Abstract
Using a genome-wide CRISPR screen, we identified CDK9, DHODH, and PRMT5 as synthetic lethal partners with gilteritinib treatment in fms-like tyrosine kinase 3 (FLT3)–internal tandem duplication (ITD) acute myeloid leukemia (AML) and genetically and pharmacologically validated their roles in gilteritinib sensitivity. The presence of FLT3-ITD is associated with an increase in anaerobic glycolysis, rendering leukemia cells highly sensitive to inhibition of glycolysis. Supportive of this, our data show the enrichment of single guide RNAs targeting 28 glycolysis-related genes upon gilteritinib treatment, suggesting that switching from glycolysis to oxidative phosphorylation (OXPHOS) may represent a metabolic adaption of AML in gilteritinib resistance. CDK9i/FLT3i, DHODHi/FLT3i, and PRMT5i/FLT3i pairs mechanistically converge on OXPHOS and purine biosynthesis blockade, implying that targeting the metabolic functions of these three genes and/or proteins may represent attractive strategies to sensitize AML to gilteritinib treatment. Our findings provide the basis for maximizing therapeutic impact of FLT3-ITD inhibitors and a rationale for a clinical trial of these novel combinations.
Down-regulation of OXPHOS/purine biosynthesis is a feature of FLT3i-induced synergistic interactions with CDK9i, PRMT5i, and DHODHi.
INTRODUCTION
Internal tandem duplication (ITD) and mutations within the tyrosine kinase domain of fms-like tyrosine kinase 3 (FLT3) occur in 30% of acute myeloid leukemia (AML) cases (1, 2). The presence of FLT3 mutations at high-variant allele frequency (allelic ratio of >0.5) is associated with poor survival (1). Both mutations lead to a constitutively active receptor tyrosine kinase, causing prolonged signal transduction along cell survival and proliferative axes. In addition, FLT3-ITD has been shown to mediate metabolic reprograming by elevating anaerobic glycolysis through up-regulation of the mitochondrial hexokinase 2 (HK2). Therefore, FLT3-ITD leukemia cells are addicted to glycolysis and susceptible to pharmacological inhibition of glycolytic activity (3).
Despite improved survival seen in FLT3-mutant AML patients treated with approved FLT3 inhibitors (FLT3is), midostaurin and gilteritinib, patients frequently experience relapse. The optimal use of these inhibitors in the upfront, relapse, and maintenance settings remains to be established (4). We previously used genome-wide CRISPR screening to identify the essentiality of XPO1 and BCL2 genes in AML with the first-generation FLT3i, midostaurin, further demonstrating the efficacy of midostaurin/selinexor and midostaurin/venetoclax combination therapies (5, 6). Given the success of the highly selective FLT3i, gilteritinib, in the ADMIRAL study (4), and its increasing adoption in the clinic, we aimed to nominate coessential genes whose knockout may confer gilteritinib sensitivity by CRISPR screening.
To this end, we identified protein arginine N-methyltransferase 5 (PRMT5), cyclin-dependent kinase 9 (CDK9), and dihydroorotate dehydrogenase (DHODH) as novel synthetic lethal partners with gilteritinib treatment in AML. Using both genetic and pharmacologic approaches, we recapitulated the coessential nature of these genes in combination with gilteritinib treatment in FLT3-ITD cell lines and patient samples. By using RNA sequencing (RNA-seq) and metabolomics, we showed that the knockdown of CDK9, PRMT5, or DHODH plus gilteritinib treatment each had in common the ability to cooperatively shut down oxidative phosphorylation (OXPHOS), and purine biosynthesis and supplementation of purine nucleosides rescued synergistic effect of gilteritinib and CDK9, PRMT5, or DHODH inhibition. We further showed the trend of enrichment of single guide RNAs (sgRNAs) targeting 28 glycolytic genes in cells treated with gilteritinib in positive selection screen. This suggests a metabolic adaption of the leukemic cells whereby they switch to OXPHOS from anaerobic glycolysis to develop resistance to gilteritinib. Therefore, OXPHOS and purine synthesis are central metabolic pathways targeted by different synthetic lethal treatments to resensitize leukemic cells to gilteritinib treatment.
Last, as a proof of concept, we examined in vivo combinatorial approaches using the 1/2/5/9 CDK inhibitor, dinaciclib, which has been used in conjunction with venetoclax in clinical trials for relapsed/refractory AML, DHODH inhibitor (DHODHi), brequinar, and a novel PRMT5 inhibitor (PRMT5i), PRT811/PRT808 (7). Our human FLT3-ITD AML xenograft model showed a promising survival benefit provided by combination therapies of dinaciclib/gilteritinib, brequinar/gilteritinib, or PRT808/gilteritinib over monotherapies, suggesting that the three combinations may improve the outcome of AML patients with FLT3 mutations.
RESULTS
Genome-wide CRISPR screen reveals novel synthetic lethal partners with gilteritinib in FLT3-ITD AML
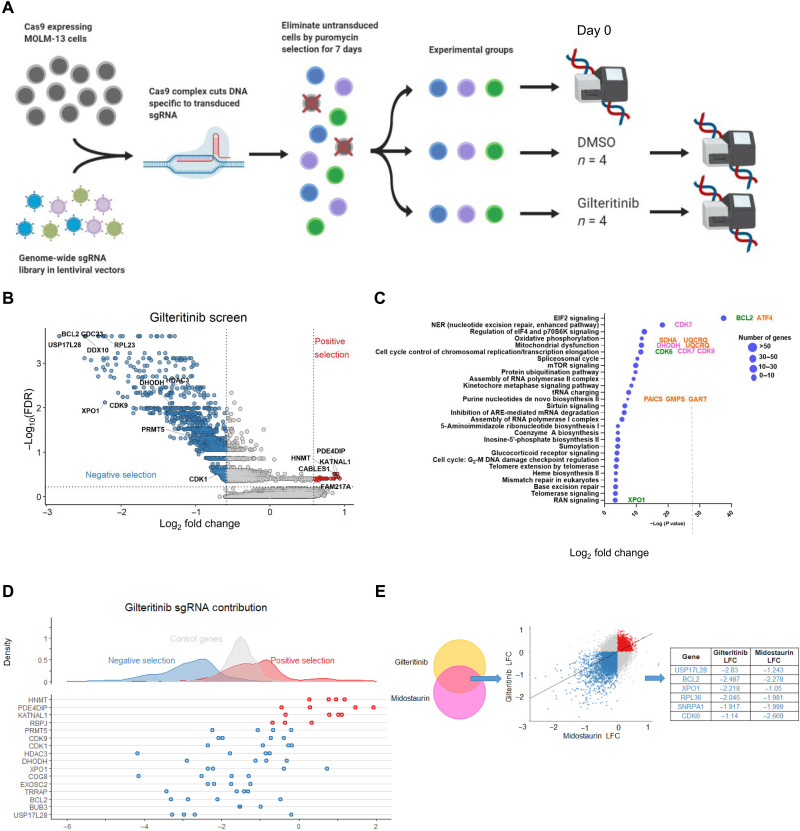
To identify coessential genes and pathways that sensitize FLT3-ITD AML cells to gilteritinib, we conducted a genome-wide CRISPR screening on MOLM-13 cells (Fig. 1A). Potential targets were highlighted on the basis of statistically significant thresholds for negative selection (synergistic) and positive selection (antagonistic) following a robust rank aggregation analysis (Fig. 1B and data file S1) (8). Across all four replicates, the screens demonstrated low Gini indices (a metric to account for the heterogeneity of sgRNA reads), a low quantity of missed sgRNAs, and a small percentage of unmapped reads (fig. S1A) (9). The top 1191 genes whose targeting sgRNAs were depleted by at least 1.5-fold (log2 fold change ≤ 0.6) were selected to identify top enriched pathways using Ingenuity Pathways Analysis (IPA) based on their low false discovery rates (FDRs)/P values (cutoff: FDR = 0.25/P = 0.03) (fig. S1B). These genes were significantly enriched in AML-related biological processes that regulate OXPHOS, mitochondria dysfunction, kinetochore metaphase signaling, cell cycle control of chromosomal replication/transcriptional elongation, and purine de novo biosynthesis (Fig. 1C and data file S2). Subsequently, one to two representative genes from each of top pathways were selected for validation based on their high expression levels in mixed-lineage leukemia (MLL)–rearranged AML compared with normal tissues, their reported properties as leukemogenesis drivers, and availability of clinical grade inhibitors: BCL2 [EIF2 (Eukaryotic Initiation Factor 2) signaling], CDK7 [nucleotide excision repair (NER)], DHODH (mitochondrial metabolism), CDK9 (transcription), and XPO1 (RAN signaling) (Fig. 1C and fig. S1B) (5, 6, 10–14). These hits also showed the high degrees of consistency of dropout, implying strong synergistic interactions with gilteritinib (Fig. 1D) (5, 6). The other top-ranked hits with small FDR/P values were either not enriched in top pathways, core essential genes therefore considered not targetable (i.e., RPL11, RPL23, and RPL28) or not relevant leukemia targets. Other top hits like EIF2 component, EIF2S1, and adenosine 5′-triphosphate (ATP) synthetase, ATP5F1A, were core essential genes for healthy cell survival. Therefore, we excluded them from our selection pool for validation. In line with our previous findings, BCL2 and XPO1 also stood out among the top-ranked hits as synthetic lethal targets with gilteritinib (Fig. 1E) (15, 16). However, because the synergistic effect of targeting the two genes with FLT3i was previously described by us and others (1–4), they were not chosen for downstream studies.
Fig. 1. CRISPR knockout screen reveals potential synergistic partners with gilteritinib.
(A) Schematic overview of genome-wide CRISPR screen design. (B) Volcano plot segregating candidate hits into positively (red) and negatively selected (blue) genes (four biological replicates per condition). (C) Pathway enrichment analysis of the top hits in negative selection by IPA. The locations of genes of interest in pathways are highlighted. Pink: Hits prioritized for validation here. Green: Hits that have high ranks in both gilteritinib and midostaurin screens but were not validated here. Orange: Hits shared by CRISPR screen and two RNA-seq datasets. (D) Contribution of each sgRNA to the top hits. (E) Selected overlapping hits shared by gilteritinib and midostaurin screens. DMSO, dimethyl sulfoxide.
To evaluate whether the fitness of other cancer cell lines is altered by loss of CDK9 or DHODH, we used the DepMap dataset (https://depmap.org/portal/), which is composed of genome-scale CRISPR knockout screens performed on 1054 cell lines. Although these genes are not core essential genes to all cell types, the dependency scores of CDK9 or DHODH in human AML cells were ≤0.5, suggesting that their depletions are particularly lethal to FLT3-ITD AML cell lines, MOLM-13 and MV4-11 (fig. S1C) (15). Clinical grade inhibitors are available for each of these gene products (10, 12), facilitating rapid clinical application. The Kyoto Encyclopedia of Genes and Genomes pathway analysis of the top hits also revealed that perturbation of multiple genes in FLT3 and cKIT signaling pathways confers vulnerability to FLT3is (fig. S2).
shRNA-mediated knockdown validates screen predictions of CDK9 and DHODH as synthetic lethal targets with FLT3i
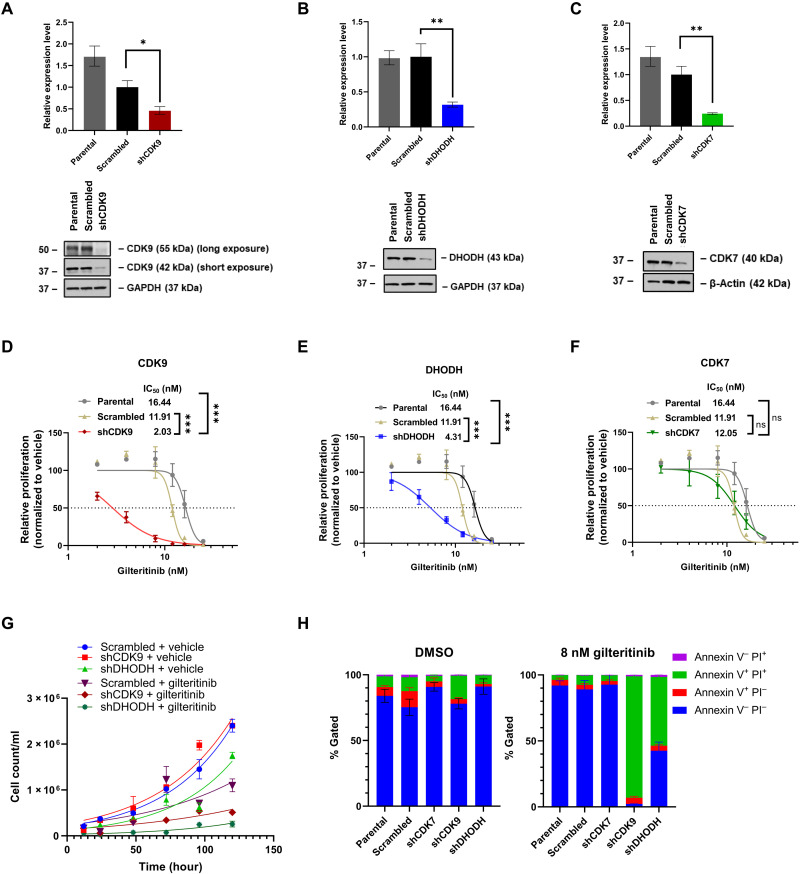
To validate the selected CRISPR screen hits, CDK9, DHODH, and CDK7, MOLM-13 cells were transduced with short hairpin RNA (shRNA) targeting each of these genes, and knockdown efficiencies were confirmed via quantitative polymerase chain reaction (qPCR) and Western blotting (Fig. 2, A to C). Scrambled or gene-targeting shRNA-transduced cell lines were treated with gilteritinib (2 to 25 nM dose range) for 120 hours before cell viability and proliferation were measured. Cells with knockdown of CDK9 or DHODH had a notably lower median inhibitory concentration (IC50) of gilteritinib compared with scrambled or the parental (untransduced) controls (Fig. 2, D and E). In contrast, knockdown of CDK7 only had a marginal impact on cell viability compared to scrambled control, suggesting that it is likely a false-positive hit (Fig. 2F). Gilterinib-treated shCDK9 or shDHODH cells had a markedly slower growth kinetics compared with vehicle-treated cell lines (Fig. 2G). In agreement with this, knockdowns of CDK9 or DHODH, but not CDK7, substantially increased the percentages of necroptotic/apoptotic cells by 9- or 2.6-fold, respectively, compared to scrambled control cells in response to gilteritinib treatment (Fig. 2H). These findings support our screen’s predictions that inactivation of CDK9 or DHODH could sensitize AML cells to FLT3-ITD inhibitors.
Fig. 2. Genetic depletion of CRISPR screen top hits, CDK9 or DHODH, but not CDK7, sensitizes AML cells to gilteritinib treatment.
(A to C) Knockdown efficiency of selected targets by shRNA as detected by qPCR and Western blotting; GAPDH or β-actin serves as loading control. n = 2. *P < 0.05 and **P < 0.01. (D to F) Dose-response curves of shCDK9-, shDHODH-, and shCDK7-stable MOLM-13 cells in response to 120-hour gilteritinib treatment. Cell viability was measured with MTS. Results are shown as means ± SEM of four technical replicates and three to four biological replicates. ***P < 0.0001; ns, not significant. (G) Cell counts of MOLM-13 cells at different time points in different treatment groups. Results are shown as means ± SEM of three biological replicates. (H) Knockdown of CDK9 or DHODH but not CDK7 with shRNA increases the frequency of apoptotic AML cell lines with gilteritinib. Parental, scrambled, or gene-targeting shRNA-stable MOLM-13 cells were treated with 8 nM gileritinib for 120 hours and stained with annexin V/PI for flow cytometry analysis. Data are shown as means ± SEM of % population from triplicates.
shCDK9 or shDHODH-mediated synthetic lethality rewires the transcriptional programs of gilteritinib-treated AML
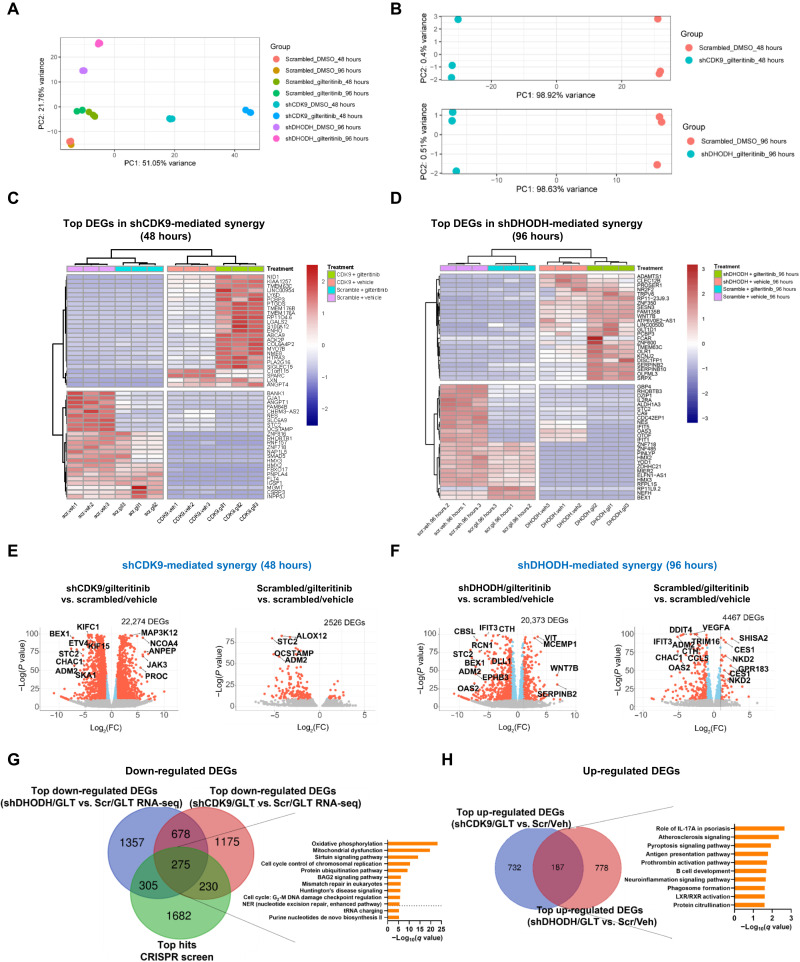
To dissect the underlying mechanism of synergy, we performed RNA-seq on scrambled, shCDK9, and shDHODH-stable MOLM-13 cells treated with a sublethal concentration of gilteritinib (8 nM) for 48 and 96 hours. To provide evidence that the assays were performed at time points where the expression of target gene of interest was sufficiently altered but before any evidence of significant cell death, kinetics of cell death [annexin/PI (Propidium Iodide)] and target gene expressions (GMPS) at different time points (e.g., 24, 48, and 96 hours after treatments) were shown. At the 48-hour time point for shCDK9 condition and 96-hour time point for shDHODH condition, the expression of purine biosynthesis gene, GMPS, was sufficiently reduced but before significant cell death (fig. S3, A to C). Therefore, 48 and 96 hours were selected as the time points for mechanistic studies. After removal of the low abundant genes and normalization, principal components analysis (PCA) plots highlighted distinctive transcriptional features of the combination treatments with respect to single agent–treated samples (Fig. 3, A and B). On the basis of heatmaps derived from hierarchical clustering, the transcriptional patterns of the top differentially expressed genes (DEGs) in shCDK9/gilteritinib and shDHODH/gilteritinib combination groups showed stark differences in expression directionality when compared to their respective controls (Fig. 3, C and D). As depicted by volcano plots, 48- and 96-hour gilteritinib treatment alone yielded 2526 and 4467 DEGs, respectively (Fig. 3, E and F, right). shCDK9/gilteritinib and shDHODH/gilteritinib combinations increased the numbers of DEGs to 22,274 and 20,373, respectively (Fig. 3, E and F, left). In comparison to shCDK9, shDHODH, or gilteritinib treatment alone, shCDK9/gilteritinib or shDHODH/gilteritinib combined treatment also differentially modulated the expressions of a number of genes, suggesting that DHODH or CDK9 knockdown orchestrates drastic transcriptome alterations in gilteritinib-treated AML cells.
Fig. 3. RNA-seq analysis reveals distinct transcriptional signatures conferred by CDK9 or DHODH inactivation in combination with gilteritinib treatment.
(A) PCA of transcriptomes of all combination and single arms over replicates. (B) Top: PCA plots of three biological replicates of gilteritinib-treated shCDK9-stable, and 48-hour vehicle-treated scrambled shRNA-stable cells. Bottom: PCA plots of three biological replicates of gilteritinib-treated shDHODH-stable and 96-hour vehicle-treated scrambled shRNA-stable cells. (C and D) Heatmap representations of normalized read counts of the top 25 down-regulated and the top 25 up-regulated DEGs in shCDK9/gilteritinib and shDHODH/gilteritnib combination treatments. Different treatment groups are color-coded (purple: scrambled + vehicle; cyan: scrambled + gilteritinib; pink: shCDK9 + vehicle or shDHODH + vehicle; and green: shCDK9 + gilteritinib or shDHODH + gilteritinib). (E and F) Volcano plots of selected treatment groups with respect to the corresponding scrambled/vehicle controls. Significantly down-regulated and up-regulated DEGs are highlighted in red. FC, fold change. (G) Venn diagram showing the overlaps among the top coessential genes in CRISPR screen negative selection, the top down-regulated DEGs in shCDK9/gilteritinib combination RNA-seq, and the top down-regulated DEGs in shDHODH/gilteritinib combination RNA-seq (CRISPR screen: FDR < 0.25; RNA-seq: Padj < 0.05). Pathway enrichment analysis of overlapped genes in three datasets is shown. (H) Up-regulated DEGs and enriched pathways shared by shCDK9/gilteritinib combination RNA-seq and shDHODH/gilteritinib combination RNA-seq. GLT, gilteritinib; Veh, vehicle; Scr, scrambled.
An overlay of the top depleted hits from the CRISPR screen and the top down-regulated DEGs from RNA-seq is shown in Fig. 3G. A total of 810 top hits from CRISPR screen negative selection were also differentially down-regulated in shCDK9/gilteritinib (CDK9 knockdown effect; 580 including 305 and 275 genes) or shDHODH/gilteritinib (DHODH knockdown effect; 505 including 230 and 275 genes) combination datasets, respectively, while 275 genes were shared by all three datasets (data file S3). Pathway analysis of the 275 overlapping genes demonstrates alterations in OXPHOS, purine de novo biosynthesis, and cell cycle control of chromosomal replication pathways, suggesting that the synergistic effect of knockout of CDK9 or DHDOH and FLT3 inhibition is dependent on functional suppression of these pathways (data file S4) (Fig. 3G). Among all differentially up-regulated genes, 187 were shared by shCDK9/gilteritinib RNA-seq and shDHODH/gilteritinib RNA-seq datasets (Fig. 3H). These shared DEGs were enriched in the role of interleukin-17A in psoriasis and pyroptosis pathways. Notably, PRMT5 transcripts were reduced in both shCDK9/gilteritinib and shDHODH/gilteritinib DEG datasets (data files S5 and S6). PRMT5 is a methyltransferase, which methylates histone 4 arginine residue and transcriptionally activates FLT3 (16). Although PRMT5 was not one of the highly ranked hits of the study, it was identified as a synergistic hit in our screen (Fig. 1, B and D). These data suggest that regulation of PRMT5 transcription may be a common downstream mechanism of actions of both shCDK9/gilteritinib and shDHODH/gilteritinib treatments. To study whether down-regulation of PRMT5 itself may be synergistic with gilteritinib, MOLM-13 cells were transduced with shRNA-targeting PRMT5, and knockdown efficiencies were confirmed (fig. S4A). Upon 120-hour treatment, cells with knockdown of PRMT5 had a significantly lower IC50 of gilteritinib compared with scrambled or the parental controls (fig. S4B). In the presence of gilteritinib, shPRMT5 cells had a considerably slower growth kinetics in comparison to scrambled cell line (fig. S4C). Last, knockdowns of PRMT5 elevated the percentages of necroptotic/apoptotic cells by twofold compared to scrambled control cells in response to gilteritinib treatment (fig. S4D).
shCDK9 or shDHODH confers sensitivity to gilteritinib treatment by down-regulating metabolic and proliferation pathways
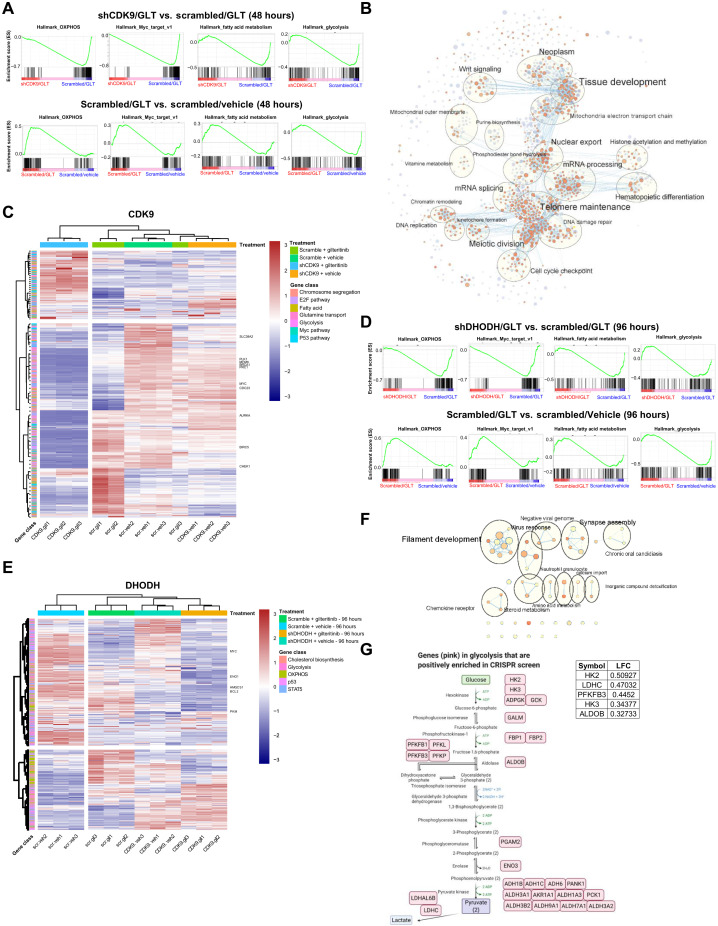
To facilitate comparative examination of pathway modulations by the different treatments, fully annotated Gene Set Enrichment Analysis (GSEA) hallmark gene sets for each comparison were analyzed by unsupervised hierarchical clustering on the basis of their normalized enrichment scores (fig. S5, A and B). As a single agent, gilteritinib down-regulated genes in glycolysis, Wnt signaling (17), and Kras signaling while concomitantly activated OXPHOS, Myc pathway, and fatty acid metabolism (Fig. 4A and figs. S5, A and B, S6, and S7). On the other hand, combination treatments substantially altered the landscapes of enriched gene sets in comparison with gilteritinib or gene-targeting shRNA alone, implying that drug combination profoundly disturbed the directionality of gene expression and pathway activities.
Fig. 4. Genetic inhibition of CDK9 or DHODH in combination with gilteritinib alters multiple signature pathways.
(A) GSEA plots of representative significantly down-regulated and up-regulated pathways in hallmark gene sets for shCDK9/gilteritinib versus scrambled/gilteritinib and scrambled/gilteritinib versus scrambled/vehicle comparisons. Combination treatment suppresses these pathways that are activated by gilteritinib treatment alone. (B) Cytoscape enrichment map of the top gene programs in shCDK9/gilteritinib combination. Enriched GSEA gene sets are predicted with Enrichment Map in Cytoscape and depicted by orange and purple nodes, where purple nodes represent significantly up-regulated pathways in combination treatment and orange nodes represent significantly down-regulated pathways in combination treatment. Node size is proportional to the number of genes in each node, line thickness indicates the overlap of genes between nodes, and the theme of genes in each cluster is specified. Clustered gene programs are labeled. (C) Heatmap showing normalized read counts of genes in selected top enriched pathways predicted by GSEA across different treatment groups of shCDK9-mediated synergy. Selected genes are labeled. The hierarchical clustering of genes and samples was performed with Euclidean distance matrix and Ward’s clustering method. (D) GSEA plots of representative significantly down-regulated and up-regulated pathways in hallmark gene sets for shDHODH/gilteritinib versus scrambled/vehicle and scrambled/gilteritinib versus scrambled/vehicle comparisons. Combination treatment suppresses these pathways that are activated by gilteritinib alone. (E) Heatmap showing normalized read counts of genes in selected top enriched pathways predicted by GSEA across different treatment groups of shDHODH-mediated synergy. Selected genes are labeled. The hierarchical clustering of genes and samples was performed with Euclidean distance matrix and Ward’s clustering method. (F) Cytoscape enrichment map of the top gene programs in shDHODH/gilteritinib combination. (G) Top glycolysis-related hits in positive selection of CRISPR screen upon gilteritinib treatment are highlighted in glycolysis pathway. Log(fold change) values of the top five hits are listed.
To gain insights into signaling network dynamics, GSEA C5 Oncology gene set and IPA analyses were conducted. In the shCDK9/gilteritinib group, both analyses robustly predicted cell division–related processes, such as kinetochore metaphase pathway, mitotic roles of polo-like kinase, DNA replication, and chromosome segregation, as top affected pathways (fig. S8, A and B), echoing the observed large fold depletion of the abundance of gene transcripts in mitosis and kinetochore formation, including SKA1, KIFC1, and KIF15 (Fig. 3E). The DNA replication, kinetochore formation, and mitochondrial electron transport chain pathways were also identified when gene sets involved in the synergistic effect of shCDK9 and gilteritinib were functionally categorized with Cytoscape enrichment maps (Fig. 4B and fig. S8C). Heatmap analysis of transcripts in the top GSEA gene sets demonstrated that the transcriptional profile of combination therapy is considerably different from those of monotherapies or vehicle control (Fig. 4C). These findings may suggest that shCDK9/gilteritinib combination treatments predominantly blunt mitosis and cell cycle progression in addition to metabolic rewiring.
In shDHODH/gilteritinib group, we found that the gilteritinib-induced OXPHOS, fatty acid metabolism, and Myc pathways were among the top pathways down-regulated by shDHODH/gilteritinib treatment (Fig. 4D and figs. S5B and S7). Cholesterols and steroids are produced from the mevalonate pathway (18). The steroid metabolic process was found enriched by IPA analysis and Cytoscape analysis (Fig. 4, E and F; and fig. S8, D to F). Leading-edge analysis of the GSEA cholesterol homeostasis gene set highlighted genes that were strongly down-regulated in combination-treated cells, including CBS, ALDOC, and HMGCS1 (fig. S7). This finding provides strong evidence that shDHODH and gilteritinib cooperatively weaken steroid biosynthesis by suppressing the mevalonate pathway.
There are some common features altered by both shDHODH/gilteritinib and shCDK9/gilteritinib combinations. For instance, the expression of key OXPHOS-related genes (FH, which encodes fumarase, and SDHA, which encodes succinate dehydrogenase) and Myc pathway–related genes (PLK1, PLK4, and Myc) was up-regulated by gilteritinib treatment yet markedly reduced by both shCDK9/gilteritinib and shDHODH/gilteritinib combination treatments (Fig. 4, A and D; and figs. S5, A and B, S6, and S7).
Together, genetic deletion of CDK9 or DHODH sensitizes AML cells to gilteritinib treatment by transcriptionally suppressing Myc pathway, OXPHOS, and related biosynthetic metabolism. A previous study suggested that depletion of HK2, a hexokinase isoform highly expressed in cancer, elevates OXPHOS, sensitizing tumor cells to cell death mediated by growth factor deprivation (19). In line with this, sgRNAs for 28 genes in glycolysis (such as HK2, HK3, and PFKFB3) tended to be positively enriched in CRISPR screen of cell fitness to gilteritinib (although changes did not reach statistical significance) (Fig. 4G and table S1) (20), implying that silencing these genes may provide survival benefit for gilteritinib-treated cells, which are adapted to OXPHOS and biosynthetic metabolism.
The synergistic interactions of gilteritinib and ablation of coessential genes converge on inactivation of purine biosynthesis
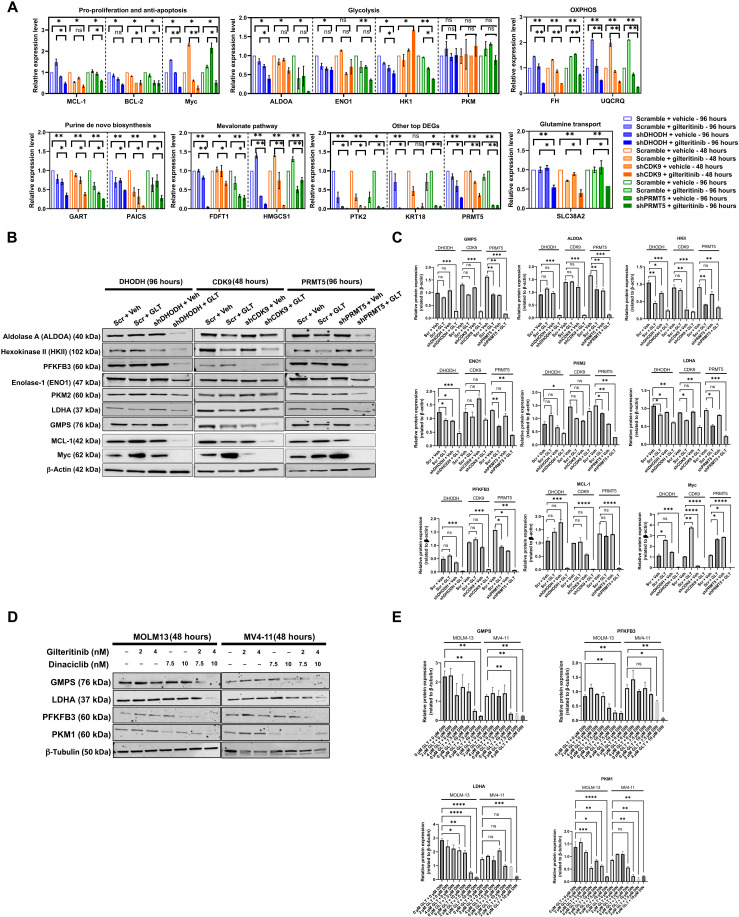
To further understand the mechanism of synergy, we measured the expression of genes involved in apoptotic and mitochondria-related metabolic pathways shared by the CRISPR screen, shCDK9/gilteritinib RNA-seq and shDHODH/gilteritinib RNA-seq datasets by real-time PCR (Fig. 3G). The expressions of MCL-1, BCL-2, and Myc were notably attenuated by shRNA-mediated knockdown of CDK9, DHODH, or PRMT5, suggesting that CDK9, DHODH, and PRMT5 promote anti-apoptotic signaling (Fig. 5A). De novo biosynthesis, mevalonate pathway, and OXPHOS are the primary mitochondria-associated metabolic pathways (21). Hallmarks of these pathways—phosphoribosylaminoimidazole carboxylase (PAICS), phosphoribosylglycinamide formyltransferase (GART), farnesyl-diphosphate farnesyltransferase 1 (FDFT1), hydroxymethylglutaryl-CoA synthase (HMGCS1), fumarase (FH), and ubiquinol-cytochrome C reductase complex III subunit VII (UQCRQ)—consistently underwent depletion in diverse combined treatment conditions (Fig. 5A).
Fig. 5. FLT3-ITD inhibition and ablation of identified coessential genes synergistically inhibit the expressions of anti-apoptotic/pro-proliferative genes and cause metabolic rewiring.
(A) The relative expressions of selected genes in pro-proliferation/anti-apoptosis, OXPHOS, purine de novo biosynthesis, mevalonate metabolism, glycolysis, and glutamine transport pathways and PTK2, KRT18, and PRMT5 in scrambled, shCDK9-, shPRMT5-, and shDHODH-stable MOLM-13 in response to vehicle or 8 nM gilteritinib treatment were measure by real-time PCR with respect to GAPDH. Values are expressed as fold changes (means ± SEM, n = 3) relative to vehicle-treated scrambled cells. *P < 0.05 and **P < 0.01. Treatment durations are indicated. (B and C) Relative expressions of selected metabolic genes in scrambled, shCDK9, shPRMT5, and shDHODH-stable MOLM-13 in response to vehicle or 8 nM gilteritnib treatment at protein levels as revealed by Western blotting. β-Actin serves as loading control. Results are representative of duplicates. Densitometric quantification of blotting band intensities was shown in (C). Treatment durations are indicated. (D and E) Relative expressions of selected metabolic genes in MOLM-13 and MV4-11 cells treated with gilteritinib and dinaciclib at different dose combinations at protein levels as revealed by Western blotting. β-Tubulin serves as loading control. Results are representative of duplicates. Densitometric quantification of blotting band intensities was shown in (E). Results are shown as means ± SEM. * P < 0.01, **P < 0.05, ***P < 0.0001, and ****P < 0.00001. DIN, dinaciclib.
Notably, knockdown of CDK9, DHODH, or PRMT5 significantly decreased SLC38A2 expression in gilteritinib-treated MOLM-13 cells while having negligible effects in untreated cells (Fig. 5A). Previous reports showed that glutamine metabolism, via its capacity of supporting both mitochondria function and cell redox metabolism, is a metabolic dependency of FLT3-ITD AML (22). In agreement with this, the glutaminase inhibitor, telaglenastat, shows synergistic cytotoxic effect with midostaurin or gilteritinib on AML cells (fig. S9). FLT3-ITD inhibition reduces glutaminolysis by blocking glutamine influx through SLC1A5, the primary glutamine transporter (22, 23). SLC38A2 can serve as a redundant glutamine transporter to compensate the deficiency of SLC1A5 (24). Therefore, our results suggest a mechanism of action where simultaneous inhibition of FLT3-ITD and co-essential targets (CDK9, PRMT5, and DHODH) can induce AML cell starvation due to the elimination of SLC1A5- and SLC38A2-mediated amino acid transport.
At the protein level, knockdown of CDK9, DHODH, or PRMT5 consistently decreased the expressions of GMPS (guanine monophosphate synthase), aldolase A, LDHA (Lactate Dehydrogenase A), and PFKFB3 (6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3) in gilteritinib-treated cells compared with scrambled control (Fig. 5, B and C). The expression of hexokinase II was predominantly down-regulated by gilteritinib treatment alone. Knockdown of CDK9 did not significantly alter the expressions of enolase-1 and pyruvate kinase 2 (PKM2) in cells being exposed to vehicle or gilteritinib (Fig. 5, B and C). At protein levels, shCDK9 alone or in combination with gilteritinib also abrogated the expressions of MCL-1 and Myc, suggesting an on-target effect of shCDK9 (Fig. 5, B and C). Combined treatment of dinaciclib and gilteritinib reduced GMPS, LDHA, PFKFB3, and PKM1 expression in a dose-dependent manner, phenocopying the genetic knockout effect (Fig. 5, D and E).
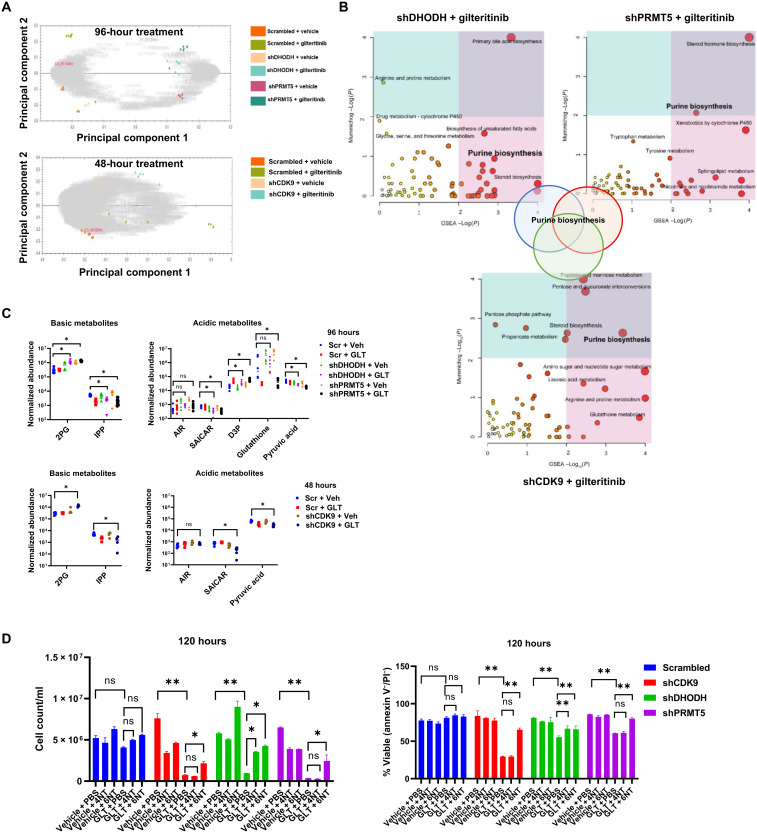
We further dissected the metabolic rewiring associated with three combination treatments by using metabolomic profiling. PCA of metabolites and heatmap analysis of the different treatment groups reveal that cells that received combined treatment exhibited distinct metabolic profiles (Fig. 6A and fig. S10, A to D). GSEA and Mummichog pathway analysis revealed that combined treatments led to negative enrichment of purine biosynthesis, amino acid metabolism, bile acid biosynthesis, and biosynthesis of unsaturated fatty acids (Fig. 6B). Notably, among the top pathways, purine biosynthesis was commonly down-regulated by all three combined treatments (Fig. 6B). Bioanalyzer results also corroborated metabolomics analysis, suggesting that gilteritinib drastically decreased glucose uptake and lactate production (fig. S10E). In agreement with previous report, knockdown of DHODH eliminated metabolites in pyrimidine biosynthesis pathway (fig. S10F) (25). In response to combined treatments, metabolic intermediates in glycolysis, 2-phospho-d-glyceric acid and d-glyceraldehyde 3-phosphate, were accumulated, and the final product pyruvic acid was concurrently depleted (Fig. 6C), echoing the down-regulation of key enzyme expression (aldolase and enolase) as revealed by RNA-seq. Combination treatments also resulted in remarkable reduction in the abundance of mevalonate pathway metabolites, isopentenyl pyrophosphate, which may arrest downstream cholesterol and steroid synthesis reaction. Our RNA-seq results indicated that purine metabolism gene, PAICS, was down-regulated by combination treatments. In agreement with this, we observed a reduction in SAICAR levels, which is the product of PAICS-mediated catalysis, in response to all three combined treatments. To further evaluate the roles of pyrimidine and purine biosynthesis in these synergistic interactions, nucleoside rescue experiments were conducted. A six-nucleoside cocktail (adenosine/guanosine/uridine/inosine/cytosine/thymidine, 20 μM each) rescued shCDK9/gilteritinib-, shDHODH/gilteritinib-, and shPRMT5/gilteritinib-mediated cell proliferation defect and cell apoptosis (Fig. 6D). A four-nucleoside cocktail (uridine/inosine/cytosine/thymidine) only partially rescued shDHODH/gilteritinib-mediated reduction in cell proliferation. This provides evidence that the synergistic anti-AML effects mediated by combined treatments of gilteritinib and CDK9 inhibitor (CDK9i), DHODHi, or PRMT5i are primarily attributed to nucleotide deficiency.
Fig. 6. FLT3-ITD inhibition and ablation of identified coessential genes synergistically inhibit purine biosynthesis.
(A) PCA of metabolomics datasets of all combination and single groups over replicates. (B) Top enriched metabolic pathways as predicted by Mummichog analysis and GSEA analysis. The size of the circle is correlated with the amounts of metabolites being identified in the pathway. Three combined treatments share purine biosynthesis pathways. (C) Normalized abundances of selected basic and acidic metabolites across different treatment groups. The normalization is performed by multiplying compound ion abundance with scalar factor calculated with a median and mean deviation approach based on all the detected abundance. *P < 0.05. AIR, aminoimidazole ribotide; SAICAR, phosphoribosylaminoimidazolesuccinocarboxamide; D3P, d-glycerol 3-phosphate; 2PG, 2-phosphoglycerate; IPP, isopentenyl diphosphate. (D) Six NT (nucleosides: A/T/C/G/U/I) cocktail but not four NT (nucleotides: T/C/U/I) cocktail protected shCDK9-, shDHODH-, or shPRMT5-stable MOLM-13 cells from gilteritinib-induced cell growth defects and apoptosis (measured by annexin V/PI staining) at 120 hours. *P < 0.05 and **P < 0.01. Values are expressed as means ± SEM from triplicates.
Pharmacologic inhibition confirms synergy of several targets with gilteritinib
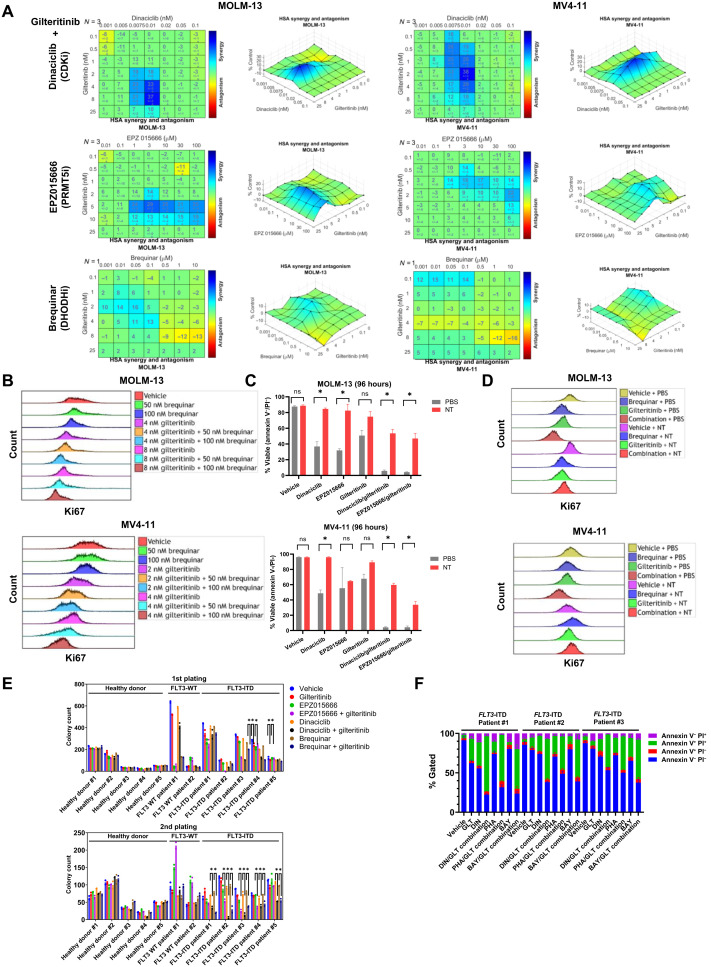
Next, we determined whether pharmacologic inhibition of CDK9, DHODH, or PRMT5 using commercially available inhibitors [brequinar for DHODH, dinaciclib, P276-00, fadraciclib (CYC065), PHA-767491, BAY-1143572 (atuveciclib) and NVP-2 for CDK9, and EPZ015666 and PRT808 for PRMT5] is synergistic with gilteritinib in AML cells. For MOLM-13 cells, synergy ranges were determined to be 2 to 10 nM gilteritinib with 0.0075 to 0.01 nM dinaciclib or 1 to 100 μM EPZ015666 (the synergy scores: dinaciclib/gilteritinib = 37; EPZ015666/gilteritinib = 29) (Fig. 7A). For MV4-11 cells, maximum synergy was observed at 0.1 to 8 nM gilteritinib in combination with 0.0075 to 0.01 nM dinaciclib or 3 to 100 μM EPZ015666 (the synergy scores: dinaciclib/gilteritinib = 38; EPZ015666/gilteritinib = 22) (Fig. 7A). P276-00, PHA-767491, BAY-1143572, or NVP-2 in combination with gilteritinib exhibited synergistic effect on both MOLM-13 and MV4-11 with variable maximum synergy scores (MOLM-13: P276-00/gilteritinib = 19, PHA-767491/gilteritinib = 25, BAY-1143572/gilteritinib = 22 and NVP-2/gilteritinib = 31; MV4-11: P276-00/gilteritinib = 19, PHA-767491/gilteritinib = 26, BAY-1143572/gilteritinib = 25, and NVP-2/gilteritinib = 21) (fig. S11, A to C). The pairwise dose combination of fadraciclib and gilteritinib lacked distinguishable synergistic dose pairs, possibly due to fadraciclib’s lower potency against CDK9 (fig. S11, A and B). Another PRMT5-specific inhibitor, PRT808, and gilteritinib produced the highest synergy scores of 13 in MV4-11 and 34 in MOLM-13, respectively (fig. S11D). Synergy ranges for each drug are physiologically achievable and represent promising opportunities for future combination treatments (26, 27).
Fig. 7. In vitro pharmacologic validation of synthetic lethal targets with gilteritinib.
(A) Synergistic effect of pairwise dose combinations of gilteritinib and dinaciclib, EPZ015666 or brequinar on MOLM-13 and MV4-11 cells. For dinaciclib, cells were treated for 2 hours with the drug before it was washed off, and all other drugs were incubated with cells for 96 hours. Cell viability was measured with MTS. HSA analysis was used to determine regions of synergy. (B) Brequinar and gilteritinib synergistically suppress cell proliferation. MOLM-13 and MV4-11 cells were treated with vehicle, 50 nM or 100 nM brequinar in combination with vehicle, 4 or 8 nM gilteritinib for 96 hours before cells were fixed and permeabilized for intracellular BV421–anti-Ki67 staining and flow cytometry analysis. (C and D) Nucleoside cocktail rescued drug combination–induced apoptosis and proliferation deficiencies of MOLM-13 and MV4-11 cells at 96 hours. Cell apoptosis and proliferation were measured with annexin V/PI staining and BV421–anti-Ki67 staining, respectively. The percentage of viable cells is expressed as means ± SEM. Results are representative of triplicates. *P < 0.05. NT, nucleoside cocktail consisting of adenosine, uridine, inosine, cytosine, guanosine, and thymidine (20 μM each). (E) Gilteritinib in combination with either EPZ015666, dinaciclib, or brequinar eliminate self-renewal potentials of FLT3-ITD primary AML cells while sparing healthy hematopoietic stem cells. Results are shown as means of two biological replicates. *P < 0.05 in comparison with single agents. (F) Synergistic effect of gilteritinib in combination with CDK9 selective inhibitors (DIN, dinaciclib; PHA, PHA-767491; BAY, BAY-1143572) on apoptosis of FLT3-ITD primary AML cells. Primary cells were treated with vehicle, single agents, or drug combinations for 96 hours with stroma coculture before AML cell apoptosis was measured with annexin V/PI staining. Results are shown as means ± SEM of three biological replicates.
Brequinar and gilteritinib combination showed modest synergy with regard to MOLM-13 and MV4-11 viability (the synergy scores of brequinar/gilteritinib: MOLM-13 = 16 and MV4-11 = 15) (Fig. 7A). Instead, brequinar and gilteritinib synergistically suppressed cell proliferation as indicated by decreased Ki67 staining (Fig. 7B). A six-nucleoside cocktail supplementation rescued the synergistic proapoptotic effect of dinaciclib/gilteritinib and EPZ015666/gilteritinib on MOLM-13 and MV4-11 cells (Fig. 7C). In the presence of pyrimidine and purine nucleosides, the combination of brequinar and gilteritinib could no longer induce cell proliferation deficiency for both MOLM-13 and MV4-11 (Fig. 7D).
To determine the effect of diverse inhibitor combinations on self-renewal of primary cells, colony-forming unit (CFU) assays were conducted on primary cells. Bone marrow cells from five FLT3-ITD patients, two FLT3–wild-type (WT) patients, and five healthy donors (the mutations and cytogenetics for each AML patient that are summarized in Table 1) were seeded into semisolid media in the presence of vehicle, 8 nM gilteritinib, 100 μM EPZ015666, 0.1 nM dinaciclib, 100 nM brequinar, the combination of gilteritinib and EPZ015666, the combination of gilteritinib and dinaciclib, or the combination of gilteritinib and brequinar. At the first plating, only FLT3-ITD patients #3 and #4 grew significantly fewer colonies in response to combination therapies (Fig. 7E). However, at the secondary plating, combination treatments robustly abolished colony formation of cells of all FLT3-ITD patients but not cells of FLT3-WT patients. Given that serial replating is a functional assay for leukemia stem and progenitor cells, we measured the frequencies of CD34+CD38− leukemia stem-like cells after CFU first plating. The combination of gilteritinib and dinaciclib or brequinar significantly decreased the frequencies of CD34+CD38− leukemia stem-like cells derived from all FLT3-ITD patients (fig. S12). The reduction in frequencies of CD34+CD38− leukemia stem-like cells was observed in three of five FLT3-ITD patients, echoing the colony-forming capacities regulated by treatments. Conversely, these combination treatments did not affect CD34+CD38− cells of FLT3-WT patients. Combination treatments did not lead to significant alterations in colony-forming properties of CD34+ healthy hematopoietic cells in primary and secondary platings, implying that the on-target cytotoxic effect is specific for AML cells. Similar to MLL-rearranged cell lines, non–MLL-rearranged FLT3-ITD patient cells were vulnerable to FLT3i/CDK9i combination treatment: gilteritinib and CDK9 inhibitors (dinaciclib, PHA-767491, and BAY-1143572) synergistically induced cell death by overcoming stroma protection (Fig. 7F).
Table 1. Mutation status of primary AML samples.
VAF, variant allele frequency; WT, wild type; N/A, not applicable.
| Patient no. | Mutation status | Cytogenetics | FLT3-ITD VAF | Age | Gender | Prior therapy |
| FLT3-ITD patient #1 |
DNMT3A/NPM1/ NRAS/TET2/FLT3-ITD |
46,XY[19]/ nonclonal[1] |
10.7% | 63 | Male | None |
| FLT3-ITD patient #2 |
NPM1/TET2/ FLT3-ITD |
46,XY[20] | 23% | 67 | Male | None |
| FLT3-ITD patient #3 |
NRAS/RUNX1/SRSF2/ TET2/FLT3-ITD |
46,XX[20] | 22.7% | 65 | Female | None |
| FLT3-ITD patient #4 |
NF1/NPM1/PTPN11/ SMC3/FLT3-ITD |
46,XY[19]/4n[1] | 47.6% | 60 | Male | None |
| FLT3-ITD patient #5 |
BIRC6/NPM1/ FLT3-ITD |
Insufficient Metaphases (46,XY[5]) |
43.7% | 68 | Male | None |
| FLT3-WT patient #1 | RUNX1/TET2/SRSF2 | 46,XY[20] | N/A | 79 | Male | None |
| FLT3-WT patient #2 | NPM1/TET2/GATA2 | 46,XY[20] | N/A | 66 | Male | None |
| Healthy donor #1 | N/A | N/A | N/A | 70 | Male | N/A |
| Healthy donor #2 | N/A | N/A | N/A | 60 | Male | N/A |
| Healthy donor #3 | N/A | N/A | N/A | 63 | Female | N/A |
| Healthy donor #4 | N/A | N/A | N/A | 65 | Male | N/A |
| Healthy donor #5 | N/A | N/A | N/A | 66 | Male | N/A |
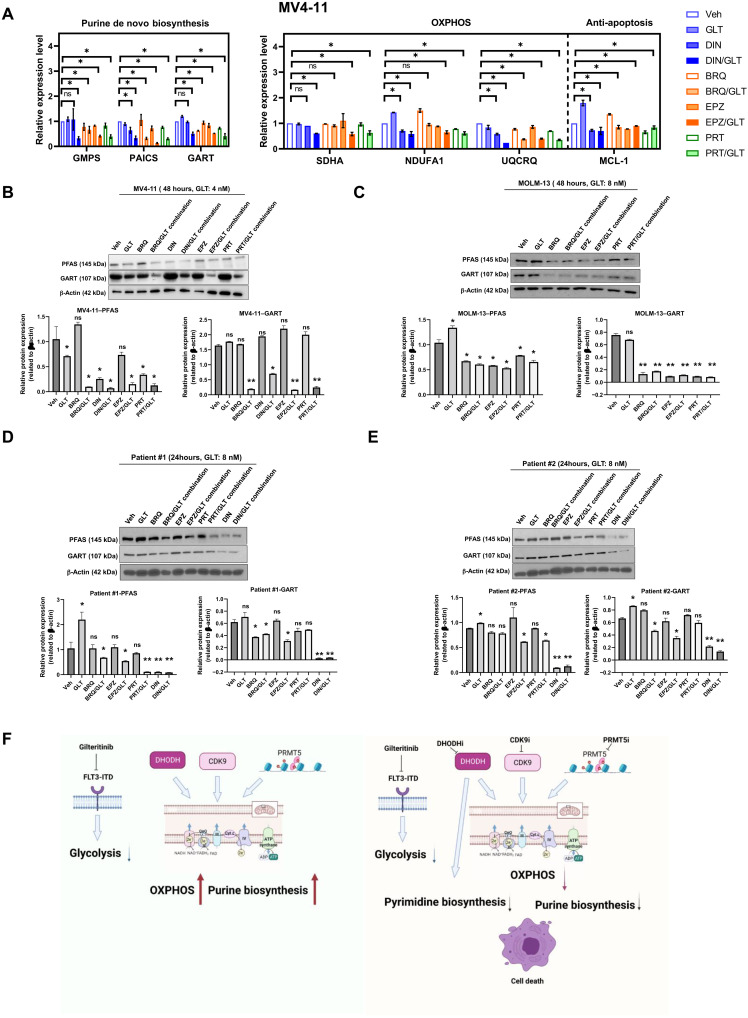
We then measured gene expressions of purine biosynthesis and OXPHOS pathways in cell lines and primary patient cells in response to a variety of treatments. Dinaciclib as a single agent reduced the expression of PAICS, NDUFA1, UQCRQ, and MCL-1 at RNA levels in MV4-11 cells (Fig. 8A). Combination of dinaciclib and gilteritinib resulted in a more robust decrease in GMPS, PACIS, GART, SDHA, NDUFA1, and UQCRQ levels than dinaciclib alone. GMPS, PAICS, GART, SDHA, NDUFA1, and UQCRQ were also drastically reduced by brequinar, EPZ015666, or PRT808 in combination with gilteritinib. At protein levels, brequinar, dinaciclib, EPZ015666, or PRT808 in combination with gilteritinib eliminated purine biosynthesis enzymes, PFAS (Phosphoribosylformylglycinamidine Synthase) and GART, in MV4-11 cells (Fig. 8B). Likewise, brequinar, EPZ015666, or PRT808 in combination with gilteritinib abolished the expressions of these enzymes in MOLM-13 cells (Fig. 8C). Down-regulation of purine biosynthesis by combination treatments was also observed on FLT3-ITD patient samples (Fig. 8, D and E). Brequinar, EPZ015666, PRT808, or dinaciclib in combination with gilteritinib decreased the expressions of PFAS and GART in these patient samples. Therefore, pharmacologic inhibition of CDK9, DHODH, or PRMT5 could recapitulate the effects of the genetic knockdown to confer sensitivity to gilteritinib treatment through purine nucleotide depletion and OXPHOS suppression (Fig. 8F).
Fig. 8. Combination of FLT3-ITD and CDK9, DHODH, or PRMT5 inhibitors attenuated the expressions of components in purine biosynthesis and OXPHOS pathways.
(A) Relative gene expressions in purine biosynthesis and OXPHOS pathways in MV4-11 in response to different treatment conditions as quantified by qPCR. *P < 0.05. (B and C) Expressions of selected metabolic enzymes in MV4-11 and MOLM-13 cells being treated with vehicle, gilteritinib, dinaciclib, brequinar, EPZ015666, or PRT808 at different combinations for 48 hours as revealed by Western blotting. β-Actin serves as loading control. Results are representative of duplicates. Densitometric quantification of blotting band intensities was shown. *P < 0.01 and **P < 0.05. (D and E) Expressions of selected metabolic enzymes and anti-apoptotic proteins in FLT3-ITD primary patient cells being treated with vehicle, gilteritinib, dinaciclib, brequinar, EPZ015666, or PRT808 at different combinations for 24 hours as revealed by Western blotting. β-Actin serves as loading control. Results are representative of duplicates. Densitometric quantification of blotting band intensities was shown. *P < 0.01 and **P < 0.05. BRQ, brequinar; EPZ, EPZ015666; PRT, PRT808. (F) Schematic illustration of the mechanism of CDK9i/FLT3i, DHODHi/FLT3i, and PRMT5i/FLT3i synergism.
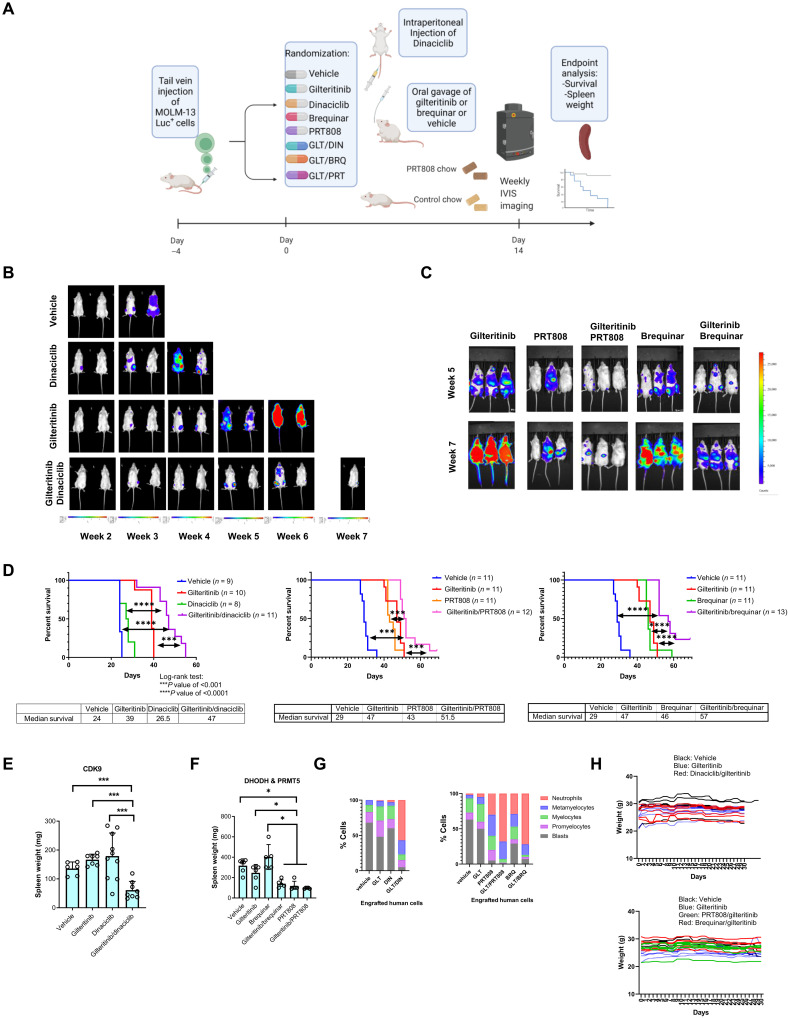
Gilteritinib in combination with dinaciclib, brequinar, or PRT808 prolongs survival in an aggressive xenograft mouse model
To assess the translational relevance of our findings, we evaluated the gilteritinib/dinaciclib and gilteritinib/brequinar synergies as a proof of concept using a MOLM-13-Luc+ engraftment mouse model (Fig. 9A). In addition, we tested the efficacy of a novel PRMT5i, PRT808 (Prelude Therapeutics), which was formulated as chow diet in combination with gilteritinib. At each given time point, AML disease burden in the combination groups was significantly lower than that of the monotherapy or vehicle group (Fig. 9, B and C). As single agents, dinaciclib provided marginal survival advantage to leukemic mice compared with vehicle control. Gilteritinib, brequinar, or PRT808 treatment significantly extended the life spans of mice. In sharp contrast, the combination arms displayed a significant survival benefit relative to vehicle or respective single-agent arms (mean survival time: the first cohort: 24 days for vehicle, 27.5 days for dinaciclib, 39 days for gilteritinib, and 47 days for gilteritinib/dinaciclib combination; the second cohort: 29 days for vehicle + control chow, 47 days for gilteritinib + control chow, 43 days for PRT808 chow, 46 days for brequinar + control chow, 51.5 days for gilteritinib/PRT808 combination, and 57 days for gilteritinib/brequinar combination; P value of <0.001 in comparison with gilteritinib alone) (Fig. 9D). At the end point, a substantial reduction in spleen weight in the combination therapy groups over all other treatment arms was observed (Fig. 9, E and F). The isolated leukemia cells from mice receiving vehicle mostly consist of immature blasts, promyelocytes, and myelocytes (Fig. 9G). Gilteritinib or dinaciclib alone did not lead to enhanced cell differentiation. Cells from mice with brequinar or PRT808 became more differentiated, yielding more granulocytes with segmented nuclei. In sharp contrast, all three combination treatments profoundly altered the cellular composition by producing morphologically differentiated cells, resembling metamyelocytes or neutrophils. The appreciable reduction of mouse weight loss was absent in any combination treatment groups, suggesting favorable tolerability of these combinations (Fig. 9H). These experiments provide strong evidence that the combination therapies of gilteritinib/dinaciclib, gilteritinib/brequinar, and gilteritinib/PRT808 at clinically relevant doses show appealing in vivo efficacy.
Fig. 9. The combination therapies of dinaciclib (CDK9i), PRT808 (PRMT5i) or brequinar (DHODHi), and gilteritinib manifest superior efficacy in a FLT3-ITD AML xenograft model.
(A) NCG mice were engrafted with MOLM-13 cells expressing luciferase and received different treatments. In the first cohort, mice received vehicle, dinaciclib (10 mg/kg) weekly, gilteritinib (30 mg/kg) daily, and dinaciclib/gilteritinib combination. In the second cohort, mice received vehicle, gilteritinib (15 mg/kg) daily, brequinar (25 mg/kg) for a total of six doses, PRT808 (5 mg/kg) chow daily, brequinar/gilteritinib combination, or PRT808/gilteritinib combination. Control high-fat chow was given to all non-PRT808 mouse groups. (B and C) IVIS imaging shows changes in luciferase signal over study periods. (D) Kaplan-Meier curves of the mouse survival times in different treatment groups. The number of mice per group was indicated. ***P < 0.001 and ****P < 0.0001. (E and F) Combination therapy significantly reduced spleen weight of engrafted mice. Statistical significance of differences in spleen weights between groups were estimated using ANOVA methods. P values have been adjusted for multiple comparisons using Holm’s procedure. *P < 0.05 and ***P < 0.001. Mice in (F) received high-fat chow. (G) Morphology scores of cytospins of enriched human cells from mouse spleens in different treatment groups. (H) Weight changes of mice in different treatment groups over 30 days.
DISCUSSION
In this study, we identified CDK9, DHODH, and PRMT5 as novel synergistic lethal partners with gilteritinib in FLT3-ITD AML. We showed that genetic deletion and pharmacological inhibition of these targets sensitize AML cell lines and primary patient samples to gilteritinib treatment. The CDK inhibitor, (dinaciclib), DHODHi (brequinar), or PRMT5i, (PRT808), in combination with gilteritinib, reduces disease burden and provides survival benefits for an AML xenograft mouse model. In addition, gilteritinib-treated AML cells are addicted to OXPHOS rather than anaerobic glycolysis for energy production and biosynthesis reactions. All three synergistic interactions converge on blocking purine de novo synthesis and OXPHOS to trigger AML cell apoptosis (Fig. 8F).
PRMT5, CDK9, and DHODH play different roles in activating proliferation and inhibiting apoptosis. DHODH is the rate limiting enzyme of the de novo pyrimidine synthesis pathway, converting dihydroorotate (DHO) to orotate (11, 25). Inhibition of DHODH induces differentiation of diverse AML subtypes (10). Direct inhibition of DHODH-mediated pyrimidine biosynthesis showed preclinical promise but lacked sufficient clinical benefit at the clinical trial stage (28). PRMT5 catalyzes symmetric demethylation of histone arginine to induce gene silencing (29). PRMT5 also methylates and regulates proteins involved in diverse cellular processes, including transcription, translation, and apoptosis. Overexpression of PRMT5 has been observed in leukemia, being involved in FLT3 gene transcription (16). PRMT5 inhibition has been shown to kill AML cells (30–32). PRMT1 and PRMT5 are both protein arginine methyltransferases that methylate FLT3 but differ in their products: PRMT1 generates ω-NG, NG-asymmetric dimethylarginine, while PRMT5 produces ω-NG, N′G-symmetric dimethylarginine. Zhu et al. (33) found that in acute lymphoblastic leukemia (ALL), PRMT1-mediated methylation of FLT3 is critical to its function as an oncogene. In a patient-derived mouse xenograft model of ALL, midostaurin and PRMT1 inhibition were more efficacious than either monotherapy alone. Given the mechanistic similarities between PRMT1 and PRMT5, this study supports our finding that PRMT5 inhibition enhances the antileukemic activity of gilteritinib. Radzisheuskaya et al. (34) showed that loss of PRMT5 causes changes in alternative splicing of multiple essential genes in AML. This work indicates that PRMT5 inhibition could be synergistic with disruption of essential genes, such as FLT3, in FLT3-ITD cancers.
CDK9 inhibitors down-regulate MCL-1 to induce cell death in AML, overcoming MCL-1–dependent drug resistance (35, 36). In addition, CDK9 inhibition suppresses the expression of relevant MYB target genes including BCL2 and CCNB1 (37). CDK9 inhibitors were also shown to inhibit active phospho-TEFb and the expression of E2F target genes necessary for the G1-S transition, DNA replication, and mitotic activity. Myc, a critical downstream transcriptional target of phospho-TEFb, was shown to be responsible for CDK9-mediated cell proliferation and survival. Inhibition of CDK9 induces MLL apoptosis by down-regulating HOXA9 and Myc (38). In agreement with these observations, our data show that kinetochore mitotic spindle, chromosome remodeling, Myc pathway, and G2-M cell cycle checkpoint gene sets are all significantly down-regulated with CDK9 knockout and that these shCDK9-induced effects are further strengthened by FLT3 inhibition. In support of our findings, a previous study found that loss of FLT3 sensitizes ER-HoxA9 conditionally immortalized murine GMP (granulocyte-macrophage progenitor) cell line to brequinar treatment (39). In addition, in an in vitro study, a combination of PRMT5 inhibition and FLT3i (AC220) resulted in an enhanced inhibition of cell proliferation compared with single agents (32). However, none of these studies provided mechanistic insights into the combination effect.
Previous work on FLT3-ITD has revealed important metabolic dependencies. Cells harboring FLT3-ITD demonstrated a highly glycolytic phenotype and had central carbon metabolism elevated by regulating FOXO activity (40). Our work builds upon the concept that metabolic vulnerabilities can be targeted to overcome anti-FLT3 therapy resistance in FLT3-ITD leukemia by inhibiting the upstream regulators of essential metabolic pathways whose activities are elevated in FLT3i-treated cells. FLT3-ITD leukemias have been shown to be dependent on serine biosynthesis (41) and creatine biosynthesis (42). Targeting each of these pathway dependencies via inhibition of their respective rate-limiting enzymes, phosphoglycerate dehydrogenase and glycine amidinotransferase, produces an antileukemic effect (41, 42). The role of metabolic targeting in treatment for FLT3-ITD AML is further supported by a study, which found that the mitochondrial oxidative consumption rate of FLT3-ITD MOLM-13 cells can be reduced by using pyrvinium that causes an amino acid deprivation–like scenario and elicits metabolic rewiring toward amino acid synthesis, transport, and glutathione metabolism (43). Our study revealed that gilteritinib-treated FLT3-ITD AML cells are addicted to OXPHOS rather than glycolysis for energy production and biosynthesis reactions, which renders these metabolically adapted cells extremely vulnerable to transcriptional silencing of the components of mitochondrial electron transport chain complexes. This is in line with previous reports suggesting that therapy-resistant AML cells increase their mitochondrial mass and have high OXPHOS (44). Our GSEA reveals that glycolysis is predominantly down-regulated by gilteritinib monotherapy. This is supported by previous studies showing that glycolytic enzymes were pronouncedly suppressed by the FLT3i, AC220 (22). Our CRISPR screen analysis shows that knockout of each of 28 glycolytic genes (including HK2) makes cells grow better in response to gilteritinib treatment, implying the development of drug resistance. Upon HK2 depletion, glucose flux to pyruvate and lactate is suppressed, but tricarboxylic acid (TCA) fluxes and OXPHOS are maintained (19). Coupling glycolysis deficiency with elevated OXPHOS promotes leukemia growth. The mechanism of CDK9, DHODH, and PRMT5 controlling OXPHOS remains unknown and warrants further exploration. However, our data suggests that it may be at least partially Myc dependent, because the Myc pathway is activated by gilteritinib treatment, which can act by transcriptionally up-regulating the genes in OXPHOS to provide drug resistance (45, 46). CDK9 and PRMT5 may drive OXPHOS and purine biosynthesis through different mechanisms. The link between CDK9 and activating transcription factor 4 (ATF4) is established in that CDK9-cyclin T1 complex phosphorylates and mediates ATF4 activation (47). It is widely known that increased expression of ATF4 is associated with up-regulated purine biosynthesis and OXPHOS (48–50). Therefore, CDK9 may drive purine biosynthesis and OXPHOS through ATF4 activation, providing resistance to FLT3i. We also found that knockdown of PRMT5 affected the mitochondria-associated metabolic pathways in such a way that leukemic cells were more susceptible to gilteritinib treatment. PRIMT5 is involved in controlling metabolic gene transcriptions, such as lipid pathway, cholesterol synthesis, and glycolysis (51). PRMT5 may also simulate gene expressions in OXPHOS/purine pathway via increases in histone H2 methylation, which enhances chromatin accessibility at gene promoters or silence negative regulators of OXPHOS/purine pathways. Another possibility is that PRMT5 may control the activities of metabolic pathways by modulating ATF4 function. Szewczyk et al.’s (52) transcriptomic analysis of poor prognostic AML unveiled that PRMT5 regulates the ATF4 pathway, which, in turn, controls the expression of genes associated with metabolic processes. In addition, PRMT5 may induce methylation of ATF4, which may in turn modulates the activities of ATF4, leading to enhanced purine biosynthesis (52).
We demonstrated that inhibitions of CDK9, PRMT5, and DHODH in combination with gilteritinib treatment converge on inhibition of the purine biosynthesis and purine supplementation protects AML cells from apoptotic effect of combination treatments. This is consistent with literature showing that blocking the purine de novo synthesis inhibits AML growth (53). It was also reported that combination of the OXPHOS inhibitor IACS-010759 with the FLT3i AC220 synergistically reduces glucose and glutamine enrichment, leading to impaired energy production and nucleotide synthesis (54). Joshi et al. (55) studied the stepwise evolution of gilteritinib resistance using RNA-seq, metabolomics, and proteomics. They found that early gilteritinib resistance is dependent on NRAS signaling, metabolic reprograming, and Aurora kinase A. They found that a 48-hour gilteritinib treatment rapidly decreased central energy metabolism and altered glycerophospholipid metabolism. In addition, the long-term presence of gilteritinib blocked sphingolipid metabolism and carnitine/fatty acid metabolism. Our own RNA-seq data suggested that cells treated with gilteritinib increased fatty acid metabolism by up-regulating CPT1A transcription. shCDK9 and shDHODH down-regulated the expression of genes in fatty acid pathway. Our metabolomics data also indicated that glutathione and steroid biosynthesis metabolisms were altered by our combination treatments. Notably, several nucleotide levels were decreased in early resistant cells (16). This is consistent with our findings that purine biosynthesis blockade is synergistic with gilteritinib. In agreement with our RNA-seq and metabolomics data, Gallipoli et al. (22) showed that glucose uptake was almost completely blocked upon FLT3 inhibition via AC220, glutamine uptake was only modestly decreased by 48-hour treatment. In addition, they revealed that AC220 treatment resulted in a more pronounced reduction of glycolysis gene LDHA than glutamine metabolism gene GLS. Our Western blotting result corroborated the findings of reduction of LDHA by FLT3i treatment. Our RNA-seq results suggested that GLS was reduced in the presence of the combination of shDHODH and gilteritinib.
Glutaminolysis, which is primarily mediated by glutaminase GLS (Glutaminase) and transporters (SLC1A5 and SLC38A2), provides metabolites to replenish the TCA cycle intermediates. AML cells shunt carbon from glutaminolysis into citrate, feeding de novo fatty acid biosynthesis in the mitochondria and providing lipids for proliferating AML cells (56). In addition, glutamine levels control OXPHOS in AML (57). Up-regulated glutamine metabolism was shown to mediate resistance to FLT3i therapies (22), and the use of a GLS inhibitor in combination with either an FLT3i or a BCL-2 inhibitor effectively eliminates AML cells (23, 56, 57). Our findings suggest that SLC38A2 expression can be down-regulated by gilteritinib in combination with inhibitors of CDK9, PRMT5, and DHODH, highlighting the metabolic plasticity of AML. SLC38A2 ranked no. 6 of 19,115 total hits in a CRISPR screen with venetoclax in AML (58), suggesting that its inactivation is synthetically lethal with BLC-2 inhibition. Therefore, it is likely that any therapies that concurrently induce glutaminolysis inhibition and BCL-2 down-regulation may enhance gilteritinib sensitivity by suppressing OXPHOS activity.
It should be noted that the top hits other than CDK9, DHODH, PRMT5, and CDK7 were not validated because of low sgRNA representation and inaccessibility of specific inhibitors. Therefore, it may worth further validations of other hits in the future studies. In addition, although we mechanistically identified purine biosynthesis and OXPHOS as the converged pathways shared by all synergistic interactions in in vitro systems, cell lines and isolated primary patient cells may not precisely reflect the complexity of tumor microenvironment responses to inhibitors in patients. Last, because of restriction of accessibility of cell lines, we only tested FLT3-ITD cell lines with MLL rearrangement, although our data in patient’s derived FLT3-ITD cells without MLL rearrangement showed similar results to the MLL-positive cell lines. It is unknown whether AML cells with other abnormalities will respond differently to these combination treatments. All these argue for cautions being taken to interpret the data. Besides dinaciclib, there are other more CDK9 selective inhibitors available, such as NVP-2 (3). NVP-2 is a highly selective aminopyrimidine-derived CDK9 inhibitor, developed by Novartis. In vitro IC50 of NVP-2 against CDK9 was <0.5 nM as compared to 584 nM(CDK1), 706 nM (CDK2), 1050 nM (CDK5), and >10 μM (CDK7). As has been observed with other CDK9 inhibitors, treatment with 10 mg/kg of NVP-2 produced a reduction in white blood cell counts, including neutrophils (13, 25), which was detectable after 3 weeks. Although NVP-2 has demonstrated tremendous preclinical potentials, no clinical studies have been reported. In the current study, we showed that NVP-2 sensitizes AML cell lines to gilteritinib treatment. Dinaciclib inhibited CDKs 1, 2, 5, and 9 with respective in vivo IC50 of 3, 1, 1, and 4 nM. It has been reported to trigger apoptosis and arrest cell growth. On-target CDK9 inhibition by dinaciclib has been shown to play major roles in in vivo efficacy on Eμ-Myc lymphomas (4). This is evident that dinaciclib treatment caused reduction in p-Rab1 and total MCL-1. We did not assess whether the observed in vivo activity of dinaciclib was completely due to on-target CDK9 inhibition, given that dinaciclib has affinities for CDK1, CDK2, and CDK5. It was reported that dinaciclib suppressed the expression of CDK2/CDK5/CDK9 in an in vivo model of cholangiocarcinoma. In addition, it was documented that simultaneous inhibitions of CDK2 and CDK9 block neuroblastoma development in vivo (5). Therefore, we cannot exclude the possibility that the synergistic in vivo elimination of leukemia by dinaciclib and gilteritinib combination was at least partially mediated by CDK1, CDK2, or CDK5 inhibition. Further studies are necessary to dissect the in vivo targets for dinaciclib in AML treatment. A cell line engraftment model with inducible shRNA-mediated CDK9 knockdown would provide insight into the in vivo efficacy of this combination in the future. The effect of different treatment strategies on dynamic metabolite flux into different pathways, mitochondrial functions, and glycolysis warrants further exploration with C13-glucose isotope tracing experiments and Seahorse technology.
In summary, our study shows that the identified synthetic lethal targets, CDK9, DHODH, and PRMT5, have high translational values in FLT3-ITD AML, because there are clinical grade inhibitors available for these targets to ensure rapid clinical applications. Inhibition of CDK9, DHODH, or PRMT5 causes metabolic reprograming and potentiates gilteritinib sensitivity in AML. Alongside their own independent activities, these target-mediated synergistic interactions share common vulnerabilities, namely, OXPHOS and purine biosynthesis. The discovery of these shared pathways adds another dimension to dissection of genetic interactions for FLT3i sensitivity. Our findings provide a rational for development of combined treatments involving CDK9, PRMT5, DHODH, or OXPHOS inhibitors to improve efficacy of FLT3is, thereby maximizing the alternative strategies for overcoming resistance associated with long-term single-treatment approaches.
MATERIALS AND METHODS
Experimental design
The overall goal of our study was to develop therapeutic strategies to enhance gilteritinib sensitivity. We conducted a whole-genome CRISPR screen to identify druggable targets that, upon inhibition, conferred sensitivity to gilteritinib-mediated FLT3 inhibition in FLT3-mutant AML. Then, genetic and pharmacologic tools were used to evaluate the underlying mechanisms by which inactivation of the nominated gene targets was related to the resistance of gilteritinib. We conducted in vitro and in vivo experiments to determine whether combined treatments were more effective in eliminating AML in comparison to single agents. For in vitro assays, at least three independent experiments were performed. The sample size was calculated to ensure 80% power to detect a 50% survival difference (pharmacological treatment group versus control group) at α = 0.05. Gender- and age-matched mice were randomly assigned into treatment groups. The pathological and biochemical outcomes were analyzed by the investigators who were blinded to the treatment groups. The primary end point was the duration of survival.
Cell culture
Cells were cultured at 37°C with 5% CO2 in RPMI 1640 (Gibco) for MOLM-13 and MV4-11 (DSMZ, Germany) or Dulbecco’s minimum essential medium (Gibco) for human embryonic kidney 293FT (Life Technologies, Carlsbad, CA), all supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin/l-glutamine (Gibco). Cell lines were validated via short tandem repeat analysis by The Ohio State University Genomic Services Core, routinely tested for mycoplasma contamination (Universal Mycoplasma Detection Kit, American Type Culture Collection 30-1012 K), and were discarded after passage 20.
Genome-wide loss-of-function screening
The human Brunello CRISPR knockout library was a gift from D. Root and J. Doench (Addgene, #73179). The library was amplified, and lentiviral particles were produced as we previously described (6, 59). The detailed description about cell transduction, puromycin selection, inhibitor treatment, library construction, and sequencing is available in the Supplementary Materials.
CFU assay of primary AML sample
Cryopreserved primary patient bone marrow cells with and without FLT3-ITD and CD34+ healthy bone marrow cells were obtained from The Ohio State University Comprehensive Cancer Center Leukemia Tissue Bank. For the first and subsequent platings, viable CD34+ cells were plated at optimal densities (50,000 cells per condition) in MethoCult H4435 (STEMCELL Technologies, Vancouver, Canada) in the presence of vehicle, single agents, or combination treatments. Formed colonies were counted blindly after 7 to 14 days after seeding. The images of colonies were captured with an Echo revolving microscope. To quantify frequencies of CD34+CD38− leukemia stem cells, after the first plating, MethoCult from each treatment cohort was melted with culture medium, and the remaining cells were dissociated and washed with phosphate-buffered saline before being fixed and stained for lineage markers (Pacific Blue), CD45 (allophycocyanin-Cy7), CD34 (phycoerythrin), and CD38 (fluorescein isothiocyanate). The frequency of leukemia stem cells (Lin−CD45+CD34+CD38−) was analyzed on a Cytomics FC 500 Flow Cytometer (Beckman Coulter).
shRNA knockdown
Lentiviral particles containing shRNAs against human CDK9, DHODH, PRMT5, and CDK7 were purchased from Sigma-Aldrich (PRMT5: TRCN0000303447; DHODH: TRCN0000025868; CDK7: TRCN0000230910; and CDK9: TRCN0000199892). Viral particles were transduced into cells by spinofecting at 32,000 rpm for 90 min at 32°C. Thereafter, puromycin was added to the culture to select shRNA-stable clones. Single-cell clones for each shRNA were established by using limited dilution.
Western blotting and qPCR
Western blotting and qPCR were conducted as previously described (59). Anti-CDK7 (MO1), CDK9 (C12F7), PRMT5 (D5P2T), DHODH (E9X8R), adolase A (D73H4), hexokinase I (C35C4), hexokinase II (C64G5), PFKFB3 (D7H4Q), enolase-1, enolase-2 (E2H9X), PKM1 (D30G6), PKM2 (D78A4), LDHA (C4B5), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (D16H11), PFAS, SDHA, UQCRQ, and GMPS antibodies were purchased from Cell Signaling Technologies (Danvers, MA). GART and PAICS antibodies were obtained from Abcam. TaqMan primers for PRMT5 (Hs01047345), DHODH (Hs00361406), CDK9 (Hs00977896), CDK7 (Hs00361486), BCL2 (Hs00608023), MCL1 (Hs06626047_g1), Myc (Hs00905030), ALDOA (Aldolase A) (Hs00605108), ENO1 (enolase 1) (Hs00361415), PKM (Hs00761782), FH (Hs00264683), UQCRQ (Hs00429571), GART (Hs00894582), PAICS (Hs00935017), FDFT1(Hs00926054), HMGCS1 (Hs00940429), PTK2 (Hs01056457), KRT18 (keratin 18) (Hs01920599), SLC38A2 (Hs01089954), and HK1 (Hs00175976) were obtained from Thermo Fisher Scientific (Waltham, MA).
Animal studies
All animal studies were carried out under protocols approved by The Ohio State University Institutional Animal Care and Use Committee. A total of 1 × 105 MOLM-13 luciferase cells were injected via tail vein into male NOD-Prkdcem26Cd52Il2rgem26Cd22/NjuCrl (NCG) from the Charles River Laboratory. On day 4 after engraftment, mice were randomized to treatment arms. Mice received the following drugs and doses: the first cohort: weekly intraperitoneal injections of dinaciclib (10 mg/kg ) (MedChemExpress, HY-10492) in 20% cyclodextrin (CTD THPB-P) diluted just before injection, daily oral gavage of gilteritinib (30 mg/kg) (MedChemExpress, HY-1432) in 6% drug (w/w) gelucire 44/14 (Gattefosse, France) aliquoted and mixed with drug weekly, oral gavage of vehicle control (gelucire 44/14), or combination therapy (dinaciclib/gilteritinib); the second cohort: oral gavage of brequinar (25 mg/kg) (MedChemExpress, HY-108325) on days 1 and 4 on a 7-day schedule for a total of six doses, PRT808 at 5 mg/kg formulated in high-fat chow (Prelude Therapeutics, Wilmington, DE), daily oral gavage of gilteritinib (15 mg/kg) (MedChemExpress, HY-1432) in 5% ethanol and 10% Cremophor EL, oral gavage of vehicle control (5% ethanol and 10% Cremophor EL), or combination therapies (brequinar/gilteritinib and PRT808/gilteritinib) at single agent regimens. Mice in non-PRT808 chow groups also received control high-fat chow.
RNA-seq and bioinformatics pipeline
RNA was extracted with an RNAeasy mini kit (QIAGEN). The quality of RNA was assessed with the Agilent 2100 BioAnalyzer and RNA 6000 Nano Kit, and the amount was quantified with the Qubit RNA HS Assay Kit. RNA-seq libraries were generated in triplicates per treatment per biological group. The RNA libraries were generated using the NEBNext Ultra II Directional (stranded) RNA Library Prep Kit for Illumina (NEB, #E7760L) and the NEBNext Polyadenylate mRNA Magnetic Isolation Module (NEB, #E7490) with the NEBNext Multiplex Oligos for Illumina Unique Dual Index Primer Pairs (NEB, #6442S/L) using an input amount of 200 ng of total RNA (quantified using a Qubit fluorometer) according to the manufacturer’s protocol.
Statistical analysis
For mouse survival experiments, the primary end point was overall survival (OS). Median survival time in each group was estimated using Kaplan-Meier methods, and the differences in OS between groups were assessed using the log-rank test. Statistical analyses were performed with GraphPad Prism 9.0 (GraphPad Prism) or SAS/STATA software (version 9.4) with analysis of variance (ANOVA) with Tukey post hoc correction for multigroup comparisons or a two-tailed Student t test for two-group comparisons, unless otherwise specified in the figure legends.
Acknowledgments
We are grateful to the patients with AML who provided samples for the above studies and to the OSU Comprehensive Cancer Center Leukemia Tissue Bank (supported by NCIP30CA016058) for sample procurement. We thank the Ohio State University Comprehensive Cancer Center Genomics Shared Resource for RNA-seq. We thank W. Dalton (H. Lee Moffitt Cancer Center) for HS5-GFP cells and R. Garzon (Ohio State University) for MOLM-13 luciferase cells.
Funding: This work was supported by the NIH, NCI (R01 CA223165), and the Ohio State University Comprehensive Cancer Center Pelotonia Foundation funds. . L.B. was supported by TRTH award from EHA and ASH.
Author contributions: P.Z. designed the research, performed the research, analyzed the data, and wrote the paper. L.T.B. designed the research, performed the research, and analyzed the data. M.G., S.S., K.W., M.C., J.S.W., D.C., L.B., C.B.C., X.C., D.S., H.P., P.Y., R.W., A.L., D.S., and R.B. performed the research. P.Y., L.A., P.S., M.W., and K.V. contributed vital new reagents or analytical tools. A.L. provided material support and reviewed the manuscript. J.S.B. supervised the study and reviewed the manuscript. R.L. designed the research and wrote the paper, supervised the study, provided experimental design, and wrote and/or reviewed the manuscript.
Competing interests: R.L. and J.B. are inventors on a provisional patent application (#63/281,934, filed 22 November 2021) related to this work filed by The Ohio State Innovation Foundation. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Data are available in the Gene Expression Omnibus via accession numbers GSE143314 and GSE185519.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S12
Table S1
Other Supplementary Material for this manuscript includes the following:
Data files S1 to S6
REFERENCES AND NOTES
- 1.Quentmeier H., Reinhardt J., Zaborski M., Drexler H. G., FLT3 mutations in acute myeloid leukemia cell lines. Leukemia 17, 120–124 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Daver N., Schlenk R. F., Russell N. H., Levis M. J., Targeting FLT3 mutations in AML: Review of current knowledge and evidence. Leukemia 33, 299–312 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ju H. Q., Zhan G., Huang A., Sun Y., Wen S., Yang J., Lu W.-H., Xu R.-H., Li J., Li Y., Garcia-Manero G., Huang P., Hu Y., ITD mutation in FLT3 tyrosine kinase promotes Warburg effect and renders therapeutic sensitivity to glycolytic inhibition. Leukemia 31, 2143–2150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perl A. E., Martinelli G., Cortes J. E., Neubauer A., Berman E., Paolini S., Montesinos P., Baer M. R., Larson R. A., Ustun C., Fabbiano F., Erba H. P., di Stasi A., Stuart R., Olin R., Kasner M., Ciceri F., Chou W.-C., Podoltsev N., Recher C., Yokoyama H., Hosono N., Yoon S.-S., Lee J.-H., Pardee T., Fathi A. T., Liu C., Hasabou N., Liu X., Bahceci E., Levis M. J., Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N. Engl. J. Med. 381, 1728–1740 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Brinton L. T., Sher S., Williams K., Canfield D., Orwick S., Wasmuth R., Cempre C., Skinner J., Lehman A., Blachly J. S., Byrd J. C., Lapalombella R., Cotargeting of XPO1 enhances the antileukemic activity of midostaurin and gilteritinib in acute myeloid leukemia. Cancers (Basel) 12, 1574 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinton L. T., Zhang P., Williams K., Canfield D., Orwick S., Sher S., Wasmuth R., Beaver L., Cempre C., Skinner J., Cannon M., Govande M., Harrington B., Lehman A., Byrd J. C., Lapalombella R., Blachly J. S., Synergistic effect of BCL2 and FLT3 co-inhibition in acute myeloid leukemia. J. Hematol. Oncol. 13, 139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., Lin H., Wang M., Angelis D., Hawkins M., Rominger D., Emm T., Luengo J., Ruggeri B., Scherle P., Vaddi K., Abstract 2919: Discovery of PRT811, a potent, selective, and orally bioavailable brain penetrant PRMT5 inhibitor for the treatment of brain tumors. Cancer Res. 80, 2919–2919 (2020). [Google Scholar]
- 8.Kolde R., Laur S., Adler P., Vilo J., Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics 28, 573–580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang B., Wang M., Zhang W., Xiao T., Chen C.-H., Wu A., Wu F., Traugh N., Wang X., Li Z., Mei S., Cui Y., Shi S., Lipp J. J., Hinterndorfer M., Zuber J., Brown M., Li W., Liu X. S., Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat. Protoc. 14, 756–780 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sykes D. B., Kfoury Y. S., Mercier F. E., Wawer M. J., Law J. M., Haynes M. K., Lewis T. A., Schajnovitz A., Jain E., Lee D., Meyer H., Pierce K. A., Tolliday N. J., Waller A., Ferrara S. J., Eheim A. L., Stoeckigt D., Maxcy K. L., Cobert J. M., Bachand J., Szekely B. A., Mukherjee S., Sklar L. A., Kotz J. D., Clish C. B., Sadreyev R. I., Clemons P. A., Janzer A., Schreiber S. L., Scadden D. T., Inhibition of dihydroorotate dehydrogenase overcomes differentiation blockade in acute myeloid leukemia. Cell 167, 171–186.e15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boukalova S., Hubackova S., Milosevic M., Ezrova Z., Neuzil J., Rohlena J., Dihydroorotate dehydrogenase in oxidative phosphorylation and cancer. Biochim. Biophys. Acta Mol. Basis Dis. 1866, 165759 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Baker A., Gregory G. P., Verbrugge I., Kats L., Hilton J. J., Vidacs E., Lee E. M., Lock R. B., Zuber J., Shortt J., Johnstone R. W., The CDK9 inhibitor dinaciclib exerts potent apoptotic and antitumor effects in preclinical models of MLL-rearranged acute myeloid leukemia. Cancer Res. 76, 1158–1169 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Anshabo A. T., Milne R., Wang S., Albrecht H., CDK9: A comprehensive review of its biology, and its role as a potential target for anti-cancer Agents. Front. Oncol. 11, 678559 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minzel W., Venkatachalam A., Fink A., Hung E., Brachya G., Burstain I., Shaham M., Rivlin A., Omer I., Zinger A., Elias S., Winter E., Erdman P. E., Sullivan R. W., Fung L., Mercurio F., Li D., Vacca J., Kaushansky N., Shlush L., Oren M., Levine R., Pikarsky E., Snir-Alkalay I., Ben-Neriah Y., Small molecules co-targeting CKIα and the transcriptional kinases CDK7/9 control AML in preclinical models. Cell 175, 171–185.e25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tzelepis K., Koike-Yusa H., de Braekeleer E., Li Y., Metzakopian E., Dovey O. M., Mupo A., Grinkevich V., Li M., Mazan M., Gozdecka M., Ohnishi S., Cooper J., Patel M., McKerrell T., Chen B., Domingues A. F., Gallipoli P., Teichmann S., Ponstingl H., McDermott U., Saez-Rodriguez J., Huntly B. J. P., Iorio F., Pina C., Vassiliou G. S., Yusa K., A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 17, 1193–1205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tarighat S. S., Santhanam R., Frankhouser D., Radomska H. S., Lai H., Anghelina M., Wang H., Huang X., Alinari L., Walker A., Caligiuri M. A., Croce C. M., Li L., Garzon R., Li C., Baiocchi R. A., Marcucci G., The dual epigenetic role of PRMT5 in acute myeloid leukemia: Gene activation and repression via histone arginine methylation. Leukemia 30, 789–799 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou P., Wu C., Wang Y., Qi R., Bhavanasi D., Zuo Z., dos Santos C., Chen S., Chen Y., Zheng H., Wang H., Perl A., Guo D., Huang J., A genome-wide CRISPR screen identifies genes critical for resistance to FLT3 inhibitor AC220. Cancer Res. 77, 4402–4413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H. Y., Appelbaum F. R., Willman C. L., Zager R. A., Banker D. E., Cholesterol-modulating agents kill acute myeloid leukemia cells and sensitize them to therapeutics by blocking adaptive cholesterol responses. Blood 101, 3628–3634 (2003). [DOI] [PubMed] [Google Scholar]
- 19.DeWaal D., Nogueira V., Terry A. R., Patra K. C., Jeon S.-M., Guzman G., Au J., Long C. P., Antoniewicz M. R., Hay N., Hexokinase-2 depletion inhibits glycolysis and induces oxidative phosphorylation in hepatocellular carcinoma and sensitizes to metformin. Nat. Commun. 9, 446 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith J. R., Hayman G. T., Wang S.-J., Laulederkind S. J. F., Hoffman M. J., Kaldunski M. L., Tutaj M., Thota J., Nalabolu H. S., Ellanki S. L. R., Tutaj M. A., de Pons J. L., Kwitek A. E., Dwinell M. R., Shimoyama M. E., The year of the rat: The rat genome database at 20: A multi-species knowledgebase and analysis platform. Nucleic Acids Res. 48, D731–D742 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panina S. B., Pei J., Kirienko N. V., Mitochondrial metabolism as a target for acute myeloid leukemia treatment. Cancer Metab. 9, 17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallipoli P., Giotopoulos G., Tzelepis K., Costa A. S. H., Vohra S., Medina-Perez P., Basheer F., Marando L., di Lisio L., Dias J. M. L., Yun H., Sasca D., Horton S. J., Vassiliou G., Frezza C., Huntly B. J. P., Glutaminolysis is a metabolic dependency in FLT3ITD acute myeloid leukemia unmasked by FLT3 tyrosine kinase inhibition. Blood 131, 1639–1653 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregory M. A., Nemkov T., Reisz J. A., Zaberezhnyy V., Hansen K. C., D’Alessandro A., DeGregori J., Glutaminase inhibition improves FLT3 inhibitor therapy for acute myeloid leukemia. Exp. Hematol. 58, 52–58 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broer A., Rahimi F., Broer S., Deletion of amino acid transporter ASCT2 (SLC1A5) reveals an essential role for transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to sustain glutaminolysis in cancer cells. J. Biol. Chem. 291, 13194–13205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bajzikova M., Kovarova J., Coelho A. R., Boukalova S., Oh S., Rohlenova K., Svec D., Hubackova S., Endaya B., Judasova K., Bezawork-Geleta A., Kluckova K., Chatre L., Zobalova R., Novakova A., Vanova K., Ezrova Z., Maghzal G. J., Novais S. M., Olsinova M., Krobova L., An Y. J., Davidova E., Nahacka Z., Sobol M., Cunha-Oliveira T., Sandoval-Acuña C., Strnad H., Zhang T., Huynh T., Serafim T. L., Hozak P., Sardao V. A., Koopman W. J. H., Ricchetti M., Oliveira P. J., Kolar F., Kubista M., Truksa J., Dvorakova-Hortova K., Pacak K., Gurlich R., Stocker R., Zhou Y., Berridge M. V., Park S., Dong L., Rohlena J., Neuzilb J., Reactivation of dihydroorotate dehydrogenase-driven pyrimidine biosynthesis restores tumor growth of respiration-deficient cancer cells. Cell Metab. 29, 399–416.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Her N.-G., Oh J.-W., Oh Y. J., Han S., Cho H. J., Lee Y., Ryu G. H., Nam D.-H., Potent effect of the MDM2 inhibitor AMG232 on suppression of glioblastoma stem cells. Cell Death Dis. 9, 792 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gullà A., Hideshima T., Bianchi G., Fulciniti M., Kemal Samur M., Qi J., Tai Y.-T., Harada T., Morelli E., Amodio N., Carrasco R., Tagliaferri P., Munshi N. C., Tassone P., Anderson K. C., Protein arginine methyltransferase 5 has prognostic relevance and is a druggable target in multiple myeloma. Leukemia 32, 996–1002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christian S., Merz C., Evans L., Gradl S., Seidel H., Friberg A., Eheim A., Lejeune P., Brzezinka K., Zimmermann K., Ferrara S., Meyer H., Lesche R., Stoeckigt D., Bauser M., Haegebarth A., Sykes D. B., Scadden D. T., Losman J. A., Janzer A., The novel dihydroorotate dehydrogenase (DHODH) inhibitor BAY 2402234 triggers differentiation and is effective in the treatment of myeloid malignancies. Leukemia 33, 2403–2415 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Zhu F., Rui L., PRMT5 in gene regulation and hematologic malignancies. Genes Dis. 6, 247–257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He X., Zhu Y., Lin Y.-C., Li M., du J., Dong H., Sun J., Zhu L., Wang H., Ding Z., Zhang L., Zhang L., Zhao D., Wang Z., Wu H., Zhang H., Jiang W., Xu Y., Jin J., Shen Y., Perry J., Zhao X., Zhang B., Liu S., Xue S.-L., Shen B., Chen C.-W., Chen J., Khaled S., Kuo Y.-H., Marcucci G., Luo Y., Li L., PRMT1-mediated FLT3 arginine methylation promotes maintenance of FLT3-ITD+ acute myeloid leukemia. Blood 134, 548–560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ando K., Hamard P.-J., Liu F., Martinez C., Ban Y., Chen X., Nimer S. D., The AML-associated FLT3-ITD kinase regulates histone modifications and cytokine signaling, via effects on PRMT5. Blood 128, 2696–2696 (2016). [Google Scholar]
- 32.Dhir A., Paterson A. J., Qiu S., Mullen A. K., Anderson N. R., Bhatia R., An epigenetic screen identifies PRMT5 as a target for inhibition of FLT3-ITD AML cell growth in combination with tyrosine kinase inhibitors. Blood 134, 2524–2524 (2019). [Google Scholar]
- 33.Zhu Y., He X., Lin Y.-C., Dong H., Zhang L., Chen X., Wang Z., Shen Y., Li M., Wang H., Sun J., Nguyen L. X., Zhang H., Jiang W., Yang Y., Chen J., Müschen M., Chen C.-W., Konopleva M. Y., Sun W., Jin J., Carlesso N., Marcucci G., Luo Y., Li L., Targeting PRMT1-mediated FLT3 methylation disrupts maintenance of MLL-rearranged acute lymphoblastic leukemia. Blood 134, 1257–1268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radzisheuskaya A., Shliaha P. V., Grinev V., Lorenzini E., Kovalchuk S., Shlyueva D., Gorshkov V., Hendrickson R. C., Jensen O. N., Helin K., PRMT5 methylome profiling uncovers a direct link to splicing regulation in acute myeloid leukemia. Nat. Struct. Mol. Biol. 26, 999–1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tibes R., Bogenberger J. M., Transcriptional silencing of MCL-1 through cyclin-dependent kinase inhibition in acute myeloid leukemia. Front. Oncol. 9, 1205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips D. C., Jin S., Gregory G. P., Zhang Q., Xue J., Zhao X., Chen J., Tong Y., Zhang H., Smith M., Tahir S. K., Clark R. F., Penning T. D., Devlin J. R., Shortt J., Hsi E. D., Albert D. H., Konopleva M., Johnstone R. W., Leverson J. D., Souers A. J., A novel CDK9 inhibitor increases the efficacy of venetoclax (ABT-199) in multiple models of hematologic malignancies. Leukemia 34, 1646–1657 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitra P., Yang R. M., Sutton J., Ramsay R. G., Gonda T. J., CDK9 inhibitors selectively target estrogen receptor-positive breast cancer cells through combined inhibition of MYB and MCL-1 expression. Oncotarget 7, 9069–9083 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Cuellar M. P., Füller E., Mäthner E., Breitinger C., Hetzner K., Zeitlmann L., Borkhardt A., Slany R. K., Efficacy of cyclin-dependent-kinase 9 inhibitors in a murine model of mixed-lineage leukemia. Leukemia 28, 1427–1435 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Sexauer A. N., Do B., Sykes D. B., Loss of FLT3 sensitizes myeloid cells to differentiation via DHODH inhibition. Blood 134, 2712–2712 (2019). [Google Scholar]
- 40.Gross D. N., van den Heuvel A. P., Birnbaum M. J., The role of FoxO in the regulation of metabolism. Oncogene 27, 2320–2336 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Bjelosevic S., Gruber E., Newbold A., Shembrey C., Devlin J. R., Hogg S. J., Kats L., Todorovski I., Fan Z., Abrehart T. C., Pomilio G., Wei A., Gregory G. P., Vervoort S. J., Brown K. K., Johnstone R. W., Serine biosynthesis is a metabolic vulnerability in FLT3-ITD-driven acute myeloid leukemia. Cancer Discov. 11, 1582–1599 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y., Newsom K. J., Zhang M., Kelley J. S., Starostik P., GATM-mediated creatine biosynthesis enables maintenance of FLT3-ITD–mutant acute myeloid leukemia. Mol. Cancer Res. 20, 293–304 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Fu Y. H., Tseng C.-Y., Lu J.-W., Lu W.-H., Lan P.-Q., Chen C.-Y., Ou D.-L., Lin L.-I., Deciphering the role of pyrvinium pamoate in the generation of integrated stress response and modulation of mitochondrial function in myeloid leukemia cells through transcriptome analysis. Biomedicine 9, 1869 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farge T., Saland E., de Toni F., Aroua N., Hosseini M., Perry R., Bosc C., Sugita M., Stuani L., Fraisse M., Scotland S., Larrue C., Boutzen H., Féliu V., Nicolau-Travers M.-L., Cassant-Sourdy S., Broin N., David M., Serhan N., Sarry A., Tavitian S., Kaoma T., Vallar L., Iacovoni J., Linares L. K., Montersino C., Castellano R., Griessinger E., Collette Y., Duchamp O., Barreira Y., Hirsch P., Palama T., Gales L., Delhommeau F., Garmy-Susini B. H., Portais J. C., Vergez F., Selak M., Danet-Desnoyers G., Carroll M., Récher C., Sarry J. E., Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov. 7, 716–735 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amaya M., Inguva A., Pei S., Jones C., Krug A., Ye H., Minhajuddin M., Winters A., Furtek S. L., Gamboni F., Stevens B., D’Alessandro A., Pollyea D. A., Reigan P., Jordan C. T., The STAT3-MYC axis promotes survival of leukemia stem cells by regulating SLC1A5 and oxidative phosphorylation. Blood 139, 584–596 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee K. M., Giltnane J. M., Balko J. M., Schwarz L. J., Guerrero-Zotano A. L., Hutchinson K. E., Nixon M. J., Estrada M. V., Sánchez V., Sanders M. E., Lee T., Gómez H., Lluch A., Pérez-Fidalgo J. A., Wolf M. M., Andrejeva G., Rathmell J. C., Fesik S. W., Arteaga C. L., MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metab. 26, 633–647.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiozaki Y., Okamura K., Kohno S., Keenan A. L., Williams K., Zhao X., Chick W. S., Miyazaki-Anzai S., Miyazaki M., The CDK9-cyclin T1 complex mediates saturated fatty acid-induced vascular calcification by inducing expression of the transcription factor CHOP. J. Biol. Chem. 293, 17008–17020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herrera K. N. G., Zaganjor E., Ishikawa Y., Spinelli J. B., Yoon H., Lin J. R., Satterstrom F. K., Ringel A., Mulei S., Souza A., Gorham J. M., Benson C. C., Seidman J. G., Sorger P. K., Clish C. B., Haigis M. C., Small-molecule screen identifies de novo nucleotide synthesis as a vulnerability of cells lacking SIRT3. Cell Rep. 22, 1945–1955 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quirós P. M., Prado M. A., Zamboni N., D’Amico D., Williams R. W., Finley D., Gygi S. P., Auwerx J., Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216, 2027–2045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yin J., Ren W., Huang X., Deng J., Li T., Yin Y., Potential mechanisms connecting purine metabolism and cancer therapy. Front. Immunol. 9, 1697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan H. F., Zhao M., Zhao L. N., Yun H. L., Yang G., Geng Y., Wang Y. F., Zheng W., Yuan Y., Song T. Q., Niu J. Q., Zhang X. D., PRMT5 confers lipid metabolism reprogramming, tumour growth and metastasis depending on the SIRT7-mediated desuccinylation of PRMT5 K387 in tumours. Acta Pharmacol. Sin. 10.1038/s41401-021-00841-y (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szewczyk M. M., Luciani G. M., Vu V., Murison A., Dilworth D., Barghout S. H., Lupien M., Arrowsmith C. H., Minden M. D., Barsyte-Lovejoy D., PRMT5 regulates ATF4 transcript splicing and oxidative stress response. Redox Biol. 51, 102282 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamauchi T., Miyawaki K., Semba Y., Takahashi M., Izumi Y., Nogami J., Nakao F., Sugio T., Sasaki K., Pinello L., Bauer D. E., Bamba T., Akashi K., Maeda T., Targeting leukemia-specific dependence on the de novo purine synthesis pathway. Leukemia 36, 383–393 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Lu X., Han L., Busquets J., Collins M., Lodi A., Marszalek J. R., Konopleva M., Tiziani S., The combined treatment with the FLT3-inhibitor AC220 and the complex I inhibitor IACS-010759 synergistically depletes Wt- and FLT3-mutated acute myeloid leukemia cells. Front. Oncol. 11, 686765 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joshi S. K., Nechiporuk T., Bottomly D., Piehowski P. D., Reisz J. A., Pittsenbarger J., Kaempf A., Gosline S. J. C., Wang Y.-T., Hansen J. R., Gritsenko M. A., Hutchinson C., Weitz K. K., Moon J., Cendali F., Fillmore T. L., Tsai C.-F., Schepmoes A. A., Shi T., Arshad O. A., McDermott J. E., Babur O., Watanabe-Smith K., Demir E., D’Alessandro A., Liu T., Tognon C. E., Tyner J. W., McWeeney S. K., Rodland K. D., Druker B. J., Traer E., The AML microenvironment catalyzes a stepwise evolution to gilteritinib resistance. Cancer Cell 39, 999–1014.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin K. H., Xie A., Rutter J. C., Ahn Y.-R., Lloyd-Cowden J. M., Nichols A. G., Soderquist R. S., Koves T. R., Muoio D. M., MacIver N. J., Lamba J. K., Pardee T. S., McCall C. M., Rizzieri D. A., Wood K. C., Systematic dissection of the metabolic-apoptotic interface in AML reveals heme biosynthesis to be a regulator of drug sensitivity. Cell Metab. 29, 1217–1231.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacque N., Ronchetti A. M., Larrue C., Meunier G., Birsen R., Willems L., Saland E., Decroocq J., Maciel T. T., Lambert M., Poulain L., Hospital M. A., Sujobert P., Joseph L., Chapuis N., Lacombe C., Moura I. C., Demo S., Sarry J. E., Recher C., Mayeux P., Tamburini J., Bouscary D., Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood 126, 1346–1356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen X., Glytsou C., Zhou H., Narang S., Reyna D. E., Lopez A., Sakellaropoulos T., Gong Y., Kloetgen A., Yap Y. S., Wang E., Gavathiotis E., Tsirigos A., Tibes R., Aifantis I., Targeting mitochondrial structure sensitizes acute myeloid leukemia to venetoclax treatment. Cancer Discov. 9, 890–909 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang P., Brinton L. T., Williams K., Sher S., Orwick S., Tzung-Huei L., Mims A. S., Coss C. C., Kulp S. K., Youssef Y., Chan W. K., Mitchell S., Mustonen A., Cannon M., Phillips H., Lehman A. M., Kauffman T., Beaver L., Canfield D., Grieselhuber N. R., Alinari L., Sampath D., Yan P., Byrd J. C., Blachly J. S., Lapalombella R., Targeting DNA damage repair functions of two histone deacetylases, HDAC8 and SIRT6, sensitizes acute myeloid leukemia to NAMPT inhibition. Clin. Cancer Res. 27, 2352–2366 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S12
Table S1
Data files S1 to S6