Abstract
To maintain homeostasis, the body, including the brain, reprograms its metabolism in response to altered nutrition or disease. However, the consequences of these challenges for the energy metabolism of the different brain cell types remain unknown. Here, we generated a proteome atlas of the major central nervous system (CNS) cell types from young and adult mice, after feeding the therapeutically relevant low-carbohydrate, high-fat ketogenic diet (KD) and during neuroinflammation. Under steady-state conditions, CNS cell types prefer distinct modes of energy metabolism. Unexpectedly, the comparison with KD revealed distinct cell type–specific strategies to manage the altered availability of energy metabolites. Astrocytes and neurons but not oligodendrocytes demonstrated metabolic plasticity. Moreover, inflammatory demyelinating disease changed the neuronal metabolic signature in a similar direction as KD. Together, these findings highlight the importance of the metabolic cross-talk between CNS cells and between the periphery and the brain to manage altered nutrition and neurological disease.
Differential metabolic flexibility of brain cells facilitates managing nutritional or pathologic challenges.
INTRODUCTION
In the brain, neurons use the majority of imported energy substrates for signal processing and propagation—energetically expensive processes. To meet this high energy demand, the brain predominantly uses glucose as energy substrate (1, 2), which is partly replaced by ketone bodies when consuming a low-carbohydrate, high-fat ketogenic diet (3, 4). According to the current view, not neurons but rather glial cells use imported glucose; they produce energy via the relatively inefficient glycolysis pathway and release glycolytic products such as lactate (5, 6). Neurons then benefit from this pool of extracellular monocarboxylates and fuel them into the mitochondrial respiratory chain/oxidative phosphorylation (OXPHOS) that produces energy with a high yield (7, 8). Accordingly, genetic studies showed that an intact OXPHOS is indispensable for neurons but not for astrocytes or oligodendrocytes (7, 9, 10), implying that glial cells follow an obligatory glycolytic energy metabolism. However, several fundamental questions remain unaddressed. The absence of pathology in glial OXPHOS mutants can also result from metabolic flexibility, which is supported by the fact that every cell in the mammalian central nervous system (CNS) is equipped with almost the entire set of metabolic pathways. Moreover, brain metabolism reacts to altered availability of energy substrates, which occur during normal postnatal development or after dietary changes, but the mechanisms in each of the CNS cell types through which nutritional changes induce the metabolic adaptation in the brain are unknown. Here, we dissect these questions by integrating proteomic and transcriptomic screens of acutely isolated brain cells in addition to tissue metabolomics and expression analyses from young and adult mice that were challenged by feeding a ketogenic diet (KD) or by neuroinflammatory disease. Together, our results reveal key mechanisms of cell type–specific metabolic remodeling that lead to an advantage in neuroinflammatory disease.
RESULTS
Proteome analysis of the cortical cell types reveals preferred pathways of energy metabolism
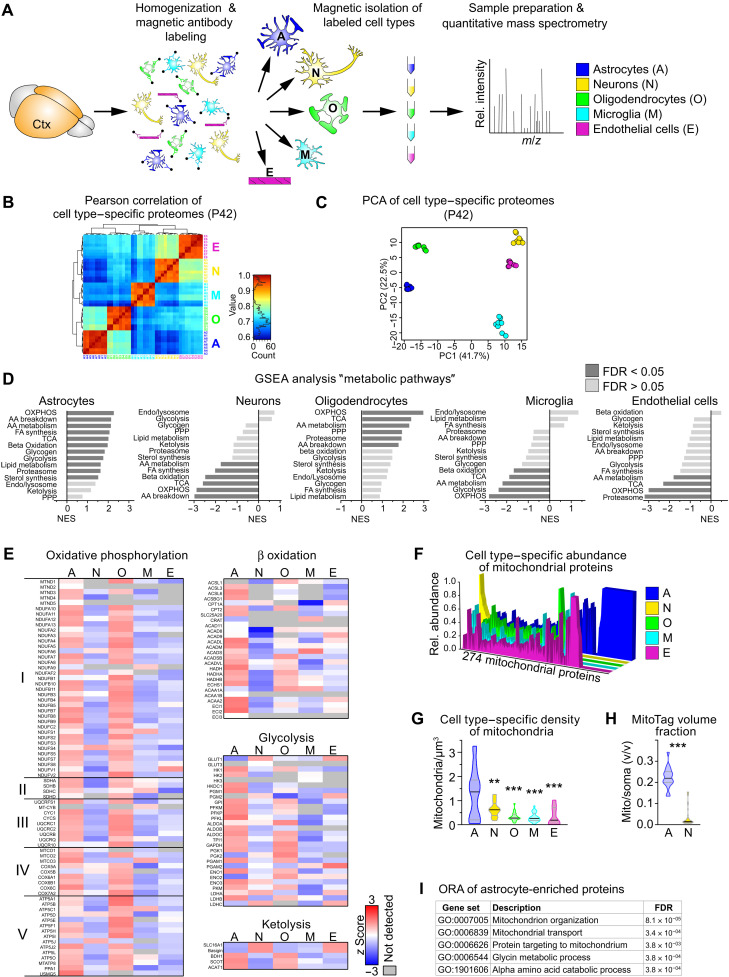
Glucose is the main energy substrate for the adult brain, but inferred from cell culture data and genetic studies, brain cell types differ in the relative utilization of energy substrates and pathways of energy metabolism (2). The capacity of a metabolic pathway is reflected in the equipment with effector enzymes irrespective of the cellular source, as shown in mitochondria ex vivo (11, 12). Therefore, the relative abundance of proteins was used as a proxy for the preferred mode of energy metabolism in the major cell types that were acutely isolated from cortices of individual adult mice followed by cell type–specific proteome profiling (Fig. 1A). We quantified a total of 3541 proteins, ranging from 1506 proteins in oligodendrocytes to 2110 proteins in endothelial cells (table S1). Pearson correlation and principal components analysis (PCA) validated the accuracy of our experimental workflow (Fig. 1, B and C). Strongly increased protein abundance of marker genes and gene set enrichment analysis (GSEA) across all five cell types using 11 custom-select gene sets related to cell function confirmed cellular identities (fig. S1 and table S2). The set related to synaptic transmission was enriched in the neuronal fraction, and myelin proteins were predominantly found in oligodendrocytes. Astrocytes enriched the gene set “homeostatic astrocyte” comprising astrocyte genes derived from recent studies (13, 14). The gene set “homeostatic microglia” comprising known markers (e.g., P2ry12, CX3CR1, TMEM190, and HexB) was significantly enriched in microglial fractions. In contrast, the gene sets related to activated glia comprising proteins associated with A1-, M1-, or disease-associated signatures, such as glial fibrillary acidic protein (GFAP) (astrocytes) or colony-stimulating factor 2 (CSF2) (microglia), were only moderately expressed in either cell type, indicating no obvious ex vivo effect. Thus, this dataset genuinely reflects the in vivo proteome of the major CNS cell types.
Fig. 1. Preferred metabolic pathways in cortical cell types.
(A) Scheme of the experimental approach. Cell types were isolated from the cortex (Ctx) of individual P42 mice (n = 4) using magnetic beads and analyzed by proteomic profiling. m/z, mass/charge ratio. (B) Pearson correlation of cell type–specific proteome datasets as in (A). (C) PCA of cell type–specific proteomes calculated from 722 complete cases of a total of 3541 proteins. (D) Normalized enrichment score (NES) of the gene sets “metabolic pathways” by gene set enrichment analysis (GSEA), comparing z scores of cell type–specific proteomes from P42 mice (AA, amino acid; FA, fatty acid; FDR, false discovery rate; OXPHOS, oxidative phosphorylation; PPP, pentose phosphate pathway; TCA, tricarboxylic acid cycle). (E) Heatmaps showing the mean cell type–specific abundance of proteins related to metabolic pathways (n = 4). (F) Cell type–specific abundance of mitochondrial proteins plotted as the relative abundance of each of the 274 identified mitochondrial proteins. (G) Violin plot of the density of mitochondria per cell soma volume, by quantification of SCOT+ mitochondria colabeled with cellular markers [n = 10 to 20 visual fields with 20 to 28 cells from four animals; one-way analysis of variance (ANOVA) with Tukey’s posttest showing significant differences to astrocytes, **P < 0.01 and ***P < 0.001]. (H) Violin plot of the relative mitochondrial volume (calculated from genetically labeled GFP fluorescence in the outer mitochondrial membrane) per cell soma volume in astrocytes (GFAP-Cre*MitoTag) and neurons (Rbp4-Cre*MitoTag) colabeled with cellular markers (n = 18 to 34 visual fields with 27 to 52 cells from three animals; Mann-Whitney test, ***P < 0.001). (I) Top five GO term processes of the 67 mitochondrial proteins found exclusively in astrocytes by overrepresentation analysis.
We next used our proteome dataset to determine the relative cell type–specific abundance of metabolic pathways by GSEA using 14 custom-select gene sets of the main metabolic pathways that contain the major enzymes but not regulatory or accessory proteins (table S3). Although this restriction could result in overlooking important differences between experimental groups regarding regulatory proteins, it focuses the analysis on the proteins directly involved in metabolic reactions. Unexpectedly, astrocytes and oligodendrocytes strongly enriched energy metabolism pathways, whereas neurons, microglia, and endothelial cells showed remarkably low normalized enrichment scores (Fig. 1D). Astrocytes contained high levels of proteins related to glycolysis (Fig. 1E), as expected (1, 2). Moreover, proteins related to the respiratory chain/OXPHOS, to the tricarboxylic acid (TCA) cycle, and to fatty acid β oxidation were also most abundant in astrocytes (Fig. 1, D and E), in accord with robust oxidative metabolism in this cell type (9, 11, 12, 15). These pathways are all located in mitochondria, and, hence, the majority of the 274 mitochondrial proteins identified in any of the proteome datasets were most abundant in astrocyte fractions (Fig. 1F). The magnetic-activated cell sorting (MACS) procedure preferentially enriches cell bodies while losing cell processes. As neurons contain the most mitochondria in their neurites (16), this likely explains the low level of mitochondrial pathways in neuronal fractions. Accordingly, astrocyte cell bodies contained the highest densities of mitochondria and the highest cumulative mitochondrial volume, compared to all other cell types (Fig. 1, G and H, and fig. S1C). These findings are consistent with a recent three-dimensional (3D) ultrastructural study that shows similar mitochondria densities in astrocytes and neuronal processes (17). Overrepresentation analysis (ORA) of the 67 mitochondrial proteins chiefly found in astrocytes implied active mitochondrial remodeling and transport especially in these glial cells (Fig. 1I). Although we did not quantify the cell type–specific contribution of cortical OXPHOS, our findings suggest a considerable contribution of astrocytic mitochondria to cortical energy metabolism.
Cortical cells adjust metabolic pathways preferences during postnatal brain maturation
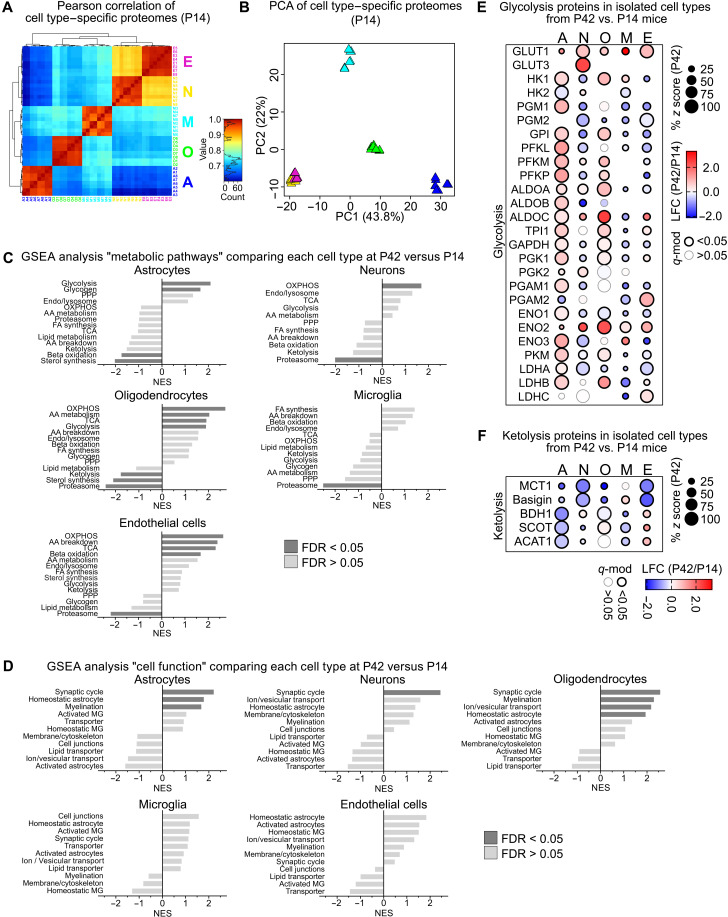
The function and energy metabolism of a given cell are closely linked by anterograde and retrograde regulations (1). When brain function matures during the first postnatal weeks, neuronal circuits and glial cell networks develop to achieve new tasks. Oligodendrocytes accomplish myelination and microglia clear surplus material from synapses and myelin. To interrogate whether brain cells adapt their metabolism in the process of postnatal brain maturation, we compared the cellular proteomes from postnatal day 42 (P42) mice with equivalent datasets obtained from mice at P14 (Fig. 2, A and B, and table S1).
Fig. 2. Metabolic changes during postnatal brain maturation.
(A) Pearson correlation of cell type–specific proteome datasets from P14 mice comprising a total of 3787 proteins, ranging from 1770 proteins in oligodendrocytes to 2664 proteins in neurons. (B) PCA of cell type–specific proteomes from P14 mice calculated from 1144 complete cases. (C) NES of the gene sets “metabolic pathways” by GSEA, shown as log2 fold changes (LFC) of cell type–specific proteomes from P42 versus P14 mice. (D) NES of gene set “cell function” by GSEA, comparing LFC of cell type–specific proteomes from P42 versus P14 mice. (E) Bubble plot of proteins related to glycolysis showing percent z scores at P42 (size), LFC of P42 versus P14 (color) with significant differences marked by the outline (q-mod values from moderated t statistics with FDR-based correction for multiple comparisons). (F) Bubble plot of proteins related to ketolysis in isolated cell types showing the percent z scores, LFC with significant differences marked by the outline as in (E).
As determined by GSEA of datasets at P42 versus P14, astrocytes and oligodendrocytes significantly increased gene sets related to carbohydrate metabolism such as glycolysis (Fig. 2, C to E). Astrocytes also increased enzymes related to glycogen synthesis and degradation in accordance with the increased “homeostatic astrocyte” cell function gene set (Fig. 2D). Oligodendrocytes also increased gene sets of the TCA cycle, OXPHOS, and amino acid metabolism but decreased sterol synthesis, together reflecting the still high anabolic demand during the advanced-state myelination at P42. In endothelial cells, the postnatal period was associated with increased abundance of endothelial glucose transporter GLUT1 in addition to unchanged glycolysis enzymes, suggestive of enhanced CNS import of glucose (Fig. 2E). Microglia that contained rather low levels of metabolic proteins showed only minor changes between P14 and P42 metabolism (reduced “proteasome” gene set) that suggested attenuating turnover of phagocytosed material, in accordance with a recent RNA-profiling resource (18). Neurons significantly increased the OXPHOS gene set, together with enhanced functional changes related to synaptic transmission (“synaptic cycle”). The gene set synaptic cycle was also increased in astrocytes and oligodendrocytes, likely reflecting cellular adaptations to active neuronal impulse propagation.
During this critical phase of postnatal brain development between P14 and P42, mice are weaned. Before weaning, the main energy metabolites in the periphery and also in the CNS are ketone bodies, predominantly β-hydroxybutyrate (3HB) and acetoacetate, which derive from consuming lipid-rich breast milk (19). Upon weaning and the switch to carbohydrate-based chow, ketone bodies are replaced by glucose as main energy substrate (20). This was paralleled by reduced levels of monocarboxylate transporter 1 (MCT1), which transports monocarboxylates, including ketone bodies, and down-regulation of the gene set “ketolysis” in almost all cell types with the strongest decreases in astrocytes and oligodendrocytes (Fig. 2, C and F). These findings show that all cell types undergo considerable metabolic adaptations during postnatal development and imply preponderant utilization of ketone bodies by astrocytes and oligodendrocytes before weaning.
Weaning mice to KD induces cortical ketolysis and maintains brain maturation
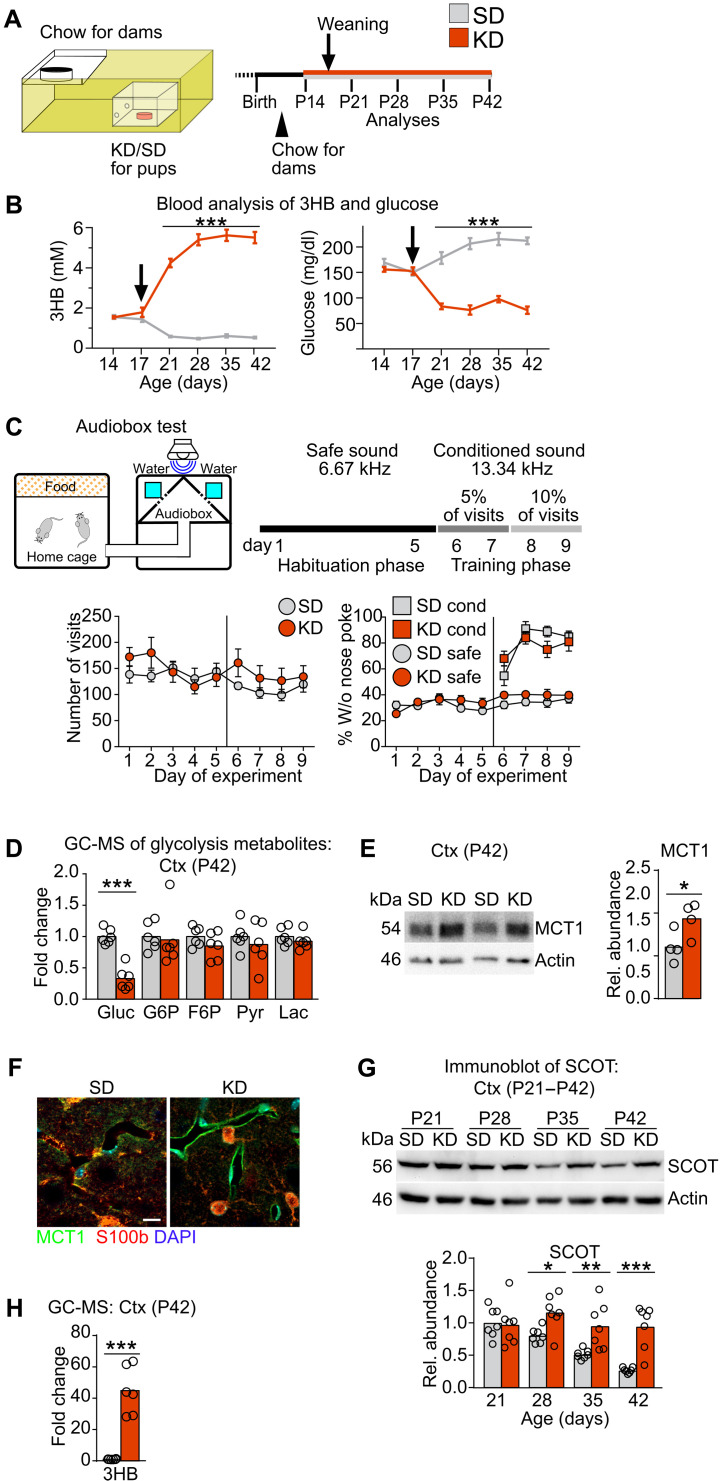
We next asked whether postweaning brain maturation is triggered by, counteracted by, or independent of the metabolic adaptation of brain cells to the altered nutritional input (ketone bodies to glucose). To uncouple brain maturation from the nutritional switch at weaning, we used a paradigm of prolonged breastfeeding by weaning mice to a classical low-carbohydrate, high-fat KD or to regular chow (standard diet, SD) (Fig. 3A). In agreement with reinforced ketosis, blood levels of 3HB significantly increased and glucose levels decreased in KD- compared to SD-fed offspring without causing liver or brain toxicity (Fig. 3B and fig. S2, A to E). Liver transcripts reflected the altered lipid metabolism, e.g., by strongly up-regulating Fgf21 expression in KD-fed mice (table S4). Despite the expected slower body weight gain of KD-fed animals (21), the brain weight was comparable in both dietary conditions (fig. S2, F and G), showing that brain growth was not delayed in KD-fed animals. In a longitudinal analysis from P21 to P42, we monitored cortical mRNA expression of genes related to brain development, testing growth factors (e.g., Bdnf), synapses (e.g., Nr2a), and myelin (e.g., Mbp). All transcript levels in KD-fed mice were similar to littermate SD-fed controls (fig. S2, H to M, and table S5). When we evaluated neurocognitive abilities using an auditory discrimination task in the Audiobox (22), mice of both treatment groups showed equivalent performance (Fig. 3C). These findings show normal brain development irrespective of nutritional paradigm.
Fig. 3. Cortical changes in mice weaned to a KD.
(A) Experimental setup. At P14, SD or KD was administered in boxes with small holes rendering this food only accessible for pups. Mice were weaned at P17 (arrow) and kept on the respective diet. (B) Blood 3HB and glucose in mice treated as in (A) (n = 20, two-way ANOVA with Sidak’s posttest, ***P < 0.001). (C) Schematic of the Audiobox test (top left) and the learning paradigm (top right) (n = 5). Mean total number of visits ± SEM shows comparable activity of mice in both treatments (bottom left). Mean percentage of visits without nose poke ± SEM under safe (circle) and conditioned (square) conditions of mice fed KD or SD at 5 weeks of age reveal similar neurocognitive abilities (bottom right). (D) Mean levels of glycolysis metabolites with individual data points, by GC-MS in cortex of KD-fed P42 mice normalized to SD-fed controls (Gluc, glucose; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; Pyr, pyruvate; Lac, lactate; t statistics with Benjamini-Hochberg correction, ***Padj < 0.001). (E) Representative immunoblot for MCT1 in P42 cortex of mice fed SD or KD with quantification normalized to reprobed actin signals (unpaired Student’s t test, *P < 0.05). (F) Representative immunolabeling of MCT1 (green), S100b (red, astrocyte marker), and 4′,6-diamidino-2-phenylindole (DAPI) (blue, nuclei) in cortex of SD and KD-fed P42 mice. Scale bar, 10 μm. (G) Representative immunoblot for SCOT in cortex of P21 to P42 mice fed SD or KD with quantification normalized to reprobed actin signals relative to P21 SD values (two-way ANOVA with Sidak’s posttest, *P < 0.05, **P < 0.005, and ***P < 0.001). (H) Mean cortical 3HB levels with individual data points measured by GC-MS (t statistics with Benjamini-Hochberg correction, ***Padj < 0.001). (D to H) Each ring represents an individual data point (one mouse).
To investigate whether brain metabolism attuned to the nutritional paradigm, we analyzed key metabolic enzymes and metabolite transporters in the cortex of KD- versus SD-fed mice. Transcripts and protein levels of the glucose transporter GLUT1 (Slc2a1 gene) were similar in both dietary regimes (fig. S2, N and O). Consistent with the reduced blood glucose in KD-fed mice, cortical glucose levels were strongly reduced as determined by metabolic profiling (Fig. 3D and table S6). In contrast, cortical levels of glycolysis intermediates and mRNA expression of glycolysis enzymes, including the rate-limiting enzyme phosphofructokinase (Pfkm) remained unchanged in KD-fed mice (Fig. 3D and fig. S2, P and Q), potentially reflecting cortical glycolysis in the presence of reduced substrate levels. MCT1 (Slc16a1) mRNA expression almost doubled in KD-fed mice, causing an approximately 1.5-fold increased MCT1 protein abundance at P42 (fig. S2R and Fig. 3E). This was particularly obvious in blood vessels (Fig. 3F), reflecting sustained ketone body uptake into the brain. In contrast, expression of the monocarboxylate transporters MCT2 and MCT4 remained unchanged (fig. S2S). Transcript levels of the rate-limiting enzyme of ketolysis, SCOT [Oxct1 gene, succinyl–coenzyme A (CoA):3-ketoacid-CoA transferase], were not influenced by age or diet (fig. S2T), consistent with the nontranscriptional regulation of ketolysis (23). In SD-fed mice, SCOT protein abundance progressively decreased with age (Fig. 3G). In contrast, SCOT persisted at high levels, resulting in an almost fourfold increased SCOT level at P42 in KD-fed mice compared to SD-fed controls (Fig. 3G). Transcript and protein abundance of other enzymes of ketolysis were similar in both dietary groups throughout the time course (fig. S2, U to X). Together with the ~40-fold increase of cortical 3HB levels (Fig. 3H), these data show robust ketolysis in the cortex of KD-fed mice at P42. Of note, the KD formulation was validated in a long-term feeding paradigm that induced chronic ketosis in the brain without causing brain damage (fig. S3).
Cortical cell types follow specific strategies to adapt their metabolism to ketosis
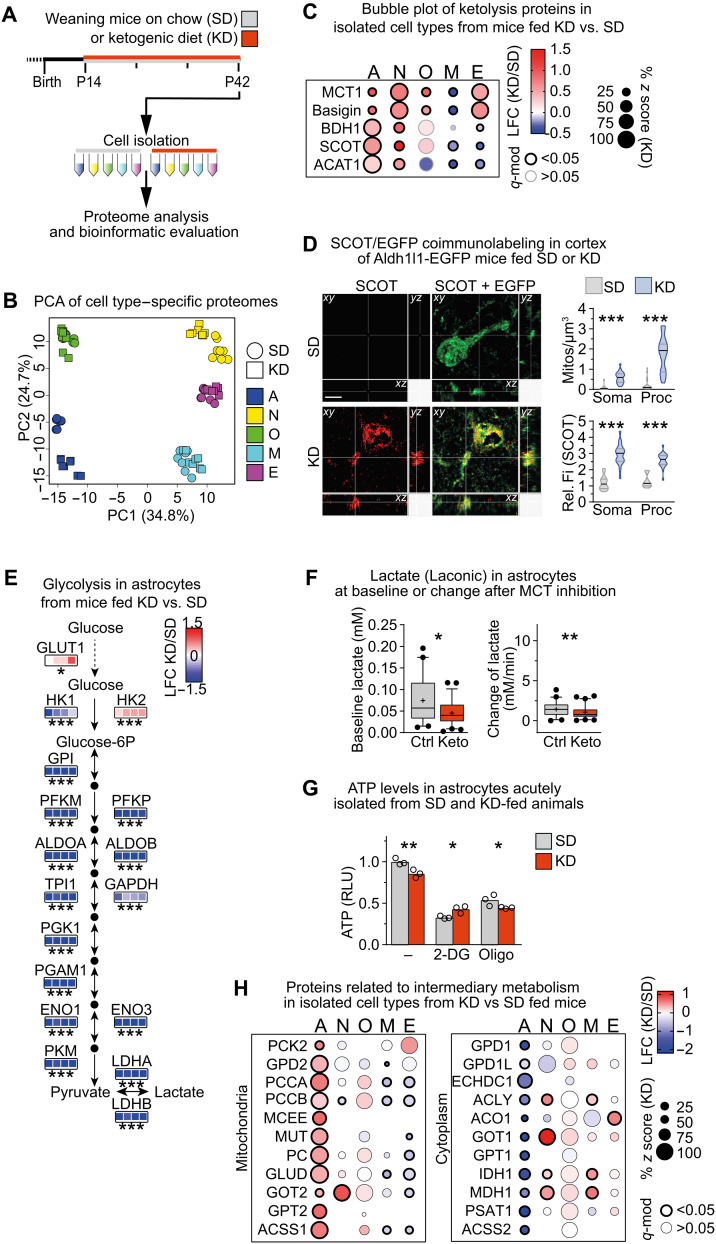
To investigate cellular adaptations to the sustained cortical ketolytic metabolism, we compared proteomes of cell types isolated from the cortex of KD- versus SD-fed mice at P42 (Fig. 4A). PCA, Pearson correlation, and volcano plots (Fig. 4B and fig. S4, A and B) showed that KD strongly altered the proteome profile of astrocytes, neurons, and microglia compared to SD. In contrast, KD and SD samples from oligodendrocytes and endothelial cells largely overlapped, implying that the degree of diet-induced adaptations was minor in these cell types. In accordance with our findings in tissue of KD-fed mice, endothelial cells strongly increased expression of MCT1 and its interaction partner basigin but not of ketolytic enzymes (Fig. 4C and fig. S4C), and the density of SCOT+ mitochondria in endothelial cells was similar in both dietary paradigms (fig. S4D). These data implicate that cerebral vessels supported cortical ketolysis in KD-fed mice by importing ketone bodies into the CNS.
Fig. 4. Cell type–specific strategies of metabolic adaptations to feeding a KD.
(A) Experimental paradigm. (B) PCA of cell type–specific proteomes from mice fed SD or KD, calculated from 546 complete cases from a total of 3621 detected proteins. (C) Bubble plot of proteins related to ketolysis in isolated cell types from KD- versus SD-fed mice. (D) Maximum intensity projection (1-μm optical section in all dimensions) of SCOT (red) and enhanced green fluorescent protein (EGFP) (green) colabeling of brain sections from ALDH1L1-EGFP transgenes fed KD or SD with quantification of the density and relative fluorescence intensity (Fi) of SCOT+ mitochondria per volume in soma and processes (Mann-Whitney test, ***P < 0.001). ALDH1L1-EGFP mice express EGFP under the astrocyte Aldh1l1 promotor. Scale bar, 5 μm. (E) LFC of proteins related to glycolysis in astrocytes from mice fed KD versus SD (n = 4). (F) Median intracellular lactate levels (Laconic nanosensor) with 5 to 95 percentiles in astrocytes that had been exposed to SD-mimicking (5 mM glucose, Ctrl) or KD-mimicking (0.4 mM glucose and 4 mM 3HB, Keto) conditions for 1 week before measuring in 2 mM glucose, showing baseline lactate levels and the rate of lactate production after inhibiting MCT transporters (AR-C 155858) (n = 41 to 60 cells from three independent experiments; Mann-Whitney test, *P < 0.05 and **P < 0.01). (G) Mean intracellular ATP relative light units (RLU) with individual values in acutely isolated astrocytes from KD- and SD-fed animals, measured in 2 mM glucose with or without inhibition of glycolysis [2-deoxyglucose (2-DG)] or OXPHOS by oligomycin (Oligo) (n = 3 measurements pooled from two mice per group). (H) Bubble plot of proteins related to intermediary metabolism in cell types isolated from mice fed KD versus SD. (C, E, and F) Significant changes in protein abundance were determined by moderated t statistics with FDR-based correction for multiple comparisons (*q mod < 0.05, **q-mod < 0.005, and ***q-mod < 0.001).
Microglia that expressed only low levels of proteins related to ketolysis in SD-fed animals further down-regulated these enzymes in KD-fed mice (Fig. 4C). The increased proteasome gene set suggested enhanced tissue remodeling, but the lipid-rich diet did not reprogram the microglial metabolism and microglia remained in a homeostatic state (fig. S4, C and E).
In oligodendrocytes of KD-fed compared to SD-fed mice, proteins of ketone body transport but not utilization were significantly enhanced (Fig. 4C), and the density of SCOT+ mitochondria did not change (fig. S4D), suggesting that ketolysis remained active in oligodendrocytes of KD-fed mice, as these cells were already adapted to the utilization of ketone bodies. We originally expected that enhanced peripheral lipid metabolism in KD-fed mice could support myelination by CNS import of precursors for lipid synthesis as cholesterol is essential for myelination (24, 25). However, GSEA revealed that feeding KD did not significantly alter any of the metabolic pathways in oligodendrocytes, including fatty acid and cholesterol synthesis (fig. S4C). By gas chromatography–mass spectrometry (GC-MS), cortical levels of cholesterol and sterol synthesis intermediates, whose levels reflect ongoing sterol synthesis, remained unchanged (table S6), and myelination was similar in both dietary conditions (fig. S5, A to C). Accordingly, proteomic profiling of oligodendrocytes revealed only a slight, but nonsignificant increase in the “myelination” gene set (fig. S4E). Of note, oligodendrocytes of KD-fed mice increased the gene set “ion/vesicular transport” (figs. S4E and S5D). This was confirmed by enhanced transcript levels related to transport and ion transport processes identified by gene ontology (GO) analysis of oligodendrocyte transcriptomes from KD- versus SD-fed animals (fig. S5, E and F, and table S7), pointing to enhanced membrane traffic in oligodendrocytes and myelin remodeling. These findings show that oligodendrocytes follow their intrinsic postnatal metabolic transformation irrespective of the altered CNS metabolism.
Consistent with increased fatty acid import into the brain of KD-fed mice (26), GSEA of astrocytes revealed enrichment of proteins related to β oxidation of fatty acids (fig. S4C). Moreover, these glial cells strongly elevated the ketolytic pathway in KD- versus SD-fed mice (Fig. 4C and fig. S4C). The density and fluorescence intensity of SCOT+ mitochondria strongly increased in astrocyte cell bodies, and even more so in astrocyte processes, compared to SD-fed mice (Fig. 4D and movie S1). In agreement with the conversion of ketone bodies to acetyl-CoA for oxidative degradation, astrocytes strongly increased the TCA and OXPHOS gene sets (fig. S4C). While the rate-limiting enzyme of the pentose phosphate pathway (PPP) was increased, the other enzymes of this pathway were decreased (fig. S4, C and F), potentially reflecting reinforced utilization of the PPP. In contrast, astrocytes strongly down-regulated glycogen metabolism and glycolysis (Fig. 4E and fig. S4, C and F). Moreover, when we accustomed astrocytes in vitro to energy metabolite levels mimicking conditions in SD [control (Ctrl)] and KD (Keto) brains, intracellular baseline lactate levels and the rate of lactate production were reduced in the Keto group (Fig. 4F). Next, we measured adenosine triphosphate (ATP) levels in acutely isolated astrocytes from KD- compared to SD-fed animals treated with inhibitors of glycolysis (2-deoxyglucose) and OXPHOS (Oligo). Regardless of the nutrition, astrocytes depended on both glycolysis and OXPHOS (Fig. 4G). The reduced glycolytic ATP and increased ATP from mitochondrial OXPHOS in astrocytes from KD-fed animals suggest attenuated glycolysis and a significant shift to OXPHOS for ATP production, in agreement with the proteome data. These alterations demonstrate increased mitochondrial fatty acid/ketone body metabolism and reduced cytosolic glycolysis in astrocytes of KD-fed mice. In combination with the increased abundance of astrocytic GLUT1, this could lead to increased intracellular glucose feeding into the PPP. Although not shown directly, this scenario is also compatible with the concept of glucose transit to other brain cells in ketotic mice.
As a consequence of attenuated glucose degradation in astrocytes, TCA anaplerosis has to be enhanced to maintaining intermediary metabolism. TCA anaplerosis can derive from amino acids involving mitochondrial glutamate dehydrogenase generating α-ketoglutarate and transaminases, such as alanine aminotransferase (GPT2) generating pyruvate followed by pyruvate carboxylase, in line with increased pyruvate consumption in 3HB-treated astrocyte cultures (27). In addition, oxidation of odd chain fatty acids followed by mitochondrial propionyl-CoA carboxylase (PCCA and PCCB subunits), methylmalonyl-CoA epimerase, and methylmalonyl-CoA mutase can give rise to TCA intermediates. Notably, all mitochondrial enzymes involved in these processes and mitochondrial transporters such as the oxoglutarate-malate carrier SLC25A11 were strongly increased in astrocytes of KD- versus SD-fed mice (Fig. 4H and table S9). In contrast, the cytoplasmic counterparts of amino acid intermediary metabolism such as GOT1 were markedly reduced (Fig. 4H). Together, these findings are compatible with the concept that astrocytes in KD-fed mice degrade proteins/amino acids, lipids, and ketone bodies and maintain the TCA cycle for subsequent oxidative energy generation, thereby sparing glucose for neurons. In contrast to these marked metabolic alterations, KD did not affect cell function pathways in astrocytes, as determined by GSEA (fig. S4E). In a corresponding transcriptome analysis, GO analysis revealed attenuated inflammatory processes in isolated astrocytes of KD- versus SD-fed mice (fig. S5, G and H, and table S8). These findings demonstrate that predominantly astrocytes sense the composition of imported energy substrates and adapt their metabolism accordingly.
Cortical neurons of KD-fed mice increase energy metabolism pathways
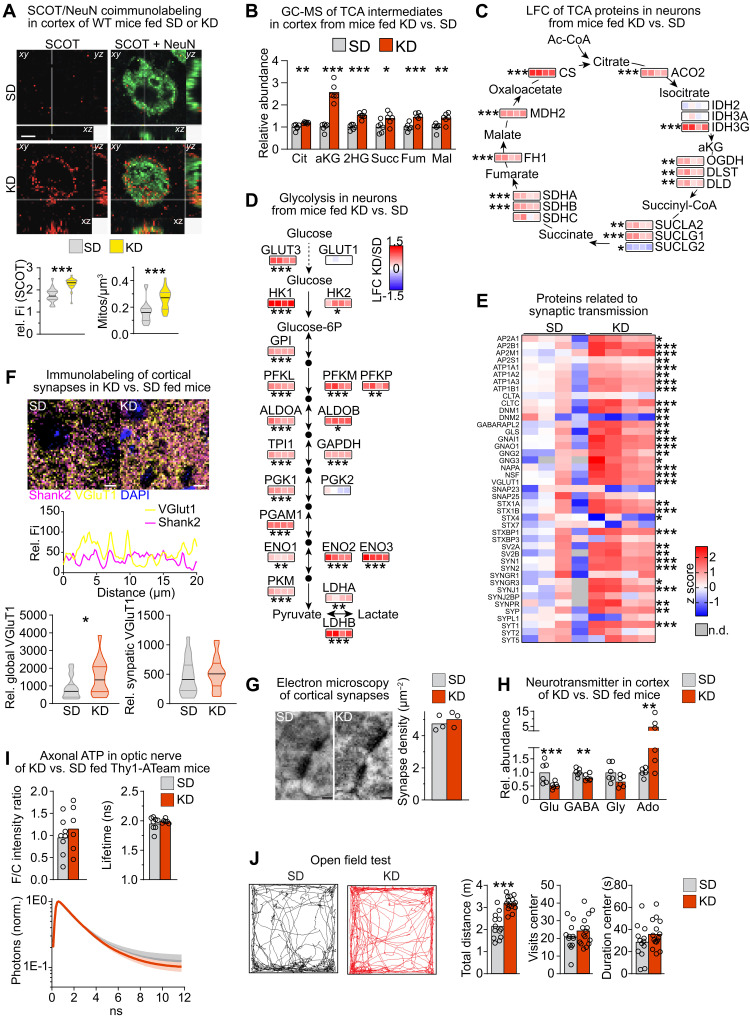
Similar to astrocytes, neurons from KD-fed mice strongly elevated protein levels related to ketone body utilization and increased the density of SCOT+ mitochondria (Figs. 4C and 5A, and fig. S4C), suggesting that neurons and astrocytes were the chief consumers of ketone bodies in KD-fed mice. Gene sets related to the catabolism of ketone bodies, TCA cycle and OXPHOS, were robustly enriched in neurons from KD-fed mice, which are also reflected by significantly increased TCA cycle enzymes and intermediates in cortical tissue (Fig. 5, B and C, and fig. S4C), in agreement with increased TCA cycle activity in the brain of rats fed a KD (28). Notably, neurons slightly reduced the PPP but strongly up-regulated glycolytic enzymes (Fig. 5D and fig. S4, C and G), likely reinforced by glucose uptake via the high-affinity GLUT3 glucose transporter (29).
Fig. 5. Feeding KD alters synapse remodeling.
(A) Maximum intensity projection (1 μm) of SCOT (red) and NeuN (green) colabeling with quantification of the density and relative fluorescence intensity of SCOT+ mitochondria per soma volume (Mann-Whitney test, ***P < 0.001; scale bar, 5 μm). (B) Mean TCA intermediates by GC-MS (n = 6; Cit, citrate; aKG, α-ketoglutarate; Succ, succinate; Fum, fumarate; Mal, malate). (C) LFC of neuronal TCA proteins (n = 4). (D) LFC of proteins related to neuronal glycolysis (n = 4). (E) Heatmap of proteins related to synaptic transmission in neurons (n = 4 mice). (F) Maximum intensity projection of Shank2 (magenta) and VGluT1 (yellow) colabeling (scale bars, 5 μm) with fluorescence intensity scan of the dashed line. Global or synaptic (colocalized with the postsynaptic marker Shank2) VGluT1 immunofluorescence with median and interquartile ranges (n = 20 images of four mice; Mann Whitney test, *P > 0.05). (G) Representative image and quantification of synapse density on electron microscopic images (n = 3 mice; scale bars, 100 nm). (H) Mean cortical neurotransmitter levels measured by GC-MS (n = 5 to 6 mice). (I) Steady-state axonal ATP levels in optic nerves from Thy1-ATeam ATP sensor mice fed KD or SD, showing the F/C intensity ratio, the sensor lifetime, and the mean normalized fluorescence decay time ± SEM [n = 6 (KD) and n = 8 (SD) nerves from three to four mice]. (J) Open-field test. Representative traces of P42 mice fed SD and KD with quantification shown as mean with individual values (n = 12 to 15 mice; two-way ANOVA with Sidak’s posttest, ***P < 0.001). (C to E) Moderated t statistics with FDR-based correction for multiple comparisons (*q mod < 0.05, **q mod < 0.005, and ***q mod < 0.001). (B and H) T statistics with Benjamini-Hochberg correction; *Padj < 0.05, **Padj < 0.01, and ***Padj < 0.001. ns, not significant. n.d., not detected.
Next, we investigated whether the increased equipment with proteins related to energy metabolism affects synapses, as the most energy-consuming process in neurons is the restoration of resting potentials after impulse generation. Neurons strongly increased the synaptic cycle gene set in KD-fed mice (fig. S4E). Although gene transcription of synaptic proteins including Vglut1 was similar in both dietary regimens (table S5), many synaptic proteins were twofold or more increased in neuronal fractions (Fig. 5E). Increased VGluT1 immunolabeling of brain sections from KD-fed mice corroborated the proteome data. However, colabeling of pre- and postsynaptic markers and counting synaptic boutons on electron micrographs revealed equivalent density of synaptic junctions in KD- and SD-fed mice (Fig. 5, F and G), implying normal synapse density but potentially with higher turnover of their components. Unexpectedly, the steady-state levels of neurotransmitters, both the excitatory glutamate and the inhibitory γ-aminobutyric acid, were reduced in KD-fed mice (Fig. 5H and table S6). Adenosine levels, which are known to dampen synaptic activity (30), were elevated 4.9-fold. Of note, the increased adenosine in the KD-fed mice was possibly in part achieved by attenuated abundance of the adenosine-metabolizing adenosine kinase that was reduced to ~30% of levels in astrocytes of SD-fed mice (table S1). When we determined neuronal steady-state energy levels in optic nerve axons using Thy1-ATeam sensor mice (31), unchanged ATP sensor signals and fluorescence lifetime suggested similar ATP levels in both dietary paradigms (Fig. 5I). To address whether the alterations of neurotransmitter levels and synaptic proteins in mice fed KD influenced behavior, we performed open field tests. Mice fed KD showed increased locomotor activity but lacked any signs of anxiety (Fig. 5J). These findings show complex adaptations of physical activity, neuronal signal transmission, and brain metabolism in KD-fed mice that could influence each other.
Feeding KD provides the required energy substrate for the disease-adapted neuronal metabolism in EAE mice
Next, we explored whether the increased equipment with proteins related to energy metabolism in neurons of KD-fed mice could also invigorate neurons in the context of neurodegenerative disease. Permanent disabilities in demyelinating disease such as multiple sclerosis (MS) are caused by progressive neuron damage. Together with adverse effects of the proinflammatory environment, neurons are at risk of suffering energy deficits. This is because of the strongly increased energy cost of impulse conduction when axons are demyelinated (32). In addition, several entities of neurodegenerative disease are associated with perturbed brain import of energy metabolites such as glucose (33, 34). To investigate the relationship between demyelinating disease and the peripheral nutrient status, we induced experimental autoimmune encephalomyelitis (EAE) in mice and monitored blood glucose values. At 7 days after induction, before the onset of clinical symptoms, glucose levels dropped to about 55% of control values and remained at low levels throughout the peak of EAE clinical scores (fig. S6A).
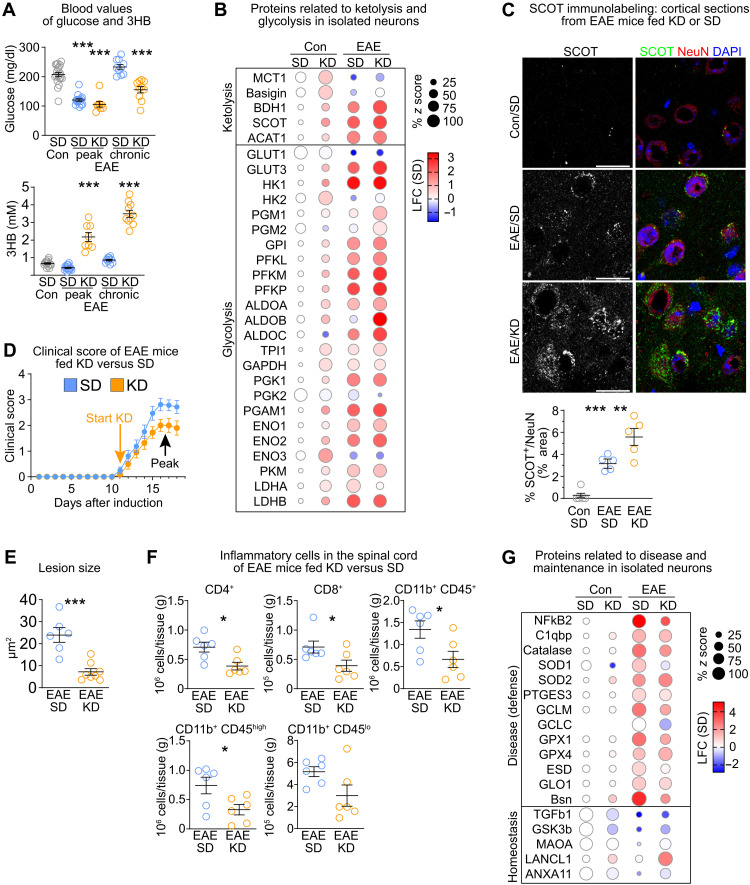
In EAE, immune cell infiltration and demyelination manifest in the spinal cord and to a lesser extent in the cortex (fig. S6, B and C). To explore, how EAE affects cortical neurons, we compared proteomes of isolated neurons from EAE mice and controls (fig. S6D). Ongoing EAE disease was reflected in increased levels of disease markers such as nuclear factor κB2 and scavenging systems for reactive oxygen species (ROS) such as superoxide dismutase 1 (SOD1), proteins of the glutathione family and catalase (fig. S6E), also confirmed by immunolabeling of cortical sections (fig. S6F). In contrast, markers of neuronal health, such as transforming growth factor–β1 or glycogen synthase kinase 3β, were markedly reduced (fig. S6E). GSEA revealed increased expression related to glycolysis and OXPHOS in neurons from EAE mice (fig. S6, G and H). Moreover, abundance of ketolysis enzymes, including the rate-limiting enzyme SCOT, was also strongly enhanced (fig. S6H). These findings show that EAE leads to increased equipping of cortical neurons, unlike spinal cord neurons (35), with enzymes involved in energy metabolism pathways, reminiscent of feeding KD (fig. S4C).
Potentially, EAE could lead to the induction of ketosis in the brain, provoked by lipolysis of degenerating myelin, and in the periphery, provoked by lipolysis of adipose tissue mass (36). However, liver abundance of HMGCS2 (3-hydroxy-3-methylglutaryl-CoA synthase 2), the rate-limiting enzyme of ketogenesis, and MCT1, involved in releasing ketone bodies into the circulation, was similar in EAE mice and non-EAE controls (fig. S7A). Liver transcripts related to lipid metabolism and ketosis, such as Fgf21, were unchanged or even decreased compared to non-EAE control mice (table S10). Moreover, at the peak of EAE, blood 3HB levels were reduced to ~60% of control levels (fig. S7B). These findings demonstrate that EAE itself does not boost peripheral ketosis, potentially restricted by the limited adipose fat stores. However, irrespective of the scarcity of corresponding energy substrates, EAE appears to interfere with the neuronal energy metabolism and induces neuronal ketolysis.
We next explored whether the induction of peripheral ketosis could rescue the shortage of circulating nutrients (due to depletion of adipose tissue) in EAE mice. When we fed KD to EAE mice therapeutically, commencing with the onset of symptoms, blood ketone body levels strongly increased at the peak of EAE disease, whereas blood glucose remained at levels found in SD-fed EAE mice (Fig. 6A). Liver transcripts of Fgf21 strongly increased, reflecting metabolic adaptations to the lipid-rich ketogenic nutrition (table S10). Peripheral ketosis was also evident by the increased protein abundance of HMGCS2 and MCT1 in the liver of KD compared to SD-fed EAE mice (fig. S7A). We then compared the proteomes of cortical neurons from EAE and control mice fed KD or SD (figs. S6D and S7C). KD-fed EAE mice had only slight increases in the energy metabolism pathways (fig. S7D). This included glycolysis and ketolysis (Fig. 6B), also confirmed by SCOT immunolabeling (Fig. 6C). Consistent with counterbalancing of peripheral ketotic with neuronal ketolytic metabolism, feeding KD to EAE mice mitigated clinical symptoms, attenuated densities of inflammatory cells, and reduced the mean lesion size (Fig. 6, D to F, and fig. S7E). In accord with reduced disease expression, protein expression of ROS scavengers such as catalase was attenuated in neurons from KD-fed versus SD-fed EAE mice (Fig. 6G). These findings demonstrate that EAE alters the neuronal metabolism in a similar direction as feeding KD without raising peripheral ketosis. Feeding KD to EAE mice provides the required energy substrate for neuronal ketolysis, which contributed to disease amelioration.
Fig. 6. Feeding KD supports neuronal metabolism in EAE.
(A) Mean blood glucose and 3HB with individual values of EAE mice fed SD or KD (n = 8 to 16 mice; one-way ANOVA with Dunnet’s posttest compared to control mice). (B) Bubble plot of ketolysis and glycolysis proteins in neurons from EAE mice and controls. (C) Representative immunolabeling of SCOT and NeuN in cortex of EAE mice and controls with quantification (n = 5 mice, one-way ANOVA with Tukey’s posttest; scale bars, 20 μm). (D) Mean clinical score ± SEM of EAE mice. KD was applied therapeutically starting with disease onset (n = 8 SD, n = 16 KD mice). (E) Mean lesion size ± SEM in lumbar spinal cord of EAE mice (n = 6 to 8 mice; unpaired two-sided Student’s t test). (F) Mean density of inflammatory cells ± SEM in spinal cord of EAE mice fed KD or SD analyzed by flow cytometry (n = 6 mice; unpaired two-sided Student’s t test). (G) Bubble plot of disease defense and homeostatic markers in cortical neurons from EAE mice and controls. *P < 0.05, **P < 0.01, and ***P < 0.001.
DISCUSSION
Detailed knowledge of the metabolic cross-talk between CNS cell types as well as between the periphery and the brain has key implications for the understanding of the mechanisms through which functional homeostasis is maintained; this becomes especially relevant during development, altered nutrition, and even disease pathogenesis. Here, we generated a comprehensive atlas of the proteome of the major cell types acutely isolated from the cortex of young and adult mice. Our study used cell type–specific protein abundances as a proxy for the activity of metabolic pathways. The quantification of effectors of metabolic pathways unravels substantial differences between cell type–specific preferences of energy metabolism, in line with the concept of metabolic coupling (2, 37). In vitro and ex vivo functional assays support this methodical approach.
We confirm high glycolytic rates in astrocytes (9). Previous studies established a nonessential role of OXPHOS in astrocytes (9, 11, 12, 15) and, in culture, lower oxygen consumption rates compared to neurons (12). In agreement with a transcriptomic study (38), we show that astrocyte fractions, but not neuronal fractions, contain highest levels of proteins related to mitochondrial oxidative energy–generating pathways, particularly OXPHOS. Our functional assays demonstrate ATP generation not only by glycolysis but also by OXPHOS in astrocytes, which is reflected in the astroglial sensitivity to changes in the partial pressure of oxygen (39, 40). Accordingly, astrocyte cell bodies contain a higher density and volume fraction of mitochondria compared to neurons, as reported in a 3D ultrastructural study (17).
Approximately 5% of the energy expenditure in the brain derives from ketone bodies in healthy adults, which can reach ~20% after starvation, dietary intervention, or preweaning (41). Our study implies ketone body utilization in all cell types before weaning and shows that postnatal brain development is accompanied by metabolic remodeling in all cell types. A prolonged breastfeeding paradigm by weaning mice to KD showed that altered brain metabolism is not achieved through equivalent regulations in all CNS cells. Rather, distinct cell type–specific strategies mediate the metabolic adaptation of the cortex to brain maturation and the altered nutritional input. Oligodendrocytes show lowest metabolic flexibility, as these glial cells followed their intrinsic metabolic reprogramming during postnatal maturation irrespective of the nutritional paradigm. In contrast, astrocytes show the highest metabolic plasticity in response to the altered nutritional input. KD increased the levels of ketone bodies and enhanced ketone body oxidation and subsequent OXPHOS in astrocytes. The reduced availability of carbohydrates resulted in diminished astrocyte glycolysis, decreasing the production and, presumably, the release of the glycolytic product lactate. Glial synthesis is a major physiological source of CNS lactate, which fuels energy production in neuronal mitochondria (5). In addition to local synthesis, lactate is also imported into the brain via endothelial MCT1 transporters (42) and there partially replaces glucose as energy metabolite during ketosis (3). Circulating lactate remains at control levels in human subjects following a KD (43). In mice, KD increased MCT1 abundance in brain vessels and, despite reduced astrocyte synthesis, maintained cortical lactate levels, contributing to the sustained mitochondrial OXPHOS in cortical neurons of KD-fed mice. Notably, KD increased not only neuronal OXPHOS but also the other major energy metabolism pathways, although it is unlikely that all pathways increased in every neuron simultaneously. It is well known that breastfeeding improves many aspects of postnatal development in children including cognitive abilities (44, 45). Mimicking prolonged breastfeeding duration by weaning mice to KD, privileged brain growth over body weight gain, and reinforced energy metabolism in neurons, suggesting that mammalian breastfeeding serves to support brain development.
Our study shows that inflammatory demyelinating disease affects peripheral glucose metabolism. Peripheral and cerebral glucose metabolism is often perturbed in individuals with MS (46–48). Designated as “brain energy gap,” this is also a recognized feature of other neurodegenerative diseases (33, 34). It is possible that proinflammatory cytokines such as the tumor necrosis factor that rise during presymptomatic EAE (49, 50) trigger lipolysis from fat stores with concomitant increase in serum leptin via hypothalamic dysregulation, as observed in mouse models and patients with MS (34, 51–53). Increased leptin attenuates food intake, thereby diminishing blood glucose, which is further aggravated by the enhanced glucose demand of proliferating immune cells (33). Despite increased adipose tissue lipolysis, blood triglyceride levels drop in EAE mice (36), suggesting their utilization by hepatic fatty acid β oxidation and ketogenesis. It is conceivable that this triggered ketolysis in the brain and in other organs, causing the depletion of circulating ketone bodies at the peak of EAE. Together, these findings demonstrate that inflammatory disease decreases blood nutrient levels that probably do not suffice to meet the energy demand of CNS neurons. This could lead to mitochondrial abnormalities in axons (54) and induce a vicious cycle of cachexia, neuronal starvation, and damage. Filling the nutrient gap by increased glucose intake as, e.g., in Western diets, however, even aggravates clinical symptoms in EAE mice and patients with MS by promoting inflammatory processes (55–57).
An unexpected finding of our study was that EAE induced similar changes in neuronal energy metabolism as feeding KD. Cortical neurons of EAE mice increased ketolysis, consistent with elevated SCOT abundance in lesions of patients with MS (58), despite the low levels of circulating ketone bodies. We hypothesize that matching ketone body supply to the neuronal metabolism contributed to the rapid amelioration of disease severity in KD-fed mice. Notably, we previously showed that feeding KD to other models of demyelinating disease normalized mitochondrial pathomorphology in axons (54). Increasing circulating levels of ketone bodies by a variety of approaches has beneficial effects in a variety of disease conditions, including inflammatory demyelinating disease in animal models and in human patients (54, 59–65). The underlying mechanisms are multifactorial involving effects on neurotransmission, epigenetic gene regulation, gut microbiota, and antioxidative as well as anti-inflammatory activities (66–68). Our data show that KD rebalances CNS and peripheral metabolism in EAE. Although potentially effective, strict dietary regimens often lack the compliance of patients. Future studies will identify alternative treatments to raise steady-state levels of circulating ketone bodies, their CNS import, and utilization, which could have implications for the management of a variety of neurodegenerative diseases.
There are some limitations to our study. It is possible that bioenergetic efficiencies differ between cell types beyond the abundance of effector enzymes of the metabolic pathways (12). We did not determine metabolic fluxes and substrate preferences in vivo, in favor of obtaining cell type–specific information from acutely isolated cells. Analyses of metabolic pathways in vitro reveal the activity of metabolic pathways but come with the drawback of lacking the local microenvironment and the complexity of the natural interplay between cell types. Our experimental paradigm does not allow discriminating between subpopulations of the different cell types but reflects the sum of the isolated cellular fractions. It is possible that some of the observed effects were even stronger in individual cells. Future studies applying spatial single-cell analysis will define neuroenergetic processes at the cellular level and reveal the local cross-talk and metabolic coupling.
In summary, our study provides a map of the protein equipment of the five major CNS cell classes acutely isolated from young and adult mouse cortex. This study provides a powerful resource for analyzing protein profiles under physiological conditions during development, in response to ketosis and challenged with neuroinflammatory disease.
MATERIALS AND METHODS
Ethical compliance
All animal experiments were performed in compliance with the ARRIVE and Institutional Animal Care and Use Committee guidelines and animal policies of the Max Planck Institute of Experimental Medicine and were approved by the German Federal State of Lower Saxony (Lower Saxony State Office for Consumer Protection and Food Safety). Animals were group-housed (three to five mice) in individually ventilated cages with 12-hour dark/light cycle and had access to food and water ad libitum. Male mice were used in this study. Animals were randomly assigned to the different treatment groups. Wild-type (WT) C57BL/6, Aldh1l1–enhanced green fluorescent protein (RRID:MMRRC_011015-UCD), ThyAT (MGI:5882597), MitoTag-floxed (RRID:IMSR_JAX:032675), GFAP-Cre (RRID:IMSR_JAX:024098), and Rpb4-Cre (RRID:MMRRC_037128-UCD) mouse strains were used in this study.
Mouse treatments
Each foster mother received five male C57BL/6N mice. To maintain normal lactogenesis in dams, commencing at the age of P10 of pups, dams received liquefied standard chow (65% ssniff V1124 and 35% ssniff E15712-10) on a shelf inaccessible for pubs. At P14, ketogenic (KD; ssniff E15149-30) or normal chow (SD; ssniff V1124) was provided in boxes with small holes, making it only accessible for pups. Mice were weaned at P17 and kept on the respective diet. Measurement of blood glucose, ketone bodies, and bodyweight was performed between 10:00 and 12:00 a.m. Blood was taken from tail tip and measured using the FreeStyle Precision System with respective strips for glucose or 3HB (Abbott Diabetes Care). Serum was prepared after clotting by centrifugation. Quantification of liver enzyme activities was done using the Architect II system (Abbott Diagnostics). Analyses were done at indicated ages.
Magnetic-activated cell sorting
Cells were isolated from cortex according to the adult brain dissociation protocol (Miltenyi Biotec, catalog no. 130-107-677). Antibody labeling was done according to the Microbead kit protocols using the antibodies specific to markers for the following cell types: oligodendrocytes (O4 1:40; 130-096-670), astrocytes (ACSA-2 1:10; 130-097-679), microglia (CD11b 1:10; 130-093-636), and endothelial cells (CD31 1:10; 130-091-935). Neurons were isolated by negative selection. The purity of cell populations was routinely determined by quantitative polymerase chain reaction (qPCR) on extracted and reverse-transcribed RNA.
Proteome analysis
Isolated cell fractions from adult mouse cortex (n = 4 per condition) were lysed in 200 μl of lysis buffer [2% CHAPS, 7 M urea, 2 M thiourea, 10 mM dithiothreitol (DTT), and 0.1 M tris (pH 8.5)] with a teflon pestle fitting into a 1.5-ml microvial. Samples were incubated in a cooled ultrasonic bath for 3 min, followed by a freeze (−20°C)/thaw cycle. This treatment was performed three times in total. After the final thaw, samples were incubated in a cooled ultrasonic bath for 10 min and centrifuged (16,100g, 4°C) to remove insoluble particles. Supernatant fractions were subjected to automated in-solution digestion by filter-aided sample preparation as previously described in detail for synaptic protein fractions (69). Briefly, samples were loaded on centrifugal filter units (30 kDa molecular weight cutoff, Merck Millipore), detergents were removed with wash buffer [8 M urea, 10 mM DTT, and 0.1 M tris (pH 8.5)], proteins were alkylated with 50 mM iodoacetamide in 8 M urea and 0.1 M tris (pH 8.5), and excess reagent was removed with wash buffer. After buffer exchange with 50 mM ammonium bicarbonate (ABC) containing 10% acetonitrile, proteins were digested overnight at 37°C with 400 ng of trypsin in 40 μl of the same buffer. Tryptic peptides were recovered by centrifugation and extracted with 40 μl of 50 mM ABC and 40 μl of 1% trifluoroacetic acid (TFA), respectively. For quantification according to the TOP3 approach (70), aliquots of combined flow-throughs were spiked with Hi3 EColi standard (10 fmol/μl; Waters Corporation), containing a set of quantified synthetic peptides derived from the Escherichia coli chaperone protein ClpB. Samples were directly subjected to liquid chromatography mass spectrometry (LC-MS) analysis using nanoscale reversed-phase ultraperformance LC separation (nanoAcquity system, Waters Corporation) coupled to quadrupole time-of-flight MS with ion mobility option (Synapt G2-S, Waters Corporation). Peptides were trapped for 4 min at a flow rate of 8 μl/min 0.1% TFA on Symmetry C18 5 μm, 180 μm by 20 mm trap column and then separated at 45°C on an HSS T3 C18 1.8 μm, 75 μm by 250 mm analytical column over 180 min at a flow rate of 300 nl/min, with a gradient comprising two linear steps of 3 to 35% mobile phase B in 165 min and 35 to 60% mobile phase B in 15 min (A, water/0.1% formic acid; B, acetonitrile/0.1% formic acid). Mass spectrometry analysis was performed in the ion mobility–enhanced data-independent acquisition mode with drift time-specific collision energies (referred to as ultradefinition mass spectrometry, UDMSE) as introduced by Distler and colleagues (71) and adapted by us to synaptic protein fractions (69). Continuum LC-MS data were processed using Waters ProteinLynx Global Server (PLGS) and searched against a custom database compiled by adding the sequence information for E. coli chaperone protein ClpB and porcine trypsin to the UniProtKB/Swiss-Prot mouse proteome and by appending the reversed sequence of each entry to enable the determination of false discovery rate (FDR). Precursor and fragment ion mass tolerances were automatically determined by PLGS and were typically below 5 parts per million (ppm) for precursor ions and below 10 ppm (root mean square) for fragment ions. Carbamidomethylation of cysteine was specified as fixed and oxidation of methionine as variable modification. One missed trypsin cleavage was allowed. Minimal ion matching requirements were two fragments per peptide, five fragments per protein, and one peptide per protein. The FDR for protein identification was set to 1% threshold.
Four animals each for the different age/diet conditions (P14, P42_SD, and P42_KD) were processed as biological replicates with a technical replicate at data acquisition level (double injection), resulting in a total of 24 LC-MS runs per cell type. EAE neurons were from two animals each at two different diets (P42_SD and P42_KD), resulting in a total of eight LC-MS runs per condition. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (72) partner repository with the identifier PXD000000.
Expression analysis
Expression analyses were done as described (54, 73). RNA was extracted from dissected tissue or isolated cells using QIAshredder and RNeasy protocols (Qiagen). Concentration and quality of RNA were evaluated using a NanoDrop spectrophotometer and RNA Nano (Agilent). cDNA was synthesized with Superscript III (Invitrogen), and qPCRs were done in triplicates using the GoTaq qPCR Master Mix (Promega) on a 7500 Fast Real-Time PCR System (Applied Biosystems) or a LightCycler 480 (Roche Diagnostics). Expression values are normalized to the mean of two housekeeping genes: Rps13 (40S ribosomal protein S13) and Rplp0 (60S acidic ribosomal protein P). Quantification was done by applying the ΔΔCt method, normalized to age matched untreated controls (set to 1). All primers are intron-spanning. All primer sequences are listed in table S11.
Antibodies
The following antibodies were used in this study: 3PGDH (Frontier Institute, catalog no. 3PGDH-GP, RRID:AB_2571654), ACAT1 (Proteintech, catalog no.16215-1-AP, RRID:AB_2220210), ACO2 (Sigma-Aldrich, catalog no. HPA001097, RRID:AB_1078092), BDH1 (Proteintech, catalog no. 15417-1-AP, RRID:AB_2274683), Actin (Sigma-Aldrich, catalog no. A3853, RRID:AB_262137), carbonic anhydrase II (M.S. Ghandour, RRID:AB_2314062), CD31 (Dianova, catalog no. DIA-310, RRID:AB_2631039), CNP (Atlas Antibodies, catalog no. AMAb91072, RRID:AB_2665789), GFAP (Chemicon, catalog no. MAB3402; RRID:AB_94844), GFP (Rockland, catalog no.600-101-215, RRID:AB_218182), GLUT1 (87), HMGCS2 (Abcam, catalog no. ab137043, RRID:AB_2749817), Iba1 (Wako, catalog no. 019-19741; RRID:AB_839504), myelin basic protein (88), MCT1 (54), NeuN (Chemicon, catalog no. MAB377B, RRID:AB_177621; Millipore, catalog no. ABN91, RRID:AB_11205760), SCOT (Proteintech, catalog no.12175-1-AP, RRID:AB_2157444), S100b (Abcam, catalog no. ab52642, RRID:AB_882426), SHANK2 (Synaptic systems, catalog no. 162204; RRID:AB_2619861), VGLUT1 (Synaptic systems, catalog no. 135511; RRID:AB_887879), Vinculin (Sigma-Aldrich, catalog no. V9131, RRID:AB_477629), CD3 (Abcam, catalog no. ab11089, RRID:AB_2889189), CD3e (BioLegend, catalog no. 100312, clone 145-2C11; RRID:AB_312677), CD4 (Becton Dickinson, catalog no. 553730, clone GK 1.5; RRID:AB_395014), CD8 (Becton Dickinson, catalog no. 550281; RRID:AB_2275792), CD11b (BioLegend, catalog no. 101201; RRID:AB_312784), CD45.2 (BioLegend, catalog no. 109805; RRID:AB_313442), ACSA-2 (Miltenyi Biotec, catalog no. 130–097-678), O4 (Miltenyi Biotec catalog no. 130–096-670; RRID:AB_2847907), CD11b/c (Miltenyi Biotec, catalog no. 130-105-634, RRID:AB_2783886), and CD31 (Miltenyi Biotec, catalog no.130-097-418, RRID:AB_2814657).
Pathway analysis
For GSEA and ORA on the biological process category of GO processes (GO term), we used WebGestalt, a WEB-based gene set analysis toolkit (74). GSEA was done with calculated z scores of cell type–specific proteins found in ≥four biological replicates from P42 mice or log2 fold change (LFC) of cellular proteomes. For ORA analysis, mitochondrial candidates exclusively found in astrocytes (67 proteins) were used against the background (all 3821 identified proteins). Standard parameters for the enrichment analysis were used with the following settings: 5, minimum number of gene IDs in the category; 2000, maximum number of gene IDs in the category; Benjamini-Hochberg correction as multiple test adjustment; and top 20 significance level.
For pathway analysis of transcriptome data, the functional annotation tool DAVID was used (75). The predefined Affymetrix background from DAVID was used as background to achieve more reliable P value results. The threshold for target genes was set to >0.5 LFC and adjusted P value (Padj) < 0.05. Analysis was performed on the basis of GO_BP direct terms. Only distinct pathways with a minimum count of five genes were considered significant. Pathways are displayed using VANTED (76) with LFCs of individual sample values.
Histochemistry
Mice were sacrificed by cervical dislocation and immersion-fixed, fresh-frozen, or perfused with 4% formaldehyde as described (54, 73). Tissue blocks were either embedded in paraffin and cut on a microtome (HMP 110, Microm) or cryoconserved and cut on a cryostat (Leica Jung Cryocut CM3000). Paraffin sections were deparaffinized followed by antigen retrieval in sodium citrate buffer [0.01 M (pH 6.0)]. For immunohistological staining, blocking was performed with 20% goat serum in phosphate-buffered saline (PBS)/bovine serum albumin (BSA) (paraffin sections) or 4% horse serum, 0.2% Triton X-100 in PBS (cryosections). Primary antibodies were diluted in 2% BSA/PBS and incubated for 24 to 48 hours. Detection was done by fluorophor coupled secondary antibodies. Specimens were analyzed on an Axio Imager.Z1 (Zeiss) equipped with an AxioCam MRc3, 0.63× camera adapter and the ZEN 2012 blue edition software using 10× objective (Plan Apochromat 10×/0.45 M27) or 20× objective (Plan-Apochromat 20×/0.8) and evaluated with ImageJ software. Confocal images were obtained using an SP5 or SP8 microscope (Leica). Cellular mitochondria were quantified on 10 images from two animals (endothelia) to 21 images from four animals (astrocytes, neurons, oligodendrocytes, and microglia) per condition. Neuronal catalase area of five randomly chosen visual fields per animal were measured by automated threshold with Fiji software (NeuN, Otsu; catalase, Yen). Electron microscopic analysis was done as previously described (25). Briefly, tissue was fixed in 4% formaldehyde, 2.5% glutaraldehyde, and 0.1 M phosphate buffer. Tissue blocks were embedded in Epon (LynxII, EMS). Of each of the three mice per treatment group, at least 15 digital pictures using an on-axis 2048 × 2048 charge-coupled device camera (×12,000 magnification, TRS, Moorenweis) of uranyl acetate contrasted ultrathin sections were taken with the LEO912 electron microscope (Zeiss, Oberkochen).
For evaluation of synapses, tissue sections were postfixed with methanol for 5 min; blocked/permeabilized with 10% normal goat serum, 3% BSA, and 0.3% Triton X-100 in PBS for 1 hour; and stained against VGluT1, Shank2, and DAPI. Leica SP8 Lightning confocal z-stack images were acquired using white light laser at 488 and 561 nm. VGluT1 and Shank2 structures were modeled using Imaris. Synaptic VGluT1 fluorescence intensity was measured from VGluT1 puncta colocalizing with Shank2 puncta.
Mitochondria morphometry
Mitochondrial morphometry and intensities were measured in different cell types, on the basis of the respective cell type–specific antibody fluorescence stain. First, low magnification overviews of the respective microscope slide were acquired with a Zeiss laser scanning confocal microscope 880 (Carl Zeiss Microscopy GmbH, Jena, Germany) equipped with a 5× objective to identify the tissue section locations and specific regions for subsequent high-resolution acquisitions. Then, a 40× [numerical aperture (NA) = 1.4] oil immersion objective was used to record oversampled (x/y/z = 26/26/100 nm), multichannel z-stacks, which were then deconvolved with Huygens Professional 21.04 (Scientific Volume Imaging B.V., Hilversum, Netherlands) to recover fluorescence signal, resolution, and sample geometry. In case of MitoTag samples, complete datasets were acquired with a Leica SP8 laser scanning confocal microscope (Leica Microsystems GmbH, Wetzlar, Germany) with the above workflow using a 2.5× air and a 63× (NA =1.4) oil immersion objective, resulting in final images oversampled at 30/30/130 nm (x/y/z). Ilastik 1.3.3 (77) was used segment the respective cell body and cell processes, allowing for later differentiation of mitochondrial morphometry in those cellular subcompartments. Cell type–specific creation parameters were implemented in Imaris 9.9.1 batch processes (Bitplane AG, Zurich, Switzerland) to automatically generate 3D surfaces of processes and cell bodies, within which the mitochondrial signal was quantified. Customized scripts automating file conversion and multichannel image assembly were written with the Fiji image processing package (78). The resulting data were exported as comma-separated values and automatically assembled and analyzed with custom-built Konstanz Information Miner pipelines (79).
Immunoblot
Tissue samples were lysed in sucrose buffer [320 mM sucrose, 10 mM tris-HCl (pH 7.4), 1 mM sodium hydrogen carbonate, 1 mM magnesium chloride, 1% Triton X-100, 2% lithium dodecyl sulphate, and 1% sodium deoxycholate] with protease inhibition (Roche) using a Precellys 24 homogenizer (Bertin Technologies) or an Ultra Turrax T8 homogenizer (Kinematika) as described (54). SDS gel electrophoresis, semidry blotting on polyvinylidene difluoride membranes (Hybond P, Biosciences), and antibody incubations were done using standard procedures. Detection of immunolabeled proteins was performed with Western Lightning Plus-ECL Reagent (PerkinElmer) or WesternBright Chemiluminescence Substrate (Biozym) using ChemoCam Imager (Intas).
Metabolome analysis
Mice were sacrificed by cervical dislocation and decapitated. After exactly 12 s, the head was transferred into a microwave (Biowave Pro, Pelco) and exposed to microwave irradiation at 750 W for 16 s. The cortex was dissected on ice, snap-frozen, and stored at −80°C until further processing. Tissue was lyophilized, and dry weight was measured. Cortices were ground to a fine powder using a beatmill (Retsch) and glass beads (5 mm, Carl Roth) and extracted with 500 μl of methanol:chloroform:water 129:50:25 (v/v/v). Phase separation was induced by addition of the internal standard (250 μl of 5 μg/ml allo-inositol in water). For the analysis of small metabolites, 10 or 100 μl of the upper aqueous phase was evaporated, and metabolites were transformed into their methoxyimino and trimethylsilyl (TMS) derivatives by incubating overnight after addition of 15 μl of methoxyamine hydrochloride and for 1 to 8 hours after addition of 30 μl of N-metyl-N-(TMS) trifluoroacetamide (MSTFA). Derivatized samples were analyzed using a split of 1:10 to 1:50 on an Agilent 5977N mass selective detector connected to an Agilent 7890B gas chromatograph equipped with a capillary HP5-MS column (30 m by 0.25 mm, 0.25-μm coating thickness, Agilent Technologies). The inlet temperature was set to 250°C, and a temperature gradient was applied (70°C for 2 min, 70° to 325°C at 5 K/min, 325°C for 2 min). A transfer line of 280°C was used. Spectra recording was performed in the range of mass/charge ratio = 71 to 600. GC-MS data were analyzed using the Agilent MSD ChemStation data analysis software (Agilent Technologies). Abundance of analytes was quantified via a target ion; however, in case that derivatization of metabolites led to several ion peaks of interest, abundance of the total ion count of the individual analytes was added. The abundance of identified metabolites was normalized to the internal standard and to the dry weight of the sample. Sterols were extracted from the organic phase by adding 800 μl of MTBE:MeOH (v/v), 200 μl of water, and 40 μl of 17:0 fatty acid (0.25 mg/ml in chloroform). Each sample was measured twice. One time, 10 μl of the organic phase was used and run later with a 1:50 split to measure cholesterol. The second time, 50 μl of the organic phase was used and run later with a 1:10 split to measure other sterols. All samples were dried by nitrogen stream and redissolved in 15 μl of pyridine. MSTFA (30 μl) was added followed by an incubation time of 1 to 8 hours. Sterols were measured by GC-MS using a similar setup as described for small metabolites except for the inlet temperature (280°C) and temperature gradient, which was run at 180°C for 1 min, 180° to 320°C at 5 K/min, and 320°C for 5 min. Quantification of sterols was based on their target ion after normalization to the internal standard and the dry weight of the sample.
Transcriptome analysis
Sequencing was performed using four to five biological replicates for each condition. RNA was extracted using the Qiagen Micro-Rneasy Kit (Qiagen, catalog no. 74004) according to the manufacturer’s protocol. As external control, 1 μl of ERCC RNA Spike-In Mix (Thermo Fisher Scientific, catalog no. 4456740) diluted at 1:5000 was added to the isolated RNA. cDNA was synthesized with the Ovation RNA-Seq system v2 (NuGEN, catalog no. 7102). As input, 100 ng of cDNA was used for library preparation using the IonXpress plus gDNA and Amplicon library preparation kit (Thermo Fisher Scientific, catalog no. 4471269). To achieve size selection of the library, a 2% E-Gel was used, and the samples were barcoded and subsequently amplified. Further quantification for each sample library was performed using the KAPA Library Quantification Kit (KAPA, catalog no. KK4827). Of each sample, equal amounts were sequenced on an Ion Torrent Sequencer. On the basis of the barcodes, the raw reads (Fastq) were split into sample-specific reads and subsequently checked for sequence quality and sequence repeats. Low-quality bases and short reads were trimmed or excluded from further analysis. The reads were mapped to Mus musculus genome (mm10) using TMAP Aligner and quantified using Partek Flow software. Genes had to have at least five reads in at least 80% of the samples to be considered for further analysis.
Behavioral assessments
Audiobox
To assess learning ability of ketogenic fed mice, sound frequency discrimination was carried out in an Audiobox (TSE, Germany) on the basis of an established protocol (22). Briefly, a sterile transponder (ISO-compliant 11784 transponder, 12 mm long, TSE, Germany) was implanted subcutaneously in the back of the isoflurane-anaesthetized mice. The injection wound was closed using topical skin adhesive (Histoacryl, Braun). This enables individual identification of mice, and the behavior (nose-poking and licking) of each mouse can be detected. The Audiobox consists of the living area, where food can be accessed ad libitum, and water corner, where water is supplied and frequency discrimination is performed. Both parts were connected through a corridor. Entrance into the corner (visit) was detected by an antenna that reads the implanted transponder and a temperature sensor. The mouse can access water by nose-poking into one of the two ports, which can be opened or closed according to the demand of the experiment. For the presentation of different sounds, a loudspeaker was positioned behind the corner. Sounds were generated using MATLAB (MathWorks). Each visit to the corner was coupled to a sound that was played for its duration. During the whole experiment, one frequency (6.67 kHz) was considered “safe”: If this frequency was presented during a given visit, then mice could nose-poke and gain access to water. In visits in which the “conditioned frequency” (13.34 kHz) was played, a nose poke was followed by an air puff and no access to water. Therefore, mice had to learn to avoid nose-poking during a “conditioned visit.” During the habituation phase, only the safe sound was presented. On days 1 and 2, ports were open, and mice were drinking freely. The ports were closed on days 3 to 5 and only opened after the mouse nose-poked into the port. On days 6 to 9 (conditioning phase), the conditioned sound was presented in a small percentage of visits, 5% (days 6 and 7) or 10% (days 8 and 9).
Open-field test
The open-field test was performed as previously described (80). Briefly, exploratory activity in a novel environment was tested in a gray arena (50 cm by 50 cm, surrounded by a wall of 50-cm height) at 20-lux light intensity. Individual animals were placed into one corner of periphery of the open field and allowed to explore it for 10 min. The exploratory behavior of the mouse was recorded using an overhead camera system and scored automatically using the Viewer software (Biobserve). The overall traveled distance was analyzed as a parameter of general activity. Time, distance, and visits in the center area were analyzed to measure anxiety behavior. Results were displayed as histograms, and two-way analysis of variance (ANOVA) with Sidak’s posttest was used for statistical significance using GraphPad Prism.
Lactate imaging in astroglia-rich primary culture
Primary cultures of cortical astrocytes were prepared from newborn mice (C57BL/6 background; P0 to P1; either sex) and imaged as described before (81). One week before imaging, cells were cultured in Dulbecco’s modified Eagle medium with 5 mM glucose (Ctrl) or with 0.4 mM glucose and 4 mM 3HB (Keto condition). Cells were transfected with 2.5 μg of plasmid encoding the lactate nanosensor Laconic (82) using Lipofectamine (Invitrogen, Carlsbad, CA, USA). Cells were mounted in a custom-built perfusion chamber and imaged using an AxioObserverZ1 microscope (Zeiss, Oberkochen, Germany), a Plan-Apochromat 20×/0.8 objective and an Axiocam 506 camera (688 × 522 pixels, 4 × 4 binning, pixel size 0.91 μm by 0.91 μm) with a time resolution of 10 s using the following filters and exposure times: mTFP (monomeric teal fluorescent protein): excitation at 436/25 nm, beam splitter at 455 nm, emission at 480/40 nm, and 90 ms; fluorescence resonance energy transfer: excitation at 436/20 nm, beam splitter at 455 nm, emission at 535/30 nm, and 70 ms; and Venus: excitation at 500/25 nm, beam splitter at 515 nm, emission at 535/30 nm, and 40 ms. During imaging, cells were superfused with incubation medium containing 116 mM NaCl, 5.4 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 0.8 mM Na2HPO4, 24 mM NaHCO3, 20 mM Hepes, and 2 mM glucose (pH 7.4). The medium was continuously bubbled with 5% CO2, 20% O2, and 75% N2. Monocarboxylate transporters were blocked using AR-C155858 (10 μM, MedChemExpress, Sollentuna, Sweden) to measure lactate production rate as a proxy for glycolytic activity (82).
ATP measurement of acutely isolated astrocytes
Astrocytes were isolated by MACS from adult male WT mice fed KD or SD and incubated for 2 hours in artificial CSF (124 mM NaCl, 2 mM CaCl2, 2 mM MgSO4, 1.25 mM NaH2PO4, 23 mM NaHCO3, 2 mM glucose, and 8 mM sucrose) with 1.25 μM oligomycin, 22.25 mM 2-deoxy-glucose, or left untreated before quantification of intracellular ATP according to the manufacturer’s instruction (Dojindo Laboratories, Japan).
Experimental autoimmune encephalomyelitis
Myelin oligodendrocyte glycoprotein (MOG)–EAE was induced in mice purchased from Charles River by immunizing subcutaneously with 200 mg of MOG 35–55 peptide in complete Freund’s adjuvant (Mycobacterium tuberculosis at 3.75 mg ml−1; Becton Dickinson, catalog no. 231141) and intraperitoneal injection twice with 500 ng of pertussis toxin (Sigma-Aldrich, catalog no. P7208) as described (73). Animals were examined daily and scored for clinical signs of the disease. If disease did not start within 15 days after induction or the clinical score rose above 4, then animals were excluded from the analysis. The clinical score was as follows: 0, normal; 0.5, loss of tail tip tone; 1, loss of tail tone; 1.5, ataxia, mild walking deficits (slip off the grid); 2, mild hindlimb weakness, severe gait ataxia, twist of the tail causes rotation of the whole body; 2.5, moderate hindlimb weakness, cannot grip the grid with hind paw but able to stay on a upright tilted grid; 3, mild paraparesis, falls down from a upright tiled grid; 3.5, paraparesis of hindlimbs (legs strongly affected but move clearly); 4, paralysis of hindlimbs, weakness in forelimbs; 4.5, forelimbs paralyzed; and 5, moribund/dead. KD was provided commencing at the first appearance of EAE symptoms.
Flow cytometry
Single-cell suspensions from spinal cords were obtained via mechanical dissociation on a cell strainer. Immune cells were separated over a two-phase Percoll density gradient by centrifugation. Blood was collected by cardiac puncture in EDTA (80 mM), and single-cell suspension was obtained by centrifugation over lymphocyte separation medium (PAA Laboratories). Staining of CD3+CD4+ T cells, CD3+CD8+ T cells, and CD11b+CD45+ cells (microglia/macrophages) was performed using the following antibodies at a 1:200 dilution: anti-CD3e (BioLegend, 100312; clone 145-2C11), anti-CD4 (Becton Dickinson, 553730; clone GK 1.5), anti-CD8 (Becton Dickinson, 100706; clone 53-6.7), anti-CD11b (BioLegend, 101207; clone M1/70), and anti-CD45.2 (BioLegend, 109814; clone 104). The addition of CaliBRITE APC (allophycocyanin) beads (BD) allowed for cell quantification. Flow cytometry was performed using a CytoFLEX S operated by CytExpert v2.4 software (Beckman Coulter v2.3). Small debris was removed with preliminary FSC/SSC (forward scatter/side scatter) gating. Single living cells were obtained by doublet exclusion. Analyzed cells were defined by marker expression, and positively and negatively stained populations were defined using isotype controls if necessary.
ATP sensor quantification
Optic nerves were acutely isolated from Thy1-ATeam ATP sensor mice (31) fed KD or SD from 8 to 12 weeks of age. Nerve ends were pinned to the bottom of the incubation chamber (ALA Scientific) using 3% agarose, and nerves were constantly superfused with carbogen-bubbled artificial CSF (124 mM NaCl, 2 mM CaCl2, 2 mM MgSO4, 1.25 mM NaH2PO4, 23 mM NaHCO3, 2 mM glucose, 4 mM 3HB, and 6 mM sucrose) at 37°C. The fluorescence lifetime imaging (FLIM) and intensity measurements of the ATP sensor ATeam1.03YEMK were performed by a two-photon laser scanning microscope (LaVision TriMscope II setup) equipped with a pulsed laser (Cronus), a 20× immersion objective (Zeiss 20×/1.0 W Plan-Apochromat), and a FLIM X1 module (LaVision Biotech). The lifetime signal of the donor fluorophore of the ATeam1.03YEMK (mseCFP, excitation at 840 nm) was acquired via a GaAsP/hybrid detector (Hamamatsu) in time-correlated single-photon counting mode (TCSPC), while the acceptor fluorescence intensity was detected using a second GaAsP detector. Both nerves of each animal were scanned subsequently by xyz tile scanning over a volume of 491 μm by 196 μm by 10 μm (voxel size of 0.192 μm by 0.192 μm by 1.000 μm). Using a 495-nm beam splitter, photons were simultaneously directed to the FLIM detector (λ < 495 nm, donor lifetime) and to the GaAsP detector (>495 nm, acceptor intensity). To perform temporal deconvolution, the instrument response function was acquired using second harmonic signals at 420 nm. Data were stitched, and the lifetime was calculated following temporal deconvolution by a double exponential decay fitting the fluorescence decay at each z plane. The lifetime reported as average-weighted lifetime (aw-LT) was determined using a custom MATLAB script (83). The aw-LT for each group (SD versus KD) was calculated for each nerve by averaging over all z planes. The low photon yield in our samples discouraged pixel-wise analysis or region of interest–based approaches. Each z plane was binned to one decay (assuming double exponential decay). The ratio of fluorescence resonance energy transfer to cyan fluorescence protein fluorescence (F/C-ratio) of acceptor and donor fluorophore intensities was determined using sum projection of the TCSPC for each z plane followed by averaging over all z planes.
Statistical analysis
Sample size for each experiment is shown in the figure legends. No inclusion or exclusion criteria were used. Studies were conducted blinded to investigators and/or formally randomized. Statistical evaluation was done using GraphPad Prism (GraphPad Inc.). Normality was tested using the Kolmogorov-Smirnov test. We applied either unpaired two-sided Student’s t test for pairwise comparisons, ANOVA for comparisons of more than two groups, or Mann-Whitney test for nonparametric comparisons, as stated in the figure legends. ANOVA was combined with a posttest to evaluate individual groups. For all statistical tests, significance was measured against an alpha value of 0.05. All error bars show means ± SEM or median ± interquartile range as indicated in the figure legends. P values are shown as *P < 0.05, **P < 0.01, and ***P < 0.001.
For label-free protein quantification of proteomic data, the freely available software ISOQuant (www.isoquant.net) was used for post-identification analysis including retention time alignment, exact mass and retention time and ion mobility clustering, peak intensity normalization, isoform/homology filtering, and calculation of absolute in-sample amounts for each detected protein (71). Only peptides with a minimum length of six amino acids that were identified with scores above or equal to 5.5 in at least two runs were considered. FDR for both peptides and proteins was set to 1% threshold, and only proteins reported by at least two peptides (one of which unique) were quantified using the TOP3 method (70). The parts per million abundance values [i.e., the relative amount (w/w) of each protein in respect to the sum over all detected proteins] were log2-transformed and normalized by subtraction of the median derived from all data points for the given protein. As described recently (69), significant changes in protein abundance were detected by moderated t statistics across all technical replicates using an empirical Bayes approach and FDR-based correction for multiple comparisons, realized in the Bioconductor R packages limma (84) and q value (85). The relative abundance of a protein was accepted as altered for q < 0.05. The exact q values are reported in table S1. PCA calculation was based on complete cases, i.e., proteins quantified in each replicate of all cell types to prevent potential clustering effects of missing values.
The statistical evaluation of metabolome and transcriptome data was performed in R with the DESeq2 package (86). Metabolites or genes with at least 0.5 LFC and Padj of less than 0.05 were considered as differentially regulated. The exact values are reported in tables S6 to S8.
Acknowledgments
We are grateful to K.-A. Nave and N. Brose for constant support. We are indebted to J. Edgar for helpful comments on the manuscript. We cordially thank A. Fahrenholz, N. Hoffmeister, S. Röglin, B. Sadowski, J. Winchenbach, and S. Thüne for technical support. We thank W. Möbius, head of the Electron Microscopy Core Unit for support. We cordially thank F. Barros (CECS, Valdivia, Chile) for providing the lactate nanosensor Laconic.
Funding: This work was supported by the following: German Research Foundation grants SPP1757 and SA2114/2 (G.S.), Alzheimer Forschung Initiative 19070 (G.S.); Wilhelm-Sander Stiftung 2019.138.1 (G.S.); German Research Foundation grants OD87/1-1, OD87/3-1, and TRR274/1 (F.O.); German Research Foundation grant TRR274/1 2020 B03/C02 ID 408885537 (T.M.); German Research Foundation grant SyNergy EXC 2145 ID 390857198 (T.M.); German Research Foundation grant IS273/10-1 (T.I.); German Research Foundation grant RO 4076/3-2 (M.J.R.); German Research Foundation grants Hi1414/6-1 and Hi1414/7-1 (J.H.); Klaus Faber Stiftung (L.H.); and Studienstiftung des deutschen Volkes (P.S.).
Author contributions: Conceptualization: T.D., L.S., S.A.B., and G.S. Methodology: T.D., L.P., D.H., M.M., A.T., T.M., T.I., L.d.H., J.H., O.J., and G.S. Investigation: T.D., L.S., S.A.B., A.M.S., N.K., L.H., P.S., A.H.S., L.V.S., D.H., T.R., M.M., T.S., L.L., A.T., S.K., I.H., T.I., L.d.H., O.J., and G.S. Visualization: T.D., L.S., S.A.B., M.M., and G.S. Resources: A.T., M.J.R., A.M.P., T.M., M.S., F.O., and J.H. Supervision: T.M. and G.S. Writing—original draft: G.S. Writing—review and editing: all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The mass spectrometry proteomics data have been deposited to ProteomeXchange Consortium via the PRIDE (72) partner repository with the identifier PXD031376 (www.ebi.ac.uk/pride/archive/projects/PXD031376). The transcriptome raw data from isolated cortical astrocytes and oligodendrocytes of KD- and SD-fed mice have been deposited at GEO:GSE206577.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S11
Movie S1
REFERENCES AND NOTES
- 1.Dienel G. A., Brain glucose metabolism: Integration of energetics with function. Physiol. Rev. 99, 949–1045 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Bonvento G., Bolanos J. P., Astrocyte-neuron metabolic cooperation shapes brain activity. Cell Metab. 33, 1546–1564 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Hui S., Cowan A. J., Zeng X., Yang L., TeSlaa T., Li X., Bartman C., Zhang Z., Jang C., Wang L., Lu W., Rojas J., Baur J., Rabinowitz J. D., Quantitative fluxomics of circulating metabolites. Cell Metab. 32, 676–688.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puchalska P., Crawford P. A., Metabolic and signaling roles of ketone bodies in health and disease. Annu. Rev. Nutr. 41, 49–77 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mächler P., Wyss M. T., Elsayed M., Stobart J., Gutierrez R., von Faber-Castell A., Kaelin V., Zuend M., San Martin A., Romero-Gomez I., Baeza-Lehnert F., Lengacher S., Schneider B. L., Aebischer P., Magistretti P. J., Barros L. F., Weber B., In vivo evidence for a lactate gradient from astrocytes to neurons. Cell Metab. 23, 94–102 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Pellerin L., Pellegri G., Martin J. L., Magistretti P. J., Expression of monocarboxylate transporter mRNAs in mouse brain: Support for a distinct role of lactate as an energy substrate for the neonatal vs. adult brain. Proc. Natl. Acad. Sci. U.S.A. 95, 3990–3995 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fünfschilling U., Supplie L. M., Mahad D., Boretius S., Saab A. S., Edgar J., Brinkmann B. G., Kassmann C. M., Tzvetanova I. D., Möbius W., Diaz F., Meijer D., Suter U., Hamprecht B., Sereda M. W., Moraes C. T., Frahm J., Goebbels S., Nave K. A., Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature 485, 517–521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edmond J., Robbins R. A., Bergstrom J. D., Cole R. A., de Vellis J., Capacity for substrate utilization in oxidative metabolism by neurons, astrocytes, and oligodendrocytes from developing brain in primary culture. J. Neurosci. Res. 18, 551–561 (1987). [DOI] [PubMed] [Google Scholar]
- 9.Supplie L. M., Duking T., Campbell G., Diaz F., Moraes C. T., Gotz M., Hamprecht B., Boretius S., Mahad D., Nave K. A., Respiration-deficient astrocytes survive as glycolytic cells in vivo. J. Neurosci. 37, 4231–4242 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz F., Garcia S., Padgett K. R., Moraes C. T., A defect in the mitochondrial complex III, but not complex IV, triggers early ROS-dependent damage in defined brain regions. Hum. Mol. Genet. 21, 5066–5077 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fecher C., Trovo L., Muller S. A., Snaidero N., Wettmarshausen J., Heink S., Ortiz O., Wagner I., Kuhn R., Hartmann J., Karl R. M., Konnerth A., Korn T., Wurst W., Merkler D., Lichtenthaler S. F., Perocchi F., Misgeld T., Cell-type-specific profiling of brain mitochondria reveals functional and molecular diversity. Nat. Neurosci. 22, 1731–1742 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Fabuel I., Le Douce J., Logan A., James A. M., Bonvento G., Murphy M. P., Almeida A., Bolanos J. P., Complex I assembly into supercomplexes determines differential mitochondrial ROS production in neurons and astrocytes. Proc. Natl. Acad. Sci. U.S.A. 113, 13063–13068 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bayraktar O. A., Bartels T., Holmqvist S., Kleshchevnikov V., Martirosyan A., Polioudakis D., Ben Haim L., Young A. M. H., Batiuk M. Y., Prakash K., Brown A., Roberts K., Paredes M. F., Kawaguchi R., Stockley J. H., Sabeur K., Chang S. M., Huang E., Hutchinson P., Ullian E. M., Hemberg M., Coppola G., Holt M. G., Geschwind D. H., Rowitch D. H., Astrocyte layers in the mammalian cerebral cortex revealed by a single-cell in situ transcriptomic map. Nat. Neurosci. 23, 500–509 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheeler M. A., Clark I. C., Tjon E. C., Li Z., Zandee S. E. J., Couturier C. P., Watson B. R., Scalisi G., Alkwai S., Rothhammer V., Rotem A., Heyman J. A., Thaploo S., Sanmarco L. M., Ragoussis J., Weitz D. A., Petrecca K., Moffitt J. R., Becher B., Antel J. P., Prat A., Quintana F. J., MAFG-driven astrocytes promote CNS inflammation. Nature 578, 593–599 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiebig C., Keiner S., Ebert B., Schaffner I., Jagasia R., Lie D. C., Beckervordersandforth R., Mitochondrial dysfunction in astrocytes impairs the generation of reactive astrocytes and enhances neuronal cell death in the cortex upon photothrombotic lesion. Front. Mol. Neurosci. 12, 40 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misgeld T., Schwarz T. L., Mitostasis in neurons: Maintaining mitochondria in an extended cellular architecture. Neuron 96, 651–666 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aten S., Kiyoshi C. M., Arzola E. P., Patterson J. A., Taylor A. T., Du Y., Guiher A. M., Philip M., Camacho E. G., Mediratta D., Collins K., Boni K., Garcia S. A., Kumar R., Drake A. N., Hegazi A., Trank L., Benson E., Kidd G., Terman D., Zhou M., Ultrastructural view of astrocyte arborization, astrocyte-astrocyte and astrocyte-synapse contacts, intracellular vesicle-like structures, and mitochondrial network. Prog. Neurobiol. 213, 102264 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q., Cheng Z., Zhou L., Darmanis S., Neff N. F., Okamoto J., Gulati G., Bennett M. L., Sun L. O., Clarke L. E., Marschallinger J., Yu G., Quake S. R., Wyss-Coray T., Barres B. A., Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron 101, 207–223.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotter D. G., d’Avignon D. A., Wentz A. E., Weber M. L., Crawford P. A., Obligate role for ketone body oxidation in neonatal metabolic homeostasis. J. Biol. Chem. 286, 6902–6910 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nehlig A., Age-dependent pathways of brain energy metabolism: The suckling rat, a natural model of the ketogenic diet. Epilepsy Res. 37, 211–221 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Vining E. P., Pyzik P., McGrogan J., Hladky H., Anand A., Kriegler S., Freeman J. M., Growth of children on the ketogenic diet. Dev. Med. Child Neurol. 44, 796–802 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Chen C., Krueger-Burg D., de Hoz L., Wide sensory filters underlie performance in memory-based discrimination and generalization. PLOS ONE 14, e0214817 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puchalska P., Crawford P. A., Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metab. 25, 262–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koper J. W., Lopes-Cardozo M., Van Golde L. M., Preferential utilization of ketone bodies for the synthesis of myelin cholesterol in vivo. Biochim. Biophys. Acta 666, 411–417 (1981). [DOI] [PubMed] [Google Scholar]
- 25.Saher G., Brügger B., Lappe-Siefke C., Möbius W., Tozawa R., Wehr M. C., Wieland F., Ishibashi S., Nave K. A., High cholesterol level is essential for myelin membrane growth. Nat. Neurosci. 8, 468–475 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Pifferi F., Laurent B., Plourde M., Lipid transport and metabolism at the blood-brain interface: Implications in health and disease. Front. Physiol. 12, 645646 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valdebenito R., Ruminot I., Garrido-Gerter P., Fernandez-Moncada I., Forero-Quintero L., Alegria K., Becker H. M., Deitmer J. W., Barros L. F., Targeting of astrocytic glucose metabolism by beta-hydroxybutyrate. J. Cereb. Blood Flow Metab. 36, 1813–1822 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy M., Beauvieux M.-C., Naulin J., El Hamrani D., Gallis J.-L., Cunnane S. C., Bouzier-Sore A.-K., Rapid adaptation of rat brain and liver metabolism to a ketogenic diet: An integrated study using 1H- and 13C-NMR spectroscopy. J. Cereb. Blood Flow Metab. 35, 1154–1162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang H., Bourdon A. K., Chen L. Y., Phelix C. F., Perry G., Gibbs free-energy gradient along the path of glucose transport through human glucose transporter 3. ACS Chem. Nerosci. 9, 2815–2823 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruskin D. N., Kawamura M., Masino S. A., Adenosine and ketogenic treatments. J. Caffeine Adenosine Res. 10, 104–109 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trevisiol A., Saab A. S., Winkler U., Marx G., Imamura H., Mobius W., Kusch K., Nave K. A., Hirrlinger J., Monitoring ATP dynamics in electrically active white matter tracts. eLife 6, e24241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neishabouri A., Faisal A. A., Saltatory conduction in unmyelinated axons: Clustering of Na+ channels on lipid rafts enables micro-saltatory conduction in C-fibers. Front. Neuroanat. 8, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cunnane S. C., Trushina E., Morland C., Prigione A., Casadesus G., Andrews Z. B., Beal M. F., Bergersen L. H., Brinton R. D., de la Monte S., Eckert A., Harvey J., Jeggo R., Jhamandas J. H., Kann O., la Cour C. M., Martin W. F., Mithieux G., Moreira P. I., Murphy M. P., Nave K. A., Nuriel T., Oliet S. H. R., Saudou F., Mattson M. P., Swerdlow R. H., Millan M. J., Brain energy rescue: An emerging therapeutic concept for neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 19, 609–633 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Candia P., Matarese G., Leptin and ghrelin: Sewing metabolism onto neurodegeneration. Neuropharmacology 136, 307–316 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Schattling B., Engler J. B., Volkmann C., Rothammer N., Woo M. S., Petersen M., Winkler I., Kaufmann M., Rosenkranz S. C., Fejtova A., Thomas U., Bose A., Bauer S., Trager S., Miller K. K., Bruck W., Duncan K. E., Salinas G., Soba P., Gundelfinger E. D., Merkler D., Friese M. A., Bassoon proteinopathy drives neurodegeneration in multiple sclerosis. Nat. Neurosci. 22, 887–896 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Bernardes D., Oliveira-Lima O. C., da Silva T. V., Juliano M. A., dos Santos D. M., Carvalho-Tavares J., Metabolic alterations in experimental autoimmune encephalomyelitis in mice: Effects of prior physical exercise. Neurophysiology 48, 117–121 (2016). [Google Scholar]
- 37.Magistretti P. J., Allaman I., A cellular perspective on brain energy metabolism and functional imaging. Neuron 86, 883–901 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Lovatt D., Sonnewald U., Waagepetersen H. S., Schousboe A., He W., Lin J. H., Han X., Takano T., Wang S., Sim F. J., Goldman S. A., Nedergaard M., The transcriptome and metabolic gene signature of protoplasmic astrocytes in the adult murine cortex. J. Neurosci. 27, 12255–12266 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butt U. J., Steixner-Kumar A. A., Depp C., Sun T., Hassouna I., Wustefeld L., Arinrad S., Zillmann M. R., Schopf N., Fernandez Garcia-Agudo L., Mohrmann L., Bode U., Ronnenberg A., Hindermann M., Goebbels S., Bonn S., Katschinski D. M., Miskowiak K. W., Nave K. A., Ehrenreich H., Hippocampal neurons respond to brain activity with functional hypoxia. Mol. Psychiatry 26, 1790–1807 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Angelova P. R., Kasymov V., Christie I., Sheikhbahaei S., Turovsky E., Marina N., Korsak A., Zwicker J., Teschemacher A. G., Ackland G. L., Funk G. D., Kasparov S., Abramov A. Y., Gourine A. V., Functional oxygen sensitivity of astrocytes. J. Neurosci. 35, 10460–10473 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cahill G. F. Jr., Fuel metabolism in starvation. Annu. Rev. Nutr. 26, 1–22 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Wang J., Cui Y., Yu Z., Wang W., Cheng X., Ji W., Guo S., Zhou Q., Wu N., Chen Y., Chen Y., Song X., Jiang H., Wang Y., Lan Y., Zhou B., Mao L., Li J., Yang H., Guo W., Yang X., Brain endothelial cells maintain lactate homeostasis and control adult hippocampal neurogenesis. Cell Stem Cell 25, 754–767.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Cox P. J., Kirk T., Ashmore T., Willerton K., Evans R., Smith A., Murray A. J., Stubbs B., West J., McLure S. W., King M. T., Dodd M. S., Holloway C., Neubauer S., Drawer S., Veech R. L., Griffin J. L., Clarke K., Nutritional ketosis alters fuel preference and thereby endurance performance in athletes. Cell Metab. 24, 256–268 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Lopez D. A., Foxe J. J., Mao Y., Thompson W. K., Martin H. J., Freedman E. G., Breastfeeding duration is associated with domain-specific improvements in cognitive performance in 9-10-year-old children. Front. Public Health 9, 657422 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R. D. Martin, The fundamental importance of breastfeeding for health and development, in Health in Transition: Translating Developmental Origins of Health and Disease Science to Improve Future Health in Africa, A. J. Macnab, A. Daar, C. Pauw, Eds. (Stellenbosch: SUN PReSS, 2020), chap. 6, pp. 67–101. [Google Scholar]
- 46.Wens I., Dalgas U., Deckx N., Cools N., Eijnde B. O., Does multiple sclerosis affect glucose tolerance? Mult. Scler. 20, 1273–1276 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Ruiz-Arguelles A., Mendez-Huerta M. A., Lozano C. D., Ruiz-Arguelles G. J., Metabolomic profile of insulin resistance in patients with multiple sclerosis is associated to the severity of the disease. Mult. Scler. Relat. Disord. 25, 316–321 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Blinkenberg M., Rune K., Jensen C. V., Ravnborg M., Kyllingsbaek S., Holm S., Paulson O. B., Sørensen P. S., Cortical cerebral metabolism correlates with MRI lesion load and cognitive dysfunction in MS. Neurology 54, 558–564 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Yang G., Parkhurst C. N., Hayes S., Gan W. B., Peripheral elevation of TNF-α leads to early synaptic abnormalities in the mouse somatosensory cortex in experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U.S.A. 110, 10306–10311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grabner G. F., Xie H., Schweiger M., Zechner R., Lipolysis: Cellular mechanisms for lipid mobilization from fat stores. Nat. Metab. 3, 1445–1465 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Kim M. S., Yan J., Wu W., Zhang G., Zhang Y., Cai D., Rapid linkage of innate immunological signals to adaptive immunity by the brain-fat axis. Nat. Immunol. 16, 525–533 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanna V., Di Giacomo A., La Cava A., Lechler R. I., Fontana S., Zappacosta S., Matarese G., Leptin surge precedes onset of autoimmune encephalomyelitis and correlates with development of pathogenic T cell responses. J. Clin. Invest. 111, 241–250 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burfeind K. G., Yadav V., Marks D. L., Hypothalamic dysfunction and multiple sclerosis: Implications for fatigue and weight dysregulation. Curr. Neurol. Neurosci. Rep. 16, 98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stumpf S. K., Berghoff S. A., Trevisiol A., Spieth L., Duking T., Schneider L. V., Schlaphoff L., Dreha-Kulaczewski S., Bley A., Burfeind D., Kusch K., Mitkovski M., Ruhwedel T., Guder P., Rohse H., Denecke J., Gartner J., Mobius W., Nave K. A., Saher G., Ketogenic diet ameliorates axonal defects and promotes myelination in Pelizaeus-Merzbacher disease. Acta Neuropathol. 138, 147–161 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang D., Jin W., Wu R., Li J., Park S.-A., Tu E., Zanvit P., Xu J., Liu O., Cain A., Chen W., High glucose intake exacerbates autoimmunity through reactive-oxygen-species-mediated TGF-β cytokine activation. Immunity 51, 671–681.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mannino A., Lithander F. E., Dunlop E., Hoare S., Shivappa N., Daly A., Phillips M., Pereira G., Sherriff J., Lucas R. M., Ponsonby A. L., Hebert J. R., van der Mei I., Black L. J.; Ausimmune Investigator Group , A proinflammatory diet is associated with an increased likelihood of first clinical diagnosis of central nervous system demyelination in women. Mult. Scler. Relat. Disord. 57, 103428 (2022). [DOI] [PubMed] [Google Scholar]
- 57.Bosch-Queralt M., Cantuti-Castelvetri L., Damkou A., Schifferer M., Schlepckow K., Alexopoulos I., Lutjohann D., Klose C., Vaculciakova L., Masuda T., Prinz M., Monroe K. M., Di Paolo G., Lewcock J. W., Haass C., Simons M., Diet-dependent regulation of TGFβ impairs reparative innate immune responses after demyelination. Nat. Metab. 3, 211–227 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han M. H., Hwang S. I., Roy D. B., Lundgren D. H., Price J. V., Ousman S. S., Fernald G. H., Gerlitz B., Robinson W. H., Baranzini S. E., Grinnell B. W., Raine C. S., Sobel R. A., Han D. K., Steinman L., Proteomic analysis of active multiple sclerosis lesions reveals therapeutic targets. Nature 451, 1076–1081 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Choi I. Y., Piccio L., Childress P., Bollman B., Ghosh A., Brandhorst S., Suarez J., Michalsen A., Cross A. H., Morgan T. E., Wei M., Paul F., Bock M., Longo V. D., A diet mimicking fasting promotes regeneration and reduces autoimmunity and multiple sclerosis symptoms. Cell Rep. 15, 2136–2146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bahr L. S., Bock M., Liebscher D., Bellmann-Strobl J., Franz L., Pruss A., Schumann D., Piper S. K., Kessler C. S., Steckhan N., Michalsen A., Paul F., Mahler A., Ketogenic diet and fasting diet as nutritional approaches in multiple sclerosis (NAMS): Protocol of a randomized controlled study. Trials 21, 3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neumann B., Baror R., Zhao C., Segel M., Dietmann S., Rawji K. S., Foerster S., McClain C. R., Chalut K., van Wijngaarden P., Franklin R. J. M., Metformin restores CNS remyelination capacity by rejuvenating aged stem cells. Cell Stem Cell 25, 473–485.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim D. Y., Hao J., Liu R., Turner G., Shi F. D., Rho J. M., Inflammation-mediated memory dysfunction and effects of a ketogenic diet in a murine model of multiple sclerosis. PLOS ONE 7, e35476 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brenton J. N., Banwell B., Bergqvist A. G. C., Lehner-Gulotta D., Gampper L., Leytham E., Coleman R., Goldman M. D., Pilot study of a ketogenic diet in relapsing-remitting MS. Neurol. Neuroimmunol. Neuroinflamm. 6, e565 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bock M., Karber M., Kuhn H., Ketogenic diets attenuate cyclooxygenase and lipoxygenase gene expression in multiple sclerosis. EBioMedicine 36, 293–303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Newman J. C., Covarrubias A. J., Zhao M., Yu X., Gut P., Ng C. P., Huang Y., Haldar S., Verdin E., Ketogenic diet reduces midlife mortality and improves memory in aging mice. Cell Metab. 26, 547–557.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simeone T. A., Simeone K. A., Stafstrom C. E., Rho J. M., Do ketone bodies mediate the anti-seizure effects of the ketogenic diet? Neuropharmacology 133, 233–241 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olson C. A., Vuong H. E., Yano J. M., Liang Q. Y., Nusbaum D. J., Hsiao E. Y., The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Cell 173, 1728–1741.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Youm Y. H., Nguyen K. Y., Grant R. W., Goldberg E. L., Bodogai M., Kim D., D’Agostino D., Planavsky N., Lupfer C., Kanneganti T. D., Kang S., Horvath T. L., Fahmy T. M., Crawford P. A., Biragyn A., Alnemri E., Dixit V. D., The ketone metabolite β-hydroxybutyrate blocks NLRP3 inflammasome-mediated inflammatory disease. Nat. Med. 21, 263–269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ambrozkiewicz M. C., Schwark M., Kishimoto-Suga M., Borisova E., Hori K., Salazar-Lazaro A., Rusanova A., Altas B., Piepkorn L., Bessa P., Schaub T., Zhang X., Rabe T., Ripamonti S., Rosario M., Akiyama H., Jahn O., Kobayashi T., Hoshino M., Tarabykin V., Kawabe H., Polarity acquisition in cortical neurons is driven by synergistic action of Sox9-regulated Wwp1 and Wwp2 E3 ubiquitin ligases and intronic miR-140. Neuron 100, 1097–1115.e15 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., Geromanos S. J., Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 (2006). [DOI] [PubMed] [Google Scholar]
- 71.Distler U., Kuharev J., Navarro P., Tenzer S., Label-free quantification in ion mobility-enhanced data-independent acquisition proteomics. Nat. Protoc. 11, 795–812 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., Tiwary S., Cox J., Audain E., Walzer M., Jarnuczak A. F., Ternent T., Brazma A., Vizcaino J. A., The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berghoff S. A., Spieth L., Sun T., Hosang L., Schlaphoff L., Depp C., Duking T., Winchenbach J., Neuber J., Ewers D., Scholz P., van der Meer F., Cantuti-Castelvetri L., Sasmita A. O., Meschkat M., Ruhwedel T., Mobius W., Sankowski R., Prinz M., Huitinga I., Sereda M. W., Odoardi F., Ischebeck T., Simons M., Stadelmann-Nessler C., Edgar J. M., Nave K. A., Saher G., Microglia facilitate repair of demyelinated lesions via post-squalene sterol synthesis. Nat. Neurosci. 24, 47–60 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liao Y., Wang J., Jaehnig E. J., Shi Z., Zhang B., WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–W205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang D. W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 76.Rohn H., Junker A., Hartmann A., Grafahrend-Belau E., Treutler H., Klapperstuck M., Czauderna T., Klukas C., Schreiber F., VANTED v2: A framework for systems biology applications. BMC Syst. Biol. 6, 139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Berg S., Kutra D., Kroeger T., Straehle C. N., Kausler B. X., Haubold C., Schiegg M., Ales J., Beier T., Rudy M., Eren K., Cervantes J. I., Xu B., Beuttenmueller F., Wolny A., Zhang C., Koethe U., Hamprecht F. A., Kreshuk A., ilastik: Interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.M. R. Berthold, N. Cebron, F. Dill, T. R. Gabriel, T. Kötter, T. Meinl, P. Ohl, C. Sieb, K. Thiel, B. Wiswedel. KNIME: The Konstanz Information Miner, in Data Analysis, Machine Learning and Applications - Proceedings of the 31st Annual Conference of the Gesellschaft für Klassifikation e.V (Springer Berlin Heidelberg, 2008), pp. 319–326. [Google Scholar]
- 80.Babaev O., Botta P., Meyer E., Muller C., Ehrenreich H., Brose N., Luthi A., Krueger-Burg D., Neuroligin 2 deletion alters inhibitory synapse function and anxiety-associated neuronal activation in the amygdala. Neuropharmacology 100, 56–65 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Köhler S., Winkler U., Sicker M., Hirrlinger J., NBCe1 mediates the regulation of the NADH/NAD+ redox state in cortical astrocytes by neuronal signals. Glia 66, 2233–2245 (2018). [DOI] [PubMed] [Google Scholar]
- 82.San Martin A., Ceballo S., Ruminot I., Lerchundi R., Frommer W. B., Barros L. F., A genetically encoded FRET lactate sensor and its use to detect the Warburg effect in single cancer cells. PLOS ONE 8, e57712 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trevisiol A., Kusch K., Steyer A. M., Gregor I., Nardis C., Winkler U., Köhler S., Restrepo A., Möbius W., Werner H. B., Nave K.-A., Hirrlinger J., Structural myelin defects are associated with low axonal ATP levels but rapid recovery from energy deprivation in a mouse model of spastic paraplegia. PLoS Biol. 18, e3000943 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., Smyth G. K., limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Storey J. D., Tibshirani R., Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Berghoff S. A., Düking T., Spieth L., Winchenbach J., Stumpf S. K., Gerndt N., Kusch K., Ruhwedel T., Möbius W., Saher G., Blood-brain barrier hyperpermeability precedes demyelination in the cuprizone model. Acta Neuropathol. Commun. 5, 94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meschkat M., Steyer A. M., Weil M. T., Kusch K., Jahn O., Piepkorn L., Agui-Gonzalez P., Phan N. T. N., Ruhwedel T., Sadowski B., Rizzoli S. O., Werner H. B., Ehrenreich H., Nave K. A., Mobius W., White matter integrity in mice requires continuous myelin synthesis at the inner tongue. Nat. Commun. 13, 1163 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S11
Movie S1