Abstract
The aim of this study was to find the application value of selective polyadenylation in immune cell infiltration, biological transcription function and risk assessment of survival and prognosis in lung adenocarcinoma (LUAD). The processed original mRNA expression data of LUAD were downloaded, and the expression profiles of 594 patient samples were collected. The (APA) events in TCGA-NA-SEQ data were evaluated by polyadenylation site use Index (PDUI) values, and the invasion of stromal cells and immune cells and tumor purity were calculated to group and select the differential genes. Lasso regression and stratified analysis were used to examine the role of risk scores in predicting patient outcomes. The study also used the GDSC database to predict the chemotherapeutic sensitivity of each tumor sample and used a regression method to obtain an IC50 estimate for each specific chemotherapeutic drug treatment. Then CIBERSORT algorithm was used to conduct Spearman correlation analysis, immune regulatory factor analysis and TIDE immune system function analysis for gene expression level and immune cell content. Finally, the Kaplan–Meier curve was used to analyze the correlation between stromal score and the immune score of LUAD. In this study, APA’s LUAD risk score prognostic model was constructed. KM survival analysis showed that immune score affected the prognosis of LUAD patients (P = 0.027) but the matrix score was not statistically significant (P = 0.1). We extracted 108 genes with APA events from 827 different genes and based on PUDI clustering and heat map, the survival rate of patients in the four groups was significantly different (P = 0.05). Multiple omics studies showed that risk score was significantly positively correlated with Macrophages M0, T cells Follicular helper, B cells naive and NK cells resting. It is significantly negatively correlated with dendritic cells resting, mast cells resting, monocyte, T cells CD4 memory resting and B cells memory. We further explored the relationship between the expression of immunosuppressor genes and risk score and found that ADORA2A, BTLA, CD160, CD244, CD274, CD96, CSF1R and CTLA4 genes were highly correlated with the risk score. Selective poly adenylation plays an important role in the development and progression of LUAD, immune invasion, tumor cell invasion and metastasis and biological transcription, and affects the survival and prognosis of LUAD patients.
Keywords: CIBERSORT algorithm, immunotherapy, lung adenocarcinoma, multiple omics research, selective poly adenylation
Introduction
Lung cancer remains the world’s leading cause of death. About 85% of lung cancers are nonsmall cell lung cancers, which can be further divided into three subtypes: large cell carcinoma, squamous cell carcinoma and lung adenocarcinoma (LUAD) [1]. LUAD is the most common histological subtype in most countries, as a result of the increased number of nonsmokers [2]. The current treatment of LUAD includes surgical resection, radiotherapy and chemotherapy, and great progress has been made. However, tumor recurrence and drug resistance are still inevitable [3]. Therefore, there is an urgent need to identify new biomarkers to help elucidate the pathological mechanisms of LUAD and develop therapeutic strategies for treating LUAD. The tumor microenvironment (TME) has attracted more and more attention as a potential therapeutic target for LUAD [4]. Various studies have demonstrated the relationship between LUAD and TME, suggesting that a better understanding of TME research can promote the progress of immunotherapy for LUAD. TME generally consists of a variety of cell types, including immune cells, extracellular matrix and stromal cells. Several algorithms have been developed to estimate stromal and immune cells in malignant tumor tissues using estimate and TIMER algorithms to estimate the abundance of invasive immune cells and predict tumor purity based on gene expression profiles [5]. Several studies have investigated the role of immune infiltration in different types of cancer, such as glioblastoma, breast cancer and melanoma. Identification of immune-related genes can also provide a better understanding of the LUAD microenvironment [6].
Selective polyadenylation is an important step in mRNA maturation. Alternative polyadenylation (APA) is highly present in more than 70% of human genes and is involved in a variety of biological processes, such as cell differentiation and proliferation and immune responses [7]. APA events have a strong ability to predict clinical outcomes in a variety of cancers, indicating their potential as new prognostic biomarkers [8]. APA events are also involved in reshaping cellular pathways and regulating specific gene expression in many cancers, providing new insights into the pathologic mechanisms of cancer development. However, the role of APA events in LUAD has not been fully elucidated [9].
In this study, we applied the estimated estimate algorithm to calculate matrix and immune scores and to determine the prognostic IRG of LUAD in the TCGA-LUAD dataset. Further, we investigated the role of APA events in IRG in RNA expression and clinical prognosis. Risk characteristics were constructed based on IRG expressions with APA events to identify the prognostic and predictive value of risk scores. CIBERSORT algorithm was used to evaluate the immune invasion of LUAD, and the relationship between risk score and immune invasion was also discussed [10]. In addition, we found that risk stratification can predict the efficacy of immunotherapy and provide reference for the treatment of LUAD.
Materials
Data acquisition
TCGA database (https://portal.gdc.cancer.gLUAD/) is the biggest cancer gene information database, including gene expression data, miRNA expression data and copy number variation, DNA methylation, SNPs and other data. We downloaded the processed original mRNA expression data of LUAD, including 594 patient sample data expression profiles. Series Matrix File of GSE37745 was downloaded from NCBI GEO public database, annotated on the GPL570 platform, and data of 106 LUAD patients with complete expression profiles and survival information were downloaded. APA events in TCGA-RNA-SEQ data were evaluated by polyadenylation site use Index (PDUI) values. PDUI values represent the frequency of APA events (on a scale of 0–1), where the larger the PDUI, the farther the polyadenylation site using the transcript, and vice versa. PDUI values for all genes per patient in the TCGA-LUAD dataset were downloaded from TC3A (http://tc3a.org/).
Differential gene expression map
The R package ‘Estimate’ was used to calculate stromal and immune cell invasion and tumor purity. All patients according to immune score and score matrix group respectively, divided into the high/low immune group and high/low matrix, and then, using ‘limma’ package | analysis of two groups of patients who were differentially expressed logFC | > 1 & adj. P < 0.05).
GO and KEGG function analysis
R package ‘ClusterProfliter’ was used for functional annotation of differential genes to comprehensively explore the functional correlation of these differential genes. Then “GOSemSim” was used to cluster the enriched pathways, and the semantic similarity between GO terms was calculated to draw the enrichment analysis results.
Model construction and prognosis
Differential genes were selected and lasso regression was used to construct prognostic correlation models. After incorporating expression values for each specific gene, a risk score formula was constructed for each patient and weighted with its estimated regression coefficients in lasso regression analysis. According to the risk scoring formula, patients were divided into a low-risk group and a high-risk group with the median risk scoring value as the cutoff point. Kaplan–Meier was used to evaluate survival differences between the two groups, and the log-rank statistical method was used for comparison. Lasso regression and stratified analysis were used to examine the role of risk scores in predicting patient outcomes. ROC curve was used to study the accuracy of the model prediction.
Drug sensitivity analysis
Genomics database based on the largest drug (GDSC cancer drugs sensitivity genomics database, https://www.cancerrxgene.org/), we use R software package ‘pRRophetic’ to predict chemotherapy sensitivity of each tumor samples; the IC50 estimates for each specific chemotherapeutic agent were obtained using a regression method, and the regression and prediction accuracy were tested using 10 cross-validation tests using the GDSC training set. Default values were selected for all parameters, including ‘combat’ for batch removal and the average of duplicate gene expression.
Immune cell infiltration analysis
CIBERSORT algorithm was used to analyze RNA-SEQ data of LUAD patients in different subgroups to infer the relative proportion of 22 immune infiltrating cells, and spearman correlation analysis was performed on gene expression and immune cell content. P < 0.05 was considered statistically significant.
GSEA enrichment analysis
Gene set enrichment analysis (GSEA) was performed on the expression profile of LUAD patients (GSEA; http://www.broadinstitute.org/gsea) to determine the high risk and low risk of differentially expressed genes between the group of patients. The genes were ordered by their degree of differential expression in the two types of samples and then tested to see whether the set of genes was enriched at the top or bottom of the sequence. In this study, GSEA was used to compare the difference in signal pathways between the high-risk group and the low-risk group, to explore the possible molecular mechanism of the difference in prognosis between the two groups, in which the number of replacements was set to 1000, and the replacement type was set to phenotype.
TISIDB analysis
TISIDB is an online site for tumor and immune system interactions that integrates multiple heterogeneous data types. The data were combined into 10 categories of information for each gene. TISIDB integrates data from multiple databases (TCGA, UniProt, GO and DrugBank, etc.) and is a valuable resource for cancer immunology research and treatment. This is used to study the interaction between tumor and immune system genes data are downloaded from the TISIDB website (http://cis.hku.hk/TISIDB/index.php).
Regulatory network analysis of key genes
The eukaryotic transcription initiation process is very complex, and often requires the assistance of a variety of protein factors, transcription factors and RNA polymerase ii form a transcription initiation complex, together participate in the process of transcription initiation. According to the functional characteristics of transcription factors can be divided into two types; the first is the universal transcription factor, which, together with RNA polymerase ii, forms a transcription initiation complex so that transcription can begin at the correct location. Cis-acting elements exist in peripheral sequences that can affect gene expression. Cis-acting elements include promoters, enhancers, regulatory sequences and inducible elements, which participate in the regulation of gene expression. Cis-acting elements themselves do not encode any protein, but only provide a site of action to interact with trans-acting factors. This analysis is mainly performed through R package cisTarget, where we use rcistarget.hg19.motifdb.cisbPond.500bp for the gene-Motif rankings database.
Statistical methods
Survival curves were generated by Kaplan–Meier method and compared by the log-rank method. Cox proportional risk model was used for multivariate analysis. All statistical analyses were performed using the R language (version 3.6). All statistical tests were bilateral, and P < 0.05 was statistically significant.
Results
Lung adenocarcinoma matrix and immune score and prognosis
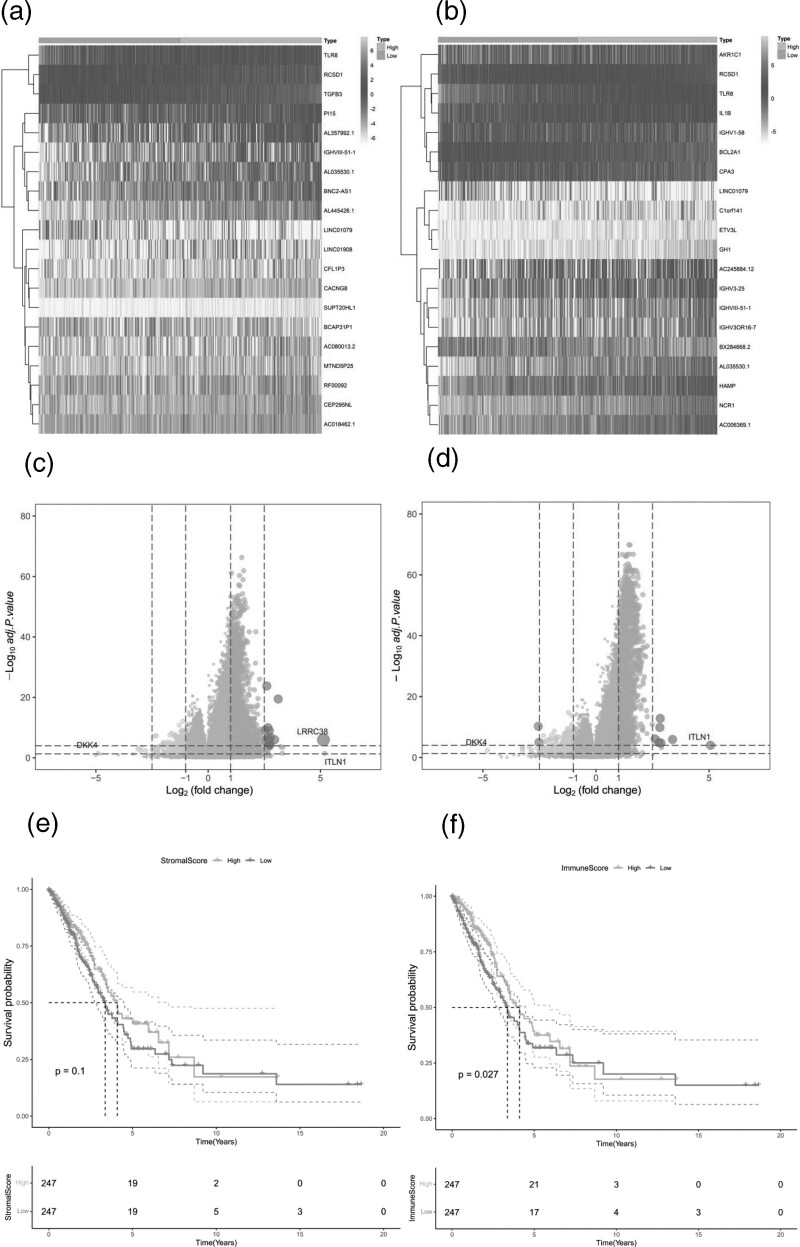
We downloaded the raw mRNA expression data (FPKM) of LUAD processed from the TCGA database and calculated the patient’s stroma score and immune score using the ‘Estimate’ package after removing the normal sample. The stroma score ranged from −1783.99 to 2107.56. Immunity scores ranged from −936.19 to 3453.01; LUAD patients were divided into low group and high group according to median values of matrix score and immune score. KM survival analysis showed that immune scores influenced the prognosis of LUAD patients. In this study, according to matrix score and immune score, the infiltration of stromal cells and immune cells showed that the TLR8 gene ranked first in the heat map of differential expression of stromal cells, but ranked third in the heat map of differential expression of immune cells, showing certain differences between the two genes (Fig. 1a and b). The volcanic map of matrix score and immune score indicated that the upregulated mutation genes accounted for the majority, whereas the downregulated genes were few. LRRC38 and ITLN1 were significantly upregulated in matrix score, ITLN1 was significantly upregulated in immune score and DKK4 was significantly down-regulated in both of them (Fig. 1c,d). Kaplan–Meier curve showed that stromal cell score had no significant difference in 5-year survival rate of LUAD (P = 0.1), but immune cell score had significant difference in 5-year survival rate of LUAD (P= 0.027) (Fig. 1e,f).
Fig. 1.
Differential expression analysis of patients with matrix and immune scores (AC) volcanic map and heat map of patients with matrix score; (BD) Analysis of differences in patients with immune score volcano map and heat map. In volcanoes, red means up and green means down. In the heat map, green means up, yellow means down. Relationship between stromal and immune scores and prognosis in patients with LUAD; (EF) kaplan–meier analysis based on grouping of stromal and immune scores in patients with LUAD.
Explore the expression and pathway enrichment of differential genes
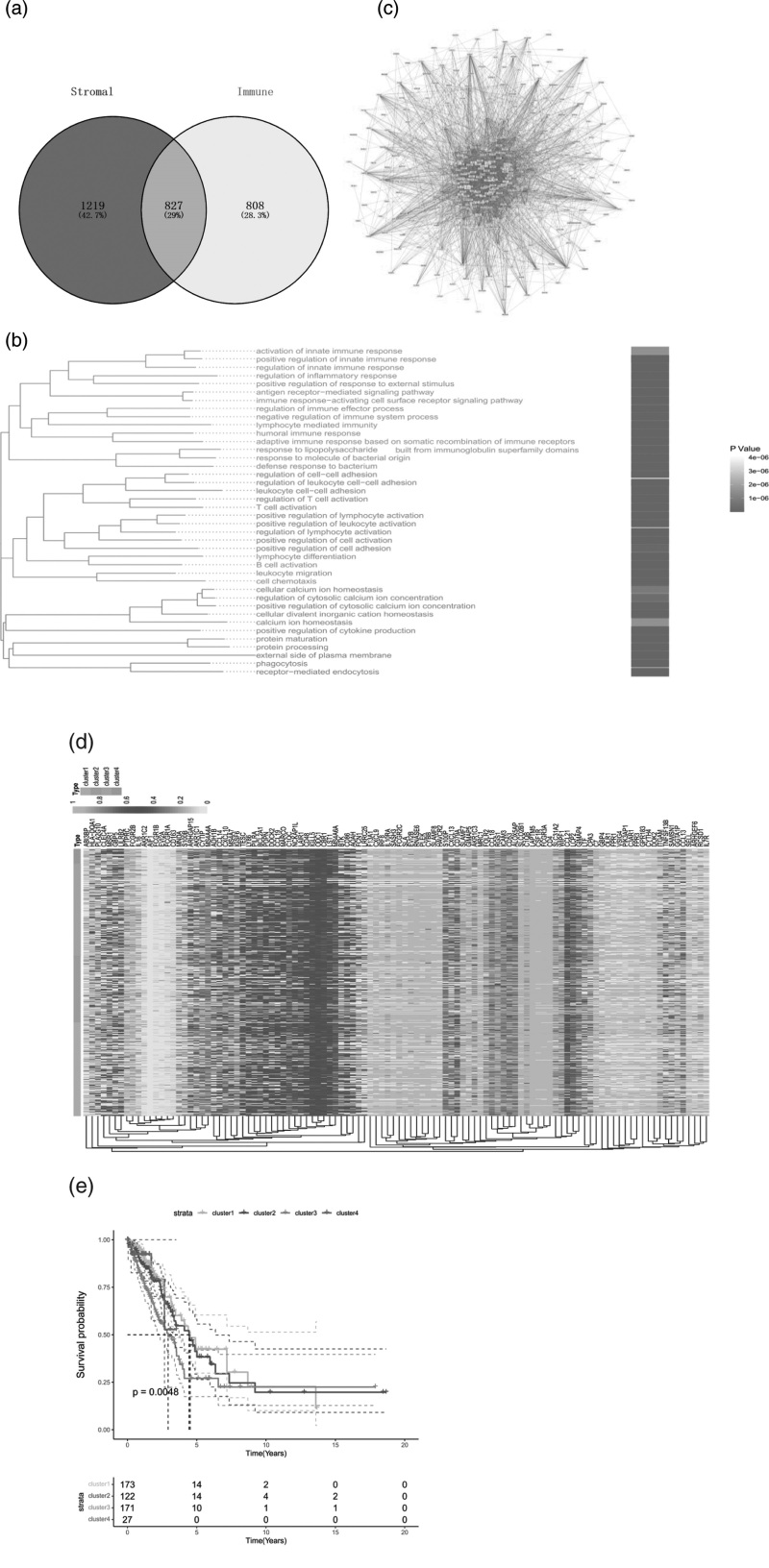
We by ‘limma’ bag of two groups of patients with variance analysis, gene screening conditions for | logFC | >1 & adj. P < 0.05, the difference matrix group identified 2046 genes, there are 1800 genes, 370 genes; A total of 1635 differential genes were identified by immunological grouping, of which 1270 genes were up-=regulated and 365 genes were down-regulated(Fig. 2a). We conducted an enrichment analysis of 827 differential genes with the intersection, and the results showed that differential genes were mainly enriched in adaptive immune response, immunoglobulin complex, lymphocyte activation and hematopoietic cell (Fig. 2b). On lineage and other pathways, cytoscape software was used to conduct protein interaction network analysis for intersecting differential genes (note: The heat map randomly showed the expression of 20 differential genes)(Fig. 2c).
Fig. 2.
Enrichment analysis of differential genes. (a)Venn of matrix score and immune score; (b) cytoscape analysis of protein interaction network of genes with differential gene concentration; (c) GO enrichment analysis of differential genes. Studies of selective polyadenylation in LUAD; (d) heat maps of PDUI values of genes in patients with APA events in the TCGA-LUAD dataset. Patients were divided into four categories according to PDUI values; (e) Kaplan–Meier analysis of the overall survival rate of the four clusters. APA, alternative polyadenylation; LUAD, lung adenocarcinoma.
Alternative polyadenylation events in differential genes
To investigate the role of APA events in LUAD prognosis, we extracted APA event data from The TCGA-LUAD dataset from The Cancer 3 ‘UTR Atlas (TC3A) database. The frequency of APA events is expressed as a percentage of the remote PDUI, which is based on the DaPars algorithm. The PDUI value represents the frequency of APA events, ranging from 0 to 1. The larger the PDUI, the farther the polyadenylation site of the transcript, and vice versa. We extracted 108 genes with APA events from 827 different genes and made cluster heat maps (clustering according to the PUDI value of each sample) (Fig. 2d). KM survival analysis showed that the survival rates of patients in the four groups were significantly different. To remove the multicollinearity of variables, we calculated the variance inflation factor (VIF) of 108 genes and selected 96 genes with VIF<10 for model construction (Fig. 2e).
Prognostic genes were obtained and prognostic models were constructed
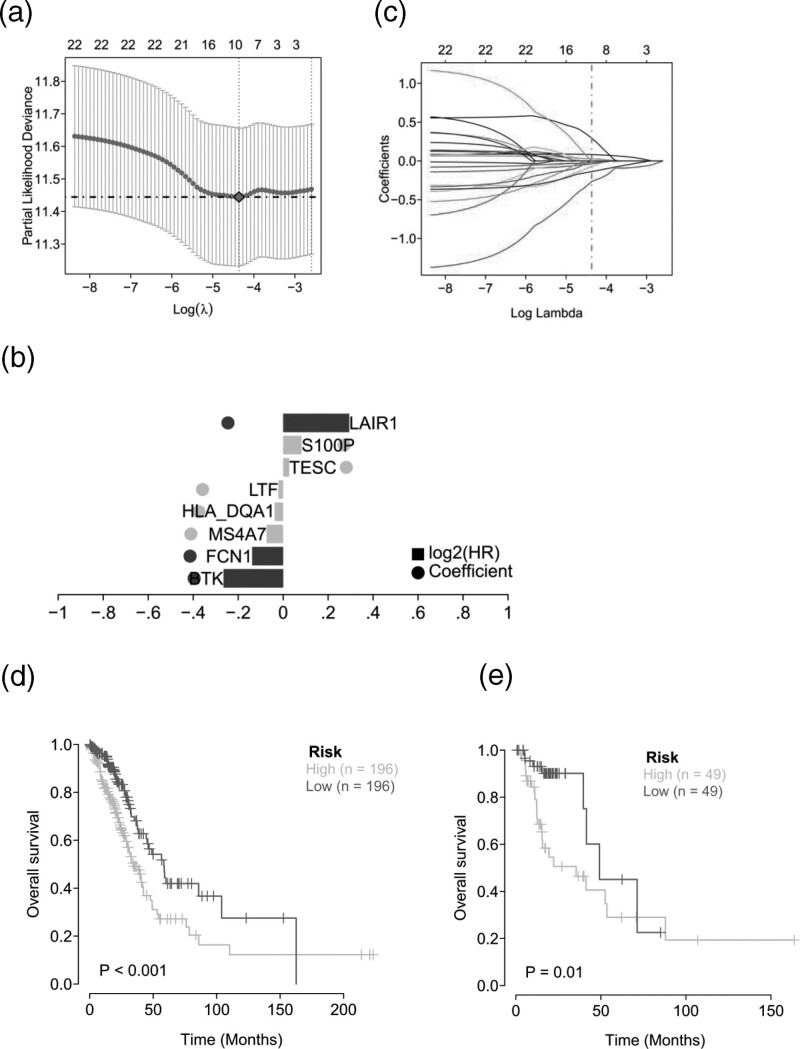
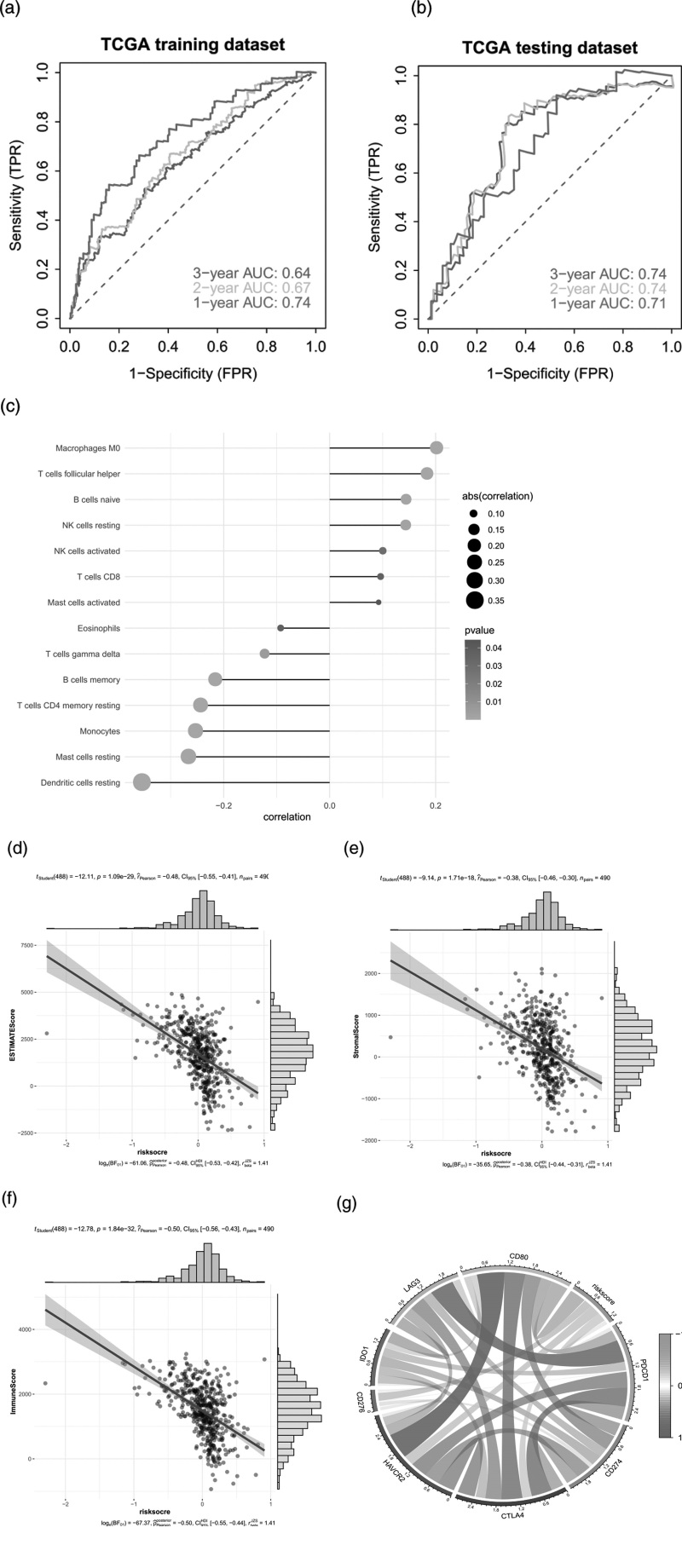
To further identify the key genes in the differential gene concentration, clinical information of LUAD patients was collected. Cox univariate regression and Lasso regression feature selection algorithm were used to screen out the characteristic genes in LUAD (Fig. 3a). The results showed that a total of eight prognostic genes were screened by Cox univariate regression (P value <0.05) (Fig. 3b). TCGA patients were randomly divided into training set and validation set in a ratio of 4:1. The best riskScore (Risk Score = BTK × (−0.26524) + FCN1 × (−0.13769) + MS4A7 × (−0.07342) + HLA_DQA1 × (−0.03788) after Lasso regression analysis + LTF × (−0.02093) + TESC × 0.02685 + S100P × 0.08172 + LAIR1 × 0.29365) for subsequent analysis(Fig. 3c). Patients were divided into high-risk and low-risk groups based on median risk scores and Kaplan–Meier curve analysis was used. The OS of the high-risk group in the training set and test set was significantly lower than that of the low-risk group. In addition, ROC curve results showed that both the training set and the test set had good validation efficiency (Fig. 3d,e).
Fig. 3.
Construction and validation of risk characteristic model. (a,c) Lasso regression constructed apa-related prognostic model; (b) Model gene coefficient diagram; (d) Survival analysis of TCGA training set model; (e) Survival analysis of TCGA test set model.
Multi-omics study to explore the clinical predictive value of the model
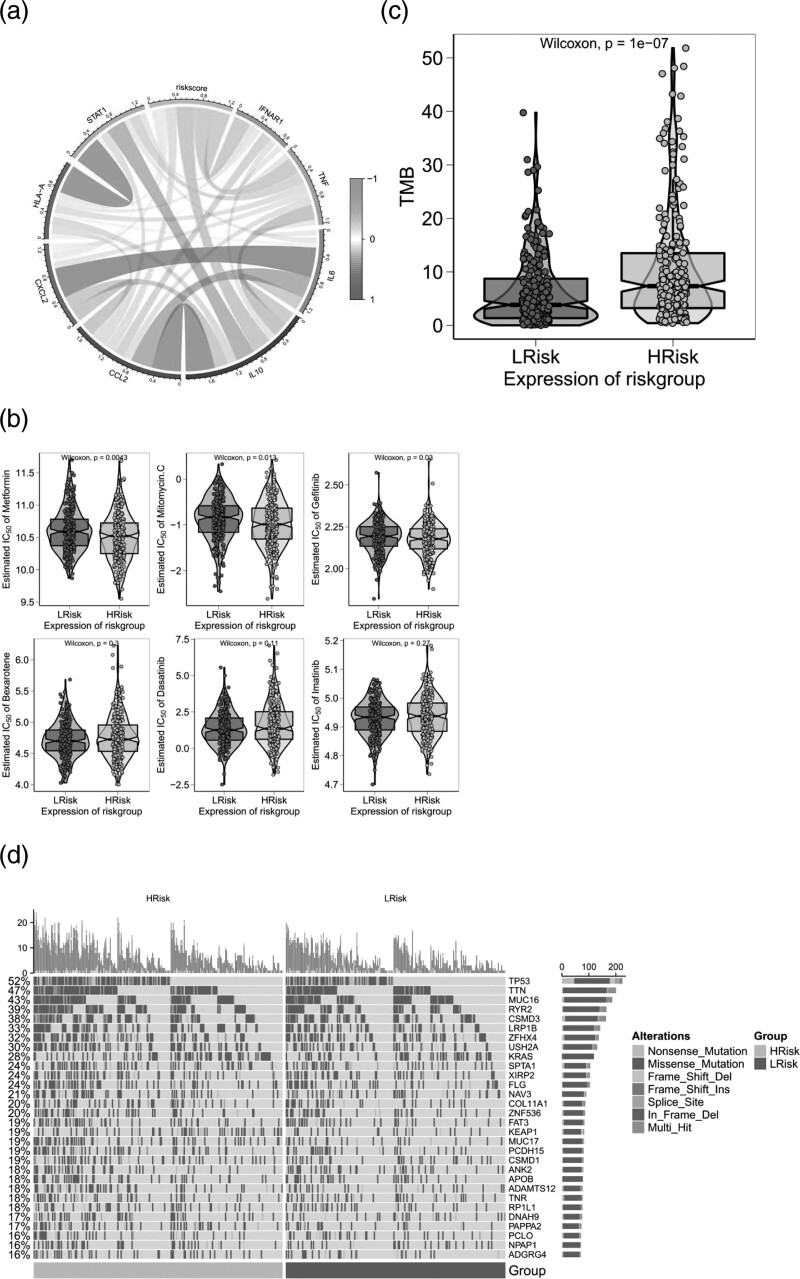
The tumor microenvironment is mainly composed of tumor-associated fibroblasts, immune cells, extracellular matrix, a variety of growth factors, inflammatory factors, special physical and chemical characteristics and cancer cells themselves, etc. the tumor microenvironment significantly affects tumor diagnosis, survival outcome and clinical treatment sensitivity. The relationship between risk score and tumor immune invasion was analyzed to further explore the potential molecular mechanism of risk score influencing the progression of LUAD. The results showed that risk score was significantly positively correlated with Macrophages M0, T cells follicular helper, B cells naive and NK cells resting. It is significantly negatively correlated with dendritic cells resting, mast cells resting, monocyte, T cells CD4 memory resting and B cells memory(Fig. 4c–f). In addition, we explored correlations between risk score and matrix score, immune score, composite score, immune checkpoint and inflammatory cytokines (Fig. 4g). The results showed that risk score was significantly negatively correlated with matrix score, immunity score and overall score. In addition, risk scores were negatively correlated with immune checkpoints and inflammatory cytokine expression. The effect of operation combined with chemotherapy on early LUAD is clear. Based on the drug sensitivity data from the GDSC database, we used R package ‘pRRophetic’ to predict the chemotherapy sensitivity of each tumor sample and further explored the risk score and the sensitivity of common chemotherapy drugs. The results showed that risk score significantly affected the sensitivity of patients to metformin, mitomycine C and gefitinib (Fig. 5b). We further analyzed the genetic mutations of patients in the high and low-risk groups, and the results were presented in the form of mutation maps(Fig. 5d). We also found that the tumor mutation load was significant between the high and low-risk groups(Fig. 5c).
Fig. 4.
ROC analysis of the prognostic model. (a) the ROC curve related to TCGA training set model (1-2-3 years). (b) the ROC curve related to TCGA test set model (1-2-3 years), and the clinical predictive value of the model explored in multiple omics studies; (c) Correlation between risk score and tumor immune invasion; (d–g) Correlation between risk score and ESTIMATES, Stromal, ImmuneImmune, checkpoint.
Fig. 5.
(a) Correlation of risk scores of cytokines. (b) Risk score and sensitivity to common chemotherapeutic drugs. (c) Tumor mutation load was significant between high and low risk groups. (d) Mutation profiles of patients at high and low risk.
Study on specific signal mechanism related to prognosis model
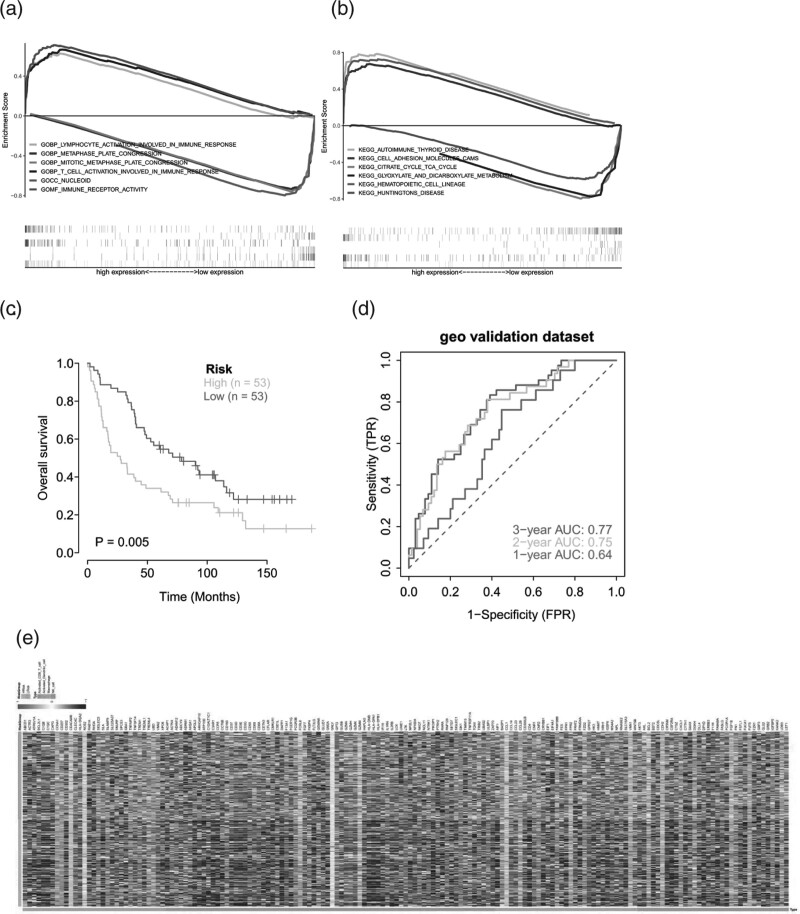
We study the specific signaling pathways involved in the high-low risk correlation model and explore the potential molecular mechanisms by which risk score influences tumor progression. Finally, we found significant enrichment in many related pathways through GSEA analysis. For example, GOBP LYMPHOCYTE ACTIVATION INVOLVED IN IMMUNE RESPONSE, GOMF IMMUNE RECEPTOR ACTIVITY and KEGG CELL ADHESION MOLECULES CAMS, KEGG HEMATOPOIETIC CELL LINEAGE. Some of these highly significant pathways were shown separately (Fig. 6a). These results suggest that the disturbance of these signaling pathways in patients with high and low-risk groups affects the prognosis of patients with LUAD(Fig. 6b).
Fig. 6.
GSEA pathway analysis between high and low risk groups. (a) GSEA-Go enrichment analysis related to high and low risk groups; (b) GSEA-KEGG enrichment analysis related to high and low risk groups. External data sets verify the predictive efficacy of risk models; (c) Kaplan–meier curves of GEO external data sets; (d) ROC curve of GEO external data set in 1-2-3; (e) Immunoregulation factor analysis: heat map of expression of immunoregulation related genes in the high and low-risk groups.
External data sets validated the robustness of the prognostic model
In this study, the data of LUAD patients with survival data processed in the GEO database (GSE37745) were downloaded, and the clinical classification of LUAD patients in the GEO database was predicted according to the model. The survival difference between the two groups was evaluated by Kaplan–Meier to explore the stability of the prediction model. The results showed that the OS of the high-risk group of the GEO external validation set was significantly lower than that of the low-risk group (Fig. 6c). To verify the accuracy of the model, we used external data sets to perform the ROC curve analysis on the model, and the results showed that the model had a strong predictive efficacy for patient prognosis prediction(Fig. 6d).
Immunoregulatory factor analysis and TIDE immune system function analysis
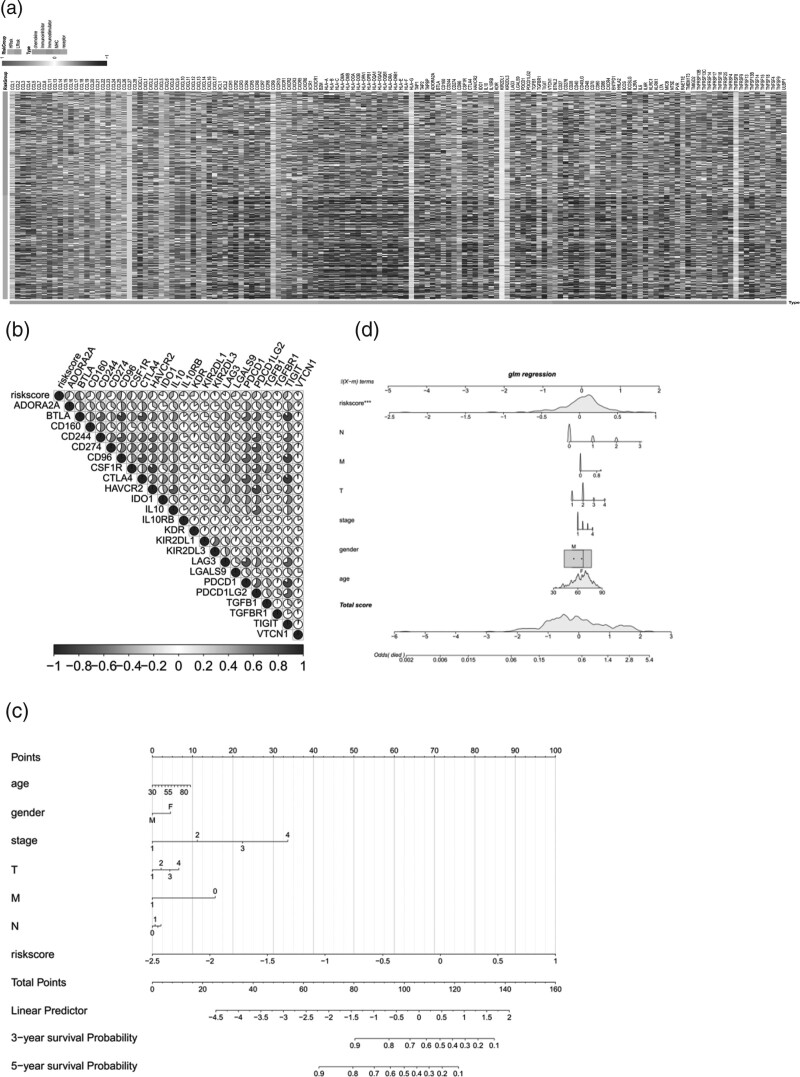
We further analyzed the interaction between tumor and immune system with the help of the website TISIDB and found that the differences of genes related to immune regulation and chemokines between the high and low-risk groups (Fig. 6e). We further explored the relationship between the expression of immunosuppressor genes and risk score (Fig. 7a). Correlation analysis showed that multiple genes such as ADORA2A, BTLA, CD160, CD244, CD274, CD96, CSF1R and CTLA4 were highly correlated with risk score (Fig. 7b).
Fig. 7.
Analysis of immune regulatory factors. (a) heat map of expression of immune regulation-related genes in the high and low-risk groups; (b) Correlation between risk score and immunomodulator; (c) Construction and efficacy evaluation of a nomogram model for risk scoring: logistic regression line graph model; (d) GLM regression line graph model.
Risk analysis, independent prognostic analysis and correlation analysis of several clinical indicators
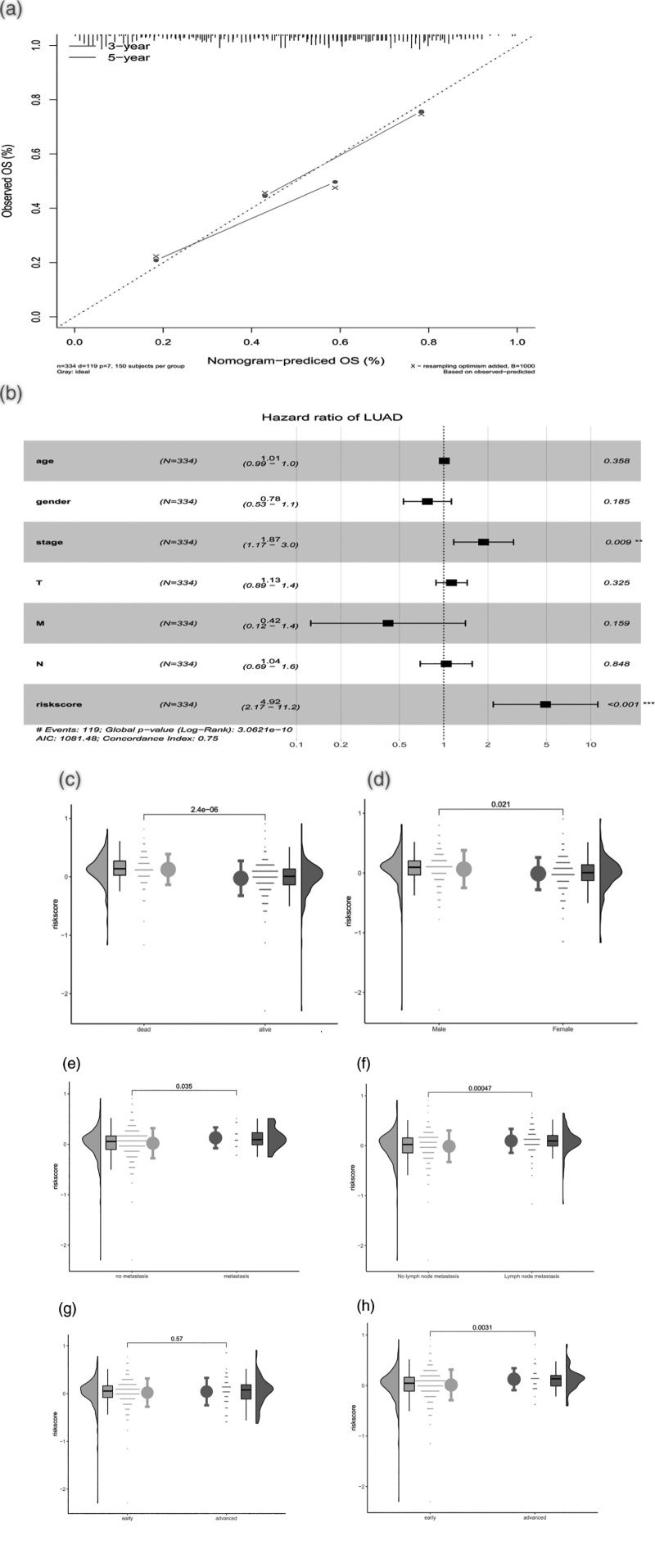
According to the median value of riskScore, the samples were divided into high-risk and low-risk groups, and the results of regression analysis were presented in the form of a column plot. The results of logistic regression analysis showed that in all of our samples, the distribution of different clinical indicators of LUAD and riskScore contributed to the whole scoring process to varying degrees. The distribution of riskScore values contributed to the scoring process in each period of cancer (Fig. 7c,d). At the same time, we also conducted a forecast analysis on OS situation in 3 years and 5 years(Fig. 8a). Multivariate analysis showed that RiskScore and stage were independent prognostic factors for LUAD patients (Fig. 8b). According to the size of the clinical index values, we divided the riskScore values corresponding to the samples into different groups, and showed the results of grouping each clinical index in the form of a violin diagram. Furthermore, kruskal.test showed that the distribution of RiskScore values in fuSTAT, GENDER, T, M, N and other clinical indicators was significant (P < 0.05) (Fig. 8c–f).
Fig. 8.
(a) Correction curve of the nomogram model for 3 years/5 years. Independent prognostic analysis of risk scoring models: (b) multi-factor analysis of risk scoring models. Relationship between risk score and clinical symptoms:(c–h) relationship between risk score and multiple clinical symptoms (fustat, gender, T, M, N, stage).
The prognostic model correlated with tumor stage
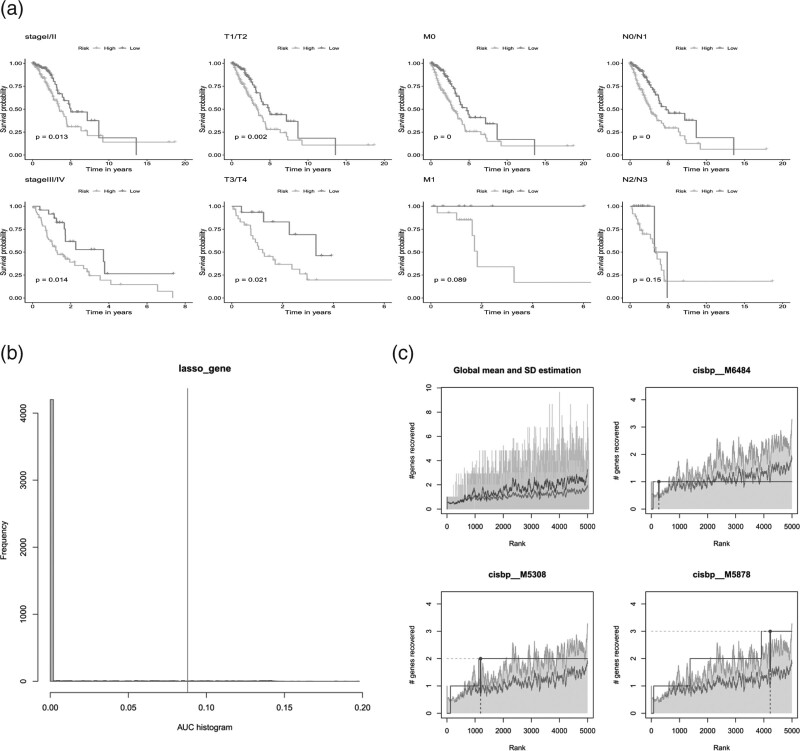
Subsequently, we discussed the significance of the prognostic model in various stages of LUAD. The survival analysis results of KM subgroup showed that the model could predict the survival of patients with different stages, T stages and M0, N0/N1 Stage, suggesting that the prognostic model had good applicability (Fig. 9a).
Fig. 9.
Prediction efficacy of prognostic models in different subgroups. (a) Prognostic models showed strong prognostic efficacy in different subgroups (Stage, T, M, N); (b) Transcriptional regulatory factor analysis of model genes: cumulative recovery curve of transcriptional factors; (c) Enrichment analysis of cumulative recovery curves of transcription factors.
Model gene transcriptional regulatory network
We applied eight model genes to the gene set for this analysis and found that they are regulated by common mechanisms, such as multiple transcription factors. Therefore, the accumulative recovery curve of these transcription factors utilization was analyzed for enrichment (Fig. 9b). Motif-tf annotation, and selection of important genes. The analysis results showed that transcription factor NFAT5 was the main regulator of gene concentration. MOTIF annotation was cisbp-M6366. A total of 2 model genes were enriched in this motif. The standardized enrichment score (NES) was 6.02. All the motifs and corresponding transcription factors enriched in the modeled genes were presented (Fig. 9c.).
Discussion
Lung cancer is one of the leading causes of tumor-related death. LUAD is the most common histological type of lung cancer, accounting for 40%, posing a great threat to human health [11–13]. In recent years, due to the rapid development of immunosuppressive therapy, the prognosis of patients with LUAD has been greatly improved [14]. However, only about one-third of patients with cancer receive a stable response from immunotherapy [15]. Therefore, how to predict the effect of immunotherapy has become a focus of clinical attention. A single biomarker can no longer meet the needs of practical application. The scoring system formed by integrating multiple data types is attracting more and more attention [16]. Tumor stem cells are a group of highly heterogeneous tumor cells in different states of differentiation. Although tumor stem cells are a small cell population in tumor tissue, they affect the efficacy of free treatment for cancer patients. Tumor stem cells can induce the production of more immunosuppressive M2 macrophages, and tumor stem cells can directly impair the activity of cytotoxic T cells to suppress the immune response [17]. In addition, cancer stem cells also play an important role in the development of cancer vaccines [18–20]. Therefore, tumor stem cells may be used as predictors of the therapeutic efficacy of immune checkpoint inhibition.
In this study, 106 differentially expressed tumor stem cell-related genes in LUAD and paracancer tissues were identified and their copy numbers were significantly changed in LUAD samples [21–25]. This may be one of the important reasons for the change of its expression in the tumor. The results of univariate Cox regression analysis showed that eight genes had a significant influence on the prognosis of LUAD, and there was a complex expression correlation network among the eight differentially expressed genes [26], suggesting that these genes may be closely related to the degree of tumor stem cell invasion. Our results showed that risk score was significantly positively correlated with Macrophages M0, T cells follicular helper, B cells naive and NK cells resting [27–29]. It is significantly negatively correlated with dendritic cells resting, mast cells resting, monocyte, T cells CD4 memory resting and B cells memory. Our study was based on drug sensitivity data from the GDSC database to further explore the risk score and sensitivity to common chemotherapeutic agents [30]. We found that risk score significantly affected the sensitivity of patients to metformin, mitomycine C and gefitinib.
Tumor dry scoring system for the treatment of immune checkpoint inhibition effect evaluation in a number of independent immunotherapy in the queue is good forecast results were obtained and compared with the existing immunotherapy predictor (such as the expression of immune checkpoints, immune cell infiltration phenotype and cytotoxicity score, antigen presented, et al.) to present a consistent correlation [31–33]. Therefore, the matrix score and the immune score are expected to be a new predictors of the efficacy of immunotherapy [34]. Although this study reveals the predictive value of the matrix score and immune score system for immune checkpoint inhibition treatment to some extent, there are still some deficiencies in the research. First, this study is based on the mRNA expression data of TCGA and GEO, which has obvious racial specificity, and whether it can be applied to other races remains to be further verified [35]. Second, the sample size of the immunotherapy validation cohort of immune checkpoint inhibition therapy is small, so the immunotherapy cohort with a larger sample size is still needed for validation [36].
In summary, eight model genes were used in the gene set for this analysis, and it was found that they were regulated by multiple transcription factors and other common mechanisms [37–39]. Transcription factor NFAT5 was the main regulator of gene concentration, and there was statistical significance in the difference of survival prognosis and immune cell infiltration in LUAD (P < 0.05). This study also constructed a matrix score and immune score model, which has potential value in predicting the efficacy of immunocheckpoint inhibition therapy in patients with LUAD [40]. Therefore, research of APA is expected to provide a theoretical basis for immune checkpoint inhibition therapy in LUAD patients, but more multicenter sample sizes are needed for further experimental verification.
Acknowledgements
The authors would like to express their gratitude to AJE for the expert linguistic services provided.
This work was supported by Scientific Research Project of Education Department of Anhui Province(YJS20210324), the National Natural Science Foundation of China (81972829), the Science and Technology Innovation Committee of Shenzhen Municipality (Grant No. JCYJ20180228162607111, JCYJ20190809104601662), the Health and Family Planning Commission of Shenzhen Municipality Research Project (Grant No. SZBC2018018), China Scholarship Council (CSC, 201908470124), Peking University-University of Michigan JI Project 2018 [2019020(PUSH)-r1], Science, Technology and Innovation Commission of Shenzhen Municipality (JCYJ20180228175531145) and Shenzhen Huada Life Sciences Open Fund Project (BGIRSZ20200003).
X.L. designed the research; J.L. guided the research; D.W., Y.Z., X.Y., Le.Lv., X.R., Q.L., Y.F. and P.X. collected and downloaded the data of our research; L.W. analyzed the data and wrote the article. All the authors revised it critically for important intellectual content, gave final approval of the version to be published and agreed to be accountable for all aspects of the work.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Liusheng Wu and Xin Ruan Contributed equally for this study.
References
- 1.Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine 2018; 34:267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szkop KJ, Cooke PIC, Humphries JA, Kalna V, Moss DS, Schuster EF, Nobeli I. Dysregulation of alternative poly-adenylation as a potential player in autism spectrum disorder. Front Mol Neurosci 2017; 10:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aphasizheva I, Yu T, Suematsu T, Liu Q, Mesitov MV, Yu C, et al. Poly(A) binding KPAF4/5 complex stabilizes kinetoplast mRNAs in Trypanosoma brucei. Nucleic Acids Res 2020; 48:8645–8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hojo H, Yashiro Y, Noda Y, Ogami K, Yamagishi R, Okada S, et al. The RNA-binding protein QKI-7 recruits the poly(A) polymerase GLD-2 for 3’ adenylation and selective stabilization of microRNA-122. J Biol Chem 2020; 295:390–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engel BJ, Grindel BJ, Gray JP, Millward SW. Purification of poly-dA oligonucleotides and mRNA-protein fusions with dT25-OAS resin. Bioorg Med Chem Lett 2020; 30:126934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yalamanchili HK, Alcott CE, Ji P, Wagner EJ, Zoghbi HY, Liu Z. PolyA-miner: accurate assessment of differential alternative poly-adenylation from 3’Seq data using vector projections and non-negative matrix factorization. Nucleic Acids Res 2020; 48:e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickard MAG, Brylow KB, Cisco LA, Anecelle MR, Pershun ML, Chandrasekaran AR, et al. Parallel poly(A) homo- and hetero-duplex formation detection with an adapted DNA nanoswitch technique. RNA 2020; 26:1118–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kashiwabara SI, Tsuruta S, Okada K, Yamaoka Y, Baba T. Adenylation by testis-specific cytoplasmic poly(A) polymerase, PAPOLB/TPAP, is essential for spermatogenesis. J Reprod Dev 2016; 62:607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dejene EA, Li Y, Showkatian Z, Ling H, Seto E. Regulation of poly(a)-specific ribonuclease activity by reversible lysine acetylation. J Biol Chem 2020; 295:10255–10270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamanaka K, Fukumoto H, Takehara M, Hamano Y, Oikawa T. The stereocontrolled biosynthesis of mirror-symmetric 2,4-diaminobutyric acid homopolymers is critically governed by adenylation activations. ACS Chem Biol 2020; 15:1964–1973. [DOI] [PubMed] [Google Scholar]
- 11.Nilsen TW. Measuring the length of poly(A) tails. Cold Spring Harb Protoc 2015; 2015:413–418. [DOI] [PubMed] [Google Scholar]
- 12.Rodríguez-Vargas JM, Rodríguez MI, Majuelos-Melguizo J, García-Diaz Á, González-Flores A, López-Rivas A, et al. Autophagy requires poly(adp-ribosyl)ation-dependent AMPK nuclear export. Cell Death Differ 2016; 23:2007–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawazaki R, Imai S, Yokogawa M, Hosoda N, Hoshino SI, Mio M, et al. Characterization of the multimeric structure of poly(A)-binding protein on a poly(A) tail. Sci Rep 2018; 8:1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng L, Zhang H, Ma J, Liu J, Ma S, Wang M, Huo Y. Phylogenetic and biological characterizations of a GI.3 norovirus. Infect Genet Evol 2020; 85:104554. [DOI] [PubMed] [Google Scholar]
- 15.Mesitov MV, Yu T, Suematsu T, Sement FM, Zhang L, Yu C, et al. Pentatricopeptide repeat poly(A) binding protein KPAF4 stabilizes mitochondrial mRNAs in Trypanosoma brucei. Nat Commun 2019; 10:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim J, Lee M, Son A, Chang H, Kim VN. mTAIL-seq reveals dynamic poly(A) tail regulation in oocyte-to-embryo development. Genes Dev 2016; 30:1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maruyama C, Niikura H, Takakuwa M, Katano H, Hamano Y. Colorimetric detection of the adenylation activity in nonribosomal peptide synthetases. Methods Mol Biol 2016; 1401:77–84. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz-Cordero R, Devine WP. Targeted therapy and checkpoint immunotherapy in lung cancer. Surg Pathol Clin 2020; 13:17–33. [DOI] [PubMed] [Google Scholar]
- 19.Schabath MB, Cote ML. Cancer progress and priorities: lung cancer. Cancer Epidemiol Biomarkers Prev 2019; 28:1563–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu L, Zheng Y, Liu J, Luo R, Wu D, Xu P, et al. Comprehensive evaluation of the efficacy and safety of LPV/r drugs in the treatment of SARS and MERS to provide potential treatment options for COVID-19. Aging (Albany NY) 2021; 13:10833–10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu L, Zheng Y, Ruan X, Wu D, Xu P, Liu J, et al. Long-chain noncoding ribonucleic acids affect the survival and prognosis of patients with esophageal adenocarcinoma through the autophagy pathway: construction of a prognostic model. Anticancer Drugs 2021; 33:e590–e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doroshow DB, Sanmamed MF, Hastings K, Politi K, Rimm DL, Chen L, et al. Immunotherapy in non-small cell lung cancer: facts and hopes. Clin Cancer Res 2019; 25:4592–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suresh K, Naidoo J, Lin CT, Danoff S. Immune checkpoint immunotherapy for non-small cell lung cancer: benefits and pulmonary toxicities. Chest 2018; 154:1416–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braun W, Rega MJ. Adenyl cyclase-stimulating catecholamines as modifiers of antibody formation. Immunol Commun 1972; 1:523–532. [DOI] [PubMed] [Google Scholar]
- 25.Régnier P, Hajnsdorf E. The interplay of Hfq, poly(A) polymerase I and exoribonucleases at the 3’ ends of RNAs resulting from Rho-independent termination: a tentative model. RNA Biol 2013; 10:602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji T, Chen Q, Tao S, Shi Y, Chen Y, Shen L, et al. The research progress of circular RNAs in hematological malignancies. Hematology 2019; 24:727–731. [DOI] [PubMed] [Google Scholar]
- 27.Boele J, Persson H, Shin JW, Ishizu Y, Newie IS, Søkilde R, et al. PAPD5-mediated 3’ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc Natl Acad Sci U S A 2014; 111:11467–11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jänicke A, Vancuylenberg J, Boag PR, Traven A, Beilharz TH. ePAT: a simple method to tag adenylated RNA to measure poly(A)-tail length and other 3’ RACE applications. RNA 2012; 18:1289–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sau SP, Larsen AC, Chaput JC. Automated solid-phase synthesis of high capacity oligo-dT cellulose for affinity purification of poly-A tagged biomolecules. Bioorg Med Chem Lett 2014; 24:5692–5694. [DOI] [PubMed] [Google Scholar]
- 30.Szymanski JJ, Jamison JT, DeGracia DJ. Texture analysis of poly-adenylated mRNA staining following global brain ischemia and reperfusion. Comput Methods Programs Biomed 2012; 105:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slomovic S, Schuster G. Circularized RT-PCR (cRT-PCR): analysis of the 5’ ends, 3’ ends, and poly(A) tails of RNA. Methods Enzymol 2013; 530:227–251. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, Sement FM, Suematsu T, Yu T, Monti S, Huang L, et al. PPR polyadenylation factor defines mitochondrial mRNA identity and stability in trypanosomes. EMBO J 2017; 36:2435–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Zhu B, Zeng Y, Shen H, Zhang J, Wang X. Clinical lipidomics in understanding of lung cancer: opportunity and challenge. Cancer Lett 2020; 470:75–83. [DOI] [PubMed] [Google Scholar]
- 34.Katoh M, Nakagama H. FGF receptors: cancer biology and therapeutics. Med Res Rev 2014; 34:280–300. [DOI] [PubMed] [Google Scholar]
- 35.Ye Y, Jing Y, Li L, Mills GB, Diao L, Liu H, Han L. Sex-associated molecular differences for cancer immunotherapy. Nat Commun 2020; 11:1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Y, Jiang Y, Li Y, Ma S. Identification of cancer omics commonality and difference via community fusion. Stat Med 2019; 38:1200–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang S. APA makes a short cut for ramping up HSC metabolism. Cell Stem Cell 2020; 26:615–616. [DOI] [PubMed] [Google Scholar]
- 38.Vallejos Baier R, Picao-Osorio J, Alonso CR. Molecular regulation of alternative polyadenylation (APA) within the Drosophila Nervous System. J Mol Biol 2017; 429:3290–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Feng W, Xu S, He B. Extensive involvement of alternative polyadenylation in single-nucleus neurons. Genes (Basel) 2020; 11:709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Travaglino A, Raffone A, Gencarelli A, Zullo F, Di Spiezio Sardo A, Insabato L. Significance of stromal markers in atypical polypoid adenomyoma. Pathol Res Pract 2020; 216:153133. [DOI] [PubMed] [Google Scholar]