Abstract
Light provides energy for photosynthesis and also acts as an important environmental signal. During their evolution, plants acquired sophisticated sensory systems for light perception and light-dependent regulation of their growth and development in accordance with the local light environment. Under natural conditions, plants adapted by using their light sensors to finely distinguish direct sunlight and dark in the soil, deep grey shade under the upper soil layer or litter, green shade under the canopy and even lateral green reflectance from neighbours. Light perception also allows plants to evaluate in detail the weather, time of day, day length and thus the season. However, in artificial lighting conditions, plants are confronted with fundamentally different lighting conditions. The advent of new light sources — light-emitting diodes (LEDs), which emit narrow-band light — allows growing plants with light of different spectral bands or their combinations. This sets the task of finding out how light of different quality affects the development and functioning of plants, and in particular, their photosynthetic apparatus (PSA), which is one of the basic processes determining plant yield. In this review, we briefly describe how plants perceive environment light signals by their five families of photoreceptors and by the PSA as a particular light sensor, and how they use this information to form their PSA under artificial narrow-band LED-based lighting of different spectral composition. We consider light regulation of the biosynthesis of photosynthetic pigments, photosynthetic complexes and chloroplast ATP synthase function, PSA photoprotection mechanisms, carbon assimilation reactions and stomatal development and function.
Keywords: LED lighting, Light quality, Photosynthetic apparatus regulation, Photosynthetic pigment synthesis, Photosynthetic carbon assimilation, Stomata
Introduction
Plants, as phototrophic organisms, heavily rely on light as a source of energy for photosynthesis. Light that is used in photosynthesis is absorbed by photosynthetic pigments — chlorophylls (Chls) and carotenoids (Cars). Chls absorb red and blue light most efficiently; Cars absorb blue light. Green light, although less efficient, can also be a source of energy in photosynthesis. Its absorption efficiency by the leaf depends on leaf structure which determines light scattering inside the leaf. Light is also an important source of information about the environment. Plants are able to evaluate the spectral composition, intensity or dose, duration and direction of light, photoperiod and its changes, which allows them to regulate their development in accordance with current light conditions. The signal, or regulatory, role of light is carried out by a system of photoreceptors that specifically perceive different ranges of the spectrum and evaluate their ratios: plants sense red and far-red light by phytochromes; blue and ultraviolet A (UV-A) light — by cryptochromes, phototropins, Zeitlupes and partly by phytochromes; ultraviolet B (UV-B) — by the photoreceptor UV-B; green — partly by cryptochromes and phototropins; and partly by phytochromes (Galvão and Fankhauser 2015). The photosynthetic apparatus (PSA) is also a kind of photoreceptor, the signals from which affect the expression of nuclear and plastid genes — primarily associated with photosynthesis, but not exclusively (Szechynska-Hebda and Karpinski 2013). Another role of light is biosynthesis, as photons directly take part in the synthesis of chemicals crucial for organism functioning. For example, one of the reactions in the Chl biosynthesis pathway — conversion of protochlorophyllide into chlorophyllide — requires a photon, and in angiosperms, it is the only pathway of Chl synthesis (Yuan et al. 2017; Solymosi and Mysliwa-Kurdziel 2021). Light also may be harmful for all living organisms — excessive light, particularly high-energy blue and ultraviolet, causes photodamage due to nonspecific absorption of these photons by various cell components (Yadav et al. 2020).
Studies of the physiology and productivity of plants grown with light of different spectral bands have a long history. In earlier works, light of different spectral quality was yielded by full-spectrum light sources (natural sunlight, incandescent, white fluorescent, high pressure sodium-vapour lamps or others) in combination with broad-band filters, or by fluorescent lamps of a certain colour (Voskresenskaya et al. 1968; Lichtenthaler et al. 1980; Leong et al. 1985; Deng et al. 1989; Bukhov et al. 1995).
During the last three decades, there has been a new interest in such works because of the advent of new light sources — light-emitting diodes (LEDs), which emit narrow-band light (with half bandwidth ~ 10–30 nm) of different wavebands and have a light intensity high enough for long-term plant growth (Goins et al. 1997; Wang et al. 2009; Berkovich et al. 2017; Tarakanov et al. 2022). With such light sources, researchers can treat plants with narrow-band light of different wavelengths, which is important for photobiological research. Also, LEDs are increasingly used in horticultural lighting (greenhouses, vertical farms and space greenhouses) due to their beneficial technical characteristics: electrical efficiency, small mass and volume, solid state construction, low heat production, longevity and safety (Goins et al. 1997; He et al. 2015; Berkovich et al. 2017). Knowledge gained through fundamental research can help build LED-based horticultural light sources with an optimal spectrum. By manipulating the spectral composition of the lighting source, it is possible to create plants with certain growth characteristics, induce accumulation of important chemicals or increase plant yield (Landi et al. 2020).
The first works involving LEDs studied the effects of individual spectral bands and their combinations on plant growth and development, photosynthetic carbon assimilation, metabolite accumulation and yield. The spectral bands studied were mostly red and blue, due to their primary importance for photosynthesis; later studies included green, far-red, orange, yellow and violet spectral bands. The aim of these works was to find a spectrum that would allow optimal plant performance (Goins et al. 1997; Matsuda et al. 2007; Wang et al. 2009; Hogewoning et al. 2010; Liu et al. 2011a, b; Xiaoying 2012). Works aimed at optimising LED horticultural lighting are still being carried out. At the same time, more works focused on understanding the physiological, biochemical and genetic mechanisms behind the effects of narrow-band light on plants (Avercheva et al. 2009; 2010; 2016; Savvides et al. 2012; Muneer et al. 2014; Su et al. 2014; Miao et al. 2016; He et al. 2017; Kochetova et al. 2018, 2022; Lanoue et al. 2018; Hamdani et al. 2019; Gao et al. 2020; Karlický et al. 2021; Tantharapornrerk et al. 2021; Trojak and Skowron 2021; Tarakanov et al. 2022). Many of them focus on the PSA as the source of organic compounds for plant growth and development. The PSA is itself a complex, multi-component structure, and its components can be affected by light spectrum in different ways (summarised in Fig. 1).
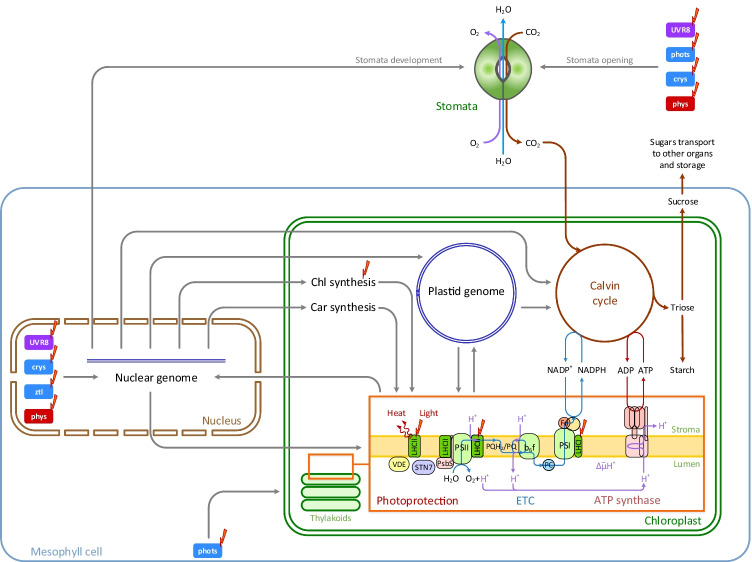
Fig. 1.
The scheme of the photosynthetic apparatus components and their interactions. Chlorophyll (Chl) and carotenoids (Car) are synthesised in plastids by nuclear coded enzymes (Yuan et al. 2017; Sun et al. 2018) and used for photosynthetic pigment-protein complex assembly. Electron transport chain (ETC) consists of the two photosystems, PSII and PSI, cytochrome b6f complex (b6f) and mobile electron carriers: plastoquinone pool (PQH2/PQ) in the thylakoid membrane, plastocyanin (PC) in the lumen and ferredoxin (Fd) in the stroma. Light harvesting antennae, LHCII and LHCI, help to harvest light for PSII and PSI, respectively. Linear electron transport through ETC from H2O to NADPH provides H+ cross-membrane transport and produces the proton gradient. ATP synthase uses this gradient energy for ATP synthesis. Calvin cycle of carbon assimilation from CO2 to sugars consumes NADPH and ATP, produced by ETC. Generated trioses may be stored transiently in the chloroplast as starch or are exported and transported to other organs as sucrose and other sugars (Raines 2003). A part of the mobile pool of LHCII can disconnect from PSII with the help of serine/threonine kinase STN7 and PsbS subunit and move to PSI or dissipate absorbed light energy to heat (red curly arrows). Violaxanthin de-epoxidase (VDE) converts violaxanthin to zeaxanthin which quenches light energy in LHCII and PSII minor antennae. The major light input targets for photosynthetic apparatus formation, function and regulation are marked with red lightning symbols. Pigment-protein complexes containing Chls and Cars capture light for photosynthesis. Photoreceptors percept light and regulate gene expression and cell metabolism. In particular, photoreceptors control stomata development and opening. The light-dependent step in the Chl biosynthesis pathway is also marked
Despite there being a large body of literature concerning LEDs as light sources for growing plants, the results of these works are often hard to compare. This is, in part, because plants of different species and age are used in these studies. Light spectrum, irradiance, photoperiod and length of plant exposure to narrow-band light may also vary. Control plants can be grown with light sources with different emission spectra. It has also been shown that effects of narrow-band light on plants are often specific to species, ecotype and cultivar (Landi et al. 2020; Yavari et al. 2021), which further complicates data systematisation. However, the accumulated knowledge allows putting together a general, though incomplete, scheme of the interaction between light of different spectral wavebands and plant physiological state. It is especially important to understand the interaction between the role of light as a source of energy and as an environmental signal.
In this review, we aim to analyse the current data on PSA development and function under narrow-band light of different spectral wavebands — red (600–700 nm), green (500–570 nm), blue (420–500 nm) and near ultraviolet (UV-A, 315–400 nm). This requires considering two topics: (1) what is the structure and function of the photosynthetic apparatus and its components in plants grown with light of different spectral bands; (2) what regulatory mechanisms light can use to control photosynthetic apparatus development. Here, we summarise the recent literature on these topics, considering separately the effects of light spectrum on different PSA components (as shown in Fig. 1) and attempt to integrate them where possible. As the photosynthetic apparatus relies on stomata for sufficient CO2 uptake, the regulation of their development and function by light spectrum is also considered.
Photoregulatory role of light
Plant photoreceptors
Plants get information about the spectral quality of the surrounding light via photoreceptors. Five groups of plant photoreceptors are known, which perceive specific wavebands from ultraviolet B to far red and, according to the signal received, regulate many physiological processes (Christie et al. 2015; Galvão and Fankhauser 2015; Kong and Okajima 2016; Yadav et al. 2020). All photoreceptor genes are located in the nucleus, and the photoreceptors themselves operate in the nucleus and/or the cytosol (Yadav et al. 2020).
Phytochromes are photoreceptors that perceive red (540–690 nm, maximum sensitivity at 640–670 nm) and far-red (695–780 nm, maximum sensitivity at 720–750 nm) light (Borthwick et al. 1954; Shinomura et al. 1996). The phytochrome chromophore is a linear tetrapyrrole phytochromobilin. Upon absorbing a red photon, it cis–trans isomerizes, which leads to conformational changes of the apoprotein from an initial inactive Pr form to a physiologically active Pfr form (Butler et al. 1964; Tu and Lagarias 2005; Rockwell and Lagarias 2020). The Pfr form interacts with partner proteins, autophosphorylates and transphosphorylates, its transduction chain partners, and most of the photoactivated phytochrome is transported from the cytosol into the nucleus, where it regulates the expression of multiple light-dependent nuclear genes (Viczián et al. 2016; Inoue et al. 2017). Dark reversion or the absorption of a far-red photon by the Pfr form inactivates the photoreceptor, changing it back into the inactive Pr form, attenuating the signal induced by the photoreceptor (Viczián et al. 2016; Klose et al. 2020). Phytochromes are a small family of photoreceptors which, in most angiosperms, includes four (in dicots) or three (in monocots) members (Li et al. 2015) with distinct and partly overlapping functions. All angiosperms possess two major phytochromes: A and B. Phytochrome A (phyA) is photolabile. It accumulates in the dark and is the predominant phytochrome in etiolated plants, determining their high sensitivity to light. The level of phyA decreases quickly and dramatically upon illumination. Its main role is to perceive the first photons during dark to light transition, upon emergence from the soil, and low lighting during seed germination and seedling development inside the soil or under litter, when the plant is under the conditions of short pulses and/or very low light intensities of any light. phyA also plays the dominant role in deep green shade under the canopy, under the conditions of relatively high intensity of continuous irradiance by light with a high content of far-red photons that have not been absorbed by the above situated green leaves (Chen and Chory 2011; Casal et al. 2014; Menon et al. 2016). Phytochrome B (phyB) is photostable — its content decreases upon illumination but less drastically than that of phyA. It is the main phytochrome of a de-etiolated adult plant growing in direct sunlight. Its role is to assess the ratio of red light suitable for photosynthesis and far-red light unsuitable for photosynthesis (Chen and Chory 2011; Casal et al. 2014; Menon et al. 2016). Minor phytochromes (phytochrome C (phyC) in monocots or phytochromes C (phyC) and E (phyE) in dicots) are also photostable, but their content and role in regulatory processes are secondary to major phytochromes A and B (Alba et al. 2000). Some plant species have lost one or all of minor phytochromes (Zostera marina — monocot without phyC (Olsen et al. 2016), Pisum and Populus nigra — dicots without phyC and phyE (Platten et al. 2005)) or acquired additional copies of phytochromes due to recent gene duplication (Arabidopsis, tomato — Alba et al. 2000, Ranunculales — Li et al. 2015).
In the dark, phytochromes are located in the cytosol. Photoactivated phytochromes initiate two major signalling chains. The rapid directional cytosolic signal branch, activated mainly by phyA, affects events on the plasma membrane (polarisation and activation of ion fluxes and the distribution of the PIN-FORMED 3 (PIN3) auxin efflux transporter) and in the cytosol (translation regulation) (Long and Iino 2001; Hughes 2013; Galvão and Fankhauser 2015). The main signalling pathway involves the import of photoactivated phytochromes into the nucleus, where they alter gene expression via two pathways. The first nuclear signalling pathway involves inhibiting the CONSTITUTIVELY PHOTOMORPHOGENIC 1/SUPPRESSOR OF phyA-105 (COP1/SPAs) complex — a component of CULLIN 4/DAMAGED DNA BINDING 1 (CUL4-DDB1COP1/SPAs) E3 ubiquitin ligase complex, a key repressor of photomorphogenesis that facilitates ubiquitylation and degradation of many transcription factors — positive regulators of photomorphogenesis, such as ELONGATED HYPOCOTYL 5 (HY5), HY5 HOMOLOGUE (HYH), LONG HYPOCOTYL IN FAR-RED 1 (HFR1), CONSTANS (CO), GIGANTEA (GI), EARLY FLOWERING 3 (ELF3). The second nuclear signalling pathway is more characteristic of phyB rather than phyA. It involves the basic helix-loop-helix (bHLH) transcription factors of the PHYTOCHROME INTERACTING FACTORs (PIFs) family, which are negative regulators of photomorphogenesis. Photoactivated phytochromes inhibit the binding capacity of PIFs with their target promoters and induce PIFs proteolytic degradation (Casal et al. 2014; Menon et al. 2016; Viczián et al. 2016; Inoue et al. 2017).
Phytochromes are involved in the regulation of germination and de-etiolation — inhibition of hypocotyl elongation, opening of the apical hook and expansion of the cotyledons. Phytochromes also define the architecture of adult plants — they modulate shoot and root gravitropism, regulate development of rosette, determine branching and apical dominance. In addition, they induce PSA and chloroplast development, shade avoidance response, play a role in modulating signalling induced by biotic and abiotic stresses, thermosensing and senescence, entrainment of the circadian clock and flowering transition (Galvão and Fankhauser 2015; Kong and Okajima 2016; Viczián et al. 2016). Thus, phytochromes are the major family of plant photoreceptors, which “function as a master regulator of photomorphogenesis to control germination, flowering, and almost everything in between” (Rockwell and Lagarias 2020). The functions of two major phytochromes, phyA and phyB, are most thoroughly studied. Some responses are induced by only one phytochrome — for example, phyA is solely responsible for germination, de-etiolation and anthocyanin synthesis under far-red light, and phyB provides thermosensing. While other responses are the result of the joint action of both phytochromes. Their effects can be additive, like in de-etiolation under red light, or opposite, like in flowering time and shade avoidance response (Shinomura et al. 1996; Casal et al. 2014; Viczián et al. 2016).
Cryptochromes are photoreceptors of blue and UV-A light (340–500 nm, maximal activity 400–500 nm with an additional peak at 360–380 nm) (Christie et al. 2015). They have a main chromophore, flavin adenine dinucleotide (FAD) (absorption maximum 450 nm with subsidiary shoulders at 430 and 470 nm), and a supplementary chromophore, pterin derivative 5,10-methenyltetrahydrofolate (MTHF) (absorption maximum at 380 nm) (Ahmad and Cashmore 1993; Christie et al. 2015; Mishra and Khurana 2017). Photoexcited FAD enters an intramolecular redox reaction with apoprotein amino acids, which changes the conformation of the apoprotein, and leads to its oligomerization, (auto)phosphorylation and interaction with partner proteins (Christie et al. 2015; Galvão and Fankhauser 2015; Mishra and Khurana 2017). Dark reversion, as well as absorption of a green photon (500–600 nm), returns the cryptochrome into the inactive form, attenuating the signal (Christie et al. 2015; Mishra and Khurana 2017). Angiosperms have two cryptochromes with overlapping and partially redundant functions: the photostable cryptochrome 1 (cry1) and the photolabile cryptochrome 2 (cry2) (Menon et al. 2016; Mishra and Khurana 2017; Wang et al. 2018). Some species have more than one copy of these cryptochromes (Mishra and Khurana 2017). In the dark, cryptochromes are located in the nucleus. In their light-activated form, they regulate the expression of many light-dependent nuclear genes via two pathways. The first pathway overlaps with phytochrome signalling and inhibits a key repressor of photomorphogenesis, the COP1/SPAs complex (explained in section “Phytochromes”), which stops the proteolysis of transcription factors — positive photomorphogenesis regulators, such as HY5, CO, and others (Christie et al. 2015; Menon et al. 2016; Wang et al. 2018; Yadav et al. 2020). The second pathway, unique to cry2, involves its direct interaction with, stabilisation and activation of cryptochrome interacting basic helix-loop-helix (CIBs) transcription factors — positive regulators of some photomorphogenesis components (Christie et al. 2015; Galvão and Fankhauser 2015; Menon et al. 2016; Wang et al. 2018). Both cryptochromes directly interact with and inhibit PIF4/5, negative photomorphogenesis regulators (Mishra and Khurana 2017; Wang et al. 2018). Also, some of the photoactivated cry1 exits the nucleus and launches the cytosolic signalling branch, activating anion channels on the plasma membrane, among other targets (Christie et al. 2015; Mishra and Khurana 2017).
Like phytochromes, cryptochromes play a global role in the regulation of growth and development, including seed dormancy and germination; de-etiolation; synthesis of Chls, Cars, anthocyanins and flavonoids; protein synthesis; circadian clock regulation; flowering time control; shade avoidance and modulation of stress responses, suppressing leaf senescence and root growth (Xu et al. 2009; Menon et al. 2016; Mishra and Khurana 2017; Wang et al. 2018; Yadav et al. 2020; Griffin et al. 2020). Functions of both cryptochromes are partially overlapping and partially distinct due to differences in their expression pattern, photolability and signallings. Photolabile cry2 predominates in germinating embryos and roots and acts under low light, and cry1 predominates in leaves, especially in green leaves, and acts under high light (Xu et al. 2009). cry1 plays a predominant role in seed dormancy (in barley), de-etiolation and circadian clock entrainment. cry2 regulates leaf senescence (in soybean) and photoperiod sensing (Galvão and Fankhauser 2015; Mishra and Khurana 2017).
Phototropins are photoreceptors of blue/UV-A light (~ 340–500 nm, maximal activity at 410–480 nm with an additional broad, less effective peak at 360–370 nm) (Liscum and Briggs 1995; Okajima et al. 2012; Christie et al. 2015). Each of the two flavin mononucleotide (FMN) chromophores, upon absorbing a blue/UV-A photon, forms covalently bound thioadducts with the polypeptide chain, which changes the conformation of the adjacent apoprotein. The photoconversion of the second FMN plays an essential role in light perception, as it liberates the apoprotein kinase activity, its autophosphorylation and interaction with partner proteins with phosphorylation of some of them. The photoconversion of the first FMN has a modulating function (Okajima et al. 2012; Kong and Wada 2014; Galvão and Fankhauser 2015). Dark reversion of the chromophore and the apoprotein shuts down the signal induced by the photoactivation (Okajima et al. 2012). Phototropins are cytosolic proteins and are predominantly localised to the plasma membrane. In darkness, they are associated with the inner face of the plasma membrane, and under blue light, they are partially internalised into the cytoplasm, relocating to the soluble fraction of the cytosol (phototropin 1 (phot1)) or associating with endomembranes (Golgi apparatus and chloroplast outer membrane for phototropin 2 (phot2)) (Kong and Wada 2014; Galvão and Fankhauser 2015; Liscum 2016). In contrast to other plant photoreceptors, which primarily act in the nucleus, phototropins affect plasma membrane and cytoplasmic proteins. They induce the opening of Ca2+-channels, activate the plasma membrane H+-ATPases and aquaporins via a kinase phosphorylation cascade, inhibit auxin transporters and induce cytoskeleton dynamic reorganisations, including cortical microtubules and specific cp-actin filaments responsible for chloroplasts attachment to the plasma membrane and their movement (Kong and Wada 2014; Christie et al. 2015; Ishka 2022). Angiosperms have two phototropins: phot1 and phot2. Their functions are largely redundant, but the light sensitivity of phot1 is much larger than phot2, firstly due to slower dark reversion (Okajima et al. 2012) and secondly due to a decrease of phot1 levels and increase of phot2 levels upon illumination (Kong and Wada 2014).
Phototropins orient plant photosynthetic organs and organelles to capture light energy efficiently by controlling phototropic stem bending, leaf expansion and flattening, petiole and leaf orientation, palisade mesophyll cells growth, chloroplast positioning and movements (accumulation or avoidance, depending on light intensity) and stomatal opening (Liscum and Briggs 1995; Kong and Wada 2014; Christie et al. 2015; Liscum 2016; Yadav et al. 2020). Thus, phototropins highly overlap in function, optimise photosynthetic efficiency and provide avoidance of damage from high light intensity in response to variable and changing light conditions. They also regulate defensive mechanisms, such as negative root phototropism, nuclear avoidance movements (mediated by the avoiding plastids), regulating resistance protein–mediated viral defence, improving plant performance (Christie et al. 2015).
The UV-B photoreceptor, UV RESISTANCE LOCUS 8 (UVR8), perceives light between 250 and 300 nm (with a maximum at 280 nm). It does not have a prosthetic chromophore. A tryptophan cluster of the polypeptide chain serves as a chromophore (Rizzini et al. 2011). Other tryptophans of the polypeptide chain act as antennae, increasing light harvesting efficiency (Li et al. 2020). In the dark, UVR8 as an inactive dimer is mainly localised in the cytosol. Upon UV-B photon absorption, a local transfer of electron and proton occurs between tryptophans forming the chromophore and other amino acid residues of the protein, and the dimer monomerizes into active monomers. The monomer of UVR8 is imported to the nucleus, where it regulates gene expression by two pathways (Rizzini et al. 2011; Yin and Ulm 2017). In the first pathway, UVR8 binds to COP1, disconnecting (COP1/SPA)2 from CUL4/DDB1COP1/SPAs multisubunit E3 ubiquitin ligase complex, thus inhibiting its E3 ubiquitin ligase activity against target proteins, which include transcription factors such as HY5, HYH and others (Rizzini et al. 2011; Ulm and Jenkins 2015; Menon et al. 2016). The rescued HY5 and HYH accumulate and induce transcription of a subset of UV-B-regulated genes, including HY5 itself (Menon et al. 2016; Yin and Ulm 2017; Liang et al. 2019; Tossi et al. 2019). In the second pathway, nuclear-localised UVR8 monomer interacts with several transcription factors, inhibiting their binding to promoters of genes which they activate or repress. UVR8 is inactivated with the help of Repressor of UV-B Photomorphogenesis 1/2 (RUP1/2) proteins, whose expression is induced by photoactivated UVR8. These proteins provide negative feedback regulation, facilitating redimerization of the photoreceptor (Liang et al. 2019; Tossi et al. 2019). Many angiosperm species have more than one copy of UVR8 gene, but most UVR8 copies from the same species show a high level of sequence identity, and it is not known if they behave as a gene family or have fully redundant roles (Fernandez et al. 2016). On the other hand, the seagrass Zostera marina has lost the UVR8 gene (Olsen et al. 2016).
The UVR8 photoreceptor, through changes in gene expression, inhibits hypocotyl elongation and other components of shade- and high temperature–promoted plant growth responses. It inhibits leaf growth and promotes leaf thickening, downward leaf curling, UV-B-dependent phototropism and entrainment of the circadian clock. To prevent damage caused by short-wavelength light, this photoreceptor also activates accumulation of screening of excessive and high-energy light flavonoids and anthocyanins and other secondary metabolites, essential for damaging light and defence responses, and induces DNA repair (photolyases expression) and protection against oxidative stress and photoinhibition (Galvão and Fankhauser 2015; Yin and Ulm 2017; Tossi et al. 2019; Yadav et al. 2020). Some physiological responses to UV-B light, high-intensity damaging and low-intensity non-stressful are independent of UVR8. They are induced by photochemical damage to cellular biomolecules that launches downstream signal cascades, including ROS production and stress hormones. The targets of these cascades are enzyme activity and gene expression (Tossi et al. 2019; Yadav et al. 2020).
Zeitlupes (ZTLs) are a family of blue/UV-A photoreceptors (the absorption spectrum is the same as in phototropins) and components of the nuclear multi-subunit SCF E3-ubiquitin-ligases. In the SCFZTL/LKP2/FKF1 complex, they play a role of substrate receptors. They specifically recognize target proteins in a light-dependent manner, bind and destabilise (via ubiquitylation and subsequent proteasomal degradation) or conversely stabilise them, thus regulating the abundance of target proteins. Their chromophore, FMN, has a photocycle like that in phototropins but with a much longer dark reversion. Photoconversion of the chromophore changes the conformation of the apoprotein and activates the E3-ligase activity of the complex (Pudasaini and Zoltowski 2013; Galvão and Fankhauser 2015; Feke et al. 2021). The photocycle of ZTL is photoreversible — shorter wavelength light (UV-A with maximal effectiveness at 382 nm) — reverses the photoactivated chromophore to the ground state, providing sensitivity of the photoreceptor to light fluence and spectral quality (Pudasaini and Zoltowski 2013; Pudasaini et al. 2017). Different members of the ZTLs protein family have highly similar primary amino acid sequences and partially redundant functions, but their major functions vary between individual proteins. ZTL/LKP2 (LOV KELCH PROTEIN 2) mostly entrains the circadian clock by targeting some of circadian oscillator components to proteolysis in a light-dependent manner. These include repressor transcription factors TIMING OF CAB EXPRESSION 1 (TOC1) and PSEUDORESPONSE-REGULATOR 5 (PRR5). FKF1 (FLAVIN-BINDING, KELCH REPEAT, F-BOX 1) regulates photoperiodism in a light-dependent manner, by targeting CYCLING DOF FACTORs (CDFs) transcription factors to proteolysis (these transcription factors suppress florigen expression) and stabilising CO transcription factor that induces florigen expression (Somers et al. 2000; Christie et al. 2015; Galvão and Fankhauser 2015; Pudasaini et al. 2017).
Phenotypically, effects of ZTL manifest in the correct expression rhythm of clock-controlled genes, including photosynthetic genes, and leaf movement rhythm, which increases plant growth, photosynthesis and productivity (Somers et al. 2000). The circadian clock independently operates and is entrained by the photoreceptor ZTL in the mesophyll and stomata guard cells. Thus, blue/UV-A light via ZTL controls circadian rhythms of CO2 fixation and stomatal conductance (Dodd et al. 2004). Since photosynthesis is only possible during daytime, when there is light, chloroplast functioning is strongly regulated by circadian clocks: up to 70% of total chloroplast-encoded protein-coding genes, including many photosynthetic genes, are dependent on the circadian clock (Atkins and Dodd 2014). This regulation is carried out mostly via SIG5 — a circadian-regulated gene, one of the six sigma-factors of plastid-encoded plastid RNA polymerase (Noordally et al. 2013). Many nuclear genes of the PSA are also rhythmic, and there are strong circadian rhythms of photosynthesis (Atkins and Dodd 2014). The measurement of the day length and its changes allow plants to sense the season and to regulate in concordance with it the time of photoperiodic flowering, and thus the time and abundance of seed production, and storage organ formation (Feke et al. 2021).
Photoreceptor interaction and its significance for plant development. Signalling pathways from photoreceptors interact on different levels. Direct interaction between some photoreceptors has been shown to modulate their activity. Photoreceptors also interact with common partner proteins. The light-dependent regulation of gene expression of photoreceptors and their signalling partners is mediated by other photoactivated photoreceptors. Signalling pathways share key components such as the nuclear COP1–HY5 module and the inhibition of PIF4/5 for phytochromes, cryptochromes and UVR8 and the interaction of phytochromes and phototropins via common protein partners PHYTOCHROME KINASE SUBSTRATEs (PKSs) in cortical cytoplasm layer. Promoters of many light-dependent genes (for example, light harvesting chlorophyll binding proteins LHCBs, chalcone isomerase (CHI)) contain regulatory elements targeted by signalling from different photoreceptors (Casal et al. 2014; Mishra and Khurana 2017; Galvão and Fankhauser 2015; Menon et al. 2016; Yadav et al. 2020). Due to this complex interaction between elements of the whole photoreceptor system, photoreceptors often have overlapping functions, synergistic or additive effects. Many signalling components are common for photoreceptors and phytohormones, which further complicates the network of plant growth regulation with light. COP1/SPAs, HY5 and PIFs are major hubs where light and phytohormone signalling pathways converge. Some aspects of these interactions have been recently reviewed (Jing and Lin 2020; Kusnetsov et al. 2020). Because of this, plants are highly sensitive to light cues and their changes, even minor ones, and some photoreceptors can partially compensate for others, at the same time affecting and being affected by phytohormones. Natural sunlight has a continuous spectrum and contains, in various proportions, photons that excite all photoreceptors to some extent. The resulting physiological response is the output of a complex interaction between all components of the photoreceptor system. When some elements of this system are artificially eliminated — for example, by using mutants lacking photoreceptors or their signalling components, or by growing plants with light lacking certain essential spectral bands — many elements of the photoreceptor system may be affected to a certain extent, and the resulting effect on plant physiological responses may be difficult to predict. Due to variations in the photoreceptor system, some physiological responses to changes in light spectrum can differ between species, ecotypes and cultivars (Casal et al. 2014).
Photoregulatory role of the photosynthetic apparatus
Photosynthetic pigments and electron transport chain also play a photoregulatory role. Through events that are termed retrograde signalling, chloroplastic signals affect the expression of many nuclear genes, mostly photosynthesis-associated and stress-related (reviewed in Singh et al. 2015; Yurina and Odintsova 2019). Multiple chloroplast components are considered signal transductors for the retrograde signal, among them tetrapyrrole biosynthesis intermediates, the redox-state of electron transport chain components (mainly the plastoquinone pool) and soluble redox compounds, reactive oxygen species and various plastid metabolites. For example, the redox state of the plastoquinone pool controls the expression of LHCB1.1 (encoding a chlorophyll binding protein of the light harvesting complex), RBCS (encoding the small Rubisco subunit) and PETE (encoding plastocyanin) among other genes (Oswald et al. 2000; Pfannschmidt et al. 2001), and methylerythritol cyclodiphosphate, a chloroplast metabolite from the isoprenoid biosynthetic pathway, regulates the expression of stress-responsive genes but not photosynthesis-associated genes (Xiao et al. 2012).
The full mechanism of signal transduction from chloroplasts to the nucleus is not yet known, but Mg-protoporphyrin-IX (Strand et al. 2003) and H2O2 (Borisova-Mubarakshina et al. 2015) apparently play a significant role. Plastid-to-nucleus signals involving tetrapyrroles, ROS and redox signals from the electron transport chain converge on GENOMES UNCOUPLED1 (GUN1), a pentatricopeptide-repeat chloroplast protein (Yuan et al. 2017). The ABA INSENSITIVE4 (ABI4) transcription factor acts downstream of GUN1 (Koussevitsky et al. 2007), providing a link with abscisic acid metabolism. Retrograde signals from chloroplasts also affect photoreceptor-mediated responses. Signals from the GUN1 pathway mediate signalling from cry1 via HY5 (Ruckle et al. 2007). Redox signals from the NADPH-thioredoxin reductase control the expression of genes for cry2 and FAR1-RELATED SEQUENCE 3 (FRS3), a protein that mediates phyA-controlled responses to far-red light (Lepistö et al. 2009).
In this way, chloroplasts can be considered another photoreceptor, with photosynthetic pigments being the chromophore. Chloroplast retrograde signals interact with photoreceptors and hormones and form a complex regulatory network that fine-tunes the PSA and cellular metabolism in response to light cues. Chloroplasts react to light in the whole PAR range, but most efficiently to red and blue light, as in the action spectrum of photosynthesis (McCree 1972). It has been shown that in rice seedlings, high-intensity red but not blue light induced retrograde signalling to repress LHCB and GOLDEN2-LIKE (GLK1) expression (Duan et al. 2020), indicating that red and blue light, while both efficiently exciting the photosynthetic photochemical reactions, can have a different role in inducing retrograde signalling.
Light reactions of photosynthesis under narrow-band light
Light reactions of photosynthesis are the processes that are needed to capture light energy and use it to synthesise ATP and NADPH which are then used to make carbohydrates from CO2 (Allen et al. 2011) and mechanisms that protect the PSA from excess light energy (Fig. 1). When the substantively unnatural narrow-band LED light is used for growing plants, it leads to changes in the PSA on many levels. These include changes in the content and ratio of photosynthetic pigments, pigment-protein complexes of thylakoid membranes and their interaction, thylakoid membrane architecture, functional parameters of PSI and PSII and electron transport activity. Plants grown with such narrow-band light have altered capability for photoprotection and plasticity in adapting to the changing environment.
Effects on photosynthetic pigment biosynthesis, content and pigment-protein complexes
Light regulation of chlorophyll biosynthesis
Light plays a significant role in the regulation of Chl biosynthesis (Fig. 2). The expression of most Chl biosynthesis genes (~ 2/3, including some isoforms of protochlorophyllide oxidoreductase (POR), a key enzyme in Chl biosynthesis) is induced by white light (Yuan et al. 2017). The mechanism of this induction involves the COP1–HY5 module and PIFs (Yuan et al. 2017), the components of signalling chains of three groups of photoreceptors — phys (red light), crys (blue/UV-A light) and UVR8 (UV-B light) (Fig. 2; Yadav et al. 2020), which can all react to components of white light. phyA has been shown to take part in the regulation of POR synthesis (Sineshchekov and Belyaeva 2019). The photoreceptor ZTL, acting via regulation of circadian clock components, also controls the biosynthesis of Chls at the transcriptional level (Fig. 2; Yuan et al. 2017). Light also plays a key role in the regulation of Chl biosynthesis at the translational and posttranslational levels (Yuan et al. 2017). If the highly orchestrated synthesis of Chls and their interaction with Chl-binding proteins is disrupted, phototoxic tetrapyrrole intermediates and unbound Chls may accumulate, leading to ROS production (Yuan et al. 2017; Solymosi and Mysliwa-Kurdziel 2021).
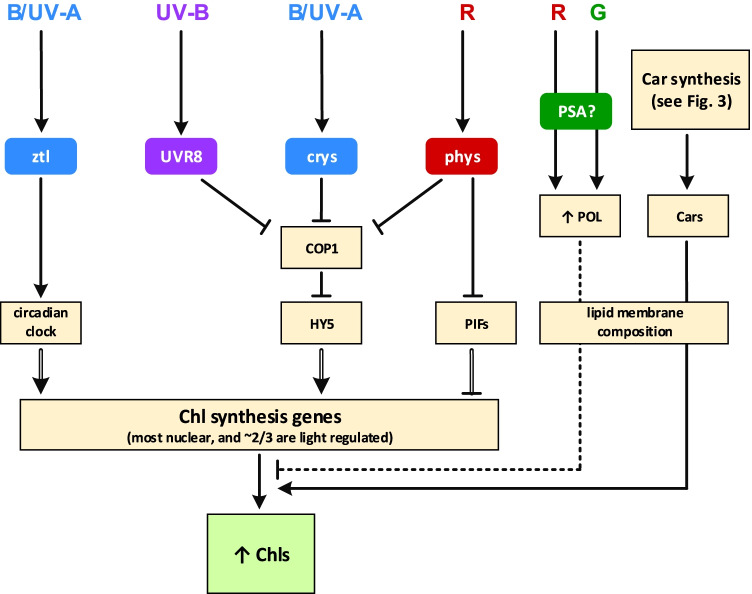
Fig. 2.
Photoregulation of chlorophyll (Chl) biosynthesis by blue (B), red (R), green (G), ultraviolet A (UV-A) and B (UV-B) light. Light-activated UV-B receptor (UVR8), cryptochromes (crys) and phytochromes (phys) inhibit COP1-based E3-ubiquitin-ligase and thus rescue HY5 transcription factor from proteolysis. Light-activated phys also inhibit PIF transcription factors. HY5 induces and PIFs repress Chl biosynthesis-related nuclear genes. Photoreceptor Zeitlupe (ztl) controls some of these genes via regulation of circadian clock. Chl biosynthesis depends on local lipid membrane composition. R and G promote lipid peroxidation (POL) in thylakoid membranes, probably via imperfectly formed PSA, which produces an excessive ROS amount. Dashed arrow — unknown mechanism, probably negative influence. Carotenoids (Cars) modulate Chl synthesis, because proper functioning of some Chl synthesis enzymes also needs the presence of carotenoids in their environment. Double line matches regulation on transcription level
Light is also required for the operation of the Chl biosynthesis pathway in plants. This pathway includes only one light-activated photochemical reaction — the reduction of protochlorophyllide to chlorophyllide, which is catalysed by POR. This step is the most well-studied and is crucial for conversion from dark-formed etioplast to chloroplast, possessing grana and mature PSA. POR enzyme reaction is strictly light-dependent in angiosperms. The light-absorbing chromophore is the substrate itself, protochlorophyllide, which accumulates in the dark as a part of a ternary complex with the enzyme POR and the second substrate NADPH. According to the absorption spectrum of protochlorophyllide, the most effective light for executing this photocatalytic reaction is blue and red light. Some angiosperms contain only one POR isoform, but several species contain two or more different isoforms, with different expression patterns and regulation (Sineshchekov and Belyaeva 2019; Solymosi and Mysliwa-Kurdziel 2021).
Light quality used for growing plants is known to affect photosynthetic pigment content. Red light, as compared to blue and white, decreased Chl and Car content in leaves of rice (Hamdani et al. 2019), cucumber (Su et al. 2014) and barley (Kochetova et al. 2018). The Chl a/b ratio did not change in barley, decreased in cucumber and increased in Arabidopsis, in all three ecotypes tested (Yavari et al. 2021). However, in tomato, no notable changes of the photosynthetic pigment content in red light were observed, as compared to green and blue. Green light notably decreased pigment content in tomato leaves (Trojak and Skowron 2021). Acclimation of Arabidopsis plants to green light also led to a decrease in total Chl and Car content, as well as a decrease in the Chl a/b ratio (Karlický et al. 2021). In blue light, total chlorophyll content decreased in cucumber (Su et al. 2014) and barley (Kochetova et al. 2018) and increased in tomato (Trojak and Skowron 2021). In rice (Hamdani et al. 2019) and barley (Kochetova et al. 2018) seedlings, blue light, as compared to white light, increased the Chl a/b ratio, which can coincide with decreased PSII antenna size and PSI/PSII ratio (Brestic et al. 2015), as well as with PSII heterogeneity (Mehta et al. 2010).
Blue light stimulates the development of “sun-type” leaves and chloroplasts, which show high photosynthetic activity and high Chl a/b ratio (Lichtenthaler et al. 2007; Zivcak et al. 2014). Using different combinations of red and blue LEDs in light sources has shown that blue light is essential to the organisation and activity of the PSA. In works with spinach (Matsuda et al. 2007), cucumber (Hogewoning et al. 2010), lettuce (Wang et al. 2016) and Mesembryanthemum crystallinum (He et al. 2017), the lowest Chl content and Chl a/b ratios were observed in pure red light. Adding even a small amount of blue light (7% for cucumber, 10% for spinach, ~ 8% for lettuce, 10% for Mesembryanthemum crystallinum) increased these parameters, and no significant change was observed upon further increase of the proportion of blue quanta. In spinach leaves, growing with 10% blue light increased LHCII content per leaf area. In Chinese cabbage grown with a red to blue ratio of 7:1 (Avercheva et al. 2009), no differences in the pigment content were found as compared to white light (high pressure sodium-vapour lamp); however, a higher Chl a/b ratio in plants grown under LEDs indicated a different proportion of functional complexes in thylakoid membranes.
An unusual effect of green light on Chl, described in Karlický et al. (2021), is that it led to accumulation of Chl containing geranylgeranyl instead of phytol (up to 50% of total Chl content), due to an incomplete hydrogenation of phytyl chains. Such Chls have been found in all major pigment-protein complexes, but preferentially in LHCII, and this hindered the formation of PSII and PSI supercomplexes and their ordered macro-arrays in the thylakoid membranes and increased the fraction of free LHCIIs. Thermal stability of LHCII trimers decreased in these conditions, which facilitates decomposition of trimers to monomers (Karlický et al. 2021). Instability of geranylgeranyl-Chl-containing PSI and especially PSII in green light apparently led to an increased probability of photodamage to reaction centre core complexes and to the decrease in the proportion of core complexes to LHCII. Incomplete hydrogenation of Chl phytyl chains was probably caused by reduced activity of geranylgeranyl reductase (GGR). Similar problems can be caused by impaired import of this enzyme into chloroplasts in high light or when the association of the enzyme with the membrane is disrupted. As GGR reduces geranylgeranyl to phytyl, not only for Chl synthesis but tocopherol and phylloquinone as well (Karlický et al. 2021), the reduced activity of this enzyme may also lead to reduced ROS scavenging by tocopherol in thylakoid membranes and to reduced activity of PSI due to phylloquinone deficit.
Another factor affecting Chl biosynthesis and function is the lipid composition of thylakoid membranes. Many enzymes of the Chl biosynthesis pathway depend on their lipid environment (Solymosi and Mysliwa-Kurdziel 2021). Lipids surrounding the LHCII trimers are important in determining their functions (Akhtar et al. 2019). Trojak and Skowron (2021) have shown that green and red light activate lipid peroxidation, which can affect the properties of LHCII by altering their lipid-protein interactions and lead to changes in Chl accumulation (Fig. 2). However, Karlický et al. (2021) show that decreased thermostability of LHCII in green light was caused by the presence of geranylgeranyl-Chl and not changes in lipid composition. Proper functioning of some of Chl synthesis enzymes, including POR, also needs the presence of Cars in their environment (Solymosi and Mysliwa-Kurdziel 2021), and Cars may thus modulate Chl biosynthesis as well (Fig. 2).
Light regulation of carotenoid biosynthesis
Light-dependent control of Car biosynthesis in plants is mediated by photoreceptors as well as by the photosynthetic electron transport chain (Fig. 3). Light-activated cryptochromes and phytochromes, acting via the COP1–HY5 signalling module, induce the expression of phytoene synthase (PSY; Fig. 3) — the first and main rate-determining enzyme in the Car biosynthesis pathway. Phytochromes also induce this enzyme by inhibiting PIF1, which represses the phytoene synthase expression in the dark or deep green shade (Fig. 3). Interestingly, both positive HY5 and negative PIF1 transcription regulators bind to the same element of the promoter, which serves as a relatively simple switch for inducing Car synthesis and accumulation upon illumination similar to direct sunlight, rich in blue and red. UV-B light, possibly at least partially mediated by the photoreceptor UVR8 acting via the same COP1–HY5 module, also induces Car synthesis, especially of zeaxanthin — a major Car for photodamage protection in the PSA (Fig. 3; Stanley and Yuan 2019). The final reactions of polyisoprenoid desaturation — from phytoene to phytofluene and from phytofluene to lycopene — are regulated by the photosynthetic electron transport chain, where oxidised plastoquinone serves as an oxidising agent in these reactions (Fig. 3; Norris et al. 1995; Ruiz-Sola and Rodríguez-Concepción 2012). Because of this, light quality that affects the PSII/PSI ratio or the activity of photosystems (Miao et al. 2016) can alter the redox state of the plastoquinone pool and, as a consequence, the biosynthesis of lycopene, the precursor of all photosynthetic Cars. High-intensity light differently modulates the expression of the enzymes of different branches and steps in Car biosynthetic pathways, leading to preferential accumulation of zeaxanthin (Fig. 3). However, mechanisms responsible for these gene expression changes remain unknown (Stanley and Yuan 2019).
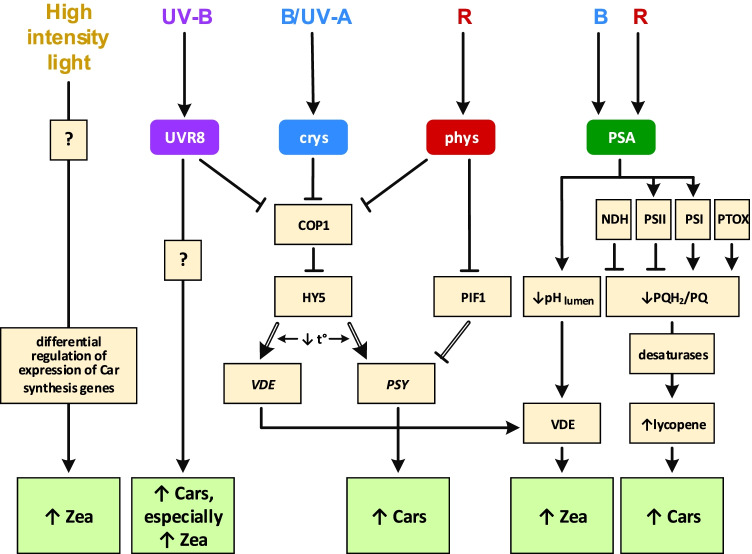
Fig. 3.
Photoregulation of carotenoid (Car) biosynthesis by blue (B), red (R), ultraviolet A (UV-A) and B (UV-B) light. Light-activated UV-B receptor (UVR8), cryptochromes (crys) and phytochromes (phys) inhibit COP1-based E3-ubiquitin-ligase and thus rescue HY5 transcription factor from proteolysis. Light-activated phys also inhibit PIF1 transcription factor. Phytoene synthase (PSY), the first and main rate-determining enzyme in the carotenoid biosynthesis pathway, is regulated by HY5 and PIF1 on the transcription level. Violaxanthin de-epoxidase (VDE) transcription is activated by HY5 as well. Low temperature increases HY5 binding with VDE and PSY promoters (Stanley and Yuan 2019). Light-dependent photosynthetic electron transport chain (ETC) decreases pH in lumen, increasing VDE activity. In addition, ETC differently changes the balance of reduced and oxidised plastoquinones (PQs): photosystem II (PSII) and plastidial NADH dehydrogenase (NDH) reduce PQ, and photosystem I (PSI) and plastidial terminal oxidase (PTOX) oxidise PQH2. Desaturases (two desaturases, phytoene desaturase and ζ-carotene desaturase, catalyse four desaturations from phytoene to lycopene) use oxidised plastoquinone as an electron acceptor for the desaturation reactions (Ruiz-Sola and Rodríguez-Concepción 2012). Zeaxanthin (Zea) is a major Car for photodamage protection in the photosynthetic apparatus. Zea level depends both on the activity of β-carotene hydroxylase, which converts β-carotene to zeaxanthin in the common Car biosynthetic pathway, and on the activity of VDE, which converts violaxanthin back to zeaxanthin (Stanley and Yuan 2019). Light quality regulates the expression of both these enzymes, and light, via lumen acidification, activates VDE post-translationally. Gene names in italics and double lines — regulation at transcription level, regular — regulation at post-translation level — protein level and/or activity
Zeaxanthin level in the leaf depends both on the activity of β-carotene hydroxylase, which converts β-carotene to zeaxanthin, and the activity of violaxanthin de-epoxidase (VDE), which converts violaxanthin back to zeaxanthin (Stanley and Yuan 2019). Light quality regulates the expression of both these enzymes: the amount of β-carotene hydroxylase increased in blue light (Fig. 3; Tran et al. 2021), and the amount of VDE increased in the series green < white < red < blue (Fig. 4; Trojak and Skowron 2021). The photosynthetic electron transport chain also post-translationally activates VDE, thus increasing zeaxanthin content (Fig. 3; Demmig-Adams 1990; we describe this in detail in the section “Photoprotection in narrow-band light”).
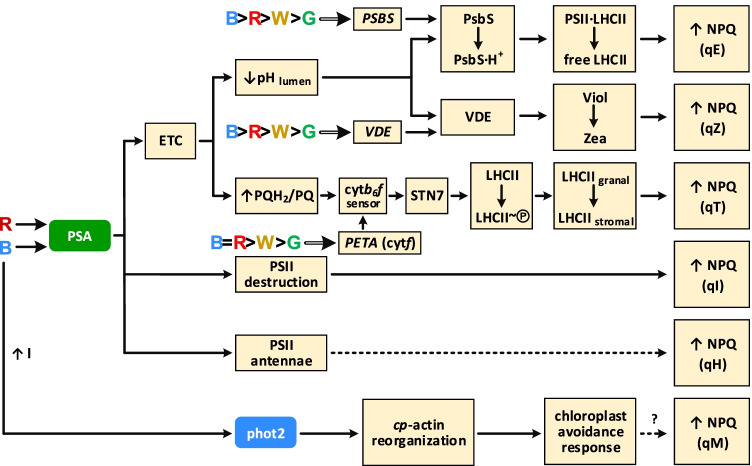
Fig. 4.
Light-dependent components of non-photochemical quenching (NPQ) of chlorophyll a fluorescence. Photoregulation of NPQ by blue (B), red (R), green (G) and white (W) light. Photosynthetic electron transport chain (ETC) affects NPQ via lumen acidification (↓pHlumen) and via change of plastoquinone redox state (↑PQH2/PQ). Lumen acidification promotes PsbS protein protonation following disconnection of LHCII trimers from PSII and energy dissipation — this is the qE component of NPQ. Lumen acidification also promotes protonation and activation of the violaxanthin de-epoxidase (VDE) following violaxanthin (Viol) to zeaxanthin (Zea) conversion and energy dissipation by Zea — this is the qZ component of NPQ. Reduced plastoquinones via cytochrome b6f complex activate the serine/threonine kinase STN7, which phosphorylates the mobile antenna LHCII and thus induces its dissociation from PSII and relocation from granal to stromal thylakoids, reducing energy transfer to PSII — this is the qT component of NPQ. Light-induced destruction of PSII (primarily D1 protein) is responsible for the photoinhibition component qI. The qH component of NPQ is connected with the PSII antenna, and its mechanism is yet to be elucidated (dashed arrow). High-intensity (↑I) blue light induces chloroplast avoidance response, decreasing light absorption by Chls. However, the contribution of this process to NPQ and the existence of qM component are questionable (dashed arrow). Several protein effectors of NPQ (PSBS, VDE and PETA for cytf subunit of cytochrome b6f complex) are regulated by light at the transcriptional level, and the light of different quality induces these genes differently (shown based on data from Trojak and Skowron 2021). The mechanism of light regulation of VDE expression is shown in Fig. 3. Gene names in italics and double lines — regulation at transcription level, regular — regulation at post-translation level — protein level and/or activity
Although different spectral bands of light can induce Car biosynthesis via their respective photoreceptors, their final effect on Car content in plant leaves is not the same. Red light, as compared to white light, decreased Car content in leaves of cucumber (Miao et al. 2016), rice (Hamdani et al. 2019), barley (Kochetova et al. 2018) and Arabidopsis (Yavari et al. 2021). Blue light increased Car content in Mesembryanthemum crystallinum (He et al. 2017) and in Arabidopsis (Yavari et al. 2021). This can increase the photoprotection capacity of plants grown with blue light. In cucumber plants, Car content per unit leaf area increased when blue light was added to red light (up to 50%) (Hogewoning et al. 2010). However, barley seedlings showed a decrease in Car content in blue light as compared to white light (Kochetova et al. 2018). In tomato, Car content in red light was the same as in blue light but higher than in green light (Trojak and Skowron 2021).
Reaction centres and electron transport chain in narrow-band light
The function of the PSA in vivo is usually studied with variable Chl fluorescence (Maxwell and Johnson 2000; Baker 2008; Murchie and Lawson 2013; Goltsev et al. 2016). It is also widely used to characterise photosynthesis of plants grown with narrow-band light.
Plants grown with narrow-band blue light usually show a more functional PSA than in red and green light, as reflected by Chl fluorescence parameters (Hogewoning, et al. 2010; Savvides et al. 2012; Miao et al. 2016; He et al. 2017; Tantharapornrerk et al. 2021; Tran et al. 2021; Trojak and Skowron 2021; Kochetova et al. 2022). In most cases, in plants grown with red light, parameters of functional activity of PSII and electron transport chain — Fv/Fm, ΦPSII and qP — were lower than in plants grown with white light or did not differ from them (cherry tomato — Xiaoying et al. 2012; cucumber — Su et al. 2014; rice — Hamdani et al, 2019; rice — Tran et al. 2021; tomato — Trojak and Skowron 2021; barley — Kochetova et al. 2022). In blue light, these parameters were higher or the same as in white light (cherry tomato — Xiaoying et al. 2012; cucumber — Su et al. 2014; rice — Tran et al. 2021; tomato — Trojak and Skowron 2021; barley — Kochetova et al. 2022). However, in other cases, blue light decreased Fv/Fm and ΦPSII (rice — Hamdani et al. 2019).
When different amounts of blue light were added to red light, Fv/Fm and ΦPSII increased until saturation: Fv/Fm was saturated at 7% of blue light and ΦPSII was saturated at 22% blue light (cucumber — Hogewoning et al. 2010). In a similar experiment, qP (reflecting the amount of open PSII reaction centres) saturated at 10% blue light, and ETR (electron transport rate) increased monotonously with increasing proportion of blue photons (Mesembryanthemum crystallinum — He et al. 2017). Thus, in these experiments, blue light increased the efficiency of photosynthesis. Chinese cabbage plants grown with red and blue LEDs with a ratio of 7:1 showed Fv/Fm and qP similar to that under white light (high pressure sodium lamps; Avercheva et al. 2009). This corresponded well with coupled electron transport rate in isolated chloroplasts, which also did not differ between red-blue light and white light (Avercheva et al. 2010).
Green light, as red light, usually decreases chlorophyll fluorescence parameters, which points to an impaired functionality of PSII and electron transport (cherry tomato — Xiaoying et al. 2012; lettuce — Muneer et al. 2014; cucumber — Su et al. 2014; rice — Tran et al. 2021; tomato — Trojak and Skowron 2021). Green light is also known to induce the accumulation of geranylgeranyl-Chl in pigment-protein complexes, which led to destabilisation of PSII and disrupted the connection between the PSII reaction centre and antenna Chls (as reflected by an increased F0), decreased Fv/Fm and increased PSII sensitivity to photoinhibition (Karlický et al. 2021).
Photoprotection in narrow-band light
Photoprotection includes dissipation of excess light energy as heat (Messant et al. 2021) and an antioxidant system (including antioxidant enzymes and low-molecular-weight antioxidants) that scavenges ROS which can be generated by the PSA when it cannot process all the absorbed energy into a useful product (Hamdani et al. 2019; Tran et al. 2021; Trojak and Skowron 2021). In greenhouses, plants are usually grown with moderate light intensities. However, narrow-band lighting creates unusual conditions for the photoreceptor system controlling PSA development, as well as the PSA itself. This kind of lighting may lead to an imbalanced pigment-protein complex ratio, which can require extra photoprotection. Photoprotective mechanisms which dissipate excess absorbed energy are often characterised by non-photochemical quenching (NPQ) of excited Chl in PSII (Goltsev et al. 2016), kinetics of its induction and relaxation.
NPQ consists of several components (shown in Fig. 4, boxes on the right). Most of them depend on processes in the PSA, which are activated most efficiently by blue and red light absorbed by photosynthetic pigments (Fig. 4; McCree 1972). qE, energy-dependent quenching, depends on the electrochemical proton gradient and acidification of the lumen, which in turn are the result of light reactions in the chloroplast electron transport chain (Fig. 4). Light-induced lumen acidification promotes the protonation of PsbS protein, a peripheral subunit of PSII. Protonated PsbS induces disconnection of LHCII trimers from PSII (Fig. 4), thus reducing energy transfer from LHCII to the reaction centre and increasing the thermal dissipation of this energy. These processes are induced in the first 1–3 min and relaxed in the first 1–3 min after dark. Lumen acidification also activates VDE. Its activity leads to zeaxanthin formation, which increases energy dissipation in PSII minor antennae. This component takes 10–15 min to induce and the same amount of time to relax. This part of qE has also been termed qZ (Fig. 4; Li et al. 2000; Dall’Osto et al. 2005; Kalituho et al. 2007; Johnson and Ruban 2009; Kereïche et al. 2010; Haniewicz et al. 2013; Sylak-Glassman et al. 2014; Ware et al. 2015; Kress and Jahns 2017; Demmig-Adams et al. 2020; Wang et al. 2020; Nicol and Croce 2021). Increased reduction of the plastoquinone pool in high light activates serine/threonine protein kinase STN7 (Bellafiore et al. 2005), which reversibly phosphorylates LHCII and leads to its dissociation from PSII and transfer from granae to stromal thylakoids (Fig. 4). This process, termed state transitions, reduces energy flow to PSII and leads to another, even slower NPQ component, qT (Fig. 4). The cytochrome b6f complex serves as the sensor for the redox state of the plastoquinone pool in this process (Chuartzman et al. 2008; Minagawa 2011; Grieco et al. 2012). NPQ can also occur due to photoinhibition of photosynthesis (qI), which usually involves photodamage to PSII (Fig. 4). Photoinhibition can also occur in dark-adapted plants right after the onset of illumination during photosynthesis induction (Malnoë 2018). qI is a slowly reversible process, requiring hours to relax.
qE, qZ, qT and qI are considered the most important mechanisms for protecting the plant from excess light. Apart from them, chloroplast avoidance movement can also contribute to NPQ, as it decreases light absorption by chloroplasts (Ptushenko et al. 2016). This component is termed qM and is activated by blue light via phototropins (Fig. 4). However, its contribution to NPQ requires re-evaluation (Wilson and Ruban 2020). Another, newly described NPQ component is qH (Fig. 4; Brooks et al. 2013; Malnoë 2018; Malnoë et al. 2018). It is the slowest NPQ component, connected with the PSII antenna, whose mechanism is yet to be elucidated.
Narrow-band LED light is known to affect the photoprotective capacity of plants, especially the faster NPQ components. In rice seedlings, blue light increased NPQ as compared to white light, and also increased the rate of initial NPQ induction (Hamdani et al. 2019). Increased NPQ amplitude and induction rate in blue light has also been observed in tomato (Trojak and Skowron 2021). This is probably due to the increased content of NPQ protein effectors: PsbS, PROTON GRADIENT REGULATION-LIKE1 (PGRL1), cytochrome b6f subunit f (cytf) and VDE (Fig. 4; Trojak and Skowron 2021). Increased PsbS content in blue light was also observed by Steen et al. (2020). Tran et al. (2021) have shown in rice seedlings that blue light not only increases NPQ and zeaxanthin content but also induces expression of genes for β-carotene hydroxylase (converts β-carotene to zeaxanthin) and VDE (Fig. 3), as well as anthocyanin accumulation. In barley plants (Kochetova et al. 2022) grown with blue light, NPQ had not just a higher amplitude but also different induction kinetics, as compared to red and white light. For the first 1–3 min, NPQ rapidly increased, and then decreased, not reaching a stationary state by 5.5 min after onset of illumination. In plants grown with red and white light, NPQ increased in the first 2–3 min, reaching a stationary state. He et al. (2017) have shown that when blue light is added to red light, NPQ increases monotonously with the increase in the proportion of blue quanta. Trojak and Skowron (2021) conclude that in plants grown in a combined red–green–blue light source, green light contributes to decreasing the amplitude of NPQ, and blue light increases the photosynthetic and photoprotective capacity of plants. Blue light appears to be beneficial to photoprotection mechanisms in most cases.
Red light also increased NPQ in rice seedlings (Hamdani et al. 2019). However, in Mesembryanthemum crystallinum (He et al. 2017) and cucumber (Miao et al. 2016), red light decreased NPQ. Tomatoes also showed lower NPQ in red light, as well as different induction kinetics (Trojak and Skowron 2021). This appears to be a more widespread reaction of NPQ to red light. Green light has been shown to decrease NPQ (tomato — Trojak and Skowron 2021). This decrease is probably due to a decrease in the amount of NPQ protein effectors. In Arabidopsis, accumulation of geranylgeranyl-Chl in green light led to a substantial increase of PSII susceptibility to photoinhibition (the rapid phase of NPQ relaxation was impaired). The mechanism of NPQ in plants grown with white and green light apparently has a different nature. In green light–grown plants, when LHCII dissociates from trimers to monomers, the ability to rapidly dissipate excess energy in free LHCII increases (Karlický et al. 2021).
Not just the overall NPQ but also its separate components can be affected differently by narrow-band light. In rice (Hamdani et al 2019), the most rapid qE phase, regulated by the transthylakoid proton gradient but independent of violaxanthin de-epoxidation to zeaxanthin (Fig. 4; Johnson et al. 2008), was faster in both red and blue light as compared to white light. The middle qZ phase, dependent on zeaxanthin accumulation and its association with minor proteins of the PSII antenna (CP26 and CP29) (Fig. 4; Dall’Osto et al. 2005; Nilkens et al. 2010), was also higher both in blue and red light. The slowest phase (200–600 s) had the highest amplitude in blue light (22.5% of white light), then in red light (6% of white light), which probably led to a decreased ΦPSII observed at this time (Hamdani et al. 2019).
Effects of light spectrum on the ATP synthase system
ATP synthesis in chloroplasts is carried out by the ATP synthase, a multi-subunit protein complex. It is integrated into the thylakoid membrane and has two components: F0, the lipid-soluble integral complex, and F1, the water-soluble peripheral complex. F0 consists of four types of subunits (I, II, III, IV14) and transfers protons through the thylakoid membrane. F1 is made up of five types of subunits (α3, β3, γ, δ, ε) and contains catalytic sites for reversible ATP synthesis (Walker 2013). Genes encoding subunits of the complex are located both in the nuclear and plastid genomes. The plastid genes of ATP synthase subunits are organised into two operons, large, AtpI/H/F/A (genes of IV, III, I, α subunits), and small, AtpB/E (genes of β and ε subunits, respectively). Genes for γ, δ, and II subunits (ATPC, ATPG and ATPD) are nuclear-encoded; the ATPC gene has been shown to have two copies in dicots (Inohara et al. 1991) and monocots (barley — Mascher et al. 2021). Because two genomes are involved in encoding ATP synthase genes, and because of a complex stoichiometry of subunits, the ATP synthase requires a strict mechanism that regulates its assembly. Thus, it has been shown that the β-subunit activates translation of α-subunit mRNA, and αβ heterodimers bind β-subunit mRNA, inhibiting its translation. Re-activation of translation occurs in the presence of γ-subunit proteins that are imported into the chloroplast stroma from the cytosol. The γ-subunit helps form (αβ)3-hexamers and releases the repression of β-subunit mRNA translation (Drapier et al. 2007).
Photoreceptors affect the chloroplast ATP synthase mainly on the level of gene expression. Phytochromes and cryptochromes regulate the expression of nuclear genes whose products take part in transcription, translation and post-translational regulation of plastid-encoded genes (Griffin et al. 2020). Genes of subunits encoded in the chloroplast genome are transcribed by the plastid-encoded RNA-polymerase (PEP) containing σ-factors which recognize their respective promoters (Ghulam et al. 2012). The expression of all σ-factor genes is activated by phytochromes and cryptochromes in red and blue light. By this mechanism, light induces the expression of plastid-encoded PEP-transcribed genes, including genes of ATP synthase subunits (Lysenko 2007; Ghulam et al. 2012; Chi et al. 2015). It has been shown that phyB increases the expression of IV, III and I ATP synthase subunits in plastids (Griffin et al. 2020). Light also takes part in the post-transcriptional regulation of plastid gene expression (Deng et al. 1989). It has been shown to affect AtpB/E-mRNA stability in spinach cotyledons. In red light, the AtpB/E-mRNA level was 50% of that in yellow and white light, while the transcriptional activity of the AtpB/E operon was similar in light of different spectrum.
Light also activates the expression of nuclear genes encoding ATP synthase subunits. In spinach, the promoter of the ATPC gene, encoding the γ-subunit, contains regulatory elements responsible for light-dependent expression (Bolle et al. 1996); illumination with red light (650 nm) led to a substantial increase of ATPC1 transcription in Arabidopsis (Yavari et al. 2021).
ATP synthase is also regulated by light on post-translational level. Differences in light intensity can lead to post-translational modification of ATP synthase proteins, which affects the rate of ATP synthesis in chloroplasts (Romanowska et al. 2008; Reiland et al. 2009; Schonberg and Baginsky 2012). The PSA itself, as a sensor of light, can induce post-translational modifications of ATP synthase subunits. When the chloroplast electron transport chain is working, it reduces stroma components, including thioredoxin. Reduced thioredoxin reduces γ-subunit thiol groups and thus activates ATP synthase (reviewed in Kang et al. 2019). Chloroplasts also have their own kinase system whose activity depends on the redox state of the plastoquinone pool and stromal redox components (Schonberg and Baginsky 2012). This kinase system affects a wide range of targets, among them σ-factors of the plastid RNA polymerase (Reiland et al. 2009; Schweer et al. 2010) and ATP synthase subunits α, β, and γ (Reiland et al. 2009). These subunits have phosphorylation sites for chloroplast kinases which are important for regulating ATP synthase activity upon changes in light intensity (Reiland et al. 2009; Schonberg and Baginsky 2012). As the PSA absorbs and utilises quanta with different efficiency, depending on their position in the visible spectrum (McCree 1972), it is reasonable to expect that light of different wavelengths will affect ATP synthesis differently.
Our results support the idea that photophosphorylation in chloroplasts is regulated by light intensity and spectrum. Thus, in Chinese cabbage plants grown with light sources made with red and blue LEDs with a ratio of 7:1, photophosphorylation rate in isolated chloroplasts decreased with an increase in light intensity. The opposite was observed in control plants grown with high-pressure sodium lamps (Avercheva et al. 2010). However, the details of ATP synthesis regulation by light spectrum remain to be investigated.
CO2 assimilation and carbon fixation reactions
The carbon fixation reactions use light energy stored in ATP and NADPH to fix CO2 and synthesise carbohydrates (Fig. 1). CO2 assimilation rate can be viewed as an integral characteristic of photosynthesis rate, incorporating rate of electron transport, Rubisco and Calvin cycle activity and triose phosphate utilisation (Busch and Sage 2017).
In most cases, blue light increases (or does not change) the net assimilation rate and maximum assimilation rate as compared to broad-spectrum white light (cucumber — Wang et al. 2009, tomato — Xiaoying et al. 2012; cucumber — Su et al. 2014, lettuce — Tarakanov et al. 2022), and growing with red light decreases these parameters as compared to white or blue light (Bukhov et al. 1995; Wang et al. 2009; Xiaoying et al. 2012; Su et al. 2014; Tarakanov et al. 2022). The same pattern was observed for starch, sucrose and soluble sugar content (Wang et al. 2009; Xiaoying et al. 2012; Hu et al. 2016) and Rubisco activity in vitro (Wang et al. 2009). Rubisco content, however, was the same in plants grown with blue, red and white light (Su et al. 2014). Other works, on the contrary, showed different patterns. Net photosynthetic rate can be higher in red light than in blue light (lettuce — Amoozgar et al. 2017; Arabidopsis — Yavari et al. 2021); the same pattern for soluble sugar content was also observed (lettuce — Chen et al. 2014). Net photosynthesis has also been observed to be lower in blue light than in white light (Hu et al. 2016). It has been shown for various species that low net photosynthetic assimilation under red light can be increased by adding blue light to the spectrum. In some cases, a small amount of blue quanta in addition to red significantly increased photosynthetic assimilation, and further increase in the amount of blue quanta did not increase this parameter (spinach — Matsuda et al. 2007, Mesembryanthemum crystallinum — He et al. 2017). In other cases, the increase in assimilation correlated well with the amount of blue quanta in the spectrum (cucumber — Hogewoning et al. 2010, Brassica alboglabra — He et al. 2015). For cucumber, this correlation was observed up to 50% blue light, for B. alboglabra — up to 16% blue light; at higher amounts of blue quanta, a decline in assimilation was observed in both cases.
The Calvin cycle is activated by thioredoxin which, in chloroplasts, is reduced primarily by ferredoxin and may also be reduced by NADPH (Kang et al. 2019), both of which are products of the photosynthetic light reactions. Chls and Cars, light-harvesting pigments in higher plants, absorb mainly red and blue light; Chl can also absorb green light to some extent, which can also activate light reactions (McCree 1972). From the action spectrum of photosynthesis, we can expect the highest activity of Calvin cycle enzymes under red and blue light, with slightly lower activity under green light.
The expression of RbcL, the gene for the large Rubisco subunit (as well as RCA, gene for Rubisco activase), is increased in blue light (cucumber — Wang et al. 2009; Su et al. 2014). For red light, the data are less certain. In different experiments, red light can decrease or not affect RCA expression and decrease or increase RbcL expression. Blue light has the same ambiguous effect on RBCS expression (gene for the small Rubisco subunit), while red light tends to decrease its expression (Wang et al. 2009; Su et al. 2014). Light spectrum affects the expression of genes for Rubisco subunits using the whole network of plant photoreceptors. It has been shown for the large Rubisco subunit that cryptochromes and phototropins either do not affect its amount in the leaf or are completely interchangeable (Weston et al. 2000). The same was shown for phototropins and RBCS (Lopez-Juez et al. 2007). The authors suggest that chloroplast-derived signals play a crucial role in the regulation of Rubisco subunit gene expression. However, phyA has also been shown to take part in the regulation of RbcL transcription, among other photosynthetic genes (plastid-encoded), in red as well as blue/UV-A light (Chun et al. 2001).
Genes of most Calvin cycle enzymes (apart from Rubisco) decrease their expression (as measured with RT-PCR) in red light as compared to broad-spectrum white light (Wang et al. 2009). Blue light increases the expression of Calvin cycle genes, except for triose-phosphate isomerase (TPI) and sedoheptulose bisphosphatase (SBPase) (Wang et al. 2009). The expression of the TPI gene in blue light is lower or equal to the control. The effect on the expression of the SBPase gene depends on the wavelength of blue light. A shorter wavelength (394.6 nm) decreased its expression, and a longer wavelength (452.5 nm) increased it (Wang et al. 2009).
Studies of carbon isotope ratios have shown that blue light increases the fraction of 12C and red light increases the fraction of 13C (Tarakanov et al. 2022). This indicates that blue light activates the Calvin cycle while red light activates photorespiration. The presence of both red and blue in the light spectrum evens out these effects.
Overall, we can conclude that blue light is favourable and red light is unfavourable for the biosynthesis of carbon assimilation enzymes, which corresponds well with data on sugar content. This is probably because cryptochrome (cry2 as well as cry1) enhances the expression of multiple photosynthetic genes, including some Calvin cycle enzymes (reviewed in Mishra and Khurana 2017). However, for some genes, the spectrum-dependent regulation of their expression can be ambiguous and depend on other factors. Light that combines red and blue light would probably be more favourable to carbon metabolism than sole red or blue light (Matsuda et al. 2007; Hogewoning et al. 2010; Savvides et al. 2012; He et al. 2015, 2017; Hu et al. 2016).
Stomata
Stomata are crucial for photosynthesis as they play a fundamental role in gas exchange regulation in plants. Gas exchange includes CO2 uptake for photoassimilation and transpiration — water vapour outward diffusion (Fig. 1). Transpiration is crucial for water balance, water and nutrient transport in plants and thermoregulation — leaf cooling, necessary to prevent overheating which can result in photoinhibition and yield penalties (von Groll et al. 2002; Hetherington and Woodward 2003; Wei et al. 2020). Stomata optimise plant gas exchange to maximise the photosynthetic rate, while avoiding starvation for CO2 or excessive water loss and drought stress. Both guard cell development and mature stomata function are regulated by many external and internal environmental factors: light, CO2, temperature, air humidity, water status, hormones and sugars play an important role (Driesen et al. 2020). One of the most important signals is lighting conditions. Long-term lighting conditions during leaf development determine mutual patterning of stomata and stomatal density (Croxdale 1998; von Groll et al. 2002; Wei et al. 2020), while current, local lighting conditions determine optimal stomatal opening (Hetherington and Woodward 2003; Driesen et al. 2020).
Light regulation of stomatal development
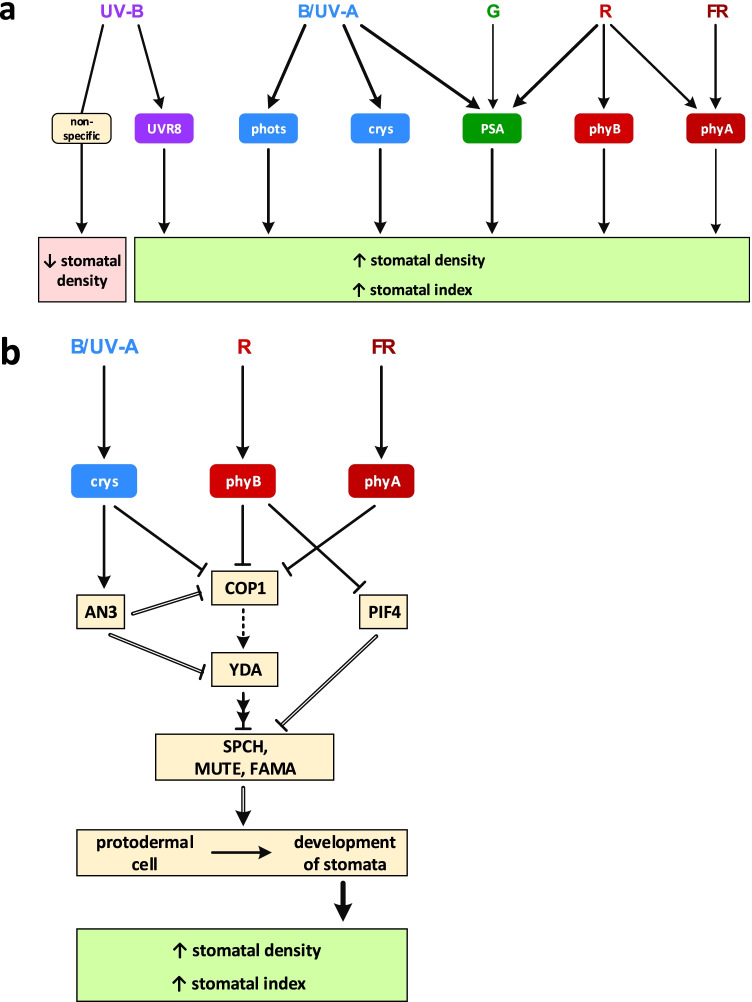
Stomata initiation and development in the leaf epidermis are affected by endogenous as well as exogenous factors, such as location on the leaf (upper or lower side, middle or edge of the leaf plate) (Croxdale 1998); phytohormones, CO2 and O2 concentrations, temperature, relative humidity, water deficit, lighting regime and intensity (Croxdale 1998; von Groll et al. 2002; Hetherington and Woodward 2003; Driesen et al. 2020); interaction with surrounding epidermal cells (Serna and Fenoll 2000) and mesophyll cells (Hetherington and Woodward 2003; Wei et al. 2020). Increasing light intensity to a certain level during leaf development increases stomata number, which enables sufficient gas exchange for a high photosynthetic rate (Driesen et al. 2020; Wei et al. 2020). In this regard, not only the irradiance illuminating the leaf directly, but also irradiance detected by phyB in mature leaves via a systemic signal has a significant impact on the stomatal index of developing leaves (Driesen et al. 2020). It has been shown that dark-grown Arabidopsis plants have only few mature stomata on the cotyledons, forming occasional stomatal clusters, and many of them are retained in the stomatal precursor stage (Wei et al. 2020).
All major spectral bands of white light take part in regulation of stomatal density and stomatal index (Fig. 5a). Red light, perceived by phyB, via PIF4 increases stomatal index and stomatal density (Fig. 5b; Arabidopsis — Boccalandro et al. 2009; Casson et al. 2009). Blue light via crys and far red light via phyA also increase stomatal index. phyA, phyB and crys work together to promote light-induced stomatal development via COP1, YODA (YDA) and transcription factors (SPEECHLESS (SPCH), MUTE and FAMA — closely related bHLH transcription factors, which regulate consecutive steps in the differentiation pathway of stomata). This pathway is critical for the development of stomata from the protodermal cell (Fig. 5b; Boccalandro et al. 2009; Kang et al. 2009). YDA is a MAPK kinase kinase, heading MAPK kinase cascade. It might be positively regulated by E3-ubiquitin ligase COP1 through yet unknown mechanisms and inhibits downstream transcription factors probably via their targeting by phosphorylation (Casson et al. 2009). Blue light via phototropins also increases stomatal density, and both phototropins work additively (Fig. 5a; Arabidopsis — Rusaczonek et al. 2021). In response to UV-B, stomatal density is increased by a UVR8-dependent mechanism but decreased in a UVR8-independent manner (Fig. 5a; Arabidopsis — Gustavsson 2018). Supplementary UV-B exposure (together with WL) via UVR8 increases stomatal index (Arabidopsis — Wargent et al. 2009). Photosynthesis in underlying mesophyll, driven mostly by red and blue light, regulates stomatal index, and stomatal density (Fig. 5a; Hronková et al. 2015).When plants are grown with light of different spectrum and the same intensity, blue LED light usually yields higher stomatal density in the developing epidermis than red LED light (cucumber — Wang et al. 2009; cucumber — Hogewoning et al. 2010; tomato — Liu et al. 2011a, b; lettuce — Muneer et al. 2014; Morus alba — Hu et al. 2016; barley — Kochetova et al. 2022).
Fig. 5.
Photoregulation of stomatal development which determines protodermal cell divisions and resulting stomatal index and stomatal density at epidermis by ultraviolet A (UV-A) and B (UV-B), blue (B), green (G), red (R), and far red (FR) light. a Light spectral ranges and the groups of photoreceptors that participate in the regulation of stomatal pattern. UV-B light acts via UVR8 photoreceptor and via nonspecific absorption by various cell components, regulating stomatal development in an opposite manner. Blue light via phots and crys, red light via phyB and far red light via phyA increase stomatal index. Photosynthetic apparatus (PSA) in underlying mesophyll regulates stomatal index and stomatal density via photosynthetic products and/or carbon dioxide concentration changes. b Key components of the most well-studied signalling pathways involved in photoregulation of stomatal development. Photoreceptors phyA, phyB and crys (cry1/2) act in concert to promote light-induced stomatal development via COP1 (E3-ubiquitin ligase), YDA (MAPK kinase kinase, heading MAPK kinase cascade) and transcription factors (SPCH, MUTE and FAMA — closely related bHLH transcription factors, which regulate consecutive steps in the differentiation pathway of stomata), critical for the development of stomata from the protodermal cell. YDA might be positively regulated by COP1 through yet unknown mechanisms (dashed arrow) and inhibits downstream transcription factors probably via their targeting by phosphorylation. PIF4 transcription factor represses SPCH expression, and phyB, inhibiting PIF4, derepresses SPCH, increasing stomatal development. Besides, light promotes ANGUSTIFOLIA3 (AN3) at transcriptional and post-translational levels, likely through the photoreceptor-mediated pathways (crys and probably phys as well), which then downregulates the expression of COP1 and YDA (Wei et al. 2020). Light-green rectangular matches promoting, and pink rectangular matches restricting conditions for photosynthesis. Double line matches regulation on transcription level
Stomata are distributed non-randomly in the plane of the epidermis: their distribution and density are strictly controlled (Croxdale 1998; von Groll et al. 2002; Wei et al. 2020). Stomatal density (the number of stomata per unit area) and stomatal index (the number of stomata per total number of epidermal cells (pavement and stomatal)) may change differently, depending on both the number and size of pavement cells (Casson et al. 2009; Wargent et al. 2009). This is achieved by regulating the number of cells entering the stomatal pathway, the number and the correct placement of unequal cell divisions during the development of stomatal complex (Serna and Fenoll 2000), when necessary — arrested differentiation of some stomatal precursors (Croxdale 1998; Kang et al. 2009; Driesen et al. 2020). This leads to a more orderly and even distribution of stomata, which must be separated by a number (at least one) of non-stomatal cells (Serna and Fenoll 2000; von Groll et al. 2002; Driesen et al. 2020). Thus, stomatal initiation is an imperfect mechanism, but stomatal development is strictly controlled and adjusted on later stages (Croxdale 1998; Driesen et al. 2020; Wei et al. 2020).
Light regulation of stomatal function
Photoregulation of stomatal movement involves most known photoreceptors, as well as the PSA as a light-perceiving system (Fig. 6). Illumination with photosynthetically active radiation, which is most efficient in the red and blue range but also operates in the green range, opens stomata by launching photosynthesis in guard cells as well as in adjacent mesophyll cells (Fig. 6). Activating photosynthetic assimilation in the mesophyll leads to a decrease in intercellular CO2 concentration, and stomatal opening is triggered by this signal (Fig. 6; Vavasseur and Raghavendra 2005). When photosynthesis is activated in the guard cells themselves, it generates soluble sugars and energy necessary for stomatal movement (Schwartz and Zeiger 1984). Carbohydrates imported from the mesophyll can also help increase the osmotic pressure in guard cells, which is necessary for stomatal opening in response to light (Fig. 6; Shimazaki et al. 2007). Stomatal opening is also induced by photoreceptors via their specific signallings. These responses are much more sensitive and can be already saturated at a low fluence rate (5 to 10 µmol m–2 s–1 for blue light response) (Driesen et al. 2020). From the PAR range, blue light induces the strongest stomatal opening (Frechilla et al. 2000; Talbott et al. 2002), and this opening is mediated by phototropins through phosphorylation of downstream signalling kinases (Fig. 6). This leads to the activation of the plasma membrane H+-ATPase AHA2 and 14–3-3 proteins, opening of potential-driven inward K+-channels in plasma membrane and accumulation of K+, followed by anions and water in guard cells’ vacuoles (Fig. 6). Starch degradation provides malate and sucrose synthesis and accumulation in vacuoles (Fig. 6; Kinoshita et al. 2001; Shimazaki et al. 2007; Kostaki et al. 2020; Ishka 2022). Less pronounced opening in response to blue/UV-A light is mediated by cryptochromes via COP1 signalling (Fig. 6; Mao et al. 2005). Blue/UV-A light also acts via ZTL and its entrainment of circadian clock (Fig. 6), which forms circadian rhythms of PSA function and thus CO2 fixation and stomatal opening (Dodd et al. 2004). UV-B light of low fluence rate induces stomatal opening, with maximum efficiency at 284 nm. This response is nearly three times greater than the response in the blue region (450 nm) (Vicia faba – Eisinger et al. 2000) and is probably provided by the photoreceptor of UV-B UVR8 (Fig. 6). Phytochromes mediate stomatal opening in response to red light (Fig. 6; Paphiopedilum — Talbott et al. 2002; Pisum – Sokolskaya et al. 2003; Arabidopsis – Wang et al. 2010). Green light, given after dark, induces the smallest stomatal opening among all visible spectral bands (Vicia faba — Frechilla et al. 2000), and this opening is mediated by phytochromes (Fig. 6; Paphiopedilum — Talbott et al. 2002). In some conditions, light may induce partial stomatal closure. Green light reverses blue/UV-A-induced stomatal opening (Fig. 6; Vicia faba — Frechilla et al. 2000; Paphiopedilum — Talbott et al. 2002), far red light reverses opening induced by red light (Fig. 6; Pisum — Sokolskaya et al. 2003) and by green light (Fig. 6; Paphiopedilum — Talbott et al. 2002). High intensity-UV-B light closes stomata via UVR8 through the COP1–HY5 signalling pathway inducing production of ethylene (Fig. 6), as well as via a UVR8- and ethylene-independent non-specific pathway (Fig. 6). Both pathways converge on H2O2 and NO production (Arabidopsis — Tossi et al. 2014; Ge et al. 2020).
Fig. 6.
Photoregulation of stomatal opening which determines stomatal conductance and transpiration rate by ultraviolet A (UV-A) and B (UV-B), blue (B), green (G), red (R) and far-red (FR) light. Light spectral ranges, the groups of photoreceptors and key components in their signalling pathways regulating stomatal opening. UV-B light acts via UVR8 photoreceptor and via nonspecific absorption by various cell components, regulating stomatal opening in an opposite manner, depending on light intensity: high UV-B light (↑I) induces stomatal closure and low UV-B light (↓I) induces stomatal opening. UVR8 closure signalling contains COP1–HY5 module and ethylene, and both closure pathways converge on H2O2 and nitric oxide (NO) production. Photoactivated crys act via COP1 as well. Photoactivated phots act via activation of the AHA2 (Arabidopsis H+-ATPase 2) in plasma membrane, following ion and water fluxes across plasma membrane and accumulation in vacuoles. phots also induce starch degradation to malate and its accumulation in vacuoles. Photosynthetic apparatus (PSA) in guard cells generates soluble sugars (also accumulating in vacuoles) and energy for all these transmembrane translocators. PSA in mesophyll produces sugars imported into guard cells and decreases intercellular CO2 concentration, which triggers stomata to open. Blue/UV-A light via ZTL and its entrainment of circadian clock forms circadian rhythms of CO2 fixation and stomatal opening. The thickness of arrows approximately symbolises relative intensity of the response. Light-green rectangular matches promoting and pink rectangular matches restricting conditions for photosynthesis
Stomatal conductance — CO2 influx through stomata inside the leaf — depends both on stomatal density and current opening state. With lighting of different spectra, especially at medium and high light intensities, stomatal conductance in blue LED light is higher than in red LED light (cucumber — Wang et al. 2009; cucumber — Hogewoning et al. 2010; cucumber — Savvides et al. 2012; cucumber — Su et al. 2014; lettuce — Muneer et al. 2014; Brassica alboglabra — He et al. 2015; Morus alba — Hu et al. 2016; tomato — Lanoue et al. 2018; Digitalis purpurea — Verma et al. 2018; Allium fistulosum — Gao et al. 2020; lettuce — Tarakanov et al. 2022) and in white or mixed red and blue light is usually in between (Wang et al. 2009; Hogewoning et al. 2010; Hu et al. 2016; Lanoue et al. 2018; Gao et al. 2020; Tarakanov et al. 2022). When the fraction of blue light increases, stomatal conductance gradually increases too (Hogewoning et al. 2010; He et al. 2015).
Conclusions
After more than 30 years of research, the effects of LED lighting on plant photosynthesis are still not fully understood, and many aspects of photosynthesis in these lighting conditions remain poorly studied. While there is ample evidence on the effects of spectral quality on net photosynthesis, photosynthetic pigment content and electron transport chain, other aspects, such as photophosphorylation rate and activities of Calvin cycle enzymes, remain under-studied. This is probably due to the lack of widely known, easy-to-use and non-invasive laboratory techniques. The regulatory mechanisms behind the observed effects are also poorly understood. While there is ample information about regulation of stomata by photoreceptors, similar information about the PSA is scarce. At the moment, it is not possible to construct a regulatory network that would link light, photoreceptors and PSA and explain the observed effects of LED lighting on plants.
The available data show that while certain patterns of red and blue light effects on photosynthetic parameters can be elucidated, the effects often vary between species, cultivar and experimental conditions. The reason for this is probably the presence of a fine-tuned, multi-component regulatory system that remains poorly understood. One way for shedding light on this regulation is comparative transcriptomic studies which allow observing how multiple genes are regulated by light. A recent analysis of microarray data (Griffin et al. 2020) showed that in Arabidopsis, multiple nuclear genes encoding chloroplast proteins are up- or downregulated by red or blue light. Phytochromes and cryptochromes also regulate nuclear genes that are involved in plastome gene expression. Among them, genes involved in transcriptional and post-transcriptional regulation are regulated both by phys and crys, while genes affecting translational activity are regulated predominantly by crys. This is probably the reason why blue light apparently yields a more competent PSA than red light in most experiments, and why adding even a small portion of blue light to red greatly improves photosynthesis (Hogewoning et al. 2010; Savvides et al.2012; He et al. 2015; Hu et al. 2016; Tarakanov et al. 2022). Such works may, in the future, help us understand this regulatory network and increase the predictability of LED lighting effects on plant photosynthesis and growth. On the other hand, a lot of the regulation of PSA assembly is post-transcriptional (Deng et al. 1989) and translational (Drapier et al. 2007), and we also need to take into account these aspects, which are still poorly studied.
Author contribution
All the authors contributed to the study conception and design. GK wrote “Plant photoreceptors” and “Stomata” and created the figures. OA wrote “Photoregulatory role of the photosynthetic apparatus”, “CO2 assimilation and carbon fixation reactions” and “Conclusions”. EB wrote “Effects of light spectrum on the ATP synthase system”. TZ wrote “Light reactions of photosynthesis under narrow-band light” (except “Effects of light spectrum on the ATP synthase system”). All the authors wrote “Introduction” and edited and approved the final manuscript.
Funding
This work was supported by the State Research Program (state registration No. 121032300068–8).
Declarations
Ethics approval
Not applicable (study did not involve animal or human subjects).
Consent to participate
Not applicable (study did not involve human subjects).
Consent for publication
Not applicable (study did not involve human subjects).
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- Akhtar P, Görföl F, Garab G, Lambrev PH. Dependence of chlorophyll fluorescence quenching on the lipid-to-protein ratio in reconstituted light-harvesting complex II membranes containing lipid labels. Chem Phys. 2019;522:242–248. doi: 10.1016/j.chemphys.2019.03.012. [DOI] [Google Scholar]
- Alba R, Kelmenson PM, Cordonnier-Pratt M-M, Pratt LH. The phytochrome gene family in tomato and the rapid differential evolution of this family in Angiosperms. Mol Biol Evol. 2000;17(3):362–373. doi: 10.1093/oxfordjournals.molbev.a026316. [DOI] [PubMed] [Google Scholar]
- Allen JF, de Paula WBM, Puthiyaveetil S, Nield J. A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci. 2011;16(12):645–655. doi: 10.1016/j.tplants.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Amoozgar A, Mohammadi A, Sabzalian MR. Impact of light-emitting diode irradiation on photosynthesis, phytochemical composition and mineral element content of lettuce cv. Grizzly. Photosynthetica. 2017;55(1):85–95. doi: 10.1007/s11099-016-0216-8. [DOI] [Google Scholar]
- Atkins KA, Dodd AN. Circadian regulation of chloroplasts. Curr Opin Plant Biol. 2014;21:43–50. doi: 10.1016/j.pbi.2014.06.008. [DOI] [PubMed] [Google Scholar]
- Avercheva OV, Berkovich YA, Erokhin AN, Zhigalova TV, Pogosyan SI, Smolyanina SO. Growth and photosynthesis of Chinese cabbage plants grown under light-emitting diode-based light source. Russ J Plant Physiol. 2009;56(1):14–21. doi: 10.1134/S1021443709010038. [DOI] [Google Scholar]
- Avercheva OV, Bassarskaya EM, Zhigalova TV, Berkovich YA, Smolyanina SO, Leont’eva MR, Erokhin AN (2010) Photochemical and photophosphorylation activities of chloroplasts and leaf mesostructure in Chinese cabbage plants grown under illumination with light-emitting diodes. Russ J Plant Physiol 57(3):382–391. 10.1134/S1021443710030106
- Avercheva OV, Berkovich YA, Konovalova IO, Radchenko SG, Lapach SN, Bassarskaya EM, Kochetova GV, Zhigalova TV, Yakovleva OS, Tarakanov IG. Optimizing LED lighting for space plant growth unit: Joint effects of photon flux density, red to white ratios and intermittent light pulses. Life Sci Space Res. 2016;11:29–42. doi: 10.1016/j.lssr.2016.12.001. [DOI] [PubMed] [Google Scholar]
- Baker NR. Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev Plant Biol. 2008;59:89–113. doi: 10.1146/annurev.arplant.59.032607.092759. [DOI] [PubMed] [Google Scholar]
- Bellafiore S, Barneche F, Peltier G, Rochaix J-D. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature. 2005;433:892–895. doi: 10.1038/nature03286. [DOI] [PubMed] [Google Scholar]
- Berkovich YA, Konovalova IO, Smolyanina SO, Erokhin AN, Avercheva OV, Bassarskaya EM, Kochetova GV, Zhigalova TV, Yakovleva OS, Tarakanov IG. LED crop illumination inside space greenhouses. REACH Rev Hum Space Explor. 2017;6:11–24. doi: 10.1016/j.reach.2017.06.001. [DOI] [Google Scholar]
- Boccalandro HE, Rugnone ML, Moreno JE, Ploschuk EL, Serna L, Yanovsky MJ, Casal JJ. Phytochrome B enhances photosynthesis at the expense of water-use efficiency in Arabidopsis. Plant Physiol. 2009;150:1083–1092. doi: 10.1104/pp.109.135509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C, Kusnetsov VV, Herrmann RG, Oelmuller R. The spinach AtpC and AtpD genes contain elements for light-regulated, plastid-dependent and organ-specific expression in the vicinity of the transcription start sites. Plant J. 1996;9:21–30. doi: 10.1046/j.1365-313X.1996.09010021.x. [DOI] [PubMed] [Google Scholar]
- Borisova-Mubarakshina MM, Ivanov BN, Vetoshkina DV, Lubimov VY, Fedorchuk TP, Naydov IA, Kozuleva MA, Rudenko NN, Dall’Osto L, Cazzaniga S, Bassi R (2015) Long-term acclimatory response to excess excitation energy: evidence for a role of hydrogen peroxide in the regulation of photosystem II antenna size. J Exp Bot 66:7151–7164. 10.1093/jxb/erv410 [DOI] [PubMed]
- Borthwick HA, Hendricks SB, Toole EH, Toole VK. Action of light on lettuce-seed germination. Bot Gaz. 1954;115(3):205–225. doi: 10.1086/335817. [DOI] [Google Scholar]
- Brestic M, Zivcak M, Kunderlikova K, Sytar O, Shao H, Kalaji HM, Allakhverdiev SI (2015) Low PSI content limits the photoprotection of PSI and PSII in early growth stages of chlorophyll b-deficient wheat mutant lines. Photosynth Res 125(1–2):151–166. 10.1007/s11120-015-0093-1 [DOI] [PubMed]
- Brooks MD, Sylak-Glassman EJ, Fleming GR, Niyogi KK. A thioredoxin-like/β-propeller protein maintains the efficiency of light harvesting in Arabidopsis. Proc Natl Acad Sci USA. 2013;110(29):2733–2740. doi: 10.1073/pnas.1305443110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhov NG, Drozdova IS, Bondar VV. Light response curves of photosynthesis in leaves of sun-type and shade-type plants grown in blue or red light. J Photochem Photobiol. 1995;30:39–41. doi: 10.1016/1011-1344(95)07124-K. [DOI] [Google Scholar]
- Busch FA, Sage RF. The sensitivity of photosynthesis to O2 and CO2 concentration identifies strong Rubisco control above the thermal optimum. New Phytol. 2017;213:1036–1051. doi: 10.1111/nph.14258. [DOI] [PubMed] [Google Scholar]
- Butler WL, Siegelman HW, Miller CO. Denaturation of phytochrome. Biochemistry. 1964;3(6):851–857. doi: 10.1021/bi00894a022. [DOI] [PubMed] [Google Scholar]
- Cackett L, Luginbuehl LH, Schreier TB, Lopez-Juez E, Hibberd JM. Chloroplast development in green plant tissues: the interplay between light, hormone, and transcriptional regulation. New Phytol. 2022;233:2000–2016. doi: 10.1111/nph.17839. [DOI] [PubMed] [Google Scholar]
- Casal JJ, Candia AN, Sellaro R. Light perception and signalling by phytochrome A. J Exp Bot. 2014;65(11):2835–2845. doi: 10.1093/jxb/ert379. [DOI] [PubMed] [Google Scholar]
- Casson SA, Franklin KA, Gray JE, Grierson CS, Whitelam GC, Hetherington AM. phytochrome B and PIF4 regulate stomatal development in response to light quantity. Cur Biol. 2009;19:229–234. doi: 10.1016/j.cub.2008.12.046. [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011;21(11):664–671. doi: 10.1016/j.tcb.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X-L, Guo W-Z, Xue X-Z, Wang L-C, Qiao X-J (2014) Growth and quality responses of ‘Green Oak Leaf’ lettuce as affected by monochromic or mixed radiation provided by fluorescent lamp (FL) and light-emitting diode (LED). Scientia Horticulturae 172:168–175
- Chi W, He B, Mao J, Jiang J, Zhang L. Plastid sigma factors: their individual functions and regulation in transcription. Biochim Biophys Acta - Bioenerg. 2015;1847:770–778. doi: 10.1016/j.bbabio.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Christie JM, Blackwood L, Petersen J, Sullivan S. Plant flavoprotein photoreceptors. Plant Cell Physiol. 2015;56(3):401–413. doi: 10.1093/pcp/pcu196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuartzman SG, Nevo R, Shimoni E, Charuvi D, Kiss V, Ohad I, Brumfeld V, Reicha Z. Thylakoid membrane remodeling during state transitions in Arabidopsis. Plant Cell. 2008;20(4):1029–1039. doi: 10.1105/tpc.107.055830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun L, Kawakami A, Christopher DA. Phytochrome A mediates blue light and UV-A-dependent chloroplast gene transcription in green leaves. Plant Physiol. 2001;125:1957–1966. doi: 10.1104/pp.125.4.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxdale J. Stomatal patterning in monocotyledons: Tradescantia as a model system. J Exp Bot. 1998;49(320):279–292. doi: 10.1093/jxb/49.Special_Issue.279. [DOI] [Google Scholar]
- Dall’Osto L, Caffarri S, Bassi R (2005) A mechanism of nonphotochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. Plant Cell 17:1217–1232. 10.1105/tpc.104.030601 [DOI] [PMC free article] [PubMed]
- Demmig-Adams B. Carotenoids and photoprotection in plants: a role for the xanthophyll zeaxanthin. Biochim Biophys Acta. 1990;1020:1–24. doi: 10.1016/0005-2728(90)90088-L. [DOI] [Google Scholar]
- Demmig-Adams B, Stewart JJ, López-Pozo M, Polutchko SK, Adams WW. Zeaxanthin, a molecule for photoprotection in many different environments. Molecules. 2020;25(24):5825 . doi: 10.3390/molecules25245825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Tonkyn JC, Peter GF, Thornber JP, Gruissem W. Post-transcriptional control of plastid mRNA accumulation during adaptation of chloroplasts to different light quality environments. Plant Cell. 1989;1:645–654. doi: 10.1105/tpc.1.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Parkinson K, Webb AAR. Independent circadian regulation of assimilation and stomatal conductance in the ztl-1 mutant of Arabidopsis. New Phytol. 2004;162(1):63–70. doi: 10.1111/j.1469-8137.2004.01005.x. [DOI] [Google Scholar]
- Drapier D, Rimbault B, Vallon O, Wollman FA, Choquet Y. Intertwined translational regulations set uneven stoichiometry of chloroplast ATP synthase subunits. EMBO J. 2007;26:3581–3591. doi: 10.1038/sj.emboj.7601802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driesen E, Van den Ende W, De Proft M, Saeys W. Influence of environmental factors light, CO2, temperature, and relative humidity on stomatal opening and development: a review. Agronomy. 2020;10:1975. doi: 10.3390/agronomy10121975. [DOI] [Google Scholar]
- Duan L, Ruiz-Sola MÁ, Couso A, Veciana N, Monte E. Red and blue light differentially impact retrograde signalling and photoprotection in rice. Phil Trans R Soc B. 2020;375:20190402. doi: 10.1098/rstb.2019.0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger W, Swartz TE, Bogomolni RA, Taiz L. The ultraviolet action spectrum for stomatal opening in broad bean. Plant Physiol. 2000;122:99–105. doi: 10.1104/pp.122.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feke A, Vanderwall M, Liu W, Gendron JM. Functional domain studies uncover novel roles for the ZTL Kelch repeat domain in clock function. PLoS ONE. 2021;16(3):e0235938 . doi: 10.1371/journal.pone.0235938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández MB, Tossi V, Lamattina L, Cassia R. A comprehensive phylogeny reveals functional conservation of the UV-B photoreceptor UVR8 from green algae to higher plants. Front Plant Sci. 2016;7:1698. doi: 10.3389/fpls.2016.01698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frechilla S, Talbott LD, Zeiger E. The CO2 response of Vicia guard cells acclimates to growth environment. J Exp Bot. 2002;53(368):545–550. doi: 10.1093/jexbot/53.368.545. [DOI] [PubMed] [Google Scholar]
- Galvão VC, Fankhauser C. Sensing the light environment in plants: photoreceptors and early signaling steps. Curr Opin Neurobiol. 2015;34:46–53. doi: 10.1016/j.conb.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Gao S, Liu X, Liu Y, Cao B, Chen Z, Xu K (2020) Photosynthetic characteristics and chloroplast ultrastructure of welsh onion (Allium fistulosum L.) grown under different LED wavelengths. BMC Plant Biol 20(1):78. 10.1186/s12870-020-2282-0 [DOI] [PMC free article] [PubMed]
- Ge X-M, Hu X, Zhang J, Huang Q-M, Gao Y, Li Z-Q, Li S, He J-M. UV RESISTANCE LOCUS8 mediates ultraviolet-B-induced stomatal closure in an ethylene-dependent manner. Plant Sci. 2020;301:110679 . doi: 10.1016/j.plantsci.2020.110679. [DOI] [PubMed] [Google Scholar]
- Ghulam MM, Zghidi-Abouzid O, Lambert E, Lerbs-Mache S, Merendino L. Transcriptional organisation of the large and the small ATP synthase operons, atpI/H/F/A and atpB/E, in Arabidopsis thaliana chloroplasts. Plant Mol Biol. 2012;79:259–272. doi: 10.1007/s11103-012-9910-5. [DOI] [PubMed] [Google Scholar]
- Goins GD, Yorio NC, Sanwo MM, Brown CS (1997) Photomorphogenesis, photosynthesis, and seed yield of wheat plants grown under red light-emitting diodes (LEDs) with and without supplemental blue lighting. J Exp Bot 48(312):1407–1413. http://jxb.oxfordjournals.org/ [DOI] [PubMed]
- Goltsev VN, Kalaji HM, Paunov M, Bąba W, Horaczek T, Mojski J, Kociel H, Allakhverdiev SI. Variable chlorophyll fluorescence and its use for assessing physiological condition of plant photosynthetic apparatus. Russ J Plant Physiol. 2016;63:869–893. doi: 10.1134/S1021443716050058. [DOI] [Google Scholar]
- Grieco M, Tikkanen M, Paakkarinen V, Kangasjärvi S, Aro EM. Steady-state phosphorylation of light-harvesting complex II proteins preserves photosystem I under fluctuating white light. Plant Physiol. 2012;160(4):1896–1910. doi: 10.1104/pp.112.206466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JHC, Prado K, Sutton P, Toledo-Ortiz G. Coordinating light responses between the nucleus and the chloroplast, a role for plant cryptochromes and phytochromes. Physiol Plant. 2020;169:515–528. doi: 10.1111/ppl.13148. [DOI] [PubMed] [Google Scholar]
- Gustavsson M (2018) The effects of ultraviolet B radiation and high temperatureon stomatal aperture and development in Arabidopsis thaliana. MSc dissertation, University of Bristol, School of Life Sciences
- Hamdani S, Khan N, Perveen S, Qu M, Jiang J, Govindjee, Zhu XG (2019) Changes in the photosynthesis properties and photoprotection capacity in rice (Oryza sativa) grown under red, blue, or white light. Photosyn Res 39(1–3):107–121. 10.1007/s11120-018-0589-6 [DOI] [PubMed]
- Haniewicz P, de Sanctis D, Büchel C, Schröder WP, Loi MC, Kieselbach T, Bochtler M, Piano D. Isolation of monomeric photosystem II that retains the subunit PsbS. Photosyn Res. 2013;118(3):199–207. doi: 10.1007/s11120-013-9914-2i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Qin L, Liu Y, Choong T. Photosynthetic capacities and productivity of indoor hydroponically grown Brassica alboglabra bailey under different light sources. Am J Plant Sci. 2015;6:554–563. doi: 10.4236/ajps.2015.64060. [DOI] [Google Scholar]
- He J, Qin L, Chong ELC, Choong T-W, Lee SK. Plant growth and photosynthetic characteristics of Mesembryanthemum crystallinum grown aeroponically under different blue- and red-LEDs. Front Plant Sci. 2017;8:361. doi: 10.3389/fpls.2017.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington AM, Woodward FI. The role of stomata in sensing and driving environmental change. Nature. 2003;424(6951):901–908. doi: 10.1038/nature01843. [DOI] [PubMed] [Google Scholar]
- Hogewoning SW, Trouwborst G, Maljaars H, Poorter H, van Ieperen W, Harbinson J. Blue light dose–responses of leaf photosynthesis, morphology, and chemical composition of Cucumis sativus grown under different combinations of red and blue light. J Exp Bot. 2010;61:3107–3117. doi: 10.1093/jxb/erq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hronková M, Wiesnerová D, Šimková M, Skůpa P, Dewitte W, Vráblová M, Zažímalová E, Šantrůček J. Light-induced STOMAGEN-mediated stomatal development in Arabidopsis leaves. J Exp Bot. 2015;66:4621–4630. doi: 10.1093/jxb/erv233Kegge. [DOI] [PubMed] [Google Scholar]
- Hu J, Dai X, Sun G. Morphological and physiological responses of Morus alba seedlings under different light qualities. Not Bot Horti Agrobot Cluj-Napoca. 2016;44(2):382–392 . doi: 10.15835/nbha44210486. [DOI] [Google Scholar]
- Hughes J. Phytochrome cytoplasmic signaling. Annu Rev Plant Biol. 2013;64:377–402. doi: 10.1146/annurev-arplant-050312-120045. [DOI] [PubMed] [Google Scholar]
- Inohara N, Iwamoto A, Moriyama Y, Shimomura S, Maeda M, Futai M. Two genes, atpC1 and atpC2, for the γ subunit of Arabidopsis thaliana chloroplast ATP synthase. J Biol Chem. 1991;266:7333–7338. doi: 10.1016/S0021-9258(20)89450-2. [DOI] [PubMed] [Google Scholar]
- Inoue K, Nishihama R, Kohchi T. Evolutionary origin of phytochrome responses and signaling in land plants. Plant Cell Environ. 2017;40(11):2502–2508. doi: 10.1111/pce.12908. [DOI] [PubMed] [Google Scholar]
- Ishka MR (2022) A surprising feature of the blue light: regulation of leaf hydraulic conductance via an autonomous phototropin-mediated blue light signaling pathway in bundle-sheath cells. Plant Cell koac088. 10.1093/plcell/koac088 [DOI] [PMC free article] [PubMed]
- Jing Y, Lin R. Transcriptional regulatory network of the light signaling pathways. New Phytol. 2020;227(3):683–697. doi: 10.1111/nph.16602. [DOI] [PubMed] [Google Scholar]
- Johnson MP, Ruban AV. Photoprotective energy dissipation in higher plants involves alteration of the excited state energy of the emitting chlorophyll(s) in the light harvesting antenna II (LHCII) J Biol Chem. 2009;284(35):23592–23601. doi: 10.1074/jbc.M109.013557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MP, Davison PA, Ruban AV, Horton P. The xanthophyll cycle pool size controls the kinetics of non-photochemical quenching in Arabidopsis thaliana. FEBS Lett. 2008;582(2):262–266. doi: 10.1016/j.febslet.2007.12.016. [DOI] [PubMed] [Google Scholar]
- Kalituho L, Beran KC, Jahns P. The transiently generated nonphotochemical quenching of excitation energy in Arabidopsis leaves is modulated by zeaxanthin. Plant Phisiol. 2007;143(4):1861–1870. doi: 10.1104/pp.106.095562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C-Y, Lian H-L, Wang F-F, Huang J-R, Yang H-Q. Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell. 2009;21:2624–2641. doi: 10.1105/tpc.109.069765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Z, Qin T, Zhao Z. Thioredoxins and thioredoxin reductase in chloroplasts: a review. Gene. 2019;706:32–42. doi: 10.1016/j.gene.2019.04.041. [DOI] [PubMed] [Google Scholar]
- Karlický V, Kmecová Materová Z, Kurasová I, Nezval J, Štroch M, Garab G, Špunda V. Accumulation of geranylgeranylated chlorophylls in the pigment-protein complexes of Arabidopsis thaliana acclimated to green light: effects on the organization of light-harvesting complex II and photosystem II functions. Photosyn Res. 2021;149(1–2):233–252. doi: 10.1007/s11120-021-00827-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kereïche S, Kiss AZ, Kouřil R, Boekema EJ, Horton P. The PsbS protein controls the macro-organisation of photosystem II complexes in the grana membranes of higher plant chloroplasts. FEBS Lett. 2010;584(4):759–764. doi: 10.1016/j.febslet.2009.12.031. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K. PHOT1 and PHOT2 mediate blue light regulation of stomatal opening. Nature. 2001;414:656–660 . doi: 10.1038/414656a. [DOI] [PubMed] [Google Scholar]
- Klose C, Nagy F, Schäfer E. Thermal reversion of plant phytochromes. Mol Plant. 2020;13:386–397. doi: 10.1016/j.molp.2019.12.004. [DOI] [PubMed] [Google Scholar]
- Kochetova GV, Belyaeva OB, Gorshkova DS, Vlasova TA, Bassarskaya EM, Zhigalova TV, Avercheva OV. Long-term acclimation of barley photosynthetic apparatus to narrow-band red and blue light. Photosynthetica. 2018;56(3):851–860. doi: 10.1007/s11099-017-0736-x. [DOI] [Google Scholar]
- Kochetova GV, Avercheva OV, Bassarskaya EM, Kushunina MA, Zhigalova TV (2022) Effects of red and blue LED light on the growth and photosynthesis of barley (Hordeum vulgare L.) seedlings. J Plant Growth Regul. 10.1007/s00344-022-10661-x (in press)
- Kong S-G. Wada M (2014) Recent advances in understanding the molecular mechanism of chloroplast photorelocation movement. Biochim Biophys Acta Bioenerg. 1837;4:522–530. doi: 10.1016/j.bbabio.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Kong S-G, Okajima K. Diverse photoreceptors and light responses in plants. J Plant Res. 2016;129:111–114. doi: 10.1007/s10265-016-0792-5. [DOI] [PubMed] [Google Scholar]
- Kostaki K-I, Coupel-Ledru A, Bonnell VC, Gustavsson M, Sun P, McLaughlin FJ, Fraser DP, McLachlan DH, Hetherington AM, Dodd AN, Franklin KA. Guard cells integrate light and temperature signals to control stomatal aperture. Plant Physiol. 2020;182(3):1404–1419. doi: 10.1104/pp.19.01528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. doi: 10.1126/science.1140516. [DOI] [PubMed] [Google Scholar]
- Kress E, Jahns P. The dynamics of energy dissipation and xanthophyll conversion in Arabidopsis indicate an indirect photoprotective role of zeaxanthin in slowly inducible and relaxing components of non-photochemical quenching of excitation energy. Front Plant Sci. 2017;8:2094. doi: 10.3389/fpls.2017.02094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnetsov VV, Doroshenko AS, Kudryakova NV, Danilova MN. Role of phytohormones and light in de-etiolation. Russ J Plant Physiol. 2020;67(6):971–984. doi: 10.1134/S1021443720060102. [DOI] [Google Scholar]
- Landi M, Zivcak M, Sytar O, Brestic M, Allakhverdiev SI (2020) Plasticity of photosynthetic processes and the accumulation of secondary metabolites in plants in response to monochromatic light environments: a review. Biochim Biophys Acta Bioenerg 1861(2):148131. 10.1016/j.bbabio.2019.148131 [DOI] [PubMed]
- Lanoue J, Leonardos ED, Grodzinski B. Effects of light quality and intensity on diurnal patterns and rates of photo-assimilate translocation and transpiration in tomato leaves. Front Plant Sci. 2018;9:756. doi: 10.3389/fpls.2018.00756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong TY, Goodchild DJ, Anderson JM (1985) Effect of light quality on the composition, function, and structure of photosynthetic thylakoid membranes of Asplenium australasicum (Sm.) hook. Plant Physiol 78(3):561–567. 10.1104/pp.78.3.561 [DOI] [PMC free article] [PubMed]
- Lepistö A, Kangasjärvi S, Luomala EM, Brader G, Sipari N, Keränen M, Keinänen M, Rintamäki E (2009) Chloroplast NADPH-thioredoxin reductase interacts with photoperiodic development in Arabidopsis. Plant Physiol 149:1261–1276. 10.1104/pp.108.133777 [DOI] [PMC free article] [PubMed]
- Li X-P, Björkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK. A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature. 2000;403:391–395. doi: 10.1038/35000131. [DOI] [PubMed] [Google Scholar]
- Li FW, Melkonian M, Rothfels C, Villarreal JC, Stevenson DW, Graham SW, Wong GK, Pryer KM, Mathews S. Phytochrome diversity in green plants and the origin of canonical plant phytochromes. Nat Commun. 2015;6:7852. doi: 10.1038/ncomms8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ren H, Kundu M, Liu Z, Zhong F, Wang L, Gao J, Zhong D. A leap in quantum efficiency through light harvesting in photoreceptor UVR8. Nat Commun. 2020;11:4316. doi: 10.1038/s41467-020-17838-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Yang Y, Liu H. Signal transduction mediated by the plant UV-B photoreceptor UVR8. New Phytol. 2019;221(3):1247–1252. doi: 10.1111/nph.15469. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H, Buschmann C, Rahmsdorf U. The importance of blue light for the development of sun-type chloroplasts. In: Senger H, editor. The blue light syndrome. Berlin: Springer-Verlag; 1980. pp. 485–494. [Google Scholar]
- Lichtenthaler HK, Ač A, Marek MV, Kalina J, Urban O. Differences in pigment composition, photosynthetic rates and chlorophyll fluorescence images of sun and shade leaves of four tree species. Plant Physiol Biochem. 2007;45(8):577–588. doi: 10.1016/j.plaphy.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Liscum E. Blue light-induced intracellular movement of phototropins: functional relevance or red herring? Front Plant Sci. 2016;7:827. doi: 10.3389/fpls.2016.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Briggs WR. Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell. 1995;7(4):473–485. doi: 10.1105/tpc.7.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XY, Chang TT, Guo SR, Xu ZG, Li J. Effect of different light quality of LED on growth and photosynthetic character in cherry tomato seedling. Acta Hortic. 2011;907:325–330 . doi: 10.17660/ActaHortic.2011.907.53. [DOI] [Google Scholar]
- Liu XY, Guo SR, Xu ZG, Jiao XL, Tezuka T. Regulation of chloroplast ultrastructure, cross-section anatomy of leaves, and morphology of stomata of cherry tomato by different light irradiations of light-emitting diodes. HortScience. 2011;46(2):217–221 . doi: 10.21273/HORTSCI.46.2.217. [DOI] [Google Scholar]
- Long C, Iino M (2001) Light-dependent osmoregulation in pea stem protoplasts. Photoreceptors, tissue specificity, ion relationships, and physiological implications. Plant Physiol 125:1854–1869. 10.1104/pp.125.4.1854 [DOI] [PMC free article] [PubMed]
- López-Juez E, Bowyer JR, Sakai T. Distinct leaf developmental and gene expression responses to light quantity depend on blue-photoreceptor or plastid-derived signals, and can occur in the absence of phototropins. Planta. 2007;227:113–123. doi: 10.1007/s00425-007-0599-7. [DOI] [PubMed] [Google Scholar]
- Lysenko EA. Plant sigma factors and their role in plastid transcription. Plant Cell Rep. 2007;26:845–859. doi: 10.1007/s00299-007-0318-7. [DOI] [PubMed] [Google Scholar]
- Malnoë A. Photoinhibition or photoprotection of photosynthesis? Update on the (newly termed) sustained quenching component qH. Environ Exp Bot. 2018;154:123–133. doi: 10.1016/j.envexpbot.2018.05.005. [DOI] [Google Scholar]
- Malnoë A, Schultink A, Shahrasbi S, Rumeau D, Havaux M, Niyogi KK. The plastid lipocalin LCNP is required for sustained photoprotective energy dissipation in Arabidopsis. Plant Cell. 2018;30:196–208. doi: 10.1105/tpc.17.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Zhang Y-C, Sang Y, Li Q-H, Yang H-Q. A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. PNAS. 2005;102(34):12270–12275. doi: 10.1073/pnas.0501011102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M, Wicker T, Jenkins J, Plott C, Lux T, Koh CS, Ens J, Gundlach H, Boston LB, Tulpova Z, Holden S, Hernandez-Pinzon I, Scholz U, Mayer KFX, Spannagl M, Pozniak CJ, Sharpe AG, Simkova H, Moscou MJ, Grimwood J, Schmutz J, Stein N. Long-read sequence assembly: a technical evaluation in barley. Plant Cell. 2021;33:1888–1906. doi: 10.1093/plcell/koab077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda R, Ohashi-Kaneko K, Fujiwara K, Kurata K (2007) Analysis of the relationship between blue-light photon flux density and the photosynthetic properties of spinach (Spinacia oleracea L.) leaves with regard to the acclimation of photosynthesis to growth irradiance. Soil Sci Plant Nutr 53(4):459–465. 10.1111/j.1747-0765.2007.00150.x
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence—a practical guide. J Exp Bot 51(345):659–668. 10.1093/jexbot/51.345.659 [DOI] [PubMed]
- McCree KJ. The action spectrum, absorptance and quantum yield of photosynthesis in crop plants. Agric Meteorol. 1972;9:191–216. doi: 10.1016/0002-1571(71)90022-7. [DOI] [Google Scholar]
- Mehta P, Allakhverdiev SI, Jajoo A (2010) Characterization of photosystem II heterogeneity in response to high salt stress in wheat leaves (Triticum aestivum). Photosynth Res 105:249–255. 10.1007/s11120-010-9588-y [DOI] [PubMed]
- Menon C, Sheerin DJ, Hiltbrunner A. SPA proteins: SPAnning the gap between visible light and gene expression. Planta. 2016;244(2):297–312. doi: 10.1007/s00425-016-2509-3. [DOI] [PubMed] [Google Scholar]
- Messant M, Krieger-Liszkay A, Shimakawa G. Dynamic changes in protein-membrane association for regulating photosynthetic electron transport. Cells. 2021;10(5):1216. doi: 10.3390/cells10051216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y-X, Wang X-Z, Gao L-H, Chen Q-Y, Qu M. Blue light is more essential than red light for maintaining the activities of photosystem II and I and photosynthetic electron transport capacity in cucumber leaves. J Integr Agric. 2016;15(1):87–100. doi: 10.1016/S2095-3119(15)61202-3. [DOI] [Google Scholar]
- Minagawa J. State transitions-the molecular remodeling of photosynthetic supercomplexes that controls energy flow in the chloroplast. Biochim Biophys Acta Bioenerg. 2011;1807(8):897–905. doi: 10.1016/j.bbabio.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Mishra S, Khurana JP. Emerging roles and new paradigms in signaling mechanisms of plant cryptochromes. CRC Crit Rev Plant Sci. 2017;36(2):89–115. doi: 10.1080/07352689.2017.1348725. [DOI] [Google Scholar]
- Muneer S, Kim E, Park J, Lee J. Influence of green, red and blue light emitting diodes on multiprotein complex proteins and photosynthetic activity under different light intensities in lettuce leaves (Lactuca sativa L.) Int J Mol Sci. 2014;15:4657–4670. doi: 10.3390/ijms15034657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie EH, Lawson T. Chlorophyll fluorescence analysis: a guide to good practice and understanding some new applications. J Exp Bot. 2013;64(13):3983–3998. doi: 10.1093/jxb/ert208. [DOI] [PubMed] [Google Scholar]
- Nicol L, Croce R. The PsbS protein and low pH are necessary and sufficient to induce quenching in the light-harvesting complex of plants LHCII. Sci Rep. 2021;11(1):7415. doi: 10.1038/s41598-021-86975-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilkens M, Kress E, Lambrev P, Miloslavina Y, Müller M, Holzwarth AR, Jahns P. Identification of a slowly inducible zeaxanthin-dependent component of non-photochemical quenching of chlorophyll fluorescence generated under steady-state conditions in Arabidopsis. Biochim Biophys Acta Bioenerg. 2010;1797(4):466–475. doi: 10.1016/j.bbabio.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Noordally ZB, Ishii K, Atkins KA, Wetherill SJ, Kusakina J, Walton EJ, Kato M, Azuma M, Tanaka K, Hanaoka M, Dodd AN. Circadian control of chloroplast transcription by a nuclear-encoded timing signal. Science. 2013;339(6125):1316–1319. doi: 10.1126/science.1230397. [DOI] [PubMed] [Google Scholar]
- Norris SR, Barrette TR, DellaPenna D. Genetic dissection of carotenoid synthesis in arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell. 1995;7(12):2139–2149. doi: 10.1105/tpc.7.12.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okajima K, Kashojiya S, Tokutomi S. Photosensitivity of kinase activation by blue light involves the lifetime of a cysteinyl-flavin adduct intermediate, S390, in the photoreaction cycle of the LOV2 domain in phototropin, a plant blue light receptor. J Biol Chem. 2012;287(49):40972–40981. doi: 10.1074/jbc.M112.406512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J, Rouzé P, Verhelst B, et al. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature. 2016;530:331–335. doi: 10.1038/nature16548. [DOI] [PubMed] [Google Scholar]
- Oswald O, Martin T, Dominy PJ, Graham IA. Plastid redox state and sugars: interactive regulators of nuclear-encoded photosynthetic gene expression. Proc Natl Acad Sci USA. 2001;98:2047–2052. doi: 10.1073/pnas.98.4.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Schütze K, Brost M, Oelmüller R. A novel mechanism of nuclear photosynthesis gene regulation by redox signals from the chloroplast during photosystem stoichiometry adjustment. J Biol Chem. 2001;276:36125–36130. doi: 10.1074/jbc.M105701200. [DOI] [PubMed] [Google Scholar]
- Platten JD, Foo E, Elliott RC, Hecht V, Reid JB, Weller JL. Cryptochrome 1 contributes to blue-light sensing in pea. Plant Physiol. 2005;139:1472–1482. doi: 10.1104/pp.105.067462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptushenko VV, Ptushenko OS, Samoilova OP, Solovchenko AE. An exceptional irradiance-induced decrease of light trapping in two Tradescantia species: an unexpected relationship with the leaf architecture and zeaxanthin-mediated photoprotection. Biol Plant. 2016;60:385–393. doi: 10.1007/s10535-016-0593-7. [DOI] [Google Scholar]
- Pudasaini A, Zoltowski BD. Zeitlupe senses blue-light fluence to mediate circadian timing in Arabidopsis thaliana. Biochemistry. 2013;52:7150–7158. doi: 10.1021/bi401027n. [DOI] [PubMed] [Google Scholar]
- Pudasaini A, Shim JS, Song YH, Shi H, Kiba T, Somers DE, Imaizumi T, Zoltowski BD. Kinetics of the LOV domain of ZEITLUPE determine its circadian function in Arabidopsis. eLife. 2017;6:e21646 . doi: 10.7554/eLife.21646.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines CA. The Calvin cycle revisited. Photosynth Res. 2003;75(1):1–10. doi: 10.1023/A:1022421515027. [DOI] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 2009;150:889–903. doi: 10.1104/pp.109.138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O'Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, Ulm R. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332(6025):103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC. Phytochrome evolution in 3D: deletion, duplication, and diversification. New Phytol. 2020;225(6):2283–2300. doi: 10.1111/nph.16240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowska E, Powikrowska M, Zienkiewicz M, Drozak A, Pokorska B. High light induced accumulation of two isoforms of the CF1 α-subunit in mesophyll and bundle sheath chloroplasts of C4 plants. Acta Biochim Pol. 2008;55:175–182 . doi: 10.18388/abp.2008_3110. [DOI] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM (2007) Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19(12):3944–3960. 10.1105/tpc.107.054312 [DOI] [PMC free article] [PubMed]
- Ruiz-Sola MÁ, Rodríguez-Concepción M. Carotenoid Biosynthesis in Arabidopsis: a colorful pathway. Arabidopsis Book. 2012;10:e0158 . doi: 10.1199/tab.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusaczonek A, Czarnocka W, Willems P, Sujkowska-Rybkowska M, Van Breusegem F, Karpiński S. Phototropin 1 and 2 influence photosynthesis, UV-C induced photooxidative stress responses, and cell death. Cells. 2021;10(2):200. doi: 10.3390/cells10020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvides A, Fanourakis D, van Ieperen W. Co-ordination of hydraulic and stomatal conductances across light qualities in cucumber leaves. J Exp Bot. 2012;63(3):1135–1143. doi: 10.1093/jxb/err348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonberg A, Baginsky S. Signal integration by chloroplast phosphorylation networks: an update. Front Plant Sci. 2012;3:1–7. doi: 10.3389/fpls.2012.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz A, Zeiger E. Metabolic energy for stomatal opening Roles of photophosphorylation and oxidative phosphorylation. Planta. 1984;161(2):129–136. doi: 10.1007/bf00395472. [DOI] [PubMed] [Google Scholar]
- Schweer J, Turkeri H, Kolpack A, Link G. Role and regulation of plastid sigma factors and their functional interactors during chloroplast transcription – recent lessons from Arabidopsis thaliana. Eur J Cell Biol. 2010;89:940–946. doi: 10.1016/j.ejcb.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Serna L, Fenoll C. Stomatal development in Arabidopsis: how to make a functional pattern. Trends Plant Sci. 2000;5(11):458–460. doi: 10.1016/s1360-1385(00)01782-9. [DOI] [PubMed] [Google Scholar]
- Shimazaki K, Doi M, Assmann SM, Kinoshita T. Light regulation of stomatal movement. Annu Rev Plant Biol. 2007;58:219–247. doi: 10.1146/annurev.arplant.57.032905.105434. [DOI] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Plant Biol. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sineshchekov VA, Belyaeva OB (2019) Regulation of chlorophyll biogenesis by phytochrome A. Biochem Mosc 84(5):491–508. 10.1134/S0006297919050043 [DOI] [PubMed]
- Singh R, Singh S, Parihar P, Singh VP, Prasad SM. Retrograde signaling between plastid and nucleus: a review. J Plant Physiol. 2015;181:55–66. doi: 10.1016/j.jplph.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Sokolskaya SV, Sveshnikova NV, Kochetova GV, Solovchenko AE, Gostimski SA, Bashtanova OB. Involvement of phytochrome in regulation of transpiration: red-/far red-induced responses in the chlorophyll-deficient mutant of pea. Funct Plant Biol. 2003;30:1249–1259. doi: 10.1071/fp03013. [DOI] [PubMed] [Google Scholar]
- Solymosi K, Mysliwa-Kurdziel B. The role of membranes and lipid-protein interactions in the Mg-branch of tetrapyrrole biosynthesis. Front Plant Sci. 2021;12:663309 . doi: 10.3389/fpls.2021.663309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. 2000;101:319–329. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- Stanley L, Yuan YW. Transcriptional regulation of carotenoid biosynthesis in plants: so many regulators, so little consensus. Front Plant Sci. 2019;10:1017. doi: 10.3389/fpls.2019.01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen CJ, Morris JM, Short AH, Niyogi KK, Fleming GR. Complex roles of psbs and xanthophylls in the regulation of nonphotochemical quenching in Arabidopsis thaliana under fluctuating light. J Phys Chem. 2020;124(46):10311–10325. doi: 10.1021/acs.jpcb.0c06265. [DOI] [PubMed] [Google Scholar]
- Strand Å, Asami T, Alonso J, Ecker JR, Chory J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature. 2003;421:79–83. doi: 10.1038/nature01204. [DOI] [PubMed] [Google Scholar]
- Su N, Wu Q, Shen Z, Xia K, Cui J. Effects of light quality on the chloroplastic ultrastructure and photosynthetic characteristics of cucumber seedlings. Plant Growth Regul. 2014;73:227–235. doi: 10.1007/s10725-013-9883-7. [DOI] [Google Scholar]
- Sun T, Yuan H, Cao H, Yazdani M, Tadmor Y, Li L. Carotenoid metabolism in plants: the role of plastids. Mol Plant. 2018;11(1):58–74. doi: 10.1016/j.molp.2017.09.010. [DOI] [PubMed] [Google Scholar]
- Sylak-Glassman EJ, Malnoë A, Re ED, Brooks MD, Fischer AL, Niyogi KK, Fleming GR. Distinct roles of the photosystem II protein PsbS and zeaxanthin in the regulation of light harvesting in plants revealed by fluorescence lifetime snapshots. Proc Natl Acad Sci USA. 2014;111(49):17498–17503. doi: 10.1073/pnas.1418317111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szechynska-Hebda M, Karpinski S. Light intensity-dependent retrograde signalling in higher plants. J Plant Physiol. 2013;170:1501–1516. doi: 10.1016/j.jplph.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Talbott LD, Zhu JX, Han SW, Zeiger E (2002) Phytochrome and blue light-mediated stomatal opening in the orchid, Paphiopedilum. Plant Cell Physiol 43(6):639–646. 10.1093/pcp/pcf075 [DOI] [PubMed]
- Tantharapornrerk N, Vichitsoonthonkul T, Techavuthiporn C, Photchanachai S (2021) Growth and antioxidant system of Chinese kale microgreens in response to different illumination of light sources. N Z J Crop Hortic Sci. 10.1080/01140671.2021.1958876
- Tarakanov IG, Tovstyko DA, Lomakin MP, Shmakov AS, Sleptsov NN, Shmarev AN, Litvinskiy VA, Ivlev AA (2022) Effects of light spectral quality on photosynthetic activity, biomass production, and carbon isotope fractionation in lettuce, Lactuca sativa L., plants. Plants 11(3):441. https://doi.org/10.3390/plants11030441 [DOI] [PMC free article] [PubMed]
- Tossi V, Lamattina L, Jenkins GI, Cassia RO. Ultraviolet-B-induced stomatal closure in Arabidopsis is regulated by the UV RESISTANCE LOCUS8 photoreceptor in a nitric oxide-dependent mechanism. Plant Physiol. 2014;164:2220–2230. doi: 10.1104/pp.113.231753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossi VE, Regalado JJ, Iannicelli J, Laino LE, Burrieza HP, Escandón AS, Pitta-Álvarez SI. Beyond Arabidopsis: differential UV-B response mediated by UVR8 in diverse species. Front Plant Sc. 2019;10:780. doi: 10.3389/fpls.2019.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LH, Lee DG, Jung S. Light quality-dependent regulation of photoprotection and antioxidant properties in rice seedlings grown under different light-emitting diodes. Photosynthetica. 2021;59(1):12–22 . doi: 10.32615/ps.2020.082. [DOI] [Google Scholar]
- Trojak M, Skowron E. Light quality-dependent regulation of non-photochemical quenching in tomato plants. Biology. 2021;10(8):721. doi: 10.3390/biology10080721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouwborst G, Hogewoning SW, van Kooten O, Harbinson J, van Ieperen W. Plasticity of photosynthesis after the “red light syndrome” in cucumber. Environ Exp Bot. 2016;121:75–82. doi: 10.1016/j.envexpbot.2015.05.002. [DOI] [Google Scholar]
- Tu S-L, Lagarias JC. The phytochromes. In: Briggs R, Spudich JL, editors. Handbook of photosensory receptors. Weinheim: Wiley-VCH; 2005. pp. 121–149. [Google Scholar]
- Ulm R, Jenkins GI. Q&A: how do plants sense and respond to UV-B radiation? BMC Biol. 2015;13:45. doi: 10.1186/s12915-015-0156-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavasseur A, Raghavendra AS. Guard cell metabolism and CO2 sensing. New Phytol. 2005;165(3):665–682. doi: 10.1111/j.1469-8137.2004.01276.x. [DOI] [PubMed] [Google Scholar]
- Verma SK, Gantait S, Jeong BR, Hwang SJ. Enhanced growth and cardenolides production in Digitalis purpurea under the influence of different LED exposures in the plant factory. Sci Rep. 2018;8:18009. doi: 10.1038/s41598-018-36113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viczián A, Klose C, Ádám É, Nagy F. New insights of red light-induced development. Plant Cell Environ. 2017;40(11):2457–2468. doi: 10.1111/pce.12880. [DOI] [PubMed] [Google Scholar]
- von Groll U, Berger D, Altmann T. The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell. 2002;14(7):1527–1539. doi: 10.1105/tpc.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskresenskaya NP, Nechayeva EP, Vlasova MP, Nichiporovich AA. The significance of blue light and kinetin for restoration of the photosynthetic apparatus in aging nearly leaves. Soviet Plant Physiol. 1968;15:890. [Google Scholar]
- Walker JE. The ATP synthase: the understood, the uncertain and the unknown. Biochem Soc Trans. 2013;41:1–16. doi: 10.1042/bst20110773. [DOI] [PubMed] [Google Scholar]
- Wang H, Gu M, Cui J, Shi K, Zhou Y, Yu J. Effects of light quality on CO2 assimilation, chlorophyll-fluorescence quenching, expression of Calvin cycle genes and carbohydrate accumulation in Cucumis sativus. J Photochem Photobiol. 2009;96:30–37. doi: 10.1016/j.jphotobiol.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Wang F-F, Lian H-L, Kang C-Y, Yang H-Q. Phytochrome B is involved in mediating red light-induced stomatal opening in Arabidopsis thaliana. Mol Plant. 2010;3(1):246–259. doi: 10.1093/mp/ssp097. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zuo Z, Wang X, Liu Q, Gu L, Oka Y, Lin C. Beyond the photocycle — how cryptochromes regulate photoresponses in plants? Curr Opin Plant Biol. 2018;45:120–126. doi: 10.1016/j.pbi.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Yan J, Ahammed GJ, Wang X, Bu X, Xiang H, Li Y, Lu J, Liu Y, Qi H, Qi M, Li T. PGR5/PGRL1 and NDH mediate far-red light-induced photoprotection in response to chilling stress in tomato. Front Plant Sci. 2020;11:669. doi: 10.3389/fpls.2020.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware MA, Belgio E, Ruban AV. Comparison of the protective effectiveness of NPQ in Arabidopsis plants deficient in PsbS protein and zeaxanthin. J Exp Bot. 2015;66(5):1259–1270. doi: 10.1093/jxb/eru477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargent JJ, Gegas VC, Jenkins GI, Doonan JH, Paul ND. UVR8 in Arabidopsis thaliana regulates multiple aspects of cellular differentiation during leaf development in response to ultraviolet B radiation. New Phytol. 2009;183(2):315–326. doi: 10.1111/j.1469-8137.2009.02855.x. [DOI] [PubMed] [Google Scholar]
- Wei H, Kong D, Yang J, Wang H. Light regulation of stomatal development and patterning: shifting the paradigm from Arabidopsis to grasses. Plant Comm. 2020;1:100030 . doi: 10.1016/j.xplc.2020.100030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston E, Thorogood K, Vinti G, López-Juez E. Light quantity controls leaf-cell and chloroplast development in Arabidopsis thaliana wild type and blue-light-perception mutants. Planta. 2000;211:807–815. doi: 10.1007/s004250000392. [DOI] [PubMed] [Google Scholar]
- Wilson S, Ruban AV. Rethinking the influence of chloroplast movements on non-photochemical quenching and photoprotection. Plant Physiol. 2020;183:1213–1223. doi: 10.1104/pp.20.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EEK, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K (2012) Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149:1525–1535. 10.1016/j.cell.2012.04.038 [DOI] [PubMed]
- Xiaoying L, Shirong G, Taotao C, Zhigang X, Tezuka T. Regulation of the growth and photosynthesis of cherry tomato seedlings by different light irradiations of light emitting diodes (LED) Afr J Biotechnol. 2012;11(22):6169–6177. doi: 10.5897/AJB11.1191. [DOI] [Google Scholar]
- Xu P, Xiang Y, Zhu H, Xu H, Zhang Z, Zhang C, Zhang L, Ma Z. Wheat cryptochromes: subcellular localization and involvement in photomorphogenesis and osmotic stress responses. Plant Physiol. 2009;149(2):760–774. doi: 10.1104/pp.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A, Singh D, Lingwan M, Yadukrishnan P, Masakapalli SK, Datta S. Light signaling and UV-B-mediated plant growth regulation. J Integr Plant Biol. 2020;62(9):1270–1292. doi: 10.1111/jipb.12932. [DOI] [PubMed] [Google Scholar]
- Yavari N, Tripathi R, Wu BS, MacPherson S, Singh J, Lefsrud M. The effect of light quality on plant physiology, photosynthetic, and stress response in Arabidopsis thaliana leaves. PLoS ONE. 2021;16:e0247380 . doi: 10.1371/journal.pone.0247380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Ulm R. How plants cope with UV-B: from perception to response. Curr Opin Plant Biol. 2017;37:42–48. doi: 10.1016/j.pbi.2017.03.013. [DOI] [PubMed] [Google Scholar]
- Yuan M, Zhao YQ, Zhang ZW, Chen YE, Ding CB, Yuan S. Light regulates transcription of chlorophyll biosynthetic genes during chloroplast biogenesis. CRC Crit Rev Plant Sci. 2017;36(1):35–54. doi: 10.1080/07352689.2017.1327764. [DOI] [Google Scholar]
- Yurina NP, Odintsova MS. Chloroplast retrograde signaling system. Russ J Plant Physiol. 2019;66:509–520. doi: 10.1134/S1021443719040149. [DOI] [Google Scholar]
- Zivcak M, Brestic M, Kalaji HM, Govindjee (2014) Photosynthetic responses of sun- and shade-grown barley leaves to high light: Is the lower PSII connectivity in shade leaves associated with protection against excess of light? Photosyn Res 119(3):339–354. 10.1007/s11120-014-9969-8 [DOI] [PMC free article] [PubMed]
- Zubo YO, Yamburenko MV, Selivankina SY, Shakirova FM, Avalbaev AM, Kudryakova NV, Zubkova NK, Liere K, Kulaeva ON, Kusnetsov VV, Borner T. Cytokinin stimulates chloroplast transcription in detached barley leaves. J Plant Physiol. 2008;148:1082–1093. doi: 10.1104/pp.108.122275. [DOI] [PMC free article] [PubMed] [Google Scholar]