells were also co-stained with MafA, a marker of mature β-cells, and PCSK2, an enzyme responsible for insulin processing (Figure 5I and J). Some insulin+ cells were bihormonal and contained 5HT or somatostatin immunoreactivity (Figure 5K and L) or glucagon. Monohormonal cells were also present (Figure 5M). Together, these findings demonstrate that FOXO1 deletion triggers the production of insulin-immunoreactive stomach cells that can be bihormonal and express markers of mature β-cells.
FOXO1 Controls Parietal Cell Abundance
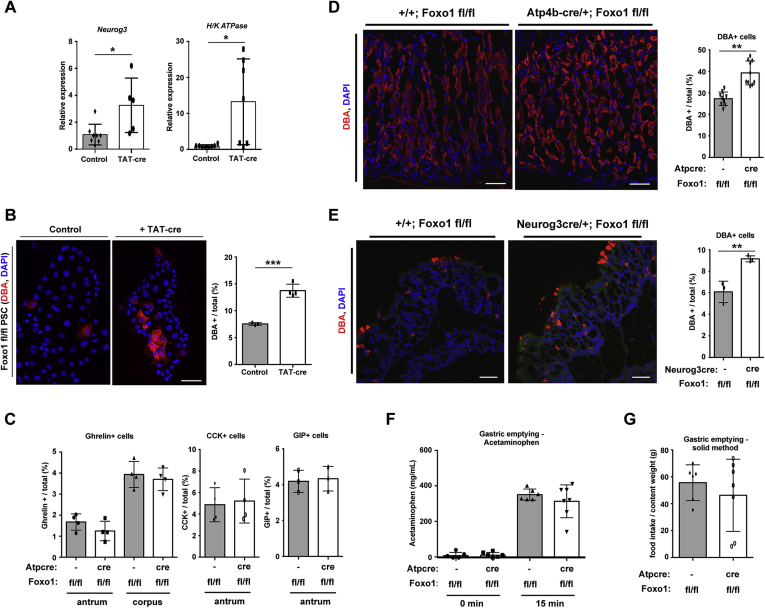
We next determined the impact of FOXO1 deletion on other stomach cell types. Similar to intestinal cells,11 FOXO1 ablation in primary stomach cells with TAT-cre also increased Neurog3 mRNA (Figure 6A). In addition, it increased H/K ATPase expression (Figure 6A), as well as numbers of DBA+ parietal cells (Figure 6B).
Figure 6.
Deletion of Foxo1 in primary culture and mice increases parietal cells. (A) Neurogenin3 (Neurog3) and parietal cell-specific H/K ATPase gene expression in Foxo1 fl/fl primary stomach culture (PSC) after overnight treatment with TAT-cre to delete Foxo1. (B) Fluorescent image with quantification of parietal cells (demarcated with DBA) after elimination of Foxo1 with TAT-cre in PSC. (C) Quantification of endocrine cell markers in stomach tissue lacking Foxo1 in parietal cells (Atp4b-cre/+; Foxo1 fl/fl). (D, E) Fluorescent staining and quantification of Rhodamine-labeled DBA lectin (parietal cells) in stomach tissue lacking Foxo1 in parietal (panel D, Atp4b-cre/+; Foxo1 fl/fl) and Neurog3+ (panel E, Neurog3cre/+; Foxo1 fl/fl at 4 days) cells. (F) Acetaminophen plasma levels before and after oral gavage of acetaminophen (100 mg/kg) and glucose (1.5 g/kg) in Foxo1 fl/fl and Atp4bcre/+; Foxo1fl/fl mice. (G) Gastric emptying measurements as calculated by 100 × (1 – [food content in stomach/1-hour food intake]). Gene expression is normalized to the epithelial marker E-cadherin. Each dot represents an individual mouse or tissue preparation. White arrow denotes positive cell. DAPI counterstains nuclei. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 vs control. Scale bar is 50 μm. Data are presented as mean ± standard deviation. Gene expression data shown are ≥4 independent experiments and histological quantification data represent N ≥ 3 mice.
Tissue-specific FOXO1 ablation in parietal cells also resulted in a 30% increase in parietal cell number in vivo, without affecting ghrelin, CCK, or gastric inhibitor polypeptide cells (Figure 6C and D). We also observed more parietal cells following FOXO1 ablation in Neurog3+ cells (Figure 6E). These changes did not alter gastric emptying as measured by either stomach clearance of solid contents after a meal or stomach clearance of acetaminophen (Figure 6F and G).
FOXO1 Regulates Cyclin CCNE1 in the Stomach
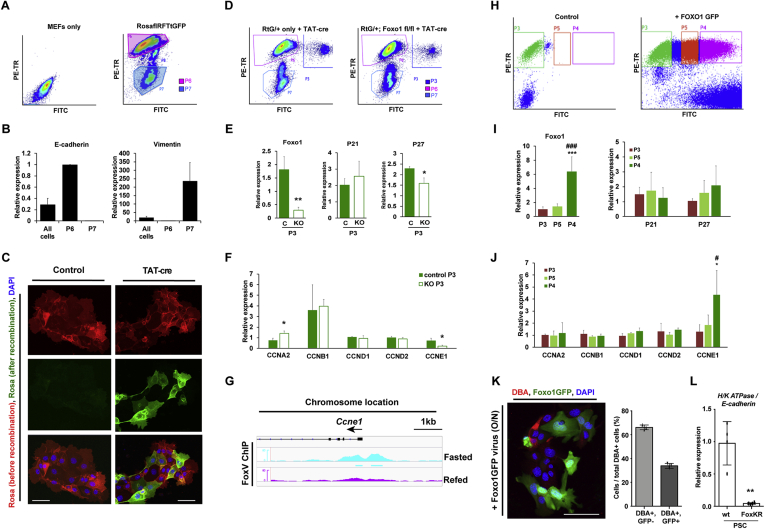
To identify potential FOXO1 targets regulating cell fate or proliferation, we used FACS to separate E-cadherin-expressing epithelial cells (P6, stomach) from vimentin-expressing mesenchymal cells (P7, MEF; Figure 7A and B). We also isolated cre-recombined stomach cells by leveraging the property of cells isolated from RosaflRFPtGFP27 (RtG) mice to undergo red-to-green fluorescence transition upon treatment with TAT-cre (Figure 7C).27 We introduced the RtG reporter allele in mice-bearing conditional Foxo1 alleles to mark Foxo1-deleted cells with green fluorescence and flow-sorted Foxo1-deleted cells (Figure 7D), assessed by measuring FOXO1 mRNA in green cells (Figure 7E).
Figure 7.
Foxo1 controls transcription of the cell cycle regulator CCNE1. (A) FACS dot plot of mouse embryonic fibroblasts (MEF) only or MEFs co-cultured with primary stomach cells isolated from RosaRFPtGFP (RtG) mice. (B) Gene expression of cells collected as in panel A. (C) RFP and GFP fluorescence of RtG primary stomach cells treated with TAT-cre. DAPI counterstains nuclei. (D) FACS dot plot of indicated cells. P3 cells show successful recombination. (E, F) Gene expression in control (RtG/+) or KO (RtG/+; Foxo1 fl/fl) primary stomach cells. ∗P < .05, ∗∗P < .01. (G) ChIP-seq tracks of Foxo1 binding to the CCNE1 promoter in fasting and refed mouse liver. (H) FACs dot plot of RtG/+ primary stomach cells transduced with Foxo1GFP adenovirus. (I, J) Gene expression in cells from panel H normalized by GAPDH. ∗P < .05 vs P3, ∗∗∗P < .001 vs P3. #P < .05 vs P4, and ###P < .001 vs P4. (K) DBA and Foxo1GFP fluorescence and quantification of parietal cells after transduction. (L) Gene expression of PSC isolated from homozygous knock-in mice with active Foxo1 (FoxKR). ∗∗P < .01. Scale bar is 50 μm. Data are presented as mean ± standard deviation. Gene expression data shown are ≥3 independent experiments and histological quantification data represent ≥3 separate primary stomach culture preparations.
As FOXO1 ablation results in changes to the cellular composition of the stomach, we analyzed changes to cell cycle genes. P21 and P27, known FOXO1 targets,28, 29, 30 were only minimally altered (Figure 7E). In contrast, we found significantly increased mRNA encoding cyclin A2 (CCNA2), a regulator of the S and G2 phase of the cell cycle, and a notable decrease of cyclin E1 (CCNE1), which controls the G1 to S phase transition (Figure 7F). To understand whether FOXO1 binds to the CCNE1 promoter, we analyzed ChIP-seq data from liver of fasted and fed mice31 and found several peaks near marks of active transcription (Figure 7G). These data are consistent with the possibility that FOXO1 controls CCNE1 in stomach cells.
To investigate whether FOXO1 regulates cyclin expression in primary stomach cells, we transduced a FOXO1-GFP adenovirus and collected fluorescent cells by FACS (Figure 7H). Gene expression analysis confirmed that Foxo1 mRNA increased 6-fold in the most fluorescent cell population (Figure 7I). Of note, only CCNE1 mRNA increased in this population, whereas other cyclins remained unchanged (Figure 7J). Furthermore, primary stomach cells overexpressing GFP-tagged Foxo1 had fewer DBA+ cells (Figure 7K). Primary stomach cells isolated from mice with active FOXO1 also had less H/K ATPase gene expression (Figure 7L). These data are consistent with the possibility that FOXO1 regulates CCNE1 transcription.
Discussion
Using our FoxV reporter mice, we found that FOXO1 is present in gastric Neurog3+ and parietal cells. The presence of FOXO1 in stomach Neurog3 cells is consistent with prior findings in other regions of the gastrointestinal tract.11,12 Interestingly, FOXO1 colocalizes with 5HT in the intestine, but not in the stomach, consistent with the finding that intestinal 5HT cells are endocrine, whereas gastric serotonin cells are derived from bone marrow mucosal mast cells.32 Furthermore, we found FOXO1 immunoreactivity in parietal cells. The function of FOXO1 in acid-secreting cells remains unclear and will be addressed in future studies.
Our studies suggest that one function of FOXO1 in stomach cells is to control the distribution of gastric cells, including acid-secreting parietal cells. There are few studies on differentiation of parietal cells in the stomach. However, a recent report indicated that differentiation of these cells increased after induction of AMP-activated protein kinase (AMPK) by metformin.33 In part, metformin and AMPK were thought to work via transcription factor KLF4 to inhibit stem cell proliferation and trigger differentiation to either foveolar or parietal cell fate. Interestingly, FOXO1 is posttranslationally regulated by AMPK.34, 35, 36 In addition, KLF4 has also been documented as a Foxo1 target.37 More studies are needed to determine a potential intersection of FOXO1 with the previously mentioned pathway in regulating parietal cell differentiation.
Although there were minimal changes in differentiated endocrine cell types in the stomach in mice harboring genetic ablation of Foxo1, removal of FOXO1 increases the gene expression of the endocrine progenitor marker Neurog3. This is analogous to FOXO1 deletion in other tissues. For example, FOXO1 deletion in murine Neurog3+ cells11 or human intestinal organoids12 increases Neurog3 levels. Similarly, Neurog3+ cells increase when Foxo1 is knocked down using small interfering RNA in cells from human fetal pancreas.19
Our data suggest the possibility that FOXO1 regulates the progression of stomach cells through the cell cycle by direct transcriptional activation of CCNE1, also known as cyclin E1. CCNE1 expression is induced when FOXO1 is overexpressed and blunted when FOXO1 is deleted. Our data indicate that Foxo1 controls CCNE1 mRNA levels in stomach cells by directly regulating gene transcription. Our evidence for this is 2-fold. First, FOXO1 directly binds to the promoter region of CCNE1 in liver (and Foxo1-binding sites are highly conserved38,39), and this promoter binding only occurs in cells isolated from fasted mice. This is important as FOXO1 is transcriptionally active under fasted conditions.40 Second, Foxo1 binding to CCNE1 occurs in DNA regions that are also modified by H3K27 acetylation and H3K4 trimethylation. The former is indicative of active transcription, whereas the latter suggests this DNA is structurally open and accessible to binding by transcription factors. Consistent with our findings, it has been suggested that FOXO1 alters CCNE1 expression in pancreatic β-cells to maintain functional β-cell mass with diabetes.41 More research will be needed to determine if a FOXO1-CCNE1 program exists in other regions of the gastrointestinal tract.
Similar to what is seen in the gut, it is possible to detect insulin-immunoreactive cells when FOXO1 is deleted in vitro and in vivo. It is beyond the scope of this article to determine if these insulin+ cells are functional “β-like” cells. However, it is interesting to note that stomach insulin+ cells co-express C-peptide, consistent with insulin processing, and MafA, a marker of mature β-cells, and are similar to intestinal insulin-positive cells generated by FOXO1 deletion in Neurog3+.11 Based on these data, it will be important to determine FOXO1 expression and distribution in human stomach, given the substantial differences with the murine stomach.42
Materials and Methods
All authors had access to the study data and had reviewed and approved the final article.
Animals
Mice were backcrossed >6 generations onto a C57BL/6J background. FoxV,21 Foxo1+/–,43 Foxo1fl/fl,44 NgnGFP,45 Ngncre,46 Ins2GFP11 mice have been described; Lgr5GFP,47 RosaYFP, and RosaflRFPtGFP27 (RtG) mice were from Jackson Laboratories; and Atp4bcre48 mice were a gift from Jason Mills at Washington University. Unless otherwise indicated, organs and cells were collected from 10- to 12-week-old male and female mice. Male and female mice were used in equal numbers. All studies were approved by, and done in accordance with, the Columbia University Institutional Animal Care and Utilization Committee.
Primary Cultures
Stomach was dissected, serosa removed, and washed extensively with ice-cold phosphate buffered saline (PBS). Tissue was separated into glands by agitation and incubation in 5 mM EDTA/PBS buffer (Invitrogen, cat. no. 15575-038) for 90 min on horizontal shaker at 4 °C. Glands were dissociated into single cells by TrypLE/DNaseI digestion (TrypLE [Invitrogen, cat. no. 12604013]) supplemented with ∼2 U/μL DNAse-1 (Sigma-Aldrich, cat. no. 10104159001) and 10 μM Y-27632 (Rock Inhibitor, Sigma-Aldrich, cat. no. Y0503-1MG). Digestion was completed with tubes placed horizontally, shaking vigorously at 37 °C for ∼30 m until tissue began to visibly dissociate. Cells were washed extensively with ice-cold Advanced DMEM/F12 (Invitrogen, cat. no. 12634010) supplemented with 10 mM HEPES (Invitrogen, cat. no. 15630080), 1× Glutamax (Invitrogen, cat. no. 35050061), 1% pen/strep (Gibco, cat. no. 15140-122), and 1% bovine serum albumin, fraction V (Fisher, cat. no. BP1600-100) ) and purified by filtration through a 40 μM cell strainer (Fisher, cat. no. 22363547).
Primary stomach cells were cultured on a layer of mouse embryonic fibroblasts (Gibco, cat. no. A34180) seeded at a density of 5 × 105 cells/6-well gelatin-coated plate (Millipore, cat. no. ES-006-B) in the same medium supplemented with 1× N2 (Invitrogen, cat. no. 17502048), 1× B-27 (Invitrogen, cat. no. 17504044), 1 mM N-Acetyl-L-cysteine (Sigma-Aldrich, cat. no. A9165-5G), 1× Primocin (Invitrogen, cat. no. NC9141851), and Rock inhibitor. After overnight recovery, cultures were treated overnight with 0.4 U/mL TAT-cre (Millipore, cat. no. SCR508) diluted in medium without rock inhibitor. Cultures were transduced overnight in serum-free complete media with adenoviruses encoding FOXO1GFP49 and FOXO1DN.50
Recombination at the Foxo1 locus by TAT-cre was detected using PCR. Genomic DNA was amplified using the KAPA2G Fast PCR kit (Sigma-Aldrich, cat. no. 2GFKB) and denatured at 95 °C for 30 seconds, annealed at 61 °C for 30 seconds, and elongated at 72 °C for 30 seconds for 34 cycles. Primers to detect recombination of Foxo1 floxed allele were as follows: 5′-GCT TAG AGC AGA GAT GTT CTC ACA TT-3′, 5′-CCA GAG TCT TTG TAT CAG GCA AAT AA-3′, and 5′-CAA GTC CAT TAA TTC AGC ACA TTG A-3′. Expected products are 115, 149, and 190 bp for the WT, floxed, and deleted Foxo1 allele.
Fluorescence-Activated Cell Sorting
Primary stomach cells were isolated as described previously and resuspended in basic culture media. For DBA staining, cells were collected, fixed for 5 min in 2% paraformaldehyde, and incubated with fluorescently conjugated lectin in the dark on a rotator at 4 °C for 1 hour before sorting. Stained cells were resuspended in ALDEFLUOR Assay Buffer (Stem Cell Technologies, cat. no. 01701). Cells were sorted using a BD Influx with a blue (488-nm) laser and 530/40 band pass filter to detect endogenous Venus fluorescence and green fluorescent protein and a green (561 nm) laser and 610/20 band pass filter to detect red fluorescent protein and Rhodamine-labeled DBA.
RNA Isolation and Quantitative PCR
RNA was isolated from 5000 to 10,000 sorted cells using the Arcturus PicoPure RNA Isolation Kit (Applied Biosystems, cat. no. 12204-01). After DNAse treatment (Qiagen, cat. no. 79254), complementary DNA (cDNA) was synthesized using qScript cDNA SuperMix (Quanta Biosciences, cat. no. 95048-100). RNA quality was confirmed with an Agilent Bioanalyzer 2100nusing Pico chip. Antral stomach was flash frozen in liquid nitrogen and RNA extracted using TRIzol (Invitrogen, cat. no. 15596026), loaded onto an RNeasy Mini Kit column (Qiagen, cat. no. 74106), and cDNA prepared using qScript cDNA SuperMix (Quanta Biosciences, cat. no. 95048-100). Quantitative PCR was performed using GoTaq qPCR Master Mix (Promega) in a Bio-Rad CFX96 system, and gene expression levels were determined by the ΔΔCt method with β-actin, GAPDH, or E-cadherin as controls. Primers are listed in the Table A1.
Chromatin Immunoprecipitation and ChIP-Seq Library Construction
The ChIP-IT High Sensitivity kit (Active Motif, Carlsbad, CA) was used for chromatin immunoprecipitation (ChIP) as recommended by the manufacturer. Mice aged 8–12 weeks were sacrificed after 4-hour fasting, 1-hour refeeding, and perfusion with 10 μM orthovanadate through the inferior vena cava Chromatin from 300 mg liver was sheared using a S220 Focused-ultrasonicator (Covaris). Four microgram of anti-GFP antibody for 10 μg of sheared chromatin was used for immunoprecipitation. The specificity of GFP antibody was confirmed by Western blot. ChIP-seq libraries were constructed using KAPA Hyper Prep Kit (KAPA Biosystems) as recommended by manufacturer. ChIP-seq libraries were quantified by Tapestation (Agilent) and sequenced on an Illumina NEXTseq (Illumina, San Diego, CA) with 75-base single-end mode.
Reads were aligned to mouse genome mm10 using Bowtie2 software.51 Only reads that passed Illumina’s purity filter, aligned with no more than 2 mismatches, and mapped uniquely to the genome were used. Duplicate reads were removed with Picard tools. The tags were extended at their 3′-ends to 200 bp. Peak calling was performed by MACS 2.1.052 with the P value cutoff of 10−7 for narrow peaks and q value cutoff of 10−1 for broad peaks against the input control sample. The transcription start site determined on mouse genome mm10 was used to measure the distance of each peak. HOMER software suite53 was used to perform motif analysis, annotate peaks, such as promoter/transcription start site, introns, exons, intergenic, 5′ UTR, noncoding RNA, and 3′ UTR, merge files, and quantify data to compare peaks. For the detection of active enhancers, we used bedtools54 by collecting the intersection of the peaks of transcription factor and histone marks. Data have been deposited in GEO [NCBI GEO [GSE151546]].31
Histopathology
Stomachs were dissected along the greater curvature, pinned flat with lumen side facing up, and fixed on ice for 90 min in 4% paraformaldehyde (Electron Microscopy Sciences, cat no. 15710). Tissue was embedded in Tissue Tek OCT compound (Sakura, Cat. no. 4583), flash frozen, and sectioned at 5 μm. For imaging, cells were cultured on gelatin-coated chamber slides (Corning, cat. no. 354114), fixed in 4% paraformaldehyde for 10 minutes at room temperature and permeabilized in PBS with 0.1% Triton X-100 (Acros Organics, Cat. no. 42235-5000) for 10 minutes.
For immunohistochemistry, samples were blocked in 10% donkey (Jackson Immuno Research Labs, cat. no. 017000121) or goat serum (Vector, Cat. no. S-1000) for 1 hour at room temperature and incubated with primary antibodies diluted in blocking solution. Primary antibodies used were GFP (rabbit; Molecular Probes, 1:400, cat. no. A-6455), GFP (goat; Abcam, 1:250, cat. no. ab6662), Serotonin (5HT-H209; Novus Biologicals, 1:50, cat. no. NB120-16007), FOXO1 (C29H4; Cell Signaling Technology, 1:100, cat. no. 2880S), Chromogranin A (rabbit; LsBio, 1:200), Somatostatin (D-20; Santa Cruz, 1:250, sc-7819), Neurogenin3 (Abcam, 1:50, cat. no. ab38548), H/K ATPase (Abcam, 1:100, cat. no. ab2866), Ki67 (Abcam, 1:500, cat. no. ab15580), pHH3 (Abcam, 1:100, cat. no. ab5176), Ecadherin (BD biosciences, 1:50, cat. no. 610181), Smooth Muscle Actin (Abcam, 1:100, cat. no. ab7817), Insulin (Dako, 1:250, A056401-2), C-peptide (Cell Signaling Technology, 1:400, cat. no. 4593S), MafA (Bethyl Laboratories, 1:100, cat. no. IHC-00352), Ghrelin (Novus Biologicals, 1:100, MAB8200), CCK8 (Thermo Fisher, 1:400, PA5-32348), and gastric inhibitor polypeptide (Abcam, 1:100, cat. no. ab30679). For immunofluorescence, primary antibodies were detected using Alexa Fluor 488, 568, and 647 secondary antibodies (1:1000, Molecular Probes) of the corresponding species. DBA (Vector Laboratories, 1:200, cat. no. RL-1032) staining was performed for 2 hours at room temperature. Slides were covered using VECTASHIELD Vibrance Antifade Mounting Medium with DAPI (Vector Laboratories, cat. no. H-1800-10) to counterstain nuclei. Images were taken with a Zeiss LSM 510/710 confocal microscope (Zeiss).
Staining for β-galactosidase was performed as described,55 and tissue was counterstained using nuclear fast red (Vector Laboratories) according to manufacturer’s protocol. Vital cell staining was performed with propidium iodide (Sigma-Aldrich, cat no. P4170-25MG, 1 μg/mL) and Hoechst 33342 (Molecular Probes, 1:1000, cat. no. H3570) for 5 minutes at 37 °C in the dark followed by cell imaging.56
For cell counts, at least 5 fields/samples in at least 3 mice were quantified. Expression and co-localization of FOXO1-positive cells were quantified by averaging 3–5 microscopic fields/sample in at least 5 independent samples. To count stomach cell types after FOXO1 deletion, a minimum of 5 fields/sample (representing at least 3000–5000 cells) were counted, as indicated in the figures. For primary cultures, stomach cells in 6–8 microscopic fields in at least 3 independent cultures were quantified.
Gastric Emptying
Liquid gastric emptying was assessed by the acetaminophen absorption test.57 Mice were fasted 5 hours and gavaged with 100 mg/kg 15% glucose, 1% acetaminophen solution (Sigma-Aldrich). 50 μL blood was collected 15 m after gavage, and plasma acetaminophen levels were measured by enzymatic assay (Sekisui Diagnostics).
To measure solid gastric emptying, mice were fasted for 16 hours with free access to water, allowed access to chow for 1 hour, and food-deprived again for 2 hours before killing and measuring stomach food content. Food intake was measured using a food dispenser, and emptying was calculated as percentage of stomach food content/food intake.
Statistics
Data are presented as means ± standard deviation. Comparisons were done by 2-tailed Student’s t-test between 2 groups and by analysis of variance followed by Tukey’s post hoc test between multiple groups. Statistical analyses were completed using GraphPad Prism 7 (La Jolla, CA), and a P < .05 was used to declare a statistical difference.
Acknowledgments
Authors' Contributions:
Wendy M. McKimpson: study concept and design; acquisition of data; analysis and interpretation of data; drafting of the manuscript; obtained funding. Taiyi Kuo: acquisition of data; analysis and interpretation of data; critical revision of the manuscript for important intellectual content. Takumi Kitamoto: acquisition of data; analysis and interpretation of data; critical revision of the manuscript for important intellectual content. Sei Higuchi: acquisition of data; analysis and interpretation of data. Jason C. Mills: acquisition of data; analysis and interpretation of data; critical revision of the manuscript for important intellectual content. Rebecca A. Haeusler: analysis and interpretation of data; critical revision of the manuscript for important intellectual content. Domenico Accili: study concept and design; analysis and interpretation of data; critical revision of the manuscript for important intellectual content; obtained funding.
Footnotes
Conflicts of Interest: These authors disclose the following: D.A. was a founder, director, stock holder, and chair of board of Forkhead Biotherapeutics Corp. The remaining authors disclose no conflicts.
Funding: Supported by a Berrie Fellow in Diabetes Research Award, T32-HL-007343-38 and 1K01DK121873-01 to W.M.M., R01DK094989 and P30DK056338 to J.C.M., HL125649 to R.A.H., and DK-57539 to D.A. Supported by S10RR027050 and P30DK063608.
Ethical Statement: The corresponding author, on behalf of all authors, jointly and severally, certifies that their institution has approved the protocol for any investigation involving humans or animals and that all experimentation was conducted in conformity with ethical and humane principles of research.
Data Transparency Statement: The data, analytic methods, and materials used in this study will be made available to other researchers on reasonable request.
Material associated with this article can be found in the online version at https://doi.org/10.1016/j.gastha.2022.05.005.
Supplementary Materials
References
- 1.Ahlman H., Nilsson The gut as the largest endocrine organ in the body. Ann Oncol. 2001;12 Suppl 2:S63–S68. doi: 10.1093/annonc/12.suppl_2.s63. [DOI] [PubMed] [Google Scholar]
- 2.Kojima M., Hosoda H., Date Y., et al. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature. 1999;402:656–660. doi: 10.1038/45230. [DOI] [PubMed] [Google Scholar]
- 3.Holst J.J. From the incretin concept and the discovery of GLP-1 to today's diabetes therapy. Front Endocrinol (Lausanne) 2019;10:260. doi: 10.3389/fendo.2019.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haber A.L., Biton M., Rogel N., et al. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egerod K.L., Engelstoft M.S., Grunddal K.V., et al. A major lineage of enteroendocrine cells coexpress CCK, secretin, GIP, GLP-1, PYY, and neurotensin but not somatostatin. Endocrinology. 2012;153:5782–5795. doi: 10.1210/en.2012-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Habib A.M., Richards P., Cairns L.S., et al. Overlap of endocrine hormone expression in the mouse intestine revealed by transcriptional profiling and flow cytometry. Endocrinology. 2012;153:3054–3065. doi: 10.1210/en.2011-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson C.A., DeLaForest A., Battle M.A. Patterning the gastrointestinal epithelium to confer regional-specific functions. Dev Biol. 2018;435:97–108. doi: 10.1016/j.ydbio.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gehart H., van Es J.H., Hamer K., et al. Identification of enteroendocrine regulators by real-time single-cell differentiation mapping. Cell. 2019;176:1158–1173.e16. doi: 10.1016/j.cell.2018.12.029. [DOI] [PubMed] [Google Scholar]
- 9.Tsuchiya K., Ogawa Y. Forkhead box class O family member proteins: the biology and pathophysiological roles in diabetes. J Diabetes Investig. 2017;8:726–734. doi: 10.1111/jdi.12651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buteau J., Accili D. Regulation of pancreatic beta-cell function by the forkhead protein FoxO1. Diabetes Obes Metab. 2007;9 Suppl 2:140–146. doi: 10.1111/j.1463-1326.2007.00782.x. [DOI] [PubMed] [Google Scholar]
- 11.Talchai C., Xuan S., Kitamura T., et al. Generation of functional insulin-producing cells in the gut by Foxo1 ablation. Nat Genet. 2012;44:406–412. doi: 10.1038/ng.2215. S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouchi R., Foo K.S., Hua H., et al. FOXO1 inhibition yields functional insulin-producing cells in human gut organoid cultures. Nat Commun. 2014;5:4242. doi: 10.1038/ncomms5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y.J., Finkbeiner S.R., Weinblatt D., et al. De novo formation of insulin-producing “neo-beta cell islets” from intestinal crypts. Cell Rep. 2014;6:1046–1058. doi: 10.1016/j.celrep.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ariyachet C., Tovaglieri A., Xiang G., et al. Reprogrammed stomach tissue as a renewable source of functional beta cells for blood glucose regulation. Cell Stem Cell. 2016;18:410–421. doi: 10.1016/j.stem.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murtaugh L.C. Pancreas and beta-cell development: from the actual to the possible. Development. 2007;134:427–438. doi: 10.1242/dev.02770. [DOI] [PubMed] [Google Scholar]
- 16.Kitamura T., Kitamura Y.I., Kobayashi M., et al. Regulation of pancreatic juxtaductal endocrine cell formation by FoxO1. Mol Cell Biol. 2009;29:4417–4430. doi: 10.1128/MCB.01622-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitamura T., Nakae J., Kitamura Y., et al. The forkhead transcription factor Foxo1 links insulin signaling to Pdx1 regulation of pancreatic beta cell growth. J Clin Invest. 2002;110:1839–1847. doi: 10.1172/JCI200216857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitamura Y.I., Kitamura T., Kruse J.P., et al. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2005;2:153–163. doi: 10.1016/j.cmet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Al-Masri M., Krishnamurthy M., Li J., et al. Effect of forkhead box O1 (FOXO1) on beta cell development in the human fetal pancreas. Diabetologia. 2010;53:699–711. doi: 10.1007/s00125-009-1632-0. [DOI] [PubMed] [Google Scholar]
- 20.Hosaka T., Biggs W.H., 3rd, Tieu D., et al. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci U S A. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo T., Kraakman M.J., Damle M., et al. Identification of C2CD4A as a human diabetes susceptibility gene with a role in beta cell insulin secretion. Proc Natl Acad Sci U S A. 2019;116:20033–20042. doi: 10.1073/pnas.1904311116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noguchi T.K., Kurisaki A. Formation of stomach tissue by organoid culture using mouse embryonic stem cells. Methods Mol Biol. 2017;1597:217–228. doi: 10.1007/978-1-4939-6949-4_16. [DOI] [PubMed] [Google Scholar]
- 23.McCracken K.W., Howell J.C., Wells J.M., et al. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat Protoc. 2011;6:1920–1928. doi: 10.1038/nprot.2011.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCracken K.W., Cata E.M., Crawford C.M., et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartfeld S., Bayram T., van de Wetering M., et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–136.e6. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCracken K.W., Aihara E., Martin B., et al. Wnt/beta-catenin promotes gastric fundus specification in mice and humans. Nature. 2017;541:182–187. doi: 10.1038/nature21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muzumdar M.D., Tasic B., Miyamichi K., et al. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 28.Dijkers P.F., Medema R.H., Pals C., et al. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1) Mol Cell Biol. 2000;20:9138–9148. doi: 10.1128/mcb.20.24.9138-9148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medema R.H., Kops G.J., Bos J.L., et al. AFX-like forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 30.Seoane J., Le H.V., Shen L., et al. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 31.Kitamoto T., Kuo T., Okabe A., et al. An integrative transcriptional logic model of hepatic insulin resistance. bioRxiv. 2021 doi: 10.1101/2021.03.15.435438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H.J., Johnston B., Aiello D., et al. Distinct cellular origins for serotonin-expressing and enterochromaffin-like cells in the gastric corpus. Gastroenterology. 2014;146:754–764.e3. doi: 10.1053/j.gastro.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miao Z.F., Adkins-Threats M., Burclaff J.R., et al. A metformin-responsive metabolic pathway controls distinct steps in gastric progenitor fate decisions and maturation. Cell Stem Cell. 2020;26:910–925.e6. doi: 10.1016/j.stem.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saline M., Badertscher L., Wolter M., et al. AMPK and AKT protein kinases hierarchically phosphorylate the N-terminus of the FOXO1 transcription factor, modulating interactions with 14-3-3 proteins. J Biol Chem. 2019;294:13106–13116. doi: 10.1074/jbc.RA119.008649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yun H., Park S., Kim M.J., et al. AMP-activated protein kinase mediates the antioxidant effects of resveratrol through regulation of the transcription factor FoxO1. FEBS J. 2014;281:4421–4438. doi: 10.1111/febs.12949. [DOI] [PubMed] [Google Scholar]
- 36.Greer E.L., Oskoui P.R., Banko M.R., et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 37.Yusuf I., Kharas M.G., Chen J., et al. KLF4 is a FOXO target gene that suppresses B cell proliferation. Int Immunol. 2008;20:671–681. doi: 10.1093/intimm/dxn024. [DOI] [PubMed] [Google Scholar]
- 38.Shin D.J., Joshi P., Hong S.H., et al. Genome-wide analysis of FoxO1 binding in hepatic chromatin: potential involvement of FoxO1 in linking retinoid signaling to hepatic gluconeogenesis. Nucleic Acids Res. 2012;40:11499–11509. doi: 10.1093/nar/gks932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalvisa A., Siersbaek M.S., Praestholm S.M., et al. Insulin signaling and reduced glucocorticoid receptor activity attenuate postprandial gene expression in liver. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2006249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langlet F., Haeusler R.A., Linden D., et al. Selective inhibition of FOXO1 activator/repressor balance modulates hepatic glucose handling. Cell. 2017;171:824–835.e18. doi: 10.1016/j.cell.2017.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., Deng S., Peng J., et al. MicroRNA-223 is essential for maintaining functional beta-cell mass during diabetes through inhibiting both FOXO1 and SOX6 pathways. J Biol Chem. 2019;294:10438–10448. doi: 10.1074/jbc.RA119.007755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee E.R., Trasler J., Dwivedi S., et al. Division of the mouse gastric mucosa into zymogenic and mucous regions on the basis of gland features. Am J Anat. 1982;164:187–207. doi: 10.1002/aja.1001640302. [DOI] [PubMed] [Google Scholar]
- 43.Nakae J., Biggs W.H., 3rd, Kitamura T., et al. Regulation of insulin action and pancreatic beta-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat Genet. 2002;32:245–253. doi: 10.1038/ng890. [DOI] [PubMed] [Google Scholar]
- 44.Paik J.H., Kollipara R., Chu G., et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee C.S., Perreault N., Brestelli J.E., et al. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 2002;16:1488–1497. doi: 10.1101/gad.985002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schonhoff S.E., Giel-Moloney M., Leiter A.B. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev Biol. 2004;270:443–454. doi: 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 47.Barker N., van Es J.H., Kuipers J., et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 48.Syder A.J., Karam S.M., Mills J.C., et al. A transgenic mouse model of metastatic carcinoma involving transdifferentiation of a gastric epithelial lineage progenitor to a neuroendocrine phenotype. Proc Natl Acad Sci U S A. 2004;101:4471–4476. doi: 10.1073/pnas.0307983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frescas D., Valenti L., Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 50.Nakae J., Kitamura T., Kitamura Y., et al. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell. 2003;4:119–129. doi: 10.1016/s1534-5807(02)00401-x. [DOI] [PubMed] [Google Scholar]
- 51.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng J., Liu T., Qin B., et al. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinz S., Benner C., Spann N., et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quinlan A.R. BEDTools: the Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;47:11.12.1–11.12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiao X.T., Ziel J.W., McKimpson W., et al. Prospective identification of a multilineage progenitor in murine stomach epithelium. Gastroenterology. 2007;133:1989–1998. doi: 10.1053/j.gastro.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McKimpson W.M., Weinberger J., Czerski L., et al. The apoptosis inhibitor ARC alleviates the ER stress response to promote beta-cell survival. Diabetes. 2013;62:183–193. doi: 10.2337/db12-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heading R.C., Nimmo J., Prescott L.F., et al. The dependence of paracetamol absorption on the rate of gastric emptying. Br J Pharmacol. 1973;47:415–421. doi: 10.1111/j.1476-5381.1973.tb08339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.