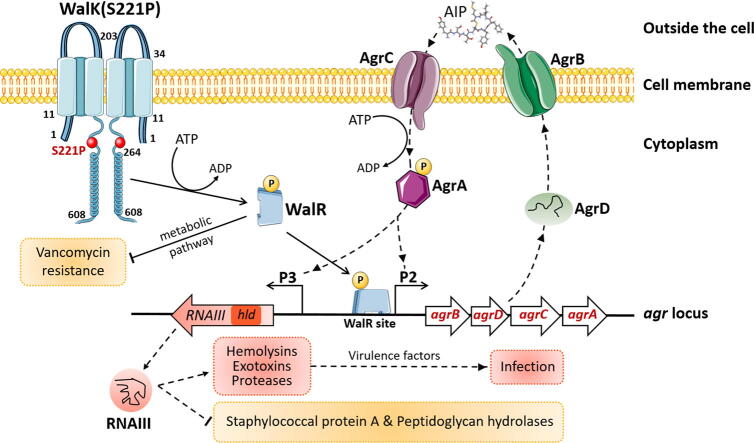
Graphical abstract
Keywords: Staphylococcus aureus, Vancomycin resistance, WalKR, WalK(S221P), Virulence, Agr system
Highlights
-
•
WalK(S221P) enhanced vancomycin resistance in S. aureus of diverse backgrounds.
-
•
WalK(S221P)-reverted VISA exacerbated murine infections.
-
•
Reversion of WalK(S221P) in VISA increased bacterial virulence.
-
•
WalK(S221P)-bearing S. aureus presented reduced virulence factor expression.
-
•
WalK(S221P) attenuated VISA virulence by contacting the agr promoter region.
Abstract
Introduction
Vancomycin-intermediate Staphylococcus aureus (VISA) is typically associated with a decline in virulence. We previously reported a WalK(S221P) mutation that plays an important role in mediating vancomycin resistance in VISA XN108. Whether this mutation is implicated in bacterial virulence remains unknown.
Objectives
This study aimed to investigate the effect of WalK(S221P) mutation on the virulence of VISA and the underlying mechanism of this effect.
Methods
The influence of WalK(S221P) mutation on VISA virulence and its underlying mechanism were explored using animal models, RNA-seq analysis, RT-qPCR, hemolytic assay, slide coagulase test, Western blot, β-galactosidase assay, and electrophoresis mobility shift assay (EMSA).
Results
Compared with XN108, WalK(S221P)-reverted strain XN108-R exacerbated cutaneous infections with increased lesion size and extensive inflammatory infiltration in mouse models. The bacterial loads of S. aureus XN108-R in murine kidney increased compared with those of XN108. RNA-seq analysis showed upregulation of a set of virulence genes in XN108-R, which exhibited greater hemolytic and stronger coagulase activities compared with XN108. Introduction of WalK(S221P) to methicillin-resistant S. aureus USA300 and methicillin-susceptible strain Newman increased the vancomycin resistance of the mutants, which exhibited reduced hemolytic activities and decreased expression levels of many virulence factors compared with their progenitors. WalK(S221P) mutation weakened agr promoter-controlled β-galactosidase activity. EMSA results showed that WalK-phosphorylated WalR could directly bind to the agr promoter region, whereas WalK(S221P)-activated WalR reduced binding to the target promoter. Inactivation of agr in S. aureus did not affect their vancomycin susceptibility but mitigated the virulence alterations caused by WalK(S221P) mutation.
Conclusion
The results of our study indicate that WalK(S221P) mutation can enhance vancomycin resistance in S. aureus of diverse genetic backgrounds. WalK(S221P)- bearing S. aureus strains exhibit reduced virulence. WalK(S221P) mutation may directly impair the activation of the agr system by WalR, thereby decreasing the expression of virulence factors in VISA.
Introduction
Staphylococcus aureus is a notorious pathogen that poses a great threat to the community and healthcare settings [1], [2]. This bacterium has evolved into a successful pathogen, largely because it features metabolic versatility, ability to subvert host immune responses, and acquisition of antibiotic resistance [2]. Vancomycin has been used as a last-resort drug to treat serious infections caused by S. aureus [3]. However, the increased use of vancomycin has led to the emergence of S. aureus with decreased vancomycin susceptibility [4]. The Clinical Laboratory Standards Institute has categorized staphylococcal isolates according to their level of vancomycin resistance as vancomycin-susceptible S. aureus (VSSA, MIC ≤ 2 μg/mL), vancomycin-intermediate S. aureus (VISA, MIC 4–8 μg/mL), and vancomycin-resistant S. aureus (VRSA, MIC ≥ 16 μg/mL) [5]. Whereas the prevalence of VRSA strains is very low [3], [6], VISA has been reported to show increased prevalence worldwide, thus raising a considerable concern for human health [3], [7].
Hitherto, the genetic basis of VISA is not completely understood. Genome sequencing and comparison of selected isogenic VSSA/VISA pairs demonstrate that VISA strains show diverse mutations in genes responsible for cell wall synthesis, cell hydrolysis, cell wall remodeling, and transcription regulation [3]. Moreover, different mutations present diverse contributions to vancomycin resistance in VISA; a single mutation may contribute variable vancomycin-resistant levels to VISA strains of different genetic lineages [8], [9]. Mutations in transcription-regulatory genes encoding two-component systems, such as WalKR, VraSR, and GraSR, are mostly found in VISA isolates [10], [11]. Although varied mutations in some genes are associated with VISA development, VISA strains exhibit several common characteristics in addition to the vancomycin-intermediate phenotype, including thickened cell walls, reduced autolysis, and attenuated virulence [3], [10], [12].
The majority of VISA cases reported in the literature involve in methicillin-resistant S. aureus (MRSA), but reduced vancomycin susceptibility in methicillin-susceptible S. aureus (MSSA) has also been documented [13]. As a bacterium, VISA may suffer a serious fitness cost when developing resistance to vancomycin, and this fitness cost can be a disadvantage to bacterial virulence [14], [15]. Several studies have demonstrated that VISA strains show decreased virulence compared with their parental isolates [16], [17], [18]. However, how VISA regulates its virulence remains unclear. Thus, whether the reduced virulence of VISA is associated with an identical altered gene that is responsible for increased vancomycin resistance deserves further investigation.
We previously reported an ST239 MRSA strain (XN108) with reduced vancomycin susceptibility (vancomycin MIC = 12 μg/mL) [19]. The allelic replacement experiments indicated that a WalK(S221P) mutation plays a crucial role in XN108 phenotypes [9]. In this study, the virulence of XN108 was compared with that of WalK(S221P) mutation- reverted strain XN108-R in mouse models. RNA-seq showed that a set of virulence factors and regulators are upregulated in XN108-R, with significantly altered genes of spa and accessory gene regulator (agr). Introduction of WalK(S221P) to MRSA USA300 and MSSA Newman strains resulted in increased vancomycin MIC and decreased hemolytic activities of the mutants. WalK(S221P) mutation affected bacterial virulence via targeting the agr system of S. aureus.
Materials and methods
Strains and plasmids
Bacterial strains and plasmids used in the present study were listed in Table S1. The VISA strain XN108 was isolated on 4 March 2004 from a burn patient who underwent skin operation on 23 February 2004 and suffered an acute S. aureus infection after the operation [19]. S. aureus XN108-R (K65) is a derivative of XN108 by curing WalK(S221P) mutation [20]. DP65 is a clinical VSSA isolate kept in our laboratory. The reference S. aureus strains USA300 (ATCC-BAA-1556), Newman (NCTC 8178), and RN4220 (NCTC 8325–4) were kindly provided by Dr. Hao Zeng (Army Medical University, China), Prof. Yu Lu (Jilin University, China), and Prof. Baolin Sun (University of Science and Technology of China), respectively. S. aureus strains were stored as frozen stocks in media containing 80% brain heart infusion (BHI) and 20% glycerol and kept at −80 °C. When preparation of working cultures, the bacterial stocks were streaked on the brain heart infusion agar (BHIA) plates and grown at 37 °C for 16 h, respectively. On the next day, a single colony of a strain of interest was inoculated in 2 mL BHI medium and grown at 37 °C for 16 h with shaking, and then the culture was performed in the subsequent experiments. The Escherichia coli strains (Table S1) in 80% Luria-Bertani (LB) medium and 20% glycerol were stored as frozen stocks. The working culture was prepared with LB agar plate or LB medium as described for S. aureus. When needed, the agar plate or broth medium was supplemented with chloramphenicol (20 μg/mL) or ampicillin (100 μg/mL) to culture strains carrying certain plasmids.
Genome sequencing and comparison of S. aureus XN108 and XN108-R
Bacterial cells of S. aureus strains XN108 and XN108-R overnight culture were collected and sent to Novogene C., LTD (Beijing, China) to perform whole-genome sequencing (WGS). Briefly, bacterial genomic DNA was prepared using a TIANamp Bacteria DNA Kit (TianGen, China), and the DNA concentration and quality were determined with a NanoDrop Spectrophotometer (Thermo Scientific, USA). WGS was performed on the Illumina HiSeqTM2500 platform (Illumina Inc., USA) using insert lengths of ∼ 350 bp. Raw data were modified and filtered as described [21], and the clean reads were mapped to the reference genome of XN108 (GenBank accession no. CP007447.1) by Samtools (0.1.19–44428 cd), and the duplicates were eliminated with Picard 1.107. The potential genomic variations were detected by using Breakdancer 1.3.7. Single nucleotide polymorphisms (SNPs) were detected by comparing the sequence of S. aureus strain XN108-R with that of strain XN108. The primary SNPs were further evaluated as previously described [21].
Bacterial growth curve
The growth curves of S. aureus XN108 and its derivative XN108-R were determined as previously described [15]. Briefly, S. aureus strains were cultured in BHI medium at 37 °C overnight with shaking, and 0.2 mL of each culture was added to 20 mL of fresh BHI broth in a sterile 50-mL flask. The optical density readings at 600 nm (OD600) were obtained every hour for 13 h after inoculation. Growth curve was drawn using the OD600 values over cultivating times.
Animal experiments
Female BALB/c mice aged 6–8 weeks were purchased from the Animal Center of Army Medical University (Third Military Medical University). All animal experiments were approved by the Laboratory Animal Welfare and Ethics Committee of the Third Military Medical University (SYXK-PLA-20120031). The experimental procedures for animals were performed according to the Regulations for Administration of Affairs Concerning Experimental Animals approved by Chinese State Council. Cervical dislocation was applied as euthanasia way of the experimental animals.
S. aureus strain XN108 or its WalK(S221P)-reverted strain XN108-R was cultivated in BHI broth and grown overnight at 37 °C with shaking. On the next day, the culture was inoculated to fresh BHI medium (1:100 dilution) and cultivated at 37 °C for 6 h (OD600 ≈ 1.0). Then, the bacterial cells were pelleted, washed two times, and resuspended in phosphate buffer saline (PBS, pH 7.2). The viable cells were counted by inoculation of 2-fold serially diluted bacterial suspension on BHIA plates and a bacterial stock was prepared by adjusting the cell concentration to 1 × 109 colony forming unit (CFU) per mL. For the formation of skin abscesses, BALB/c mice (n = 10) were treated as previously described [22]. Briefly, mice were pretreated with 6% Na2S to remove the hair on back, and then subcutaneously inoculated with 1 × 108 CFU of bacteria cells (100 μL of stock) or PBS alone in one flank of the murine back. Skin abscess formation was monitored, and abscess areas were measured daily by the maximal length × the width of the developing lumps. The body weights of mice were also determined daily. For histopathological examination, skin lesions of the infected mice (n = 2) on 5 d post-infection (pi) were prepared and fixed by 4 % paraformaldehyde, then embed with paraffin, sliced, stained with hematoxylin-eosin (HE), and observed under a Microscope (Olympus BX53, Japan).
For bacterial dissemination, BALB/c mice (n = 7) were challenged with 5 × 107 CFU of the GFP-carrying plasmid transformed XN108 or XN108-R (namely, XN108-pGFP and XN108-R-pGFP) through tail vein injection and sacrificed on 5 d pi. Organs of the infected mice were obtained for the determination of fluorescence efficiency of GFP with the IVIS® Lumina LT system (PerkinElmer, USA). The bacterial loads in murine organs were counted with plate dilution method as previously described [23].
Proinflammatory cytokine determination
Blood samples were collected from BALB/c mice (n = 3) challenged via tail vein injection with 5 × 107 CFU of XN108-pGFP or XN108-R-pGTP on 5 d pi, and mouse sera were obtained after blood clotting. Livers and kidneys of the infected mice were isolated and homogenized by adding PBS (pH 7.2) to achieve homogenate of 1 mL/organ. The supernatants were collected by centrifugation at 5,000 × g at 4 °C for 10 min to remove tissue debris. Then, the levels of IL-6, IL-1β, and TNF-α were determined with an ELISA kit according to the manufacturer’s instructions (R&D Systems, USA). The PBS-injected mice (n = 3) served as negative control.
RNA-seq analysis
S. aureus strains were grown in BHI broth at 37 °C for 16 h. Then, the culture of XN108 or XN108-R was inoculated into fresh BHI medium and cultured at 37 °C. Bacterial cells at the logarithmic phase (6 h post inoculation, OD600 ≈ 1.0) were harvested by centrifugation at 5,000 × g at 4 °C for 10 min, and total RNA was extracted with the RNAprep Pure Cell/Bacteria Kit (TianGen, China). Three biological repeated samples were prepared for each strain. The 16S and 23S rRNAs were removed using the Ribo-ZeroTM Gold Kit (Illumina, USA). The cDNA libraries were prepared from mRNA samples with a TruSeq Illumina Kit as described [24]. RNA-seq was performed by Novogene Co., LTD (Beijing, China) with Illumina HiSeqTM2500 sequencing system. The numbers of successful reads were quantified and assessed by Novogene Co., LTD. The differentially expressed genes (DEGs) were analyzed by software DESeq version 1.10.1. The RNA-seq data were deposited in the GEO DataSets under ID codes GSE127706.
RT-qPCR
S. aureus strains of interest at the logarithmic phase were prepared. Total RNA and the relevant cDNA were obtained using the RNAprep Pure Cell/Bacteria Kit (TianGen, China) and the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA), respectively. Quantitative polymerase chain reaction (qPCR) was performed, and the relative expression level of each gene was calculated as a ratio of its threshold cycles (Ct value) relative to that of the reference gene gyrB [25]. The relative expression levels of genes in XN108 were adjusted to 1.0, and the relative levels of genes in XN108-R were calculated. The primers used were listed in Table S2.
Hemolysis assay
To evaluate the hemolytic activities of strains of interest, the overnight cultures of S. aureus XN108, USA300, Newman, and their derivatives were diluted at 1:1,000 with BHI medium, and 20 μL aliquots were streaked onto Columbia sheep blood agar plate (Pengtong Medical, China), and cultured at 37 °C for 16 h. On the next day, the cultured plates were kept at 4 °C in a refrigerator for 24 h, and then photographed. The cross-streak assay was used to qualitatively evaluate the production of α-, β-, and δ-hemolysin (Hla, Hlb, and Hld) in S. aureus strains as previously described [26]. For the tube test method, S. aureus strains of interest were cultivated in BHI medium at 37 °C for 16 h (OD600 ≈ 2.5). The culture supernatant (100 μL) of each bacterial strain was mixed with 900 μL PBS carrying 3% rabbit erythrocytes (Pengtong Medical, China), followed by incubation at 37 °C for 20 min. After centrifugation at 1,000 × g at 4 °C for 10 min, OD543 values of the supernatants were measured. The 3% rabbit erythrocytes in 1,000 μL ddH2O and 1,000 μL PBS were used as the positive and negative controls, respectively. The percentage of hemolytic activity was calculated relative to the positive control, which represented 100% hemolytic activity.
Western blot
S. aureus strains of interest were cultured in BHI medium at 37 °C overnight. Bacterial cells in 2 mL culture were harvested, washed two times with PBS, and resuspended in 1 mL of PBS on ice. Then, bacterial cells were broken by the addition of 2 g zirconia-silica beads and treated with Minibeadbeater (Biospec, USA) for three times (5 min for each). The protein concentration of the solution was determined using the Bradford protein assay kit (Beyotime Biotechnology, China). The proteins of Hla and Hlb in S. aureus strains of interest were separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene difluoride membrane (GE, USA). A first antibody against Hla or Hlb (murine polyclonal antibodies, kind gifts from Dr. Hao Zeng, Department of Microbiology and Biochemical Pharmacy, Army Medical University) and a secondary sheep anti-mouse antibody coupled with horseradish peroxidase (Boster, China) were used in the experiment. The target proteins were visualized by the enhanced chemiluminescence system (Amersham Pharmacia Biotech, USA) and photographed.
Slide coagulase test
The bound coagulases of S. aureus strains of interest were detected using slide coagulase tests as previously described [27]. Briefly, S. aureus strains were cultured on the sheep blood plates at 37 °C overnight. A drop of sheep plasma containing ethylene diamine tetraacetic acid (EDTA) anticoagulant and several S. aureus colonies were mixed on a clean glass slide, and the results were observed within 10 s. Bacterial cells in PBS were used as the negative control.
Vancomycin E-test
S. aureus strains of interest were cultured in BHI medium overnight with shaking. On the next day, the vancomycin E-test was determined on BHIA plates with vancomycin strips according to the instruction of manufacturer (bioMérieux, France).
Construction of allelic replacement and gene deletion mutants
To introduce the WalK(S221P) mutation to S. aureus strain USA300, the genomic DNA template of VISA strain XN108 was prepared using a TIANamp Bacteria DNA Kit (TianGen, China), and two ∼ 900 bp flanking DNA fragments were obtained using PCR with primers US-WalK-up/US-S221P-down and US-S221P-up/US-WalK-down (Table S2). Then, the PCR fragments were ligated by overlap-PCR with primers US-WalK-up US-WalK-down, and the fusion fragment was digested by EcoR I and BamH I, cloned into the pBT2 shuttle vector to construct the pBT2-WalK(S221P) plasmid. The resultant plasmid was sequentially transformed into strains RN4220 and USA300. The plasmid-carried USA300 was firstly cultured at 30 °C for 16 h, followed by inoculation into fresh BHI medium and cultivation at 42 °C to achieve the integration of the pBT2-WalK(S221P) plasmid into the bacterial genomic DNA, and then cultured at 25 °C to eliminate the plasmid. The final WalK(S221P) mutation-carried strain (K-USA300) was verified by PCR detection and nucleic acid sequencing. The WalK(S221P) mutation-carried strain K-Newman was constructed with the same allelic replacement strategy described above and confirmed by DNA sequencing.
To generate gene deletion mutant XN108/Δagr, two ∼ 900 bp DNA fragments were amplified by PCR with primer pairs AgrA up-for/AgrA up-rev and AgrA down-for/AgrA down-rev (Table S2). Next, the PCR fragments were ligated by overlap-PCR with primers AgrA-for/AgrA-rev. The fusion fragment was digested by EcoR I and BamH I and cloned into the pBT2 vector to achieve the pBT2-ΔagrA plasmid. The resultant plasmid was sequentially transformed into strains RN4220 and XN108. The pBT2-ΔagrA plasmid-carried XN108 was subjected to screen XN108/Δagr mutant by sequentially cultivating at 30 °C, 42 °C, and 25 °C as described above. The final mutant strain (XN108/Δagr) was verified by PCR detection and nucleic acid sequencing. Construction of XN108-R/Δagr and USA300/Δagr was performed using the same strategy as for XN108/Δagr. USA300/Δagr was transformed with pBT2-WalK(S221P) plasmid to obtain K-USA300/Δagr using the aforementioned procedure.
To construct Newman/Δagr and K-Newman/Δagr, a plasmid pBT2-ΔagrBDC was generated with the strategy identical to that for pBT2-ΔagrA construction. After electrotransformed into S. aureus Newman or K-Newman, the desired mutant was screened following the method described above. The agr deletion mutants were finally characterized by PCR amplification and nucleic acid sequencing.
Construction of a reporter strain carrying the agr-promoter controlled LacZ
The DNA fragment spanning −235 to + 18 of the agr gene was obtained by PCR amplification from XN108 genomic DNA template using primers pOS1-p2p3-5(EcoR I)N and pOS1-p2p3-3(BamH I)N (Table S2). The amplified PCR products were digested with EcoR I and BamH I and inserted into pOS1 plasmid carrying a promoterless gene encoding LacZ. The resultant plasmid (pOS1-agrP) was transformed to E. coli DH5α and characterized by DNA sequencing, then transformed into S. aureus RN4220 for restriction modification, and subsequently transformed into S. aureus XN108 and XN108-R, respectively.
β-Galactosidase assay
The pOS1-agrP plasmid-transformed S. aureus XN108 or XN108-R was cultured in BHI medium at 37 °C overnight, then the culture was diluted in fresh BHI (200-fold) and cultivated at 37 °C for 7 h with shaking (OD600 ≈ 1.2). Bacterial cells were collected by centrifugation at 5,000 × g at 4 °C for 10 min, washed two times with PBS, and resuspended in 100 μL of AB buffer (100 mM KH2PO4, 100 mM NaCl, pH 7.0). The β-galactosidase assay was performed as previously described [22]. Briefly, bacterial cells in 100 μL of AB buffer were lysed by the addition of lysostaphin (Sigma-Aldrich, USA) to a final concentration of 1 µg/mL, and incubation at 37 °C for 15 min. The suspension was added with another 900 μL of ABT solution (AB buffer added with 0.1% TritonX-100) and mixed. Then, 50 μL of the sample was taken and mixed with 10 μL of 4-methylumbelliferyl -β-D-galactoside (4 mg/mL, Sigma-Aldrich, USA) and incubated at room temperature for 1 h. Finally, 20 μL of the solution was mixed with 180 μL of ABT buffer, and the reaction was monitored at 445 nm with an excitation wavelength of 365 nm. All bacterial samples were determined at least three times.
Expression and purification of recombinant proteins
To achieve protein expression in S. aureus, a shuttle vector pXR was constructed by insertion of xyl regulon into plasmid pLI50 at the sites of EcoR I and Kpn I [28], [29]. Genes encoding WalK(S221P) and WalR were amplified from S. aureus XN108 genomic DNA template by PCR with primer pairs pXR-walK-5́/pXR-walK-3́ and pXR-walR-5́/pXR- walR-3́ (Table S2), respectively. The gene encoding WT WalK was obtained from S. aureus XN108-R genomic DNA template by PCR with primer pairs pXR-walK-5́/pXR-walK-3́. A 6 × His tag was fused with the C-terminal of target proteins and introduced by primers pXR-walK-3́ and pXR-walR-3́, respectively. PCR products were digested with the restriction enzymes BamH I and Hind III, then purified with the Wizard®SV Gel and PCR Clean-up System (Promega, USA) and cloned into the pXR vector to generate pXR-WalK(S221P), pXR-WalR, and pXR-WalK, respectively. Next, the recombinant plasmids were individually transformed into S. aureus RN4220 and a chloramphenicol-resistant colony was picked up for each transformation and cultivated at 37 °C in 100 mL of BHI medium with 0.5% xylose for 15 h to induce the expression of recombinant proteins. For protein purification, bacterial cells after induction with xylose were harvested by centrifugation at 5,000 × g at 4 °C for 30 min and washed twice with buffer A (20 mM Tris, 100 mM NaCl, pH 8.0). Then, the pellet was resuspended with 100 mL of buffer A containing 1 mM phenylmethylsulfonyl fluoride (PMSF) (Beyotime Biotechnology, China), DNase (10 µg/mL), and lysostaphin (30 µg/mL) and incubated at 37 °C for 3 h to disrupt the cell wall. After centrifugation at 16,000 × g at 4 °C for 30 min to remove cell debris, the supernatant was collected and centrifuged again at 10,000 × g for 30 min, filtered through a 0.22 µm filter unit (Beyotime Biotechnology, China), and applied to a Ni–NTA resin column (GE, USA) pre-equilibrated with buffer A. The column was washed with buffer A, followed by buffer A mixed with a linear gradient of 5%–100% (v/v) buffer B (buffer A containing 500 mM imidazole) to remove the nonspecifically bound proteins and elute the target proteins. Recombinant proteins in the elute fractions were identified by SDS-PAGE analysis, then were dialyzed five times against buffer A to remove imidazole. The concentration of recombinant WalK, WalK(S221P), or WalR proteins in buffer A containing 25% glycerol was detected with the Bradford protein assay kit (Beyotime Biotechnology, China).
Electrophoresis mobility shift assay (EMSA)
A biotinylated DNA probe of S. aureus agr gene P2 promoter region (agr-P2) was obtained by PCR from XN108 genomic DNA using primer pair Agr-p1-5′-biotin/Agr-p2-3′ (Table S2). One similar fragment without the predicted WalR-binding site (agr-P2M) was also amplified with PCR using primer pair Agr-p1-5′-biotin/Agr-p2M-3′ (Table S2) and served as negative control. The unlabelled DNA fragment of the mecA ORF amplified by PCR with primers mecA-5′/mecA-3′ (Table S2) was added as a nonspecific competitor. The promoter region of sle1 gene was obtained by PCR from XN108 genomic DNA using primers sle1-5′-biotin/sle1-3′ (Table S2) and served as positive probe.
The EMSA was performed with a LightShift® Chemiluminescent EMSA Kit (Thermo Scientific, USA) as previously described [22]. Briefly, 10 picomoles (pM) of DNA probe was incubated with a variable amount of recombinant WalR (0 to 320 pM) activated with 20 µM of ATP and 0.5 µM of WalK or WalK(S221P) in a 20 µL reaction solution containing 1 × binding buffer, 5 mM MgCl2, 0.05% (v/v) NP40, 2.5% (v/v) glycerol, and 50 ng/µL poly[dI-dC]. Binding reaction was performed at room temperature for 30 min. Reaction mixtures were then separated with a 6% (m/v) native polyacrylamide gel and electrophoresed at 90 V at 4 °C for 2 h in the 0.5 × TBE (Tris-boric acid-EDTA) buffer. Gels were stained by GelRed dye (Biotium, USA) and observed under UV light. To compare the probe binding ability between WalK- and WalK(S221P)-activated WalR, the gray value of the bound DNA fragments in each lane was analyzed using ImageJ software. The value of bound probe in each lane (0 to 320 pM WalR) relative to that of loading control (free probe in the lane of 0 pM WalR) was calculated and compared.
Statistical analysis
The results were statistically analyzed with GraphPad Prism 7. Results derived from samples between two groups were treated with unpaired two-tailed student's t-test, and the difference between groups was compared by analysis of variance (ANOVA) test. Each experiment was conducted at least three times, and the relevant results were presented as mean ± standard deviations (SD). The statistically significant was considered as a P value < 0.05.
Results
Reversion of WalK(S221P) in XN108 resulted in enhanced virulence of XN108-R in mouse models
XN108 is a VISA strain with a vancomycin MIC of 12 μg/mL, while the WalK(S221P)- reverted strain XN108-R exhibits a vancomycin MIC of 4 μg/mL [19], [20]. Reversion of WalK(S221P) did not affect the growth rate of XN108-R (Fig. S1). Complete genome sequencing and comparative genomic analysis identified a total of 15 mutations in five genes of XN108-R relative to those of XN108 (Table S3). Except for the WalK(S221P) site reversion, other mutations occurring in genes encoding phage proteins, cassette chromosome recombinase C, and transposase in XN108-R may not be associated with the vancomycin resistance of S. aureus [3].
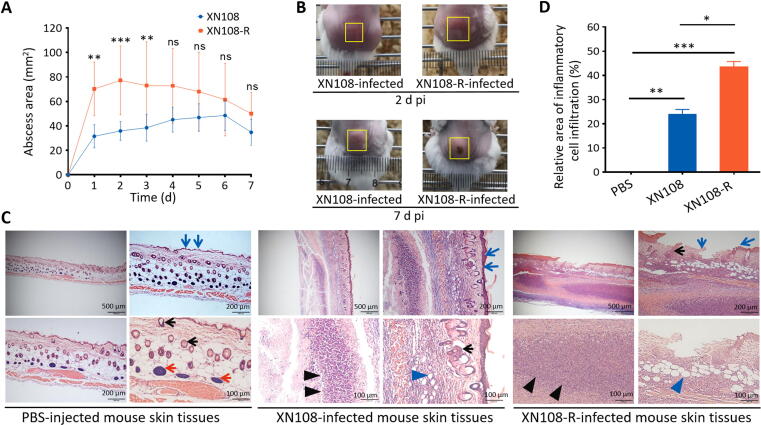
A mouse infection model was established by subcutaneously injecting the back flank of mice with 1 × 108 CFU of S. aureus XN108 or XN108-R. The course of infection was then monitored for 7 d to determine whether WalK(S221P) reversion affects the virulence of VISA. Abscesses caused by XN108-R between 1 and 3 d pi were significantly larger than those caused by XN108 (Fig. 1A). Photographs of mouse skin abscesses obtained on 2 d pi confirmed this observation. However, the abscess sizes observed on 7 d pi were comparable, and more skin lesions were observed in XN108-R-infected mice relative to those in XN108-challenged ones (Fig. 1B and Fig. S2). The weights of mice challenged with XN108 and XN108-R were comparable (Fig. S3A). Histological examination indicated that XN108-infected mouse skin presents subcutaneous inflammatory necrosis, abscess formation, gland vanish, inflammatory cell infiltration, congestion, hair follicle degeneration, and fat vacuole formation; by comparison, PBS-injected murine skin presented a normal structure with abundant subcutaneous tissue glands, including light-colored sebaceous glands and sweat glands (Fig. 1C). S. aureus XN108-R-infected mouse skin exhibited more serious subcutaneous inflammatory necrosis, larger abscesses, heavier inflammatory cell infiltration, larger degenerated fat vacuoles, incomplete dermis structure, and a partially broken epidermis compared with XN108-infected mouse skin (Fig. 1C). The severity of skin lesions was numerically classified, and the relative inflammatory cell infiltration area observed in the sections of XN108-R-infected mice was significantly larger than that of XN108-infected animals (P < 0.05) (Fig. 1D).
Fig. 1.
Reversion of WalK(S221P) enhanced VISA virulence. (A) Skin abscess areas of the S. aureus XN108- and XN108-R-infected mice. Mice were injected subcutaneously with 1 × 108 CFU of bacterial cells. Abscess sizes were measured daily. The number of animals used, n = 10. Statistical significance was calculated by 2Way ANOVA. ** P < 0.01, *** P < 0.001, and ns indicates no significance. (B) Representative abscesses in mice on 2 and 7 d pi. The abscess size for each animal was indicated by a yellow box. (C) Histological observation. The skin of infected mice was obtained on 5 d pi. The histological sections were prepared, stained with HE, and observed under a microscope (Olympus BX53). The bar represented 100, 200 or 500 µm was indicated. The blue arrows indicated mouse epidemis, the red arrows showed subcutaneous tissue glands, the black arrows indicated hair follicles, the black tangles pointed to abscesses and the blue tangles showed the degenerated fat vacuoles. (D) Severity of the mouse lesions. The severity of the skin lesions in histological sections of XN108-, XN108-R-, and mock (PBS)-infected mice was numerically classified with ImageJ software, and the relative inflammatory cell infiltration area (%) was calculated and indicated. Three sections with the bars of 100 µm were used. Statistical significance was calculated by One-Way ANOVA. * P < 0.05, ** P < 0.01, and *** P < 0.001.
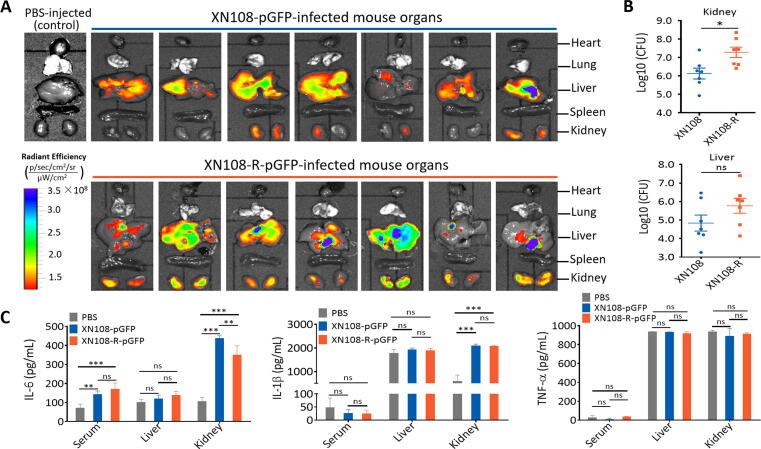
We investigated whether XN108-R, which shows reduced vancomycin resistance relative to XN108, is associated with bacterial loads and inflammatory responses in a murine systemic infection model. Mice were intravenously challenged with pGFP plasmid-carrying XN108 or XN108-R. The weights of XN108-R-pGFP-infected mice significantly decreased compared with those of XN108-pGFP-infected animals from 2 d pi (Fig. S3B). All experimental mice were sacrificed on 5 d pi, and the GFP intensity in various organs (i.e., heart, lung, liver, spleen, and kidney) was examined via an animal-imaging system. The radiant efficiency in the kidneys and livers of both XN108-R-pGFP- and XN108-pGFP-infected mice was higher than that of PBS-injected animals (Fig. 2A). Obvious abscesses were observed on the kidney surface of 57.1% (4/7) of the XN108-R-pGFP-infected mice compared with the gross specimens of XN108-pGFP-infected and PBS-injected animals (Fig. S3C). Mouse kidney and liver homogenates were prepared, and the bacteria were counted by the plate-dilution method as described previously [23]. The bacterial loads in the kidneys of XN108-R-pGFP- infected mice were significantly higher than those of XN108-pGFP-challenged animals, but no difference in bacterial burden was observed in the livers of both groups (Fig. 2B).
Fig. 2.
Reversion of WalK(S221P) in VISA XN108 contributed to bacterial dissemination. (A) Dissemination of XN108 and XN108-R in organs of the infected mice. BALB/c mice (n = 7 in each group) were challenged via tail vein injection with 5 × 107 CFU of the pGFP plasmid-transformed S. aureus XN108 and XN108-R, respectively. The infected mice were sacrificed on 5 d pi and their organs were isolated. The radiant efficiency of the murine organs was determined using the IVIS® Lumina LT system. The organs of the PBS-injected mice served as negative control. (B) Bacterial burdens in organs of the infected mice. After radiant efficiency determination, the kidney and liver of the infected mice were grinded to homogenate and bacteria in the tissue homogenates were counted by plate dilution method. Statistical significance was calculated by Student’s t-test. * P < 0.05 and ns represents no significance. (C) Detection of the levels of proinflammatory cytokines. Sera and organs (i.e., liver and kidney) of the XN108-pGFP-, XN108-R-pGFP-infected, and PBS-injected mice (n = 3 for each group) were respectively collected on 5 d pi, and the levels of IL-6, IL-1β, and TNF-α in the sera and organ homogenates were determined by ELISA as described in Materials and methods. Statistical significance was calculated by 2way ANOVA. ** P < 0.01, *** P < 0.001, and ns represents no significance.
Levels of proinflammatory cytokines, such as IL-6, IL-1β, and TNF-α, in the sera and organs (i.e., liver and kidney) of XN108-R-pGFP- and XN108-pGFP-infected mice were determined to evaluate whether WalK(S221P) mutation reversion enhances the virulence of VISA. As shown in Fig. 2C, IL-6 levels in the sera and kidneys of XN108-R-pGFP- and XN108-pGFP-infected mice were significantly higher than those of PBS-injected animals. Higher IL-6 levels were also noted in the kidneys of XN108-pGFP-infected mice relative to XN108-R-pGFP-challenged animals, but no difference in IL-6 and IL-1β levels in the livers of both groups was observed. S. aureus-challenged mice exhibited serum, liver, and kidney TNF-α and IL-1β levels comparable with those of the mock-infected group except for IL-1β levels in the mouse kidney. These data are consistent with the results of bacterial burden counting (Fig. 2B), thereby indicating that the severity of abscess formation in the kidneys of XN108-R-pGFP-infected mice may affect IL-6 production. Taken together, our results thus far suggest that reversion of WalK(S221P) mutation results in the enhanced virulence of VISA.
Virulence-associated gene expression changed after WalK(S221P) mutation reversion in XN108
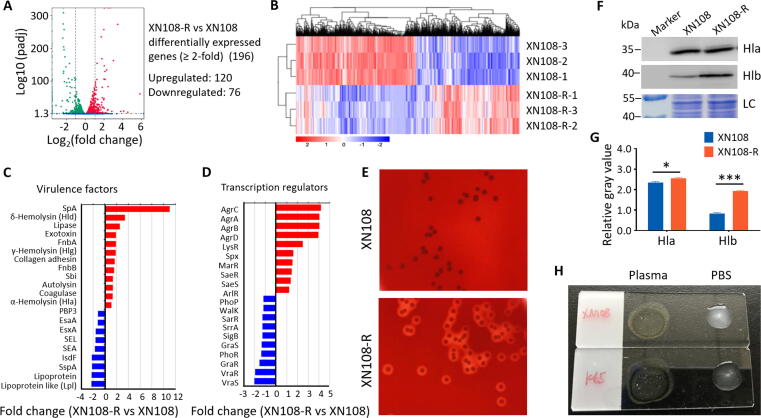
S. aureus is characterized by a large array of virulence factors, which are the basis of its virulence [30]. RNA-seq was performed to explore whether the observed increased virulence of WalK(S221P)-reverted XN108-R is associated with alterations in the expression of virulence factors. The RNA-seq data showed that XN108-R and XN108 have markedly distinct gene expression profiles (Fig. 3A). Euclidean distance calculation was used to illustrate DEGs among biological replicates (Fig. 3B). Ten altered genes, including significantly (i.e., spa,hla, fnbB,sbi, RNAIII, sigB, and saeR) and slightly (i.e., isdA, graR, and rot) altered genes, were noted in XN108-R relative to those in XN108. These results were confirmed by RT-qPCR, and the findings obtained were consistent with those of RNA-seq analysis (Fig. S4A).
Fig. 3.
Transcriptomic analysis and virulence factor determination of XN108-R and XN108. (A) RNA-seq analysis showed DEGs between XN108-R and XN108. (B) Transcriptional profiles for each sample analyzed with Pairwise Euclidean distance (heat map). (C) Virulence gene set and (D) transcription regulator gene set analysis, showing upregulated genes (red) and downregulated genes in XN108-R (blue). (E) Hemolytic experiments. Sheep blood plates were inoculated with XN108 and XN108-R, cultured at 37 °C overnight, then kept at 4 °C for 24 h and photographed. (F) Western blot analysis of Hla and Hlb in S. aureus strains indicated. The total bacterial proteins served as loading control (LC). Molecular weights of the protein marker are indicated on the left. (G) Evaluation of gray values of the expressed Hla and Hlb using ImageJ software. The relative values of Hla/LC and Hlb/LC were indicated. Statistical significance was calculated by 2way ANOVA. * P < 0.05 and *** P < 0.001. (H) Slide coagulase test. More agglutinated particles were presented in XN108-R (K65) relative to those of XN108.
Compared with XN108, WalK(S221P)-reverted XN108-R with reduced vancomycin resistance showed 120 upregulated genes and 76 downregulated genes (changes ≥ 2-fold, Fig. 3A). Of these 196 DEGs, 29.6% (58/196) encoded proteins with hypothetical functions (Table S4). We performed virulence gene set enrichment analysis to explore the pathogenic basis of VISA. The expression levels of SpA, δ-hemolysin (Hld, encoded by RNAIII), γ-hemolysin (Hlg), α-hemolysin (Hla), lipase, FnbA, FnbB, Sbi, Agr, LysR, SaeRS, LysR, and MarR were upregulated in XN108-R when compared with XN108. However, the expressions of Lpl, IsdA, IsdF, enterotoxins (i.e., SEA and SEL), type-VII secretion system-associated effect and apparatus molecules (i.e., EsxA and EsaA), and regulatory molecules (i.e., VraSR, GraRS, SigB, SarR, and WalK) were downregulated in XN108-R (Fig. 3C–D). The hemolytic activity of XN108-R increased in comparison with that of XN108, as tested by blood-agar plates (Fig. 3E) and the tube hemolysis method (Fig. S4B–C). Western blot indicated that the expression levels of Hla and β-hemolysin (Hlb) were significantly increased in XN108-R compared with those in XN108 (Fig. 3F–G). Coagulase activity was higher in XN108-R than in XN108 (Fig. 3H), consistent with the increased expression level of coagulase observed during RNA-seq (Fig. 3C). Overall, these observed alterations in virulence factors and regulatory profiles may contribute to the increased virulence of XN108-R.
Introduction of WalK(S221P) mutation to S. Aureus enhanced vancomycin resistance and attenuated bacterial virulence
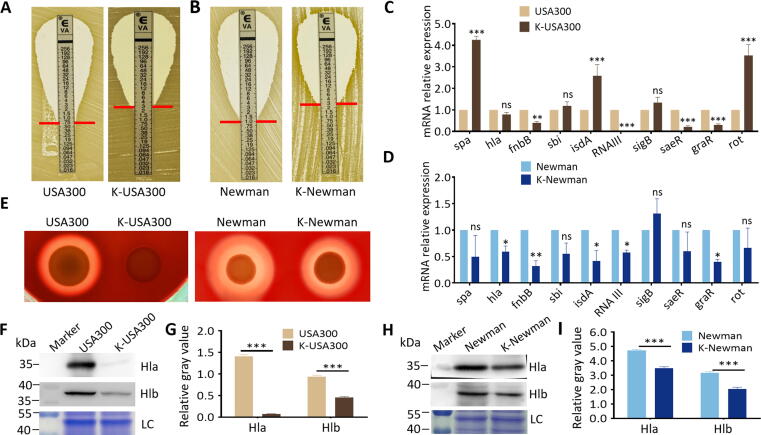
WalK(S221P) mutation contributes to vancomycin resistance in VISA XN108 [9]. Allelic replacement experiments were performed by substituting walK in the vancomycin-susceptible strains MRSA USA300 and MSSA Newman with the walK from XN108 to determine the effects of WalK(S221P) mutation on the vancomycin resistance and virulence of S. aureus further (Fig. S5A–C). WalK(S221P)-carrying K-USA300 exhibited increased vancomycin resistance (MIC = 3.0 μg/mL) when compared with WT USA300 (MIC = 0.75 μg/mL). The resultant MSSA strain K-Newman also presented increased vancomycin resistance (MIC = 3.0 μg/mL) compared with its parent strain (MIC = 1.0 μg/mL) (Fig. 4A–B). RT-qPCR showed that the expression levels of some virulence factors, including those encoding hemolysin (hla), adhesion factor (fnbB), and virulence regulators (i.e., RNAIII, saeR, and graR), decreased in K-USA300 compared with those in USA300. By contrast, the expression of immune evasion factors (i.e., spa and sbi), adhesion factor (isdA), and virulence regulators (i.e., sigB and rot) increased in K-USA300 compared with those in USA300 (Fig. 4C). In comparison with WT Newman, WalK(S221P)-carrying K-Newman exhibited decreased expression levels of all tested virulence factors except for sigB, which slightly increased in K-Newman (Fig. 4D). Therefore, the effect of WalK(S221P) mutation on the expression of S. aureus virulence factors may be strain-dependent, and this effect could be associated with the strain-based virulence regulatory network. However, both K-USA300 and K-Newman presented decreased hemolytic activity in comparison with their progenitors (Fig. 4E and Fig. S6). Western blot revealed that the protein expression levels of Hla and Hlb in the WalK(S221P)-introduced mutants decreased (Fig. 4F–I). Taken together, these results indicate that a single WalK(S221P) mutation can enhance vancomycin resistance in S. aureus and affect bacterial virulence by regulating virulence factors and regulatory profiles.
Fig. 4.
Introduction of WalK(S221P) to S. aureus increased vancomycin resistance and attenuated bacterial virulence. E-test of vancomycin susceptibilities in (A) MRSA strains USA300 and K-USA300, and (B) MSSA strains Newman and K-Newman. RT-qPCR detection of the expression levels of virulence factors in (C) USA300 and K-USA300, and (D) Newman and K-Newman. Statistical significance was calculated by 2way ANOVA. * P < 0.05, ** P < 0.01, *** P < 0.001, and ns indicates no significance. (E) Hemolytic experiments. Sheep blood plates were inoculated with USA300 and K-USA300 or strains Newman and K-Newman, cultured at 37 °C overnight, then kept at 4 °C for 24 h and photographed. Western blot analysis of Hla and Hlb levels in (F) USA300 and K-USA300, and (H) Newman and K-Newman. Molecular weights of the protein marker are indicated on the left. LC, loading control. Evaluation of gray values of the expressed Hla and Hlb using ImageJ software. The relative values of Hla/LC and Hlb/LC in (G) USA300 and K-USA300, and (I) Newman and K-Newman were indicated. Statistical significance was calculated by 2way ANOVA. *** P < 0.001.
The effect of WalK(S221P) mutation on S. Aureus virulence depended on agr activity
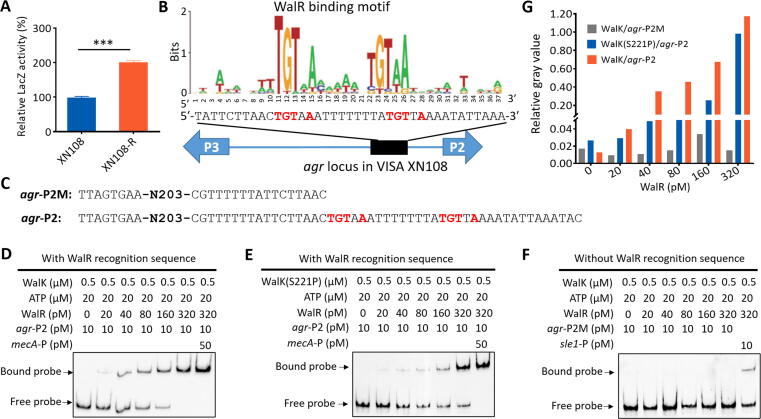
Given the upregulation of agr genes in WalK(S221P)-reverted XN108-R and corresponding downregulation in WalK(S221P)-carrying K-USA300 and K-Newman (Fig. 3D, Fig. S4A, and Fig. 4C–D), we hypothesized that WalK(S221P) mutation may affect agr activity in S. aureus. A reporter vector (pOS1-agrP) (Table S1) was generated, and β-galactosidase assay was performed by transforming pOS1-agrP into XN108 and its recovery strain XN108-R to test this hypothesis. The results demonstrated significantly lower β-galactosidase activity in XN108 compared with that in XN108-R (Fig. 5A), which suggests that WalKR can affect agr activity in S. aureus.
Fig. 5.
WalK-activated WalR bound to agr promoter region to control the expression of agr in S. aureus. (A) β-galactosidase assay. The pOS1-agrP reporter plasmid was transformed into S. aureus XN108 and XN108-R. The LacZ activity was detected and represented as mean ± SD (n = 3). Statistical significance was calculated by Student’s t-test. ***P < 0.001. (B) Predicted WalR-binding site in the promoter regions of agr P2. (C) Mutation of the predicted WalR-binding site in agr promoter regions for EMSA experiment. Interaction between agr promoter region (agr-P2) and (D) WalK-activated WalR proteins or (E) WalK(S221P)-activated WalR proteins was detected. The unlabeled DNA fragment of mecA promoter region (50 pM) was used for non-specific competition of the interaction. (F) EMSA with agr promoter mutant (agr-P2M), the sle1 promoter DNA fragment (10 pM) was used as positive control. (G) Evaluation of gray value of the bound probe in each lane using ImageJ software. The value of the free probe in the first lane (0 pM WalR protein) was used as loading control (LC), and the relative gray values of the bound probe to LC in other lanes were calculated and indicated.
We subsequently searched the possible binding site of the WalR response regulator [28], [31] between the promoter regions of agr P2 and P3, and a typical WalR-binding site was predicted (Fig. 5B). The His-tagged proteins of WalK, WalK(S221P), and WalR were prepared from the engineered S. aureus strains (Fig. S7). EMSA showed that WalK-activated WalR could bind to the putative WalR-binding site-carrying agr promoter DNA fragment (Fig. 5C–D and 5G) but not the fragment without the WalR-binding site (Fig. 5C and 5F). The promoter binding capability of WalK(S221P)-activated WalR was weaker than that of WalK-activated WalR (Fig. 5C–E and 5G). Overall, the WalKR system can directly control the activity of the agr P2 promoter, and the mutant WalK-mediated WalR exhibited decreased control of P2, which affected Agr activity and led to the reduced virulence of VISA XN108 in comparison with that of its isogenic XN108-R with WalK(S221P) mutation reversion.
Inactivation of agr in S. Aureus resulted in comparable vancomycin resistance and mitigated WalK(S221P)-mediated virulence
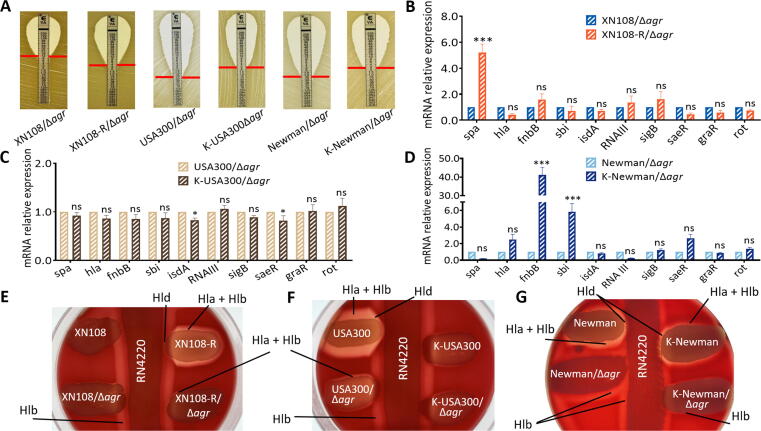
Agr activity was independently inactivated by deletion of the agrA gene in strains XN108, XN108-R, USA300, and K-USA300 or by knockout the agrBDC genes in Newman and K-Newman to confirm the Agr-mediated effect of WalK(S221P) mutation on the virulence of S. aureus (Fig. S5D–K). E-test showed that inactivation of agr does not affect the vancomycin susceptibility of the S. aureus strains tested in comparison with that of their progenitors (Fig. 6A and Fig. 4A-B) [20]. However, RT-qPCR revealed significant changes in the expression levels of several virulence factors, including hla, fnbB, sbi, RNAIII, sigB, and rot, in XN108-R relative to those in WT XN108 (Fig. S4A) were vanished in XN108-R/Δagr compared with those in the XN108/Δagr (Fig. 6B). This phenomenon was also observed in the strain pairs USA300/Δagr–K-USA300/Δagr and Newman/Δagr–K-Newman/Δagr (Fig. 6C–D) compared with USA300–K-USA300 and Newman–K-Newman (Fig. 4C–D), respectively. Agr-inactivated strains displayed similarly weak hemolytic activities on blood agar plates (Fig. S8A–C). Among the S. aureus-produced hemolysins, Hla and Hld were strongly upregulated whereas Hlb was weakly regulated by agr [26]. We used cross-streak assay to evaluate the production of Hla, Hlb, and Hld in the S. aureus strains of interest qualitatively. As shown in Fig. 6E–G, XN108-R presented higher Hla, Hlb, and Hld activities compared with XN108; by contrast, the agr-inactivated strains (XN108/Δagr and XN108-R/Δagr) exhibited comparable hemolytic activity. The WalK(S221P) mutation-bearing strain K-USA300 revealed lower production of Hla, Hlb, and Hld than USA300; however, the agr-deletion strains (USA300/Δagr and K-USA300/Δagr) showed remarkably decreased expression of hemolysins and similar hemolytic activity (Fig. 6F). WalK(S221P)-bearing strain K-Newman presented decreased expression levels of Hla, Hlb, and Hld compared with Newman, and Newman/Δagr showed reduced expressions of Hla and Hld but not Hlb. K-Newman/Δagr presented decreased Hlb activity relative to Newman/Δagr (Fig. 6G). Subsequent determination of Hla and Hlb by Western blot revealed results consistent with those of the cross-streak assay (Fig. S8D–I). Taken together, the data indicate that the contribution of WalK(S221P) to the vancomycin resistance of VISA may be independent of agr activity, which is required in the effect of WalK(S221P) on the reduced virulence of diverse S. aureus.
Fig. 6.
Agr inactivation resulted in comparable vancomycin resistance and bacterial virulence. (A) E-test of vancomycin susceptibilities in XN108/Δagr, XN108-R/Δagr, USA300/Δagr, K-USA300/Δagr, Newman/Δagr, and K-Newman/Δagr. RT-qPCR detection of the expression levels of virulence factors in (B) XN108/Δagr and XN108-R/Δagr, (C) USA300/Δagr and K-USA300/Δagr, and (D) Newman/Δagr and K-Newman/Δagr. Statistical significance was calculated by 2way ANOVA. * P < 0.05, *** P < 0.001, and ns indicates no significance. Cross streak assay for (E) XN108 and its derivatives, (F) USA300 and its derivatives, and (G) Newman and its derivatives. S. aureus RN4220 was used to represent Hlb production. The production of other hemolysins was indicated.
Discussion
As the most prevalent Gram-positive pathogen worldwide, S. aureus is characterized by antibiotic resistance and numerous virulence factors that could cause various diseases [1], [26]. The World Health Organization has listed VISA as a superbug, and the discovery and development of new control strategies for this bacterium have been prioritized [32]. S. aureus strains that develop resistance to vancomycin often result in decreased virulence; however, the mechanism underlying this effect is unknown [16], [17], [18], [33]. Peleg et al. [16] used Galleria mellonella as a model insect and demonstrated that VISA strains show decreased pathogenicity in comparison with their parental isolates. One VISA strain exhibited decreased infectivity in a rat endocarditis model and rapid clearance in the blood compared with the WT strain [17]. These previous studies used laboratory-derived VISA from a VSSA or clinical isolate [3]. In the present work, we showed that the natural WalK(S221P) mutation in the clinical VISA isolate XN108 can enhance vancomycin resistance in MRSA and MSSA strains. Using mouse cutaneous and systemic infection models, we demonstrated that WalK(S221P)-reverted XN108-R exhibits increased virulence compared with its progenitor XN108 (Fig. 1 and Fig. 2). Compared with XN108, S. aureus XN108-R caused larger abscesses, especially between 1 and 3 d pi. XN108-R-infected mouse also presented more serious subcutaneous inflammatory necrosis and greater bacterial burdens in the kidney compared with XN108-challenged animals. These data are consistent with the previous finding that the USA300-derived VISA strain SG-R exhibits reduced ability to cause tissue necrosis and skin abscess in mice compared with its isogenic progenitor SG-S [18].
The genetic determinants of increased vancomycin resistance in VISA have been intensively evaluated by genomic comparison of isogenic VSSA/VISA strains, and hundreds of mutations in certain genes have been determined in VISA strains compared with those in VSSA isolates [3], [10]. VISA can possess several mutations in certain genes, and a cumulative effect of these mutated genes may exist and contribute to the formation of VISA. For instance, the first characterized VISA strain, Mu50, carries six mutations, and sequential introduction of these mutations to six genes of the S. aureus N315ΔIP could completely reconstitute the VISA phenotype [8]. By contrast, the contribution of a single mutation to vancomycin resistance in S. aureus is varied. Allele-swapping experiments revealed that WalK(G223D) increases the vancomycin MICs of strains JKD6009 and MW2 from 1.5 μg/mL to 3 μg/mL and from 2 μg/mL to 4 μg/mL, respectively [10], [34]. In this work, we showed that WalK(S221P) can enhance vancomycin resistance in MRSA USA300 and MSSA Newman. WalK(S221P)-bearing strains K-USA300 and K-Newman presented similar MICs, which, at 3 μg/mL, is lower than that of WalK(S221P)-bearing N315 (MIC = 8 μg/mL) [20]. This difference may be attributed to the diverse genetic backgrounds of S. aureus isolates.
Agr is a delicate regulatory element of S. aureus that controls the expression of most genes encoding surface-anchored proteins and secreting factors that contribute to S. aureus virulence [35], [36]. VISA strains exhibiting defective agr function have been frequently reported [37], [38]; however, the reasons behind the dysfunction of VISA agr are unclear. Dai et al. [35] reported that an increase in vraR expression in a laboratory-induced VISA is accompanied by a gradual decrease in its agr expression. Our RNA-seq data indicated that agr expression increases in XN108-R relative to that in XN108 (Fig. 3D). A set of virulence factors involved in bacterial adhesion (i.e., FnbA, FnbB), host innate immune evasion (i.e., SpA, Sbi), and toxin and virulence enzymes (i.e., hemolysins, lipase, and autolysin) were upregulated in XN108-R relative to those in XN108 (Fig. 3C). These data may explain why VISA XN108 shows reduced virulence. In contrast to the upregulation of VraSR in VISA strains, VISA-associated mutation impaired WalKR function [3], [9], [39]. We performed LacZ reporter assay and found that WalK(S221P) mutation could repress the promoter activity of agr (Fig. 5A). EMSA also indicated that the effect of WalKR on agr expression is direct and that WalK(S221P)-activated WalR mitigates the control of WalKR on agr promotor. These findings indicate that vancomycin resistance-associated WalK(S221P) mutation may contribute to S. aureus virulence through the agr system.
The negative regulatory effects of agr on spa and fnbAB have been reported [40]. However, in this study, the expression of spa and fnbAB was significantly upregulated and accompanied by agr upregulation in XN108-R (Fig. 3C–D and Fig. S4A). This result is consistent with a previous study that the expression of both spa and agrA in VISA SG-R decreased compared with that in VSSA SG-S [18]. This phenomenon may be ascribed to a coordinated regulation network in S. aureus. Some regulatory molecules, such as Spx and SaeRS, can eliminate the agr function in S. aureus to increase surface-associated protein expression [41], [42]. XN108 features the agr-I group, which may result in variable effects on target genes, including those encoding surface-anchored proteins in S. aureus [43]. As the most important virulence factors, at least four hemolysins (i.e., Hla, Hlb, Hld, and Hlg) could be produced by S. aureus [26]. In some S. aureus strains, Hlb is truncated because of the insertion of a prophage into the hlb gene, such as φSa3 in XN108 and φNM3 in Newman [9], [44]. However, lysogeny is reversible, and excision of the prophage activates the expression of Hlb via a phage regulatory switch mechanism [45], [46]. The activities of Hla, Hlb, and Hld could be scored directly using blood agar plates, but that of Hlg cannot because it is inhibited by sheep blood agar [26]. We used two well-characterized agr-positive strains, namely, USA300 (agr-IV) and Newman (agr-I), and found that WalK(S221P)-bearing strains K-USA300 and K-Newman exhibit increased vancomycin MICs and decreased hemolytic activities; these effects were accompanied by significantly reduced Hla and Hlb levels in the mutants (Fig. 4E–I and Fig. S6). Deletion of agr resulted in decreased Hla, Hlb, and Hld production in strains USA300/Δagr, K-USA300/Δagr, Newman/Δagr, and K-Newman/Δagr, although the Hlb level in Newman/Δagr was higher than that in K-Newman/Δagr (Fig. 6E–G and Fig. S8). Adhikari et al. [26] reported that Hlb is weakly controlled by agr and generates a relatively turbid zone of hemolysis whereas Hla and Hld are controlled strongly by agr. Our data suggest that WalK(S221P) mutation may influence Hlb expression through an agr-independent pathway. However, the exact regulatory role of WalKR on Hlb expression requires further investigation.
Conclusions
Our research highlights the balance between vancomycin resistance and bacterial virulence in VISA strains. The data collected in the present study suggest a naturally existing WalK(S221P) mutation that is not only associated with vancomycin resistance in VISA but also attenuates VISA virulence via an agr-dependent pathway. We propose that WalKR can increase agr activity whereas WalK(S221P) mutation results in weak WalKR activity, which downregulates agr expression and contributes to decreased virulence in VISA.
Credit author statement
Xiancai Rao and Renjie Zhou designed the study; Yifan Rao, Huagang Peng, Weilong Shang, and Li Tan performed the experiments. Yifan Rao wrote the manuscript with the assistance of Ming Li and Yi Yang. Zhen Hu and Yi Yang collected the samples and assisted in data analysis. Xiancai Rao and Ming Li provided their invaluable contributions in critically revising the manuscript.
Compliance with ethics requirements
All animal experiments were approved by the Laboratory Animal Welfare and Ethics Committee of the Third Military Medical University (SYXK-PLA-20120031). The institutional and national guidelines for the care and use of animals (the Third Military Medical University Laboratory Animal Welfare and Ethics Committee) were followed.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank Hao Zeng from Army Medical University for supplying S. aureus strain USA300 and antibodies against Hla and Hlb. We acknowledge Yu Lu from Jilin University for kindly providing S. aureus strain Newman and Baolin Sun from University of Science and Technology of China for his gift of S. aureus strain RN4220.
The present work was supported by the National Natural Science Foundation of China (grant numbers 82102405, 82071857). The funders played no role in the study design, data collection, and manuscript preparation.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.11.015.
Contributor Information
Ming Li, Email: sotx7080@163.com.
Renjie Zhou, Email: zhou_rj@aliyun.com.
Xiancai Rao, Email: raoxiancai@126.com.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Chen J., Zhou H., Huang J., Zhang R., Rao X. Virulence alterations in Staphylococcus aureus upon treatment with the sub-inhibitory concentrations of antibiotics. J Adv Res. 2021;31:165–175. doi: 10.1016/j.jare.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das S., Lindemann C., Young B.C., Muller J., Österreich B., Ternette N., et al. Natural mutations in a Staphylococcus aureus virulence regulator attenuate cytotoxicity but permit bacteremia and abscess formation. Proc Natl Acad Sci USA. 2016;113(22):E3101–E3110. doi: 10.1073/pnas.1520255113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu Q., Peng H., Rao X. Molecular events for promotion of vancomycin resistance in vancomycin intermediate Staphylococcus aureus. Front Microbiol. 2016;7:1601. doi: 10.3389/fmicb.2016.01601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiramatsu K., Hanaki H., Ino T., Yabuta K., Oguri T., Tenover F.C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40(1):135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 5.Patel J.B. CLSI; Wayne, PA, USA: 2014. Performance standards for antimicrobial susceptibility testing: twenty-fourth informational supplement. [Google Scholar]
- 6.Cong Y., Yang S., Rao X. Vancomycin resistant Staphylococcus aureus infections: A review of case updating and clinical features. J Adv Res. 2020;21:169–176. doi: 10.1016/j.jare.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rishishwar L., Kraft C.S., Jordan I.K. Population genomics of reduced vancomycin susceptibility in Staphylococcus aureus. mSphere. 2016;1(4):e00094–e116. doi: 10.1128/mSphere.00094-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katayama Y., Sekine M., Hishinuma T., Aiba Y., Hiramatsu K. Complete reconstitution of the vancomycin-intermediate Staphylococcus aureus phenotype of strain Mu50 in vancomycin-susceptible S. aureus. Antimicrob Agents Chemother. 2016;60(6):3730–3742. doi: 10.1128/AAC.00420-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng H, Hu Q, Shang W, Yuan J, Zhang X, Liu H, et al. WalK(S221P), a naturally occurring mutation, confers vancomycin resistance in VISA strain XN108. J Antimicrob Chemother. 2017; 72(4): 1006–13. doi: 10.1093/jac/dkw518. [DOI] [PubMed]
- 10.Howden B.P., McEvoy C.R.E., Allen D.L., Chua K., Gao W., Harrison P.F., et al. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog. 2011;7(11):e1002359. doi: 10.1371/journal.ppat.1002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Li X., Jiang L., Han W., Xie X., Jin Y., et al. Novel mutation sites in the development of vancomycin-intermediate resistance in Staphylococcus aureus. Front Microbiol. 2017;7:2163. doi: 10.3389/fmicb.2016.02163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui L., Ma X., Sato K., Okuma K., Tenover F.C., Mamizuka E.M., et al. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J Clin Microbiol. 2003;41(1):5–14. doi: 10.1128/JCM.41.1.5-14.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pillai S.K., Wennersten C., Venkataraman L., Eliopoulos G.M., Moellering R.C., Karchmer A.W. Development of reduced vancomycin susceptibility in methicillin-susceptible Staphylococcus aureus. Clin Infect Dis. 2009;49(8):1169–1174. doi: 10.1086/605636. [DOI] [PubMed] [Google Scholar]
- 14.Andersson D.I., Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8(4):260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 15.Shang W., Hu Q., Yuan W., Cheng H., Yang J., Hu Z., et al. Comparative fitness and determinants for the characteristic drug resistance of ST239-MRSA-III-t030 and ST239-MRSA-III-t037 strains isolated in China. Microb Drug Resist. 2016;22(3):185–192. doi: 10.1089/mdr.2015.0226. [DOI] [PubMed] [Google Scholar]
- 16.Peleg A.Y., Monga D., Pillai S., Mylonakis E., Moellering R.C., Jr, Eliopoulos G.M. Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J Infect Dis. 2009;199(4):532–536. doi: 10.1086/596511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majcherczyk P.A., Barblan J.L., Moreillon P., Entenza J.M. Development of glycopeptide-intermediate resistance by Staphylococcus aureus leads to attenuated infectivity in a rat model of endocarditis. Microb Pathog. 2008;45(5–6):408–414. doi: 10.1016/j.micpath.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Gardete S., Kim C., Hartmann B.M., Mwangi M., Roux C.M., Dunman P.M., et al. Genetic pathway in acquisition and loss of vancomycin resistance in a methicillin resistant S. aureus strain of clonal type USA300. PLoS Pathog. 2012;8(2) doi: 10.1371/journal.ppat.1002505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X., Hu Q., Yuan W., Shang W., Cheng H., Yuan J., et al. First report of a sequence type 239 vancomycin-intermediate Staphylococcus aureus isolate in Mainland China. Diagn Microbiol Infect Dis. 2013;77(1):64–68. doi: 10.1016/j.diagmicrobio.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Peng H., Rao Y., Yuan W., Zheng Y., Shang W., Hu Z., et al. Reconstruction of the vancomycin-susceptible Staphylococcus aureus phenotype from a vancomycin-intermediate S. aureus XN108. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y., Liu C., Li W.G., Xu J.L., Zhang W.Z., Dai Y.F., et al. Independent Microevolution Mediated by Mobile Genetic Elements of Individual Clostridium difficile Isolates from Clade 4 Revealed by Whole-Genome Sequencing. mSystems. 2019;4(2):e00252–e318. doi: 10.1128/mSystems.00252-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H., Shang W., Hu Z., Zheng Y., Yuan J., Hu Q., et al. A novel SigB(Q225P) mutation in Staphylococcus aureus retains virulence but promotes biofilm formation. Emerg Microbes Infec. 2018;7(1):1–12. doi: 10.1038/s41426-018-0078-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burts M.L., Williams W.A., DeBord K., Missiakas D.M. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc Natl Acad Sci USA. 2005;102(4):1169–1174. doi: 10.1073/pnas.0405620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu T., Wang X.Y., Cui P., Zhang Y.M., Zhang W.H., Zhang Y. The agr quorum sensing system represses persister formation through regulation of phenol soluble modulins in Staphylococcus aureus. Front Microbiol. 2017;8:2189. doi: 10.3389/fmicb.2017.02189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Liu Q., Liu Q., Gao Q., Lu H., Meng H., et al. Phylogenetic analysis and virulence determinant of the host-adapted Staphylococcus aureus lineage ST188 in China. Emerg Microbes Infect. 2018;7(1):45. doi: 10.1038/s41426-018-0048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adhikari R.P., Arvidson S., Novick R.P. A nonsense mutation in agrA accounts for the defect in agr expression and the avirulence of Staphylococcus aureus 8325–4 traP::kan. Infect Immun. 2007;75(9):4534–4540. doi: 10.1128/IAI.00679-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Cai M, Yan H, Fu J, Wu G, Zhao Z, et al. Yunnan Baiyao reduces hospital-acquired pressure ulcers via suppressing virulence gene expression and biofilm formation of Staphylococcus aureus. Int J Med Sci. 2019;16(8):1078–88. doi: 10.7150/ijms.33723. eCollection 2019. [DOI] [PMC free article] [PubMed]
- 28.Sizemore C., Buchner E., Rygus T., Witke C., Gotz F., Hillen W. Organization, promoter analysis and transcriptional regulation of the Staphylococcus xylosus xylose utilization operon. Mol Gen Genet. 1991;227(3):377–384. doi: 10.1007/BF00273926. [DOI] [PubMed] [Google Scholar]
- 29.You Y., Xue T., Cao L., Zhao L., Sun H., Sun B. Staphylococcus aureus glucose-induced biofilm accessory proteins, GbaAB, influence biofilm formation in a PIA-dependent manner. Int J Med Microbiol. 2014;304(5–6):603–612. doi: 10.1016/j.ijmm.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Zecconi A., Scali F. Staphylococcus aureus virulence factors in evasion from innate immune defenses in human and animal diseases. Immunol Lett. 2013;150(1–2):12–22. doi: 10.1016/j.imlet.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Dubrac S., Bisicchia P., Devine K.M., Msadek T. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol. 2008;70(6):1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- 32.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 33.Gao W, Cameron DR, Davies JK, Kostoulias X, Stepnell J, Tuck KL, et al. The RpoB H481Y rifampicin resistance mutation and an active stringent response reduce virulence and increase resistance to innate immune responses in Staphylococcus aureus. J Infect Dis. 2013; 207(6): 929–39. doi: 10.1093/infdis/jis772. [DOI] [PMC free article] [PubMed]
- 34.Hu J., Zhang X., Liu X., Chen C., Sun B. Mechanism of reduced vancomycin susceptibility conferred by walK mutation in community-acquired methicillin-resistant Staphylococcus aureus strain MW2. Antimicrob Agents Chemother. 2015;59(2):1352–1355. doi: 10.1128/AAC.04290-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai Y., Chang W., Zhao C., Peng J., Xu L., Lu H., et al. VraR binding to the promoter region of agr inhibits its function in vancomycin-intermediate Staphylococcus aureus (VISA) and heterogeneous VISA. Antimicrob Agents Chemother. 2017;61(5):e02740–e2816. doi: 10.1128/AAC.02740-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunman P.M., Murphy E., Haney S., Palacios D., Tucker-Kellogg G., Wu S., et al. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol. 2001;183(24):7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakoulas G., Eliopoulos G.M., Moellering R.C., Wennersten C., Venkataraman L., Novick R.P., et al. Accessory gene regulator agr locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob Agents Chemother. 2002;46(5):1492–1502. doi: 10.1128/AAC.46.5.1492-1502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harigaya Y., Ngo D., Lesse A.J., Huang V., Tsuji B.T. Characterization of heterogeneous vancomycin-intermediate resistance, MIC and accessory gene regulator agr dysfunction among clinical bloodstream isolates of Staphylococcus aureus. BMC Infect Dis. 2011;11:287. doi: 10.1186/1471-2334-11-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCallum N., Meier P.S., Heusser R., Berger-Bächi B. Mutational analyses of open reading frames within the vraSR operon and their roles in the cell wall stress response of Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55(4):1391–1402. doi: 10.1128/AAC.01213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamber S., Cheung A.L. SarZ promotes the expression of virulence factors and represses biofilm formation by modulating SarA and agr in Staphylococcus aureus. Infect Immun. 2009;77(1):419–428. doi: 10.1128/IAI.00859-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jousselin A., Kelley W.L., Barras C., Lew D.P., Renzoni A. The Staphylococcus aureus thiol/oxidative stress global regulator Spx controls trfA, a gene implicated in cell wall antibiotic resistance. Antimicrob Agents Chemother. 2013;57(7):3283–3292. doi: 10.1128/AAC.00220-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh V., Phukan U.J. Interaction of host and Staphylococcus aureus protease-system regulates virulence and pathogenicity. Med Microbiol Immunol. 2019;208(5):585–607. doi: 10.1007/s00430-018-0573-y. [DOI] [PubMed] [Google Scholar]
- 43.Geisinger E., Chen J., Novick R.P. Allele-dependent differences in quorum-sensing dynamics result in variant expression of virulence genes in Staphylococcus aureus. J Bacteriol. 2012;194(11):2854–2864. doi: 10.1128/JB.06685-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baba T., Bae T., Schneewind O., Takeuchi F., Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol. 2008;190(1):300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feiner R., Argov T., Rabinovich L., Sigal N., Borovok I., Herskovits A.A. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol. 2015;13(10):641–650. doi: 10.1038/nrmicro3527. [DOI] [PubMed] [Google Scholar]
- 46.Tran P.M., Feiss M., Kinney K.J., Salgado-Pabón W. φSa3mw prophage as a molecular regulatory switch of Staphylococcus aureus β-toxin production. J Bacteriol. 2019;201(14):e00766–e818. doi: 10.1128/JB.00766-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.