Summary
Human macrophages are a natural host of many mycobacterium species, including Mycobacterium abscessus (M. abscessus), an emerging pathogen affecting immunocompromised and cystic fibrosis patients with few available treatments. The search for an effective treatment is hindered by the lack of a tractable in vitro intracellular infection model. Here, we established a reliable model for M. abscessus infection using human pluripotent stem cell-derived macrophages (hPSC-macrophages). hPSC differentiation permitted reproducible generation of functional macrophages that were highly susceptible to M. abscessus infection. Electron microscopy demonstrated that M. abscessus was present in the hPSC-macrophage vacuoles. RNA sequencing analysis revealed a time-dependent host cell response, with differing gene and protein expression patterns post-infection. Engineered tdTOMATO-expressing hPSC-macrophages with GFP-expressing mycobacteria enabled rapid image-based high-throughput analysis of intracellular infection and quantitative assessment of antibiotic efficacy. Our study describes the first to our knowledge hPSC-based model for M. abscessus infection, representing a novel and accessible system for studying pathogen-host interaction and drug discovery.
Keywords: pluripotent stem cells, macrophage, mycobacterium abscessus, infection, highthroughput, genetic modification
Graphical abstract
Highlights
-
•
A simplified chemically defined and serum-free protocol for the generation of functional macrophages from hPSCs
-
•
An efficient human model recapitulating intracellular infection of Mycobacterium abscessus in hPSC-macrophages
-
•
A high-throughput system testing antibiotic sensitivity with fluorescent hPSC-macrophages and M. abscessus
In this article, Ed Stanley, Sohinee Sarkar, and colleagues develop a simplified method for producing functional macrophages from hPSCs. They use these macrophages to examine intracellular infection with M. abscessus, host cell responses, and antibiotic sensitivity. This model may serve as a system for studying macrophage-pathogen interactions and screening for antimicrobial compounds.
Introduction
The human macrophage is an important innate immune cell type, playing key roles in development, regeneration, inflammation, and infection (Wynn et al., 2013). Macrophages are responsible for the detection, uptake, and processing of infectious microbes, setting up a sequence of immune responses to eliminate potential pathogens (Tosi, 2005). However, human macrophages are also a natural host of some pathogens, including most types of mycobacteria, which circumvent the macrophage’s normal strategy for microbe elimination (Diacovich and Gorvel, 2010). This results in difficult-to-treat chronic infections, a situation that is further exacerbated in immune-compromised individuals and those with chronic pulmonary disorders. One such pathogen is Mycobacterium abscessus, an environmentally ubiquitous non-tuberculosis species that opportunistically infects individuals with lung diseases, such as cystic fibrosis, and also causes skin and soft tissue infections in people with immunodeficiencies (Bryant et al., 2013; Johansen et al., 2020; Lee et al., 2015).
M. abscessus is highly resistant to antibiotics, leaving infected people with few treatment options (Lee et al., 2015; Nessar et al., 2012). Moreover, the search for new antibiotics has been hindered by the lack of tractable experimental human models suitable for high-throughput phenotypic drug screening. Although animal models including Drosophila, zebrafish embryos, and immunocompromised mice are reported to be susceptible to M. abscessus infection, platforms for studying M. abscessus in a human context are limited (Bernut et al., 2017; Ordway et al., 2008). An ideal system would employ functional human macrophages; however, such cells are difficult to obtain routinely from human blood and are likely to display variability associated with different donors. These limitations make primary macrophages a less-than-ideal platform for studying pathogen-host interactions and drug screening.
Human pluripotent stem cells can be grown in large numbers and differentiated to produce any cell type found within the human body. hPSC-derived blood cells have been used for studying hematopoietic development, modeling diseases, and developing new therapies (Ivanovs et al., 2017). Physiologically, hPSC-macrophages resemble their counterparts isolated from human donors, making them a preferable system in comparison to transformed cell lines that often harbor multiple uncharacterized genetic mutations (Cao et al., 2019; Han et al., 2019). In addition, hPSCs can be easily genetically modified, potentially enabling mechanistic studies that could provide avenues for the development of new therapeutic strategies (Hendriks et al., 2016). More recently, hPSC-macrophages have been shown to support infection of a variety of viral and bacterial pathogens, including Mycobacterium tuberculosis (Mtb) (Han et al., 2019; Lang et al., 2018; Neehus et al., 2018; Vaughan-Jackson et al., 2021). In this study, we show that genetically modified hPSC-macrophages support M. abscessus infection and represent a tractable and scalable system for studying host-microbe interactions.
Results
Generation of functional human macrophages from pluripotent stem cells
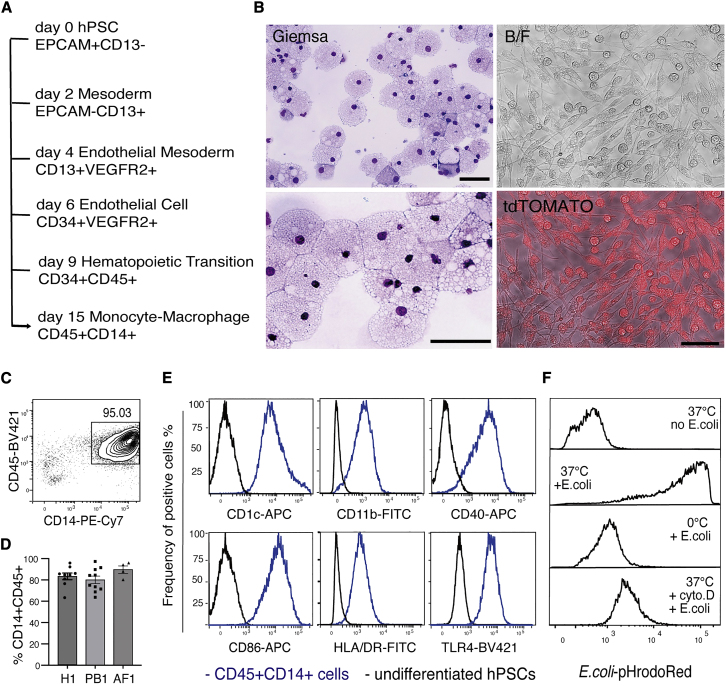
We established a simple and serum-free chemically defined method to generate functional macrophages from hPSCs, including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs). This method directs the stepwise differentiation of hPSCs through critical stages of hematopoietic development, recapitulating key events in the genesis of early myeloid cells (Figures 1A and S1A). Flow cytometry analysis for cell surface markers was used to monitor the differentiation steps transiting key development stages, including mesoderm (CD13+EPCAM–), endothelial progenitor (CD13+VEGFR2+), endothelial cell (CD34+CD45−), and hematopoietic progenitor (CD34+CD45+) (Figure S1B) (Ivanovs et al., 2017; Ng et al., 2016). The frequency of CD34+CD45+ hematopoietic progenitor cells increased from day 9 to day 12 (Figures S1C and S1D), providing a substantial population for the subsequent efficient generation of macrophages at later timepoints (Figure S1E). At day 15, May-Grunwald-Giemsa hematologic-stained cytospin preparations revealed a uniform population of large round monocytic cells with vacuolated cytoplasm, with the latter characteristic also evident by bright field and fluorescence microscopy (Figure 1B). At this stage, flow cytometry analysis showed that CD45+CD14+ myeloid cells accounted for approximately 70%–95% of the total suspension cells (Figures 1C and 1D). This result was reproduced using three independent hiPSC and hESC lines (Figure 1D), attesting to the efficiency and robustness of this protocol for generating myeloid cells.
Figure 1.
Generation of functional human macrophages from pluripotent stem cells
(A) Roadmap of hPSC differentiation to functional macrophages in vitro.
(B) Morphological analysis for hPSC-derived macrophages. Panels show cyto-spin preparations of May-Grunwald-Giemsa-stained macrophages (left) and a bright field (BF) and a BF fluorescence-merged (tdTOMATO) image of macrophages generated from an hPSC line that constitutively expresses a tdTOMATO transgene. Scale bar, 50 μm.
(C) Flow cytometric analysis showing the expression of CD45 and CD14 on cells from differentiation cultures at day 15. The frequency of CD45+CD14+ cells is indicated.
(D) Histograms summarizing the frequency of CD45+CD14+ cells obtained from three independent hPSC lines over more than four independent experiments: H1, n = 9; PB1, n = 10; AF1, n = 4. Data shown as mean ± SEM, non-significant, examined by one-way ANOVA test.
(E) Flow cytometry analysis of CD45+CD14+ cells (blue line) for the expression of surface markers typically associated with functional macrophages. Undifferentiated hPSCs were used as negative controls (black line).
(F) Flow cytometry analysis of hPSC-derived macrophages incubated with pHrodoRed-conjugated E. coli bioparticles under the conditions indicated. Incubation of E. coli bioparticles with hPSC-macrophages was conducted at 37°C (37°C + E. coli). Control groups: hPSC-derived macrophages at 37°C (37°C no E. coli), incubation of E. coli bioparticles with hPSC-derived macrophages at 0°C (0°C + E. coli), incubation of E. coli bioparticles with macrophages with cytochalasin D treatment at 37°C (37°C + cyto. D + E. coli).
CD14+ blood cells produced by this method had typical characteristics of macrophages. Flow cytometry analysis showed that the CD45+CD14+ cells expressed functional surface proteins of macrophages, which are critical for phagocytosis (CD11b), bacterial pathogen responsiveness (TLR4), antigen presentation (CD1c and HLA-DR), and T cell co-stimulation (CD40 and CD86) (Figure 1E). Furthermore, hPSC-macrophages showed robust phagocytic activity, evidenced by their capacity to uptake fluorescence-labeled E. coli bioparticles, activity that was largely absent at 0°C and inhibited by treatment with the actin polymerization inhibitor cytochalasin D (Figure 1F). Functional macrophages could be maintained in cultures until at least day 30 (Figures S2A–S2E), a characteristic that has logistical advantages for in vitro applications.
Establishment of an infection model for M. abscessus
Next, we examined whether our hPSC-macrophages could be infected by M. abscessus. To easily monitor the infection process, we generated a GFP-tagged M. abscessus variant using the pTEC15 GFP-expressing plasmid (Takaki et al., 2013). The GFP-expressing strain displayed identical growth kinetics to the genetically unmodified wild-type strain (Figure S2F). Preliminary experiments indicated that a multiplicity of infections (MOI) of 5 provided a robust level of infection after 3 h, as assessed by flow cytometry analysis of GFP fluorescence (Figure S2G). This result was also consistent with prior studies showing that an MOI of 5 was efficient for M. tuberculosis infection of hPSC-macrophages (Han et al., 2019). Furthermore, we found that the initial uptake of mycobacteria was inhibited by cytochalasin D or incubation at 0°C (Figure S2H), the former confirming the infection process is dependent on phagocytosis (Roux et al., 2016).
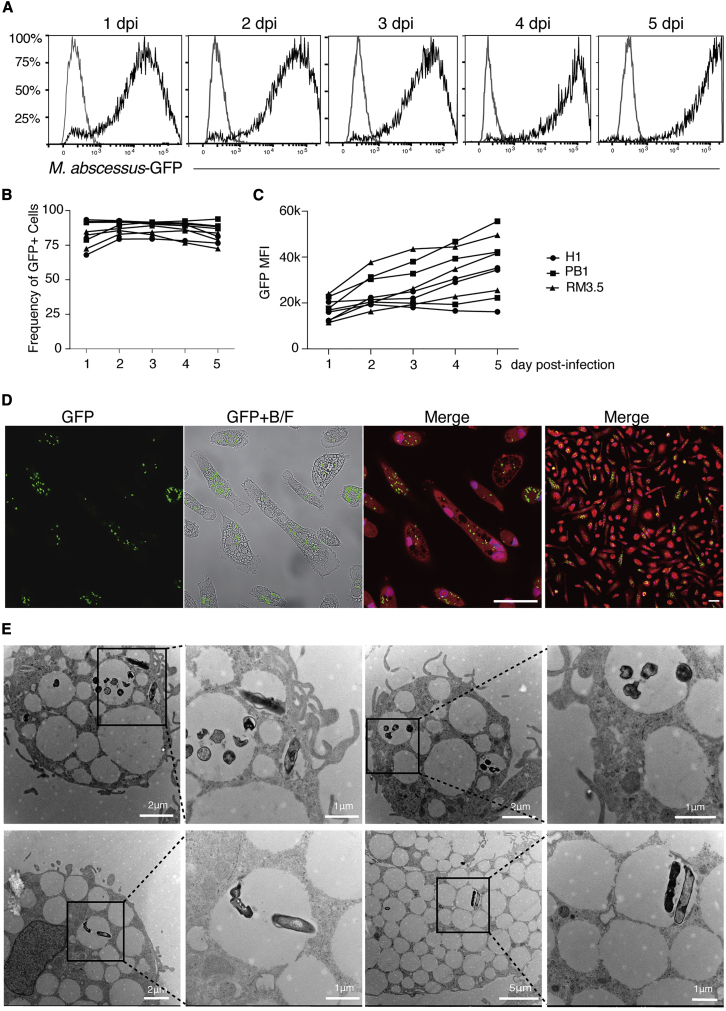
By 1 day post-infection (dpi), more than 70% of cells were infected (Figures 2A and 2B). Over the ensuing days, GFP intensity increased, demonstrating a progressive accumulation of mycobacteria in each cell (Figures 2A and 2C). Although it is unclear if this accumulation was due to increased uptake or intracellular replication, M. abscessus is known to replicate within macrophages and can even escape the phagosomal compartment to infect neighboring cells (Kim et al., 2019). Indeed, confocal microscopy of 2-dpi cultures revealed multiple GFP+ M. abscessus in individual macrophages and that nearly all macrophages were infected (Figure 2D). Furthermore, transmission electron microscopy showed that M. abscessus was present in macrophage vacuoles in groups or as individual cells (Figure 2E), reminiscent of the previously reported rough or smooth variants, respectively (Johansen et al., 2020; Roux et al., 2016). Taken together, these results demonstrate that hPSC-derived macrophages are a tractable model for M. abscessus infection.
Figure 2.
Infection of hPSC-macrophages by M. abscessus
(A) Flow cytometry analysis of M. abscessus infection (GFP) of CD14+CD45+ hPSC-macrophages from 1 day post-infection (dpi) to 5 dpi.
(B) Summary of infection kinetics of macrophages from three independent hPSC lines from 1 to 5 dpi using an MOI of 5. H1, n = 3; PB001, n = 3, RM3.5, n = 3. Data are shown in individual values, non-significant tested by two-way ANOVA. “n,” number of independent experiments.
(C) Summary of the average of intensity of GFP fluorescence in infected hPSC-macrophages from 1 to 5 dpi. MFI, mean of fluorescent intensity. H1, n = 3; PB1, n = 3, RM3.5, n = 3. Data shown are shown in individual values, significance tested by two-way ANOVA, P(RM3.5 d1 versus d5) = 0.0284. “n,” number of independent experiments.
(D) Confocal fluorescence images of tdTOMATO-expressing hPSC-macrophages infected with GFP-labelled M. abscessus at 4 dpi. Macrophage nuclei were identified by DAPI staining (blue). Scale bar, 50 μm.
(E) Transmission electron micrographs of hPSC-macrophages infected with M. abscessus presenting in groups or by individual after 2 days of infection. The length of scale bars associated with each micrograph is indicated.
M. abscessus induces time-dependent responses in infected macrophages
To probe the impact of M. abscessus on hPSC-derived macrophages, we examined transcriptomic changes over the course of infection at 3, 24, and 48 h by RNA sequencing. Multidimensional scaling (MDS) analysis showed that uninfected and 3-h-infected samples were separated from 24- to 48-h-infected samples in the first dimension (Figure 3A). In addition, the 24-h-infected samples were more tightly clustered than those representing other time points, suggesting that mycobacterial infection induced a convergent transcriptomic response program (Figure 3A). Gene ontology analysis showed that the major changes in the top 100 differentially expressed genes related to cytoplasmic translation, cell cycle and DNA damage, and inflammation (Figure 3B). A heatmap of the top 100 genes based on FDR-corrected p values showed clear distinctions between infected and uninfected conditions and revealed distinct upregulated gene signatures between 3-h-infected versus 24- and 48-h-infected macrophages (Figure 3C). This analysis separated samples into an uninfected and 3-h-infected group, and a late infected group (24 and 48 h), consistent with the grouping pattern shown in the MDS plot (Figure 3C), suggesting that mycobacterial infection of hPSC-macrophages triggered time-dependent transcriptomic responses.
Figure 3.
Host cell responses of hPSC-macrophages to M. abscessus
(A) Multidimensional scaling (MDS) plot showing the relationship between samples representing uninfected (n = 4) and infected macrophages were derived from the cell lines PB1 at 3 h (3h, n = 4), 24 h (24h, n = 3) and 48 h (48h, n = 3) post-infection. “n,” number of independent experiments.
(B) Gene ontology analysis for top 100 differentially expressed genes between samples representing 48 h post-infection and 3 h post-infection.
(C) Heatmap and hierarchical clustering analysis of the top 100 differentially expressed genes across the samples shown in (A). Hierarchy relationship between samples is shown across the top, and individual genes are shown on the right. Heatmap color represents Log2 fold change from the mean row expression.
(D) ELISA assay of secreted human IL8 for PB1 hPSC-macrophages infected with M. abscessus. The table shows concentrations of IL8 in the indicated experiments (Exp 1–3), and lower panel shows fold change in IL8 concentration using data from the table above. Statistics were calculated by a one-way ANOVA test. “n” = 3, independent experiments.
In addition to the independent experiments performed with the PB1 line described above, we also tested macrophages representing additional independent genetic backgrounds over an extended infection period time to 4 dpi (see experimental procedures). MDS analysis showed that uninfected and 3-h-infected samples were separated from 2-dpi and 4-dpi samples on the first dimension, and the 2-dpi samples were more tightly clustered than those representing other time points, confirming our conclusion that mycobacterial infection induced a common transcriptomic response program that developed irrespective of genetic background variability of the host cells (Figures S3A and S3C). By 4 dpi, this uniformity was decreased, perhaps reflecting degradation of the culture by the infection and potential donor variability in the immune response at the late infection stage. Similar to the results shown in Figure 3, gene ontology analysis showed that the major changes in the top 100 differentially expressed genes related to infection and immune responses (Figure S3B). This analysis also separated samples into an uninfected and 3-h-infected group, and a late infected group (2 and 4 dpi), consistent with the grouping pattern shown in the MDS plot (Figure S3C). Using the self-organizing maps technique (Tamayo et al., 1999), we classified time-variable cytokine genes further into three clusters based on their expression pattern over the course of infection (Figure S3D), which also showed a degree of variability of donors in the regulation of cytokine gene expression. However, overall, these analyses indicate that the immediate effect of M. abscessus infection is to induce a common transcriptomic response in the infected host cells.
To further understand cytokine induction by M. abscessus, we examined the kinetics of IL8 secretion, a well-documented response of macrophages to microbial infection (Lévêque et al., 2017; Meera et al., 2004; Khan et al., 1995). Using ELISA, we found that IL8 secretion was an immediate response of hPSC-macrophages to M. abscessus infection, with elevated levels of IL8 detected in the culture media within 24 h (Figure 3D). A similar pattern of IL8 secretion was observed with the three independent PSC lines tested (Figures 3D and S3E). Overall, these results suggest that our hPSC-macrophages were able to develop a host response to M. abscessus in a time-dependent manner.
hPSC-macrophage-resident M. abscessus as a model for assessing antibiotic sensitivity
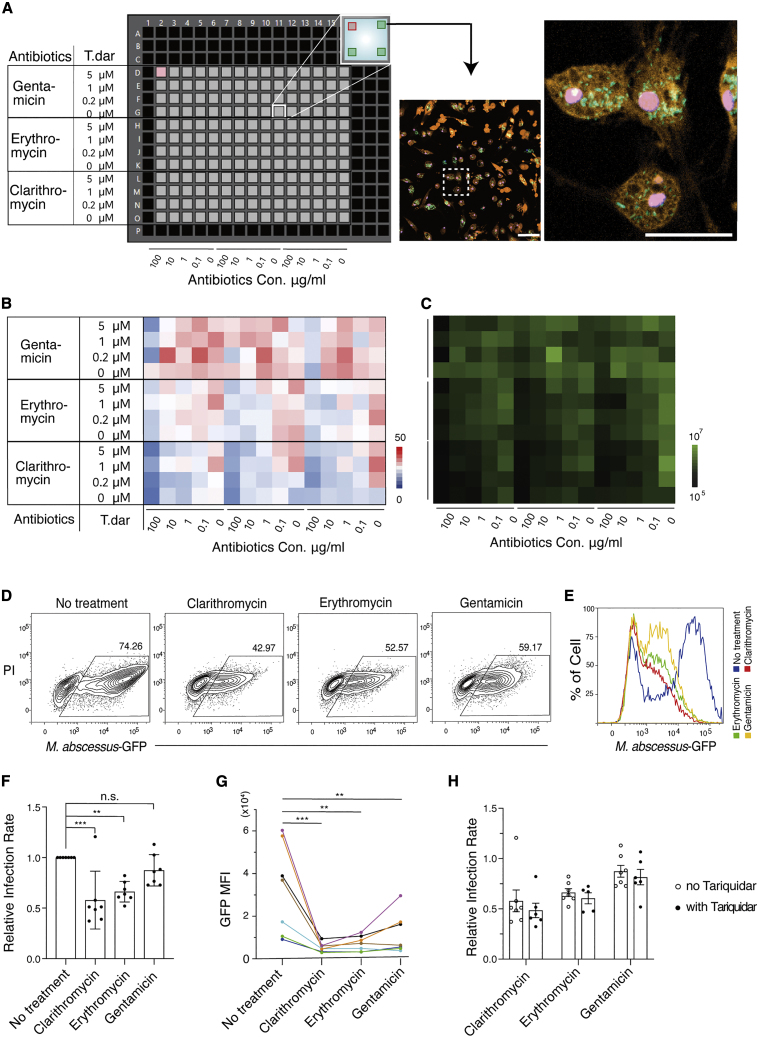
Antibiotic resistance is a defining characteristic of mycobacteria with important clinical ramifications (Johansen et al., 2020). A key challenge for such treatments is the ability to kill these bacteria when they are contained within host cells, especially macrophages. We therefore examined whether M. abscessus-infected hPSC-macrophages could be used to test the efficacy of antibiotic treatments. For these experiments we utilized generic antibiotics that are used clinically for treating M. abscessus infection, including clarithromycin, erythromycin, and gentamicin. However, there is limited knowledge concerning the ability of these antibiotics to target intracellular M. abscessus in human macrophages. Such knowledge could underpin further research into the development of antibiotic resistance, especially to clarithromycin, and whether this resistance is influenced by M. abscessus’ intracellular location (Mougari et al., 2016; Rubio et al., 2015). In this experiment, we also examined the consequence of antibiotic treatments in the presence of tariquidar, a p-glycoprotein drug efflux pump inhibitor previously shown to increase the antibiotic sensitivity of macrophage-resident M. tuberculosis (Figure 4A) (Hartkoorn et al., 2007).
Figure 4.
Analysis of M. abscessus antibiotic sensitivity
(A) Experimental setup of a 384-well plate format for a cross titration of the indicated antibiotics and the efflux inhibitor tariquidar (T. dar). Concentrations of tariquidar are shown on the left-hand side, and concentrations of antibiotics are shown below. Four fields of view per well were acquired for fluorescent imaging using high-speed confocal microscopy as indicated in the inset. A representative confocal fluorescent image showing merged channels of GFP+ M. abscessus, tdTOMATO+ macrophages, and DAPI+ nuclei. Scale bar left: 100 μm, right: 50 μm.
(B) Heatmap representation indicating the average number of intracellular GFP+ M. abscessus per tdTOMATO+ macrophages in each well under the conditions indicated. Concentrations of T. dar are shown on the left-hand side, and concentrations of antibiotics are shown below.
(C) Heatmap representation indicating the average GFP intensity of each well under the conditions indicated in (B).
(D) Representative flow cytometry plots assessing the degree of M. abscessus infection in the presence of 100 ug/ml of the indicated antibiotic. Cells shown in each plot were pre-gated for CD45 and CD14 double positivity.
(E) Representative histogram of GFP mean fluorescence intensity (MFI) of infected cultures treated with/without antibiotics at 100 ug/ml. Cells shown in the histogram were pre-gated for CD45 and CD14 double positivity.
(F) Histogram summarizing quantitative analyses of antibiotic efficacy as determined by flow cytometry. Relative infection rate was calculated as the frequency of infected cells in antibiotic-treated samples relative to the corresponding untreated control sample. Data show the mean ± SEM of replicates representing seven independent experiments (n = 7). Statistical analysis was calculated by a one-way ANOVA test, P(clarithromycin) = 0.0003, P(erythromycin) = 0.0031, P(gentamicin) = 0.3917.
(G) Summary of GFP intensity of GFP+ M. abscessus-infected macrophages. Each colored data point and line represents a replicate of seven independent experiments (n = 7). Statistical analysis was calculated by a one-way ANOVA test, P(clarithromycin) = 0.0006, P(erythromycin) = 0.0012, P(gentamicin) = 0.0080.
(H) Histogram summarizing quantitative analyses of antibiotic sensitivity in combination with (closed circles) or without (open circles) tariquidar, as determined by flow cytometry. Relative infection rate was calculated by normalizing the frequency of infected cells in antibiotic-treated samples to their untreated control samples. Data show the mean ± SEM of replicates of six independent experiments (n = 6). Statistical analysis was calculated by a two-way ANOVA test.
Our experimental setup incorporated macrophages and mycobacteria expressing distinct fluorescent proteins, enabling the use of high-resolution and high-throughput fluorescence confocal microscopy to visualize and quantify the extent of infection (Figure 4A). After 4 days of treatment with clinically relevant concentrations of antibiotics, hPSC-macrophages retained intracellular GFP+ mycobacteria, consistent with the widely documented multidrug resistance of M. abscessus (Figures 4B and 4C). Nevertheless, higher concentrations of antibiotics (100 ug/ml) reduced the frequency of intracellular mycobacteria (Figures 4B, 4C, and S4A). In contrast to results obtained from M. tuberculosis infection of THP1 leukemic cells, our experiments showed that the efflux inhibitor, tariquidar, did not improve antibiotic sensitivity against M. abscessus infection. Although these experiments involved different species of mycobacteria, we believe this discrepancy is most likely due to the lack of ABCB1 expression on hPSC-macrophages, a phenotype that they share with peripheral blood-derived macrophages. In this context, it is noteworthy that ABCB1 is upregulated in leukemic cells (Drach et al., 1992; Moreau et al., 2011), potentially making them amenable to targeting with ABCB1 inhibitors, particular in the context of intracellular mycobacterium antibiotic resistance (Hartkoorn et al., 2007). Furthermore, the number of hPSC-macrophages was not significantly affected by tariquidar or high concentration of antibiotics, suggesting neither of these agents substantially reduced macrophage viability (Figures S4B and S4C).
We used flow cytometry to validate the efficacy of the highest concentration of antibiotic (100 ug/ml) in the presence or absence of tariquidar. Our results showed that clarithromycin and erythromycin were able to significantly reduce the frequency of M. abscessus infection (Figures 4D and 4F). Furthermore, all three antibiotics were able to reduce the M. abscessus-GFP fluorescence intensity of infected macrophages, suggesting the ability to inhibit the intracellular growth of M. abscessus (Figures 4E and 4G). These side-by-side comparisons also suggested that clarithromycin outperformed erythromycin and gentamicin in the clearance and/or growth inhibition of intracellular M. abscessus in human macrophages. Consistent with the results of the high-throughput analysis, we found that tariquidar did not improve the efficacy of these three antibiotics (Figure 4H). Collectively, these results demonstrate the hPSC-macrophage model can be used to assess the ability of antibiotics to kill intracellular M. abscessus, and this model could be readily adapted for high-throughput drug testing and screening.
Discussion
In this study, we describe a simple pluripotent stem cell-based model for M. abscessus infection using hPSC-derived macrophages, permitting studies of host cell immune responses and drug testing. In our differentiation protocol, M-CSF was the only myeloid growth factor required to induce functional macrophages from hPSCs (see experimental procedures) (Figure 1). In distinction to previous methods that often include multiple myeloid growth factors, such as interleukin (IL) 1, IL3, IL6, granulocyte-macrophage colony stimulating factor (GM-CSF), and tissue necrosis factor (Lyadova et al., 2021), we found that using M-CSF as the sole myeloid factor was sufficient to efficiently induce functional macrophages from the endothelial cell stage onward, and it maintained hPSC-macrophages over a prolonged period of in vitro culture (Figure S2). An optimized version of the basal medium free of albumin also enabled the derivation of endodermal cell lineages from hPSCs and primary human tissue (Sun et al., 2022), potentially facilitating co-culture studies. Our streamlined differentiation protocol efficiently generated macrophages that resembled those created with more complex methods (as reviewed in Lyadova et al., 2021), forming a robust platform upon which to undertake functional studies. This simplified cell culture condition negated the need to include pro-inflammatory cytokines, which may have advantages for dissecting downstream infection-associated events.
Using this chemically defined system, we were able to examine key aspects of M. abscessus intracellular infection, an environmentally common non-TB species that is an emerging pathogen in specific clinical settings. Our results showed that M. abscessus robustly infected hPSC-macrophages through macrophage’s phagocytotic activity and then stably resided in vacuoles in groups or as individual cells (Figure 2D), consistent with previous findings from animal models (Bernut et al., 2017). The macrophage phagosome represents an ideal intracellular habitat, providing a physical barrier that may serve to protect mycobacterium from extracellular antibiotics (Queval et al., 2017). Our system enables intracellular infection, potentially allowing future investigation of mycobacterial replication inside host cells. Employing genetically engineered fluorescent macrophages and mycobacteria enabled us to easily identify and quantify the mycobacterium present within infected cells, creating a system that could be readily adapted to high-throughput image-based phenotypic examination of sensitivities for three antibiotics (Figure 4). Our proof-of-concept experiments confirmed the superiority of clarithromycin for targeting intracellular M. abscessus in human macrophages compared with erythromycin and gentamicin, laying the foundation for future large-scale high-throughput screens and the quantitative comparison of anti-mycobacterial chemical compounds. In addition, the ability of our system to differentiate the potency of specific treatments suggests it could be used to optimize new formulations of clinal antibiotic treatments and to study the development of antibiotic resistance inside host cells.
Infected hPSC-macrophages showed dynamic host responses to M. abscessus infection, highlighting the potential of our model for studying pathogen-host interactions. In this regard, our observation of IL8 induction in response to M. abscessus infection (Figures 3D and S3E) mirrors the upregulation of this cytokine seen in genetically susceptible individuals, such as patients with cystic fibrosis (Johansen et al., 2020; Lévêque et al., 2017; Khan et al., 1995). We anticipate that our fluorescence-based screening system would be also compatible with GFP-expressing Mtb and complement previous platforms for tuberculosis drug discovery (Han et al., 2019). In summary, our pluripotent stem cell-based human model provides a flexible platform for studying human macrophage biology and host-pathogen interactions.
Experimental procedures
Full details of experimental procedures are provided in supplemental information.
hPSC lines and macrophage differentiation
Work related to pluripotent stem cell lines was conducted in accordance with RCH Human Research Ethics Committee 33001A. The four human PSC lines used in this study have been described previously: ESC line H1 and the iPSC lines PB1 (PB001), PB4 (PB004), PB5 (PB005), RM (RM3.5), AF1. Methods for hPSC culture and macrophage differentiation in detail are provided in supplemental information.
Generation of GFP-expressing Mycobacterium abscessus
M. abscessus ATCC 19977 was obtained from the American Type Culture Collection (In Vitro Technologies, L948). The GFP-expressing variant was generated using the pTEC-15 GFP-expressing plasmid previously constructed by the Ramakrishnan Lab. Full details are included in the supplemental information.
Mycobacterium abscessus infection
Approximately 30,000–50,000 hPSC-derived macrophages were plated to each well of a 96-well plate 1 day before infection. On the day of infection, M. abscessus-GFP culture was harvested and washed twice in PBS. Single cell suspensions were prepared and counted in a hemocytometer to determine bacterial cell numbers. hPSC-derived macrophages were inoculated with M. abscessus-GFP at the MOI as indicated. For flow cytometry analysis of infected macrophages, cells were enzymatically detached and stained with conjugated anti-CD14-PE/Cy7 and anti-CD45-BV421 antibodies. To avoid contamination in the flow cytometer by live M. abscessus-GFP (ATCC, 19977), samples were fixed with 4% paraformaldehyde solution in PBS prior to flow cytometry analysis.
High-throughput confocal imaging
TdTOMATO-expressing hPSC-derived macrophages were plated into a 384-well plate at the density of 2,500 cells/well. Cells were adherent to the plate surface overnight, which was followed by inoculation of GFP-expressing M. abscessus at the MOI of 5 (0 dpi). On 1 dpi, antibiotics were added to the indicated wells at the indicated concentrations, in the presence or absence of the efflux inhibitor tariquidar. On 5 dpi, cultures were fixed with 3.7% paraformaldehyde and stained with DAPI (4′,6-diamidino-2-pheylindole) for high-throughput confocal imaging analysis. Cells were imaged using an automated spinning disc confocal microscope (CV8000, Yokogawa). Four fields of view (FOV) were acquired for each condition, and a single maximum intensity projection (MIP) image from five Z planes with a 2-μm Z step (10-μm range) was generated for each FOV. Individual images were captured using a 20x air objective and 250-ms laser exposure time. MIP images were used for downstream image analysis using particle analysis detection protocol (CellPathfinder, Yokogawa).
Statistical analysis
Statistical analyses were performed in Prism (Graphpad, Version 8.0.2) by one-way ANOVA or two-way ANOVA tests. Analyses are indicated in the figure legends. The data are reported as mean ± SEM or SD as indicated. Statistical significance was indicated in the figure legends.
Data availability
RNA sequencing data are available in the GEO data repository with the identification numbers GSE183865 and GSE207456.
Author contributions
Conceptualization, E.G.S., P.S., S. Sarker, and S. Sun. Methodology, S. Sun., S. Sarker, and E.G.S. Investigation, S. Sun, S. Sarker, E.N., A.H., and K.S. Formal analysis, S. Sun, S. Sarker, A.H., M.S., H.N., and M.R. Supervision, A.G.E., E.G.S., and P.S. Writing – original draft, S. Sun and E.G.S. Writing – review & editing, all authors. Funding acquisition, A.G.E., E.G.S., M.R., and P.S. All authors contributed to the article and approved the submitted version.
Acknowledgments
Eric Hanssen and Zlatan Trifunovic for electron microscopy; Tanya Labonne for the PB001-tdTOMATO hiPSC line; Steve Graham, Freya Bruveris, and Holly Voges for helpful discussions; Melbourne Ian Holmes Imaging Center (Bio21 Institute) and the Stafford Fox Disease Modeling Facility (Murdoch Children’s Research Institute).
This study was funded by the National Health & Medical Research Council of Australia through research fellowships awarded to A.G.E. (GNT1117596) and E.G.S (GNT1079004), project grants awarded to A.G.E. and E.G.S. (GNT1068866, GNT1129861) and a new investigator grant awarded to E.S.N (GNT1164577), by the Australian Research Council Special Research Initiative in Stem Cells (Stem Cells Australia), and by the Stafford Fox Medical Research Foundation. M.R. is funded by an NHMRC Ideas Grant (APP1180905). Additional infrastructure funding to the Murdoch Children's Research Institute was provided by the Australian Government National Health and Medical Research Council Independent Research Institute Infrastructure Support Scheme and the Victorian Government’s Operational Infrastructure Support Program. The Australian Regenerative Medicine Institute is supported by grants from the State Government of Victoria and the Australian Government. The Novo Nordisk Foundation Center for Stem Cell Medicine is supported by Novo Nordisk Foundation grants (NNF21CC0073729).
Conflicts of interest
E.G.S. is member of the editorial board of Stem Cell Reports.
Published: August 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.07.013.
Contributor Information
Sohinee Sarkar, Email: sohinee.sarkar@mcri.edu.au.
Edouard G. Stanley, Email: ed.stanley@mcri.edu.au.
Supplemental information
References
- Bernut A., Herrmann J.L., Ordway D., Kremer L. The diverse cellular and animal models to decipher the physiopathological traits of Mycobacterium abscessus infection. Front. Cell. Infect. Microbiol. 2017;7:100–108. doi: 10.3389/fcimb.2017.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant J.M., Grogono D.M., Greaves D., Foweraker J., Roddick I., Inns T., Reacher M., Haworth C.S., Curran M.D., Harris S.R., et al. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet. 2013;381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Yakala G.K., Van Den Hil F.E., Cochrane A., Mummery C.L., Orlova V.V. Stem cell Reports article differentiation and functional comparison of Monocytes and macrophages from hiPSCs with peripheral blood Derivatives. Stem Cell Rep. 2019;12:1282–1297. doi: 10.1016/j.stemcr.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diacovich L., Gorvel J.P. Bacterial manipulation of innate immunity to promote infection. Nat. Rev. Microbiol. 2010;8:117–128. doi: 10.1038/nrmicro2295. [DOI] [PubMed] [Google Scholar]
- Drach D., Zhao S., Drach J., Mahadevia R., Gattringer C., Huber H., Andreeff M. 1992. Subpopulations of Normal Peripheral Blood and Bone Marrow Cells Express a Functional Multidrug Resistant Phenotype. [PubMed] [Google Scholar]
- Han H.-W., Seo H.-H., Jo H.-Y., Han H.J., Falcão V.C.A., Delorme V., Heo J., Shum D., Choi J.-H., Lee J.-M., et al. Drug discovery platform targeting M. tuberculosis with human embryonic stem cell-derived macrophages. Stem Cell Rep. 2019;13:980–991. doi: 10.1016/j.stemcr.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartkoorn R.C., Chandler B., Owen A., Ward S.A., Bertel Squire S., Back D.J., Khoo S.H. Differential drug susceptibility of intracellular and extracellular tuberculosis, and the impact of P-glycoprotein. Tuberculosis. 2007;87:248–255. doi: 10.1016/j.tube.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Hendriks W.T., Warren C.R., Cowan C.A. Genome editing in human pluripotent stem cells: Approaches, Pitfalls, and solutions. Cell Stem Cell. 2016;18:53–65. doi: 10.1016/j.stem.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovs A., Rybtsov S., Ng E.S., Stanley E.G., Elefanty A.G., Medvinsky A. Human haematopoietic stem cell development: from the embryo to the dish. Development. 2017;144:2323–2337. doi: 10.1242/dev.134866. [DOI] [PubMed] [Google Scholar]
- Johansen M.D., Herrmann J.L., Kremer L. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat. Rev. Microbiol. 2020;18:392–407. doi: 10.1038/s41579-020-0331-1. [DOI] [PubMed] [Google Scholar]
- Kim B.-R., Kim B.-J., Kook Y.-H., Kim B.-J. Phagosome escape of rough Mycobacterium abscessus strains in Murine macrophage via phagosomal Rupture can Lead to type I Interferon Production and their cell-to-cell Spread. Front. Immunol. 2019;10:125. doi: 10.3389/fimmu.2019.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang J., Cheng Y., Rolfe A., Hammack C., Vera D., Kyle K., Wang J., Meissner T.B., Ren Y., Cowan C., Tang H. An hPSC-derived tissue-resident macrophage model Reveals differential responses of macrophages to ZIKV and DENV infection. Stem Cell Rep. 2018;11:348–362. doi: 10.1016/j.stemcr.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.R., Sheng W.H., Hung C.C., Yu C.J., Lee L.N., Hsueh P.R. Mycobacterium abscessus complex infections in humans. Emerg. Infect. Dis. 2015;21:1638–1646. doi: 10.3201/eid2109.141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévêque M., le Trionnaire S., del Porto P., Martin-Chouly C. The impact of impaired macrophage functions in cystic fibrosis disease progression. J. Cyst. Fibros. 2017;16:443–453. doi: 10.1016/j.jcf.2016.10.011. [DOI] [PubMed] [Google Scholar]
- Lyadova I., Gerasimova T., Nenasheva T. Macrophages derived from human induced pluripotent stem cells: the Diversity of protocols, future Prospects, and Outstanding Questions. Front. Cell Dev. Biol. 2021;9:640703. doi: 10.3389/fcell.2021.640703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M., Eidelman O., Zhang J., Paweletz C., Caohuy H., Yang Q., Jacobson K.A., Heldman E., Huang W., Jozwik C., et al. Digitoxin mimics gene therapy with CFTR and suppresses hypersecretion of IL-8 from cystic fibrosis lung epithelial cells. Proc. Natl. Acad. Sci. USA. 2004;101:7693–7698. doi: 10.1073/pnas.0402030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau A., le Vee M., Jouan E., Parmentier Y., Fardel O. Drug transporter expression in human macrophages. Fundam. Clin. Pharmacol. 2011;25:743–752. doi: 10.1111/j.1472-8206.2010.00913.x. [DOI] [PubMed] [Google Scholar]
- Mougari F., Bouziane F., Crockett F., Nessar R., Chau F., Veziris N., Sapriel G., Raskine L., Cambau E. Selection of resistance to clarithromycin in Mycobacterium abscessus Subspecies. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.00943-16. e00943-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neehus A.L., Lam J., Haake K., Merkert S., Schmidt N., Mucci A., Ackermann M., Schubert M., Happle C., Kühnel M.P., et al. Impaired IFNγ-Signaling and mycobacterial clearance in IFNγR1-Deficient human iPSC-derived macrophages. Stem Cell Rep. 2018;10:7–16. doi: 10.1016/j.stemcr.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nessar R., Cambau E., Reyrat J.M., Murray A., Gicquel B. Mycobacterium abscessus: a new antibiotic nightmare. J. Antimicrob. Chemother. 2012;67:810–818. doi: 10.1093/jac/dkr578. [DOI] [PubMed] [Google Scholar]
- Ng E.S., Azzola L., Bruveris F.F., Calvanese V., Phipson B., Vlahos K., Hirst C., Jokubaitis V.J., Yu Q.C., Maksimovic J., et al. Differentiation of human embryonic stem cells to HOXA + hemogenic vasculature that resembles the aorta-gonad-mesonephros. Nat. Biotechnol. 2016;34:1168–1179. doi: 10.1038/nbt.3702. [DOI] [PubMed] [Google Scholar]
- Ordway D., Henao-Tamayo M., Smith E., Shanley C., Harton M., Troudt J., Bai X., Basaraba R.J., Orme I.M., Chan E.D. Animal model of Mycobacterium abscessus lung infection. J. Leukoc. Biol. 2008;83:1502–1511. doi: 10.1189/jlb.1007696. [DOI] [PubMed] [Google Scholar]
- Queval C.J., Brosch R., Simeone R. The macrophage: a Disputed Fortress in the Battle against Mycobacterium tuberculosis. Front. Microbiol. 2017;8:2284. doi: 10.3389/fmicb.2017.02284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A.L., Viljoen A., Bah A., Simeone R., Bernut A., Laencina L., Deramaudt T., Rottman M., Gaillard J.L., Majlessi L., et al. The distinct fate of smooth and rough Mycobacterium abscessus variants inside macrophages. Open Biol. 2016;6:160185. doi: 10.1098/rsob.160185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio M., March F., Garrigó M., Moreno C., Español M., Coll P. Inducible and acquired clarithromycin resistance in the Mycobacterium abscessus complex. PLoS One. 2015;10:e0140166. doi: 10.1371/journal.pone.0140166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Li J.Y., Nim H.T., Piers A., Ramialison M., Porrello E.R., Konstantinov I.E., Elefanty A.G., Stanley E.G. CD90 Marks a Mesenchymal program in human Thymic epithelial cells in vitro and in Vivo. Front. Immunol. 2022;13:846281. doi: 10.3389/fimmu.2022.846281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan T.Z., Wagener J.S., Bost T., Martinez J., Accurso F.J., Riches D.W. Early pulmonary inflammation in infants with cystic fibrosis. Am. J. Respir. Crit. Care Med. 1995;151:1075–1082. doi: 10.1164/ajrccm/151.4.1075. [DOI] [PubMed] [Google Scholar]
- Takaki K., Davis J.M., Winglee K., Ramakrishnan L. Evaluation of the pathogenesis and treatment of Mycobacterium marinum infection in zebrafish. Nat. Protoc. 2013;8:1114–1124. doi: 10.1038/nprot.2013.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo P., Slonim D., Mesirov J., Zhu Q., Kitareewan S., Dmitrovsky E., Lander E.S., Golub T.R. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci. USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi M.F. Innate immune responses to infection. J. Allergy Clin. Immunol. 2005;116:241–249. doi: 10.1016/j.jaci.2005.05.036. [DOI] [PubMed] [Google Scholar]
- Vaughan-Jackson A., Stodolak S., Ebrahimi K.H., Browne C., Reardon P.K., Pires E., Gilbert-Jaramillo J., Cowley S.A., James W.S. Differentiation of human induced pluripotent stem cells to authentic macrophages using a defined, serum-free, open-source medium. Stem Cell Rep. 2021;16:1735–1748. doi: 10.1016/j.stemcr.2021.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn T.A., Chawla A., Pollard J.W. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data are available in the GEO data repository with the identification numbers GSE183865 and GSE207456.