Abstract
Background:
Epigenetics, and especially DNA methylation, contributes to the pathogenesis of sporadic amyotrophic lateral sclerosis (SALS). This study aimed to investigate the role of DNA methylation in SALS using whole blood of SALS patients.
Methods:
In total, 32 SALS patients and 32 healthy controls were enrolled in this study. DNA was isolated from whole blood collected from the participants. DNA methylation profiles were generated using Infinium MethylationEPIC BeadChip.
Results:
We identified 34 significant differentially methylated positions (DMPs) in whole blood from SALS patients, compared with the healthy controls. Of these DMPs, five were hypermethylated and 29 were hypomethylated; they corresponded to 13 genes. For the DMPs, ATAD3B and BLK were hypermethylated, whereas DDO, IQCE, ABCB1, DNAH9, FIGN, NRP1, TMEM87B, CCSAP, ST6GALNAC5, MYOM2, and RUSC1-AS1 were hypomethylated. We also identified 12 differentially methylated regions (DMRs), related to 12 genes (NWD1, LDHD, CIS, IQCE, TNF, PDE1C, LGALS1, CSNK1E, LRRC23, ENO2, ELOVL2, and ELOVL2-AS1). According to data from the Kyoto Encyclopedia of Genes and Genomes database, DNAH9 and TNF are involved in the amyotrophic lateral sclerosis (ALS) pathway. Correlation analysis between clinical features and DNA methylation profiling indicated that the methylation level of ELOVL2 and ARID1B was positively associated with the age of onset (r = 0.86, adjust P = 0.001) and disease duration (r = 0.83, adjust P = 0.01), respectively.
Conclusions:
We found aberrant methylation in DMP- and DMR-related genes, implying that many epigenetic alterations, such as the hypomethylation of DNAH9 and TNF, play important roles in ALS etiology. These findings can be helpful for developing new therapeutic interventions.
Keywords: Amyotrophic lateral sclerosis, DNA methylation, Differentially methylated positions, Differentially methylated regions, Whole blood
Introduction
Amyotrophic lateral sclerosis (ALS) is a rare neurodegenerative disease that is characterized by the progressive loss of upper and lower motor neurons.[1] Most patients die within 3 to 5 years after the onset of symptoms because of respiratory failure.[2] However, the pathophysiological mechanisms underlying ALS are still not fully understood. It has been suggested that both familial ALS (FALS) and sporadic ALS (SALS) must be addressed to comprehensively explain the disease.[3] Many novel genes have been identified as targets for next-generation sequencing developments.[1,2] However, mutations in these genes account for approximately two-thirds of all FALS cases and approximately 10% of SALS cases. In this regard, genetic factors can only explain a small proportion of patients with SALS.
A number of population-based and case-control studies have revealed that environmental risk factors are associated with ALS.[4–6] Thus, focus has recently been directed into ALS environmental research to better elucidate the disease mechanisms.[7] Though epigenetic processes are modified by the environment, they have a genetic component and can regulate genetic signals. Thus, they are considered to be links between genetic risks, environmental exposure, and diseases.[8,9] Growing evidence suggests that in addition to genetic mechanisms, epigenetic mechanisms also play a critical role in ALS. Indeed, ALS has been posited to be caused by complex interactions between genes, environmental exposure, and impaired molecular pathways.[9] DNA methylation is one of the major and most widely studied epigenetic mechanisms. It refers to the post-transcriptional covalent addition of a methyl group to cytosine residues in DNA, leading to the formation of 5-methylcytosine (5mC).[9] This epigenetic phenomenon commonly occurs in genomic regulatory regions, such as at promoter element CpG dinucleotides, or is clustered in CpG islands or on CpG island shores.[9,10] Moreover, a recent study revealed that DNA methylation signatures are unique patterns of DNA methylation alterations that can be defined for rare disorders caused by pathogenic variants.[11] Considering that ALS is a rare disorder for which a number of related pathogenic variants have been discovered, it is essential to explore the association between DNA methylation and ALS, particularly regarding SALS.
Several case-control studies have used postmortem nervous tissues or blood to reveal differential DNA methylation patterns. For instance, an investigation of whole genome methylation in brain DNA suggested that SALS patients exhibited either hypermethylation or hypomethylation at 38 methylation sites, of which 23 were associated with genes and three were associated with CpG islands.[10] Figueroa-Romero et al[12] discovered alterations in global methylation in postmortem SALS spinal cord. However, they failed to find a differential percentage in global 5mC in the whole blood of SALS patients. In contrast, Tremolizzo et al[13] not only found that DNA methylation was increased in the whole blood of ALS patients but also considered this to be a possible marker of epigenetic dysfunction in ALS, independent of onset age. Notably, global DNA methylation levels in blood have been shown to be significantly higher in ALS patients than in asymptomatic and paucisympomatic carriers or in family members who are non-carriers of SOD1 mutations.[14] Together, these findings suggest that altered DNA methylation contributes to neurodegeneration in ALS patients.
In this study, therefore, we profiled epigenome-wide DNA methylation in 64 blood-derived DNA samples from 32 SALS patients and 32 healthy controls. We used Infinium MethylationEPIC BeadChip (Illumina, San Diego, CA, USA), which is a highly efficient tool for DNA methylation research that contains >850,000 CpG probes, to identify DNA methylation patterns associated with SALS.
Methods
Ethical approval
The study was approved by the Ethics Committee of Peking Union Medical College Hospital (No. JS-2624) and written informed consent was obtained from each participant.
Patients and controls
All patients were enrolled between December 2020 and May 2021 at Peking Union Medical College Hospital. Patients were diagnosed with SALS according to the revised El Escorial criteria.[15] Healthy relatives who had lived with these patients for a long time and accompanied them to the clinic were enrolled as the control group.
Clinical information, including sex, age, age of onset, site of onset, disease duration, Revised ALS Functional Rating Scale (ALSFRS-R) score, King's clinical staging, and diagnostic categories, were recorded for all patients. “ΔALSFRS-R,” defined here as (48 - ALSFRS-R at baseline)/duration from symptom onset to baseline clinical diagnosis in months, was used as a measure of the disease progression rate. Sex and age information were also recorded for the control group.
Sample collection, DNA extraction, and measurement of DNA methylation
At the time of diagnosis, peripheral blood samples were collected from patients and controls. DNA was isolated from whole blood using a DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). A Nanodrop 2000 (Thermo Scientific, Wilmington, DE, USA) was used to estimate the purity and concentration of DNA. Approximately, 500 ng of genomic DNA from each sample was bisulfite-converted using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA), according to the manufacturer's standard protocol. DNA methylation profiles were measured using the Infinium MethylationEPIC BeadChip (Illumina), following the manufacturer's instructions.
Data analysis
R software (R Foundation, Vienna, Austria) was used for data analysis. The array data (.IDAT files) were read with the “minfi” package and analyzed using the “ChAMP” package to determine the methylation level. For all probes, the methylation status is denoted here as b value, which comprises the ratio of the methylated probe intensity to the overall probe intensity (ie, the sum of methylated and unmethylated probe intensities plus the constant a, where a = 100). CpG sites having |Δβ| ≥ 0.1 (in patients vs. controls) and P < 0.05 were considered to be differentially methylated positions (DMPs). If Δβ ≥ 0.1, the CpG site in question was considered to be hypermethylated; if Δβ ≤ −0.1, the site was considered to be hypomethylated. When a region contained more than seven CpG sites within a distance of 1000 bp, and the false discovery rate was <0.05, the said region was defined as a differentially methylated region (DMR). The averageβ values of promoters and CpG islands were compared between patients and controls. Promoters and CpG islands for which |Δβ| ≥ 0.10 and P < 0.05 were considered for further analysis. DMPs were identified using the “limma” package and DMRs were identified using the “DMRcate” package. To investigate the significances of DMP and DMR target genes in biological processes and functions, the Gene Ontology (GO) annotations and functions of the said genes were obtained from UniProt (https://www.uniprot.org/). The pathways involved with genes were searched for in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database at https://www.kegg.jp/kegg/pathway.html. To explore the potential genetic and epigenetic relationships in ALS, network string interactions were performed between established ALS genes[2,3] and the DMP/DMR-related genes identified in our study. All of the genes were ranked using the degree method in Cytoscape.
Clinical data are shown here as means ± standard deviation if they were normally distributed, or as medians (interquartile range) if they were not normally distributed. The Wilcoxon test was used to compare DNA methylation levels between patients and controls, as well as between patients at different stages. The correlation between DNA methylation level and clinical features was calculated by Spearman correlation analysis using the Weighted Correlation Network Analysis function “corAndPvalue”. Statistical significance was set at P < 0.05.
Results
Demographic characteristics of patients and controls
The demographic and clinical characteristics of the patients are summarized in Table 1. The patient group comprised nine individuals with clinically definite ALS, nine with clinically probable ALS, and 14 with clinically laboratory-supported probable ALS. There were 19 males and 13 females; the group's mean age was 54.7 ± 10.2 years at the time of sample collection. The control group comprised 14 males and 18 females, with a mean age of 38.6 ± 10.0 years. In the patient group, the mean age of onset was 53.5 ± 10.2 years. Ten patients had bulbar onsets, with the remaining 22 having limb onsets; the group's mean disease duration was 11.5 months. The mean ALSFRS-R score was 37, and the mean DALSFRS-R was 0.9. The number of patients at each King's stage is also shown in Table 1.
Table 1.
Characteristics of ALS patients and healthy controls.
| Variables | ALS patients (n = 32) | Controls (n = 32) |
| Male, n | 19 | 14 |
| Age at sample collection, mean (SD) | 54.7 (10.2) | 38.6 (10.0) |
| Age of onset, mean (SD) Site of onset, n | 53.5 (10.2) | – |
| Bulbar | 10 | – |
| Limb | 22 | – |
| Disease duration (months), median (IQR) | 11.5 (11.0) | – |
| ALSFRS-R score, median (IQR) | 37 (11) | – |
| DALSFRS-R, median (IQR) King's stage, n | 0.9 (1.0) | – |
| 2 | 14 | – |
| 3 | 6 | – |
| 4 | 12 | – |
ΔALSFRS-R: (48 - ALSFRS-R at baseline)/duration from symptom onset to baseline clinical diagnosis in months. ALS: Amyotrophic lateral sclerosis; ALSFRS-R: Revised ALS Functional Rating Scale; IQR: Interquartile range; SD: Standard deviation.
Comparison of methylation status between ALS patient and healthy control groups
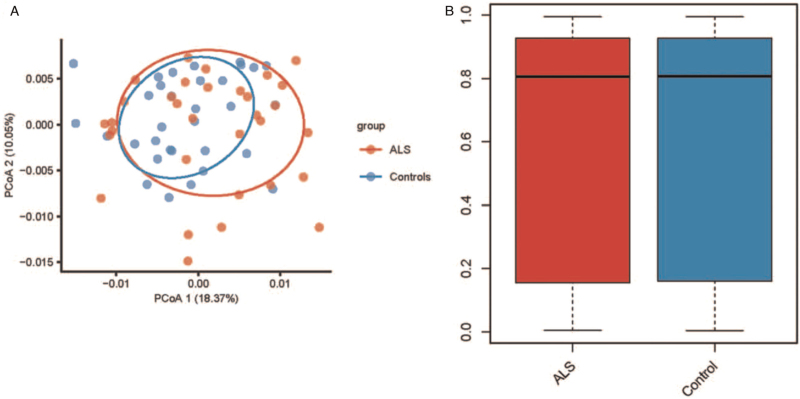
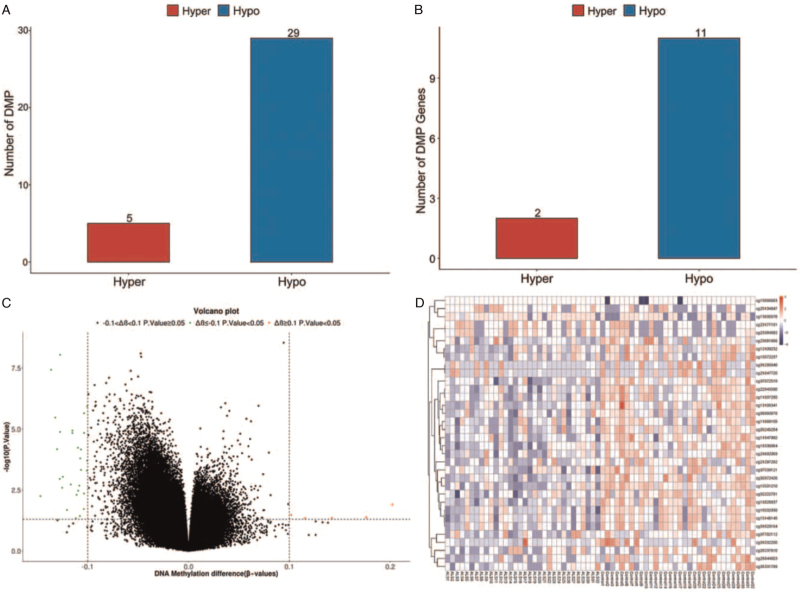
The principal coordinates analysis of samples from the two groups is shown in Figure 1A. There was no significant difference between the ALS patient and control groups regarding the whole DNA methylation level (P > 0.05, Figure 1B). A total of 34 DMPs were identified, among which five were hypermethylated and 29 were hypome-thylated [Figure 2A]. These DMPs were distributed across 13 genes, including two hypermethylated genes and 11 hypomethylated genes [Figure 2B]. Among all DMPs, cg01295646, cg21847720, and cg01032200 were located in CpG islands [Supplementary Table 1]. The two hypermethylated genes were ATAD3B and BLK, and the 11 hypomethylated genes were DDO, IQCE, ABCB1, DNAH9, FIGN, NRP1, TMEM87B, CCSAP, ST6GALNAC5, MYOM2, and RUSC1-AS1. Detailed information on the DMPs and their related genes is provided in Supplementary Table 1. By comparing the average β values of DMPs between the two groups, we found that β value was significantly lower in the ALS group than in the control group (P = 0.002). Furthermore, 12 DMRs were identified to be related to 12 genes: NWD1, LDHD, C1S, IQCE, TNF, PDE1C, LGALS1, CSNK1E, LRRC23, ENO2, ELOVL2, and ELOVL2-AS1. Among these 12 DMPs, ten regions overlapped with promoters. Detailed information regarding the DMRs and their related genes or promoters is provided in Supplementary Table 2.
Figure 1.
(A) PCoA of DNA methylation signature in ALS patients and controls. (B) Whole level of DNA methylation in ALS patients and controls. ALS: Amyotrophic lateral sclerosis; PCoA: Principal coordinates analysis.
Figure 2.
(A) Number of hypermethylated and hypomethylated DMPs. (B) Number of hypermethylated and hypomethylated DMPs related genes. (C) Volcano plot of DMPs. (D) Heatmap of DMPs. DMPs: Differentially methylated positions.
Pathway and GO analysis
Considering the limited number of genes that were identified as being related to DMPs and DMRs, we directly research for these genes in UniProt and KEGG to explore the potential functions and pathways associated with ALS. Detailed information is shown in Supplementary Tables 3 and 4. The molecular functions, biological processes, and cellular components of DMP- and DMR- related genes were identified through GO analysis (via UniProt).
Considering the neurodegenerative nature of ALS, among the 13 identified DMP- related genes, DNAH9 may be the most related to ALS, as it is active in the axoneme, involved in the biological processes of cilium and microtubule (MT)-based movements, and enables dynein light intermediate/ intermediate chain binding, minus-end-directed MT motor activity, and protein-adenosine triphosphate (ATP)-nucle-otide binding. Similarly, among the 12 identified DMR-related genes, TNF may be the most significant as it participates in numerous biological processes, including the positive regulation of neuron apoptotic processes and the positive or negative regulation of gene expression, and is widely distributed across cell surface and extracellular regions. Consistent with the GO analysis results, KEGG analysis revealed that DNAH9 and TNF participated in the pathways of both ALS and neurodegeneration [Supplementary Tables 3 and 4].
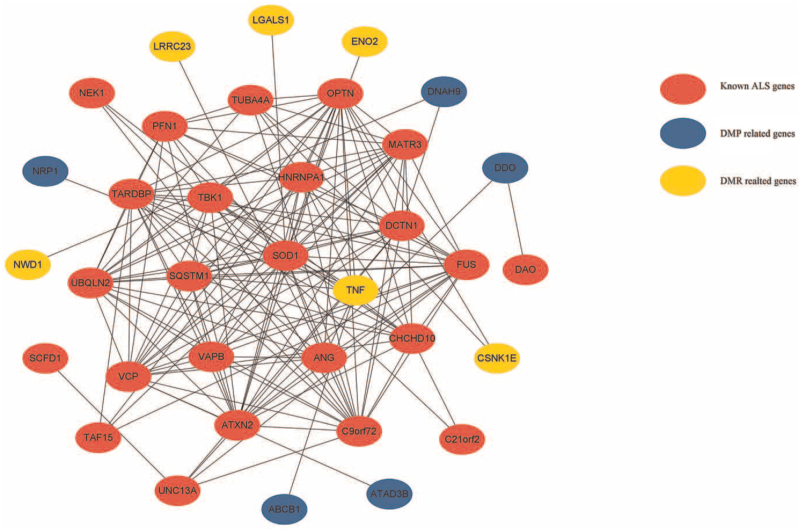
Network string interactions between established ALS genes and DMP/DMR-related genes
Figure 3 shows the top 35 causative or significant aberrant methylated gene network string interactions, as ranked by the degree method. SOD1, OPTN, and C9orf72 were ranked first. SOD1 was ranked first, with a score of 19 for all genes. TNF was ranked first among all DMP/ DMR-related genes, with a score of eight. DNAH9 was shown to be capable of interacting with the established ALS gene DCTN1 and with DMR-related genes such as NWD1. TNF was found to be capable of interacting with known ALS genes such as ANG, SOD1, OPTN, TBK1, HNRNPA1, and SQSTM1 and with DMP-related genes such as NRP1 and ABCB1. Detailed information is provided in Supplementary Table 5.
Figure 3.
Network string interactions between already known ALS genes and DMP/DMR related genes. ALS: Amyotrophic lateral sclerosis; DMP: Differentially methylated position; DMR: Differentially methylated region.
Association of DNA methylation with clinical features in SALS patients
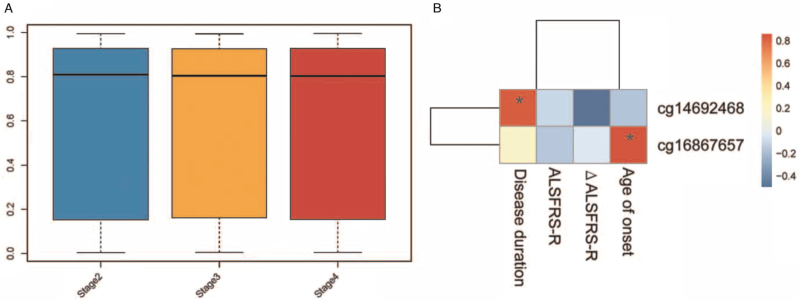
Comparing the DNA methylation levels between SALS patient groups at various stages indicated that there were no significant differences between the three groups (P > 0.05; Figure 4A). The methylation level of ELOVL2 (cg16867657) and ARID1B (cg14692468) was positively associated with age of onset (r = 0.86, adjust P = 0.001; Figure 4B), and disease duration (r = 0.83, adjust P = 0.01; Figure 4B), respectively. There were no significant CpG sites associated with ALSFRS-R or ΔALSFRS-R.
Figure 4.
(A) Whole level of DNA methylation in ALS patients with different King's stages. (B) Significant CpG sites associated with clinical features. ALS: Amyotrophic lateral sclerosis; ALSFRS-R: Revised ALS Functional Rating Scale.
Discussion
To the best of our knowledge, this is the first study to use Infinium MethylationEPIC BeadChip to investigate epigenome-wide DNA methylation changes in whole blood from SALS patients, compared with healthy controls. Although we did not find significant differences in DNA methylation between the two groups, our results suggest that SALS patients exhibited significant DMPs and DMRs compared with controls and that more hypomethylated CpG sites appear in the DMPs/DMRs of SALS patients than in those of the controls. A number of significant DMP- and DMR-related genes were identified to be involved in the pathological process of ALS, indicating that DNA methylation at specific sites plays an important role in ALS.
The etiology of ALS has not been fully elucidated. In approximately 90% of individuals who do not have any family history of ALS, the contribution of heritability or genetic variability to the disease is estimated to be 20% to 60%. Furthermore, some individuals can carry a known genetic mutation for their entire lifetime without developing ALS. Environmental triggers, when superimposed on genetic risks and cellular changes related to aging, are widely believed to play a role in ALS.[7] The controls in our study were all healthy relatives of patients, who had lived with them for a long time and thus could have undergone exposure to the same environmental factors. We found that the control group as a whole had no significant difference in DNA methylation compared to the patient group but did exhibit significant differences at certain CpG sites. This may therefore suggest that external factors influence local epigenetic modifications, rather than the whole genome.
Oates et al[16] examined methylation in the promoters of SOD1 and VEGF in white cell DNA and brain DNA from ALS patients. They found that the promoter regions were largely unmethylated. In contrast, Coppedè et al[14] observed that the promoters of SOD1, FUS, TARDBP, and C9orf72 were demethylated in ALS. Garton et al[17] investigated the DNA methylation signatures of nine C9orf72 hexanucleotide repeat expansion-positive ALS cases, identifying two as being methylation outliers (in the two probes in the CpG island upstream of the transcriptional start site and in the probe nearest to the hexanucleotide repeat expansion). They also found that neither SOD1 nor TARDBP presented compelling evidence that rare singlenucleotide variants were associated with differential DNA methylation in blood. A study into the genome-wide methylation profiles of blood DNA from monozygotic twins suggested that the similarity in these profiles was far greater than that observed in profiles extracted from the blood DNA of other types of siblings or unrelated individuals, thus demonstrating that epigenetic changes are genetically controlled.[18] Here, we also found no significant differences in methylation levels between ALS patients and controls regarding known common ALS causative genes such as SOD1, TARDBP, FUS, and C9orf72 (data not shown). However, interaction network analysis of the identified DMP/DMR-related genes and known ALS genes implied that there were potential associations between abnormal methylation and the expressions of causative genes. Thus, more mechanistic studies are needed to confirm the interactions between genetics and epigenetics in ALS.
Previous studies have concentrated on nerve-tissue derived DNA methylation. Oates et al[16] performed bisulfite polymerase chain reaction to analyze brain DNA from six SALS patients, focusing on DNA methylation in the promoter regions of SOD1 and VEGF. They discovered that these regions were largely unmethylated in all patients, which was confirmed in white cell DNA extracted from peripheral blood. Morahan et al[10] found 38 differentially methylated sites in ten brain samples from SALS patients using Affymetrix GeneChip Human Tiling 2.0R Arrays. Figueroa-Romero et al,[12] meanwhile, identified 3574 methylation genes in postmortem SALS spinal cord using Illumina Infinium HumanMethylation27 BeadArrays. However, through using enzyme-linked immunosorbent assays, they only observed global 5mC alterations in spinal cord, but not in whole blood. In our study into blood-tissue derived DNA, we found that global methylation levels were not significantly different between the SALS patient and control groups, but identified 34 DMPs and 12 DMRs in SALS patients. Differences in DNA methylation profiles between nerve and blood tissues may result from various detection methods, diverse ancestries, and different sample sizes. Thus, the examination of both blood and brain DNA may be important when exploring methylation in neurological disorders.[16]
We found no significant difference in the whole DNA methylation levels between patient groups for different ALS stages. There were no significant CpG sites associated with ALSFRS-R or ΔALSFRS-R. This suggests that aberrant DNA methylation may trigger the initial pathological process of ALS and become relatively stable in the progression and advanced stages. An in vivo study revealed that DNA methylation controlled by DNA methyltransferases (Dnmts) was strictly associated with motor neuron death during mouse brain and spinal cord maturation.[19] In adulthood, when Dnmt3a is abundant in synapses and mitochondria, Dnmt1 and Dnmt3a are expressed differently. During the apoptosis of these neurons, nuclear and cytoplasmic 5mC immunoreactivity, Dnmt3a protein levels, and Dnmt enzyme activity all increased preapoptotically.[19] The inhibition of Dnmts completely blocked the increase in 5mC and the apoptosis of motor neurons in mice.[19]
DNAH9 and TNF are both already known to be linked to the ALS pathway and to neurodegeneration in multiple diseases, according to KEGG. DNAH9 encodes a large multi-subunit molecular motor named the heavy chain subunit of axonemal dynein. Axonemal dynein attaches to MTs and hydrolyzes ATP to mediate the movement of cilia and flagella. Mutations in this protein have been shown to be involved in axonal transport defects and motoneuron degeneration.[20] Recently, Maimon et al[21] found that the interaction between dynein and another ALS-related protein, collapsin response mediator protein 4 (CRMP4), may contribute to ALS. By blocking the CRMP4-dynein interaction, motor neuron loss was shown to be reduced in human-derived motor neurons and ALS model mice.[21] Comley et al[22] found that vulnerable motor neurons displayed higher dynein protein levels. In our study, we found that hypomethylation mainly occurs at the CpG opensea of the DNAH9 gene body. This may have an impact on the protein levels of dynein, or may regulate the expressions of other genes such as DCTN1 and NWD1, consequently leading to axon injury or other pathological changes. TNF encodes tumor necrosis factor, which is mainly secreted by macrophages and can induce cell death in tumor cell lines. It is traditionally considered to be related to cancer or autoimmune diseases. However, in ALS animal models and patients, TNFα as well as its receptors have been shown to be progressively upregulated from the presymptomatic stage until the end stage. This suggests that the TNF system plays a key role in ALS.[23] Our results revealed not only aberrant methylation in TNF but also the complex interactions between ALS causative gene and other differentially methylated genes. In addition, special attention should be paid to another gene ELOVL2, as it is not only located in a DMR but is also closely linked to the age of onset. ELOVL2 encodes the elongation of very long chain fatty acid protein 2; it is mainly involved in the process of fatty acid metabolism. Although the role of ELOVL2 in ALS has not yet been studied, considering that metabolic factors are related to ALS development and prognosis, it is also essential to clarify the role of ELOVL2 methylation in ALS.
Here, we have found aberrant methylation in DMP- and DMR- related genes, implying that many epigenetic alterations such as the hypomethylation of DNAH9 and TNF play important roles in ALS etiology. These findings could help to develop new therapeutic interventions. Future studies can aim to replicate the methodology used to arrive at theses findings to determine the functional consequences of the observed DNA methylation changes on ALS pathophysiology.
Funding
This work was supported by the National Key Research and Development Program of China (No. 2016YFC0905103), the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (CIFMS) (No. 2016-I2M-1-004), and the Strategic Priority Research Program of the Chinese Academy of Sciences (Nos. XDB39000000, XDB39040100).
Conflicts of interest
None.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Footnotes
How to cite this article: Cai Z, Jia X, Liu M, Yang X, Cui L. Epigenome-wide DNA methylation study of whole blood in patients with sporadic amyotrophic lateral sclerosis. Chin Med J 2022;135:1466–1473. doi: 10.1097/CM9.0000000000002090
Supplemental digital content is available for this article.
References
- 1.van Es MA, Hardiman O, Chio A, Al-Chalabi A, Pasterkamp RJ, Veldink JH, et al. Amyotrophic lateral sclerosis. Lancet 2017; 390:2084–2098. doi: 10.1016/S0140-6736(17)31287-4. [DOI] [PubMed] [Google Scholar]
- 2.Chia R, Chiò A, Traynor BJ. Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol 2018; 17:94–102. doi: 10.1016/S1474-4422 (17) 30401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown RH, Al-Chalabi A. Amyotrophic lateral sclerosis. N Engl J Med 2017; 377:162–172. doi: 10.1056/NEJMra1603471. [DOI] [PubMed] [Google Scholar]
- 4.Lian L, Liu M, Cui L, Guan Y, Liu T, Cui B, et al. Environmental risk factors and amyotrophic lateral sclerosis (ALS): a case-control study of ALS in China. J Clin Neurosci 2019; 66:12–18. doi: 10.1016/j. jocn.2019.05.036. [DOI] [PubMed] [Google Scholar]
- 5.Dickerson AS, Hansen J, Thompson S, Gredal O, Weisskopf MG. A mixtures approach to solvent exposures and amyotrophic lateral sclerosis: a population-based study in Denmark. Eur J Epidemiol 2020; 35:241–249. doi: 10.1007/s10654-020-00624-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellavia A, Dickerson AS, Rotem RS, Hansen J, Gredal O, Weisskopf MG. Joint and interactive effects between health comorbidities and environmental exposures in predicting amyotrophic lateral sclerosis. Int J Hyg Environ Health 2021; 231:113655.doi: 10.1016/j. ijheh.2020.113655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goutman SA, Feldman EL. Voicing the need for amyotrophic lateral sclerosis environmental research. JAMA Neurol 2020; 77:543–544. doi: 10.1001/jamaneurol.2020.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease. Nature 2019; 571:489–499. doi: 10.1038/s41586-019-1411-0. [DOI] [PubMed] [Google Scholar]
- 9.Paez-Colasante X, Figueroa-Romero C, Sakowski SA, Goutman SA, Feldman EL. Amyotrophic lateral sclerosis: mechanisms and therapeutics in the epigenomic era. Nat Rev Neurol 2015; 11:266–279. doi: 10.1038/nrneurol.2015.57. [DOI] [PubMed] [Google Scholar]
- 10.Morahan JM, Yu B, Trent RJ, Pamphlett R. A genome-wide analysis of brain DNA methylation identifies new candidate genes for sporadic amyotrophic lateral sclerosis. Amyotroph Lateral Scler 2009; 10:418–429. doi: 10.3109/17482960802635397. [DOI] [PubMed] [Google Scholar]
- 11.Chater-Diehl E, Goodman SJ, Cytrynbaum C, Turinsky AL, Choufani S, Weksberg R. Anatomy of DNA methylation signatures: emerging insights and applications. Am J Hum Genet 2021; 108:1359–1366. doi: 10.1016/j.ajhg.2021.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figueroa-Romero C, Hur J, Bender DE, Delaney CE, Cataldo MD, Smith AL, et al. Identification of epigenetically altered genes in sporadic amyotrophic lateral sclerosis. PLoS One 2012; 7:e52672.doi: 10.1371/journal.pone.0052672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tremolizzo L, Messina P, Conti E, Sala G, Cecchi M, Airoldi L, et al. Whole-blood global DNA methylation is increased in amyotrophic lateral sclerosis independently of age of onset. Amyotroph Lateral Scler Frontotemporal Degener 2014; 15:98–105. doi: 10.3109/21678421.2013.851247. [DOI] [PubMed] [Google Scholar]
- 14.Coppedè F, Stoccoro A, Mosca L, Gallo R, Tarlarini C, Lunetta C, et al. Increase in DNA methylation in patients with amyotrophic lateral sclerosis carriers of not fully penetrant SOD1 mutations. Amyotroph Lateral Scler Frontotemporal Degener 2018; 19:93–101. doi: 10.1080/21678421.2017.1367401. [DOI] [PubMed] [Google Scholar]
- 15.Brooks BR, Miller RG, Swash M, Munsat TL. World Federation of Neurology Research Group on Motor Neuron Diseases. El Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 2000; 1:293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 16.Oates N, Pamphlett R. An epigenetic analysis of SOD1 and VEGF in ALS. Amyotroph Lateral Scler 2007; 8:83–86. doi: 10.1080/17482960601149160. [DOI] [PubMed] [Google Scholar]
- 17.Garton FC, Benyamin B, Zhao Q, Liu Z, Gratten J, Henders AK, et al. Whole exome sequencing and DNA methylation analysis in a clinical amyotrophic lateral sclerosis cohort. Mol Genet Genomic Med 2017; 5:418–428. doi: 10.1002/mgg3.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang M, Xi Z, Ghani M, Jia P, Pal M, Werynska K, et al. Genetic and epigenetic study of ALS-discordant identical twins with double mutations in SOD1 and ARHGEF28. J Neurol Neurosurg Psychiatry 2016; 87:1268–1270. doi: 10.1136/jnnp-2016-313592. [DOI] [PubMed] [Google Scholar]
- 19.Chestnut BA, Chang Q, Price A, Lesuisse C, Wong M, Martin LJ. Epigenetic regulation of motor neuron cell death through DNA methylation. J Neurosci 2011; 31:16619–16636. doi: 10.1523/JNEUROSCI.1639-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kieran D, Hafezparast M, Bohnert S, Dick JR, Martin J, Schiavo G, et al. A mutation in dynein rescues axonal transport defects and extends the life span of ALS mice. J Cell Biol 2005; 169:561–567. doi: 10.1083/jcb.200501085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maimon R, Ankol L, Gradus Pery T, Altman T, Ionescu A, Weissova R, et al. A CRMP4-dependent retrograde axon-to-soma death signal in amyotrophic lateral sclerosis. EMBO J 2021; 40:e107586.doi: 10.15252/embj.2020107586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comley L, Allodi I, Nichterwitz S, Nizzardo M, Simone C, Corti S, et al. Motor neurons with differential vulnerability to degeneration show distinct protein signatures in health and ALS. Neuroscience 2015; 291:216–229. doi: 10.1016/j.neuroscience.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Guidotti G, Scarlata C, Brambilla L, Rossi D. Tumor necrosis factor alpha in amyotrophic lateral sclerosis: Friend or foe? Cells 2021; 10:518.doi: 10.3390/cells10030518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.