Graphical abstract
Keywords: Plant-associated microbiome; Root exudates; Synergistic microbiomes, rhizospheric microbiomes; Endosphere; Phyllosphere
Highlights
-
•
Plant-microbiome interaction occurs at the rhizosphere, endosphere, and phyllosphere.
-
•
Root exudates can favor the recruitment of a beneficial microbiome in the rhizosphere.
-
•
Plant topology and phytochemistry influence the recruitment of the phyllosphere microbiome.
-
•
Diverse plant strategies selectively recruit beneficial microbiomes.
-
•
Multiple plant mechanisms displace potential pathogens from the rhizosphere.
-
•
The beneficial microbiome helps plants to recruit other beneficial microbiota.
Abstract
Background
Research on beneficial mechanisms by plant-associated microbiomes, such as plant growth stimulation and protection from plant pathogens, has gained considerable attention over the past decades; however, the mechanisms used by plants to recruit their microbiome is largely unknown.
Aim of Review
Here, we review the latest studies that have begun to reveal plant strategies in selectively recruiting beneficial microbiomes, and how they manage to exclude potential pathogens.
Key Scientific concepts of Review: We examine how plants attract beneficial microbiota from the main areas of interaction, such as the rhizosphere, endosphere, and phyllosphere, and demonstrate that such process occurs by producing root exudates, and recognizing molecules produced by the beneficial microbiota or distinguishing pathogens using specific receptors, or by triggering signals that support plant-microbiome homeostasis. Second, we analyzed the main environmental or biotic factors that modulate the structure and successional dynamics of microbial communities. Finally, we review how the associated microbiome is capable of engaging with other synergistic microbes, hence providing an additional element of selection. Collectively, this study reveals the importance of understanding the complex network of plant interactions, which will improve the understanding of bioinoculant application in agriculture, based on a microbiome that interacts efficiently with plant organs under different environmental conditions.
Introduction
The association between plants and microorganisms is over 400 million years old; there exists fossil record that supports the establishment of interactions between plants and certain fungi, such as arbuscular mycorrhiza. It is possible that there are no fossil records of plant-bacteria associations due to their fragile unicellular nature [1], [2]. Thus, this association is expected, since microorganisms occurred before plants and colonized a major portion of the planet's ecosystems. Therefore, plants perhaps encountered microorganisms, and since then, formed symbiotic and commensal associations [3]. In this regard, both groups of organisms began establishing chemical and physical communications, where multicellular and larger plants enabled selected microbes to establish and colonize their internal compartments, such as roots, stems, and leaves [4]. Certain groups even generated organelles such as chloroplasts and mitochondria, according to the symbiotic theory of Margulis [5].
However, initially, not all associations are beneficial, since certain microorganisms, such as bacteria, fungi, or viruses, exploited their genetic resources to evolve, attack, and infect systems to survive, at the expense of harming their host (i.e., pathogenesis) [6]. In contrast, when non-pathogenic microorganisms thrive in association with plants, both benefit, since the plant secretes compounds that are produced during photosynthesis, while heterotrophic microbes provide protection against pathogens, in addition to stimulating their growth and fitness through the synthesis and excretion of compounds of diverse origin and function [7], [8], [9], [10]. For example, multiple species of avirulent fungi of the genus Trichoderma or bacteria of the genus Bacillus or Pseudomonas, are capable of synthesizing hundreds of compounds that have a fundamental role during interaction with plants, either by stimulating their growth or antagonizing fungal pathogens or nematodes [11], [12]. Similarly, other groups of mycorrhizal fungi, which inhabit and are associated with biogeochemical cycles, are capable of improving soil fertility conditions, thereby improving the nutritional status of the plant [13]. This plant-associated microbiota with beneficial metabolic capacities have been used as bioinoculants (i.e., biofungicides/biofertilizers or both) to improve the growth and production of grains, vegetables, and fruits in various agricultural systems [14], [15], [16]. Thus, they are an excellent replacement to using agrochemicals that pollute the environment and are toxic to human health [17], [18].
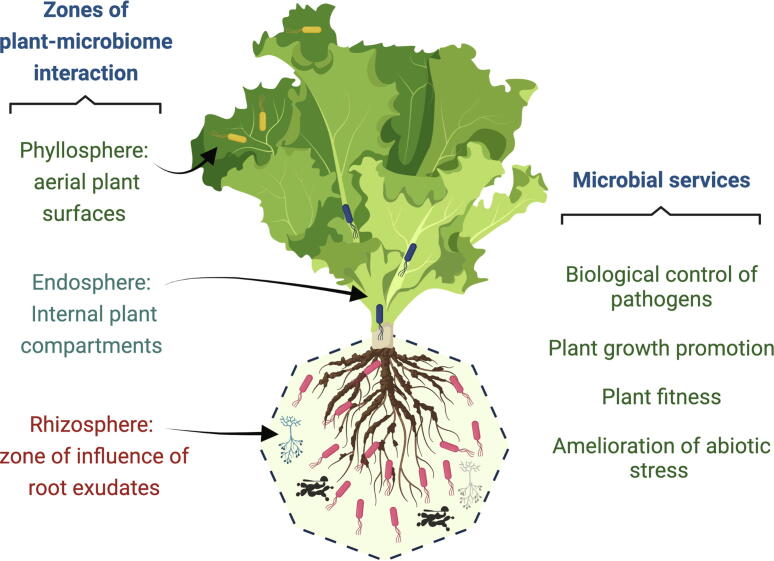
These diverse ecological interactions between plants and microorganisms raises a question: how do plants recruit their associated microbiota? This has been examined in the last few decades via field and laboratory approaches. However, this question remains central to the field of molecular plant–microbe interaction [10], [19]. Recently, various omics techniques have unveiled a global panorama of stimulus–response in both directions of plant-microorganism or vice versa [20], [21], [22]. In this study, we review the most recent and important literature to determine how plants recruit beneficial microbiota, mainly from zones such as the rhizosphere, endosphere, and phyllosphere. The strong interaction of beneficial microbiome in these zones provides several beneficial services, such as plant protection and growth stimulation under different environmental conditions (see Fig. 1). Several recent reviews have illustrated these beneficial microbial activities [20], [23], [24]. In addition, we will discuss the selection system that enables plant to preferentially associate with the microbiota, which provides certain adaptive advantage (versus opportunistic pathogens), and the possible factors aiding this adaptation. Finally, we propose that the same plant-associated microbiome favors the recruitment of other beneficial organisms through diverse mechanisms.
Fig. 1.
Main areas of interaction and recruitment of the plant microbiome, and some of the main services provided by the microbiome to the plant.
Plant-microbiome interactions
The vast majority of microorganisms existing in the environment does not interact with when encountering plants, which indicates a neutral interaction between them [25]. In contrast, certain microorganisms form parasitic interactions (pathogens), which affects the host plant’s health and development [26], [27], [28]. In commensalism relationship, which is a relationship between individuals of two species, one species benefits and the other is neither positively nor negatively affected [29]. Finally, in symbiotic or mutualistic relationships, both plants and microorganisms obtain certain advantage and/or receive some benefit. In certain cases, the symbiosis can be extremely narrow and obligate, where neither organism can survive without the other. To the best of our knowledge, obligate symbiosis are rare between plants of agronomic interest and associated organisms, but are beneficial to both, if discovered [30], [31], [32]. One such bacteria identified as rhizobia (free-living saprophytes), inhabit the rhizosphere, which communicates specifically with certain species of legumes by forming nodules in the roots and fixing atmospheric nitrogen. The obtained ammonia after nitrogen fixing can be used by the plant, while the symbiont receives a “home to live” and nutrients to survive [33], [34]. Such reciprocal interactions between the rhizobiome (and non-rhizobial bacteria associated with root nodules) and the plant defines what few authors call the “MicrobiHome” [35], [36].
Factors affecting plant-microbiome short- and long-term associations
In general, the type of interactions can be defined depending on their duration, that is, the plant can casually (short-term) associate with certain microbes, and stops interacting in due course [37], [38]. In contrast, certain associations are long-term, perhaps during the life cycle of plants and beyond [39]. The microorganisms with long-term association are called the core microbiome [1], [40], which can be defined as the total microbiota associated with the plant (and its genomic reserves) that presents long-term interactions, which can survive, thrive, and interact with different tissues such as roots, shoots, leaves, flowers, and locations [7], [41], [42], [43]. Defining the core microbiome of each plant is essential to better understand the species-specific relationships and recognize the advantage of applied aspects, such as in agrobiotechnology, which increases the efficiency of bioinoculants applied for various environmental situations.
Various abiotic and biotic factors affect the short- and long-term association between plants and their microbiome [44]. Abiotic factors include soil pH, supposedly for the microbiome- plant interactions in the rhizosphere, in addition to the availability of resources, such as nutrients, oxygen, temperature, water, presence of contaminating metals or metalloids, salinity, and soil type [45], [46], [47]. In a study using rhizoboxes, Blossfeld et al. [48] performed non-invasive analyses of the presence of oxygen and pH standards in the rhizosphere of plants (Juncus spp.). Interestingly, the study concluded that the dynamic interaction of O2 concentration gradients and pH, and the organic acids exuded, were key parameters in the physicochemical environment of the rhizosphere. It is important to evaluate the population dynamics of the rhizospheric microbiome through different gradients of O, pH, and the presence of root exudates, and observe how these factors, in the presence of certain microbial species, can modulate (or be modulated) and influence the growth and suitability of the plant.
A review by Philippot et al. [49] also showed that, in general, the bacterial phylum Proteobacteria was dominant in the rhizosphere, while the fungal phyla Ascomycota and Glomeromycota were also the most abundant and regularly observed in the plant rhizosphere. Their study concluded that the microbiome in natural environments can be distinctly influenced by the agricultural ecosystems. For example, the main drivers of microbiota diversity in an agricultural ecosystem are agricultural practices, followed by environmental conditions, soil type, and plant species, whereas in natural ecosystems, mainly biotic interactions, plant species, and plant diversity play a relevant role in modulating microbial communities.
However, biotic factors can also affect plant-microbiome dynamics. For example, the plant can interact specifically depending on its developmental stage. The type of tissue or part that interacts include the root (rhizosphere), the stem or the leaves, and an internal zone (endosphere) or external/superficial (phyllosphere) region of the plant. Furthermore, Chaparro et al. [50] reported that the rhizospheric microbiome of Arabidopsis plants varies depending on their stage of development (seedling, vegetative, bolting, and flowering), and that certain phyla such as Acidobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, and specific genera were strongly correlated with the types of root exudates. Their work showed that plants exude different compounds depending on their developmental stage, and in return, these exudates aid in assembling a specific microbiome for the plant. Studies by Bai and colleagues [51] showed that the microbiome of Arabidopsis leaves differes from that of the rhizosphere, but there exists a huge functional overlap of more than 400 analyzed genomes. This suggests that the microbiomes in both niches may perform similar functions when interacting with the plant, although not at the level of species diversity. Multiple species (gram-positive or gram-negative) such as various plant growth-promoting strains of the genera Pseudomonas spp. and Bacillus spp., can perform similar beneficial functions.
From beneficial to pathogenic interactions
Importantly, the interactions between the plant and its microbiome can notably change, that is, they can proceed from a neutral or commensal state to a pathogenic state. Newton et al. [29] reviewed certain factors that ignite a state of pathogenesis in few plant-associated organisms, such as bacteria or fungi. For example, the authors highlight the importance of its trophic requirements and environmental conditions, or a simple exposure of the host can transform a mutualist organism to a parasite/pathogen. In particular, the fungus Pectobacterium atrosepticum, can transform from a symbiont to a pathogen, depending on its interaction with the plant host, or resembling the opportunistic bacterium Pseudomonas syringae, whose ability to infect depends on quorum sensing. Similarly, certain plant species are more susceptible to attack by certain opportunistic pathogens than the others, depending on their chemical defenses and immune response, and the associated microbiome itself that stimulates their defense and helps in their protection [52].
Plant call to recruit the microbiome
Plants, being sessile organisms, are restricted to associating with surrounding organisms, as compared to migrating animals, which can look for new interactions [53]. However, the offspring of some plants achieve this by managing to colonize new areas through seed dispersal mechanisms or reproductive structures. Some seeds can travel a long distance before finding suitable conditions for germination [54]. However, once the seeds germinate, they interact with the resident soil microbiota through the roots (rhizosphere) [55]. The aerial zones of the plant (phyllosphere) are also prone to colonization by microbes that reach their tissues, such as leaves, stems, flowers, and fruits, via air [56]. Furthermore, plants have an internal compartment known as the endosphere, which are also predisposed to colonization by rhizospheric and phyllospheric microorganisms [57], [58]. Thus, the interaction (or recruitment) of microbiome with plants occurs at three main zones: the rhizosphere, phyllosphere, and endosphere.
Recruitment in the rhizosphere
Root activities modify microbial activity, along with the physiochemical properties of the surrounding soil or rhizosphere [59], [60]. The rhizosphere is defined as a narrow part of the soil that is influenced by root exudates, which is the habitat of thousands of microbial species, including bacteria, fungi, oomycetes, viruses, and archaea, and few other macro-organisms such as nematodes or insects [61]. These microbiota can interact with the plant through the roots, which not only functions as a soil anchoring system, but also perceives and communicates with abiotic elements. Some of these abiotic factors have been previously described. A plant initiates recruitment or “call” to generate its own microbiome from seeds (endophyte reservoirs), or through the planting of cuttings, bulbs, rhizomes, or stolons [62], [63]. These structures are known to contain diverse endophytic microorganisms that are inherited vertically from mother plants, including seeds [39], [64].
The number of endophytic cells may be low compared to mature plants; the roots emerge and interact with the microbiota of the bulk soil, and forms the rhizosphere, where the exudates first attract the future associated microbiome. Exudates can contain amino acids (i.e. α-Alanine, β-alanine, γ-aminobutyric, α-aminoadipic, arginine, asparagine, aspartic, citrulline, cystathionine, cysteine, cystine, deoxymugineic, 3-epihydroxymugineic, glutamine, glutamic, glycine, histidine, and homoserine), sugars (arabinose, fructose, galactose, glucose, maltose, mannose, mucilages of various compositions, oligosaccharides, rafnose, rhamnose, ribose, sucrose, xylose, deoxyribose), growth factors and vitamins (p-amino benzoic acid, biotin, choline, inositol, N-methyl nicotinic acid, niacin, pathothenic, pantothenate, pyridoxine ribofavin, strigolactones, thiamine), fatty acids (linoleic, linolenic, oleic, palmitic, stearic), organic acids (acetic, aconitic, ascorbic, aldonic, benzoic, butyric, cafeic, citric, p-coumaric, erythronic, ferulic, formic, fumaric, glutaric, glycolic, lactic, glyoxilic, malic, malonic, oxalacetic, oxalic), and other chemoattractants [65]. Additionally, some exudates such as flavonoids, citrate, malate, oxalate coumarins, malic acid, camalexin, benzoxazinoids, or even ethylene, can be specific taxon recruiters [7].
Under certain conditions of environmental stress, such as phosphate or iron limitation, some plants tend to enhance the secretion of citrate, malate, or oxalate to enrich
the rhizosphere with organic carbon, which attracts beneficial microorganisms [65]. This hypothesis was confirmed in an interesting study published by Kost et al. [66], who reported a strict association between beneficial Burkholderia species and their ability to use oxalate as a carbon source. Interestingly, the authors when analyzing other species of the genus Burkholderia, such as B. glumae and B. plantarii, which are characterized as plant pathogens, or even human opportunistic pathogens, such as Burkholderia cepacia, they were unable to degrade oxalate. Therefore, the oxalotrophy or ability to use oxalate as a carbon source was strongly associated with the ability of the beneficial strain B. phytofirmans PsJN to colonize lupine (Lupinus albus L.) and maize (Zea mays). Finally, a mutant with deleted oxc gene was impaired during the early colonization of lupine and maize seedlings, confirming the role of oxalotrophy in colonizing the plant, and in turn, demonstrating that certain root exudates selectively favor a beneficial microbiome.
Multiple factors such as identically associated-microbiota and their volatile organic compounds (VOCs), influence the diversity of root exudates excreted by the plant. For example, a recent study showed that inoculation with PGPR Bacillus amyloliquefaciens GB-03 stimulates the production of volatiles in tomato plants (i.e., β-caryophyllene), which are perceived in the neighboring plants. By detecting these VOCs, the “receiver” plants modify the production of their root exudates (i.e., salicylic acid), in turn modifying the rhizospheric microbiota. Interestingly, the receiver plants and emitting plants display a broad similarity in their rhizosphere-associated microbial communities. This was the first study to demonstrate the involvement of microbe-induced plant volatiles (MIPVs) in generating signals that modify (and synchronize) the rhizosphere-associated microbial communities of the neighboring plants [67].
In contrast, depending on the bulk soil type surrounding the plant and the resident microbiota, a specific rhizospheric microbiome can be formed (along with the seed microbiome) [39], [47], [62], [68]. In particular, the ability of a plant to recruit an associated microbiome can be restricted by the native soil microbiota. This was demonstrated by Bakker et al. [69], who observed that the microbiome formation is dynamic and largely depends on the soil type, and the input of resources (cocktail of exudates and carbon content) in corn plants. Similarly, in the microbiota that inhabited each of the four analyzed soil types (sandy loam, clay 1, silt loam, and clay 2), certain microbial communities were more sensitive to variations in nutrient availability, which decreased their diversity, while others handled the change better. Certain amendments, such as ferulic acid, reportedly stimulated the abundance of a few phyla, and decreased the diversity of the rhizospheric microbiome (bacteria and fungi) of Cucumis sativus L. (cucumber) plants [70].
Furthermore, plant species can specifically interact with certain groups of microorganisms. For example, legume plants excrete phenolic compounds, such as flavonoids and isoflavonoids, which induces the expression of nodulation (nod) genes in nitrogen-fixing symbiotic bacteria, prominently known as rhizobia. In response, rhizobia produces nod factors, which are lipoquitooligosaccharides, whose function is to induce a series of physiological changes in the plant root to subsequently form nitrogen-fixing nodules. This type of rather specific communication between certain species of rhizobia and legume species (with exceptions), occurred approximately 75–80 million years ago [71]. A few examples of this type of symbiotic and extremely specific interactions are Rhizobium spp. that are symbionts of Phaseolus spp. (e.g., common bean), Sinorhizobium spp. of Medicago species (e.g., alfalfa), Mesorhizobium spp. of Lotus spp. (e.g., lotus, chickpea), and Bradyrhizobium spp. of Glycine spp. (e.g., soybean, cowpea) [72], [73], [74], [75]. Endosymbiosis of rhizobia within plant roots suggest a shared signaling pathway in plants for fungi (e.g., arbuscular mycorrhizal fungi) and symbiotic proteobacteria, which has been termed as the common symbiotic pathway (CSP) [76], [77]. Recently Genre and Russo [76] reviewed works suggesting the role of at least ten proteins, such as HMGR1 (3-Hydroxy-3-Methylglutaryl CoA Reductase 1), a key enzyme in the mevalonate biosynthetic pathway, and kinases like SYMRK (receptor-like kinase) and CCaMK (nuclear calcium- and calmodulin-dependent kinase), which participate in common signaling pathways in mutualistic symbioses of plants with species of rhizobia and glomeromycetes, the so-called CSP [76].
However, recent work with rice plants suggests that the plant responds to the endophytic colonization of Azoarcus sp., inducing metabolic changes and Ca2+-dependent signaling, without CSP [78]. This further highlights the exclusivity of plant-endophyte bacterial interactions and recognition. Similarly, Liu et al. [79] recently observed that the genotype of soybean plants (Glycine max) is important in structuring the rhizospheric microbiome. Regardless of the soil type, specific bacterial genera such as Rhizobium, Novosphingobium, Phenylobacterium, Streptomyces, and Nocardioides were found in abundance. Finally, their results predicted a convergent metabolic capacity between soil types and genotypes in the rhizosphere of Glycine max, and pathways related to xenobiotic degradation, plant–microbe interactions, and nutrient transport. Studies by Micallef et al. [80] suggested that the plant age and genotype impact the successional progression of the rhizospheric bacterial community in model plants such as Arabidopsis thaliana. In conclusion, their results imply that variations in exudation during plant development are highly specific and depend on the genotype, suggesting the local and unique co-evolution of certain communication processes, which developed because of the accessions between Arabidopsis and its endemic microbiota.
A recent work by Son et al. [81] involved the screening of Arabodopsis thaliana mutants, where the identified FERONIA (RES) receptor kinase mutant (fer-8) “allowed” the enrichment of bacterial species Pseudomonas fluorescens in the rhizosphere. Interestingly, the fer-8 mutant line showed basal levels of reactive oxygen species (ROS) in roots, suggesting that under these conditions, pseudomonads took advantage by increasing their abundance, concluding that FER-mediated ROS production regulates the levels of beneficial pseudomonads in the rhizospheric microbiome. P. fluorescens is widely known for its direct and indirect plant growth promoting capabilities [82]; therefore, this work provides a novel approach in communicating and recruiting beneficial bacteria in the rhizosphere, and possibly facilitating endophytic colonization.
Successional conformation of the rhizosphere microbiome
The rhizosphere, being a dynamic ecosystem, poses a challenge due to its microbial diversity and the complex interactions that occur within, such as competing for nutrients and spaces, survival in the presence of antibiotics, and other activities that shape the community structure [83], [84]. Certain succession studies have shown interest in establishing the role of certain plant species in reshaping the microbial communities that inhabit the soil. Recently, the work done by Sun et al. [85] demonstrated the important role of pioneer plants and their impact on the diversity and structure of bacterial communities in the rhizosphere and bulk soil of copper mine tailings in China. A taxonomic analysis using Illumina MiSeq sequencing showed that Alphaproteobacteria, Deltaproteobacteria, Chloroflexi, Acidobacteria, and Gemmatimonadetes significantly increased their relative abundance in the rhizosphere and bulk soil. In contrast, Gammaproteobacteria and Firmicutes were abundant in bare tailings, in which Bacillus, Pseudomonas, and Lactococcus constituted 63.04% of the total bacterial community.
Similarly, Shi et al. [59] analyzed the succession response of soil bacterial communities during the onset of roots of the common annual grass Avena fatua via 16 s rRNA gene sequencing over two growth periods. The results were as follows: the roots of Avena fatua caused a successional change in the conformation of the rhizospheric bacterial community, which was characterized by the overall reduction in its diversity by decreasing the relative abundance of Acidobacteria, Actinobacteria, and Firmicutes, which was evaluated using taxonomic and phylogenetic analysis; in other cases, plant roots selectively stimulated the relative abundance of bacterial groups such as Alphaproteobacteria, Betaproteobacteria, and Bacteroidetes.
Another interesting work performed recently [38], involved the authors growing modern and wild tomato genotypes, Solanum lycopersicum var. During four successive cycles of Moneymaker (modern) and the wild relative S. pimpinellifolium LA1578, the assembly of the microbiome differed between the two genotypes, and these differences significantly amplified over time. The authors analyzed certain core genera of the rhizospheric microbiome of tomato, and highlighted that the wild tomato increased the abundance of the genera Acidovorax, Massilia, and Rhizobium, whereas the modern tomato plants were enriched by bacterial species of the genus Pseudomonas.
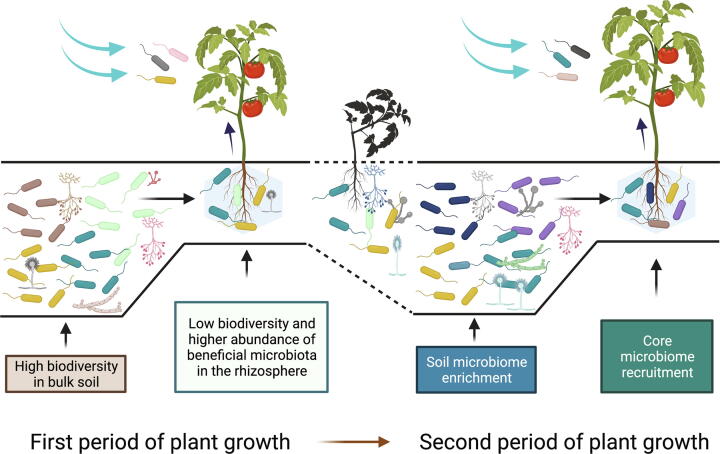
Together, these results suggest and corroborate the dynamic aspect of the plant microbiome assembly, in addition to the fact that certain bacterial taxa may be selected (i.e., selection of a beneficial microbiome from bulk soil), and plant genotype plays the role of another selection factor. Fig. 2 illustrates the dynamic aspects of plant microbiome recruitment during different periods of plant growth (i.e., an agricultural ecosystem).
Fig. 2.
Recruitment dynamics of the plant microbiome during two stages of plant growth in a potential agricultural ecosystem. Initially, bulk soil diversity is high; however, planting seeds promotes the recruitment of a beneficial microbiome that may include a few taxa, decreasing biodiversity. The microbiota recruited from the rhizosphere could also colonize the endosphere and travel to other plant tissues, such as aerial areas. The plant phyllosphere is also susceptible to recruiting new members of the microbiome. Once agricultural production ends in the first period, the soil can regain its biodiversity through the input of organic matter, crop rotation, etc. In a second period, the plant can have a microbiome made up of endophytic bacteria that were inherited horizontally, characterized by long-term relationships or that are part of the core microbiome of the plant host. Additionally, the plant can recruit other members of the rhizosphere and the phyllosphere. See the text for the references that support the illustration.
Colonizing the endosphere
Plants can recruit microbiome from above-ground or below-ground areas. As previously mentioned, rhizodeposits fundamentally influence the gradual recruitment of rhizospheric microbiome [86]. A significant progress in understanding the approach by which plant roots recruit the endophytic microbiome from the rhizosphere is in the work of Edwards et al. [87]. Through a detailed analysis, conducted under different experimental conditions, either in greenhouse or in field, and in various plant models, the authors detected gradual changes in the microbial composition at three interacting root zones: rhizosphere (soil close to and influenced by the root surface), rhizoplane (root surface), and the endosphere (root interior). The results observed gradual changes in the microbiota composition that inhabited the three microecosystems associated with the roots, rhizosphere, rhizoplane, and endosphere. This work is consistent with the two-step model, although with certain variations, proposed by [88], [89], where soil abiotic properties (first step) and plant rhizodeposits (second step) are selective factors that filter (and recruit) certain microbial diversity from the rhizosphere. This model has recently been tested in wild tomato plants and domesticated varieties, indicating that the associated microbiome can be influenced by plant domestication trade-offs [90].
Once the rhizosphere microbiota manage to pass the rhizoplane and penetrate the root tissues, these endophytes will have the opportunity to colonize other plant compartments. The trip includes a walk through the vascular system of the plant toward aerial tissues, such as stem or leaves, along with the flowers and fruits (once the plant enters these developmental stages) [91]. The rhizosphere, which is below-ground and in close contact with the soil microbiota, may also contain unexpected entry zones into the internal tissues of the roots, including root wounds or cracks, which may not require an active system to penetrate tissues. In contrast, rhizospheric bacteria desiring an entry must exhibit more active colonization determinants in the rhizosphere, with characteristics such as the ability to hydrolyze the cells of the plant host, produce biofilms for better attachment, have motility, quorum sensing, and synthesize antibiotics to displace other competitors, in addition to producing antioxidant enzymes [57], [92], [93]. Although no evident genetic differences exist between rhizospheric and endophytic bacteria, a list of genes coding for the aforementioned functions has been suggested.
A pioneering work [94], proposed certain differences between rhizobacteria and endophytes by comparing the genomes of nine endophytic species, including Burkholderia phytofirmans PsJN, Burkholderia spp. strain JK006, Azospirillum lipoferum 4B, Enterobacter cloacae ENHKU01, Klebsiella pneumoniae 342, Pseudomonas putida W619, Enterobacter spp. 638, Azoarcus spp. BH72, and Serratia proteamaculans 568. The authors found a match for approximately 40 genes that could be involved in endophyte behavior, such as transporters, secretion and delivery systems, detoxification and redox maintenance potential, and plant polymer degradation, among other transcriptional regulators. Unfortunately, for decades, only few genes have been identified as important factors in endophytic colonization and existence. However, some important studies have been previously reviewed [57]. For example, Meneses et al. [95] generated mutants in the gumD gene of the N2-fixing bacterium, Gluconacetobacter diazotrophicus, for the production of exopolysaccharides, which are required to generate biofilms and for the endophytic colonization of rice plants. Similarly, in the same bacterial species, Alquéres et al. [96] characterized relevant activities of glutathione reductase (gr::Tn5) and superoxide dismutase (sod::Tn5) in endophytic colonization. In the case of Azoarcus sp. strain BH72, mutations in pilA and eglA, affecting twitching motility and endoglucanase activities, respectively, were described as essential for endophytic colonization of rice plant roots [97]. Furthermore, Buschart et al. [98] generated mutants containing three copies of fliC genes, which encode the major structural protein flagellin. However, only the fliC3 copy was functional and necessary for motility, and was associated with rice roots. Endophytic colonization of rice roots was significantly reduced in the fliC3 mutant. Interestingly, other activities in the rhizosphere, such as the establishment of microcolonies on the root surface remained unaffected, which further confirmed its important role in the endophyte lifecycle.
In contrast, mutant strains affecting swimming motility (fliI) or siderophore pyoverdine production (pvdI) in the rhizobacterium Pseudomonas fluorescens PICF7, and the facultative endophytes in the olive tree roots (Olea europaea L.) did not significantly differ from the wild strain PICF7 in its endophytic colonization capacity Therefore, these characteristics were rejected as being relevant to the endophytic lifestyle of this strain. However, as a facultative endophyte, it reveals that certain functions, such as motility, may be relevant in other non-facultative endophytic strains. Accordingly, in the case of Azoarcus olearius BH72, where mechanisms such as motility are relevant for endophytic colonization of rice, plant defenses are not induced (i.e. bacterial molecular components or MAMPs) [99], [100].
More recently, the importance of motility activities and the production of cyclic di-GMP (c-di-GMP) have been reported in the bacterium Azoarcus sp. CIB strains to endophytically colonize rice plants [101]. Previously, the significant role of c-di-GMP for endophytic lifestyle in Azoarcus sp. species has also been suggested through transcriptional analysis [100]. Interestingly, c-di-GMP in bacteria is involved in modulating the synthesis of exopolysaccharides, adhesins, and the formation of biofilms. As previously mentioned, these functions regulate various mechanisms, including colony morphology, quorum sensing processes, cell motility, nodulation, and virulence [101].
Role of plant defense responses and phytohormones in controlling endophyte colonization
Pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) are one of the main mechanisms recognizing the invasive pathogens [99], [102]. Plants can also detect other “non-pathogenic” microorganisms, including bacteria, from microbe-associated molecular patterns (MAMPs), which regulate the plant defense responses through its innate immune system. These PRRs are the first recognition filter, the second being the intracellular immune receptors, also known as the NOD-like receptor (NLR) type. These receptors directly or indirectly recognize virulence effectors secreted by pathogens within host cells, thereby inducing effector-triggered immunity (ETI) [99], [103]. Although considerable progress has been made in studying these microbial recognition mechanisms by plants, only a few examples of PRRs have been studied in Arabidopsis and rice [104], and other plant models need to be investigated and compared.
Systems, such as MAMPs, individually recognize and detect colonizing microorganisms, which attempt to form beneficial associations with their plant hosts. Other active players include phytohormones, such as salicylic acid (SA), jasmonate acid (JA), abscisic acid (ABA), and ethylene (ET) which may be related to defense functions [105], [106], [107], [108]. In general, SA responds to the interaction between abiotic stimuli, and biotrophic and hemobiotrophic organisms, inducing the defense response in plants, called systemic acquired resistance or SAR [109]. In contrast, JA/ET is an alternative plant pathway that respond to the interaction of beneficial organisms, such as PGPBs [110]. However, some studies [111], reportedly observed concentration changes in the SA and JA content in Metarhizium (an entomopathogenic fungi)-treated maize (Zea mays) plants, which corresponds to plant responses related to the known SA and JA/ET pathways.
Another work performed by Sheoran et al. [112] mentions that the endophytic bacterium Pseudomonas putida BP25, capably alters the root architecture and triggers signaling using SA as a feedback loop in regulating endophytic colonization in Arabidopsis. The same working group has previously reported the beneficial characteristics of BP25 strain, such as the production of a broad spectrum of volatile compounds that suppress a broad range of plant pathogens [113]. Moreover, Lebeis et al. (2015) used mutants in the immune system of Arabidopsis thaliana Col-0 plants, and observed that SA plays an important role in the selection of certain bacterial taxa and structuring their root microbiome, including those that colonize the rhizosphere and endosphere. The authors also reported that endophytic communities were less abundant than rhizosphere inhabitants. Interestingly, the cell densities of some isolates of genera such as Terracoccus sp., Streptomyces sp., and Mitsuaria sp., were modulated in culture media supplemented with or without SA.
More recently, Chen et al. [104] found contradictory results, where they observed that JA, and not SA, is the pathway that restricts endophytic colonization of the beneficial endophyte Azoarcus olearius BH72 in rice plants, while the pathogen Xanthomonas oryzae pv. oryzae PXO99 induces a response to the SA pathway only in roots. As expected, the PX099 strain induced a considerably higher defense response than BH72, which is non-pathogenic. Interestingly, both endophytes induced the expression of genes encoding several enzymes involved in phytoalexin biosynthesis, ROS production, and pathogenesis-related (PR) proteins. To elucidate the detailed response pathways in both endophytes, rice mutant lines cpm2 deficient in jasmonate synthesis, and NPR1 (by RNAi), which mediates responses by SA were used. The authors found that only BH72 increased its endophytic colonization in cpm2, while PX099 was unaffected. The authors concluded that the JA pathway regulates the selective interaction with beneficial endophytic species, modulating their beneficial microbiome.
ET, conversely, is a gaseous plant hormone that is involved in other plant responses such as senescence or fruit ripening. At low concentrations, it can participate as a signal for seed germination, elongation of plant roots, the formation of leaf and root primordia, and flowering initiation [115]. In addition, ET responds or increases its levels when the plant experiences different abiotic or biotic stress, such as attack by pathogens. Therefore, lowering their levels under stress is important for maintaining growth and fitness [116]. To achieve this, the plant can also associate with 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase-producing bacteria. ACC deaminase is widely studied for its ability to reduce ethylene levels in plants through the hydrolysis of ACC, a precursor of ET synthesis, generating α-ketobutyrate and ammonia [117], [118]. Multiple bacterial species have been reported to inhabit the rhizosphere, endosphere, and phyllosphere that contain this function, which helps the plant reduce its ethylene levels under stress conditions, stimulating its growth. In the case of plants of agricultural interest, it has been observed that the application of biofertilizers based on ACC-producing bacteria increases the production and protection of crops [119].
Reaching the phyllosphere
The phyllosphere is an ecosystem that is particularly different from the rhizosphere and endosphere. It is perhaps the most challenging habitat for microorganisms that “desire” to establish long associations with their hosts. This is due to abiotic factors such as insolation, UV rays, drastic temperature variations, rain, and wind [56]. Additionally, the microbiota that is relatively new to the epiphytic life must compete for space and nutrients with the resident and well-established biota. Therefore, adventitious microbes that reach the rhizosphere must be resilient during extremely stressful situations [120], [121]. However, in certain predominantly studied bacteria, certain determinants such as exopolysaccharides, biolfilm and/or adhesion compounds, the flagellum, antibiotics, biosufactant compounds, free radical detoxifying proteins (catalases and superoxide dismutases), versatile metabolism, and quorum sensing signal molecules that reportedly aid in colonizing habitats such as the rhizosphere or the endosphere, can also be of significance in colonizing the phyllosphere [122], [123]. In contrast, the production of pigments by epiphytic bacteria displays a protective mechanism against ultraviolet radiation, which is essential for survival in an epiphytic lifestyle. Jacobs et al. [124] demonstrated the use of mutants in the synthesis of pigments (orange, cream, and pink) in Clavibacter michiganensis, which was highly sensitive when exposed to solar UV radiation (UVR), with lower leaf survival rate, compared to wild strains.
Microscopy analyses have revealed that the vast majority of bacterial cells may be located in the grooves formed by the junctions of the epidermal or apoplast of plant cells, where water and certain nutrients can flow and serve as a reserve for the microbiota. Other structures, such as the presence of veins, stomata, trichomes, and hydathodes, can also alter nutrient availability (i.e., sugars such as fructose and sucrose) [125].
The phyllosphere of plants can be considered as one of the most abundant areas or ecosystems on earth. Some data suggest that such an environment, measured as global leaf area, can reach figures of more than 500 million km2 [40], [123]. These habitats are considered highly abundant, since a square centimeter of leaf surface can host between 106 and 107 bacterial cells [126]. In an excellent review documented by Vorholt [123], the phyllosphere can be widely diverse, where groups such as Proteobacteria (alpha, beta, and gamma), Actinobacteria, and Bacteroidetes are among the most abundant in rice, clover, soybean, and Arabidopsis plants. However, differences are observed between these four plant models, since genera such as Agrobacterium, Methylobacterium (Alphaproteobacteria), and Mycobacterium (Actinobacteria), are the commonly reported epiphytes of rice plants, whereas Pseudomonas is a common inhabitant of soybean, clover, and Arabidopsis. These data are based on the frequency reads of predicted protein coding and rRNA genes within the metagenomes of microbial communities obtained from MG ‑ RAST.
Driving plant factors allowing/restricting microbiome phyllosphere establishment
To date, notably the plant's immune system and signaling pathways that consider the action of phytohormones, among other determinants, can be factors that encourage the intimate interaction and selection of a specific microbiome in ecosystems such as the rhizosphere and the endosphere [19], [127], [128]. However, defense systems in plant aerial zones can act in a systematic and localized manner. Therefore, some authors, such as Schlechter et al. [56], suggest that “to better understand this impact (immune responses of plants), it is necessary to study bacterial communities at a micrometric resolution, which is a scale that is relevant for members of the community.” The microenvironments present on the plant leaf surface can be defined at the micrometer level. Thus, it has been suggested that plant topology and phytochemistry may influence the recruitment of a particular microbiome [129], [130].
Microscopic analysis has revealed the presence of vast majority of bacterial cells in the grooves formed by the junctions of the plant cell epidermis or apoplast. This is where water and some nutrients flow and serve as a reserve for the microbiota. Other structures, such as the presence of veins, stomata, trichomes, and hydathodes, can also alter the availability of nutrients (e.g., sugars such as fructose and sucrose) [125].
Therefore, there may be local and systemic responses in leaves. According to [131], plants use internal immune receptors that recognize specific bacterial effectors, which leads to effector-activated immunity (ETI), which can detect and distinguish the presence of potential pathogens. These immune responses are located at the site of interaction (or infection), which causes the early entry of calcium ions (Ca2+) and MAP kinase, and activation of calcium-dependent protein kinase. This cascade of signals subsequently comprises callose deposition and stomatal closure as defense against pathogens, in addition to cell death (for more information, see [56], [131]).
As aforementioned, certain factors can successfully colonize environments such as the phyllosphere [56]. However, stochastic events also influence the composition of a plant-associated microbiome. As demonstrated by Maignien et al. [132], when analyzing more than 260,000 sequences from hypervariable regions of the 16S rRNA gene, they managed to characterize the phyllosphere bacterial community structure on 32 plant and 21 air samples over 73 days. Their results confirmed that in addition to ecological successions, a stochastic variation influences the assembly of bacterial communities associated with the microenvironments of the A. thaliana phyllosphere. Alternatively, the phyllospheric microbiome of plants can be colonized by those microbes that reside not only in the air but also that originate from the rhizosphere [133], as previously mentioned.
In an extensive study of the fungal and bacterial communities of the Arabidopsis thaliana phyllosphere, the host genotype was also a contributing factor that determined the presence or absence of certain microbial groups. This result was achieved through genome-wide association studies (GWAS), which revealed that certain genes responsible for defense, kinase-related activities, and cell wall integrity (comprising the polysaccharides cellulose, callose, and pectin) impact the community variations [134].
In an interesting study, the availability of O2 in the marine plant phyllosphere was considered an important factor in the conformation of the associated microbiome, which modulates the microbial metabolic processes that occur within epiphytic biofilms [135]. Although O2 is available in terrestrial environments, other gases such as CO2 can affect the diversity of the phyllosphere (and endosphere) of rice plants [136]. However, it would be interesting to observe their influence in the colonization and microbial activity in the resident plant phyllosphere in regions with high levels of environmental pollution (with gases such as CO2 or other elements).
Can the plant microbiome help to recruit other beneficial microbes?
The plant associated-microbiome provides different services to the plant, such as protection against pathogens, stimulation of its growth, and increased levels of resistance against different abiotic stresses. The examples are diverse, and the literature is filled with these reports (see [137], [138], [139]). However, few reports illustrate the beneficial action by the associated microbiome in recruiting new members to provide beneficial services to the plant. One way to evaluate this hypothesis is by analyzing the inoculation of a “microbial agent” in the rhizosphere, endosphere or phyllosphere, and observe a rearrangement in the associated-microbial composition; hence, improvement in health, plant growth or fitness will be indirectly observed compared to those plants that were not previously inoculated with the “microbial agent.”
This type of work was conducted by Zhang et al. [140], who observed that the pre-inoculation of pepper seedlings with the PGPR Bacillus velezensis NJAU-Z9 induced changes in the composition of the rhizospheric microbiome in a field experiment over two seasons. This pre-inoculation significantly increased the abundance of bacterial genera, such as Sphingomonas, Sphingopyxis, Bradyrhizobium, Chitinophaga, Dyadobacter, Streptomyces, Lysobacter, Pseudomonas, and Rhizomicrobium, and the fungal genera such as Aspergillus, Cladorrhinum, and Cladorrhinum. Similarly, the transplanted seedlings pre-colonized with NJAU-Z9 significantly increased pepper yield by 11% and 24%, respectively, compared to non-inoculated plants.
Another study by [141] also showed that the nodule-endophyte Agrobacterium sp. 10C2 affects the richness and structure of rhizosphere bacterial communities of common bean plants (Phaseolus vulgaris). The inoculation also induced plant nodulation and growth. Some of the bacterial taxa that increased their relative abundance were mainly members of the genus Bacillus, such as Bacillus licheniformis, Bacillus pumilus, Bacillus senegalensis, Bacillus subtilis, Bacillus firmus, and Paenibacillus koreensis. These Bacillus species have been widely reported as PGPB.
In addition, to the beneficial agents modulating the beneficial microbiome of plants, their presence or infection by fungal pathogens can induce changes in the microbiota. In some cases, the plant detects the presence of the pathogen (through unknown mechanisms) and increases the population of beneficial endophytic microbiota that help protect the host. This was demonstrated by the excellent work of Carrión et al. [142], who inoculated an endophytic consortium of Flavobacterium and Chitinophaga in sugar beet seedling plants, previously identified by their population increase and stimulation, in the presence of the fungal pathogen Rhizoctonia solani, which also increased plant protection. Another work conducted by Solís-García et al. [143] also suggest that pathogens such as Phytophtora oomycete can cause disease in avocado plants and modify the rhizosphere microbiota. Interestingly, root rot increased the proportion of Pseudomonadales and Burkholderiales, which are PGPB agents, while the abundance of other opportunistic fungal pathogens, such as Mortierella sp., Fusarium spp., Lasiodiplodia sp., and Scytalidium sp. was also increased. The previous inoculation of one, two, or an entire microbiome can also prohibit the establishment or colonization of pathogens in plant rhizosphere, including those that cause disease in humans, such as E.coli [144].
Previous studies have shown that the inoculation of unique species can modify an entire plant-associated microbiome, that is, they stimulate the abundance of certain genera that may also exert direct and indirect mechanisms that influences plant growth and protection. However, there remains a question about how they stimulate the presence of other beneficial taxa. Certain new evidence may arise from the synergistic interaction between species, as shown in the study by Baas et al. [145] where, through the inoculation of a phosphate-solubilizing consortium, which includes the species Comamonas testosteroni, Pseudomonas putida, Enterobacter cloacae, and Citrobacter freundii, the growth and productivity increased twice as that of the uninoculated cultures. The cultures that benefited from the consortium were red winter wheat (Triticum aestivum), fescue turf grass (Festuca arundinacea; Kentucky 31 variety), jalapeño (Capsicum annuum; early jalapeño variety), cherry tomato (Solanum lycopersicum; Sweetie variety), and basil (Ocimum basilicum; Italian Genovese variety).
Another study on the beneficial interaction between PGPB is that of [146], who observed that Bacillus thuringiensis and Pseudomonas fluorescens strains can interact physically, which additively and/or synergistically stimulate the growth of husk plants tomato (Physalis ixocarpa Brot. ex Horm.). Notably, even certain PGP mechanisms in a single PGPB can exhibit synergistic action to stimulate the growth of tomato plants under salinity conditions, such as the production of ACC deaminase and trehalose in PGPB Pseudomonas sp. UW4. However, there are other studies, where inoculation through microbial strains or consortia have improved the growth or production of different cultures [147], but the effect on the associated microbiota was not evaluated in the plant, and its effect is unknown. Therefore, it is advisable to incorporate this analysis with new bioinoculant agents, to understand if the beneficial interactions with another associated microbiota can also indirectly benefit the plant.
Microbial communication plays a significant role in promoting the abundance of certain plant-associated microbial taxa. Therefore, quorum sensing (QS) molecules, such as N-acyl homoserine lactones (AHL), play an important role in this communication to regulate and sense bacterial populations in environments, such as the rhizosphere. Similarly, plants can sense AHL and produce different signals or defense pathways. Although Rodriguez et al. [148] mentioned that AHLs are also produced by pathogens, these molecules do not provide sufficient information for plants to provide a defense response signal, but they do suggest that “ Possibly the combinations and concentrations of QS molecules may indicate an imbalanced microbial composition.”
Other signal molecules, with functions similar to AHL, have been reported in studies such as those by Orozco-Mosqueda et al., Velázquez-Becerra et al., and Martínez-Cámara et al. [149], [150], [151]. These studies have identified and characterized volatile molecules such as dimethylhexadecylamine, produced by the curl-and endobacterium Arthrobacter agilis UMCV2, which may play an important role in communicating with other beneficial taxa in the rhizosphere or the endosphere of plants, since this molecule has an antifungal action and modulates the growth and swarming motility of other bacterial species, among other stimulating activities of plant growth and nutrition. However, the modulating role of microbial communities in the rhizosphere or endosphere of plants is still unknown.
Other molecules that are involved in microbial communication are lipopolysaccharides, which have been widely studied in beneficial bacteria, and whose functions are diverse, including stimulating plant growth, biofilms function, communication with the environment, recognition between microorganisms, and antagonism towards pathogens [152]. It would be important to know whether the mutant strains in the synthesis of polysaccharides, such as Bacillus velezensis NJAU-Z9 [140] or Agrobacterium sp. 10C2 [141], increase the abundance and diversity of beneficial taxa in environments such as the rhizosphere.
Restricting the recruitment of harmful microbes
The microbiota not only helps the plant recruit other beneficial microorganisms, but it can also restrict access to others, including potential pathogens [153]. The mechanisms by which certain microbes restrict growth are mainly antibiosis [154]. For example, PGPB of the genus Pseudomonas is widely known for producing compounds such as phenazine-1-carboxylic acid, phenazine-1-carboxyamide, 2,4-diacetyl phloroglucinol, pyoluteorin, pyrrolnitrin, volatile hydrogen cyanide, and dimethyl disulfide (DMDS) [155], [156]. In addition, PGPB can synthesize lytic enzymes, such as glucanases or cellulals, which are capable of degrading the membranes of plant pathogenic fungi. In addition, Bacillus species can restrict the growth of pathogens through the production of lipopeptides such as fengycin, surfactin, and iturin [157], [158]. Other beneficial mechanisms that interact with plant hosts, such as Trichoderma, can restrict growth in the rhizosphere of plants through physical mechanisms, by occupying spaces, in addition to producing antibiotics [159], [160]. In general, there are multiple plant protection services provided by the plant-associated microbiota from different microecosystems, such as the rhizosphere, endosphere, and phyllosphere, thus avoiding harmful interactions with the host.
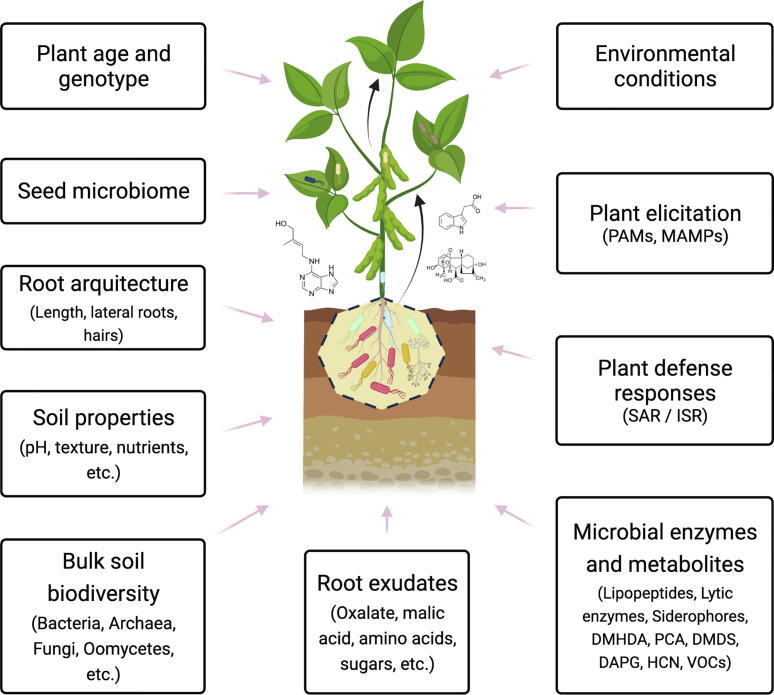
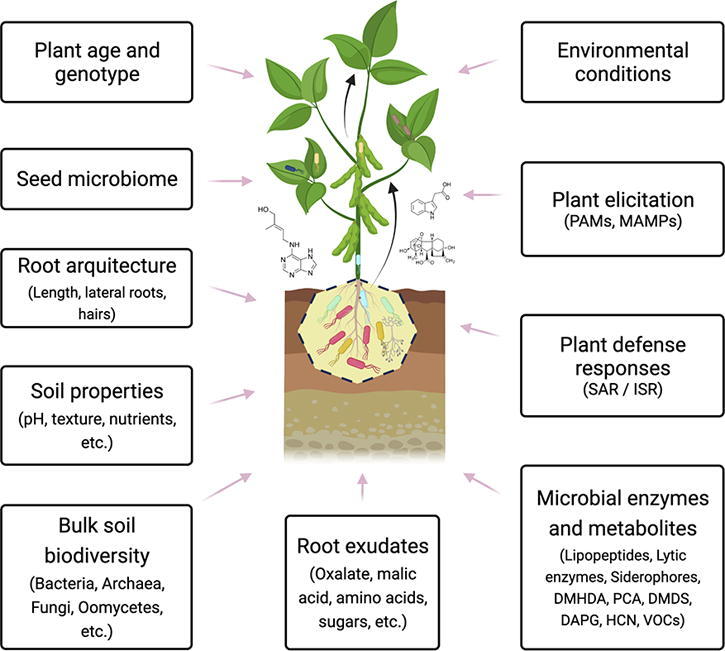
Evidently, there are still sufficient gaps in microbe-microbe interaction, and the plant social network [161], and in understanding how certain stimulating strains of the beneficial plant microbiome can achieve this function. Therefore, it is necessary to further investigate the aspects that regulate these functions, including the important abiotic factors [162], such as pH and soil type, salinity, or even the type of root exudate. Fig. 3 shows several abiotic and biotic factors affecting the recruitment of the plant microbiome into the three different interaction zones reviewed here: the rhizosphere, endosphere, and phyllosphere.
Fig. 3.
Various factors that modulate the recruitment of a microbiome and interactions with the plant. See text for further details. Abbreviations meaning: Pathogen-Associated –Molecular- Patterns (PAMs); Microbe-Associated Molecular Patterns (MAMPs); Systemic Acquired Resistance (SAR); Induced Systemic Resistance (ISR); N, N-dimethyl-1-hexadecylamine (DMHDA); Phenazine-1-carboxylic acid (PCA); Dimethyl disulfide (DMDS); 2,4-diacetyl phloroglucinol (DAPG); Hydrogen cyanide (HCN).
Conclusions and perspectives
Plant-host interactions, whether beneficial or pathogenic, are complex and involve a wide variety of stimuli and connections that affect the type of final association [76], [161]. In particular, the question of how plants recruit their beneficial microbiota/microbiome, and how they differentiate them from pathogens desiring to infect them, is only just being answered [19], which undoubtedly requires different approaches (biochemical, microbiological, microscopic, omics techniques, etc.) to identify the molecules, metabolites, interaction networks, signaling pathways, genes, and the physical interactions involved in recruiting the associated microbiome.
In addition, the vast majority of plant-host interaction analyses use model plants such as Arabidopsis [89], [114], [132] (with undeniable genetic advantages); however, other plant species of agricultural importance still lack data. In addition, more than 300,000 plant species have been proposed to exist worldwide [163], which is expected to have broad and diverse modes of interaction and recruitment of their associated microbiome. Therefore, future research is needed to provide new insights into the molecular interactions and recruitment of beneficial organisms, whose function is still particularly unknown in natural ecosystems.
Finally, different agricultural systems should consider the use of beneficial microorganisms as a requirement for sustainable agricultural practices. For example, the inoculation of microbial agents or consortia in seeds prior to sowing should be considered a routine activity. In some regions, synthetic fertilizers or fungicides (i.e., captan or benomyl) are applied to avoid subsequent fungal infection by pathogens. However, the collateral damage exerted by these agrochemicals on the environment, human, and animal health is widely documented. Therefore, the application of microbial biocontrols or biostimulants, previously evaluated as active hosts of the crops, will allow agricultural products that provide a sustainable future.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
CRediT authorship contribution statement
Gustavo Santoyo: Conceptualization, Visualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by CONACYT-México (Proposal: A1-S-15956). The author very much appreciates the support of Mariana Orozco Mosqueda, Montserrat Orozco Mosqueda and Ma. del Carmen Orozco Mosqueda during the writing of this review.
Biography

Dr. Gustavo Santoyo obtained his Doctor of Science degree in Biomedical Sciences at the National Autonomous University of Mexico (UNAM) in 2005 and did postdoctoral studies during 2005-2007 at the Center for Cancer Research, NIH, USA, as well as a sabbatical year (2015) at the Wilfrid Laurier University, Canada. He is currently a full-time research professor in the Institute of Chemical and Biological Research of the Universidad Michoacana de San Nicolás de Hidalgo, in Morelia, Mexico. His research interest is led towards plant-microbe interactions, biocontrol of fungal pathogens, development of bioinoculants for agricultural crops, and basic research on microbial diversity of extreme environments. In these research areas has published 80 scientific publications with more than 3,300 citations and a h-factor 27. In the Academy, He has supervised 30 doctoral, master and undergrad thesis. Dr. Santoyo is a member of the Mexican Academy of Sciences and the National Research System (SNI) of the National Council of Science and Technology (CONACYT) in Mexico.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Lemanceau P, Barret M, Mazurier S, Mondy S, Pivato B, Fort T, et al. Plant Communication With Associated Microbiota in the Spermosphere, Rhizosphere and Phyllosphere. vol. 82. Elsevier Ltd; 2017. 10.1016/bs.abr.2016.10.007.
- 2.Krings M., Taylor T.N., Hass H., Kerp H., Dotzler N., Hermsen E.J. Fungal endophytes in a 400-million-yr-old land plant: Infection pathways, spatial distribution, and host responses. New Phytol. 2007;174:648–657. doi: 10.1111/j.1469-8137.2007.02008.x. [DOI] [PubMed] [Google Scholar]
- 3.Javaux E.J. Extreme life on Earth - Past, present and possibly beyond. Res Microbiol. 2006;157:37–48. doi: 10.1016/j.resmic.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Upson JL, Zess EK, Białas A, Wu C hang, Kamoun S. The coming of age of EvoMPMI: evolutionary molecular plant–microbe interactions across multiple timescales. Curr Opin Plant Biol 2018;44:108–16. 10.1016/j.pbi.2018.03.003. [DOI] [PubMed]
- 5.Lazcano A., Peretó J. On the origin of mitosing cells: A historical appraisal of Lynn Margulis endosymbiotic theory. J Theor Biol. 2017;434:80–87. doi: 10.1016/j.jtbi.2017.06.036. [DOI] [PubMed] [Google Scholar]
- 6.Badet T., Croll D. The rise and fall of genes: origins and functions of plant pathogen pangenomes. Curr Opin Plant Biol. 2020;56:65–73. doi: 10.1016/j.pbi.2020.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Arif I., Batool M., Schenk P.M. Plant Microbiome Engineering: Expected Benefits for Improved Crop Growth and Resilience. Trends Biotechnol. 2020;38:1385–1396. doi: 10.1016/j.tibtech.2020.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Bakker P.A.H.M., Berendsen R.L., Van Pelt J.A., Vismans G., Yu K., Li E., et al. The Soil-Borne Identity and Microbiome-Assisted Agriculture: Looking Back to the Future. Mol Plant. 2020;13:1394–1401. doi: 10.1016/j.molp.2020.09.017. [DOI] [PubMed] [Google Scholar]
- 9.Hassan M.K., McInroy J.A., Kloepper J.W. The interactions of rhizodeposits with plant growth-promoting rhizobacteria in the rhizosphere: A review. Agric. 2019;9 doi: 10.3390/agriculture9070142. [DOI] [Google Scholar]
- 10.Haney C.H., Samuel B.S., Bush J., Ausubel F.M. Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat Plants. 2015;1 doi: 10.1038/nplants.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santoyo G, Guzmán-Guzmán P, Parra-Cota FI, Santos-Villalobos S de los, Orozco-Mosqueda M del C, Glick BR. Plant Growth Stimulation by Microbial Consortia. Agron 2021, Vol 11, Page 219 2021;11:219. 10.3390/AGRONOMY11020219.
- 12.Méndez-Santiago E.W., Gómez-Rodríguez O., Sánchez-Cruz R., Folch-Mallol J.L., Hernández-Velázquez V.M., Villar-Luna E., et al. Serratia sp., an endophyte of Mimosa pudica nodules with nematicidal, antifungal activity and growth-promoting characteristics. Arch Microbiol. 2020 doi: 10.1007/s00203-020-02051-2. [DOI] [PubMed] [Google Scholar]
- 13.Bona E., Cantamessa S., Massa N., Manassero P., Marsano F., Copetta A., et al. Arbuscular mycorrhizal fungi and plant growth-promoting pseudomonads improve yield, quality and nutritional value of tomato: a field study. Mycorrhiza. 2017;27:1–11. doi: 10.1007/s00572-016-0727-y. [DOI] [PubMed] [Google Scholar]
- 14.Sarma B.K., Yadav S.K., Singh S., Singh H.B. Microbial consortium-mediated plant defense against phytopathogens: Readdressing for enhancing efficacy. Soil Biol Biochem. 2015;87:25–33. doi: 10.1016/j.soilbio.2015.04.001. [DOI] [Google Scholar]
- 15.Díaz-Rodríguez A, Alejandra Salcedo Gastelum L, María Félix Pablos C, Isela Parra-Cota F, Santoyo G, Laura Puente M, et al. The Current and Future Role of Microbial Culture Collections in Food Security Worldwide 2021;4:614739. 10.3389/fsufs.2020.614739.
- 16.Orozco-Mosqueda M.C., Flores A., Rojas-s B., Urtis-flores C.A., Morales-cedeño L.R., Valencia-marin M.F., et al. Plant Growth-Promoting Bacteria as Bioinoculants : Attributes and Challenges for Sustainable. Crop Improvement. 2021:1–15. [Google Scholar]
- 17.Walters D.R., Havis N.D., Paterson L., Taylor J., Walsh D.J., Sablou C. Control of foliar pathogens of spring barley using a combination of resistance elicitors. Front Plant Sci. 2014;5:1–9. doi: 10.3389/fpls.2014.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adesemoye A.O., Torbert H.A., Kloepper J.W. Plant growth-promoting rhizobacteria allow reduced application rates of chemical fertilizers. Microb Ecol. 2009;58:921–929. doi: 10.1007/s00248-009-9531-y. [DOI] [PubMed] [Google Scholar]
- 19.Thoms D., Liang Y., Haney C.H. Maintaining Symbiotic Homeostasis: How Do Plants Engage With Beneficial Microorganisms While at the Same Time Restricting Pathogens? Mol Plant-Microbe Interact. 2021;34:462–469. doi: 10.1094/mpmi-11-20-0318-fi. [DOI] [PubMed] [Google Scholar]
- 20.Orozco-Mosqueda M del C, Santoyo G. Plant-microbial endophytes interactions: Scrutinizing their beneficial mechanisms from genomic explorations. Curr Plant Biol 2021;25:100189. 10.1016/j.cpb.2020.100189.
- 21.Bosamia TC, Barbadikar KM, Modi A. Genomic insights of plant endophyte interaction: prospective and impact on plant fitness. Elsevier Inc.; 2020. 10.1016/b978-0-12-819654-0.00009-0.
- 22.Levy A., Salas Gonzalez I., Mittelviefhaus M., Clingenpeel S., Herrera Paredes S., Miao J., et al. Genomic features of bacterial adaptation to plants. Nat Genet. 2018;50:138–150. doi: 10.1038/s41588-017-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khatoon Z., Huang S., Rafique M., Fakhar A., Kamran M.A., Santoyo G. Unlocking the potential of plant growth-promoting rhizobacteria on soil health and the sustainability of agricultural systems. J Environ Manage. 2020;273 doi: 10.1016/j.jenvman.2020.111118. [DOI] [PubMed] [Google Scholar]
- 24.Morales-Cedeño LR, Orozco-Mosqueda M del C, Loeza-Lara PD, Parra-Cota FI, de los Santos-Villalobos S, Santoyo G. Plant growth-promoting bacterial endophytes as biocontrol agents of pre- and post-harvest diseases: Fundamentals, methods of application and future perspectives. Microbiol Res 2021;242. 10.1016/j.micres.2020.126612. [DOI] [PubMed]
- 25.Morgan J.A.W., Bending G.D., White P.J. Biological costs and benefits to plant-microbe interactions in the rhizosphere. J Exp Bot. 2005;56:1729–1739. doi: 10.1093/jxb/eri205. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H., Tang M., Chen H., Tian Z., Xue Y., Feng Y. Communities of arbuscular mycorrhizal fungi and bacteria in the rhizosphere of Caragana korshinkii and Hippophae rhamnoides in Zhifanggou watershed. Plant Soil. 2010;326:415–424. doi: 10.1007/s11104-009-0022-1. [DOI] [Google Scholar]
- 27.Tian L., Lin X., Tian J., Ji L., Chen Y., Tran L.S.P., et al. Research advances of beneficial microbiota associated with crop plants. Int J Mol Sci. 2020;21:1–18. doi: 10.3390/ijms21051792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choudhary D.K., Johri B.N. Interactions of Bacillus spp. and plants - With special reference to induced systemic resistance (ISR) Microbiol Res. 2009;164:493–513. doi: 10.1016/j.micres.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Newton A.C., Fitt B.D.L., Atkins S.D., Walters D.R., Daniell T.J. Pathogenesis, parasitism and mutualism in the trophic space of microbe-plant interactions. Trends Microbiol. 2010;18:365–373. doi: 10.1016/j.tim.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Lima P.T., Faria V.G., Patraquim P., Ramos A.C., Feijó J.A., Sucena É. Plant-microbe symbioses: New insights into common roots. BioEssays. 2009;31:1233–1244. doi: 10.1002/bies.200800177. [DOI] [PubMed] [Google Scholar]
- 31.Massa N., Cesaro P., Todeschini V., Capraro J., Scarafoni A., Cantamessa S., et al. Selected autochthonous rhizobia, applied in combination with AM fungi, improve seed quality of common bean cultivated in reduced fertilization condition. Appl Soil Ecol. 2020;148 doi: 10.1016/j.apsoil.2020.103507. [DOI] [Google Scholar]
- 32.Sasse J., Martinoia E., Northen T. Feed Your Friends: Do Plant Exudates Shape the Root Microbiome? Trends Plant Sci. 2018;23:25–41. doi: 10.1016/j.tplants.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Das K., Prasanna R., Saxena A.K. Rhizobia: a potential biocontrol agent for soilborne fungal pathogens. Folia Microbiol (Praha) 2017;62:425–435. doi: 10.1007/s12223-017-0513-z. [DOI] [PubMed] [Google Scholar]
- 34.Orozco-Mosqueda M d. C, Altamirano-Hernandez J, Farias-Rodriguez R, Valencia-Cantero E, Santoyo G. Homologous recombination and dynamics of rhizobial genomes. Res Microbiol 2009;160. 10.1016/j.resmic.2009.09.011. [DOI] [PubMed]
- 35.Hakim S, Mirza BS, Imran A, Zaheer A, Yasmin S, Mubeen F, et al. Illumina sequencing of 16S rRNA tag shows disparity in rhizobial and non-rhizobial diversity associated with root nodules of mung bean (Vigna radiata L.) growing in different habitats in Pakistan. Microbiol Res 2020;231:126356. 10.1016/j.micres.2019.126356. [DOI] [PubMed]
- 36.Song C., Jin K., Raaijmakers J.M. Designing a home for beneficial plant microbiomes. Curr Opin Plant Biol. 2021;62 doi: 10.1016/j.pbi.2021.102025. [DOI] [PubMed] [Google Scholar]
- 37.Orozco-Mosqueda M del C, Santoyo G. Plant-microbial endophytes interactions: Scrutinizing their beneficial mechanisms from genomic explorations. Curr Plant Biol 2020:100189. 10.1016/j.cpb.2020.100189.
- 38.Cordovez V., Rotoni C., Dini-Andreote F., Oyserman B., Carrión V.J., Raaijmakers J.M. Successive plant growth amplifies genotype-specific assembly of the tomato rhizosphere microbiome. Sci Total Environ. 2021;772 doi: 10.1016/j.scitotenv.2020.144825. [DOI] [PubMed] [Google Scholar]
- 39.Kong H.G., Song G.C., Ryu C.M. Inheritance of seed and rhizosphere microbial communities through plant–soil feedback and soil memory. Environ Microbiol Rep. 2019;11:479–486. doi: 10.1111/1758-2229.12760. [DOI] [PubMed] [Google Scholar]
- 40.Massoni J., Bortfeld-Miller M., Jardillier L., Salazar G., Sunagawa S., Vorholt J.A. Consistent host and organ occupancy of phyllosphere bacteria in a community of wild herbaceous plant species. ISME J. 2020;14:245–258. doi: 10.1038/s41396-019-0531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orozco-Mosqueda M del C, Rocha-Granados M del C, Glick BR, Santoyo G. Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol Res 2018;208:25–31. 10.1016/j.micres.2018.01.005. [DOI] [PubMed]
- 42.Hanin M., Ebel C., Ngom M., Laplaze L., Masmoudi K. New insights on plant salt tolerance mechanisms and their potential use for breeding. Front Plant Sci. 2016;7:1–17. doi: 10.3389/fpls.2016.01787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y., Zhu A., Tan H., Cao L., Zhang R. Engineering banana endosphere microbiome to improve Fusarium wilt resistance in banana. Microbiome. 2019;7:1–15. doi: 10.1186/s40168-019-0690-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu Q., Zhang Z., Peijnenburg W.J.G.M., Liu W., Lu T., Hu B., et al. Rhizosphere Microbiome Assembly and Its Impact on Plant Growth. J Agric Food Chem. 2020;68:5024–5038. doi: 10.1021/acs.jafc.0c00073. [DOI] [PubMed] [Google Scholar]
- 45.Lozupone C.A., Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci U S A. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fierer N., Jackson R.B. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zarraonaindia I., Owens S.M., Weisenhorn P., West K., Hampton-Marcell J., Lax S., et al. The soil microbiome influences grapevine-associated microbiota. MBio. 2015;6 doi: 10.1128/mBio.02527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blossfeld S., Gansert D., Thiele B., Kuhn A.J., Lösch R. The dynamics of oxygen concentration, pH value, and organic acids in the rhizosphere of Juncus spp. Soil Biol Biochem. 2011;43:1186–1197. doi: 10.1016/j.soilbio.2011.02.007. [DOI] [Google Scholar]
- 49.Philippot L., Raaijmakers J.M., Lemanceau P., Van Der Putten W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat Rev Microbiol. 2013;11:789–799. doi: 10.1038/nrmicro3109. [DOI] [PubMed] [Google Scholar]
- 50.Chaparro J.M., Badri D.V., Vivanco J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014;8:790–803. doi: 10.1038/ismej.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bai Y., Müller D.B., Srinivas G., Garrido-Oter R., Potthoff E., Rott M., et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature. 2015;528:364–369. doi: 10.1038/nature16192. [DOI] [PubMed] [Google Scholar]
- 52.Aoun M. Host defense mechanisms during fungal pathogenesis and how these are overcome in susceptible plants: A review. Int J Bot. 2017;13:82–102. doi: 10.3923/ijb.2017.82.102. [DOI] [Google Scholar]
- 53.Zhu J.K. Abiotic Stress Signaling and Responses in Plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wright S.J., Trakhtenbrot A., Bohrer G., Detto M., Katul G.G., Horvitz N., et al. Understanding strategies for seed dispersal by wind under contrasting atmospheric conditions. Proc Natl Acad Sci U S A. 2008;105:19084–19089. doi: 10.1073/pnas.0802697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Compant S., Clément C., Sessitsch A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem. 2010;42:669–678. doi: 10.1016/j.soilbio.2009.11.024. [DOI] [Google Scholar]
- 56.Schlechter R.O., Miebach M., Remus-Emsermann M.N.P. Driving factors of epiphytic bacterial communities: A review. J Adv Res. 2019;19:57–65. doi: 10.1016/j.jare.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santoyo G., Moreno-Hagelsieb G., del Carmen Orozco-Mosqueda M, Glick B.R. Plant growth-promoting bacterial endophytes. Microbiol Res. 2016;183 doi: 10.1016/j.micres.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 58.Compant S., Cambon M.C., Vacher C., Mitter B., Samad A., Sessitsch A. The plant endosphere world – bacterial life within plants. Environ Microbiol. 2020;23:1812–1829. doi: 10.1111/1462-2920.15240. [DOI] [PubMed] [Google Scholar]
- 59.Shi S., Nuccio E., Herman D., Rijkers R., Estera K., Li J., et al. Successional Trajectories of Rhizosphere Bacterial Communities over. MBio. 2015;6:13–20. doi: 10.1128/mBio.00746-15.Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X., Rui J., Mao Y., Yannarell A., Mackie R. Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol Biochem. 2014;68:392–401. doi: 10.1016/j.soilbio.2013.10.017. [DOI] [Google Scholar]
- 61.Hartmann A., Rothballer M., Schmid M. Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil. 2008;312:7–14. doi: 10.1007/s11104-007-9514-z. [DOI] [Google Scholar]
- 62.Girsowicz R., Moroenyane I., Steinberger Y. Bacterial seed endophyte community of annual plants modulated by plant photosynthetic pathways. Microbiol Res. 2019;223–225:58–62. doi: 10.1016/j.micres.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 63.Bodhankar S, Grover M, Hemanth S, Reddy G, Rasul S, Yadav SK, et al. Maize seed endophytic bacteria: dominance of antagonistic, lytic enzyme-producing Bacillus spp. 3 Biotech 2017;7. 10.1007/s13205-017-0860-0. [DOI] [PMC free article] [PubMed]
- 64.Rahman M.M., Flory E., Koyro H.W., Abideen Z., Schikora A., Suarez C., et al. Consistent associations with beneficial bacteria in the seed endosphere of barley (Hordeum vulgare L.) Syst Appl Microbiol. 2018;41:386–398. doi: 10.1016/j.syapm.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 65.Vives-Peris V., de Ollas C., Gómez-Cadenas A., Pérez-Clemente R.M. Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep. 2020;39:3–17. doi: 10.1007/s00299-019-02447-5. [DOI] [PubMed] [Google Scholar]
- 66.Kost T., Stopnisek N., Agnoli K., Eberl L., Weisskopf L. Oxalotrophy, a widespread trait of plant-associated Burkholderia species, is involved in successful root colonization of lupin and maize by Burkholderia phytofirmans. Front Microbiol. 2013;4:1–9. doi: 10.3389/fmicb.2013.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kong H.G., Song G.C., Sim H.J., Ryu C.M. Achieving similar root microbiota composition in neighbouring plants through airborne signalling. ISME J. 2021;15:397–408. doi: 10.1038/s41396-020-00759-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramakrishnan B., Kaur S., Prasanna R., Ranjan K., Kanchan A., Hossain F., et al. Microbial inoculation of seeds characteristically shapes the rhizosphere microbiome in desi and kabuli chickpea types. J Soils Sediments. 2017;17:2040–2053. doi: 10.1007/s11368-017-1685-5. [DOI] [Google Scholar]
- 69.Bakker M.G., Chaparro J.M., Manter D.K., Vivanco J.M. Impacts of bulk soil microbial community structure on rhizosphere microbiomes of Zea mays. Plant Soil. 2015;392:115–126. doi: 10.1007/s11104-015-2446-0. [DOI] [Google Scholar]
- 70.Zhou X., Wu F. Effects of amendments of ferulic acid on soil microbial communities in the rhizosphere of cucumber (Cucumis sativus L.) Eur J Soil Biol. 2012;50:191–197. doi: 10.1016/j.ejsobi.2012.03.001. [DOI] [Google Scholar]
- 71.Bonfante P., Anca I.A. Plants, mycorrhizal fungi, and bacteria: A network of interactions. Annu Rev Microbiol. 2009;63:363–383. doi: 10.1146/annurev.micro.091208.073504. [DOI] [PubMed] [Google Scholar]
- 72.Abd-Alla M.H., El-Enany A.W.E., Nafady N.A., Khalaf D.M., Morsy F.M. Synergistic interaction of Rhizobium leguminosarum bv. viciae and arbuscular mycorrhizal fungi as a plant growth promoting biofertilizers for faba bean (Vicia faba L.) in alkaline soil. Microbiol Res. 2014;169:49–58. doi: 10.1016/j.micres.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 73.Li R., Feng Y., Chen H., Zhang C., Huang Y., Chen L., et al. Whole-Genome Sequencing of Bradyrhizobium diazoefficiens 113–2 and Comparative Genomic Analysis Provide Molecular Insights Into Species Specificity and Host Specificity. Front Microbiol. 2020;11:1–17. doi: 10.3389/fmicb.2020.576800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lopez-Gomez M., Sandal N., Stougaard J., Boller T. Interplay of flg22-induced defence responses and nodulation in Lotus japonicus. J Exp Bot. 2012;63:393–401. doi: 10.1093/jxb/err291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peralta H., Mora Y., Salazar E., Encarnación S., Palacios R., Mora J. Engineering the nifH promoter region and abolishing poly-β -hydroxybutyrate accumulation in Rhizobium etli enhance nitrogen fixation in symbiosis with Phaseolus vulgaris. Appl Environ Microbiol. 2004;70:3272–3281. doi: 10.1128/AEM.70.6.3272-3281.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Genre A., Russo G. Does a common pathway transduce symbiotic signals in plant–microbe interactions? Front Plant Sci. 2016;7:1–8. doi: 10.3389/fpls.2016.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Genre A., Lanfranco L., Perotto S., Bonfante P. Unique and common traits in mycorrhizal symbioses. Nat Rev Microbiol. 2020;18:649–660. doi: 10.1038/s41579-020-0402-3. [DOI] [PubMed] [Google Scholar]
- 78.Chen X., Miché L., Sachs S., Wang Q., Buschart A., Yang H., et al. Rice responds to endophytic colonization which is independent of the common symbiotic signaling pathway. New Phytol. 2015;208:531–543. doi: 10.1111/nph.13458. [DOI] [PubMed] [Google Scholar]
- 79.Liu F., Hewezi T., Lebeis S.L., Pantalone V., Grewal P.S., Staton M.E. Soil indigenous microbiome and plant genotypes cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiol. 2019;19:1–19. doi: 10.1186/s12866-019-1572-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Micallef S.A., Channer S., Shiaris M.P., Colón-Carmona A. Plant age and genotype impact the progression of bacterial community succession in the Arabidopsis rhizosphere. Plant Signal Behav. 2009;4:777–780. doi: 10.4161/psb.4.8.9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song Y., Wilson A.J., Zhang X.-C., Thoms D., Sohrabi R., Song S., et al. FERONIA restricts Pseudomonas in the rhizosphere microbiome via regulation of reactive oxygen species. Nat Plants. 2021;7:644–654. doi: 10.1038/s41477-021-00914-0. [DOI] [PubMed] [Google Scholar]
- 82.Garrido-Sanz D, Meier-Kolthoff JP, Göker M, Martín M, Rivilla R, Redondo-Nieto M. Genomic and genetic diversity within the Pseudomonas fluoresces complex. PLoS One 2016;11. 10.1371/journal.pone.0150183. [DOI] [PMC free article] [PubMed]
- 83.Gamalero E., Bona E., Todeschini V., Lingua G. Saline and arid soils: Impact on bacteria, plants, and their interaction. Biology (Basel) 2020;9:1–27. doi: 10.3390/biology9060116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Backer R., Rokem J.S., Ilangumaran G., Lamont J., Praslickova D., Ricci E., et al. Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front Plant Sci. 2018;871:1–17. doi: 10.3389/fpls.2018.01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun X., Zhou Y., Tan Y., Wu Z., Lu P., Zhang G., et al. Restoration with pioneer plants changes soil properties and remodels the diversity and structure of bacterial communities in rhizosphere and bulk soil of copper mine tailings in Jiangxi Province. China. Environ Sci Pollut Res. 2018;25:22106–22119. doi: 10.1007/s11356-018-2244-3. [DOI] [PubMed] [Google Scholar]
- 86.Reinhold-Hurek B., Bünger W., Burbano C.S., Sabale M., Hurek T. Roots Shaping Their Microbiome: Global Hotspots for Microbial Activity. Annu Rev Phytopathol. 2015;53:403–424. doi: 10.1146/annurev-phyto-082712-102342. [DOI] [PubMed] [Google Scholar]
- 87.Edwards J., Johnson C., Santos-Medellín C., Lurie E., Podishetty N.K., Bhatnagar S., et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A. 2015;112:E911–E920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bulgarelli D., Schlaeppi K., Spaepen S., Van Themaat E.V.L., Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 89.Bulgarelli D., Rott M., Schlaeppi K., Loren Ver, van Themaat E., Ahmadinejad N., et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 90.Barajas H.R., Martínez-Sánchez S., Romero M.F., Álvarez C.H., Servín-González L., Peimbert M., et al. Testing the Two-Step Model of Plant Root Microbiome Acquisition Under Multiple Plant Species and Soil Sources. Front Microbiol. 2020;11:1–17. doi: 10.3389/fmicb.2020.542742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dini-Andreote F. Endophytes: The Second Layer of Plant Defense. Trends Plant Sci. 2020;25:319–322. doi: 10.1016/j.tplants.2020.01.007. [DOI] [PubMed] [Google Scholar]
- 92.Brader G., Compant S., Mitter B., Trognitz F., Sessitsch A. Metabolic potential of endophytic bacteria. Curr Opin Biotechnol. 2014;27:30–37. doi: 10.1016/j.copbio.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu H., Carvalhais L.C., Crawford M., Singh E., Dennis P.G., Pieterse C.M.J., et al. Inner plant values: Diversity, colonization and benefits from endophytic bacteria. Front Microbiol. 2017;8:1–17. doi: 10.3389/fmicb.2017.02552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ali S., Duan J., Charles T.C., Glick B.R. A bioinformatics approach to the determination of genes involved in endophytic behavior in Burkholderia spp. J Theor Biol. 2014;343:193–198. doi: 10.1016/j.jtbi.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 95.Meneses C.H.S.G., Rouws L.F.M., Simões-Araújo J.L., Vidal M.S., Baldani J.I. Exopolysaccharide production is required for biofilm formation and plant colonization by the nitrogen-fixing endophyte Gluconacetobacter diazotrophicus. Mol Plant-Microbe Interact. 2011;24:1448–1458. doi: 10.1094/MPMI-05-11-0127. [DOI] [PubMed] [Google Scholar]
- 96.Alquéres S., Meneses C., Rouws L., Rothballer M., Baldani I., Schmid M., et al. The bacterial superoxide dismutase and glutathione reductase are crucial for endophytic colonization of rice roots by Gluconacetobacter diazotrophicus PAL5. Mol Plant-Microbe Interact. 2013;26:937–945. doi: 10.1094/MPMI-12-12-0286-R. [DOI] [PubMed] [Google Scholar]
- 97.Böhm M., Hurek T., Reinhold-Hurek B. Twitching motility is essential for endophytic rice colonization by the N2-fixing endophyte Azoarcus sp. strain BH72. Mol Plant-Microbe Interact. 2007;20:526–533. doi: 10.1094/MPMI-20-5-0526. [DOI] [PubMed] [Google Scholar]
- 98.Buschart A., Sachs S., Chen X., Herglotz J., Krause A., Reinhold-Hurek B. Flagella mediate endophytic competence rather than act as MAMPs in rice-Azoarcus sp. strain BH72 interactions. Mol Plant-Microbe Interact. 2012;25:191–199. doi: 10.1094/MPMI-05-11-0138. [DOI] [PubMed] [Google Scholar]
- 99.Macho A.P., Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol Cell. 2014;54:263–272. doi: 10.1016/j.molcel.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 100.Shidore T., Dinse T., Öhrlein J., Becker A., Reinhold-Hurek B. Transcriptomic analysis of responses to exudates reveal genes required for rhizosphere competence of the endophyte Azoarcus sp. strain BH72. Environ Microbiol. 2012;14:2775–2787. doi: 10.1111/j.1462-2920.2012.02777.x. [DOI] [PubMed] [Google Scholar]
- 101.Fernández-Llamosas H., Díaz E., Carmona M. Motility, adhesion and c-di-gmp influence the endophytic colonization of rice by azoarcus sp. cib. Microorganisms. 2021;9:1–17. doi: 10.3390/microorganisms9030554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu S., Shan L., He P. Microbial signature-triggered plant defense responses and early signaling mechanisms. Plant Sci. 2014;228:118–126. doi: 10.1016/j.plantsci.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tsuda K., Katagiri F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol. 2010;13:459–465. doi: 10.1016/j.pbi.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 104.Chen X., Marszałkowska M., Reinhold-Hurek B. Jasmonic Acid, Not Salicyclic Acid Restricts Endophytic Root Colonization of Rice. Front Plant Sci. 2020;10:1–15. doi: 10.3389/fpls.2019.01758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kloepper J.W., Ryu C.M., Zhang S. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology. 2004;94:1259–1266. doi: 10.1094/PHYTO.2004.94.11.1259. [DOI] [PubMed] [Google Scholar]
- 106.Jung H.W., Tschaplinski T.J., Wang L., Glazebrook J., Greenberg J.T. Priming in systemic plant immunity. Science (80-) 2009;324:89–91. doi: 10.1126/science.1170025. [DOI] [PubMed] [Google Scholar]
- 107.Ueda H., Kikuta Y., Matsuda K. Plant communication. Plant Signal Behav. 2012;7:222–226. doi: 10.4161/psb.18765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Naseem H., Ahsan M., Shahid M.A., Khan N. Exopolysaccharides producing rhizobacteria and their role in plant growth and drought tolerance. J Basic Microbiol. 2018;58:1009–1022. doi: 10.1002/jobm.201800309. [DOI] [PubMed] [Google Scholar]
- 109.Syed Ab Rahman S.F., Singh E., Pieterse C.M.J., Schenk P.M. Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 2018;267:102–111. doi: 10.1016/j.plantsci.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 110.Khan N., Bano A., Ali S., Babar M.A. Crosstalk amongst phytohormones from planta and PGPR under biotic and abiotic stresses. Plant Growth Regul. 2020;90:189–203. doi: 10.1007/s10725-020-00571-x. [DOI] [Google Scholar]
- 111.Rivas-Franco F., Hampton J.G., Narciso J., Rostás M., Wessman P., Saville D.J., et al. Effects of a maize root pest and fungal pathogen on entomopathogenic fungal rhizosphere colonization, endophytism and induction of plant hormones. Biol Control. 2020;150 doi: 10.1016/j.biocontrol.2020.104347. [DOI] [Google Scholar]
- 112.Sheoran N., Kumar A., Munjal V., Nadakkakath A.V., Eapen S.J. Pseudomonas putida BP25 alters root phenotype and triggers salicylic acid signaling as a feedback loop in regulating endophytic colonization in Arabidopsis thaliana. Physiol Mol Plant Pathol. 2016;93:99–111. doi: 10.1016/j.pmpp.2016.01.008. [DOI] [Google Scholar]
- 113.Sheoran N., Valiya Nadakkakath A., Munjal V., Kundu A., Subaharan K., Venugopal V., et al. Genetic analysis of plant endophytic Pseudomonas putida BP25 and chemo-profiling of its antimicrobial volatile organic compounds. Microbiol Res. 2015;173:66–78. doi: 10.1016/j.micres.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 114.Lebeis S.L., Paredes S.H., Lundberg D.S., Breakfield N., Gehring J., McDonald M., et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science (80-) 2015;349:860–864. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- 115.Gamalero E., Glick B.R. Bacterial modulation of plant ethylene levels. Plant Physiol. 2015;169:13–22. doi: 10.1104/pp.15.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Glick B.R. Bacterial ACC deaminase and the alleviation of plant stress. Adv Appl Microbiol. 2004;56:291–312. doi: 10.1016/S0065-2164(04)56009-4. [DOI] [PubMed] [Google Scholar]
- 117.Orozco-Mosqueda M del C, Glick BR, Santoyo G. ACC deaminase in plant growth-promoting bacteria (PGPB): An efficient mechanism to counter salt stress in crops. Microbiol Res 2020;235. 10.1016/j.micres.2020.126439. [DOI] [PubMed]
- 118.Gamalero E., Lingua G., Berta G., Glick B.R. Beneficial role of plant growth promoting bacteria and arbuscular mycorrhizal fungi on plant responses to heavy metal stress. Can J Microbiol. 2009;55:501–514. doi: 10.1139/W09-010. [DOI] [PubMed] [Google Scholar]
- 119.Glick B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res. 2014;169:30–39. doi: 10.1016/j.micres.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 120.Ryffel F., Helfrich E.J.N., Kiefer P., Peyriga L., Portais J.C., Piel J., et al. Metabolic footprint of epiphytic bacteria on Arabidopsis thaliana leaves. ISME J. 2016;10:632–643. doi: 10.1038/ismej.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chaudhry V., Runge P., Sengupta P., Doehlemann G., Parker J.E., Kemen E. Shaping the leaf microbiota: Plant-microbe-microbe interactions. J Exp Bot. 2021;72:36–56. doi: 10.1093/jxb/eraa417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hernandez M.N., Lindow S.E. Pseudomonas syringae increases water availability in leaf microenvironments via production of hygroscopic syringafactin. Appl Environ Microbiol. 2019;85 doi: 10.1128/AEM.01014-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vorholt J.A. Microbial life in the phyllosphere. Nat Rev Microbiol. 2012;10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 124.Jacobs J.L., Carroll T.L., Sundin G.W. The role of pigmentation, ultraviolet radiation tolerance, and leaf colonization strategies in the epiphytic survival of phyllosphere bacteria. Microb Ecol. 2005;49:104–113. doi: 10.1007/s00248-003-1061-4. [DOI] [PubMed] [Google Scholar]
- 125.Leveau J.H.J., Lindow S.E. Appetite of an epiphyte: Quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc Natl Acad Sci U S A. 2001;98:3446–3453. doi: 10.1073/pnas.061629598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lindow S.E., Brandl M.T. Microbiology of the phyllosphere. Appl Environ Microbiol. 2003;69:1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Prieto P., Schilirò E., Maldonado-González M.M., Valderrama R., Barroso-Albarracín J.B., Mercado-Blanco J. Root Hairs Play a Key Role in the Endophytic Colonization of Olive Roots by Pseudomonas spp. with Biocontrol Activity. Microb Ecol. 2011;62:435–445. doi: 10.1007/s00248-011-9827-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oukala N, Aissat K, Pastor V. Bacterial endophytes : The hidden actor in plant immune re- sponses against biotic stress 2021:1–24. [DOI] [PMC free article] [PubMed]
- 129.Sivakumar N, Sathishkumar R, Selvakumar G, Shyamkumar R, Arjunekumar K. Phyllospheric Microbiomes: Diversity, Ecological Significance, and Biotechnological Applications. Springer International Publishing; 2020. 10.1007/978-3-030-38453-1_5.
- 130.Povolotsky T.L., Keren-Paz A., Kolodkin-Gal I. Metabolic Microenvironments Drive Microbial Differentiation and Antibiotic Resistance. Trends Genet. 2021;37:4–8. doi: 10.1016/j.tig.2020.10.007. [DOI] [PubMed] [Google Scholar]
- 131.Henry E., Yadeta K.A., Coaker G. Recognition of bacterial plant pathogens: Local, systemic and transgenerational immunity. New Phytol. 2013;199:908–915. doi: 10.1111/nph.12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Maignien L, DeForce EA, Chafee ME, Murat Eren A, Simmons SL. Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. MBio 2014;5. 10.1128/mBio.00682-13. [DOI] [PMC free article] [PubMed]
- 133.Wagner M.R., Lundberg D.S., Del Rio T.G., Tringe S.G., Dangl J.L., Mitchell-Olds T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat Commun. 2016;7 doi: 10.1038/ncomms12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Beilsmith K., Thoen M.P.M., Brachi B., Gloss A.D., Khan M.H., Bergelson J. Genome-wide association studies on the phyllosphere microbiome: Embracing complexity in host–microbe interactions. Plant J. 2019;97:164–181. doi: 10.1111/tpj.14170. [DOI] [PubMed] [Google Scholar]
- 135.Elgetti Brodersen K., Kühl M., Trampe E., Koren K. Imaging O2 dynamics and microenvironments in the seagrass leaf phyllosphere with magnetic optical sensor nanoparticles. Plant J. 2020;104:1504–1519. doi: 10.1111/tpj.15017. [DOI] [PubMed] [Google Scholar]
- 136.Ren G., Zhu C., Alam M.S., Tokida T., Sakai H., Nakamura H., et al. Response of soil, leaf endosphere and phyllosphere bacterial communities to elevated CO2 and soil temperature in a rice paddy. Plant Soil. 2015;392:27–44. doi: 10.1007/s11104-015-2503-8. [DOI] [Google Scholar]
- 137.Santoyo G., Gamalero E., Glick B.R. Mycorrhizal-Bacterial Amelioration of Plant Abiotic and Biotic Stress. Front Sustain Food Syst. 2021;5 doi: 10.3389/fsufs.2021.672881. [DOI] [Google Scholar]
- 138.Kumar A., Verma J.P. Does plant—Microbe interaction confer stress tolerance in plants: A review? Microbiol Res. 2018;207:41–52. doi: 10.1016/j.micres.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 139.Alori E.T., Glick B.R., Babalola O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front Microbiol. 2017;8:1–8. doi: 10.3389/fmicb.2017.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang Y., Gao X., Shen Z., Zhu C., Jiao Z., Li R., et al. Pre-colonization of PGPR triggers rhizosphere microbiota succession associated with crop yield enhancement. Plant Soil. 2019;439:553–567. doi: 10.1007/s11104-019-04055-4. [DOI] [Google Scholar]
- 141.Chihaoui S.A., Trabelsi D., Jdey A., Mhadhbi H., Mhamdi R. Inoculation of Phaseolus vulgaris with the nodule-endophyte Agrobacterium sp. 10C2 affects richness and structure of rhizosphere bacterial communities and enhances nodulation and growth. Arch Microbiol. 2015;197:805–813. doi: 10.1007/s00203-015-1118-z. [DOI] [PubMed] [Google Scholar]
- 142.Carrión V.J., Perez-Jaramillo J., Cordovez V., Tracanna V., De Hollander M., Ruiz-Buck D., et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science (80-) 2019;366:606–612. doi: 10.1126/science.aaw9285. [DOI] [PubMed] [Google Scholar]
- 143.Solís-García I.A., Ceballos-Luna O., Cortazar-Murillo E.M., Desgarennes D., Garay-Serrano E., Patiño-Conde V., et al. Phytophthora Root Rot Modifies the Composition of the Avocado Rhizosphere Microbiome and Increases the Abundance of Opportunistic Fungal Pathogens. Front Microbiol. 2021;11:1–15. doi: 10.3389/fmicb.2020.574110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Van Elsas J.D., Chiurazzi M., Mallon C.A., Elhottova D., Krištůfek V., Salles J.F. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc Natl Acad Sci U S A. 2012;109:1159–1164. doi: 10.1073/pnas.1109326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Baas P., Bell C., Mancini L.M., Lee M.N., Conant R.T., Wallenstein M.D. Phosphorus mobilizing consortium Mammoth PTM enhances plant growth. PeerJ. 2016;2016:1–16. doi: 10.7717/peerj.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rojas-Solis D., Hernandez-Pacheco C.E., Santoyo G. Evaluation of Bacillus and Pseudomonas to colonize the rhizosphere and their effect on growth promotion in tomato (Physalis ixocarpa Brot. ex Horm.) Rev Chapingo, Ser Hortic. 2016;22:45–57. doi: 10.5154/r.rchsh.2015.06.009. [DOI] [Google Scholar]
- 147.De Corato U. Disease-suppressive compost enhances natural soil suppressiveness against soil-borne plant pathogens: A critical review. Rhizosphere. 2020;13 doi: 10.1016/j.rhisph.2020.100192. [DOI] [Google Scholar]
- 148.Rodriguez P.A., Rothballer M., Chowdhury S.P., Nussbaumer T., Gutjahr C., Falter-Braun P. Systems Biology of Plant-Microbiome Interactions. Mol Plant. 2019;12:804–821. doi: 10.1016/j.molp.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 149.del Carmen Orozco-Mosqueda M, Velázquez-Becerra C., Macías-Rodríguez L.I., Santoyo G., Flores-Cortez I., Alfaro-Cuevas R., et al. Arthrobacter agilis UMCV2 induces iron acquisition in Medicago truncatula (strategy I plant) in vitro via dimethylhexadecylamine emission. Plant Soil. 2013;362:51–66. doi: 10.1007/s11104-012-1263-y. [DOI] [Google Scholar]
- 150.Velázquez-Becerra C., Macías-Rodríguez L.I., López-Bucio J., Flores-Cortez I., Santoyo G., Hernández-Soberano C., et al. The rhizobacterium Arthrobacter agilis produces dimethylhexadecylamine, a compound that inhibits growth of phytopathogenic fungi in vitro. Protoplasma. 2013;250 doi: 10.1007/s00709-013-0506-y. [DOI] [PubMed] [Google Scholar]
- 151.Martínez-Cámara R., Montejano-Ramírez V., Moreno-Hagelsieb G., Santoyo G., Valencia-Cantero E. The volatile organic compound dimethylhexadecylamine affects bacterial growth and swarming motility of bacteria. Folia Microbiol (Praha) 2020 doi: 10.1007/s12223-019-00756-6. [DOI] [PubMed] [Google Scholar]
- 152.Abdalla AK, Ayyash MM, Olaimat AN, Osaili TM, Al-Nabulsi AA, Shah NP, et al. Exopolysaccharides as Antimicrobial Agents: Mechanism and Spectrum of Activity. Front Microbiol 2021;12. 10.3389/fmicb.2021.664395. [DOI] [PMC free article] [PubMed]
- 153.Compant S., Duffy B., Nowak J., Clément C., Barka E.A. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Appl Environ Microbiol. 2005;71:4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Carmona-Hernandez S., Reyes-Pérez J.J., Chiquito-Contreras R.G., Rincon-Enriquez G., Cerdan-Cabrera C.R., Hernandez-Montiel L.G. Biocontrol of postharvest fruit fungal diseases by bacterial antagonists: A review. Agronomy. 2019;9 doi: 10.3390/agronomy9030121. [DOI] [Google Scholar]
- 155.Wang Z., Mei X., Du M., Chen K., Jiang M., Wang K., et al. Potential modes of action of Pseudomonas fluorescens ZX during biocontrol of blue mold decay on postharvest citrus. J Sci Food Agric. 2020;100:744–754. doi: 10.1002/jsfa.10079. [DOI] [PubMed] [Google Scholar]
- 156.Hernández-León R., Rojas-Solís D., Contreras-Pérez M., Orozco-Mosqueda M.D.C., Macías-Rodríguez L.I., Reyes-de la Cruz H., et al. Characterization of the antifungal and plant growth-promoting effects of diffusible and volatile organic compounds produced by Pseudomonas fluorescens strains. Biol Control. 2015;81 doi: 10.1016/j.biocontrol.2014.11.011. [DOI] [Google Scholar]
- 157.Fira D., Dimkić I., Berić T., Lozo J., Stanković S. Biological control of plant pathogens by Bacillus species. J Biotechnol. 2018;285:44–55. doi: 10.1016/j.jbiotec.2018.07.044. [DOI] [PubMed] [Google Scholar]
- 158.Lahlali R., Peng G., McGregor L., Gossen B.D., Hwang S.F., McDonald M. Mechanisms of the biofungicide Serenade (Bacillus subtilis QST713) in suppressing clubroot. Biocontrol Sci Technol. 2011;21:1351–1362. doi: 10.1080/09583157.2011.618263. [DOI] [Google Scholar]
- 159.Abbey J.A., Percival D., Abbey Lord, Asiedu S.K., Prithiviraj B., Schilder A. Biofungicides as alternative to synthetic fungicide control of grey mould (Botrytis cinerea)–prospects and challenges. Biocontrol. Sci Technol. 2019;29:241–262. doi: 10.1080/09583157.2018.1548574. [DOI] [Google Scholar]
- 160.Bae H., Roberts D.P., Lim H.S., Strem M.D., Park S.C., Ryu C.M., et al. Endophytic Trichoderma isolates from tropical environments delay disease onset and induce resistance against Phytophthora capsici in hot pepper using multiple mechanisms. Mol Plant-Microbe Interact. 2011;24:336–351. doi: 10.1094/MPMI-09-10-0221. [DOI] [PubMed] [Google Scholar]
- 161.Park Y.S., Ryu C.M. Understanding plant social networking system: Avoiding deleterious microbiota but calling beneficials. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22073319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Santoyo G., Hernández-Pacheco C., Hernández-Salmerón J., Hernández-León R. The role of abiotic factors modulating the plant-microbe-soil interactions: Toward sustainable agriculture. A review. Spanish. J Agric Res. 2017;15 doi: 10.5424/sjar/2017151-9990. [DOI] [Google Scholar]
- 163.Smith S.A., Tank D.C., Boulanger L.A., Bascom-Slack C.A., Eisenman K., Kingery D., et al. Bioactive endophytes warrant intensified exploration and conservation. PLoS ONE. 2008;3:1–4. doi: 10.1371/journal.pone.0003052. [DOI] [PMC free article] [PubMed] [Google Scholar]