Abstract
Sphingomonas paucimobilis SYK-6 is able to grow on various dimeric lignin compounds, which are converted to vanillate and syringate by the actions of unique lignin degradation enzymes in this strain. Vanillate and syringate are degraded by the O-demethylase and converted into protocatechuate (PCA) and 3-O-methylgallate (3MGA), respectively. PCA is further degraded via the PCA 4,5-cleavage pathway, while the results suggested that 3MGA is degraded through another pathway in which PCA 4,5-dioxygenase is not involved. In a 10.5-kb EcoRI fragment carrying the genes for PCA 4,5-dioxygenase (ligAB), 2-pyrone-4,6-dicarboxylate hydrolase (ligI), and a portion of 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase (ligC), we found the ligJ gene encoding 4-oxalomesaconate (OMA) hydratase, which catalyzes the conversion of OMA into 4-carboxy-4-hydroxy-2-oxoadipate. The ligJ gene is transcribed in the same direction as ligABC genes and consists of an 1,023-bp open reading frame encoding a polypeptide with a molecular mass of 38,008 Da, which is located 73-bp upstream from ligA. The ligJ gene product (LigJ), expressed in Escherichia coli, was purified to near homogeneity and was estimated to be a homodimer (69.5 kDa) by gel filtration chromatography. The isoelectric point was determined to be 4.9, and the optimal temperature is 30°C. The Km for OMA and the Vmax were determined to be 138 μM and 440 U/mg, respectively. LigJ activity was inhibited by the addition of thiol reagents, suggesting that some cysteine residue is part of the catalytic site. The ligJ gene disruption in SYK-6 caused the growth defect on and the accumulation of common metabolites from both vanillate and syringate, indicating that the ligJ gene is essential to the degradation of these two compounds. These results indicated that syringate is converted into OMA via 3MGA, and it enters the PCA 4,5-cleavage pathway.
Lignin is the most abundant aromatic compound on the earth, and its mineralization is a fundamental step in the terrestrial carbon cycle. It is expected that lignin can be used as biomass by converting it to valuable materials. Bacterial enzyme systems for lignin degradation and modification are of great use for this purpose. Sphingomonas paucimobilis SYK-6 is able to grow on various dimeric lignin compounds, including β-aryl ether, biphenyl, and diarylpropane, as sole carbon and energy sources (18). We have characterized the enzymes and genes involved in β-aryl ether cleavage (16, 17) and biphenyl degradation (23, 24), which include essential and late limiting steps of lignin degradation, respectively. These unique specific lignin degradation enzymes in SYK-6 would be suitable tools for conversion of lignin to useful intermediate metabolites.
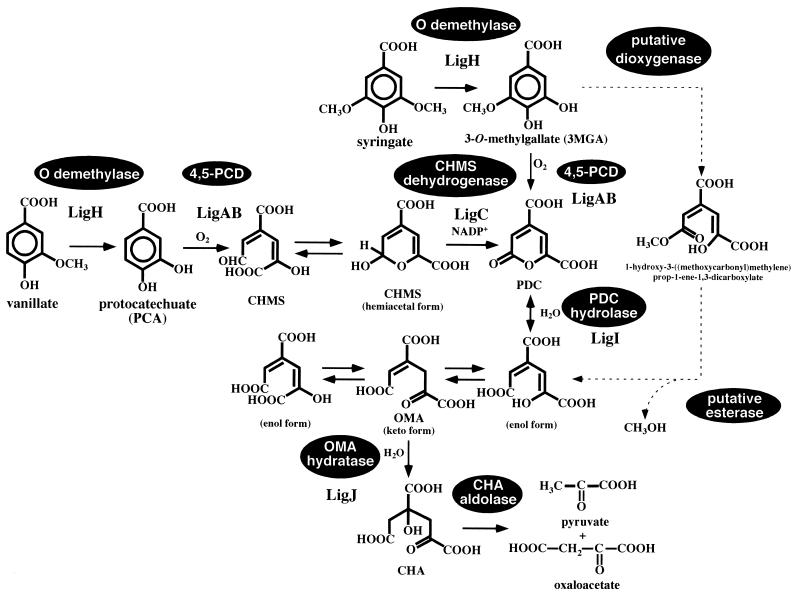
Vanillate and syringate are important intermediate metabolites from lignin, having guaiacyl and syringyl moieties, respectively. In SYK-6, vanillate and syringate are converted to protocatechuate (PCA) and 3-O-methylgallate (3MGA), respectively, by the O-demethylase encoded by ligH (21). PCA is a key intermediate metabolite among various aromatic degradation pathways. Three kinds of dioxygenases are involved in the aromatic ring cleavage of PCA: PCA 3,4-dioxygenase (5, 6, 37), PCA 4,5-dioxygenase (22, 31), and PCA 2,3-dioxygenase (35). In the case of SYK-6, PCA is metabolized through the PCA 4,5-cleavage pathway (Fig. 1), which was enzymatically characterized in 1980s by Kersten et al. (8) and Maruyama and colleagues (11–15). PCA is initially transformed to 4-carboxy-2-hydroxymuconate-6-semialdehyde (CHMS) by PCA 4,5-dioxygenase (LigAB). CHMS is nonenzymatically converted to an intramolecular hemiacetal form and then oxidized by CHMS dehydrogenase (11, 15). The resulting intermediate, 2-pyrone-4,6-dicarboxylate (PDC), is hydrolyzed by PDC hydrolase to yield the keto form and enol form (4-carboxy-2-hydroxymuconate) of 4-oxalomesaconate (OMA), which are in equilibrium (8, 12, 19). OMA is converted to 4-carboxy-4-hydroxy-2-oxoadipate (CHA) by OMA hydratase (13). Finally, CHA is cleaved by CHA aldolase to produce pyruvate and oxaloacetate (14, 32). We previously characterized the PCA 4,5-dioxygenase gene (ligAB) (22, 31) and PDC hydrolase gene (ligI) (19); recently, the CHMS dehydrogenase gene (ligC) was also characterized (E. Masai, K. Momose, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda, submitted for publication). However, the PCA 4,5-cleavage pathway has not been genetically characterized in detail.
FIG. 1.
The proposed degradation pathway of vanillate and syringate via the PCA 4,5-cleavage pathway in S. paucimobilis SYK-6. LigA and LigB, the small and large subunits of PCA 4,5-dioxygenase (4,5-PCD) (22); LigH, a gene product essential for vanillate and syringate O-demethylations (21); LigC, CHMS dehydrogenase (Masai et al., submitted); LigI, PDC hydrolase (19); LigJ, OMA hydratase (this study). The PCA 4,5-cleavage pathway is illustrated according to findings from previous studies (11–15, 19, 22). The degradation pathway for syringate indicated by a dashed line was suggested on the basis of the results obtained in this and a previous (4) study.
On the other hand, the pathway for 3MGA degradation is ambiguous. PCA 4,5-dioxygenase was reported to catalyze the ring cleavage of 3MGA to form PDC, and metabolism of 3MGA through the PCA 4,5-cleavage pathway was suggested (8). However, two mutant strains of SYK-6, in which the ligAB and ligI genes were insertionally inactivated could grow on syringate but not on vanillate (19; H. Aoshima, E. Masai, S. Nishikawa, Y. Katayama, and M. Fukuda, Abstr. 8th Int. Symp. Microb. Ecol., abstr. 93, 1998). These results indicated that 3MGA generated from syringate is predominantly metabolized via a pathway other than the PCA 4,5-cleavage pathway.
In this study, we characterized the OMA hydratase gene and the enzymatic properties of the gene product to obtain detailed genetic information on the PCA 4,5-cleavage pathway and insight into the metabolism of syringate in SYK-6. We also present evidence that the OMA hydratase gene is essential to the metabolism of both vanillate and syringate and that OMA is the common intermediate metabolite of vanillate and syringate.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. S. paucimobilis SYK-6 was grown at 30°C in W minimal salt medium (23) containing 0.2% (wt/vol) vanillate or syringate or in Luria-Bertani (LB) medium (Bacto Tryptone, 10 g/liter; yeast extract, 5 g/liter; NaCl, 5 g/liter).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| Sphingomonas paucimobilis | ||
| SYK-6 | Wild type; Nalr Smr | 7 |
| DLJ | Mutant derivative of SYK-6; Kmr gene insertion mutant of ligJ; Nalr Smr Kmr | This study |
| Escherichia coli | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′[traD36 proAB+ lacIq lacZΔM15] | 36 |
| BL21(DE3) | hsdS gal(λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) | 30 |
| Plasmids | ||
| pUC18 and pUC19 | Cloning vectors; Apr | 36 |
| pBluescript II KS(+) | Cloning vector; Apr | 29 |
| pT7-blue(R) | TA cloning vector; Apr | 3 |
| pET21(+) | Expression vector; Apr T7 promoter | Novagen |
| pUC4K | Source of Kmr cassette; Apr Kmr | 33 |
| pK19mobsacB | oriT sacB Kmr | 27 |
| pHN139F | pUC18 with a 10.5-kb EcoRI fragment of pVA01 carrying ligJAB, a part of ligC, and ligI | 19 |
| pHN139R | pUC18 carrying the same fragment as pHN139F in the opposite direction | 19 |
| pSS30F | pBluescript II KS(+) with a 3.0-kb SalI fragment carrying ligJ of pHN139F | This study |
| pSS30R | pBluescript II KS(+) carrying the same fragment as pSS30F in the opposite orientation | This study |
| pSS73F, pSS73R, pSS50F, pSS50R, pSS17F, pSS17R, pSS20F, pSS20R | pBluescript II KS(+) with deletion fragments of pHN139F and pHN139R (Fig. 1) | 19, this study |
| pSXB17 | pBluescript II KS(+) with a 1.7-kb SmaI-XbaI fragment carrying a portion of ligJ | This study |
| pETJ | pET21(+) with a 1.1-kb PCR-amplified fragment carrying ligJ | This study |
| pSXB17K | pSXB17 with an insertion of the Kmr gene of pUC4K replacing a 500-bp Eco47III fragment | This study |
| pLJD | pK19mobsacB with a 2.4-kb SmaI-XbaI fragment of pSXB17K | This study |
Preparation of substrate.
PDC was prepared from PCA by using cells of Pseudomonas putida PpY1100 harboring pVAD4, which conferred transformation activity from PCA to PDC as described earlier (19). To obtain OMA, 1 mmol of PDC was hydrolyzed by 0.057 N NaOH at room temperature for 3 h and then neutralized with 0.5 N HCl by the method of Maruyama (12). In a previous study, we identified the trimethylsilyl (TMS) derivatives of two isomeric enol forms of OMA (19). In this study, the OMA preparation was derivatized with methoxyamine hydrochloride to identify the keto form of OMA because α-keto acids are difficult to analyze by gas chromatography-mass spectrometry (GC-MS). Methoxyamine hydrochloride (final concentration, 10 mg/ml) was added to 200 μM OMA solution. The resultant solution was alkalinized (pH 11 to 12) and kept at 60°C for 1 h (34). This solution was acidified by 2 N HCl, and the derivatized OMA was extracted with ethyl acetate. The extract was dried in vacuo and trimethylsilylated. The gas chromatogram of the sample showed two peaks with retention times of 29.5 min (compound I) and 30.2 min (compound II). The mass spectrum of compound I corresponded to that of the enol form OMA (19). On the other hand, the mass spectrum of compound II showed the major fragments at m/z 447, 432, 416, and 330, which seemed to correspond to the molecular ions of methoxime-TMS derivatives of the keto form OMA (M), M-CH3, M-OCH3, and M-COOTMS, respectively. Thus, the OMA preparation contains the two isomeric enol forms and the keto form as shown in Fig. 1.
Vanillate, syringate, and other chemicals were purchased from Tokyo Kasei Kogyo Co. (Tokyo, Japan) or Wako Pure Chemical Industries (Osaka, Japan).
DNA manipulations and nucleotide sequencing.
DNA manipulations were carried out essentially as described elsewhere (1, 25). A Kilosequence kit (Takara Shuzo Co., Ltd., Kyoto, Japan) was used to construct a series of deletion derivatives, whose nucleotide sequences were determined by the dideoxy termination method with an ALFexpress DNA sequencer (Pharmacia Biotech, Milwaukee, Wis.).
A Sanger reaction (26) was carried out by using a Thermosequenase fluorescent labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). Sequence analysis and homology alignment were carried out with the GeneWorks programs (IntelliGenetics, Inc., Mountain View, Calif.). The DDBJ database was used for searching homologous proteins. Southern hybridization analysis of SYK-6 and its OMA hydratase gene (ligJ) insertion mutants were performed with the DIG system (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) according to the procedure recommended by the manufacturer.
Subcloning of ligJ gene.
To overexpress the ligJ gene, the ligJ coding region in pHN139F was amplified by PCR using the Ex Taq polymerase (Takara Shuzo), pHN139F as a template, and primers ligJF and ligJR. Primers ligJF (forward; GAGACGATCACGAGAGGTAACC) and ligJR (reverse; GAAATCACGGGAAACAAAGC) were designed from the pHN139F ligJ coding sequence.
The 1.1-kb PCR product was inserted into pT7-blue(R) (3). Then the EcoRI-HindIII fragment of the resulting plasmid was ligated to pET21(+) to generate pETJ.
Enzyme assay.
Using the method of Maruyama (13), OMA hydratase activity was spectrophotometrically determined by measuring the decrease in the absorbance at 265 nm (ɛ265 = 2.6 × 103 M−1 cm−1; pH 8.0) with a DU-7500 spectrophotometer (Beckman, Fullerton, Calif.). The enzyme reaction was carried out at 30°C in a cuvette. The 1-ml reaction mixture contained 200 μM OMA and the enzyme in 0.1 M Tris-acetate buffer (pH 8.0). The OMA preparation contained the keto form and enol form (4-carboxy-2-hydroxymuconate), which are in equilibrium. One unit of enzyme activity was defined as the amount that degrades 1 μmol of substrate per min at 30°C. Specific activity was expressed as units per milligram of protein. Km and Vmax values were obtained from Hanes-Woolf plots and expressed as means from at least three independent experiments.
Enzyme purification.
Enzyme purification was performed according to the method described below by using a BioCAD700E apparatus (PerSeptive Biosystems, Framingham, Mass.).
(i) Preparation of cell extract. Cells were grown in 100 ml of 2×YT medium (Bacto Tryptone, 20 g/liter; yeast extract, 10 g/liter; NaCl, 5 g/liter) containing 100 mg of ampicillin/liter. Expression of ligJ was induced for 12 h at 30°C by the addition of isopropyl-β-d-thiogalactopyranoside (final concentration, 1 mM). Cells were harvested by centrifugation and resuspended in 20 mM Tris-HCl buffer (pH 8.0) (buffer A). The cells were broken by two passages through a French pressure cell. The cell lysate was centrifuged at 15,000 × g for 15 min. Streptomycin (final concentration, 1%) was added to the supernatant, which was recentrifuged at 15,000 × g for 15 min to remove nucleic acids. The supernatant was recovered and then centrifuged again at 170,000 × g for 60 min at 4°C. The crude extract was obtained after concentration by ultrafiltration using a Minicon B15 (Amicon, Beverly, Mass.).
(ii) POROS PI anion-exchange chromatography.
The crude extract was applied to a POROS PI (polyethyleneimine) column (7.5 by 100 mm; PerSeptive Biosystems) previously equilibrated with buffer A. The enzyme was eluted with 88 ml of linear gradient of 0 to 0.5 M NaCl. The OMA hydratase was eluted at approximately 0.34 M.
(iii) POROS HQ anion-exchange chromatography.
The fractions containing OMA hydratase activity eluted from a PI column were pooled, desalted, and concentrated by ultrafiltration using a Minicon B15. The resulting solution was applied to a POROS HQ (quaternized PI) column (4.6 by 100 mm; PerSeptive Biosystems) previously equilibrated with buffer A. The enzyme was eluted with 33 ml of a linear gradient of 0 to 0.5 M NaCl. The fractions containing OMA hydratase activity that eluted at approximately 0.28 M were pooled.
(iv) POROS PE hydrophobic interaction chromatography.
The fractions containing OMA hydratase activity eluted from a HQ column were pooled, desalted, and concentrated. Ammonium sulfate was added to the enzyme solution to a final concentration of 2 M. After centrifugation at 15,000 × g for 10 min, the supernatant was recovered and applied to a POROS PE column (4.6 by 100 mm; PerSeptive Biosystems) equilibrated with buffer B (buffer A containing 2 M ammonium sulfate). The enzyme was eluted with 25 ml of a linear gradient of 2.0 to 0 M ammonium sulfate. The fractions containing OMA hydratase activity that eluted at approximately 1.1 M were pooled, desalted, and concentrated as described above. Glycerol was added to a final concentration of 10%, and the purified enzyme was stored at −80°C until use.
Analytical method.
The protein concentration was determined by the method of Bradford (2). The purity of the enzyme preparation was examined by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis (SDS-PAGE) (10). The molecular mass of the native enzyme was determined by Superdex 200 HR10/30 (Pharmacia Biotech) gel filtration column chromatography using a BioCAD700E apparatus. Elution was performed with 50 mM potassium phosphate buffer (pH 7.0) containing 0.15 M NaCl at a flow rate of 0.8 ml/min. The molecular weight was estimated on the basis of calibration curve of reference proteins.
To determine the N-terminal amino acid sequence, a cell extract of Escherichia coli BL21(DE3) harboring pETJ was subjected to SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane (Bio-Rad, Hercules, Calif.). The area at 35 kDa was cut out and analyzed on a PPSQ-21 protein sequencer (Shimadzu, Kyoto, Japan). The isoelectric point of LigJ was determined by isoelectric point focusing on an Ampholine PAG plate (pH 3.5 to 9.5; Pharmacia Biotech) using a model Multiphor II electrophoresis system (Pharmacia Biotech).
The substrate and the reaction products were detected and identified by GC-MS using model 5971A with an Ultra-2 capillary column (50 m by 0.2 mm; Hewlett-Packard Co., Palo Alto, Calif.) and electrospray ionization (ESI)-MS using HP1100 series LC-MSD (Hewlett-Packard Co.). The analytical conditions for GC-MS were the same as described previously (19). In ESI-MS analysis, mass spectra were obtained by negative-mode ESI, with a needle voltage of −3.5 kV and a source temperature at 350°C. The sample was injected into the flow system; the water/methanol ratio was 90:10 (vol/vol), and the flow rate was 0.2 ml/min.
Identification of the reaction product.
OMA was incubated with purified LigJ (0.5 μg) in 0.1 M Tris-acetate buffer (pH 8.0) for 10 min. The reaction mixture was acidified and extracted with ethyl acetate, and then the extract was trimethylsilylated. The resultant TMS derivatives are analyzed by GC-MS as described above.
In the case of ESI-MS analysis, the reaction mixture was diluted to 1/10 with 10 mM Tris-acetate buffer (pH 8.0), and 5 μl of the mixture was injected into the flow system.
Insertional inactivation of the ligJ gene.
The 1.7-kb SmaI-XbaI fragment carrying a portion of ligJ was cloned into pBluescript II KS(+) to generate pSXB17. pSXB17 was digested with Eco47III, and the 500-bp fragment in the middle of ligJ was deleted. The 1.2-kb PstI fragment containing the kanamycin resistance (Kmr) gene from pUC4K (33) was inserted into this Eco47III site. The resultant plasmid, pSXB17K, was digested with EcoRI and KpnI, and the insert containing the inactivated ligJ gene was cloned into pK19mobsacB (27) to generate pLJD.
pLJD was introduced into SYK-6 cells by electroporation as described previously (19). Kmr transformants were selected on an LB agar plate containing 50 mg of kanamycin/liter. They were cultured for 12 h in LB liquid medium containing 50 mg of kanamycin/liter and 10% sucrose. The candidates for mutants were isolated on an LB agar plate containing 10% sucrose and kanamycin. Southern hybridization analyses of the SalI digests of total DNA prepared from the candidates for mutants were carried out with the 1.2-kb SalI-XbaI and 1.2-kb PstI fragment probes containing a portion of ligJ and the Kmr gene, respectively.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper was deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AB035121.
RESULTS
Nucleotide sequence of the OMA hydratase gene.
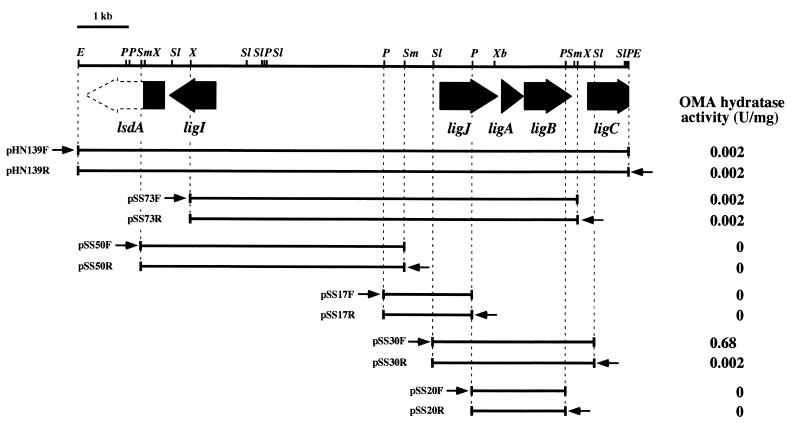
We have already isolated the SYK-6 10.5-kb EcoRI fragment carrying ligAB (22), ligI (19), and a part of ligC (18; Masai et al., submitted). We examined the OMA hydratase activity on this fragment to see whether the OMA hydratase gene is located in it. The OMA hydratase activity was observed in E. coli JM109 harboring pHN139F, which contained this 10.5-kb EcoRI fragment (Fig. 2). In the deletion analysis, the DNA region that conferred OMA hydratase activity was limited to the 2.7-kb SalI-XhoI fragment, which spanned the most of the insert of pSS30F and pSS30R. The ligAB gene was included in this 2.7-kb fragment as indicated in Fig. 2. Therefore, the OMA hydratase gene seemed to be located upstream from the ligA gene. Based on the difference of the activity between pSS30F and pSS30R, the direction of transcription of OMA hydratase gene was suggested to be the same as that of ligAB. The deletion derivatives of pSS30F were constructed, and the nucleotide sequence of the region except for the ligAB gene, whose sequence had been established previously (22), was determined. The only open reading frame (ORF) found was 73 bp upstream from ligA and designated ligJ. The start codon of ligJ could not be deduced because the 5′ end of the ligJ sequence has three consecutive ATG codons. The ligJ gene spans bp 1017 to 1023 and has a putative ribosome binding sequence upstream.
FIG. 2.
Deletion analysis of the 10.5-kb EcoRI fragment and lig gene organization. The OMA hydratase activity of cells containing each subclone is presented on the right. Arrows indicate the direction of transcription from the lac promoters. The ligI, ligJ, ligA, ligB, and ligC genes are indicated by filled arrows; a partly filled arrow represents the part of the ORF of the lignostilbene α, β-dioxygenase homolog (lsdA). Each plasmid was introduced in E. coli JM109. E, EcoRI; P, PstI; Sl, SalI; Sm, SmaI; X, XhoI; Xb, XbaI.
Expression of ligJ in E. coli.
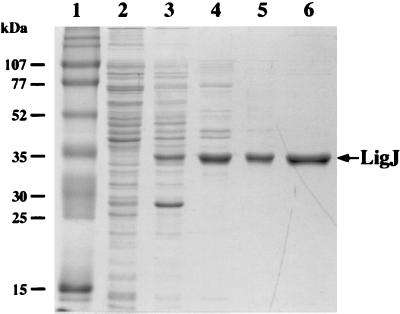
The ligJ gene was amplified by PCR and subcloning in pET21(+) to generate plasmid pETJ as described in Materials and Methods. The ligJ gene was expressed in E. coli BL21(DE3) harboring plasmid pETJ with the aid of its T7 promoter. OMA hydratase activity of the cell extract of E. coli BL21(DE3) harboring pETJ was 24.3 U/mg, indicating the high level of expression of ligJ. In SDS-PAGE analysis, the molecular mass of a subunit of the ligJ product (LigJ) was estimated to be 35 kDa (Fig. 3, lane 3), this 35-kDa protein was subjected to the N-terminal amino acid sequencing. The sequence of the first 15 residues was determined to be Met-Met-Met-Ile-Ile-Asp-Xaa-His-Gly-Xaa-Tyr-Thr-Val-Leu-Pro, which corresponded to the deduced amino acid sequence of ligJ translated from the first ATG of the three consecutive ATG codons. Therefore, the ligJ gene was estimated to be an ORF of 1,023 bp, encoding 341 amino acid residues. The molecular mass deduced from the amino acid sequence of LigJ (Mr, 38,008) is close to the value estimated by SDS-PAGE (35 kDa). A homology search with the deduced amino acid sequence of ligJ in the SwissProt and DAD databases showed identity only with LigY (37%), which is a hydrolase for the meta-cleavage compound 2,2′,3-trihydroxy-3′-methoxy-5,5′-dicarboxybiphenyl, (OH-DDVA), involved in the catabolism of the lignin-related biphenyl by S. paucimobilis SYK-6 (24).
FIG. 3.
SDS-PAGE analysis of protein fractions. Proteins were separated on an SDS–12% polyacrylamide gel and stained with Coomassie brilliant blue. Lanes: 1, molecular weight markers; 2, crude extract of E. coli BL21(DE3) harboring pET21(+) (10 μg of protein); 3, crude extract of E. coli BL21 (DE3) harboring pETJ (10 μg of protein); 4, PI fraction (5 μg of protein); 5, HQ fraction (3 μg of protein); 6, PE fraction (2 μg of protein). Molecular masses are given on the left.
Purification of OMA hydratase.
LigJ protein expressed in E. coli BL21(DE3) harboring pETJ was purified to near homogeneity (Fig. 3) by a series of column chromatographyies with PI, HQ, and PE (Table 2 and Fig. 3). LigJ was purified approximately 13-fold, with a recovery of 20%.
TABLE 2.
Purification of OMA hydratase from E. coli BL21(DE3) harboring pETJ
| Fraction | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Fold purification |
|---|---|---|---|---|---|
| Crude extract | 72.0 | 3,100 | 43.1 | 100 | 1.0 |
| PI | 8.21 | 1,380 | 168 | 45 | 3.9 |
| HQ | 3.00 | 921 | 307 | 30 | 7.1 |
| PE | 1.10 | 613 | 557 | 20 | 13 |
Identification of the reaction product.
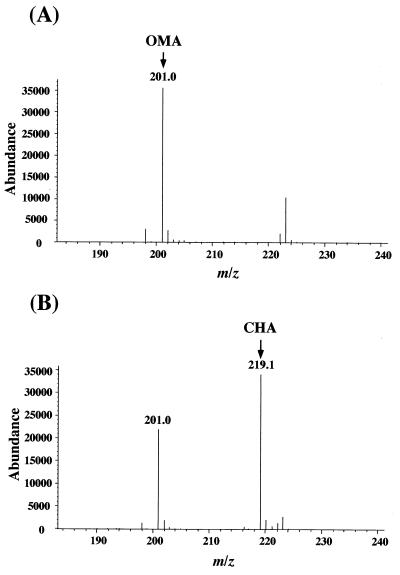
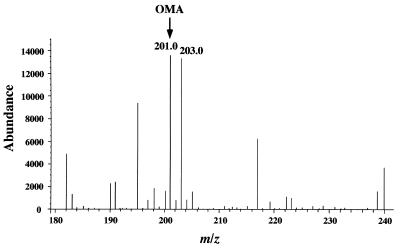
To identify the reaction product of OMA catalyzed by the purified LigJ, the reaction mixture was analyzed by ESI-MS. Figure 4A shows the mass spectrum of the substrate. The major fragment at m/z 201 in Fig. 4A was estimated to be the deprotonated molecular ion ([M-H]−) of OMA (where M is a molecular ion of OMA). In the mass spectrum of the reaction mixture shown in Fig. 4B, the fragment at m/z at 201 of OMA decreased to 65% of its initial intensity, and the new major fragment at m/z 219 corresponding to [M-H]− of CHA (where M is the molecular ion of CHA) was observed. Thus, the results strongly suggested that OMA was converted to CHA by incorporation of a water molecule.
FIG. 4.
Identification of the reaction product from OMA catalyzed by LigJ. (A) Negative-ion ESI-MS spectrum of OMA. The peak at m/z 201 was assigned to the deprotonated molecular ion [M-H]− of OMA. (B) Negative-ion ESI-MS spectrum of the reaction product generated from OMA catalyzed by purified LigJ. The peak at m/z 201 derived from OMA reduced, and the generation of the product peak at m/z 219 was observed.
Enzyme properties.
Gel filtration column chromatography using Superdex 200 indicated that the molecular mass of the native LigJ was 69.5 kDa. This result suggested that LigJ is a homodimer. The isoelectric point of LigJ was determined by isoelectric focusing gel electrophoresis to be 4.9. The optimal temperature for LigJ hydratase activity on OMA was determined to be 30°C; the optimal pH was not determined since OMA was unstable in at high pH. The Km for OMA and the Vmax were determined to be 138 μM and 440 U/mg, respectively.
The influence of thiols and thiol reagents on LigJ was also examined. Purified LigJ (0.5 μg) was preincubated with 1 mM cysteine, reduced glutathione, and dithiothreitol individually at 30°C for 10 min, and the remaining activities were determined. OMA hydratase activity was activated to 147, 135, and 120% in the presence of cysteine, reduced glutathione, and dithiothreitol, respectively. On the other hand, the addition of 1 mM HgCl2 completely inhibited the activity. These results suggested that some cysteine residue in LigJ is involved in the enzyme reaction.
Disruption of the ligJ gene in S. paucimobilis SYK-6.
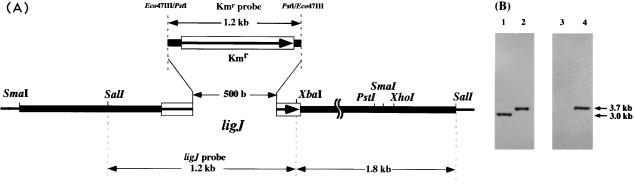
The ligJ gene was disrupted to investigate its role in the catabolism of vanillate and syringate by SYK-6. Gene inactivation was carried out using the ligJ disruption plasmid pLJD, which was constructed by replacing the internal segment of ligJ inserted in pK19mobsacB by a Kmr gene. The ligJ insertional mutation was confirmed by Southern hybridization analysis using the 1.2-kb SalI-XbaI fragment carrying a portion of ligJ and the 1.2-kb PstI fragment carrying the Kmr gene as probes (Fig. 5). The mutant strain DLJ obtained completely lost the ability to grow on both vanillate and syringate.
FIG. 5.
The ligJ disruption in S. paucimobilis SYK-6. (A) Schematic representation of the insertional inactivation of ligJ by the Kmr gene. Thick arrows indicate orientations of transcription of the ligJ and Kmr genes. (B) Southern hybridization analysis of the ligJ insertion mutant (DLJ). Lanes 1 and 3, total DNA of SYK-6 digested with SalI; lanes 2 and 4, total DNA of DLJ digested with SalI. The 1.2-kb SalI-XbaI fragment carrying a portion of ligJ (lanes 1 and 2) and the 1.2-kb PstI fragment of the Kmr gene (lanes 3 and 4) were used as probes.
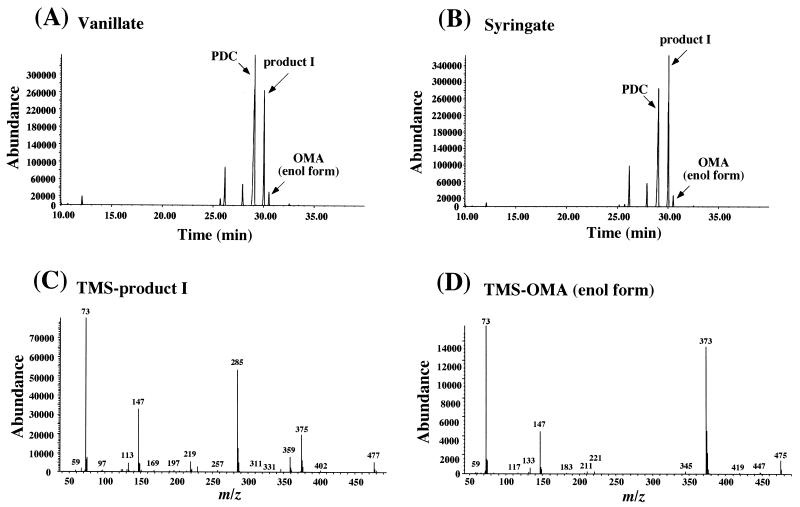
Vanillate and syringate (0.2 %) were independently incubated with whole cells of strain DLJ in W minimal medium. The metabolites produced from vanillate and syringate were examined and determined by GC-MS and ESI-MS. As shown in the gas chromatogram (Fig. 6), vanillate and syringate, detected with retention times of 21.2 and 25.2 min, respectively, disappeared completely, and accumulation of the enol form of OMA, PDC, and product I with a retention time of 30.5 min was observed in both cultures. The major fragments at m/z 475 and m/z 373 were identified in the mass spectrum of the TMS derivative of the enol form OMA as reported previously (Fig. 6D) (19). These fragments agree with M-CH3 and M-COOTMS, respectively, where M is a molecular ion of TMS-OMA. Similarly, the fragments at m/z 477 and m/z 375 of product I are thought to be M-CH3, and M-COOTMS, respectively (Fig. 6C). On the other hand, ESI-MS analysis indicated the accumulation of the ions at m/z 183 and 201 in the metabolites from vanillate and syringate, which corresponded to the deprotonated molecular ions of PDC and OMA, respectively. In addition to these ions, another ion at m/z 203 accumulated from both vanillate and syringate. Thus, this ion seemed to be the deprotonated molecular ion of product I. A ligJ inactivated mutant, DLJ should have accumulated a significant amount of OMA. However, the amount of OMA accumulated was smaller than expected. In our previous study, we found that the keto form of OMA, which is an α-keto acid, could not be detected by GC-MS in the condition that we used (19). Thus, product I did not correspond to the keto form of OMA. Possibly, product I was generated from OMA by an unknown reaction in SYK-6. To examine this hypothesis, OMA was incubated with the DLJ cell extract prepared from cells grown in LB. The reaction was carried out in 0.1 M Tris-acetate buffer (pH 8.0) containing 100 μg DLJ cell extract, 1 mM NADPH, 200 μM OMA, and 1 mM ZnSO4 at 30°C. Zn2+ was added to inhibit the reverse reaction of the PDC hydrolase, which catalyzes the conversion of OMA to PDC (19). The reaction product after 10 min of incubation was analyzed by ESI-MS and GC-MS. In the ESI-MS analysis, the peak at m/z 201 derived from OMA was converted to the peak at m/z 203 only in the presence of NADPH (Fig. 7). GC-MS analysis showed the generation of the product with the same retention time (30.5 min) as product I. Although the ion at m/z 477 observed with product I was not detected, the relative intensities of the mass fragments at m/z 375, 285, 147, and 73 were identical to those of product I (data not shown). These results strongly suggested that product I was produced from accumulated OMA possibly by the addition of two hydrogen atoms catalyzed by an unknown NADPH-dependent reductase in SYK-6 and that the accumulation of product I represents the accumulation of OMA. Thus, the results indicate that ligJ encodes OMA hydratase, which is essential for catabolism of both vanillate and syringate.
FIG. 6.
Identification of accumulated products from vanillate and syringate by DLJ. (A and B) Gas chromatograms of TMS derivatives of the accumulated products from vanillate and syringate, respectively. In both cultures, PDC, the enol form of OMA, and the unidentified product I were observed. (C and D) Mass spectra of the TMS derivatives of product I and the enol form of OMA, respectively.
FIG. 7.
Conversion of OMA by DLJ cell extract. OMA was converted to the product of which [M-H]− is m/z 203 only in the presence of NADPH.
DISCUSSION
The OMA hydratase gene, designated as ligJ, encoding a protein of 38,008 Da (341 amino acids), was characterized in this study. LigJ showed no similarity to the functionally related 2-hydroxypent-2,4-dienoate hydratases of catechol (9, 28) and biphenyl (20) degradation pathways. However, LigJ showed 37% identity only with the OH-DDVA meta-cleavage compound hydrolase (LigY) involved in the degradation of 5,5′-dehydrodivanillate (DDVA) by S. paucimobilis SYK-6. LigY showed no similarity to other aromatic compound hydrolases involved in benzene, toluene, xylene, and biphenyl degradation and did not contain a lipase box (Gly-Xaa-Ser-Gly motif), which constitutes an active site in serine hydrolases. Alignment of the deduced amino acid sequences of ligJ and ligY is shown in Fig. 8. Similarity is distributed throughout the whole sequence; striking identity is found in their amino-terminal sequences, Met-Ile-Ile-Asp-Cys-His-Gly-His. It is interesting that these two proteins in the successive degradation pathway are highly similar. LigJ and LigY seem to evolve from the same ancestral origin. The structural similarity between the LigJ substrate (OMA) and the organic acid moiety of the LigY substrate might have contributed to the evolution of these proteins.
FIG. 8.
Alignment of amino acid sequences between LigJ and LigY of S. paucimobilis SYK-6. A BLAST search indicated that the most similar protein whose function has been clarified is the OH-DDVA meta-cleavage compound hydrolase (LigY) of S. paucimobilis SYK-6 (24). Identical and similar amino acids are indicated by asterisks and colons, respectively.
Only the OMA hydratase of P. ochraceae had been characterized. The OMA hydratases of both S. paucimobilis SYK-6 and P. ochraceae are dimeric proteins, and their subunit molecular masses and pIs are very similar. Various thiols and thiol reagents affected the activities of both enzymes. However, their kinetic parameters are considerably different. The Km and Vmax values of SYK-6 OMA hydratase are approximately 10 and 3.7 times higher than those of the P. ochraceae enzyme, respectively, indicating that the P. ochraceae enzyme has significantly higher affinity toward OMA than SYK-6 enzyme.
The production of CHA from OMA catalyzed by OMA hydratase was suggested by Maruyama (13). Kersten et al. suggested that OMA is in equilibrium among the two isomeric enol forms and keto form. Both enol forms of OMA (4-carboxy-2-hydroxymuconate) were easily detected as TMS derivatives by GC-MS; however, the keto form of OMA could be detected only when it was modified to the methoxime form as described in Materials and Methods. In the case of CHA, it could not be detected by GC-MS even in its methoxime form. Therefore, we used ESI-MS to detect these compounds and demonstrate the production of CHA from OMA by LigJ. It is difficult to specify which form of OMA is the real substrate; however, considering the chemical structure of the LigJ reaction product, CHA, the keto form is the most likely candidate.
The ligJ gene of SYK-6 was inactivated by gene replacement to clarify the catabolic role of the ligJ gene. In a previous study, each disruption of ligB, ligC, and ligI did not affect syringate metabolism, while these mutants could not grow on vanillate (19; Aoshima et al., Abstr. 8th Int. Symp. Microb. Ecol., 1998; Masai et al., submitted). On the other hand, the ligJ insertion mutant DLJ could not grow on vanillate and syringate. Thus, it appears that the syringate catabolic pathway adjoined the PCA 4,5-cleavage pathway at the LigJ reaction step. DLJ accumulated the enol form of OMA, PDC, and the unknown product I from vanillate and syringate (Fig. 6). Accumulation of PDC seemed to be a result of the reverse reaction of PDC hydrolase, which converts OMA to PDC (19). Product I was suggested to be generated from OMA probably by addition of two atoms of hydrogen by NADPH-dependent reductase in DLJ cells. Product I is estimated to be 4-hydroxybut-1-ene-1,2,4-tricarboxylate. Taken together, the results suggest that the ligJ gene is essential for both vanillate and syringate degradation and that the syringate degradation pathway joins to the PCA 4,5-cleavage pathway at OMA.
Donnelly and Dagley reported that P. putida TMC degrades 3MGA to oxaloacetate and pyruvate via OMA, with the release of methanol (4). They proposed the involvement of a 3MGA dioxygenase and an esterase in the transformation of 3MGA to OMA. Characterization of a putative 3MGA dioxygenase and an esterase is necessary for a better understanding of syringate catabolism in S. paucimobilis SYK-6.
ACKNOWLEDGMENTS
H.H. was financially supported by research fellowship 2068 from the Japan Society for the Promotion of Science for Young Scientists. This work was supported in part by Grant-in-Aid for Encouragement of Young Scientists 11760057 from the Ministry of Education, Science, Sports and Culture, Japan.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1990. [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Brownstein M J, Carpten J D, Smith J R. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. BioTechniques. 1996;20:1004–1010. doi: 10.2144/96206st01. [DOI] [PubMed] [Google Scholar]
- 4.Donnelly M I, Dagley S. Bacterial degradation of 3,4,5-trimethoxycinnamic acid with production of methanol. J Bacteriol. 1981;147:471–476. doi: 10.1128/jb.147.2.471-476.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frazee R W, Livingston D M, LaPorte D C, Lipscomb J D. Cloning, sequencing, and expression of the Pseudomonas putida protocatechuate 3,4-dioxygenase genes. J Bacteriol. 1993;175:6194–6202. doi: 10.1128/jb.175.19.6194-6202.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartnett C, Neidle E L, Ngai K L, Ornston L N. DNA sequences of genes encoding Acinetobacter calcoaceticus protocatechuate 3,4-dioxygenase: evidence indicating shuffling of genes and of DNA sequences within genes during their evolutionary divergence. J Bacteriol. 1990;172:956–966. doi: 10.1128/jb.172.2.956-966.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katayama Y, Nishikawa S, Nakamura M, Yano K, Yamasaki M, Morohoshi N, Haraguchi T. Cloning and expression of Pseudomonas paucimobilis SYK-6 genes involved in the degradation of vanillate and protocatechuate in P. putida. Mokuzai Gakkaishi. 1987;33:77–79. [Google Scholar]
- 8.Kersten P J, Dagley S, Whittaker J W, Arciero D M, Lipscomb J D. 2-Pyrone-4,6-dicarboxylic acid, a catabolite of gallic acids in Pseudomonas species. J Bacteriol. 1982;152:1154–1162. doi: 10.1128/jb.152.3.1154-1162.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kukor J J, Olsen R H. Genetic organization and regulation of a meta cleavage pathway for catechols produced from catabolism of toluene, benzene, phenol, and cresols by Pseudomonas pickettii PKO1. J Bacteriol. 1991;173:4587–4594. doi: 10.1128/jb.173.15.4587-4594.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Maruyama K. Isolation and identification of the reaction product of α-hydroxy-γ-carboxymuconic-ɛ-semialdehyde dehydrogenase. J Biochem. 1979;86:1671–1677. doi: 10.1093/oxfordjournals.jbchem.a132687. [DOI] [PubMed] [Google Scholar]
- 12.Maruyama K. Purification and properties of 2-pyrone-4,6-dicarboxylate hydrolase. J Biochem. 1983;93:557–565. doi: 10.1093/oxfordjournals.jbchem.a134210. [DOI] [PubMed] [Google Scholar]
- 13.Maruyama K. Purification and properties of γ-oxalomesaconate hydratase from Pseudomonas ochraceae grown with phthalate. Biochem Biophys Res Commun. 1985;128:271–277. doi: 10.1016/0006-291x(85)91674-2. [DOI] [PubMed] [Google Scholar]
- 14.Maruyama K. Purification and properties of 4-hydroxy-4-methyl-2-oxo-glutarate aldolase from Pseudomonas ochraceae grown on phthalate. J Biochem. 1990;108:327–333. doi: 10.1093/oxfordjournals.jbchem.a123201. [DOI] [PubMed] [Google Scholar]
- 15.Maruyama K, Ariga N, Tsuda M, Deguchi K. Purification and properties of α-hydroxy-γ-carboxymuconic-ɛ-semialdehyde dehydrogenase. J Biochem. 1978;83:1125–1134. doi: 10.1093/oxfordjournals.jbchem.a132002. [DOI] [PubMed] [Google Scholar]
- 16.Masai E, Katayama Y, Kawai S, Nishikawa S, Yamasaki M, Morohoshi N. Cloning and sequencing of the gene for Pseudomonas paucimobilis enzyme that cleaves β-aryl ether. J Bacteriol. 1991;173:7950–7955. doi: 10.1128/jb.173.24.7950-7955.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masai E, Katayama Y, Kubota S, Kawai S, Yamasaki M, Morohoshi N. A bacterial enzyme degrading the model lignin compound β-etherase is a member of the glutathione-S-transferase superfamily. FEBS Lett. 1993;323:135–140. doi: 10.1016/0014-5793(93)81465-c. [DOI] [PubMed] [Google Scholar]
- 18.Masai E, Katayama Y, Nishikawa S, Fukuda M. Characterization of Sphingomonas paucimobilis SYK-6 genes involved in degradation of lignin-related compounds. J Ind Microbiol Biotech. 1999;23:364–373. doi: 10.1038/sj.jim.2900747. [DOI] [PubMed] [Google Scholar]
- 19.Masai E, Shinohara S, Hara H, Nishikawa S, Katayama Y, Fukada M. Genetic and biochemical characterization of 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J Bacteriol. 1999;181:55–62. doi: 10.1128/jb.181.1.55-62.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masai E, Sugiyama S, Iwashita N, Shimizu S, Hauschild J E, Hatta T, Kimbara K, Yano K, Fukuda M. The bphDEF meta-cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphACB genes in Rhodococcus sp. strain RHA1. Gene. 1997;187:141–149. doi: 10.1016/s0378-1119(96)00748-2. [DOI] [PubMed] [Google Scholar]
- 21.Nishikawa S, Sonoki T, Kasahara T, Obi T, Kubota S, Kawai S, Morohoshi N, Katayama Y. Cloning and sequencing of Sphingomonas (Pseudomonas) paucimobilis gene essential for the O demethylation of vanillate and syringate. Appl Environ Microbiol. 1998;64:836–842. doi: 10.1128/aem.64.3.836-842.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noda Y, Nishikawa S, Shiozuka K, Kadokura H, Nakajima H, Yoda K, Katayama Y, Morohoshi N, Haraguchi T, Yamasaki M. Molecular cloning of the protocatechuate 4,5-dioxygenase gene of Pseudomonas paucimobilis. J Bacteriol. 1990;172:2704–2709. doi: 10.1128/jb.172.5.2704-2709.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng X, Egashira T, Hanashiro K, Masai E, Nishikawa S, Katayama Y, Kimbara K, Fukuda M. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl Environ Microbiol. 1998;64:2520–2527. doi: 10.1128/aem.64.7.2520-2527.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng X, Masai E, Katayama Y, Fukuda M. Characterization of the meta-cleavage compound hydrolase gene involved in degradation of lignin-related biphenyl structure by Sphingomonas paucimobilis SYK-6. Appl Environ Microbiol. 1999;65:2789–2793. doi: 10.1128/aem.65.6.2789-2793.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 28.Shingler V, Powlowski J, Marklund U. Nucleotide sequence and functional analysis of the complete phenol/3,4-dimethylphenol catabolic pathway of Pseudomonas sp. strain CF600. J Bacteriol. 1992;174:711–724. doi: 10.1128/jb.174.3.711-724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Short J M, Fernandez J M, Sorge J A, Huse W. λZAP: a bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res. 1988;16:7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 31.Sugimoto K, Senda T, Aoshima H, Masai E, Fukuda M, Mitsui Y. Crystal structure of an aromatic-ring-opening dioxygenase LigAB which is a protocatechuate 4,5-dioxygenase. Struct Fold Des. 1999;15:953–965. doi: 10.1016/s0969-2126(99)80122-1. [DOI] [PubMed] [Google Scholar]
- 32.Tack B F, Chapman P J, Dagley S. Purification and properties of 4-hydroxy-4-methyl-2-oxoglutarate aldolase. J Biol Chem. 1972;247:6444–6449. [PubMed] [Google Scholar]
- 33.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primer. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 34.Williams V P, Ching D K, Cederbaum S D. Adsorption of organic acids from amniotic fluid and urine onto silica gel before analysis by gas chromatography and combined gas chromatography/mass spectrometry. Clin Chem. 1979;25:1814–1820. [PubMed] [Google Scholar]
- 35.Wolgel S A, Dege J E, Perkins-Olson P E, Jaurez-Garcia C H, Crawford R L, Münck E, Lipscomb J D. Purification and characterization of protocatechuate 2,3-dioxygenase from Bacillus macerans: a new extradiol catecholic dioxygenase. J Bacteriol. 1993;175:4414–4426. doi: 10.1128/jb.175.14.4414-4426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13 mp 18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 37.Zylstra G J, Olsen R H, Ballou D P. Genetic organization and sequence of the Pseudomonas cepacia genes for the alpha and beta subunits of protocatechuate 3,4-dioxygenase. J Bacteriol. 1989;171:5915–5921. doi: 10.1128/jb.171.11.5915-5921.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]