Abstract
The product of bacteriophage φ29 early gene 6, protein p6, is a double-stranded-DNA binding protein and one of the more abundant proteins during viral infection. We have studied the role of protein p6 in vivo through the infection of suppressor and nonsuppressor Bacillus subtilis strains with a phage carrying a nonsense mutation in gene 6, sus6(626). In the absence of functional protein p6, the two major processes of the viral cycle, transcription and DNA replication, were affected. Viral DNA synthesis was practically abolished, and early transcription was remarkably delayed and, in addition, underregulated at late times of the infection. The amount of protein p6 synthesized after infection with mutant phage sus6(626) under suppressor conditions was sixfold lower than that produced after wild-type infection. Nonetheless, phage production was as high as that obtained after wild-type infection. These results indicate that p6 is synthesized in amounts higher than those needed for most of its functions. However, the concentration of protein p6 appeared to be important for repression of the early promoter C2.
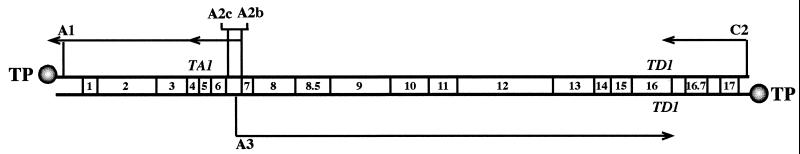
The bacteriophage φ29 genome encodes at least 20 proteins whose genes have been divided into two groups, early and late, based on the time during infection when they are first expressed (19) (see Fig. 1). Early genes are transcribed from three main promoters: C2, A2b, and A2c. The transcript originating from the C2 promoter encodes proteins p17 and p16.7, which are involved in viral DNA replication (5, 11), and in addition, it contains four open reading frames. Transcripts starting at promoters A2b and A2c give rise to proteins p6 to p1. Protein p1 is involved in viral DNA replication, and it becomes attached to the bacterial membrane (4). Genes 2 and 3 encode, respectively, the viral DNA polymerase and the protein that primes the initiation of replication and becomes covalently linked to the 5′ termini of the phage DNA (18). The product of gene 4, protein p4, is the transcriptional regulator of the promoters located in the central region of the genome. Protein p4 is responsible for the switch from early to late transcription, repressing early promoters A2b and A2c and activating late promoter A3 (17). Gene 5 encodes a single-stranded-DNA binding protein, and the product of gene 6, protein p6, is a double-stranded-DNA binding protein (18). Late genes encode structural proteins and proteins involved in viral morphogenesis and bacterial lysis, and they are transcribed from the late A3 promoter. Finally, the RNA transcribed from the other main promoter, A1, is required for the encapsidation of the phage genome (7).
FIG. 1.
Transcription map of the bacteriophage φ29 genome. Locations of promoters A1, A2c, A2b, A3, and C2 are indicated by vertical bars. Arrows indicate the direction of transcription, with the arrowheads at the termination sites (TA1 and TD1). The genetic map is depicted. The phage terminal protein (TP) is shown attached to the 5′ ends of the genome.
Protein p6 is a 103-amino-acid, non-sequence-specific DNA binding protein, able to recognize the phage genome; binding yields a multimeric complex in which the DNA adopts a right-handed toroidal conformation (9). The in vitro formation of multiple protein p6-DNA complexes, scattered through virtually the entire phage genome, led to the proposal that protein p6 plays a structural role in the organization of the viral genome into a compact nucleoprotein complex (8). Multimeric complexes adopt a variety of dynamic structures that can provide an adequate frame for multiple processes ranging from DNA unwinding to the interaction of proteins with DNA. The formation of the p6-DNA complex at the phage genome ends, where the origins of replication and promoter C2 are located, is required in vitro for activation of the initial step of φ29 DNA replication (24) and for repression of the early promoter C2 (2, 25; A. Camacho and M. Salas, unpublished data). Furthermore, it is also through the formation of a nucleoprotein complex that p6 is involved in the regulation of the central promoter region complementing the transcriptional regulatory function of protein p4 (6). Hence, protein p6 is an interesting candidate for the study of the function of architectural DNA binding proteins.
In this work we have analyzed viral development in the absence of p6 and in the presence of different amounts of p6 by using a φ29 mutant with a nonsense mutation in gene 6 under nonsuppressor and suppressor conditions. The results indicate that, in vivo, protein p6 function is essential both for viral DNA synthesis and for the correct regulation of the promoters expressed early in infection. Furthermore, results obtained after infection of suppressor bacteria with mutant sus6(626), where a functional but reduced synthesis of protein p6 takes place, indicated that p6 is synthesized in amounts higher than those needed for most of its functions. However, early promoter C2 repression was dependent on p6 concentration.
MATERIALS AND METHODS
Bacterial strains and bacteriophages.
Bacillus subtilis 110NA Trp− SpoA− su− (15) was used to grow wild-type phage φ29, while conditional lethal mutants sus6(626) (16) and sus14(1242) (10) were propagated in the suppressor strain MO-101-P SpoA− [Met−]+ Thr− su+44 (12). Bacteria were grown in Luria-Bertani medium (20) with 5 mM MgSO4 in phage infection assays. Phage stocks were prepared essentially as described elsewhere (15).
Isolation and analysis of the viral RNA.
Cultures of B. subtilis su− and su+ strain exponentially grown to a density of 5 × 108 cells/ml were infected at the multiplicities of infection (MOIs) indicated below, with mutant sus6(626) or sus14(1242). Total RNA was isolated from 20 ml of culture at the times indicated below and purified as previously described (14). Each RNA species was identified by the extension of specific primers designed to hybridize downstream from the transcription start sites of the promoter under study. Primer positions were 98 nucleotides (nt) from promoter C2, 87 nt from promoter A2b, 77 nt from promoter A2c, 69 nt from promoter A1, and 68 nt from promoter A3 (Fig. 1). Mixtures of primers (50 μmol each) were end labeled with 20 U of T4 polynucleotide kinase and 20 μCi of [γ-32P]ATP for 1 h at 37°C, precipitated with ethanol, and resuspended in H2O to a final concentration of 0.2 pM. One microgram of RNA was mixed with 10 pmol of each 32P-labeled primer in 40 mM Tris-HCl (pH 7.5) and 100 mM NaCl in a final volume of 20 μl. After denaturation at 85°C for 2 min, hybridization was carried out by allowing the DNA mixture to cool slowly to 30°C. Samples were then put on ice, and 120 μl of ice-cooled reverse transcriptase buffer (Promega) was added. Primers were extended with 5 U of avian myeloblastosis virus reverse transcriptase (Promega) for 1 h at 42°C. Samples were then filtered through 1-ml Sephadex G-50 spun columns, and the eluted cDNA was precipitated with ethanol. Truncated transcripts were analyzed by electrophoresis in 6% polyacrylamide–urea gels and quantified using a Fuji Bas-IIIs image analyzer.
In vitro transcription assays.
Runoff transcription assays (10 μl) contained 25 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 2 mM dithiothreitol, 100 μM (each) CTP, GTP, and ATP, 10 μM UTP, 0.5 μCi of [α-32P]UTP, 1 μg of poly(dI-dC), 4 U of RNasin, and 50 mM KCl. Each reaction mixture also contained a 4 nM concentration of a 268-bp DNA fragment containing promoter C2, 25 nM RNA polymerase (RNAP), and protein p6 in the amounts indicated below. Reaction mixtures containing DNA, RNAP, and p6 were incubated for 10 min at 37°C before nucleoside triphosphate addition, and transcription was allowed to proceed for 20 min at 37°C. Reactions were stopped by the addition of 0.15% sodium dodecyl sulfate (SDS) and 2.5 mM EDTA. Nonincorporated radioactivity was removed with Sephadex-G50 spun columns. Transcripts were precipitated with ethanol, resolved on 6% polyacrylamide denaturing gels, and quantified by using a Fuji Bas-IIIs image analyzer.
DNA purification and analysis.
DNA was purified from 1.5 ml of a culture of B. subtilis infected at an MOI of 5 with mutant sus6(626) or control phage. Cells were pelleted by centrifugation and resuspended in 500 μl of buffer BBA (10 mM Tris-HCl [pH 8], 10 mM EDTA, 50 mM NaCl, 20% sucrose) with 1 mg of lysozyme and 50 μg of RNase A per ml. After 10 min of incubation at room temperature, SDS (1.5%) and proteinase K (50 μg/ml) were added and the samples were incubated for 5 h at 27°C. DNA was extracted with phenol, precipitated with ethanol, and resuspended in 100 μl of TE (10 mM Tris-HCl [pH 8], 1 mM EDTA). Aliquots of 20 μl were run in 0.6% agarose gels containing ethidium bromide. Pictures of the gels were taken with a Polaroid machine, and the amount of viral DNA in each sample was quantified by scanning the picture negative of the gel with a Molecular Dynamics 300A densitometer.
Analysis of the virus-induced proteins.
Exponentially grown cultures of 5 × 108 cells of the B. subtilis su− or su+ strain per ml were infected with the mutant sus6(626) or sus14(1242) at the MOI indicated for each experiment. At the times indicated below, cells from 1.5-ml cultures were collected by centrifugation and resuspended in 250 μl of SDS buffer (625 mM Tris-HCl [pH 6.8], 2% [wt/vol] SDS, 5% [vol/vol] 2-mercaptoethanol, 10% glycerol). Proteins were separated in a 10-to-20% SDS-polyacrylamide gel electrophoresis gradient and stained with Coomassie blue or analyzed by Western blotting. Protein p6 was quantified from the wet Coomassie blue-stained gel by scanning the band with a Molecular Dynamics 300A densitometer. For Western blot analysis, proteins were transferred to an Immobilon-P membrane (Millipore) for 2 h at 200 mA and the assay was carried out following the instructions of the supplier.
Purification of protein p6.
Exponentially growing B. subtilis su+ cells were infected at an MOI of 5 with mutant sus6(626). Cells were collected by centrifugation after 45 min of infection and ground with alumina, and proteins were suspended in buffer B6 (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 7 mM β-mercaptoethanol) with 0.8 M NaCl. DNA was removed with 0.3% polyethylenimine, and protein p6 was precipitated by adjusting the salt concentration to 0.15 M NaCl with buffer B6 containing 0.004% polyethylenimine. A further step of precipitation was done by addition of ammonium sulfate to 70% saturation. Proteins were solubilized in buffer B6 with 50 mM NaCl and 25% glycerol and passed through a phosphocellulose column (5 ml) equilibrated in buffer B6 with 50 mM NaCl. The fraction that had been eluted with 100 mM NaCl was passed through a DEAE-cellulose column (2 ml), and protein p6 was eluted with 0.5 M NaCl. The protein was dialyzed in buffer B6 containing 50% glycerol and stored at −70°C. The purity and concentration of p6 were assayed by gel electrophoresis and Coomassie blue staining.
RESULTS
The function of protein p6 was studied by analyzing the viral development of a mutant with a nonsense mutation in gene 6, sus6(626), in suppressor and nonsuppressor B. subtilis strains. In the nonsuppressor bacteria (su−) the phage development has to cope with the absence of the gene function, whereas in the suppressor strain (su+), functionality is at least partially restored. With the aim of producing viral development without lysis of the infected bacteria beyond min 40 of the infection, another nonsense mutant, sus14(1242), was used instead of the wild-type phage for the control infection. Mutant sus14(1242) does not lyse the bacteria because it has a termination codon in the coding sequence of the holin protein; otherwise, it undergoes a normal phage development (10).
Synthesis of protein p6 in mutant sus6(626) infection.
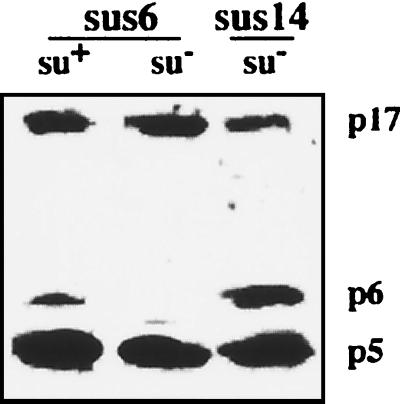
The location of the mutation of mutant sus6(626) was determined by sequencing of its PCR-amplified gene 6. A single base substitution in codon 30 was found, which changes the CAA triplet coding for glutamine (Gln30) to the nonsense triplet TAA (data not shown). Thus, when mutant sus6(626) infects the B. subtilis nonsuppressor strain 110NA (su−), a truncated peptide containing only the 29 N-terminal amino acids of p6 should be synthesized. Figure 2 shows that full-size protein p6 was produced when the sus6(626) mutant infected the suppressor su+44 strain (su+), as shown by Western blot analysis; however, the amount of protein was sixfold lower than that obtained after sus14(1242) phage infection. On the other hand, as expected, after infection of su− bacteria with mutant sus6(626), no full-size protein p6 was recognized by the monospecific anti-p6 serum. Two other early induced proteins were analyzed in this experiment, p5 and p17, and whereas the amounts of protein p5 were similar after infection with sus14 and sus6 phage, the amount of p17 in either su+ or su− cells infected with mutant sus6(626) was threefold higher than that in control cells infected with sus14(1242). Results obtained with proteins p2 and p4 were similar to those for p5. Since genes 2, 3, 5, and 6 are transcribed from promoters A2b and A2c, while gene 17 is transcribed from promoter C2, this result could reflect differences in early promoter regulation induced by the mutation of gene 6.
FIG. 2.
Analysis of protein p6 in B. subtilis infected with mutant sus6(626). Polypeptides present 40 min after the infection of suppressor (su+) and nonsuppressor (su−) B. subtilis with φ29 mutants carrying nonsense mutations in gene 6 (sus6) and gene 14 (sus14) were separated by electrophoresis in a 10-to-20% gradient polyacrylamide gel in the presence of SDS. Proteins were transferred to a Millipore polyvinylidene difluoride membrane, and proteins p5, p6, and p17 were detected with a mixture of the corresponding monospecific antisera.
Viral DNA synthesis and transcription in bacteria infected with mutant sus6(626).
To get further insight into the function(s) of p6 in vivo, we analyzed the development of mutant sus6(626). Infection of the nonsuppressor strain (su−) with mutant sus6(626) gave rise to a remarkable delay of bacterial lysis and a burst size of about 10 phage particles per bacterium, whereas the burst size was about 300 virus progeny per cell when mutant sus14(1242) was used instead. Infection of the suppressor strain (su+) with mutant sus6(626) produced normal lysis and phage progeny (results not shown). These results indicate that several steps of the viral development are affected by the absence of p6.
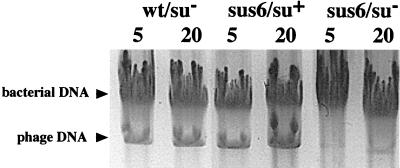
Previous results have shown that mutant sus6(626) does not incorporate labeled thymidine when infected su− bacteria are treated with 6-(p-hydroxyphenylazo)-uracil, indicating that the mutant is defective in DNA synthesis (5). As shown in Fig. 3, the amount of DNA synthesized by mutant sus6(626) in su− bacteria infected at an MOI of 5 or 20, analyzed by agarose gel electrophoresis, was very low after 35 min of infection compared with the amount synthesized after infection with wild-type phage or after infection of su+ bacteria with the sus6 mutant. In addition, two- and threefold increases in viral DNA levels were obtained by increasing the MOI from 5 to 20 in the control wild-type infection and in su+ bacteria infected with mutant sus6(626), respectively. Therefore, protein p6 is required for the correct synthesis of phage DNA, and the protein synthesized by the mutant in the suppressor bacteria fully rescued the DNA synthesis capacity.
FIG. 3.
DNA synthesis in B. subtilis infected with mutant sus6(626). Agarose electrophoresis of the DNA present after 60 min of infection of nonsuppressor (su−) and suppressor (su+) B. subtilis with φ29 mutants carrying nonsense mutations in gene 6 (sus6) or gene 14 (sus14). The MOI (phage per bacterium) is indicated on top of each lane.
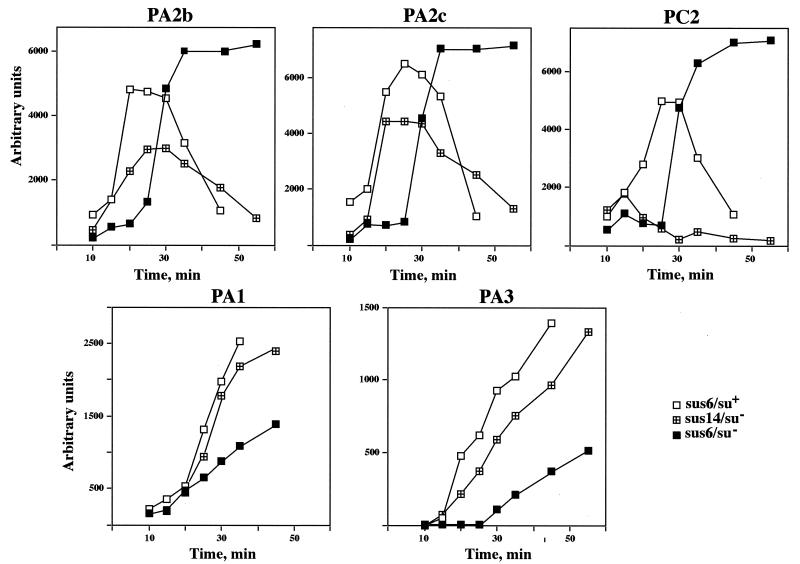
The analysis of protein p6 function in transcription regulation was followed by analysis of the transcripts produced from each of the five main viral promoters in a restrictive infection with phage mutant sus6(626) (Fig. 4). Since RNA synthesis was quantified by primer extension assays, the results indicate the amount of RNA accumulated but not the turnover rate for each transcript. During the development of mutant sus14(1242), as it occurs in a wild-type infection, early transcription is driven from promoters C2, A2c, and A2b. Promoter C2 can be considered the earliest transcribed one, since its transcripts are detected first and reached the maximum by 15 min postinfection, while the accumulation of the transcripts derived from the other two early promoters, A2b and A2c, did not reach the maximum before 20 to 25 min postinfection. Without functional protein p6 [mutant sus6(626) infecting su− bacteria], a quite different picture was observed; the early transcription derived from promoters A2b, A2c, and C2 was low in the first 20 min of the infection, with a dramatic increase at 30 min, reaching a plateau by 35 to 40 min. Since no significant DNA synthesis was observed in the absence of p6 (Fig. 3), the template for transcription in this case should be mainly the input DNA, while in a wild-type infection the newly synthesized DNA molecules are transcribed, too. Hence, in the absence of p6, the amount of early transcripts late in infection is severalfold higher than when p6 is functional, indicating a failure of the repression of the early promoters A2b, A2c, and C2 at this stage of the infection. Development of mutant sus6(626) in the su+ bacteria restored the level of the transcripts derived from promoters A2b and A2c late in infection and, therefore, the regulation of those promoters. However, the amount of transcripts from promoter C2 was fivefold higher than after wild-type infection and the repression was delayed by about 15 min.
FIG. 4.
Primer extension analysis of the transcripts produced throughout the infection cycle in B. subtilis infected with mutant sus6(626). RNA synthesized by mutant sus6(626) in nonsuppressor (su−) or suppressor (su+) B. subtilis or by the control mutant sus14(1242) at an MOI of 5 was isolated at the time after the infection indicated at the bottom of each graph. Transcripts derived from each of the promoters (indicated above each graph) were analyzed by the extension of radioactively labeled specific primers and quantitated as described in Materials and Methods. The graphs show the amount of mRNA observed during an infection cycle, where each point represents the average of at least three independent experiments. Arbitrary units are comparable between graphs.
Transcription from the other two main promoters, the late A3 promoter and the constitutively expressed A1 promoter, was also analyzed (Fig. 4). In the control infection, transcripts derived from promoter A1 were detectable after 10 min of infection, and from then on they accumulated almost linearly, while the expression of promoter A3 started after 15 min of infection, when the virus-encoded transcription activator, protein p4, is synthesized (14). There was not much difference in the level of transcription of promoter A1 in the absence or presence of p6 synthesis, with about twofold fewer transcripts in the sus6(626)-infected nonsuppressor bacteria than in those with the wild-type infection. This does not seem to be significant, taking into account the above-mentioned defect in DNA synthesis in the su− bacteria infected with the sus6 mutant; this defect could be also responsible for the reduced rate of transcription from the late promoter A3. Transcription from both promoters (A1 and A3) was restored when the mutant sus6(626) was grown in the suppressor bacteria.
Repression activity of the protein p6 produced in suppressor bacteria infected with mutant sus6(626).
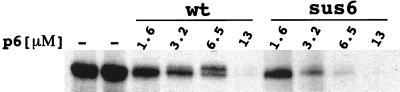
Most nonsense mutants can recover the activity of the protein if the tRNA of the suppressor strain introduces an amino acid homologous to the one originally mutated, or if the changed amino acid is located in a position which does not interfere with the structure or activity of the protein. However, even under favorable conditions of amino acid substitution, the amount of protein produced in the suppressor strain could be insufficient if the protein has to function in stoichiometric amounts. Protein p6, together with the single-stranded-DNA binding protein p5, is the most abundant protein after φ29 infection, the estimated amount of protein p6 being about 7 × 105 molecules per cell after 30 min of infection (1). Hence, the deficient repression of promoter C2 in the suppressor strain infected with mutant sus6(626) could be due either to the synthesis of a not-fully-functional protein or to a small amount of synthesis of the protein. To analyze these possibilities we purified the p6 synthesized in the su+ strain infected with the sus6(626) mutant and assayed its ability to repress promoter C2 in vitro. As shown in Fig. 5, the protein expressed from the mutant was found to have repressor function similar to or even slightly better than that of the wild-type protein. Thus, the loss of functionality of protein p6 as a repressor of the C2 promoter in the sus6-infected suppressor bacteria could be ruled out.
FIG. 5.
Effect of protein p6 on in vitro transcription of promoter C2. Runoff assays were performed in the presence of the indicated amounts of purified protein p6 from a wild-type (wt) phage infection or purified p6 from B. subtilis su+ infected with the mutant sus6(626) (sus6).
Relation between concentration of protein p6 and repression of promoter C2.
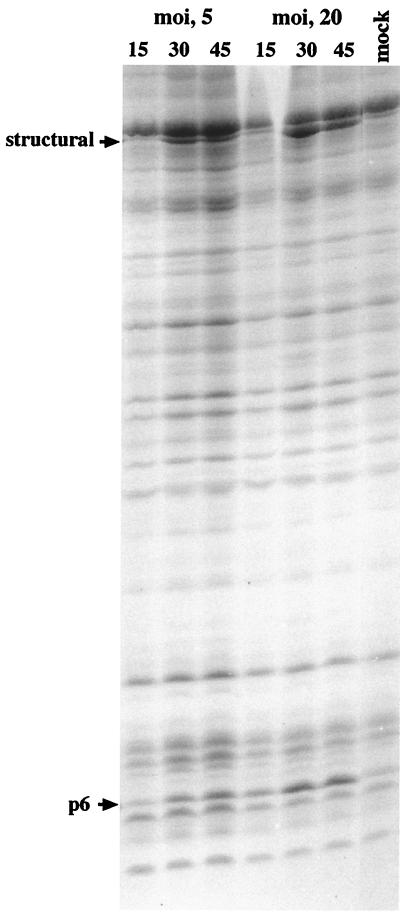
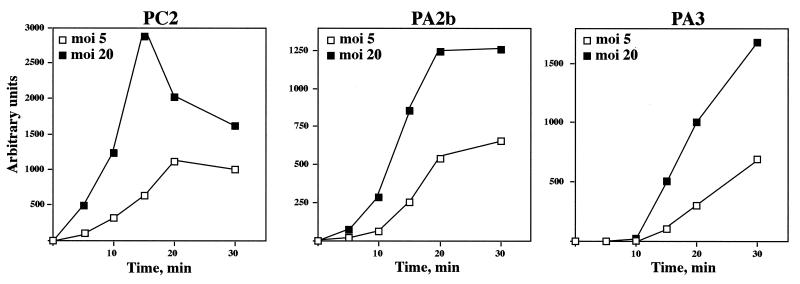
The full functionality of protein p6, isolated from the sus6-infected su+ strain, in the repression of promoter C2 in vitro indicated that the deficient repression of this promoter during the development of mutant sus6(626) in su+ bacteria is not due to the amino acid changed by the mutated tRNA. On the other hand, the amount of protein synthesized after sus6(626) infection of su+ bacteria was smaller than in the wild-type infection (Fig. 2). Both data suggest that p6 could function in a dose-dependent manner in the repression of promoter C2. If this is the case, since promoter C2 is repressed as early as 15 min postinfection and p6 is an early protein, infection by mutant sus6(626) at various MOIs should produce different levels of p6 at early times of the infection, which would influence the degree of repression of promoter C2. To test this hypothesis, we infected suppressor bacteria with mutant sus6(626) at an MOI of 5 or 20, and the levels of synthesis of protein p6 and of repression of promoter C2 were analyzed in the same experiment. As shown in Fig. 6, the MOI affected the amount of protein p6 present in the cells in the first 15 min of the infection, that is, at the time when promoter C2 is silenced in a wild-type infection (Fig. 4). At 15 min, about fivefold more p6 was present in the extract from cells infected with the higher MOI. On the other hand, the amount of the transcripts derived from promoters C2, A2b, and A3 increased with the genome dose, with about threefold more transcripts at an MOI of 20 than at an MOI of 5 (Fig. 7). The time kinetics showed analogous curves for the transcripts derived from promoters A2b and A3 at both MOIs, in contrast to the different kinetics obtained for promoter C2. At an MOI of 20, repression of the C2 promoter was stronger and took place by 15 min postinfection, which is what occurs in wild-type infection (Fig. 3) and which is in agreement with the increase of p6 synthesis (Fig. 6). Thus, the amount of p6 seems to be important in repressing promoter C2. This result suggests a fine tuning of the molecular ratio between the viral genome and protein p6.
FIG. 6.
Analysis of the amount of protein p6 induced as a function of the MOI with mutant sus6(626). The suppressor B. subtilis strain was infected with mutant sus6(626) at an MOI of 5 or 20, and at the times indicated (in minutes), proteins were analyzed by SDS-polyacrylamide gel electrophoresis and stained with Coomassie blue.
FIG. 7.
Viral RNA synthesis as a function of the MOI with mutant sus6(626). RNA derived from the early promoters C2 and A2b or the late promoter A3 was analyzed by the extension of specific primers hybridized to transcripts purified from extracts of the suppressor strain infected with mutant sus6(626) at an MOI of 5 or 20 at the indicated times.
DISCUSSION
From the results presented in this paper, together with previous ones, we can envision in more detail the role of protein p6 during the life cycle of bacteriophage φ29. Early transcription, driven from promoters C2, A2b, and A2c, starts as soon as the DNA is injected into the cell and interacts with the host RNAP. The transcripts expressed from promoters A2b and A2c end at the termination site TA1, located in gene 4, or at the DNA end (3) (Fig. 1). Therefore, genes 6 and 5 are expressed from all transcripts derived from these promoters, while genes 4 to 1 are expressed only from the transcripts passing through the TA1 terminator. This fact probably contributes to the large amount of proteins 6 and 5 in the infected cells relative to the level of the proteins encoded only by the transcripts passing through the TA1 terminator, which include, among others, the transcriptional regulator, protein p4, and the phage-encoded DNA polymerase. Upon its synthesis, protein p6 binds in a non-sequence-specific manner but with some preference for certain sequences of the phage DNA, such as the nucleation sites located at the genome ends (24). This interaction of p6 with the DNA might produce a restructuring more suitable for early transcription, which could explain the delayed induction of the early promoters in the absence of p6, shown in Fig. 4. Binding of p6 to the nucleation sites facilitates further binding of p6 dimers to DNA, and a multimeric complex is formed that can extend far from the nucleation site as the synthesis of protein p6 progresses. Our current model holds that the DNA at these protein-DNA complexes forms a right-handed coil wrapped around a multimeric protein p6 core (23). The multimeric complexes formed at the genome ends activate DNA synthesis (22) and repress promoter C2 (2) located 160 bp from the right DNA end by impairing the stability of the closed complex (Camacho and Salas, unpublished). However, they do not affect the transcription complex formed at promoter A1 located 321 bp from the left DNA end. The different outcomes in the C2 and A1 promoters may depend on the orientation of the transcription unit relative to the right-handed toroidal conformation of the DNA in the p6-DNA complex. Transcription from promoter C2 is codirectional with p6-DNA complex growth, while the direction of transcription from promoter A1 is opposite to that of p6-DNA complex formation. In addition, the distance between the transcription start site of each promoter and the corresponding nucleation site is three times larger at promoter A1 than at promoter C2. Results presented here show that the amount of protein p6 present during the first 15 min of the infection is crucial for promoter C2 repression. Since phage-encoded DNA synthesis starts by 15 min of infection (19), this result suggests that the DNA/p6 ratio is essential for repression of promoter C2 to start.
Protein p4 is believed to play a major role in the control of promoters A2b, A2c, and A3. The protein binds upstream of RNAP at promoters A2c and A3, and by direct interaction of p4 with the RNAP α subunit it represses promoter A2c, impeding promoter clearance, and activates transcription from promoter A3, stabilizing the RNAP as a close complex (17). In addition, p4 plays a major role in the repression of promoter A2b through its binding to the site upstream of PA3 which overlaps with the −35 box of PA2b. It has been recently shown that protein p6 promotes p4-mediated repression of promoter A2c and activation of promoter A3 by enhancing the capacity of p4 to bind to the DNA (6). Figure 4 shows that in the wild-type phage infection, accumulation of transcripts derived from promoters A2b and A2c is arrested by 25 min and decreases afterwards; in contrast, no decrease in such transcripts was observed after infection of su− bacteria with the mutant sus6(626). The impairment on repression of promoters A2b and A2c seems to be due to deficient synthesis of protein p6, since activation of promoter A3 indicated the presence of functional protein p4.
Large amounts of protein p6 accumulate throughout wild-type infection, suggesting a need for such amounts of the protein for at least some of its functions. The results presented in this paper suggest that a high concentration of protein p6 is critical for repressing promoter C2 but not for its other functions, such as regulation of promoters A2c and A2b or activation of DNA synthesis. Since for repression of promoters A2b and A2c and activation of DNA synthesis, other proteins in addition to p6 are required, protein p6 could function by saturating the nonspecific DNA sequence, thereby ensuring that the concentration of the corresponding proteins (the transcriptional regulator p4 or DNA polymerase) remains high enough to successfully find its targets even when the amount of DNA increases. We cannot exclude an architectural role of p6 in transcription regulation through the transient flexing of the DNA upon its binding, which most probably increases the interaction between the two p4 dimers, and between these and the DNA backbone, thereby enhancing p4 binding affinity.
Prokaryotic cells contain proteins involved in the organization and compaction of their chromosomal DNA, among them, proteins HU and H-NS of Escherichia coli (21) and HBsu of B. subtilis (13). Bacteriophage φ29 protein p6 shares several properties with these proteins; like them, it is small and basic, it forms dimers in solution and binds double-stranded DNA depending on structural features rather than by sequence recognition, and it is a global transcriptional regulator which represses its own promoter both in vivo (this paper) and in vitro (6). Therefore, protein p6 is an excellent tool for analyzing and elucidating the nonspecific but precise function of the so-called histone-like prokaryotic proteins.
ACKNOWLEDGMENTS
This investigation was aided by research grants 2R01 GM27242-20 from the National Institutes of Health and PB98-0645 from the Dirección General de Investigación de Ciencia y Tecnología and an institutional grant from the Fundación Ramón Areces.
We thank F. Rojo for critical reading of the manuscript and J. M. Lázaro and L. Villar for purification of proteins.
REFERENCES
- 1.Abril A, Salas M, Andreu J M, Hermoso J M, Rivas G. Phage φ29 protein p6 is in a monomer-dimer equilibrium that shifts to higher association states at the millimolar concentration found in vivo. Biochemistry. 1997;36:11901–11908. doi: 10.1021/bi970994e. [DOI] [PubMed] [Google Scholar]
- 2.Barthelemy I, Mellado R P, Salas M. In vitro transcription of bacteriophage φ29 DNA: inhibition of early promoter by the viral replication protein p6. J Virol. 1989;63:460–462. doi: 10.1128/jvi.63.1.460-462.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barthelemy I, Salas M, Mellado R P. In vivo transcription of bacteriophage φ29 DNA: transcription termination. J Virol. 1987;61:1751–1755. doi: 10.1128/jvi.61.5.1751-1755.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bravo A, Salas M. Initiation of bacteriophage φ29 DNA replication in vivo: assembly of a membrane-associated multiprotein complex. J Mol Biol. 1997;269:102–112. doi: 10.1006/jmbi.1997.1032. [DOI] [PubMed] [Google Scholar]
- 5.Carrascosa J L, Camacho A, Moreno F, Jiménez F, Mellado R P, Viñuela E, Salas M. Bacillus subtilis bacteriophage φ29. Characterization of gene products and functions. Eur J Biochem. 1976;66:229–241. doi: 10.1111/j.1432-1033.1976.tb10512.x. [DOI] [PubMed] [Google Scholar]
- 6.Elías-Arnanz M, Salas M. Functional interactions between a phage histone-like protein and a transcriptional factor in regulation of φ29 early-late transcriptional switch. Genes Dev. 1999;13:2502–2513. doi: 10.1101/gad.13.19.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo P, Erikson S, Anderson D. A small RNA is required for in vivo packaging of bacteriophage φ29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 8.Gutiérrez C, Freire R, Salas M, Hermoso J M. Assembly of phage φ29 genome with viral protein p6 into a compact complex. EMBO J. 1994;13:169–176. doi: 10.1002/j.1460-2075.1994.tb06257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermoso J M, Freire R, Bravo A, Gutiérrez C, Serrano M, Salas M. DNA structure in the nucleoprotein complex that activates replication of phage φ29. Biophys Chem. 1994;50:183–189. doi: 10.1016/0301-4622(94)85030-5. [DOI] [PubMed] [Google Scholar]
- 10.Jiménez F, Camacho A, Torre J, Viñuela E, Salas M. Assembly of Bacillus subtilis phage φ29: mutants in the cistrons coding for the non-structural proteins. Eur J Biochem. 1977;73:57–72. doi: 10.1111/j.1432-1033.1977.tb11291.x. [DOI] [PubMed] [Google Scholar]
- 11.Meijer W, Lewis P, Errington J, Salas M. Dynamic relocalization of phage φ29 DNA during replication and the role of the viral protein p16.7. EMBO J. 2000;19:4182–4190. doi: 10.1093/emboj/19.15.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellado R P, Viñuela E, Salas M. Isolation of a strong suppressor of nonsense mutations in Bacillus subtilis. Eur J Biochem. 1976;65:213–223. doi: 10.1111/j.1432-1033.1976.tb10408.x. [DOI] [PubMed] [Google Scholar]
- 13.Micka B, Groch N, Heinemann U, Marahiel M A. Molecular cloning, nucleotide sequence, and characterization of the Bacillus subtilis gene encoding the DNA-binding protein HBsu. J Bacteriol. 1991;173:3191–3198. doi: 10.1128/jb.173.10.3191-3198.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monsalve M, Mencía M, Rojo F, Salas M. Transcription regulation in Bacillus subtilis phage φ29: expression of the viral promoters throughout the infection cycle. Virology. 1995;20:23–31. doi: 10.1006/viro.1995.1048. [DOI] [PubMed] [Google Scholar]
- 15.Moreno F, Camacho A, Viñuela E, Salas M. Suppressor-sensitive mutants and genetic map of Bacillus subtilis bacteriophage φ29. Virology. 1974;62:1–16. doi: 10.1016/0042-6822(74)90298-0. [DOI] [PubMed] [Google Scholar]
- 16.Reilly B E, Zeece V M, Anderson D L. Genetic study of suppressor-sensitive mutants of the Bacillus subtilis bacteriophage φ29. J Virol. 1973;11:756–760. doi: 10.1128/jvi.11.5.756-760.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rojo F, Mencía M, Monsalve M, Salas M. Transcription activation and repression by interaction of a regulator with the α subunit of RNA polymerase: the model of phage φ29 protein p4. Prog Nucleic Acid Res Mol Biol. 1998;60:29–46. doi: 10.1016/s0079-6603(08)60888-0. [DOI] [PubMed] [Google Scholar]
- 18.Salas M, Miller J T, Leis J, DePamphilis M L. Mechanisms for priming DNA synthesis. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 131–176. [Google Scholar]
- 19.Salas M, Rojo F. Replication and transcription of bacteriophage φ29 DNA. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 843–857. [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Schmid B M. More than just “histone-like” proteins. Cell. 1990;63:451–453. doi: 10.1016/0092-8674(90)90438-k. [DOI] [PubMed] [Google Scholar]
- 22.Serrano M, Gutiérrez C, Salas M, Hermoso J M. Superhelical path of the DNA in the nucleoprotein complex that activates the initiation of phage φ29 DNA replication. J Mol Biol. 1993;230:248–259. doi: 10.1006/jmbi.1993.1140. [DOI] [PubMed] [Google Scholar]
- 23.Serrano M, Salas M, Hermoso J M. A novel nucleoprotein complex at a replication origin. Science. 1990;248:1012–1016. doi: 10.1126/science.2111580. [DOI] [PubMed] [Google Scholar]
- 24.Serrano M, Gutiérrez J, Prieto I, Hermoso J M, Salas M. Signals at the bacteriophage φ29 DNA replication origins required for protein p6 binding and activity. EMBO J. 1989;8:1879–1885. doi: 10.1002/j.1460-2075.1989.tb03584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whiteley H R, Ramey W D, Spiegelman G B, Holder R D. Modulation of in vivo and in vitro transcription of bacteriophage φ29 early genes. Virology. 1986;155:392–401. doi: 10.1016/0042-6822(86)90202-3. [DOI] [PubMed] [Google Scholar]