Abstract
Chondromyxoid fibroma (CMF) is a rare benign bone neoplasm that manifests histologically as a lobular proliferation of stellate to spindle-shaped cells in a myxoid background, exhibiting morphological overlap with other cartilaginous and myxoid tumors of bone. CMF is characterized by recurrent genetic rearrangements that place the glutamate receptor gene GRM1 under the regulatory control of a constitutively active promoter, leading to increased gene expression. Here, we explore the diagnostic utility of GRM1 immunohistochemistry as a surrogate marker for GRM1 rearrangement using a commercially available monoclonal antibody in a study of 230 tumors, including 30 CMF cases represented by 35 specimens. GRM1 was positive by immunohistochemistry in 97% of CMF specimens (34/35), exhibiting moderate to strong staining in more than 50% of neoplastic cells; staining was diffuse (>95% of cells) in 25 specimens (71%). Among 9 CMF specimens with documented exposure to acid decalcification, 4 (44%) exhibited diffuse immunoreactivity (>95%) for GRM1, whereas all 15 CMF specimens (100%) with lack of exposure to decalcification reagents were diffusely immunoreactive (P=0.003). High GRM1 expression at the RNA level was previously observed by quantitative reverse transcription PCR (RT-qPCR) in 9 CMF cases that were also positive by immunohistochemistry; low GRM1 expression was observed by RT-qPCR in the single case of CMF that was negative by immunohistochemistry. GRM1 immunohistochemistry was negative (<5%) in histological mimics of CMF, including conventional chondrosarcoma, enchondroma, chondroblastoma, clear cell chondrosarcoma, giant cell tumor of bone, fibrous dysplasia, chondroblastic osteosarcoma, myoepithelial tumor, primary aneurysmal bone cyst, brown tumor, phosphaturic mesenchymal tumor, CMF-like osteosarcoma, and extraskeletal myxoid chondrosarcoma. These results indicate that GRM1 immunohistochemistry may have utility in distinguishing CMF from its histologic mimics.
Keywords: GRM1, chondromyxoid fibroma, promoter swapping, immunohistochemistry, bone tumors
INTRODUCTION
Chondromyxoid fibroma (CMF) is a rare benign bone tumor that most commonly arises in the metaphysis of long bones and typically affects adolescents or young adults, although it can be seen in a wide range of ages and in a variety of anatomic sites.1 Histologically, CMF consists of lobules of stellate to spindle-shaped cells in a predominantly myxoid background. The lobules generally are more cellular at their periphery, imparting a characteristic zonal architecture. Whereas myofibroblastic spindle-shaped cells tend to occupy the peripheral zone, stellate and chondroid-appearing cells are found in the center.2 Hyaline cartilage is identified in a minority of cases.3 Enlarged, hyperchromatic, and pleomorphic nuclei, likely degenerative in nature, are sometimes present, mimicking malignancy. In keeping with the morphological features of both cartilaginous and myofibroblastic differentiation, by immunohistochemistry the lesional cells express S100 protein and SOX9,4,5 along with smooth muscle actin.2 ERG expression has also been observed in CMF.6
CMF exhibits recurrent rearrangements involving chromosome arm 6q,7 resulting in translocation or chromoplexy-mediated fusion of the glutamate receptor gene GRM1, a G protein-coupled receptor primarily expressed in neurons of the central nervous system.8,9 This recurrent genetic abnormality places the entire protein-coding sequence of GRM1 downstream of any one of several strongly active gene promoters, such as COL12A1, BCLAF1, or MEF2A. The result of this promoter swapping is an up to 1400-fold increase in GRM1 expression in CMF. While GRM1 rearrangement and overexpression have been observed in 90% of CMF cases, GRM1 expression was found to be negligible in 174 non-CMF mesenchymal tumors, suggesting that aberrantly increased GRM1 expression is a distinctive feature of CMF among mesenchymal neoplasms.8
Immunohistochemical markers are increasingly being used as efficient and cost-effective surrogates for recurrent, diagnostically relevant genetic events in bone and soft tissue tumors. For instance, immunohistochemistry can be used to detect aberrant expression of components of fusion oncoproteins, such as CAMTA1 in epithelioid hemangioendothelioma and STAT6 in solitary fibrous tumor.10–14 Additionally, immunohistochemistry enables identification of protein overexpression secondary to either gene amplification, as observed with MDM2 in low-grade central, parosteal, and dedifferentiated osteosarcoma,15–17 or dysregulation of transcript and protein degradation, as occurs with FOS in osteoid osteoma and osteoblastoma.18–20 Here, we explore the diagnostic utility of GRM1 immunohistochemistry as a surrogate for GRM1 overexpression resulting from recurrent promoter-swapping rearrangements in CMF.
MATERIALS AND METHODS
Cases were retrieved from the surgical pathology archives of Stanford Medical Center, Dartmouth-Hitchcock Medical Center, and Leiden University Medical Center under Institutional Review Board-approved protocols. Representative H&E-stained slides were reviewed to confirm the diagnostic classification. A combination of whole tissue sections and tissue microarrays (TMA) were used to evaluate 230 cases altogether: chondromyxoid fibroma (30 total, 4 TMA; 5 cases had two specimens, representing primary tumor and recurrence), primary aneurysmal bone cyst (35 total, 25 TMA), giant cell tumor of bone (27 total, 17 TMA), chondroblastoma (24 total, 11 TMA), conventional chondrosarcoma with myxoid stroma (20 total, 0 TMA), fibrous dysplasia (15 total, 0 TMA), chondroblastic osteosarcoma (15 total, 0 TMA), extraskeletal myxoid chondrosarcoma (15 total, 3 TMA), myoepithelial tumor (13 total, 0 TMA), enchondroma (10 total, 0 TMA), chordoma (10 total, 0 TMA), clear cell chondrosarcoma (5 total, 0 TMA), phosphaturic mesenchymal tumor (5 total, 0 TMA), brown tumor of hyperparathyroidism (4 total, 4 TMA), and chondromyxoid fibroma-like osteosarcoma (2 total, 0 TMA). The tissue microarrays (TMAs) were constructed using a tissue arrayer (Beecher Instruments, Silver Spring, MD, USA) as previously described.21 Tissues were evaluated as single cores, ranging from 0.6 to 2.0 mm in diameter, taken from representative areas of each formalin-fixed paraffin-embedded block. Cores were not considered if targeted tissue was not included on the array, as assessed morphologically for each core.
Immunohistochemistry for GRM1 was performed on 4-μm-thick formalin-fixed paraffin-embedded tissue sections following pressure cooker antigen retrieval (0.01M citrate buffer, pH 6.0) using a rabbit monoclonal antibody directed against an epitope within amino acids 280–420 of human GRM1 (1:500; clone JM11–61; Thermo Fisher Scientific Inc., Waltham, MA, USA). Immunodetection was completed using the VECTASTAIN ABC kit (Vector Laboratories, Inc., Burlingame, CA, USA) and DAB chromogen (Abcam, Cambridge, UK), according to the manufacturers’ specifications. Appropriate positive control (cerebellum; Supplemental Figure 1) and negative control were employed throughout, including independent controls for each iteration of immunohistochemistry. The extent of immunoreactivity was graded according to the percentage of positive tumor cells and the intensity of staining was graded as weak, moderate, or strong.
Quantitative reverse transcription polymerase chain reaction (RT-qPCR) for GRM1 was previously performed on freshly frozen tissue in ten cases.8 In brief, the TaqMan Gene Expression assay (Hs00168250_m1, Thermo Fisher Scientific Inc.) was used with TBP (4333769-F), ACTB (4333762-T), and HPRT1 (Hs02800695_m1) housekeeping genes as endogenous RNA controls. Control tissues for comparison of GRM1 expression levels included three cases each of extraskeletal myxoid chondrosarcoma, central conventional chondrosarcoma, chondroma, and osteochondroma, in addition to two chondroblastic osteosarcomas and two synovial chondromatoses. RT-qPCR reactions were performed in triplicate using the 7500 RT-PCR system (Thermo Fisher Scientific Inc.). Relative gene expression levels were calculated using the comparative Ct (ΔΔCt) method. Assessment of GRM1 immunohistochemistry was performed by A.M.S.T. and G.W.C. while blinded to the results of RT-qPCR. Fisher’s exact test was used to assess the association between GRM1 immunostaining intensity and tissue decalcification.
RESULTS
GRM1 immunohistochemistry was tested in a cohort of 30 CMF cases (Table 1). Clinical information was available for 26 patients, including 14 females and 12 males. Patients ranged from 9 to 80 years old at presentation (median 26 years old). Tumors were localized to the tibia (n=7), ilium (n=6), metatarsals (n=3), femur (n=2), and phalanges of the foot (n=2), with one case each involving sternum, rib, scapula, metacarpal, radius, and nasal septum. There were five patients with two specimens available for analysis, representing primary and recurrent tumor, yielding 35 CMF specimens altogether. These specimens were derived from curettage (n=21), resection (n=8), or biopsy (n=6) procedures.
Table 1.
Summary of immunohistochemical staining for GRM1.
| Tumor type | Total cases | GRM1 positive (%)a | GRM1 negative (%) |
|---|---|---|---|
| Chondromyxoid fibroma | 30 | 29 (97) | 1 (3) |
| Primary aneurysmal bone cyst | 35 | 0 (0) | 35 (100) |
| Giant cell tumor of bone | 27 | 0 (0) | 27 (100) |
| Chondroblastoma | 24 | 0 (0) | 24 (100) |
| Conventional chondrosarcoma | 20 | 0 (0) | 20 (100) |
| Chondroblastic osteosarcoma | 15 | 0 (0) | 15 (100) |
| Fibrous dysplasia | 15 | 0 (0) | 15 (100) |
| Extraskeletal myxoid chondrosarcoma | 15 | 0 (0) | 15 (100) |
| Myoepithelial tumor | 13 | 0 (0) | 13 (100) |
| Chordoma | 10 | 0 (0) | 10 (100) |
| Enchondroma | 10 | 0 (0) | 10 (100) |
| Clear cell chondrosarcoma | 5 | 0 (0) | 5 (100) |
| Phosphaturic mesenchymal tumor | 5 | 0 (0) | 5 (100) |
| Brown tumor | 4 | 0 (0) | 4 (100) |
| Chondromyxoid fibroma-like osteosarcoma | 2 | 0 (0) | 2 (100) |
Positivity was defined as presence of cytoplasmic staining in greater than 5% of cells.
GRM1 was positive for expression by immunohistochemistry in 29 of 30 tumors (97%) and in 34 of 35 specimens (97%). Anti-GRM1 immunostaining was invariably localized to the cytoplasm. The extent of anti-GRM1 immunoreactivity ranged from ~50% to >95% of neoplastic cells; 25 specimens (25/35; 71%) showed staining in >95%. Likewise, the intensity of immunoreactivity in positive tumors ranged from moderate to strong, with strong staining intensity observed in 27 specimens (27/35; 77%). GRM1 expression was present within cells across the entire spectrum of cytomorphology observed in CMF, including spindle-shaped, stellate, and pleomorphic cells (Figure 1).
Figure 1.
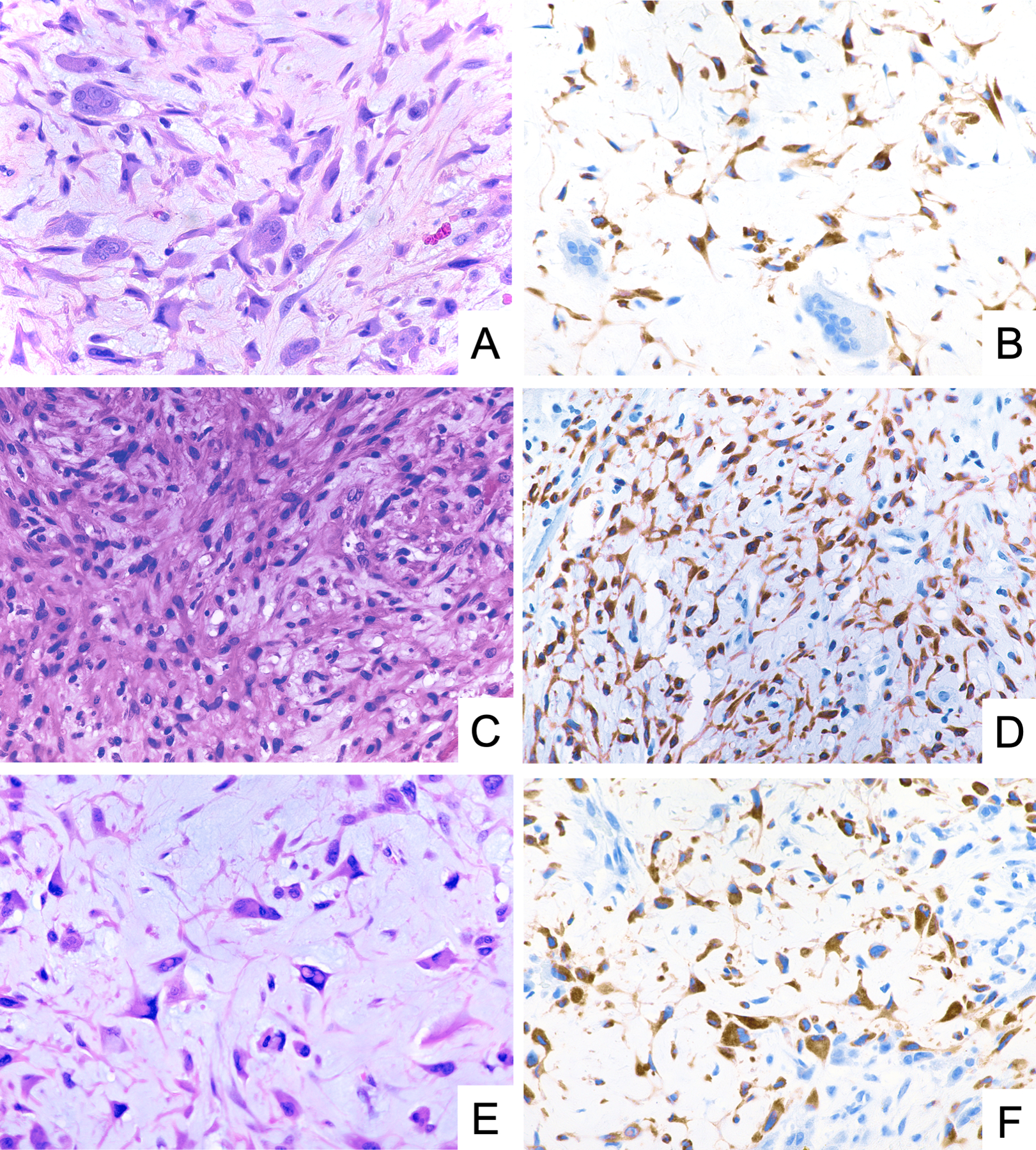
Representative photomicrographs of H&E stain and GRM1 immunohistochemistry demonstrating diffuse and strong GRM1 expression in neoplastic cells of chondromyxoid fibroma, including stellate cells (A, B), spindle-shaped cells (C, D), and pleomorphic cells (E, F).
For all five CMF cases with two separate specimens available for analysis, representing primary and recurrent tumor in each case, GRM1 immunohistochemistry was positive in both specimens. While the intensity of GRM1 staining was similar when comparing each pair of specimens, two of the pairs showed variation in the extent of staining. In both specimen pairs, the loss of immunostaining selectively occurred in the center of tissue fragments, suggesting that the variation in extent of staining was caused by incomplete fixation.
GRM1 RNA expression levels were previously analyzed by RT-qPCR in ten tumors.8 Nine tumors (90%) showed high levels of GRM1 expression, defined as more than 100-fold increased expression relative to other cartilaginous tumors. All nine tumors (100%) with high levels of GRM1 expression by RT-qPCR were positive for GRM1 by immunohistochemistry. The single case with low levels of GRM1 by RT-qPCR, which involved the metatarsal of a 10-year-old female patient, was also the only tumor in the CMF cohort that was negative for GRM1 by immunohistochemistry (Figure 2).
Figure 2.
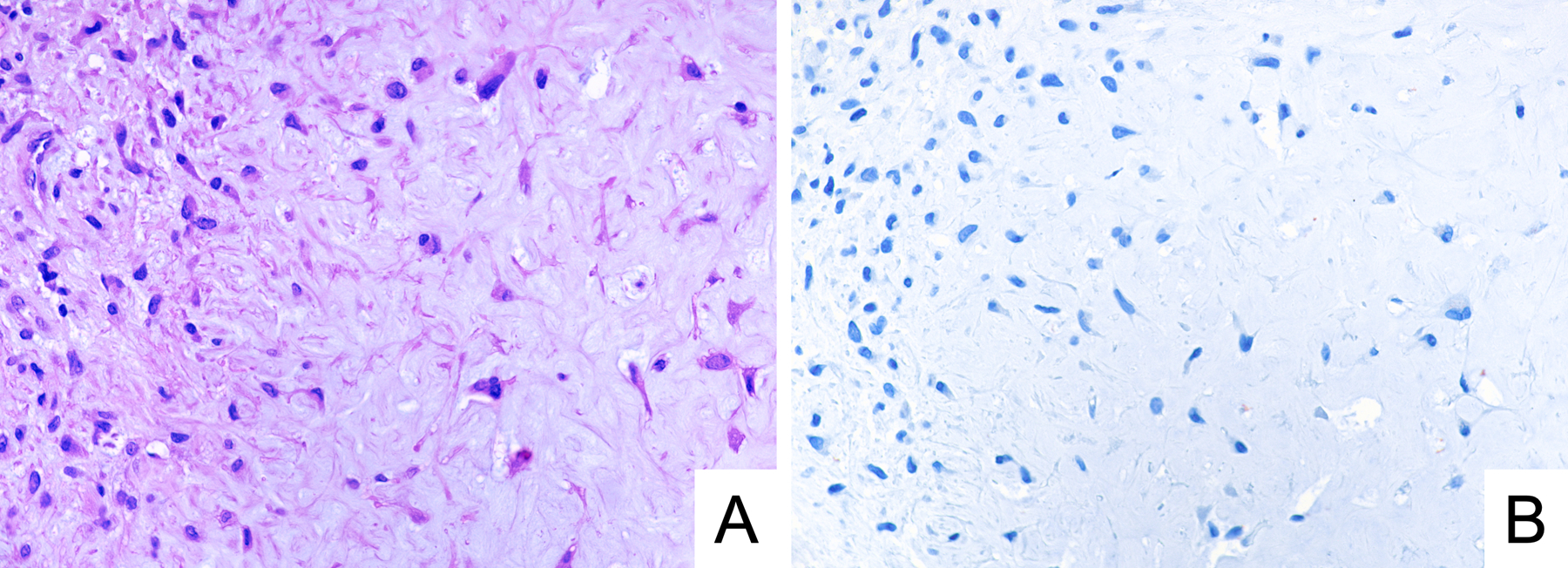
Representative photomicrographs of H&E stain (A) and GRM1 immunohistochemistry (B) in a case of GRM1-negative chondromyxoid fibroma. The tumor involved the first metatarsal in a 10-year-old female patient, and it showed a low level of GRM1 expression by RT-qPCR.
Records related to tissue processing were available for 24 specimens. Acid decalcification was used in nine cases (9/24; 38%; Figure 3). The intensity of staining was strong in 4 decalcified specimens (4/9; 44%) and in all 15 non-decalcified specimens (15/15; 100%; P=0.003). Similarly, whereas immunoreactivity in >95% of neoplastic cells was observed in 13 non-decalcified specimens (13/15; 87%), such extensive staining was seen in only 4 decalcified specimens (4/9; 44%; P=0.06). The single specimen that was negative for GRM1 expression by immunohistochemistry was decalcified; a non-decalcified freshly frozen sample of the same tumor showed low GRM1 levels by RT-qPCR. There were three additional decalcified cases of CMF in which paired non-decalcified fresh-frozen tissue was analyzed by RT-qPCR – all three exhibited high GRM1 expression at the RNA level on non-decalcified freshly tissue and were positive for GRM1 by immunohistochemistry on decalcified formalin-fixed paraffin-embedded tissue.
Figure 3.
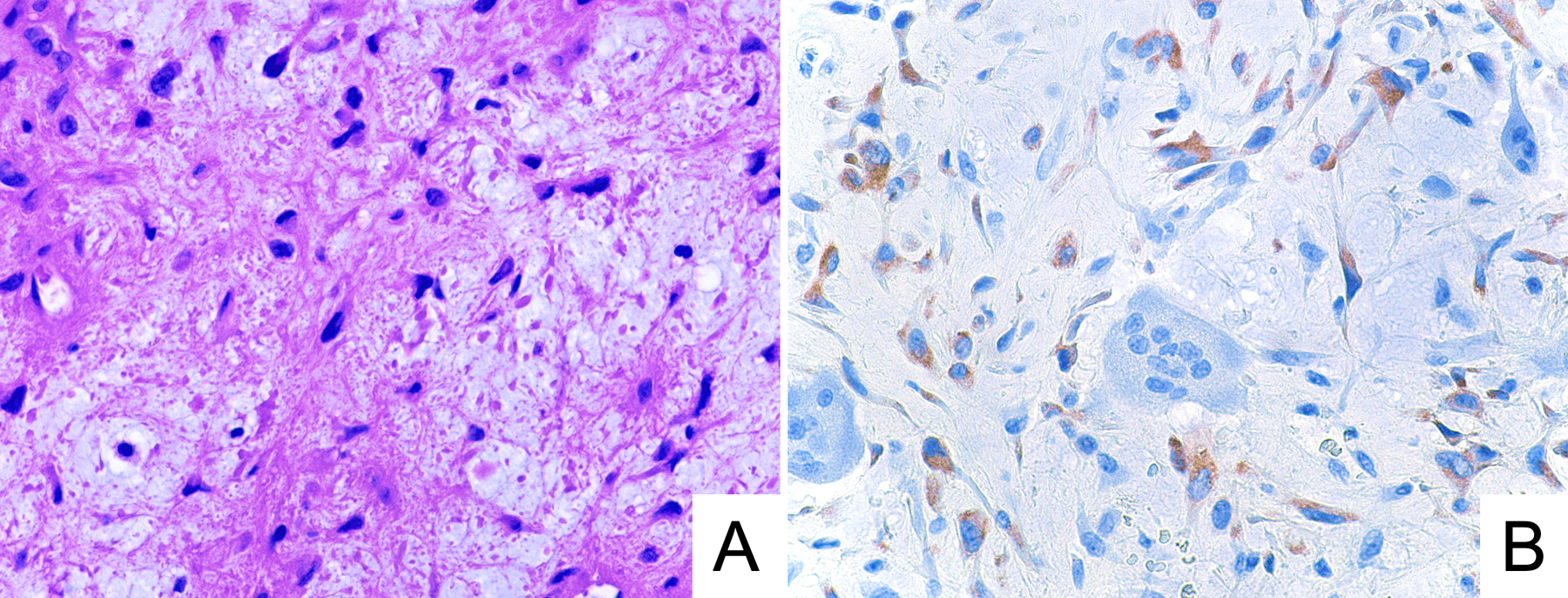
Representative photomicrographs of H&E stain (A) and GRM1 immunohistochemistry (B) in a case of chondromyxoid fibroma exhibiting moderate anti-GRM1 staining intensity. This specimen underwent acid decalcification.
To determine the specificity of GRM1 immunohistochemistry, we tested 200 samples representing potential histological mimics of CMF (Table 1). Among these samples, which encompassed cartilaginous (Figure 4), giant cell-rich (Figure 5), and myxoid tumors (Figure 6), we found no cases that were positive for GRM1 expression at a threshold of staining in 5% of neoplastic cells. Rare, weakly immunoreactive cells, accounting for less than 5% of the lesional cell population, were identified in two cases of chordoma, two cases of chondroblastoma, and one case of chondroblastic osteosarcoma. In addition, we observed no anti-GRM1 immunoreactivity in background non-neoplastic tissue, including bone, cartilage, hematopoietic marrow, blood vessels, and adipose tissue.
Figure 4.
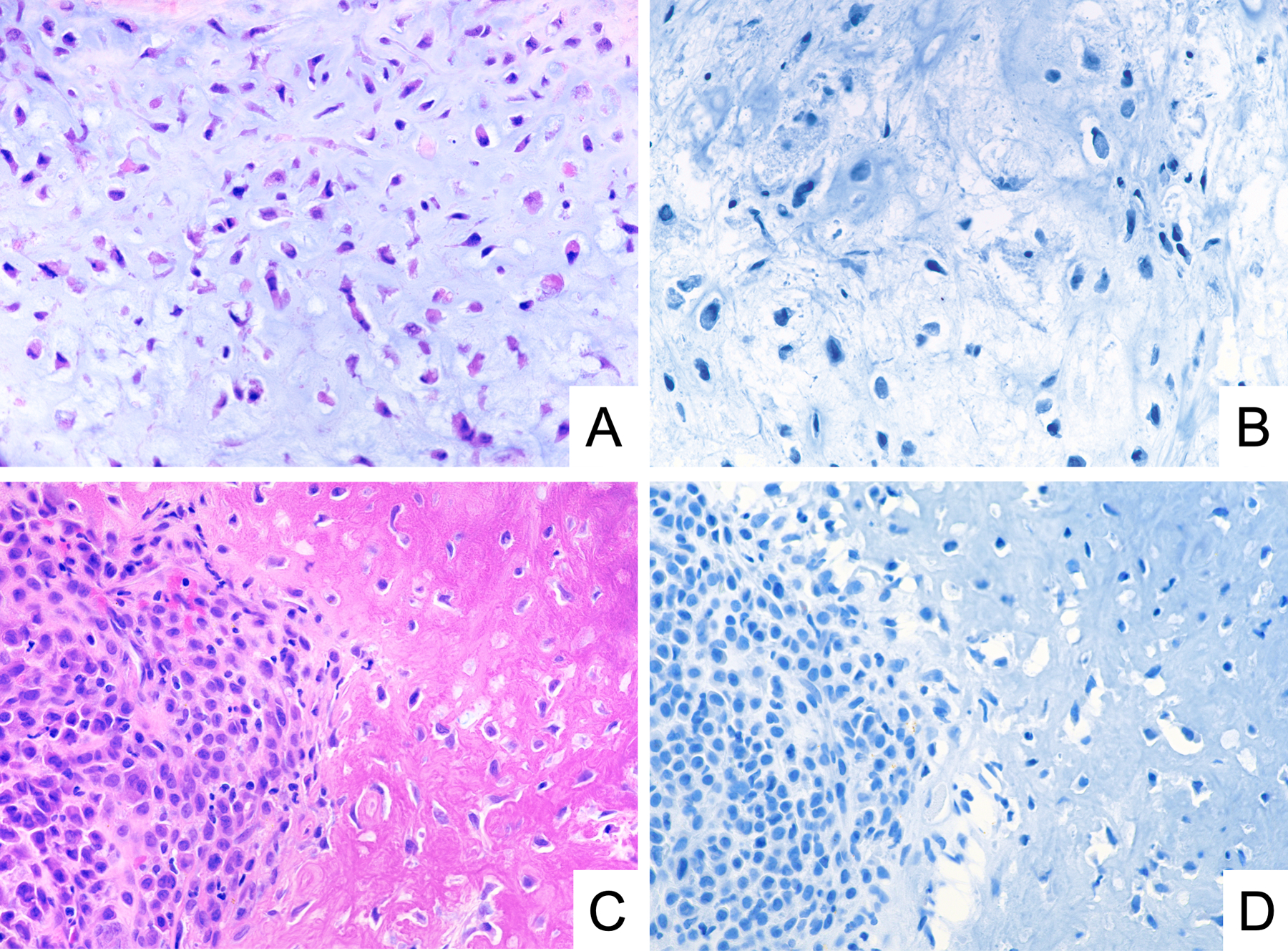
Representative photomicrographs of H&E stain and GRM1 immunohistochemistry demonstrating lack of GRM1 expression in conventional chondrosarcoma with myxoid stroma (A, B) and chondroblastoma (C, D).
Figure 5.
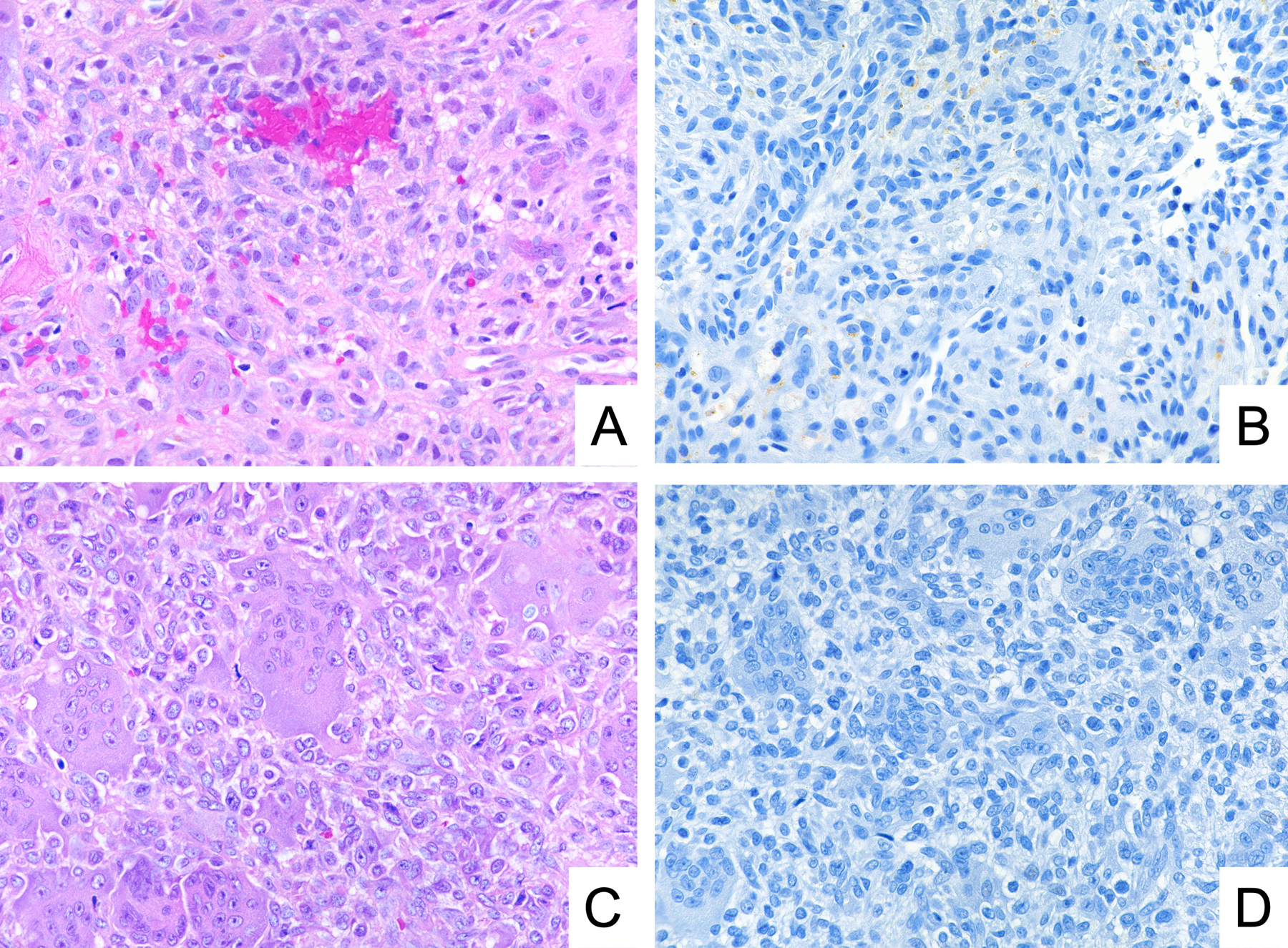
Representative photomicrographs of H&E stain and GRM1 immunohistochemistry demonstrating lack of GRM1 expression in primary aneurysmal bone cyst (A, B) and giant cell tumor of bone (C, D).
Figure 6.
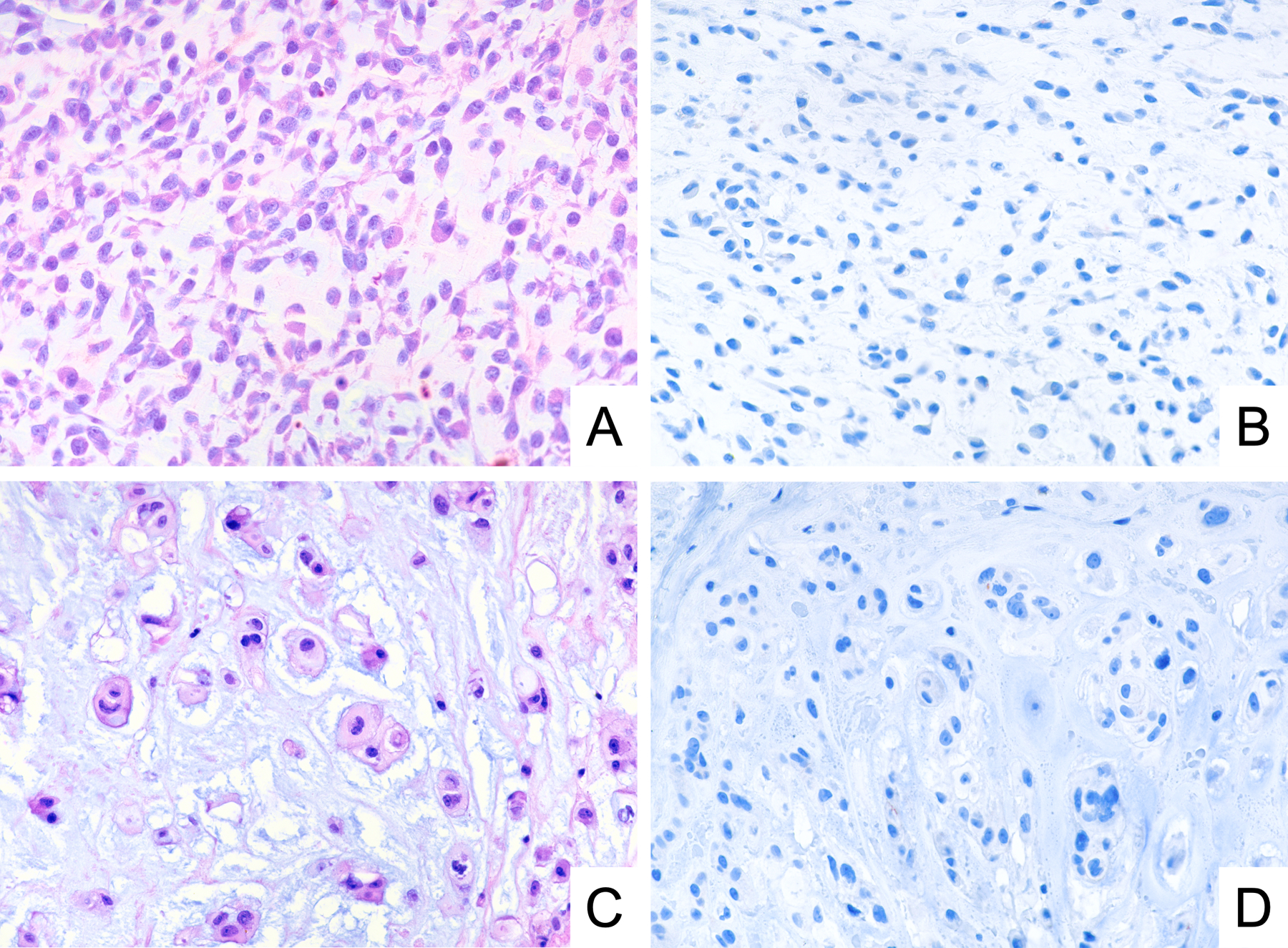
Representative photomicrographs of H&E stain and GRM1 immunohistochemistry demonstrating lack of GRM1 expression in extraskeletal myxoid chondrosarcoma (A, B) and chordoma (C, D).
DISCUSSION
Our data suggest that GRM1 immunohistochemistry is a useful ancillary technique for the diagnosis of CMF, serving as a surrogate marker of recurrent promotor-swapping GRM1 rearrangements. As with other immunohistochemical surrogates of recurrent molecular alterations, GRM1 immunohistochemistry offers practical advantages relative to alternative cytogenetic and molecular techniques, including accessibility, cost, and turnaround time. We anticipate that GRM1 immunohistochemistry may have particular utility in scant or fragmented biopsy specimens in which characteristic morphological features, such as distinctive lobular architecture, are difficult to discern, or when there is a discrepancy with the radiological imaging. Assessment of GRM1 may also prove to be useful in tumors with unusual features, such as cytologic atypia. Thus, GRM1 may complement other immunohistochemical markers for recurrent molecular alterations in primary bone tumors, such as histone H3.3 G34W for giant cell tumor of bone and H3.3 K36M for chondroblastoma.22–25
CMF can be difficult to distinguish from its histologic mimics, especially chondrosarcoma of bone with myxoid changes. Previous studies have used immunohistochemistry to compare CMF with high-grade chondrosarcoma showing decreased expression of CCND1 (67% vs. 20%) and p16INK4A (67% vs 27.5%) in chondrosarcoma.26 However, these results could not be easily translated to routine diagnostics. Here, we use GRM1 immunohistochemistry as a surrogate histologic tool to detect GRM1 rearrangement in CMF, and show GRM1 expression in 29 of 30 CMF (97%) versus 0 out of 20 (0%) conventional chondrosarcomas with myxoid stroma. Therefore, GRM1 immunohistochemistry is a highly specific and sensitive tool to make this distinction.
The histologic differential diagnosis of CMF also includes the extremely rare CMF-like variant of osteosarcoma, which is characterized by spindle-shaped or stellate tumor cells in a background of myxoid stroma.27–29 We observed no GRM1 expression in two cases of CMF-like osteosarcoma, suggesting that GRM1 immunohistochemistry may aid in this challenging diagnostic distinction. However, given the limited number of CMF-like osteosarcoma cases in this series, additional studies are warranted. Thorough morphological examination remains the key to diagnosing CMF-like osteosarcoma, which shows more cytologic atypia and infiltrative growth than CMF. CMF-like osteosarcoma also characteristically exhibits osteoid production by the malignant cells, albeit inconspicuous in some cases.27–29 In addition, correlation with imaging studies is required when considering this differential diagnosis, as CMF-like osteosarcoma will be more aggressive-appearing radiographically with ill-defined margins.
One case of CMF (1/30; 3%) was negative for GRM1 by immunohistochemistry. This tumor involved the first metatarsal in a 10-year-old female patient. It also was the only one of ten examined cases that had low levels of GRM1 mRNA expression by RT-qPCR.8 From a practical perspective, the finding of this GRM1-negative case indicates that an absence of anti-GRM1 immunoreactivity does not entirely exclude a diagnosis of CMF. This finding also raises the possibility that there is a small subset of CMF that arise via pathogenic mechanisms other than GRM1 promoter-swapping rearrangements. Alternatively, given the substantial morphological similarity between CMF and several other cartilaginous and myxoid neoplasms, it may be that rare GRM1-negative CMF cases in fact represent “CMF-like” variants of another entity. However, review of the histology of the GRM1-negative case in our series did not reveal any unusual histological features (Figure 2). Identification and analysis of additional GRM1-negative CMF cases will allow for a better understanding of the molecular pathogenesis of this rare subset of tumors.
We found that GRM1 is susceptible to diminished immunoreactivity secondary to acid decalcification, similar to other immunohistochemical markers. Overall, the effect of decalcification on GRM1 immunoreactivity was fairly modest, with all decalcified specimens exhibiting at least moderate staining intensity in more than 50% of neoplastic cells, except for the one case that was completely negative for GRM1. Given that the GRM1-negative CMF case also showed low expression of GRM1 by RT-qPCR in a non-decalcified fresh-frozen sample, the lack of GRM1 staining in this case is likely to reflect the absence of an underlying GRM1 rearrangement, rather than an effect of decalcification. Still, we recommend that results of GRM1 immunohistochemistry be interpreted with caution in the setting of a previously acid-decalcified specimen. Whenever possible, decalcification should be avoided in order to optimize epitope preservation.
Our findings suggest that GRM1 expression by immunohistochemistry is a specific feature that characterizes the vast majority of CMFs. Although our study aimed to assess the specificity of GRM1 immunohistochemistry by evaluating a wide variety of tumors that reasonably could be included in a broad differential diagnosis of CMF, we would recommend caution when implementing and interpreting GRM1 immunostaining, given that this marker has yet to be applied extensively to the vast landscape of human neoplasms. Moreover, while we relied mostly on whole tissue sections to analyze 230 unique tumors, the use of TMAs to test a subset of tumors represents a limitation of our study. Reassuringly, our GRM1 immunohistochemistry data corroborate previous RT-qPCR data demonstrating that increased GRM1 expression is highly sensitive and specific for CMF.8 Therefore, we conclude that GRM1 immunohistochemistry, when combined with both careful histomorphological assessment and consideration of radiographic imaging features, may be a useful ancillary tool for the diagnosis of CMF.
Supplementary Material
Representative photomicrographs of GRM1 immunohistochemistry in the cerebellum. A) Protein expression is present in the cytoplasm and dendritic processes of Purkinje cells (straight arrows). Patchy expression is also present within the processes extending into the granular cell layer (arrowhead). Bergman glial cell cytoplasm (bent arrow) is not highlighted by the GRM1 immunostain. B) Expression is also present within the neurons and processes of the dentate nucleus (arrows) while surrounding white matter and glial cells (asterisk) are negative.
Funding:
G.W.C. is supported in part by the Stanford University School of Medicine Clinical and Translational Science Award Program (National Center for Advancing Translational Sciences, KL2TR003143).
Footnotes
Declaration of conflicting interests: The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics declarations: The manuscript is an original work of all authors. All authors made a significant contribution to this study. All authors have read and approved the final version of the manuscript. This study was performed in accordance with research policies approved by the Institutional Review Boards of the authors’ affiliated institutions.
REFERENCES
- 1.WHO Classification of Tumours Editorial Board. Soft Tissue and Bone Tumours. vol. 3 (International Agency for Research on Cancer, 2020). [Google Scholar]
- 2.Nielsen GP, Keel SB, Dickersin GR, Selig MK, Bhan AK & Rosenberg AE Chondromyxoid fibroma: a tumor showing myofibroblastic, myochondroblastic, and chondrocytic differentiation. Mod. Pathol 12, 514–517 (1999). [PubMed] [Google Scholar]
- 3.Wu CT, Inwards CY, O’Laughlin S, Rock MG, Beabout JW & Unni KK Chondromyxoid fibroma of bone: a clinicopathologic review of 278 cases. Hum. Pathol 29, 438–446 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Konishi E, Nakashima Y, Iwasa Y, Nakao R & Yanagisawa A Immunohistochemical analysis for Sox9 reveals the cartilaginous character of chondroblastoma and chondromyxoid fibroma of the bone. Hum. Pathol 41, 208–213 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Bleiweiss IJ & Klein MJ Chondromyxoid fibroma: report of six cases with immunohistochemical studies. Mod. Pathol 3, 664–666 (1990). [PubMed] [Google Scholar]
- 6.Shon W, Folpe AL & Fritchie KJ ERG expression in chondrogenic bone and soft tissue tumours. J. Clin. Pathol 68, 125–129 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Romeo S, Duim RAJ, Bridge JA, Mertens F, de Jong D, Dal Cin P et al. Heterogeneous and complex rearrangements of chromosome arm 6q in chondromyxoid fibroma: delineation of breakpoints and analysis of candidate target genes. Am. J. Pathol 177, 1365–1376 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nord KH, Lilljebjörn H, Vezzi F, Nilsson J, Magnusson L, Tayebwa J et al. GRM1 is upregulated through gene fusion and promoter swapping in chondromyxoid fibroma. Nat. Genet 46, 474–477 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Anderson ND, de Borja R, Young MD, Fuligni F, Rosic A, Roberts ND et al. Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumors. Science 361, eaam8419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibuya R, Matsuyama A, Shiba E, Harada H, Yabuki K & Hisaoka M CAMTA1 is a useful immunohistochemical marker for diagnosing epithelioid haemangioendothelioma. Histopathology 67, 827–835 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Doyle LA, Fletcher CDM & Hornick JL Nuclear Expression of CAMTA1 Distinguishes Epithelioid Hemangioendothelioma From Histologic Mimics. Am. J. Surg. Pathol 40, 94–102 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Cheah AL, Billings SD, Goldblum JR, Carver P, Tanas MZ & Rubin BP STAT6 rabbit monoclonal antibody is a robust diagnostic tool for the distinction of solitary fibrous tumour from its mimics. Pathology 46, 389–395 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Doyle LA, Vivero M, Fletcher CD, Mertens F & Hornick JL Nuclear expression of STAT6 distinguishes solitary fibrous tumor from histologic mimics. Mod. Pathol 27, 390–395 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Yoshida A, Tsuta K, Ohno M, Yoshida M, Narita Y, Kawai A et al. STAT6 immunohistochemistry is helpful in the diagnosis of solitary fibrous tumors. Am. J. Surg. Pathol 38, 552–559 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Yoshida A, Ushiku T, Motoi T, Shibata T, Beppu Y, Fukayama M et al. Immunohistochemical analysis of MDM2 and CDK4 distinguishes low-grade osteosarcoma from benign mimics. Mod. Pathol 23, 1279–1288 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Yoshida A, Ushiku T, Motoi T, Beppu Y, Fukayama M, Tsuda H et al. MDM2 and CDK4 immunohistochemical coexpression in high-grade osteosarcoma: correlation with a dedifferentiated subtype. Am. J. Surg. Pathol 36, 423–431 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Dujardin F, Binh MBN, Bouvier C, Gomez-Brouchet A, Larousserie F, Muret A. de et al. MDM2 and CDK4 immunohistochemistry is a valuable tool in the differential diagnosis of low-grade osteosarcomas and other primary fibro-osseous lesions of the bone. Mod. Pathol 24, 624–637 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Amary F, Markert E, Berisha F, Ye H, Gerrand C, Cool P et al. FOS Expression in Osteoid Osteoma and Osteoblastoma: A Valuable Ancillary Diagnostic Tool. Am. J. Surg. Pathol 43, 1661–1667 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Fittall MW, Mifsud W, Pillay N, Ye H, Strobl A-C, Verfaillie A et al. Recurrent rearrangements of FOS and FOSB define osteoblastoma. Nat. Commun 9, 2150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam SW, Cleven AHG, Kroon HM, Briaire-de Bruijn IH, Szuhai K & Bovée JVMG Utility of FOS as diagnostic marker for osteoid osteoma and osteoblastoma. Virchows Arch. 476, 455–463 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med 4, 844–847 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Kerr DA, Brcic I, Diaz-Perez JA, Shih A, Wilky BA, Pretell-Mazzini J et al. Immunohistochemical Characterization of Giant Cell Tumor of Bone Treated With Denosumab: Support for Osteoblastic Differentiation. Am. J. Surg. Pathol 45, 93–100 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Amary F, Berisha F, Ye H, Gupta M, Gutteridge A, Baumhoer D et al. H3F3A (Histone 3.3) G34W Immunohistochemistry: A Reliable Marker Defining Benign and Malignant Giant Cell Tumor of Bone. Am. J. Surg. Pathol 41, 1059–1068 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaefer I-M, Fletcher JA, Nielsen GP, Shih AR, Ferrone ML, Hornick JL et al. Immunohistochemistry for histone H3G34W and H3K36M is highly specific for giant cell tumor of bone and chondroblastoma, respectively, in FNA and core needle biopsy. Cancer Cytopathol. 126, 552–566 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amary MF, Berisha F, Mozela R, Gibbons R, Guttridge A, O’Donnell P et al. The H3F3 K36M mutant antibody is a sensitive and specific marker for the diagnosis of chondroblastoma. Histopathology 69, 121–127 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Romeo S, Oosting J, Rozeman LB, Hameetman L, Taminiau AHM, Cleton-Jansen AM et al. The role of noncartilage-specific molecules in differentiation of cartilaginous tumors. Cancer 110, 385–394 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Chow LT, Lin J, Yip KM, Kumta SM, Ahuja AT, King WW et al. Chondromyxoid fibroma-like osteosarcoma: a distinct variant of low-grade osteosarcoma. Histopathology 29, 429–436 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Derqaoui S, Marbouh O, Madhi T, Najat L & Rouas L Chondromyxoid Fibroma-Like Osteosarcoma in a 13 Years Old Girl: A Report of a New Case. Clin. Pathol 14, 2632010X211057555 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong J, Si L, Geng J, Xing Y, Hu Y, Jiao Q et al. Chondromyxoid fibroma-like osteosarcoma: a case series and literature review. BMC Musculoskelet. Disord 21, 53 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative photomicrographs of GRM1 immunohistochemistry in the cerebellum. A) Protein expression is present in the cytoplasm and dendritic processes of Purkinje cells (straight arrows). Patchy expression is also present within the processes extending into the granular cell layer (arrowhead). Bergman glial cell cytoplasm (bent arrow) is not highlighted by the GRM1 immunostain. B) Expression is also present within the neurons and processes of the dentate nucleus (arrows) while surrounding white matter and glial cells (asterisk) are negative.


