Abstract
Observations of comorbidity in heart diseases, including cardiac dysfunction (CD) are increasing, including and cognitive impairment, such as Alzheimer’s disease and dementia (AD/D). This comorbidity might be due to a pleiotropic effect of genetic variants shared between CD and AD/D. Here, we validated comorbidity of CD and AD/D based on diagnostic records from millions of patients in Korea and the University of California, San Francisco Medical Center (odds ratio 11.5 [8.5–15.5, 95% Confidence Interval (CI)]). By integrating a comprehensive human disease–SNP association database (VARIMED, VARiants Informing MEDicine) and whole-exome sequencing of 50 brains from individuals with and without Alzheimer's disease (AD), we identified missense variants in coding regions including APOB, a known risk factor for CD and AD/D, which potentially have a pleiotropic role in both diseases. Of the identified variants, site-directed mutation of ADIPOQ (268 G > A; Gly90Ser) in neurons produced abnormal aggregation of tau proteins (p = 0.02), suggesting a functional impact for AD/D. The association of CD and ADIPOQ variants was confirmed based on domain deletion in cardiac cells. Using the UK Biobank including data from over 500000 individuals, we examined a pleiotropic effect of the ADIPOQ variant by comparing CD- and AD/D-associated phenotypic evidence, including cardiac hypertrophy and cognitive degeneration. These results indicate that convergence of health care records and genetic evidences may help to dissect the molecular underpinnings of heart disease and associated cognitive impairment, and could potentially serve a prognostic function. Validation of disease–disease associations through health care records and genomic evidence can determine whether health conditions share risk factors based on pleiotropy.
Subject terms: Personalized medicine, Prognostic markers
Introduction
Adequate identification and risk stratification of patients based on substantial evidence including genetic contributions is a central theme in precision medicine [1]. For example, the identification of a pleiotropic effect as a risk factor shared between diseases is a central premise for beneficial care outcomes including co-occurring (or sequential) diagnosis of diseases in an individual [2, 3], and for predicting mortality [4]. While mapping disease relationships has a long history, the advent of digitalized health records has led to a rapid increase in ability to analyze health care data. This allows for the recapitulation of known temporal disease correlations, such as illness trajectories in Denmark [5]. For example, in our previous study, we traced sequential diagnosis patterns of millions of patients, then identified an unexpected risk of patients with schizophrenia that would change clinical care meaningfully [6]. However, the validation of newly identified genomic risk factors underpinning the comorbidity between diseases using functional evidence has shown limited success.
Pleiotropy is a phenomenon whereby one gene affects multiple traits and appears to be pervasive in biology. Over decades, genome-wide association studies (GWAS) have illustrated that trait-associated genetic variants are substantially shared across different traits, which may lead to comorbid diseases [7]. Detection of shared genetic risks between traits and diseases have been mainly presented based on a large-scale analysis of GWAS data [8]. Li et al. determined that corpuscular volume is elevated before diagnosis of acute lymphoblastic leukemia by associated variants in the gene for IKZF1 through the integration of electronic medical records (EMRs) and a comprehensive GWAS database, VARIMED [9]. In other previous attempts, the beneficial results of clinical application of analysis of pleiotropy effect have been demonstrated via subtyping of patients with type 2 diabetes using their personal genotypes and aligning medical records, including those for disease comorbidities [10].
Dementia is a spectrum of neurological conditions and is an increasingly prevalent issue, covering a wide range of medical conditions, including Alzheimer's disease (AD), dementia with Lewy bodies, and vascular dementia. The risk factors for Alzheimer's disease, and dementia (AD/D) have been suggested in previous attempts to identify them, which are mainly based on the analysis of clinical epidemiology, such as smoking pattern [11] and hemodynamic monitoring [12]. Various etiologies of cardiac dysfunction (CD), such as heart failure, obesity, and myocardial infarction (heart attack), have been ranked [13]. Multimorbid conditions (i.e. multiple diseases in a single patient) are a major clinical issue in patients with CD [14, 15]. Interestingly, patients with CD present a higher risk for AD/D [16]. Although it is suspected that heart disease and AD/D share similar genetic backgrounds and risk factors, such as ApoE polymorphisms [17], many aspects of the genetic architecture shared between AD/D and CD remain unknown.
In this study, we recapitulate the comorbidity of CD and AD/D by repurposing the digital health records of a large number of patients from different countries. The purpose of this is to unravel the shared genetic risk factors of these diseases based on functional evidence, which comprises a genetic associations database, in vitro experiments, medical images, and associated cognitive behavior.
Methods
Use of administrative health care records
The dataset used in this analysis was the National Inpatients Set of Korea (NISK) from the Health Insurance Review and Assessment Service (HIRA; www.hira.or.kr). The HIRA database contains deidentified inpatient and outpatient billing information from all medical facilities in Korea covered by insurance, including primary care and academic medical centers. We used three of the annual builds of HIRA (2009–2011) and merged these iterations. To avoid redundancy in the merged HIRA dataset, we only used records of deceased individuals and their diagnostic records in the 2009–2010 versions, and merged these with the most recent 2011 version. All diagnostic codes were reported using the International Classification of Disease, 10th Revision (ICD-10) and grouped to the 3-letter code level to minimize overlap and subclassification of diagnoses. To validate the identified diagnostic patterns, we utilized electronic medical records (EMRs) from the University of California, San Francisco (UCSF) Medical Center, collected using Epic (Verona, WI) between 2011 and 2017, which includes inpatient and outpatient records for 816504 unique individuals. The records were deidentified and contained no direct patient identifiers as defined by the Health Insurance Portability and Accountability Act (HIPAA).
Disease trajectory (temporal diagnoses of diseases) by directed acyclic graph modeling
The primary diagnosis code was used to determine the primary disease state of each patient. We used a method identical to that used in our previous work to trace the temporal order of disease diagnoses for each patient [6, 18]. In summary, a trajectory consists of the first node for patients diagnosed as being with disease i and nodes for subsequent diagnoses presenting in patients who were diagnosed in a prior diagnosis node, which are connected via directed acyclic edges showing that subsequent diagnoses occurred more frequently than at random. To determine the association between a pair of diseases that were co-diagnosed in one patient, we first calculated the relative association (RA) measurement of all disease pairs (Disease i – Disease j) that occurred within 1 year for each patient. Then, we quantified the overall temporal directionality of disease co-occurrence (δi→j for Disease i → Disease j) using mean difference of dates of diagnoses or associated admissions for each individual. The statistical significance was determined by a binomial test (Benjamin–Hochberg false discovery rate [FDR] < 0.1). Finally, we used selected pairs of disease co-occurrences (RA > 1, FDR < 0.1) with the directional order of diagnoses (δi→j ≠ 0, FDR < 0.1) in further analysis.
Based on the shared patients between two pairs of diagnoses steps (Disease 1→2 and Disease 2→3, FDR < 0.1), we joined multiple steps of disease-to-disease trajectories by concatenating the progressions of the two diseases into three or multiple steps of diseases (Disease 1→2→3). All details of methods utilized are presented in Additional file 1.
Analysis of whole-exome sequencing of AD/D samples
Tissue collection and genomic DNA extraction
In the present study, we used the same samples and case-control criteria as used in our previous work [19]. In summary, two brain banks (the Netherlands Brain Bank [NBB] and the Human Brain and Spinal Fluid Resource Center [HBSFRC]) provided fresh frozen human postmortem hippocampus and matched whole blood samples. Samples were classified into AD-affected, and unaffected age-matched controls based on clinical and neuropathological findings. Genomic DNA (gDNA) was isolated from 6 to 10 Nissl-stained tissue slides by laser capture microdissection (LCM). Cryosectioning of a frozen hippocampal tissue block (20 μm depth) was performed by cryostat (CM1850, Leica Germany) and attached to ultraviolet (UV)-treated 1.0 mm PEN-membrane slides (Zeiss, 415190-9041-000). The slides were stained with 1% cresyl violet-75% EtOH solution right before LCM. After confirming the proper staining of sub-regions of the hippocampal formation (HIF), slides were mounted on the stage of an LCM instrument (PALM MicroBeam, Zeiss Germany). HIFs from the slides were captured and stored in a lysis buffer (56304, Qiagen, Germany). Mechanical crushing of the acquired tissue was performed using a bead-beating homogenizer (FastPrep-24, MP Biomedicals USA). The homogenized tissue was then lysed at 56 °C for 12 h, using a column-based QIAamp DNA Micro Kit (56304, Qiagen). Whole blood cells were processed using QIAamp DNA Blood Midi Kit (51183, Qiagen), according to the manufacturer’s instructions. Quantification of extracted gDNA was performed using a Bioanalyzer 2100 (Agilent, USA), and its integrity was checked by running it on 1% agarose gel. The volume of the LCM-captured region from the PALM Robo software and the concentration of gDNA from the Bioanalyzer was used to determine the average gDNA yield from the HIF. The process of estimating the number of neurons in the LCM-captured region is as follows: first, the average number of observed neurons in each sub-region of the HIF is determined by finding the number of maximum signals from 8-bit converted Nissl-stained images using ImageJ software. Thereafter, the area of the LCM-captured region is multiplied by the average neuron count.
Whole-exome sequencing
Each exome library was prepared based on the manufacturer’s instructions (Human All Exon V4/V5 + UTR 50 Mb Kit, Agilent USA) using about 1 μg of gDNA. Paired-end sequencing with an Illumina HiSeq 2000/2500 sequencing system was performed according to the manufacturer’s protocol using quality control-passed exome libraries (http://nextgen.mgh.harvard.edu/attachments/Paired-End_SamplePrep_Guide_1005063_D.pdf).
To identify germline variants from the samples, we applied the GATK’s best practice workflows. First, germline variants of each sample were identified with a GATK HaplotypeCaller (release 3.5.0) in GVCF mode. All GVCFs generated from each sample were jointly genotyped by using the GATK GenotypeGVCFs tool (release 3.5.0) and we recalibrated the variant quality score of the jointly genotyped vcf with the GATK VariantRecalibrator. Next, we applied a hard filter with the criteria of QD < 2.0, FS > 60.0, MQ < 20.0, HaplotypeScore >13.0, MappingQualityRankSum < −12.5, or ReadPosRankSum < −8.0 on the recalibrated vcf as recommended in GATK’s best practice workflows. Finally, variants that passed the hard filter were annotated with an Ensemble Variant Effect Predictor [20].
Identification of newly identified pleiotropic variants
By integrating a comprehensive human disease–SNP association database (VARIMED), Exome Aggregation Consortium (ExAC), and whole-exome sequencing (WES) of 50 brains from individuals with and without AD, we identified missense variants. In summary, we first selected 92 genes including APOE shared between the relevant genetic variants of CD and AD/D from VARIMED. Among those of genes, we selected 183 pathogenic genetic variants harbored in 58 genes. The pathogenicity of germline variants was determined based on the averaged functional impact of missense variants (mean score >0) from 16 algorithms, including SIFT [21] and Polyphen2 [22]. In Additional File 5, the list of 16 used algorithms is presented. We selected those 16 scoring methods based on the coverage (>90%) and the median score (range 0.2–0.3) criteria, and averaged their scores for each missense mutation. Among those 183 variants, we finally selected three variants in three genes as candidates for newly identified pleiotropic variants that might contribute to both CD and AD/D. Those three variants are (i) among the top 10 ranked pathogenic score (>0.7), (ii) not detected in individuals without AD in our WES samples, (iii) rare variants based on ExAC (allele frequency <1%) [23], and (iv) not reported in ClinVar, implying that they were not identified as mutations in human phenotypes [24].
Validation of the pleiotropic variants for CD and AD/D
Molecular cloning of the selected variants and expression of the human protein, tau
Three of the selected germline mutations (MTHFD1L c.1688G>A; p.Arg563His, DPP10 c.2254C>A; p.Gln752Lys, ADIPOQ c.268G>A; p.Gly90Ser) are reconfirmed in identical brain samples through Sanger sequencing. After the reconfirmation of germline mutations, full-length cDNA of human MTHFD1L (2937 bp, ENST00000367321.7) was chemically synthesized and mutant form MTHFD1L (c.1688G>A), ADIPOQ (735 bp, ENST00000320741.6; c.268G>A), and DPP10 (2391 bp, ENST00000410059.5; c.2254C>A) were generated by mutagenesis. To append a 3×FLAG tag on either N-, C-terminal of wild-type and mutant form, we synthesized 3×FLAG DNA fragments by annealing oligos (Table 1).
Table 1.
Oligos used to synthesize 3×FLAG DNA fragments.
| sense 5′–3′ | anti-sense 5′–3′ | |
|---|---|---|
| (i) N-3×FLAG | ATGGACTACAAAGACCA | CTTGTCATCGTCATCCTTGTA |
| TGACGGTGATTATAAAGAT | GTCGATGTCATGATCTTTATAA | |
| CATGACATCGACTACAAG | TCACCGTCATGGTCTT | |
| GATGACGATGACAAG | TGTAGTCCAT | |
| (ii) 3×FLAG-C | GACTACAAAGACCATGACGG | CTACTTGTCATCGTCATC |
| TGATTATAAAGATCATGACAT | CTTGTAGTCGATGTCATG | |
| CGACTACAAGGATGACGATG | ATCTTTATAATCACCGTC | |
| ACAAGTAG | ATGGTCTTTGTAGTC |
Then, we performed overlap PCR with 3×FLAG and each cDNA as templates to make the final insert. We assembled each insert and linearized Xho1/EcoR1 double-digested form of pCIG2-C1 plasmid (modified from pCIG2) by using an EZ-fusion cloning kit (EZ016S, Enzynomics South Korea) to obtain pCIG-3×FLAG-hMTHFD1L, pCIG-3×FLAG-hADIPOQ, and pCIG-3×FLAG-hDPP10. For an expression test, 3 μg of pCIG2-3×FLAG (Empty), pCIG2-3×FLAG-WT, and pCIG2-3×FLAG-mutant plasmid DNA were transiently expressed in Neuro-2a cell line (American type Culture Collection [ATCC], CCL-131) by using iN-fect reagent (15081, iNtRON South Korea) and cells were harvested after 72-h transfection. Neuro-2a is a fast-growing mouse neuroblastoma cell line and has been widely used to study Alzheimer’s disease [25]. Therefore, we used this cell line for the functional validation.
The expression levels of human proteins, such as tau (a known phenotype signature of AD), were estimated using western blotting. The cells were washed with phosphate-buffered saline (PBS) and lysed with 1× RIPA buffer (9806, Cell signaling Technology) in combination with protease inhibitor cocktail (P8465, Sigma USA). The lysates were cleared by centrifugation at 13,000 × g for 30 min and the protein concentration of the supernatant was determined using a Bradford protein assay (Bio-Rad). Total proteins were separated by NuPage 4–12% bis Tris–polyacrylamide gel electrophoresis in MES SDS running buffer (ThermoFisher Scientific), and then transferred to a PVDF membrane (GE Healthcare USA). The membranes were probed with specific antibodies. Immunocomplexes were detected using horseradish peroxidase-conjugated anti-rabbit, mouse antibodies followed by chemiluminescence detection (ECL, Amersham). The band intensities were quantified using NIH ImageJ software. We used specific primary antibody against CP13 (Anti-CP13, phospho-tau ser202) (39357, Cell Signaling Technology). Polyclonal tau (A0024) antibody was purchased from Dako, and PHF1 (phospho ser396/404) antibody was kindly provided by Dr. P. Davies. Anti-β-actin antibody was obtained from Sigma (A2228).
Analysis of the functional role of ADIPOQ in a cardiomyoblast cell (H9c2) and associated disease pathway
To evaluate the functional role of ADIPOQ on CD, we cloned ADIPOQ knockout (ADIPOQ-KO) cells using cardiomyoblasts (H9c2). Using a CRISPR-Cas9 system (px 330 all-in-one vector), we constructed ADIPOQ-KO cells [26], single-cell RNA-seq data of these ADIPOQ-KO cells were analyzed using the Chromium platform (10X Genomics). The single-cell RNA-seq reads were filtered based on the percentage of mitochondrial genes (<30%). All of doublet artifacts are removed using DoubletFinder [27]. From 15,423 cells and 800,233,426 reads of transcripts, we calculated the pseudobulk expression of all genes [28] to identify differentially expressed genes (DEGs) by comparing the reads with those from conventional RNA-seq of H9c2 cells (GSE89130). After quantile normalization between our pseudobulk expressions of genes and transcript abundance of H9c2, we selected DEGs using R software. The associated pathways and Gene Ontology (GO) of the selected DEGs were determined based on the p value of the gene set enrichment test [29] using the R software package clusterProfiler (https://git.bioconductor.org/packages/clusterProfiler).
Structural and functional differences of hearts and brains by the germline variants of ADIPOQ
We also examined anatomical and functional differences by the selected germline variant in ADIPOQ, such as volumes of heart, trends in numeric memory function, and asymmetricity of brains in a longitudinal manner from the UK Biobank. Of 502,591 individuals in the UK Biobank study, germline variants of 49,960 of individuals were sequenced using WES. Of these 49,960 of participants, we selected 310 who had a rare variant in ADIPOQ (ENST00000320741.6; c.268A), then selected 49,642 individuals who have a major allele in ADIPOQ (ENST00000320741.6; c.268G). To show the contribution of the rare variant of ADIPOQ, we selected 69 individuals from a minor allele group, and 275 from a major allele group based on a propensity score analysis [30], implying that other confounders such as age, level of obesity, and sex are similar between compared sets. The volume of hearts including the thickness of the heart wall, volume of ventricles, and aorta were estimated based on the cardiac magnetic resonance images (MRIs) using deep learning-based auto-segmentations [31]. Likewise, the longitudinal trends of numeric memory function of the individuals from the UK Biobank were also compared according to the selected allele groups of ADIPOQ.
Results
Overview of study
The exploration and discovery phases of the present study are depicted in Fig. 1. The merged HIRA covered 55,291,171 diagnoses for 2,182,356 individuals with 43,545 death outcomes. The mean age of patients in the merged HIRA was 54.17 ± 23.26 years. We also utilized deidentified EMRs of UCSF Medical Center consisting of 685,200 patients for the validation of the identified diagnosis patterns from NISK of HIRA (Table 2).
Fig. 1. Identifying shared genetic architecture by repurposing scaled digital health data.
Schematic workflow of a developing hypothesis in the observation phase based on digital health data (gray boxes) to study, using genomic data (white boxes). In the gray boxes, the database of HIRA and the electronic health records (EHR) of UCSF Medical Center were used to suggest the association of cardiac dysfunction (CD), such as hypertensive cardiac disease, with dementia and Alzheimer’s disease (AD/D); the public database of HIRA (Health Insurance Review and Assessment service) was used as a nationwide inpatient/outpatient diagnoses observation database in South Korea; the EHR of UCSF was used as the validation database. After presenting the propensity of CD amongst AD/D diagnoses, we examined the genetic associations between CD and AD/D in a discovery phase (i.e., white boxes).
Table 2.
Summary of the datasets used.
| Datasets and features | Frequency | |
|---|---|---|
| Observation phase | National inpatient/outpatient set of HIRAa | |
| Total patients | 2,182,356 | |
| Selected patientsb | 763,892 | |
| No. of men in the selected set | 341,788 (44.7%) | |
| No. of women in the selected set | 420,104 (55.2%) | |
| No. of diagnoses | 13,459,583 | |
| No. of unique diagnosis codes (ICD-10)c | 5859 (5–3-letter level) | |
| Unique diagnosis high-level (3 letter)d | 1151 | |
| Mean of diagnoses age | 54.19 (±23.26) | |
| Outcome of diagnoses | ||
| Deceasede | 43,545 (5.7%) | |
| Alivef | 1,252,114 | |
| UCSF Medical Centerg (2012.01 to 2017.01) | ||
| Total patients | 6,852,000 | |
| Total cases of diagnoses | 44,545,038 | |
| No. of unique diagnoses codes (ICD-10-CM) | 29,893 | |
| Unique diagnosis codes with 3 letters | 1831 | |
| Mean of diagnoses age | 48.67 (±23.41), | |
| Outcome of diagnoses | ||
| Deceased | 2961 | |
| Not deceased | 143,996 | |
| Pending | 21,248,546 | |
| Discovery phase | VARIMED (VARiant Informing MEDicine) | |
| No. of reviewed publication | 10,331 | |
| No. of SNPs (dbSNP IDs) | 130,426 (129,890) | |
| No. of traits (disease/non-disease traits)h | 4 223 (1 489/2374) | |
| No. of associations between SNPs and traits | 135,410 | |
| NBB (Netherlands Brain Bank) | ||
| Total patients (AD/non-AD)i | 50 (AD 43/non-AD 7) | |
| Hippocampal formation (HF) samples | 50 | |
| Blood (BL) samples | 50 | |
| Mean deceased age (AD/non-AD deceased) | 83.5±8/71.4±12.6 | |
| Sex (AD deceased) | Male = 14; Female = 29 | |
| Sex (non-AD deceased) | Male = 3; Female = 4 | |
| Braak staging (AD/non-AD deceased) | 5.02±1.1/0.71 ± 0.48 | |
| UK Biobank | ||
| No. of total participants | 502,543 | |
| Participants with whole-exome seq (WES)j | 49,960 |
aThe Health Insurance Review and Assessment Service of Korea (HIRA). We utilized non-longitudinal sets consisting of randomly sampled in/outpatient sets built annually from 2009 to 2011 (www.hira.or.kr).
bTo minimize the re-enrollment of patients into the 2011 set from the 2009 and 2010 sets, we selected only deceased patients from the 2009 and 2010 sets. In addition, we excluded records of non-disease-related diagnoses, including injuries, poisoning, and childbirth, using diagnosis codes.
cInternational Statistical Classification of Disease and Related Health Problems 10th Revision (ICD-10).
dBased on the hierarchical structure of ICD-10 codes, which consists of a 5-letter level for a disease with familial history and a 3-letter level for general disease classification, we used transformed diagnosis codes at the 3-digit level in this study.
eDetected outcomes in health insurance reviews (HIRA).
fOther non-deceased outcomes included ongoing patients, transferred, sent back, others, and discharged while alive.
gDeidentified electronic medical records (EMRs) from the University of California, San Francisco (UCSF) Medical Center (a tertiary-care university hospital).
hCounted based on MeSH terms (Medical Subject Headings, the National Library of Medicine’s controlled vocabulary) for traits including eye color and diseases such as asthma.
iAll collected samples were of Western European ancestry.
jWe analyzed participants with WES data to validate the phenotypic effect of the germline variant of interest.
Bold characters emphasize the numbers of the table.
We traced the temporal relationships between diseases diagnosed within individuals to investigate the possibility of a pleiotropic effect on genetic risk factors. To systematically trace the disease diagnosis trajectories, we simplified the timeline for each patient by recording the first instance of each primary diagnosis, such that any later diagnosis of the same disease was removed. We used our previous approach, directed acyclic graph (DAG) modeling approach to recapitulate the pattern of disease diagnoses by time [32]. In DAG modeling, the trajectory consists of the first node for a set of patients diagnosed with a disease i, and nodes are connected via directed acyclic edge presenting subsequent diagnoses occurred frequently than random. The details of our DAG-based approach were published previously [6, 18]. The concept of our DAG-based approach is presented graphically in Additional file 2. The significance of sequential patterns of diagnoses and directionalities were determined by the p-value of the binomial test (FDR < 0.1). In our model, there were 405 disease trajectories traced in total, and our model showed 3.24 steps of disease diagnoses after the initial diagnosis on average (Table 3). After excluding co-occurring diagnoses, which may be associated with the milieu of the 405 disease trajectories, we selected a diagnosis pattern starting from CD to AD/D to examine a possible pleiotropic effect between the diseases through shared genetic risk factors.
Table 3.
Statistics of diagnosis trajectory analysis using the set of HIRA.
| Features | Frequency | |
|---|---|---|
|
Trajectorya = the first and followed nodes linked via edges |
No. of trajectories | 405 |
|
Node = Diagnosed patients as a disease i |
Total type of diagnosis | 604 |
| The 1st diagnosis | 170 | |
| Patients in the 1st diagnosis | 1,023,171 | |
| Fatal outcomes of diagnosis | 2 134 | |
|
Edge (Directed acyclic edge) = Sequence between diagnosis i and j (FDR < 0.1)b |
Subsequent diagnoses after the 1st diagnosis | 80,316 |
| Mean of steps diagnoses in a trajectory | 3.24 ± 1.12 |
aThe trajectory consists of the first node, representing a set of patients diagnosed with disease i, succeeding nodes are connected via a directed acyclic edge, representing subsequent diagnoses that occur more frequently than randomly.
bAll the presented edges were statistically significant (Relative association (RA) for the co-occurrence of diagnosis i and j > 1 and FDR of the binomial test for the co-diagnosis of diagnosis i and j < 0.1; FDR of the binomial test for the sequential directionality of diagnosis dates <0.1). The details of the diagnosis trajectory model are presented on the following website (https://www.youtube.com/watch?v=jJMds31-e2g).
Bold characters to emphasize the numbers of the table.
After we validated a sequential pattern of disease diagnoses, including CD and AD/D, based on the HIRA and UCSF datasets, the shared genetic risk contributions (i.e., a pleiotropic effect) were examined using population-wide evidence from the UK Biobank data and experimental validations.
Validation of the association between CD and AD/D
The 405 disease trajectories modeled based on the HIRA dataset are presented in Additional file 3. Our DAG model demonstrates the real-world phenomena, including the known clinical burden of hospital deaths, and regional pattern of diagnoses prevalence. For example, chronic obstructive pulmonary disease (COPD) patterns in patients with dementia and the elderly were captured in our model with 854 fatal outcomes via the sequential diagnoses including pneumonia, which reinforces the findings of previous studies (Additional file 2) [33, 34].
The CD to AD/D diagnostic path identified in patients from South Korea is shown in Fig. 2A. In total, 425 patients were diagnosed as having hypertensive heart and renal disease, and then presented diverse disease status including aging-related diseases (senile cataracts) and known comorbidity of cardiovascular diseases (hyperlipidemia) [35] within 1 year. Of those 425 patients, 6.8% (n = 26) were diagnosed with AD/D. Patients who were diagnosed with dementia following hypertensive heart and kidney disease were enriched with those having hypertensive heart disease (p = 1.08E–04, hypergeometric test; Fig. 2C). We note that diagnoses of vascular dementia were not included among these 26 patients. Although there is a growing number of reports for the coexistence of heart failure and dementia, the shared risk factors are unclear [36] and warrant further analysis.
Fig. 2. The preceding diagnosis of heart disease before dementia.
A, B Traced models of disease diagnosis patterns using the directed acyclic graph (DAG) model. Our DAG model consisted of a node for the disease diagnosis and an edge for subsequent diagnoses presented that were not random (FDR < 0.1). A From the HIRA dataset, we found that 6.1% of the patients suffering from hypertensive heart and renal disease (n = 425) were diagnosed with unspecified dementia and Alzheimer’s disease (a major subtype of dementia). B We validated whether the identical pattern was repeated in the independent dataset obtained from the electronic health records (EHR) of the UCSF Medical Center. Of the 672 patients with hypertensive heart and kidney disease, 6.8% (n = 46) were diagnosed with dementia. C, D Due to the vague definition of the “hypertensive heart and kidney disease” diagnosis, we looked into a more detailed diagnosis code at the 5-digit level of ICD-10. C In the HIRA dataset, approximately 46% of dementia diagnoses made in patients who already had hypertensive heart and renal diseases were identified after heart failure. Therefore, the diagnosis of dementia after the diagnosis of hypertensive heart and renal disease was significantly enriched by cardiac disease (CD) (p = 1.08E–04, hypergeometry test). D Likewise, the EHR from the UCSF Medical Center also showed an identical pattern. Of the 46 dementia diagnoses made in patients with pre-existing hypertensive heart and renal disease, 33 were identified after the diagnosis of heart failure. The diagnosis of dementia after the diagnosis of hypertensive heart and kidney disease was enriched by cardiac diseases, such as heart failure (p = 4.66E–02, hypergeometric test).
To examine confounders for this propensity for nonvascular AD/D among patients with hypertensive heart failure, we scrutinized independent medical records. We hypothesized that repeated representation of co-occurring diagnoses from an independent dataset shows shared genetic risk factors between diseases. Indeed, the same pattern of diagnoses is observed in the EMR of UCSF Medical Center (Fig. 2B). The UCSF Medical Center EMR provided deidentified data from 816504 patients from 2011 to 2018. In summary, among 672 patients with hypertensive heart and kidney disease, mostly those with hypertensive heart failure (p = 4.66E–02, hypergeometric test; Fig. 2C), 6.8% (n = 46) were also diagnosed with dementia. HIRA and UCSF Medical Center EMR were gathered for nonbiomedical research purposes, indicating a possibility of diagnoses primarily for the billing of health insurance. However, for confirmation of CD and AD/D, general guidelines from the American Heart Association (AHA) [37] and the Diagnostic and Statistical Manual of Mental Disorders: 5th Edition (DSM-5) [38] require evidence-based evaluation of patients, such as MRIs and Mini-Mental State Examination (MMSE) score. Therefore, we were confident of the categorization of the disease patients and the sequential pattern of their diseases. Moving forward to the discovery phase of our study, we sought to verify the shared underlying mechanisms in disease trajectories.
Identification of pleiotropic variants contributing to AD/D and CD
We analyzed WES of 50 samples from deceased individuals consisting of 43 patients with AD and seven individuals from normal group. Since we were mainly interested in the genetic risk of AD/D and CD in the elderly (Fig. 2), we included only the group of subjects with AD are those with late-onset cases (age of diagnosis ≥65, mean deceased age 83.5 ± 8 years). A summary of the demography of patients whose samples were analyzed by WES is shown in Table 4.
Table 4.
Candidates with pleiotropic features (i.e., shared genetic risks) between CD and AD/D.
| Genomic locationa | Gene symbol | Ref/Alt2b | Average of pathogenic scorec | No. of ADd | Allele frequency | Clinical significance (ClinVar) | |
|---|---|---|---|---|---|---|---|
| ExAC (all) | ExAC (Finnish cohort) | ||||||
| Chr6, 150949095 | MTHFD1L | G/A | 0.815 | 3 | 0.01264 | 0.0065 | NS |
| Chr2, 115840821 | DPP10 | C/A | 0.792 | 2 | 0.00308 | 0.00076 | NS |
| Chr3, 186854237 | ADIPOQ | G/A | 0.861 | 1 | 0.00302 | 0.00045 | NS |
None of the selected variants were detected in the non-AD samples.
NS not significant.
aGenomic assembly version of GRCh 38.
bReference nucleotides (Ref) and alternative nucleotides (Alt).
cAverage pathogenic scores from 16 algorithms (SIFT, Polyphen2_HDIV, Polyphen2_HVAR, MutationTester, PROVEAN, REVEL, CADD, fathmm-MKL, Eigen-PC-raw, GERP++, PolyP100way_vertebrate, PolyP20way_Mammalian, phastCons100way_vertebrate, phastCons20way_mammalian, VEST3, and Siphy_29way_logOdds).
dNumber of patientswith Alzheimer’s disease (AD) in our NBB WES dataset (43 AD and seven non-ADsamples).
We analyzed 1,327,403 bp of exonic regions of 88 genes of CD- and AD/D-associated genes (Fig. 3A). Those 88 genes were selected from our comprehensive human disease–SNP association database (VARIMED), a manually curated database of disease–SNP associations, containing over 100 features of association studies from 8962 human genetics papers covering 2376 diseases and traits [9]. The identified set of 88 genes commonly associated with CD and AD/D is shown in Additional file 4. For example, it is known that rare variants of APOB elevate the level of low-density lipoprotein cholesterol (LDL-C) and then ultimately increase the risk of the AD [39]. Meanwhile, genetic mutations of APOB are also associated with the risk of coronary artery diseases and CD [40, 41]. Similarly, other notorious shared genetic risks, such as variants of lipoprotein lipase (LPL), shared between CD and AD/D were detected [42, 43]. In addition, variants with lower pathogenicity score including one variant in ADIPOQ (score = 0.227) were excluded from further analysis (Additional file 4). We filtered 184 germline variants of 58 genes as shared genetic risks of CD and AD/D, and then finally selected three variants in three genes (MTHFD1L, DPP10, ADIPOQ) as candidates with pleiotropic features for the comorbidity of CD and AD/D (Additional file 4). Those three variants were selected based on the newly identified and rare variants in ExAC (<1%) and ClinVar [24], and the averaged pathogenicity scores (>0.7; range [0,1]) from 16 algorithms including SIFT [21] and Polyphen [22], which quantify the functional impact of missense variants (Fig. 3A). The overall distribution of pathogenicity scores across the 16 methods we used are presented in Additional file 5. The three variants were detected in six samples from patients with AD, but in no samples from those without. The average of pathogenicity scores of the variants are presented in Table 4. The variant of MTHFD1L was detected in samples from 3 patients with AD in our NBB WES data (c.1688G>A; p.Arg563His) with pathogenicity score of 0.81, and rarely detected in the ExAC database covering over 60706 individuals. Similarity, variants of DPP10 and ADIPOQ were detected in two and one of our AD samples with high pathogenic scores (0.79 and 0.86, respectively), and rarely detected in the ExAC database. Therefore, we selected those variants as newly identified candidates, which may be associated with CD and AD/D.
Fig. 3. Identification of the candidates of the pleiotropic variants for CD and AD/D.
A Integration of the variant–disease associations metaset from our VARIMED and the whole-exome sequencing (WES) dataset. We selected 3 variants in MTHFD1L, DPP10, and ADIPOQ for further validation. B Reconfirmation of selected germline variants using Sanger sequencing.
Because our WES data were generated from brain samples of patients with AD, we confirmed the variants of the matched blood samples experimentally, if possible, to exclude the possibility of somatic mutations. Three selected germline mutations (MTHFD1L c.1688G>A; p.Arg563His, DPP10 c.2254C>A; p.Gln752Lys, ADIPOQ c.268G>A; p.Gly90Ser) were reconfirmed in brain samples and available blood samples from identical individuals through Sanger sequencing (Fig. 3B).
The rare variants MTHFD1L (c.1688G>A), DPP10 (c.2254C>A), and ADIPOQ (c.268G>A) were selected as candidates for pleiotropic variants for CD and AD/D.
Functional validation of the pleiotropic variants using in vitro experiments
The degree of abundance of CP13 (phosphorylated tau Ser202), PHF1 (phosphorylated tau Ser396/404), and tau proteins in the neuronal cell lines by transfected genes is shown in Fig. 4A. We assessed the abundance of CP12, PHF1, and tau proteins for three biological replicates for each cell line. The abnormal aggregation of CP13, PHF1, and tau is a well-known neuropathology of AD/D [44–46]. We quantified the aggregation of CP13, PHF1, and tau across the status of genetic variants that we selected (three repeats per condition). Although the variants of MTHFD1L (MTHFD1L-M) and DPP10 (DPP10-M) showed similar levels of expression for CP13, PHF1, and tau, the site-directed mutant ADIPOQ (ADIPOQ-M; ADIPOQ c.268G>A) displayed abnormal aggregation of tau and CP13 (Fig. 4B–D; p < 0.05, t-test). Therefore, the identified ADIPOQ variant (c.268G>A) seems to contribute the aggregation of tau and CP13 (phosphorylated tau Ser202).
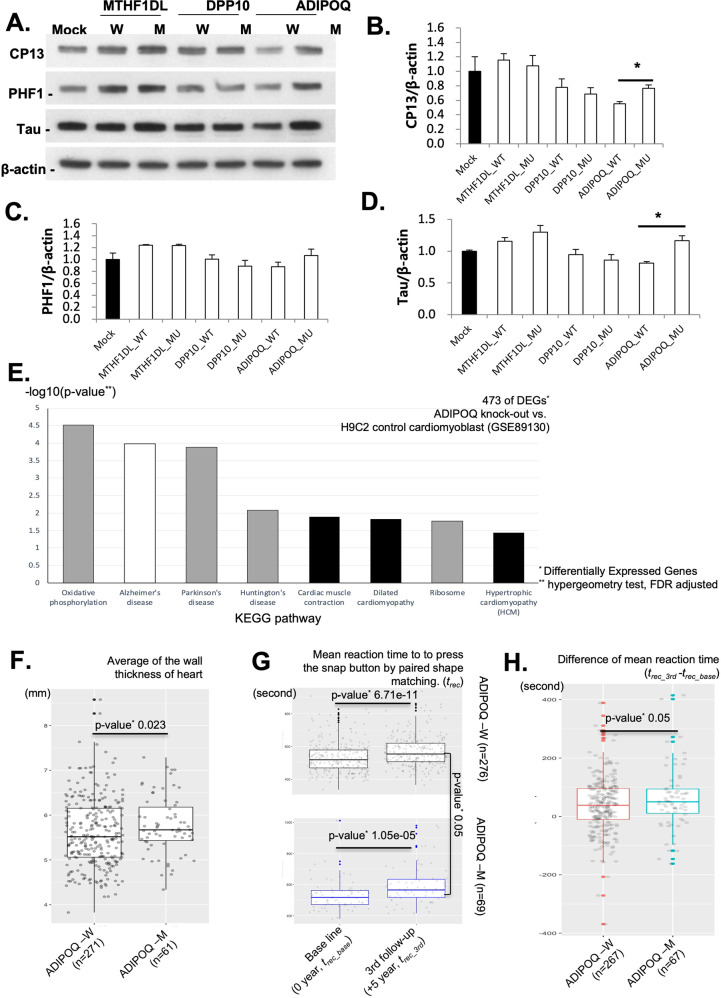
Fig. 4. Validation of pleiotropic effects for CD and AD/D.
A–D Validation of the functional impact of the 3 identified AD/D variants. We constructed site-directed mutant Neuro-2a cell lines (MTHFD1L-M, DPP10-M, and ADIPOQ-M). We assessed the abundance of each cell line of CP12, PHF1, and tau proteins via three biological replicates. A Loading abundance of CP13, PHF1, and tau proteins, as well as β-actin in neuronal cell lines by transfected genes. ‘M’ = mutated. ‘W’ = wild type. B–D Levels of aggregation of CP13, PHF1, and tau normalized by β-actin. ADIPOQ-M displayed abnormal aggregation of tau and CP13 (D) (p < 0.05, t-test). E Analysis of an enriched pathway of 473 selected differentially expressed genes (DEGs) (FDR p < 0.05, log2 fold change >5) in ADIPOQ knockout cells, H9c2 (rat cardiac cell). Cardiac dysfunction and cognition impairment pathways were enriched in 473 DEGs (FDR p < 0.05, hypergeometric test). F–H Functional impact of the ADIPOQ variant on a population scale using the UK Biobank. We selected 69 individuals from the minor allele group of ADIPOQ (c.268G>A) and 276 from the major allele group based on the propensity score matching analysis. F The average heart wall thickness across 16 sites was significantly increased in the ADIPOQ-M group (ADIPOQ c.268A) (p = 0.0023, Wilcoxon test). G Differences in mean reaction time (RT), calculated over 12 rounds, to press a “snap” button when both cards presented matched correctly. The X-axis represents the difference in the measured RTs between the initial assessment (0 years) and the third assessment (5–10 years later). Each plot shows the RTs by allele group (ADIPOQ-W and ADIPOQ-M). H Difference in the mean RTs between the baseline and the third assessment. The minor allele group (ADIPOQ-M) showed a longer mean RT than the major allele group in the third assessment compared to the first assessment (p < 0.05, t-test).
Owing to the poor transfection efficacy of ADIPOQ-M (ADIPOQ c.268G>A) in a cardiac cell line (H9c2), we constructed an ADIPOQ knockout (ADIPOQ-KO) cardiac cell using a CRISPR-Cas9 system (px 330 all-in-one vector, Additional file 6). Then, we constructed the pseudobulk expression of ADIPOQ-KO across 14116 cells by aggregating the count matrix of single-cell RNA-seq. The pseudobulk expression of 15791 genes was calculated based on aggregating the raw read count as depicted in the Methods section and a previous attempt [28]. By comparing the RNA-seq result from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/, accession number: GSE89130), 473 DEGs in ADIPOQ-KO were determined (FDR p < 0.05 and log2 of fold change >5). We conducted a quantile normalization followed by a comparison of the pseudobulk expression to a cardiomyoblast control (GSE89130, bulk RNA-seq results). Here, we note that the identified variant (ADIPOQ 268G>A) is a missense variant with a high pathogenicity score (0.86, Additional File 4), meaning the protein of the ADIPOQ variant would be damaging or truncated. Therefore, we examined the overall impact of DEGs of ADIPOQ-KO to prioritize associated biological function for further validation. The overall gene expression between ADIPOQ-KO and GSE89130 was similar (Pearson correlation coefficient 0.721, p = 1.54E–125) indicating that the batch variation by sequencing technology was negligible (Additional file 6). The 473 DEGs are enriched with CD and cognition impairment pathways including AD, cardiac muscle contraction, and hypertrophic cardiomyopathy (FDR p < 0.05, hypergeometry test; Fig. 4E). The enriched gene ontology (GO) terms of those DEGs show favorable support such as sarcomere (GO:0030017; FDR p = 0.0049), muscle system process (GO:0003012, FDR p = 1.23E–05), and contractile fiber (GO:0043292, FDR p = 0.00049, Additional file 7). The functional impact of ADIPOQ deletion shows that the alteration of overall gene expression is associated with cardiac function, including hypertrophic cardiomyopathy.
In summary, we confirmed that the variant of ADIPOQ (c.268G>A) contributes to abnormal aggregation of tau, and that gene expression altered by the knockout of ADIPOQ is peculiarly associated with dysfunction of cardiac muscle.
Functional validation of the pleiotropic variants using population-wide data
We scrutinized phenotypic differences by the germline variant of ADIPOQ (ADIPOQ c.268G>A). Of 502,591 individuals in the UK Biobank, 49,960 participants have a matched WES data (Table 2). Of them, we selected 310 who have the rare variant in ADIPOQ (ENST00000320741.6; c.268A), then selected 49,642 individuals who have the major allele in ADIPOQ (ENST00000320741.6; c.268G). We selected 69 individuals from the minor allele group, and 276 individuals in the major allele group based on the propensity score matching analysis [47], thereby minimizing differences in other confounders, such as age, level of obesity (measured by the body mass index), and sex, between the compared sets (Additional file 8).
The volume of hearts, including the thickness of heart wall, volume of ventricles, and aorta, were estimated based on cardiac magnetic resonance images (MRIs) using deep learning-based auto segmentation [31]. The average of the wall thickness of the heart across 16 sites is significantly increased in the ADIPOQ-M group (ADIPOQ c.268A, p = 0.0023 in one-sided Wilcoxon test for whether the thickness of ADIPOQ-M group is greater than W); mean of the wall thickness in ADIPOQ-W = 5.60 ± 0.76 mm; mean of the wall thickness in ADIPOQ-M = 5.77 ± 0.64 mm (Fig. 4F). The wall thickness measured from 16 different sites in the heart between ADIPOQ-W (major genotype GG) and ADIPOQ-M (minor genotype AG) is shown in Additional file 9. However, other screened phenotypes from heart MRIs, such as ventricular volume and atrial volume, are similar in the allele group of ADIPOQ, indicating that a thick heart wall associated with the germline variant of ADIPOQ (ADIPOQ c.268G>A) is independent of heart volume.
We also examined the genetic contribution of the ADIPOQ variant (ADIPOQ c.268G>A) for AD/D. Based on the reliability of the cognitive tests in the UK Biobank [48], we selected mean reaction time (RT) of participants. Within an assessment, the RT was measured for 12 rounds by pressing a snap button for which both cards presented matched correctly. We compared the mean RTs between the results of the initial assessment (0 year) and the third assessment (5–10 years later). We omitted the second assessment due to the lack of assessment results. The longitudinal trends of cognition and processing functions are decreased in both ADIPOQ allele groups with the participants’ aging (Fig. 4G). The mean age of all participants was 53.95 years (ADIPOQ-W) and 53.45 years (ADIPOQ-M) in the initial assessment, respectively (Additional file 8). Meanwhile, the mean RTs between ADIPOQ-W and ADIPOQ-M were found to be similar from the initial assessment (ADIPOQ-W = 531.94 s; ADIPOQ-M = 531.60 s, p = 0.4, t-test), however the minor allele group (ADIPOQ-M) showed a significant increase at the third assessment (ADIPOQ-W = 569.87 s; ADIPOQ-M = 596.92 s, p < 0.05, t-test). Similarly, the difference of the mean RT alterations between allele groups is shown in Fig. 4H. The Y-axis of Fig. 4H indicates the difference between the mean RTs at the baseline and third assessment; the larger value of Y-axis of Fig. 4H indicates degeneration of the cognitive processes and the slowing of RT of participants after the first assessment. At the third assessment time, the minor allele group ADIPOQ (ADIPOQ-M, ADIPOQ c.268G>A, Minor AG) showed an almost 155.24% longer mean RT after the first assessment than the major allele group (mean difference of the RT between the first and third test; major GG = 39.16 ± 95.7 s, minor AG = 60.70 ± 108.44 s, p < 0.05, t-test; Fig. 4H). Although the onset of AD/D was undetected from the health care records of these individuals owing to their relatively young age (mid-50s), our examination shows the contribution of the germline variants of ADIPOQ (c.268G>A) for the phenotype of AD/D, such as degeneration of the processing and response.
These results indicated that we successfully validate potential pleiotropic effects of the variant of ADIPOQ (c.268G>A) for the phenotype of AD/D (aggregation of tau, and the longitudinal trend of cognitive degeneration; p < 0.05), and the phenotype of CD (thickened muscle in the wall of the heart; p < 0.05) (Fig. 4A–H). The repeated patterns of paired diagnoses (i.e., CD and AD/D) (Figs. 1–3) from the population-scale EMRs are based on the pleiotropy of ADIPOQ. Our identified germline variant of ADIPOQ (c.268G>A) is a genetic risk shared between CD and AD/D.
Discussion
In the present study, we hypothesized that a repeated diagnosis of two diseases within a single patient is associated with a shared biological mechanism, such as a pleiotropic variant, and can be validated via big data. Using the systematic approach of our previous work (i.e., DAG modeling) [6], we observed from the NISK of the HIRA and UCSF data that a CD diagnosis, such as hypertensive heart disease or heart failure, is particularly frequent amongst patients with AD/D (odds ratio 11.5 [8.5–15.5, 95% confidence interval (CI)]). We identified three missense variants, including ADIPOQ, which might have a pleiotropic role in both diseases. Functional evidence covering tau aggregation, transcriptional impact of ADIPOQ knockout for cardiomyoblasts, and the quantitative assessment of cognition and cardiac muscle structures persistently indicate the pleiotropy of the ADIPOQ variant (c.268G>A) in CD and AD/D.
Although the expression ADIPOQ has been mainly observed in fat tissue, genetic variants of ADIPOQ showed association with diverse disease traits, such as heart failure and dementia. For example, the release of adiponectin, encoded by ADIPOQ, in epicardial adipose tissues contributes to the defense mechanism for myocardial oxidative stress [49]. Moreover, chronic adiponectin deficiency causes cerebral insulin resistance, leading to AD-like cognition impairments and Aβ deposition in aged mice [50]. Functional contributions of ADIPOQ for both of CD and AD/D been suggested previously, whereas confirmation of pleiotropic loci of ADIPOQ has hitherto been lacking.
Although a plethora of GWAS studies shed light on the genetic risks shared between diseases and traits, they are mainly based on statistical support from a population, calling for independent functional validation. However, analysis of multifunctional genes like that for ADIPOQ, which is involved in regulating fat metabolism and obesity-associated cognitive decline [51], requires multiple evidences for identification of their functional roles. Here, the power of big data analytics, including next-generation sequencing analysis and our disease trajectory modeling, catalyze the identification and validation of newly identified pleiotropic variants for CD and AD/D and showed that significant genetic evidences is indispensable to interpret data-driven analysis in medicine. Therefore, investigation of national projects to exploit the human genome aggregating phenotype data, such as AllofUs (https://allofus.nih.gov/) and FinnGen (https://www.finngen.fi/en), would accelerate the identification of genetic architecture underlying human diseases.
The present study has several limitations. Although we validated phenotypic impact for cardiac muscle alongside the variant of ADIPOQ (c.268G>A) using the UK Biobank, underlying transcriptional evidence remained unclear because of the poor transfection efficacy of ADIPOQ (c.268G>A) in H9c2 cells. The suggested pleiotropic locus of ADIPOQ shows functional impact for CD- and AD/D-asociated phenotypes (i.e., tau aggregation and hypertrophic cardiac muscle). The chronologic order of these diseases (CD followed by AD/D) and late-onset propensities are open questions. We also acknowledge that the suggested pleiotropic locus of ADIPOQ (c.268G>A) is a rare variant (minor allele frequency [MAF] <1% in ExAC), whereas the known incidence of CD and AD/D is substantial. While it is possible that common variants (MAF > 5%) are also associated with CD and AD/D, the functional impact of rare variants has been shown to have more substantial contributions [52]. The influence of confounders, including aging, as well as the subtle effects of common variants, require further study. Therefore, the presented Additional file 4 would pave the way to identify additional pleiotropic loci of CD and AD/D by conjugating genomic data and linked phenotype data, such as that in the UK Biobank, AllofUs, and FinnGen.
Supplementary information
Acknowledgements
This research was supported by the National Institute of General Medical Sciences of the National Institutes of Health (R01GM079719), the Post Genome Multi-Ministerial Genome Project (3000-3031-317) and Korea Institute of Science and Technology Information (KISTI) (K21-L02-C10-S1, K-22-L02-C04-S01). This research was also supported by the project of Building and Operating a Bio Data Station (Platform building and tool development) (NRF-2021M3H9A2030520) funded by the Korean government. This research was also supported by Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (2021M3H9A203052011), and Gil Medical Center (FRD2020-08, funder: Gachon Gil Medical Center).
Author contributions
SBC, and HJP designed the study. HJP, JHL, CSJ, NR, YHK, and JMK analyzed the data. JSP, JHL, HYS, and CMJ performed the experiments. HJP and SBC wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-022-02144-0.
References
- 1.Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–35. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blair DR, Lyttle CS, Mortensen JM, Bearden CF, Jensen AB, Khiabanian H, et al. A nondegenerate code of deleterious variants in Mendelian loci contributes to complex disease risk. Cell. 2013;155:70–80. doi: 10.1016/j.cell.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park J, Lee D-S, Christakis NA, Barabási A-L. The impact of cellular networks on disease comorbidity. Mol Syst Biol. 2009;5:262. doi: 10.1038/msb.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganna A, Ingelsson E. 5 year mortality predictors in 498 103 UK Biobank participants: a prospective population-based study. Lancet. 2015;386:533–40. doi: 10.1016/S0140-6736(15)60175-1. [DOI] [PubMed] [Google Scholar]
- 5.Jensen AB, Moseley PL, Oprea TI, Ellesøe SG, Eriksson R, Schmock H, et al. Temporal disease trajectories condensed from population-wide registry data covering 6.2 million patients. Nat Commun. 2014;5:4022. doi: 10.1038/ncomms5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paik H, Kan MJ, Rappoport N, Hadley D, Sirota M, Chen B, et al. Tracing diagnosis trajectories over millions of patients reveal an unexpected risk in schizophrenia. Sci Data. 2019. 10.1038/s41597-019-0220-5. [DOI] [PMC free article] [PubMed]
- 7.Watanabe K, Stringer S, Frei O, Umićević Mirkov M, de Leeuw C, Polderman TJC, et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet. 2019. 10.1038/s41588-019-0481-0. [DOI] [PubMed]
- 8.Pickrell JK, Berisa T, Liu JZ, Ségurel L, Tung JY, Hinds DA. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet. 2016. 10.1038/ng.3570. [DOI] [PMC free article] [PubMed]
- 9.Li L, Ruau DJ, Patel CJ, Weber SC, Chen R, Tatonetti NP, et al. Disease risk factors identified through shared genetic architecture and electronic medical records. Sci Transl Med. 2014;6:234ra57. doi: 10.1126/scitranslmed.3007191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Cheng WY, Glicksberg BS, Gottesman O, Tamler R, Chen R, et al. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci Transl Med. 2015. 10.1126/scitranslmed.aaa9364. [DOI] [PMC free article] [PubMed]
- 11.Prince M, Cullen M, Mann A. Risk factors for Alzheimer's disease and dementia: a case-control study based on the mrc elderly hypertension trial. Neurology. 1994. 10.1212/wnl.44.1.97. [DOI] [PubMed]
- 12.Seshadri S, Beiser A, Selhub J, Jacques PF, Rosenberg IH, D’Agostino RB, et al. Plasma homocysteine as a risk factor for dementia and Alzheimer’s disease. N Engl J Med. 2002. 10.1056/NEJMoa011613. [DOI] [PubMed]
- 13.Poirier P, Giles TD, Bray GA, Hong Y, Stern JS, Pi-Sunyer FX, et al. Obesity and cardiovascular disease: Pathophysiology, evaluation, and effect of weight loss: an update of the 1997 American Heart Association Scientific Statement on obesity and heart disease from the Obesity Committee of the Council on Nutrition, Physical Activity, and Metabolism. Circulation. 2006;113:898–918. doi: 10.1161/CIRCULATIONAHA.106.171016. [DOI] [PubMed] [Google Scholar]
- 14.Buddeke J, Bots ML, Van Dis I, Visseren FLJ, Hollander M, Schellevis FG, et al. Comorbidity in patients with cardiovascular disease in primary care: a cohort study with routine healthcare data. Br J Gen Pract. 2019;69:E398–E406. doi: 10.3399/bjgp19X702725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buddeke J, Bots ML, van Dis I, Liem A, Visseren FLJ, Vaartjes I. Trends in comorbidity in patients hospitalised for cardiovascular disease. Int J Cardiol. 2017;248:382–8. doi: 10.1016/j.ijcard.2017.06.106. [DOI] [PubMed] [Google Scholar]
- 16.Qiu C, Winblad B, Marengoni A, Klarin I, Fastbom J, Fratiglioni L. Heart failure and risk of dementia and Alzheimer disease: a population-based cohort study. Arch Intern Med. 2006;166:1003–8. doi: 10.1001/archinte.166.9.1003. [DOI] [PubMed] [Google Scholar]
- 17.Kivipelto M, Helkala EL, Laakso MP, Hänninen T, Hallikainen M, Alhainen K, et al. Midlife vascular risk factors and Alzheimer’s disease in later life: Longitudinal, population based study. Br Med J. 2001. 10.1136/bmj.322.7300.1447. [DOI] [PMC free article] [PubMed]
- 18.Paik H, Kim J. Condensed trajectory of the temporal correlation of diseases and mortality extracted from over 300,000 patients in hospitals. PLoS ONE. 2021;16:e0257894. doi: 10.1371/journal.pone.0257894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park JS, Lee J, Jung ES, Kim MH, Kim IB, Son H, et al. Brain somatic mutations observed in Alzheimer’s disease associated with aging and dysregulation of tau phosphorylation. Nat Commun. 2019. 10.1038/s41467-019-11000-7. [DOI] [PMC free article] [PubMed]
- 20.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The ensembl variant effect predictor. Genome Biol. 2016. 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed]
- 21.Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat Protoc. 2016. 10.1038/nprot.2015.123. [DOI] [PubMed]
- 22.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010. 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed]
- 23.Karczewski KJ, Weisburd B, Thomas B, Solomonson M, Ruderfer DM, Kavanagh D, et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 2017. 10.1093/nar/gkw971. [DOI] [PMC free article] [PubMed]
- 24.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014. 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed]
- 25.Liu N, Yu Z, Xun Y, Li M, Peng X, Xiao Y, et al. TNFAIP1 contributes to the neurotoxicity induced by Aβ25–35 in Neuro2a cells. BMC Neurosci. 2016;17:51. doi: 10.1186/s12868-016-0286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013. 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed]
- 27.McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019. 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed]
- 28.Hou W, Ji Z, Ji H, Hicks SC. A systematic evaluation of single-cell RNA-sequencing imputation methods. Genome Biol. 2020. 10.1186/s13059-020-02132-x. [DOI] [PMC free article] [PubMed]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed]
- 30.Zhao Q-Y, Luo J-C, Su Y, Zhang Y-J, Tu G-W, Luo Z. Propensity score matching with R: conventional methods and new features. Ann Transl Med. 2021. 10.21037/atm-20-3998. [DOI] [PMC free article] [PubMed]
- 31.Bai W, Sinclair M, Tarroni G, Oktay O, Rajchl M, Vaillant G, et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks 08 Information and Computing Sciences 0801 Artificial Intelligence and Image Processing. J Cardiovasc Magn Reson. 2018. 10.1186/s12968-018-0471-x. [DOI] [PMC free article] [PubMed]
- 32.Paik H, Kan MJ, Rappoport N, Hadley D, Sirota M, Chen B, et al. Tracing diagnosis trajectories over millions of patients reveal an unexpected risk in schizophrenia. Sci Data. 2019. 10.1038/s41597-019-0220-5. [DOI] [PMC free article] [PubMed]
- 33.Lowery EM, Brubaker AL, Kuhlmann E, Kovacs EJ. The aging lung. Clin Interv Aging. 2013. 10.2147/CIA.S51152. [DOI] [PMC free article] [PubMed]
- 34.Manabe T, Fujikura Y, Mizukami K, Akatsu H, Kudo K. Pneumonia-associated death in patients with dementia: A systematic review and meta-analysis. PLoS ONE. 2019. 10.1371/journal.pone.0213825. [DOI] [PMC free article] [PubMed]
- 35.Sander GE, Giles TD. Hypertension and lipids: Lipid factors in the hypertension syndrome. Curr Hypertens Rep. 2002. 10.1007/s11906-002-0026-1. [DOI] [PubMed]
- 36.Ng JB, Turek M, Hakim AM. Heart disease as a risk factor for dementia. Clin Epidemiol. 2013.10.2147/CLEP.S30621. [DOI] [PMC free article] [PubMed]
- 37.Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE, Drazner MH, et al. 2013 ACCF/AHA Guideline for the Management of Heart Failure. J Am Coll Cardiol. 2013. 10.1016/j.jacc.2013.05.019.
- 38.Apostolova LG. Alzheimer Disease. Continuum (Minneap Minn). 2016. 10.1212/CON.0000000000000307. [DOI] [PMC free article] [PubMed]
- 39.Wingo TS, Cutler DJ, Wingo AP, Le NA, Rabinovici GD, Miller BL, et al. Association of early-onset Alzheimer disease with elevated low-density lipoprotein cholesterol levels and rare genetic coding variants of APOB. JAMA Neurol. 2019. 10.1001/jamaneurol.2019.0648. [DOI] [PMC free article] [PubMed]
- 40.Ludwig EH, Hopkins PN, Allen A, Wu LL, Williams RR, Anderson JL, et al. Association of genetic variations in apolipoprotein B with hypercholesterolemia, coronary artery disease, and receptor binding of low density lipoproteins. J Lipid Res. 1997. 10.1016/s0022-2275(20)37419-8. [PubMed]
- 41.Tybjærg-Hansen A, Steffensen R, Meinertz H, Schnohr P, Nordestgaard BG. Association of mutations in the apolipoprotein B gene with hypercholesterolemia and the risk of ischemic heart disease. N Engl J Med. 1998. 10.1056/nejm199805283382203. [DOI] [PubMed]
- 42.Baum L, Chen L, Masliah E, Chan YS, Ng HK, Pang CP. Lipoprotein lipase mutations and Alzheimer’s disease. Am J Med Genet. 1999. 10.1002/(SICI)1096-8628(19990416)88:2<136::AID-AJMG8>3.0.CO;2-D. [DOI] [PubMed]
- 43.Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators, Stitziel NO, Stirrups KE, Masca NG, Erdmann J, Ferrario PG, et al. Coding Variation in ANGPTL4, LPL, and SVEP1 and the risk of coronary disease. N Engl J Med. 2016. 10.1056/nejmoa1507652. [DOI] [PMC free article] [PubMed]
- 44.Janocko NJ, Brodersen KA, Soto-Ortolaza AI, Ross OA, Liesinger AM, Duara R, et al. Neuropathologically defined subtypes of Alzheimer’s disease differ significantly from neurofibrillary tangle-predominant dementia. Acta Neuropathol. 2012. 10.1007/s00401-012-1044-y. [DOI] [PMC free article] [PubMed]
- 45.Torres AK, Jara C, Olesen MA, Tapia-Rojas C. Pathologically phosphorylated tau at S396/404 (PHF-1) is accumulated inside of hippocampal synaptic mitochondria of aged wild-type mice. Sci Rep. 2021. 10.1038/s41598-021-83910-w. [DOI] [PMC free article] [PubMed]
- 46.Iqbal K, Liu F, Gong C-X, Grundke-Iqbal I. Tau in Alzheimer disease and related tauopathies. Curr Alzheimer Res. 2010. 10.2174/156720510793611592. [DOI] [PMC free article] [PubMed]
- 47.Austin PC An introduction to propensity score methods for reducing the effects of confounding in observational studies. Multivariate Behav Res. 2011. 10.1080/00273171.2011.568786. [DOI] [PMC free article] [PubMed]
- 48.Lyall DM, Cullen B, Allerhand M, Smith DJ, Mackay D, Evans J, et al. Cognitive test scores in UK biobank: Data reduction in 480,416 participants and longitudinal stability in 20,346 participants. PLoS ONE. 2016. 10.1371/journal.pone.0154222. [DOI] [PMC free article] [PubMed]
- 49.Antonopoulos AS, Margaritis M, Verheule S, Recalde A, Sanna F, Herdman L, et al. Mutual regulation of epicardial adipose tissue and myocardial redox state by PPAR-γ/adiponectin signalling. Circ Res. 2016. 10.1161/CIRCRESAHA.115.307856. [DOI] [PMC free article] [PubMed]
- 50.Ng RCL, Cheng OY, Jian M, Kwan JSC, Ho PWL, Cheng KKY, et al. Chronic adiponectin deficiency leads to Alzheimer’s disease-like cognitive impairments and pathologies through AMPK inactivation and cerebral insulin resistance in aged mice. Mol Neurodegener. 2016. 10.1186/s13024-016-0136-x. [DOI] [PMC free article] [PubMed]
- 51.Forny-Germano L, De Felice FG, Do Nascimento Vieira MN. The role of leptin and adiponectin in obesity-associated cognitive decline and Alzheimer’s disease. Front Neurosci. 2019. 10.3389/fnins.2018.01027. [DOI] [PMC free article] [PubMed]
- 52.Bomba L, Walter K, Soranzo N. The impact of rare and low-frequency genetic variants in common disease. Genome Biol. 2017;11:8308.. doi: 10.1186/s13059-017-1212-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.