Abstract
Ubiquinone is an essential component of the electron transfer system in both prokaryotes and eukaryotes and is synthesized from chorismate and polyprenyl diphosphate by eight steps. p-Hydroxybenzoate (PHB) polyprenyl diphosphate transferase catalyzes the condensation of PHB and polyprenyl diphosphate in ubiquinone biosynthesis. We isolated the gene (designated ppt1) encoding PHB polyprenyl diphosphate transferase from Schizosaccharomyces pombe and constructed a strain with a disrupted ppt1 gene. This strain could not grow on minimal medium supplemented with glucose. Expression of COQ2 from Saccharomyces cerevisiae in the defective S. pombe strain restored growth and enabled the cells to produce ubiquinone-10, indicating that COQ2 and ppt1 are functional homologs. The ppt1-deficient strain required supplementation with antioxidants, such as cysteine, glutathione, and α-tocopherol, to grow on minimal medium. This suggests that ubiquinone can act as an antioxidant, a premise supported by our observation that the ppt1-deficient strain is sensitive to H2O2 and Cu2+. Interestingly, we also found that the ppt1-deficient strain produced a significant amount of H2S, which suggests that oxidation of sulfide by ubiquinone may be an important pathway for sulfur metabolism in S. pombe. Ppt1-green fluorescent protein fusion proteins localized to the mitochondria, indicating that ubiquinone biosynthesis occurs in the mitochondria in S. pombe. Thus, analysis of the phenotypes of S. pombe strains deficient in ubiquinone production clearly demonstrates that ubiquinone has multiple functions in the cell apart from being an integral component of the electron transfer system.
Ubiquinone is known to be an electron transporter in the respiratory chain in prokaryotes and eukaryotes. It varies among organisms in the length of its isoprenoid side chain. For example, Saccharomyces cerevisiae uses ubiquinone-6 (UQ-6), Escherichia coli uses UQ-8, and Schizosaccharomyces pombe uses UQ-10 (9, 16, 37). It has been shown that the type of ubiquinone in organisms is determined by the polyprenyl diphosphate synthase enzyme, which catalyzes the condensation reaction of isopentenyl diphosphate with allylic diphosphate to give a defined length of the isoprenoid (22, 26). When polyprenyl diphosphate synthase genes from other sources were expressed in S. cerevisiae and E. coli, the ubiquinone generated was of the same type as that expressed in the donor organism (22–26). By this method, we successfully produced various ubiquinone species (UQ-5 to UQ-10) in the S. cerevisiae COQ1 mutant (22), which in turn indicates that p-hydroxybenzoate (PHB) polyprenyl diphosphate transferase, which catalyzes the condensation reaction between the isoprenoid side chain and PHB, has a broad substrate specificity. This is supported by consistent observations showing that purified PHB polyprenyl diphosphate transferases from Pseudomonas putida (12, 40) and E. coli (17) have fairly wide substrate specificities in terms of polyprenols. In contrast, PHB geranyltransferase, which is responsible for the synthesis of shikonin, is highly specific, as it uses only geranyl diphosphate as a substrate (21). Studying the PHB polyprenyl diphosphate transferases from different sources may enhance our understanding of this type of enzyme.
Ubiquinone appears to play roles in addition to acting as a component of the electron transfer system. One such role is that of an antioxidant, as indicated by a number of studies (1, 5, 6, 7, 8, 14). A strain of S. cerevisiae unable to produce ubiquinone is sensitive to lipid peroxide, suggesting that ubiquinone protects against oxidants (5). Similarly, an S. pombe strain which does not produce ubiquinone because of a deficiency of decaprenyl diphosphate synthase is sensitive to H2O2 and requires an antioxidant to grow on glucose-containing medium (37). Antioxidant roles of ubiquinone in E. coli also have been reported recently (18, 36). Furthermore, physiological concentrations of ubiquinone act as antioxidants on human low-density lipoprotein (1, 7). Another role of ubiquinone is that it can accept electrons from sources other than the respiratory chain. Recently, it was elegantly shown that ubiquinone (or menaquinone) will accept electrons generated by the formation of protein disulfide in E. coli (3). Sulfide-ubiquinone oxidoreductase, previously thought to occur mainly in photobiosynthetic bacteria as a component in energy metabolism, has been shown to be present in S. pombe and other eukaryotic organisms (42). This suggests that there may be a link between sulfide metabolism and ubiquinone in eukaryotes.
To increase our knowledge of the ubiquinone biosynthetic pathway and the various functions of ubiquinone, we have characterized in this study a strain of S. pombe that cannot produce ubiquinone because of a defect in its PHB polyprenyl transferase gene. We show clearly that ubiquinone can act both as an antioxidant and as an acceptor of electrons from sulfide.
MATERIALS AND METHODS
Materials.
Restriction enzymes and other DNA-modifying enzymes were purchased from Takara Shuzo Co. Ltd. and New England Biolabs, Inc.
Strains, plasmids, and media.
E. coli strains DH10B and DH5α were used for the general construction of plasmids (32). Plasmids pBluescript II KS+/−, pT7Blue-T (Novagen), pREP1 (15), and pREP1-GFPS65A (39) were used as vectors. The S. pombe homothallic haploid wild-type strain SP870 (h90 leu1-32 ade6-M210 ura4-D18) (13) and the diploid strains SP826 (h+ leu1-32 ade6-M210 ura4-D18/h+ leu1-32 ade6-M216 ura4-D18) (13) and TP4-1D/TP4-5A (h+ leu1-32 ura4-D18 his2 ade6-M216/h− leu1-32 ura4-D18 ade6-M210) (37) were used to produce Δppt1::ura4 strains by homologous recombination. KS10 (h+ leu1-32 ade6-M216 ura4-D18 Δdps::ura4) was previously described (37). JV5 (h− ura4-294 leu1-32 Δhmt2::URA3+) was obtained from D. W. Ow (42). Yeast cells were grown in YE (0.5% yeast extract, 3% glucose) or PM minimal medium (111 mM glucose, 93.5 mM NH4Cl, 15.5 mM Na2HPO, 14.7 mM potassium hydrogen phthalate, 5.2 mM MgCl2 · 6H2O, 13.4 mM KCl, 0.28 mM Na2SO4, 0.1 mM CaCl2 · 2H2O, 81.2 μM nicotinic acid, 55.5 μM myo-inositol, 40.8 μM biotin, 4.2 μM calcium pantothenate, 8.1 μM boric acid, 2.37 μM MnSO4, 1.39 μM ZnSO4 · 7H2O, 0.74 μM FeCl3 · 6H2O, 0.25 μM MoO4 · 2H2O, 0.6 μM KI, 0.16 μM CuSO4 · 5H2O, 4.76 μM citric acid) with appropriate supplements as described by Moreno et al. (19). YEA and PMA contain 75 μg of adenine per ml in YE and PM, respectively. The concentration of supplemented amino acids was 100 μg/ml. Yeast transformation was performed according to the method described by Rose et al. (29).
DNA manipulations.
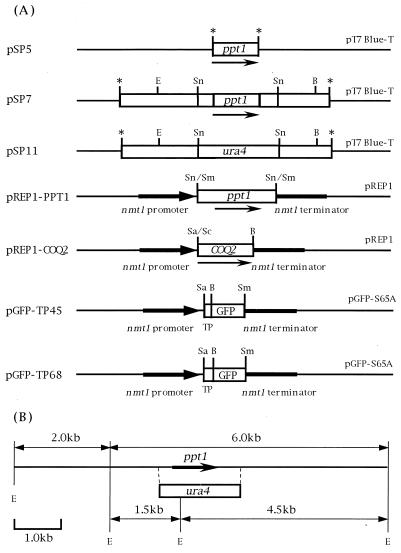
Cloning, restriction enzyme analysis, and preparation of plasmid DNAs were performed essentially as described previously (32). PCR was done according to the procedure described before (31). DNA sequences were determined by the dideoxynucleotide chain termination method (33) using an ABI377 DNA sequencer. To clone the ppt1 gene, the following four primers were designed. Two oligonucleotides, 5′-TGAATTCGATGATAATTAAGCCTATAGCGT-3′ (creates an EcoRI site) and 5′-TCCAAGACTGCAGTAGAACGTTTAAGAATC-3′, were used to amplify the ppt1 gene. The amplified fragment was then cloned into pT7Blue-T to yield pSP5. The two additional oligonucleotides 5′-TGATGAACCACATTTACTTGATTTAGTCGA-3′ and 5′-TCGAGCTCTTCTGACACCTCAACCTTTAAA-3′ were used to amplify the 4.5-kb fragment containing the ppt1 gene and the surrounding region. The amplified fragment was then cloned into pT7Blue-T to yield pSP7. To make pSP11, pSP7 was digested with SnaBI and ligated with the ura4 cassette derived from pHSG398-ura4 (39). The 1.8-kb SnaBI fragment containing ppt1 was cloned into the SmaI site of pREP1 to yield pREP1-PPT1. The SacI-BamHI fragment containing COQ2 (38) was cloned into pREP1 to yield pREP1-COQ2. Putative mitochondrial transit sequences of ppt1 were amplified by PCR using the oligonucleotides 5′-AGGTCGACAGATTAGCATGTAAATAG-3′ (sense primer; creates a SalI site) and 5′-ATGGATCCGGGGGTTACAGAGTTTGA-3′ (antisense primer; creates a BamHI site) or 5′-TAGGATCCTTCAGCGTAGTATTGCCA-3′ (antisense primer; creates a BamHI site). The PCR products were cloned into the SalI and BamHI sites of pREP1-GFPS65A (39), which contains the GFPS65A gene (20) in pREP1, to yield pGFP-TP45 and pGFP-TP68.
Gene disruption.
The one-step gene disruption technique was performed according to the procedure of Rothstein (30). Plasmid pSP11 was linearized by appropriate restriction enzymes, and the linearized plasmid was used to transform SP870 and SP826 to uracil prototrophy. Southern hybridization was performed as described before (32).
Ubiquinone extraction and measurement.
Ubiquinone was extracted as previously described (37, 41). S. pombe cells were grown in a PMA-based medium (20 ml) until the mid-log phase. After harvesting, the cells were lysed with 3 mg of Novozyme, and ubiquinone was extracted with 3 ml of hexane-acetone (1:1, vol/vol), followed by evaporation of the organic solution to dryness. Samples were then redissolved in 1 ml of chloroform-methanol (1:1, vol/vol) and the solution was washed with 0.5 ml of 0.7% NaCl. After evaporation to dryness, the residue was taken up in 30 μl of chloroform-methanol (2:1, vol/vol) and analyzed by normal-phase thin-layer chromatography on a Kiesel gel 60 F254 plate (Merck) with benzene-acetone (93:7, vol/vol). A UQ-10 standard (Kaneka) was also applied. The UV-visualized band containing ubiquinone was collected from the thin-layer chromatography plate and extracted with chloroform-methanol (1:1, vol/vol). The solution was evaporated to dryness and the residue was redissolved in ethanol. The purified ubiquinone was further analyzed by high-pressure liquid chromatography using ethanol as a solvent (41).
Measurement of sulfide.
Hydrogen sulfide was detected by production of PbS from lead acetate. A quantitative determination of sulfide was performed by the methylene blue method as previously described (28). Briefly, S. pombe cells were grown in YEA medium (50 ml) until the late log phase. The cells were collected and disrupted by glass beads, and cell extracts were resuspended in 0.1 ml of 0.1% dimethylphenylenediamine (in 5.5 N HCl) and 0.1 ml of 23 mM FeCl3 (in 1.2 N HCl). The samples were incubated at 37°C for 5 min, after which the absorbance at 670 nm was determined using a blank consisting of the reagents alone.
Staining of mitochondria and fluorescence microscopy.
Mitochondria were stained by the mitochondrion-specific dye MitoTracker Green FM (Molecular Probes, Inc.). Cells were suspended in 10 mM HEPES, pH 7.4, containing 5% glucose, and MitoTracker Green FM was added to yield a final concentration of 100 nM. After standing for 15 min at room temperature, cells were visualized by fluorescence microscopy at 490 nm. Fluorescence microscopy was carried out with a Zeiss Axioskop microscope at a magnification of ×1,000. GFPS65A fluorescence was observed by illumination at 485 nm. Images were captured by a Hamamatsu C5985 CCD camera.
RESULTS
Cloning of the ppt1 gene and construction of strains with a defective ppt1 gene.
We found a putative gene for PHB polyprenyl diphosphate transferase in the S. pombe genomic DNA sequence determined by the Sanger Center. This gene (SPAC56F8.04c) shows high sequence similarity with the COQ2 gene from S. cerevisiae and was designated ppt1 (for PHB polyprenyl diphosphate transferase). ppt1 and putative PHB polyprenyl diphosphate transferases from other species could also be found in the National Center for Biotechnology Information database (Fig. 1). Of these genes, only ubiA from E. coli and COQ2 from S. cerevisiae have been functionally characterized (2, 16, 17, 35, 38).
FIG. 1.
Comparison of the amino acid sequences of PHB-polyprenyl diphosphate transferases. EC, E. coli (accession no. X66619); SY, Synechocystis sp. strain PCC6803 (D64006); SC, S. cerevisiae (M81698); SP, S. pombe (Z69728); CE, Caenorhabditis elegans (U13876); Dm, Drosophila melanogaster (AE003678). A putative substrate recognition sequence is indicated by the underline. Conserved amino acids of at least three in six sequences are highlighted.
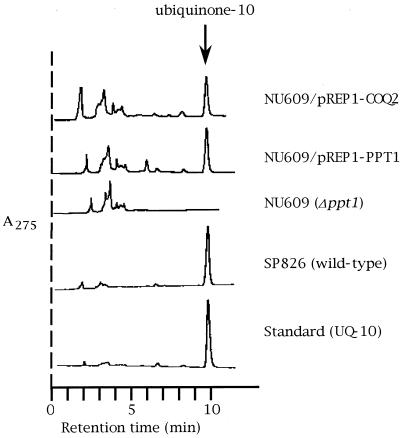
To analyze the function of the ppt1 gene, we amplified the ppt1 gene from S. pombe genomic DNA by PCR to yield the 1-kb DNA fragment containing ppt1 and the 4.5-kb fragment containing the surrounding DNA. To make S. pombe strains containing a defective ppt1 gene, we constructed the plasmid pSP11, in which the ppt1 gene is disrupted by the ura4 gene (Fig. 2A). This plasmid was then linearized by the appropriate restriction enzymes and the fragment initially used to transform the S. pombe wild-type haploid strain SP870. However, although some Ura+ transformants were obtained, no strains with disruptions in the ppt1 gene could be isolated. Thus, we decided to transform the diploid SP826 and TP4-1D/TP4-5A strains with the pSP11 fragment. When SP826 was transformed, 30 colonies of Ura+ transformants could be picked and grown on YEA-rich medium. The stability of the Ura+ phenotype was examined by replica plating, and nine stable Ura+ transformants were thus obtained. One of these strains, designated SP826Δppt1, was allowed to make spores. Germinated haploid cells were plated in replicates on plates containing YEA and PMA-Leu. While all cells grew well on YEA medium some grew only very slowly on the PMA-Leu plate, and these were examined for ubiquinone synthesis. As none synthesized ubiquinone (Fig. 3), these strains were considered to potentially have a disruption in ppt1. One such haploid strain, designated NU609, was used for further experiments. Transformation of TP4-1D/TP4-5A similarly generated a strain with a putative disruption in ppt1 that was designation TP4-1D/TP4-5AΔppt1.
FIG. 2.
Plasmid constructions used in this study (A) and EcoRV restriction map of the ppt1 and the ppt1::ura4 regions (B). Asterisks indicate the sites of TA ligation with the T-tailed vector pT7Blue-T. pREP1-PPT1 and pREP1-COQ2 contain the entire lengths of the ppt1 and COQ2 genes, respectively, and are under the control of the strong nmt1 promoter. pGFP-TP45 and pGFP-TP68 contain putative mitochondrial transit peptides (TP) of Ppt1. Thin arrows indicate the direction and the length of open reading frames. Abbreviations for restriction enzymes: B, BamHI; E, EcoRV; Sa, SalI; Sc, SacI; Sn, SnaBI; Sm, SmaI.
FIG. 3.
Detection of UQ-10. Ubiquinone was extracted from the wild-type SP826, NU609 (Δppt1::ura4), NU609 harboring plasmid pREP1-PPT1, and NU609 harboring plasmid pREP1-COQ2. Ubiquinone was first separated by thin-layer chromatography and then further analyzed by high-pressure liquid chromatography.
Verification of ppt1 disruption by Southern hybridization analysis.
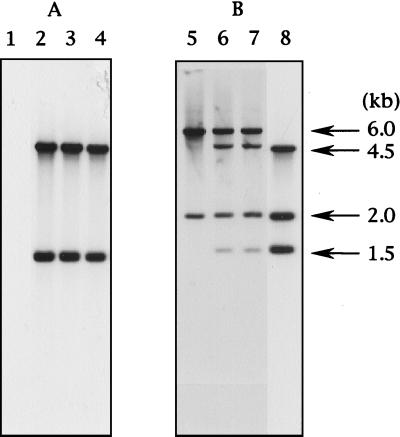
Genomic DNAs from SP826, SP826Δppt1, NU609, and TP4-1D/TP4-5AΔppt1 were subjected to Southern hybridization analysis to confirm the disruption of ppt1 by ura4. The genomic DNAs were first digested with EcoRV and run on an agarose gel. The ura4 cassette and the ppt1 gene were used as probes. In lanes containing SP826Δppt1 and TP4-1D/TP4-5AΔppt1 DNAs, 1.5- and 4.5-kb bands appeared with both probes (Fig. 2B and Fig. 4, lanes 2, 3, 6, and 7), because SP826Δppt1 and TP4-1D/TP4-5AΔppt1 contain both the complete ppt1 gene and the ura4-disrupted ppt1 gene. When the ura4 cassette was used as a probe, no band appeared with DNA from SP826 (Fig. 4, lane 1), but 1.5- and 4.5-kb bands appeared with the DNAs from SP826Δppt1 and TP4-1D/TP4-5AΔppt1 strains (Fig. 4, lanes 2 and 3, respectively) as well as with NU609 DNA (Fig. 4, lane 4). When the ppt1 fragment was used as a probe, four bands of 1.5, 2.0, 4.5, and 6.0 kb appeared with SP826Δppt1 and TP4-1D/TP4-5AΔppt1 DNAs (Fig. 4, lanes 6 and 7), and three bands of 1.5, 2.0, and 4.5 kb appeared with NU609 DNA (Fig. 4, lane 8). Thus, the ppt1 gene is properly disrupted in SP826Δppt1, TP4-1D/TP4-5AΔppt1, and NU609.
FIG. 4.
Southern hybridization analysis. Genomic DNAs of SP826, SP826Δppt1, TP4-1D/TP4-5AΔppt1, and NU609 were prepared, separated on an agarose gel, and probed with the ura4 gene (A) and the ppt1 gene from pSP7 (B). Lanes 1 and 5, wild-type SP826 (diploid); lanes 2 and 6, SP826Δppt1 (diploid); lanes 3 and 7, TP4-1D/TP4-5AΔppt1 (diploid); lanes 4 and 8, NU609 (haploid). The EcoRV restriction map is shown in Fig. 2B.
Complementation of ppt1 disruption-containing cells with COQ2.
In the disruption of the ppt1 gene in NU609 by homologous recombination, it is possible that the upstream and downstream deletion of ppt1 could have damaged other genes. To eliminate this possibility, the plasmid pREP1-PPT1, which includes only the ppt1 gene, was used in a complementation assay. We also constructed pREP1-COQ2, in which only the COQ2 region is expressed under the control of the strong promoter nmt1, to test the functional conservation between Coq2 and Ppt1. Thus, NU609 harboring either or both of the vectors pREP1-PPT1 and pREP1-COQ2 were plated on PM-based medium and growth was observed. A few days later, NU609 harboring only the pREP1 vector formed only a very tiny colony, while NU609 harboring pREP1-PPT1 and pREP1-COQ2 grew as well as the wild-type strain. Thus, only the ppt1 function was abolished in NU609. That pREP1-PPT1 is as competent as pREP1-COQ2 in correcting the poor growth of the ppt1-defective strain indicates that ppt1 is a functional homologue of COQ2. When we extracted ubiquinone from each strain, UQ-10 was detected in the wild-type strain, in NU609 harboring pREP1-PPT1, and in NU609 harboring pREP1-COQ2, but not in NU609 alone (Fig. 3). That COQ2 complements the ppt1 disruptant and allows the production of UQ-10 in S. pombe is consistent with the idea that PHB polyprenyl diphosphate transferase has a broad substrate specificity.
Phenotypes of the NU609 ppt1 disruptant.
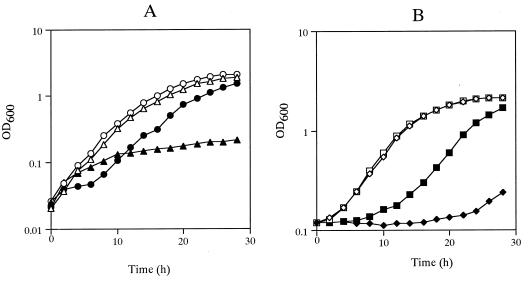
It was previously reported that KS10 (Δdps::ura4), a strain of S. pombe with a disruption in the dps (decaprenyl diphosphate synthase) gene, is unable to produce ubiquinone and has some notable phenotypes, including H2O2 and Cu2+ sensitivity and a requirement of cysteine or glutathione for growth on minimal medium (37). Thus, NU609 was tested for these phenotypes. NU609 (Δppt1) was first grown on PM-based medium with and without supplementation with 200 μg of cysteine per ml or 200 μg of glutathione per ml. The addition of cysteine or glutathione effectively caused a recovery of NU609 growth similar to that of the dps disruptant KS10 when it was treated similarly (data not shown). NU609 was next tested for growth on PM-based liquid medium supplemented with α-tocopherol, a well-known lipid antioxidant. NU609 cells did not grow on the minimal medium, but interestingly, NU609 cells grew well when 1 mM α-tocopherol was added (data not shown). This suggests that ubiquinone may act as an antioxidant in S. pombe. If this is so, it follows that the ppt1-deficient strain might be susceptible to oxygen radical producers, and we duly noted that NU609 growth is severely inhibited by the presence of 2.5 mM H2O2 (Fig. 5A) or 0.5 mM Cu2+ (Fig. 5B). The oxidants at these concentrations did not, however, affect the growth of wild-type cells (Fig. 5). These results suggest strongly that ubiquinone can serve as an antioxidant in normal fission yeast cells.
FIG. 5.
Sensitivity of NU609 to oxygen radical producers. Wild-type (▵, ○, ◊, and □) and NU609 (Δppt1::ura4) (▴, ●, ⧫, and ■) strains were pregrown in YEA liquid medium until saturation and then placed in 40-fold dilutions in fresh YEA medium with 2.5 mM H2O2 (A) (▵ and ▴), 0.5 mM Cu2+ (B) (◊ and ⧫), or neither (○, ●, □, and ■). Cell growth was measured at 2-h intervals by optical density at 600 nm (OD600).
Production of hydrogen sulfide in S. pombe strains unable to produce ubiquinone.
We found that when the S. pombe strains with disruptions in ppt1 or dps were cultivated, they smelled unpleasant. This was found to be due to their production of H2S when we tested for the formation of PbS by the chemical reaction of H2S with lead acetate (data not shown). Strains deficient in either ppt1 or dps could produce H2S, but the wild-type cells could not. Since HMT2 catalyzes sulfide oxidation by concomitant reduction of ubiquinone in S. pombe (42), we tested for H2S production in hmt2 mutants, but H2S could not be detected using this method. The production of H2S was also observed in NU609 cells grown on liquid minimal medium supplemented with α-tocopherol, indicating that the antioxidant function of α-tocopherol could not overcome the production of H2S. We measured the amount of acid-labile sulfide present in the cells and found that while JV5 (Δhmt2) produced a 2.5-fold-larger amount of S2− than wild-type cells (82.1 and 33.7 nmol/106 cells, respectively), KS10 (Δdps) and NU609 (Δppt1) produced 9-fold-larger amounts of S2− than the Δhmt2 strain (758.1 and 718.6 nmol/109 cells, respectively). This surprisingly high level of S2− production presumably leads to the production of H2S in the ppt1- and dps-deficient strains. This unexpected phenotype suggests that ubiquinone may be important in sulfide oxidation in S. pombe.
Mitochondrial localization of Ppt1.
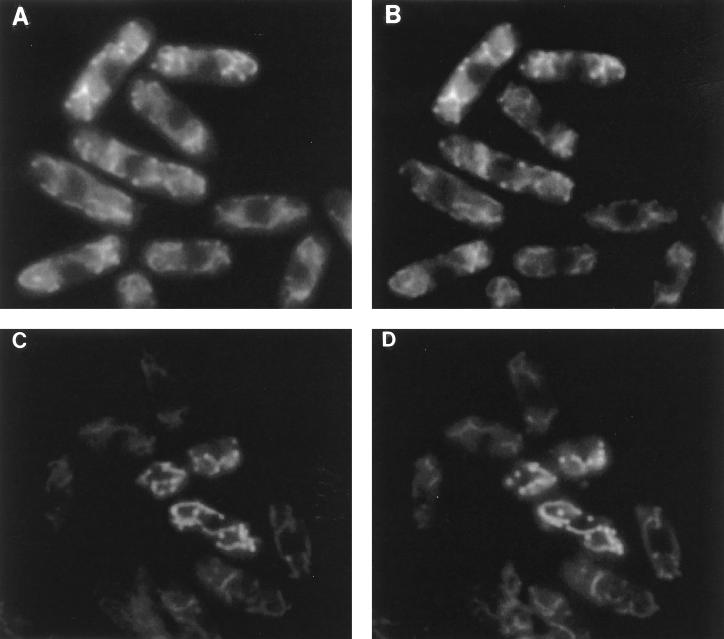
Since ubiquinone biosynthetic enzymes are localized to the mitochondria of S. cerevisiae (4, 11, 27), it has been suggested that ubiquinone biosynthesis occurs in the mitochondria. To assess the case for homologous enzymes from S. pombe, the localization of S. pombe Ppt1 was examined by Ppt1-green fluorescent protein (GFP) fusions. Thus, genes expressing putative Ppt1 mitochondrial transit peptides of either 45 (pGFP-TP45) or 68 (pGFP-TP68) amino acids fused with GFP were constructed. pGFP-TP45 and pGFP-TP68 were used to transform the S. pombe wild-type strain, and Leu+ transformants were selected. When selected transformants were examined by fluorescence microscopy, accumulation of the fusion proteins in the mitochondria was observed (Fig. 6). The transformants were simultaneously stained with MitoTracker Green FM, which stains mitochondria. The dye stained the cells in exactly the same pattern produced by fluorescing of the Ppt1-GFP fusions, indicating that Ppt1 localizes to the mitochondria.
FIG. 6.
Colocalization of Ppt1-GFP fusion proteins with a mitochondrion-specific dye. The patterns of fluorescence produced by Ppt1-GFP fusion proteins (A and C) and by MitoTracker Green FM (mitochondrion-specific dye) (B and D) in pGFP-TP45 (A and B) and in pGFP-TP68 (C and D).
DISCUSSION
In this study, we examined the ppt1 gene, which encodes a 358-amino-acid protein with high homology to E. coli UbiA and S. cerevisiae Coq2 (34 and 48% identity, respectively). In S. cerevisiae, Coq2 acts to transfer six isoprenoid units to PHB to produce UQ-6. If, however, the COQ2 gene is expressed in an E. coli ubiA mutant, the cells produce UQ-8 (38). Similarly, as shown in this study, expression of the COQ2 gene in an S. pombe ppt1 disruptant resulted in production of UQ-10. Thus, COQ2 can transfer both octaprenyl diphosphate and decaprenyl diphosphate to PHB. S. cerevisiae can also generate various ubiquinone species (UQ-5 to UQ-10) when polyprenyl diphosphate synthases from other species are expressed in the COQ1 mutant (22). Those observations all indicate that PHB polyprenyl diphosphate transferases can act with a broad range of different polyprenyl diphosphate substrates.
Sequence alignment of the various PHB polyprenyl transferases suggests a putative substrate binding site constituted by an aspartic acid-rich motif (NDXXDXXXD) (35). This motif is well conserved in homolog proteins from Providencia stuartii, Neisseria meningitidis, Pasteurella haemolytica, and Arabidopsis thaliana as well as in the proteins listed in Fig. 1. However, the assumption that this motif is the substrate binding site is based merely on its similarity with the substrate recognition site (DDXXD) in polyprenyl diphosphate synthases (23, 24). The exact substrate recognition sequence in PHB polyprenyl diphosphate transferases remains to be determined.
While eight COQ genes (COQ1 to COQ8) are known to be involved in ubiquinone biosynthesis in S. cerevisiae (2, 4, 10, 11, 27), the ppt1 gene was only the second gene found to be involved in S. pombe biosynthesis of ubiquinone. When we subsequently examined the database from the S. pombe genome project for more genes, we found several COQ homologs. Besides dps (COQ1 homolog) and ppt1 (COQ2 homolog), there are also COQ3, COQ4, COQ5, COQ6, and COQ7 homologs in the S. pombe genome, with amino acid sequence identities of 40, 42, 54, 37, and 51%, respectively (the sequence of COQ8 is not public). Thus, the entire gene set known to be involved in S. cerevisiae ubiquinone biosynthesis is also preserved in S. pombe. However, the enzymatic activities of Coq4 and Coq8 have, as yet, not been determined, and it is also not clear if all eight S. cerevisiae COQs are necessary and sufficient for the biosynthesis of ubiquinone.
Coq1 (our unpublished observations), Coq3 (27), Coq5 (4), and Coq7 (11) have all been localized to the mitochondria of S. cerevisiae, indicating that ubiquinone biosynthesis occurs in mitochondria. When we examined Ppt1 localization in this study, we found that it also localized to the mitochondria, suggesting that biosynthesis of ubiquinone in S. pombe also occurs in the mitochondria.
S. pombe strains whose ppt1 gene had been disrupted had several interesting phenotypes. First, while the ppt1 disruption strain did not grow well on PMA-glucose, growth was greatly improved by the presence of cysteine or glutathione. The addition of the lipid antioxidant α-tocopherol also improved growth. A requirement for cysteine, glutathione, or α-tocopherol for growth on minimal medium is interesting and is consistent with the concept that ubiquinone acts as an antioxidant. Supporting this idea further is that the ppt1-deficient strain is sensitive to active oxygen-producing reagents, such as H2O2 and Cu2+. These phenotypes of ppt1-deficient S. pombe are essentially equivalent to those observed for the dps-deficient S. pombe (37), confirming that these phenotypes arise as a consequence of not being able to produce ubiquinone. A role for ubiquinone as an antioxidant has also been reported for E. coli, S. cerevisiae, and mammalian cells (1, 7, 14, 18, 36). In E. coli, a ubiquinoneless mutant is more susceptible to H2O2 and Cu2+ (36). In S. cerevisiae, strains unable to produce ubiquinone are more susceptible to lipid peroxide and show lower stabilities of extracellular ascorbate (34). In mammalian cells, ubiquinone works synergistically with α-tocopherol to reduce lipid peroxide or low-density lipoprotein (1, 7). It is deduced that ubiquinone in S. pombe also has the role of suppressing lipid peroxidation of the membrane, although more direct evidence will be necessary to prove this point.
This study also detected an additional, and very interesting, phenotype of fission yeast strains unable to produce ubiquinone. The ppt1 and dps mutants both produced large amounts of H2S. This observation could not be explained by ordinary metabolic pathways. However, the recent finding that sulfide-ubiquinone reductase exists in S. pombe (42) suggested to us that there may be a metabolic link between ubiquinone and H2S production. We speculate that in the absence of ubiquinone in the cell, sulfide-ubiquinone reductase cannot function and thus the cell accumulates H2S. Since sulfide-ubiquinone reductase is not present in S. cerevisiae, mutants of S. cerevisiae that are unable to produce ubiquinone do not produce H2S (our unpublished observation). Interestingly, humans as well as some other higher eukaryotes possess sulfide-ubiquinone reductases similar to the one found in photosynthetic bacteria (42). HMT2 catalyzes sulfide oxidation by concomitant reduction of ubiquinone in S. pombe. That the hmt2 mutant does not release H2S in equivalent quantities as strains unable to produce ubiquinone is perhaps due to the gradual oxidization of sulfide by ubiquinone that occurs despite the absence of sulfide-ubiquinone reductase.
All the observed phenotypes of S. pombe strains that are unable to produce ubiquinone serve to emphasize the fact that ubiquinone does not function solely as a component of the electron transfer system, as is generally believed. Ubiquinone appears to also be important in the oxidative stress response and the sulfide oxidation pathways, at least in S. pombe. The former role seems to be common in eukaryotes, while the latter role may occur in the majority of eukaryotes that have sulfide-ubiquinone reductases. Further investigation into the importance of the alternative functions of ubiquinone in other species can be carried out by the construction of ubiquinone-deficient organisms.
ACKNOWLEDGMENTS
This study was supported by a grant-in-aid from the Ministry of Education, Science, and Culture of Japan.
JV5 was kindly provided by D. W. Ow.
REFERENCES
- 1.Alleva R, Tomasetti M, Battino M, Curatola G, Littarru G P, Folkers K. The roles of coenzyme Q10 and vitamin E on the peroxidation of human low density lipoprotein subfractions. Proc Natl Acad Sci USA. 1995;92:9388–9391. doi: 10.1073/pnas.92.20.9388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashby M N, Kutsunai S Y, Ackerman S, Tzagoloff A, Edwards P A. COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate:polyprenyltransferase. J Biol Chem. 1992;267:4128–4136. [PubMed] [Google Scholar]
- 3.Bader M, Muse W, Ballou D P, Gassner C, Bardwell J C. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–227. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- 4.Barkovich R J, Shtanko A, Shepherd J A, Lee P T, Myles D C, Tzagoloff A, Clarke C F. Characterization of the COQ5 gene from Saccharomyces cerevisiae. Evidence for a C-methyltransferase in ubiquinone biosynthesis. J Biol Chem. 1997;272:9182–9188. doi: 10.1074/jbc.272.14.9182. [DOI] [PubMed] [Google Scholar]
- 5.Do T Q, Schultz J R, Clarke C F. Enhanced sensitivity of ubiquinone-deficient mutants of Saccharomyces cerevisiae to products of autoxidized polyunsaturated fatty acids. Proc Natl Acad Sci USA. 1996;93:7534–7539. doi: 10.1073/pnas.93.15.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ernster L, Dallner G. Biochemical, physiological and medical aspects of ubiquinone function. Biochim Biophys Acta. 1995;1271:195–204. doi: 10.1016/0925-4439(95)00028-3. [DOI] [PubMed] [Google Scholar]
- 7.Frei B, Kim M C, Ames B N. Ubiquinol-10 is an effective lipid-soluble antioxidant at physiological concentrations. Proc Natl Acad Sci USA. 1990;87:4879–4883. doi: 10.1073/pnas.87.12.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant C M, MacIver F H, Dawes I W. Mitochondrial function is required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. FEBS Lett. 1997;410:219–222. doi: 10.1016/s0014-5793(97)00592-9. [DOI] [PubMed] [Google Scholar]
- 9.Grünler J, Ericsson J, Dallner G. Branch-point reactions in the biosynthesis of cholesterol, dolichol, ubiquinone and prenylated proteins. Biochim Biophys Acta. 1994;1212:259–277. doi: 10.1016/0005-2760(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 10.Hsu A Y, Do T Q, Lee P T, Clarke C F. Genetic evidence for a multi-subunit complex in the O-methyltransferase steps of coenzyme Q biosynthesis. Biochim Biophys Acta. 2000;1484:287–297. doi: 10.1016/s1388-1981(00)00019-6. [DOI] [PubMed] [Google Scholar]
- 11.Jonassen T, Proft M, Randez-Gil F, Schultz J R, Marbois B N, Entian K D, Clarke C F. Yeast Clk-1 homologue (Coq7/Cat5) is a mitochondrial protein in coenzyme Q synthesis. J Biol Chem. 1998;273:3351–3357. doi: 10.1074/jbc.273.6.3351. [DOI] [PubMed] [Google Scholar]
- 12.Kawahara K, Koizumi N, Kawaji H, Oishi K, Aida K, Uchida K. Partial purification and characterization of 4-hydroxybenzoate-polyprenyltransferase in ubiquinone biosynthesis of Pseudomonas putida. Agric Biol Chem. 1991;55:2307–2311. [Google Scholar]
- 13.Kawamukai M, Gerst J, Field J, Riggs M, Rodgers L, Wigler M, Young D. Genetic and biochemical analysis of the adenylyl cyclase-associated protein, cap, in Schizosaccharomyces pombe. Mol Biol Cell. 1992;3:167–180. doi: 10.1091/mbc.3.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kontush A, Hubner C, Finckh B, Kohlschutter A, Beisiegel U. Antioxidative activity of ubiquinol-10 at physiologic concentrations in human low density lipoprotein. Biochim Biophys Acta. 1995;1258:177–187. doi: 10.1016/0005-2760(95)00115-s. [DOI] [PubMed] [Google Scholar]
- 15.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 16.Meganathan R. Biosynthesis of the isoprenoid quinones menaquinone (vitamin K2) and ubiquinone (coenzyme Q) In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 642–656. [Google Scholar]
- 17.Melzer M, Heide L. Characterization of polyprenyl diphosphate:4-hydroxybenzoate polyprenyltransferase from Escherichia coli. Biochim Biophys Acta. 1994;1212:93–102. doi: 10.1016/0005-2760(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 18.Messner K R, Imlay J A. The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J Biol Chem. 1999;274:10119–10128. doi: 10.1074/jbc.274.15.10119. [DOI] [PubMed] [Google Scholar]
- 19.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 20.Moriyoshi K, Richards L J, Akazawa C, O'Leary D D, Nakanishi S. Labeling neural cells using adenoviral gene transfer of membrane targeted GFP. Neuron. 1996;16:255–260. doi: 10.1016/s0896-6273(00)80044-6. [DOI] [PubMed] [Google Scholar]
- 21.Muhlenweg A, Melzer M, Li S M, Heide L. 4-Hydroxybenzoate 3-geranyltransferase from Lithospermum erythrorhizon: purification of a plant membrane-bound prenyltransferase. Planta. 1998;205:407–413. doi: 10.1007/s004250050337. [DOI] [PubMed] [Google Scholar]
- 22.Okada K, Kainou T, Matsuda H, Kawamukai M. Biological significance of the side chain length of ubiquinone in Saccharomyces cerevisiae. FEBS Lett. 1998;431:241–244. doi: 10.1016/s0014-5793(98)00753-4. [DOI] [PubMed] [Google Scholar]
- 23.Okada K, Kainou T, Tanaka K, Nakagawa T, Matsuda H, Kawamukai M. Molecular cloning and mutational analysis of the ddsA gene encoding decaprenyl diphosphate synthase from Gluconobacter suboxydans. Eur J Biochem. 1998;255:52–59. doi: 10.1046/j.1432-1327.1998.2550052.x. [DOI] [PubMed] [Google Scholar]
- 24.Okada K, Kamiya Y, Zhu X, Suzuki K, Tanaka K, Nakagawa T, Matsuda H, Kawamukai M. Cloning of the sdsA gene encoding solanesyl diphosphate synthase from Rhodobacter capsulatus and its functional expression in Escherichia coli and Saccharomyces cerevisiae. J Bacteriol. 1997;179:5992–5998. doi: 10.1128/jb.179.19.5992-5998.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okada K, Minehira M, Zhu X, Suzuki K, Nakagawa T, Matsuda H, Kawamukai M. The ispB gene encoding octaprenyl diphosphate synthase is essential for growth of Escherichia coli. J Bacteriol. 1997;179:3058–3060. doi: 10.1128/jb.179.9.3058-3060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okada K, Suzuki K, Kamiya Y, Zhu X, Fujisaki S, Nishimura Y, Nishino T, Nakagawa T, Kawamukai M, Matsuda H. Polyprenyl diphosphate synthase essentially defines the length of the side chain of ubiquinone. Biochim Biophys Acta. 1996;1302:217–223. doi: 10.1016/0005-2760(96)00064-1. [DOI] [PubMed] [Google Scholar]
- 27.Poon W W, Barkovich R J, Hsu A Y, Frankel A, Lee P T, Shepherd J N, Myles D C, Clarke C F. Yeast and rat Coq3 and Escherichia coli UbiG polypeptides catalyze both O-methyltransferase steps in coenzyme Q biosynthesis. J Biol Chem. 1999;274:21665–21672. doi: 10.1074/jbc.274.31.21665. [DOI] [PubMed] [Google Scholar]
- 28.Rabinowitz J C. Analysis of acid-labile sulfide and sulfhydryl groups. Methods Enzymol. 1978;53:275–277. doi: 10.1016/s0076-6879(78)53033-4. [DOI] [PubMed] [Google Scholar]
- 29.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 30.Rothstein R J. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 31.Saiki R K, Gelf D H, Stoffel I S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sanger F, Coulson R, Barrel B G, Smith J H, Roe B A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980;143:161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- 34.Santos-Ocana C, Cordoba F, Crane F L, Clarke C F, Navas P. Coenzyme Q6 and iron reduction are responsible for the extracellular ascorbate stabilization at the plasma membrane of Saccharomyces cerevisiae. J Biol Chem. 1998;273:8099–8105. doi: 10.1074/jbc.273.14.8099. [DOI] [PubMed] [Google Scholar]
- 35.Soballe B, Poole R K. Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology. 1999;145:1817–1830. doi: 10.1099/13500872-145-8-1817. [DOI] [PubMed] [Google Scholar]
- 36.Soballe B, Poole R K. Ubiquinone limits oxidative stress in Escherichia coli. Microbiology. 2000;146:787–796. doi: 10.1099/00221287-146-4-787. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki K, Okada K, Kamiya Y, Zhu X, Nakagawa T, Kawamukai M, Matsuda H. Analysis of the decaprenyl diphosphate synthase (dps) gene in fission yeast suggests a role of ubiquinone as an antioxidant. J Biochem. 1996;121:496–505. doi: 10.1093/oxfordjournals.jbchem.a021614. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki K, Ueda M, Yuasa M, Nakagawa T, Kawamukai M, Matsuda H. Evidence that Escherichia coli ubiA product is a functional homolog of yeast COQ2, and the regulation of ubiA gene expression. Biosci Biotechnol Biochem. 1994;58:1814–1819. doi: 10.1271/bbb.58.1814. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka K, Nishide J, Okazaki K, Kato H, Niwa O, Nakagawa T, Matsuda H, Kawamukai M, Murakami Y. Characterization of a fission yeast SUMO-1 homologue, Pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol Cell Biol. 1999;19:8660–8672. doi: 10.1128/mcb.19.12.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uchida K, Koizumi N, Kawaji H, Kawahara K, Aida K. Solubilization of 4-hydroxybenzoate-polyprenyltransferase from cell membrane of Pseudomonas putida and its properties. Agric Biol Chem. 1991;55:2299–2305. [Google Scholar]
- 41.Wallace B J, Young I G. Role of quinones in electron transport to oxygen and nitrate in Escherichia coli. Studies with a ubiA− menA− double quinone mutant. Biochim Biophys Acta. 1977;461:84–100. doi: 10.1016/0005-2728(77)90071-8. [DOI] [PubMed] [Google Scholar]
- 42.Weghe J G V, Ow D W. A fission yeast gene for mitochondrial sulfide oxidation. J Biol Chem. 1999;274:13250–13257. doi: 10.1074/jbc.274.19.13250. [DOI] [PubMed] [Google Scholar]