Abstract
The cellular prion protein PrPC partners with caveolin-1 (CAV1) in neurodegenerative diseases but whether this interplay occurs in cancer has never been investigated. By leveraging patient and cell line datasets, we uncover a molecular link between PrPC and CAV1 across cancer. Using cell-based assays, we show that PrPC regulates the expression of and interacts with CAV1. PrPC additionally controls the expression of the amyloid precursor protein APP and of the Aβ generating enzyme BACE1, and regulates the levels of Aβ, whose accumulation is a central event in Alzheimer’s disease. We further identify DKK1 and DKK3, involved in both Alzheimer’s disease and cancer progression, as targets of the PrPC-dependent axis. Finally, we establish that antibody-mediated blocking of the Aβ-PrPC interaction delays the growth of prostate cancer cell line-derived xenografts and prevents the development of metastases. Our data additionally support an enrichment of the Aβ-PrPC-dependent pathway in the basal subtype of prostate cancer, associated with anti-hormonal therapy resistance, and in mesenchymal colon cancer, associated with poor prognosis. Thus, based on a parallel with neurodegenerative diseases, our results bring to light an Aβ-PrPC axis and support the potential of targeting this pathway in patients with selected subtypes of prostate and colon cancer.
Subject terms: Targeted therapies, Prognostic markers
Introduction
Since its discovery in 1985, the cellular prion protein PrPC has been extensively studied for its involvement in a group of fatal neurodegenerative disease known as transmissible spongiform encephalopathies (TSEs) [1]. PrPC is ubiquitously expressed and has since been ascribed a plethora of functions according to the cellular context [2]. Via its GPI anchorage at the outer plasma membrane and its interaction with diverse partners, PrPC can act as a receptor or co-receptor, leading to the mobilization of intracellular signalling cascades [2]. A major PrPC partner in the recruitment of cell signalling pathways is the membrane protein caveolin 1 (CAV1) [2, 3]. In 2009, the discovery that PrPC acts as a receptor for the amyloid Aβ peptide [4], which is produced from the amyloid precursor protein APP following endoproteolytic cleavage and whose accumulation is a central event in Alzheimer’s disease (AD) [5], sparked renewed interest in the signalling function of PrPC. It is now well-established that Aβ oligomers hijack PrPC-dependent signalling to foster synaptic impairment [2].
A set of studies have allowed to uncover a link between PrPC and cancer [6]. For instance, we recently showed that PrPC is overexpressed in the poor-prognosis subtype of colon cancer and that it controls the expression of a set of genes associated with a mesenchymal phenotype [7]. Along the same line, CAV1 was identified as a specific marker of epithelial to mesenchymal transition (EMT) across cancer cell lines [8]. Altogether, this converging set of data led us to make the provocative assumption that an Aβ-PrPC-CAV1 platform may operate in cancer cells and recruit pro-tumorigenic downstream signalling cascades. Here, by combining in silico analysis of several cancer datasets, cell-based assays and mouse xenografts, we delineate an Aβ-PrPC-CAV1 axis that sustains cancer growth and metastasis. We further highlight the enrichment of this axis in basal prostate cancer and mesenchymal colon cancer, suggesting its targeting may be beneficial in these specific prostate and colon cancer subtypes.
Results and discussion
PRNP is highly correlated to CAV1 across cancer types and PrPC controls caveolin-1 expression in cancer cells
To probe for a potential link between PrPC and caveolin-1 in cancer, we interrogated published datasets for potential associations between levels of transcripts encoding PrPC (PRNP) and caveolin-1 (CAV1). We first leveraged the cancer cell line encyclopaedia (CCLE) [9], which provides a unique resource to perform queries on a pan-cancer scale. As shown in Fig. 1A, CAV1 represented the second most correlated transcript with PRNP across the CCLE, suggesting a strong association between the expression of these two genes. We then mined published data sets from The Cancer Genomic Atlas (TCGA) via the Morpheus platform at the Broad Institute (https://software.broadinstitute.org/morpheus/) and found significant correlations between PRNP and CAV1 expression in several cancer types (Supplementary Fig. 1A). The strong correlation between PRNP and CAV-1 was also recovered in two cell line panels (Supplementary Fig. 1A). Of note, the levels of PRNP transcripts also strongly correlated with those of CAV1 protein in several of the dataset studied, most notably prostate carcinoma (PRAD) and colon adenocarcinoma (COAD) (Supplementary Fig. 1B).
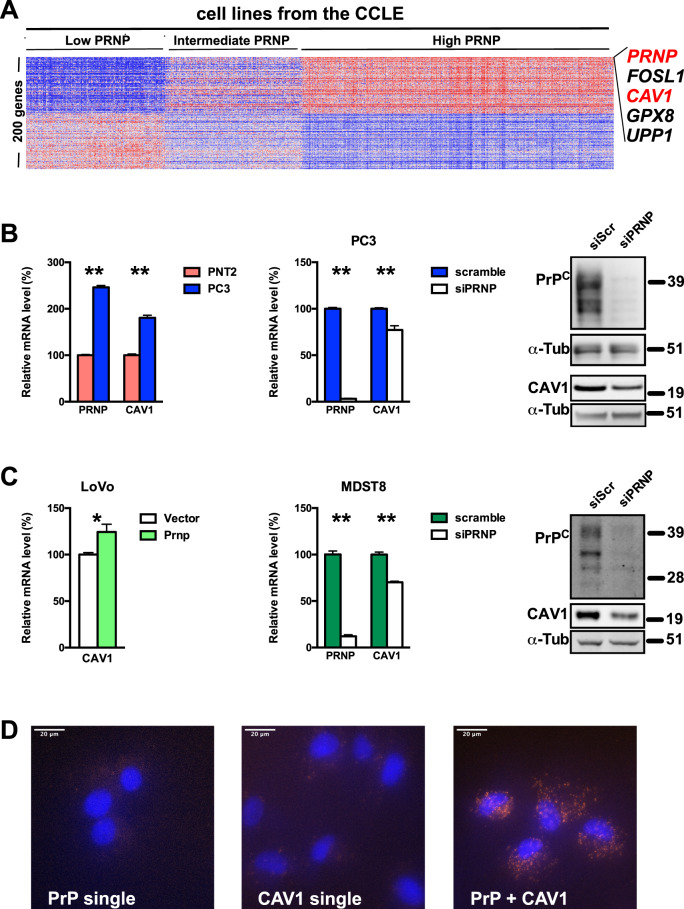
Fig. 1. PrPC and Caveolin-1 relationship across cancer.
A Heatmap showing the top genes most positively and negatively correlated with the expression on PRNP in the CCLE. B qRT-PCR analysis of the relative expression of PRNP and CAV1 in PC3 prostate cancer cells versus PNT2 normal prostate cells (left panel); relative expression of PrPC and CAV1 mRNA (middle panel) or protein (right panel) in PRNP-silenced versus control PC3 prostate cancer cells. C qRT-PCR analysis of the relative expression of CAV1 PrPC-overexpressing versus control LoVo colon cancer cells (day 3 post-transfection) (left panel); relative expression of PrPC and CAV1 mRNA (middle panel) or protein (right panel) in PRNP-silenced versus control MDST8 colon cancer cells. Results are expressed as means of n = 2 independent triplicates of cell preparations ± s.e.m. *p < 0.05, ***p < 0.001 vs. control (Mann-Whitney test). Quantification for western blots is provided in Supplementary Fig. S2, with protein levels normalized to α-tubulin (α-tub). D Proximity ligation assay showing co-localization of PrPC and CAV1 in MDST8 cells (orange spots, right panel). Cells incubated with anti-PrPC antibody alone (left panel) or anti-Caveolin-1 antibody alone (middle panel) were used as controls. Nuclei were stained with DAPI.
Next, we turned to cell-based approaches to probe for a functional link between PrPC and CAV1. We measured higher PRNP and CAV1 levels in the PC3 prostate cancer cell line as compared to the PNT2 normal epithelial prostatic cell line (Fig. 1B), and found reduced levels of CAV1 mRNA and protein when PrPC expression was silenced in PC3 cells (Fig. 1B and Supplementary Fig. 2A). We further exploited the LoVo and MDST8 colon cancer cell lines that express PrPC at low and high levels, respectively [7]. In line with the data obtained in prostate cell lines, overexpression of PrPC in LoVo cells led to an upregulation of CAV1 mRNA (Fig. 1C) while PrPC knockdown in MDST8 cells caused a reduction in CAV1 mRNA and protein (Fig. 1C and Supplementary Fig. 2B). Finally, PrPC and CAV1 were found to co-localize in MDST8 cells, as evidenced with the Proximity Ligation Assay (Fig. 1D). Altogether, these data highlight regulatory and physical relationships between PrPC and CAV1 in the context of cancer, reminiscent of that described in neuronal cells [2, 3].
PrPC controls Aβ levels in prostate and colon cancer cells
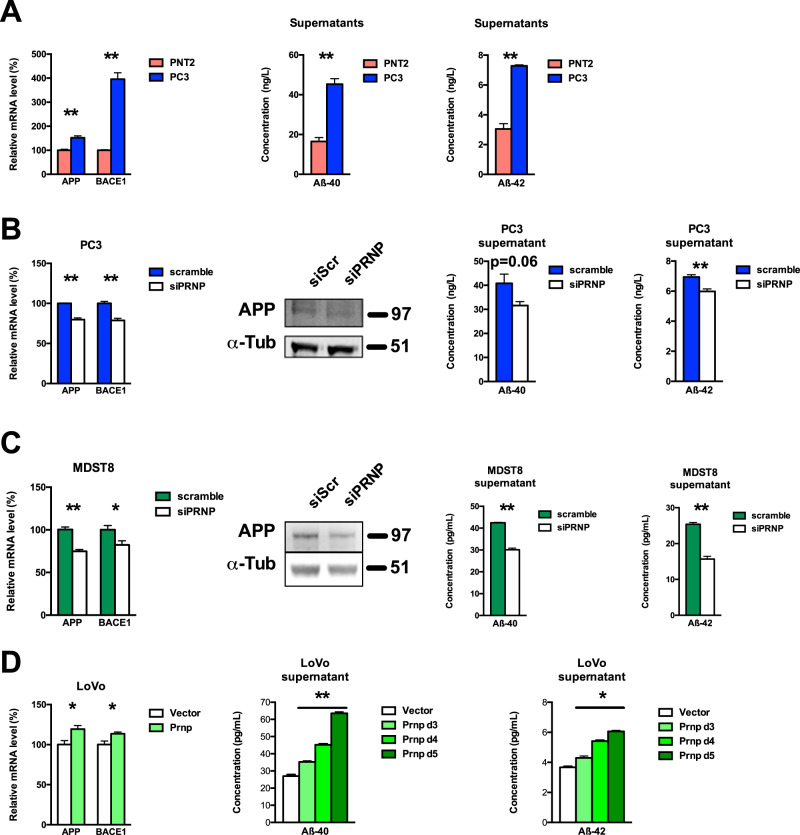
Since PRNP mRNA and CAV1 mRNA and protein levels consistently correlated with those of transcripts encoding APP in the CCLE (Supplementary Fig. 1C), we next examined a potential link between PrPC and APP and its proteolytic fragments in prostate and colon cancer cells. As shown in Fig. 2A, PC3 cells expressed higher levels of mRNAs encoding APP as well as BACE1, a protease involved in the processing of APP and the generation of Aβ peptides [10], when compared to PNT2 cells. Accordingly, we measured higher levels of the Aβ-40 and Aβ-42 peptides in the supernatants of PC3 cells, when compared to PNT2 cells (Fig. 2A). Silencing of PrPC (48 h) in the PC3 cell line was associated with a reduction in APP mRNA and protein, BACE1 mRNA, as well as extracellular Aβ-40 and Aβ-42 concentrations (Fig. 2B and Supplementary Fig. 2C). As observed with PC3 cells, APP mRNA and protein, BACE1 mRNA, and extracellular Aβ-40 and Aβ-42 concentrations were all reduced in MDST8 cells after PrPC knockdown (Fig. 2C and Supplementary Fig. 2D). Finally, in the LoVo colon cell line, PrPC overexpression promoted an upregulation of APP and BACE1 transcripts (Fig. 2D). Of note, we found that the concentrations of Aβ-40 and Aβ-42 peptides in the supernatants of LoVo cells progressively increased along a 3 to 5-day window post-transfection with the PrPC-encoding plasmid (Fig. 2D and Supplementary Fig. 2E). Altogether, these data indicate that PrPC controls the expression of APP, as well as that of BACE1, the main sheddase responsible for the production of Aβ peptides [10]. These observations differ with several studies reporting a negative correlation between PrPC and BACE1 or Aβ levels [11–14]. Yet, they are in agreement with the observed positive correlation between PrPC and APP levels in the brain of transgenic mice [15]. They also fully fit in with the reported promotion of APP cleavage by PrPC in the brain through the upregulation of BACE1 expression [16], as well as the recently described PrPC-dependent secretion of Aβ in cell lines [17]. Accordingly, our gain and loss of function experiments strongly support a control of PrPC over the soluble concentrations of Aβ peptides in cancer cells.
Fig. 2. PrPC controls Aβ levels in prostate and colon cancer cells.
A qRT-PCR analysis of the relative expression of APP and BACE1 in PC3 prostate cancer cells versus PNT2 normal prostate cells (left panel). Extracellular Aβ40 and Aβ42 levels were measured in the supernatants of PNT2 and PC3 cells (right panels). B APP and BACE1 mRNA levels, APP protein levels and extracellular Aβ40 and Aβ42 levels were measured in PRNP-silenced versus control PC3 prostate cancer cell extracts and supernatants. C APP and BACE1 mRNA levels, APP protein levels and extracellular Aβ40 and Aβ42 levels were measured in PRNP-silenced versus control MDST8 colon cancer cell extracts and supernatants. D APP and BACE1 mRNA levels and extracellular Aβ40 and Aβ42 levels were measured in PrPC-overexpressing versus control LoVo colon cancer cell extracts (day 3 post-transfection) and supernatants (days 3, 4 and 5 post-transfection). In B and C, protein levels were normalized to α-tubulin (α-tub). Results are expressed as means of n = 2 independent triplicates of cell preparations ± s.e.m. *p < 0.05, **p < 0.01 vs. control (Mann-Whitney test except for D middle and right panels Kruskal-Wallis and posthoc Wilcoxon rank-sum test with Holm’s correction). Quantification for western blots is provided in Supplementary Fig. S2C, D.
Targeting the Aβ-PrPC interaction in vitro reduces the expression of a set of genes associated with mesenchymal features of cancer cells and lowers the levels of extracellular Aβ40, Aβ42 and TGFβ1
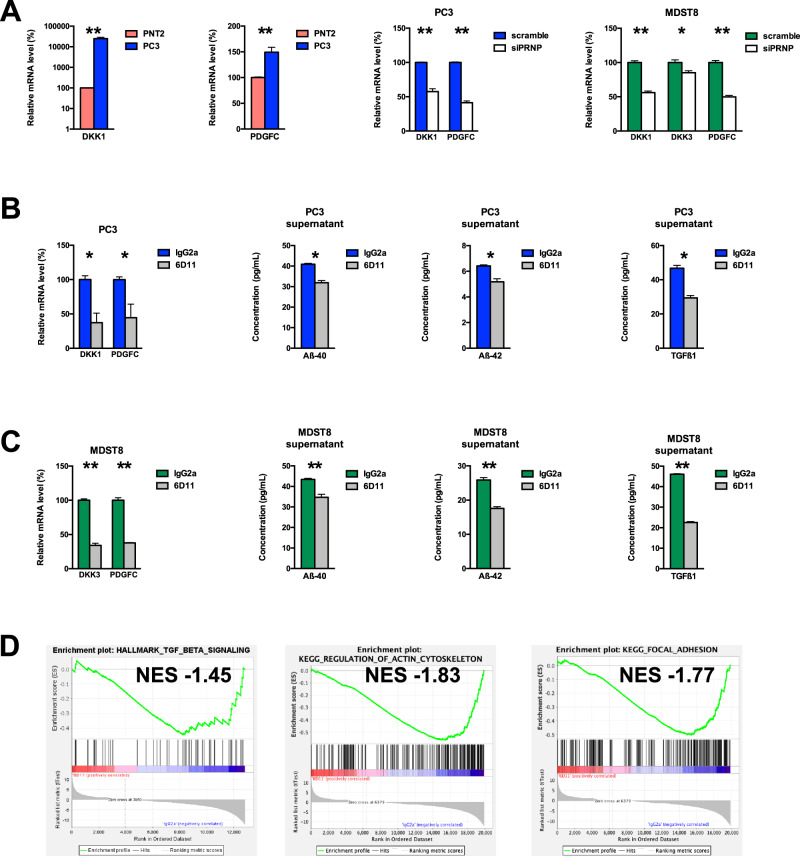
Next, we searched for candidate targets downstream of the PrPC-CAV1 module that may potentially be recruited upon binding of Aβ to PrPC. Among the genes whose levels significantly correlated with those of PRNP mRNA, APP mRNA and CAV1 mRNA and protein in the CCLE, we selected DKK1, DKK3 and PDGFC for further analysis (Supplementary Fig. 3), in view of the potential link between DKK1/DKK3 and Alzheimer’s disease [18, 19], their contribution to tumour progression by activating the tumour microenvironment [20, 21] and the PrPC-dependent regulation of PDGFC in colon cancer cells [7]. We measured increased levels of DKK1 and PDGFC mRNA in PC3 versus PNT2 cells, which were strongly reduced after PrPC knockdown in PC3 cells (Fig. 3A). In line with this, PrPC-silenced MDST8 cells exhibited reduced mRNA levels of DKK1, DKK3 and PDGFC (Fig. 3A).
Fig. 3. Identification of potential Aβ-PrPC targets and impact of antibody-mediated blockade of the Aβ-PrPC interaction.
A qRT-PCR analysis of the relative expression of DKK1 and PDGFC in PC3 prostate cancer cells versus PNT2 normal prostate cells (left panels) as well as in PRNP-silenced versus control PC3 cells (middle panel) and qRT-PCR analysis of the relative expression of DKK1, DKK3 and PDGFC in PRNP-silenced versus control MDST8 cells (right panel). B DKK1 and PDGFC mRNA levels and extracellular Aβ40, Aβ42 and TGFβ levels were measured in cell extracts and supernatants of PC3 prostate cancer cells exposed to 6D11 antibodies versus control isotype antibodies. C DKK3 and PDGFC mRNA levels and extracellular Aβ40, Aβ42 and TGFβ levels were measured in cell extracts and supernatants of MDST8 colon cancer cells exposed to 6D11 antibodies versus control isotype antibodies. Results are expressed as means of n = 2 independent triplicates of cell preparations (except for A and B n = 2 independent duplicates of cell preparations) ± s.e.m. *p < 0.05, ***p < 0.001 vs. control (Mann-Whitney test). D GSEA analysis showing the downregulation of the TGFβ signalling, regulation of actin cytoskeleton and focal adhesion signatures in 6D11-treated versus control MDST8 cells. NES normalized enrichment score.
Next, we probed whether blockade of the Aβ-PrPC interaction would impact on DKK1/3 and PDGFC transcript levels. To this end, cells were exposed to the 6D11 antibody (10 µg/mL, 72 h) targeting the 92-110 epitope within PrPC that binds Aβ [4]. Treatment of PC3 cells with 6D11 induced a significant reduction in DKK1 and PDGFC transcript levels (Fig. 3B). We further observed a decrease in the levels Aβ-40 and Aβ-42 in the supernatants of PC3 cells treated with 6D11, as well as that of TGFβ1, which we previously showed to be controlled by PrPC in colon cancer cells [7] (Fig. 3B). In line with the above data, DKK3 and PDGFC transcripts were reduced in MDST8 cells after treatment with 6D11 (Fig. 3C). We further explored some PrPC-regulated genes described above (BACE1, CAV1) or that we previously showed to be associated with the mesenchymal phenotype of MDST8 cells (IDO1, TGFB1, ZEB1) [7, 22], and found that expression of this set of genes was robustly decreased upon blockade of the PrPC- Aβ interaction with 6D11 antibodies (Supplementary Fig. 4A). As with PC3 cells, the levels of Aβ-40, Aβ-42 and TGFβ1 were decreased in the supernatants of MDST8 treated with 6D11 (Fig. 3C). In contrast to the effects observed with 6D11, treatment of PC3 cells with the Sha31 antibody that does not interfere with the Aβ-PrPC interaction induced only a mild decrease in DKK1 mRNA and was neutral to PDGFC mRNA (Supplementary Fig. S4B). As for MDST8 cells, exposure to the Sha31 antibody promoted an increase in DKK3 and PDGFC levels (Supplementary Fig. 4C), in agreement with Sha31 mimicking a ligand-induced activation of PrPC [2]. A more global profiling of the transcriptome of MDST8 cells treated with 6D11 through RNAseq followed by GSEA analysis revealed that 3 gene signatures, TGFβ signalling, regulation of actin cytoskeleton and focal adhesion, which are significantly correlated with PRNP gene expression in colon cancer [7] and data not shown), were downregulated in response to 6D11 treatment (Fig. 3D). Whether 6D11 promotes the internalization of PrPC, as described by Pankiewicz et al. [23] or its shedding, as depicted by Linsenmeier et al. [24], or operates through another mode of action warrants further investigation. As a whole, our data suggest that Aβ-PrPC signalling controls the expression of DKK1/3 and PDGFC as well as Aβ and TGFβ levels in prostate and colon cancer cells and regulates pathways involved in the mesenchymal subtype of colon cancer.
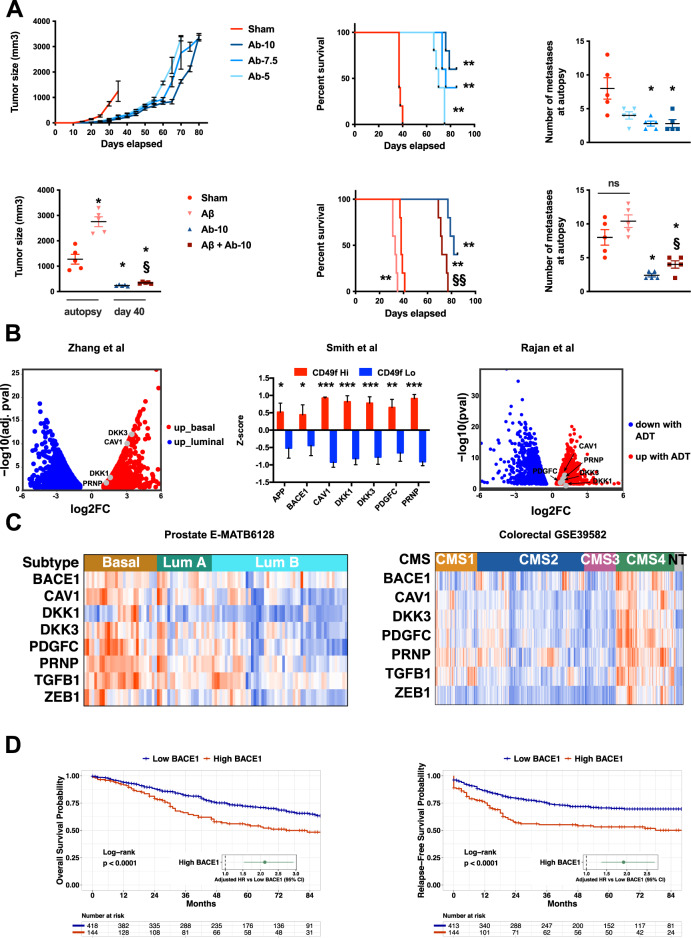
Blockade of the Aβ-PrPC interaction reduces the growth of prostate cancer cells in vivo and the Aβ-PrPC connection has translational relevance in prostate and colon cancer patients
The above results prompted us to target the Aβ-PrPC interaction in an in vivo model of tumorigenesis. To this end, we studied the effect of 6D11 on the growth of PC3 xenografts in mice (see Materials and Methods). Castrated mice bearing palpable tumours were treated twice weekly with PBS or 6D11 at various doses (5, 7.5, or 10 mg per kg of body weight). As shown in Fig. 4A and Supplementary Fig. 5A, B, the 6D11 antibody markedly reduced tumour growth at all doses, when compared to controls. This effect was prolonged beyond 60 days at the highest dose. While the growth of tumours strongly resumed around day 60 depending on the dose received, prolonged administration of 6D11 promoted a > 100% improvement in the mean survival time of PC3-grafted mice (Fig. 4A). Importantly, the metastatic burden in xenografted mice was strongly reduced upon 6D11 treatment (Fig. 4A and Supplementary Fig. 5C). Furthermore, we found that treating PC3-grafted mice with recombinant Aβ boosted tumour growth and shortened survival (Fig. 4A). Remarkably, these effects were counteracted upon concomitant treatment with 6D11 antibodies, which also drastically reduced metastatic burden (Fig. 4A). Thus, these data establish a tumour-promoting effect of Aβ, which has to be brought together with the recent identification of melanoma-secreted Aβ as a promoter of brain metastasis [25]. They also demonstrate that abrogating the Aβ-PrPC interaction is beneficial in a pre-clinical prostate cancer model. Next, we interrogated public datasets to evaluate the potential translational relevance of the PrP-dependent axis in prostate cancer. Interestingly, we found an enrichment of PRNP, CAV1, as well as DKK1 and DKK3 mRNA levels in basal versus luminal benign prostate biopsies in the Zhang dataset [26] (Fig. 4B). Of note, the basal cell signature was shown to be associated with aggressive prostate cancer [26]. Along the same line, PRNP, CAV1, DKK1, DKK3 and PDGFC were all significantly enriched in the basal cell population from benign and cancer prostate tissue, characterized by high expression of the cell surface marker CD49f [27] (Fig. 4B). As in [26], Smith et al. argued that the CD49fHi-associated gene signature is a hallmark of aggressive prostate cancer [27]. Furthermore, analysis of matched samples before and after androgen-deprivation therapy (ADT) revealed an increase in the same set of genes after ADT [28] (Fig. 4B). Finally, we sought to assess the distribution of the various genes of interest according to the recently described molecular classification of prostate cancer samples into Luminal A, Luminal B or Basal subtypes [29]. Applying the PAM50 classifier designed by [29] to the dataset of Kamoun et al. [30], we observed an enrichment of the various genes of the pathway in tumours of the basal subtype compared to those of the luminal B subtype (Fig. 4C). PRNP, PDGFC and TGFB1 mRNAs were also significantly higher in basal versus luminal A tumours (Fig. 4C and Supplementary Fig. 6). The enrichment of the PrPC-dependent axis in the basal versus luminal B subtype was also recovered in TCGA prostate cancer samples (Supplementary Fig. 7). Of note, although patients with basal and luminal A prostate cancer have a better prognosis than those with luminal B prostate cancer, they were shown to respond poorly to ADT treatment [29]. As for colon cancer, we had previously analysed the expression of PRNP according to the consensus molecular classification defined by Guinney et al. [31] and we had documented an enrichment of PRNP transcripts in the poor-prognosis mesenchymal subtype referred to as CMS4 [7], associated with poor prognosis [31]. Here, in line with the above findings, we monitored a significant enrichment of the PrPC-dependent network in tumours of the CMS4 subtype (Fig. 4C and Supplementary Fig. 8). Finally, the expression of the BACE1 gene was highly prognostic for overall survival and relapse-free survival in the GSE39582 cohort both as a continuous variable in univariate (Hazard Ratio HR = 1.85, 95% CI = 1.29 to 2.66, p < 0.001 for OS and HR = 2.39, 95% CI = 1.65–3.46, p < 0.001 for RFS) and multivariate (HR = 1.88, 95% CI = 1.27–2.78, p < 0.01 for OS and HR = 1.99, 95% CI = 1.34–2.95, p < 0.001 for RFS) analysis and categorical variable (dataset GSE39582) (Fig. 4D), as well as in a large validation cohort (Supplementary Fig. 9). In summary, the Aβ-PrPC-dependent axis appears to represent a hallmark of basal prostate cancer and mesenchymal colon cancer, thus opening new avenues for the development of therapeutic strategies specifically targeting these cancer molecular subgroups.
Fig. 4. The Aβ-PrPC axis can be targeted in vivo and has clinical relevance in basal prostate cancer and mesenchymal colon cancer.
A Top panels. Tumour growth, survival curves and number of metastases in mice bearing PC3 xenografts and treated with 5, 7.5 or 10 mg per kg of 6D11 antibody (Ab-5, Ab-7.5, Ab-10, respectively) versus control mice (Sham). Bottom panels. Tumour size (measured at autopsy for the sham and Aβ groups, and at day 40 for the Ab-10 and the Aβ + Ab-10 groups), survival curves and number of metastases in mice bearing PC3 xenografts and treated with recombinant Aβ, 6D11 antibody (at 10 mg per kg, Ab-10) or both Aβ and 6D11 antibody (Aβ + Ab-10) versus control mice (Sham). Data for tumour size and number of metastases are expressed as means ± s.e.m. of n = 5 values (*p < 0.05 and **p < 0.01 versus Sham, §p < 0.05 and §§p < 0.01 versus Aβ alone, Kruskal-Wallis and posthoc Wilcoxon rank-sum test with Holm’s correction for tumour size and number of metastases, log-rank test for survival curves. ns not significant). B Volcano plot showing the enrichment of CAV1, DKK1, DKK3 and PRNP transcripts in basal versus luminal benign prostate cancers from the Zhang study [26] (left panel); Enrichment of APP, BACE1, CAV1, DKK1, DKK3, PDGFC and PRNP transcripts in CD49fHi versus CD49fLo prostate tissue from the Smith study [27] (middle panel) (*p < 0.05 and ***p < 0.001 versus CD49fLo, two-tailed t-test); Volcano plot showing the upregulation of CAV1, DKK1, DKK3, PDGFC and PRNP transcripts after ADT in the Rajan study [32] (right panel). C Heatmaps showing the distribution of PRNP-associated genes in prostate cancer patients (E-MATB6128 data set) according to the PAM50 classification by Zhao [29] (left panel) or in colon cancer patients (GSE39582 dataset) according to the CMS classification by Guinney [31] (right panel). LumA: Luminal A. Lum B: Luminal B. NT: non tumour. See Supplementary Fig. S5 and S7 for statistics. D Kaplan-Meier overall survival (OS) (left panel) and relapse free survival (RFS) (right panel) according to high and low BACE1 gene expression was determined in colon cancer patients of the GSE39582 dataset. Hazard ratios were adjusted for TNM stage, MMR status and adjuvant chemotherapy.
Taken together, our study brings compelling evidence supporting the activation of an Aβ-PrPC-CAV1 dependent signalling pathway in specific subtypes of prostate and colon cancer. It provides a proof of concept that targeting the Aβ-PrPC interaction has beneficial effect and warrants testing the impact of Aβ-PrPC binding blockade in combination with standard chemotherapy.
Materials and Methods
See Supplementary Information for further details.
Supplementary information
Acknowledgements
Grant support was provided by the following: Cancéropôle Ile de France (grant number 2016-1-EMERG-36-UP 5-1), Association pour la Recherche sur le Cancer (grant number PJA 20171206220), SIRIC CARPEM (CAncer Research for PErsonalized Medicine, INCa-DGOS Inserm_12561), Labex Immuno-oncology, SATT Ile de France (grant number 415) as well as INSERM and Groupement des Entreprises Françaises dans la Lutte contre le Cancer (GEFLUC). M. Sroussi was supported by a fellowship from Fondation pour la Recherche Médicale (FRM grant FDM202006011237). The authors acknowledge N. Pierron and F. d’Agostini for expert methodological assistance.
Author contributions
SMR, JML, and PLP contributed to the conception and design of this study. SMR, SML, DLC, TZH, AG, MS, CP, and JML performed the experiments and acquired the data. SMR, AdR, FD, NV, JML, and PLP analysed and interpreted the data. SMR, JML, and PLP wrote and edited the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41388-022-02430-7.
References
- 1.Aguzzi A, Calella AM. Prions: Protein aggregation and infectious diseases. Physiol Rev. 2009;89:1105–52. doi: 10.1152/physrev.00006.2009. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch TZ, Martin-Lannerée S, Mouillet-Richard S. Functions of the prion protein. Prog Mol Biol Transl Sci. 2017;150:1–34. doi: 10.1016/bs.pmbts.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Mouillet-Richard S, Ermonval M, Chebassier C, Laplanche JL, Lehmann S, Launay JM, et al. Signal transduction through prion protein. Science. 2000;289:1925–8. doi: 10.1126/science.289.5486.1925. [DOI] [PubMed] [Google Scholar]
- 4.Lauren J, Gimbel DA, Nygaard HB, Gilbert JW, Strittmatter SM. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers. Nature. 2009;457:1128–32. doi: 10.1038/nature07761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karran E, De, Strooper B. The amyloid hypothesis in Alzheimer disease: New insights from new therapeutics. Nat Rev Drug Disco. 2022;21:306–18. doi: 10.1038/s41573-022-00391-w. [DOI] [PubMed] [Google Scholar]
- 6.Mouillet-Richard S, Ghazi A, Laurent-Puig P. The cellular prion protein and the hallmarks of cancer. Cancers. 2021;13:5032. doi: 10.3390/cancers13195032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Corre D, Ghazi A, Balogoun R, Pilati C, Aparicio T, Martin-Lannerée S, et al. The cellular prion protein controls the mesenchymal-like molecular subtype and predicts disease outcome in colorectal cancer. EBioMedicine. 2019;46:94–104. doi: 10.1016/j.ebiom.2019.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Zhao W, Akbani R, Liu W, Ju Z, Ling S, et al. Characterization of human cancer cell lines by reverse-phase protein arrays. Cancer Cell. 2017;31:225–39. doi: 10.1016/j.ccell.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hampel H, Vassar R, De Strooper B, Hardy J, Willem M, Singh N, et al. The β-Secretase BACE1 in Alzheimer’s Disease. Biol Psychiatry. 2021;89:745–56. doi: 10.1016/j.biopsych.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkin ET, Watt NT, Hussain I, Eckman EA, Eckman CB, Manson JC, et al. Cellular prion protein regulates beta-secretase cleavage of the Alzheimer’s amyloid precursor protein. Proc Natl Acad Sci USA. 2007;104:11062–7. doi: 10.1073/pnas.0609621104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths HH, Whitehouse IJ, Baybutt H, Brown D, Kellett KA, Jackson CD, et al. Prion protein interacts with BACE1 protein and differentially regulates its activity toward wild type and Swedish mutant amyloid precursor protein. J Biol Chem. 2011;286:33489–500. doi: 10.1074/jbc.M111.278556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitehouse IJ, Brown D, Baybutt H, Diack AB, Kellett KAB, Piccardo P, et al. Ablation of prion protein in wild type human Amyloid Precursor Protein (APP) transgenic mice does not alter the proteolysis of APP, levels of Amyloid-β or pathologic phenotype. PloS One. 2016;11:e0159119. doi: 10.1371/journal.pone.0159119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitehouse IJ, Miners JS, Glennon EBC, Kehoe PG, Love S, Kellett KAB, et al. Prion protein is decreased in Alzheimer’s brain and inversely correlates with BACE1 activity, amyloid-β levels and Braak stage. PloS One. 2013;8:e59554. doi: 10.1371/journal.pone.0059554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kralovicova S, Fontaine SN, Alderton A, Alderman J, Ragnarsdottir KV, Collins SJ, et al. The effects of prion protein expression on metal metabolism. Mol Cell Neurosci. 2009;41:135–47. doi: 10.1016/j.mcn.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 16.McHugh PC, Wright JA, Williams RJ, Brown DR. Prion protein expression alters APP cleavage without interaction with BACE-1. Neurochem Int. 2012;61:672–80. doi: 10.1016/j.neuint.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Qin K, Zhao L, Gregory C, Solanki A, Mastrianni JA. “Dual Disease” TgAD/GSS mice exhibit enhanced Alzheimer’s disease pathology and reveal PrPC-dependent secretion of Aβ. Sci Rep. 2019;9:8524. doi: 10.1038/s41598-019-44317-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruggink KA, Kuiperij HB, Gloerich J, Otte-Holler I, Rozemuller AJ, Claassen JA, et al. Dickkopf-related protein 3 is a potential Abeta-associated protein in Alzheimer’s Disease. J Neurochem. 2015;134:1152–62. doi: 10.1111/jnc.13216. [DOI] [PubMed] [Google Scholar]
- 19.Killick R, Ribe EM, Al-Shawi R, Malik B, Hooper C, Fernandes C, et al. Clusterin regulates beta-amyloid toxicity via Dickkopf-1-driven induction of the wnt-PCP-JNK pathway. Mol Psychiatry. 2014;19:88–98. doi: 10.1038/mp.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Amico L, Mahajan S, Capietto A-H, Yang Z, Zamani A, Ricci B, et al. Dickkopf-related protein 1 (Dkk1) regulates the accumulation and function of myeloid derived suppressor cells in cancer. J Exp Med. 2016;213:827–40. doi: 10.1084/jem.20150950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrari N, Ranftl R, Chicherova I, Slaven ND, Moeendarbary E, Farrugia AJ, et al. Dickkopf-3 links HSF1 and YAP/TAZ signalling to control aggressive behaviours in cancer-associated fibroblasts. Nat Commun. 2019;10:130. doi: 10.1038/s41467-018-07987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghazi A, Le Corre D, Pilati C, Taieb J, Aparicio T, Didelot A, et al. Prognostic value of the PrPC-ILK-IDO1 axis in the mesenchymal colorectal cancer subtype. Oncoimmunology. 2021;10:1940674. doi: 10.1080/2162402X.2021.1940674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pankiewicz JE, Sanchez S, Kirshenbaum K, Kascsak RB, Kascsak RJ, Sadowski MJ. Anti-prion protein antibody 6D11 restores cellular proteostasis of prion protein through disrupting recycling propagation of PrPSc and targeting PrPSc for lysosomal degradation. Mol Neurobiol. 2019;56:2073–91. doi: 10.1007/s12035-018-1208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linsenmeier L, Mohammadi B, Shafiq M, Frontzek K, Bär J, Shrivastava AN, et al. Ligands binding to the prion protein induce its proteolytic release with therapeutic potential in neurodegenerative proteinopathies. Sci Adv. 2021;7:eabj1826. doi: 10.1126/sciadv.abj1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleffman K, Levinson G, Rose IVL, Blumenberg LM, Shadaloey SAA, Dhabaria A, et al. Melanoma-secreted amyloid beta suppresses neuroinflammation and promotes brain metastasis. Cancer Disco. 2022;12:1314–35. doi: 10.1158/2159-8290.CD-21-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D, Park D, Zhong Y, Lu Y, Rycaj K, Gong S, et al. Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer. Nat Commun. 2016;7:10798. doi: 10.1038/ncomms10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith BA, Sokolov A, Uzunangelov V, Baertsch R, Newton Y, Graim K, et al. A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc Natl Acad Sci USA. 2015;112:E6544–6552. doi: 10.1073/pnas.1518007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajan P, Sudbery IM, Villasevil MEM, Mui E, Fleming J, Davis M, et al. Next-generation sequencing of advanced prostate cancer treated with androgen-deprivation therapy. Eur Urol. 2014;66:32–9. doi: 10.1016/j.eururo.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao SG, Chang SL, Erho N, Yu M, Lehrer J, Alshalalfa M, et al. Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy. JAMA Oncol. 2017;3:1663–72. doi: 10.1001/jamaoncol.2017.0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamoun A, Cancel-Tassin G, Fromont G, Elarouci N, Armenoult L, Ayadi M, et al. Comprehensive molecular classification of localized prostate adenocarcinoma reveals a tumour subtype predictive of non-aggressive disease. Ann Oncol. 2018;29:1814–21. doi: 10.1093/annonc/mdy224. [DOI] [PubMed] [Google Scholar]
- 31.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajan P, Sudbery I, Villasevil M, Mui E, Fleming J, Davis M, et al. Next-generation sequencing of advanced prostate cancer treated with androgen-deprivation therapy. Eur Urol. 2014 [cited 2021 Jun 18];66. Available from: https://pubmed-ncbi-nlm-nih-gov.proxy.insermbiblio.inist.fr/24054872/ [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.