ABSTRACT
Profilin 4 (Pfn4) is expressed during spermiogenesis and localizes to the acrosome-acroplaxome-manchette complex. Here, we generated PFN4-deficient mice, with sperm displaying severe impairment in manchette formation. Interestingly, HOOK1 staining suggests that the perinuclear ring is established; however, ARL3 staining is disrupted, suggesting that lack of PFN4 does not interfere with the formation of the perinuclear ring and initial localization of HOOK1, but impedes microtubular organization of the manchette. Furthermore, amorphous head shape and flagellar defects were detected, resulting in reduced sperm motility. Disrupted cis- and trans-Golgi networks and aberrant production of proacrosomal vesicles caused impaired acrosome biogenesis. Proteomic analysis showed that the proteins ARF3, SPECC1L and FKBP1, which are involved in Golgi membrane trafficking and PI3K/AKT pathway, are more abundant in Pfn4−/− testes. Levels of PI3K, AKT and mTOR were elevated, whereas AMPK level was reduced, consistent with inhibition of autophagy. This seems to result in blockage of autophagic flux, which could explain the failure in acrosome formation. In vitro fertilization demonstrated that PFN4-deficient sperm is capable of fertilizing zona-free oocytes, suggesting a potential treatment for PFN4-related human infertility.
KEY WORDS: Profilin 4, Acrosome biogenesis, PI3K/AKT signaling, Cis-Golgi, Trans-Golgi, Infertility, In vitro fertilization, Morula, Manchette, Spermiogenesis
Summary: PFN4-deficient male mice exhibit impaired acrosome formation and malformation of the manchette, leading to amorphous sperm head shape, flagellar abnormalities and sterility.
INTRODUCTION
Fertilization is the process of fusing two gametes for sexual reproduction. Male gametes are produced through spermatogenesis whereby the germ cells undergo a series of various differentiation steps, including condensation of chromatin, development of the manchette, and flagellar and acrosome formation. In mice, formation of the caudal manchette starts during spermiogenesis at step 8, which is present along the base of the developing spermatid nucleus (Long and Cook, 1991; O'Donnell and O'Bryan, 2014). At the beginning of nuclear elongation, the microtubular manchette extends in a posterior direction from the nuclear marginal ring to the cytoplasmic lobe at the distal end, creating a ring-like structure that surrounds the spermatid head underneath the acrosome. Protein synthesis and storage occur in the cytoplasmic lobe and the proteins responsible for the strengthening of the head-tail attachment are transported along manchette microtubules by intra-manchette transport (IMT).
In addition to shaping the sperm head, the manchette is also responsible for trafficking proteins to the developing sperm flagella (Teves et al., 2020). Transport of proteins and vesicles occurs between the cytoplasm and nucleus via nuclear pores, developing centrosomes and sperm flagella. Protein trafficking occurs via IMT using the manchette (Teves et al., 2020). A number of proteins localize to the manchette and contribute to IMT, such as Hook1 (Mendoza-Lujambio et al., 2002), Gopc (Yao et al., 2002), Pfn3 (Umer et al., 2021), Spag17 (Kazarian et al., 2018), Ift20 (Li et al., 2016), Lrguk1 (also known as Lrguk) (Okuda et al., 2017), Spef2 (Lehti et al., 2017) and Cdc42 (Chapin et al., 2001), and mutation results in a mis-shaped sperm head. However, the molecular mechanisms of trafficking proteins responsible for sperm head shaping and flagella development via IMT are not clear (Teves et al., 2020).
During fertilization, the acrosomal reaction takes place, which results in release of the acrosomal content. This is instrumental in breaking down the zona pellucida of the oocyte and enables the penetration and fusion of sperm and oocyte (Sun et al., 2011). Therefore, structural or functional abnormalities of the acrosome are potentially able to interfere with sperm-egg fusion, eventually resulting in sub- or infertility.
Acrosome biogenesis is brought about by several endoplasmic reticulum (ER)- and Golgi-associated proteins and Golgi-mediated vesicle trafficking (Khawar et al., 2019). This vesicle trafficking is in part regulated by the autophagy machinery. Autophagy is the intracellular catabolic process responsible for the degradation and recycling of organelles and cytosolic proteins by autophagosomes (Mizushima and Levine, 2010; Zhu et al., 2019). Autophagy is crucial for the proliferation and differentiation of spermatogonia stem cells, meiotic progression and spermiogenesis. During spermiogenesis, autophagy plays a role in acrosome development, head shaping, flagellum formation and cytoplasmic removal. During the process of autophagy, initially a cup-like structure termed phagophore develops, engulfing cytoplasmic components and then closing as a double-membrane structure named the autophagosome. This process consists of three key steps: initiation, elongation and closure of the membrane (Yang et al., 2017).
Autophagy is a highly specialized and tightly regulated process involving autophagy-related proteins (ATGs). To date, 41 ATGs are known and ATG7 has been demonstrated to be crucial for autophagosome biogenesis (Wang et al., 2014). ATG7 activates LC3 (microtubule-associated protein 1/11-light chain 3, MAP1LC3A), which is transported to Golgi-derived vesicles via phagophores during auto-lysosomal or acrosomal biogenesis. There, the activated LC3 protein facilitates the fusion and transportation of Golgi-derived vesicles in the phagophore-like structures towards the nucleus for acrosome biogenesis (Wang et al., 2014). The major regulator of autophagy, mTOR, is a downstream component of the phosphatidylinositol 3 kinase (PI3K) and AKT-kinase (AKT) pathway (Kaleağasıoğlu et al., 2020). mTOR is the main intracellular autophagic suppressor and is positively regulated by the PI3K/AKT pathway (Li et al., 2018).
The autophagy-related proteins and caspases interplay to regulate autophagy and apoptosis under basal conditions. Recently, the role of caspases in the regulation of autophagy has begun to be described. Core ATGs interact, recruit and activate several caspases by various autophagosomal membranes. Caspases 3 and 9 are markers for apoptosis known to play a role in autophagy (Tsapras and Nezis, 2017). In addition, autophagy is also thought be involved in phagocytosis of the apoptotic cells. It is known that under some physiological and pathological conditions Sertoli cells play a role in phagocytosis of apoptotic germ cells (Dong et al., 2016).
Profilins are small proteins of 15 kDa molecular weight. There are four known profilin genes in mouse and human. The functions of the various profilins seem to have diverged in mammals to either regulate membrane trafficking or regulate the cytoskeleton (Sun et al., 2013). Profilin 1 is ubiquitously expressed (Tariq et al., 2016), localized in the trans-Golgi network and required for the formation of constitutive vesicle transport at the trans-Golgi network (Dong et al., 2000). Profilin 2, expressed in brain, associates with a number of proteins involved in endocytosis and membrane trafficking (Gareus et al., 2006). The testis-specific PFN3 is localized to the cis- and trans-Golgi networks and plays a role in autophagy regulation during acrosome biogenesis (Umer et al., 2021).
Profilin 4 (Pfn4, PFN4) is highly expressed in testes and localized to the acrosome-acroplaxome-manchette complex (Behnen et al., 2009). Hence, it is tempting to speculate that PFN4 plays a role in acrosome and manchette biogenesis. Of note, Pfn4 shows only 30% homology to other profilin family members, and it does not encode for the actin and poly-L-proline binding sites that are present in PFN1-3 (Behnen et al., 2009), suggesting that Pfn4 has functions in vivo that are independent of the regulation of actin dynamics.
Here, we generated PFN4-deficient mice by CRISPR/Cas9-mediated gene editing in zygotes. PFN4-deficient males show disturbed spermatogenesis, resulting in male infertility. Development of the manchette in Pfn4−/− mice was greatly disturbed leading to defects in the sperm head and flagella defects impinging on sperm motility. Furthermore, acrosome formation was impaired and abnormalities in the cis- and trans-Golgi network and disrupted proacrosomal vesicles were observed. Proteomic analyses indicated deregulation of the PI3K/AKT and mTOR/AMPK pathways, suggestive of inhibited autophagy. Taken together, our results show that PFN4 is required for proper manchette and acrosome formation.
RESULTS
Generation and phenotypic characterization of PFN4-deficient mouse lines
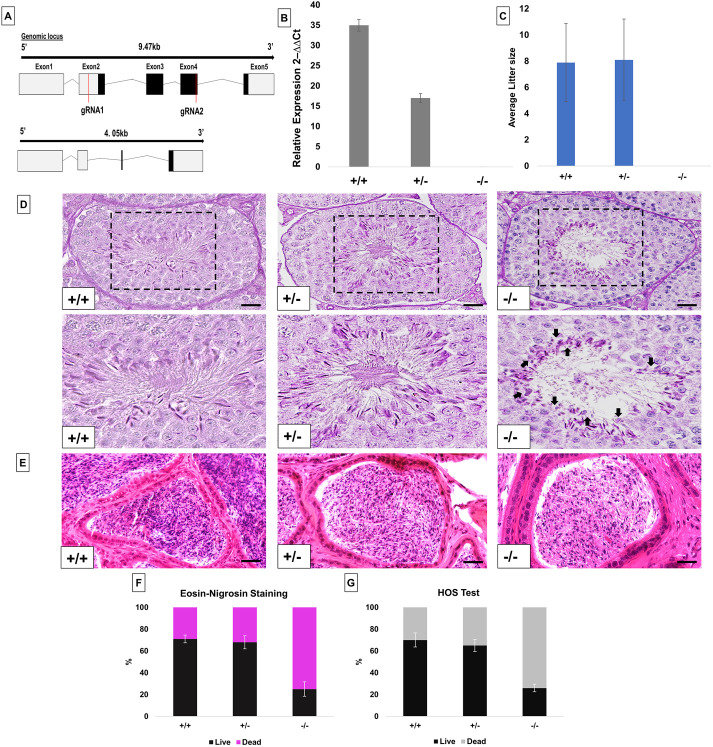
To delete the Pfn4 allele, two single guide RNAs annealing to exon 2 and exon 4 of the Pfn4 locus (Fig. 1A) and Cas9 mRNA were injected into fertilized oocytes. Successful gene editing in the offspring was detected by PCR and confirmed by sequencing (Fig. S1A,B). Two PFN4-deficient mouse lines harboring identical deletions of 5417 bp were established, backcrossed to C57BL/6 mice and named Pfn4Δa and Pfn4Δb. The deletion causes a frame shift in the PFN4 reading frame, leading to premature translational termination giving rise to a hypothetical truncated protein of 25 amino acids.
Fig. 1.
Generation and characterization of PFN4-deficient mice (Pfn4Δa). (A) Schematic of the Pfn4 locus and established PFN4-deficient mouse line. Red lines mark positions of the guide RNAs used for gene editing. (B) Validation of PFN4-deficient mice (Pfn4Δa). qRT-PCR was performed to check the relative expression of Pfn4 mRNA in murine testis of WT, Pfn4+/− and Pfn4−/− mice for the Pfn4Δa mutation. (C) Mating statistics of WT, Pfn4+/− and Pfn4−/− male mice (n=9/genotype). (D) PAS staining on WT, Pfn4+/− and Pfn4−/− testes sections. Dashed boxes indicate the areas shown at higher magnification below. Arrows indicate abnormal head morphology. (E) Hematoxylin and Eosin staining on WT, Pfn4+/− and Pfn4−/− cauda epididymis sections. (F) Quantification of E&N staining (n=3 biological replicates/genotype) of mature WT, Pfn4+/− and Pfn4−/− sperm isolated from cauda epididymis. (G) Hypo-osmotic swelling (HOS) test (n=3 biological replicates/genotype) of mature WT, Pfn4+/− and Pfn4−/− sperm isolated from cauda epididymis. Error bars represent mean±s.d. Scale bars: 10 μm.
For Pfn4Δa, real-time quantitative PCR (qRT-PCR) showed that the level of Pfn4 mRNA is reduced by half in Pfn4+/− compared with WT mice. No Pfn4 transcript was detected in Pfn4−/− mice confirming the Pfn4Δ allele as a null allele (Fig. 1B). Results for Pfn4Δb line are shown in the Fig. S1C.
After generating and validating the PFN4-deficient lines, we performed fertility analysis on males from Pfn4Δa and Pfn4Δb lines. Adult males heterozygous and homozygous for the Pfn4 deletion mated normally with females as vaginal plugs were clearly seen. Males heterozygous for the Pfn4 mutation retained normal fertility, with an average litter size comparable to that of wild-type (WT) littermates. This suggests that the reduced level of Pfn4 mRNA observed in heterozygous males does not affect fertility. In contrast, Pfn4−/− mice of both Pfn4Δa and Pfn4Δb lines were infertile as none of the males (n=9) produced any pregnancy or offspring (Fig. 1C, Pfn4Δa; Fig. S1D, Pfn4Δb). Of note, female Pfn4−/− mice did not show any alterations in fertility (Fig. S1E).
For both PFN4-deficient lines (Pfn4Δa, Pfn4Δb), body weight (Fig. S1F), and the absolute and relative weight of testes and epididymis in Pfn4−/− males were comparable with Pfn4+/− and WT littermates (Fig. S1F-J). Periodic acid Schiff (PAS) and Hematoxylin and Eosin staining was performed on testes and cauda epididymis and showed that the overall morphology of the seminiferous tubules and cauda epididymis is not affected (Fig. 1D,E), but elongating spermatozoa of Pfn4−/− mice appeared mis-shapen with seemingly smaller nuclei (Fig. 1D, arrows). These results suggest that loss of Pfn4 affects spermiogenesis.
In order to assess the viability of spermatozoa, Eosin and Nigrosin (E&N) staining was performed on mature sperm cells isolated from cauda epididymis. E&N staining differentiates live (white heads) from dead (pink heads) sperm (Fig. S1K). In mice, a percentage of 60-80% live sperm is considered normal, 40-60% borderline and less than 40% abnormal. Using males from the Pfn4Δa line, E&N staining showed that Pfn4+/− males mice display a normal percentage of viable sperm whereas in Pfn4−/− mice an abnormally low percentage of viable sperm (25%) was observed (Fig. 1F). To confirm this, a hypo-osmotic swelling test was performed on cauda epididymal sperm, wherein intact (live) sperm show tail curling (Fig. S1M). The percentage of hypo-osmotic reactive sperm in Pfn4−/− mice was abnormally low (24%) (Fig. 1G). The results were confirmed with the Pfn4Δb animals (Fig. S1L,N). In conclusion, loss of Pfn4 affects sperm viability. As noted before, we observed that sperm from Pfn4−/− display an amorphous and smaller head shape (black arrows) (Fig. S1K).
Loss of PFN4 results in aberrant sperm head morphology
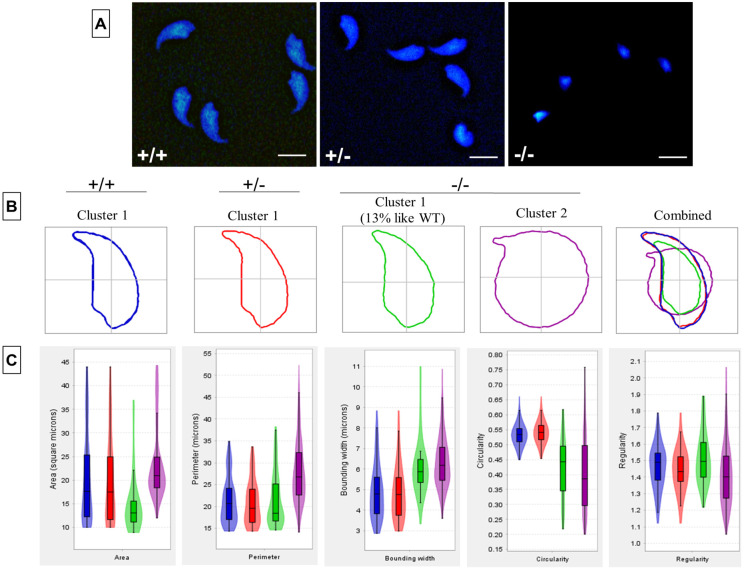
In order to determine the shape of the sperm head, we next isolated mature sperm from cauda epididymis, and performed nuclear morphology analysis (Fig. 2A). Here, 77% of Pfn4−/− sperm head showed a dramatically disturbed, irregular/round and smaller head size (Fig. 2B, Cluster 2). Of note, 16% (Cluster 1) of Pfn4−/− sperm showed a less irregular head shape, which was more comparable to WT and Pfn4 heterozygous sperm (Fig. 2B). Detailed analysis revealed that in Pfn4−/− sperm nuclear area, perimeter, regularity, bounding width and circularity were significantly affected compared with WT and Pfn4+/− sperm (Fig. 2C). Details of parameters are given in Table S1.
Fig. 2.
Sperm head morphology analysis. Sperm head morphology analysis on WT, Pfn4+/− and Pfn4−/− sperm cells (n=3 biological replicates/genotype). In total, 901 nuclei were analyzed: 309 for WT (cluster 1), 276 for Pfn4+/− (cluster 1) and 541 for Pfn4−/− (86 sperm nuclei in cluster 1 and 455 nuclei in cluster 2). (A) Sperm cells stained with DAPI. (B) Head morphology of WT, Pfn4+/− and Pfn4−/− sperm cells. (C) Area, perimeter, regularity, bounding width, and circularity of WT, Pfn4+/− and Pfn4−/− sperm cells. Bonferroni's corrected P-values were calculated using the non-parametric Mann–Whitney–Wilcoxon test. Error bars show the mean±s.d. Box boundaries indicate consensus nucleus (overall examination of the population). Horizontal lines indicate mean of the respective parameter and error bars indicate coefficient of variability, standard error and standard deviation per parameter.
Defective manchette development in Pfn4−/− mice
The manchette is a temporary microtubular structure that surrounds the sperm nucleus during spermatid elongation and is thought to play a role in shaping of the sperm head (Kierszenbaum and Tres, 2004). As PFN4 is localized to the manchette (Behnen et al., 2009), we investigated whether the loss of PFN4 affects manchette formation. α-Tubulin staining demonstrated that Pfn4+/− and WT spermatids showed proper manchette development and structural organization, exemplified by a manchette microtubular array forming a tight junction in the caudal region of spermatids (Fig. 3A,B). In contrast, Pfn4−/− mice showed only marginal staining for α-tubulin, presenting as punctate and dispersed. Hence, the developing Pfn4−/− spermatids completely lack the typical parallel microtubular array structure of the manchette (Fig. 3C).
Fig. 3.
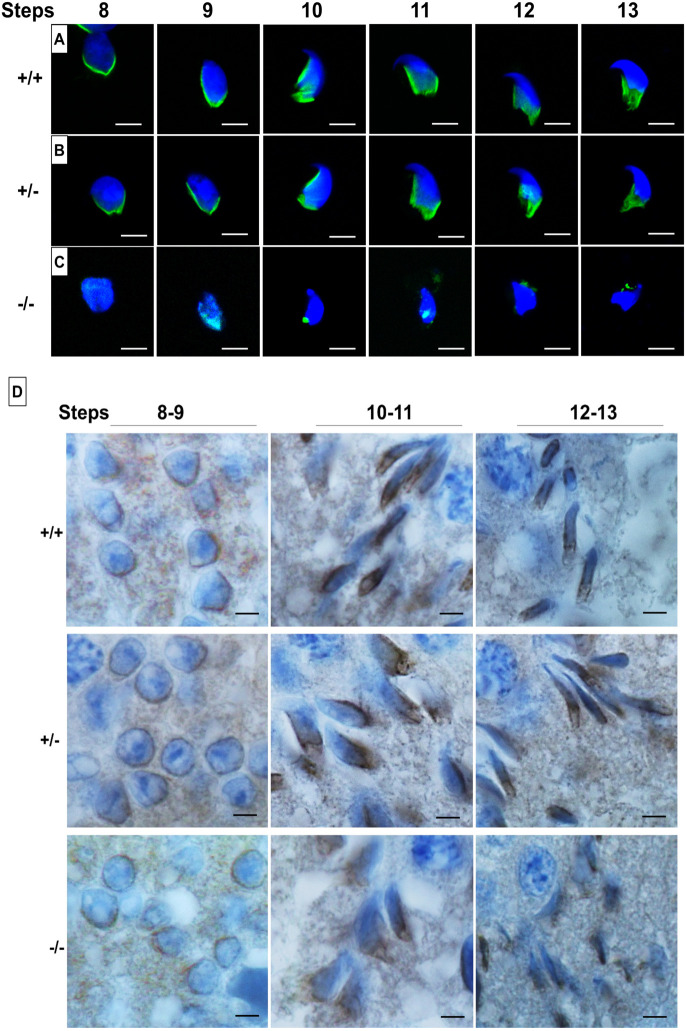
Manchette staining using α-tubulin and HOOK1 antibodies. (A-C) Immunofluorescence staining for the manchette using an α-tubulin antibody (green) on a germ cell population isolated from WT (A), Pfn4+/− (B) and Pfn4−/− (C) testes (n=3/genotype). Nuclei were stained with DAPI (blue). (D) IHC using an anti-HOOK1 antibody on WT, Pfn4+/− and Pfn4−/− testes sections. Scale bars: 20 µm.
In order to understand this defect at the morphological level, we used TEM to analyze the ultrastructure. Pfn4−/− spermatids showed mislocalization (steps 8-9) and an angular shape (step 10) of the microtubular manchette, whereas in Pfn4+/+ spermatids the microtubular manchette was properly aligned (steps 8-9) and had a crescent shape (step 10) in the middle of the posterior region of the developing spermatid (Fig. S2A). The manchette defect was more prominent in later steps (11-13) of development. In steps 11-13, Pfn4−/− spermatids lacked the microtubular array structure, and appeared scraggy and rugged in shape, compared with Pfn4+/+ spermatids, in which the microtubular mantle was aligned in arrays (Fig. S2A). These data indicate that deletion of PFN4 disrupts manchette formation, which, in turn, leads to the deformed sperm heads observed in Fig. 2C and Fig. S2A.
Furthermore, we performed immunohistochemistry (IHC) using an α-tubulin antibody on WT, Pfn4+/− and Pfn4−/− testis sections. IHC staining was abnormal at steps 8-11 in Pfn4−/− spermatids (Fig. S2B, stars), compared with WT and Pfn4+/− testis sections, which showed a properly formed, sickle-shaped microtubular manchette in the posterior region of developing spermatids. Furthermore, at steps 12-16 a complete loss of the manchette and amorphous shape of elongated sperm heads were observed (indicated by stars) in Pfn4−/− spermatids compared with WT and Pfn4+/− spermatids (Fig. S2B).
In order to examine further the structural origin of the manchette malformation, we performed IHC using HOOK1 (hook microtubule tethering protein 1) and ARL3 (ADP-ribosylation factor like 3) antibodies on testes sections for all three genotypes to see whether the localization of these proteins is affected in Pfn4−/− sperm. HOOK1 is localized in the manchette as well in the nuclear ring between the manchette and the nucleus. HOOK1 directs correct positioning and elongation of the microtubular manchette (Mendoza-Lujambio et al., 2002; Schwarz et al., 2017). ARL3 is localized in the manchette and essential for its development (Qi et al., 2013). In addition, ARL3 plays a role in ciliary trafficking and intraflagellar transport (Li et al., 2010). Interestingly in Pfn4−/− spermatids, HOOK1 is correctly localized in the nuclear ring in steps 8-9 sperm (Fig. 3D). In steps 10-13, HOOK1 expression appears disturbed, reflecting the staining pattern of α-tubulin. This suggests that the manchette starts to form (including the microtubule mantle) with the development of the sperm head, but the manchette structure becomes progressively more abnormal and, in some cases, appears to be absent. Interestingly, ARL3 expression seems to be absent up to steps 12-13, where the ARL3 antibody appears to stain random regions of the sperm heads (Fig. S2C); by contrast, in WT and Pfn4+/− testes sections, the microtubular manchette is properly aligned from steps 8-13.
Flagellar defects in Pfn4−/− sperm
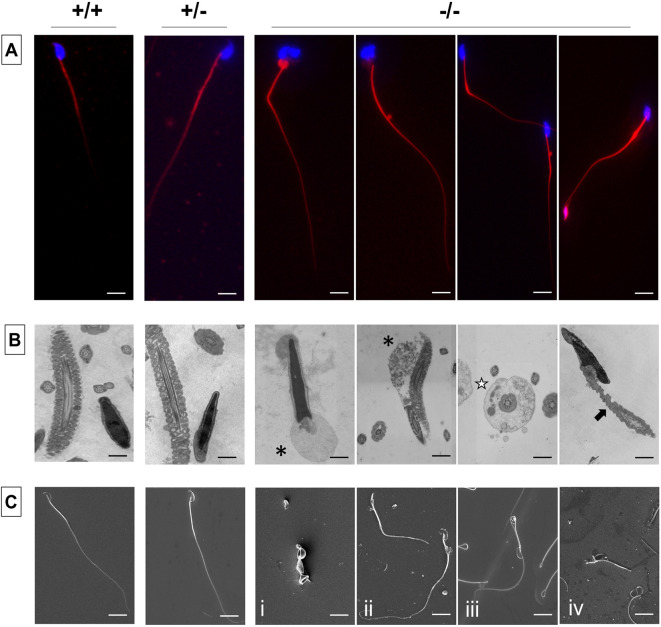
It has been reported that during flagellar development sperm tail proteins are also delivered through the manchette, aside from the intraflagellar transport (IFT) (Mochida et al., 1991; Mendoza-Lujambio et al., 2002). Given that loss of Pfn4 disturbs development of the manchette, we next examined midpiece and flagellar structures in sperm cells. First, Mito Red dye was used to stain the sperm mitochondria in the midpiece of the flagella and DAPI was used to stain sperm heads. Here, Pfn4−/− sperm revealed numerous deformities including cytoplasmic droplets in the connecting piece, and a bent midpiece and abnormally thick midpiece (Fig. 4A). This suggests that the disturbed manchette seen in Pfn4−/− males causes the mitochondria abnormalities in sperm flagellum.
Fig. 4.
Flagellar staining and ultrastructural analysis. (A-C) Flagellar staining and ultrastructural analysis using MitoRed (A), TEM (B) and SEM (C) of WT, Pfn4+/− and Pfn4−/− sperm isolated from the cauda epididymis (n=3 biological replicates/genotype). Black asterisks indicate cytoplasmic droplets in the midpiece, white star indicates a defective fibrous sheath, and black arrow indicates mitochondrial structural defects. Scale bars: 10 μm.
Next, transmission electron microscopy (TEM) was performed on cauda epididymis sperm. TEM on Pfn4−/− sperm revealed disorganized flagellar components such as cytoplasmic droplets in the midpiece (black asterisks), a defective fibrous sheath (white star), and mitochondrial structural defects (black arrow) (Fig. 4B). Scanning electron microscopy (SEM) revealed morphological anomalies of Pfn4−/− sperm, including coiled flagella (Fig. 4Ci), a shortened thick midpiece (Fig. 4Cii), cytoplasmic droplets in the midpiece (Fig. 4Ciii) and a bent connecting piece (Fig. 4Civ).
Reduced sperm motility in Pfn4−/− mice
Flagellar deformities might affect the sperm motility parameters, so we performed computer-assisted semen analysis (CASA), to analyze the swimming properties of Pfn4−/− sperm. Compared with Pfn4+/+ and Pfn4+/−, sperm motility parameters [curvilinear velocity (VCL), straight-line velocity (VSL), average path velocity (VAP) and progressive and total motility] were significantly reduced in Pfn4−/− sperm (Table S1). These results indicate that abnormalities in sperm flagella lead to reduced overall sperm motility in Pfn4−/− mice.
PFN4 is essential for acrosome biogenesis
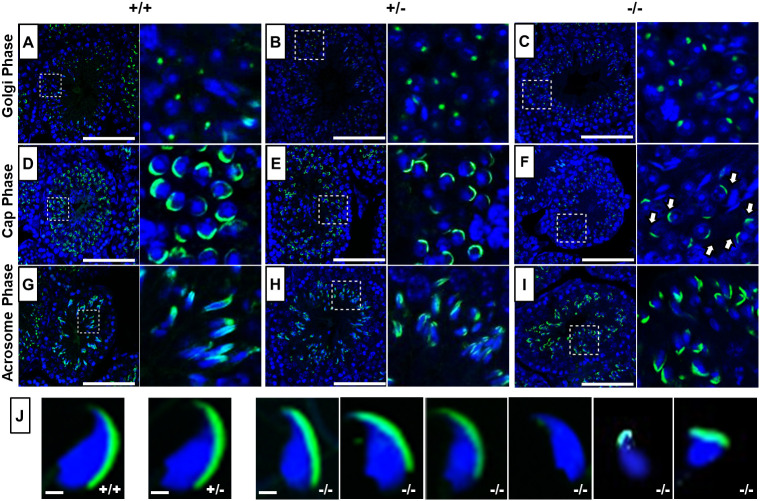
In addition to the manchette, PFN4 is also localized to the acroplaxome (Behnen et al., 2009), suggesting a role in biogenesis/function of the acrosome. In order to examine acrosome development, PNA-FITC fluorescence labeling was performed on testes sections to visualize the different steps of acrosome development. In seminiferous tubules of WT (Fig. 5A) and Pfn4+/− (Fig. 5B) male mice, the developing acrosome forms a single homogenous cluster present on the anterior face of nuclei in Golgi-phase spermatids, whereas in Pfn4−/− spermatozoa PNA-FITC staining showed a non-uniform shape of the acrosomal vesicle (Fig. 5C). This abnormal acrosome development was further seen in the next step, known as the cap phase. Here, a cap-like covering can be seen on the apical surface of the WT and Pfn4+/− spermatozoa (Fig. 5D,E). In Pfn4−/− sperm cells, the cap-like structures were partially impaired (white arrows) (Fig. 5F). In the acrosomal phase, when elongation and head remodeling of spermatids starts, this abnormal acrosome development was most prominent. In Pfn4−/− sperm, the acrosome structure failed to develop into an arrow-like structure (Fig. 5I), as seen in the WT (Fig. 5G) and Pfn4+/− sperm (Fig. 5H). These suggest that loss of PFN4 partially impairs biogenesis of the acrosome starting from the first phase (Golgi phase) of acrosome development. Furthermore, PNA-FITC labeling on mature sperm isolated from cauda epididymis of Pfn4−/− mice showed malformed acrosomes and aberrant head morphology (Fig. 5J). In addition, PAS staining further confirmed the abnormalities in acrosome development (Fig. S3A).
Fig. 5.
Acrosome analysis using PNA-FITC fluorescence staining. (A-I) Immunofluorescence staining for acrosome biogenesis on testes sections and mature sperm of WT, Pfn4+/− and Pfn4−/− mice (n=3 biological replicates/genotype). In the Golgi phase, the proacrosomal granule (green) was labeled by PNA-FITC in WT (A), Pfn4+/− (B) and Pfn4−/− (C) round spermatozoa. In the cap phase, acrosomal caps (green) were stained in WT (D), Pfn4+/− (E) and Pfn4−/− (F) round spermatozoa (white arrows show abnormal cap structures). In the acrosomal phase, PNA-FITC-labeled the acrosomal area in WT (G), Pfn4+/− (H) and Pfn4−/− (I) elongated spermatids. Dashed boxes indicate the areas enlarged to the right. (J) Immunofluorescence staining using PNA-FITC (green) on epididymal sperm cells of WT, Pfn4+/− and Pfn4−/− mice (n=3/genotype) to show the variety of Pfn4−/− sperm such as malformed acrosome and abnormal head morphology. Scale bars: 20 μm.
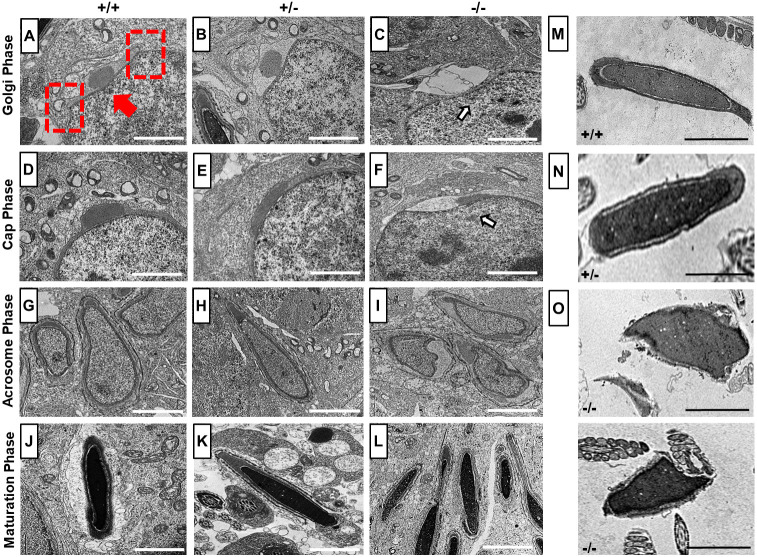
Next, TEM was used for ultrastructural analysis of the developing sperm cells. In the Golgi phase, the trans-Golgi domain releases proacrosomal vesicles, which fuse to form a single, large and dense acrosomal granule in the center at the marginal ring (red dashed boxes) of the acroplaxome membrane (red arrow) on the nuclear surface as seen in WT and Pfn4+/− sperm (Fig. 6A,B). In Pfn4−/− mice, the proacrosomal vesicles released from the trans-Golgi network did not seem to gather and fuse as a single, large and dense acrosomal vesicle on the apical face of developing spermatozoa (Fig. S3B). This suggests that the acrosomal vesicle formation is greatly delayed retarding acrosome biogenesis (Fig. 6C, arrow indicates large acrosomal vesicle) in subsequent phases. In the cap phase, the proacrosomal vesicle develops and flattens into a cap-like structure over the nucleus in WT (Fig. 6D) and Pfn4+/− spermatids (Fig. 6E). However, in Pfn4−/− spermatids, the cap-like structure did not spread out over the nucleus, but remained as a pinched granule (Fig. 6F, arrow shows abnormal cap structure). In the acrosomal and maturation phase, this defect became more prominent, and the cap developed further and formed a proper acrosome in WT (Fig. 6G,J) and Pfn4+/− (Fig. 6H,K) spermatids, whereas in Pfn4−/− sperm proacrosomal vesicles failed to develop into a polarized cap and acrosome (Fig. 6I,L).
Fig. 6.
TEM analysis. (A-L) Ultrastructural analysis using TEM on testes sections of WT, Pfn4+/− and Pfn4−/− mice (n=3 biological replicates/genotype). (A-C) Golgi phase spermatozoa of WT (A), Pfn4+/− (B) Pfn4−/− (C) mice (arrow indicates deformed developing acrosome). Red arrow indicates the acroplaxome, red dashed rectangles indicate the marginal ring. (D-F) Cap phase spermatozoa of WT (D), Pfn4+/− (E) and Pfn4−/− (F) mice (arrow indicates abnormal cap-like structures). (G-I) Acrosomal phase spermatids of WT (G), Pfn4+/− (H) and Pfn4−/− (I) mice. (J-L) Maturation phase spermatids of WT (J), Pfn4+/− (K) and Pfn4−/− (L) mice. (M-O) Ultrastructural analysis of cauda epididymal sperms of WT (M), Pfn4+/− (N) and Pfn4−/− (O) mice. Scale bars: 2 μm.
To confirm the acrosomal defects, we performed IHC using Sp56 (Zp3r) and acrosin antibodies on testes sections of WT, Pfn4+/− and Pfn4−/− mice. Sp56 and acrosin are markers of acrosomal vesicle and acrosomal matrix, respectively (Xin et al., 2020). Sp56 is detectable as early as the start of acrosome biogenesis starts (Golgi phase) (Kim et al., 2001). IHC showed that Sp56 and acrosin staining were not uniformly distributed in developing acrosomes in Pfn4−/− compared with WT and Pfn4+/− testes sections (Fig. S3C,D). These results further validate the observations of impaired acrosome biogenesis in Pfn4−/− mice.
In order to investigate the acrosomal defect in mature sperm, we used TEM to analyze mature sperm cells isolated from cauda epididymis. Ultrastructural analysis showed a lack of properly formed acrosomes in epididymal sperm of Pfn4−/− mice (Fig. 6O), whereas WT and Pfn4+/− sperm cells showed proper and intact acrosomes (Fig. 6M,N).
Loss of Pfn4 results in a fragmented Golgi network
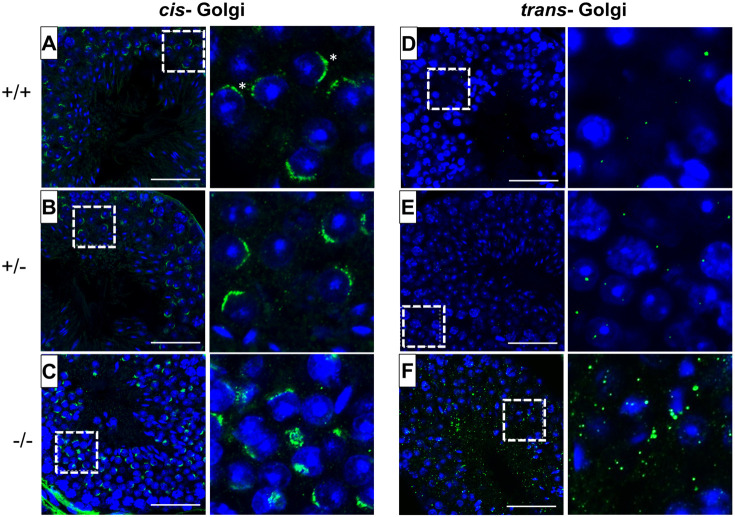
The Golgi apparatus is responsible for releasing proacrosomal vesicles for the formation of the acrosome during the Golgi phase of acrosome biogenesis (Tang et al., 1982). We therefore examined the orientation and morphology of the Golgi network domains. Immunofluorescence (IF) staining was performed using GM130 (also known as Golga2) and TGN46 (trans-Golgi network protein) as markers for the cis- and trans-Golgi networks, respectively. GM130 plays a crucial role in vesicle tethering and fusion and in maintaining cis-Golgi structural integrity (Tiwari et al., 2019), whereas TGN46 is important for the formation of exocytic vesicles and secretion from the trans-part of the Golgi network (Huang et al., 2019). Compared with WT (Fig. 7A,D) and Pfn4+/− (Fig. 7B,E) spermatids, IF staining showed defective and fragmented cis- (Fig. 7C) and trans- (Fig. 7F) Golgi network in Pfn4−/− testis sections. In Pfn4−/− testis sections, structural disruption and mild mislocalization of the cis-Golgi network was observed, and the trans-Golgi was fragmented and dispersed throughout the cytoplasm indicating impaired vesicle trafficking and transport, unlike in WT and Pfn4+/− spermatids, where cis- and trans-Golgi were properly formed and aligned. These observations indicate that loss of PFN4 results in disorganization of the Golgi sub-domains, leading to malformed proacrosomal vesicles that seem to impair acrosome biogenesis.
Fig. 7.
Fluorescence staining of the Golgi network. Cis- and trans-Golgi immunofluorescence staining using GM130 (green) and TGN46 (green) antibodies and nuclei stained with DAPI (blue) on WT, Pfn4+/− and Pfn4−/− (n=3/genotype) testes sections. (A-C) WT (A), Pfn4+/− (B) and Pfn4−/− (C) spermatozoa stained for the cis-Golgi compartment (asterisks). (D-F) WT (D), Pfn4+/− (E) and Pfn4−/− (F) spermatozoa stained for the trans-Golgi network. Dashed boxes indicate the areas enlarged to the right. Scale bars: 50 μm.
Furthermore, we checked the localization of COPA (coatomer protein complex subunit α), PICK1 (protein interacting with C kinase 1) and GOPC (Golgi associated PDZ and coiled-coil motif containing) using IHC on testes sections of WT, Pfn4+/− and Pfn4−/− mice. COPA is essential for cargo transport of Golgi-derived proteins (Lepelley et al., 2020), and PICK1 and GOPC play an important role in vesicle release from Golgi network and is further associated with proacrosomal granule formation (Xiao et al., 2009). Our IHC showed that COPA is mislocalized (arrowheads) in Pfn4−/− spermatids, indicating disrupted vesicle transport for acrosome biogenesis, whereas in WT and Pfn4+/− developing spermatids COPA is properly localized in the Golgi apparatus (black arrows) (Fig. S4A). IHC using anti-PICK1 and anti-GOPC antibodies showed mislocalization of Golgi was observed in Pfn4−/− testes sections, whereas in WT and Pfn4+/− testes sections (black arrows) staining showed proper Golgi localization (Fig. S4B,C). These results further indicate that vesicle trafficking is affected in Pfn4−/− mice, ultimately resulting in abnormal formation of proacrosomal granules.
Disruption of PI3K/AKT and AMPK/mTOR signaling pathways leads to inhibited autophagy in Pfn4−/− mice
In order to gain insight into the molecular mechanisms affecting the process of Golgi vesicle trafficking, liquid chromatography-mass spectrometry (LC-MS) was performed on protein lysates extracted from testes. A principal component analysis (PCA) performed on the top 5% proteins with highest variance across samples showed an overall biological difference between all three genotypes (WT, Pfn4+/− and Pfn4−/−) and the knockout replicates clustered apart from the controls (Fig. S5A).
Eleven proteins were differentially abundant when comparing Pfn4−/− samples with Pfn4+/− samples; however, significantly differentially abundant proteins were not detected in Pfn4−/− samples compared with the WT (Fig. S5B). The effect of PFN4 deficiency is likely restricted to the round spermatid stages. Because proteins were extracted from the whole testis, the samples include proteins from a very heterogeneous cell population. This could lead to difficulties in detecting signals of differential abundance due to PFN4 deficiency.
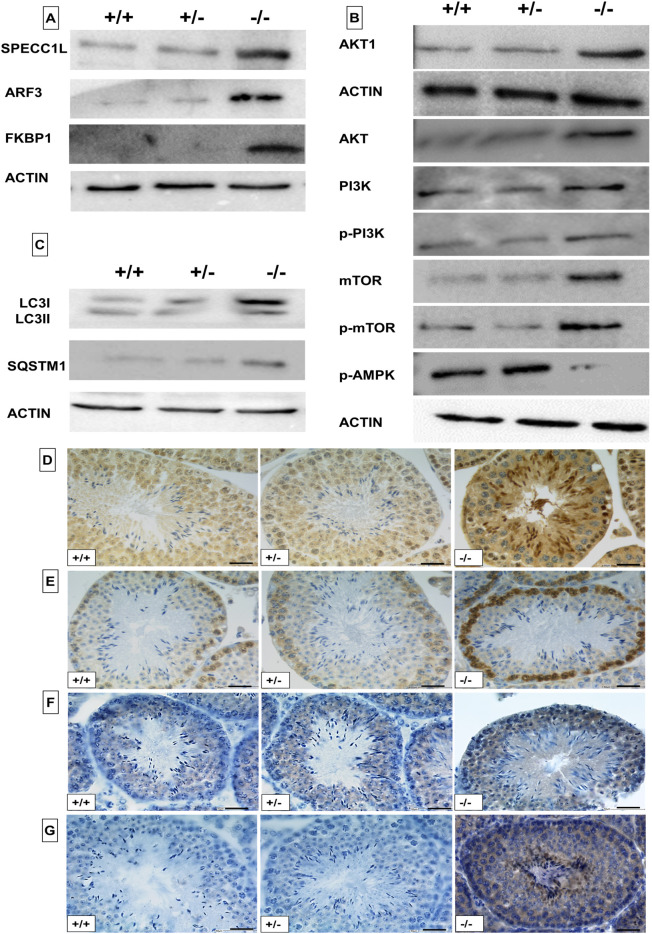
However, visualization of the data by heatmap showed that in Pfn4−/− extracts three proteins ARF2:3 (ADP-ribosylation factor), SPECC1L (sperm antigen with calponin homology and coiled-coil domains 1 like) and FKBP1A (FKBP prolyl isomerase 1a) were moderately more abundant compared with Pfn4+/+ and Pfn4+/− (Fig. S5C). These proteins are involved in the trans-Golgi network, vesicle tethering and autophagy regulation. To validate these results, western blots on protein lysates from testes were performed. Here, enhanced levels of ARF3, SPECCL1L and FKBP1 were detected in Pfn4−/− testes (Fig. 8A). Furthermore, pathways for autophagy, PI3K/AKT and mTOR signaling, protein processing in the ER and Golgi vesicle transport were enriched in Pfn4−/− (Fig. S5D). Next, we aimed to validate this upregulation of P13K/AKT and AMPK/mTOR signaling in PFN4-deficient mice. PI3K/AKT and mTOR/AMPK regulate autophagic flux and are negative and positive regulators of autophagy, respectively. Western blot analyses showed increased protein levels for AKT (total), AKT1 (phosphorylated), PI3K (total) and phospho-P13K (phosphorylated), mTOR and phospho-mTOR, whereas the level of phospho-AMPKα was reduced in Pfn4−/− testes (Fig. 8B). These results together indicate that loss of PFN4 affects the autophagy-related signaling pathways PI3K/AKT and mTOR as well as protein processing in the ER–trans-Golgi network (ER-TGN).
Fig. 8.
Analysis of autophagy regulators. Western blot analysis and immunohistochemical staining on WT, Pfn4+/− and Pfn4−/− mice (n=3/genotype). (A) Western blot using SPECC1L, ARF3 and FKBP1 antibodies on protein extracts from WT, Pfn4+/− and Pfn4−/− testes. (B) Western blot using AKT, AKT1, PI3K, p-PI3K, mTOR, p-mTOR and p-AMPK antibodies on protein extracts from WT, Pfn4+/− and Pfn4−/− testes. (C) Western blot using LC3I/II and SQSTM1 antibodies on protein extracts from WT, Pfn4+/− and Pfn4−/− testes. (D-G) IHC using LC3I/II (D), SQSTM1 (E), AKT1 (F) and ARF3 (G) antibodies on WT, Pfn4+/− and Pfn4−/− testes sections. Scale bars: 50 μm.
Next, we investigated whether the altered levels of PI3K/AKT and mTOR/AMPK affect formation of the autophagosome. Therefore, levels of LC3-I/II, a marker for autophagy activation, and SQSTM1 (also known as p62), a marker for autophagic/lysosomal degradation, were analyzed by western blot. The immunoblot showed elevated protein levels of LC3I/II and SQSTM1/p62 in PFN4-deficient testis (Fig. 8C). Furthermore, increased protein levels of LC3II (Fig. 8D), SQSTM1 (Fig. 8E), AKT1 (Fig. 8F) and ARF3 (Fig. 8G) were also confirmed by IHC. Quantification of the western blots showed increased protein levels for LC3BI/II, SQSTM1, ARF3, FKBP1, SPECC1L, AKT1, AKT, mTOR, p-mTOR, p-PI3K and PI3K, and decreased levels for AMPK (Fig. S5E). These results suggest that the autophagic flux is impaired leading to inhibition of autophagy.
The acrosomal reaction is significantly reduced in Pfn4−/− sperm
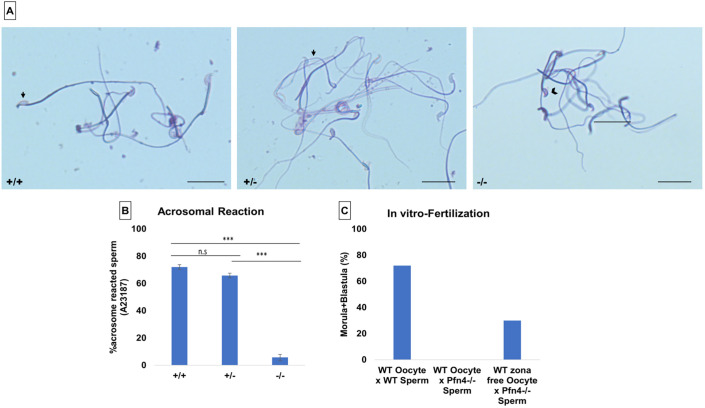
As a result of the impaired acrosome biogenesis, we expected the acrosomal reaction to be affected. To induce the acrosomal reaction, we used A23187 and Coomassie staining to differentiate the acrosome-reacted sperm (in which the acrosome is not present on the sperm head; Fig. 9A, black arrows) from the non-acrosomal-reacted sperm [in which the acrosome is detectable as a crescent-like shape (dark blue) on the sperm head; Fig. 9A, black arrowhead]. Bright-field microscopy was used to assess 200 spermatozoa in triplicate for each genotype. The acrosomal reaction was assessed by the absence of the acrosome (a dark-blue crescent shape on the sperm head). Upon exposure to A23187, more than 72% of sperm from WT mice and 65% of sperm from Pfn4+/− mice showed acrosomal exocytosis, whereas only 5-6% sperm of the Pfn4−/− mice underwent the acrosome reaction (Fig. 9B). This finding demonstrates that sperm of Pfn4−/− males display a significantly reduced acrosome reaction.
Fig. 9.
Acrosomal reaction and in vitro fertilization. Calcium ionophore-induced acrosomal reaction in WT, Pfn4+/− and Pfn4−/− sperm cells (n=3/genotype). (A) Coomassie-stained sperm cells of WT, Pfn4+/− and Pfn4−/−. Black arrows indicate that a successful acrosomal reaction took place. Black arrowhead indicates a crescent-shaped acrosome on a sperm head, indicating that the acrosomal reaction did not take place. Scale bars: 20 μm. (B) Percentage of acrosomal-reacted sperm WT, Pfn4+/− and Pfn4−/− (n=3 biological replicates/genotype). ***P<0.0005, one-tailed, paired Student's t-test. P-value relates to acrosomal-reacted WT and Pfn4+/− sperm compared with Pfn4−/− sperm. (C) Percentage of oocytes developing into blastocysts after fertilization of oocytes with WT and Pfn4−/− sperm and zona-free oocytes with Pfn4−/− sperm (n=1/genotype).
PFN4-deficient sperm are able to fertilize zona-free oocytes
The acrosome contains digestive enzymes that degrade the zona pellucida and enable fusion of the sperm with the egg (Saldívar-Hernández et al., 2015). Because PFN4-deficient sperm display defects in acrosome function, we performed in vitro fertilization on oocytes with and without the zona pellucida using Pfn4−/− sperm. Although Pfn4−/− sperm were not capable of fertilizing oocytes with zonae, using zona-free oocytes resulted in successful fertilization with 30% of oocytes developing to the morula/blastocyst stage (Fig. 9C). In comparison, 70% of oocytes were successfully fertilized using WT sperm. This indicates that the acrosome defects in Pfn4−/− sperm contribute to male infertility in PFN4-deficient mice.
Actin cytoskeletal organization is normal in Pfn4−/− seminiferous epithelium
In order to check whether loss of PFN4 also affects actin cytoskeletal organization in testicular seminiferous tubules, we performed phalloidin staining on testes sections of WT, Pfn4+/− and Pfn4−/− mice. The actin cytoskeleton is important for migration of germ cells from the apical to basal regions of seminiferous tubules, and maintenance of cell–cell (Sertoli–spermatid) interactions (Kanatsu-Shinohara et al., 2008). Interestingly, we found that the dynamic actin cytoskeleton is comparable for all three genotypes (Fig. S6). This result suggests that loss of PFN4 does not impede actin assembly in seminiferous epithelium of testes.
Apoptosis and phagocytosis are not observed in Pfn4−/− mice
To determine whether apoptosis was induced in Pfn4−/− germ cells, IHC was performed for the apoptotic markers cleaved caspase 3, cleaved caspase 9 and active caspase 3. Upon apoptosis induction, caspases are actively transported into the nucleus resulting in cell death. Interestingly, IHC revealed no indication of germ cell death in Pfn4−/− testes, with staining comparable to WT and Pfn4−/− sections (Fig. S7A). Furthermore, we performed TEM analysis on testes sections to detect phagocytosis of germ cells by Sertoli cells. Surprisingly, phagocytosis was not observed in Pfn4−/− Sertoli cells, which is comparable to WT and Pfn4+/− sections (Fig. S7B).
DISCUSSION
Here, we have established and analyzed mouse lines deficient in PFN4 and demonstrate that lack of PFN4 leads to male infertility. We show that PFN4-null spermatids display malformed formation of the manchette and flagella. Disturbed manchette development is the main cause of abnormal formation of sperm heads and flagellar defects, which lead to reduced sperm motility. We further reveal that acrosome biogenesis is impaired. Defects in acrosome biogenesis seems to originate from a malfunctioning Golgi network, which is incapable of forming a proper acrosomal vesicle. Proteomic analysis revealed an enrichment of the PI3K/AKT and mTOR pathways in Pfn4−/− mice, which was confirmed by western blot. Therefore, LC3I/II and SQSTM1 protein levels were increased, indicating inhibition of autophagy. Finally, we demonstrate that zona-free oocytes treated with Pfn4−/− sperm develop up to morula stage, suggesting that PFN4 is required for proper acrosome biogenesis and function.
The phenotype observed in PFN4-deficient mice correlates with PFN4 protein subcellular localization in the acrosome and manchette (Behnen et al., 2009). We observed defects in manchette development that seem to result in the amorphous head shape of Pfn4−/− sperm. Furthermore, we provide evidence that the flagellar structure is affected by the impaired manchette. In addition, acrosome biogenesis is partially aberrant, starting from the Golgi phase in Pfn4−/− mice. Hence, we show that PFN4 contributes to manchette formation, protein transport, flagellar structural integrity, sperm motility, acrosome biogenesis and sperm head morphology.
Pfn4−/− mice display greatly reduced sperm viability as a result of severe defects in sperm membrane integrity. The infertility seems to be mainly caused by perturbation of manchette development and acrosome biogenesis.
PFN4 is localized in the manchette of spermatids during elongation steps (Behnen et al., 2009). We observed defects in manchette development in Pfn4−/− sperm. Several studies on spermiogenesis-related genes, e.g. Hook1 (Mendoza-Lujambio et al., 2002), Ift20 (Li et al., 2016), Pfn3 (Umer et al., 2021), Spag17 (Kazarian et al., 2018), Lrguk1 (Okuda et al., 2017), Spef2 (Lehti et al., 2017), Gopc (Yao et al., 2002) and Cdc42 (Chapin et al., 2001), showed aberrant manchette development resulting in altered sperm head shape. The manchette and nucleus are connected with linkers, suggesting a structural relationship between manchette and nucleus. Furthermore, clutching forces exerted by the manchette are responsible for shaping of the sperm head (Kierszenbaum and Tres, 2004).
In addition, we observed disrupted localization of HOOK1 and ARL3 in the manchette of Pfn4−/− spermatids. Interestingly HOOK1-deficient males display a similar phenotype (impaired manchette and flagellar development, abnormal sperm head shape, reduced motility and malformed acrosome formation) as we observed in our Pfn4−/− males (Schwarz et al., 2017). Recently, it has been reported that HOOK1 interacts with CCDC181 (coiled-coil domain containing protein 181) and plays a role in flagellar development and regulation (Schwarz et al., 2017). HOOK1 acts as cargo protein for CCDC181 protein in the IMT process. We observed perinuclear ring staining using HOOK1, suggesting that perinuclear ring formation is not affected in Pfn4−/− spermatids; however, in later steps (10-13), we did not observe formation of the microtubular manchette in Pfn4−/− spermatids. ARL3 plays a role in cargo transport for axoneme formation and is essential for ciliogenesis (Alkanderi et al., 2018). We speculate that PFN4 plays a role as a structural and cargo protein, whereby it might participate in binding motor proteins and transporting their cargo to specific locations during sperm development. We further hypothesize that PFN4 might interact directly or in a complex with HOOK1 and ARL3. Unfortunately, owing to a lack of PFN4 antibody, the exact localization of PFN4 protein (at the ultrastructural level) in sperm flagellar is not known; in addition, the co-immunoprecipitation experiments cannot be performed to find the interacting partners of PFN4. The localization of PFN4 to the manchette suggests a role in its organization and/or remodeling. Indeed, we find that loss of PFN4 results in defective manchette development and a smaller sperm head with severely altered shape. PFN3 also localized in the manchette and Pfn3−/− mice show amorphous sperm head morphology as a result of abnormal manchette development, which is comparable to the phenotype of Pfn4−/− mice.
As described previously, the manchette also contributes to the cargo of motor proteins before transporting them for tail formation through IMT. We observed flagellar defects that contribute to the reduced sperm motility in PFN4-deficient mice. The phenotypic similarities observed in other knockout mouse models, e.g. LRGUK (Liu et al., 2015), MEIG1 (Li et al., 2015), PACRG (Li et al., 2015), SUN4 (SPAG4) (Calvi et al., 2015), HOOK1 (Mendoza-Lujambio et al., 2002) and GOPC (Yao et al., 2002), emphasize the significance of the IMT mechanism in sperm flagellar development. This underlies the movement of proteins via acrosome-acroplaxome-manchette-tail association for manchette development, sperm head shaping, acrosome biogenesis, maintaining Golgi sub-domains, and flagella formation. However, further studies are needed to elucidate the role of PFN4 in IFT during late steps of development of sperm tail.
Ultrastructural examination of Pfn4−/− sperm revealed that round spermatids fail to properly fuse proacrosomal granules/vesicles to form the acrosome. Further analysis using cis- and trans-Golgi markers indicates that the structural disorganization of Golgi sub-domains might be the reason for aberrant proacrosomal vesicle formation and fusion during acrosome biogenesis. IHC using Sp56 and acrosin also showed disturbed acrosome biogenesis in PFN4-deficient mice. These results suggest a role of PFN4 in vesicle trafficking and protein transport from the Golgi network for acrosome biogenesis. Interestingly, PICK1, GM130 and PFN3 proteins have been shown to localize to the Golgi apparatus as well. Mice deficient for these genes also displayed a fragmented Golgi network resulting in impaired acrosome development due to failure of proacrosomal granule fusion, as observed in PFN4 knockouts.
In addition, several proteins, such as COPA, PICK1 and GOPC, play a role in vesicle trafficking and transport. It has been reported that mutations in COPA lead to aberrant protein trafficking (Lepelley et al., 2020). We observed mislocalization of COPA, PICK1 and GOPC in PFN4-deficient mice, suggesting disturbed vesicle trafficking and transport and resulting in arrested acrosome formation. We hypothesize that PFN4 might interact directly or indirectly with COPA, PICK1 and GOPC, which are crucial for protein transport and trafficking, and loss of PFN4 might disturb this complex. However, further experiments need to be done to find the interacting partners of PFN4.
We reported previously (Umer et al., 2021) that PFN3 is located in the Golgi apparatus, acrosome and manchette, and hence is similar to PFN4 in subcellular localization (Behnen et al., 2009). Both Pfn3−/− and Pfn4−/− male mice display impaired acrosome biogenesis with disrupted Golgi sub-domains. However, Pfn4−/− mice show a more dramatic phenotype in terms of sperm viability, acrosome biogenesis and fertility compared with Pfn3−/− mice. We speculate that this is due to PFN4 affecting the main signaling pathway of autophagy (PI3K/AKT), whereas PFN3 affects only downstream targets (mTOR/AMPK) of autophagy (Umer et al., 2021).
Protein trafficking from the Golgi network is vital throughout the process of sperm differentiation. During acrosome development, proacrosomal vesicles are released from the trans-Golgi network and fuse to form the acrosome (Tang et al., 1982). In acrosome biogenesis, components of the autophagy network play a role (Wang et al., 2014). They modulate proacrosomal formation, and support transport and fusion towards the nucleus for acrosome development (Shang et al., 2016). P13K/AKT and mTOR are key regulators of autophagy and an imbalance in these factors leads to inhibition of autophagy (Jia et al., 2022). We demonstrate that protein levels of P13K, p-PI3K, AKT and AKT1 were increased in PFN4-deficient mice, likely leading to the activation of mTOR and p-mTOR and suppression of AMPK, resulting in autophagy inhibition. The increase of LC3BI/II and SQSTM1 protein levels in Pfn4−/− males suggests that autophagosomes accumulate. Ultimately, this seems to result in a blockage of autophagic flux and impaired acrosome biogenesis in PFN4-deficient mice.
SIRT1 (Liu et al., 2017) and ATG7 (Wang et al., 2014) deficient mice show inhibited autophagy, resulting in LC3BI/II and SQSTM1 accumulation and failure of proacrosomal granule formation and fusion. Our study on Pfn3−/− shows a disturbance in the mTOR/AMPK pathway along with the accumulation of autophagosomes (Umer et al., 2021). This suggests that SIRT1 and ATG7 proteins play a role in the downstream cascade, whereas PFN3 protein is essential for regulating the central player (mTOR) of autophagy. However, PFN4 seems to play a role in the upstream PI3K/AKT signaling mechanism of autophagy.
Proteomics analysis of Pfn4−/− mice indicated moderate deregulation of SPECC1L, ARF2:3 and FKBP1 proteins. They play a role in PI3K signaling, the trans-Golgi network and vesicle tethering and transport. SPECC1L is known to be a modulator of PI3K/AKT signaling (Wilson et al., 2016), which is essential for autophagy regulation. Specc1l mutants exhibit reduced PI3K/AKT signaling (Wilson et al., 2016). We observed increased protein levels of SPECC1L in Pfn4−/− mice, which seem to lead to the activation of PI3K/AKT signaling, as confirmed by enrichment and western blot analysis (Fig. 6; Fig. S5D). Increased protein levels of AKT leads to activation of mTOR, which results in the inhibition of autophagy (Hahn-Windgassen et al., 2005). ARF2:3 and FKBP1A localize in the trans-Golgi network, play a role in post-Golgi trafficking and act as a sorting station from Golgi to plasma membrane (Manolea et al., 2010). Enrichment analysis further showed that biological processes for protein transport are altered in Pfn4−/− mice. This suggests that loss of PFN4 might disturb the autophagy mechanism responsible for acrosome biogenesis, Golgi structural organization, and vesicle formation, transport and fusion.
Aberrant acrosome biogenesis is likely responsible for the disrupted acrosome reaction and inability of sperm to fertilize oocytes. Indeed, Pfn4−/− sperm showed a very low acrosomal exocytosis rate compared with controls, but Pfn4−/− sperm were still able to fertilize oocytes that had been treated to remove the zona pellucida. Thus, impaired acrosome biogenesis seems to be responsible, at least in part, for the Pfn4−/− male infertility. In contrast to PFN3-deficient mice (Umer et al., 2021), acrosomal exocytosis was affected more severely in the PFN4-deficient mouse model.
In addition, we did not observe any organizational defects in the actin cytoskeleton in Pfn4−/− testes. This could be because PFN4 does not retain the actin-binding site, and actin organization and polymerization are affected only when F-actin capping or cross-linking protein junctions are disturbed.
Interestingly, we did not observe male germ cell apoptosis and phagocytosis by Sertoli cells in our PFN4-deficient mice. This suggests that inhibition of autophagy caused by disrupted signaling pathways does not contribute to apoptosis in testicular germ cells of Pfn4−/− males.
Our results suggest that PFN4 plays a role in manchette development and vesicle trafficking from the Golgi network to developing spermatozoa for acrosome biogenesis. In addition, defects in manchette development were observed, which seems to result in abnormal sperm head morphology. Furthermore, we demonstrated the deregulation of PI3K/AKT and mTOR/AMPK signaling pathways, which results in the autophagy inhibition during acrosome biogenesis in Pfn4−/− mice. This unusual member of the profilin family is essential for mouse male fertility and is an intriguing target for studies concerning human male factor infertility.
MATERIALS AND METHODS
Ethical statement
Animal care, breeding setup and all experimental experiments were approved according to German laws of animal protection and in agreement with the approval of the local institutional animal care committees (Landesamt für Natur, Umwelt und Verbraucherschutz, North Rhine-Westphalia, approval ID: AZ84- 02.04.2013.A429).
Generation of Pfn4 knockout mice
Superovulation was achieved by intraperitoneal injection of C57BL/6J female mice with pregnant mare serum (5 IU) and human chorionic gonadotropin (5 IU); two females were then mated with one C57BL/6J male. Zygotes were isolated and injected as described previously (Umer et al., 2021). Pfn4 knockout mice were generated by co-injection of Cas9 mRNA (100 ng/μl) (Sigma-Aldrich), and in vitro-transcribed single guide RNAs (50 ng/μl each guide) (Table S2) into the cytoplasm of zygotes. The zygotes surviving after microinjection were kept for 3 days in KSOM medium in a CO2 incubator. The resulting blastocysts were transferred into the uteri of pseudo-pregnant foster recipients as described previously (Umer et al., 2021).
Genotyping PCR
Genomic DNA was isolated using phenol:chloroform methodology from mouse tails and subjected to PCR. Gene-specific primers were designed and used for genotyping PCR. PCR products were sequenced to identify the locus-specific mutations mediated by CRISPR/Cas9 genome editing. Primers are listed in Table S1.
Fertility analysis
For fertility analysis, a total of seven to nine WT and knockout male mice aged 10-12 weeks were individually mated with sexually mature C57BL/6J WT females in a controlled breeding experiment. Females were observed for the vaginal plugs and pregnancies. The average litter size from each pregnant female was calculated.
Morphological analysis
Pfn4−/−, Pfn4+/− and WT littermate male mice were used for phenotypic characterization (body, testes and cauda epididymis weights) as previously reported (Umer et al., 2021) (n=9 animals/genotype). Male mice aged 10-12 weeks were used in this study.
Epididymis sperm assessment
Epididymal sperm were extracted by multiple incisions into the cauda epididymis followed by a swim-out for 30-60 min in M2 medium. Sperm viability analysis was performed using E&N staining and sperm membrane integrity was accessed by the hypo-osmotic swelling test (Umer et al., 2021).
Nuclear morphology analysis
For nuclear morphology analysis, after swim-out sperm cells were washed three times followed by fixation in 2% paraformaldehyde. Sperm cells were diluted in fixative and spread evenly on a glass slide and allowed to air dry. Slides were counterstained with DAPI (Carl Roth) as described previously (Skinner et al., 2019). Analysis was performed on three independent experiments in total (n=3/genotype). Images were taken using a Leica DM5500 B/JVC KY-F75U digital camera. Images were analyzed using the ImageJ plugin ‘Nuclear morphology analysis v1.15.3’ according to the developer's instructions.
Protein extraction and western blot analysis
For total protein extracts from the testes, tissue was homogenized using a Dounce homogenizer (Sartorius) into 500-900 μl RIPA buffer (Thermo Fisher Scientific). The homogenized tissue was kept for 15 min on ice followed by 15 min centrifugation on 4°C at 13,000 rpm (17,383 g). In order to load equal amounts of protein, the concentration of supernatant was measured using the Pierce BCA Protein-Assay Kit (Thermo Fisher Scientific). SDS gel followed by western blot was performed as described previously (Umer et al., 2021) with primary antibodies against LC3I/II (1:1000; ab58610, Abcam), p-mTOR (1:1000; 2971, Cell Signaling Technology), mTOR (1:1000; 2972, Cell Signaling Technology), SQSTM1 (1:1000; 5114, Cell Signaling Technology), AMPK (1:1000; 5831, Cell Signaling Technology), β-actin (1:10,000; A5441, Sigma-Aldrich), as described previously (Umer et al., 2021), ARF3 (1:1000; 10800-1-AP), FKBP1 (1:1000; 10273-1-AP) (Hou et al., 2017), SPECC1L (1:1000; PA5-71632; Thermo Fisher Scientific), p-AKT (1:2000; 4060, Cell Signaling Technology), p-PI3K (1:1000; 17366, Cell Signaling Technology), PI3K (1:1000; 4263, Cell Signaling Technology) and AKT (1:1000; 4691, Cell Signaling Technology), as described previously (Chen et al., 2022).
cDNA synthesis and qRT-PCR
After removal of the tunica albuginea from testes tissue, RNA isolation using TRIzol reagent according to manufacturer's protocol (Life Technologies) was performed. The concentration and purity of isolated RNA was measured using NanoDrop (Peqlab). After DNaseI treatment of RNA, cDNA synthesis was performed using the RevertAid First Strand cDNA Synthesis Kit (Fermentas, now Thermo Fisher Scientific). qRT-PCR was performed using Maxima SYBR Green qPCR Master Mix (Life Technologies) on ViiA 7 Real Time PCR System (Applied Biosystems, distributed by Life Technologies) as described previously (Umer et al., 2021). At the end of each PCR run, a melting point analysis was performed. Gapdh was used as reference gene for data normalization.
TEM
Testes tissue and epididymal mature sperm cells were washed in PBS and fixed in 1.5% glutaraldehyde and 0.1 M cacodylate buffer (pH 7.4). Sperm cells were washed with 0.1 M cacodylate buffer, followed by fixation in 2% osmium tetroxide with an additional wash in 0.1 M cacodylate buffer. Further steps were performed as described previously (Umer et al., 2021). Images of ultra-thin sections were taken using a Verios 460L microscope (FEI) with a STEM III-detector.
Immunohistochemistry
Bouin's fixed 5-μm thick sections were processed for immunohistochemistry. Staining was carried out using a Lab Vision PT module (Thermo Fisher Scientific) and Autostainer 480S (medac) as described previously (Umer et al., 2021). Primary antibodies against HOOK1 (1:200; 10871-1-AP) (Zhou et al., 2009), LC3B (1:100; ab58610, Abcam), ARL3 (1:200; 10961-1-AP) (Qi et al., 2013), GOPC (1:200; 12163-1-AP, Thermo Fisher Scientific), COPA (1:200; BS-12464R, Thermo Fisher Scientific), PICK1 (1:200; PA5-76084, Thermo Fisher Scientific), SP56 (1:200; MA1-10866, Thermo Fisher Scientific), acrosin (1:200; BS-5151R, Thermo Fisher Scientific), AKT1 (1:100; 4060, Cell Signaling Technology), SQSTM1 (1:200; 5114, Cell Signaling Technology) and ARF3 (1:200; 10800-1-AP, Proteintech) were used.
Isolation of germ cell population from testes
Testes were collected in ice-cold PBS and the tunica albuginea was removed carefully. Testicular tissue was minced in 200 μl of digestion medium [1 mg/ml collagenase/dispase (10269638001, Roche), 1 mg/ml hyaluronidase (H3506; Sigma-Aldrich), 1 mg/ml DNAse I (DN-25; Sigma-Aldrich) in DMEM] until all seminiferous tubules seemed to be digested. Then 800 μl of digestion medium was added and incubated for 25 min at 37°C with slow continuous rotation. Additional mechanical disruption was performed by pipetting the solution five times using recovery tips. The suspension was filtered through a 35 mm pore size filter to achieve a single-cell suspension. The cell suspension was pipetted slowly to avoid clogging and cell damage. Cells were collected by centrifugation at 400 g for 10 min at 37°C, the supernatant was discarded, and the pellet was resuspended in 1 ml of PBS (Umer et al., 2021).
Immunofluorescence staining
Immunofluorescence staining was performed on testes sections using PNA-FITC Alexa Fluor 488 conjugate (Molecular Probes, Invitrogen), anti-mouse GM130 (1:250; 610823, BD Biosciences) and anti-rabbit TGN46 (1:100; JF1-024, Thermo Fisher Scientific) to assess acrosome biogenesis, cis- and trans-Golgi structural organization, respectively. Immunofluorescence staining of α-tubulin to detect the manchette was performed as described (Umer et al., 2021) on testes sections, epididymal sperm cells and the isolated germ cell population. Images were taken within 24 h using an LSM 710 confocal microscope (Zeiss).
PAS staining
Bouin's fixed 5-μm thick testes sections were deparaffinized and rehydrated followed by incubation for 10 min with periodic acid (0.5% in H2O). Slides were washed with H2O and incubated for 20 min with Schiff reagent. Finally, slides were rinsed for 7 min with H2O, followed by counterstaining with Hematoxylin, dehydration and mounting with a coverslip. Images were taken using a panoramic 3DHistech™ slide scanner.
Mass spectrometry
To prepare tissue for mass spectrometry, 20 mg of testes tissue was suspended in 200 µl LYSE buffer from the iST-NHS sample preparation kit (PreOmics) supplemented with 1× HALT1/2 protease inhibitors (Thermo Fisher Scientific). To each sample, 50 mg glass beads with 0.5 mm diameter (VWR International) were added. Tissues were disrupted in a Bioruptor ultrasound device (Diagenode SA) at 4°C. Samples were reduced by incubation at 95°C for 10 min shaking in a thermomixer at 1000 rpm. Insoluble matter was separated by centrifugation (10 min, 14,000 g, 10°C) and the supernatant was transferred to a new reaction tube. An aliquot was diluted tenfold in water for measurement of protein content with a BCA assay (Pierce Thermo Scientific) according to the manufacturer's instructions.
Solution containing 50 µg protein was added to 50 µl of DIGEST buffer (iST-NHS kit) for digestion of proteins (3 h, 37°C) and 0.4 mg of TMT10plex isobaric Mass Tag Labeling reagent was added to each sample and incubated at room temperature for 1 h. The reaction was quenched with 10 µl hydroxylamine. The preparation procedure was continued according to the iST-NHS kit instructions. Pooled peptides were dried in a vacuum concentrator, dissolved in IPG buffer pH 3-10 (GE Healthcare) and fractionated with an Off Gel device (Agilent) according to the manufacturer's instructions. Dried peptide fractions were desalted using ZipTip C18 tips (Sigma-Aldrich Chemie). Eluates were dried and re-dissolved in 10 µl 0.1% formic acid (FA).
Peptide separation was performed on a Dionex Ultimate 3000 RSLC nano HPLC system. The autosampler was operated in ‘μl-pickup’ mode. Peptides were dissolved in 10 µl 0.1% FA (solvent A) and 1 µl of this solution was injected onto a C18 analytical column (self-packed 300 mm length, 75 µm inner diameter, ReproSil-Pur 120 C18-AQ, 1.9 µm, Dr. Maisch). Peptides were separated over a linear gradient from 5% to 35% of solvent B (90% acetonitrile, 0.1% FA) at 300 nl/min. The nanoHPLC was coupled online to an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). Gradient length was 120 min. Peptide ions between 330 and 1600 m/z were scanned in the Orbitrap detector every 3 s with a resolution of 120,000 (maximum fill time 50 ms, AGC target 100%). Polysiloxane (445.12002 Da) was used for internal calibration (typical mass error ≤1.5 ppm). Using a top-speed method, peptides were subjected to collision-induced dissociation for identification (CID: 0.7 Da isolation, normalized energy 35%) and fragments analyzed in the linear ion trap with target 10,000 and maximum fill time 35 ms, turbo mode. Fragmented peptide ions were excluded from repeat analysis for 25 s. The top eight fragment ions were chosen for synchronous precursor selection and fragmented with higher energy CID (HCD: 2 Da MS2 isolation, 65% collision energy) for detection of reporter ions in the Orbitrap analyzer (resolution 50,000, maximum fill time 86 ms, target 100,000).
Raw data processing and database search were performed using Proteome Discoverer software 2.5.0.400 (Thermo Fisher Scientific). Peptide identification was carried out with an in-house Mascot server version 2.6.1 (Matrix Science). MS data were searched against Mus musculus sequences from the SwissProt database including isoforms (2019/06, 560,459+40,403 sequences) and contaminants database (cRAP) (Mellacheruvu et al., 2013). Precursor ion m/z tolerance was 10 ppm, fragment ion tolerance 0.5 Da (CID). Tryptic peptides with up to two missed cleavages were searched. C6H11NO-modification of cysteines (delta mass of 113.08406) and tandem mass tag (TMT) on N-termini and lysines were set as static modifications. Oxidation was allowed as dynamic modification of methionine. Mascot results were evaluated using the Percolator algorithm version 3.02.1 (The et al., 2016) as implemented in Proteome Discoverer. Spectra with identifications above 1% q-value were sent to a second round of database search with semi tryptic enzyme specificity (one missed cleavage allowed). Protein N-terminal acetylation, methionine oxidation, TMT and cysteine alkylation were then set as dynamic modifications. Actual false discovery rate values were 0.6% (peptide spectrum matches) and 1.0% (peptides and proteins). Reporter ion intensities (most confident centroid) were extracted from the MS3 level, with co-isolation <90% and SPS mass match >65%.
The statistical analyses of the MS data was performed using an in-house developed workflow in R environment (R version 3.6) (https://www.gbif.org/tool/81287/r-a-language-and-environment-for-statistical-computing). Prior to the statistical analysis, non-unique peptides and single-shot proteins were removed. From all available fractions, only those with a minimum number of missing values per peptide-spectrum match (PSM) and across all TMT channels were selected. When this filter returned more than one fraction per PSM, the one with the highest average intensity across all TMT channels was selected. The PSM-level data were then transformed and variance-stabilized using the VSN package (Huber et al., 2002) and then aggregated from PSM-level to protein-level intensities using Tukey's median polish method. The statistical inference analysis was performed using the R package Limma (Ritchie et al., 2015), in which the peptide pools were modeled as fixed effects. The resulting P-values for each statistical contrast were adjusted for multiple testing using the Benjamini–Hochberg method and the false discovery rates were calculated. The volcano plots, heatmaps and PCA plots were generated using ggplot2, ComplexHeatmap (Gu et al., 2016) and FactoMineR (Lê et al., 2008) packages, respectively.
Acrosomal reaction
After sperm swim-out in M2 medium, capacitation was induced by incubating in HTF medium (Millipore) for 90 min at 37°C, 5% CO2. To induce the acrosome reaction, the calcium ionophore A23187 (10 µM; c7522, Sigma-Aldrich) was added and the experiment was performed according to Umer et al. (2021). Spermatozoa for each genotype (n=3/genotype) were assessed by bright-field microscope.
Statistics
Statistical analysis was performed using Student's t-test (two-tailed unpaired and one-tailed paired), and pairwise comparisons between populations using Mann–Whitney U-test and ANOVA (Tukey's post-hoc) to ascertain the significance of the variability in the data. The mean of all values for a particular data set is represented in the graphical form. Error bars are used to denote standard deviation. P<0.05 was considered statistically significant.
In vitro fertilization
Oocytes from B6D2F1 females were super-ovulated by intraperitoneal injection of 5 IU pregnant mare serum and human chorionic gonadotropin. Oocytes were extracted from the oviducts after 15 h of the last hormone injection and freed from cumulus cells by treatment with 0.5-1 ml hyaluronidase for 30 s to 1 min at 37°C and 5% CO2. Oocytes and sperm were co-cultured in M2 medium (Sigma-Aldrich) for 2-4 h. Surviving oocytes were cultivated in KSOM in micro-drops (Gynemed) under mineral oil (Gynemed) at 37°C and 5% CO2.
Supplementary Material
Acknowledgements
We kindly thank the Core Facilities for Mass Spectrometry and Bioinformatics Data Analysis (CUBA) of the Medical Faculty, University of Bonn for their support. The mass spectrometer was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Projektnummer 386936527. We also thank Angela Egert and Andrea Jäger for excellent technical assistance, and Prof. Gielen and Dr Lea for their support in the scanning of slides.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: H.S., N.U.; Methodology: N.U., K.L., A.B.; Formal analysis: N.U., S.P., F.S., M.S., A.B.; Investigation: N.U., S.P., G.K., M.S., A.B.; Resources: H.S.; Data curation: F.S., A.B., M.S.; Writing - original draft: N.U.; Writing - review & editing: L.A., G.K., H.S.; Visualization: F.S., L.A., M.S.; Supervision: L.A., H.S.; Project administration: H.S.; Funding acquisition: H.S., N.U.
Funding
This study was funded by grants by the German Academic Exchange Program (DAAD; Deutscher Akademischer Austauschdienst) (91646787 to N.U.). Open Access funding provided by Medizinische Fakultät, Rheinische Friedrich-Wilhelms-Universität Bonn. Deposited in PMC for immediate release.
Data availability
Mass spectrometry proteomics data have been deposited to the ProteomeXchange ConsoSrtium via the PRIDE partner repository with the dataset identifier PXD030394.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/lookup/doi/10.1242/dev.200499.reviewer-comments.pdf.
References
- Alkanderi, S., Molinari, E., Shaheen, R., Elmaghloob, Y., Stephen, L. A., Sammut, V., Ramsbottom, S. A., Srivastava, S., Cairns, G., Edwards, N.et al. (2018). ARL3 mutations cause Joubert syndrome by disrupting ciliary protein composition. Am. J. Hum. Genet. 103, 612-620. 10.1016/j.ajhg.2018.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnen, M., Murk, K., Kursula, P., Cappallo-Obermann, H., Rothkegel, M., Kierszenbaum, A. L. and Kirchhoff, C. (2009). Testis-expressed profilins 3 and 4 show distinct functional characteristics and localize in the acroplaxome-manchette complex in spermatids. BMC Cell Biol. 10, 34. 10.1186/1471-2121-10-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi, A., Wong, A. S. W., Wright, G., Wong, E. S. M., Loo, T. H., Stewart, C. L. and Burke, B. (2015). SUN4 is essential for nuclear remodeling during mammalian spermiogenesis. Dev. Biol. 407, 321-330. 10.1016/j.ydbio.2015.09.010 [DOI] [PubMed] [Google Scholar]
- Chapin, R. E., Wine, R. N., Harris, M. W., Borchers, C. H. and Haseman, J. K. (2001). Structure and control of a cell-cell adhesion complex associated with spermiation in rat seminiferous epithelium. J. Androl. 22, 1030-1052. 10.1002/j.1939-4640.2001.tb03444.x [DOI] [PubMed] [Google Scholar]
- Chen, C., Bao, H., Lin, W., Chen, X., Huang, Y., Wang, H., Yang, Y., Liu, J., Lv, X. and Teng, L. (2022). ASF1b is a novel prognostic predictor associated with cell cycle signaling pathway in gastric cancer. J. Cancer 13, 1985-2000. 10.7150/jca.69544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, J., Radau, B., Otto, A., Müller, E. C., Lindschau, C. and Westermann, P. (2000). Profilin I attached to the Golgi is required for the formation of constitutive transport vesicles at the trans-Golgi network. Biochim. Biophys. Acta Mol. Cell Res. 1497, 253-260. 10.1016/S0167-4889(00)00056-2 [DOI] [PubMed] [Google Scholar]
- Dong, Y.-S., Hou, W.-G., Li, Y., Liu, D.-B., Hao, G.-Z., Zhang, H.-F., Li, J.-C., Zhao, J., Zhang, S., Liang, G.-B.et al. (2016). Unexpected requirement for a binding partner of the syntaxin family in phagocytosis by murine testicular Sertoli cells. Cell Death Differ. 23, 787-800. 10.1038/cdd.2015.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareus, R., di Nardo, A., Rybin, V. and Witke, W. (2006). Mouse profilin 2 regulates endocytosis and competes with SH3 ligand binding to dynamin 1. J. Biol. Chem. 281, 2803-2811. 10.1074/jbc.M503528200 [DOI] [PubMed] [Google Scholar]
- Gu, Z., Eils, R. and Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847-2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Hahn-Windgassen, A., Nogueira, V., Chen, C.-C., Skeen, J. E., Sonenberg, N. and Hay, N. (2005). Akt activates the mammalian target of rapamycin by regulating cellular ATP level and AMPK activity. J. Biol. Chem. 280, 32081-32089. 10.1074/jbc.M502876200 [DOI] [PubMed] [Google Scholar]
- Hou, X., Zhang, J., Wang, Y., Xiong, W. and Mi, J. (2017). TGFBR-IDH1-Cav1 axis promotes TGF-β signalling in cancer-associated fibroblast. Oncotarget 8, 83962-83974. 10.18632/oncotarget.20861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., Ma, T., Lau, P. K., Wang, J., Zhao, T., Du, S., Loy, M. M. T. and Guo, Y. (2019). Visualization of protein sorting at the trans-golgi network and endosomes through super-resolution imaging. Front. Cell Dev. Biol. 7, 1-13. 10.3389/fcell.2019.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, W., von Heydebreck, A., Sültmann, H., Poustka, A. and Vingron, M. (2002). Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics (Oxford, England) 18 Suppl. 1, S96-S104. 10.1093/bioinformatics/18.suppl_1.S96 [DOI] [PubMed] [Google Scholar]
- Jia, M., Qiu, H., Lin, L., Zhang, S., Li, D. and Jin, D. (2022). Inhibition of PI3K/AKT/mTOR signalling pathway activates autophagy and suppresses peritoneal fibrosis in the process of peritoneal dialysis. Front. Physiol. 13, 778479. 10.3389/fphys.2022.778479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaleağasıoğlu, F., Ali, D. M. and Berger, M. R. (2020). Multiple Facets of Autophagy and the Emerging Role of Alkylphosphocholines as Autophagy Modulators. Front. Pharmacol. 11, 547. 10.3389/fphar.2020.00547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara, M., Takehashi, M., Takashima, S., Lee, J., Morimoto, H., Chuma, S., Raducanu, A., Nakatsuji, N., Fässler, R. and Shinohara, T. (2008). Homing of mouse spermatogonial stem cells to germline Niche depends on β1-integrin. Cell Stem Cell 3, 533-542. 10.1016/j.stem.2008.08.002 [DOI] [PubMed] [Google Scholar]
- Kazarian, E., Son, H. Y., Sapao, P., Li, W., Zhang, Z., Strauss, J. F., III and Teves, M. E. (2018). SPAG17 is required for male germ cell differentiation and fertility. Int. J. Mol. Sci. 19, 1252. 10.3390/ijms19041252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khawar, M. B., Gao, H. and Li, W. (2019). Mechanism of acrosome biogenesis in mammals. Front. Cell Dev. Biol. 7, 195. 10.3389/fcell.2019.00195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum, A. L. and Tres, L. L. (2004). The acrosome-acroplaxome-manchette complex and the shaping of the spermatid head. Arch. Histol. Cytol. 67, 271-284. 10.1679/aohc.67.271 [DOI] [PubMed] [Google Scholar]
- Kim, K.-S., Cha, M. C. and Gerton, G. L. (2001). Mouse sperm protein sp56 is a component of the acrosomal matrix. Biol. Reprod. 64, 36-43. 10.1095/biolreprod64.1.36 [DOI] [PubMed] [Google Scholar]
- Lê, S., Josse, J. and Husson, F. (2008). FactoMineR: an R package for multivariate analysis. J. Stat. Softw. 25, 1-18. 10.18637/jss.v025.i01 [DOI] [Google Scholar]
- Lehti, M. S., Zhang, F.-P., Kotaja, N. and Sironen, A. (2017). SPEF2 functions in microtubule-mediated transport in elongating spermatids to ensure proper male germ cell differentiation. J. Cell Sci. 130, e1.2. 10.1242/jcs.208710 [DOI] [PubMed] [Google Scholar]
- Lepelley, A., Martin-Niclós, M. J., le Bihan, M., Marsh, J. A., Uggenti, C., Rice, G. I., Bondet, V., Duffy, D., Hertzog, J., Rehwinkel, J.et al. (2020). Mutations in COPA lead to abnormal trafficking of STING to the Golgi and interferon signaling. J. Exp. Med. 217, e20200600. 10.1084/jem.20200600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Wei, Q., Zhang, Y., Ling, K. and Hu, J. (2010). The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J. Cell Biol. 189, 1039-1051. 10.1083/jcb.200912001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Tang, W., Teves, M. E., Zhang, Z., Zhang, L., Li, H., Archer, K. J., Peterson, D. L., Williams, D. C., Strauss, J. F.et al. (2015). A MEIG1/PACRG complex in the manchette is essential for building the sperm flagella. Development (Camb.) 142, 921-930. 10.1242/dev.119834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Zhang, Y., Zhang, L., Tevesb, M. E., Liu, H., Strauss, J. F., Pazour, G. J., Foster, J. A., Hess, R. A., Zhang, Z.et al. (2016). Intraflagellar transport protein IFT20 is essential for male fertility and spermiogenesis in mice. Mol. Biol. Cell 27, 3705-3716. 10.1091/mbc.e16-05-0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Hu, X., Wang, J. H., Xu, W., Yi, C., Ma, R. and Jiang, H. (2018). Inhibition of autophagy via activation of PI3K/Akt/mTOR pathway contributes to the protection of hesperidin against myocardial ischemia/reperfusion injury. Int. J. Mol. Med. 42, 1917-1924. 10.3892/ijmm.2018.3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., DeBoer, K., de Kretser, D. M., O'Donnell, L., O'Connor, A. E., Merriner, D. J., Okuda, H., Whittle, B., Jans, D. A., Efthymiadis, A.et al. (2015). LRGUK-1 is required for basal body and Manchette function during spermatogenesis and male fertility. PLoS Genet. 11, e1005090. 10.1371/journal.pgen.1005090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J.et al. (2017). Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development (Camb.) 144, 441-451. 10.1242/jcs.202085 [DOI] [PubMed] [Google Scholar]
- Long, F. J. and Cook, S. (1991). Formation of the perinuclear theca in spermatozoa of diverse mammalian species: relationship of the manchette and multiple band polypeptides. Mol. Reprod. Dev. 28, 380-393. 10.1002/mrd.1080280411 [DOI] [PubMed] [Google Scholar]
- Manolea, F., Chun, J., Chen, D. W., Clarke, I., Summerfeldt, N., Dacks, J. B. and Melançon, P. (2010). Arf3 is activated uniquely at the trans-golgi network by Brefeldin A-inhibited guanine nucleotide exchange factors. Mol. Biol. Cell 21, 1836-1849. 10.1091/mbc.e10-01-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu, D., Wright, Z., Couzens, A. L., Lambert, J.-P., St-Denis, N. A., Li, T., Miteva, Y. V., Hauri, S., Sardiu, M. E., Low, T. Y.et al. (2013). The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730-736. 10.1038/nmeth.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Lujambio, I., Burfeind, P., Dixkens, C., Meinhardt, A., Hoyer-Fender, S., Engel, W. and Neesen, J. (2002). The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh) mutant mouse. Hum. Mol. Genet. 11, 1647-1658. 10.1093/hmg/11.14.1647 [DOI] [PubMed] [Google Scholar]
- Mizushima, N. and Levine, B. (2010). Autophagy in mammalian development and differentiation. Nat. Cell Biol. 12, 823-830. 10.1038/ncb0910-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida, K., Tres, L. L. and Kierszenbaum, A. L. (1991). Structural and Biochemical Features of Fractionated Spermatid Manchettes and Sperm Axonemes of the Azh/Azh Mutant Mouse. Mol. Reprod. Dev. 52, 434-444. [DOI] [PubMed] [Google Scholar]
- O'Donnell, L. and O'Bryan, M. K. (2014). Microtubules and spermatogenesis. Semin. Cell Dev. Biol. 30, 45-54. 10.1016/j.semcdb.2014.01.003 [DOI] [PubMed] [Google Scholar]
- Okuda, H., DeBoer, K., O'Connor, A. E., Merriner, D. J., Jamsai, D. and O'Bryan, M. K. (2017). LRGUK1 is part of a multiprotein complex required for manchette function and male fertility. FASEB J. 31, 1141-1152. 10.1096/fj.201600909R [DOI] [PubMed] [Google Scholar]
- Qi, Y., Jiang, M., Yuan, Y., Bi, Y., Zheng, B., Guo, X., Huang, X., Zhou, Z. and Sha, J. (2013). Adp-ribosylation factor-like 3, a manchette-associated protein, is essential for mouse spermiogenesis. Mol. Hum. Reprod. 19, 327-335. 10.1093/molehr/gat001 [DOI] [PubMed] [Google Scholar]
- Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W. and Smyth, G. K. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldívar-Hernández, A., González-González, M. E., Sánchez-Tusié, A., Maldonado-Rosas, I., López, P., Treviño, C. L., Larrea, F. and Chirinos, M. (2015). Human sperm degradation of zona pellucida proteins contributes to fertilization. Reprod. Biol. Endocrinol. 13, 99. 10.1186/s12958-015-0094-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, T., Prieler, B., Schmid, J. A., Grzmil, P. and Neesen, J. (2017). Ccdc181 is a microtubule-binding protein that interacts with Hook1 in haploid male germ cells and localizes to the sperm tail and motile cilia. Eur. J. Cell Biol. 96, 276-288. 10.1016/j.ejcb.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, Y., Wang, H., Jia, P., Zhao, H., Liu, C., Liu, W., Song, Z., Xu, Z., Yang, L., Wang, Y.et al. (2016). Autophagy regulates spermatid differentiation via degradation of PDLIM1. Autophagy 12, 1575-1592. 10.1080/15548627.2016.1192750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner, B. M., Rathje, C. C., Bacon, J., Johnson, E. E. P., Larson, E. L., Kopania, E. E. K., Good, J. M., Yousafzai, G., Affara, N. A. and Ellis, P. J. I. (2019). A high-throughput method for unbiased quantitation and categorization of nuclear morphology. Biol. Reprod. 100, 1250-1260. 10.1093/biolre/ioz013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T. T., Chung, C. M. and Chan, H. C. (2011). Acrosome reaction in the cumulus oophorus revisited: involvement of a novel sperm-released factor NYD-SP8. Protein Cell 2, 92-98. 10.1007/s13238-011-1022-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T., Li, S. and Ren, H. (2013). Profilin as a regulator of the membrane-actin cytoskeleton interface in plant cells. Front. Plant Sci. 4, 512. 10.3389/fpls.2013.00512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X. M., Lalli, M. F. and Clermont, Y. (1982). A cytochemical study of the Golgi apparatus of the spermatid during spermiogenesis in the rat. Am. J. Anat. 163, 283-294. 10.1002/aja.1001630402 [DOI] [PubMed] [Google Scholar]
- Tariq, N., Basharat, Z., Butt, S. and Baig, D. N. (2016). Distribution analysis of profilin isoforms at transcript resolution with mRNA-seq and secondary structure in various organs of Rattus norvegicus. Gene 589, 49-55. 10.1016/j.gene.2016.05.023 [DOI] [PubMed] [Google Scholar]
- Teves, M. E., Roldan, E., Krapf, D., Strauss, J. F., III, Bhagat, V. and Sapao, P. (2020). Sperm differentiation: the role of trafficking of proteins. Int. J. Mol. Sci. 21, 3702. 10.3390/ijms21103702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The, M., MacCoss, M. J., Noble, W. S. and Käll, L. (2016). Fast and accurate protein false discovery rates on large-scale proteomics data sets with Percolator 3.0. J. Am. Soc. Mass Spectrom. 27, 1719-1727. 10.1007/s13361-016-1460-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, N., Graham, M., Liu, X., Yue, X., Zhu, L., Meshram, D., Choi, S., Qian, Y., Rothman, J. E. and Lee, I. (2019). Golgin45-Syntaxin5 interaction contributes to structural integrity of the Golgi stack. Sci. Rep. 9, 12465. 10.1038/s41598-019-48875-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsapras, P. and Nezis, I. P. (2017). Caspase involvement in autophagy. Cell Death Differ. 24, 1369-1379. 10.1038/cdd.2017.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umer, N., Arévalo, L., Phadke, S., Lohanadan, K., Kirfel, G., Sons, D., Sofia, D., Witke, W. and Schorle, H. (2021). Loss of Profilin3 impairs spermiogenesis by affecting acrosome biogenesis, autophagy, manchette development and mitochondrial organization. Front. Cell Dev. Biol. 9, 749559. 10.3389/fcell.2021.749559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Wan, H., Li, X., Liu, W., Chen, Q., Wang, Y., Yang, L., Tang, H., Zhang, X., Duan, E.et al. (2014). Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 24, 852-869. 10.1038/cr.2014.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, N. R., Olm-Shipman, A. J., Acevedo, D. S., Palaniyandi, K., Hall, E. G., Kosa, E., Stumpff, K. M., Smith, G. J., Pitstick, L., Liao, E. C.et al. (2016). SPECC1L deficiency results in increased adherens junction stability and reduced cranial neural crest cell delamination. Sci. Rep. 6, 17735. 10.1038/srep17735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, N., Kam, C., Shen, C., Jin, W., Wang, J., Lee, K. M., Jiang, L. and Xia, J. (2009). PICK1 deficiency causes male infertility in mice by disrupting acrosome formation. J. Clin. Invest. 119, 802-812. 10.1172/JCI36230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, A., Qu, R., Chen, G., Zhang, L., Chen, J., Tao, C., Fu, J., Tang, J., Ru, Y., Chen, Y.et al. (2020). Disruption in ACTL7A causes acrosomal ultrastructural defects in human and mouse sperm as a novel male factor inducing early embryonic arrest. Sci. Adv. 6, eaaz4796. 10.1126/sciadv.aaz4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., Ahmed, N., Wang, L., Chen, H., Waqas, Y., Liu, T., Haseeb, A., Bangulzai, N., Huang, Y. and Chen, Q. (2017). In vivo autophagy and biogenesis of autophagosomes within male haploid cells during spermiogenesis. Oncotarget 8, 56791-56801. 10.18632/oncotarget.18221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, R., Ito, C., Natsume, Y., Sugitani, Y., Yamanaka, H., Kuretake, S., Yanagida, K., Sato, A., Toshimori, K., Noda, T.et al. (2002). Lack of acrosome formation in mice lacking a Golgi protein, GOPC. Proc. Natl. Acad. Sci. USA 99, 11211-11216. 10.1073/pnas.162027899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Du, Y.-R., Qin, W.-H., Hu, Y.-G., Huang, Y.-N., Bao, L., Han, D., Mansouri, A. and Xu, G.-L. (2009). RIM-BP3 is a manchette-associated protein essential for spermiogenesis. Development (Cambridge, England) 136, 373-382. 10.1242/dev.030858 [DOI] [PubMed] [Google Scholar]
- Zhu, Z., Yang, C., Iyaswamy, A., Krishnamoorthi, S., Sreenivasmurthy, S. G., Liu, J., Wang, Z., Tong, B. C.-K., Song, J., Lu, J.et al. (2019). Balancing mTOR signaling and autophagy in the treatment of Parkinson's disease. Int. J. Mol. Sci. 20, 728. 10.3390/ijms20030728 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.