Graphical abstract
Keywords: CCNA, Cell wall remodeling, AdZAT5, AdPL5, Adβ-Gal5
Research Highlights
-
•
CCNA was advanced by introducing physiological traits.
-
•
Six cell wall genes and four transcription factors were identified for pectin degradation.
-
•
A series of experiments validated the regulations of AdZAT5 on AdPL5 and Adβ-Gal5.
-
•
CCNA would be powerful for phishing the unknown regulators with higher efficiency and accuracy.
Abstract
Introduction
Cell wall degradation and remodeling is the key factor causing fruit softening during ripening.
Objectives
To explore the mechanism underlying postharvest cell wall metabolism, a transcriptome analysis method for more precious prediction on functional genes was needed.
Methods
Kiwifruits treated by ethylene (a conventional and effective phytohormone to accelerate climacteric fruit ripening and softening as kiwifruits) or air were taken as materials. Here, Consensus Coexpression Network Analysis (CCNA), a procedure evolved from Weighted Gene Co-expression Network Analysis (WGCNA) package in R, was applied and generated 85 consensus clusters from twelve transcriptome libraries. Advanced and comprehensive modifications were achieved by combination of CCNA and WGCNA with introduction of physiological traits, including firmness, cell wall materials, cellulose, hemicellulose, water soluble pectin, covalent binding pectin and ionic soluble pectin.
Results
As a result, six cell wall metabolisms related structural genes AdGAL1, AdMAN1, AdPL1, AdPL5, Adβ-Gal5, AdPME1 and four transcription factors AdZAT5, AdDOF3, AdNAC083, AdMYBR4 were identified as hub candidate genes for pectin degradation. Dual-luciferase system and electrophoretic mobility shift assays validated that promoters of AdPL5 and Adβ-Gal5 were recognized and trans-activated by transcription factor AdZAT5. The relatively higher enzyme activities of PL and β-Gal were observed in ethylene treated kiwifruit, further emphasized the critical roles of these two pectin related genes for fruit softening. Moreover, stable transient overexpression AdZAT5 in kiwifruit significantly enhanced AdPL5 and Adβ-Gal5 expression, which confirmed the in vivo regulations between transcription factor and pectin related genes.
Conclusion
Thus, modification and application of CCNA would be powerful for the precious phishing the unknown regulators. It revealed that AdZAT5 is a key factor for pectin degradation by binding and regulating effector genes AdPL5 and Adβ-Gal5.
Introduction
Ripening is a growth phase of fruits, in which stage the appearance, texture, aroma volatiles and nutrients of fruits undergo significant changes to be more attractive for potential consumers [1]. However, over-ripe is accompanied by symptoms of senescence, such as cellular disruption, rot, pathogen infection and so on, that causes detrimental damage to fruit industry. Thus, regulation on fruit ripening is indispensable and strategically important. As a model plant, the ripening mechanisms of tomato (Solanum lycopersicum) have been comprehensively studied. A series of ripening mutants, Never-ripe (Nr) [2], Ripening-inbibitor (Rin) [3]and Colorless non-ripening (Cnr) [4], revealed several regulators in concert with ethylene perception and/or signaling related ripening. Furthermore, silencing a pectate lyase gene produced firmer fruits without sacrificing other aspects of ripening, but having prolonged shelf life and improved resistance to grey mould [5], [6]. However, due to morphological structural difference and evolutionary time divergence, some conclusions from model systems may be not totally applied in other species of fruits. For example, the softening of tomato fruit is regulated by β-galactosidase 4 (TBG4, EC 3.2.1.23) and pectate lyase (PL, EC 4.2.2.2) rather than polygalacturonase (PG, EC 3.2.1.15) according to observations in last decades [6], [7], [8], while PG is a crucial candidate of fruit softening for strawberry [9], pear [10] and apple [11]. The discrepancy of underlying ripening mechanism between tomato and other kinds of fruits emphasizes the necessary of researches in different species of fruits.
Texture change, usually with softening and limited shelf life, causes severe and direct economic deterioration during ripening of fleshy fruits [12]. It is shown that this process can be affected by several factors, including N-glycan processing [13], water loss and cellular turgor [14], [15], hydroxyl radicals attack [16], cell wall degradation and remodeling [17]. During ripening, cell walls undergo most significant changes, which involves xyloglucan-cellulose network loosening, depolymerization of polyuronide and matrix glycans, loss of pectic galactan side chains and pectic arabinan side chains, solubilisation of pectin and so on [17]. Overexpressed the key genes of cell wall loosening, xyloglucan endotransglucosylase/hydrolase (FvXTH9 and FvXTH6, EC 2.4.1.207) which were isolated from Fragaria × vesca, by infiltration in fruits of Fragaria × ananassa produced less firm fruits [18]. In kiwifruit, the transcripts of AdXTH5 and enzyme activity of XTH accumulated in a considerable abundance in ripe fruit [19], [20]. As described by recent studies, pectin plays a more important role in mechanical support for plant cell wall through interacting extensively with cellulose and filling the space left by twining cellulose microfibrils and hemicellulose [21], [22], [23]. Transgenic antisense lines of PG, degrading unesterfied pectin, exhibited reduced pectin depolymerisation, higher intercellular adhesion and firmer fruits in apple [11] and decreased cell wall disassembly, increased ionically bound and covalently bound pectin, firmer fruits and extended shelf life in strawberry [24]. Whether these identified cell wall related genes play the same role in other fruits or different mechanisms exist need to be elucidated.
Kiwifruit (Actinidia spp.) is a world-wide circulated, economic fruit crop. Soften to eaten firmness is essential for kiwifruit consumption [25]. At the commencement of softening, starch converting to sugars initiated the process, followed by pectin solubilization and deploymerization [26]. Our previous study indicated that AdDof3 is an activator for AdBAM3L, which contributed to starch degradation [27]. Consequent to starch degradation, the cell wall components, including cellulose, hemicellulose, covalent binding pectin, decreased in parallel with softening, while water soluble pectin increased on the contrary [27]. Due to the importance of softening regulation for kiwifruit, a large number of research efforts have focused on biochemical and molecular mechanisms of kiwifruit texture improvement. At physiological level, the volume of intercellular air spaces was proven to be enlarged while cell wall material shrinked together with reduction of xyloglucan molecular weight [28], [29]. Furthermore, the activities of critical enzyme, such as XTH, PG, PME, β-Gal were investigated by several reports [19], [30], [31], [32], [33], although there seem to be some conflict among them. At molecular level, it was reported that AdXTH5 can be actively regulated by AdEIL2 (EIN3-Like) and AdEIL3 to promote softening [34]. Knocking down ethylene synthesized enzyme 1-aminocyclopropane-1-carboxylic acid oxidase (ACO) retarded softening [26]. Overexpressing a zinc finger protein AdDof3 lead to an elevated expression of AdBAM3L as well as a decrease in starch content in kiwifruit leaves [27]. Although pectin underwent extraordinary changes during kiwifruit softening, compared to above, the researches about pectin-related transcriptional regulation and/or transgenic implementation were rare, which claims more exploration in the pectin related genes.
In this study, molecular basis for cell wall degradation in ripening kiwifruit were investigated. In order to increase the reliability of gene screening from transcriptome data (generated from ethylene treated and non-treated fruits), Consensus Co-Expression Network Analysis (CCNA) [35] was conducted, with the adding of consequent characteristics joint analysis. As a result, the 85 robust gene clusters were interacted with the content of cell wall materials and firmness. After intersection with differently expressed genes (DEGs) and hierarchical filtering by trait-related cluster correlations, top 10 hub genes were selected for gene function and transcriptional regulation confirmation. Dual-luciferase assay and electrophoretic mobility shift assays (EMSAs) indicated that a zinc finger transcription factor AdZAT5 positively regulated the promoters of pectin degradation related genes AdPL5 and Adβ-Gal5, which code for PL and b-Gal enzyme in kiwifruit. Transient overexpressing AdZAT5 in kiwifruit ‘Xuxiang’ (Actinidia deliciosa) resulted in the increase of AdPL5 and Adβ-Gal5 transcripts, as well as an increase of PL and β-Gal activities in transgenic kiwifruit core.
Material and methods
Plant material and treatments
Kiwifruit (Actinidia deliciosa [A. Chev.] C.F. Liang et A.R. Ferguson var. deliciosa cv. Hayward), harvested from a commercial orchard in 2015 at Shanxi, China, were subjected to two treatments, which has been described in detail in our previous study by Zhang et al., 2019 [27] Briefly, the ethylene treatment was designated as ETH; air control treatment was designated as CK. At each sampling point, ethylene production, firmness, cell wall materials (CWM), cellulose, hemicellulose, water soluble pectin (WSP), covalent binding pectin (CBP) and ionic soluble pectin (ISP) were extracted and detected previously by Zhang et al., 2019 [27]. The end of sampling in each treatment was determined according to the ethylene production peak. Each treatment and physiological trait examination was conducted with three biological replicates with five fruits in each replicate at each sampling point. The following analysis and experiments were performed based on these samples.
Kiwifruit (Actinidia deliciosa [A. Chev.] C.F. Liang et A.R. Ferguson var. deliciosa cv. Xuxiang), harvested from a commercial orchard in 2019 at Shanxi, China, were adopted as transient overexpression materials with mean firmness of 90 N. Seven fruits were used in which one fruit was injected twice as a replicate.
RNA and genomic DNA extraction
Total RNA and genomic DNA were extracted according to the CTAB (hexadecyl trimethyl ammonium bromide) methods by Wang et al., 2020 [36]. Total RNA and genomic DNA of each biological replicate were extracted independently.
cDNA synthesis and RT-qPCR
Potential genomic DNA contamination was removed before reverse transcribe by PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Beijing, China). All the operations were executed according to the instruction manual.
Real time-quantitative PCR (RT-qPCR) was conducted with LightCycler® 480 (Roche) and LightCycler® 480 SYBR Green I Master (Roche). The primers of each gene were designed referring to ‘Hongyang’ (Actinidia chinensis) kiwifruit genome [37] and checked both by melting curves and products resequencing. The compositions of PCR mix and following procedure were set according to the instruction manual. Kiwifruit Actin (GenBank no. EF063572) was employed as internal control of transcripts abundance. Primers for RT-qPCR are listed in Table S5.
Consensus Co-expression Network Analysis (CCNA)
Twelve samples of ethylene treatment and air control at 1-day storage and 4-day storage were collected to construct 12 libraries. RNA-seq was conducted on Illumina HiSeq 4000 sequencing platform with paired-end reads. The genome ‘Hong Yang’ (Actinidia chinensis, http://kiwifruitgenome.org/organism/) was adopted as reference genome. Gene expression level were estimated by Fragments Per Kilobase of transcript per Million mapped reads (FPKM) with false discovery rate. Genes were annotated from four databases: Nr (nonredundant protein)), GO (Gene Ontology), KO (KEGG ortholog database) and Swiss-Prot (a manually annotated and reviewed protein sequence database). Transcriptome data are available at GenBank SRR6885590–SRR6885601.
The scripts of consensus co-expression network analysis were obtained from Shahan et al., 2018 [35] with modifications. First of all, 80% genes except for FPKM < 1 at 12 samples (four libraries each with three replicates) were subsampled 1000 times each following by a standard Weighted Gene Co-expression Network Analysis (WGCNA) with randomized parameters (power transformation [1,2,4,8,12,16], minModuleSize [40, 60, 90, 120, 150, 180, 210], merge on eigengenes [true/false]). The according scripts are make_subsamp_wgcna.py and subsamp_wgcna.R. The number of times each gene pair clustered together were defined as matrix A, the number of times each gene pair subsampled together were defined as matrix B with scripts seq_adding_mat.R and seq_indicator_mat.R respectively. The adjacency matrix C, representing connection strength between gene pairs, was calculated by matrix A dividing matrix B with scripts merge_mats.R. Then, the matrix C as an input to replace gene pair connections of standard WGCNA and correlated with physiological trait data. Principal component analysis (PCA) was conducted by packages FactoMineR and factoextra. The R packages ggpubr and ggcor selected candidate genes.
Genes and promoters cloning
The sequences of candidate genes were mapped to genome ‘Hongyang’ (Actinidia chinensis). The primers of full-length coding sequences of AdZAT5 (Achn248441, Acc20475), AdNAC083 (Achn169681, Acc06142), AdMYBR4 (Achn217431, Acc29824, MG581952) were designed according to ‘Hongyang’ (Actinidia chinensis) reference genome. Next, candidate genes were amplified with these primers and kiwifruit ‘Hayward’ cDNA as template.
Promoters of AdPL5 (Achn315151, Acc29689, MG835626), Adβ-Gal5 (Achn294421, Acc10736, MG857556), AdPME1 (Achn102711, Acc27094) were isolated referring to ‘Red 5’ (A. chinensis) genome. Since the coding sequences (CDS) of these genes in ‘Red 5’ and 'Hayward' (A. deliciosa) were almost identical, the CDS of these genes from 'Hayward' transcriptome were aligned to 'Red 5’ genome to find the promoters of each gene. Then, the cloning of promoters was conducted by two rounds. In the first round, reverse primers were designed behind the initiation codon about 20–40 bp to make sure the promoters were from correct target genes with gDNA from ‘Hayward’ as template. In the second round, reverse primers were designed just before the initiation codon to exclude the coding sequence in final construct with plastids from first round as template. The cloning of AdGAL1 (Achn155581, Acc23330, HQ10811) promoter failed even with three pairs of primers. All the primers are listed in Table S5.
Dual-luciferase assays
The full-length coding sequences of AdZAT5 (Achn248441, Acc20475, MZ676710), AdNAC083 (Achn169681, Acc06142, MZ676711), AdMYBR4 (Achn217431, Acc29824, MG581952), cloned from last step, were inserted into pGreen II 0029 62-SK vector (SK). In the meanwhile, the promoters of AdPL5 (Achn315151, Acc29689, MG835626), Adβ-Gal5 (Achn294421, Acc10736, MG857556), AdPME1 (Achn102711, Acc27094, MZ676712) were fused with pGreen II 0800-LUC vector (LUC). Primers used for vectors constructions are listed in Table S5. The SK vector of AdDOF3 (Achn014701, Acc23702, MH105003) and LUC vectors of AdMAN1 (Achn197721, Acc04619, MH105013), AdPL1 (Achn039701, Acc18073, HQ108112) have been constructed previously by Zhang et al., 2019 [27]. All of the vectors were transferred into Agrobacterium tumefaciens GV3101 before the strains grown on Luria-Bertani (LB) medium plates with 50 µg mL−1 kanamycin and 25 µg mL−1 gentamycin. After another time of reactivation of these strains, they were resuspended in the infiltration buffer (10 mM MES, 10 mM MgCl2, 150 µM acetosyringone, pH = 5.6) and the concentration adjusted to OD600 ≈ 0.75 with UV–VIS spectrophotometer (UV-2600, Shimadzu, Japan). The suspensions of SK vectors and LUC vectors were mixed (10:1, v/v) and injected into tobacco (Nicotiana benthamiana) leaves by syringes without needles. The empty SK vector was taken as control. Three lines of tobacco were infected as three biological replicates. After three days of injection, the leaf discs near the lesion were taken and scrunched in 1 × PBS buffer (KH2PO4 2 mM, Na2HPO4 8 mM, NaCl 136 mM, KCl 2.6 mM, pH = 7.4) to detect firefly luciferase and Renilla luciferase with the Dual-Luciferase Reporter Assay System (Promega). The regulatory effects higher than 2-fold were confirmed by three independent experiments.
PL activity and β-Gal activity
The methods of extraction and enzyme activity assay of pectate lyase were referring Marín-Rodríguez et al., 2003 [38] with modifications. First, 0.5 g (0.1 g for leaves) frozen kiwifruit pulp were dissolved in 4 °C 1 mL extraction buffer (0.5 M mannitol, 0.05 M sodium phosphate (pH = 7.0), 2% β-mercaptoethanol, 1% w/v polyvinyl pyrrolidone (PVP K30), 1 mM PMSF, 10 mg L−1 leupeptin), the mixture was vortexed and extracted on ice constantly for 1 h. Then, the samples were centrifuged at 18,514 rcf for 20 min at 4 °C, the supernatant was loaded onto a Sephadex® G-25 column (Sigma) in BeyoGold™ 63 mm × 8.9 mm affinity chromatography column empty column tube (3 mL). The filtration was used for pectate lyase detection. The substrate was composed of 0.24% w/v Macklin® polygalacturonic acid (PGA), 60 mM Tris-HCl (pH = 8.5), 0.3 mM CaCl2, 1 mM PMSF, 10 mg L−1 leupeptin. To prevent the formation of insoluble calcium-PGA complexes, a 2 × PGA/ Tris-HCl (0.48% w/v) was mixed with identical volume of 2 × CaCl2 (0.6 mM). Finally, 20 µL extract or 20 µL water as control was added to 200 µL detection substrate and was incubated at 40 °C for 30 min, the absorbance at 235 nm was read with BioTek® synergy™ H1 microplate reader and Corning® UV-transparent microplate. The molar extinction coefficient was assumed to be 5200 M−1 cm−1 for unsaturated oligogalacturonates. One unit PL enzyme activity (U) is defined as the amount of kiwifruit pulp, which produces 1 mM of 4, 5-unsaturated product in 1 min under the assay conditions.
To investigate the enzyme activity of β-galactosidase, 0.5 g (0.1 g for leaves) frozen kiwifruit pulp were dissolved in 4 °C 1 mL 0.1 M citrate-sodium citrate buffer (pH = 4.6), which consist of 1 M NaCl, 13 mM EDTA, 1.5% w/v PVP, 5 mM β-mercaptoethanol, 1 mM PMSF and 10 mg L−1 leupeptin [39]. The samples were vortexed adequately and extracted on ice. After one hour, the mixture was centrifuged at 18,514 rcf for 20 min at 4 °C and the supernatant was retained for β-Gal activity examination. For detection, 80 µL crude extract was mixed with 82.5 µL substrate buffer (0.1 M citrate-sodium citrate buffer (pH = 4.6), 0.1% w/v bovine serum albumin (BSA), 13 mM p-nitrophenyl-β-galactoside (pNPG), 1 mM PMSF and 10 mg L−1 leupeptin) and reacted at 37 °C for 15 min. The reaction was terminated by adding of 125 µL 0.2 M Na2CO3. The liberated p-nitrophenol was measured at 410 nm with BioTek® synergy™ H1 microplate reader and Costar® 96 well cell culture plate. One unit of β-Gal activity (U) was defined as the amount of p-nitrophenol (µmol) produced for 1 g frozen kiwifruit pulp in 1 min, and it expressed as U/g. The p-nitrophenol was taken as standards. The calibration curve was in Table S6.
Recombinant protein and EMSA analysis
The binding sites of AdZAT5 on AdPL5 and Adβ-Gal5 promoters were predicted on PlantRegMap [40]. According to the cis-elements predicted, two pairs of primers with 3′ biotin end-labeled and two pairs of competing probes without labeled (cold probe) were designed by HuaGene (Shanghai, China) and double-stranded DNA probes were obtained by annealing complementary oligonucleotides. The probes used in EMSA are listed in Table S5.
Meanwhile, the coding sequence of AdZAT5 was combined with pET-32a vector (Novagen) and transformed to Escherichia coli Rosetta (DE3) pLysS (Novagen) by heat shock. The protein expressed by the recombinant vector was induced by 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 16 °C for 18 h. After centrifugation at 6000 rcf for 20 min at room temperature, the precipitate was lysed with 1 × PBS buffer and frozen-thawed from −80 °C to 4 °C. The cells were crushed further by sonication on ice at 200 W with 3 s intervals for about 20 min. The supernatant obtained from centrifugation with 7000 rcf for 10 min was filtered by 0.45 µm Millpore Express® PES Membrane, subsequently by His-tag Affinity Chromatographic Column (Transgene). The eluted protein was confirmed by SDS-PAGE and residual protein was used in the following EMSA experiment. LightShift Chemiluminescent EMSA kit (ThermoFisher Scientific) together with AdZAT5 recombinant protein and labeled probes as well as cold probes were employed to identify the binding sites on promoters.
Subcellular localization of AdZAT5
The coding sequence without stop codon of AdZAT5 was amplified and inserted into 35S-eGFP vector. The construct was transformed to Agrobacterium tumefaciens GV3101 and transiently expressed in transgenic N. benthamiana leaves with nucleus-located mCherry. After three days of infiltration, the GFP and RFP fluorescence was imaged with a Zeiss LSM710NLO confocal laser-scanning microscope. The primers are listed in Table S5.
Transient overexpression in ‘Xuxiang’ kiwifruit
The full length of AdZAT5 was inserted into pSAK277 vector [41] and transformed into Agrobacterium tumefaciens EHA105. The infiltrations were adjusted to OD600 ≈ 0.75 with the same buffer in dual-luciferase assays. After two hours culturing in dark at 28 °C, blue or red ink was added separately in empty pSAK277 suspension or AdZAT5 suspension to indicate the diffusion area. Either 0.1 mL of AdZAT5 or empty vector suspension was injected in separate end of ‘Xuxiang’ kiwifruit core tissue. Then, injected fruits were stored at 28 °C for three days and samples were collected in liquid nitrogen with each fruit as a replicate and stored at −80 °C for RNA extraction.
Statistical analysis
GraphPad Prism (Version 8.0.0 (131)) was used to compute least-significant difference (LSD) and t-test. Softewares R with related packages and Adobe Photoshop CC 2017, Adobe Illustrator CC 2017 were employed to handle with pictures.
Accession numbers
Sequence data from this article have been deposited in the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) under the following accession numbers: AdZAT5, MZ676710; AdNAC083, MZ676711; AdPME1, MZ676712. The accession numbers of other genes can be found in Table S4.
Results
CCNA comprehensive analysis of kiwifruit transcriptome data
Twelve libraries (ETH1d, CK1d, ETH4d, CK4d, with three replicates in each) were constructed and sequenced [27]. To affirm the effectiveness of ethylene treatment on gene expression, FPKM from these libraries were dimensionality reduced and compared by PCA and a preliminary clustering (Fig. S1). On the second principal component (1.6% and 7.9%, respectively), ETH1d vs CK1d and ETH4d vs CK4d were both separated apparently, meanwhile, the clustering of samples based on their Euclidean distance split ETH and CK into two disparate clades. All these indicated that the differences between ethylene treatment and air control are sufficient for further analysis.
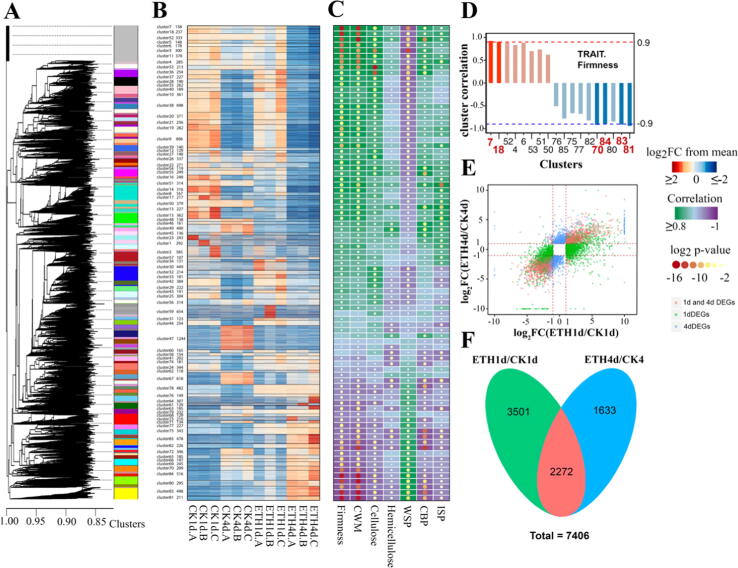
Genes with FPKM < 1 at 12 libraries were excluded, the remain 24191genes were put into the procedure of CCNA, starting by subpopulation before standard WGCNA [42] with random parameters (Table S1). After 1000 times iterations, the weighted adjacency matrix was calculated then used as an input to WGCNA to obtain the consensus clusters (Fig. 1A, B and Table S1). To precisely predict the candidate genes underlying kiwifruit softening, a modification was conducted from this step compared to original CCNA. The gene clusters with narrowing down expression profiles were specifically distributed to cell wall related trait data from our previous report [27], including firmness, cell wall materials, cellulose, hemicellulose, water soluble pectin, covalent binding pectin, ionic soluble pectin, and corresponding correlations were calculated (Fig. 1C and Table S1). For each trait, specific correlation coefficient was set to ±0.9 for firmness and cell wall materials, and ±0.8 for the others (Fig. 1D and Fig. S2). When foldchange was set as two, 5773 DEGs in ETH1d vs CK1d and 3905 DEGs in ETH4d vs CK4d were identified (Fig. 1, E and F, and Table S1).
Fig. 1.
Transcriptome analysis and cluster-trait associations. (A) Dendrogram and CCNA clusters of transcriptome data. (B) Gene expression profiles of 85 clusters in 12 transcriptome libraries. (C) Interactions between 85 clusters and traits, including firmness, cell wall materials (CWM), cellulose, hemicellulose, water soluble pectin (WSP), covalent binding pectin (CBP) and ionic soluble pectin (ISP). (D) Cluster correlations of firmness trait. The others were listed in Fig. S2. (E and F) Different expressed genes in one day after ethylene treatment /one day after air control (ETH1d/CK1d) and four days after ethylene treatment /four days after air control (ETH4d/CK4d).
Candidate genes screened by the association of consensus clusters and cell wall related traits
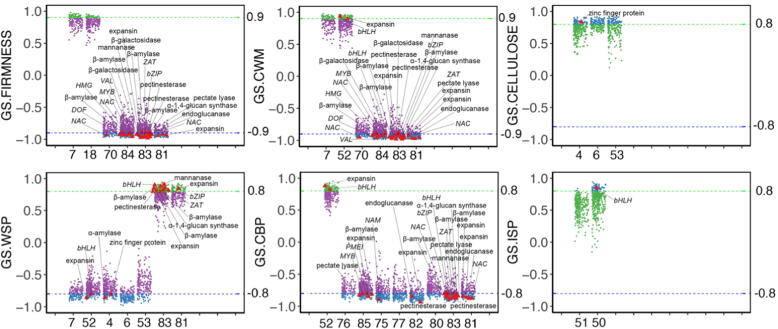
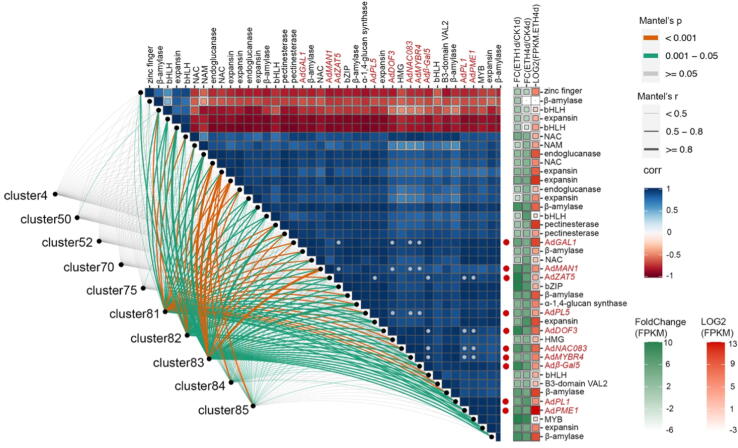
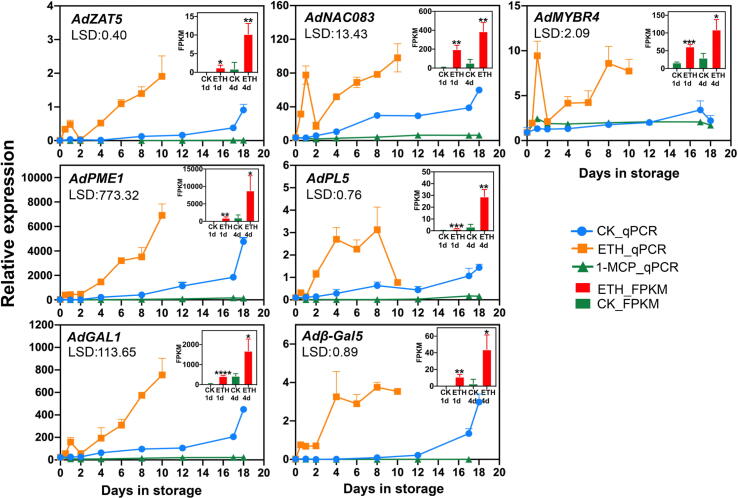
To further locate candidate genes accurately, more stringent rules were set to individual gene in each candidate clusters. Under the condition of absolute value of GS higher than 0.9 for firmness and cell wall materials and 0.8 for the other traits, 40 candidate genes were selected (Fig. 2 and Table S3). Again, the trait data we employed are one-orientation directed, cell wall associated fruit softening. Genes involved in cell wall metabolism and with more abundant transcripts in ETH4d, as well as higher foldchange in ETH4d vs CK4d were picked first, that are β-D-galactosidase 1 (AdGAL1, Achn155581, Acc23330, HQ10811), endo-β-mannanase1 (AdMAN1, Achn197721, Acc04619, MH105013), pectate lyase 1 (AdPL1, Achn039701, Acc18073, HQ108112), pectate lyase 5 (AdPL5, Achn315151, Acc29689, MG835626), β-galactosidase 5 (Adβ-Gal5, Achn294421, Acc10736, MG857556), pectinesterase 1 (AdPME1, Achn102711, Acc27094) (Fig. 3 and Table S4). Furthermore, the transcription factors potentially regulated these genes were also screened with the same rules. They are C2H2 zinc finger protein 5 (AdZAT5, Achn248441, Acc20475), Dof zinc finger protein 3 (AdDOF3, Achn014701, Acc23702, MH105003), AdNAC083 (Achn169681, Acc06142), AdMYBR4 (Achn217431, Acc29824, MG581952) (Fig. 3 and Table S4). The expression profiles of these genes in ethylene treatment and in air control were verified by qRT-PCR (Fig. 4) except for AdMAN1, AdPL1, and AdDOF3 which were included in our previous research [27]. At the same time, the relative expression of candidate genes in 1-Methylcyclopropene (1-MCP, 1 µL L−1, 20 °C), an antagonist of the ethylene receptor, were identified to verify the effect of repressing treatment (Fig. 4).
Fig. 2.
Gene significance of individual gene in candidate clusters with physiological traits. Physiological traits include firmness, cell wall materials (CWM), cellulose, water soluble pectin (WSP), covalent binding pectin (CBP) and ionic soluble pectin (ISP), with one for each subplot. The x axis in each subplot indicates the number of consensus cluster, while y axis indicates gene significance (GS). In GS.FIRMNESS, GS.CWM, GS.WSP and GS.CBP subplots, the red triangles refer to candidate genes, green dots refer to genes with GS higher than 0.9 or 0.8, blue dots refer to genes with GS lower than −0.9 or −0.8, purple dots refer to genes with GS between −0.8 to 0.8 or between −0.9 to 0.9. In GS.CELLULOSE and GS.ISP subplots, the red triangles refer to candidate genes, blue dots represent genes with GS higher than 0.8, green dots represent genes with GS lower than 0.8. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Criteria of candidate genes screening. Including correlation coefficients among 40 candidate genes, their belongings to clusters, fold-change of one day after ethylene treatment /one day after air control (FC(ETH1d/CK1d)) and four days after ethylene treatment /four days after air control (FC(ETH4d/CK4d)), FPKM in ETH 4d. Gene names with red color and highlighted by red dots indicate 10 candidate genes. The grey dots refer to correlation coefficients between six candidate transcription factors and four candidate structural genes. The values are indicated by color bars. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Relative expression profiles and FPKM of candidate genes in ethylene treatment (ETH, 100 µL L−1 for 24 h at 20 °C), in air control (CK, 20 °C) and in 1-Methylcyclopropene treatment (1-MCP, 1 µL L−1, 20 °C). The expression profiles of AdDof3 and AdPL1, AdMAN1 were verified by Zhang et al., 2019. Error bars represent SE based on three replicates. Least Significant Difference (LSD) values represent LSD at p < 0.05.
In vivo and in vitro regulations of AdZAT5 on AdPL5 and Adβ-Gal5
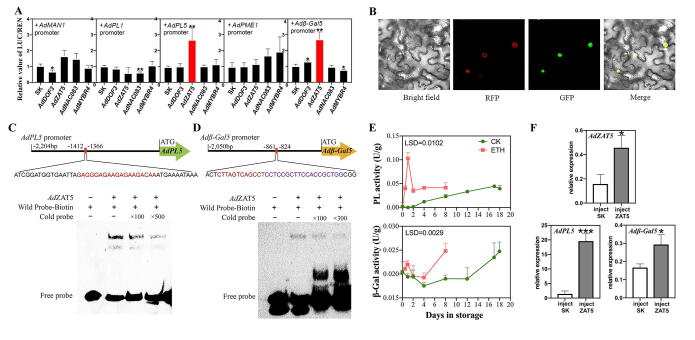
To assess the potential effects of candidate transcription factors on candidate cell wall related genes, dual-luciferase reporter assay system was taken up with pGreen II 0029 62-SK vectors fused into coding sequences of four transcription factors, pGreen II 0800-LUC vectors recombined to promoters of five structural genes except for AdGAL1. Three pairs of primers designed based on the genome sequence of ‘Red 5’ failed to clone the promoter of AdGAL1 (Table S5). It turned out that 2.7- and 2.7-fold inductions were observed from AdZAT5 on AdPL5 and Adβ-Gal5 promoters, respectively (Fig. 5A). The regulations of AdDOF3 on AdMAN1 and AdPL1 were reported by our previous work [27], which were also repeated (Fig. 5A, grey bars).
Fig. 5.
Investigation and confirmation of regulations between candidate transcription factors and structural genes. (A) Regulatory verified by dual-luciferase assay. (B) Subcellular localization of AdZAT5. (C and D) Electrophoretic mobility shift assays (EMSA) of recombinant AdZAT5 protein binding cis-elements on AdPL5 and Adβ-Gal5 promoters. (E) Enzyme activities of pectate lyase and β-galactosidase. (F) Relative expression of AdZAT5, AdPL5, Adβ-Gal5 in transient overexpression ‘Xuxiang’ kiwifruit core tissue. The two ends of core tissue were injected with empty vector as control or AdZAT5 recombinant vector. Error bars in A indicate SE from three replicates (*, p < 0.05; **, p < 0.01; and ***, p < 0.001). Error bars represent SE based on three replicates. Least Significant Difference (LSD) values represent LSD at p < 0.05.
Subcellular localization revealed that AdZAT5 is expressed in nucleus (Fig. 5B). Then, the promoter sequences of AdPL5 and Adβ-Gal5 were put into PlantRegMap [40] database to predict cis-elements. It came out that GAGGGAGAAGAGAAGACAA (−1412 bp to −1366 bp) in AdPL5 and CTTAGTCAGCCTCCTCCGC and CCTCCGCTTCCACCGCTGG (−861 bp to −824 bp) in Adβ-Gal5 are potential binding domains of C2H2 zinc finger protein. Thus, corresponding biotin probes were designed and prokaryotic expressed AdZAT5 protein was purified. Finally, EMSA assay indicated that AdZAT5 could physically bind to AdPL5 and Adβ-Gal5 promoters (Fig. 5C and D).
In addition of the molecular level, the enzyme activity of PL and β-Gal were examined (Fig. 5E). The results indicated that the PL activity was significantly enhanced by ethylene treatment and raced up sharply to 0.1 U/g at 1 d, then dropped down and maintained at 0.03–0.04 U/g from 2 d to 8 d. In control fruit, PL activity increased gradually and peaked parallel with ethylene burst peak. On the other hand, β-Gal activity also showed the transient responses to exogenous ethylene treatment and later enhancement (at 8 d), which is earlier than that in control fruit (Fig. 5E). Thus, molecular (mRNA abundance) and biochemical (enzyme activities) evidences pointed out the importance of PL and β-Gal for kiwifruit postharvest ripening.
Transient overexpression analysis of AdZAT5 indicated regulations on AdPL5 and Adβ-Gal5
Transient overexpression of AdZAT5 in mature ‘Xuxiang’ kiwifruit core tissue was analyzed. It resulted that the gene expression of AdPL5 was successfully provoked about 13-fold higher (p < 0.001) by injection of AdZAT5. Similarly, the expression profile of Adβ-Gal5 exhibited 70% (p < 0.05) increase in samples injecting AdZAT5 (Fig. 5F).
Discussion
RNA-sequencing (RNA-seq) is one of the widely-used omics and with the development of sequencing techniques, it becomes more attractive for the gradually lower cost and the emergence of third sequencing technology PacBio. RNA-seq has been applied in a wide range in analyzing critical genes of fruit ripening, such as apricot [43], persimmon [44], strawberry [45] and kiwifruit [27], [46]. In our earlier work, the selection of candidate genes was mainly depending on foldchange [27] or standard WGCNA [46]. Although several regulated genes were identified, most of the candidate genes, produced by the analysis procedures, did not possess functions [27], [46]. Similarly, the same situation was also found in other fruit development and ripening research [47], [48]. So, it is quite meaningful and necessary to explore how to modify RNA-seq analysis procedure with more precise prediction and is extremely vital for researches using RNA-seq. It has been demonstrated that CCNA is an optimal protocol to propose a hypothesis with efficiency and accuracy [35]. We adopted and improved this method by combining CCNA with physiological trait in kiwifruit ripening research.
Attempt and advantage of combination of CCNA and WGCNA analysis on the transcriptome data
The attempts to minimize the effects of user-picked parameters and increase the stability of gene modules in WGCNA have been tried several times [48], [49]. In similarity across bootstrap re-sampling (SARE) procedure, a re-sampling strategy was adopted to assess gene module stability of WGCNA [50]. Re-sampling and repeat runs were also used in Consensus Clustering to strengthen the robustness of clusters [49]. In CCNA, 80% genes of original dataset were sub-sampled for subsequent WGCNA with random parameters, such as minimum module size, power transformation, and merging on eigengenes [49], [50]. Constantly, this process was iterated for 1000 times for the purpose of more stable clusters without sacrificing the high intracluster similarity or low intercluster similarity produced by WGCNA. Similarly, this methodology was compared with standard WGCNA and proved to be more reliable with higher bootstrap confidence interval, which assure the accuracy of genes assignment [35]. However, such analysis stopping at gene pairs correlations, such as the explorations on critical candidate genes using developmental flowers, without corresponding phenomenon- or physiology- data. It is worth to emphasize that the applications of transcriptomic analyses were usually employed to unveil the underlying regulatory genes for specific phenomenon- or physiology- data. With the introduction of physiological data in CCNA in this research, the candidate regulator and effector gene sets were narrowed down to 10 genes, while there are 388 cell wall related genes (Table S6) and enormous transcription factors in kiwifruit genome. Besides, consequent dual-luciferase assay, EMSA and transient overexpression system verified the hypothesis experimentally with 3/10 accuracy (Fig. 5A). If the candidate AdBAM3L, which was reported in our previous article [27] was counted, the percentage will be 4/11. However, the accuracy of our previous simple and rough analysis method was 5/26 [27]. As for workload, there were 24 gene-interactions (six transcription factors × four structural genes) needed to be verified, while it was 168 gene-interactions (14 transcription factors × 12 structural genes) in previous work [27], which was 7-fold labors consuming. Moreover, the attempts on combination of CCNA and WGCNA analysis on the transcriptome data were rarely reported. Thus, application of CCNA and/or WGCNA required further attempts and modifications.
Here, 85 consensus clusters were further correlated with several cell walls associated traits, and specific candidate hub genes with close interactions with fruit softening were selected (Fig. 3). In principle, the interactions between gene modules and trait dataset is calculated by the built-in function of WGCNA with two respective matrixes. The gene module matrix in WGCNA and consensus clusters in CCNA are essentially matrixes representing correlations of gene pairs. Thus, the advanced steps in present analysis used the consensus clusters in CCNA to replace the gene module matrix in WGCNA. With such modification, the candidate clusters were specifically correlated with related to cell wall metabolism. Again, the combination of CCNA and WGCNA analysis were limited, such experiment design could be applied extensively in other organisms with different physiological traits, development phases or various tissues.
Characterized the key genes for kiwifruit cell wall remodeling
Fruit softening is a complex trait controlled by multiple quantitative trait loci [51]. Application of CCNA and WGCNA centered the key candidate genes for kiwifruit postharvest softening, including AdGAL1, AdMAN1, AdPL1, AdPL5, Adβ-Gal5, AdPME1 (Fig. 3 and Table S4). The characterized six genes were far less than the existing cell wall related genes that annotated within kiwifruit genome (388 genes, Table S6). Taken together physiology data [27] and the following transcriptional regulation investigations (Fig. 5), two key enzymes (PL and β-Gal) for kiwifruit pectin solubilization were also investigated. The results indicated that both PL and β-Gal activities were positively correlated with kiwifruit softening and were enhanced by exogenous ethylene treatment (Fig. 5). Gene family analysis with Arabidopsis and tomato demonstrated that AdPL1, AdPL4 and AdPL5 were the major members among PL family, AdGAL1 and Adβ-Gal5 were the major members among β-Gal family. Among β-Gal family, PSS15514.1 (Achn155581) AdGAL1 (Achn155581) share the same ID in ‘Hongyang’ genome and share the same FPKM in transcriptome (Fig. S3). Similar changes on transcripts and enzyme activities were observed in various fruits. For example, the high expression of PL was observed earlier in strawberry [52], banana [53] and grape berry [54]. While hindered by technology and few attentions, PL enzyme activity was detected successfully first in banana fruit at 2003 and proved to be ripening-related [38]. Similarly, β-galactosidase II activity increased markedly during tomato ripening and the increment contributed to net galactosyl loss [38], [55]. The mRNA level of TBG4 corresponding to β-galactosidase II protein peaked at turning stage [56] and analogous phenomena were also observed in pear [57]. What’s more, the same enzyme seems to act in a different way in various fruits. According to the reports, PL and β-Gal play more crucial roles in tomatoes [5], [55], while PG and XTH affected the strawberry and apple firmness most [9], [11], [18]. Due to the difficulty of the stable transformation for fruit researches, most of these investigations mainly focused on gene expression and biochemical level.
Recently, Uluisik et al., 2016 [5] silenced a PL gene by antisense approach and produced significantly firmer tomatoes with reduced PL activity. Meanwhile, the same PL gene was reported by Yang et al., 2017 [6] and proved to prolong shelf-life. Then, CRISPR mutant lines of PL exhibited firmer fruits compared with that of PG2a and TBG4 [58]. At this point, the importance of PL in cell wall remodeling and fruit softening has been aware by a growing number of researchers. However, the application of stable transgenic transformation in perennial fruits is limited in a range of extend by technological difficulty. Till now, only a few species, such as papaya [59], apple [60] and kiwifruit [27], [36], [61], could adopt this technique. Here, AdPL and Adβ-Gal expressions were enhanced in ox-AdZAT5 with 13-fold (p < 0.001) and 0.7-fold (p < 0.05), respectively (Fig. 5). Thus, the precious prediction on softening related genes would guide the further transgenic works in kiwifruit.
Transcription factors AdZAT5 coordinated AdPL5 and Adβ-Gal5 for fruit softening and ripening regulation
The transcriptional regulatory mechanisms underlying fruit softening has been widely investigated in various fruits. It is believed that there are master regulators to switch on/off downstream structural genes of fruit texture [62]. For example, starch degradation enzymes could be activated by MabHLH6 in banana [63] and AdDof3 in kiwifruit [27], DkERF8/16/19 could bind DxXTH9 and promote its expression to accelerate persimmon softening [44], cell wall modifying enzymes PG were regulated by MdEIL3 and MdCBF2 in apple [64]. RNA interference and overexpression lines of FaRIF (a NAC transcription factor) indicated that this transcription factor extensively regulated many aspects of strawberry fruit ripening, including cell wall degradation and modification [45]. Through above reports, it is easy to find that the phenomena of several transcription factors regulating one structural gene or several structural genes affected by one transcription factor are prevalent.
Again, using CCNA and WGCNA analysis, only four transcription factors were raised, including newly characterized AdZAT5, AdNAC083 and AdMYBR4 (Fig. 4) and previously reported AdDof3 [27]. Among them, AdZAT5 showed trans-activations on promoters of AdPL5 and Adβ-Gal5 (Fig. 5). Meanwhile, transient overexpression lines of AdZAT5 exhibited upregulations of AdPL5 and Adβ-Gal5 (Fig. 5), which confirmed the in vivo regulatory links in kiwifruit and also suggested the AdZAT5 is a potential regulator for kiwifruit softening by controlling the two structural genes within pectin degradation pathway. Moreover, PL gene was recently characterized as the key gene for tomato fruit texture regulation [5], [6]. However, the upstream regulators for PL gene were rarely investigated. Thus, the finding of AdZAT5 provided the one of the uncharacterized master regulators for PL gene and fruit softening.
Conclusions
In order to perform more precious prediction on candidate genes for quantitative traits (eg. fruit softening), combination and modification on CCNA and WGCNA were performed in present research. This methodology was modified with combination of physiological trait data so as to expand the application scope to multiple organisms in various treatments or different development phases. The output of this analysis centered on very limited numbers of structural genes and transcription factors for kiwifruit softening. With gene expression, enzyme activity, transcriptional regulatory analysis, as well as the stable transformation in kiwifruit, AdZAT5 was characterized as a positive regulator of kiwifruit softening by binding to AdPL5 and Adβ-Gal5 promoters as well as promoting their transcriptions. These analyses and results not only provided the insights for kiwifruit softening regulations, but also showed the powerful of precious predictions with modified approach for transcriptome analysis, which would be useful for more extensive applications in other plants and phenomena.
Compliance with Ethics Requirement
There was no use of any animals or human patients.
CRediT authorship contribution statement
Qiu-yun Zhang: Conceptualization, Methodology, Investigation, Writing – original draft, Visualization. Jun Ge: Software, Formal analysis. Xin-cheng Liu: Validation, Formal analysis. Wen-qiu Wang: Methodology, Resources. Xiao-fen Liu: Methodology, Resources. Xue-ren Yin: Conceptualization, Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We would like to appreciate Dr. Andrew Allan (Plant&Food Research, NZ) for providing pSAK277 vector. This research was supported by the National Key Research and Development Program (2018YFD1000200), the National Natural Science Foundation of China (32072635), the Key Research and Development Program of Zhejiang Province (2021C02015), China Postdoctoral Science Foundation (2020107), Fruit New Varieties Breeding Project of Zhejiang Province (2021C02066-8).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.11.019.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Grierson D. Ethylene and the control of fruit ripening. In: Seymour GB, Poole M, Giovannoni JJ, Tucker GA (eds.), The Molecular Biology and Biochemistry of Fruit Ripening. John Wiley & Sons, Inc. First Edition; 2013. p. 43–73.
- 2.Lanahan M.B., Yen H.C., Giovannoni J.J., Klee H.J. The Never Ripe mutation blocks ethylene perception in tomato. Plant Cell. 1994;6:521–530. doi: 10.1105/tpc.6.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vrebalov J., Ruezinsky D., Padmanabhan V., White R., Medrano D., Drake R., et al. A MADS-box gene necessary for fruit ripening at the tomato Ripening-Inhibitor (Rin) Locus. Science. 2002;296(5566):343–346. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- 4.Manning K., Tör M., Poole M., Hong Y., Thompson A.J., King G.J., et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38(8):948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 5.Uluisik S., Chapman N.H., Smith R., Poole M., Adams G., Gillis R.B., et al. Genetic improvement of tomato by targeted control of fruit softening. Nat Biotechnol. 2016;34(9):950–952. doi: 10.1038/nbt.3602. [DOI] [PubMed] [Google Scholar]
- 6.Yang L., Huang W., Xiong F., Xian Z., Su D., Ren M., et al. Silencin of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol J. 2017;15:1544–1555. doi: 10.1111/pbi.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giovannoni J.J., DellaPenna D., Bennett A.B., Fischer R.L. Expression of a chimeric Polygalacturonase gene in transgenic rin (Ripening Inhibitor) tomato fruit results in polyuronide degradation but not fruit softening. Plant Cell. 1989;1:53–63. doi: 10.1105/tpc.1.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith D.L., Abbott J.A., Gross K.C. Down-regulation of tomato β-Galactosidase 4 results in decreased fruit softening. Plant Physiol. 2002;129:1755–1762. doi: 10.1104/pp.011025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paniagua C., Ric-Varas P., García-Gago J.A., López-Casado G., Blanco-Portales R., Muñoz-Blanco J., et al. Elucidating the role of polygalacturonase genes in strawberry fruit softening. J Exp Bot. 2020;71:7103–7117. doi: 10.1093/jxb/eraa398. [DOI] [PubMed] [Google Scholar]
- 10.Zhang S., Ma M., Zhang H., Zhang S., Qian M., Zhang Z., et al. Genome-wide analysis of polygalacturonase gene family from pear genome and identification of the member involved in pear softening. BMC Plant Biol. 2019;19(1) doi: 10.1186/s12870-019-2168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atkinson R.G., Sutherland P.W., Johnston S.L., Gunaseelan K., Hallett I.C., Mitra D., et al. Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus x domestica) fruit. BMC Plant Biol. 2012;12(1) doi: 10.1186/1471-2229-12-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seymour G.B., Chapman N.H., Chew B.L., Rose J.K.C. Regulation of ripening and opportunities for control in tomato and other fruits. Plant Biotechnol J. 2013;11(3):269–278. doi: 10.1111/j.1467-7652.2012.00738.x. [DOI] [PubMed] [Google Scholar]
- 13.Meli V.S., Ghosh S., Prabha T.N., Chakraborty N., Chakraborty S., Datta A. Enhancement of fruit shelf life by suppressing N-glycan processing enzymes. Proc Natl Acad Sci USA. 2010;107(6):2413–2418. doi: 10.1073/pnas.0909329107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saladié M., Matas A.J., Isaacson T., Jenks M.A., Goodwin S.M., Niklas K.J., et al. A reevaluation of the key factors that influence tomato fruit softening and integrity. Plant Physiol. 2007;144:1012–1028. doi: 10.1104/pp.107.097477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romero P., Rose J.K.C. relationship between tomato fruit softening, cuticle properties and water availability. Food Chem. 2019;295:300–310. doi: 10.1016/j.foodchem.2019.05.118. [DOI] [PubMed] [Google Scholar]
- 16.Fry S.C., Dumville J.C., Miller J.G. Fingerprinting of polysaccharides attacked by hydroxyl radicals in vitro and in the cell walls of ripening pear fruit. Biochem J. 2001;357(3):729–737. doi: 10.1042/0264-6021:3570729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brummell D.A. Cell wall disassembly in ripening fruit. Funct Plant Biol. 2006;33(2):103. doi: 10.1071/FP05234. [DOI] [PubMed] [Google Scholar]
- 18.Witasari L.D., Huang F.C., Hoffmann T., Rozhon W., Fry S.C., Schwab W. Higher expression of the strawberry xyloglucan endotransglucosylase/hydrolase genes FvXTH9 and FvXTH6 accelerates fruit ripening. Plant J. 2019;100:1237–1253. doi: 10.1111/tpj.14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redgwell R.J., Fry S.C. Xyloglucan endotransglycosylase activity increases during kiwifruit ripening. Plant Physiol. 1993;103:1399–1406. doi: 10.1104/pp.103.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atkinson R.G., Johnston S.L., Yauk Y.-K., Sharma N.N., Schröder R. Analysis of xyloglucan endotransglucosylase/hydrolase (XTH) gene families in kiwifruit and apple. Postharvest Biol Technol. 2009;51(2):149–157. [Google Scholar]
- 21.Zykwinska A.W., Ralet M.-C.J., Garnier C.D., Thibault J.-F.J. Evidence for in vitro binding of pectin side chains to cellulose. Plant Physiol. 2005;139:397–407. doi: 10.1104/pp.105.065912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dick-Pérez M., Zhang Y., Hayes J., Salazar A., Zabotina O.A., Hong M. Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry. 2011;50(6):989–1000. doi: 10.1021/bi101795q. [DOI] [PubMed] [Google Scholar]
- 23.Wang T., Park Y.B., Cosgrove D.J., Hong M. Cellulose-pectin spatial contacts are inherent to never-dried Arabidopsis primary cell walls: evidence from solid-state nuclear magnetic resonance. Plant Physiol. 2015;168(3):871–884. doi: 10.1104/pp.15.00665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posé S., Paniagua C., Cifuentes M., Blanco-Portales R., Quesada M.A., Mercado J.A. Insights into the effects of polygalacturonase FaPG1 gene silencing on pectin matrix disassembly, enhanced tissue integrity, and firmness in ripe strawberry fruits. J Exp Bot. 2013;64:3803–3815. doi: 10.1093/jxb/ert210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ilina N., Alem H.J., Pagano E.A., Sozzi G.O. Suppression of ethylene perception after exposure to cooling conditions delays the progress of softening in ‘Hayward’ kiwifruit. Postharvest Biol Technol. 2010;55(3):160–168. [Google Scholar]
- 26.Atkinson R.G., Gunaseelan K., Wang M.Y., Luo L., Wang T., Norling C.L., et al. Dissecting the role of climacteric ethylene in kiwifruit (Actinidia chinensis) ripening using a 1-aminocyclopropane-1-carboxylic acid oxidase knockdown line. J Exp Bot. 2011;62:3821–3835. doi: 10.1093/jxb/err063. [DOI] [PubMed] [Google Scholar]
- 27.Zhang A.-d., Wang W.-Q., Tong Y., Li M.-J., Grierson D., Ferguson I., et al. Transcriptome analysis identifies a zinc finger protein regulating starch degradation in kiwifruit. Plant Physiol. 2018;178(2):850–863. doi: 10.1104/pp.18.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redgwell R.J., Melton L.D., Brasch D.J. Cell-wall polysaccharides of kiwifruit (Actinidia deliciosa): effect of ripening on the structural features of cell-wall materials. Carbohydr Res. 1991;209:191–202. [Google Scholar]
- 29.Hallett I.C., Macrae E.A., Wegrzyn T.F. Changes in kiwifruit cell wall ultrastructure and cell packing during postharvest ripening. Int J Plant Sci. 1992;153(1):49–60. [Google Scholar]
- 30.Wegrzyn T.F., MacRae E.A. Pectinesterase, polygalacturonase, and β-galactosidase during softening of ethylene-treated kiwifruit. HortScience. 1992;27(8):900–902. [Google Scholar]
- 31.Bonghi C., Pagni S., Vidrih R., Ramina A., Tonutti P. Cell wall hydrolases and amylase in kiwifruit softening. Postharvest Biol Technol. 1996;9(1):19–29. [Google Scholar]
- 32.Fullerton C.G., Prakash R., Ninan A.S., Atkinson R.G., Schaffer R.J., Hallett I.C., et al. Fruit from two kiwifruit genotypes with contrasting softening rates show differences in the xyloglucan and pectin domains of the cell wall. Front Plant Sci. 2020;11:964. doi: 10.3389/fpls.2020.00964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang W., Chen M., Zhao T., Han F., Zhang Q.i., Liu X., et al. Genome-wide identification and expression analysis of polygalacturonase gene family in kiwifruit (Actinidia chinensis) during fruit softening. Plants. 2020;9(3):327. doi: 10.3390/plants9030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin X.R., Allan A.C., Chen K.S., Ferguson I.B. Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol. 2010;153:1280–1292. doi: 10.1104/pp.110.157081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shahan R., Zawora C., Wight H., Sittmann J., Wang W., Mount S.M., et al. Consensus coexpression network analysis identifies key regulators of flower and fruit development in wild strawberry. Plant Physiol. 2018;178(1):202–216. doi: 10.1104/pp.18.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W.-Q., Wang J., Wu Y.-Y., Li D.-W., Allan A.C., Yin X.-R. Genome-wide analysis of coding and non-coding RNA reveals a conserved miR164-NAC regulatory pathway for fruit ripening. New Phytol. 2020;225(4):1618–1634. doi: 10.1111/nph.16233. [DOI] [PubMed] [Google Scholar]
- 37.Pilkington S.M., Crowhurst R., Hilario E., Nardozza S., Fraser L., Peng Y.Y., et al. A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Bioinf. 2018;19:257. doi: 10.1186/s12864-018-4656-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marín-Rodríguez M.C., Smith D.L., Manning K., Orchard J., Seymour G.B. Pectate lyase gene expression and enzyme activity in ripening banana fruit. Plant Mol Biol. 2003;51:851–857. doi: 10.1023/a:1023057202847. [DOI] [PubMed] [Google Scholar]
- 39.Pressey R. β-Galactosidases in Ripening Tomatoes. Plant Physiol. 1983;71(1):132–135. doi: 10.1104/pp.71.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian F., Yang D.C., Meng Y.Q., Jin J., Gao G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 2020;48:1104–1113. doi: 10.1093/nar/gkz1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng Y.Y., Lin-Wang K., Cooney J.M., Wang T.C., Espley R.V., Allan A.C. Differential regulation of the anthocyanin profile in purple kiwifruit (Actinidia species) Hortic Res. 2019;6:3. doi: 10.1038/s41438-018-0076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langfelder P., Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Q.Y., Feng C., Li W.H., Qu Z.H., Zeng M., Xi W.P. Transcriptional regulatory networks controlling taste and aroma quality of apricot (Prunus armeniaca L.) fruit during ripening. BMC Genomics. 2019;20:45. doi: 10.1186/s12864-019-5424-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang M.-M., Zhu Q.-G., Deng C.-l., Luo Z.-R., Sun N.-J., Grierson D., et al. Hypoxia-responsive ERFs involved in post deastringency softening of persimmon fruit. Plant Biotechnol J. 2017;15(11):1409–1419. doi: 10.1111/pbi.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martín-Pizarro C., Vallarino J.G., Osorio S., Meco V., Urrutia M., Pillet J., et al. The NAC transcription factor FaRIF controls fruit ripening in strawberry. Plant Cell. 2021;33:1574–1593. doi: 10.1093/plcell/koab070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang A., Zhang Q., Li J., Gong H., Fan X., Yang Y., et al. Transcriptome co-expression network analysis identifies key genes and regulators of ripening kiwifruit ester biosynthesis. BMC Plant Biol. 2020;20(1) doi: 10.1186/s12870-020-2314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hollender C.A., Kang C., Darwish O., Geretz A., Matthews B.F., Slovin J., et al. Floral transcriptomes in woodland strawberry uncover developing receptacle and anther gene networks. Plant Physiol. 2014;165(3):1062–1075. doi: 10.1104/pp.114.237529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L.N., Zhang Q.Y., Li W.H., Zhang S.K., Xi W.P. Identification of key genes and regulators associated with carotenoid metabolism in apricot (Prunus armeniaca) fruit using weighted gene coexpression network analysis. BMC Genomics. 2019;20:876. doi: 10.1186/s12864-019-6261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monti S., Tamayo P., Mesirov J., Golub T. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Machine Language. 2003;52:91–118. [Google Scholar]
- 50.Shannon C.P., Chen V., Takhar M., Hollander Z., Balshaw R., McManus B.M., et al. SABRE: a method for assessing the stability of gene modules in complex tissues and subject populations. BMC Bioinf. 2016;17(1) doi: 10.1186/s12859-016-1319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seymour G.B., Østergaard L., Chapman N.H., Knapp S., Martin C. Fruit development and ripening. Annu Rev Plant Biol. 2013;64(1):219–241. doi: 10.1146/annurev-arplant-050312-120057. [DOI] [PubMed] [Google Scholar]
- 52.Medina-Escobar N., Cárdenas J., Moyano E., Caballero J.L., Muñoz-Blanco J. Cloning, molecular characterization and expression pattern of a strawberry ripening-specific cDNA with sequence homology to pectate lyase from higher plants. Plant Mol Biol. 1997;34:867–877. doi: 10.1023/a:1005847326319. [DOI] [PubMed] [Google Scholar]
- 53.Domínguez-Puigjaner E., LLop I., Vendrell M., Prat S. A cDNA clone highly expressed in ripe banana fruit shows homology to pectate lyases. Plant Physiol. 1997;114:1071–1076. doi: 10.1104/pp.114.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nunan K.J., Davies C., Robinson S.P., Fincher G.B. Expression patterns of cell wall-modifying enzymes during grape berry development. Planta. 2001;214(2):257–264. doi: 10.1007/s004250100609. [DOI] [PubMed] [Google Scholar]
- 55.Carrington C.M.S., Pressey R. ß-Galactosidase II activity in relation to changes in cell wall galactosyl composition during tomato ripening. J Am Soc Hortic Sci. 1996;121(1):132–136. [Google Scholar]
- 56.Smith D.L., Gross K.C. A family of at least seven β-galactosidase genes is expressed during tomato fruit development. Plant Physiol. 2000;123:1173–1183. doi: 10.1104/pp.123.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitagawa Y., Kanayama Y., Yamaki S. Isolation of β-galactosidase fractions from Japanese pear: activity against native cell wall polysaccharides. Physiol Plant. 1995;93:545–550. [Google Scholar]
- 58.Wang D.D., Samsulrizal N.H., Yan C., Allcock N.S., Craigon J., Blanco-Ulate B., et al. Characterization of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol. 2018;179:544–557. doi: 10.1104/pp.18.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fitch M.M.M., Manshardt R.M., Gonsalves D., Slightom J.L., Sanford J.C. Stable transformation of papaya via microprojectile bombardment. Plant Cell Rep. 1990;9:189–194. doi: 10.1007/BF00232177. [DOI] [PubMed] [Google Scholar]
- 60.Yao J.-L., Cohen D., Atkinson R., Richardson K., Morris B. Regeneration of transgenic plants from the commercial apple cultivar Royal Gala. Plant Cell Rep. 1995;14:407–412. doi: 10.1007/BF00234044. [DOI] [PubMed] [Google Scholar]
- 61.Varkonyi-Gasic E., Wang T.C., Voogd C., Jeon S., Drummond R.S.M., Gleave A.P., et al. Mutagenesis of kiwifruit CENTRORADIALIS-like genes transforms a climbing woody perennial with long juvenility and axillary flowering into a compact plant with rapid terminal flowering. Plant Biotechnol J. 2019;17:869–880. doi: 10.1111/pbi.13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tucker G., Yin X.R., Zhang A.D., Wang M.M., Zhu Q.G., Liu X.F., et al. Ethylene† and fruit softening. Food Quality Safety. 2017;1:253–267. [Google Scholar]
- 63.Xiao Y.-y., Kuang J.-F., Qi X.-n., Ye Y.-J., Wu Z.-X., Chen J.-y., et al. A comprehensive investigation of starch degradation process and identification of a transcriptional activator MabHLH6 during banana fruit ripening. Plant Biotechnol J. 2018;16(1):151–164. doi: 10.1111/pbi.12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tacken E., Ireland H., Gunaseelan K., Karunairetnam S., Wang D., Schultz K., et al. The role of ethylene and cold temperature in the regulation of the apple POLYGALACTURONASE1 gene and fruit softening. Plant Physiol. 2010;153:294–305. doi: 10.1104/pp.109.151092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.