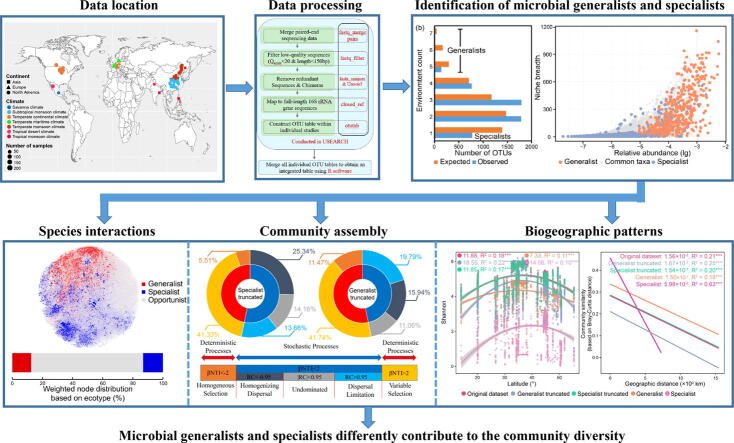
Graphical abstract
Keywords: Biogeographical pattern, Community assembly, Community diversity, Niche breadth, Species interaction
Highlights
-
•
Microbial generalists and specialists were investigated at the global scale.
-
•
Generalists and specialists simultaneously contributed to species interactions.
-
•
Generalists had higher diversification and transition rates than specialists.
-
•
Generalists were more characterized by stochastic processes than specialists.
-
•
Generalists dampened microbial biogeographical patterns.
Abstract
Introduction
Microbial generalists and specialists are thought to have distinct impacts on community dynamics, while there have been limited efforts to estimate their contribution to microbial diversity.
Objectives
We aimed to resolve this research gap in microbial ecology to strengthen our understanding of the biogeography of microbial diversity, with implications for global-scale biodiversity mapping.
Methods
Herein, we identified the ecological characteristics of microbial generalists and specialists across over 3,000 farmland soil samples from eleven countries that encompassed seven climate types.
Results
Considering the distinct distributions of generalists and specialists in degree of connexions, betweenness and as key species in network topology, both generalists and specialists contributed to species interactions, though through different modalities. A stronger signature of deterministic processes in specialists indicated their lower tolerance to environment fluctuations. Generalists, in contrast, were more characterized by stochastic processes with higher diversification and transition rates that suggested more important roles in maintaining community stability when exposed to environmental disturbances. The relationship between latitude and diversity combining with distance-decay effects showed that generalists dampened microbial biogeographical patterns, with contrasting impacts by specialists.
Conclusion
By demonstrating the ecological characteristics of microbial generalists and specialists, this study deepens our understanding of microbial diversity and highlights the need to impart systematic distinctions among different categories of species when modelling and predicting the fate of ecosystems in the face of global climate change, rather than assuming that species are functionally equivalent.
Introduction
Microorganisms are ubiquitous and perform crucial functions in various ecosystems, such as the Earth’s biogeochemical cycles [1], human metabolism [2] and biotechnological processes [3]. Broad interest has arisen in terms of understanding and modeling how microbes influence ecosystem functions [4]. It is thought that microbial communities that exhibit higher diversity are more functionally stable. The broad assumption in these studies, which use microbial diversity as a predictor of ecosystem service and function stability, is that distant taxa are functionally equivalent [5]. However, there is mounting evidence that bias may be introduced by using this assumption [6]. Microbial generalists and specialists impart different impacts on microbial community dynamics. Generalists exhibit broad environmental tolerances, while specialists have narrower range of habitats and specific environment fitness [7]. Therefore, there is a need to parse these broad microbial groups to estimate their contribution to diversity (e.g., structure and population size), and further to better understand and predict how ecosystem functions may respond to changes in global climate.
Rare taxa have been found to greatly contribute to community stability and play a more important role in species interactions as compared to abundant taxa. This was previously indicated by the assignment of the majority of microbial network hubs to rare taxa [8]. This is counter-intuitive, considering that abundant taxa with more individuals have a higher probability of interaction with others and appear to impart stronger impacts on population dynamics. The contribution of generalists/specialists may be analogous with that of abundant/rare microbes in terms of species interactions. Herein, we hypothesized that (i) although existing in limited habitats, specialists may play a broader role in species interactions. This would be measurable in co-occurrence network topologies by higher degree of connexions (i.e., more links with other species in the network) and a larger number of keystone species than that exhibited by generalists. This is based on the idea that specialists are more dependent on species interactions (e.g., auxotrophy) [9]. Specialists are also expected to have stricter growth conditions that may include specific metabolic requirements while generalists are less affected or better buffered to environmental filtering [10]. Thus, we also hypothesized that (ii) in the microbial assembly, the specialists are expected to contribute more to deterministic processes (e.g., environmental filtering), while the generalists are expected to mainly contribute to stochastic processes. In addition to these expected distinct species interactions and contrasting impacts on microbial community assembly, we also aimed to decipher the spatial biodiversity patterns impacted by generalists and specialists. Within the biogeography context, the distance–decay relationship allows to assess the community similarity changes with increasing geographical distance [11] and indicates the impacts of both dispersal limitation and spatial autocorrelation [12]. Due to a limited impact of dispersal limitation and environmental filtering, generalists are expected to minimize species turnover, the β-diversity, which reflects the loss or replacement of species across space or time. Hence, we further hypothesized that (iii) the presence of microbial generalists will result in a flatter slope of the distance–decay relationship while microbial specialists are expected to increase the slope.
To test these hypotheses, we used high-resolution community profiling to determine the spatial distribution of bacterial communities across farmland soil ecosystems. While there are various concepts and definitions to describe ecological specialization [13], [14], herein, species were defined as ‘habitat generalists’ and ‘habitat specialists’ based on their spatial distribution [10], [15], rather than by functional traits. >2,000 samples from eleven countries across seven climate types were collected to determine the characteristics (e.g., roles in species interactions, community assemblies and biogeographical patterns) of microbial generalists and specialists in community dynamics. This allowed us to better understand the ecological roles of microbial generalists and specialists, laying a foundation for imparting systematic distinctions among different categories of species when modelling and predicting the fate of ecosystems.
Materials and methods
Data collection and species table generation
Bacterial sequencing datasets related to farmland soils were collected from 66 studies, published within 2015.01 to 2018.12, including a total of 3,086 samples (Supplementary Data) by searching terms “microbe” and “farmland soil” in the Web of Science database. The locations of farmlands in each study were also collected for analysis related to biogeographic patterns. Publicly accessible bacterial sequencing data, primarily from the NCBI SRA database, for the studies were downloaded. Nineteen primer pairs (8F:556R; 8F:533R; 8F:343R; 9F:530R; 28F:219R; 27F:533R; 784F:1046R; 577F:926R; 519F:926R; 515F:907R; 515F:806R; 502F:802R; 479F:888R; 341F:907R; 341F:805R; 341F:785R; 341F:534R; 338F:806R; 1106F:1378R) were identified from the datasets to perform sequencing based on Roche 454 (30.8%) or Illumina technologies (69.2%). All the data were obtained from published articles in public database (NCBI) using Sratoolkit tool. We listed all the data sources and detailed environment information in the metadata.xlsx file on Github (https://github.com/Qicheng-Xu/Metadata-for-Generalists-and-Specialists-in-Global-Farmland-Soils).
Raw paired-end (forward and reverse) sequences in each study were merged with the “fastq_mergepairs” function and low-quality sequences (length < 150 or quality score < 20) were filtered with “fastq_filter” function in USEARCH software [16]. The “fastx_uniques” and “unosie3” functions were then used to perform the dereplication and denoising (error-correction) of sequences. Singletons were discarded, as they may result from erroneous sequencing or prediction.
As the datasets include sequences targeted at different regions of the 16S rRNA gene, it’s impossible to perform the analysis at the single nucleotide difference level, such as zOTU nor ASV level, and the sequences after quality control cannot be compared directly. Hence, a closed-reference workflow was employed to map these fragments to full-length 16S rRNA gene sequences. The “closed_ref” function was used to perform the mapping processes at a 97% identify threshold with RDP database (http://www.drive5.com/usearch/manual/sintax_downloads.html), whereas sequences that could not be mapped were assigned to unknown parts. This RDP training set containing high quality 16S rRNA gene sequences is recommended for USEARCH by the recent study [17]. When a fragment could be matched to multiple full-length 16S rRNA sequences, the Occum’s Razor principle was followed and the fragment was assigned to the candidate full-length sequence that contained the most fragment hits. The corresponding full-length 16S rRNA gene sequence and its annotation were taken as representative sequence and taxonomic classification for further study. Mapped results of each study were merged into an integrated table with the “merge” function in R software. This workflow (Fig. S1) shared similar idea with a previous study (18) and can solve the problem that sequences with different primers cannot be compared directly.
Samples with > 90% unassigned sequences or < 1000 sequencing depth were discarded. The remaining 2,079 samples from eleven countries were further classified into seven climate types according to their locations, namely temperate maritime climate, temperate continental climate, subtropical monsoon climate, temperate monsoon climate, tropical monsoon climate, savanna climate and tropical desert climate (Fig. S2).
The identification of microbial generalists and specialists
The generalists and specialists were classified according to a recent study [19]. The ANOSIM showed that the climate (global R = 0.21) was the best factor to divide the samples, compared with country (global R = 0.13) and continent (global R = 0.14) (Table S1). Similar results were obtained when only considering the absence/presence of species. Hence, each climate was considered as a unique environment to explore the distribution of species. The species table was randomly shuffled 10,000 times by preserving the richness in each sample to obtain a random background distribution. An enrichment of the observed species compared with the random distribution indicated that the species were selected by the environment or had higher ability to adapt to the environment than expected. If the species enrichment occurred in narrow environments, these species were defined as specialists (e.g., enriched in limited environments). Accordingly, generalists were species enriched in wide environments. The niche breadth of species was calculated by the “niche.width” function in the “spaa” package in R.
Construction of co-occurrence networks and detection of keystone species
Microbial co-occurrence networks were constructed based on a Spearman correlation [20]. The P-values were multiple-testing-corrected with the Benjamini-Hochberg FDR controlling procedure [21]\. Direct correlation dependencies were determined using the network enhancement method [22]. Edges with an adjusted P-value < 0.05 and a score above the threshold determined by the random matrix theory (RMT) method [23] were retained. Networks were graphed in Gephi.
The modules, within which nodes were highly connected while with few connections outside [24], were defined with the greedy modularity optimization [25]. To assess the topological roles of taxa in the networks, the nodes were classified into four categories based on the within-module connectivity (Zi) and among-module connectivity (Pi), including module hubs (Zi > 2.5), network hubs (Zi > 2.5 and Pi > 0.62), connectors (Pi > 0.62) and peripherals (Zi < 2.5 and Pi < 0.62) [26].
The structural robustness (i.e., natural connectivity) of each network was calculated to compare the stability of the networks. It is an average eigenvalue derived from network spectrum, which describes the redundancy of alternative paths [27]. A higher robustness indicates a more stable network structure.
Bacterial assembly processes based on the null-model-framework
The estimation of assembly processes based on the null model and the calculation of nearest taxon index (NTI) were based on a subset of total samples. The samples with top 10 mapping ratios (i.e., samples with lower proportion of unknown parts) in each climate type were selected and a total of 70 samples was obtained. The NTI was calculated by the “ses.mntd” function in “picante” package in R.
Significant positive correlations (P < 0.05) across short phylogenetic distances (i.e., phylogenetic signals) were detected in all groups, ensuring the further use of null-model-framework [28]. The null model was conducted as described by recent studies [29], [30]. This framework is based on the phylogenetic turnover, which is the evolutionary distance separating species in one community from species in another community. A null distribution of β-mean nearest taxon distance (βMNTDnull) was obtained by shuffling the species across the tips of the phylogeny. This randomization was repeated 999 times. The β-nearest taxon index (βNTI) indicates the difference between observed βMNTD and the βMNTDnull. |βNTI| > 2 indicates that the observed βMNTD deviated significantly from the βMNTDnull distribution (i.e., the control of deterministic processes). In this case, βNTI > +2 and βNTI < -2 represent heterogeneous selection and homogeneous selection, respectively. When |βNTI| < 2, the community diversity results from a stochastic assembly process. In this case, the Raup–Crick (RCvalue) index was calculated based on the contingency matrix for further assessment in stochastic processes. The contingency matrices were randomly shuffled by maintaining the observed species richness and the number of sequences for each sample. This random shuffle was performed for 999 times to generate a null distribution. RCvalue > 0.95 and RCvalue < − 0.95 indicate significant departures from the null distribution, and represent dispersal limitation and homogenizing dispersal, respectively. |RCvalue| < 0.95 represents the undominated scenario, which indicates that community turnover is dominated by multiple processes consisting of weak selection and dispersal, diversification, and/or drift (see [31] for details).
Estimation of the stochastic ratio in community assembly
The modified stochasticity ratio (MST) was also one of null model based indexes and assumes that deterministic processes drive the community to be more similar or dissimilar than the null expectation. Different from the null-model-based framework mentioned above, MST reflects the contribution of stochastic processes based on relative differences between the observed situation and the null expectation, rather than the significance of the difference, and therefore can better quantitatively measure the stochasticity in assembly [32], [33]. The MST index defines 0.5 as the boundary point to determine whether the community assembly is more deterministic (<0.5) or more stochastic (>0.5). However, the phylogenetic diversity was not considered as the above-mentioned null model approach. The MST was calculated based on both Bray-Curtis and Jaccard distance by using the “NST” package in the R software environment.
Estimated contribution of the neutral processes based on Sloan neutral model
The Sloan neutral model is a neutral-theory-based approach and assumes that species are ecologically functionally equivalent and could be randomly lost and then replaced by other members in the local community and/or supplemented from the metacommunity via dispersal [34], [35]. The relationship between the occurrence frequency of species in local communities and their relative abundance in the metacommunity was examined to estimate the potential contribution of neutral processes in assembly. Herein, the species associated with independent samples were taken to be local communities that are a part of a broader metacommunity consisting of the species associated with all of the samples in datasets. The parameters R2 and m represent the goodness of fit and migration rate, respectively. The species with an occurrence frequency that deviated from the neutral distribution are considered to be selected for or against by environment, or have a distinct dispersal ability. The model fitting was performed in R software as previously described [35].
Evolutionary trends of specialists and generalists
The binary-state speciation and extinction (BiSSE) model was performed to explore the ecological roles of generalists and specialists as described [18]. The model considers generalists and specialists as distinct evolutionary states and calculates evolutionary rate parameters (the speciation and extinction rates), allowing for the estimation of the transformation from one ecological state to another (the transition rate between generalists and specialists). The BiSSE method creates a model in which specialist or generalist is able to become more or less abundant in one of two ways: either by differences in character state transition (i.e., specialist species evolving to become generalists or vice-versa), or through relative diversification (specialist species give rise to more or fewer descendent species than generalist species do). BiSSE then selects a combination of rate parameters that with the highest likelihood to generate the data that it is given. The phylogenetic tree is the most critical input for BiSSE model. The Living Tree Project datasets provide curated entries and the best quality sequences with manually checked alignment. The identified generalists and specialists were mapped to the All-species Living Tree (LTP tree from this well-established Living Tree Project) according to their accession number. A subtree was obtained by retaining the branches with mapped taxa. The subtree was linearized and reconstructed to be an ultrametric tree using the “ape” package in R. BiSSE model was performed with the “diversitree” package in R software [36]. The “starting.point.bisse” function was used to determine the starting point for the simulation by setting identical speciation and extinction rates. Then, a maximum likelihood method was used to estimate the evolutionary rate parameters. A Markov chain Monte Carlo (MCMC) simulation with 10,000 permutations was performed to ensure the stability of the final estimate. MCMC simulations with 1,000 and 5,000 permutations obtained similar results.
Other statistical analyses
The α-diversity (including the Shannon and Chao index) was calculated using “vegan” package. The ecological community thresholds were identified using threshold indicator species analyses (TITAN) in the “TITAN2” package [37]. TITAN divides the community into two groups: Z- taxa negatively respond to the increased gradient, and Z + taxa positively respond to the increased gradient. The taxa with no response to the environment gradient were not considered. Then, TITAN tracks cumulative responses of declining [sum(Z-)] and increasing [sum(Z + )] taxa in the community. The ecological thresholds were defined as points where the maximum aggregate change in the frequency and relative abundance of responding taxa occurs. When the environmental values reach by and exceed the ecological threshold, the abundance and occurrence frequency of species will decrease in group Z-, while increase in group Z+. Hence, the range of niche optima of the community is defined as the gradient below sum(Z-) and above sum(Z + ). The curve fitting (linear and quadratic) was performed with “lm” function in “vegan” package. The heatmap was plotted with “pheatmap” package. The phylogenetic tree was constructed using Neighbor-Joining method in R based on default parameters with the multiple alignment matrix produced in Muscle, and plotted with iTOL (http://itol.embl.de/).
Results
The composition of generalist and specialist groups
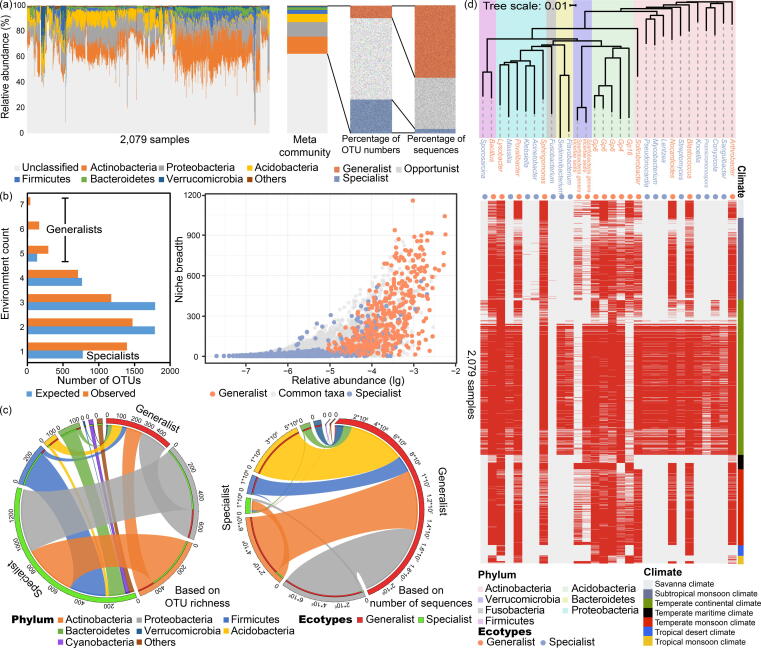
The merged datasets provided a contingency table matching to 5,266 annotated bacterial species from 2,079 samples of satisfying sequencing depth. These samples were from eleven countries and across seven climate types (Fig. S2). The bacterial communities in farmland soils were dominated by Actinobacteria (13%), Proteobacteria (11%), Acidobacteria (6%), Firmicutes (3%), Bacteroidetes (2%) and Verrucomicrobia (1%) (Fig. 1a).
Fig. 1.
The composition of microbial communities in farmland soils and the definition of generalist and specialist groups. (a) The distribution of communities at phylum level. The assigned species were further classified into generalist, specialist and opportunist groups. The richness and relative abundance composition of different groups were provided. (b) Classification of generalists and specialists. By comparing the expected and observed distribution, species observed in one environment and more than five environments were defined as specialists and generalists, respectively. The remaining were opportunists. The niche breadth, which indicates the occurrence frequency of species, and the relative abundance of the specialists and generalists are shown. Relative abundance is shown on a logarithmic (lg) scale on the x axis. (c) Taxonomic distribution of generalists and specialists at the phylum level, based on richness (left) and relative abundance (right). The thickness of each ribbon (i.e., the links in the middle area) in the circos-plot represents the richness or relative abundance of generalist and specialist groups assigned to different phyla. (d) Phylogenetic tree of the most abundant generalists and specialists (i.e., taxa with top 15 relative abundance in generalist and specialist groups) with their occurrence in different samples. Red and grey represents the existence and absence of species, respectively. The sample climate type, taxonomic information and species ecotypes are indicated by different colours.
By comparing the random and observed species distribution patterns (see methods for details), species existing in only one climate type were defined as specialists, species existing in more than five climate types were defined as generalists, and the remaining were considered as opportunists (Fig. 1b). In total, 1,396 species (∼27%) were classified as specialists and 495 species (∼9%) were classified as generalists (Fig. 1a). Specialists and generalists accounted for ∼ 4% and 56% of the total relative abundance, respectively. While the richness of the generalists was approximately one-third that of specialists, the generalists were ∼ 16 times higher in relative abundance. In general, generalists were distributed across a wider range of samples and exhibited a higher relative abundance than the specialists. However, some specialists were found to have a high relative abundance and were identified in a wide range of samples (across one climate type), while some generalists were only found in a limited number of samples (from various climate types). Hence, the frequency of occurrence across samples and relative abundances do not necessarily dictate the ecotype. As such, the environmental heterogeneity of the species should be considered. The proportions of generalists and specialists varied across different climate types (Table S2).
Whether considering species richness or relative abundance, both the generalist and specialist groups were principally within the Actinobacteria, Proteobacteria, Acidobacteria, Firmicutes, Bacteroidetes, Verrucomicrobia and Cyanobacteria (Fig. 1c). More generalists were identified within the Proteobacteria, Acidobacteria and Verrucomicrobia, while more specialists were related to the Actinobacteria, Firmicutes, Bacteroidetes and Cyanobacteria. Dominant generalists (species within the top 15, based on relative abundance) were classified as Arthrobacter, Blastococcus, Nocardioides, Solirubrobacter, Gp16, Gp4, Gp6, Spartobacteria genera incertae sedis, Sphingomonas, Povalibacter, Lysobacter and Bacillus (Fig. 1d). Dominant specialists were Sanguibacter, Conyzicola, Promicromonospora, Knoellia, Streptomyces, Lentzea, Mycobacterium, Pseudonocardia, Flavobacterium, Sediminibacterium, Fusobacterium, Acinetobacter, Klebsiella, Massilia and Sporosarcina (Fig. 1d). Although there may be preferences within the generalist or specialist groups to a specific phylum, it was not possible to determine ecotypes at this coarse phylogenetic level due to the high similarity of dominant phylum. However, at the genus level, there was no overlap in the dominant generalists and specialists.
The contribution of generalist and specialist groups to the co-occurrence networks
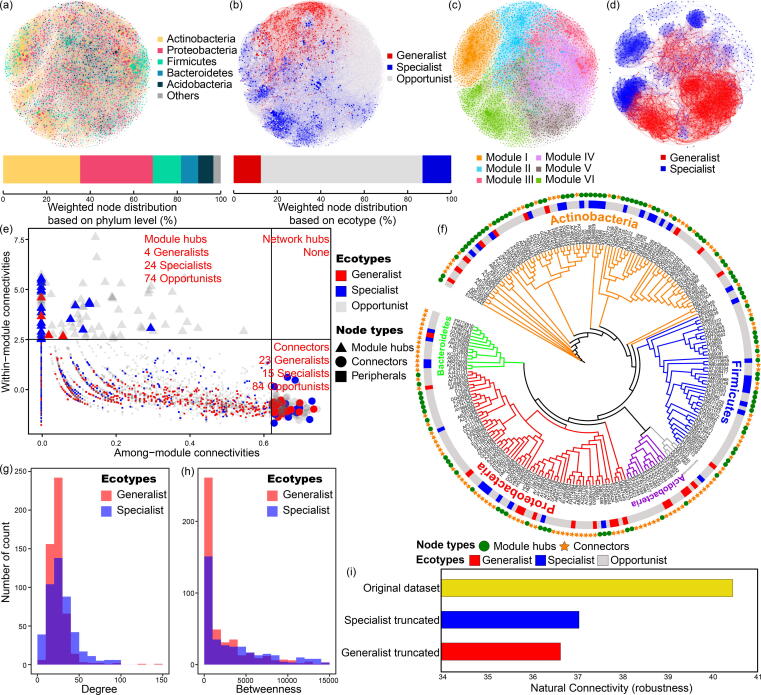
The co-occurrence network consisted of 3,619 nodes and 51,594 edges with the average degree of connexions (i.e., average links per node) at 28.51. Actinobacteria (35%), Proteobacteria (33%), Firmicutes (13%), Bacteroidetes (8%) and Acidobacteria (7%) harboured the majority (>96% totally) of links in the network (Fig. 2a). Compared to the mixed distribution of phyla across the network topology, there was greater separation between generalists and specialists (Fig. 2b). With the exception of module I, in which 27% generalist nodes and 17% specialist nodes were detected, generalists and specialists were more biased to exist in different modules (Fig. 2b and 2c). Modules II and III were comprised of 49% and 11% generalists, respectively. In contrast, modules IV, V, and VI contained 10%, 16% and 49% specialists, respectively. When constructing a new network only containing generalists and specialists, this phenomenon of topological separation was confirmed (Fig. 2d). In addition, generalists and specialists simultaneously contributed to the overall network topology with similar contributions (13%, Fig. 2b).
Fig. 2.
The co-occurrence networks, network properties and keystone species of generalist and specialist groups. The networks are based on entire communities with different colors indicating (a) phyla distribution, (b) ecotype distribution and (c) module distribution of species. (d) The network constructed with the identified generalist and specialist species (i.e., opportunists were not included). (e) Classification of keystone species. The nodes were classified into four categories including module hubs (Zi > 2.5), network hubs (Zi > 2.5 and Pi > 0.62), connectors (Pi > 0.62) and peripherals (Zi < 2.5 and Pi < 0.62). Shapes and colours indicate the node types and ecotypes, respectively. (f) The phylogenetic tree of keystone species. Branch colours indicate the phyla distribution. Shapes and colours outside the tree indicate the node types and ecotypes, respectively. (g) The distribution of degree of connexions and (h) betweenness in generalist and specialist groups. The ecotypes are represented by colours. (i) The robustness of network before or after the removal of generalists/specialists to measure the importance of the contribution of generalists/specialists to the network structure. The original dataset represented the entire community, the specialist truncated represented the dataset after removing specialists, and the generalist truncated represented the dataset after removing generalists.
Specialists were more likely module hubs, with a six times larger number over generalists. In contrast, generalists were more likely to be connectors (23 vs. 15). There were no network hubs detected in this network (Fig. 2e). These keystone species (module hubs and connectors) principally belonged to the Actinobacteria, Proteobacteria and Firmicutes (Fig. 2f).
Generalists exhibited higher distribution at medium (20–30) and high (130–150) degree of connexions than the specialists (Fig. 2g). Generalists were also in higher distribution in low (0–4,000) and medium (8,000–12,000) betweenness values (Fig. 2h). These differences in degree of connexions and betweenness distributions between the generalists and specialists indicated their different contributions to network topology.
The removal of generalist or specialist groups from the original dataset caused a decline in network robustness. The decrease in the generalists truncated dataset (from 40.5 to 36.4) is slightly higher than when the specialists were removed (from 40.5 to 37.1) (Fig. 2i). This indicated that the removal of generalists resulted in a more sensitive, less robust network compared with the removal of specialists.
Overall, microbial generalists and specialists simultaneously contributed to the species interactions, and the absence of these two groups would decrease the robustness of the co-occurrence networks.
The roles of generalist and specialist groups in assembly processes
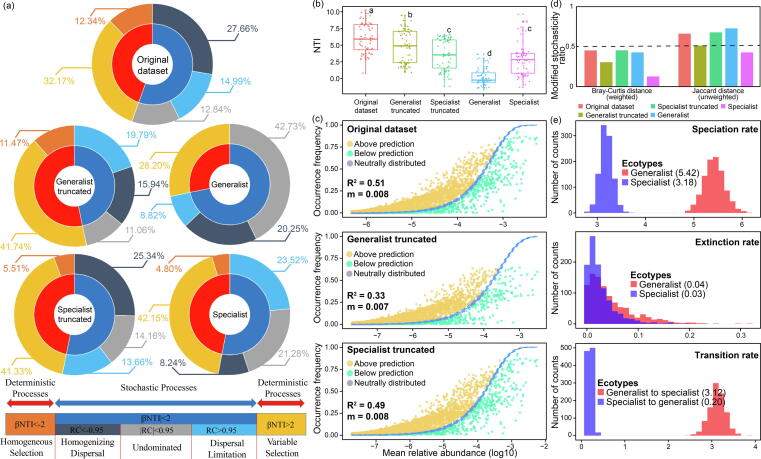
The null-model-based framework (Fig. 3a) showed that both deterministic and stochastic processes were important in microbial community assembly in these farmland soils. Stochastic processes (dispersal limitation: ∼15%, homogenizing dispersal: ∼28%, undominated: ∼13%, in total ∼55%) contributed more than deterministic processes (heterogeneous selection: ∼32%, homogeneous selection: ∼12%, in total ∼46%) at the continental scale. The removal of generalists from the original dataset resulted in an increased contribution of deterministic processes (from 45% to 53%) and dispersal limitation (from 15% to 20%) that was accompanied by a decrease in homogenizing dispersal (from 28% to 16%). Together, this indicates that the removal of generalists increased the environmental selection but limited the microbial dispersal in the community. The removal of specialists only imparted minor changes. Furthermore, compared with specialists (deterministic processes: ∼47%, dispersal limitation: ∼24%), the generalists (deterministic processes: ∼28%, dispersal limitation: ∼9%) were less controlled by deterministic processes and exhibited a higher dispersal ability (less controlled by dispersal limitation). In addition, the nearest taxon index (NTI) suggested that the specialists were more phylogenetically clustered than the generalists. The removal of specialists caused a larger decrease in NTI, leading the original community to become more phylogenetically random than with the removal of the generalists (Fig. 3b).
Fig. 3.
The assembly and evolutionary roles of generalist and specialist groups. (a) Assembly of different groups based on the null-model-based framework. The inner circle represents the contribution of stochastic and deterministic processes to community assembly. The outer circle represents the percentage of detailed ecological processes statistically assigned to stochastic or deterministic processes. The original dataset represents the entire community, the specialist truncated represents the dataset after removing specialists, and the generalist truncated represents the dataset after removing generalists. (b) The nearest taxon index (NTI) to assess the phylogenetic structure of different groups. A higher NTI indicates that the community exhibits more phylogenetic clustering and a NTI close to 0 indicates the community is phylogenetically random. (c) Estimation of the neutral processes based on the Sloan neutral model. The parameters R2 and m represents the goodness of fitting and migration rate, respectively. The species that occur more and less frequently than predicted are shown in yellow and green, respectively. Dashed lines represent 95% confidence intervals and the species falling within the confidence intervals are considered neutrally distributed. (d) The modified stochasticity ratio (MST) of different groups based on Bray-Curtis and Jaccard distance. The higher MST indicated the community assembly was more stochastic. (e) Evolutionary characteristics of the generalists and specialists based on the binary-state speciation and extinction (BiSSE) model. The distribution of speciation rate, extinction rate and transition rate are shown, and their average values among the specialist and generalist groups were provided.
The relationship between the distribution and relative abundances of microbial taxa was well described by the Sloan neutral model (Fig. 3c). As such, neutral processes played an important role in microbial community assembly (R2 = 0.51). The removal of specialists slightly decreased the impacts of neutral processes (R2 decreased from 0.51 to 0.49), whereas the removal of generalists led to a larger decrease in R2 (from 0.51 to 0.33) and a slight decrease in estimated migration rates.
When considering the relative abundance of taxa, the modified stochasticity ratio (MST) showed (Fig. 3d) that the generalists (0.43) exhibited higher stochasticity than specialists (0.13). Removal of generalists caused a decrease (from 0.45 to 0.30) in stochasticity, whereas, the removal of specialists resulted in a small increase. Similar trends among groups was observed when only considering the presence/absence of taxa, while stochasticity indexes were higher overall.
Compared with specialists (speciation rate: 3.18, extinction rate: 0.03), the generalists (speciation rate: 5.42, extinction rate: 0.04) were characterized by an approximately two-fold higher speciation rate and a slightly higher extinction rate. The transition rate from generalists to specialists (3.12) was approximately 15-fold higher than that from specialists to generalists (0.20).
Overall, three different models, the null-model-based framework, Sloan neutral model, and MST, were employed to investigate the roles of generalists and specialists in microbial assembly by assessing the relative contribution of deterministic and stochastic processes (Fig. 3a, 3c and 3d). Generalists contributed more to stochastic processes while the specialists contributed more to deterministic processes of community assembly. Generalists imparted greater impacts on assembly than specialists. In addition, the higher diversification rate (speciation rate minus extinction rate) and the higher transition rate of generalists indicated their important roles in community assembly, for example in the maintenance of diversity, from an evolutionary perspective (Fig. 3e).
Geographic patterns of generalist and specialist groups
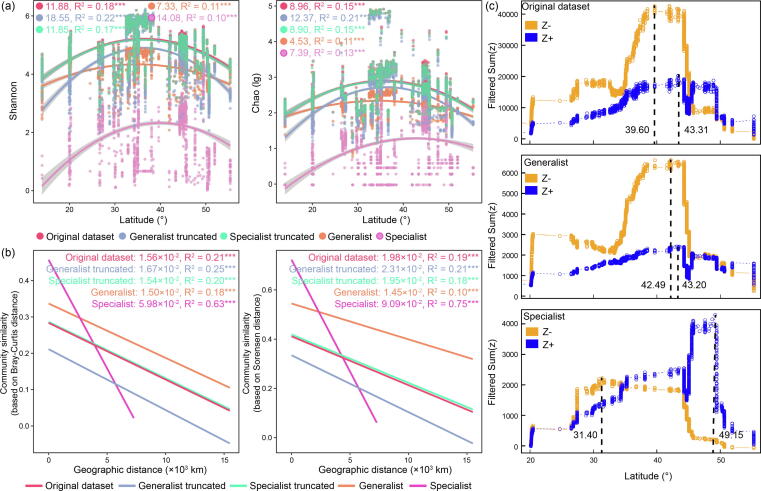
Alpha diversity indices (Shannon and Chao) showed an increasing then decreasing trend (quadratic distribution) with latitude across all groups (Fig. 4a). Diversity was highest at ∼ 35° latitude for the generalist group and the entire community and at ∼ 40° for the specialist group in our datasets. The removal of generalists or specialists did not alter the position at which the maximum points occurred. Quadratic coefficients were used to compare the trend strength. For the Shannon index, the specialists (quadratic coefficient: 14.08) had a stronger trend compared to the generalists (quadratic coefficient: 7.33). The removal of generalists increased the trend strength with an increase in the quadratic coefficient from 11.88 to 18.55, while the removal of specialists has only a slight effect of decrease on these quadratic coefficients. Results based on the Chao index were similar to Shannon.
Fig. 4.
The biogeographical patterns and ecological thresholds of generalist and specialist groups. (a) The relationship between latitude and alpha diversity (i.e., Shannon and Chao) in different groups. Quadratic coefficients (absolute value) are provided to compare the strength of trend. A higher quadratic coefficient indicates a stronger relationship. R2 represents the goodness of fitting. ***P < 0.001. The original dataset represents the entire community, the specialist truncated represents the dataset after removing specialists, and the generalist truncated represents the dataset after removing generalists. (b) The distance decay in community similarity based on Bray-Curtis distance (considering relative abundance) and Sorensen distance (considering presence/absence). Slopes (absolute value) are provided to compare the strength of distance decay. A higher slope indicates a stronger distance decay. (c) The ecological community thresholds, indicating the environmental point where the maximum aggregate change in the species frequency and relative abundance occurs, for different groups identified using threshold indicator species analyses (TITAN). Blue and orange symbols represent sum values of positive (Z+) and negative (Z-) indicator taxa along with latitude. The range of niche optima is the gradient below sum(Z-) and above sum(Z+).
A distance-decay relationship in community similarity was detected across all groups (Fig. 4b). Slopes were used to compare the strength of distance decay relationship. The specialists (slope: 5.98 × 10-2) had stronger distance-decay compared with generalists (slope: 1.50 × 10-2), and the removal of specialists slightly weakened the distance-decay of the entire community. In contrast, the removal of generalists strengthened the distance-decay with an increase in slope from 1.56 × 10-2 to 1.67 × 10-2. The distance decay based on the presence/absence of species showed similar results.
The environmental points, where the maximum aggregate change in the species frequency and relative abundance occurs (i.e., the ecological community thresholds), were identified using threshold indicator species analyses (TITAN, see methods for details). The communities were divided into two groups: Z- taxa negatively respond to the latitude, and Z + taxa positively respond to the latitude. The range of niche optima of the community is defined as the latitude gradient below sum(Z-) and above sum(Z + ). The sum(Z-) ecological thresholds were 31.40°, 42.49° and 39.60° for specialists, generalists and the entire community, respectively. The sum(Z+) ecological thresholds were 49.15°, 43.20° and 43.31° for specialists, generalists and the entire community, respectively. Hence, compared with specialists, the generalists had a wider range of niche optima, where the accumulated relative abundance was higher.
Overall, compared with specialists, the generalists demonstrated weaker biogeographic patterns, including the relationship between latitude and α-diversity and distance-decay in community similarity, and broader environment adaptation. In addition, generalists appear to dampen the biogeographical patterns of the entire community, while the specialists strengthen the biogeographical patterns.
The species interactions, assembly processes and biogeographic patterns in different climate types and the related effects of microbial generalists and specialists were also explored (Fig. S3-5). Similar results were obtained.
In addition, the generalists were predicated to have larger genomes and proteomes (Fig. S6), which is in agreement with the expectation that generalists require larger gene and protein repertoires to survive in multiple environment conditions. We also found that the G + C contents and coding density were significantly different between generalists and specialists (Fig. S6).
Discussion
When predicting the ecosystem service and functional stability, a critical assumption underlying many models is that distant taxa are functionally equivalent. If taxa are functionally equivalent, microbial communities with varying compositions will function in an identical manner when they are placed in the same environment. Given high species diversity and the rapid adaptability of microbes to new conditions, this functional redundancy seems plausible. However, this may not be the truth. For example, microbial communities that shared a common history with a given habitat were found to exhibit higher functional performance compared to communities foreign to that habitat, due to local specialization [38]. Hence, an improved predictive ability may be achieved by imparting systematic distinctions among different categories of microbial species. The most common classification schemes of microbial groups include the separation of fungal and bacterial taxa [39], active and dormant pools [40] and generalists and specialists [41]. Among these categories, the contribution of microbial generalists and specialists to diversity has been found to be significant, though it is rarely investigated. Herein, we explored the characteristics of microbial generalists and specialists in terms of species interactions, assembly rules and biogeographic patterns.
We found that (i) generalists and specialists simultaneously contributed to interactions between species through different mechanisms (Fig. 2); (ii) generalists contribute more to stochastic processes in community assembly while specialists contribute more to the deterministic processes (Fig. 3); (iii) the existence of microbial generalists dampens microbial biogeographic patterns, with contrasting impacts by specialists (Fig. 4). These results give insights into how these two groups contribute to community diversity and enrich the knowledge on better understanding and predicting ecosystem functions and diversity in the future.
The implications of generalists and specialists on occurrence networks
Species interactions may affect community composition and drive the stability and distribution patterns in microbial ecology. Patterns of species interactions were highly dynamic and contingent on community composition, species density and the environmental condition [42]. Compared with specialists, generalists are more available due to wider distribution and higher population densities, which is thought to lead to a higher probability of interaction with others. However, recent studies in microbial ecology have shown that rare taxa may play more important role in species interactions [8]. As such, high availability may not determine the strength of interactions. Based on existing knowledge, it remains difficult to determine whether generalists or specialists contribute more to microbial interactions.
Our results, based on co-occurrence network analyses, showed that both the generalists and specialists had similar degree of connexions and that the removal of generalist or specialist nodes from the network resulted in a clear decrease in network robustness (Fig. 2). Thus, both specialists and generalists were important in shaping interactions among species. However, the significance of species interactions may be different between the two groups. Microbial species specialize towards the consumption of common resources in order to escape competition and offset the energetic survival costs [9], [43]. This is explained by the Black Queen Hypothesis [43], [44] that adaptive genome reduction allows free-living organisms to become beneficiary of a common good produced by a helper (i.e., dependency interaction) [9]. Generalists are likely to be helpers (common good producers), and from the Black Queen Hypothesis corollary, their ecological status is supposed to be paired with higher genome sizes and possibly with genome expansion [43]. Thus, in the face of environmental stress, generalists are supposed to be less dependent of the presence of others to grow and may pass the environmental filter more efficiently than non-generalists. That means, species interactions seem not to be obligatory for generalists while are more likely to stabilize the community. In contrast, specialists are expected to more rely on external nutrients or metabolites produced by a different microorganism for growth, the absence of these interactions would lead to cell death.
The implications of generalists and specialists on assembly processes
Understanding the mechanisms that underlie the assembly of microbial communities are essential to unravelling the sustainability of ecological systems [45]. Recent studies have revealed that niche and neutral processes are not mutually exclusive; in contrast, they are complementary and work together in structuring microbial communities [46]. However, the assembly processes of microbial generalists and specialists remains unexplored. Generalists, inhabiting a wide range of environments, and specialists, having a narrower habitat range, are thought to be dominated by contrasting assembly processes and thus contribute differently to the overall diversity. Generalists are thought to be favoured by environments that vary frequently, while specialists evolve in environments that remain constant in space and time [47]. As such, in constantly varying natural systems, generalists appear to be more impactful on the overall community structure and function, even though they have a lower species richness.
We found that generalists contribute more to stochastic processes in community assembly while specialists contribute more to the deterministic processes (Fig. 3). This is in agreement with a previous study that showed that the distribution of generalists was primarily determined by neutral processes due to their general indifference to variations in habitat conditions, while habitat specialists are more affected by species sorting (deterministic processes) due to their “preferences” for certain environmental conditions [48]. At the community level, these results indicated that specialists may contribute more to the function of the ecosystem in stable environments, considering that the deterministically assembled community was found to have an increased function compared with the stochastically assembled one [49]. However, there may be another scenario that occurs within fluctuating environments. Community-level functional acclimatisation to environment changes could be obtained through a greater generalist physiological breadth through adjustments to new conditions or through the multitude of specialist physiologies as they become more abundant/active from dormant pools [50]. A generalist-specialist trade-off would affect the potential of the community to acclimatise to new environments. For generalists, the stochastic assembly process may be an acclimatization to future possible environment changes. The large pool of generalists may expand the range of optimal niches and lead to a greater chance of a compensatory response to alterations in the environment. However, there may be a tipping point in this wide niche breadth effect, where the negative effect of containing too high a proportion of generalists begins to reduce species richness and the resource use efficiency in the ecosystem.
The implications of generalists and specialists on biogeographic patterns
Biogeographical patterns describe the distribution of species across space or time and reveal the mechanisms maintaining and generating species diversity. Although crucial for understanding population dynamics, microbial generalists or specialists are often neglected when attempting to predict and explain spatial patterns of biodiversity. There are similarities in biogeographical patterns in macro- and microorganisms [45], whereas, it is important to emphasize that some classical biogeographical patterns observed for macro-organisms, such as the links between latitude and diversity, are generally not observed in microorganisms [45], [51]. This observation suggests differences in biogeography between macro- and microorganisms. The large populations of microbial generalists may distort the linkages between latitude and diversity of community members. We found that, when the generalists were absent from the entire community, the linkages between latitude and diversity as well as the distance decay relationship were weaker (Fig. 4a and 4b), which supports this assumption. Dispersal limitation and environmental filtering are two of the main forces shaping biogeographical patterns. For generalists, the higher tolerance, which was indicated by a wider ecological range of fitness (Fig. 4c), and more individuals within groups (Fig. 1a) may result in a higher probability of dispersal and successful survival in new environments which would lead to weaker biogeographic patterns.
Furthermore, it may be possible to quantify the dampening effect of microbial generalists on biogeographical patterns. To accomplish this, differences in biogeographical patterns for total communities and their generalist and specialist fractions could be compared and the differences quantified using a measure of effect size. This approach would bring new insights into predicting microbial biogeographic community patterns, where systematic distinctions among different categories of taxa were neglected before.
In addition, a subset was created to investigate the effects of environmental factors. We found that planted crops, climates, soil depths and soil properties had significant impacts on the bacterial communities (Fig. S7). Hence, future work should take other biotic and abiotic factors into consideration when explore the microbial generalists and specialists, such as plant physiological activity. It is well known that, under stress, plants create and excrete a broad suite of secondary metabolites which microbiomes can use for signalling and food [52], [53], [54], [55], [56]. Some proxy (e.g., LICOR measurements, photosynthetic activity) can be used to identify possible differences of plant activity that can ultimately shape the associated microbial generalists and specialists.
Conclusion
This global survey of microbiomes originating from farmland soils revealed the distinct contributions of generalists and specialists to microbial diversity from the perspective of species interactions, community assembly and biogeographical patterns. In stable environments, specialists contribute more to community diversity and function through their more robust deterministic processes and higher richness. However, in constantly fluctuating natural environments, generalists that confer a high degree of ecological resistance to altered environmental conditions and exhibit higher diversification and transition rates, potentially play a more important role in maintaining community and functional stability. Generalists and specialists simultaneously contributed to interactions between species through different mechanisms. Considering that specialists appear to have a dependency of species interactions in terms of survival, when facing an environment disturbance, conservation strategies should focus on microbial specialists to avoid a decrease in overall diversity. Lastly, our results support that the existence of microbial generalists dampens microbial biogeographic patterns and indicate that the contribution of both generalists and specialists should be taken into consideration when predicting global patterns of microbial diversity to likely increase the predictive power of the data analysis.
CRediT authorship contribution statement
Qicheng Xu: Methodology, Software, Writing – original draft, Visualization. Philippe Vandenkoornhuyse: Writing – review & editing, Project administration, Supervision. Ling Li: Investigation, Validation, Data curation. Junjie Guo: Investigation, Validation, Data curation, Resources. Chen Zhu: Investigation, Validation, Data curation, Software. Shiwei Guo: Writing – review & editing, Project administration, Supervision. Ning Ling: Conceptualization, Writing – original draft, Project administration, Supervision, Writing – review & editing. Qirong Shen: Writing – review & editing, Project administration, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA28030302), the National Natural Science Foundation of China (41977080, 31902114), the Natural Science Foundation of Jiangsu Province (BK20190543), the China Postdoctoral Science Foundation (2019M651861), the Young Elite Scientists Sponsorship Program by CAST (2019QNRC001), and a grant from the CNRS (EC2CO).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.12.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Falkowski P.G., Fenchel T., Delong E.F. The microbial engines that drive Earth's biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson J.K., Holmes E., Wilson I.D. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 3.Rittmann B.E. Microbial ecology to manage processes in environmental biotechnology. Trends Biotechnol. 2006;24:261–266. doi: 10.1016/j.tibtech.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Delgado-Baquerizo M., Maestre F.T., Reich P.B., Jeffries T.C., Gaitan J.J., Encinar D., et al. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat Commun. 2016;7:1–8. doi: 10.1038/ncomms10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence C.R., Neff J.C., Schimel J.P. Does adding microbial mechanisms of decomposition improve soil organic matter models? A comparison of four models using data from a pulsed rewetting experiment. Soil Biol Biochem. 2009;41:1923–1934. [Google Scholar]
- 6.Shade A. Diversity is the question, not the answer. ISME J. 2017;11:1–6. doi: 10.1038/ismej.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Székely A.J., Silke L. The importance of species sorting differs between habitat generalists and specialists in bacterial communities. FEMS Microbiol Ecol. 2014;87:102–112. doi: 10.1111/1574-6941.12195. [DOI] [PubMed] [Google Scholar]
- 8.Xiong C., He J.Z., Singh B.K., Zhu Y.G., Wang J.T., Li P.P., et al. Rare taxa maintain the stability of crop mycobiomes and ecosystem functions. Environ Microbiol. 2020;23:1907–1924. doi: 10.1111/1462-2920.15262. [DOI] [PubMed] [Google Scholar]
- 9.Zengler K., Zaramela L.S. The social network of microorganisms—How auxotrophies shape complex communities. Nat Rev Microbiol. 2018;16:383–390. doi: 10.1038/s41579-018-0004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Q., Luo G., Guo J., Xiao Y., Zhang F., Guo S., et al. Intraspecific variation and dormancy potential matter. Mol Ecol; Microbial generalist or specialist: 2021. Shen Q; pp. 1–13. [DOI] [PubMed] [Google Scholar]
- 11.Nekola J.C., White P.S. The distance decay of similarity in biogeography and ecology. J Biogeogr. 1999;26:867–878. [Google Scholar]
- 12.McGill B.J. Towards a unification of unified theories of biodiversity. Ecol Lett. 2010;13:627–642. doi: 10.1111/j.1461-0248.2010.01449.x. [DOI] [PubMed] [Google Scholar]
- 13.Devictor V., Clavel J., Julliard R., Lavergne S., Mouillot D., Thuiller W., et al. Defining and measuring ecological specialization. J Appl Ecol. 2010;47:15–25. [Google Scholar]
- 14.Bell T.H., Bell T. Many roads to bacterial generalism. FEMS Microbiol Ecol. 2021;97(fiaa240) doi: 10.1093/femsec/fiaa240. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y.-J., Leung P.M., Wood J.L., Bay S.K., Hugenholtz P., Kessler A.J., et al. Metabolic flexibility allows bacterial habitat generalists to become dominant in a frequently disturbed ecosystem. ISME J. 2021;1–19 doi: 10.1038/s41396-021-00988-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 17.Edgar R. Taxonomy annotation and guide tree errors in 16S rRNA databases. PeerJ. 2018;6 doi: 10.7717/peerj.5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sriswasdi S., Yang C.C., Iwasaki W. Generalist species drive microbial dispersion and evolution. Nat Commun. 2017;8:1–8. doi: 10.1038/s41467-017-01265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Q., Ling N., Chen H., Duan Y., Wang S., Shen Q., et al. Long-term chemical-only fertilization induces a diversity decline and deep selection on the soil bacteria. mSystems. 2020;5:e00337–e420. doi: 10.1128/mSystems.00337-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lima-Mendez G., Faust K., Henry N., Decelle J., Colin S., Carcillo F., et al. Determinants of community structure in the global plankton interactome. Science. 2015;348:1262073. doi: 10.1126/science.1262073. [DOI] [PubMed] [Google Scholar]
- 21.Benjamini Y., Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- 22.Feizi S., Marbach D., Médard M., Kellis M. Network deconvolution as a general method to distinguish direct dependencies in networks. Nat Biotechnol. 2013;31:726–733. doi: 10.1038/nbt.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo F., Zhong J., Yang Y., Scheuermann R.H., Zhou J. Application of random matrix theory to biological networks. Phys Lett A. 2006;357:420–423. [Google Scholar]
- 24.Newman M.E. Modularity and community structure in networks. Proc Natl Acad Sci USA. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng Y., Jiang Y.H., Yang Y., He Z., Luo F., Zhou J. Molecular ecological network analyses. BMC Bioinf. 2012;13:1–20. doi: 10.1186/1471-2105-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J., Deng Y., Luo F., He Z., Tu Q., Zhi X. Functional molecular ecological networks. mBio. 2010;1:e00169–00110. doi: 10.1128/mBio.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng G.-s., Wu J. Optimal network topology for structural robustness based on natural connectivity. Phys A. 2016;443:212–220. [Google Scholar]
- 28.Stegen J.C., Lin X., Fredrickson J.K., Chen X., Kennedy D.W., Murray C.J., et al. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013;7:2069–2079. doi: 10.1038/ismej.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou J., Ning D. Stochastic community assembly: Does it matter in microbial ecology? Microbiol Mol Biol R. 2017;81:e00002–00017. doi: 10.1128/MMBR.00002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia X., Dini-Andreote F., Salles J.F. Community assembly processes of the microbial rare biosphere. Trends Microbiol. 2018;26:738–747. doi: 10.1016/j.tim.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 31.Stegen J.C., Lin X., Fredrickson J.K., Konopka A.E. Estimating and mapping ecological processes influencing microbial community assembly. Front Microbiol. 2015;6(370) doi: 10.3389/fmicb.2015.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo X., Feng J., Shi Z., Zhou X., Yuan M., Tao X., et al. Climate warming leads to divergent succession of grassland microbial communities. Nat Clim Change. 2018;8:813–818. [Google Scholar]
- 33.Ning D., Deng Y., Tiedje J.M., Zhou J. A general framework for quantitatively assessing ecological stochasticity. Proc Natl Acad Sci USA. 2019;116:16892–16898. doi: 10.1073/pnas.1904623116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sloan W.T., Lunn M., Woodcock S., Head I.M., Nee S., Curtis T.P. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol. 2006;8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- 35.Burns A.R., Stephens W.Z., Stagaman K., Wong S., Rawls J.F., Guillemin K., et al. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 2016;10:655–664. doi: 10.1038/ismej.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.FitzJohn R.G. Diversitree: Comparative phylogenetic analyses of diversification in R. Methods Ecol Evol. 2012;3:1084–1092. [Google Scholar]
- 37.Baker ME, King RS, Kahle D, Kahle MD. Package ‘TITAN2’. 2015.
- 38.Strickland M.S., Lauber C., Fierer N., Bradford M.A. Testing the functional significance of microbial community composition. Ecology. 2009;90:441–451. doi: 10.1890/08-0296.1. [DOI] [PubMed] [Google Scholar]
- 39.Waring B.G., Averill C., Hawkes C.V. Differences in fungal and bacterial physiology alter soil carbon and nitrogen cycling: Insights from meta-analysis and theoretical models. Ecol Lett. 2013;16:887–894. doi: 10.1111/ele.12125. [DOI] [PubMed] [Google Scholar]
- 40.Wang G., Mayes M.A., Gu L., Schadt C.W. Representation of dormant and active microbial dynamics for ecosystem modeling. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0089252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moorhead D.L., Sinsabaugh R.L. A theoretical model of litter decay and microbial interaction. Ecol Monogr. 2006;76:151–174. [Google Scholar]
- 42.Ramsey M.M., Rumbaugh K.P., Whiteley M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS pathog. 2011;7 doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mas A., Jamshidi S., Lagadeuc Y., Eveillard D., Vandenkoornhuyse P. Beyond the black queen hypothesis. ISME J. 2016;10:2085–2091. doi: 10.1038/ismej.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris B.E., Henneberger R., Huber H., Moissleichinger C. Microbial syntrophy: Interaction for the common good. FEMS Microbiol Rev. 2013;37:384–406. doi: 10.1111/1574-6976.12019. [DOI] [PubMed] [Google Scholar]
- 45.Nemergut D.R., Schmidt S.K., Fukami T., O'Neill S.P., Bilinski T.M., Stanish L.F., et al. Patterns and processes of microbial community assembly. Microbiol Mol Biol R. 2013;77:342–356. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J., Liu W., Deng Y., Jiang Y.H., Xue K., He Z., et al. Stochastic assembly leads to alternative communities with distinct functions in a bioreactor microbial community. mBio. 2013;4:e00584–00512. doi: 10.1128/mBio.00584-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kassen R. The experimental evolution of specialists, generalists, and the maintenance of diversity. J Evolution Biol. 2002;15:173–190. [Google Scholar]
- 48.Liao J., Cao X., Zhao L., Wang J., Gao Z., Wang M.C., et al. The importance of neutral and niche processes for bacterial community assembly differs between habitat generalists and specialists. FEMS Microbiol Ecol. 2016;92(fiw174) doi: 10.1093/femsec/fiw174. [DOI] [PubMed] [Google Scholar]
- 49.Graham E.B., Stegen J.C. Dispersal-based microbial community assembly decreases biogeochemical function. Processes. 2017;5:65. [Google Scholar]
- 50.Hawkes C.V., Keitt T.H. Resilience vs. historical contingency in microbial responses to environmental change. Ecol Lett. 2015;18:612–625. doi: 10.1111/ele.12451. [DOI] [PubMed] [Google Scholar]
- 51.Fierer N., Jackson R.B. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan X., Hong S., Xiong W., Raza W., Shen Z., Wang B., et al. Development of fungal-mediated soil suppressiveness against Fusarium wilt disease via plant residue manipulation. Microbiome. 2021;9:1–15. doi: 10.1186/s40168-021-01133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Vries F.T., Griffiths R.I., Knight C.G., Nicolitch O., Williams A. Harnessing rhizosphere microbiomes for drought-resilient crop production. Science. 2020;368:270–274. doi: 10.1126/science.aaz5192. [DOI] [PubMed] [Google Scholar]
- 54.Huang A.C., Jiang T., Liu Y.-X., Bai Y.-C., Reed J., Qu B., et al. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science. 2019;364 doi: 10.1126/science.aau6389. [DOI] [PubMed] [Google Scholar]
- 55.Carrión V.J., Perez-Jaramillo J., Cordovez V., Tracanna V., De Hollander M., Ruiz-Buck D., et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science. 2019;366:606–612. doi: 10.1126/science.aaw9285. [DOI] [PubMed] [Google Scholar]
- 56.Hu L., Robert C.A., Cadot S., Zhang X., Ye M., Li B., et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat commun. 2018;9:1–13. doi: 10.1038/s41467-018-05122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.