Abstract
Like many other bacteria, Corynebacterium glutamicum possesses two types of l-malate dehydrogenase, a membrane-associated malate:quinone oxidoreductase (MQO; EC 1.1.99.16) and a cytoplasmic malate dehydrogenase (MDH; EC 1.1.1.37) The regulation of MDH and of the three membrane-associated dehydrogenases MQO, succinate dehydrogenase (SDH), and NADH dehydrogenase was investigated. MQO, MDH, and SDH activities are regulated coordinately in response to the carbon and energy source for growth. Compared to growth on glucose, these activities are increased during growth on lactate, pyruvate, or acetate, substrates which require high citric acid cycle activity to sustain growth. The simultaneous presence of high activities of both malate dehydrogenases is puzzling. MQO is the most important malate dehydrogenase in the physiology of C. glutamicum. A mutant with a site-directed deletion in the mqo gene does not grow on minimal medium. Growth can be partially restored in this mutant by addition of the vitamin nicotinamide. In contrast, a double mutant lacking MQO and MDH does not grow even in the presence of nicotinamide. Apparently, MDH is able to take over the function of MQO in an mqo mutant, but this requires the presence of nicotinamide in the growth medium. It is shown that addition of nicotinamide leads to a higher intracellular pyridine nucleotide concentration, which probably enables MDH to catalyze malate oxidation. Purified MDH from C. glutamicum catalyzes oxaloacetate reduction much more readily than malate oxidation at physiological pH. In a reconstituted system with isolated membranes and purified MDH, MQO and MDH catalyze the cyclic conversion of malate and oxaloacetate, leading to a net oxidation of NADH. Evidence is presented that this cyclic reaction also takes place in vivo. As yet, no phenotype of an mdh deletion alone was observed, which leaves a physiological function for MDH in C. glutamicum obscure.
Recently, we discovered the gene for a relatively unknown type of malate dehydrogenase called malate:quinone oxidoreductase (MQO; (EC 1.1.99.16) [also called “malate dehydrogenase (acceptor)”] (22). Like the NAD-dependent malate dehydrogenase (MDH; EC 1.1.1.37), MQO catalyzes the oxidation of malate to oxaloacetate. The enzyme is membrane associated, probably through weak ionic or hydrophobic interactions. Tightly bound flavin adenine dinucleotide serves as a prosthetic group, and quinones instead of NAD are the electron acceptors of the enzyme. The quinones are subsequently oxidized by the electron transfer chain. These properties place MQO, like succinate dehydrogenase (SDH), both in the electron transfer chain and in the citric acid cycle. The existence of MQO as an enzymatic entity was first proven in 1956 (8). MQO activity was since then observed in several bacteria, both gram positive and gram negative (see references cited in reference 22) but not in archaebacteria or eucaryotes. After identification (for the first time) of a DNA sequence encoding an MQO in Corynebacterium glutamicum, it was found that similar sequences with, until then, unknown function existed in many other bacteria (see Fig. 1 in references 16 and 22).
C. glutamicum possesses both MQO and MDH. Moreover, as shown below, these enzymes are simultaneously present and active. As a consequence of the fact that the electron acceptors of MDH and MQO differ, the Gibbs standard free energy differences (ΔG°′) of the reactions catalyzed by these enzymes are also very much different. The ΔG°′ of malate oxidation by MDH has a highly positive value of +28.6 kJ · mol−1, whereas the ΔG°′ of this reaction catalyzed by MQO is −55.0 or −18.9 kJ · mol−1 depending on whether ubiquinones or menaquinones, respectively, act as acceptors. A consequence of the large difference in ΔG°′ would be that MDH and MQO, when active at the same time, catalyze reactions in opposite directions as follows:
Table.
| (MDH) | NADH + H+ + oxaloacetate | → | NAD+ + malate | −28.6 | ||
| (MQO) | MQ + malate | → | oxaloacetate + MQH2 | −18.9 | ||
| + | + | |||||
|---|---|---|---|---|---|---|
| (Net) | NADH + H+ + MQ | → | NAD+ + MQH2 | −47.5 | ||
Here MQ and MQH2 represent oxidized and reduced menaquinone, respectively. The numerical values are ΔG°′ in kilojoules per mole. The net reaction equals that of a non-proton-pumping NADH:quinone oxidoreductase (type II NADH dehydrogenase or NDH). This cycle could serve as an additional NADH dehydrogenase or could be involved in regulation of malate or oxaloacetate concentrations. In the present paper, we show that this reaction cycle occurs with isolated membranes and purified MDH of C. glutamicum, and we present evidence that the cycle also occurs in vivo. Furthermore, the physiological function of MQO and MDH was assessed by studying their regulation and the effect on growth of deletion of their genes.
These studies are of general interest for bacterial physiology since genes encoding MQO and MQO activity have been found in many gram-positive and -negative bacteria.
MATERIALS AND METHODS
Strains, growth, and media.
Plasmids and strains used in this study are listed in Table 1. C. glutamicum was cultivated at 30°C.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Characteristics | Reference |
|---|---|---|
| C. glutamicum | ||
| ATCC 13032 | Wild type | 1 |
| Δmqo | ATCC 13032 Δmqo | This work |
| Δmdhint | ATCC 13032 mdh::pEMmdhint; Kmr | This work |
| Δmdh | ATCC 13032 Δmdh | This work |
| Δmdhint Δmqo | Δmqo mdh::pEMmdhint; Kmr | This work |
| Δndh | ATCC 13032 ndh::pEMndhint; Kmr | This work |
| Δmdh Δndhint | Δmdh ndh::pEMndhint; Kmr | This work |
| Δmqo Δndhint | Δmqo ndh::pEMndhint; Kmr | This work |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 29 |
| S17-1 | thi-1 F−endAR1 hsdR17 supE44 λ− pro | 34 |
| Plasmids | ||
| pK19MobSacB | Integration vector, Kmr; oriVE. coli oriT sacB | 31 |
| pBluescript II SK(+) | Ampr; lacZ | 33 |
| pEM1 | Integration vector; Kmr; oriVE. coli oriT | 32 |
| pEMmdhint | Internal fragment of mdh in pEM1 | This work |
| pEMndhint | Internal fragment of ndh in pEM1 | This work |
Tryptone-yeast extract medium (2 × TY) was described before (29). The design of the minimal medium used in this study was based on minimal media for Corynebacterium described in other studies (17, 18), and on the Neidhardt minimal medium for enterobacteria (23). It was modified to optimize growth rate, to guarantee pH stability over a long period of growth, and to prevent the formation of precipitates, thereby correcting three major disadvantages of minimal media previously used for growth of C. glutamicum. A fourth advantage of this medium, which proved instrumental in the discovery, described below, of the nicotinamide-dependent growth of the Δmqo mutant, is the fact that it reproducibly allows growth of C. glutamicum after inoculation at very low cell densities.
Two trace-element solutions, TE1 and TE2, were prepared as described previously (22). The minimal medium consisted of (per liter) 8.37 g of 3-[N-morpholino]-propanesulfonic acid (MOPS), 0.72 g of N-tris[hydroxymethyl]-methylglycine (Tricine), 4.05 g of NH4Cl, 1 g of KCl, 0.3 g of K2HPO4, 0.23 g of MgCl2 · 6H2O, 50 mg of CaCl2 · 2H2O, 0.2 g of EDTA, 50 mg of K2SO4, 15 mg of protocatechuate, and 0.2 mg of biotin. MOPS, Tricine, NH4Cl, and KCl were prepared together as a 10-fold concentrated stock solution. The other components were prepared individually as 100-fold concentrated stock solutions, except for protocatechuate, which was added in dry form during medium preparation, and biotin, which was stored at −20°C as a 1,000-fold stock solution. The EDTA stock solution was titrated to pH 7 with NaOH. The medium (1 liter) was prepared by first diluting 100 ml of the solution containing MOPS, Tricine, NH4Cl, and KCl and 10 ml of the EDTA solution in 700 ml of distilled water. Subsequently, 3 ml of TE1 and 1 ml of TE2 were added, and finally all other components were added. The medium was titrated to pH 7.0 with NaOH, the volume was adjusted to 900 ml with water, and the medium was filter sterilized. Carbon and energy sources were added from 10-fold concentrated and filter-sterilized stocks. If necessary, these stocks had been adjusted to pH 7 with NaOH. For the preparation of minimal medium agar plates, the medium was prepared twofold concentrated and was mixed with an equal volume of autoclaved liquid agar (30 g/liter of water).
For growth measurements, a colony was suspended in 2 ml of minimal medium without carbon source. A few microliters of this suspension was transferred with an inoculating loop to 2 ml of minimal medium with carbon source. These cultures were used to establish whether mutants were able to grow or were used as a preculture for measurements of growth rate on the corresponding carbon sources. In the latter case, the preculture was diluted to a final optical density at 600 nm between 0.05 and 0.1 into 10 ml of minimal medium with carbon source in a 50-ml conical flask. The air contact-surface/volume ratio in these flasks at rest was 2 cm2/ml. The culture was incubated in a rotary shaker at 200 rpm. Under these conditions, growth was exponential at least up to an optical density of 10. The growth rate was calculated by least-squares fitting of the exponential growth equation to the data.
Purification of MDH.
MDH was purified from raw cell extract in four steps: a streptomycinsulfate precipitation, an anion-exchange purification, and a dye affinity purification repeated twice. Wild-type C. glutamicum was cultivated for 16 h in 400 ml of 2× TY medium. The cells were harvested, washed twice in 50 mM K-phosphate (pH 7.5) with additional 1 mM dithiothreitol and 2 mM EDTA, and resuspended in the same buffer at a concentration of 0.25 g (wet weight)/ml (total volume approximately 20 ml). The suspension was passed three times through a French press cell at 165 MPa. Cell debris was removed by centrifugation for 10 min at 75,000 × g and 4°C. A 10% (wt/vol) streptomycinsulfate solution was slowly added, while being stirred, to the supernatant up to a final concentration of 0.75%. The extract was left on ice for 15 min and subsequently centrifuged for 10 min at 15,000 × g and 4°C. The clear supernatant was transferred to dialysis tubing and was dialyzed twice for 1.5 h at 4°C against 1 liter of 10 mM K-phosphate (pH 7.5) with additional 1 mM dithiothreitol and 2 mM EDTA (buffer A). The dialysate was cleared by centrifugation for 15 min at 75,000 × g and 4°C. It contained approximately 10 mg of protein/ml.
For anion-exchange chromatography, a Resource-Q column (Pharmacia) with a 1-ml column volume was used. Purification was performed at 4°C. The column was loaded with 15 mg of protein and washed with 4 ml of buffer A. The protein was eluted with a linear NaCl gradient, from 0 to 1 M, in buffer A and with a total volume of 15 ml. MDH activity eluted from the column at approximately 0.5 M NaCl. The fractions containing more than 10% of the total MDH activity were combined and desalted by gel filtration on a PD-10 column (Pharmacia).
Affinity chromatography was performed with the dye ligand reactive brown 10 cross-linked to agarose (Sigma). A column with a volume of 2.5 ml was loaded with 10 mg of protein or less and was subsequently washed with 6 column volumes of 10 mM K-phosphate (pH 7.5). MDH was eluted with the same buffer supplemented with 1 mM NADH. The MDH preparation was desalted by gel filtration on a PD-10 column, and the dye affinity purification and desalting were repeated once more. The resulting MDH preparation was concentrated using a centrifugal filter device with a 10-kDa cutoff filter (Amicon). The preparation contained only one protein of 33 kDa as judged by sodium dodecyl sulfate-polyacrylamide gelelectrophoresis of 50 μg of protein and subsequent staining with Coomassie brilliant blue. To stabilize the MDH activity, bovine serum albumin (1-mg/ml final concentration) was added, and the preparation was stored at −20°C. Under these conditions, the activity did not significantly decrease over at least 3 months.
Construction of site-directed mutants.
A defined chromosomal deletion of the mqo gene was constructed as follows. A 3,174-bp DNA fragment excised from pRM30 (22) using SalI and XbaI containing the 1,500-bp open reading frame (ORF) encoding MQO was inserted into the integration vector pK19MobSacB using the same enzymes. Preceding this ligation, the HindIII site had been removed from the vector by filling in using T4 DNA polymerase. The resulting plasmid, pK19MI1, was partially digested with HindIII. By religation, the 766-bp SalI/HindIII fragment containing the 5′ end of the mqo gene and the 1,341-bp HindIII/XbaI fragment containing the 3′ end of the mqo gene were joined to form an mqo ORF with a 1,067-bp internal deletion. The resulting plasmid, pK19MI3, was used to replace the mqo gene in wild-type C. glutamicum according to the procedure of Schäfer et al. (30). Success of the deletion procedure was checked by PCR of the chromosomal template. The resulting Δmqo strain did not possess detectable MQO activity as measured with 2,6-dichlorophenolindophenol (DCPIP) as the electron acceptor.
Insertion mutants of the ndh gene were constructed as follows. A 0.39-kbp fragment of the ndh gene was amplified from chromosomal DNA of the wild type by PCR with degenerative primers designed to hybridize with conserved stretches in known ndh genes. The primers used were 5′-CCGCTGCT(G/C)TACCA(A/G)GTGGC-3′ and 5′-CCGGT(G/C)GGGCC(A/C)GC(G/C)CCGAC-3′. Annealing was performed at 53°C. The fragment was cloned in vector pBluescript II SK(+) opened with EcoRV. An 0.34-kbp BamHI fragment isolated from this plasmid was cloned into pEM1. The resulting plasmid, pEMndhint, was used to create insertion mutants in C. glutamicum wild-type and Δmqo strains by the simplified transformation protocol described in the work of Van der Rest et al. (39), resulting in strains ndh::pEMndhint and Δmqo ndh::pEMndhint, respectively. Other parts of the ndh gene were cloned by plasmid rescue (24) from the ndh::pEMndhint insertion mutant (R. Yücel, unpublished results). These fragments were sequenced on both strands.
Insertion mutants of the mdh gene encoding cytoplasmic MDH (EC 1.1.1.37) were constructed as follows. A 0.47-kbp fragment of the mdh gene was amplified from chromosomal DNA of the C. glutamicum wild-type strain by PCR with degenerative primers. One primer was designed using the N-terminal amino acid sequence of the purified MDH protein from C. glutamicum (A. Drysch, unpublished results). The other primer was designed to hybridize with a conserved stretch in known mdh genes. The primers used were 5′-AA(AG)GT(CT)AC(CT)GT(CT)AC(CT)GG(CT)-3′ and 5′-CG(AG)TT(AG)TG(AG)TC(AGC)A(AG)(AG)CG-3′. Annealing was performed at 63°C. The amplified fragment was cloned in vector pBluescript II SK(+) (Stratagene) opened with EcoRV. The fragment was excised from this plasmid with BamHI and HindIII and cloned in vector pEM1 opened with the same enzymes. The resulting plasmid, pEMmdhint, was used to create insertion mutants in C. glutamicum wild-type and Δmqo strains (39), resulting in strains mdh::pEMmdhint and Δmqo mdh::pEMmdhint, respectively. The mdh insertion mutants did not possess MDH activity.
A defined chromosomal deletion of the mdh gene was constructed as follows. A 1,503-bp DNA fragment containing the 984-bp mdh ORF was amplified using the oligonucleotides 5′-GTGGATCCTGCGCTTGGACATGCCAG-3′ and 5′-CGCTCTAGATTAGAGCAAGTCGCG-3′. The first oligonucleotide introduces a BamHI site 505 bp in front of the mdh ORF, and the second introduces an XbaI site 4 bp following the TAA stop codon of the mdh ORF. The amplified fragment was digested with BamHI, XbaI, and XhoII. The XhoII digestion removes a 395-bp internal fragment from the mdh ORF. By religation, a 645-bp BamHI/XhoII DNA fragment containing the 5′ end of the mdh ORF and a 456-bp XhoII/XbaI DNA fragment containing the 3′ end of the mdh ORF were joined to form an mdh ORF with a 395-bp internal deletion. This fragment was inserted into the integration vector pK19MobSacB opened with BamHI and XbaI. The resulting plasmid pK19Δmdh was transformed by electroporation into wild-type C. glutamicum (39). Deletion mutants of mdh were selected according to the procedure described above for mqo deletion mutants. The resulting Δmdh strain did not possess detectable MDH activity. The Δndhint mutation was introduced in this strain as described above for the wild type.
Measurement of the membrane-bound dehydrogenase activities.
Cultures growing exponentially on 50 ml of minimal medium with carbon source were placed on ice for 10 min after reaching an optical density at 600 nm between 2 and 3. They were centrifuged for 5 min at 4,000 × g and 4°C. The supernatant was discarded, and the pellets either were frozen in liquid nitrogen and stored at −20°C or were processed immediately. Membrane fragments were isolated from these cells as described before (22). The activities of the dehydrogenases for NADH, d- or l-malate, d- or l-lactate, and succinate associated with these membranes were measured with DCPIP as an electron acceptor and in the presence of a 1 mM concentration of the substrate (22). NADH dehydrogenase activity was also measured by observing the decrease in absorption at 340 nm over a certain period. The absorption coefficients used were 22 and 6.3 cm−1 · mM−1 for DCPIP and NADH, respectively.
Measurement of intracellular NAD and NADH concentrations.
Cells were extracted by thoroughly mixing 1 ml of culture with either 1 M HClO4 or 1 M KOH containing 50% ethanol. The final pH of the extracts was 1 and 12.4 for the acid and alkaline extracts, respectively. The acid and alkaline extracts were incubated at 55°C for 8 and 5 min, respectively. They were subsequently cooled on ice and carefully neutralized with 2 M KOH or 1 M HCl. The concentration of the pyridine nucleotides in the extracts was measured with the cycling assay (6), and intracellular concentrations were calculated by assuming an intracellular volume of 1.5 μl · mg−1 (dry mass). The dry mass was estimated by measuring the optical density at 600 nm and assuming that an optical density equal to 1 corresponds to 0.34 mg (dry mass) · ml−1 (28).
Measurement of cytoplasmic malate and lactate dehydrogenases.
Exponentially growing cells (50 ml of medium; optical density at 600 nm between 2 and 3) were harvested and washed with 50 mM K-phosphate, pH 7.5, as described above and resuspended in 0.7 ml of this buffer. The cells were broken by sonication for 5 min on ice with a Branson B15 sonifier, equipped with a microtip at a 30% duty cycle and 30% of maximal output. Debris was removed by centrifugation for 15 min at 28,000 × g. The MDH activity in the extract was measured in the direction of malate oxidation or oxaloacetate reduction according to the method of Smith (35), except that 3-amino-1-propanol instead of diethanolamine was used as a buffer. d-or l-lactate dehydrogenase was measured in the direction of lactate oxidation as described by Wahlefeld (40).
Measurement of extracellular organic acids.
Organic acids produced by cultures were quantified by chromatography of the culture supernatant in 5 mM H2SO4 on a 300- by 8-mm organic acid resin column (Chromatography Service). The identity of the acids was confirmed by standard enzymatic assays of the culture supernatants (5).
Protein determination.
Protein was determined with bicinchoninic acid according to a protocol adapted from the work of Smith et al. (36). When protein was determined in membrane fragment preparations, the assay solution contained an additional 0.5% (mass/vol) sodium dodecyl sulfate.
Nucleotide sequence accession number.
The complete sequence of the ndh gene has been deposited in the EMBL data bank under accession no. AJ238250.
RESULTS
Activity of NADH, succinate, and malate dehydrogenases during growth on various substrates.
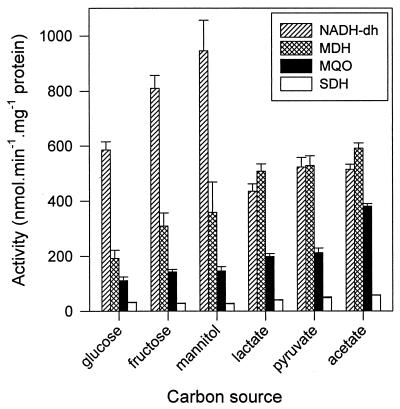
The activities of the membrane-bound dehydrogenases, NADH dehydrogenase, SDH (EC 1.3.99.1), and MQO, differ, depending on the carbon and energy source (Fig. 1). The citric acid cycle (or tricarboxylic acid [TCA] cycle) enzymes MQO and SDH are more active during growth on L-lactate, acetate, and pyruvate. For example, MQO and SDH activities are, by a factor of 3.4 (±0.4) and 1.8 (±0.1), respectively, higher during growth on acetate than on glucose. The cytoplasmic MDH is regulated in a similar manner, and a positive correlation between MDH and MQO activities is observed (Fig. 1). Thus, it is shown that MDH and MQO are present at the same time in C. glutamicum and that their activities are coordinately regulated.
FIG. 1.
Specific dehydrogenase activities in cells grown on various substrates. The NADH dehydrogenase (NADH-dh), SDH, MQO, and MDH activities were determined in cells from the exponential growth phase growing on various carbon sources. The values are the averages of two independent experiments in which the activities were determined in duplicate. The error bars indicate the overall sample standard deviations of the four measurements. Note that the values of the specific activities cannot be directly compared because MDH activity is standardized with respect to total protein in the cleared cell extract, whereas MQO activity is standardized with respect to total membrane protein. Moreover, MDH activity was assayed in the direction of malate oxidation at pH 9.2. Lactate is l-lactate.
NADH dehydrogenase is regulated differently from these TCA cycle enzymes. Its activity might be partly correlated with the degree of reduction of the substrate. The dependency on the degree of reduction of the substrate is apparent from the very high activity found in cells growing on mannitol. Complete oxidation of this substrate generates an extra reducing equivalent (NADH) compared to oxidation of glucose or fructose.
Properties of the Δmqo mutant.
An MQO-negative mutant (DM22) generated by random mutation with 1-methyl-3-nitro-1-nitrosoguanidine (NTG) was described before (22). The mutant grew more slowly than the wild type on several substrates tested (D. Molenaar, unpublished data). This phenotype allowed the cloning of the mqo gene by complementation. Sequencing of the mqo gene from DM22 confirmed the existence of a deleterious mutation, consisting of a single base-pair mutation leading to an internal stop codon. However, since the plasmid-borne mqo gene was not able to fully complement the phenotype, it was suspected that DM22 might carry additional mutations. This is not uncommon in strains generated by NTG mutagenesis. Therefore, to be able to discern the physiological effects of the deletion of mqo alone, a site-directed mutant, Δmqo, was constructed. Assaying for MQO activity confirmed its absence in this strain.
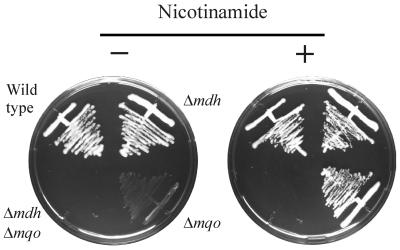
This mutant does not grow on the minimal medium described in Materials and Methods, except when nicotinamide is added (Fig. 2). Other supplements tested, but not stimulating growth, were the vitamins riboflavin, folic acid, d-pantothenic acid, thiamine, pyridoxine, and p-aminobenzoic acid and the amino acid aspartate. For growth on glucose, fructose, mannitol, pyruvate, and lactate, 1 mg of nicotinamide liter−1 is sufficient to let Δmqo attain the same optical density as that of the wild type after 48 h of incubation. Still, even in the presence of 1 mg of nicotinamide liter−1, Δmqo grows more slowly than the wild type. During growth on glucose, a small but significant reduction of the growth rate by approximately 10% was observed. The defect was very pronounced in the case of growth on acetate (75% reduction) or pyruvate (40% reduction), substrates during growth on which the MQO activity is relatively high (Fig. 1). The activities of the NADH dehydrogenase, SDH, and MDH in strain Δmqo do not differ significantly from those in the wild type when grown on the substrates referred to in Fig. 1 (data not shown).
FIG. 2.
Complementation of Δmqo by mdh when cells grow in the presence of nicotinamide. Strains were streaked on minimal medium agar with 1% (wt/vol) glucose, and where indicated, 1 mg of nicotinamide liter−1 was added. Plates were incubated for 4 days.
Properties of the Δmdh single and the Δmdh Δmqo double mutants.
Insertion mutations of the mdh gene were made in the wild type and in Δmqo. The single mutant Δmdh did not have an observable phenotype when tested for growth on glucose, fructose, lactate, acetate, or succinate, neither when growth on plates was scored nor when the growth rate in liquid medium was measured. The Δmdh Δmqo double mutant, however, was unable to grow on these substrates in defined medium, even in the presence of nicotinamide (Fig. 2). This indicates that MDH is able to complement the absence of MQO activity but only when Δmqo is supplied with nicotinamide. Apparently, the function of MQO, that is, the oxidation of malate to oxaloacetate, is taken over by MDH in these cells. An obvious reason for the nicotinamide dependence of Δmqo would be that it leads to an increased concentration of NAD in the cell, enabling MDH to oxidize malate. As can be seen in Table 2, addition of nicotinamide indeed causes an increase in intracellular NAD and NADH concentrations. Although the [NAD]/[NADH] ratio decreases from 4.4 in the absence of nicotinamide to 2.8 at 1 mg of nicotinamide liter−1, the sevenfold increase of the NAD concentration might have a favorable effect on the kinetics of MDH which suffices to enable malate oxidation by the enzyme. The low growth rates of Δmqo even in the presence of nicotinamide are, nevertheless, indicative of the difficulties the organism has with this backup solution, which are likely to be due to the unfavorable thermodynamics of the reaction.
TABLE 2.
NAD and NADH concentrations in C. glutamicum grown in the presence of different nicotinamide concentrations
| [Nicotinamide] (μg · liter−1) | Pyridine nucleotide concn (mM)
|
[NAD+]/[NADH] ratio | |
|---|---|---|---|
| [NAD+] | [NADH] | ||
| 0 | 0.22 | 0.05 | 4.4 |
| 10 | 0.60 | 0.26 | 2.3 |
| 1,000 | 1.53 | 0.54 | 2.8 |
Properties of the Δndhint single and the Δmqo Δndhint and Δmdh Δndhint double mutants.
C. glutamicum possesses highly active membrane-bound NADH dehydrogenase (Fig. 1). In order to gain evidence for the hypothesis that MQO and MDH catalyze opposite reactions in the citric acid cycle and consequently catalyze a net NADH dehydrogenase reaction, the membrane-bound NADH dehydrogenase had to be inactivated. For this purpose, we disrupted the ndh gene, which encodes a type II NADH dehydrogenase, expecting that possibly also a nuo homolog, encoding type I NADH dehydrogenase, might have to be disrupted. However, membranes isolated from strains carrying the ndh disruption, when grown on 2× TY medium, did not possess detectable NADH dehydrogenase activity, neither when assayed with DCPIP nor when NADH consumption was observed directly by measuring the absorption at 340 nm. This indicates that a type II NADH dehydrogenase (NDH), the non-proton-pumping enzyme encoded by the ndh gene, is under these conditions the only active membrane-bound NADH dehydrogenase. This mutant grows at a slightly lower rate than does the wild type (Table 3).
TABLE 3.
Growth of wild type and mutants by scoring of colonies on minimal medium agar supplied with different carbon sources after 24 or 48 h of incubationa
| Strain | Growth on substrate at time (h)b
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No addition
|
Plus nicotinamide
|
|||||||||||
| Glucose
|
Mannitol
|
Acetate
|
Glucose
|
Mannitol
|
Acetate
|
|||||||
| 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | |
| Wild type | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Δmqo | − | − | − | − | − | − | +++ | +++ | + | +++ | +/− | +/− |
| Δndh | + | +++ | +/− | + | + | ++ | + | +++ | +/− | +++ | + | ++ |
| Δmqo Δndhint | − | − | − | − | − | − | +/− | +/− | − | + | − | − |
Nicotinamide was added to a concentration of 1 mg/liter.
Symbols: +++, large colonies; ++, small colonies; +, tiny colonies; +/−, growth visible only in regions of dense inoculation; −, no growth.
Escherichia coli mutants lacking both type I and II membrane-bound NADH dehydrogenases have clear phenotypes (7, 42). Since they are unable to regenerate NAD by respiration, these mutants grow mainly fermentatively and excrete large amounts of lactate (41). They are also unable to grow on mannitol, since fermentative growth on this substrate is not possible. As a diagnostic indicator of the absence of NADH dehydrogenase activity, growth of the C. glutamicum double mutant Δmqo Δndhint was tested on mannitol. Although this mutant lacked all known NADH dehydrogenase activities, it did grow slowly on mannitol. It also did not excrete lactate during growth on glucose. An explanation for this phenomenon would be that, similar to the MQO-MDH cycle, a cyclic reaction involving lactate and pyruvate catalyzed by an NAD-dependent lactate dehydrogenase and membrane-bound quinone-dependent lactate dehydrogenase could constitute an alternative NADH dehydrogenase (see Discussion). In the Δmqo, Δndhint, and Δmqo Δndhint strains, the membrane-bound l-lactate dehydrogenase was more active than in the wild type by a factor of 1.5, 1.8, and 2.3, respectively. Neither the membrane-bound d-lactate dehydrogenase nor the cytoplasmic lactate dehydrogenase activities were increased in these strains. A significant difference of Δmqo Δndhint from the single mutants was that it excreted fumarate and malate to high concentrations (Table 4). This may be explained by assuming that the TCA cycle is almost completely blocked in the double mutant due to the fact that the MQO is absent and that the backup function of MDH is restricted by high NADH concentrations due to the absence of NADH dehydrogenase.
TABLE 4.
Extracellular products after growing for 16 h on minimal medium with 2% glucose as carbon source and containing 1 mg of nicotinamide · ml−1
| Strain | α-Ketoglutarate (mM) | Pyruvate (mM) | Malate (mM) | Fumarate (mM) |
|---|---|---|---|---|
| Wild type | 0.29 | 0.005 | 0.05 | 0.002 |
| Δmqo | 0.3 | 0.025 | 0.34 | 0.059 |
| Δndh | 0.36 | 0.059 | 0.05 | 0.019 |
| Δmqo Δndhint | 0.26 | 0.074 | 3.9 | 10.4 |
The fact that only the double mutants Δmqo Δndhint and Δmdh Δndhint display a clear temperature-sensitive phenotype (Fig. 3) is indirect evidence for the limited NADH dehydrogenase capacity in this mutant compared to the single mutants. For the interpretation of these results, an observation for Mycobacterium smegmatis is of importance. In M. smegmatis, a bacterium containing MQO as the only malate dehydrogenase, the disruption of ndh causes growth inhibition at high temperatures (21) and also causes isoniazid resistance. Isoniazid sensitivity and temperature resistance can be restored by introducing MDH from the closely related Mycobacterium bovis in these mutants, thus restoring an NADH dehydrogenase activity catalyzed by native MQO and the foreign MDH. Although corynebacteria and mycobacteria are evolutionary closely related, C. glutamicum wild type is not sensitive toward isoniazid. However, C. glutamicum mutant strains display a behavior similar to that of M. smegmatis at elevated temperatures, although in this case MQO and MDH are both innate.
FIG. 3.
Temperature-sensitive phenotype of Δmqo Δndhint and Δmdh Δndhint double mutants. The strains were streaked on 2× TY agar, and plates were incubated for 3 days at the temperatures indicated. Strains with the integration disruption Δndhint (upper halves) were streaked on plates containing an additional 25 μg of kanamycin ml−1 to prevent the growth of revertants.
It has been proposed by others that membranes of C. glutamicum also possess an NADPH dehydrogenase activity with an optimum at pH 5.5 (20). We could measure a similar activity in our assay system at pH 7. However, NADPH dehydrogenase activity was completely absent in the Δndhint mutant. This result indicates that NADPH dehydrogenase is a minor activity of NDH and is not likely to be due to a separate NADPH-specific dehydrogenase, as was proposed by Matsushita et al. (20).
Properties of purified MDH. Assaying malate oxidation by MDH is usually performed at high pH, since, the proton being a product of the reaction, the free energy difference is more favorable under these conditions than at neutral pH. Of course, for the physiological function the activity of the enzyme at neutral pH is of interest. Although malate oxidation with NAD as the electron acceptor is thermodynamically unfavorable, an MDH might still be able to catalyze malate oxidation at a substantial rate under the proper conditions, i.e., relatively low NADH and oxaloacetate concentrations. The assay of malate oxidation by MDH quickly runs into equilibrium at pH 7.5, as early as attainment of 2 μM oxaloacetate and NADH in the assay used here with starting concentrations of 1 mM malate and 0.1 mM NAD. This amount of NADH is hardly detectable, and therefore, in the simple assay at pH 7.5 no malate oxidation by purified MDH was observed. A low activity of maximally 18 U · mg−1 was observed only when a sink for oxaloacetate in the form of glutamate oxaloacetate transaminase together with glutamate was added to the reaction mixture. On the other hand, the rate of oxaloacetate reduction catalyzed by MDH at pH 7.5 is 770 U · mg−1. At pH 9.5, the specific activities were 60 and 1,020 U · mg−1 for malate oxidation and oxaloacetate reduction, respectively.
MDH from C. glutamicum was also able to catalyze the NADPH-dependent reduction of oxaloacetate. Similar observations with respect to pyridine nucleotide specificity have been made previously for other MDHs (4).
In vitro reconstitution of the cyclic reaction between MDH and MQO.
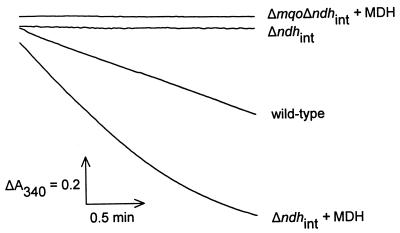
Membranes isolated from the Δndhint mutant have no observable NADH dehydrogenase activity (Fig. 4). When malate and purified MDH from C. glutamicum are added to these membranes, an apparent NADH dehydrogenase activity can be observed. This activity is dependent on the presence of MQO, since the reaction could not be reconstituted with membranes from the double mutant Δmqo Δndhint.
FIG. 4.
In vitro reconstitution of NADH dehydrogenase activity using membrane fragments and purified MDH. Membrane fragments were isolated from cells grown on 2× TY medium with 1% (wt/vol) glucose. Absorption by NADH was recorded at 340 nm. The reaction mixture contained 50 to 150 μg of membrane protein per ml, 0.2 mM NADH, and where indicated 175 ng of purified MDH ml−1. The reaction was started by adding 1 mM malate.
Since MDH also catalyzes the NADPH-dependent oxaloacetate reduction, an apparent NADPH dehydrogenase activity could also be reconstituted with the system (data not shown).
Inhibition of SDH by oxaloacetate.
Oxaloacetate is a known inhibitor of a number of enzymes in central metabolism, notably of SDH, for which examples exist in procaryotes as well as eucaryotes (2). The possible role of MQO and MDH in controlling the cytoplasmic oxaloacetate concentration prompted us to study the effect of oxaloacetate on SDH in C. glutamicum. The Michaelis constant for succinate of C. glutamicum SDH is approximately 20 to 30 μM. The modulation of SDH by oxaloacetate is complex but is basically a competitive type of inhibition (2). From the results of inhibition experiments, a competitive inhibition constant by oxaloacetate of 0.15 to 0.22 μM could be deduced. This illustrates the potential importance of oxaloacetate as a regulator of TCA cycle activity in C. glutamicum.
The inhibition of MQO by oxaloacetate was also assessed. A significant effect of oxaloacetate was observed only at very high concentrations, with the activity inhibited by 50% at 8 mM oxaloacetate. Based on the equilibrium constant for the MDH-catalyzed reaction, such high free oxaloacetate concentrations can hardly be expected in the cell (see Discussion). Therefore, the inhibition of MQO by oxaloacetate is probably not of physiological significance.
DISCUSSION
The observation of in vitro NADH dehydrogenase activity catalyzed by MQO and MDH and the observation of the temperature sensitivity of the Δmqo Δndhint double mutant support the hypothesis that MQO and MDH catalyze opposite reactions in vivo. More evidence is supplied by two observations from the literature. A study by Miesel et al. (21) indicated that isoniazid resistance in M. smegmatis is often associated with mutations which inactivate the type II NADH dehydrogenase. Isoniazid resistance is thought to be caused by the high NADH/NAD ratio in these mutants. Expressing the wild-type ndh gene from a plasmid restored isoniazid sensitivity in an NDH-negative mutant. Interestingly, isoniazid sensitivity was also restored in this mutant by expressing the heterologous mdh gene from the related species M. bovis. Bearing in mind that M. smegmatis possesses only MQO and no MDH of its own (26), we propose that the expression of a heterologous MDH in M. smegmatis causes the above-mentioned cyclic reaction to occur, resulting in a net NADH dehydrogenase reaction. The explanation by Miesel et al. (21) that MDH alone might oxidize NADH is incomplete. This would require a continuous source for oxaloacetate and a sink for malate and thereby a considerable reorganization of central metabolism.
The situation with respect to MQO and MDH is reminiscent of the d-lactate dehydrogenases found in E. coli. This organism possesses a cytoplasmic NAD-dependent enzyme (EC 1.1.1.28) and a membrane-bound d-lactate:quinone oxidoreductase (12). Also in this case, the oxidation of a 2-hydroxy acid is involved, and the standard free energies of the reactions are comparable to those of the respective malate dehydrogenases. The NAD-dependent oxidation of lactate has a positive ΔG°′ (equal to +25.0 kJ · mol−1) whereas the ubiquinone-dependent reaction has a negative ΔG°′ (equal to −58.6 kJ · mol−1). Consequently, the physiological function of the NAD-dependent lactate dehydrogenase is in most cases to reduce pyruvate, whereas the quinone-dependent enzyme is exclusively involved in the oxidation of lactate. The gene (dld) encoding d-lactate:quinone oxidoreductase in E. coli was accidentally cloned in an attempt to clone the ndh gene. When overexpressed from a plasmid, dld was able to complement the phenotype of an ndh mutation (41). The ndh mutant excretes large quantities of d-lactate since, being unable to dispose of reducing equivalents in another way, it grows in a fermentative mode. In contrast, the ndh mutant strain overexpressing d-lactate:quinone oxidoreductase did not excrete lactate. The authors concluded that the NAD-dependent d-lactate dehydrogenase and the d-lactate:quinone oxidoreductase together function as an NADH dehydrogenase.
Why do some organisms use MQO to catalyze malate oxidation, whereas others apparently use MDH (see accompanying article about E. coli)? An obvious advantage of using MQO is the highly negative ΔG°′ of malate oxidation by this enzyme compared to the highly positive ΔG°′ of the reaction catalyzed by MDH. In fact, malate oxidation by MDH has the highest ΔG°′ value of all reactions in central metabolism, that is, glycolysis and the citric acid cycle. The next highest value (ΔG°′ = +24 kJ · mol−1) is for the aldolase reaction. Since this reaction is one-sided bimolecular, a less extreme situation with respect to product concentrations exists than that for MDH. To illustrate this, it can be calculated that the ratio [oxaloacetate]/[malate] at pH 7 and 30°C has to be smaller than 1.2 × 10−5 times the [NAD]/[NADH] ratio if MDH oxidizes malate. The [NAD]/[NADH] ratio can be on the order of 1 in bacterial cells, for example, under conditions of low oxygen tension (10). Assuming a malate concentration on the order of 10−3 M, the oxaloacetate concentration under these conditions would have to be on the order of 10−8 M or less. Oxaloacetate is an important precursor in gluconeogenesis and in the synthesis of amino acids of the aspartate branch. Furthermore, it is the substrate of citrate lyase in the citric acid cycle. A very low oxaloacetate concentration might cause a growth limitation by limiting the reaction rates of one or more enzymes using oxaloacetate as a substrate. Higher levels of MDH would not improve this situation because the oxaloacetate concentration would be limited by the equilibrium of the MDH reaction. The problem of the unfavorable equilibrium of the MDH-catalyzed malate oxidation has been recognized before, and an alternative solution in the form of substrate channeling has been proposed for mitochondria. Here the reactant oxaloacetate or NADH would be transferred directly from MDH to citrate synthase or NADH dehydrogenase, respectively, thus directly coupling a reaction with an unfavorable ΔG°′ to one with a favorable ΔG°′ (9, 11, 27, 37). The fact that the biosynthetic and energy-generating pathways take place in separate compartments may also provide part of the solution for this problem in eucaryotes.
Using an MQO instead of an MDH may allow an organism to attain a high TCA cycle flux independently of the NADH/NAD and malate/oxaloacetate ratios. Thus, MQO may be of importance for a robust TCA cycle flux under adverse conditions. Such conditions could be low electron acceptor concentrations, limiting concentrations of carbon sources, or growth on carbon sources requiring a high TCA cycle flux for the generation of metabolic energy. In C. glutamicum, MQO is important during growth on organic acids, in particular, acetate, as reflected by its high activity and the severe effect on growth of an mqo deletion. Similarly, it was noticed that mqo disruption in Pseudomonas fluorescens impaired its ability to colonize tomato roots (L. C. Dekkers, personal communication). Under these circumstances, this organism probably grows on organic acids excreted by the plant (19). Nevertheless, bacteria like E. coli are able to attain a high growth rate on acetate using MDH (accompanying paper). This may, however, require more restricted and optimized conditions, in particular of aeration, to maintain a high TCA cycle flux rate.
Another situation in which an organism might benefit from the use of an MQO is when it faces continuous or occasional low intracellular pH. The MDH reaction produces a proton, and therefore its ΔG°′ becomes even more unfavorable at low pH. This situation might, for example, apply to Helicobacter pylori (16).
Energetically, the subsequent reactions of malate oxidation by MDH and NADH oxidation by NDH are equivalent to the MQO reaction alone, because the net reactions are the same. The situation would be different when an organism expresses the proton-pumping (type I) NADH dehydrogenase because then, in the net reaction, protons would be pumped by MDH and the type I NADH dehydrogenase, making in principle more metabolic energy available than with the MQO reaction. However, this energy gain might be possible only at the expense of a low oxaloacetate concentration, as explained above.
In bacteria like M. smegmatis (26), H. pylori (16), Neisseria meningitidis (13, 38), and several Pseudomonas species (25), which possess only MQO and no (known) MDH, the function of MQO seems clear. In those species, it is the only enzyme which is able to directly oxidize malate to oxaloacetate in the TCA cycle. However, the presence of two malate dehydrogenases as in C. glutamicum, E. coli, and many other bacteria, is puzzling (3, 8, 14, 15). In some cases, like that of E. coli (see accompanying paper), the activity of MQO may not be high enough to be of importance or may be induced only under special conditions. However, as shown above in C. glutamicum MDH and MQO activities are high and coordinately regulated on different carbon and energy sources. This suggests a similar or cooperative metabolic function of MDH and MQO. However, whereas the deletion of mqo is deleterious in this species, the fact that deletion of the mdh gene in C. glutamicum has no effect on growth indicates that the latter has no physiological function under the circumstances tested. Consequently, although it is very likely that MQO and MDH catalyze opposite reactions, neither the oxidation of NADH nor the potentially regulatory role of the cycle in regard to oxaloacetate and malate concentrations seems to be of importance. It was shown that in an MQO deletion strain MDH can take over the function of MQO when the cells are supplied with sufficient nicotinamide. This implies that, under circumstances where MQO does not function, MDH might serve as a backup.
ACKNOWLEDGMENTS
We thank Andreas Burkovski for discussion, Yiulia Bertsova for drawing our attention to the literature concerning E. coli lactate dehydrogenases, and Linda Dekkers for discussion and sharing unpublished information with us.
This research was funded by the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie in Germany (project 0316712).
REFERENCES
- 1.Abe S, Takayama K, Kinoshita S. Taxonomical studies on glutamic acid producing organisms. J Gen Appl Microbiol (Tokyo) 1967;13:279–301. [Google Scholar]
- 2.Ackrell B A. Metabolic regulatory functions of oxalacetate. Horiz Biochem Biophys. 1974;1:175–219. [PubMed] [Google Scholar]
- 3.Asano A, Kaneshiro T, Brodie A F. Malate-vitamin K reductase, a phospholipid-requiring enzyme. J Biol Chem. 1965;240:895–905. [PubMed] [Google Scholar]
- 4.Banaszak L J, Bradshaw R A. Malate dehydrogenases. In: Boyer P D, editor. The enzymes. XI. New York, N.Y: Academic Press; 1976. pp. 369–396. [Google Scholar]
- 5.Bergmeyer H U, Bergmeyer J, Grassl M, editors. Methods of enzymatic analysis. Weinheim, Germany: Chemie Verlag; 1985. [Google Scholar]
- 6.Bernofsky C, Swan M. An improved cycling assay for nicotinamide adenine dinucleotide. Anal Biochem. 1973;53:452–458. doi: 10.1016/0003-2697(73)90094-8. [DOI] [PubMed] [Google Scholar]
- 7.Calhoun M W, Gennis R B. Demonstration of separate genetic loci encoding distinct membrane-bound respiratory NADH dehydrogenases in Escherichia coli. J Bacteriol. 1993;175:3013–3019. doi: 10.1128/jb.175.10.3013-3019.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohn D V. The oxidation of malic acid by Micrococcus lysodeikticus. J Biol Chem. 1956;221:413–423. [PubMed] [Google Scholar]
- 9.Datta A, Merz J M, Spivey H O. Substrate channeling of oxalacetate in solid-state complexes of malate dehydrogenase and citrate synthase. J Biol Chem. 1985;260:15008–15012. [PubMed] [Google Scholar]
- 10.De Graef M R, Alexeeva S, Snoep J L, Teixeira de Mattos M J. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J Bacteriol. 1999;181:2351–2357. doi: 10.1128/jb.181.8.2351-2357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukushima T, Deckers R V, Anderson W M, Spivey H O. Substrate channeling of NADH and binding of dehydrogenases to complex I. J Biol Chem. 1989;264:16483–16488. [PubMed] [Google Scholar]
- 12.Gennis R B, Stewart V. Respiration. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology; 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 217–261. [Google Scholar]
- 13.Holten E. Pyridine nucleotide independent oxidation of l-malate in genus Neisseria. Acta Pathol Microbiol Scand B. 1976;84:17–21. doi: 10.1111/j.1699-0463.1976.tb01895.x. [DOI] [PubMed] [Google Scholar]
- 14.Jones M, King H K. Particulate malate oxidation in strictly aerobic bacteria: the respiratory chain of Moraxella lwoffi. FEBS Lett. 1972;22:277–279. doi: 10.1016/0014-5793(72)80249-7. [DOI] [PubMed] [Google Scholar]
- 15.Jurtshuk P, Bednarz A J, Zey P, Denton C H. l-Malate oxidation by the electron transport fraction of Azotobacter vinelandii. J Bacteriol. 1969;98:1120–1127. doi: 10.1128/jb.98.3.1120-1127.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kather B, Stingl K, van der Rest M E, Altendorf K, Molenaar D. Another unusual type of citric acid cycle enzyme in Helicobacter pylori: the malate: quinone oxidoreductase. J Bacteriol. 2000;182:3204–3209. doi: 10.1128/jb.182.11.3204-3209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelle R, Laufer B, Brunzema C, Weuster-Botz D, Krämer R, Wandrey C. Reaction engineering analysis of L-lysine transport by Corynebacterium glutamicum. Biotechnol Bioeng. 1996;51:40–50. doi: 10.1002/(SICI)1097-0290(19960705)51:1<40::AID-BIT5>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 18.Liebl W, Klamer R, Schleifer K H. Requirement of chelating compounds for the growth of Corynebacterium glutamicum in synthetic media. Appl Environ Microbiol. 1989;32:205–210. [Google Scholar]
- 19.Lugtenberg B, Dekkers L C. What makes Pseudomonas bacteria rhizosphere competent? Environ Microbiol. 1999;1:9–13. doi: 10.1046/j.1462-2920.1999.00005.x. [DOI] [PubMed] [Google Scholar]
- 20.Matsushita K, Yamamoto T, Toyama H, Adachi O. NADPH oxidase system as a superoxide-generating cyanide-resistant pathway in the respiratory chain of Corynebacterium glutamicum. Biosci Biotechnol Biochem. 1998;62:1968–1977. doi: 10.1271/bbb.62.1968. [DOI] [PubMed] [Google Scholar]
- 21.Miesel L, Weisbrod T R, Marcinkeviciene J A, Bittman R, Jacobs W R., Jr NADH dehydrogenase defects confer isoniazid resistance and conditional lethality in Mycobacterium smegmatis. J Bacteriol. 1998;180:2459–2467. doi: 10.1128/jb.180.9.2459-2467.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molenaar D, van der Rest M E, Petrovic S. Biochemical and genetic characterization of the membrane-associated malate dehydrogenase (acceptor) (EC 1.1.99.16) from Corynebacterium glutamicum. Eur J Biochem. 1998;254:395–403. doi: 10.1046/j.1432-1327.1998.2540395.x. [DOI] [PubMed] [Google Scholar]
- 23.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niaudet B, Goze A, Ehrlich S D. Insertional mutagenesis in Bacillus subtilis: mechanism and use in gene cloning. Gene. 1982;19:277–284. doi: 10.1016/0378-1119(82)90017-8. [DOI] [PubMed] [Google Scholar]
- 25.O'Brien R W, Taylor B L. Formation and dissimilation of oxalacetate and pyruvate Pseudomonas citronellolis grown on noncarbohydrate substrates. J Bacteriol. 1977;130:131–135. doi: 10.1128/jb.130.1.131-135.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prasada Reddy T L, Suryanarayana Murthy P, Venkitasubramanian T A. Variations in the pathways of malate oxidation and phosphorylation in different species of Mycobacteria. Biochim Biophys Acta. 1975;376:210–218. doi: 10.1016/0005-2728(75)90012-2. [DOI] [PubMed] [Google Scholar]
- 27.Robinson J B, Inman L, Sumegi B, Srere P A. Further characterization of the Krebs tricarboxylic acid cycle metabolon. J Biol Chem. 1987;262:1786–1790. [PubMed] [Google Scholar]
- 28.Ruffert S, Lambert C, Peter H, Wendisch V F, Krämer R. Efflux of compatible solutes in Corynebacterium glutamicum mediated by osmoregulated channel activity. Eur J Biochem. 1997;247:572–580. doi: 10.1111/j.1432-1033.1997.00572.x. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Schäfer A, Kalinowski J, Simon R, Seep-Feldhaus A, Pühler A. High-frequency conjugal plasmid transfer from gram-negative Escherichia coli to various gram-positive coryneform bacteria. J Bacteriol. 1990;172:1663–1666. doi: 10.1128/jb.172.3.1663-1666.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 32.Schrumpf B, Schwarzer A, Kalinowski J, Pühler A, Eggeling L, Sahm H. A functionally split pathway for lysine synthesis in Corynebacterium glutamicum. J Bacteriol. 1991;173:4510–4516. doi: 10.1128/jb.173.14.4510-4516.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Short J M, Fernandez J M, Sorge J A, Huse W D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988;16:7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 35.Smith A F. Malate dehydrogenase. In: Bergmeyer H U, Bergmeyer J, Grassl M, editors. Methods of enzymatic analysis. Weinheim, Germany: Chemie Verlag; 1985. pp. 163–171. [Google Scholar]
- 36.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olsen B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 37.Sumegi B, Srere P. Complex I binds several mitochondrial NAD-coupled dehydrogenases. J Biol Chem. 1984;259:15040–15045. [PubMed] [Google Scholar]
- 38.Tettelin H, Saunders N J, Heidelberg J, Jeffries A C, Nelson K E, Eisen J A, Ketchum K A, Hood D W, Peden J F, Dodson R J, Nelson W C, Gwinn M L, DeBoy R, Peterson J D, Hickey E K, Haft D H, Salzberg S L, White O, Fleischmann R D, Dougherty B A, Mason T, Ciecko A, Parksey D S, Blair E, Cittone H, Clark E B. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 39.Van der Rest M E, Lange C, Molenaar D. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl Microbiol Biotechnol. 1999;52:541–545. doi: 10.1007/s002530051557. [DOI] [PubMed] [Google Scholar]
- 40.Wahlefeld A W. Lactate dehydrogenase. UV-method with L-lactate and NAD. In: Bergmeyer H U, Bergmeyer J, Grassl M, editors. Methods of enzymatic analysis. Weinheim, Germany: Chemie Verlag; 1985. pp. 126–133. [Google Scholar]
- 41.Young I G, Jaworowski A, Poulis M. Cloning of the gene for the respiratory D-lactate dehydrogenase of Escherichia coli. Biochemistry. 1982;21:2092–2095. doi: 10.1021/bi00538a017. [DOI] [PubMed] [Google Scholar]
- 42.Young I G, Wallace B J. Mutations affecting the reduced nicotinamide adenine dinucleotide dehydrogenase complex of Escherichia coli. Biochim Biophys Acta. 1976;449:376–385. doi: 10.1016/0005-2728(76)90149-3. [DOI] [PubMed] [Google Scholar]