Abstract
Virulence factors of Pseudomonas aeruginosa include hydrogen cyanide (HCN). This secondary metabolite is maximally produced at low oxygen tension and high cell densities during the transition from exponential to stationary growth phase. The hcnABC genes encoding HCN synthase were identified on a genomic fragment complementing an HCN-deficient mutant of P. aeruginosa PAO1. The hcnA promoter was found to be controlled by the FNR-like anaerobic regulator ANR and by the quorum-sensing regulators LasR and RhlR. Primer extension analysis revealed two transcription starts, T1 and T2, separated by 29 bp. Their function was confirmed by transcriptional lacZ fusions. The promoter sequence displayed an FNR/ANR box at −42.5 bp upstream of T2 and a lux box centered around −42.5 bp upstream of T1. Expression of the hcn genes was completely abolished when this lux box was deleted or inactivated by two point mutations in conserved nucleotides. The lux box was recognized by both LasR [activated by N-(oxododecanoyl)-homoserine lactone] and RhlR (activated by N-butanoyl-homoserine lactone), as shown by expression experiments performed in quorum-sensing-defective P. aeruginosa mutants and in the N-acyl-homoserine lactone-negative heterologous host P. fluorescens CHA0. A second, less conserved lux box lying 160 bp upstream of T1 seems to account for enhanced quorum-sensing-dependent expression. Without LasR and RhlR, ANR could not activate the hcn promoter. Together, these data indicate that expression of the hcn promoter from T1 can occur under quorum-sensing control alone. Enhanced expression from T2 appears to rely on a synergistic action between LasR, RhlR, and ANR.
Pseudomonas aeruginosa is a gram-negative bacterium that can cause serious infections in patients suffering from cystic fibrosis, cancer, infection with human immunodeficiency virus, or severe burn wounds (16, 29). The pathogenesis of P. aeruginosa infections is due to the production of both cell-associated and extracellular virulence factors. One of these extracellular compounds, hydrogen cyanide (HCN), has been found at relatively high concentrations in patients with freshly infected burns (27). Evidence for HCN being a virulence factor comes from an experimental infection model in which an hcn insertion mutant of P. aeruginosa had a strongly reduced ability to kill the nematode Caenorhabditis elegans (L. Gallagher and C. Manoil, Abstr. Pseudomonas '99: Biotechnol. Pathog., abstr. 75, 1999). Cyanide is a potent inhibitor of cytochrome c oxidase, the terminal component of the aerobic respiratory chain in many organisms, and of several other important metalloenzymes (52).
In Pseudomonas spp., HCN biosynthesis is catalyzed by the membrane-bound enzyme HCN synthase, which forms HCN and CO2 from glycine (8). The enzyme is sensitive to molecular oxygen and has been purified only partially from a Pseudomonas species (60); hence, little is known about the biochemistry of the enzymatic reaction. In P. fluorescens, the structural hcnABC genes encoding HCN synthase are clustered and probably form an operon (34). From an analysis of nucleotide sequence data, it can be concluded that HCN synthase essentially functions as a glycine dehydrogenase/oxidase, transferring four electrons to a cyanide-resistant branch of the aerobic respiratory chain (5).
In P. aeruginosa and P. fluorescens, cyanogenesis proceeds at low oxygen concentrations (9, 11) and depends on ANR (anaerobic regulator of arginine deiminase and nitrate reductase), a transcriptional regulator, which is converted to its active form under low oxygen supply. ANR belongs to the FNR (fumarate and nitrate reductase regulator) family of transcriptional regulators (53); anr mutants of both species produce little HCN (34, 62). ANR can activate target promoters by binding to conserved sequences known as ANR boxes, with a recognition specificity which is similar but not identical to that of FNR (58).
Cell density is a second parameter that influences cyanogenesis in Pseudomonas spp. It was discovered 20 years ago that optimal expression of HCN synthase occurs during the transition from the exponential to the stationary phase (12). This expression pattern is characteristic of regulatory mechanisms which more recently have been termed quorum sensing (21). In P. aeruginosa, the production of virulence factors and secondary metabolites is under quorum-sensing control involving N-acyl-homoserine lactone signals (20). These signals are produced by an autoinduction mechanism with increasing cell density, and they are assumed to diffuse freely through the cell envelope. When reaching a threshold concentration, they activate LuxR-type transcriptional regulators which control the expression of target genes by recognizing conserved sequences termed lux boxes (17, 20). P. aeruginosa contains two interdependent quorum-sensing systems (43, 44, 57). In the lasRI system, the LasI protein directs the synthesis of N-(3-oxododecanoyl)-homoserine lactone (OdDHL), which triggers the transcriptional activator, LasR, to induce the expression of virulence factors such as elastases (LasB and LasA), exotoxin A, and alkaline protease (47). In addition, the las system autoregulates the lasI gene, leading to the production of more OdDHL (51), and upregulates the second quorum-sensing system, consisting of the transcriptional activator RhlR and the RhlI protein, which directs the synthesis of N-butanoyl-homoserine lactone (BHL). The rhlRI system regulates the production of multiple exoproducts including rhamnolipids, elastase (LasB), pyocyanin, and cyanide (7, 33, 57).
From previous studies, it appears that both autoinducers OdDHL and BHL contribute to the regulation of the hcn genes in P. aeruginosa (33, 45, 56, 57). However, it is not clear whether the OdDHL-LasR team can exert its effect on hcn gene expression directly or indirectly, i.e., by activating the rhlRI system. Furthermore, the question arises of how anaerobic control by ANR and quorum-sensing control by LasR and RhlR may cooperate during initiation of transcription of the hcn genes. In the present study, we have undertaken a systematic analysis of the hcn promoter region of P. aeruginosa. We show that the simultaneous action of three activators ANR, LasR, and RhlR ensures optimal expression of the hcnABC cluster.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used in this study are listed in Table 1. P. aeruginosa and Escherichia coli strains were routinely grown on nutrient agar plates and in nutrient yeast broth (NYB) with aeration at 37°C, whereas P. fluorescens strains were cultivated on these media at 30°C. For determination of HCN production, P. aeruginosa strains were grown with shaking at 180 rpm under mild oxygen limitation in 100-ml Erlenmeyer flasks containing 40 ml of a synthetic glycine minimal medium (MMC) described by Castric (10). Severe oxygen limitation was achieved by growing the cells in tightly closed 125-ml bottles containing 60 ml of MMC, with gentle shaking; at the end of growth, the oxygen present initially was consumed by the cells. Antibiotics were used at the concentrations (in micrograms per milliliter) indicated in parentheses: for P. aeruginosa strains, carbenicillin (250), chloramphenicol (250), gentamicin (10), tetracycline (125), and mercuric chloride (10); for P. fluorescens, gentamicin (10) and tetracycline (125); and for E. coli, ampicillin (100), chloramphenicol (25), gentamicin (10), and tetracycline (25). 5-Bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) was incorporated into solid media at 0.02% to monitor β-galactosidase expression (49).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Genotype or description; phenotype | Reference or origin |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Wild type | 30 |
| PAO2324 | met-9020 catA1 nar-9011 tyu-9009 puuD6 hcn | 36 |
| PAO6261 | Δanr | 61 |
| PAO6289 | hcnA′-′lacZ | This study (Fig. 2) |
| PAO6290 | Δanr hcnA′-′lacZ; PAO6261 derivative | This study |
| PAO6293 | rhlR::Tn501 hcnA′-′lacZ; Hgr; PDO111 derivative | This study |
| PAO6326 | lasR::pUC19 (carrying ′lasR′ on a 0.4-kb PstI fragment) hcnA′-′lacZ; Cbr; PAOR derivative | This study |
| PAO6330 | ΔlasRI | C. Reimmann, unpublished |
| PAO6351 | rhlR::Tn501 derivative of PAOJP1; Hgr Tcr | This study |
| PAOJP1 | ΔlasI::Tcr; Tcr | 43 |
| PAOJP2 | ΔlasI::Tcr ΔrhlI::Tn501; Tcr Hgr | 43 |
| PAOR | lasR::pUC19 (carrying ′lasR′ on a 0.4-kb PstI fragment); Cbr | 32 |
| PDO111 | rhlR::Tn501; Hgr | 7 |
| P. fluorescens CHA0 | Wild type | 55 |
| E. coli DH5α | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 deoR Δ(lacZYA-argF)U169 λ-(φ 80dlacZΔM15) | 49 |
| Plasmids | ||
| pBluescript II KS | Cloning vector, ColE1 replicon; Apr | Stratagene |
| pBBR1MCS | Broad-host-range cloning vector; Cmr | 31 |
| pJF118EH | Cloning vector, ColE1 replicon, lacIq, ptac; Apr | 22 |
| pKT240 | Cloning vector, IncQ replicon, Mob; Apr Kmr | 3 |
| pME3071 | pMMB67EH with a 5-kb XhoI-HindIII fragment of CHA0 containing hcnABC′ | 34 |
| pME3087ΔE | Suicide vector, ColE1 replicon, Mob; EcoRI site deleted; Tcr | 59 |
| pME3326 | pBBR1MCS with a 6.6-kb BamHI fragment of PAO1 containing the hcnABC region | This study |
| pME3333 | pBBR1MCS with a 4.25-kb SalI-XbaI fragment of PAO1 containing the hcnABC genes | This study (Fig. 2A) |
| pME3336 | pBluescript with a 0.78-kb EcoRV fragment of PAO1 containing the hcn promoter region and the first 9 codons of hcnA | This study |
| pME3823 | pKT240 with a 0.78-kb EcoRV hcnA upstream fragment and a translational hcnA′::′lacZ fusion at the EcoRV site in hcnA | This study |
| pME3825 | pME3087ΔEcoRI with a 4.8-kb EcoRV-SmaI fragment containing a translational hcnA′::′lacZ fusion | This study (Fig. 2C) |
| pME3826 | pME6010 with a 0.78-kb hcnA upstream fragment and a translational hcnA′::′lacZ fusion at the EcoRV site in hcnA | This study |
| pME3827 | pME6001 carrying lasR on a 1.1-kb PvuI-KpnI fragment; Gmr | This study |
| pME3832 | pME6010 with the 111-bp hcnA upstream fragment and a translational hcnA′::′lacZ fusion at the EcoRV site in hcnA | This study (Fig. 6) |
| pME3835 | pME6010 with the 88-bp hcnA upstream fragment and a translational hcnA′::′lacZ fusion at the EcoRV site in hcnA | This study (Fig. 6) |
| pME3837 | pME6010 with the 143-bp hcnA upstream fragment and a translational hcnA′::′lacZ fusion at the EcoRV site in hcnA | This study (Fig. 6) |
| pME3840 | pME6001 carrying rhlRI′ on a 2-kb PstI fragment; Gmr | This study |
| pME3844 | Derivative of pME3837 carrying 2 point mutations | This study (Fig. 6) |
| pME3850.1 | pME6522 with 130 bp upstream of transcriptional start site T1 | This study (Fig. 2D) |
| pME3850.2 | pME6522 with 159 bp upstream of transcriptional start site T2 | This study (Fig. 2D) |
| pME3852 | Derivative of pME3826 with 6-bp insertion specifying an SphI site | This study (Fig. 6) |
| pME6000 | Cloning vector derived from pBRR1MCS; Tcr | 37 |
| pME6001 | Gmr derivative of pME6000 | 6 |
| pME6010 | pACYC177-pVS1 shuttle vector; Tcr | 28 |
| pME6522 | pME6010 containing the promoterless lacZ gene on a 3.3-kb PstI/DraI fragment | 6 |
| pMJG1.7 | pSW200 carrying the lasR gene on a 1.7-kb EcoRI-SacI fragment of P. aeruginosa PAO1; Apr | 23 |
| pMMB67EH | Cloning vector, IncQ replicon, lacIq, ptac; Apr | 22 |
| pMP21 | pMMB190 carrying the rhlRI′ genes on a 2-kb PstI fragment; Apr | 33 |
| pNM482 | ColE1 replicon; ′lacZ; Apr | 40 |
| pRK2013 | Helper plasmid, ColE1 replicon, Tra; Kmr | 18 |
DNA techniques and nucleotide sequencing.
Small-scale preparations of plasmid DNA were carried out by the cetylmethylammonium bromide method (15), and large-scale preparations were performed using Qiagen-Tips (Qiagen). Chromosomal DNA was extracted from P. aeruginosa and purified as described elsewhere (24). Restriction enzyme digestions, ligations, and agarose gel electrophoresis were performed using standard procedures (49). Restriction fragments were purified from agarose gels using GeneClean II (Bio 101). Transformation of E. coli, P. aeruginosa, and P. fluorescens was carried out by electroporation (18). Southern blotting of P. aeruginosa DNA with Hybond N membranes (Amersham), random-primed DNA labeling of a 3.1-kb EcoRI-XhoI hcnABC fragment from pME3071 (Table 1) with digoxigenin-11-dUTP (Boehringer Mannheim), hybridization with this probe, and detection were all performed according to the protocols of the supplier. Nucleotide sequences of the hcnA gene with its upstream region and of the proximal 0.4 kb of the hcnB gene as well as of all PCR-derived constructs were determined by the dideoxy chain termination method using [35S]dATP (Amersham), 7-deaza-dGTP, and Sequenase version 2.0 (United States Biochemical Corp.). Sequence analysis was performed using the Genetics Computer Group programs FASTA (for homology searches in the Genbank/EMBL and SwissProt databases), GAP, and BESTFIT (for comparisons of pairs of sequences).
Plasmid construction and mutagenesis. (i) Construction of a translational hcnA′-′lacZ fusion.
The 0.78-kb EcoRV fragment from plasmid pME3336 containing the hcn promoter region and the first nine codons of the hcnA gene was fused in frame to the ′lacZ fragment from pNM482, in pME6010, resulting in plasmid pME3826. The same hcnA′-′lacZ fusion was also cloned on a 3.88-kb fragment into vector pKT240, giving pME3823.
(ii) Construction of the deletion derivatives pME3832, pME3835, and pME3837.
The deletion derivatives were obtained by PCR. Plasmid pME3826 was used as the template to amplify the respective hcnA′ fragments by the use of primers homologous to positions −111 (5′-GATCGAATTCACCTACCAGAATTGGCAGG-3′; pME3832), −88 (5′-GCTCGGATCCGATACCCACCTGTCATGG-3′; pME3835), and −143 (5′-GCTCGGATCCGTTCGACTTTTCCGCGCG-3′, pME3837) and of a primer annealing within the lacZ sequence (5′-TGCTGCAAGGCGATTAAGTTGG-3′). The positions are given relative to the translational start (see Fig. 2B). These primers contain a restriction site (underlined) at the 5′ end: an EcoRI site for pME3832, and a BamHI site for pME3835 and pME3837. For the amplification reactions, thermostable DNA polymerase (Eurobio) was used. Thermal cycling (15 cycles) consisted of denaturation at 95°C for 1 min, primer annealing at 58°C for 1 min, and elongation at 72°C for 1 min. The PCR fragments obtained were digested with EcoRV and EcoRI or BamHI, cloned into pBluescript II SK, and sequenced. The deleted promoter regions were fused to the ′lacZ gene (40) in pME6010, resulting in pME3832, pME3835, and pME3837 (see Fig. 6).
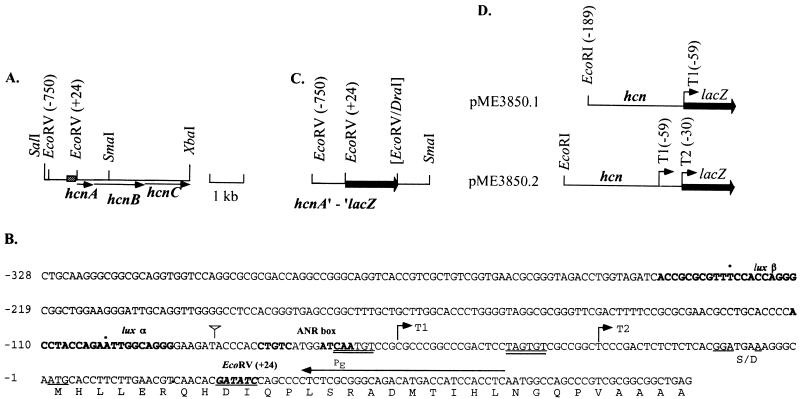
FIG. 2.
The hcnABC cluster, its promoter region, and lacZ constructs. (A) The hcnABC biosynthetic genes (indicated by arrows) were cloned on a 4.25-kb SalI/XbaI fragment into pBRR1MCS, giving pME3333. (B) The hcnA promoter region (indicated by ▩ in panel A) was mapped as shown in Fig. 4. Numbering is relative to the hcnA ATG start codon. The transcriptional start sites T1 and T2 are indicated by arrows. The putative translation initiation codon (deduced by comparison with the HcnA polypeptide of P. fluorescens [34]) and the potential ribosome binding site (S/D) are underlined. Double underlining indicates the −10 promoter regions. The ANR box and the palindromic lux boxes α and β (Fig. 5) are indicated in boldface. The primer (PE) used in the primer extension experiment (Fig. 4) is indicated by an arrow above the sequence. The insertion of an SphI restriction site in plasmid pME3852 (Fig. 6) is indicated by an inverted triangle. (C) A translational hcnA′-′lacZ fusion was integrated into the chromosome of the wild type and various mutants (Fig. 3) via homologous recombination. (D) PCR-amplified fragments, containing 130 bp upstream of T1 or 159 bp upstream of T2, were fused to the promoterless lacZ gene of pME6522, resulting in two transcriptional fusions, pME3850.1 and pME3850.2, respectively. The EcoRI site used was created by PCR.
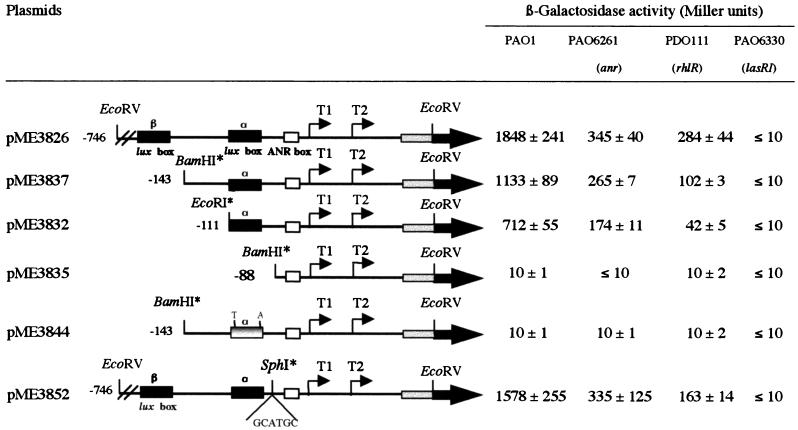
FIG. 6.
Effects of deletions and site-directed mutations on hcnA promoter activity. β-Galactosidase activities were determined in P. aeruginosa wild type PAO1 and in mutants PDO111 (rhlR), PAO6330 (lasRI), and PAO6261 (anr) containing the hcnA′-′lacZ fusion plasmids shown. Cells were grown in 40 ml of MMC with mild oxygen limitation to an OD600 of 1.0 to 1.3. Restriction sites introduced artificially are indicated by ∗. The mutated lux box α of pME3844 is indicated by two nucleotides mutated (T, A), and the hcnA gene is shown in gray. β-Galactosidase activity is provided along with the standard deviation (mean of three independent experiments).
(iii) Site-directed mutagenesis of the hcnA promoter region.
The two-base substitution in plasmid pME3844, which is otherwise identical to pME3837, was generated by the overlap extension method (39). Primer 5′-CCCACTTACCAGAATTGGCAAGGGAAG-3′ was used to generate the mutations (boldface) in the lux box (see Fig. 5). The mutation was verified by sequencing. An SphI restriction site was introduced between the ANR box and the lux box by PCR, using as template pME3336, which contains the hcn promoter on a 0.78-kb EcoRV insert. The whole 3.78-kb plasmid was amplified using primers I1 (5′GTCCGCATGCACCCACCTGTCATGGAT-3′) and I2 (5′-GTCCGCATGCATCTTCCCCTGCCAATT-3′), each containing an SphI restriction site (underlined). The PCR product was digested with SphI and ligated. The 6-bp insertion was verified by sequencing, and the 0.78-kb EcoRV insert was fused in frame with the ′lacZ reporter gene from pNM482 in vector pME6010, creating plasmid pME3852.
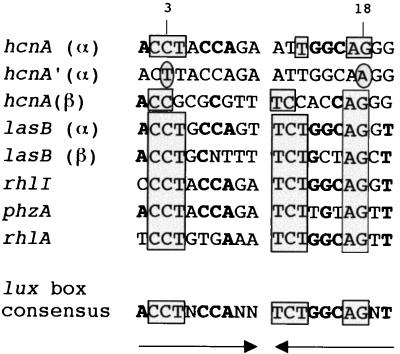
FIG. 5.
Alignment of the lux boxes found in the hcnABC promoter region with other lux boxes of P. aeruginosa. The sequences shown are found in the promoters of four autoinducible genes, rhlI (42), lasB (48), rhlA (43), and phzA (GenBank accession number AF005404). A similar palindromic (→ ←) consensus sequence has been proposed previously by Whiteley et al. (56). Highly conserved nucleotides (six out of seven) are boxed, and less conserved nucleotides (five out of seven) are indicated in boldface. Two nucleotides that were mutated in the hcnA′ α lux box of pME3844 are shown in ovals.
(iv) Construction of pME3850.1 and pME3850.2 containing a transcriptional lacZ fusion.
Plasmids pME3850.1 and pME3850.2 were obtained by PCR amplification of the hcn promoter regions using an EcoRI-tagged primer (the EcoRI site is underlined), homologous to position −188 (5′-GCTCGAATTCACGGGTGAGCCGGC-3′), and two primers which anneal at the +1 transcriptional start sites T1 (5′-GCTCCTGCAGGCGGACATTGATCC-3′ for pME3850.1) and T2 (5′-GCTCCTGCAGAGCCGGCGACACTAG-3′ for pME3850.2) and which each create an artificial PstI site (underlined). The 136- and 165-bp EcoRI/PstI-digested PCR fragments were fused to the lacZ gene of pME6522, resulting in pME3850.1 and pME3850.2, respectively.
Construction of P. aeruginosa mutant strains.
The chromosomal hcnA′-′lacZ reporter strains PAO6289 (hcnA′-′lacZ), PAO6290 (anr hcnA′-′lacZ), PAO6293 (rhlR hcnA′-′lacZ), and PAO6326 (lasR hcnA′-′lacZ) were obtained as follows. The translational hcnA′-′lacZ fusion from plasmid pME3826 was cloned into the suicide plasmid pME3087ΔE. The resulting plasmid pME3825 was mobilized by the helper plasmid pRK2013 into the different derivatives of P. aeruginosa and chromosomally integrated with selection for tetracycline resistance (Tcr). Excision of the vector by a second crossover was obtained by enrichment for tetracycline-sensitive (Tcs) cells (61). The chromosomal insertions of the hcnA′-′lacZ translational fusion were checked by Southern blotting (data not shown) and by testing their β-galactosidase-positive phenotype. The P. aeruginosa rhlR lasI double-mutant strain PAO6351 was constructed by transduction. The transducing phage E79tv2 (41) was propagated on strain PAOJP1 and then used to transduce the lasI::Tcr mutation into the rhlR mutant strain PDO111. The transductants were selected on nutrient agar plates containing tetracycline and mercuric chloride.
RNA isolation and transcriptional start site mapping.
Extraction of total RNA from P. aeruginosa cells grown in MMC (for conditions, see the legend to Fig. 4) was performed by a single-step RNA isolation method (14) using the TRIzol reagent (GIBCO-BRL). After treatment with amplification-grade DNase (Pharmacia), 10-μg aliquots of the isolated RNA were analyzed by primer extension as described elsewhere (54). The oligonucleotide PE 5′-GAGGTGGATGGTCATGTCTGCCCGCGAGAG-3′ (positions +65 to +36), which anneals to the coding strand of hcnA, was 5′-end labeled with 20 μCi of [γ-32P]dATP (Amersham) and 10 U of T4 polynucleotide kinase (Pharmacia) at 37°C for 30 min; 0.05 pmol of the [32P]dATP-labeled primer, purified with a nucleotide removal kit (Qiaquick; Qiagen) was dissolved in 20 mM Tris-HCl (pH 8.3)–200 mM NaCl–0.1 mM EDTA together with 10 μg of total RNA in a final volume of 30 μl. The solution was boiled for 3 min and incubated at 60°C for 2 h, then at 37°C for 30 min, and at room temperature for 30 min. Avian myeloblastosis virus reverse transcriptase (30 U; Pharmacia) was used to extend the primer in a reaction mixture containing 94 mM Tris-HCl (pH 8.3), 10 mM MgCl2, 10 mM dithiothreitol, 45 U of RNase inhibitor (Pharmacia), and 0.5 mM (each) deoxynucleoside triphosphate. The reaction was carried out in a total volume of 100 μl at 42°C for 1 h. After phenol-chloroform extraction, the extended product was precipitated by the addition of 0.1 volume of 3 M sodium acetate and 3 volumes of ethanol. The pellet was washed with 75% ethanol, dried, and redissolved in 10 μl of H2O. The unlabeled primer was used to generate a nucleotide sequence ladder using a T7 sequencing kit (Pharmacia) with [35S]dATP. Primer extension products were separated in an 8 M urea–8% polyacrylamide gel, in parallel with the sequencing reactions to map the transcription initiation sites.
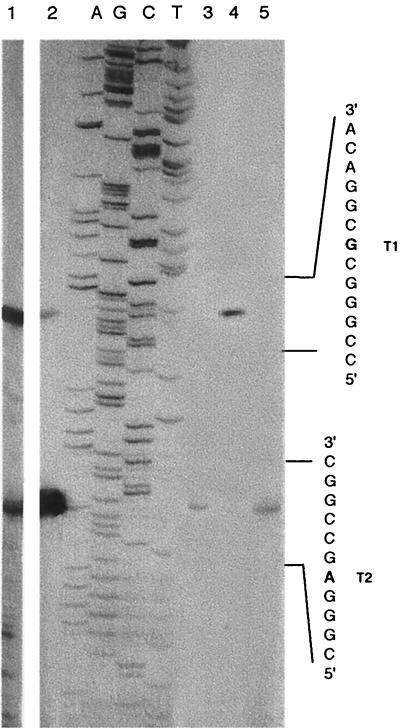
FIG. 4.
Primer extension analysis of the 5′ end of the hcnABC transcripts. The 5′-end-labeled oligonucleotide PE (Fig. 2B) was used as described in Materials and Methods. Lanes 1 and 2 show the extension products obtained with RNA from P. aeruginosa PAO1 grown semianaerobically to OD600 of 2.3 and 0.4, respectively, under the same conditions as for Fig. 1 and 3. Lane 3 contains the extension product of the hcn transcript from PAO1 grown to an OD600 of 0.9 under anaerobic conditions (in 60 ml of MMC; tightly closed 125-ml bottles). Lanes 4 and 5 show transcriptional start sites obtained with RNA from an anr mutant (PAO6261) and an rhlR mutant (PDO111), respectively, grown semianaerobically to an OD600 of 0.9. In lanes A, G, C, and T, the sequencing ladders obtained with the unlabeled oligonucleotide PE were run in parallel. Because of the 5′ phosphate, the primer extension products move 0.5 nucleotide ahead of the corresponding band in the ladder.
Assay for regulation by LasR and RhlR in a heterologous background.
To develop an RhlR/BHL assay in P. fluorescens CHA0, we cloned the 2-kb PstI fragment containing the rhlRI′ genes (33) behind the lac promoter of the vector pME6001, producing pME3840. Although the rhlI gene lacks nine codons at the 3′ end in this construct, BHL synthase activity was not affected. For a LasR/OdDHL assay, the 1.1-kb PvuI fragment containing lasR (23) was similarly expressed from the lac promoter of vector pME6001, resulting in pME3827. The two plasmids obtained were electroporated separately into P. fluorescens strain CHA0, which additionally contained an hcnA′-′lacZ translational fusion of P. aeruginosa on plasmid pME3837 or pME3844. To examine the effect of the las or rhl autoinduction system on hcnA expression, we measured β-galactosidase activities.
HCN production.
HCN was quantified in P. aeruginosa culture supernatants as described previously (26). Strains growing on plates were tested qualitatively for HCN production by an indicator paper method (9).
Assay of β-galactosidase activity.
Pseudomonas cells were routinely grown in 100-ml flasks containing 40 ml of MMC with shaking at 180 rpm. β-Galactosidase specific activities were determined by the Miller method (49).
Autoinducer estimation by thin-layer chromatography.
Cultures (200 ml) of strains PAO1, PDO111, PAO6261, PAO6330, PAOJP1, PAOJP2, PAO6351, CHA0, and CHA0 containing pME3840 were grown in NYB at 37°C (P. aeruginosa) or at 30°C (P. fluorescens) with shaking to an optical density at 600 nm (OD600) of 1.7 to 2.0. Cells were removed by centrifugation (10,000 rpm for 10 min), and the cell-free supernatants were adjusted to pH 5.0 prior to extraction with an equal volume of dichloromethane in a separating funnel. The solvent phase was treated with anhydrous MgSO4 to eliminate H2O and evaporated to dryness using a rotary evaporator. The total extract was concentrated 200-fold by dissolving it in 1 ml of 50% (vol/vol) acetonitrile and was stored at −20°C. The presence of N-acyl-homoserine lactones in 1 to 5 μl of extract was tested by C18 reverse-phase thin-layer chromatography, developed with methanol-water (60:40, vol/vol), and revealed by the indicator organisms, Chromobacterium violaceum mutant CV026 (38) for BHL and Agrobacterium tumefaciens (traG-lacZ) (13) for OdDHL. By comparison with known amounts of BHL and OdDHL standards, we estimated that P. aeruginosa wild-type strain PAO1 and the anr mutant PAO6261 produced about 5 μM BHL and 2 μM OdDHL. The rhlR mutant strain PAO6293 showed about 2 μM each BHL and OdDHL. Strains PAO6330 and PAOJP1 were defective for the production of OdDHL and produced eight times less BHL than did the wild type. Strain PAOJP2 was completely defective for both autoinducers, while the rhlR strain PAO6351 produced only 2 μM BHL. The introduction of plasmid pME3840, which contains the rhlR gene and almost the whole rhlI gene, resulted in the production of 2 μM BHL in P. fluorescens strain CHA0; without this plasmid, neither BHL nor OdDHL was detectable.
Nucleotide sequence accession number.
The sequence of the complete hcnABC cluster of P. aeruginosa is available under GenBank accession number AF208523 and at http://www.pseudomonas.com.
RESULTS
Relative importance of ANR, LasR, and RhlR for cyanogenesis in P. aeruginosa.
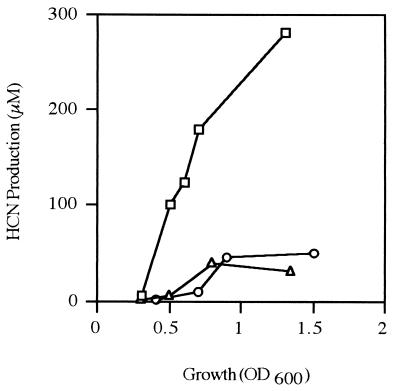
To assess quantitatively the contribution of each of the transcriptional regulators, we measured HCN formation in mildly oxygen limited P. aeruginosa cultures growing in MMC. In the wild-type strain PAO1, HCN production strongly depended on cell density (Fig. 1). Both an anr and an rhlR mutant excreted about six times less HCN than did the wild type, at 109 cells per ml (corresponding to an OD600 of 1.0) (Fig. 1). A lasR mutant did not produce any detectable quantities of HCN (data not shown). These results demonstrate that LasR is absolutely required for cyanogenesis and suggest that LasR, RhlR, and ANR do not act independently of each other.
FIG. 1.
Cell density-dependent HCN production in P. aeruginosa. The wild type PAO1 (□), the anr mutant PAO6261 (○), and the rhlR mutant PDO111 (▵) were grown with oxygen limitation in 40 ml of MMC at 37°C. HCN was measured as described in Materials and Methods. Each point is the mean of three independent experiments.
Identification and characterization of the P. aeruginosa hcnABC genes.
We used the hcnABC genes of P. fluorescens CHA0 (34) as a probe for Southern hybridization analysis to detect the homologous genes with their promoter in P. aeruginosa PAO1. A chromosomal 6.6-kb BamHI fragment of strain PAO1 gave a hybridization signal and was inserted into the broad-host-range vector pBBR1MCS, resulting in pME3326. Subcloning of a 4.25-kb SalI-XbaI fragment (Fig. 2A) into the same vector produced pME3333, which complemented the HCN-negative mutant PAO2324 for HCN production, as determined by a qualitative test. The nucleotide sequence of this fragment revealed three open reading frames showing 77% identity with the hcnABC genes of P. fluorescens CHA0. The order of the hcn genes and their organization as a putative operon are conserved in both Pseudomonas species. The deduced amino acid sequences of the HcnA, HcnB, and HcnC polypeptides of P. aeruginosa show 69, 70, and 76% identity, respectively, with the corresponding gene products of P. fluorescens. In particular, the [2Fe-2S] motif in HcnA and the dehydrogenase motifs in HcnB and HcnC were conserved. By contrast, the promoter region upstream of hcnA in P. aeruginosa (Fig. 2B) diverges from the hcnA promoter of P. fluorescens (34), showing only 40% identity on a stretch of 210 nucleotides. This suggests that there may be differences in the regulatory mechanisms that control hcn expression in these two species.
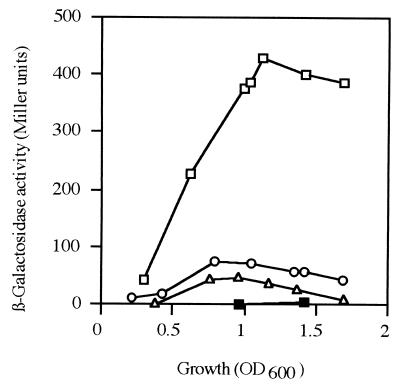
Inactivation of the chromosomal hcnABC genes of P. aeruginosa by deletion resulted in an HCN-negative phenotype (data not shown), confirming the function of these genes. A translational hcnA′-′lacZ fusion (Fig. 2C) was introduced by gene replacement into the chromosome of the wild type and the anr, lasR, and rhlR mutants. β-Galactosidase activities measured in the resulting PAO derivatives (Fig. 3) closely paralleled HCN levels (Fig. 1), confirming the regulatory roles of the three transcription activators in cyanogenesis.
FIG. 3.
Cell density-dependent expression of a translational hcnA′-′lacZ fusion in P. aeruginosa. Cultures of PAO6289 (□), the anr mutant PAO6290 (○), the rhlR mutant PAO6293 (▵), and the lasR mutant PAO6326 (■) were grown with oxygen limitation in 40 ml of MMC at 37°C. Each β-galactosidase measurement is the mean of three independent experiments.
Mapping and characterization of the P. aeruginosa hcnA promoter region.
The region upstream of hcnA was mapped by primer extension using primer PE, which anneals to positions +65 to +36 relative to the translational start (Fig. 2B). Total RNA was extracted from P. aeruginosa wild-type strain PAO1 grown in MMC under mild oxygen limitation to different cell densities (Fig. 4, lanes 1 and 2). Two hcn transcripts were found (Fig. 4), suggesting two transcriptional start sites, T1 and T2 (Fig. 2B). At high cell density, the upstream T1 transcript, which mapped to nucleotide −59 (relative to the hcnA start codon), was more abundant than was the downstream T2 transcript starting at nucleotide −30, whereas at low cell density the opposite transcript abundance was observed (Fig. 4). This suggests that quorum-sensing control might be particularly important for transcription from start site T1. In agreement with this interpretation, RNA isolated from the rhlR mutant PDO111 gave only the downstream T2 transcript (Fig. 4, lane 5). No attempt was made to map transcripts in a lasR mutant because in this background hcn gene expression was extremely low (Fig. 3). When RNA was extracted from the wild-type strain PAO1 grown under anaerobic conditions, only the downstream T2 transcript was detectable (Fig. 4, lane 3). Furthermore, this transcript was absent from the anr mutant PAO6261 (Fig. 4, lane 4). These data suggest that transcription starting at T2 is basically under the control of the anaerobic regulator ANR.
A potential ANR/FNR box (CTGTC… .ATCAA) was found centered at −42.5 bp from the transcriptional start site T2 (Fig. 2B). As shown below, this box is functional although it deviates in the left half-site from the ANR/FNR consensus box (TTGAT… .ATCAA) (53, 58). The right ANR half-site overlaps with the −10 sequence of the upstream promoter. Centered at −42.5 bp from the start site T1 of this promoter, there is a palindromic sequence resembling the Vibrio fischeri lux box (17, 21). This potential lux box (designated α in Fig. 2B) is very similar to the quorum-sensing-controlled operators of the P. aeruginosa rhlI, lasB, rhlA, and phzA promoters (1, 42, 43, 48, 56). An alignment (Fig. 5) shows that 13 out of 16 conserved nucleotides in a P. aeruginosa consensus lux box are also present in box α, which therefore could be recognized by LasR and/or RhlR. The postulated lux and ANR boxes in the hcn promoter both had a canonical distance of 42.5 bp from the respective transcription start sites, T1 and T2 (17, 53, 58). Interestingly, there is a second, less conserved lux box-like sequence (designated β in Fig. 2B) 108 bp upstream of lux box α (Fig. 5).
To confirm the function of the two promoters, P1 and P2, corresponding to the transcriptional starts T1 and T2, respectively, we constructed two transcriptional lacZ fusions. To this end, both promoters were fused precisely at their +1 transcription start sites to the lacZ reporter gene (Fig. 2D). The β-galactosidase activities of the resulting plasmids pME3850.1 and pME3850.2 were measured in P. aeruginosa wild type and in anr, rhlR, and lasRI mutants grown in MMC with mild oxygen limitation (Table 2). Plasmid pME3850.1, which contains only the quorum-sensing-dependent promoter P1, depended on RhlR and LasR for expression, whereas the presence or absence of ANR did not influence the activity significantly (Table 2). Plasmid pME3850.2, which contains both promoters, showed higher β-galactosidase activity and a clear dependence on ANR, LasR, and RhlR (Table 2). The impact of ANR appears to be greater on the translational hcnA′-′lacZ fusion (Fig. 3) than on the transcriptional hcn (P2)-lacZ fusion (Table 2), for unknown reasons. Under the conditions chosen (ca. 109 cells/ml), the promoter activity of P1 was slightly stronger than that of P2 (Table 2).
TABLE 2.
β-Galactosidase activities of two transcriptional hcnA-lacZ fusions constructed at T1 and T2 in P. aeruginosa wild type and regulatory mutantsa
| Strain (genotype) | β-Galactosidase activity (Miller units)
|
|
|---|---|---|
| pME3850.1 (P1) | pME3850.2 (P1+P2) | |
| PAO1 (wild type) | 2,299 ± 259 | 3,971 ± 183 |
| PAO6261 (anr) | 1,887 ± 80 | 2,721 ± 111 |
| PDO111 (rhlR) | 186 ± 32 | 281 ± 4 |
| PAO6330 (lasRI) | 99 ± 1 | 98 ± 1 |
Strains were grown in 40 ml of MMC in 100-ml Erlenmeyer flasks at 37°C with shaking to an OD600 of 1.0 and 1.2. β-Galactosidase activities were determined in triplicate; means ± standard deviations of three independent experiments are given. The hcnA-lacZ transcriptional fusion constructs are shown in Fig. 2D.
Mutational analysis of the hcnA promoter region.
To explore further the role of the potential operator sequences associated with the hcnA promoter, we used a translational hcnA′-′lacZ reporter with the entire upstream region (in pME3826) (Fig. 6). Progressive deletions removed the upstream lux box β and neighboring sequences (in pME3837 and pME3832) and then the downstream lux box α (in pME3835). The ANR box was not deleted, as this would also have abolished the −10 sequence of the promoter P1. All constructs were assayed in the wild type and in the anr, rhlR, and lasRI mutants at a uniform cell density (109 cells/ml) under mild oxygen limitation. None of the constructs was active in the lasRI background (Fig. 6), confirming the essential role of the las system. Deletion of the lux box α or introduction of two point mutations into this box at conserved nucleotides (pME3844 [Fig. 5]) also prevented expression completely (Fig. 6). This indicated that lux box α is required for the functioning of both promoters and that ANR alone is insufficient to activate the promoter via its (imperfect) ANR box. The region lying upstream of lux box α enhanced expression about threefold; part of this effect could be due to the poorly conserved lux box β (Fig. 6). In the anr mutant PAO6261, constructs pME3826, pME3837, and pME3832 were expressed at levels about fivefold below the levels in the wild type, indicating that ANR acts as a downstream activator at the ANR box (Fig. 6). In the rhlR mutant PDO111, expression of the same constructs was 7 to 17 times lower than in the wild type PAO1 but above the background levels in the lasRI mutant PAO6330 (Fig. 6). These data indicate that LasR, together with either RhlR or ANR, can provide some expression, but that a synergy between all three regulators is necessary for maximal expression. Spacing between the lux box and the ANR box could be altered by a 6-bp insertion (in pME3852), without significant effects on expression (Fig. 6). In conclusion, the data in Table 2 and Fig. 4 and 6 show that quorum-sensing control operates on promoter P1 and, when boosted by ANR and low oxygen availability, also on promoter P2.
Both LasR and RhlR individually can activate the hcnA promoter.
The lasI rhlI double mutant PAOJP2 expressed an hcnA′-′lacZ fusion poorly. Addition of both OdDHL and BHL restored β-galactosidase activity to near the wild-type level, whereas each autoinducer added singly had a smaller effect (Table 3), confirming the data of Whiteley et al. (56) obtained with an hcnB-lacZ fusion. Since LasR/OdDHL activates the expression of the rhlR and rhlI genes (32, 44) in the quorum-sensing cascade, the effect of OdDHL in strain PAOJP2 might have been indirect. Therefore, a lasI rhlR double mutant (PAO6351) was constructed. In this mutant, addition of OdDHL stimulated hcnA′-′lacZ expression about fivefold (Table 3). To confirm that LasR/OdDHL and RhlR/BHL can individually activate the hcnA promoters, we used P. fluorescens strain CHA0 as a heterologous host which does not synthesize N-acyl-homoserine lactones (6). (E. coli DH5α proved unsuitable for this experiment because the hcnA promoter was not expressed in this background.) In strain CHA0, the P. aeruginosa hcnA′-′lacZ fusion carried by pME3837 (Fig. 6) was expressed at a basal level (103 Miller units [Table 4]). Introduction of the P. aeruginosa rhlRI genes on pME3840 enhanced expression fourfold (Table 4). The production of BHL (and N-hexanoyl-homoserine lactone) by strain CHA0 containing pME3837 and pME3840 was verified by a bioassay (see Materials and Methods); in the vector control (CHA0 harboring pME6001 instead of pME3840), no BHL was detected. Introduction of the P. aeruginosa lasR gene on pME3827 into P. fluorescens CHA0/pME3837 also enhanced hcnA′-′lacZ expression fourfold, provided that 50 nM OdDHL was added to the culture medium (Table 4). When the crucial lux box α was mutated at two positions (pME3844 [Fig. 6]), hcnA′-′lacZ expression was stimulated by neither LasR/OdDHL nor RhlR/BHL in P. fluorescens (Table 4). Attempts to introduce both LasR and RhlR into P. fluorescens were unsuccessful. Nevertheless, the data presented in Table 4 show that LasR/OdDHL alone or RhlR/BHL alone can activate the hcn promoter to some extent.
TABLE 3.
Effects of added autoinducers OdDHL and BHL on a translational hcnA′-′lacZ fusion (pME3823) in P. aeruginosa wild type and regulatory mutantsa
| Strain (genotype) | β-Galactosidase activity (Miller units)
|
|||
|---|---|---|---|---|
| No addition | +BHL | +OdDHL | +BHL +OdDHL | |
| PAO1/pME3823 (wild type) | 3,022 ± 112 | ND | ND | ND |
| PAOJP2/pME3823 (rhlI lasI) | 24 ± 6 | 400 ± 13 | 455 ± 4 | 2,197 ± 22 |
| PAO6351/pME3823 (lasI rhlR) | 112 ± 1 | 123 ± 8 | 581 ± 70 | 766 ± 76 |
The activity of an hcnA′-′lacZ translational fusion construct (pME3823) was determined when cells grown in 40 ml of MMC (in 100-ml Erlenmeyer flasks) at 37°C with shaking reached an OD600 of 1.0 to 1.2. BHL was dissolved in 15% acetonitrile and added at a concentration of 10 μM; OdDHL was dissolved in 50% acetonitrile and added at a concentration of 10 μM. β-Galactosidase activities were determined in three independent experiments; means ± standard deviations are given. ND, not determined.
TABLE 4.
LasR- and RhlR-dependent expression of the hcnA promoter of P. aeruginosa in P. fluorescens CHA0a
| Plasmids | Regulator present | β-Galactosidase
activity (Miller units)
|
||
|---|---|---|---|---|
| No addition | +BHL | +OdDHL | ||
| pME3837 + pME6001 | 103 ± 17 | ND | ND | |
| pME3837 + pME3840 | RhlRI | 421 ± 32 | 452 ± 36 | ND |
| pME3837 + pME3827 | LasR | 94 ± 5 | ND | 441 ± 36 |
| pME3844 + pME3827 | LasR | 62 ± 3 | ND | 89 ± 7 |
| pME3844 + pME3840 | RhlRI | 60 ± 6 | 69 ± 4 | 68 ± 5 |
The hcnA′-′lacZ fusion constructs pME3837 and pME3844 (Fig. 6) were tested in strain CHA0. Cells were grown in 40 ml of MMC (in 100-ml Erlenmeyer flask) at 30°C with shaking to an OD600 of 1.0 to 1.2. BHL was dissolved in 15% acetonitrile and added at a concentration of 10 μM; OdDHL was dissolved in 50% acetonitrile and added at a concentration of 50 nM. β-Galactosidase activities were determined in triplicate; means ± standard deviations are given. ND, not determined.
DISCUSSION
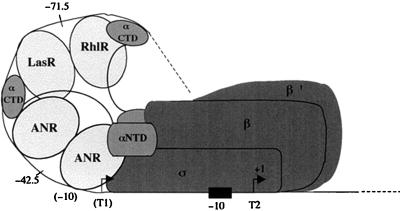
Early physiological experiments on HCN formation in P. aeruginosa, which showed that both the growth phase and reduced oxygen levels are important for induction (9, 11, 12), can now be explained in the light of this study on the hcn promoter. With increasing cell density, expression of the quorum-sensing regulators LasR and RhlR increases as does the concentration of OdDHL and BHL (32, 44, 45). As a consequence, transcription from start site T1 is enhanced in parallel (Fig. 4), presumably because both LasR/OdDHL and RhlR/BHL can bind to lux box α. Although each regulator alone can achieve partial activation of this promoter (Table 4), the combined action of both regulators is more effective (Table 3); for this reason, our model (Fig. 7) pictures a hypothetical RhlR-LasR heterodimer. The RhlR-LasR order shown is arbitrary; the opposite order could also be true. Expression of elastase in P. aeruginosa shows a similar dependence on LasR and RhlR, whereas the expression of rhamnolipid biosynthetic genes is regulated tightly by RhlR alone (7). In the hcn promoter, lux box α is centered at −42.5 bp relative to the transcription start T1. As noted previously for LuxR in V. fischeri, a 42.5-bp spacing between the center of the lux box and the transcription start site (17) is characteristic of ambidextrous transcriptional activators, such as CRP (cyclic AMP receptor protein) and FNR, which interact with the C-terminal domain of the α subunits of RNA polymerase and with ς70 (4, 46).
FIG. 7.
Model for the recognition of the hcn promoter P2 by the transcriptional regulators ANR, LasR, and RhlR and their interaction with RNA polymerase. αCTD (C-terminal domain of α) and αNTD (N-terminal domain of α) of RNA polymerase are shown as separate domains joined by a linker. Positions relative to the transcriptional start site T2 are indicated below the sequence. This model has been adapted from that proposed by Belyaeva et al. (4) for CRP-dependent promoter activation. During oxygen limitation, ANR blocks transcription from site T1 by binding to the corresponding −10 sequence (in brackets).
At low oxygen levels, the ANR protein provides additional hcn expression, by activating transcription from start site T2 (Fig. 4 and 6). The ANR protein itself appears to be expressed constitutively in P. aeruginosa (50). Interestingly, the function of promoter P2 was strictly dependent on the quorum-sensing machinery. As shown by pME3835, which lacks lux box α, and pME3844, which has a mutant variant of this lux box, ANR alone does not promote hcn transcription under mild oxygen limitation (Fig. 6). Even under severe oxygen limitation, no significant expression was observed with these constructs (data not shown). The reason for ANR not acting as an independent activator probably lies in the poorly conserved left half-site of the ANR box. The tandem arrangement of the lux and ANR boxes spaced 29 bp apart (Fig. 7) allows synergistic promoter activation by LasR, RhlR, and ANR (Fig. 6; Table 2). An analogous situation has been studied extensively by Belyaeva et al., who showed that E. coli promoters carrying tandem recognition sites for CRP at −41.5 and −71.5 bp are synergistically activated by CRP (4). The spacing between the sites can be increased with relatively minor consequences for promoter strength, suggesting a remarkable flexibility of the interactions between CRP and the subunits of RNA polymerase. It is therefore not surprising that a 6-bp insertion between the lux box and the ANR box affected hcn expression very little (Fig. 6). Because CRP, FNR, ANR, LuxR, LasR, and RhlR all have similar spacing requirements for their recognition sites, we have modeled the hcn promoter P2 (Fig. 7) after the tandem CRP promoter (−74.5/−41.5) (4). Binding of ANR to the hcn promoter not only activates transcription from T2 but also represses that from T1 (Fig. 6). The promoter region upstream of lux box α enhances hcn expression about threefold (Fig. 6). This region contains a second, suboptimal lux box (β) (Fig. 2B and 5), which could account for part of the enhancing effect. Although we did not investigate this in detail, we wish to point out the resemblance to the lasB promoter of P. aeruginosa, where two LasR recognition sites lying 60 bp apart synergistically enhance lasB promoter strength (1).
In P. aeruginosa, ANR is known to control anaerobic respiration with nitrate or nitrite as the electron acceptor (2) and the anaerobically inducible arginine deiminase pathway encoded by the arcDABC operon (62). It is interesting to compare the role of ANR as an activator of the arc and hcn promoters. At the arc promoter, ANR and the ANR box (located at −41.5 bp from the transcriptional start site) are sufficient to give strong anaerobic activation (25, 58). In the presence of arginine, the ArgR regulatory protein boosts the expression of the arc promoter by binding to the −70 region (35). Without ANR, ArgR does not activate this promoter (35). At the hcn promoter, by contrast, it is ANR that has an auxiliary role and the regulators (LasR and RhlR) binding to the upstream region can function autonomously.
In P. fluorescens CHA0, cyanogenesis also depends on ANR (34) and on cell density (6). However, in this organism the hcn promoter does not contain any lux box, and deletion of the region upstream of the ANR box does not affect hcn expression (5a). Furthermore, P. fluorescens CHA0 does not appear to produce N-acyl-homoserine lactones, although heterologous expression of the P. aeruginosa rhlI gene results in the synthesis of BHL by this strain (Table 4). A posttranscriptional mechanism of quorum-sensing regulation has been proposed for P. fluorescens CHA0 (6). Whether such a mechanism is conserved in P. aeruginosa remains to be seen. We find it striking that HCN production in one fluorescent pseudomonad, P. aeruginosa PAO, depends on the complex interactions between the las and the rhl systems, whereas in a closely related species, P. fluorescens CHA0, cyanogenesis can do without.
ACKNOWLEDGMENTS
We thank Paul Williams for generously supplying autoinducer samples, and we thank B. Iglewski, J. Pearson, D. Ohman, C. Reimmann, and A. Lazdunski for providing strains. We also thank Steve Busby, Birgit Wackwitz, Karin Heurlier, Caroline Blumer, and Laura Serino for discussion and SmithKline Beecham for generously supplying carbenicillin.
This work was supported by the Swiss National Foundation for Scientific Research (31-56608.99) and European Biotechnology project BIO4CT960119.
REFERENCES
- 1.Anderson R M, Zimprich C A, Rust L. A second operator is involved in Pseudomonas aeruginosa elastase (lasB) activation. J Bacteriol. 1999;181:6264–6270. doi: 10.1128/jb.181.20.6264-6270.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arai H, Kodama T, Igarashi Y. Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa. Mol Microbiol. 1997;25:1141–1148. doi: 10.1046/j.1365-2958.1997.5431906.x. [DOI] [PubMed] [Google Scholar]
- 3.Bagdasarian M M, Amann E, Lurz R, Rückert B, Bagdasarian M. Activity of the hybrid trp-lac (tac) promoter of Escherichia coli in Pseudomonas putida. Construction of broad-host-range, controlled-expression vectors. Gene. 1983;26:273–282. doi: 10.1016/0378-1119(83)90197-x. [DOI] [PubMed] [Google Scholar]
- 4.Belyaeva T A, Rhodius V A, Webster C L, Busby S J. Transcription activation at promoters carrying tandem DNA sites for the Escherichia colicyclic AMP receptor protein: organisation of the RNA polymerase alpha subunits. J Mol Biol. 1998;277:789–804. doi: 10.1006/jmbi.1998.1666. [DOI] [PubMed] [Google Scholar]
- 5.Blumer C, Haas D. Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch Microbiol. 2000;173:170–177. doi: 10.1007/s002039900127. [DOI] [PubMed] [Google Scholar]
- 5a.Blumer C, Haas D. Iron regulation of the hcnABC genes encoding hydrogen cyanide synthase depends on the anaerobic regulator ANR rather than on the global activator GacA in Pseudomonas fluorescensCHA0. Microbiology. 2000;146:2417–2424. doi: 10.1099/00221287-146-10-2417. [DOI] [PubMed] [Google Scholar]
- 6.Blumer C, Heeb S, Pessi G, Haas D. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescensinvolves specific ribosome binding sites. Proc Natl Acad Sci USA. 1999;96:14073–14078. doi: 10.1073/pnas.96.24.14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosais under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castric P A. Glycine metabolism by Pseudomonas aeruginosa: hydrogen cyanide biosynthesis. J Bacteriol. 1977;130:826–831. doi: 10.1128/jb.130.2.826-831.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castric P A. Hydrogen cyanide production by Pseudomonas aeruginosaat reduced oxygen levels. Can J Microbiol. 1983;29:1344–1349. doi: 10.1139/m83-209. [DOI] [PubMed] [Google Scholar]
- 10.Castric P A. Hydrogen cyanide, a secondary metabolite of Pseudomonas aeruginosa. Can J Microbiol. 1975;21:613–618. doi: 10.1139/m75-088. [DOI] [PubMed] [Google Scholar]
- 11.Castric P A. Influence of oxygen on the Pseudomonas aeruginosahydrogen cyanide synthase. Curr Microbiol. 1994;29:19–21. [Google Scholar]
- 12.Castric P A, Ebert R F, Castric K F. The relationship between growth phase and cyanogenesis in Pseudomonas aeruginosa. Curr Microbiol. 1979;2:287–292. [Google Scholar]
- 13.Cha C, Gao P, Chen Y C, Shaw P D, Farrand S K. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol Plant-Microbe Interact. 1998;11:1119–1129. doi: 10.1094/MPMI.1998.11.11.1119. [DOI] [PubMed] [Google Scholar]
- 14.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 15.Del Sal G, Manfioletti G, Schneider C. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 1988;16:9878. doi: 10.1093/nar/16.20.9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Döring G. Pseudomonas aeruginosa infection in cystic fibrosis patients. In: Campa M, Bendinelli M, Friedman H, editors. Pseudomonas aeruginosa as an opportunistic pathogen. New York, N.Y: Plenum Press; 1993. pp. 245–273. [Google Scholar]
- 17.Egland K A, Greenberg E P. Quorum sensing in Vibrio fischeri: elements of the luxlpromoter. Mol Microbiol. 1999;31:1197–1204. doi: 10.1046/j.1365-2958.1999.01261.x. [DOI] [PubMed] [Google Scholar]
- 18.Farinha M A, Kropinski A M. High efficiency electroporation of Pseudomonas aeruginosausing frozen cell suspensions. FEMS Microbiol Lett. 1990;58:221–225. doi: 10.1111/j.1574-6968.1990.tb13982.x. [DOI] [PubMed] [Google Scholar]
- 19.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuqua C, Greenberg E P. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr Opin Microbiol. 1998;1:183–189. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- 21.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 22.Fürste J P, Pansegrau W, Frank R, Blöcker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 23.Gambello M J, Iglewski B H. Cloning and characterization of the Pseudomonas aeruginosa lasRgene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gamper M, Ganter B, Polito M R, Haas D. RNA processing modulates the expression of the arcDABC operon in Pseudomonas aeruginosa. J Mol Biol. 1992;226:943–957. doi: 10.1016/0022-2836(92)91044-p. [DOI] [PubMed] [Google Scholar]
- 25.Gamper M, Zimmerman A, Haas D. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J Bacteriol. 1991;173:4742–4750. doi: 10.1128/jb.173.15.4742-4750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gewitz H S, Pistorius E K, Voss H H, Vennesland B. Cyanide formation in preparations from Chlorella vulgarisBeijerinck: effect of sonication and amygdalin addition. Planta. 1976;131:145–148. doi: 10.1007/BF00389986. [DOI] [PubMed] [Google Scholar]
- 27.Goldfarb W B, Margraf H. Cyanide production by Pseudomonas aeruginosa. Ann Surg. 1967;165:104–110. doi: 10.1097/00000658-196701000-00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, Wade J, Walsh U, O'Gara F, Haas D. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol Plant-Microbe Interact. 2000;13:232–237. doi: 10.1094/MPMI.2000.13.2.232. [DOI] [PubMed] [Google Scholar]
- 29.Holder I A. Pseudomonas aeruginosa burn infections: pathogenesis and treatment. In: Campa M, Bendinelli M, Friedman H, editors. Pseudomonas aeruginosa as an opportunistic pathogen. New York, N.Y: Plenum Press; 1993. pp. 275–295. [Google Scholar]
- 30.Holloway B W. Genetics of Pseudomonas. Bacteriol Rev. 1969;33:419–443. doi: 10.1128/br.33.3.419-443.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovach M E, Phillips R W, Elzer P H, Roop II R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 32.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosalinks the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 33.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosaPAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 34.Laville J, Blumer C, Von Schroetter C, Gaia V, Défago G, Keel C, Haas D. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescensCHA0. J Bacteriol. 1998;180:3187–3196. doi: 10.1128/jb.180.12.3187-3196.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu C D, Winteler H, Abdelal A, Haas D. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J Bacteriol. 1999;181:2459–2464. doi: 10.1128/jb.181.8.2459-2464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto H, Ohta S, Kobayashi R, Terawaki Y. Chromosomal location of genes participating in the degradation of purines in Pseudomonas aeruginosa. Mol Gen Genet. 1978;167:165–176. doi: 10.1007/BF00266910. [DOI] [PubMed] [Google Scholar]
- 37.Maurhofer M, Reimmann C, Schmidli-Sacherer P, Heeb S, Haas D, Défago G. Salicylic acid biosynthetic genes expressed in Pseudomonas fluorescensstrain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology. 1998;88:678–684. doi: 10.1094/PHYTO.1998.88.7.678. [DOI] [PubMed] [Google Scholar]
- 38.McClean K H, Winson M K, Fish L, Taylor A, Chhabra S R, Camara M, Daykin M, Lamb J H, Swift S, Bycroft B W, Stewart G S, Williams P. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 39.Mikaelian I, Sergeant A. Modification of the overlap extension method for extensive mutagenesis on the same template. Methods Mol Biol. 1996;57:193–202. doi: 10.1385/0-89603-332-5:193. [DOI] [PubMed] [Google Scholar]
- 40.Minton N P. Improved plasmid vectors for the isolation of translational lacgene fusions. Gene. 1984;31:269–273. doi: 10.1016/0378-1119(84)90220-8. [DOI] [PubMed] [Google Scholar]
- 41.Morgan A F. Transduction of Pseudomonas aeruginosawith a mutant of bacteriophage E79. J Bacteriol. 1979;139:137–140. doi: 10.1128/jb.139.1.137-140.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ochsner U A, Koch A K, Fiechter A, Reiser J. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J Bacteriol. 1994;176:2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearson J P, Pesci E C, Iglewski B H. Roles of Pseudomonas aeruginosa las and rhlquorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pesci E C, Pearson J P, Seed P C, Iglewski B H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 46.Rhodius V A, Busby S J W. Positive activation of gene expression. Curr Opin Microbiol. 1998;1:152–159. doi: 10.1016/s1369-5274(98)80005-2. [DOI] [PubMed] [Google Scholar]
- 47.Rumbaugh K P, Griswold J A, Iglewski B H, Hamood A N. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosain burn wound infections. Infect Immun. 1999;67:5854–5862. doi: 10.1128/iai.67.11.5854-5862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rust L, Pesci E C, Iglewski B H. Analysis of the Pseudomonas aeruginosa elastase (lasB) regulatory region. J Bacteriol. 1996;178:1134–1140. doi: 10.1128/jb.178.4.1134-1140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Savioz A, Zimmermann A, Haas D. Pseudomonas aeruginosapromoters which contain a conserved GG-N10-GC motif but appear to be RpoN-independent. Mol Gen Genet. 1993;238:74–80. doi: 10.1007/BF00279533. [DOI] [PubMed] [Google Scholar]
- 51.Seed P C, Passador L, Iglewski B H. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonasautoinducer PAI: an autoinduction regulatory hierarchy. J Bacteriol. 1995;177:654–659. doi: 10.1128/jb.177.3.654-659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solomonson L P. Cyanide as a metabolic inhibitor. In: Vennesland B, Conn E E, Knowles C J, Westley J, Wissing F, editors. Cyanide in biology. London, England: Academic Press Ltd.; 1981. pp. 11–28. [Google Scholar]
- 53.Spiro S. The FNR family of transcriptional regulators. Antonie Leeuwenhoek. 1994;66:23–36. doi: 10.1007/BF00871630. [DOI] [PubMed] [Google Scholar]
- 54.Vögtli M, Hütter R. Characterisation of the hydroxystreptomycin phosphotransferase gene (sph) of Streptomyces glaucescens: nucleotide sequence and promoter analysis. Mol Gen Genet. 1987;208:195–203. doi: 10.1007/BF00330442. [DOI] [PubMed] [Google Scholar]
- 55.Voisard C, Bull C T, Keel C, Laville J, Maurhofer M, Schnider U, Défago G, Haas D. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches. In: O'Gara F, Dowling D N, Boesten B, editors. Molecular ecology of rhizosphere microorganisms. Weinheim, Germany: VCH; 1994. pp. 67–89. [Google Scholar]
- 56.Whiteley M, Lee K M, Greenberg E P. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winson M K, Camara M, Latifi A, Foglino M, Chhabra S R, Daykin M, Bally M, Chapon V, Salmond G P, Bycroft B W, Lazdunski A, Stewart G S A B, Williams P. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winteler H V, Haas D. The homologous regulators ANR of Pseudomonas aeruginosa and FNR of Escherichia colihave overlapping but distinct specificities for anaerobically inducible promoters. Microbiology. 1996;142:685–693. doi: 10.1099/13500872-142-3-685. [DOI] [PubMed] [Google Scholar]
- 59.Winteler H V, Schneidinger B, Jaeger K E, Haas D. Anaerobically controlled expression system derived from the arcDABC operon of Pseudomonas aeruginosa: application to lipase production. Appl Environ Microbiol. 1996;62:3391–3398. doi: 10.1128/aem.62.9.3391-3398.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wissing F, Andersen K A. The enzymology of cyanide production from glycine by a Pseudomonas species. In: Vennesland B, Conn E E, Knowles C J, Westley J, Wissing F, editors. Cyanide in biology. London, England: Academic Press Ltd.; 1981. pp. 275–288. [Google Scholar]
- 61.Ye R W, Haas D, Ka J O, Krishnapillai V, Zimmermann A, Baird C, Tiedje J M. Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosarequires Anr, an analog of Fnr. J Bacteriol. 1995;177:3606–3609. doi: 10.1128/jb.177.12.3606-3609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zimmerman A, Reimmann C, Galimand M, Haas D. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol Microbiol. 1991;5:1483–1490. doi: 10.1111/j.1365-2958.1991.tb00794.x. [DOI] [PubMed] [Google Scholar]